Abstract
The vertebrate innate immune system confers host cells with mechanisms to protect against both evolutionarily ancient pathogens and newly emerging pathogenic strains. Innate immunity relies on the host cell’s ability to distinguish between self and pathogen-derived molecules. To achieve this, the innate immune system uses germline encoded receptors called pattern recognition receptors (PRRs), which recognize various molecular signatures, including nucleic acids, proteins, lipids, glycans and glycolipids. Among these molecules, the recognition of pathogenic, mislocalized, or damaged DNA by cellular protein receptors, commonly called DNA sensors, represents a major surveillance pathway for initiating immune signaling. The ability of cells to temporally regulate DNA sensor activation and subsequent signal termination is critical for effective immune signaling. These same mechanisms are also co-opted by pathogens to promote their replication. Therefore, there is significant interest in understanding DNA sensor regulatory networks during microbial infections and autoimmune disease. One emerging aspect of DNA sensor regulation is through post-translational modifications (PTMs), including phosphorylation, acetylation, ubiquitination, ADP-ribosylation, SUMOylation, methylation, deamidation, glutamylation. In this chapter, we discuss how PTMs have been shown to positively or negatively impact DNA sensor functions via diverse mechanisms, including direct regulation of enzymatic activity, protein-protein and protein-DNA interactions, protein translocations and protein turnover. In addition, we highlight the ability of virus-induced PTMs to promote immune evasion. We also discuss the recent evidence linking PTMs on DNA sensors with human diseases and more broadly, highlight promising directions for future research on PTM-mediated regulation of DNA sensor-dependent immune signaling.
1. Introduction
The vertebrate innate immune system is at the front line of defense against pathogens. This ancient defense mechanism confers host cells protection not only against pathogens that have existed for much of the vertebrate evolution, but also against novel pathogenic strains. A critical function of innate immunity is to distinguish between self and pathogen- or damage-derived molecules. To recognize pathogen-associated molecular patterns (PAMPs), the innate immune system employs a series of germline encoded receptors, known as pattern recognition receptors (PRRs). PRRs recognize a vast range of molecular signatures, including nucleic acids, proteins, lipids, glycans and glycolipids, and among them, the sensing of DNA has been studied for decades (Chen et al., 2016b; Gordon, 2002; Tan et al., 2018; Thompson et al., 2011).
DNA (both double- and single-stranded) is a common genetic material for many pathogens (DNA viruses, bacteria, etc.), and thus detecting pathogen-associated DNA provides a unique signature for initiating innate immune programs. In addition, host DNA that is either damaged or mislocalized due to pathological conditions can also be sensed and elicit an immune response. This gives rise to autoimmunity, where the host immune system initiates responses against healthy tissues (Chen et al., 2016b; Tan et al., 2018). To date, researchers have identified and characterized the function of many host receptors for pathogenic DNA, which collectively are classified as DNA sensors. At minimum, a protein that functions as a DNA sensor should directly bind to pathogenic DNA, and then trigger innate immune signaling through the expression of antiviral or inflammatory cytokines and chemokines (Crow et al., 2015; Liu et al., 2016). Additionally, DNA sensors can protect host cells by inducing apoptosis (Zierhut et al., 2019), autophagy (Lei et al., 2018), or repressing virus gene expression upon binding to viral DNA (Diner et al., 2016; Johnson et al., 2014).
The first protein to be characterized as a DNA sensor was Toll-like receptor 9 (TLR9). TLR9 recognizes unmethylated cytosine-phosphate-guanine (CpG) nucleotide sequences, which is rare in vertebrate genomes compared to DNA viruses and bacteria (Krieg et al., 1995; Takeshita et al., 2001). After unmethylated CpG stimulation, TLR9 moves from the endoplasmic reticulum (ER) to the Golgi apparatus and then to endo-lysosomes, where it interacts with MyD88 to trigger downstream proinflammatory cytokine responses (Chockalingam et al., 2009; Leifer et al., 2004) (Fig. 1).
Fig. 1.
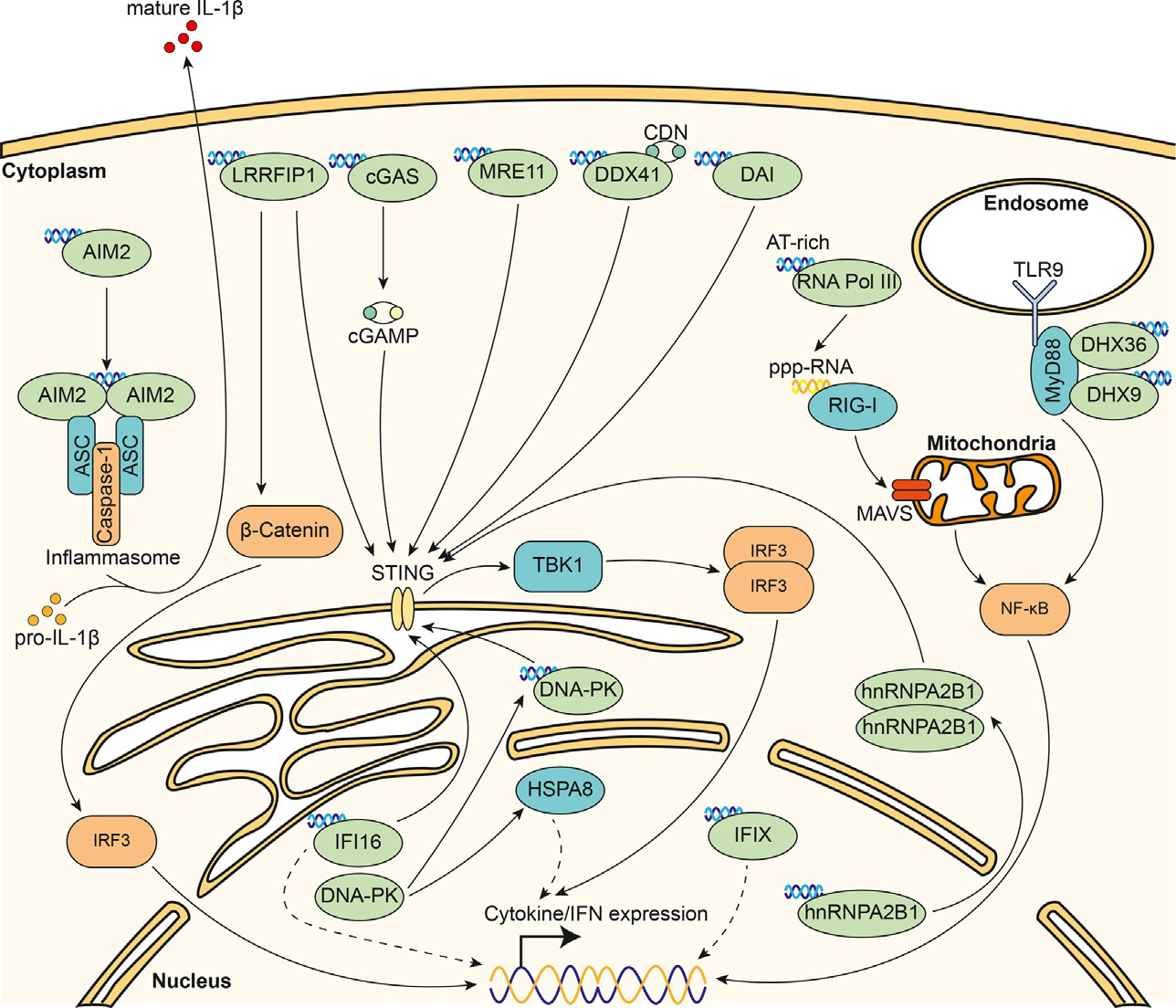
DNA sensors and their downstream signaling pathways. Known cytoplasmic (AIM2, cGAS, DAI, DDX41, DHX36, DHX9, DNAPK, LRRFIP1, MRE11, RNA Pol III) and nuclear (IFI16, IFIX, hnRNPA2B1) DNA sensors are indicated. Most DNA sensors converge on STING-TBK1-IRF3 axis, where the activated DNA sensor eventually leads to the dimerization of STING. Subsequently, this results in the phosphorylation of TBK1 and IRF3. Phosphorylated IRF3 dimerizes and translocates into the nucleus, leading to gene transcription of cytokines and interferons. Other pathways include the NF-κB pathway and the β-catenin pathway.
Unlike TLR9, many other DNA sensors do not use unique molecule features to distinguish pathogenic vs host DNA. Rather, their DNA sensing ability is intimately tied to their subcellular localization. For example, DNA sensors are often localized to the cytosol and are positioned to detect mislocalized (cytosolic) DNA, a marker for viral infection, cellular DNA damage, and mitochondrial DNA leakage (Chen et al., 2016b; Riley and Tait, 2020; Tan et al., 2018). The first cytosolic DNA sensor discovered was the cytosolic protein, DNA-dependent activator of IFN-regulatory factors (DAI), also known as Z-DNA binding protein 1 (Takaoka et al., 2007). Subsequent to the identification of DAI, many other “cytosolic” DNA sensors have been identified, including absent in melanoma 2 (AIM2) (Burckstummer et al., 2009; Hornung et al., 2009), DNA-protein kinase (DNA-PK) (Burleigh et al., 2020; Ferguson et al., 2012; Zhang et al., 2011a), DEAD box polypeptide 41 (DDX41) (Miyashita et al., 2011; Zhang et al., 2011b), DEAH box helicase 9 and 36 (DHX9 and DHX36) (Kim et al., 2010), meiotic recombination 11 homolog A (MRE11) (Kondo et al., 2013), leucine-rich repeat flightless interacting protein-1 (LRRFIP1) (Yang et al., 2010), RNApolymerase III (RNA Pol III) (Ablasser et al., 2009; Chiu et al., 2009), and cyclic-GMP-AMP synthase (cGAS) (Sun et al., 2013; Wu et al., 2013) (Table 1). Among these sensors, cGAS emerged as a dominant cytosolic DNA sensor during DNA virus or retrovirus infection (Gao et al., 2013; Sun et al., 2013), bacterial infections (Hansen et al., 2014), as well as self-DNA leakage (Harding et al., 2017; Mackenzie et al., 2017). cGAS binds double stranded DNA and catalyzes the production of a second messenger, cyclic GMP-AMP (cGAMP), using ATP and GTP as substrates (Lohofener et al., 2015; Zhang et al., 2013b, 2014). Intracellular cGAMP then binds the endoplasmic reticulum (ER)-associated adaptor STING, which recruits TANK-binding kinase 1 (TBK1) to phosphorylate the transcription factor interferon regulatory factor 3 (IRF3) (Bowie, 2012; Tanaka and Chen, 2012). The STING-TBK1-IRF3 signaling axis is critical, as most DNA sensors identified to date activate this signaling pathway (Fig. 1). Phosphorylated IRF3 then dimerizes and translocates into the nucleus, activating the expression of Type I interferons (IFNs) and other cytokines. Through both autocrine and paracrine signaling, Type I IFNs activate interferon stimulated genes (ISGs) through Jak-STAT signaling (Yan et al., 2016). ISGs possess a wide range of activities, which include stimulating cell-to-cell communication and cell death pathways, and targeting pathways required for viral entry, translation, replication, and egress. Overall, these activities convey a concerted effort to control and/or restrict the pathogenic infection process (Schneider et al., 2014).
Table 1.
Identified PTMs on DNA sensors.
| DNA sensor | Sensing location/pathway | |
|---|---|---|
| PTM: amino acid(s) | PTM characterization | References |
|
AIM2 – |
Cytoplasm/Inflammasome – |
Burckstummer et al. (2009) and Hornung et al. (2009) |
|
cGAS Acetylation: K7a, K21c, K47c, K50abc, K62c, K63c, K82c, K83c, K198b, K285b, K292b, K355b, K384ab, K392a, K394a, K414a, K432b, and K479b Deamidation: N210d, N389d, Q451dand Q454d Glutamylation: E272e, E302e Phosphorylation: S13l, S37bl, S64l, T69l, T91l, S116bl, S129l, S143lm, S201b, Y215g, S221b, S263b, and S305bfl SUMOylation: K217h, K464h Ubiquitinationk: K271i, K335j, K414i |
Cytoplasm/STING Ab-based detection Mutagenesis In-vitro assay IP-MS identification Targeted MS (PRM) |
Sun et al. (2013) and Wu et al. (2013) Dai et al. (2019)a Song et al. (2020a)b Song et al. (2020b)c Zhang et al. (2018)d Xia et al. (2016)e Seo et al. (2015)f and Liu et al. (2018)g Hu et al. (2016)h Chen et al. (2016a)i, Seo et al. (2018)j and Chen and Chen (2019)k Li et al. (2021)l and Olsen et al. (2010)m |
|
DAI – |
Cytoplasm/STING – |
Takaoka et al. (2007) |
|
DDX41 Ubiquitinationa Phosphorylation: S4b, S21b, and S23b SUMOylation: K416c and K442c |
Cytoplasm/STING Ab-based detection Mutagenesis In-vitro assay |
Zhang et al. (2011b) Zhang et al. (2013a)a Dephoure et al. (2008)b, Mayya et al.(2009)b, Olsen et al. (2010)b and Zhou et al. (2013a)b Hendriks et al. (2017)c |
|
DHX9 Methylationa: K146c and R1175c Phosphorylationb: S87d, S125d, S321d, S449d, and S506d Acetylation: K191e, K199e, and K1024e SUMOylation: K697f |
Cytoplasm/MyD88 Ab-based detection Mutagenesis In-vitro assay |
Kim et al. (2010) Smith et al. (2004)a and Sadler et al. (2009)b Guo et al. (2014)c Bian et al. (2014)d, Dephoure et al. (2008)d, Olsen et al. (2010)d and Zhou et al. (2013a)d Choudhary et al. (2009)e Hendriks et al. (2017)f |
|
DHX36 Acetylation: K947a Phosphorylation: S161b and S963b |
Cytoplasm/MyD88 – |
Kim et al. (2010)
Choudhary et al. (2009) a Zhou et al. (2013a) b |
|
DNA-PK: PRKDC Phosphorylation: S2056c, T2609abc, S2612bf, T2638bf, T2647bf, S511g, S687g, S841g, S1065g, T2535g, S2789g, S3205g, S3731g, S3821g, and S4026g Acetylation: K117h, K828h, K1209h, K1970h, K2259h, K3241h, K3260h, K3621h, K3638h, and K3642h Ku70 ADP-ribosylatione Phosphorylation: S6a, S51d, S2g, S27g, S306g, T455g, S477g, S520g, S550g, S560g SUMOylation: K287i, K317i, and K556i Ku80 ADP-ribosylatione |
Cytoplasm/HSPA8 Ab-based detection Mutagenesis In-vitro assay MS identification |
Burleigh et al. (2020), Ferguson et al. (2012) and Zhang et al. (2011a) Chan et al. (2002)a Douglas et al. (2002)b Wechsler et al. (2004)c Jin and Weaver (1997)d Beck et al. (2014)e Ismail et al. (2015)f Olsen et al. (2010)g, Rigbolt et al. (2011)g and Zhou et al. (2013a)g Choudhary et al. (2009)h Hendriks et al. (2014)i and Hendriks et al. (2017)i |
|
Phosphorylation: S577a, S579a, S580a, T715a, S255g, S258g, S318g, and T535g Ubiquitinationf Acetylation: K144h, K265h, K332h, K660h, and K665h SUMOylation: K195i, K532i, K534i, K566i, K568i, K669i, and K688i |
||
|
hnRNPA2B1 Methylation/Demethylation: Arg226a Acetylation: K168b and K173b Methylation: R203c, R213c, R228c, R238c, R266c, R325c, and R350c Phosphorylation: T4d, S29d, S85d, T140d, T159d, T176d, S189d, S201d, S212d, S225d, S231d, S236d, S259d, S324d, Y331d, S341d, S344d, and Y347d SUMOylation: K22e, K104e, K112e, K120e, K137e, K152e, K168e and K173e |
Nucleus/STING Ab-based detection Mutagenesis |
Wang et al. (2019) Wang et al. (2019)a Choudhary et al. (2009)b Guo et al. (2014)c Bian et al. (2014)d, Mayya et al. (2009)d, Olsen et al. (2010)d, Rigbolt et al.(2011)d and Zhou et al. (2013a)d Hendriks et al. (2017)e |
|
IFI16 Acetylation: K45a, K99a, K128a, K214a, K444a, K451a, K561a, K598a, and K614a Phosphorylationb: S95a, S106a, S153a, S168a, S174a, S575a, S780a Ubiquitinationc SUMOylation: K116d, K128d, K561d, and K683d |
Nucleus/STING Ab-based detection Mutagenesis In-vitro assay IP-MS identification |
Kerur et al. (2011) and Unterholzner et al. (2010) Li et al. (2012)a Dell’Oste et al. (2014)b Li et al. (2019)c Hendriks et al. (2014)d, Hendriks et al. (2017)d and Hendriks et al. (2015)d |
|
IFIX – |
Nucleus/STING – |
Diner et al. (2015a) |
|
LRRFIP1 Phosphorylation: S16a, S115a, S116a, S120a, S555a, S581a, S618a, T676a, S714a, S733a, S766a, S768a SUMOylation: K305b, K606b |
Cytoplasm/β-catenin – |
Yang et al. (2010) Bian et al. (2014)a, Dephoure et al. (2008)a, Mayya et al. (2009)a, Olsen et al. (2010)a and Zhou et al. (2013a)a Hendriks et al. (2017)b and Impens et al. (2014)b |
|
MRE11 Methylation in gly-arg-rich regiona Phosphorylation: S275b, S619b, S649b, S678b, S688b, S689b, and S701b SUMOylation: K255c, K416c, and K625c |
Cytoplasm/STING Ab-based detection Mutagenesis In-vitro assay IP-MS validation |
Kondo et al. (2013) Zhuang et al. (2009)a Bian et al. (2014)b, Dephoure et al. (2008)b, Mayya et al. (2009)b, Olsen et al. (2010)b and Zhou et al. (2013a)b Hendriks et al. (2017)c |
|
RNA Pol III Acetylation: K445 |
Cytoplasm/RIG-I – |
Ablasser et al. (2009) and Chiu et al. (2009) Choudhary et al. (2009)a |
|
TLR9 Ubiquitination |
Endosome/MyD88 Ab-based detection |
Krieg et al. (1995) and Takeshita et al. (2001) Chuang and Ulevitch (2004) |
Listed in the table are the PTM types and sites, PTM characterization approaches, DNA sensing location and pathway involved, and the papers that reported the DNA sensor identification (first line) or the identification of the PTMs.
The functionally characterized sites are labeled in bold.
Superscript letters denote the papers that reported the identification of specific PTM sites.
Although cytosolic DNA sensors are deployed to detect aberrant cytosolic DNA, many DNA viruses are known to replicate their genomes in the nucleus, including the herpesviruses, herpes simplex virus type 1 (HSV-1) and human cytomegalovirus (HCMV), as well as adenovirus (AdV), human papilloma virus (HPV) and polyomavirus (PyV) (Schmid et al., 2014). Researchers have identified several DNA sensors that reside in the host cell nucleus, including interferon-inducible protein 16 (IFI16) (Kerur et al., 2011; Unterholzner et al., 2010) and interferon-inducible protein X (IFIX) (Diner et al., 2015a). Notably, even the cytosolic DNA sensor cGAS has been detected in the nucleus (Liu et al., 2018). Like cytosolic sensors, IFN-mediated signaling of many nuclear DNA sensors is also STING-dependent (Unterholzner et al., 2010; Wang et al., 2019), although STING-independent mechanisms have also been proposed (Diner et al., 2016). As an additional layer of versatility, some DNA sensors may act synergically and thus may often compensate for each other. For example, the well-known ribonucleoprotein, hnRNPA2B1, moonlights as a nuclear DNA sensor activating cGAS-dependent and -independent pathways to stimulate type I interferon signaling (Wang et al., 2019). During HSV-1 infection, hnRNPA2B1 binds viral DNA in the nucleus, dimerizes and translocates to the cytoplasm to activate the STING-TBK1-IRF3 axis and thus initiate antiviral signaling. hnRNPA2B1 also potentiates cGAS-STING-dependent cytokine expression by facilitating m6A modification and nucleocytoplasmic trafficking of CGAS, IFI16, and STING mRNAs (Wang et al., 2019). Overall, these findings (and those from many other laboratories) highlight the importance of both cytosolic and nuclear DNA sensors and demonstrate the convergence many sensors have on the TBK1-IRF3 signaling axis. Additional studies are required to help define how these DNA sensors are regulated to coordinate a unified immune response.
The accelerated study of DNA sensors in recent years has highlighted their important roles in microbial infections and autoimmune diseases (Gao et al., 2015; Zhou et al., 2020). By understanding the regulation of these cellular sensors, there is the potential to develop more effective treatments for these conditions. However, despite a rapid expansion in our knowledge of DNA sensing mechanisms, several key questions remain unanswered. For example, it remains to be fully elucidated how host cells selectively recognize the DNA genome of the nuclear-replicating DNA viruses in the context of the host genome, and how cells maintain the DNA sensors in the inactive state during cellular homeostasis. These open questions emphasize the need to continue investigating the molecular mechanisms of DNA sensor regulation.
Given the immune activating nature of DNA sensing signal transduction, the ability of cells to temporally regulate DNA sensor activation is critical. From a fundamental perspective, many aspects of DNA sensor regulation invoke basic biochemical principles. For example, signal termination can occur via degradation of the immunostimulatory DNA, the second messenger, or the DNA sensor protein itself (Paludan, 2015). These mechanisms often involve protein-protein interactions, which may exert either negative or positive regulation depending on the context. For example, cGAS interacts with the DNA sensor IFI16 to promote interferon signaling (Orzalli et al., 2015), while its interaction with OASL, a protein that classically functions in the RNA sensing pathway, inhibits cGAS activity (Lum et al., 2018). Viral proteins have also been found to interact with DNA sensors to suppress host immune signaling, for example, Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF52 (Wu et al., 2015), HSV-1 pUL37 (Zhang et al., 2018) and VP22 (Huang et al., 2018a), and HCMV pUL31 (Huang et al., 2018b) and pUL83 (Biolatti et al., 2018) interact with and inhibit cGAS.
More recently, post-translational modifications (PTMs) of DNA sensor proteins have emerged as an additional layer of regulation. The rapid identification of potential functional PTMs has been aided by the development of high-resolution tandem mass spectrometry (MS), which confidently identifies the specific modified residue(s). Like most proteins, DNA sensors also have been found to be decorated by a diverse range of PTMs, including phosphorylation, acetylation, methylation, glutamylation, deamidation, ADP-ribosylation, SUMOylation, and ubiquitination. While only a fraction of the identified PTMs have been functionally assessed, for the sites that have been characterized, they provide key toggles of sensor functions. In this chapter, we will focus on painting the current PTM landscape of DNA sensors. Specifically, we will highlight how PTM identification using MS-based proteomics has accelerated the discovery of regulatory hubs of DNA sensor bioactivity. We will provide key examples of PTMs that impact the activity of DNA sensors. This regulation can occur directly through enzymatic activation/inhibition or protein degradation, as well as indirectly by altering the subcellular localization of DNA sensors. We will also highlight specific PTMs that are pathogen- or disease-associated. Finally, we will discuss future implications of PTM characterization studies in contributing to the development of selective therapeutics under pathogenic or autoimmune states.
2. Mass spectrometry-based proteomic characterization of PTMs
Without the aid of mass spectrometry, PTM research is largely based on prior knowledge or predictive analyses, and frequently begins with identification of an interaction between a protein of interest (POI) and a specific PTM regulatory enzyme (kinase, phosphatase, acetyltransferase, etc.) (Fig. 2, bottom). For example, cGAS was discovered to interact with tubulin polyglutamylase TTLL4 and TTLL6, two enzymes involved in protein glutamylation (Xia et al., 2016). Subsequent biochemical and molecular experiments confirmed that these enzymes were responsible for glutamylation of cGAS at two sites. Practically, PTM identification by these hypothesis-driven approaches often employs in vitro assays with purified enzymes and recombinant proteins or synthetic peptides from the POI. This approach has been invaluable for demonstrating direct modification of DNA sensors (Chan et al., 2002; Sadler et al., 2009; Zhang et al., 2018). Historically, PTM detection and visualization has employed antibody-based approaches with (2D)-SDS-PAGE and western blotting techniques. The antibodies may be PTM-specific for direct detection, or raised against the POI, which can indirectly suggest the presence of modifications through molecular weight and/or charge shifts (Fig. 2, bottom). These biochemical approaches are often followed with site-directed mutagenesis for downstream functional characterization assays (Fig. 1, bottom). One example is the IFI16 acetylation at K99 within its nuclear localization signal, which mutagenesis studies indicated plays a role in the subcellular localization of this DNA sensor (Li et al., 2012).
Fig. 2.
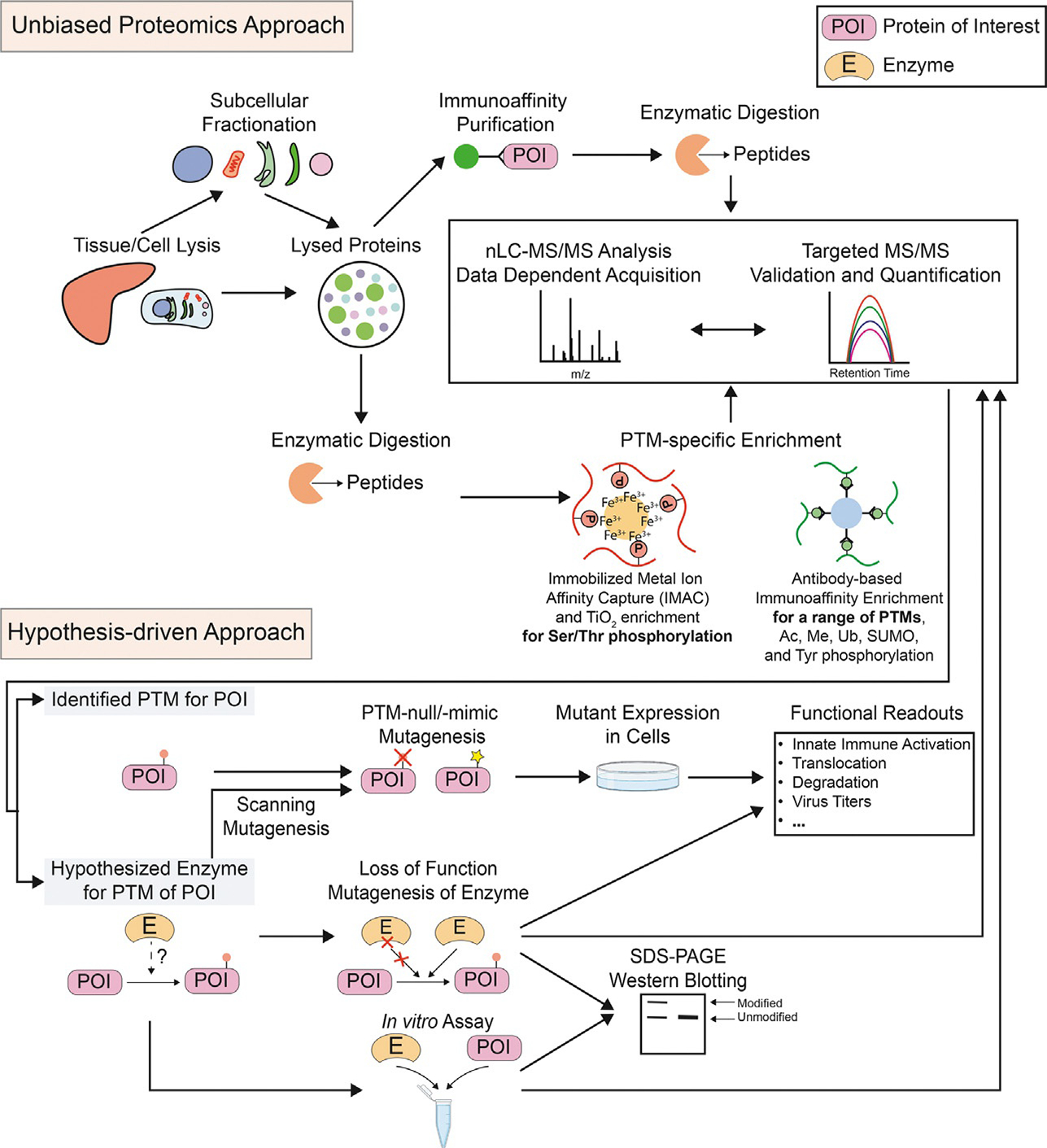
Representative biochemical and proteomic methods for identifying PTMs and modifying enzymes for DNA sensors. (Top) In unbiased proteomics approaches, following tissue/cell lysis and optional subcellular fractionation, immunoaffinity purification can be performed using antibodies that target the DNA sensor of interest. The isolated proteins will be subjected to enzymatic digestion and nano liquid chromatography-tandem mass spectrometry (nLC-MS/MS) to identify modified peptides on the DNA sensor itself, as well as any interacting protein which may serve as a potential modifying enzyme. Alternatively, one can carry out PTM-specific enrichment after enzymatic digestion of the lysed proteins. This allows for systematic detection of specific modified peptides in the proteome. Ac, acetylation; Me, methylation; SUMO, SUMOylation; Ub, ubiquitination. (Bottom) In contrast, hypothesis-driven approaches usually begin with prior knowledge of candidate PTM sites or modifying enzymes, which can be followed up by in-depth characterization using mutagenesis, in vitro enzymatic assays, and in vivo cellular assays. The impact of PTM-null or loss of function enzymes in these assays can be monitored by SDS-PAGE/western blot, functional readouts, or mass spectrometry.
More recently, with the rapid development of analytical and proteomic technologies, researchers can detect and analyze PTMs from a wide range of tissues and cells with increased specificity and sensitivity (Fig. 1, top) (Gillen et al., 2020; Greco et al., 2014). Nonetheless, as PTMs are often present at low stoichiometry, adequate depth of analysis is often achieved with biochemical fractionation or protein and peptide (immuno)affinity enrichment techniques (Fig. 2, top). For example, if the goal is to identify different types of PTMs present on a POI, the enrichment of the protein by immunoaffinity purification coupled with mass spectrometry (IP-MS) is commonly performed. IP-MS approaches benefit from high sequence coverage of the POI, and therefore, have the potential to identify the greatest number of unique PTM sites. For example, IP-MS analyses of IFI16 and cGAS have contributed the majority of the known phosphorylation and acetylation sites, accelerating the identification of PTM sites that regulate innate immune signaling (Dai et al., 2019; Li et al., 2012; Song et al., 2020a,b). In addition to studying multiple PTMs of one protein of interest, mature proteomic technologies are available to broadly detect and quantify certain types of PTMs on different proteins across diverse cellular and tissue proteome landscapes (Ke et al., 2016). Enrichment strategies have been developed for different PTM types, such as the enrichment of phosphorylated peptides by affinity-based approaches using metal oxides, e.g., titanium dioxide (TiO2) (Larsen et al., 2005; Pinkse et al., 2004) or immobilized metal ions (IMAC), e.g., Fe3+, Ti4+, or Zn4+ (Beausoleil et al., 2004; Feng et al., 2007; Nuhse et al., 2003; Zhou et al., 2013b) (Fig. 2, top). These core enrichment techniques have provided the foundation on which phosphorylation signaling networks have been studied and cataloged at the proteome scale (Hornbeck et al., 2015; Olsen et al., 2006; White and Wolf-Yadlin, 2016; Yu et al., 2019). As PTMs are exquisitely regulated in cellular space and time, proteome scale characterizations of these PTM dynamics present unique challenges. Yet, recent work shows promise in addressing some of these issues (Lun and Bodenmiller, 2020; Martinez-Val et al., 2021). For example, recently a high-throughput approach was developed to decipher spatio-temporal phospho-proteome regulation using sequential cell fractionation and titanium IMAC (Bekker-Jensen et al., 2020), coupled with rapid LC-MS/MS and data independent acquisition (Martinez-Val et al., 2021).
Although TiO2- and IMAC-based methods have been shown to provide effective enrichment of peptides containing Ser and Thr phosphorylation sites, they are known to be less suitable for enriching for Tyr phosphorylated peptides. Therefore, immunoaffinity purifications using anti-phosphotyrosine antibodies are often used as a complementary enrichment method (Rush et al., 2005). Antibody-based immunoaffinity approaches have been proven valuable for investigating a range of different PTMs, including acetylation, methylation, ubiquitination, and SUMOylation (Choudhary et al., 2009; Guo et al., 2014; Hendriks et al., 2018; Kim et al., 2011; Murray et al., 2018; Zhang et al., 2009). The reliance on antibodies comes with their own inherent drawbacks, including specificity and affinity issues. For example, motif bias has been observed with anti-acetyllysine antibodies, motivating the use of a cocktail of monoclonal antibodies to enrich acetylated lysines under different sequence contexts (Shaw et al., 2011; Svinkina et al., 2015). Specific antibodies against mono- and dimethyl arginine, and mono-, di-, and trimethyl lysine have been developed for methylation enrichment (Guo et al., 2014; Rothbart et al., 2015), while ubiquitination can be enriched by antibodies against the diglycyl lysine (K-GG) ubiquitin (Ub) remnant after trypsinization (Bustos et al., 2012). In addition, a linear-linkage-specific antibody can be applied for IP-MS analysis of polyubiquitinations (Matsumoto et al., 2012). Overall, both affinity and immunoaffinity-based approaches can introduce biases for specific motifs. Therefore, a combination of chemistries (Herring et al., 2015; Yue et al., 2015) and antibodies (Carabetta et al., 2016; Svinkina et al., 2015; Zhang et al., 2009) provide complementary methods to eliminate or reduce these biases.
In addition to identification of PTM site-localization, mass spectrometry enables quantitative analysis of PTMs. The advent of accurate mass and high-resolution mass spectrometers coupled to nano liquid chromatography separations (nLC) have facilitated global quantification of intact modified peptides by label-free methods using MS1 peak intensity or area under the curve metrics (Montoya et al., 2011). Additionally, targeted mass spectrometry approaches, such as parallel reaction monitoring (PRM) (Peterson et al., 2012), have further increased the sensitivity and specificity of detection (Manes and Nita-Lazar, 2018). These targeted approaches are designed to use signature features (nLC retention time, peptide mass) and fragmentation information to uniquely detect and quantify a peptide of interest. In contrast to global quantification, targeted-MS/MS quantification is performed at the MS/MS level, i.e., using fragment ions. These targeted approaches can be designed for the detection of hundreds of proteins, comprised of either unmodified or modified peptides. Once the signature parameters are determined, the assays are transferable between different laboratories. This method improves quantification sensitivity and reproducibility, which are particularly welcome benefits when analyzing low abundance PTMs that are modulated under different biological conditions (Song et al., 2020a). In addition, targeted-MS/MS is uniquely suited to resolve ambiguities in the sites of modification when multiple potential residues are present in the modified peptide sequence. Overall, this method provides an effective mean for detecting PTMs for which site-specific antibodies are not available and for accurately quantifying changes in a certain modification during a biological process.
3. DNA sensors are regulated by a combination of PTMs
One way that PTMs affect protein activity is by causing conformational changes to the protein structure. For DNA sensor proteins, these conformation changes may impact their DNA binding ability, enzymatic activity, and/or protein-protein interactions. PTMs may also influence the subcellular localization or turnover of DNA sensor proteins. This section highlights three prominent mechanisms by which PTMs regulate DNA sensors: enzymatic activity (Section 3.1), subcellular localization (Section 3.2), and half-life (Section 3.3). The subset of PTMs discussed below and that provide these functional molecular toggles are enumerated in Table 1 and graphically summarized in Fig. 3.
Fig. 3.
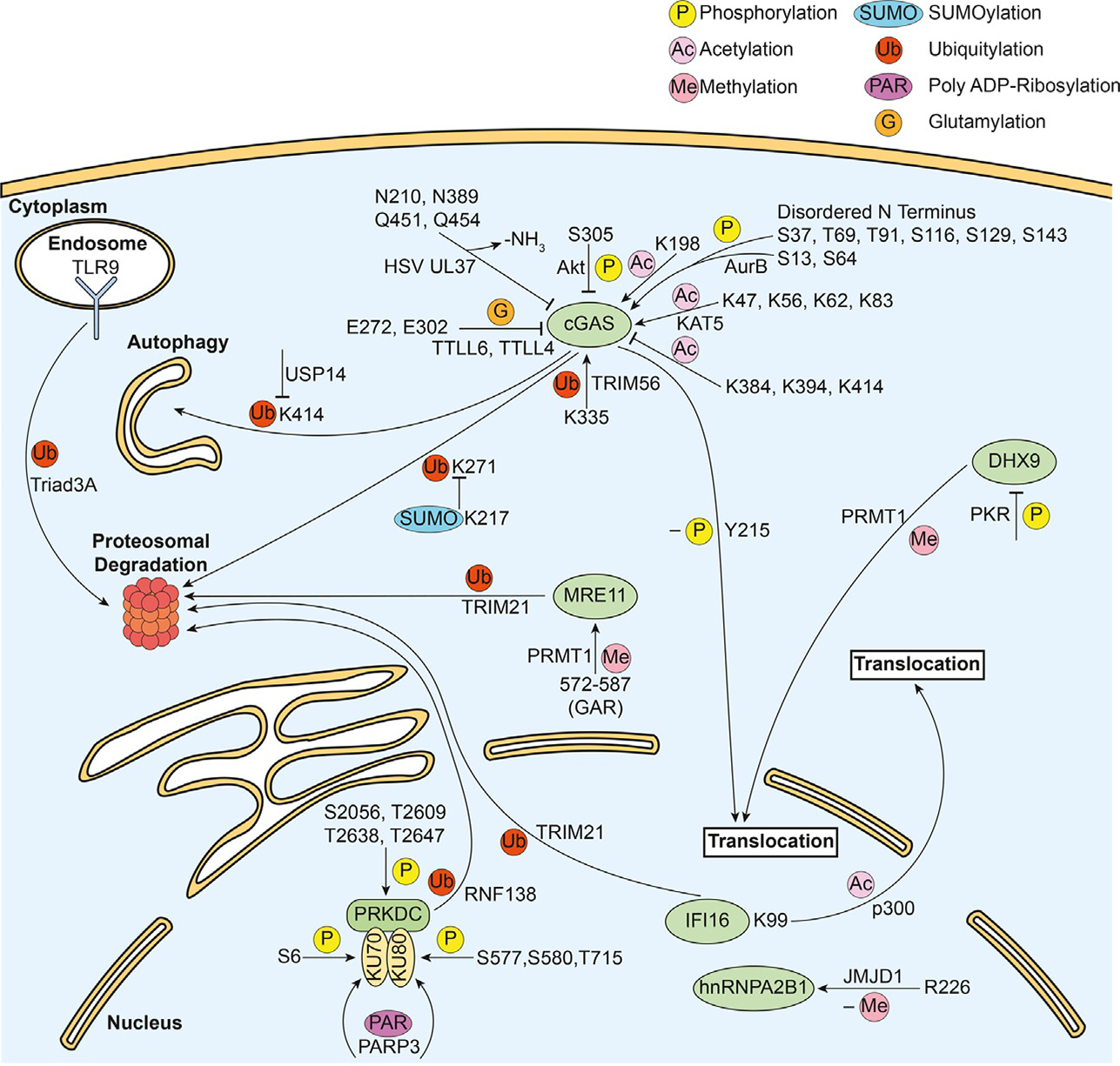
Characterized PTMs on DNA sensors and their reported functions. The corresponding modifying enzymes (if any) are presented beside each arrow. Illustrated are PTMs reported to impact DNA sensor functions by leading to activation/inhibition of sensor-induced immune signaling, intracellular shuttling of the sensor, or proteasome- or autophagy-mediated sensor degradation.
3.1. PTM-mediated activation or inhibition of DNA sensor activity
3.1.1. cGAS
Perhaps not surprisingly, the most well-studied cytosolic DNA sensor, cGAS, also has the distinction of possessing the most extensive compilation and functional characterization of PTMs, with modifications on at least 42 different amino acid residues. Interestingly, opposing regulation of cGAS functions is achieved not only through its variety of PTMs, but also for the same type of PTM, when present at different sites. The first functionally characterized PTM on cGAS was phosphorylation at Ser305 (Seo et al., 2015). During vesicular stomatitis virus (VSV) infection in L929 cells, Akt kinase mediated Ser305 phosphorylation was shown to down-regulate cGAS-dependent immune signaling (Seo et al., 2015). The inhibition of enzymatic activity is consistent with the location of Ser305 within the catalytic core of cGAS. In addition to Ser305, multiple phosphorylation sites have been identified (Ser37, Ser116, Ser201, Ser221, Ser263), however, mutational analyses at these residues indicated that none of these phosphorylations significantly impact cGAS-mediated immune signaling or apoptosis induction (Song et al., 2020a). Most recently, cGAS has been demonstrated to be hyperphosphorylated at its N-terminus during mitosis, with Ser13, Ser37, Ser64, Thr69, Thr91, Ser116, Ser129, and Ser143 found as the phosphorylated residues (Li et al., 2021). Aurora B was identified as the kinase that phosphorylates the cGAS N-terminus, and mutagenesis of these residues showed that these phosphorylations inhibit cGAS activity. The N-terminus was further characterized as being necessary for sensing chromatin. Therefore, the hyperphosphorylation likely inhibits chromatin-binding cGAS activity through the negatively charged phosphate groups, providing a regulatory mechanism to prevent an autoimmune response (Li et al., 2021).
cGAS is also a target of glutamylation and lysine ubiquitination. The interaction of cGAS with the E3 ubiquitin ligase TRIM56, promotes monoubiquitination of Lys335, resulting in an increase in cGAS dimerization, DNA-binding ability, and cGAMP production (Seo et al., 2018). E3 ubiquitin ligase TRAF6 promotes cGAS polyubiquitination, and this polyubiquitination upregulates cGAS-dependent IFN signaling (Chen and Chen, 2019). cGAS is also modified at two glutamate residues by glutamylation. Glutamylation is a reversible PTM that has been well-characterized in the regulation of microtubule dynamics (Garnham et al., 2015). Glutamylation forms a covalent, “peptide bond” between the γ-carboxy group of the target protein and the amino group of free glutamate. Similar to ubiquitination, this modification supports the addition of multiple PTM. In cGAS, two glutamylation sites were reported, both of which inhibit cGAS function, but with different mechanisms. The polyglutamylation of Glu272 by tubulin tyrosine ligase-like protein 6 (TTLL6) weakens its DNA binding ability, while monoglutamylation at Glu302 by TTLL4 dampens its synthase activity (Xia et al., 2016). Additionally, deamidation of Asn and Gln residues occurs on cGAS during HSV-1 infection. Specifically, mouse cGAS is deamidated by HSV-1 pUL37 both in cells and in vitro at Asn196, Asn377, Gln436 and Gln439 (corresponding to Asn210, Asn389, Gln451 and Gln454 of human cGAS). While single deamidations do not abolish cGAS function, three or four co-occurring deamidations led to a reduction in cGAS-dependent cytokine expression due to impaired enzymatic activity (Zhang et al., 2018).
Recently, many cGAS acetylation sites (Lys7, Lys21, Lys47, Lys50, Lys62, Lys63, Lys82, Lys83, Lys198, Lys285, Lys292, Lys355, Lys384, Lys392, Lys394, Lys414, Lys432, and Lys479) have been discovered using mass spectrometry (MS) (Dai et al., 2019; Song et al., 2020a,b). Among these sites, acetylation of Lys47, Lys56, Lys62, Lys83, and Lys198 appear to enhance cGAS immune signaling function (Dai et al., 2019; Song et al., 2020a,b). In contrast, Lys384, Lys394, and Lys414 acetylations impair cGAS enzymatic activity by inhibiting cGAMP production, while cGAS-DNA binding remains largely preserved and its dimerization is unaffected (Dai et al., 2019). In addition to inhibiting cytokine induction, Lys384 and Lys414 acetylations inhibit cGAS-dependent apoptosis (Song et al., 2020a). Interestingly, the regulation of Lys384 and Lys414 acetylation status is mediated in part through opposing acetylation and deacetylation by aspirin and histone deacetylase 3 (HDAC3), respectively (Dai et al., 2019). While the roles of lysine acetylation during active pathogen infections remains to be fully elucidated, several recent studies point to their regulatory roles. For instance, acetylation of Lys198 has been shown to be down regulated during herpesvirus infections with either HSV-1 or HCMV. As Lys198 acetylation promotes cGAS activity, the reduced PTM level during infection might reflect a viral mechanism to inhibit cGAS-dependent signaling (Song et al., 2020a). Most recently, Lys47, Lys56, Lys62, and Lys83 were shown to be acetylated by KAT5, increasing the DNA-binding ability of cGAS (Song et al., 2020b). Upon HSV-1 infection, mice with inactivated KAT5 had lower cytokine responses and survival rates, indicating these cGAS acetylations positively regulate cGAS function in vivo (Song et al., 2020b).
3.1.2. DNA-PK
The DNA-dependent protein kinase (DNA-PK) is a serine/threonine protein kinase consisting of a catalytic subunit (DNA-PKcs, also known as PRKDC), as well as a Ku heterodimer composed of Ku70 (XRCC6) and Ku80 (XRCC5) subunits (Chan et al., 1999; Jin and Weaver, 1997). While DNA-PK has been generally thought of as playing a critical role in the DNA damage response (DDR) pathways, more recent evidence suggests that it can also function as a DNA sensor during virus infection. Interestingly, it can activate IRF3-dependent innate immunity independently of its DNA-PKcs kinase activity, suggesting that its function in DNA sensing and DDR may be decoupled (Ferguson et al., 2012). DNA-PK has also been found to activate a STING-independent DNA sensing pathway (Burleigh et al., 2020). Although the exact molecular mechanisms underlying DNA-PK’s DNA sensing abilities remain to be elucidated, it is evident that this protein contributes to the regulation of innate immunity in response to foreign DNA.
The catalytic subunit of DNA-PK, PRKDC, is regulated by multiple phosphorylations. PRKDC is autophosphorylated on Ser2056, Ser2612, Thr2609, Thr2638 and Thr2647. Autophosphorylation induces a conformational change that leads to remodeling of the DNA-PK complex, a requisite for efficient end processing during DNA repair (Chan et al., 2002; Douglas et al., 2002). Protein phosphatase 5 catalyzes the dephosphorylation of PRKDC. The phosphorylation status of these residues is tightly regulated as hypo- or hyperphosphorylation of these sites leads to increased sensitivity to DNA damage induction (Wechsler et al., 2004).
The Ku70 and Ku80 subunits of DNA-PK are also regulated by PTMs. Both Ku70 and Ku80 are physiological targets of PRKDC. Researchers found Ser51 in Ku70 could be phosphorylated by PRKDC in vitro, however, this phosphorylation did not affect DNA repair (Jin and Weaver, 1997). Subsequently, it was found that Ku70 is phosphorylated at Ser6, and Ku80 is phosphorylated at Ser577, Ser579, Ser580 and Thr715, and phosphorylation can enhance the helicase activity of their respective Ku subunits (Chan et al., 1999). In addition, Ku70/Ku80 can be ADP-ribosylated by Poly(ADP-ribose) polymerase 3 (PARP3). This is functionally relevant as the depletion of PARP3 inhibits recruitment of Ku80 to sites of DNA damage and reduces double strand break repair (Beck et al., 2014). Finally, Ku80 can be ubiquitinated by RNF138, leading to the removal of Ku complex from the DNA breaks (Ismail et al., 2015).
3.1.3. DHX9
Nuclear DNA helicase II (NDH II), also known as ATP-dependent RNA helicase A (RHA), is encoded by the DHX9 gene and functions as a sensor for both viral DNA and RNA (Kim et al., 2010). Although we know DHX9 can be regulated by phosphorylations, the impact of the modifications on its DNA or RNA sensing ability is not clear. DHX9 is phosphorylated by PRKDC, the catalytic subunit of DNA-PK, and this phosphorylation can be stimulated by both viral DNA and RNA, consistent with its functions in DNA and RNA metabolism (Yang et al., 2012). In addition, DHX9 is phosphorylated by protein kinase R (PKR), which has an overall inhibitory effect on DHX9 function (Sadler et al., 2009). DHX9 is known to regulate expression of the HIV-1 transactivation response via direct binding with viral RNA. Of note, the regions of DHX9 that bind viral RNA are also targets of PKR phosphorylation, and thus present a plausible therapeutic target (Sadler et al., 2009).
3.1.4. MRE11
MRE11 has been shown to be recruited to exogenous dsDNA in the cytosol, and can induce STING-dependent Type I interferon expression (Kondo et al., 2013). In addition to its DNA sensing function, MRE11 is a DNA double-strand break repair protein, containing a DNA binding domain and a phosphodiester domain, hence functioning also as an ssDNA endonuclease and dsDNA exonuclease. The exonuclease function is involved in deletional nonhomologous end joining (D-NHEJ), contributing to DNA damage repair (Boisvert et al., 2005; Zhuang et al., 2009). The exonuclease function of MRE11 is positively regulated by arginine methylation in its glycine-arginine-rich (GAR) region, between amino acids 572 and 587 (Boisvert et al., 2005; Zhuang et al., 2009). Although methylation is required for MRE11 functions during DNA damage, it remains unknown whether the DNA sensor activity is also affected by methylation (Zhuang et al., 2009).
3.1.5. hnRNPA2B1
Prior to the characterization of hnRNPA2B1 as a DNA sensor, methylation was identified within its arginine-glycine-glycine–rich (RGG) domain (Gary and Clarke, 1998). Recent work characterizing hnRNPA2B1 functions during HSV-1 infection found that it senses nuclear DNA (Wang et al., 2019). Specifically, nuclear hnRNPA2B1 binds viral DNA, dimerizes, then becomes demethylated within the RGG domain at Arg226, upon which the homodimer translocates to the cytoplasm to initiate downstream type I interferon signaling (Wang et al., 2019). This dimerization-dependent demethylation is mediated by the arginine demethylase JMJD6, as an JMJD6 inhibitor prevented demethylation and also inhibited downstream signaling. Given that the JMJD6 inhibitor did not affect hnRNPA2B1 translocation, it is thought that demethylation at Arg226 is selectively required for hnRNPA2B1 activation (Wang et al., 2019).
3.2. PTM-controlled translocation of DNA sensors
In addition to the direct regulation of the activity of DNA sensors, PTMs can also indirectly regulate their functions by controlling their subcellular localization. This allows for a spatially restricted activation of DNA sensors. For example, IFI16, commonly regarded as a nuclear DNA sensor, has been shown to be acetylated at K99 within one of its nuclear localization sequence motifs by the acetyltransferase p300, leading to its localization to the cytoplasm (Li et al., 2012). This allows for its DNA sensing function to occur in the cytoplasm, as well as for the activation of inflammasome, leading to the expression of antiviral cytokines (Ansari et al., 2015; Li et al., 2012). In addition, the DEAH-box family protein DHX9, which was found to recognize cytosolic microbial DNA in plasmacytoid dendritic cells (pDCs) (Kim et al., 2010), can interact with and be methylated by PRMT1, which mediates its nuclear import (Smith et al., 2004). Although direct evidence is lacking, it is possible that PRMT1-mediated DHX9 methylation negatively regulates the nucleic acid sensing of DHX9 to prevent overactivation. This is supported by a previous study that reported a higher proinflammatory response in myeloid-specific PRMT1 knock-out mice (Tikhanovich et al., 2017).
Changes in DNA sensor subcellular localization can also be regulated by PTMs outside the context of pathogen invasion. cGAS is phosphorylated at Y215 by B-lymphoid tyrosine kinase (BLK), facilitating its cytoplasmic retention. Upon dephosphorylation, cGAS was shown to shuttle into the nucleus, where it inhibits PARP1-mediated homologous recombination pathways upon DNA damage (Liu et al., 2018). These initial reports of localization-dependent activities of DNA sensors provide a promising avenue for future mechanistic investigations of PTMs with currently unknown functions.
3.3. PTM-modulated protein degradation of DNA sensors
It is critical for cells to either maintain or return to a state of immune homeostasis in the absence of or after being challenged by pathogens, respectively, in order to prevent overactivation of immune responses that are linked to different human diseases. One of the major negative feedback mechanisms is the degradation of the DNA sensors that initiated innate immune signaling. Frequently, this is achieved through ubiquitination of the proteins to facilitate degradation through the proteasome. Both IFI16 and DDX41 have been shown to be ubiquitinated by the E3 ubiquitin ligase TRIM21 for subsequent degradation (Li et al., 2019; Zhang et al., 2013a). TRIM21-mediated degradation of IFI16 is dependent on STING activation, a downstream signaling protein of IFI16. This highlights the importance of negative feedback regulation in the context of DNA sensing. Similarly, the Toll-like receptor proteins TLR4 and TLR9 were found to be ubiquitinated by Triad3A and targeted for degradation to inhibit their signaling functions (Chuang and Ulevitch, 2004).
cGAS was reported to be ubiquitinated at K271 and K414 which facilitate degradation through the proteasome and p62-mediated autophagy, respectively (Chen et al., 2016a; Hu et al., 2016). In addition, both sites have mechanisms in place to inhibit their polyubiquitination. Hu et al. reported that in uninfected cells, cGAS can be SUMOylated at K217, which resides in relative proximity to the ubiquitination site K271, thereby being proposed to serve as a steric hindrance to the E3 ligase (Hu et al., 2016). Chen et al. found that TRIM14, which can be upregulated by interferons, recruits the ubiquitin carboxyl-terminal hydrolase (USP14) to K414 to cleave the polyubiquitination chains, thereby preventing the interaction of cGAS with p62 (Chen et al., 2016a). cGAS was also found to be SUMOylated at K464 early during HSV-1 infection, but later de-SUMOylated by sentrin-specific protease 2 (SENP2) to allow for polyubiquitination on the same site, promoting its degradation (Hu et al., 2016). Given the proximity of K414 and K464, it is also possible that SUMOylation on K464 could sterically inhibit the interaction between cGAS and p62 mediated by K414 polyubiquitination. Altogether, these modifications, their crosstalk, and their functional significance highlight the intricate balance cells have to maintain between activation and inhibition of DNA sensors.
4. Virus-induced PTMs for dampening or evading the host innate immune response
The abilities of PTMs to regulate DNA sensor functions are not exclusive to the host cell. Viruses often hijack host PTM modifying enzymes or alternatively encode viral enzymes that directly modify DNA sensors to promote immune evasion and facilitate viral replication. For instance, cGAS was reported to be deamidated in its Mab21 enzymatic domain by the viral deamidase pUL37 from HSV-1 (Zhang et al., 2018). Deamidation of human cGAS at one single site (N210) was shown to be sufficient to decrease the cGAMP-synthesizing activity of cGAS and inhibit downstream interferon and ISG production. Furthermore, the N210 site was found to be under strong positive selection through analysis of the protein sequences in nonhuman primates. This further supports the notion that the deamidation site in cGAS is an active target for viral evasion (Zhang et al., 2018).
IFI16 is also known to be a primary target of viral proteins. Orzalli et al. reported that, during HSV-1 infection, the RING finger domain of the viral protein ICP0 promotes the degradation of IFI16 by acting as an E3 ubiquitin ligase (Orzalli et al., 2012). Although others have reported that ICP0 is neither necessary nor sufficient for IFI16 degradation (Cuchet-Lourenco et al., 2013), the HSV-1-induced degradation of IFI16 is well recognized and shown to be mediated via the targeting of the IFI16 pyrin domain (Diner et al., 2015b). The β-herpesvirus HCMV also acquired several mechanisms to suppress IFI16’s DNA sensing ability, one of which is driven by regulatory PTM event. The viral kinase pUL97 phosphorylates IFI16, causing its mislocalization to the cytoplasm early in infection, while later in infection, trapping IFI16 into egressing virions, likely by promoting the interaction between IFI16 and the multivesicular body protein Vps4A (Dell’Oste et al., 2014). Although the specific site of pUL97-mediated IFI16 phosphorylation is currently unknown, many of the previously reported but as yet uncharacterized phosphorylation sites reside near the nuclear localization signal and are therefore candidates for regulating subcellular distribution during infection. Although not directly added to the DNA sensor, but rather impacting its function, PTM-driven regulation was also shown to counteract one of the mechanisms of HCMV inhibition of IFI16. HCMV was reported to use its tegument protein pUL83 to clamp the IFI16 pyrin domain, preventing its oligomerization and the ability of IFI16 to induce immune signaling. Host cell induced phosphorylation at Ser364 within the pyrin-associating domain of pUL83 was shown to restore the ability of IFI16 to oligomerize, likely by interfering with the ability of pUL83 to fully clamp the IFI16 pyrin domain (Li et al., 2013). To date, no systematic comparison of the PTM landscape during different types of virus infections has been performed. It is expected that continued global host PTM characterization, as well studies focused on DNA sensors, under various infection conditions will help point to unique or shared PTM signatures that underlie mechanisms of host defense or virus immune evasion.
5. PTMs on DNA sensors affect disease progression and treatment
Innate immunity is a double-edged sword: the activation of innate immune system protects us from pathogen infection and cellular damages; however, its over-activation drives autoimmune diseases and compromises human health. As self-DNA sensing is linked to autoimmune and autoinflammatory diseases, cells employ multiple DNases to cleave aberrantly located and accumulated DNA, and thus avoid self-immune activation (Motwani et al., 2019). Therefore, mutations on DNases could cause autoimmune diseases in a DNA-sensor dependent manner. For example, the loss of DNase II endonuclease activity would lead to excessive production of type I interferons, and further lead to neonatal anemia, membranoproliferative glomerulonephritis, liver fibrosis, and deforming arthropathy (Rodero et al., 2017). Another clinically relevant example is the loss-of-function of three-prime repair exonuclease 1 (TREX1; also known as DNase III), which can lead to systemic lupus erythematosus and Aicardi–Goutieres Syndrome (AGS) (Crow and Manel, 2015; Stetson et al., 2008). Aicardi-Goutières syndrome (AGS) is a genetic disorder that affects the white matter in the brain and spinal cord. AGS patients with DNA 3′ repair exonuclease 1 (TREX1) loss-of-function mutations cannot degrade mislocalized cytosolic DNA (Crow and Manel, 2015). This causes elevated levels of cGAS-dependent IFN production, contributing to autoimmune signaling that destructively targets the white matter (Gao et al., 2015). A promising treatment for AGS patients was suggested to be the inhibition of cGAS using the common anti-inflammatory drug aspirin, which directly acetylates cGAS at Lys384 and Lys414, leading to reduced interferon signaling. In patient cells and an AGS mouse model, aspirin effectively represses AGS symptoms (Dai et al., 2019). This demonstrates the potential of aspirin to broadly mitigate the impact of cGAS-dependent autoimmunity on disease progression.
6. Future perspectives
The combination of hypothesis-driven molecular approaches and discovery-based global proteomics methods has facilitated the functional characterization of many PTMs on DNA sensors. Yet, owing to the systems-wide advantages of proteomic approaches to simultaneously survey the PTM landscape, novel PTMs are being identified faster than they can be functionally characterized. For example, IFI16 has been found to be SUMOylated and to also possess many phosphorylation sites that remain to be linked to specific protein functions (Impens et al., 2014; Li et al., 2012). Moreover, we have recently reported that cGAS is decorated with numerous acetylation and phosphorylation sites that could differentially impact immune signaling and apoptosis (Song et al., 2020a), but additional characterization of these sites is needed to determine whether they reflect bona fide regulatory elements during pathogen infections.
The ability of PTMs to govern a protein’s localization, for example, by inducing shuttling events, is a fascinating post-translational regulatory mechanism, particularly in the context of DNA sensors. The core functionality of DNA sensors to surveil the intracellular environment requires precise spatial and temporal regulation of DNA recognition. Given the different cellular routes for the trafficking of pathogenic DNA, sensors must have the ability to respond to these diverse stimuli. Mechanistically, much remains to be understood about how PTMs govern protein localization and how they participate in host immune defense and viral immune evasion. However, these early reports of PTM-dependent regulation of sensor localization have shown the remarkable ability of a single modification to establish the DNA sensor’s localization. Moreover, the fact that localization-defining PTM sites use both addition and removal mechanisms suggests a mechanistic flexibility that can leverage opposing PTM modifying enzymes, e.g., kinases and phosphatases. As DNA sensor PTMs continue to be characterized, it is likely that a greater appreciation will be realized for the ability of PTMs to act as dynamic, stimulus-dependent molecular switches to control protein translocations.
Recent work suggests that DNA sensor PTMs do not exclusively regulate protein activities in isolation, but rather through a combination of co-occurring modifications that give rise to different proteoforms. For example, modifications that occur on lysine residues (acetylation, ubiquitination, and SUMOylation) are often found to target the same residue, and thus the existence of one PTM precludes the others. A more intricate mechanism of regulation occurs when proximal modified residues either always co-occur, or are present only in isolation on the same protein molecule. Given the large number of potential residues and types of modifications, characterizing the proteoforms of a single gene product is no trivial task (Schaffer et al., 2019). Toward this goal, intact, often lower molecular weight proteins are directly analyzed by mass spectrometry. This approach is commonly referred to as top-down MS. In contrast to bottom-up proteomics, top-down analysis is able to better preserve unique PTM combinations (Schaffer et al., 2019). Depending on the proteoform complexity and molecular weight of a particular gene product, a variation on top-down MS, called middle-down MS, can be used. Middle-down approaches use a selective enzymatic proteolysis that generates larger polypeptides than traditional digestion methods (e.g., using trypsin), thereby retaining regions of the protein intact for mass spectrometry analysis (Wu et al., 2012). As these approaches continue to be refined (Cristobal et al., 2017; Cupp-Sutton and Wu, 2020), they have the potential to provide a unique perspective to proteoform-specific functions of DNA sensors.
In the interim, clues to the inter-relationships between DNA sensor PTMs are emerging. For example, a mapping study of the SUMO proteome, found a significant degree of co-modification with phosphorylation (Hendriks et al., 2017), and crosstalks were also investigated for ADP-ribosylation and phosphorylation (Daniels et al., 2020), to name just a few. So far, a limited number of examples of PTM co-regulation are known for DNA sensors. For example, cGAS ubiquitination of cGAS (K271) is inhibited by its SUMOylation (K217) (Hu et al., 2016). Thus, SUMOylation provides a “brake” on ubiquitin-dependent proteasomal degradation of cGAS. The existence of histone “PTM code” is well known as a primary determinant of their epigenetic regulation (Onder et al., 2015). While a PTM code for DNA sensors is still being explored, it is likely that as their PTM landscapes become richer, a systems-level synthesis of PTM interdependence will emerge. Also, comparison of PTM codes between DNA sensors may provide novel mechanisms for how specific immune responses achieve specificity despite redundancies and convergent signaling pathways.
Globally, the link between disease states and the PTM profiles of individual patients is of great clinical interest (Thygesen et al., 2018). Analogous to patient-specific proteome profiling, the PTM status of patients may also serve to establish disease biomarkers or contribute to understanding disease progression, and therefore, assist more in the development of precise therapeutics. From the perspective of DNA sensors, their cooperation and convergence to stimulate interferon signaling suggests that future therapies may benefit from targeting multiple DNA sensors, and present selective therapeutic targets. This underscores the need to continue characterizing functional PTMs on DNA sensor proteins, and specifically, PTMs that are associated with overactivation of the immune response and self-DNA sensing states. Profiling the PTM landscape of DNA sensors in inflammatory states may be informative, for example, during virus-associated inflammation or in lung inflammatory diseases where the STING-mediated sensing pathway plays a prominent role (Benmerzoug et al., 2019; Heil and Brockmeyer, 2019). Ultimately, by increasing PTM coverage across DNA sensors, we will gain mechanistic insights into key post-translational regulatory events that fine-tune sensor activities across a wide range of physiological time scales.
Acknowledgments
Funding was provided by NIH NIGMS (GM114141) and by the Edward Mallinckrodt Foundation to I.M.C., and China Scholarship Council (CSC) scholarship (201506210052) to B.S.
Abbreviations
- AdV
adenovirus
- AGS
Aicardi-Goutières syndrome
- AIM2
absent in melanoma 2
- BLK
B-lymphoid tyrosine kinase
- cGAMP
cyclic GMP-AMP
- cGAS
cyclic-GMP-AMP synthase
- CpG
cytosine-phosphate-guanine
- DAI
DNA-dependent activator of IFN-regulatory factor
- DDR
DNA damage response
- DDX41
DEAD box polypeptide 41
- DHX36
DEAH box helicase 36
- DHX9
DEAH box helicase 9
- DNA-PK
DNA-dependent protein kinase
- DNA-PKcs/PRKDC
DNA-dependent protein kinase catalytic subunit
- D-NHEJ
deletional nonhomologous end joining
- ER
endoplasmic reticulum
- GAR
glycine-arginine-rich
- HCMV
human cytomegalovirus
- HDAC
histone deacetylase
- HIV-1
human immunodeficiency virus type 1
- hnRNPA2B1
heterogeneous nuclear ribonucleoproteins A2/B1
- HPV
human papilloma virus
- HSV-1
herpes simplex virus type 1
- IFI16
interferon-inducible protein 16
- IFIX
interferon-inducible protein X
- IFN
interferon
- IMAC
immobilized metal affinity chromatography
- IP
immunoaffinity purification
- IRF3
interferon regulatory factor 3
- ISG
interferon-stimulated gene
- Jak
Janus kinase
- JMJD6
histone arginine demethylase
- KAT5
lysine acetyltransferase 5
- KSHV
Kaposi’s sarcoma-associated herpesvirus
- LRRFIP1
leucine-rich repeat flightless interacting protein-1
- m6A
N6-methyladenosine
- MRE11
meiotic recombination 11 homolog A
- MS
mass spectrometry
- MyD88
myeloid differentiation primary response protein
- nLC
nano liquid chromatography
- OASL
2′−5′-oligoadenylate synthase-like protein
- PAMP
pathogen-associated molecular pattern
- PARP
poly ADP-ribose polymerase
- pDC
plasmacytoid dendritic cell
- PKR
protein kinase RNA-activated
- POI
protein of interest
- PRM
parallel reaction monitoring
- PRMT1
protein arginine N-methyltransferase 1
- PRR
pattern recognition receptor
- PTM
post-translational modifications
- PyV
polyomavirus
- RGG
arginine-glycine-glycine–rich
- RNA Pol III
RNA polymerase III
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SENP2
sentrin-specific protease 2
- STAT
signal transducer and activator of transcription
- STING
endoplasmic reticulum-associated adaptor
- TBK1
TANK-binding kinase 1
- TLR
Toll-like receptor
- TREX1
DNA 3′ repair exonuclease 1
- Triad3A
triad domain-containing protein 3
- TRIM14
tripartite motif-containing protein 14
- TRIM21
tripartite motif-containing protein 21
- TRIM56
tripartite motif-containing protein 56
- TTLL4
tubulin polyglutamylase 4
- TTLL6
tubulin polyglutamylase 6
- USP14
ubiquitin-specific-processing protease 14
- VSV
vesicular stomatitis virus
- XRCC5
X-ray repair cross-complementing protein 6
- XRCC6
X-ray repair cross-complementing protein 5
References
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V, 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol 10 (10), 1065–1072. 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari MA, Dutta S, Veettil MV, Dutta D, Iqbal J, Kumar B, et al. , 2015. Herpesvirus genome recognition induced acetylation of nuclear IFI16 is essential for its cytoplasmic translocation, inflammasome and IFN-beta responses. PLoS Pathog. 11 (7), e1005019. 10.1371/journal.ppat.1005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, et al. , 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U. S. A 101 (33), 12130–12135. 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C, Boehler C, Guirouilh Barbat J, Bonnet ME, Illuzzi G, Ronde P, et al. , 2014. PARP3 affects the relative contribution of homologous recombination and nonhomologous end-joining pathways. Nucleic Acids Res. 42 (9), 5616–5632. 10.1093/nar/gku174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen DB, Bernhardt OM, Hogrebe A, Martinez-Val A, Verbeke L, Gandhi T, et al. , 2020. Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries. Nat. Commun 11 (1), 787. 10.1038/s41467-020-14609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerzoug S, Ryffel B, Togbe D, Quesniaux VFJ, 2019. Self-DNA sensing in lung inflammatory diseases. Trends Immunol. 40 (8), 719–734. 10.1016/j.it.2019.06.001. [DOI] [PubMed] [Google Scholar]
- Bian Y, Song C, Cheng K, Dong M, Wang F, Huang J, et al. , 2014. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J. Proteomics 96, 253–262. 10.1016/j.jprot.2013.11.014. [DOI] [PubMed] [Google Scholar]
- Biolatti M, Dell’Oste V, Pautasso S, Gugliesi F, von Einem J, Krapp C, et al. , 2018. Human cytomegalovirus tegument protein pp65 (pUL83) dampens type I interferon production by inactivating the DNA sensor cGAS without affecting sting. J. Virol 92 (6). 10.1128/JVI.01774-17, e01774–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Dery U, Masson JY, Richard S, 2005. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 19 (6), 671–676. 10.1101/gad.1279805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, et al. , 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol 10 (3), 266–272. 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Burleigh K, Maltbaek JH, Cambier S, Green R, Gale M Jr., James RC, Stetson DB, 2020. Human DNA-PK activates a STING-independent DNA sensing pathway. Sci. Immunol 5 (43), eaba4219. 10.1126/sciimmunol.aba4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos D, Bakalarski CE, Yang Y, Peng J, Kirkpatrick DS, 2012. Characterizing ubiquitination sites by peptide-based immunoaffinity enrichment. Mol. Cell. Proteomics 11 (12), 1529–1540. 10.1074/mcp.R112.019117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabetta VJ, Greco TM, Tanner AW, Cristea IM, Dubnau D, 2016. Temporal regulation of the Bacillus subtilis acetylome and evidence for a role of MreB acetylation in cell wall growth. mSystems 1 (3). 10.1128/mSystems.00005-16, e00005–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Ye R, Veillette CJ, Lees-Miller SP, 1999. DNA-dependent protein kinase phosphorylation sites in Ku 70/80 heterodimer. Biochemistry 38 (6), 1819–1828. 10.1021/bi982584b. [DOI] [PubMed] [Google Scholar]
- Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ, 2002. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 16 (18), 2333–2338. 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chen Y, 2019. Ubiquitination of cGAS by TRAF6 regulates anti-DNA viral innate immune responses. Biochem. Biophys. Res. Commun 514 (3), 659–664. 10.1016/j.bbrc.2019.05.022. [DOI] [PubMed] [Google Scholar]
- Chen M, Meng Q, Qin Y, Liang P, Tan P, He L, et al. , 2016a. TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol. Cell 64 (1), 105–119. 10.1016/j.molcel.2016.08.025. [DOI] [PubMed] [Google Scholar]
- Chen Q, Sun L, Chen ZJ, 2016b. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol 17 (10), 1142–1149. 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ, 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138 (3), 576–591. 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chockalingam A, Brooks JC, Cameron JL, Blum LK, Leifer CA, 2009. TLR9 traffics through the Golgi complex to localize to endolysosomes and respond to CpG DNA. Immunol. Cell Biol 87 (3), 209–217. 10.1038/icb.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. , 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325 (5942), 834–840. 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Chuang TH, Ulevitch RJ, 2004. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat. Immunol 5 (5), 495–502. 10.1038/ni1066. [DOI] [PubMed] [Google Scholar]
- Cristobal A, Marino F, Post H, van den Toorn HW, Mohammed S, Heck AJ, 2017. Toward an optimized workflow for middle-down proteomics. Anal. Chem. 89 (6), 3318–3325. 10.1021/acs.analchem.6b03756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Manel N, 2015. Aicardi-Goutieres syndrome and the type I interferonopathies. Nat. Rev. Immunol. 15 (7), 429–440. 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- Crow MS, Javitt A, Cristea IM, 2015. A proteomics perspective on viral DNA sensors in host defense and viral immune evasion mechanisms. J. Mol. Biol 427 (11), 1995–2012. 10.1016/j.jmb.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuchet-Lourenco D, Anderson G, Sloan E, Orr A, Everett RD, 2013. The viral ubiquitin ligase ICP0 is neither sufficient nor necessary for degradation of the cellular DNA sensor IFI16 during herpes simplex virus 1 infection. J. Virol 87 (24), 13422–13432. 10.1128/JVI.02474-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupp-Sutton KA, Wu S, 2020. High-throughput quantitative top-down proteomics. Mol. Omics 16 (2), 91–99. 10.1039/c9mo00154a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Huang YJ, He X, Zhao M, Wang X, Liu ZS, et al. , 2019. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell 176 (6), 1447–1460. e1414. 10.1016/j.cell.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels CM, Kaplan PR, Bishof I, Bradfield C, Tucholski T, Nuccio AG, et al. , 2020. Dynamic ADP-ribosylome, phosphoproteome, and interactome in LPS-activated macrophages. J. Proteome Res 19 (9), 3716–3731. 10.1021/acs.jproteome.0c00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Oste V, Gatti D, Gugliesi F, De Andrea M, Bawadekar M, Lo Cigno I, et al. , 2014. Innate nuclear sensor IFI16 translocates into the cytoplasm during the early stage of in vitro human cytomegalovirus infection and is entrapped in the egressing virions during the late stage. J. Virol 88 (12), 6970–6982. 10.1128/JVI.00384-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP, 2008. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U. S. A 105 (31), 10762–10767. 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner BA, Li T, Greco TM, Crow MS, Fuesler JA, Wang J, Cristea IM, 2015a. The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Mol. Syst. Biol 11 (1), 787. 10.15252/msb.20145808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner BA, Lum KK, Javitt A, Cristea IM, 2015b. Interactions of the antiviral factor interferon gamma-inducible protein 16 (IFI16) mediate immune signaling and herpes simplex Virus-1 immunosuppression. Mol. Cell. Proteomics 14 (9), 2341–2356. 10.1074/mcp.M114.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner BA, Lum KK, Toettcher JE, Cristea IM, 2016. Viral DNA sensors IFI16 and cyclic GMP-AMP synthase possess distinct functions in regulating viral gene expression, immune defenses, and apoptotic responses during herpesvirus infection. MBio 7 (6). 10.1128/mBio.01553-16, e01553–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, et al. , 2002. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem. J 368 (Pt. 1), 243–251. 10.1042/BJ20020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Ye M, Zhou H, Jiang X, Jiang X, Zou H, Gong B, 2007. Immobilized zirconium ion affinity chromatography for specific enrichment of phosphopeptides in phosphoproteome analysis. Mol. Cell. Proteomics 6 (9), 1656–1665. 10.1074/mcp.T600071-MCP200. [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL, 2012. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife 1, e00047. 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, et al. , 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341 (6148), 903–906. 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Li T, Li XD, Chen X, Li QZ, Wight-Carter M, Chen ZJ, 2015. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. U. S. A 112 (42), E5699–E5705. 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham CP, Vemu A, Wilson-Kubalek EM, Yu I, Szyk A, Lander GC, et al. , 2015. Multivalent microtubule recognition by tubulin tyrosine ligase-like family glutamylases. Cell 161 (5), 1112–1123. 10.1016/j.cell.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Clarke S, 1998. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol 61, 65–131. 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- Gillen J, Bridgwater C, Nita-Lazar A, 2020. Approaching complexity: systems biology and ms-based techniques to address immune signaling. Expert Rev. Proteomics 17 (5), 341–354. 10.1080/14789450.2020.1780920. [DOI] [PubMed] [Google Scholar]
- Gordon S, 2002. Pattern recognition receptors: doubling up for the innate immune response. Cell 111 (7), 927–930. 10.1016/s0092-8674(02)01201-1. [DOI] [PubMed] [Google Scholar]
- Greco TM, Diner BA, Cristea IM, 2014. The impact of mass spectrometry-based proteomics on fundamental discoveries in virology. Annu. Rev. Virol 1 (1), 581–604. 10.1146/annurev-virology-031413-085527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, et al. , 2014. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics 13 (1), 372–387. 10.1074/mcp.O113.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K, Prabakaran T, Laustsen A, Jorgensen SE, Rahbaek SH, Jensen SB, et al. , 2014. Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 33 (15), 1654–1666. 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA, 2017. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548 (7668), 466–470. 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, Brockmeyer NH, 2019. Self-DNA sensing fuels HIV-1-associated inflammation. Trends Mol. Med 25 (11), 941–954. 10.1016/j.molmed.2019.06.004. [DOI] [PubMed] [Google Scholar]
- Hendriks IA, D’Souza RC, Yang B, Verlaan-de Vries M, Mann M, Vertegaal AC, 2014. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol 21 (10), 927–936. 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, Treffers LW, Verlaan-de Vries M, Olsen JV, Vertegaal ACO, 2015. SUMO-2 orchestrates chromatin modifiers in response to DNA damage. Cell Rep. 10 (10), 1778–1791. 10.1016/j.celrep.2015.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal AC, Nielsen ML, 2017. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol 24 (3), 325–336. 10.1038/nsmb.3366. [DOI] [PubMed] [Google Scholar]
- Hendriks IA, Lyon D, Su D, Skotte NH, Daniel JA, Jensen LJ, Nielsen ML, 2018. Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun 9 (1), 2456. 10.1038/s41467-018-04957-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring LE, Grant KG, Blackburn K, Haugh JM, Goshe MB, 2015. Development of a tandem affinity phosphoproteomic method with motif selectivity and its application in analysis of signal transduction networks. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 988, 166–174. 10.1016/j.jchromb.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E, 2015. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43 (Database issue), D512–D520. 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. , 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458 (7237), 514–518. 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MM, Yang Q, Xie XQ, Liao CY, Lin H, Liu TT, et al. , 2016. Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity 45 (3), 555–569. 10.1016/j.immuni.2016.08.014. [DOI] [PubMed] [Google Scholar]
- Huang J, You H, Su C, Li Y, Chen S, Zheng C, 2018a. Herpes simplex virus 1 tegument protein VP22 abrogates cGAS/STING-mediated antiviral innate immunity. J. Virol 92 (15), e00841–18. 10.1128/JVI.00841-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZF, Zou HM, Liao BW, Zhang HY, Yang Y, Fu YZ, et al. , 2018b.Human cytomegalovirus protein UL31 inhibits DNA sensing of cGAS to mediate immune evasion. Cell Host Microbe 24 (1), 69–80.e64. 10.1016/j.chom.2018.05.007. [DOI] [PubMed] [Google Scholar]
- Impens F, Radoshevich L, Cossart P, Ribet D, 2014. Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc. Natl. Acad. Sci. U. S. A 111 (34), 12432–12437. 10.1073/pnas.1413825111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Gagne JP, Genois MM, Strickfaden H, McDonald D, Xu Z, et al. , 2015. The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nat. Cell Biol 17 (11), 1446–1457. 10.1038/ncb3259. [DOI] [PubMed] [Google Scholar]
- Jin S, Weaver DT, 1997. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO J. 16 (22), 6874–6885. 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Bottero V, Flaherty S, Dutta S, Singh VV, Chandran B, 2014. IFI16 restricts HSV-1 replication by accumulating on the HSV-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications. PLoS Pathog. 10 (11), e1004503. 10.1371/journal.ppat.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke M, Shen H, Wang L, Luo S, Lin L, Yang J, Tian R, 2016. Identification, quantification, and site localization of protein posttranslational modifications via mass spectrometry-based proteomics. Adv. Exp. Med. Biol 919, 345–382. 10.1007/978-3-319-41448-5_17. [DOI] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B, 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe 9 (5), 363–375. 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, et al. , 2010. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U. S. A 107 (34), 15181–15186. 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, et al. , 2011. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44 (2), 325–340. 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, et al. , 2013. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc. Natl. Acad. Sci. U. S. A 110 (8), 2969–2974. 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, et al. , 1995. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374 (6522), 546–549. 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ, 2005. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4 (7), 873–886. 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- Lei Z, Deng M, Yi Z, Sun Q, Shapiro RA, Xu H, et al. , 2018. cGAS-mediated autophagy protects the liver from ischemia-reperfusion injury independently of STING. Am. J. Physiol. Gastrointest. Liver Physiol 314 (6), G655–G667. 10.1152/ajpgi.00326.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifer CA, Kennedy MN, Mazzoni A, Lee C, Kruhlak MJ, Segal DM, 2004. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J. Immunol 173 (2), 1179–1183. 10.4049/jimmunol.173.2.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Diner BA, Chen J, Cristea IM, 2012. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl. Acad. Sci. U. S. A 109 (26), 10558–10563. 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chen J, Cristea IM, 2013. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 14 (5), 591–599. 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wu R, Guo W, Xie L, Qiao Z, Chen S, et al. , 2019. STING-mediated IFI16 degradation negatively controls type I interferon production. Cell Rep. 29 (5), 1249–1260.e1244. 10.1016/j.celrep.2019.09.069. [DOI] [PubMed] [Google Scholar]
- Li T, Huang T, Du M, Chen X, Du F, Ren J, Chen ZJ, 2021. Phosphorylation and chromatin tethering prevent cGAS activation during mitosis. Science 371, eabc5386. 10.1126/science.abc5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Qian C, Cao X, 2016. Post-translational modification control of innate immunity. Immunity 45 (1), 15–30. 10.1016/j.immuni.2016.06.020. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, et al. , 2018. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563 (7729), 131–136. 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- Lohofener J, Steinke N, Kay-Fedorov P, Baruch P, Nikulin A, Tishchenko S, et al. , 2015. The activation mechanism of 2′−5’-oligoadenylate synthetase gives new insights into OAS/cGAS triggers of innate immunity. Structure 23 (5), 851–862. 10.1016/j.str.2015.03.012. [DOI] [PubMed] [Google Scholar]
- Lum KK, Song B, Federspiel JD, Diner BA, Howard T, Cristea IM, 2018. Interactome and proteome dynamics uncover immune modulatory associations of the pathogen sensing factor cGAS. Cell Syst. 7 (6), 627–642.e626. 10.1016/j.cels.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun XK, Bodenmiller B, 2020. Profiling cell signaling networks at single-cell resolution. Mol. Cell. Proteomics 19 (5), 744–756. 10.1074/mcp.R119.001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, et al. , 2017. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548 (7668), 461–465. 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manes NP, Nita-Lazar A, 2018. Application of targeted mass spectrometry in bottom-up proteomics for systems biology research. J. Proteomics 189, 75–90. 10.1016/j.jprot.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Val A, Bekker-Jensen DB, Steigerwald S, Mehta A, Tran T, Sikorski K, et al. , 2021. Spatial-proteomics reveal in-vivo phospho-signaling dynamics at subcellular resolution. bioRxiv. 10.1101/2021.02.02.425898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto ML, Dong KC, Yu C, Phu L, Gao X, Hannoush RN, et al. , 2012. Engineering and structural characterization of a linear polyubiquitin-specific antibody. J. Mol. Biol 418 (3–4), 134–144. 10.1016/j.jmb.2011.12.053. [DOI] [PubMed] [Google Scholar]
- Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, et al. , 2009. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal 2 (84), ra46. 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- Miyashita M, Oshiumi H, Matsumoto M, Seya T, 2011. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol. Cell. Biol 31 (18), 3802–3819. 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya A, Beltran L, Casado P, Rodriguez-Prados JC, Cutillas PR, 2011. Characterization of a TiO(2) enrichment method for label-free quantitative phosphoproteomics. Methods 54 (4), 370–378. 10.1016/j.ymeth.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motwani M, Pesiridis S, Fitzgerald KA, 2019. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet 20 (11), 657–674. 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
- Murray LA, Sheng X, Cristea IM, 2018. Orchestration of protein acetylation as a toggle for cellular defense and virus replication. Nat. Commun 9 (1), 4967. 10.1038/s41467-018-07179-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuhse TS, Stensballe A, Jensen ON, Peck SC, 2003. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 2 (11), 1234–1243. 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M, 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127 (3), 635–648. 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, et al. , 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal 3 (104), ra3. 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Onder O, Sidoli S, Carroll M, Garcia BA, 2015. Progress in epigenetic histone modification analysis by mass spectrometry for clinical investigations. Expert Rev. Proteomics 12 (5), 499–517. 10.1586/14789450.2015.1084231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, DeLuca NA, Knipe DM, 2012. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. U. S. A 109 (44), E3008–E3017. 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, Broekema NM, Diner BA, Hancks DC, Elde NC, Cristea IM, Knipe DM, 2015. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc. Natl. Acad. Sci. U. S. A 112 (14), E1773–E1781. 10.1073/pnas.1424637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan SR, 2015. Activation and regulation of DNA-driven immune responses. Microbiol. Mol. Biol. Rev 79 (2), 225–241. 10.1128/MMBR.00061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ, 2012. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics 11 (11), 1475–1488. 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ, 2004. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem 76 (14), 3935–3943. 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, et al. , 2011. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal 4 (164), rs3. 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- Riley JS, Tait SW, 2020. Mitochondrial DNA in inflammation and immunity. EMBO Rep. 21 (4), e49799. 10.15252/embr.201949799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero MP, Tesser A, Bartok E, Rice GI, Della Mina E, Depp M, et al. , 2017. Type I interferon-mediated autoinflammation due to DNase II deficiency. Nat. Commun 8 (1), 2176. 10.1038/s41467-017-01932-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Dickson BM, Raab JR, Grzybowski AT, Krajewski K, Guo AH, et al. , 2015. An interactive database for the assessment of histone antibody specificity. Mol. Cell 59 (3), 502–511. 10.1016/j.molcel.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, et al. , 2005. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol 23 (1), 94–101. 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Sadler AJ, Latchoumanin O, Hawkes D, Mak J, Williams BR, 2009. An antiviral response directed by PKR phosphorylation of the RNA helicase A. PLoS Pathog. 5 (2), e1000311. 10.1371/journal.ppat.1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer LV, Millikin RJ, Miller RM, Anderson LC, Fellers RT, Ge Y, et al. , 2019. Identification and quantification of proteoforms by mass spectrometry. Proteomics 19 (10), e1800361. 10.1002/pmic.201800361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Speiseder T, Dobner T, Gonzalez RA, 2014. DNA virus replication compartments. J. Virol 88 (3), 1404–1420. 10.1128/JVI.02046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM, 2014. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol 32, 513–545. 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Yang A, Tan B, Kim S, Liang Q, Choi Y, et al. , 2015. Akt kinase-mediated checkpoint of cGAS DNA sensing pathway. Cell Rep. 13 (2), 440–449. 10.1016/j.celrep.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Kim C, Shin WJ, Sklan EH, Eoh H, Jung JU, 2018. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat. Commun 9 (1), 613. 10.1038/s41467-018-02936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PG, Chaerkady R, Zhang Z, Davidson NE, Pandey A, 2011. Monoclonal antibody cocktail as an enrichment tool for acetylome analysis. Anal. Chem 83 (10), 3623–3626. 10.1021/ac1026176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WA, Schurter BT, Wong-Staal F, David M, 2004. Arginine methylation of RNA helicase a determines its subcellular localization. J. Biol. Chem 279 (22), 22795–22798. 10.1074/jbc.C300512200. [DOI] [PubMed] [Google Scholar]
- Song B, Greco TM, Lum KK, Taber CE, Cristea IM, 2020a. The DNA sensor cGAS is decorated by acetylation and phosphorylation modifications in the context of immune signaling. Mol. Cell. Proteomics 19 (7), 1193–1208. 10.1074/mcp.RA120.001981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song ZM, Lin H, Yi XM, Guo W, Hu MM, Shu HB, 2020b. KAT5 acetylates cGAS to promote innate immune response to DNA virus. Proc. Natl. Acad. Sci. U. S. A 117 (35), 21568–21575. 10.1073/pnas.1922330117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R, 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134 (4), 587–598. 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ, 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339 (6121), 786–791. 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svinkina T, Gu H, Silva JC, Mertins P, Qiao J, Fereshetian S, et al. , 2015. Deep, quantitative coverage of the lysine acetylome using novel anti-acetyl-lysine antibodies and an optimized proteomic workflow. Mol. Cell. Proteomics 14 (9), 2429–2440. 10.1074/mcp.O114.047555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. , 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448 (7152), 501–505. 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Takeshita F, Leifer CA, Gursel I, Ishii KJ, Takeshita S, Gursel M, Klinman DM, 2001. Cutting edge: role of toll-like receptor 9 in CpG DNA-induced activation of human cells. J. Immunol 167 (7), 3555–3558. 10.4049/jimmunol.167.7.3555. [DOI] [PubMed] [Google Scholar]
- Tan X, Sun L, Chen J, Chen ZJ, 2018. Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol 72, 447–478. 10.1146/annurev-micro-102215-095605. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ, 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal 5 (214), ra20. 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA, 2011. Pattern recognition receptors and the innate immune response to viral infection. Viruses 3 (6), 920–940. 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thygesen C, Boll I, Finsen B, Modzel M, Larsen MR, 2018. Characterizing disease-associated changes in post-translational modifications by mass spectrometry. Expert Rev. Proteomics 15 (3), 245–258. 10.1080/14789450.2018.1433036. [DOI] [PubMed] [Google Scholar]
- Tikhanovich I, Zhao J, Olson J, Adams A, Taylor R, Bridges B, et al. , 2017. Protein arginine methyltransferase 1 modulates innate immune responses through regulation of peroxisome proliferator-activated receptor gamma-dependent macrophage differentiation. J. Biol. Chem 292 (17), 6882–6894. 10.1074/jbc.M117.778761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, et al. , 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol 11 (11), 997–1004. 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wen M, Cao X, 2019. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 365 (6454), eaav0758. 10.1126/science.aav0758. [DOI] [PubMed] [Google Scholar]
- Wechsler T, Chen BP, Harper R, Morotomi-Yano K, Huang BC, Meek K, et al. , 2004. DNA-PKcs function regulated specifically by protein phosphatase 5. Proc. Natl. Acad. Sci. U. S. A 101 (5), 1247–1252. 10.1073/pnas.0307765100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White FM, Wolf-Yadlin A, 2016. Methods for the analysis of protein phosphorylation-mediated cellular signaling networks. Annu. Rev. Anal. Chem. (Palo Alto, Calif.) 9 (1), 295–315. 10.1146/annurev-anchem-071015-041542. [DOI] [PubMed] [Google Scholar]
- Wu C, Tran JC, Zamdborg L, Durbin KR, Li M, Ahlf DR, et al. , 2012. A protease for ‘middle-down’ proteomics. Nat. Methods 9 (8), 822–824. 10.1038/nmeth.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ, 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339 (6121), 826–830. 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Li W, Shao Y, Avey D, Fu B, Gillen J, et al. , 2015. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe 18 (3), 333–344. 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Ye B, Wang S, Zhu X, Du Y, Xiong Z, et al. , 2016. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat. Immunol 17 (4), 369–378. 10.1038/ni.3356. [DOI] [PubMed] [Google Scholar]
- Yan Z, Gibson SA, Buckley JA, Qin H, Benveniste EN, 2016. Role of the JAK/STAT signaling pathway in regulation of innate immunity in neuroinflammatory diseases. Clin. Immunol 189, 4–13. 10.1016/j.clim.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X, 2010. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat. Immunol 11 (6), 487–494. 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- Yang H, Lundback P, Ottosson L, Erlandsson-Harris H, Venereau E, Bianchi ME, 2012. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol. Med 18, 250–259. 10.2119/molmed.2011.00389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yu K, Zhang Q, Liu Z, Zhao Q, Zhang X, Wang Y, et al. , 2019. qPhos: a database of protein phosphorylation dynamics in humans. Nucleic Acids Res. 47 (D1), D451–D458. 10.1093/nar/gky1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X, Schunter A, Hummon AB, 2015. Comparing multistep immobilized metal affinity chromatography and multistep TiO2 methods for phosphopeptide enrichment. Anal. Chem 87 (17), 8837–8844. 10.1021/acs.analchem.5b01833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, et al. , 2009. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics 8 (2), 215–225. 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, et al. , 2011a. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol 186 (8), 4541–4545. 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ, 2011b. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol 12 (10), 959–965. 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Bao M, Lu N, Weng L, Yuan B, Liu YJ, 2013a. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat. Immunol 14 (2), 172–178. 10.1038/ni.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ, 2013b. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for sting. Mol. Cell 51 (2), 226–235. 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wu J, Du F, Xu H, Sun L, Chen Z, et al. , 2014. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 6 (3), 421–430. 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhao J, Xu S, Li J, He S, Zeng Y, et al. , 2018. Species-specific deamidation of cGAS by herpes simplex virus UL37 protein facilitates viral replication. Cell Host Microbe 24 (2), 234–248.e235. 10.1016/j.chom.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ, Mohammed S, 2013a. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res 12 (1), 260–271. 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
- Zhou H, Ye M, Dong J, Corradini E, Cristobal A, Heck AJ, et al. , 2013b. Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc 8 (3), 461–480. 10.1038/nprot.2013.010. [DOI] [PubMed] [Google Scholar]
- Zhou R, Xie X, Li X, Qin Z, Wei C, Liu J, Luo Y, 2020. The triggers of the cGAS-STING pathway and the connection with inflammatory and autoimmune diseases. Infect. Genet. Evol 77, 104094. 10.1016/j.meegid.2019.104094. [DOI] [PubMed] [Google Scholar]
- Zhuang J, Jiang G, Willers H, Xia F, 2009. Exonuclease function of human Mre11 promotes deletional nonhomologous end joining. J. Biol. Chem 284 (44), 30565–30573. 10.1074/jbc.M109.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C, Yamaguchi N, Paredes M, Luo JD, Carroll T, Funabiki H, 2019. The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell 178 (2), 302–315. e323. 10.1016/j.cell.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]


