Abstract
Nearly all FDA approved drugs and bioactive small molecules exert their effects by binding to and modulating proteins. Consequently, understanding how small molecules interact with proteins at an atomic level is a central challenge of modern chemical biology and drug development. Complementary to structure-guided approaches, chemoproteomics has emerged as a method capable of high-throughput identification of proteins covalently bound by small molecules. To profile noncovalent interactions, established chemoproteomic workflows typically incorporate photoreactive moieties into small molecule probes, which enable trapping of small molecule-protein interactions (SMPIs). This strategy, termed photoaffinity labelling (PAL), has been utilized to profile an array of small molecule interactions, including for drugs, lipids, metabolites, and cofactors. Herein we describe the discovery of photocrosslinking chemistries, including a comparison of the strengths and limitations of implementation of each chemotype in chemoproteomic workflows. In addition, we highlight key examples where photoaffinity labelling has enabled target deconvolution and interaction site mapping.
Graphical Abstract
Introduction
Small molecules, including chemical probes, natural products, FDA-approved drugs, and clinical candidates are widely employed tools used to manipulate and investigate biological systems. Consequently, determining the physiologically relevant protein targets and modes of action (MoA) of these bioactive molecules is a central challenge of chemical biology, pharmacology, and drug development programs. Complementary to structure-based and genetic approaches (e.g. CRISPR-Cas9 screens1, 2), mass spectrometry-based proteomic methods have emerged as favored tools for deconvolving the small molecule protein interactome, distinguished by their high throughput nature and compatibility with in cell and in vivo studies. Showcasing the utility of proteomics for target deconvolution, application of the method Cellular Thermal Shift Assay (CETSA) to target deconvolution has revealed the functional targets of anti-malarial drugs,3 cellular dynamics of cancer drugs,4, 5 and streamlined the drug discovery process using high-throughput thermal profiling methods such as System-wide Identification of Enzyme Substrates by Thermal Analysis (SIESTA).6 Affinity enrichment studies, using small molecules immobilized on resin have been successfully adopted for target deconvolution studies, notably for assaying the kinome (using kinobeads)7, 8 and chromatin-associated proteins (using BET bromodomain inhibitors bound to resin).9
Proteomics is particularly well suited to deconvolve the protein targets of small molecules that react irreversibly with amino acid side chains. Methods aimed at assaying such covalent compounds can be grouped into two general categories (1) those that rely on mechanism-based probes to assay specific classes of proteins (e.g. kinases,10, 11 proteases,12, 13 and hydrolases,14–16 and (2) those that employ highly electrophilic probes that react promiscuously with specific amino acid side chains.17–20
Activity based protein profiling (ABPP) is a hallmark example of a mechanism-based approach. ABPP implements a chemical probe that contains both a warhead (e.g. epoxide to react with mammalian histone deacetalases (HDACs),21 fluorophosphonate to react with serine hydrolases, 14 vinyl sulfone to react with the proteasome,22–24 or acyloxymethyl ketone to react with cysteine proteases25, 26) and an enrichment/visualization handle (e.g. radiolabel, biotin, azide/alkyne, or fluorophore). While these probes were initially employed primarily to fractionate the proteome based on a specific enzymatic activity, subsequent utilization in a “competitive” format has allowed for inhibitor discovery and target deconvolution, including for the clinical candidate molecule BIA 10–2474.27 For readers less familiar with the distinction between direct labeling and competition studies, in the former the probe is used to label and enrich proteins of interest, whereas in the latter samples are pre-treated with either an active molecule or vehicle. Subsequently both treated and control samples are labeled with the activity-based probes and labeling by the molecule of interest is reported by a decreased fluorescent signal in gel-based studies or a decreased MS1 signal during MS/MS analysis. Competition studies offer the advantage of delineating high and low occupancy labeling events, whereas direct labeling studies can prove particularly useful for identification of the precise sites of protein-probe interactions.
The advent of reactivity-based chemoproteomics began with the Isotope-Coded Affinity Tag (ICAT) reagents, which featured a cysteine-reactive electrophile and a biotin enrichment handle.28 First used primarily to simplify complex proteomes by fractionation based on the presence of cysteine residues, related reagents and workflows have subsequently been applied to identify redox-sensitive cysteines, hyper-reactive or pKa perturbed cysteines20 and small molecule ligandable or potentially druggable cysteines.17 With the advent of additional electrophiles, such chemoproteomic studies have been extended to a number of additional amino acid side chains, including lysine, 29 methionine,30 tyrosine,19 serine,31–33 and glutamate/aspartate.34–36 However, a key limitation of both reactivity- and activity-based profiling methods is their reliance on covalent mechanisms of labeling, which by definition exclude small molecules that interact reversibly with proteins of interest (POIs).
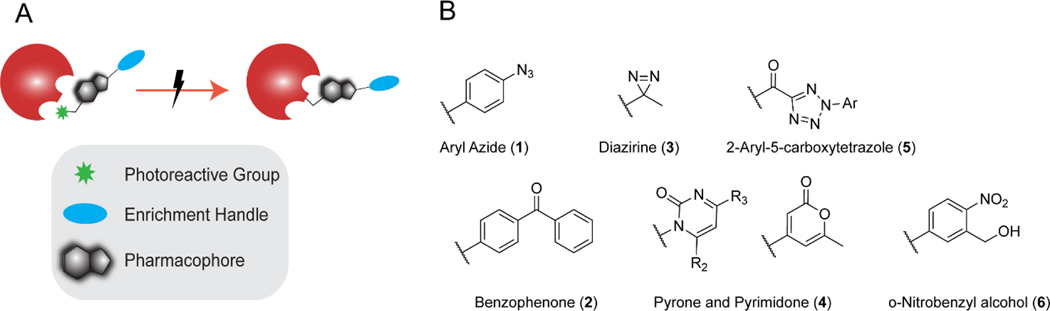
Photoaffinity groups serve as latent reactive groups unleashed only upon irradiation with light. Therefore, incorporation of a photoactive moiety allows for irreversible trapping of reversible interactions and by doing so enables the technical innovations of reactivity and activity-based profiling platforms to be extended to dynamic and transient small molecule-protein interactions (SMPIs). This method termed photoaffinity labeling (PAL) commonly employs both a photoreactive group and enrichment handle on the small molecule of interest (pharmacophore). When irradiated with light, the photoaffinity probe covalently modifies nearby proteins, which can then be identified via protein enrichment and MS analysis (Figure 1A,B). Beyond small molecules, photocrosslinking has also been applied to capture protein-protein (using unnatural amino acid incorporation),37–40 protein-oligonucleotide,41–45 protein-cofactor,46, 47 protein-lipid,48, 49 and protein-carbohydrate interactions.50, 51 For several excellent prior reviews on these areas, we would direct the readers to Murale’s review of protein-protein interactions (PPIs),52 Peng’s and Laguerre’s protein-lipid photocrosslinking review,53, 54 Trads’ review of PAL for DNA-protein interactions,55 and Wu’s review of trapping carbohydrate-protein interactions.56 These methods have been extended to many organisms, including Mycobacterium tuberculosis,57 Pseudomonas aeruginosa,58 Staphylococcus aureus,59 and Caenorhabditis elegans.60
Figure 1.
(A) Shows general workflow for photoaffinity labeling (PAL) between pharmacophore (shown in black) and protein of interest (POI, shown in red) (B) Structures of commonly used photoreactive groups.
Here we aim to provide an in-depth update on the state-of-the-art developments in PAL, including novel reactive groups and their applications to target deconvolution by chemoproteomics with particular emphasis on efforts to map the precise interaction sites and modes of action of small molecules. We also will provide a retrospective analysis of the history of the field, including the pioneering efforts that led to the first applications of photocrosslinkers to protein chemistry and biological systems. Finally, we will delve into some of limitations of photolabeling strategies and will highlight future opportunities both in the areas of new labeling strategies and mass spectrometry-based proteomic platforms, which together should unlock the full potential of PAL to map SMPIs.
1. Photoaffinity Labeling Chemistries.
1.1. Discovery of Photoactive Groups.
The scientific roots of photoaffinity labeling for studying SMPIs can be found in chemical photoregulation of enzymes and in applications of photoactivatable groups to chemical modification of proteins. In one of the earliest applications of protein photochemistry, the enzyme chymotrypsin was chemically modified with p-nitrophenyl diazoacetate to generate the photocaged inactive diazoacetyl chymotrypsin.61 Upon exposure to UV light (tungsten lamp or sunlight), chymotrypsin’s esterolytic activity was restored, supporting a mechanism whereby the reactive carbene generated upon photolysis was quenched by water and the acyl enzyme further hydrolyzed to regenerate the active enzyme and release glycolic acid. As ~25% of the enzyme activity was not restored upon photolysis, irreversible modification of chymotrypsin by the carbene was postulated. Shafer and coworkers further analyzed the photoproducts of the diazoacetyl chymotrypsin reaction and found that carboxymethyl serine and carboxymethyl tyrosine were two reaction products, further supporting irreversible enzyme modification by a carbene intermediate.62
Demonstrating the generalizability and the broad scope of amino acids compatible with this diazo labeling chemistry, covalent modification of the enzyme trypsin at an alanine residue was achieved upon photolysis of a diazoacyl-modified trypsin.63 Detailed mechanistic studies revealed that, upon exposure to UV light, ɑ-diazo carbonyl compounds, such as ethyl diazoacetate yield two major products, the carbene insertion into water and a rearranged product analogous to the Wolf rearrangement of diazo ketones.64 Despite these initial promising findings, the widespread adoption of diazo compounds for photolabeling was hindered by the notorious instability of this compound class.65 Also, the requirement for adjacent carbonyl functionality—diazo compounds are known to show light-independent protein reactivity, most notably towards cysteine thiols and the imidazole ring of histidine,66, 67—proved problematic.
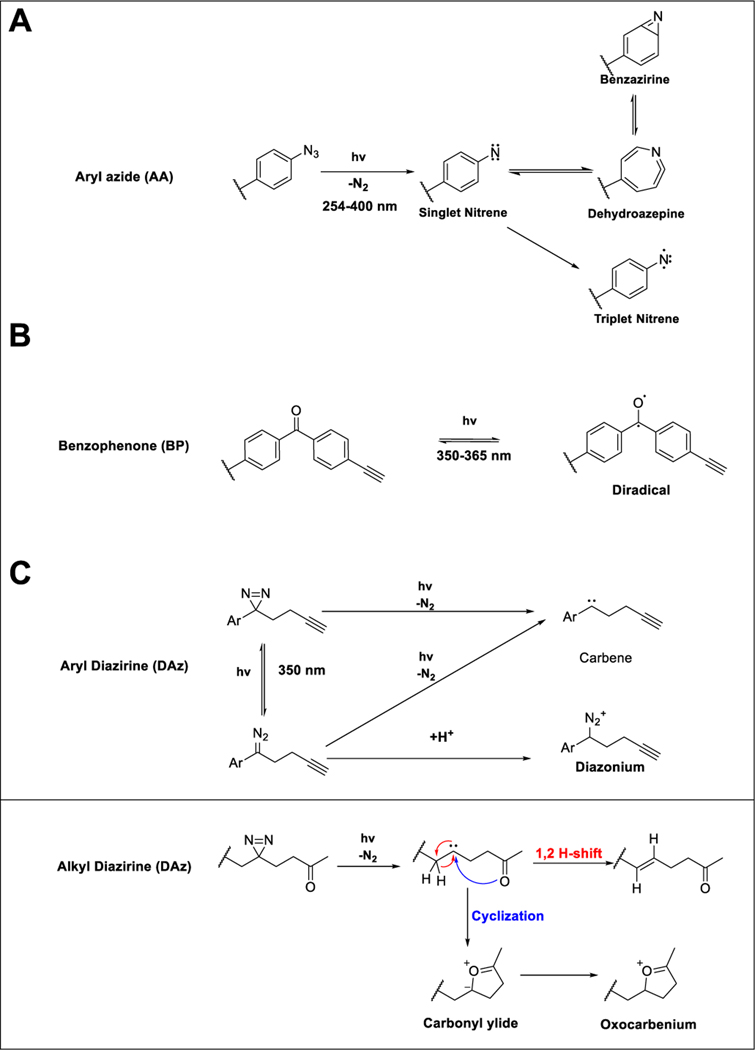
To circumvent these limitations, azides (Figure 2A) were developed as second-generation PAL reagents. Looking beyond photocaged enzymes, Fleet and coworkers chemically modified both the bovine antibody gamma globulin and human serum albumin to contain a 4-azido-2-nitrophenyl (NAP) group, with the objective of trapping interacting antibodies. Using these modified proteins, immunoglobulin G (IgG) was identified as the primary interacting antibody. Further studies utilizing radioactive NAP analogs revealed NAP localization to the antigen-binding site.68 This chemistry was then extended to small molecule-protein interaction mapping for acetylcholine, using an azide-functionalized analogue, both in red blood cell membranes and in frog sartorius muscle cells.69 In pioneering work aimed at studying protein-phospholipid interactions, Khorana demonstrated the photolysis and crosslinking of azido-modified fatty acids.70 However, the use of azides in photoaffinity labeling was hindered by the relatively short wavelength of light needed for activation and long lifetime of the nitrene intermediate.
Figure 2.
(A) Photolysis of aryl azide to a singlet nitrene, which can either relax to a triplet nitrene or isomerize to a dehydroazepine or benzazirine. (B) Photolysis of benzophenone to a triplet diradical. (C) Photolysis of diazirine to a carbene or diazo species, which can subsequently lose nitrogen to form the reactive carbene or gain a proton to form a diazonium species. The carbene from alkyl diazirines can further self-quench via a 1,2 H-shift. Alternatively, if the handle bears a carbonyl the carbene can undergo cyclization forming a carbonyl ylide or oxocarbenium species.
The emergence of the benzophenone (Figure 2B) and diazirine (Figure 2C) groups addressed many of these limitations. The preparation and photodecomposition of aryl diazirines were first evaluated by Knowles,71 and shortly thereafter 3-phenyl-3H-diazirine and adamantyl diazirine photoprobes were applied to identify small molecule-lipid interactions,72 which revealed that the lipid-labeling capacity of diazirines was not quenched by glutathione, in contrast with the thiol-sensitivity of the aryl azides. Following the promising application of diazirines to label lipid bilayers, diazirine labeling was subsequently extended to protein labeling using the probe 3H-adamantyl diazirine and the intrinsic membrane protein glycophorin A, which revealed interaction with multiple sites in the hydrophobic region of the protein.73
As the rapid quenching of carbenes by water was perceived as a limitation for protein labeling studies, aryl ketones, which form diradicals upon UV irradiation were also investigated as an alternative photolabel that demonstrated improved stability and inertness towards water.74 Acetophenone and benzophenone (Figure 2B) were benchmarked against aryl azide, using the peptide pentagastrin, with the model proteins bovine serum albumin (BSA) and lysozyme.74 Both ketone- and azide-based labels afforded comparable labeling, with the benzophenone distinguished by decreased protein damage afforded by the longer wavelength (320 nm) of light employed.
In addition to these three established photoreactive groups, considerable effort has also been made to discover additional functionalities compatible with PAL. In one key example, Battenberg and colleagues looked to nature and identified pyrones and pyrimidones (Figure 1B) as photoactivatable structural motifs naturally present in many natural products and bioactive molecules. When photoactivated with 350 nm light, pyrones yield a reactive ketene intermediate while pyrimidones afford either a bicyclic urea or isocyanate intermediate, all of which are susceptible to nucleophilic crosslinking with adjacent amino acids.75
Continuing in the theme of nature-inspired photoactivatable handles, another functional group found in many natural products is the tetrazole moiety (Figure 1B). The photolability and subsequent nucleophilic capture of the diaryltetrazole (DAT) was first reported by Rolf Huisgen in 1961.76 Nearly forty years later, the tetrazole functionality was debuted as a bioorthogonal group for bioconjugation, functioning via irreversible loss of nitrogen to form a reactive nitrile imine species that is then trapped by an alkene through a 1,3-dipolar cycloaddition.77 This reactive intermediate could also be intercepted with carboxylate functionalities and as such was proposed as a photoreactive group for PAL experiments.36, 78 A similar handle, the 2-aryl-5-carboxytetrazole (ACT; Figure 1B), was developed in parallel and was reported to have higher crosslinking yield when compared with structurally matched benzophenone and diazirine probes.79 In addition to DAT and ACT, other photoreactive groups identified within the past five years include o-nitrobenzyl alcohols (o-NBA; Figure 1B), as lysine specific photoactivatable groups, and o-(hydroxymethyl)phenols (o-HMP) as tryptophan specific groups.80, 81 Upon irradiation at 365 nm, o-NBA forms a reactive nitroso aldehyde species which can quickly react with proximal lysine side chains and o-HMP generates ortho quinone methides, which react selectively with tryptophan residues.
1.2. Pros and Cons of Photoreactive Groups.
As the early photolabeling discovery efforts revealed, general instability and side reactivity, particularly towards thiols, and requirement for protein-damaging wavelengths of light are ubiquitous liabilities of photoactive groups. Thus, the choice of photoreactive group should be guided by these potential limitations. As highlighted above, the diazo group, although highly reactive, has generally been sidelined for protein labeling due to instability and UV-independent labeling. Therefore, the primary photoactivatable groups used in protein labeling studies are benzophenones, aryl azides, and diazirines. Here we will briefly expound upon the mechanisms of labeling and the strengths and limitations of each photolabel (Table 1, respectively), with the goal of providing a template for judicious label selection for future studies. Photolysis of aryl azides (Figure 2A) using relatively short wavelength light (300 nm) generates a singlet nitrene intermediate which can react with nearby biomolecules via two pathways: (1) relaxation to a triplet nitrene and subsequent C-H bond insertion or (2) ring expansive isomerization to either an dehydroazepine or benzazirine, which are both susceptible to nucleophilic attack with lysine amines and cysteine thiols, respectively.82 The relatively long half-life of the nitrene and ring expanded species may result in an increased radius of labeling, compared with other groups. The relatively short wavelength required to generate these species is known to cause protein damage, especially since PAL experiments can require constant irradiation for a prolonged period of time (e.g. 10 minutes).83 Despite these many limitations, aryl azides are still widely used for protein labeling studies, likely in large part due to the compatibility of the comparably small azide moiety with applications that preclude bulkier substitutions.
Table 1.
Comparison of different photoreactive groups
| Photoreactive Group | Activation Wavelength | Reversibility of Excited State | Functional Group Selectivity | Refs |
|---|---|---|---|---|
| Aryl Azide (1) | 300 nm | Irreversible | Non-Specific | 68, 69 |
| Benzophenone (2) | 350 nm | Reversible | Non-specific | 74 |
| Diazirine (3) | 350 nm | Irreversible | Non-specific | 71, 73 |
| Pyrone and Pyrimidone (4) | 350 nm | Reversible | Nucleophilic | 75 |
| 2-Aryl-5-carboxytetrazole (5) | 302 nm | Irreversible | Carboxylate | 79 |
| o-Nitrobenzyl alcohol (6) | 365 nm | Irreversible | Amine | 18, 80 |
Addressing many of the limitations of aryl azides, benzophenones are widely adopted for photolabeling. In contrast with aryl azides and the structurally related acetophenone photolabel, the comparably long wavelength required for benzophenone photoactivation (350–365 nm) decreases protein destruction.74 While the diradical formed upon irradiation (Figure 2B) can react with water, this quenching is reversible, as dehydration will regenerate the ground state benzophenone. Consequently, benzophenones can be repeatedly excited over prolonged irradiation periods, affording increased protein labeling. Benzophenones are typically straightforward to incorporate synthetically by coupling 4-benzoylbenzoic acid to alcohol and amine groups. Due to its inherently large size, benzophenone-based probes are not suited to applications intolerant of bulky substitutions. In addition, because benzophenone excitation is reversible, these moieties require significantly longer irradiation times (e.g. 0.5–2 hours) and can act as photosensitizers leading to oxidative damage of proteins.84
In many ways, aryl and alkyl diazirines (Figure 2C) are ideal alternatives to both benzophenone and aryl azides. These functionalities are photoactivated with 350 nm light and, upon irradiation, irreversibly eliminate N2 gas, resulting in a highly reactive carbene species. Carbenes can non-specifically insert into R-H and C-H bonds.72 Due to their many beneficial features, diazirines are now ubiquitous in small molecule protein interaction site mapping studies. Carbenes are generally highly reactive, affording comparably efficient protein labeling after short irradiation periods, using longer wavelengths of light, which, as described for benzophenones, decreases the risk of protein damage. Photoactivated diazirines are reactive towards all twenty amino acid side chains as well as the peptide backbone, although marked bias towards labeling of polar and nucleophilic amino acids has recently been reported for alkyl diazirines.85
This biased reactivity likely stems from an alternate photodegradation pathway where, upon UV irradiation, the diazirine isomerizes to form a diazo species. This diazo species can either lose N2 and form the desired carbene or, in acidic media, become protonated to the diazonium species which are known alkylating agents (Figure 2C). Nucleophilic amino acid side chains, particularly carboxylates, are known to react with diazo and diazonium species,86–88 which rationalizes the aforementioned biased amino acid reactivity of alkyl diazirines. Whether aryl diazirines show comparable amino acid reactivity profiles to that observed for alkyl diazirines remains unexplored, though we expect that the decreased basicity of the aryl diazirines compared with their alkyl counterparts should disfavour diazonium formation and afford a decrease in the polar amino acid bias.
Recently, O’Brien and coworkers demonstrated that dialkyl diazirines can proceed through more than just these two defined pathways. Specifically, if the photoprobe contains a carbonyl, upon photolysis the resulting carbene can intramolecularly cyclize forming carbonyl ylides and oxocarbenium intermediates that can alkylate proximal biomolecules. Alternatively these species can be quenched by solvent or through a 1,2-hydride shift (Figure 2C).89 Another important liability to consider for probe development efforts is the propensity of alkyl diazirines to modify proteins in a UV-independent manner. Exemplifying this limitation, diazirine-containing analogues of S-adenosyl-L-homocysteine (SAH) were found to label proteins during CuAAC bioorthogonal labeling.47 It is intriguing to speculate that the CuAAC reaction conditions catalyze the formation of the aforementioned diazo species.
Incorporation of a difluoromethylene unit alpha to the diazirine is one notable strategy that has been pursued to simultaneously decrease the size and increase the stability of diazirine tags.90 Exemplifying this strategy, by incorporating a difluoromethylene unit connecting both the diazirine and alkyne Chang and coworkers obtained a difluoro photoreactive handle with high stability and small size (6.9 Å compared to the standard 9.4 Å handle).90 As an added benefit, the difluoro substituent also accelerates both strain-promoted azide-alkyne cycloaddition and CuAAC.91, 92 Prior mechanistic studies analyzing the photoactivation of 3-trifluoromethyl-3-phenyldiazirine revealed a product distribution that consisted of 65% carbene and 35% diazoisomer, the latter being stable in mildly acidic conditions for 12 hours. Supporting the stability of the carbene, no internal rearrangement of the carbene via fluorine migration was observed 93 However, recent benchmarking of this minimalist difluoro (DF) tag revealed poor labeling efficiency relative to other diazirine reagents In contrast, a terminal diazirine tag showed comparably efficient protein labeling with minimal background reactivity.94
2. The Advent of Affinity-Based Protein Profiling (AfBP) using Photoaffinity Chemistry
Due to an absence of suitable enrichment strategies and limited mass spectrometry capabilities, most early studies employing photoaffinity labels were restricted to in vitro tagging of recombinant proteins. The move to more complex systems was first enabled by combining radiolabeling with photoaffinity labeling,95 comparable to early ABPP studies, which incorporated radioisotopic labels into electrophilic probes.96 Modified proteins were then visualized by gel-based autoradiography and identified after fractionation by MS analysis, as was shown for thiazolidinedione photoprobes.97 With the advent of non-radioactive ABPP methods that rely on fluorophore-, biotinyl-,98 or azide/alkyne-visualization and enrichment handles, identification of the protein targets of chemical probes was vastly streamlined. While protein targets were initially identified by Edman degradation protein sequencing, advances in tandem mass spectrometry, including most notably increased data collection speed and resolution afforded by the development of the Orbitrap,99 obviated the need for substantial sample fraction and allowed for more rapid identification of labeled proteins. Hence, the fields of activity- and affinity-based chemoproteomics were born.
2.1. AfBP chemoproteomics to identify the protein targets of small molecules.
Affinity-based chemoproteomic studies using small molecule photoprobes all rely on the same general workflow (Figure 3). First cells or cell lysates are labeled with the photoactivatable reagent. The system is then subjected to ultraviolet (UV) irradiation and the modified proteins can be visualized by SDS-page and in-gel fluorescence and/or identified by MS/MS analysis after enrichment and sequence-specific proteolytic digestion. In one of the first examples of such affinity-based protein profiling, hydroxamate photoprobes featuring a benzophenone and fluorophore/enrichment handles, were used to profile matrix metalloproteinases (MMPs) in complex proteomes. Using these probes, a comparison of invasive and non-invasive cancer cell proteomes revealed differential expression of several MMPs, including neprilysin, highlighting the utility of AfBP to identify potential therapeutic targets.100 Complementary to this study, a similar report demonstrated that tetrapeptide AfBPs that featured hydroxamate functionality together with diazirine or benzophenone photoreactive groups could be used to identify MMPs, including the unique recognition sequences of individual enzymes.101 The utility of this substrate fingerprinting strategy was further demonstrated by combining a ‘cocktail’ of AfBPs with mass spectrometry identification to profile MPs in cancer proteomes.102
Figure 3.
General workflow for photoaffinity-activity-based chemoproteomics. 1) Cells are incubated with photoprobe, irradiated, and lysed. 2) Labeled proteins can be fluorescently labeled and imaged on gel. 3) Alternatively, labeled proteins can be enriched w/ a capture handle (e.g. biotin) digested and identified through MS/MS analysis. For both the gel-based and MS-based assays, blockade of probe labeling by pre-treatment with an untagged competitor molecule prior to irradiation can facilitate delineation of high and low affinity binding events.
In addition to mapping MMP interactions, affinity-based chemical proteomics has proven valuable for uncovering the protein targets of a wide range of other small molecules. For example, the anti-cancer candidate compound Arenobufagin, a steroid component of toad venom that is widely used in traditional Chinese medicine and naturally contains a pyrone group, has been studied using this approach. Similarly, Encequidar (HM30181), a P-glycoprotein inhibitor, contains a photoactive tetrazole. Capitalizing on the presence of these groups, Ma and coworkers conducted chemoproteomic analyses of the targets of both HM30181 and Arenobufagin,103 which identified Poly [ADP-ribose] polymerase 1 (PARP1) as a functional off-target of Arenobufagin potentially responsible for its toxicity. In a similar fashion, malate dehydrogenase 2 (MDH2) was identified as a functional target of a previously identified HIF-1ɑ inhibitor LW6, using a clickable diazirine probe bearing the aryloxyacetyl-amino benzoic ester scaffold of LW6.104 The utility of PAL for mode-of-action studies was further showcased by the discovery, of the FoF1-ATP synthase as the active target for the anti-trypanosomal activity of bis-tetrahydropyran 1,4-triazole (B-THP-T).105
Building upon these earlier studies, Parker and coworkers combined photoaffinity labeling with fragment based screening—a method that uses small minimalized compounds as scout molecules and initial leads for mapping potentially druggable sites on proteins106, 107—to generate a chemoproteomic map of the protein interactions of a 15-member fully functionalized fragment (FFF) library.108 A key finding from this study was the identification of >400 protein-ligand interactions, including 83% of proteins without previously identified ligands in the DrugBank,109 supporting that this form of proteome-wide affinity-based interaction mapping can discover new druggable pockets in proteins previously inaccessible to small molecule binding. This study also demonstrated that FFFs are useful for phenotypic screening applications, enabling rapid target deconvolution of hit molecules. By screening the FFF library in an adipogenesis assay, napthyl-compound 25 was identified to promote adipogenesis. Chemoproteomic enrichment studies revealed progesterone receptor membrane component 2 (PGRMC2) as the functional target responsible for this activity, which was further verified using genetic approaches. Extension of the FFF method to map the interactions of 16 chiral FFF probes (8 (R) and 8 (S)) revealed 108 enantiomer-specific protein interactions.110
In further extensions of this methodology, PAL-chemoproteomic analysis recently revealed that carbonic anhydrase is an off target of MCC950, an NLR-family inflammasome pyrin domain-containing 3 (NLRP3) inhibitor.111 A PAL-chemoproteomic approach was also recently used to confirm the cell-based on-target activity of peptidomimetic RAS ligands.112 Recent work using a pyridine photoactivatable group has been applied to assay the targets of JNJ-40411813, a metabotropic glutamate receptor subtype 2 (mGlu2) modulator that was withdrawn from clinical trials due to adverse effects.113 As shown in Table 2, the interactomes of a wide range of drugs, lipids, metabolites, and co-factors have been profiled by photoaffinity labelling, showcasing the generalizability of the method, as will be described in more detail in the following sections.
Table 2.
Examples of PAL studies for drug, lipid, metabolite, cofactor interactions.
| Entry | Probe | Photolabel | Protein Target | Site of Labeling | Refs |
|---|---|---|---|---|---|
| Drugs | |||||
| Celecoxib, Indomethacin, Naproxen | Diazirine | COX1, COX2, PTGES | For Celecoxib | 115 116 | |
| Δ8/9-THC | Diazirine | Reep5, Mtch2, Gnb1, Cox4il | N/A | 117 | |
| Arenobufagin, HM30181 | Diazirine ɑ-Pyrone | PARP1, RAB6A/6B/39A, SRI | N/A | 103 | |
| HM30181 | Diazirine Diaryltetrazole | NOP2, DDX5, ERGIC1, RAB6A/6B, and UCDH | N/A | 103 | |
| PFQX | 2-aryl-5-carboxytetrazole | AMPAR | Yes | 118 | |
| Dasatinib | 2-aryl-5-carboxytetrazole, Diazirine | Abl, BTK, Fym, Lck, SRC, PDGFRB. SIK1, TIMK2 | For BTK | 119.79 | |
| Nintedanib | Diazirine | TPP1 | No | 120 | |
| Lipids | |||||
| Sterol | Diazirine | HeLa Proteome | N/A | 48 | |
| Fatty Acid | Diazirine | PTGS1 PTGS2 IFITM3 | N/A | 121,122,60 | |
| Sphingolipid | Diazirine | Caveolin 1, PITPβ, Nicastrin, p24 | N/A | 123, 124,49, 125 | |
| Phospholipid | Benzophenone Diazirine | MDA-MB-435 Proteome A431 Proteome | N/A | 126, 127 | |
| Metabolite/Cofactor | |||||
| PIP2 | Diazirine | ARF1 | N/A | 128 | |
| iE-DAP | Diazirine | NOD1, ARF6 | N/A | 129 | |
| MDP | Diazirine | NOD2 | N/A | 129 | |
| NAD+ | Aryl azide Diazirine | IL-2, PARP1, PARP10, Ctbp1, Psmc2, AK1, AK2, TRMT10C/MRPP1, (other targets not listed) | For PARP1 | 46, 130 | |
| SAH | Aryl Azide, Benzophenone, Diazirine | MT, MT-associated proteins, and proteins known to bind onto SAH | For Probe 2 47 | 47, 131, 132 | |
| GTP | Benzophenone | BSC1L, GNL3, CSNK2A2, ATL3, OPA1, (other targets not listed) | N/A | 120 | |
| ATP | Benzophenone Aryl azide | CK2, IL-2 | Active site in CK2 | 130 133, 134 | |
| Aminoacyl-AMS | Benzophenone | Nonribosomal peptide synthetases | No | 135 |
The choice of mass spectrometry-based enrichment and quantification strategies are relevant for all AfBP analyses described in this review. While early studies relied on spectral counting to identify bona fide ligands, the advent of stable isotope labeling with amino acids in cell culture (SILAC) allowed for more precise stratification of interactions, particularly for proteins only identified by several tryptic peptides, such as low abundance proteins.114 In these studies, cells are isotopically labeled with either light (14N-, 12C-) or heavy light (15N-, 13C-) lysine and arginine and then the relative enrichment quantified on the MS1 level, comparing heavy and light chromatographic peak areas. A distinct disadvantage of SILAC is that many cell lines, most notably primary cells, are not amenable to metabolic labeling. As an alternative strategy, fully functionalized enrichment handles that incorporate an isotopically enriched azido acetate for click capture, diphenylsilane cleavable linker, and biotin moiety for enrichment, have been found to enable quantification of photo-affinity labeled peptides.115 The incorporation of isobaric tandem mass tags (TMTs) into chemoproteomic workflow has also recently enabled increased sample throughput and data reproducibility.110
2.2.1. Drugs and Clinical Candidates.
Target deconvolution and mode-of-action studies are an essential part of modern drug development pipelines that together help to establish the safety and efficacy of clinical candidates. Among the many available techniques often employed to establish the protein binders of clinical candidates (e.g. CETSA, genetic screens, and affinity capture), PAL, particularly when coupled to a chemoproteomic readout, has emerged as a favored approach for small molecule interactome mapping efforts. Application of PAL has revealed the on- and off-target activity of a number of clinical candidates and even FDA-approved drugs (Table 2).
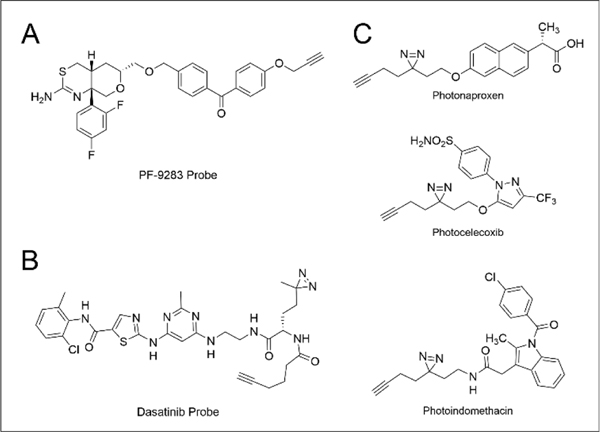
While BACE1 (β-site APP-cleaving enzyme 1 inhibitors have received considerable attention as promising candidates for treatment and prevention of Alzheimer’s disease, most BACE inhibitors, including Amgen preclinical candidate AMG-8718 and Lilly & Co. clinical candidate LY2811376, have been withdrawn from development due to toxicity and BACE1-independent activity. Zuhl and coworkers136 applied a chemoproteomic strategy, using a benzophenone probe, to inform the mechanism of ocular toxicity of the Pfizer BACE inhibitor PF-9283 (Figure 4A), designating Cathepsin D as a major off-target of PF-9283 and the primary driver of its ocular toxicity.
Figure 4.
Structure(s) of photoreactive probes of (A) PF-9283, (B) Dasatinib, and (C) NSAIDs.
PAL labeling strategies have proven informative for assessing the selectivity of kinase inhibitors, which is particularly important given the high sequence and structural homology of the 700+ members of the kinase family. For example, chemoproteomic profiles using diazirine and benzophenone analogues of Dasatinib (Figure 4B), an FDA approved Bcr-Abl inhibitor used to treat chronic myelogenous leukemia, revealed 84 Dasatinib interactors, including both kinase and non-kinase proteins.119 Consistent with this promiscuity, Dasatinib-mediated inhibition of Src and c-Kit have been implicated as contributing to Dasatinib’s anti-cancer efficacy.137 Similar studies have also been conducted to analyze the interactomes of non-steroidal anti-inflammatory drugs (NSAIDs), including celecoxib, naproxen, and indomethacin (Figure 4C). Roughly seven hundred protein interactors were identified between the three probes with an overlap of only 187 interactors between the three, supporting substantial promiscuity. Curiously, the interactomes observed for two cell lines (K562 and Jurkat) were largely distinct, supporting the importance of assaying multiple cell types, including those with disease- and side-effect relevance.115
In more recent work, Tripeptidyl-peptidase 1 (TPP1) was identified as a new target of the triple angiokinase inhibitor Nintedanib, which may in part rationalize the efficacy of this molecule for a number of indications, including lung carcinoma and interstitial lung diseases.120
Looking beyond clinical candidates, photolabeling strategies have also extended to traditional medicines and psychoactive natural products. For Δ−8 and Δ−9 tetrahydrocannabinol (THC), chemoproteomic analysis using a THC-diazirine probe captured not only the CB1 receptor but also four additional high-affinity interactors, Reep5, Mtch2, Gnb1, and Cox4il 117. These additional binders may help to rationalize some of the adverse effects of long-term THC usage, most notably memory loss, which in term should help to guide the usage of THC in indications where it has proven efficacious, such as moderating the pain of multiple sclerosis (MS) 138, cancer, and AIDS.139
2.2.2. Lipids.
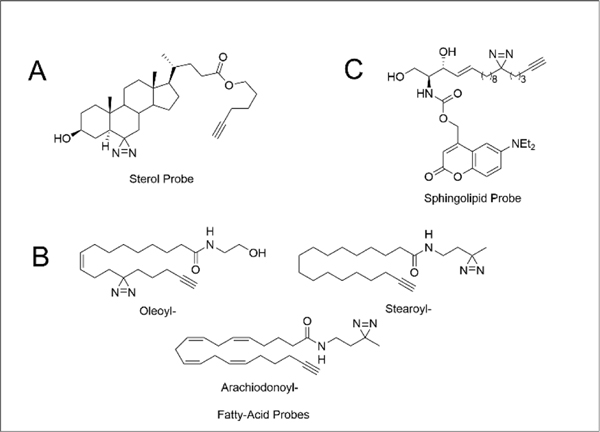
In addition to traditional small-molecule drugs, PAL has proven to be a useful method for identifying lipid-protein interactions, including for sterol,48 sphingolipids,49, 125 phospholipids,126, 127 and fatty acids.60, 121 When compared with conventional methods140–142 for mapping protein-lipid binding events (e.g. radiolabeling or limited proteolysis), PAL-lipid probes offer several unique advantages, including compatibility with cell-based studies and stratification of high- and low-affinity binders in vivo. Using three photo-sterol probes, cis-, trans-, and epi- (Figure 5A), Hulce and coworkers identified over 800 cholesterol-protein interactions in HeLa cells.48 Of these 800 interactions, 265 were also competed by pretreatment with excess cholesterol, further supporting their high affinity and cholesterol-specific nature.
Figure 5.
Structure(s) of (A) trans-sterol probe, (B) fatty acid probes and (C) sphingolipid probe.
Niphakis and coworkers next sought to extend this methodology to additional lipid-protein interactions. In this report, lipids probes featuring arachidonoyl-, oleoyl-, palmitoyl-, and stearoyl- acyl chains were prepared incorporating both diazirine and alkyne functionalities (Figure 5B).121 The authors generated a map of lipid-protein interactions by comparing the protein targets of each lipid probe both with direct enrichment and competition studies using a SILAC chemoproteomic platform. A key finding from this study was a propensity for lipid binding by proteins that have known drug-binding activity. Consequently, lipid photoaffinity probes can be used to assay target engagement of active small molecules. As a demonstration of this screening capacity, flurbiprofen, a PTGS1 and PTGS2 inhibitor was found to competitively block lipid probe labeling for both enzymes. The Lipid-Protein Interaction Profiling (LiPIP) platform expanded on this concept of lipid probes for small molecule target deconvolution, using fatty-acid based probes121 to profile the ligandability of small-molecules.143 As a demonstration of the utility of this approach, LiPIP uncovered the targets of multiple small-molecule drugs, including the antidiabetic agent KDT501. When compared with other PAL-labeling strategies, LiPIP offers the distinct advantage of eliminating the need for synthesis of custom probes for each active small molecule.
Looking to the future, one key, and as yet to be not fully addressed, challenge for the study of lipid-protein interactions is the transient nature of these binding events and relatively fast kinetics of many lipid-mediated signaling cascades. Trifunctional lipid probes that contain a photocaging group, photocrosslinker, and enrichment handle represent an intriguing option to address these challenges, allowing for sequential photo reactions in cells (first uncaging and then photocrosslinking; Figure 5C). Exemplifying this premise, trifunctional sphingosine (TFS), trifunctional diacylglycerol (TFDAG), and trifunctional fatty acid (TFFA) probes have all been employed for protein interaction identification subcellular localization analyses. Application of the TFS probe to Niemann-Pick disease type C (NPC) cells identified late endosomal/lysosomal vesicles as the site of sphingosine storage in this cell line.49 in a similar manner, photocaged phosphatidylinositol bis- and trisphosphate (PIP) probes have enabled the de novo identification of lipid binding proteins involved in PIP transport to the plasma membrane.144
2.2.3. Metabolite/Cofactor.
Much like lipids and small molecules, metabolites and protein cofactors are ripe for photoaffinity interactome analysis. The compatibility of photoaffinity labeling studies to cell-based assays sets them apart from other useful methods, including limited proteolysis,145, 146 thermal proteome profiling,147, 148 and resin-based affinity capture studies.149
The kinome is a large 700+ member family that has been extensively profiled using photoaffinity, covalent active site directed, and reversible probes. Both the desthiobiotin-ATP developed by Activx150,10 and Kinobeads151 have been used extensively for small molecule screening and functional studies. Complementary to these platforms, photoaffinity probes targeting kinases fall into several main categories. First, there are pan kinase probes, which bind promiscuously to large swaths of the kinome (e.g. staurosporine152, 153). A second group are probes that target specific classes of kinases (e.g. certain tyrosine, serine/threonine or metabolic kinases including lipid kinases 154, 155 (e.g. Dasatinib, discussed above). Placement of the photoactive group in both of these classes of probes has proven critical for achieving sufficient photocrosslinking.78, 156–158 And the final category of kinase photoprobes, which we will discuss in more detail here, are those that have modified gamma-phosphate (in ATP) that are used to capture specific phosphorylation events of kinase-interacting proteins. For a comprehensive recent review of the first two categories of kinase photoaffinity probes, we would direct the readers to Korovesis’ review.159
Given the promiscuity of many kinases and transient nature of most kinase-substrate interactions, mapping the substrates of a particular kinase remains a particularly important and challenging problem. While bump-and-hole strategies160 can address this problem, they are by definition low throughput, relying on production of an engineered kinase and non-natural ATP analogue. As an alternative approach, photoaffinity analogues of ATP, modified on the gamma-phosphate can be used to annotate kinase-substrate interactions. In the method kinase catalyzed crosslinking and streptavidin purification (K-CLASP), Dedigama-Arachchige and coworkers built upon their earlier studies that developed ATP-aryl azide (ATP-AA) and ATP-benzophenone (ATP-BP) photoprobes133, 134 to identify both the kinase and interacting proteins associated with specific phosphorylation events. In K-CLASP, cell lysates are treated with both a biotinylated synthetic substrate peptide and the ATP-AA photoprobe. After UV irradiation, the proteins bound to the peptide are enriched, proteolyzed and sequenced by LC-MS/MS. Using a well characterized protein kinase A (PKA) substrate peptide, the authors identify 324 interacting proteins, including the known interactors PKA, CHEK1, and MAP2K7.161 Low crosslinking efficiency and promiscuity of labeling are limitations of K-CLASP that have in part been addressed by conceptually related methods that rely on covalent trapping using a methacrylate ATP analogue.162, 163
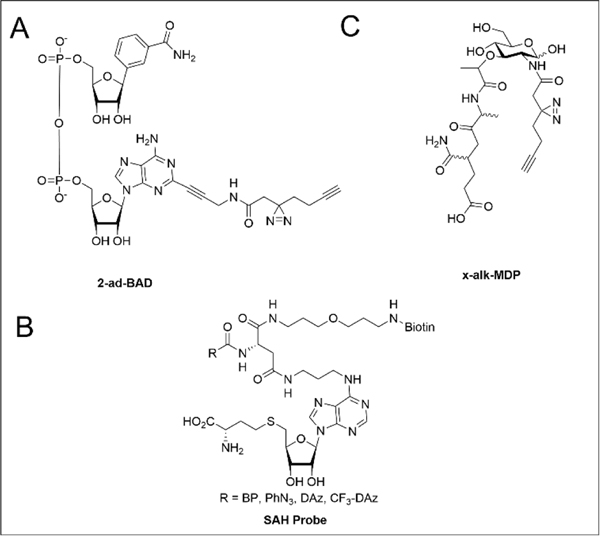
Beyond the kinome, photoaffinity capture methods have been extended to numerous other metabolites and cofactors, including nicotinamide dinucleotide (NAD), S-adenosylmethionine (SAM), and peptidoglycan fragments. NAD photoaffinity probes have been applied to capture poly-ADP−ribose polymerases (PARPs) and other NAD+/NADH binding proteins. Šileikytė and coworkers investigated the NAD interactome using two clickable benzamide-adenine-dinucleotide (BAD) diazirine probes, 2-ad-BAD and 6-ad-BAD (Figure 6A). Chemoproteomic comparison of the proteins labeled by these two probes revealed that 2-ad-BAD bound PARP 1 and 10, whereas 6-ad-BAD did not afford appreciable crosslinking with any PARPs. Collectively these probes enriched 74 total proteins consisting of NAD+/NADH/NADP+/NADPH, ATP, AMP, GTP, ADPr, and FAD binders. This study highlights the importance of the positioning of the photoreactive group (2-Ad-BAD vs 6-Ad-BAD) as shown by the relatively small overlap in targets enriched by each probe, with a combined 74 proteins enriched but only 10 shared targets.46
Figure 6.
Structures of photoreactive probes of (A) NAD+ analog, (B) SAH, and (C) MDP.
Another cofactor extensively studied using PAL is S-adenosylmethionine (SAM), a methyl donor involved in methylation of proteins, lipids, and nucleic acids. Most photoaffinity probes have been derived from S-adenosylhomocysteine (SAH), the demethylated product formed from SAM-dependent methylation. Initial affinity-labeling experiments using probes derived from SAH and methyl transferase (MT) inhibitors identified a handful of MTs.164, 165 Using a suite of four photoaffinity SAH probes to profile human cell lysates and cancer cell lines, Horning and coworkers successfully enriched ~25% of the 200+ known and predicted human MTs in cancer cell proteomes (Figure 6B).47 Nearly all (~90%) of the 50+ significantly enriched targets were MTs or MT-associated proteins. These results provide a strong basis for future development and application of optimized photoaffinity probes to functionally characterize MTs, MT complexes, and MT inhibitors.
The use of photoaffinity-based metabolite and cofactors probes has also been extended to the study of microbial metabolite-host interactions. To understand how host cell detection of peptidoglycan (PG) fragments triggers an inflammatory response, Wang and coworkers synthesized PG diazirine-photoaffinity reporters, which were based on two PG fragments common in many bacterial species: γ-D-glutamyl-meso-diaminopimelic acid (iE-DAP) or muramyl-dipeptide (MDP; Figure 6C).129 Both iE-DAP and MDP photoaffinity reporters selectively crosslinked human nucleotide-binding oligomerization domain-containing protein 1 and 2 (NOD1 and NOD2), consistent with the previously reported activation of these NOD receptors.166, 167 Unexpectedly, the MDP probe was also found to crosslink ARF GTPases, hinting at a putative function of these proteins in the innate immune response. Given the significance of the PG and other bacterial cell wall components for modulating host-pathogen interactions and for determining the efficacy, or lack thereof, of many antibiotics, we expect that future studies using both the PG and related probes should inform additional bacterial metabolite-protein interactions relevant to immune evasion and antibiotic resistance.
2.3. High resolution mapping of small molecule binding sites with photoaffinity labeling.
Mapping of precise interaction sites in proteins is a particularly important opportunity for PAL. Binding site information is useful for deciphering the mode of action of lead compounds, for informing how compound binding impacts protein function, and for guiding medicinal chemistry efforts to improve the potency and selectivity of lead compounds. At face value, mapping specific interaction sites via PAL appears straightforward, requiring simply the enrichment and MS/MS analysis of crosslinked tryptic peptides. However, only a subset of PAL probes have proven compatible with interaction site identification (Table 2). Such analyses are often complicated by the low efficiency of crosslinking, unpredictable fragmentation of crosslinked peptides, and the increased hydrophobicity of labeled peptides, which further complicates enrichment and identification. PAL interaction site identification has however functioned well for smaller probes. For example, by employing their previously developed SIM-PAL platform with a modified biotin capture handle, Miyamato and coworkers identified Prostaglandin E Synthase (PTGES) as one of eight significantly enriched interacting proteins with their photo-celecoxib probe. Then using recombinant PTGES, the binding site was uncovered through PAL and MS analysis with a total of 27 peptide spectral matches of the photo-celecoxib-conjugate. Through competitive displacement experiments, with a known PTGES inhibitor, licofelone, the authors validated the binding site of celecoxib being in proximity to His-53.116
Flaxman and coworkers then applied this same approach towards binding site identification of the macrocyclic lactone rapamycin. Rapamycin was previously known to act as a molecular glue stabilizing a protein-protein interaction between FKBP12 and the FRB domain of mTOR. Using a photoaffinity probe with a diazirine handle on C40 of rapamycin the authors found modification on residues 75–110 of FKBP12 and residues 10–22 of FRB. Predicted residues of interaction, D79 of FKBP12 and E18 of the FRB domain, were mutated to alanine which resulted in significant decrease in labeling implicating these residues as major hits of the rapamycin probe. Further molecular dynamics simulations informed by these experimental results found a 5 Å distance between rapamycin and either protein with a labeling radius of 9 Å.168
Efforts to map the cholesterol-protein binding sites further showcase the challenges associated with deciphering the complex fragmentation patterns of larger crosslinked molecules. Using a PAL strategy, Budelier and coworkers mapped the cholesterol binding site in the voltage-dependent anion channel-1 (VDAC1),169 a mitochondrial protein required for mitochondrial respiration and apoptosis.170, 171 By combining this labeling data with mutagenesis and molecular modeling, cholesterol was proposed to bind with the aliphatic tail 3.5 Å from Glu73 and the steroid ring 2.5 Å from Thr83. While a promising approach for the study of recombinant proteins in vitro, cholesterol-PAL has yet to be extended to, and is likely incompatible with, cell-wide binding studies, due to a requirement for manual interpretation of MS/MS spectra. Further showcasing the complexity of such mapping studies, Budelier and coworkers found that KK174, one of their two structurally related diazirine cholesterol probes (KK174 and LKM38), produced a long half-life reactive species, which confounded interaction site identification efforts with that probe.
In these examples, understanding how these small molecules exert their intended effects is the primary focus. However, characterizing off-target effects is an equally important criterion in the drug-development process. Li and coworkers developed and applied a photoreactive probe labeled LK2-P1 to inform the off-target activity of the Parkinson’s disease (PD) inhibitor LRRK2-IN-1. Proliferating cell nuclear antigen (PCNA) was significantly enriched by LK2-P1, confirming this protein as an off-target of LRRK2-IN-1. The interaction site was pinpointed to Glu238, using recombinant PCNA labeled with LK2-P1. Molecular docking studies suggest the aminopyridine moiety of LRRK2-IN-1 is in proximity to the interdomain connecter loop (IDCL) region which is responsible for interacting with binding proteins.172 Since PCNA plays a major role in DNA synthesis, the authors hypothesized that complexing PCNA with LRRK2-IN-1 significantly decreases its binding with DNA polymerase delta (Pol d) and epsilon (Pol e) thus reducing DNA replication and synthesis. This model was supported by immunoprecipitation studies measuring Pol δ and Pol ε binding affinity for LRRK2-IN-1 treated cells.173
Such interaction site mapping has also been successful using benzophenone probes. Using a benzophenone-functionalized analogue of the HIV drug Raltegravir, the drug’s interaction site was localized to the active site of the HIV-1 integrase enzyme. 174 Further molecular dynamics and docking studies revealed several key amino acid residues that stabilize this probe-protein interaction. As with most of the aforementioned studies, the interaction site identified was limited to in vitro studies using recombinant protein, which further highlights the technical challenges associated with identifying crosslinked peptides, particularly in more complex samples.
2.4. Other Photolabeling strategies
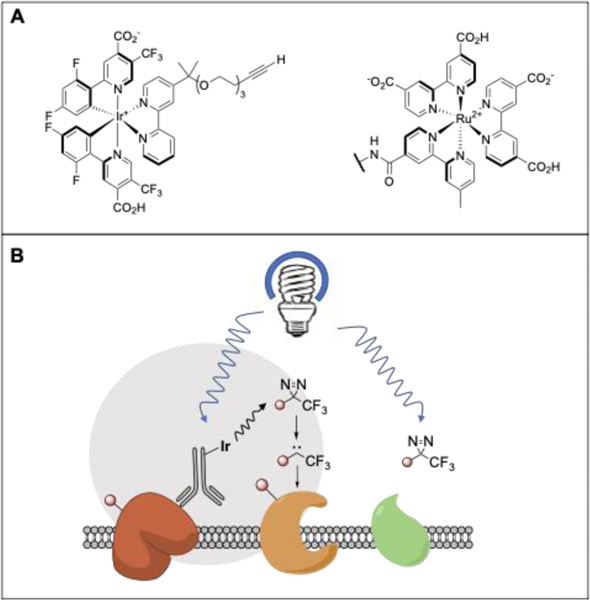
The use of photocatalysts for proximity labeling applications represents an exciting emerging strategy for small molecule target deconvolution applications. Pioneering work by Kodadek and colleagues revealed that the combination of a ruthenium photocatalyst and visible light could be harnessed to crosslink proteins through the excitation of tyrosine residues.175 Looking beyond protein-protein crosslinking, Nakamura and coworkers demonstrated the utility of photocatalysts for small molecule protein interaction mapping. In this work a ruthenium photocatalyst appended small molecule was used to catalyze the crosslinking of tyrosine residues to the dimethyl-1,4-phenylenediamine radical trap176 A recent study, Nakamura employed a ruthenium based photocatalyst (Figure 7A) to excite molecular oxygen. The photogenerated singlet oxygen labels nearby histidine residues in a [4+2] manner followed by nucleophilic attack with methyl-arylurazole (MAU). By using an enrichment handle substituted MAU, labeled His residues can then be identified via purification and MS analysis.177
Figure 7.
(A) Structures of microenvironment mapping photo catalyst (left) and singlet oxygen photocatalyst (right).177, 178 (B) Mechanism of microenvironment mapping via dexter energy transfer.
This general concept has been built upon by MacMillan and coworkers who recently demonstrated that Dexter energy transfer from an excited iridium catalyst appended to an antibody (Figure 7A) could efficiently photoactivate proximal diazirines to carbenes, affording labeling of antibody-adjacent proteins (Figure 7B). When compared with APEX-based methods, this photolabeling offers the distinct advantage of more precise microenvironment map of the target protein due to the significantly shorter half-life of a carbene as compared to a phenoxyl radical.178 This approach has recently been extended to small molecule-protein mapping.179 Looking to the future, we can envision a range of applications for these photocatalytic labeling strategies, including in small molecule screening and target deconvolution, particularly for low abundance and tough-to-detect proteins. The large size of these catalysts may prove confounding for some such studies, possibly by altering the interactomes or subcellular distribution of labeled probes.
Conclusions and Future Prospects
Deciphering the protein-small molecule interactome is an essential step for both functional biology and drug development. Photoaffinity labeling, particularly when combined with a chemoproteomic readout, is a powerful approach to gain high-throughput insight into where and when such interactions occur. As reviewed here, a number of photoactivatable groups (Table 1) have already been applied to such interactomic studies, including for mapping small molecule, drug, lipid, metabolite and cofactor interactions. As demonstrated by the breadth of interactions profiled by photoaffinity labeling (Table 2), this field is rapidly evolving with numerous recent reports of novel probes and interactomic studies.
Choice of photoaffinity label is an essential component of the design and successful execution of these studies. The workhorse reactive groups for such studies are the aryl azides, benzophenone, and aryl and alkyl diazirines, which each offer unique strengths and limitations. Aryl azides are distinguished by their relatively small size. However, the relatively short wavelength of light required for activation and propensity to rearrange to form longer half-life species has largely sidelined the aryl azide in favor of the latter two photoactivatable groups. The ubiquity of the benzophenone moiety in pharmaceutical target deconvolution campaigns is a testament to this group’s general utility for chemoproteomics. Similarly, the diazirine has seen widespread adoption in chemoproteomic studies, chosen in part due to its small size, promiscuous reactivity and the generally short half-life of the reactive carbene—importantly, recent work89 has revealed that diazirines can release both short and long half-life reactive species, which may complicate their use in studies aimed at pinpointing precise interaction sites. Additional factors that should guide photoreactive group selection include 1) the presence of naturally occurring photoactivatable moieties (e.g. pyrone, pyrimidone, or tetrazole that are often found in natural products); 2) the presence of specific amino acids in a binding site (e.g. Glu/Asp) that favour reactivity with certain photoactive groups; and 3) if a putative target shows poor reactivity towards a specific photoactive groups, alternatives should be considered, including the use of photocatalyst-based labelling strategies.179
An ideal photoactivatable group would be small, afford high labelling efficiency (potentially through repeated photoactivation), rely upon relatively long wavelength light to decrease protein damage, and react promiscuously with nearly all amino acid side chains as well as with the peptide backbone. Several intriguing alternatives to these ubiquitous photolabels have emerged, including 2-Aryl-5-carboxytetrazole (ACT)118 and thienyl-substituted α-ketoamide,180 with the former reacting with carboxylates specifically and the latter distinguished by its relatively small size, reversible activation, and low hydrophobicity.181. As shown in Table 1, the unique features of each photoaffinity label should be considered when selecting an appropriate labeling strategy for probe development efforts.
As highlighted here and previously covered extensively by Flaxman’s prior review, a key challenge for photoaffinity labeling is capturing precisely where these interactions occur.182 Low crosslinking efficiency and functional group selectivity are challenges for developing new photoreactive species. Often, there is a tradeoff between the two (as seen with ACT), which offers high crosslinking efficiency and reactivity limited to acidic side chains. In addition, recent literature suggests diazirines, the gold standard, also show a pronounced bias towards reacting towards a subset of amino acid side chains and are therefore not as promiscuous as previously thought.85 This biased reactivity almost certainly complicates pinpointing binding sites particularly when further confounded by the propensity of some diazirines to rearrange into more long half-life reactive species. Further complicating matters, the position of the crosslinker has proven repeatedly to be a key driver of the protein targets captured by PAL studies, as exemplified by the low overlap between the targets captured by 2-Ad-BAD and 6-Ad-BAD.46 It remains unclear whether these differences in interactions are due mainly to blockade of binding by the photoactivatable substituent or by poor positioning of the photoactivatable group relative to reactive to reactive side chains or the peptide backbone. The nature of the protein target can also impact the relative success of binding site mapping efforts. Interaction site identification for transmembrane proteins, distinguished by their low abundance and hydrophobicity, may be hindered by lower crosslinking efficiency and in some cases will require optimization of sample preparation workflows, including proteolytic digest using multiple or alternative sequence specific proteases. Even for relatively successful studies that yield labelled peptides, efforts to build a high confidence binding model may be confounded both by any ambiguity surrounding the precise sites of labelling within a peptide and the identification of multiple modified peptides that support multiple potential binding modes.183 184 Future studies that can more robustly capture crosslinked peptides in complex biological setting for larger molecules will likely shed light on approaches to address these important challenges.
The complicated fragmentation pattern of crosslinked peptides, particularly those modified by larger probes adds an additional layer of complexity to interaction site identification. As show for mapping cholesterol’s binding to VDAC1169 and Rapamycin’s binding to FKBP12 and the FRB domain of mTOR,168 low throughput manual interpretation of MS/MS spectra is still a mainstay of these workflows. The fragmentation of larger molecules is often unpredictable, which has largely hindered efforts towards high throughput data analysis. We expect that advances in proteomic search algorithms, including readily available open search algorithms (e.g. MSFragger185) and offset searches, such as those performed for glycoproteomics,186 likely will, at least in part address these challenges. Complementary to these computational innovations, we envision that the development of new photoaffinity handles, including those that react with higher efficiency and those that can be cleaved either chemically or during MS/MS analysis will further facilitate interaction site mapping.
Beyond small molecule-protein interactions, there is tremendous potential for photoaffinity labeling in assaying nearly all biomolecular interactions. As reviewed here, this technology has already seen widespread use in lipid-, metabolite-, and cofactor-binder identification studies. The study of protein-protein interactions using PAL labeling strategies has seen some traction using unnatural amino acid incorporation. As with small molecule interactions, pinpointing the specific sites of binding for crosslinked peptides has proven extremely challenging. We expect that the use of recently developed trifunctional amino acids that incorporate a photocrosslinker, cleavable linker and an enrichment handle and are readily incorporated into proteins through genetic code expansion, should in part address this challenge.187 Such interactomic analyses will likely also be further facilitated by the recently developed iridium and ruthenium-based photocatalytic labelling platforms.177, 178 Taken together, the rapid and widespread adoption of PAL chemistries, particularly when combined with chemoproteomics, is a testament to the significance of the approach, both for functional biology and chemical probe and drug development campaigns.
Acknowledgements
This study was supported by NIH P01HL146358-01 to KMB and NIH T32 GM067555-11 to NRB. We gratefully acknowledge all members of the Backus lab for helpful suggestions.
Footnotes
Footnotes relating to the title and/or authors should appear here.
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG and Zhang F, Science, 2014, 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neggers JE, Kwanten B, Dierckx T, Noguchi H, Voet A, Bral L, Minner K, Massant B, Kint N, Delforge M, Vercruysse T, Baloglu E, Senapedis W, Jacquemyn M. and Daelemans D, Nature Communications, 2018, 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dziekan JM, Yu H, Chen D, Dai L, Wirjanata G, Larsson A, Prabhu N, Sobota RM, Bozdech Z. and Nordlund P, Science Translational Medicine, 2019, 11, eaau3174-eaau3174. [DOI] [PubMed] [Google Scholar]
- 4.Savitski MM, Reinhard FBM, Franken H, Werner T, Savitski MF, Eberhard D, Molina DM, Jafari R, Dovega RB, Klaeger S, Kuster B, Nordlund P, Bantscheff M. and Drewes G, Science, 2014, 346. [DOI] [PubMed] [Google Scholar]
- 5.Becher I, Werner T, Doce C, Zaal EA, Tögel I, Khan CA, Rueger A, Muelbaier M, Salzer E, Berkers CR, Fitzpatrick PF, Bantscheff M. and Savitski MM, Nature Chemical Biology, 2016, 12, 908–910. [DOI] [PubMed] [Google Scholar]
- 6.Saei AA, Beusch CM, Sabatier P, Wells JA, Gharibi H, Meng Z, Chernobrovkin A, Rodin S, Näreoja K, Thorsell AG, Karlberg T, Cheng Q, Lundström SL, Gaetani M, Végvári Á, Arnér ESJ, Schüler H. and Zubarev RA, Nature Communications, 2021, 12, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golkowski M, Subba R, Vidadala R, Lombard CK, Suh W, Maly DJ and Ong S-E, J Proteome Res, 2017, 16, 1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dittus L, Werner T, Muelbaier M. and Bantscheff M, ACS Chem. Biol, 2017, 12, 29–29. [DOI] [PubMed] [Google Scholar]
- 9.Tyler DS, Vappiani J, Cañeque T, Lam EYN, Ward A, Gilan O, Chan YC, Hienzsch A, Rutkowska A, Werner T, Wagner AJ, Lugo D, Gregory R, Molina CR, Garton N, Wellaway CR, Jackson S, Macpherson L, Figueiredo M, Stolzenburg S, Bell CC, House C, Dawson SJ, Hawkins ED, Drewes G, Prinjha RK, Rodriguez R, Grandi P. and Dawson MA, Science, 2017, 356, 1397–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, Jagannathan S, Aban A, Okerberg E, Herring C, Nordin B, Weissig H, Yang Q, Lee JD, Gray NS and Kozarich JW, Chemistry and Biology, 2011, 18, 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srinivasan J, Cload ST, Hamaguchi N, Kurz J, Keene S, Kurz M, Boomer RM, Blanchard J, Epstein D, Wilson C. and Diener JL, Chemistry and Biology, 2004, 11, 499–508. [DOI] [PubMed] [Google Scholar]
- 12.Greenbaum D, Medzihradszky KF, Burlingame A. and Bogyo M, Chemistry and Biology, 2000, 7, 569–581. [DOI] [PubMed] [Google Scholar]
- 13.Hang HC, Loureiro J, Spooner E, van der Velden AW, Kim YM, Pollington AM, Maehr R, Starnbach MN and Ploegh HL, ACS Chem Biol, 2006, 1, 713–723. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Patricelli MP and Cravatt BF, Proceedings of the National Academy of Sciences of the United States of America, 1999, 96, 14694–14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S. and Cravatt BF, Proceedings of the National Academy of Sciences of the United States of America, 2010, 107, 20941–20946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Der Linden WA, Segal E, Child MA, Byzia A, Drąg M. and Bogyo M, Chemistry and Biology, 2015, 22, 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Backus KM, Correia BE, Lum KM, Forli S, Horning BD, González-Páez GE, Chatterjee S, Lanning BR, Teijaro JR, Olson AJ, Wolan DW and Cravatt BF, Nature, 2016, 534, 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zanon PRA, Yu F, Musacchio P, Lewald L, Zollo M, Krauskopf K, Mrdović D, Raunft P, Maher TE, Cigler M, Chang C, Lang K, Toste FD, Nesvizhskii Alexey I. and Hacker SM, chemRxiv, 2021, DOI: 10.26434/CHEMRXIV.14186561.V1. [DOI] [Google Scholar]
- 19.Hahm HS, Toroitich EK, Borne AL, Brulet JW, Libby AH, Yuan K, Ware TB, McCloud RL, Ciancone AM and Hsu KL, Nature Chemical Biology, 2020, 16, 150–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MBD, Bachovchin DA, Mowen K, Baker D. and Cravatt BF, Nature, 2010, 468, 790–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taunton J, Hassig CA and Schreiber SL, Science, 1996, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- 22.Bogyo M, McMaster JS, Gaczynska M, Tortorella D, Goldberg AL and Ploegh H, Proceedings of the National Academy of Sciences of the United States of America, 1997, 94, 6629–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson KD, Gan-Erdene T. and Kolli N, Methods in Enzymology, 2005, 399, 37–51. [DOI] [PubMed] [Google Scholar]
- 24.Borodovsky A, Ovaa H, Meester WJN, Venanzi ES, Bogyo MS, Hekking BG, Ploegh HL, Kessler BM and Overkleeft HS, ChemBioChem, 2005, 6, 287–291. [DOI] [PubMed] [Google Scholar]
- 25.Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KAH, Salvesen GS and Bogyo M, Journal, 2005, 1, 33–38. [DOI] [PubMed] [Google Scholar]
- 26.Berger AB, Witte MD, Denault JB, Sadaghiani AM, Sexton KMB, Salvesen GS and Bogyo M, Molecular Cell, 2006, 23, 509–521. [DOI] [PubMed] [Google Scholar]
- 27.Van Esbroeck ACM, Janssen APA, Cognetta AB, Ogasawara D, Shpak G, Van Der Kroeg M, Kantae V, Baggelaar MP, De Vrij FMS, Deng H, Allarà M, Fezza F, Lin Z, Van Der Wel T, Soethoudt M, Mock ED, Den Dulk H, Baak IL, Florea BI, Hendriks G, De Petrocellis L, Overkleeft HS, Hankemeier T, De Zeeuw CI, Di Marzo V, Maccarrone M, Cravatt BF, Kushner SA and Van Der Stelt M, Science, 2017, 356, 1084–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH and Aebersold R, Nature Biotechnology, 1999, 17, 994–999. [DOI] [PubMed] [Google Scholar]
- 29.Hacker SM, Backus KM, Lazear MR, Forli S, Correia BE and Cravatt BF, Nat Chem, 2017, 9, 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin S, Yang X, Jia S, Weeks AM, Hornsby M, Lee PS, Nichiporuk RV, Iavarone AT, Wells JA, Toste FD and Chang CJ, Science, 2017, 355, 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shenoy VM, Thompson BR, Shi J, Zhu HJ, Smith DE and Amidon GL, Molecular Pharmaceutics, 2020, 17, 1706–1714. [DOI] [PubMed] [Google Scholar]
- 32.Li W, Blankman JL and Cravatt BF, Journal of the American Chemical Society, 2007, 129, 9594–9595. [DOI] [PubMed] [Google Scholar]
- 33.Simon GM and Cravatt BF, Journal of Biological Chemistry, 2010, 285, 11051–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bach K, Beerkens BLH, Zanon PRA and Hacker SM, ACS Central Science, 2020, DOI: 10.1021/acscentsci.9b01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma N, Hu J, Zhang ZM, Liu W, Huang M, Fan Y, Yin X, Wang J, Ding K, Ye W. and Li Z, Journal of the American Chemical Society, 2020, 142, 6051–6059. [DOI] [PubMed] [Google Scholar]
- 36.Cheng K, Lee J-S, Hao P, Yao SQ, Ding K. and Li Z, Angewandte Chemie, 2017, 129, 15240–15244. [DOI] [PubMed] [Google Scholar]
- 37.Mathur S, Fletcher AJ, Branigan E, Hay RT and Virdee S, Cell Chemical Biology, 2020, 27, 74–82.e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baruah H, Puthenveetil S, Choi YA, Shah S. and Ting AY, Angewandte Chemie - International Edition, 2008, 47, 7018–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farell IS, Toroney R, Hazen JL, Mehl RA and Chin JW, Nature Methods, 2005, 2, 377–384. [DOI] [PubMed] [Google Scholar]
- 40.Tantama M, Lin WC and Licht S, Journal of the American Chemical Society, 2008, 130, 15766–15767. [DOI] [PubMed] [Google Scholar]
- 41.Buchmueller KL, Hill BT, Platz MS and Weeks KM, Journal of the American Chemical Society, 2003, 125, 10850–10861. [DOI] [PubMed] [Google Scholar]
- 42.Dziuba D, Hoffmann JE, Hentze MW and Schultz C, ChemBioChem, 2020, 21, 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Zheng W, Zhang W, Chen N, Liu Y, Chen L, Zhou X, Chen X, Zheng H. and Li X, Chemical Science, 2015, 6, 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dasovich M, Beckett MQ, Bailey S, Ong SE, Greenberg MM and Leung AKL, Journal of the American Chemical Society, 2021, DOI: 10.1021/jacs.0c12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gérard-Hirne T, Thiebaut F, Sachon E, Désert A, Drujon T, Guérineau V, Michel BY, Benhida R, Coulon S, Saintomé C. and Guianvarc’h D, Biochimie, 2018, 154, 164–175. [DOI] [PubMed] [Google Scholar]
- 46.Šileikytė J, Sundalam S, David LL and Cohen MS, Journal of the American Chemical Society, 2021, 143, 6787–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horning BD, Suciu RM, Ghadiri DA, Ulanovskaya OA, Matthews ML, Lum KM, Backus KM, Brown SJ, Rosen H. and Cravatt BF, Journal of the American Chemical Society, 2016, 138, 13335–13343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hulce JJ, Cognetta AB, Niphakis MJ, Tully SE and Cravatt BF, Nature Methods, 2013, 10, 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Höglinger D, Nadler A, Haberkant P, Kirkpatrick J, Schifferer M, Stein F, Hauke S, Porter FD and Schultz C, Proceedings of the National Academy of Sciences of the United States of America, 2017, 114, 1566–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu SH, Boyce M, Wands AM, Bond MR, Bertozzi CR and Kohler JJ, Proceedings of the National Academy of Sciences of the United States of America, 2012, 109, 4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu H, Shajahan A, Yang JY, Capota E, Wands AM, Arthur CM, Stowell SR, Moremen KW, Azadi P. and Kohler JJ, Cell Chem Biol, 2021, DOI: 10.1016/j.chembiol.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murale DP, Hong SC, Haque MM and Lee JS, Proteome Science, 2017, 15, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng T, Yuan X. and Hang HC, Journal, 2014, 21, 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.lien Laguerre A. and Schultz C, Current Opinion in Cell Biology, 2018, 53, 97–104. [DOI] [PubMed] [Google Scholar]
- 55.Trads JB, Tørring T. and Gothelf KV, Accounts of Chemical Research, 2017, 50, 1367–1374. [DOI] [PubMed] [Google Scholar]
- 56.Wu H. and Kohler J, Journal, 2019, 53, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Touchette MH, Van Vlack ER, Bai L, Kim J, Cognetta AB, Previti ML, Backus KM, Martin DW, Cravatt BF and Seeliger JC, ACS Infectious Diseases, 2017, 3, 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srinivas N, Jetter P, Ueberbacher BJ, Werneburg M, Zerbe K, Steinmann J, Van Der Meijden B, Bernardini F, Lederer A, Dias RLA, Misson PE, Henze H, Zumbrunn J, Gombert FO, Obrecht D, Hunziker P, Schauer S, Ziegler U, Käch A, Eberl L, Riedel K, Demarco SJ and Robinson JA, Science, 2010, 327, 1010–1013. [DOI] [PubMed] [Google Scholar]
- 59.Le P, Kunold E, Macsics R, Rox K, Jennings MC, Ugur I, Reinecke M, Chaves-Moreno D, Hackl MW, Fetzer C, Mandl FAM, Lehmann J, Korotkov VS, Hacker SM, Kuster B, Antes I, Pieper DH, Rohde M, Wuest WM, Medina E. and Sieber SA, Nature Chemistry, 2020, 12, 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haberkant P, Raijmakers R, Wildwater M, Sachsenheimer T, Brügger B, Maeda K, Houweling M, Gavin A-C, Schultz C, van Meer G, Heck AJR and Holthuis JCM, Angewandte Chemie International Edition, 2013, 52, 4033–4038. [DOI] [PubMed] [Google Scholar]
- 61.Singh A, Thornton ER and Westheimer FH, The Journal of biological chemistry, 1962, 237, 3006–3008. [PubMed] [Google Scholar]
- 62.Shafer J, Baronowsky P, Laursen R, Finn F. and Westheimer FH, Journal of Biological Chemistry, 1966, 241, 421–427. [PubMed] [Google Scholar]
- 63.Vaughan RJ and Westheimer FH, J. Am. Chem. Soc, 1968, 1, 217–218. [DOI] [PubMed] [Google Scholar]
- 64.Chaimovich H, Vaughan RJ and Westheimer FH, Journal of the American Chemical Society, 1968, 90, 4088–4093. [Google Scholar]
- 65.Knowles JR, Accounts of Chemical Research, 1972, 5, 155–160. [Google Scholar]
- 66.Neuberger A, Biochemical Journal, 1938, 32, 1452–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ermer A, Baumann H, Steude G, Peters K, Fittkaut S, Dolaschka P. and Genov NC, Journal of Enzyme Inhibition and Medicinal Chemistry, 1990, 4, 35–42. [DOI] [PubMed] [Google Scholar]
- 68.Fleet GWJ, Porter RR and Knowles JR, Nature, 1969, 224, 511–512. [Google Scholar]
- 69.Kiefer H, Lindstrom J, Lennox ES and Singer SJ, Proceedings of the National Academy of Sciences of the United States of America, 1970, 67, 1688–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chakrabarti P. and Khorana G, Biochemistry, 1975, 14, 5021–5033. [DOI] [PubMed] [Google Scholar]
- 71.Smith RAG and Knowles JR, Journal, 1973, 95, 5072–5073. [DOI] [PubMed] [Google Scholar]
- 72.Bayley H. and Knowles JR, Biochemistry, 1978, 2420–2423. [DOI] [PubMed] [Google Scholar]
- 73.Goldman DW, Pober JS, White J. and Bayley H, Nature, 1979, 280, 841–843. [DOI] [PubMed] [Google Scholar]
- 74.Galardy RE, Craig LC, Jamieson JD and Printz MP, Journal of Biological Chemistry, 1974, 249, 3510–3518. [PubMed] [Google Scholar]
- 75.Battenberg OA, Nodwell MB and Sieber SA, J. Org. Chem, 2011, 76, 6075–6087. [DOI] [PubMed] [Google Scholar]
- 76.Huisgen R, Sauer J. and Seidel M, Chemische Berichte, 1961, 94, 2503–2509. [Google Scholar]
- 77.Song W, Wang Y, Qu J, Madden MM and Lin Q, Angewandte Chemie - International Edition, 2008, 47, 2832–2835. [DOI] [PubMed] [Google Scholar]
- 78.Li Z, Qian L, Li L, Bernhammer JC, Huynh HV, Lee JS and Yao SQ, Angewandte Chemie International Edition, 2016, 55, 2002–2006. [DOI] [PubMed] [Google Scholar]
- 79.Herner A, Marjanovic J, Lewandowski TM, Marin V, Patterson M, Miesbauer L, Ready D, Williams J, Vasudevan A. and Lin Q, Journal of the American Chemical Society, 2016, 138, 14609–14615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo AD, Wei D, Nie HJ, Hu H, Peng C, Li ST, Yan KN, Zhou BS, Feng L, Fang C, Tan M, Huang R. and Chen XH, Nature Communications, 2020, 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin C, Du HJ, Zhao H, Yan DF, Liu NX, Sun H, Wen X. and Xu QL, Organic and Biomolecular Chemistry, 2017, 15, 3472–3478. [DOI] [PubMed] [Google Scholar]
- 82.Vodovozova EL, Biochemistry (Mosc), 2007, 72, 1–20. [DOI] [PubMed] [Google Scholar]
- 83.Katzenellenbogen JA, Johnson HJ, Carlson KE and Myers HN, Biochemistry, 1974, 13, 2986–2994. [DOI] [PubMed] [Google Scholar]
- 84.Morin B. and Cadet J, Journal of the American Chemical Society, 1995, 117, 12408–12415. [Google Scholar]
- 85.West AV, Muncipinto G, Wu H-Y, Huang AC, Labenski MT, Jones LH and Woo CM, J. Am. Chem. Soc, 2021, 143, 6700–6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McGrath NA, Andersen KA, Davis AK, Lomax JE and Raines RT, Chem Sci, 2015, 6, 752–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mix KA, Lomax JE and Raines RT, J Am Chem Soc, 2017, 139, 14396–14398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Iacobucci C, Götze M, Piotrowski C, Arlt C, Rehkamp A, Ihling C, Hage C. and Sinz A, Anal Chem, 2018, 90, 2805–2809. [DOI] [PubMed] [Google Scholar]
- 89.O’Brien JGK, Jemas A, Asare-Okai PN, Am Ende CW and Fox JM, Org Lett, 2020, 22, 9415–9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chang CF, Mfuh A, Gao J, Wu HY and Woo CM, Tetrahedron, 2018, 74, 3273–3277. [Google Scholar]
- 91.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA and Bertozzi CR, Proceedings of the National Academy of Sciences of the United States of America, 2007, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grée D. and Grée R, Tetrahedron Letters, 2010, 51, 2218–2221. [Google Scholar]
- 93.Brunner J, Senn H. and Richards FM, Journal of Biological Chemistry, 1980, 255, 3313–3318. [PubMed] [Google Scholar]
- 94.Conway LP, Jadhav AM, Homan RA, Li W, Rubiano JS, Hawkins R, Michael R. and Parker CG, Chem. Sci, 2021, DOI: 10.1039/d1sc01360b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guo L.-w., Hajipour AR, Gavala ML, Arbabian M, Martemyanov KA, Arshavsky VY and Ruoho AE, Bioconjug. Chem, 2005, 16, 685–693. [DOI] [PubMed] [Google Scholar]
- 96.Bogyo M, Verhelst S, Bellingard-Dubouchaud V, Toba S. and Greenbaum D, Chemistry and Biology, 2000, 7, 27–38. [DOI] [PubMed] [Google Scholar]
- 97.Colca JR, McDonald WG, Waldon DJ, Leone JW, Lull JM, Bannow CA, Lund ET and Mathews WR, American Journal of Physiology Endocrinology and Metabolism, 2004, 286, 252–260. [DOI] [PubMed] [Google Scholar]
- 98.Kam CM, Abuelyaman AS, Li Z, Hudig D. and Powers JC, Bioconjugate Chemistry, 1993, 4, 560–567. [DOI] [PubMed] [Google Scholar]
- 99.Zubarev RA and Makarov A, Anal. Chem, 2013, 85. [DOI] [PubMed] [Google Scholar]
- 100.Saghatelian A, Jessani N, Joseph A, Humphrey M. and Cravatt BF, Proceedings of the National Academy of Sciences of the United States of America, 2004, 101, 10000–10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chan EWS, Chattopadhaya S, Panicker RC, Huang X. and Yao SQ, Journal of the American Chemical Society, 2004, 126, 14435–14446. [DOI] [PubMed] [Google Scholar]
- 102.Sieber SA, Niessen S, Hoover HS and Cravatt BF, Nature Chemical Biology, 2006, 2, 274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ma N, Zhang ZM, Lee JS, Cheng K, Lin L, Zhang DM, Hao P, Ding K, Ye WC and Li Z, ACS Chemical Biology, 2019, 14, 2546–2552. [DOI] [PubMed] [Google Scholar]
- 104.Lee K, Ban HS, Naik R, Hong YS, Son S, Kim BK, Xia Y, Song KB, Lee HS and Won M, Angewandte Chemie International Edition, 2013, 52, 10286–10289. [DOI] [PubMed] [Google Scholar]
- 105.Tulloch LB, Menzies SK, Fraser AL, Gould ER, King EF, Zacharova MK, Florence GJ and Smith TK, PLOS Neglected Tropical Diseases, 2017, 11, e0005886-e0005886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scott DE, Coyne AG, Hudson SA and Abell C, Biochemistry, 2012, 51, 4990–5003. [DOI] [PubMed] [Google Scholar]
- 107.Hajduk PJ and Greer J, Nature Reviews Drug Discovery, 2007, 6, 211–219. [DOI] [PubMed] [Google Scholar]
- 108.Parker CG, Galmozzi A, Wang Y, Correia BE, Sasaki K, Joslyn CM, Kim AS, Cavallaro CL, Lawrence RM, Johnson SR, Narvaiza I, Saez E. and Cravatt BF, Cell, 2017, 168, 527–541.e529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, MacIejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C. and Wilson M, Nucleic Acids Research, 2018, 46, D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y, Dix MM, Bianco G, Remsberg JR, Lee HY, Kalocsay M, Gygi SP, Forli S, Vite G, Lawrence RM, Parker CG and Cravatt BF, Nature Chemistry, 2019, 11, 1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kennedy CR, Goya Grocin A, Kovačič T, Singh R, Ward JA, Shenoy AR and Tate EW, ACS Chemical Biology, 2021, DOI: 10.1021/acschembio.1c00218, acschembio.1c00218-acschembio.00211c00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hong SH, Yoo DY, Conway L, Richards-Corke KC, Parker CG and Arora PS, Proceedings of the National Academy of Sciences of the United States of America, 2021, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hellyer SD, Aggarwal S, Chen ANY, Leach K, Lapinsky DJ and Gregory KJ, ACS Chemical Neuroscience, 2020, 11, 1597–1609. [DOI] [PubMed] [Google Scholar]
- 114.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A. and Mann M, Molecular & cellular proteomics : MCP, 2002, 1, 376–386. [DOI] [PubMed] [Google Scholar]
- 115.Gao J, Mfuh A, Amako Y. and Woo CM, Journal of the American Chemical Society, 2018, 140, 4259–4268. [DOI] [PubMed] [Google Scholar]
- 116.Miyamoto DK, Flaxman HA, Wu HY, Gao J. and Woo CM, ACS Chemical Biology, 2019, 14, 2527–2532. [DOI] [PubMed] [Google Scholar]
- 117.Soethoudt M, Alachouzos G, Van Rooden EJ, Moya-Garzón MD, Van Den Berg RJBHN, Heitman LH and Van Der Stelt M, Cannabis and Cannabinoid Research, 2018, 3, 136–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miyajima R, Sakai K, Otani Y, Wadatsu T, Sakata Y, Nishikawa Y, Tanaka M, Yamashita Y, Hayashi M, Kondo K. and Hayashi T, ACS Chemical Biology, 2020, 15, 2364–2373. [DOI] [PubMed] [Google Scholar]
- 119.Shi H, Zhang CJ, Chen GYJ and Yao SQ, Journal of the American Chemical Society, 2012, 134, 3001–3014. [DOI] [PubMed] [Google Scholar]
- 120.Cisar EAG, Nguyen N. and Rosen H, Journal of the American Chemical Society, 2013, 135, 4676–4679. [DOI] [PubMed] [Google Scholar]
- 121.Niphakis MJ, Lum KM, Cognetta AB, Correia BE, Ichu TA, Olucha J, Brown SJ, Kundu S, Piscitelli F, Rosen H. and Cravatt BF, Cell, 2015, 161, 1668–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Peng T. and Hang HC, J. Am. Chem. Soc, 2015, 137, 6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Haberkant P, Schmitt O, Contreras FX, Thiele C, Hanada K, Sprong H, Reinhard C, Wieland FT and Brügger B, Journal of Lipid Research, 2008, 49, 251–262. [DOI] [PubMed] [Google Scholar]
- 124.Contreras FX, Ernst AM, Haberkant P, Björkholm P, Lindahl E, Gönen B, Tischer C, Elofsson A, Von Heijne G, Thiele C, Pepperkok R, Wieland F. and Brügger B, Nature, 2012, 481, 525–529. [DOI] [PubMed] [Google Scholar]
- 125.Haberkant P, Stein F, Höglinger D, Gerl MJ, Brügger B, Van Veldhoven PP, Krijgsveld J, Gavin AC and Schultz C, ACS Chemical Biology, 2016, 11, 222–230. [DOI] [PubMed] [Google Scholar]
- 126.Rowland MM, Bostic HE, Gong D, Speers AE, Lucas N, Cho W, Cravatt BF and Best MD, Biochemistry, 2011, 50, 11143–11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang D, Du S, Cazenave-Gassiot A, Ge J, Lee JS, Wenk MR and Yao SQ, Angewandte Chemie - International Edition, 2017, 56, 5829–5833. [DOI] [PubMed] [Google Scholar]
- 128.Huang W, Sun W, Song Z, Yu Y, Chen X. and Zhang Q, Organic and Biomolecular Chemistry, 2012, 10, 5197–5201. [DOI] [PubMed] [Google Scholar]
- 129.Wang Y-C, Westcott NP, Griffin ME and Hang HC, ACS Chem. Biol, 2019, 14, 21–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Campbell S, Kim H, Doukast M. and Haley B, Photoaffinity labeling of ATP and NAD’ binding sites on recombinant human interleukin 2 (signal transduction/autophosphorylation/glycoprotein hormone/lymphokine), 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dalhoff C, Hüben M, Lenz T, Poot P, Nordhoff E, Köster H. and Weinhold E, ChemBioChem, 2010, 11, 256–265. [DOI] [PubMed] [Google Scholar]
- 132.Brown LJ, Baranowski M, Wang Y, Schrey AK, Lenz T, Taverna SD, Cole PA and Sefkow M, Analytical Biochemistry, 2014, 467, 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Suwal S, Kay M. and Pflum H, Angew Chem Int Ed Engl, 2010, 49, 1627–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Garre S, Senevirathne C. and Pflum MKH, Bioorganic and Medicinal Chemistry, 2014, 22, 1620–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ishikawa F, Konno S, Uchida C, Suzuki T, Takashima K, Dohmae N, Kakeya H. and Tanabe G, Cell Chem Biol, 2021, DOI: 10.1016/j.chembiol.2021.05.014. [DOI] [PubMed] [Google Scholar]
- 136.Zuhl AM, Nolan CE, Brodney MA, Niessen S, Atchison K, Houle C, Karanian DA, Ambroise C, Brulet JW, Beck EM, Doran SD, O’Neill BT, Am Ende CW, Chang C, Geoghegan KF, West GM, Judkins JC, Hou X, Riddell DR and Johnson DS, Nature Communications, 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shah NP, Tran C, Lee FY, Chen P, Norris D. and Sawyers CL, Science, 2004, 305, 399–401. [DOI] [PubMed] [Google Scholar]
- 138.Baker D, Pryce G, Ludovic Croxford J, Brown P, Pertwee RG, Huffman JW and Layward L, Nature, 2000, 404, 84–87. [DOI] [PubMed] [Google Scholar]
- 139.Beal JE, Olson R, Laubenstein L, Morales JO, Bellman P, Yangco B, Lefkowitz L, Plasse TF and Shepard KV, Journal of Pain and Symptom Management, 1995, 10, 89–97. [DOI] [PubMed] [Google Scholar]
- 140.Charrin S, Manié S, Thiele C, Billard M, Gerlier D, Boucheix C. and Rubinstein E, European Journal of Immunology, 2003, 33, 2479–2489. [DOI] [PubMed] [Google Scholar]
- 141.Thiele C, Hannah MJ, Fahrenholz F. and Huttner WB, Nature Cell Biology, 2000, 2, 42–49. [DOI] [PubMed] [Google Scholar]
- 142.Motamed M, Zhang Y, Wang ML, Seemann J, Kwon HJ, Goldstein JL and Brown MS, Journal of Biological Chemistry, 2011, 286, 18002–18012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lum KM, Sato Y, Beyer BA, Plaisted WC, Anglin JL, Lairson LL and Cravatt BF, ACS Chemical Biology, 2017, 12, 2671–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mueller R, Kojic A, Citir M. and Schultz C, Angewandte Chemie International Edition, 2021, DOI: 10.1002/anie.202103599, anie.202103599-anie.202103599. [DOI] [Google Scholar]
- 145.Schopper S, Kahraman A, Leuenberger P, Feng Y, Piazza I, Müller O, Boersema PJ and Picotti P, Nature Protocols, 2017, 12, 2391–2410. [DOI] [PubMed] [Google Scholar]
- 146.Piazza I, Kochanowski K, Cappelletti V, Fuhrer T, Noor E, Sauer U. and Picotti P, Cell, 2018, 172, 358–372.e323. [DOI] [PubMed] [Google Scholar]
- 147.Sridharan S, Kurzawa N, Werner T, Günthner I, Helm D, Huber W, Bantscheff M. and Savitski MM, Nature Communications, 2019, 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huber KVM, Olek KM, Müller AC, Tan CSH, Bennett KL, Colinge J. and Superti-Furga G, Journal, 2015, 12, 1055–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Luzarowski M, Kosmacz M, Sokolowska E, Jasińska W, Willmitzer L, Veyel D. and Skirycz A, Journal of Experimental Botany, 2017, 68, 3487–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Okerberg ES, Wu J, Zhang B, Samii B, Blackford K, Winn DT, Shreder KR, Burbaum JJ and Patricelli MP, Proceedings of the National Academy of Sciences of the United States of America, 2005, 102, 4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B. and Drewes G, Nature Biotechnology, 2007, 25, 1035–1044. [DOI] [PubMed] [Google Scholar]
- 152.Fischer JJ, Graebner OY, Dalhoff C, Michaelis S, Schrey AK, Ungewiss J, Andrich K, Jeske D, Kroll F, Glinski M, Sefkow M, Dreger M. and Koester H, Journal of Proteome Research, 2010, 9, 806–817. [DOI] [PubMed] [Google Scholar]
- 153.Shi H, Cheng X, Sze SK and Yao SQ, Chemical Communications, 2011, 47, 11306–11308. [DOI] [PubMed] [Google Scholar]
- 154.Sherratt AR, Nasheri N, McKay CS, O’Hara S, Hunt A, Ning Z, Figeys D, Goto NK and Pezacki JP, ChemBioChem, 2014, 15, 1253–1256. [DOI] [PubMed] [Google Scholar]
- 155.Desrochers GF, Cornacchia C, McKay CS and Pezacki JP, ACS Infectious Diseases, 2018, 4, 752–757. [DOI] [PubMed] [Google Scholar]
- 156.Lanning BR, Whitby LR, Dix MM, Douhan J, Gilbert AM, Hett EC, Johnson TO, Joslyn C, Kath JC, Niessen S, Roberts LR, Schnute ME, Wang C, Hulce JJ, Wei B, Whiteley LO, Hayward MM and Cravatt BF, Nature Chemical Biology, 2014, 10, 760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Murphy JM, Zhang Q, Young SN, Reese ML, Bailey FP, Eyers PA, Ungureanu D, Hammaren H, Silvennoinen O, Varghese LN, Chen K, Tripaydonis A, Jura N, Fukuda K, Qin J, Nimchuk Z, Mudgett MB, Elowe S, Gee CL, Liu L, Daly RJ, Manning G, Babon JJ and Lucet IS, Biochemical Journal, 2014, 457, 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kwarcinski FE, Brandvold KR, Phadke S, Beleh OM, Johnson TK, Meagher JL, Seeliger MA, Stuckey JA and Soellner MB, ACS Chem. Biol, 2016, 38, 17–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Korovesis D, Beard HA, Mérillat C. and Verhelst SHL, ChemBioChem, 2021, DOI: 10.1002/cbic.202000874, 1–14. [DOI] [PubMed] [Google Scholar]
- 160.Shah K, Liu Y, Deirmengian C. and Shokat KM, Proceedings of the National Academy of Sciences of the United States of America, 1997, 94, 3565–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dedigama-Arachchige PM and Pflum MKH, ACS Chemical Biology, 2016, 11, 3251–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Riel-Mehan MM and Shokat KM, Chemistry and Biology, 2014, 21, 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mitchell DC, Menon A. and Garner AL, Cell Chemical Biology, 2019, 26, 980–990.e988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tam EK, Li Z, Goh YL, Cheng X, Wong SY, Santhanakrishnan S, Chai CL and Yao SQ, Chem Asian J, 2013, 8, 1818–1828. [DOI] [PubMed] [Google Scholar]
- 165.Zhu B, Zhang H, Pan S, Wang C, Ge J, Lee JS and Yao SQ, Chemistry - A European Journal, 2016, 22, 7824–7836. [DOI] [PubMed] [Google Scholar]
- 166.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K. and Girardin SE, Nature Reviews Immunology, 2014, 14, 9–23. [DOI] [PubMed] [Google Scholar]
- 167.Caruso R, Warner N, Inohara N. and Núñez G, Immunity, 2014, 41, 898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Flaxman HA, Chang CF, Wu HY, Nakamoto CH and Woo CM, Journal of the American Chemical Society, 2019, 141, 11759–11764. [DOI] [PubMed] [Google Scholar]
- 169.Budelier MM, Cheng WWL, Bergdoll L, Chen ZW, Janetka JW, Abramson J, Krishnan K, Mydock-McGrane L, Covey DF, Whitelegge JP and Evers AS, Journal of Biological Chemistry, 2017, 292, 9294–9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.McCommis KS and Baines CP, Biochimica et Biophysica Acta - Biomembranes, 2012, 1818, 1444–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Colombini M, Biochimica et Biophysica Acta Biomembranes, 2012, 1818, 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bravo R, Frank R, Blundell PA and Macdonald-bravo H, Nature, 1987, 326, 515–517. [DOI] [PubMed] [Google Scholar]
- 173.Li W, Zhou Y, Tang G, Wong NK, Yang M, Tan D. and Xiao Y, Molecular Pharmaceutics, 2018, 15, 3252–3259. [DOI] [PubMed] [Google Scholar]
- 174.Pala N, Esposito F, Tramontano E, Singh PK, Sanna V, Carcelli M, Haigh LD, Satta S. and Sechi M, ACS Medicinal Chemistry Letters, 2020, 11, 1986–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fancy DA and Kodadek T, Proc Natl Acad Sci U S A, 1999, 96, 6020–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sato S. and Nakamura H, Angew Chem Int Ed Engl, 2013, 52, 8681–8684. [DOI] [PubMed] [Google Scholar]
- 177.Nakane K, Sato S, Niwa T, Tsushima M, Tomoshige S, Taguchi H, Ishikawa M. and Nakamura H, Journal of the American Chemical Society, 2021, DOI: 10.1021/jacs.1c01626. [DOI] [PubMed] [Google Scholar]
- 178.Geri JB, Oakley JV, Reyes-Robles T, Wang T, McCarver SJ, White CH, Rodriguez-Rivera FP, Parker DL, Hett EC, Fadeyi OO, Oslund RC and MacMillan DWC, Science, 2020, 367, 1091–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Trowbridge AD, Seath CP, Rodriguez-Rivera FP, Li BX, Dul BE, Schwaid AG, Geri JB, Oakley JV, Fadeyi OO, Oslund RC, Ryu KA, White C, Reyes-Robles T, Tawa P, Parker DL and MacMillan DWC, 2021, DOI: 10.1101/2021.08.02.454797 %J bioRxiv, 2021.2008.2002.454797. [DOI] [Google Scholar]
- 180.Ota E, Usui K, Oonuma K, Koshino H, Nishiyama S, Hirai G. and Sodeoka M, ACS Chem Biol, 2018, 13, 876–880. [DOI] [PubMed] [Google Scholar]
- 181.Ota E, Usui K, Oonuma K, Koshino H, Nishiyama S, Hirai G. and Sodeoka M, ACS Chemical Biology, 2018, 13, 876–880. [DOI] [PubMed] [Google Scholar]
- 182.Flaxman HA and Woo CM, Biochemistry, 2018, 57, 8-8. [DOI] [PubMed] [Google Scholar]
- 183.Yip GM, Chen ZW, Edge CJ, Smith EH, Dickinson R, Hohenester E, Townsend RR, Fuchs K, Sieghart W, Evers AS and Franks NP, Nat Chem Biol, 2013, 9, 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lanyon-Hogg T, Ritzefeld M, Zhang L, Andrei SA, Pogranyi B, Mondal M, Sefer L, Johnston CD, Coupland CE, Greenfield JL, Newington J, Fuchter MJ, Magee AI, Siebold C. and Tate EW, Angew Chem Int Ed Engl, 2021, 60, 13542–13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D. and Nesvizhskii AI, Nature Methods, 2017, 14, 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Polasky DA, Yu F, Teo GC and Nesvizhskii AI, Nature Methods, 2020, 17, 1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lin J, Bao X. and Li XD, Molecular Cell, 2021, 0. [DOI] [PubMed] [Google Scholar]