Abstract
Objective
The objective of this study was to better understand the pathophysiology and underlying genetic mechanisms behind two abdominal aortic aneurysm (AAA) subtypes using computed tomographic imaging in combination with whole genome sequencing.
Methods
Patients with a known AAA and European ancestry were included in this investigation and underwent genetic and image analysis. Patients with AAAs and indications of descending thoracic aortic pathology (aortic dissection, penetrating aortic ulcers, intramural hematoma, atheromas, ulcerative plaque, and intramural ulceration, and intimal flaps/tears) were classified as having thoracic aortic disease, grouped together, and compared with patients with an AAA and a normal descending thoracic aorta. Whole genome sequencing was then performed on the 93 patients who had imaging features consistent with thoracic aortic disease and the 126 patients with a normal descending thoracic aorta.
Results
The results of this study suggest one variant-level, four gene-level, and one gene set-level associations in patients with thoracic aortic disease who also had an AAA. The variant rs79508780 located in TSEN54 achieved study-wide significance (P = 1.71E-06). BATF3 and SMLR1 were significantly associated and EFCAB3 and TAF4 were reached suggestive assocation with a diseased descending thoracic aorta (P = 5.23E-26, P = 1.86E-25, P = 1.54E-05, and P = 8.31E-05, respectively). Gene sets were also compiled using MSigDB and trait-based index single nucleotide variation from major genome-wide association studies. GO_DNA_DOUBLE_STRAND_BREAK_PROCESSING, a gene set related to double-stranded DNA break repair, was significantly associated with thoracic aortic disease in AAA patients (P = 1.80E-06).
Conclusions
This pilot study provides further evidence that an AAA may be the end result of multiple degenerative pathways. Genetic variations in vitamin D signaling, cholesterol metabolism, extracellular matrix breakdown, and double-stranded DNA break repair pathways were associated with European patients who had an AAA and thoracic aortic disease. Additionally, this study provides support for the application of a radiogenomic approach for the investigation of other potential pathologies that could lead to the development of an AAA or influence future management decisions. (JVS–Vascular Science.)
Clinical Relevance
In this study, we provide evidence that abdominal aortic aneurysms (AAAs) may be a result of multiple pathophysiologies rather than a single disease. We have identified genetic variants involved in vitamin D signaling, cholesterol metabolism, extracellular matrix breakdown, and double-stranded DNA break repair associated with structural defects in the aortic wall in patients with AAAs who are of European descent. Patients with AAAs and structural defects in the thoracic aorta have been previously linked to differential behavior after endovascular aneurysm repair. These patients with wall defects exhibited greater sac regression, a marker of surgical success, after endovascular aneurysm repair. Our study demonstrates the usefulness of a radiogenomic approach for elucidating mechanisms behind the formation and future behavior of AAAs that could aid surgeons in making future procedural and management decisions.
Keywords: Abdominal aortic aneurysm, Genetic association, Aortic dissection, Genetics, Morphology
Article Highlights.
-
•
Type of Research: Single-center cross-sectional study
-
•
Key Findings: This study suggests the association of rs79508780 (P = 1.71E-06), BATF3 (P = 5.23E-26), SMLR1 (P = 1.86E-25) and a set of genes related to double-stranded DNA break repair (P = 1.80E-06) were significantly associated with structural defects in the descending thoracic aorta in a study of 219 patients with abdominal aortic aneurysms.
-
•
Take Home Message: A radiogenomic approach for identifying underlying genetic mechanisms behind morphologic subtypes of abdominal aortic aneurysms can expand our knowledge of aneurysm etiology and help to direct future management decisions.
Infrarenal abdominal aortic aneurysm (AAA) is a multifactorial condition characterized by the dilation of the aorta below the level of the renal arteries. AAAs occur predominantely in elderly men and affect 2% to 8% of men over the age of 65.1 AAA has both genetic and environmental risk factors including a history of smoking, hypertension, hypercholesterolemia, and a family history of AAA.1 Thoracic aortic aneurysms and AAAs can also occur as a result of an aortic dissection (AD) involving the descending thoracic and abdominal aorta. Dissections that originate in the descending thoracic aorta may be monitored and treated with antihypertensive medication in the absence of complications.2 Approximately 40% of medically managed patients with descending thoracic AD who do not undergo emergency surgery develop aortic aneurysms. Although the aneurysmal dilation of dissected aortic segments is not uncommon, intimal and medial interruptions of the aortic wall may also be an indication of an underlying degenerative pathology in the absence of a fully dissected aorta.3 Genetic and histologic studies have also suggested overlapping mechanisms in the etiology of AAA and AD. One of the main pathways implicated in the development of both conditions is the degradation of the extracellular matrix.4,5 Matrix metalloproteases, a family of endoproteases, are capable of degrading components of the extracellular matrix. Histologic studies show elevated levels of matrix metalloprotease family members in AAA6 and AD,7 suggesting that upregulation of matrix metalloproteases may be involved in the development of both conditions. Previous genetic testing has also suggested the involvment of a number of genes and loci in the cholesterol metabolism and atherosclerosis pathways in the formation of AAA and AD. Low-density lipoprotein receptor-related protein 1 (LRP1) encodes a receptor involved in lipid homeostasis and a genetic variant in the LRP1 locus was significantly associated with both AAA8 and AD9 in multiple genome-wide association (GWA) studies.
Other computed tomography findings associated with an AD include extension of the dissection, penetrating aortic ulcer (PAU), and intramural hematoma (IMH).10 When AD, PAU, and IMH present acutely with symptoms such as severe chest or back pain, these conditions are often grouped together as part of a spectrum of disease known as acute aortic syndrome.10 However, AD, PAU, and IMH can all be clinically silent and seen incidentally on computed tomography angiography (CTA). A study evaluating 236 AD patients found that 15% of patients were asymptomatic at the time of presentation, which suggests that a subgroup of AD patients could have an AD that does not present with pain and then results in subsequent abdominal aortic dilation.11 There is currently no universally accepted nomenclature when AD, PAU, or IMH are incidentally seen. In this article, we are defining the incidental presence of a thoracic AD, PAU, IMH, atheromas, ulcerative plaque, and intramural ulceration, and intimal flaps or tears indicating structural defects in the aortic wall as having thoracic aortic disease.
A previous investigation comparing sac regression after endovascular aneurysm repair between AAA patients with and without thoracic aortic disease showed that AAA patients with imaging features suggestive of a previous AD exhibited greater sac regression following the procedure than AAA patients with a normal thoracic aorta.12 We hypothesize that different AAA morphologic subtypes may have varying genetic etiologies. Specifically, AAA associated with structural defects in the aortic wall such as a previous subclinical thoracic AD, PAU, and IMH, which are identifiable on diagnostic imaging, and could be driven by distinct genetic architecture. The objective of this investigation was to identify genetic differences between AAA patients with a fully normal thoracic aorta and AAA patients with imaging characteristic consistent with structural defects in the descending thoraicic aorta.
Methods
Study population
This single-center cross-sectional study was approved by the Institutional Review Board at the Mount Sinai Hospital. Inclusion criteria for this study were patients of European ancestry with a diagnosis of AAA confirmed on diagnostic imaging and a minimum aortic diameter of 3 cm (Supplementary Fig 1). Exclusion criteria include a past history of any syndromic connective tissue disorder such as Marfan syndrome, Ehlers-Danlos syndrome, Loeys-Dietz syndrome, or Turner syndrome. The 360 consecutive patients enrolled in the study were asked to complete a comprehensive medical and family history survey, which included a history of cardiovascular, pulmonary, and renal conditions (Supplementary Fig 1). The incidence of any cases of sudden or unexplained death were also recorded because these deaths could indicate a ruptured AAA. Patients were also asked to provide a sample of peripheral blood for sequencing and underwent a high resolution helical CTA scan as a part of their normal course of care. Electronic medical records were used to validate survey answers and complete any missing information in the patient's medical history. All study patients provided written consent to participate in this study.
Supplementary Fig 1.
Flow diagram of patient selection, quality control steps, and analysis.
Vascular measurements
All AAA patients included in this study had undergone a high-resolution helical CTA scan as part of normal clinical care. The CTA scans were reconstructed using Vital Vitrea software to fully visualize the aorta, and vascular measurements and assessments were performed by a single investigator trained in CTA imaging analysis with extensive experience with the use of three-dimensional vascular reconstruction software. The maximum diameter of the infrarenal aorta was determined on three-dimensional reconstructions. A further analysis of the CTA scans was then performed to assess the features of thoracic aortic disease, which includes the presence of AD, PAU, and structural defects in the descending thoracic aorta of the AAA patients (Fig 1, A-D). PAU was defined as blood flow into the aortic wall without proximal or distal extension resulting in outpouching of the aortic wall. The presence of other indicators of structural defects in the aortic wall such as IMH, atheromas, intimal flaps or tears, ulcerative or atherosclerotic plaque, wall ulceration, wall thickening, and calcified wall tissue projecting into the lumen of the aorta were also included in the designation of thoracic aortic disease. The AAA patients were then divided into two phenotypic groups. The thoracic aortic disease group had features indicating structural defects in the descending thoracic wall and an AAA, whereas the nonthoracic disease group had a normal thoracic aorta and an AAA (Fig 1, E and F).
Fig 1.
The presence of computed tomography (CT) features was assessed in the descending thoracic aorta of patients with an abdominal aortic aneurysm (AAA). A, A patient with a normal descending thoracic aorta. B, Descending thoracic aortic dissection (AD). C, Penetrating aortic ulcers (PAUs). D, Structural defects in the aortic wall (IMH, atheromas, ulcerative/atherosclerotic plaque, wall ulceration, wall thickening, and calcified lesions/wall tissue projecting into the lumen of the aorta). CT images of two different patients, each with an AAA. E, A patient with an AAA and thoracic aortic disease. F, A patient with an AAA and a normal thoracic aorta.
Sequencing
Genomic DNA was extracted from peripheral venous blood then paired-end whole genome sequencing was then performed on Illumina HiSeq XTen, with approximately 30× coverage. Reads were aligned to the hg19 human genome reference using BWA (version 0.7.12).13 Subsequent processing steps included insertion/deletion (INDEL) realignment, de-duplication, and base quality score recalibration. Genetic variants (single nucleotide variations [SNVs] and INDELs) were called with GATK (version 2.8). Multiple filters such as missingness and read depth, were applied to remove low quality variants and genotypes. Following variant-level QC, relatives and sex discordant samples were excluded. Finally, a principal component analysis was conducted to increase the power of the analysis by controlling for population stratification thereby minimizing differences in allele frequency that were due to non-European ancestry. Principal components were computed for the AAA patient genomes and the 1000 Genomes Project populations (Supplementary Fig 2, A and B). AAA patients more than four standard deviations from the mean of the European superpopulation and patients with low sample-level sequencing quality metrics were excluded. SnpEff (version 4.2) was used to generate HIGH, MODERATE, LOW, or MODIFIER variant impact predictions and population frequencies.14 Only variants with predicted HIGH or MODERATE impact were retained. A gene set-level association analysis was performed using Magma (version 1.07). Gene sets were obtained from MSigDB (version 6.2) and generated using the SNVs from two different phenotypically relevant GWA studies. The AAA gene set, the strongest associated SNV in each genome-wide significant locus from the largest and most recently published meta-analysis was selected.15 In the AD gene set, the top three genome wide significant SNVs were selected.9 All GENCODE v19 genes located ±1 Mb from each index SNV in the AD and AAA studies were added to the relevant gene set.
Supplementary Fig 2.
Principal component analysis. A, Final analysis of the abdominal aortic aneurysm (AAA) cohort and 1000 Genomes dataset. All AAA samples cluster with 1000 Genomes European super population. B, Scree plots showing the cumulative variation explained by the principal component and previous principal components. Red dotted line indicates the point where 95% of the variation is explained
Statistical analysis
Categorical variables were reported as the count and percentage of patients in each group and analyzed using χ2 tests. Continuous variables were reported as mean and confidence interval or median and interquartile range for nonparametric measurements and analyzed using Kruskal-Wallis tests. Association and power analyses were undertaken using the SKAT package (version 1.3.2.1). Association analysis models were adjusted for sex, hypertension, and the first five principal components. Multiple testing-corrected thresholds were used to define study-wide statistical significance (P < .05/number of tests) and a suggestive P value threshold was used to define suggestive significance (P < 1/number of tests) for the variant- and gene-level association tests. The power analysis was performed with an effect size of 0.03 for a variant with a minor allele frequency (MAF) of 0.03. All statistical analyses were conducted using R (version 3.5.1; The R Foundation, Vienna, Austria).
Results
After the initial screening, 243 patients were included in the sequencing cohort. After applying variant level and subject level filters, eight patients were excluded owing to low sequencing of imaging quality (more than six standard deviations from the mean), nine patients were excluded owing to relatedness to another patient in the cohort, one patient was excluded owing to sex discordance, and seven patients were excluded owing non-European ancestry (more than four standard deviations from the mean of the European superpopulation from the 1000 Genomes Project. The final study population consisted of 219 patients who were grouped based on the presence or absence of imaging features consistent with thoracic aortic disease. Ninety-three patients had a diseased thoracic aorta and were included in group 1. The remaining 126 patients showed no evidence of a previous AD or associated imaging features in the descending thoracic or abdominal aorta and were included in group 2 (Supplementary Fig 1). Patient demographics and comorbid conditions were compared between group 1 and group 2 patients. There were no significant differences in age, sex, smoking history, clinical comorbidities, or family history of AAA between the two groups (P = .449; Table I). There were no significant differences between the maximum diameters of the AAA or common iliac arteries (Table II).
Table I.
Demographics of study patients
| Group 1 | Group 2 | P value | |
|---|---|---|---|
| No. | 93 | 126 | |
| Age | 77.00 [70.00-83.00] | 77.00 [71.00-82.00] | .961 |
| Male sex | 74 (79.6) | 108 (85.7) | .309 |
| Smoking | 76 (81.7) | 94 (74.6) | .278 |
| Body mass index | 25.87 [23.59-28.98] | 27.34 [24.30-29.76] | .078 |
| Hypertension | 72 (77.4) | 85 (67.5) | .143 |
| Hypercholesterolemia | 69 (74.2) | 89 (70.6) | .669 |
| Kidney disease | 7 (7.5) | 12 (9.5) | .782 |
| Diabetes | 11 (11.8) | 16 (12.7) | NS |
| Hernia | 33 (35.5) | 45 (35.7) | NS |
| Myocardial infarction | 27 (29.0) | 45 (35.7) | .371 |
| Stroke | 11 (11.8) | 9 (7.1) | .341 |
| Heart defect | 2 (2.2) | 4 (3.2) | NS |
| Mitral valve prolapse | 1 (1.1) | 8 (6.3) | .082 |
| Sudden unexplained death | 13 (14.0) | 18 (14.3) | NS |
| Family history of AAA | .449 | ||
| No | 50 (53.8) | 64 (50.8) | |
| Suspected | 2 (2.2) | 7 (5.6) | |
| Yes | 41 (44.1) | 55 (43.7) |
NS, Not significant.
Values are median [IQR] or number (%).
Table II.
Summary of maximum diameters of the infrarenal aorta, and the left and right common iliac arteries
| Maximum diameter, mm | Group1 | Group 2 | P value |
|---|---|---|---|
| Infrarenal aorta | 55.50 [50.50-61.40] | 52.80 [45.40-59.70] | .409 |
| Right common iliac artery | 15.90 [11.80-20.70] | 16.50 [13.00-21.50] | .18 |
| Left common iliac artery | 15.00 [12.15-20.10] | 16.50 [12.80-21.20] | .387 |
Values are median [IQR].
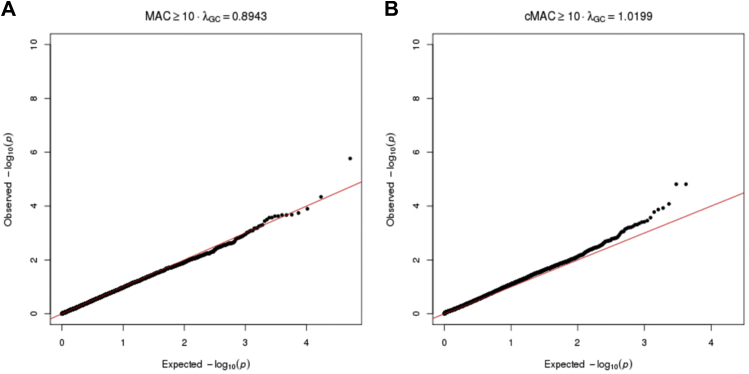
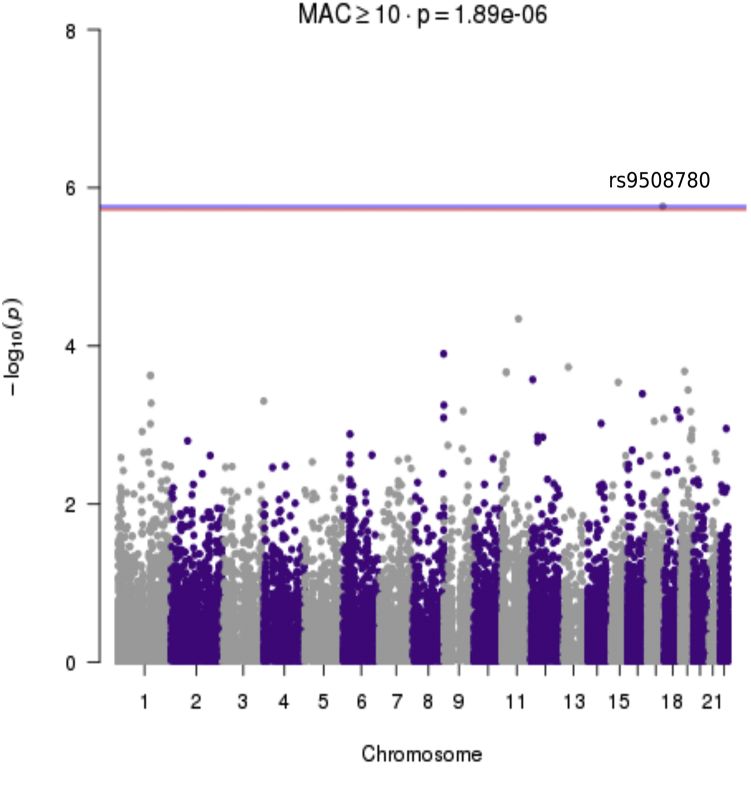
We performed a variant-level association comparing the two phenotypes of patients with AAA. The analysis model was slightly deflated with a λ of 0.8943 (Supplementary Fig 3, A). A single variant achieved study-wide significance, rs79508780 located in TSEN54 on chromosome 17 (P = 1.71E-06; Fig 2). The presence of the ALT allele of rs79508780 increases the risk of thoracic aortic disease in patients with AAA (odds ratio, 7.89). Rs79508780 is a common missense variant encoding p.Val190Met with an allele frequency of approximately 6.33% in the ExAC non-Finnish European population. The gene-level SKAT-O analysis showed the genomic model exhibited minimal inflation with a λ of 1.0199 (Supplementary Fig 3, B). Two genes, BATF3 (P = 5.23E-26) and SMLR1 (1.86E-25), achieved study-wide significance (P = 4.57E-06), whereas EFCAB3 (P = 1.54E-05) and TAF4 (P = 8.31E-05) reached the suggestive P value threshold (9.14E-05; Fig 3). The association of BATF3 with the group 1 patients was driven by both a rare in-frame deletion and a common protective variant (Table III). The association of SMLR1 with the group 1 patients had a similar architecture and was driven by a single rare variant in one group 1 patient and a more common protective variant (Table III). EFCAB3 and TAF4 exhibited more complex architecture with multiple rare variants associated with the presence of thoracic aortic disease and a common protective variant associated with patients with a normal thoracic aorta (Table III).
Supplementary Fig 3.
Quantile–quantile (QQ) plots for the association tests showing the distribution of the genomic data. A, A QQ plot of the variant-level association analysis. B, A QQ plot for the gene-level association analysis.
Fig 2.
Manhattan plot showing the results of the variant association analysis. The P values of variants with a minor allele count of greater than or equal to 10 were plotted against chromosomal location. The blue line indicates the suggested P value threshold and the red line indicates the study-wide P value threshold of 1.89–06. The analysis was adjusted for sex and the first five principal components.
Fig 3.
Manhattan plot showing the results of the study-wide gene-based association analysis. The plot shows P values genes with a cumulative minor allele count of greater than or equal to 10 plotted against the chromosomal location. The blue line indicates the suggestive association P value threshold (P = 9.14–05) and the red line indicates the study-wide P value threshold of 4.57–06. The analysis covariates included sex and the first three principal components from the principal component analysis.
Table III.
Results of the variant level tests for the four genes that reached study-wide significance in gene-level association tests between AAA patients with and without features of the thoracic aorta consistent with structural defects in the descending thoracic aorta
| Gene | Gene ID | CHR | BP | REF/ALT | ExAC_NFE_AF | MAC_A | MAC_U | MAF_A | MAF_U | IMPACT | EFFECT | HGVS.p | OR.ALL | P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BATF3 | NA | 1 | 212873019 | TGCTGCG/T | NA | 1 | 0 | 0.005376 | 0 | MODERATE | disruptive_inframe_deletion | p.Pro27_Gln28del | Inf | 0.17427451 |
| rs2202683 | 1 | 212873074 | C/T | 0.130553 | 22 | 30 | 0.1183 | 0.121 | MODERATE | missense_variant | p.Val11Ile | 0.97 | 0.55676196 | |
| SMLR1 | NA | 6 | 131148644 | G/A | 0.00333511 | 1 | 0 | 0.005376 | 0 | MODERATE | missense_variant | p.Val31Met | Inf | 0.17427451 |
| NA | 6 | 131148737 | G/A | 0.304505 | 46 | 66 | 0.2473 | 0.2619 | MODERATE | missense_variant | p.Val62Met | 0.93 | 0.66361606 | |
| EFCAB3 | rs6504103 | 17 | 60451185 | G/C | 2.66E-04 | 1 | 0 | 0.0054 | 0 | MODERATE | missense_variant | p.Lys15Asn | Inf | 0.174274505 |
| rs114592394 | 17 | 60464702 | G/A | 0.0012 | 1 | 0 | 0.0054 | 0 | MODERATE | missense_variant splice_region_variant | p.Asp78Asn | Inf | 0.211393998 | |
| rs78388447 | 17 | 60493386 | A/G | 0.07519 | 25 | 26 | 0.1344 | 0.1032 | MODERATE | missense_variant | p.Tyr390Cys | 1.35 | 0.414902518 | |
| TAF4 | rs201112070 | 20 | 60640232 | G/A | 0.00199712 | 3 | 0 | 0.01667 | 0 | MODERATE | missense_variant | p.Ala212Val | Inf | 0.03009 |
| NA | 20 | 60528844 | C/T | 1.34E-04 | 4 | 1 | 0.02151 | 0 | MODERATE | missense_variant | p.Arg71His | 5.52 | 0.03465 | |
| NA | 20 | 60640470 | T/A | 0.00197628 | 1 | 0 | 0.005814 | 0 | MODERATE | missense_variant | p.Ser133Cys | Inf | 0.19438 | |
| NA | 20 | 60640556 | C/T | 0.00866317 | 3 | 1 | 0.01667 | 0 | MODERATE | missense_variant | p.Arg104His | 4.02 | 0.23108 | |
| NA | 20 | 60639732 | T/C | 2.86E-04 | 1 | 0 | 0.005952 | 0 | MODERATE | missense_variant | p.Met379Val | Inf | 0.23956 | |
| rs201584478 | 20 | 60572616 | G/A | 6.66E-05 | 2 | 0 | 0.01075 | 0 | MODERATE | missense_variant | p.Ser1027Leu | Inf | 0.30147 | |
| NA | 20 | 60639840 | TGACCCCCGGCGCCGGCGCCGC/T | 3.11E-04 | 1 | 1 | 0.005618 | 0 | MODERATE | inframe_deletion | p.Ala336_Val342del | 1.32 | 0.78424 | |
| rs113875178 | 20 | 60640461 | A/T | 1 | 168 | 226 | 1 | 1 | MODERATE | missense_variant | p.Ser136Ala | NaN | NS |
NS, Not significant.
Neither the AAA or AD gene set reached the study-wide P value threshold of 2.71E-06 (P = .0772 and P = .2444, respectively). Only a single gene set, GO_DNA_DOUBLE_STRAND_BREAK_PROCESSING, related to the biological process of formation of a 3ʹ end single stranded DNA overhang at the site of a break in the DNA, achieved study-wide significance (P = 1.80E-06; Table IV). Within the gene set, five genes had a P value of less than .05: BARD1, BLM, RBBP8, DNA2, and RAD52 (Table V). A statistical power analysis was performed to determine the study's ability to robustly detect genetic association. The study with the current sample size of 219 patients has less than .1 power to detect an associate of effect size of 0.03 for a variant with a MAF of 0.03 at an alpha of 1.89E-06. The power analysis suggests that more than 30,000 samples would be required to detect a causal association with these parameters.
Table IV.
List of the gene sets from MSigDB with a P value of less than .05
| Gene set | No. of genes | BETA | BETA_STD | SE | P | P_CORR | SELF_P |
|---|---|---|---|---|---|---|---|
| M15933:GO_DNA_DOUBLE_STRAND_BREAK_PROCESSING | 17 | 0.95 | 0.0281 | 0.205 | 1.80E-06 | 0.0356 | 2.55E-05 |
| M14146:BOYAULT_LIVER_CANCER_SUBCLASS_G1_UP | 106 | 0.375 | 0.0276 | 0.0859 | 6.47E-06 | 0.1099 | 7.71E-05 |
| M4336:OSADA_ASCL1_TARGETS_DN | 21 | 0.805 | 0.0265 | 0.204 | 4.03E-05 | 0.4891 | 2.92E-05 |
| M12146:GO_REGULATION_OF_LEUKOCYTE_MEDIATED_CYTOTOXICITY | 44 | 0.49 | 0.0233 | 0.129 | 6.97E-05 | 0.6748 | 6.52E-04 |
| M14105:DACOSTA_LOW_DOSE_UV_RESPONSE_VIA_ERCC3_XPCS_DN | 8 | 1.15 | 0.0234 | 0.313 | 1.13E-04 | 0.8293 | 6.77E-04 |
| M15265:GO_RESPONSE_TO_INTERFERON_GAMMA | 127 | 0.284 | 0.0229 | 0.08 | 1.98E-04 | 0.945 | 4.82E-04 |
| M9277:GSE39110_DAY3_VS_DAY6_POST_IMMUNIZATION_CD8_TCELL_DN | 185 | 0.226 | 0.022 | 0.0646 | 2.34E-04 | 0.9677 | 0.001 |
| M18849:MODULE_153 | 29 | 0.571 | 0.0221 | 0.164 | 2.46E-04 | 0.9723 | 0.002 |
| M19177:GO_STRUCTURAL_MOLECULE_ACTIVITY | 609 | 0.127 | 0.0221 | 0.0364 | 2.51E-04 | 0.9739 | 4.93E-04 |
| M19218:LIU_CDX2_TARGETS_DN | 6 | 1.24 | 0.0218 | 0.358 | 2.58E-04 | 0.9764 | 0.008 |
| M1958:MORF_ACTG1 | 90 | 0.318 | 0.0216 | 0.0924 | 2.89E-04 | 0.9848 | 1.16E-04 |
| M8904:GSE37301_PRO_BCELL_VS_GRANULOCYTE_MONOCYTE_PROGENITOR_UP | 123 | 0.276 | 0.0219 | 0.0801 | 2.91E-04 | 0.9851 | 6.51E-04 |
| M12539:GO_CELLULAR_RESPONSE_TO_INTERFERON_GAMMA | 108 | 0.296 | 0.022 | 0.0868 | 3.26E-04 | 0.9898 | 4.68E-04 |
| M4011:GSE17721_LPS_VS_CPG_24H_BMDC_DN | 180 | 0.217 | 0.0208 | 0.0638 | 3.27E-04 | 0.9898 | 1.55E-04 |
| M9263:GSE40068_CXCR5NEG_BCL6NEG_CD4_TCELL_VS_CXCR5POS_BCL6NEG_TFH_DN | 188 | 0.216 | 0.0212 | 0.0643 | 3.79E-04 | 0.9948 | 4.00E-04 |
| M2602:BIOCARTA_RANKL_PATHWAY | 15 | 0.784 | 0.0218 | 0.234 | 4.04E-04 | 0.996 | 2.38E-04 |
| M8602:GSE32128_INOS_DEPENDENT_VS_INOS_INDEPENDENT_ACTIVATED_TCELL_DN | 174 | 0.217 | 0.0205 | 0.0661 | 5.15E-04 | 0.9988 | 1.80E-04 |
| M10546:REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | 9 | 0.999 | 0.0215 | 0.305 | 5.23E-04 | 0.999 | 0.0015548 |
| M651:REACTOME_CS_DS_DEGRADATION | 10 | 0.906 | 0.0206 | 0.276 | 5.24E-04 | 0.9991 | 0.039 |
| M16351:GO_REGULATION_OF_CELL_KILLING | 53 | 0.38 | 0.0199 | 0.118 | 6.34E-04 | 0.9995 | 0.002 |
| M17301:GO_CHROMATIN | 337 | 0.156 | 0.0203 | 0.0484 | 6.56E-04 | 0.9996 | 2.53E-04 |
| M11562:GO_DNA_DEALKYLATION | 18 | 0.608 | 0.0185 | 0.191 | 7.06E-04 | 0.9997 | 0.004 |
| M16566:GO_WALKING_BEHAVIOR | 30 | 0.513 | 0.0202 | 0.161 | 7.20E-04 | 0.9997 | 9.54E-05 |
| M14997:GO_REGULATION_OF_BONE_RESORPTION | 31 | 0.512 | 0.0204 | 0.161 | 7.20E-04 | 0.9997 | 0.008 |
| M15:PID_LYSOPHOSPHOLIPID_PATHWAY | 53 | 0.384 | 0.02 | 0.121 | 7.39E-04 | 0.9998 | 5.54E-04 |
| M10280:ZHAN_LATE_DIFFERENTIATION_GENES_UP | 24 | 0.57 | 0.0201 | 0.18 | 7.65E-04 | 0.9999 | 0.023 |
| M115:PID_REG_GR_PATHWAY | 65 | 0.363 | 0.021 | 0.115 | 8.37E-04 | 1 | 4.08E-04 |
| M13213:GO_INNATE_IMMUNE_RESPONSE | 506 | 0.125 | 0.0199 | 0.0399 | 8.77E-04 | 1 | 1.56E-04 |
| M5437:GSE3982_MAST_CELL_VS_BCELL_DN | 170 | 0.211 | 0.0197 | 0.0682 | 9.86E-04 | 1 | 0.001 |
| M11171:DACOSTA_UV_RESPONSE_VIA_ERCC3_UP | 267 | 0.163 | 0.0189 | 0.0526 | 9.97E-04 | 1 | 4.20E-04 |
Table V.
Summary statistics from the genes in the gene set GO_DNA_DOUBLE_STRAND_BREAK_PROCESSING
| ENSEMBLE_ID | GENE | CHR | START | STOP | NSNPS | N | ZSTAT | P |
|---|---|---|---|---|---|---|---|---|
| ENSG00000138376 | BARD1 | 2 | 215590370 | 215674428 | 14 | 219 | 2.738 | 0.0030911 |
| ENSG00000197299 | BLM | 15 | 91260558 | 91358859 | 10 | 219 | 2.3482 | 0.0094324 |
| ENSG00000101773 | RBBP8 | 18 | 20378224 | 20606451 | 5 | 219 | 2.1133 | 0.017287 |
| ENSG00000138346 | DNA2 | 10 | 70173821 | 70231879 | 11 | 219 | 2.0754 | 0.018975 |
| ENSG00000002016 | RAD52 | 12 | 1021243 | 1099219 | 9 | 219 | 2.0436 | 0.020498 |
| ENSG00000020922 | MRE11A | 11 | 94152895 | 94227074 | 6 | 219 | 1.52 | 0.06425 |
| ENSG00000104320 | NBN | 8 | 90945564 | 91015456 | 7 | 219 | 1.429 | 0.076508 |
| ENSG00000188827 | SLX4 | 16 | 3631182 | 3661599 | 31 | 219 | 0.71212 | 0.23819 |
| ENSG00000012048 | BRCA1 | 17 | 41196312 | 41277500 | 19 | 219 | 0.59316 | 0.27654 |
| ENSG00000081177 | EXD2 | 14 | 69658228 | 69709075 | 6 | 219 | 0.55103 | 0.29081 |
| ENSG00000163104 | SMARCAD1 | 4 | 95128762 | 95212443 | 5 | 219 | 0.49758 | 0.30939 |
| ENSG00000113522 | RAD50 | 5 | 131891711 | 131980313 | 6 | 219 | 0.45653 | 0.32401 |
| ENSG00000169139 | UBE2V2 | 8 | 48920960 | 48977268 | 1 | 219 | 0.16467 | 0.4346 |
| ENSG00000149311 | ATM | 11 | 108093211 | 108239829 | 27 | 219 | 0.15251 | 0.43939 |
| ENSG00000172977 | KAT5 | 11 | 65479467 | 65487075 | 2 | 219 | -0.11052 | 0.544 |
| ENSG00000134758 | RNF138 | 18 | 29671818 | 29711524 | 1 | 219 | -0.21175 | 0.58385 |
| ENSG00000170364 | SETMAR | 3 | 4344988 | 4359251 | 11 | 219 | -0.36877 | 0.64385 |
In this study, we have provided further evidence that abdominal aortic aneuryms (AAAs) may be a result of multiple pathophysiologies rather than a single disease. We have identified genetic variants involved in vitamin D signaling, cholesterol metabolism, extracellular matrix breakdown, and double-stranded DNA break repair associated with structural defects in the aortic wall in patients with AAAs who are of European descent. Patients with AAAs and structural defects in the thoracic aorta have been previously linked to differential behavior following endovascular aneurysm repair. These patients with wall defects exhibited greater sac regression, a marker of surgical success, following endovascular aneurysm repair. Our study demonstrates the utility of a radiogenomic approach for elucidating mechanisms behind the formation and future behavior of AAAs which could aid surgeons in making future procedural and management decisions.
Discussion
The primary results of this study are that it supports the application of a radiogenomic approach to elucidating the etiology of AAA and provides supporting evidence for the theory that AAAs are a result of multiple pathologies rather than a single disease. The largest AAA GWA study replicated previous associated loci (SORT1, IL6R, 9p21, DAP2IP, and LDLR) and suggested the novel association of 1q32.3 (SMYD2), 13q12.11 (LINC00540), 20q13.12 (near PCIF1/MMP9/ZNF335), and 21q22.2 (ERG) with AAA development.15 These loci implicate cell cycle regulation, inflammation, cholesterol metabolism, and breakdown of the extracellular matrix in the development of AAA; however, the definition of AAA used in these studies was based soley on diameter. This sequencing study identified novel genetic signals associated with distinct morphologic subtypes of AAA. Patients with thoracic aortic disease and an AAA were associated with the variant rs79508780 in TSEN54, two genes (BATF3 and SMLR1), and a set of genes involved in repair of double-stranded DNA breaks, as well as suggested association with the genes EFCAB3 and TAF4. None of the variants, genes, or gene set identified in this study have been previously associated with a diagnosis of either AAA or AD.
The variant rs79508780 identified in association with dissection morphology is located in the gene TSEN54. TSEN54 encodes a subunit of the tRNA splicing endonuclease complex involved in the processing of mature RNA and has been implicated in the development of pontocerebellar hypoplasia types 2, 4, and 5.16,17 Pontocerebellar hypoplasia is a rare, prenatal neurodegenerative disorder characterized by cerebellum atrophy, microcephaly, and delayed development.16 Although other mutations in the TSEN54 gene can have highly deleterious effects, rs79508780 is reported to be benign and is a low frequency common variant found in 5.8% of the non-Finnish European population (gnomAD–Non-Finnish European). Interestingly, the MAF in patients with AAA and thoracic aortic disease was higher than the reported non-Finnish European frequency, whereas the frequency in patients with a normal thoracic aorta was lower than the expected frequency for a person of European descent (MAF = 0.113 and MAF = 0.016, respectively).
BATF3 encodes a protein that is a member of the basic leucine zipper family. These proteins form a heterodimer with JUN family proteins. This heterodimer structure recognizes and binds to the 5ʹ end of the DNA sequence thus regulating the expression of target genes. The current target predictions include repression of transcription of IL-1 and matrix metalloproteinase 1.18 Matrix metalloproteinases are a family of endonucleases that regulate the turnover of various components of the aortic wall and have been widely implicated in the development of aortic aneurysms and dissections.19 SMLR1 encodes small leucine rich protein 1, which is located on chromosome 6. Although the exact function of small leucine rich protein 1 has not been characterized, it has been shown to be coexpressed with BSCL2, Berardinelli-Seip congenital lipodystrophy 2, which is a regulator of lipid catabolism.20 An atherosclerosis prone mouse model (LDLR-/-) showed an increased plaque burden when BSCL2 was knocked out.21 A high plaque burden and penetrating atherosclerotic plaques can initiate PAUs and eventual aneurysm formation.22
TAF4 encodes a TATA-box binding protein associated factor 4. TAF4 is a component of the transcription factor IID a protein that coordinates the assembly of RNA polymerase by positioning the polymerase and serves as a scaffold for the other components of the complex.18 Although not genome-wide significant, TAF4 has been reported in the NHGRI-EBI GWAS catalog with a P value of 9E-6 as a potential regulator of the level of circulating plasma factor VII activating protease also called hyaluronic acid binding protein 2 (HABP2).23 HABP2 is an extracellular protein involved in blood coagulation and fibrinolysis. Increased concentrations of HABP2 in atherosclerotic plaques made the plaques less stable, which could result in plaque rupture and downstream complications, such as coronary artery stenosis or development of PAU.24 Although the incidence of hypercholesterolemia was not different between the two morphologic groups, differences inTAF4 could result in the defects in the structure of the aortic wall observed in group 2 study patients, and the mechanism behind this and possible mediation by statin use requires further investigation. In addition, the formation of HABP2/inhibitor complex is internalized via the low-density lipoprotein receptor related protein (LRP).25 LRP1, a member of the LRP protein family, has been previously implicated in the development of both AD9 and AAA.26 In addition, TAF4 potentiates the transcription of vitamin D3 receptors.27 Low concentrations of vitamin D or vitamin deficiencies have been previously associated with both dissection and dilation of the thoracic aorta and AAA.28,29 EFCAB3 encodes an EF-hand calcium binding domain 3. EF-hand motifs are the most commonly found calcium-binding domain. In many cases, binding of calcium induces a conformational changes that can expose another active site or target binding area such as the binding of calmodulin to myosin kinase.30 Calmodulin can form a complex with many of the nitric oxide synthases. Inhibition of nitric oxide synthase in murine models has reduced aneurysm progression by inhibiting the production of matrix metalloproteases suggesting nitric oxide synthase 2 as a potential drug target for treating aortic aneurysms and dissections.31,32
The single gene set GO_DNA_DOUBLE_STRAND_BREAK_PROCESSING, which includes genes related to the repair of a double-stranded DNA break, was significantly associated with the presence of thoracic aortic disease in patients with AAA. Genes included in this gene set have also been implicated in the development of various cancers, neurological syndromes like Nijmegen breakage syndrome, DNA replication and repair, and apoptosis.33 Interestingly, three of the genes in this gene set, BARD1, BLM, and RBBP8, are associated with the BRCA1-mediated DNA repair pathway in response to oxidative stress.34 Increased expression of BRCA1 has been shown to protect vascular smooth muscle cells from oxidative stress in murine and cell-based models.35 Additionally BRCA1 over expression in murine models of atherosclerosis exhibited higher levels of endothelial nitric oxide synthetase, lower levels of reactive oxygen species, and decreased atherosclerosis.36
This study had a few limitations. Study criteria included only patients with confirmed European ancestry, and although this factor increased the study's overall power to detect genetic variation, it limited the applicability of the results to patients of European descent. Further investigation of additional populations is necessary to identify genetic variation associated with structural defects in the descending thoracic aorta in non-European populations. Additionally, the study had a limited sample size; despite the biological relevance of both BATF3 and SMLR1, the significance of each was driven by two SNVs, one common and one rare, with the rare SNV only observed in a single person. These signals could be real, but also could be statistical artifacts generated by the inclusion of a single minor allele. A larger sample size could bestow greater power, enabling us to determine whether these gene-based signals are spurious or real. The second major limitation is the lack of replication in a large independent cohort. The dual requirement of specific imaging and whole genome/exome sequencing limits the use public databases for evaluation of an independent replication cohort and also. Further replication in a larger independent cohort or model will be necessary to validate the finding of this study. The final limitation is the lack of tissue sample evaluation to determine downstream effects of these variants on transcription and translation. However, because the use of endovascular approaches to aortic aneurysm repair increases, aortic tissue becomes less available. The use of high-resolution imaging in concert with previous findings from tissue, in vivo, and in vitro studies may provide a noninvasive solution for understanding the biological mechanisms underlying AAA.
Conclusions
We propose a methodology using next generation sequencing and CTA analysis for evaluating endophenotypes possibly related to the etiology of AAA. Our pilot investigation suggests novel associations of one variant (rs79508780), four genes (BATF3, SMLR1, EFCAB3, and TAF4), and one gene set (DNA double strand break processing) with the presence of imaging features related to structural defects in the descending thoracic wall in patients with an AAA. The genes associated with the AAA and thoracic aortic disease in this pilot study are involved in pathways related to regulation of matrix metalloproteases, cholesterol metabolism, vitamin D signaling, tRNA splicing, and DNA repair in response to oxidative stress. This pilot study provides further evidence that AAA is not a single disease and is rather resultant from a number of different pathophysiologies. Although the results of this study will require further validation and replication in the future, this study suggests the application of a radiogenomic approach for identifying specific variants and genes that can differentiate two AAA endophenotypes: patients with both AAA and thoracic aortic disease and patients with isolated AAA and a normal thoracic aorta. Furthermore, the two morphologies analyzed in this study have been previously linked to differential sac regression rates following endovascular aneurysm repair, which suggests that a radiogenomic approach to AAA classification may also have usefulness in making procedural and patient management decisions.
AUTHOR CONTRIBUTIONS
Conception and design: GM, AR, ET, RT, EM, PF, MM
Analysis and interpretation: GM, AR, RL, MM
Data collection: GM, ET
Writing the article: GM, ET
Critical revision of the article: AR, RT, EM, RL, PF, MM
Final approval of the article: GM, AR, ET, RT, EM, RL, PF, MM
Statistical analysis: GM, AR, ET
Obtained funding: Not applicable
Overall responsibility: GM
Acknowledgments
We are very thankful to Shea J. Andrews, PhD, and Brian Fulton-Howard, PhD, for their assistance with the principle component analysis.
Footnotes
Supported in part through the Icahn School of Medicine at Mount Sinai and through the resources and staff expertise provided by the Charles Bronfman Institute for Personalized Medicine and The BioMeTM Biobank Program at the Icahn School of Medicine at Mount Sinai and the Office of Research Infrastructure of the National Institutes of Health under award numbers S10OD018522 and S10OD026880. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author conflict of interest: none.
The editors and reviewers of this article have no relevant financial relationships to disclose per the JVS-Vascular Science policy that requires reviewers to decline review of any manuscript for which they may have a conflict of interest.
Appendix
References
- 1.Nordon I.M., Hinchliffe R.J., Loftus I.M., Thompson M.M. Pathophysiology and epidemiology of abdominal aortic aneurysms. Nat Rev Cardiol. 2011;8:92–102. doi: 10.1038/nrcardio.2010.180. [DOI] [PubMed] [Google Scholar]
- 2.Rossella F., Piergiorgio C., Paola De R., Martin C., Arturo E., Christoph N. Interdisciplinary expert consensus document on management of Type B aortic dissection. J Am Coll Cardiol. 2013;61:1661–1678. doi: 10.1016/j.jacc.2012.11.072. [DOI] [PubMed] [Google Scholar]
- 3.Durham C.A., Cambria R.P., Wang L.J., Ergul E.A., Aranson N.J., Patel V.I. The natural history of medically managed acute type B aortic dissection. J Vasc Surg. 2015;61:1192–1199. doi: 10.1016/j.jvs.2014.12.038. [DOI] [PubMed] [Google Scholar]
- 4.Fiotti N., Calvagna C., Sgorlon G., Altamura N., Pitacco P., Zamolo F. Multiple sites of vascular dilation or aneurysmal disease and matrix metalloproteinase genetic variants in patients with abdominal aortic aneurysm. J Vasc Surg. 2017;67:1727–1735. doi: 10.1016/j.jvs.2017.09.047. [DOI] [PubMed] [Google Scholar]
- 5.Li T., Lv Z., Jing J.J.J., Yang J., Yuan Y. Matrix metalloproteinase family polymorphisms and the risk of aortic aneurysmal diseases: a systematic review and meta-analysis. Clin Genet. 2018;93:15–32. doi: 10.1111/cge.13050. [DOI] [PubMed] [Google Scholar]
- 6.Keeling B.W., Armstrong P.A., Stone P.A., Bandyk D.F., Shames M.L. An overview of matrix metalloproteinases in the pathogenesis and treatment of abdominal aortic aneurysms. Vasc Endovascular Surg. 2005;39:457–464. doi: 10.1177/153857440503900601. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X., Wu D., Choi J.C., Minard C.G., Hou X., Coselli J.S. Matrix metalloproteinase levels in chronic thoracic aortic dissection. J Surg Res. 2014;189:348–358. doi: 10.1016/j.jss.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris D.R., Biros E., Cronin O., Kuivaniemi H., Golledge J. The association of genetic variants of matrix metalloproteinases with abdominal aortic aneurysm: a systematic review and meta-analysis. Heart. 2014;100:295–302. doi: 10.1136/heartjnl-2013-304129. [DOI] [PubMed] [Google Scholar]
- 9.Guo D.-C., Grove M.L., Prakash S.K., Eriksson P., Hostetler E.M., LeMaire S.A. Genetic variants in LRP1 and ULK4 are associated with acute aortic dissections. Am J Hum Genet. 2016;99:762–769. doi: 10.1016/j.ajhg.2016.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vilacosta I., San Román J.A. Acute aortic syndrome. Heart. 2001;85:365–368. doi: 10.1136/heart.85.4.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spittell P.C., Spittell J.A., Joyce J.W., Tajik A.J., Edwards W.D., Schaff H.V. Clinical features and differential diagnosis of aortic dissection: experience with 236 cases (1980 through 1990) Mayo Clin Proc. 1993;68:642–651. doi: 10.1016/s0025-6196(12)60599-0. [DOI] [PubMed] [Google Scholar]
- 12.Miner G., Taubenfeld E., Tadros R., Han D., Marin M. Decreased abdominal aortic aneurysm size following EVAR in patients with CT evidence of subclinical thoracic aortic dissection. Ann Vasc Surg. 2020;66:95–103. doi: 10.1016/j.avsg.2019.10.091. [DOI] [PubMed] [Google Scholar]
- 13.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones G.T., Tromp G., Kuivaniemi H., Gretarsdottir S., Baas A.F., Giusti B. Meta-analysis of genome-wide association studies for abdominal aortic aneurysm identifies four new disease-specific risk loci. Cir Res. 2017;120:341–353. doi: 10.1161/CIRCRESAHA.116.308765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Namavar Y., Chitayat D., Barth P.G., van Ruissen F., de Wissel M.B., Poll-The B.T. TSEN54 mutations cause pontocerebellar hypoplasia type 5. Eur J Hum Genet. 2011;19:724–726. doi: 10.1038/ejhg.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasher P.R., Namavar Y., Tijn Pv, Fluiter K., Sizarov A., Kamermans M. Impairment of the tRNA-splicing endonuclease subunit 54 (tsen54) gene causes neurological abnormalities and larval death in zebrafish models of pontocerebellar hypoplasia. Hum Mol Genet. 2011;20:1574–1584. doi: 10.1093/hmg/ddr034. [DOI] [PubMed] [Google Scholar]
- 18.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X., Shen Y.H., LeMaire S.A. Thoracic aortic dissection: are matrix metalloproteinases involved? Vascular. 2009;17:147–157. doi: 10.2310/6670.2008.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolland T., Taşan M., Charloteaux B., Pevzner S.J., Zhong Q., Sahni N. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao J., Liu X., Gao M., Wang M., Wang Y., Wang F. Dyslipidemia, steatohepatitis and atherogenesis in lipodystrophic apoE deficient mice with Seipin deletion. Gene. 2018;648:82–88. doi: 10.1016/j.gene.2018.01.062. [DOI] [PubMed] [Google Scholar]
- 22.Gifford S.M., Duncan A.A., Greiten L.E., Gloviczki P., Oderich G.S., Kalra M. The natural history and outcomes for thoracic and abdominal penetrating aortic ulcers. J Vasc Surg. 2016;63:1182–1188. doi: 10.1016/j.jvs.2015.11.050. [DOI] [PubMed] [Google Scholar]
- 23.Olsson M., Stanne T.M., Pedersen A., Lorentzen E., Kara E., Martinez-Palacian A. Genome-wide analysis of genetic determinants of circulating factor VII-activating protease (FSAP) activity. J Thromb Haemost J Thromb Haemost. 2018;16:2024–2034. doi: 10.1111/jth.14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanse S.M., Parahuleva M., Muhl L., Kemkes-Matthes B., Sedding D., Preissner K.T. Factor VII-activating protease (FSAP): vascular functions and role in atherosclerosis. Thromb Haemost. 2008;99:286–289. doi: 10.1160/TH07-10-0640. [DOI] [PubMed] [Google Scholar]
- 25.Muhl L., Nykjaer A., Wygrecka M., Monard D., Preissner K.T., Kanse S.M. Inhibition of PDGF-BB by Factor VII-activating protease (FSAP) is neutralized by protease nexin-1, and the FSAP-inhibitor complexes are internalized via LRP. Biochem J. 2007;404:191–196. doi: 10.1042/BJ20061630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bown M.J., Jones G.T., Harrison S.C., Wright B.J., Bumpstead S., Baas A.F. Abdominal aortic aneurysm is associated with a variant in low-density lipoprotein receptor-related protein 1. Am J Hum Genet. 2011;89:619–627. doi: 10.1016/j.ajhg.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mengus G., May M., Carré L., Chambon P., Davidson I. Human TAF(II)135 potentiates transcriptional activation by the AF-2s of the retinoic acid, vitamin D3, and thyroid hormone receptors in mammalian cells. Genes Dev. 1997;11:1381–1395. doi: 10.1101/gad.11.11.1381. [DOI] [PubMed] [Google Scholar]
- 28.Vianello E., Dozio E., Barassi A., Tacchini L., Lamont J., Trimarchi S. Vitamin D deficiency is associated with increased osteocalcin levels in acute aortic dissection: a pilot study on elderly patients. Mediators Inflamm. 2017;2017:6412531. doi: 10.1155/2017/6412531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takagi H., Umemoto T., Alice g. Vitamins and abdominal aortic aneurysm. Int Angiol. 2017;36:21–30. doi: 10.23736/S0392-9590.16.03618-X. [DOI] [PubMed] [Google Scholar]
- 30.Meador W.E., Means A.R., Quiocho F.A. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 31.Pimiento J.M., Maloney S.P., Tang P.C.Y., Muto A., Westvik T.S., Fitzgerald T.N. Endothelial nitric oxide synthase stimulates aneurysm growth in aged mice. J Vasc Res. 2008;45:251–258. doi: 10.1159/000112940. [DOI] [PubMed] [Google Scholar]
- 32.Oller J., Méndez-Barbero N., Ruiz E.J., Villahoz S., Renard M., Canelas L.I. Nitric oxide mediates aortic disease in mice deficient in the metalloprotease Adamts1 and in a mouse model of Marfan syndrome. Nat Med. 2017;23:200–212. doi: 10.1038/nm.4266. [DOI] [PubMed] [Google Scholar]
- 33.Chrzanowska K.H., Gregorek H., Dembowska-Bagińska B., Kalina M.A., Digweed M. Nijmegen breakage syndrome (NBS) Orphanet J Rare Dis. 2012;7:13. doi: 10.1186/1750-1172-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi Y.W., Kang H.J., Bae I. BRCA1 and oxidative stress. Cancers (Basel) 2014;6:771–795. doi: 10.3390/cancers6020771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lovren F., Pan Y., Quan A., Singh K.K., Khan R., Gupta N. BRCA1 shields vascular smooth muscle cells from oxidative stress. J Thorac Cardiovasc Surg. 2014;147:1946–1955. doi: 10.1016/j.jtcvs.2013.09.060. [DOI] [PubMed] [Google Scholar]
- 36.Singh K.K., Shukla P.C., Quan A., Al-Omran M., Lovren F., Pan Y. BRCA1 is a novel target to improve endothelial dysfunction and retard atherosclerosis. J Thorac Cardiovasc Surg. 2013;146:949–960. doi: 10.1016/j.jtcvs.2012.12.064. [DOI] [PubMed] [Google Scholar]