Abstract
Objective
Somatic mosaicism of KRAS gene is currently recognized as the only established molecular basis of arteriovenous malformations (AVM). However, given the limitations of the current technologies, KRAS somatic mutations are detected only in a limited proportion of AVMs and tissue biopsy remains an invasive high risky, sometimes life-threatening, diagnostic procedure. Next-generation sequencing liquid biopsy using cell-free DNA (cfDNA) has emerged as an innovative noninvasive approach for early detection and monitoring of cancer. This approach overcomes the space-time profile constraint of tissue biopsies opens a new scenario for vascular malformations owing to somatic mosaicism. Here, we propose a new approach as a fast noninvasive reliable tool in order to investigate the cfDNA coming from the AVMs.
Methods
A group of five patients suffering from AVM were selected. Blood samples from peripheral vein and efferent vein from vascular malformation were collected and cfDNA was extracted. The cfDNA libraries were performed using Oncomine Pan-Cancer Cell-Free Assay. We used Ion Proton for sequencing and Ion Reporter Software for analysis (Life Technologies, Carlsbad, Calif).
Results
In all cases, either G12D or G12V mutations in KRAS were identified. The mutational load was higher in the efferent vein than in peripheral blood, confirming the causative role of the identified mutation at a somatic level.
Conclusions
We demonstrate that cfDNA next-generation sequencing liquid biopsy is able to identify the KRAS mutation detected in affected tissues. Moreover, we have shown that blood sample withdrawal at the lesion site increases variant allele frequency with an order of magnitude above the limit of detection (usually 0.05%), decreasing the risk of a false negative. Finally, the noninvasiveness of the method avoids any risk of bleeding, being easily performed also in children. We propose this technique as the method of choice to better investigate AVMs and consequently to identify the therapy tailored to the genetic defect.
Clinical Relevance
This article highlights the importance of using liquid biopsy as a new method to investigate the molecular profile of AVMs. In view of the frequent inaccessibility of vascular tissues owing to the invasiveness of solid biopsy and the relative high incidence of biopsies with low diagnostic power, here we evaluated the efficacy of detecting cfDNA fragments released into the bloodstream from the affected tissue cells. Through a simple blood draw from the efferent vein at the vascular malformation site, the liquid biopsy allowed us to identify KRAS pathogenic mutations piloting a personalized therapeutic approach and opening a new scenario for new therapeutic strategies.
Keywords: Arteriovenous malformation, Liquid biopsy, cf-DNA, KRAS mutation, Noninvasive technique
Article Highlights.
-
•
Type of Research: multicenter, prospective study
-
•
Key Findings: Cell-free DNA next-generation sequencing liquid biopsy is able to identify somatic KRAS mutations in 100% of investigated cases without the need of tissue biopsy. The mutational load is higher in the plasma sample collected in the efferent vein from vascular malformation than in peripheral vessel, confirming the causative role of the identified mutation.
-
•
Take Home Message: We propose this noninvasive technique as the method of choice for arteriovenous malformation investigation and identification of tailored therapy.
Arteriovenous malformations (AVM) are fast-flow vascular malformations composed of tangles of abnormally developed vasculature. The absence of capillaries between arteries and veins often leads to high blood pressure and rupture. They can occur in some part of the body, including the brain.1,2 In the majority of cases an activating KRAS mutation has been identified.1, 2, 3, 4, 5 These KRAS variants have been previously described as gain-of-function mutations in cancer and Nikolaev et al3 have recently shown that hot spot mutations in KRAS, namely p.G12V are associated with arteriovenous brain malformations through an increase in angiogenesis, migration and cell proliferation.6 During the last years, next-generation sequencing (NGS) liquid biopsy has emerged as an innovative noninvasive technique for the identification of key mutations that are responsible for tumor growth allowing to optimize diagnosis, monitoring, and therapeutic choice.7, 8, 9 Therefore, cell-free DNA (cfDNA) analysis has the possibility to overcome the space-time profile constraint of physical biopsies and it opens a new scenario for vascular malformations where tissue biopsy represents an invasive, high-risk, sometime life-threatening, diagnostic procedure. The use of liquid biopsy would also improve the opportunity to monitor illness evolution at a molecular level.
In the present study, we performed a comprehensive analysis of five patients with AVMs to determine if noninvasive NGS liquid biopsy from the efferent vein at the lesion site could detect the key variant bypassing the need for a high-risk, life-threatening tissue biopsy. The blood from the efferent vein at the vascular malformation site was sampled during embolization procedures before the injection of embolizing materials or liquids without causing further discomfort to the patient. For this reason, we define this technique as noninvasive. The NGS-liquid biopsy at the venous malformation site detected pathogenic mutations in KRAS gene in each patient. This study was consistent with Institutional guidelines and approved by the ethical committees of Azienda Ospedaliera Senese, Siena.
Methods
Patient enrollment and sample collection
Five patients affected by AVM were enrolled at the Medical Genetics Unit of the Azienda Ospedaliera Universitaria Senese, Siena, Italy, for a new diagnostic approach. Written informed consent for genetic analysis was obtained from all patients. Clinical information as well as genealogic trees and cancer family history were collected on a genetic consultation setting. For all patients, liquid biopsy withdrawal from the lesion efferent vein was performed by Vascular Surgery of Ospedale Maggiore di Crema. All the blood specimens were taken during embolization procedures before the injection of embolizing materials or liquids. All the patients underwent a complete hemodynamic and radiologic evaluation including computed tomography angiography or magnetic resonance angiography. According to the Yakes Classification, these shunts belong to category II or III.
Extraction of cfDNA from plasma
Blood samples (10 mL) were collected from each patient and placed into cfDNA BCT blood collection tube (Streck, Neb). The cfDNA was extracted from 4 ml of plasma using MagMAX cell-free Total Nucleic Acid Isolation Kit (ThermoFisher Scientific, Waltham, Mass), according to the manufacturer's instructions. The quality and quantity of cfDNA were verified respectively using the Agilent High Sensitivity DNA Kit (Agilent Technologies, Palo Alto, Calif) on Agilent2100 Bioanalyzer (Agilent Technologies) and Qubit dsDNA HS Assay Kits on Qubit 2.0 fluorometer (Invitrogen, Carlsbad, Calif).
NGS sequencing on cfDNA
The cfDNA sequencing was performed using Oncomine Pan-Cancer Cell-Free Assay (ThermoFisher Scientific) on Life Technologies Ion Proton sequencer (Life Technologies, Carlsbad, Calif). This technology identifies various types of alterations, including single nucleotide variants, insertions/deletions, gene fusions and copy number variations in cancer-related genes (clinical actionable mutations) with a reportable range up to 0.05%. Sequencing analysis was performed using Ion Reporter Server System (Thermo Fisher Scientific).
Results
Patient 1 is a 56-year-old man with a congenital port wine stain angioma on the lower right leg. He also suffers from varicose veins since the age of 18. At the age of 54, after meniscal injury, magnetic resonance imaging of the right knee showed an AVM with the presence of flow voids and enlargement of the arterial and venous vessels in the medial and lateral compartment, and bone involvement at femoral and tibial level; the morphology of the shunt is compatible with type III B according to the classification of Wayne Yakes
Patient 2 is a 40-year-old woman with a congenital port wine stain angioma on the external part of the lower right leg, associated with hypertrophy. Angiography and computed tomography angiography showed a complex angiodysplasia of the right limb, which presented with multiple arteriovenous fistulas, high-flow fistulas involving the tibial bone malformation, and abnormal communication between the common iliac artery and vein. Over the time, the patient underwent multiple embolizations and resection of arteriovenous fistulas. Because of limb length discrepancy, the patient underwent the elongation of the contralateral limb with the Ilizarov technique. Previous molecular analysis with a targeted panel did not identify any RASA1 pathogenic mutation, excluding Parkes-Weber syndrome.
Patient 3 is a 29-year-old man with congenital angiodysplasia of the lower left limb, extending to the groin-abdominal region. Several endovascular and surgical interventions have been performed since childhood. He underwent a below-the-knee amputation at the age of 14 because of life-threatening bleeding and experienced a recurrence of AV shunts on the stump 10 years later. The patient reported episodes of nose bleeding, also present in other family members, but he did not satisfy Curacao criteria for a clinical diagnosis of hereditary hemorrhagic telangiectasia.
Patient 4 is a 45-year-old woman with congenital red vascular malformation in the upper right limb associated with limb hypermetry. The patient underwent several devascularization procedures, both surgical and endovascular, and resections over time.
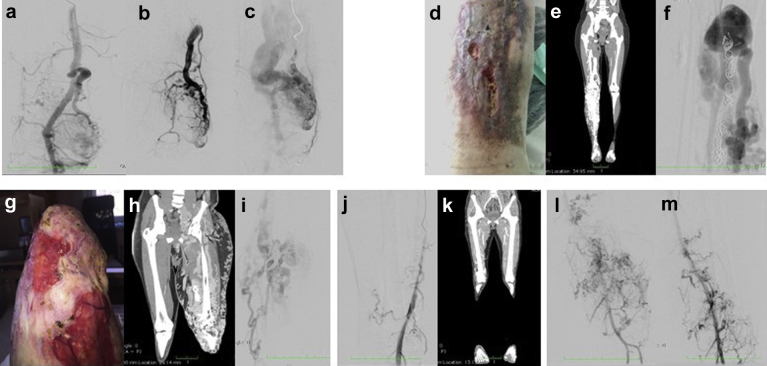
Patient 5 is a 40-year-old woman who was referred for red-purple spots on the left lower limb, with hypertrophy and dysmetry. The first clinical manifestation dates back to the age of 11 years, with deep venous thrombosis in the left lower limb. The patient underwent multiple endovascular procedures, and surgical removal of an aneurysm at the level of the femoral artery. The sampling for the cfDNA analysis was taken from the extremity AVM. Magnetic resonance imaging identified an additional hepatic arteriovenous fistula. Clinical features of all patients are shown in the Fig.
Fig.
Clinical features of the patients. a-c, Patient 1: intra-articular arteriovenous shunt and outflow vein where the blood specimen was taken. d-f, Patient 2: right lower limb arteriovenous malformation (AVM) with leg ulceration (d). Computed tomography (CT) scan (e) and angiography (f) of diffuse arteriovenous shunt at the tibial artery level. g-i, Patient 3: left lower stump AVM with leg ulceration (g). CT scan (h) and angiography (i) with huge diffuse arteriovenous shunting. j, k, Patient 4: angiography of the right lower limb (j) and CT reconstruction of arteriovenous shunt at the knee level (k). l, m, Patient 5: angiography of diffuse arteriovenous shunt.
In patients 1, 3, and 5, NGS-liquid biopsy analysis detected the same pathogenic mutation in KRAS gene c.35G>A; p.(Gly12Asp) (Table). In patients 2 and 4, NGS liquid biopsy analysis identified the mutation in KRAS gene c.35G>T; p.(Gly12Val). The variant allele frequency (VAF) ranged from 0,19% to 4,10%. In patient 1, who had the lowest VAF, a second blood draw from the efferent vein of AVMs of the knee was performed. The percentage of VAF in this experiment increased of one order of magnitude from 0.19% to 1.63%. Clinical features and molecular findings are shown in the Fig and the Table.
Table.
Patient clinical features and molecular findings
| Patient | Code number | Gender | Age, years | Phenotype | Peripheral NGS liquid biopsy | Efferent vein NGS liquid biopsy |
|---|---|---|---|---|---|---|
| 1 | 2023/19 | Male | 56 | AVM dx leg | KRAS (p.(G12D)) 0.19% | KRAS (p.(G12D)) 1.63% |
| 2 | 4417/19 | Female | 40 | AVM dx leg | na | KRAS (p.(G12V)) 4.11% |
| 3 | 4477/19 | Male | 29 | AVM sx leg | na | KRAS (p.(G12D)) 1.18% |
| 4 | 4820/19 | Female | 45 | AVM dx leg | na | KRAS (p.(G12V)) 4.19% |
| 5 | 3859/19 | Female | 40 | AVM sx leg | na | KRAS (p.(G12D)) 1.18% |
AVM, Arteriovenous malformation; dx, dextrum; na, not applicable; NGS, next-generation sequencing; sx, sinistrum.
Discussion
AVMs are a nontumor subset of vascular anomalies owing to a dysmorphogenesis in the developmental process.
In view of the increasing role of endovascular treatments, the frequent inaccessibility of vascular tissues, the invasiveness of solid biopsy, and the relative high incidence of nondiagnostic biopsies, we evaluated the efficacy of detecting cfDNA fragments that are released into the bloodstream from the affected tissue cells. Different cells, including normal healthy cells and hematopoietic cells, contribute to the cfDNA in the blood. However, in efferent venous blood, major cfDNA is released by cells; thus, this technique is highly sensitive for circulating cfDNA, allowing the identification of pathogenic mutations in KRAS genes, even with a very low VAF percentage in all patients.
In patient 1, liquid biopsy from the efferent vein detected an enrichment for the causative KRAS mutation, allowing a search for a causative role in the angiodysplastic process. Taken together, our data strengthen the idea that KRAS somatic mosaicism for gain-of-function mutations is the key genetic driver involved in the development of AVMs. Furthermore, our data suggest that our novel approach, based on the combination of NGS and liquid biopsy from the efferent vein at the vascular malformation site, allows detecting even low-grade somatic mosaicism responsible of the vascular phenotype, thus bypassing the need for a high-risk tissue biopsies.
Interestingly, the identified KRAS mutations (p.Gly12Asp and p.Gly12Val) are cancer hotspot mutations, which increase MAPK-ERK pathway activation, thus inducing endothelial cell proliferation3 and enhancing their migratory behavior.10, 11, 12, 13, 14, 15 Noteworthy, although individuals who harbor RAS mutations at a germline level present with an increased risk of tumors such a juvenile myelomonocytic leukemia, acute leukemia, neuroblastoma, and rhabdomyosarcoma,16 no increased risk for cancer development has so far been clearly shown for patients with AVMs who harbor somatic KRAS mutations. According to recent lines of evidence, drugs that specifically target a cancer driver gene represent innovative repurposing-based treatments readily available for use in different settings for hereditary conditions or somatic mosaic syndromes that carry the same driver genomic aberration.14,17 Thus, in the new era of personalized medicine, agents that inhibit the MAP-ERK pathway commonly used in phase II clinical trials for the treatment of several solid tumors18 could be likely considered as a therapy for sporadic brain AVMs caused by KRAS mutations. In conclusion, the diagnostic use of liquid biopsy for nononcologic diseases will forward the development of personalized therapeutic approach for AVMs and will open the door to new therapeutic strategies potentially able to block or slow down disease progression.
Author contributions
Conception and design: AR, MV
Analysis and interpretation: MP, LD, EF, AS, AP, AR, MV
Data collection: MP, AC, AT, LD, GD, MB, EF, AG, CF, AP, AR, MV
Writing the article: MP, AC, AT, LD, GD, MB, EF, AG, CF, AS, AP, AR, MV
Critical revision of the article: EF, AP, AR, MV
Final approval of the article: MP, AC, AT, LD, GD, MB, EF, AG, CF, AS, AP, AR, MV
Statistical analysis: Not applicable
Obtained funding: Not applicable
Overall responsibility: AR
Acknowledgments
We thank AVMs patients and ILA association (Italian association of childhood angiodysplasias and hemangiomas). The “Cell lines and DNA bank of Rett Syndrome, X-linked mental retardation and other genetic diseases”, member of the Telethon Network of Genetic Biobanks (project no. GTB12001 and GFB18001), funded by Telethon Italy, and of the EuroBioBank network provided us with specimens. We thank SienaGenTest srl, a Spin-off of the University of Siena (www.sienagentest.dbm.unisi.it) for assessment of data analysis.
Footnotes
Funded by ONG ILA.
Author conflict of interest: none.
The editors and reviewers of this article have no relevant financial relationships to disclose per the JVS-Vascular Science policy that requires reviewers to decline review of any manuscript for which they may have a conflict of interest.
Contributor Information
Alessandra Renieri, Email: alessandra.renieri@unisi.it.
Massimo Vaghi, Email: vaghim@yahoo.it.
References
- 1.Oka M., Kushamae M., Aoki T., Yamaguchi T., Kitazato K., Abekura Y. KRAS G12D or G12V mutation in human brain arteriovenous malformations. World Neurosurg. 2019;126:e1365–e1373. doi: 10.1016/j.wneu.2019.03.105. [DOI] [PubMed] [Google Scholar]
- 2.Nikolaev S.I., Vetiska S., Bonilla X., Boudreau E., Jauhiainen S., Jahromi B.R. Somatic activating KRAS mutations in arteriovenous malformations of the brain. N Engl J Med. 2018;378:250–261. doi: 10.1056/NEJMoa1709449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikolaev S.I., Vetiska S., Bonilla X., Boudreau E., Jauhiainen S., Rezai Jahromi B. Somatic activating KRAS mutations in arteriovenous malformations of the brain. N Engl J Med. 2018;378:250–261. doi: 10.1056/NEJMoa1709449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng F., Nussinov R. KRAS activating signaling triggers arteriovenous malformations. Trends Biochem Sci. 2018;43:481–483. doi: 10.1016/j.tibs.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Priemer D.S., Vortmeyer A.O., Zhang S., Chang H.Y., Curless K.L., Cheng L. Activating KRAS mutations in arteriovenous malformations of the brain: frequency and clinicopathologic correlation. Hum Pathol. 2019;89:33–39. doi: 10.1016/j.humpath.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Starke R.M., McCarthy D., Komotar R.J., Connolly E.S. Somatic KRAS mutation found in sporadic arteriovenous malformations. Neurosurgery. 2018;83:e14–e15. doi: 10.1093/neuros/nyy163. [DOI] [PubMed] [Google Scholar]
- 7.Palmieri M., Baldassarri M., Fava F., Fabbiani A., Gelli E., Tita R. Two point-NGS analysis of cancer genes in cell free-DNA of metastatic cancer patients. Cancer Med. 2020;9:2052–2061. doi: 10.1002/cam4.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowley E., Di Nicolantonio F., Loupakis F., Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 9.Corcoran R.B., Chabner B.A. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018;379:1754–1765. doi: 10.1056/NEJMra1706174. [DOI] [PubMed] [Google Scholar]
- 10.McDonald J., Bayrak-Toydemir P., Pyeritz R.E. Hereditary hemorrhagic telangiectasia: an overview of diagnosis, management, and pathogenesis. Genet Med. 2011;13:607–616. doi: 10.1097/GIM.0b013e3182136d32. [DOI] [PubMed] [Google Scholar]
- 11.Gallione C.J., Repetto G.M., Legius E., Rustgi A.K., Schelley S.L., Tejpar S. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4) Lancet. 2004;363:852–859. doi: 10.1016/S0140-6736(04)15732-2. [DOI] [PubMed] [Google Scholar]
- 12.Bayrak-Toydemir P., McDonald J., Markewitz B., Lewin S., Miller F., Chou L.S. Genotype–phenotype correlation in hereditary hemorrhagic telangiectasia: mutations and manifestations. Am J Med Genet A. 2006;140:463–470. doi: 10.1002/ajmg.a.31101. [DOI] [PubMed] [Google Scholar]
- 13.Revencu N., Boon L.M., Mulliken J.B., Enjolras O., Cordisco M.R., Burrows P.E. Parkes Weber syndrome, vein of Galen aneurysmal malformation, and other fast-flow vascular anomalies are caused by RASA1 mutations. Hum Mutat. 2008;29:959–965. doi: 10.1002/humu.20746. [DOI] [PubMed] [Google Scholar]
- 14.Luks V.L., Kamitaki N., Vivero M.P., Uller W., Rab R., Bovée J.V.M.G. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J Pediatr. 2015;166:1048–1054.e1-5. doi: 10.1016/j.jpeds.2014.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Do Prado L.B., Han C., Paul Oh S., Su H. Recent advances in basic research for brain arteriovenous malformation. Int J Mol Sci. 2019;20:5324. doi: 10.3390/ijms20215324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kratz C.P., Franke L., Peters H., Kohlschmidt N., Kazmierczak B., Finckh U. Cancer spectrum and frequency among children with Noonan, Costello, and cardio-facio-cutaneous syndromes. Br J Cancer. 2015;112:1392–1397. doi: 10.1038/bjc.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burotto M., Chiou V.L., Lee J.M., Kohn E.C. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120:3446–3456. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braicu C., Buse M., Busuioc C., Drula R., Gulei D., Raduly R. A comprehensive review on MAPK: a promising therapeutic target in cancer. Cancers (Basel) 2019;11:1618. doi: 10.3390/cancers11101618. [DOI] [PMC free article] [PubMed] [Google Scholar]