Abstract
Background
The heat shock factor (Hsf) and small heat shock protein (sHsp, also called Hsp20) complex has been identified as a primary component in the protection of plant cells from ubiquitous stresses, particularly heat stress. Our study aimed to characterize and analyze the Hsf and Hsp genes in Brachypodium distachyon, an annual temperate grass and model plant in cereal and grass studies.
Results
We identified 24 Hsf and 18 Hsp20 genes in B. distachyon and explored their evolution in gene organization, sequence features, chromosomal localization, and gene duplication. Our phylogenetic analysis showed that BdHsfs could be divided into three categories and BdHsp20s into ten subfamilies. Further analysis showed that the 3’UTR length of BdHsp20 genes had a negative relationship with their expression under heat stress. Expression analyses indicated that BdHsp20s and BdHsfs were strongly and rapidly induced by high-temperature treatment. Additionally, we constructed a complex regulatory network based on their expression patterns under heat stress. Morphological analysis suggested that the overexpression of five BdHsp20 genes enhanced the seed germination rate and decreased cell death under high temperatures.
Conclusion
Ultimately, our study provided important evolutionary and functional characterizations for future research on the regulatory mechanisms of BdHsp20s and BdHsfs in herbaceous plants under environmental stress.
Keywords: Heat shock protein, Heat shock factor, Evolution, Seed germination, Brachypodium distachyon, Correlation analysis
Background
Although plants may appear inactive, they have actually evolved a series of countermeasures to respond to various extracellular stimuli. Therefore, plants are not completely passive under external stresses and will form some complex protective mechanisms for adaptation. Abiotic stress, such as high temperature, high salt, heavy metal pollution, and drought, threaten crops’ normal growth and development (Wang, Vinocur & Altman, 2003; Mittler, 2006). Global warming resulting from the greenhouse effect has made high temperatures a greater threat to plants and the main factor affecting plant productivity. It is well-known that heat shock factors (Hsfs) (Nover et al., 2001) and heat shock proteins (Hsps) (Lindquist & Craig, 1988) are initial response genes that can be rapidly induced to repsond to high temperatures. Under external stimuli, Hsp expression can be induced by Hsfs binding the heat shock elements (HSEs; nGAAnnTTCn or nTTCnnGAAn) that exist in multiple copies upstream of the Hsp genes (Giorno et al., 2010). Hsp acts as a molecular chaperone that prevents protein misfolding and eliminates non-natural protein aggregation.
Hsps are widespread in living organisms and generally fall into five groups according to their molecular weight: Hsp100, Hsp90, Hsp70, Hsp60, and Hsp20 (Marrs et al., 1993; Schirmer, Lindquist & Vierling, 1994; Krishna et al., 1995; Mayer & Bukau, 2005; Waters, 2013). Hsp20 is a structurally conserved small heat shock protein (sHsp) with a molecular weight of 15–42 KD. It consists of a variable N-terminal region, a conserved C-terminal region, and a C-terminal extension region (Kriehuber et al., 2010; Poulain, Gelly & Flatters, 2010; Waters, 2013). The N-terminal region sequences across different subfamilies vary greatly, while their sequences in the same subfamily show high similarity (Bondino, Valle & Have, 2012). In contrast, the C-terminal sequence contains a conserved a-crystallin domain (ACD), composed of approximately 90 amino acids, and can be divided into CRI and CRII regions with a β6-loop junction in the middle region. sHsps are the “first responders” in response to cellular stress. They are ATP-independent chaperones that do not change the protein activity themselves, but instead bind to denature-modified proteins of the same molecular weight to form stable compounds and prevent their aggregation (Hartl, Bracher & Hayer-Hartl, 2011). For example, LimHSP16.45 and OsHSP18.2 are molecular chaperones that enhance cell viability under stress (Mu et al., 2011; Kaur et al., 2015). Additionally, abiotic stresses other than high temperature, such as drought, low temperature, high salt, heavy metals, anaerobic stress, and UV-light, can also induce sHsp synthesis (Wang et al., 2004; Sun & Macrae, 2005).
Unlike Hsps, Hsfs usually contain five conserved domains: the DNA binding domain (DBD), hydrophobic oligomerization domain (HR-A/B), nuclear localization signal domain (NLS), nuclear export signal (NES), and C-terminal activation domain (CTAD), which is characterized by short peptide motifs (AHA motifs). Hsfs can be divided into three subfamilies based on the number of amino acid residues inserted between HR-A/B: Class A, B, and C (Guo et al., 2008). Twenty-one amino acids were inserted in Class A, seven amino acids in Class C, and no amino acid residues in Class B (Kotak et al., 2007; Scharf et al., 2012). Hsf plays a key role in regulating plant responses to heat shock (Lin et al., 2011; Ohama et al., 2016), drought, salt, light, and cold stress (Hwang et al., 2014; Zeng et al., 2016; Wang et al., 2020) by regulating the expression of different Hsps. For example, three HSFs induced by heat stress were initially cloned and identified in tomato (Solanum lycopersicum) (Scharf et al., 1990). Although the role of Hsps and Hsfs in stress responses is well established, the regulatory network involved in stress tolerance and the many functions of Hsps need to be further studied.
Brachypodium distachyon (2n = 10) is an annual temperate grass that has been widely accepted as a model plant. Its genome is completely sequenced in order to study characteristics that are unique to cereals and grasses (International Brachypodium, 2010; Catalan et al., 2014). These characteristics include small stature, rapid life cycle, genetic tractability, self-fertility, and small diploid genome size (Garvin et al., 2008; Opanowicz et al., 2008). To obtain more detailed information, we performed correlation network and functional analysis of its Hsfs and sHsps. First, we identified and surveyed phylogenetic relationships of Hsfs and sHsps. Then, we investigated their evolutionary relationship in terms of 3D structure, chromosomal localization, gene duplication, and cis-elements. Next, the expression patterns of Hsfs and sHsps in different tissues and several abiotic stresses were evaluated, and we established the signaling regulatory network based on the expression patterns following different stress treatments. Finally, we measured the seed germination rate and trypan blue staining of five overexpressed BdsHsps in Arabidopsis thaliana after heat treatment.
Materials and Methods
Sequence retrieval
The 23 Hsp20s and 21 Hsfs in A. thaliana (Scharf, Siddique & Vierling, 2001; Sarkar, Kim & Grover, 2009) and the 19 Hsp20s and 25 Hsfs in Oryza sativa (Guo et al., 2008; Sarkar, Kim & Grover, 2009) were obtained from the Arabidopsis Information Resources database (TAIR: http://www.arabidopsis.org/) (Lamesch et al., 2012) and the TIGR Rice Genome Annotation Resources database (http://rice.plantbiology.msu.edu/) (Ouyang et al., 2007), respectively. These sequences were used to search for the Hsp20s and Hsfs in B. distachyon in the publicly available PLAZA database (http://bioinformatics.psb.ugent.be/plaza/) Bel et al., 2018). Collected sequences were scanned using InterPro software and were only considered if they possessed Hsp20 or Hsf consensus sequences, as confirmed by a previous study (Mitchell et al., 2019). Any redundant or alternative splice variants were eliminated. Ultimately, we collected 18 Hsp20s and 24 Hsfs belonging to B. distachyon. Additionally, the molecular weights (Mw) and isoelectric points (pI) of Hsps and Hsfs were obtained from the ExPASy’s pl/Mw calculation tool (https://web.expasy.org/compute_pi/). Subcellular localization predictions for each protein were performed using the CELLO server (http://cello.life.nctu.edu.tw/).
Gene structure, multiple sequence alignment, protein motif, and 3D structure analyses
The exon/intron organization of candidate Hsp20s and Hsfs were surveyed using Gene Structure Display Server (GSDS) software (http://gsds.gao-lab.org/; Hu et al., 2015). The sequence alignments were carried out using Clustal Omega with default parameters (http://www.ebi.ac.uk/Tools/msa/clustalo/). The Hsp20 and Hsf domains and motifs were identified using MEME (http://meme-suite.org/tools/meme; Bailey et al., 2015). Visualization of the protein-conserved domain of these genes was drawn using the WebLogo3 application (http://weblogo.threeplusone.com/). The 3D structure of the BdHsp20s and BdHsfs was modeled using PHYRE2 with default parameters (highest percent identity, most sequence coverage) (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index).
Chromosomal locations, duplication, synteny, and phylogenetic analyses of BdHsp20s and BdHsfs
We collected the chromosome location information of BdHsp20 and BdHsf genes from the PLAZA database and visualized them using the CorelDRAW X3 program. Synteny information of duplicate genes was based on the PlantDGD database (http://pdgd.njau.edu.cn:8080/; Qiao et al., 2019). Phylogenetic trees based on the alignment of Hsp20 and Hsf full-length nucleotide or amino acid sequences were constructed using the maximum likelihood (ML) method with the Jones-Taylor-Thornton (JTT) model, 1000 bootstrap values, and partial deletion in MEGA 5.0 software for O. sativa, A. thaliana, and B. distachyon, respectively (Tamura et al., 2011).
Cis-element analyses of BdHsp20s genes
The upstream 2 kb sequences of the identified BdHsp20 genes were downloaded from the PLAZE database. The extracted sequences were submitted to the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Lescot et al., 2002) for cis-elements analysis. The diagram of the cis-elements of BdHsp20 genes was displayed using CorelDraw software.
Plant growth, treatment, and expression analysis
Bd21 seeds were sown in 1/2 Murashige and Skoog (MS) medium in the dark for 4 d at 25 °C, transferred to a soil mix, and grown in a greenhouse with a cycle of 21 °C for 14 h of light and (>3000 lux)/18 °C for 10 h of darkness. Root, sheet, leaf, and caryopsis were collected from 2-week-old seedlings for tissue-specific expression analysis. For heat analysis, 2-week-old B. distachyon Bd21 plants were treated in liquid MS medium under 40 °C for 1 h, 3 h, 6 h, 12 h, and 24 h. For dark and UV analysis, 2-week-old B. distachyon Bd21 plants were treated in MS liquid medium for 1 h, 3 h, 6 h, 12 h, and 24 h. For abscisic acid (ABA) analysis, 2-week-old B. distachyon Bd21 plants were treated in MS liquid medium containing 100 µM ABA for 1 h, 3 h, 6 h, 12 h, and 24 h. Plants were harvested at their corresponding time points after treatment for further analyses. All samples were flash-frozen in liquid nitrogen and used for RNA extraction.
We extracted the total RNA from each sample using Trizol reagent, and reverse-transcribed 1–2 ug into cDNA using PrimeScript RT Master Mix Perfect Real-Time (TaKaRa, Kyoto, Japan) according to the manufacturer’s instructions. The total RNA was detected for quality using Nanodrop1000 and its integrity was calculated by electrophoresis in 1.5% (w/v) agarose gel. The real-time quantitative polymerase chain reaction (RT-qPCR) was executed in 10 µl reactions with 5–50 ng of first-stand cDNA products (4 µl), 5 pmol of each primer (0.4 µl), 5 µl SYBR green master mix (2X), and 0.2 µl ROX as a passive reference standard to normalize the SYBR fluorescent signal. The RT-qPCR thermal profile was as follows: initial activation at 95 °C for 5 min followed by 45 cycles of 95 °C for 30 s and 60 °C for 30 s. Afterward, PCR product specificity was monitored using a melting curve analysis (61–95 °C with fluorescence read every 0.5 °C). The B. distachyon actin (Bradi2g24070) gene was used as an internal reference for all RT-qPCR analyses. Each experiment was implemented with three independent biological replicates. The relative BdHsp and BdHsf gene expression was calculated using the 2−ΔΔCt method. The up-regulated genes were defined as a fold-change greater than 2 with a p value of < 0.05, and the down-regulated genes were less than 0.5 with a p value of < 0.05. The primer-sets are listed in Table S1.
Regulatory network and pattern analyses of BdHsp20 and BdHsf genes
The length of the mRNA untranslated region (5′UTR and 3′UTR) can significantly affect mRNA stability and protein translation efficiency (Srivastava et al., 2018). To assess the relationship of 3′UTR length and the expression levels of the BdHsp20 gene under heat stress, its correlation coefficient was investigated and the results were displayed in a linear graph. Additionally, the BdHsp20 and BdHsf expression data were detected using RT-qPCR under heat stress and were clustered together to form an integrated expression profile. BdHsf and BdHsp20 genes with correlation coefficients greater than 0.7 or less than −0.7 were screened as a set of their expression regulatory, which we submitted into Cytoscape software (Shannon et al., 2003) to construct their correlation regulatory network.
Binary vector construction, Arabidopsis transformation, heat stress treatment, and trypan blue staining of the transgenic plants
We amplified full-length cDNAs encoding BdHsp20s (including BdHsp16.9-CI, BdHsp17.2A-CI, BdHsp17.2B-CI, BdHsp18-CII, and BdHsp16.4-CI; Table S2) using a pair of specific primers (Table S3) from Bd21 plants, cloned them into pCAMBIA1300 vector at BamHI/KpnI sites, and yielded corresponding plasmid 35S:BdHsp20s that were used for Agrobacterium-mediated Arabidopsis transformation. Positive transgenic plants were screened and planted on 1/2 MS medium containing 25 µg/L hygromycin for one week. The presence of the transgenic insertion was confirmed by PCR in the transgenic plants. Homozygous T3 transgenic plants were subjected to 50 °C for 1 h during the seed germination experiment. More than 100 independent plant seeds were picked for the experiment, and Arabidopsis wild-type (WT) seed was used as the control. Each treatment was conducted in triplicate independently using three biological replicates. After two weeks, the morphological observation was recorded, and the germination rate was counted.
Dead cells were determined by trypan blue staining (Cui et al., 2018; Luan et al., 2018) using the homozygous T3 transgenic plants. The T3 generation transgenic and WT seeds were treated at 50 °C for 1 h and then placed in normal growth conditions. The trypan blue staining solution contained LPTB: 25% w/v lactic acid, 2.5 mg/ml trypan blue, 25% glycerin, and 23% phenol water. The seedlings were placed in a vacuum machine and extracted for 10 min until the dye was completely injected into the plants. Afterwards, we added an appropriate amount of Chloral hydrate solution to the hood to repeat decolorization. After complete decolorization, equilibration for 2 h using 70% glycerol, and observation and photographing with a microscope, the dead blue cells were measured. The greater the number of inactive cells or cells with incomplete membranes, the deeper the trypan blue staining.
Results
Identification and annotation of the Hsp20 and Hsf genes in B. distachyon
The identification and evolution of Hsf genes has been explored in B. distachyon (Table 1; Yang et al., 2014), but the relationship between Hsp20 and Hsf genes needs further investigation. To identify the Hsp20 genes, we used their sequences from A. thaliana and O. sativa as queries for BLASTP analyses in the B. distachyon genome database. Ultimately, a total of 18 non-redundant Hsp20 and 24 Hsf sequences were retrieved (Table 1). Moreover, the genes of Hsp20 and Hsf in B. distachyon followed the same nomenclature rule as those of A. thaliana and O. sativa based on their sequence similarities and subcellular localization, particularly BdHsp20s. For example, Bradi1g44230 was designated as BdHsp15.8-PX according to its subcellular peroxisomal (PX) localization and Mw of 15.8 kD (Table 1). The results showed that most of the BdHsps (11) were located at the cytoplasm (C), except for three BdHsps in the endoplasmic reticulum, one in the PX, one in the chloroplast (CP), and two in the mitochondria (M) (Table 1). Additionally, the protein size of all BdHsp20 proteins varied from 144aa (BdHsp15.8-PX) to 364aa (BdHsp40.0-M), while BdHsf proteins ranged from 248aa (BdHsfC1b) to 649aa (BdHsfA6a) (Table 1). Furthermore, BdHsp20s possessed one intron or were intronless, while all BdHsfs, except BdHsfA6a (4) and BdHsfA1a (2), contained one intron (Table 1). The pI of BdHsp20 proteins ranged from 4.78 (BdHsp18.7-CI) to 8.86 (BdHsp40.0-M) and the acidic proteins dominated, while the BdHsf proteins varied from 4.69 (BdHsfA2a) to 9.82 (BdHsfB1a) with the acidic proteins also occupying the main points (Table 1).
Table 1. Characteristics of BdHsp20 and BdHsf gene families.
| Gene Name | Locus ID | Orientation | ORF | No. of a.a | No. of introns | 5′–3′Coordinate | pI | Mw(kD) |
|---|---|---|---|---|---|---|---|---|
| BdHsp16.8-CIV | Bradi1g26190 | – | 459 | 152 | 1 | 21279133–21279701 | 6.06 | 16.8 |
| BdHsp15.8-PX | Bradi1g44230 | + | 435 | 144 | 0 | 42485161–42485595 | 6.92 | 15.8 |
| BdHsp17.3-CI | Bradi1g53850 | + | 471 | 156 | 0 | 52437678–52438148 | 5.79 | 17.3 |
| BdHsp16.9-CI | Bradi1g67040 | + | 456 | 151 | 0 | 65991938–65992393 | 5.98 | 16.9 |
| BdHsp17.6-CI | Bradi1g67080 | + | 477 | 158 | 1 | 66023992–66024468 | 5.98 | 17.6 |
| BdHsp18.7-CI | Bradi2g02350 | + | 537 | 178 | 1 | 1609756–1610292 | 4.78 | 18.7 |
| BdHsp17.2A-CI | Bradi2g02400 | – | 465 | 154 | 0 | 1631165–1631629 | 6.76 | 17.2 |
| BdHsp17.2B-CI | Bradi2g02410 | + | 462 | 153 | 0 | 1632105–1632566 | 6.19 | 17.2 |
| BdHsp18.0-CII | Bradi2g05374 | – | 498 | 165 | 0 | 3913040–3913537 | 5.96 | 18 |
| BdHsp16.4-CI | Bradi2g12990 | + | 444 | 147 | 0 | 11414902–11415345 | 6.19 | 16.4 |
| BdHsp20.5-ER | Bradi3g02710 | – | 585 | 341 | 0 | 1675529–1676113 | 7.94 | 20.5 |
| BdHsp22.0-ER | Bradi4g21070 | + | 615 | 204 | 0 | 24169313–24169927 | 6.25 | 22 |
| BdHsp24.2-ER | Bradi5g11110 | – | 657 | 218 | 0 | 14808972–14809628 | 5.52 | 24.2 |
| BdHsp18.3-CIII | Bradi3g60100 | + | 510 | 169 | 1 | 58879440–58880020 | 6.59 | 18.3 |
| BdHsp26.4-CP | Bradi1g68440 | + | 720 | 239 | 1 | 67159967–67160773 | 6.77 | 26.4 |
| BdHsp23.2-M | Bradi3g58590 | – | 642 | 213 | 1 | 57826850–57827614 | 7.9 | 23.2 |
| BdHsp21.0-CV | Bradi2g20767 | – | 570 | 189 | 1 | 18240585–18242132 | 4.84 | 21 |
| BdHsp40.0-M | Bradi3g07421 | – | 1095 | 364 | 1 | 57826850–57827614 | 8.86 | 40 |
| BdHsfA1a | Bradi1g01130 | + | 1866 | 622 | 2 | 758757–763160 | 4.92 | 67.716 |
| BdHsfA2a | Bradi1g37720 | – | 1047 | 349 | 1 | 33623352–33624526 | 4.69 | 38.014 |
| BdHsfA2b | Bradi2g41530 | + | 1143 | 381 | 1 | 41768002–41770456 | 5.94 | 42.777 |
| BdHsfA3a | Bradi3g44700 | – | 1536 | 512 | 1 | 46678418–46681158 | 5.52 | 56.011 |
| BdHsfA4a | Bradi2g49860 | + | 1317 | 439 | 1 | 49859988–49861908 | 5.39 | 48.86 |
| BdHsfA5a | Bradi2g18980 | – | 1095 | 365 | 1 | 16735799–16737098 | 5.38 | 40.652 |
| BdHsfA5b | Bradi3g43710 | + | 1407 | 469 | 1 | 45299059–45301814 | 4.93 | 51.677 |
| BdHsfA6a | Bradi1g08891 | + | 1947 | 649 | 4 | 6281780–6283033 | 6.43 | 70.343 |
| BdHsfA6b | Bradi1g74350 | – | 1020 | 340 | 1 | 71592465–71594730 | 5.25 | 38.841 |
| BdHsfA6c | Bradi3g26920 | – | 1071 | 357 | 1 | 27700103–27702438 | 5.16 | 40.236 |
| BdHsfA7a | Bradi1g05550 | – | 1044 | 348 | 1 | 3777047–3779557 | 5.5 | 39.021 |
| BdHsfA7b | Bradi1g55630 | – | 1374 | 458 | 1 | 54141088–54143900 | 4.88 | 50.304 |
| BdHsfA8a | Bradi1g69407 | – | 1182 | 394 | 1 | 67838063–67840368 | 4.83 | 43.389 |
| BdHsfB1a | Bradi4g32130 | + | 909 | 303 | 1 | 37847110–37850047 | 9.82 | 32.79 |
| BdHsfB2a | Bradi3g42130 | – | 1038 | 346 | 1 | 43857313–43858451 | 8.1 | 36.641 |
| BdHsfB2b | Bradi4g35780 | – | 1200 | 400 | 1 | 41115331–41116660 | 5.05 | 41.873 |
| BdHsfB3a | Bradi5g18680 | + | 924 | 308 | 1 | 21774260–21775269 | 5.29 | 32.975 |
| BdHsfB4a | Bradi1g19900 | – | 942 | 314 | 1 | 15943892–15946059 | 6.68 | 34.736 |
| BdHsfB4b | Bradi1g61620 | + | 840 | 280 | 1 | 60967400–60969034 | 6.15 | 31.752 |
| BdHsfB4c | Bradi4g32050 | – | 1215 | 405 | 1 | 37779303–37780616 | 8.97 | 42.982 |
| BdHsfC1a | Bradi2g44050 | + | 1008 | 336 | 1 | 44543658–44544857 | 5.23 | 36.787 |
| BdHsfC1b | Bradi2g48990 | + | 744 | 248 | 1 | 49157232–49158080 | 9.07 | 26.933 |
| BdHsfC2a | Bradi1g38140 | + | 759 | 253 | 1 | 34296765–34297648 | 6.11 | 26.937 |
| BdHsfC2b | Bradi3g08870 | + | 945 | 315 | 1 | 6966950–6968174 | 6 | 33.332 |
Notes.
C, Cytoplasmic; CP, Chloroplast; ER, endoplasmic reticulum; M, Mitochondrial and PX, peroxisomal.
Phylogenetic classification and gene structure of BdHsp20s and BdHsfs
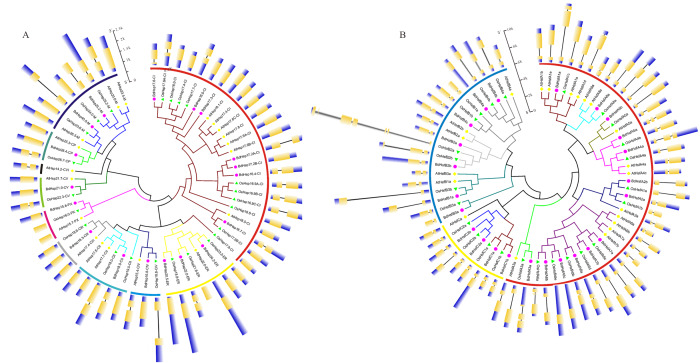
To evaluate the evolutionary relationship between BdHsp20s and BdHsfs, the phylogenetic tree was independently reconstructed based on the sequence alignments of B. distachyon, A. thaliana, and O. sativa full-length amino acids (Table S4) and using the ML method. According to our phylogenetic analysis and subcellular localization prediction results, the BdHsp20s could be divided into cytoplasmic (CI, CII, CIII, CIV, CV, and CVI), CP, ER, M, and PX subfamilies (Fig. 1A). Most Hsp20s belonged to the CI subfamily, suggesting that the cytoplasm may be the main functional region of the plant Hsp20s. The CVI subfamily had only one member and was derived from A. thaliana, while B. distachyon and O. sativa had no members (Fig. 1A), implying that CVI Hsp20 might only exist in dicotyledons. We also found a close relationship between the M and CP subfamilies (Fig. 1A), which supports the results of a previous study (Waters, Aevermann & Sanders-Reed, 2008). It was interesting that the classification of subfamilies was closely consistent with the pattern of intron-exon structure, as supported by previous research (Ouyang et al., 2009).
Figure 1. Phylogenetic relationships and gene structure analysis of Hsf (A) and Hsp20 (B) genes from O. sativa, A. thaliana, and B. distachyon, respectively.
Likewise, phylogenetic analysis divided Hsfs into three main clades, designated as classes A, B, and C (Fig. 1B). Class A Hsfs were further divided into nine groups based on their phylogenetic relationships: subgroup A1, A2, A3, A4, A5, A6, A7, A8, and A9. Of these nine groups, most of the subgroups contained genes in three plant genomes, except for subgroup A9 which was only in A. thaliana and subgroup A2 and A7 which were absent in O. sativa (Fig. 1B). Moreover, class B was divided into four subgroups (B1, B2, B3, and B4), while class C Hsfs were separated into C1 and C2. It was notable that there was only one member in A. thaliana, while class C Hsfs were amplified in B. distachyon and O. sativa (Fig. 1B), indicating that these multiple gene copies may exhibit expansion in monocots. Furthermore, we also found that the subgroup classification patterns were related to intron-exon organization, e.g., class C Hsfs had fully similar intron numbers, exon lengths, and intron phases (Fig. 1B).
Chromosomal location and gene duplication of BdHsp20s and BdHsfs
To analyze the relationship between the gene duplication and genetic divergence within BdHsp20s or BdHsfs, we investigated their chromosomal locations based on the information from the PLAZA database. The physical locations and CpG island distributions of the BdHsp20 and BdHsf genes on B. distachyon chromosomes were drawn (Fig. 2). The results showed that the 18 BdHsp20s and 24 BdHsfs were mapped on five B. distachyon chromosomes: 16 genes were located on chromosome 1, 11 genes on chromosome 2, nine genes on chromosome 3, four genes on chromosome 4, and only one Hsf and Hsp20 member on chromosome 5 (Fig. 2). Interestingly, all BdHsp20 and BdHsf gene members were encoded at the low-density region of CpG islands on all five chromosomes, suggesting that the expression of these genes may be regulated by other genes, which was consistent with previous studies (Bird, 1986; Hahn et al., 2011).
Figure 2. Chromosomal location and gene duplication analyses for B. distachyon Hsp20 and Hsf genes.
The chromosomal position of each BdHsf and BdHsp20 gene was mapped according to the B. distachyon genome. The chromosome number is indicated at the top of each chromosome. The CpG island distribution maps shown in each chromosome depended on the CpG density in the B. distachyon genome. The colored lines and characters represent the subgroup to which proteins in each clade belong. The dotted line shows the gene duplication events across these BdHsf and BdHsp20 genes. The scale bar is shown on the left.
Gene duplication events, such as tandem duplication and segmental duplication, might generate new members of a gene family because of the important role that base substitution, deletion, and insertion into different chromosomes play in the expansion of gene families (Cannon et al., 2004). Therefore, we surveyed the duplication events of BdHsp20s and BdHsfs. We found that five pairs of duplicates underwent tandem duplication and 12 pairs of gene duplicates underwent segmental duplication (Fig. 2), indicating that tandem duplication and segmental duplication might contribute to the amplification of these gene families. These results showed that the greater number of gene family members might be the result of genomic rearrangement and expansion during the evolution process.
Protein structure and tissue-specific expression profile of BdHsp20s and BdHsfs
To better understand BdHsp20 and BdHsf characteristics, we further analyzed their sequence features, specifically the ACD of BdHsp20s and the HR-A/B of BdHsfs (Fig. 3A and 3B). As expected, all BdHsp20s contained an ACD consensus sequence that was composed of two regions: CRI and CRII (Fig. 3A). CRI was formed by β2, β3, β4, and β5, while CRII was formed by β6, β7, β8, and β9. However, BdHsp16.8-CIV and BdHsp23.2-M lacked a β6 structure that played an important role in the normal formation of dimers (Siddique et al., 2008), which was also supported by the 3D structure of the two proteins (Fig. 3C), indicating that they might be replaced by another BdHsp20 member in performing biological functions. Additionally, the typical BdHsf HRA/B contained approximately 100 conserved amino acids. Using the difference of the flexible linkers between the HR-A and B regions, BdHsfs were divided into three categories: Class A (13), Class B (7), and Class C (4) (Fig. 3B). The HR-A containing the most conserved amino acids was L-K/R-R-R-D/E-X3-L-X2-E-L/V, and M-MX-F-L-X-K/R contained the most in HR-B (Fig. 3B). Indeed, the 3D structure of the 24 BdHsfs constructed using phyre2 showed several conserved motifs, including DBD, NLS, and NES (Fig. 3D).
Figure 3. Multiple-sequence alignment analysis and three-dimensional structural model of BdHsfs and BdHsp20s.
(A–B) Alignment of selected conserved domains of BdHsf and BdHsp20 amino acid sequences. (C–D) Three-dimensional structural model of BdHsf and BdHsp20 proteins.
Moreover, the phylogenetic tree of full-length BdHsf and BdHsp20 proteins was reconstructed using MEGA 5.0 (Fig. 4A and 4B). The clustering result was slightly different from the clustering based on the protein sequences of B. distachyon, A. thaliana, and O. sativa. Remarkably, although the gene coding regions had different lengths, the BdHsf domains were highly identical. All the BdHsf members contained DBD, HR-A/B, and NLS domains, while class B BdHsfB3a shared a NES motif and class A BdHsfs also contained an AHA domain that was different from BdHsfA5a (Fig. 4C). In addition, seven class A BdHsf members also had a NES domain similar to BdHsfA6a’s (Fig. 4C). A schematic representing the structure of all BdHsp20 members was drawn using the analysis results from the MEME server (Fig. 4D). We found that all BdHsp20s contained an ACD conserved domain, which was composed of a CRI region (β2, β3, β4, and β5) formed by motif 1 and motif 4, and a CRII region (β6, β7, β8, and β9) formed by motif 2 and motif 3 (Fig. S1). Interestingly, the clustered BdHsp20 pairs localizing the same cellular compartments also exhibited highly similar motif organization, such as BdHsp18.3-CIII/BdHsp18-CII and BdHsp24.2-ER/BdHsp22-ER (Fig. 4D), indicating that the protein architecture was helpful in the distribution and aggregation of the protein.
Figure 4. Phylogenetic relationships and protein structure analysis of BdHsf and BdHsp20 proteins with tissue-specific expression profiles.
(A–B) The phylogenetic tree was constructed from the amino acid sequences using the ML program from MEGA 5, and represents relationships between BdHsf and BdHsp20 proteins. (C–D) Domain analysis of BdHsf and BdHsp20 proteins. (E–F) Expression patterns of BdHsf and BdHsp20 genes in different tissues including root, sheet, leaf, and caryopsis, respectively.
We also investigated the expression patterns of BdHsf and BdHsp20 genes in four tissues or organs (root, sheet, leaf, and caryopsis) using the RT-qPCR method. We found that BdHsfB2a and BdHsfB2b were highly expressed in four tissues; BdHsfB1a, BdHsfC1b, and BdHsfA7a had a relatively high expression level in all tissues except for root; and BdHsfA6a was highly expressed in root, sheet, and caryopsis (Fig. 4E). Other BdHsf genes displayed a relatively low expression level in all tissues (Fig. 4E). Additionally, most BdHsp20 gene members also showed low expression in all tissues, except for a few genes with moderate or high expression levels (Fig. 4F). For example, BdHsp18-CII showed high expression in root and sheet, BdHsp18.3-CIII showed high expression in caryopsis, and BdHsp17.3-CI and BdHsp15.8-PX were moderately expressed in the root (Fig. 4F). These results suggested that BdHsf and BdHsp20 genes may not be expressed during normal growth and development, but are rapidly and highly expressed in response to stresses.
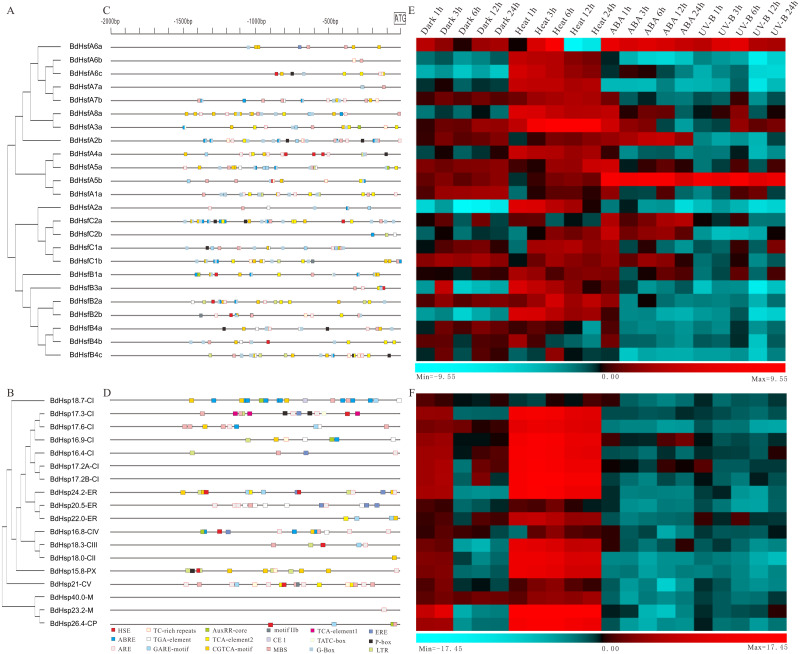
Cis-elements and expression analysis of BdHsp20s and BdHsfs under multiple abiotic stresses and ABA treatment
To explore the function of BdHsp20s’ and BdHsfs’ response to abiotic stresses and hormones, the Bd21 seedling expression profiles were examined using RT-qPCR under dark, heat, UV-B, and ABA treatments. We first investigated the cis-elements in the -2 kb promoter region of the BdHsf and BdHsp20 genes, except for BdHsp17.2A-CI, BdHsp17.2B-CI, and BdHsp40.0-M on account of their lack of a promoter region. The clustering result was obtained based on the phylogenetic tree of the full-length CDS sequence (Fig. 5A and 5B). We found that BdHsp20s and BdHsfs contained various cis-elements involved with defense and hormone and stress responsiveness (Fig. 5C and 5D). For instance, almost all BdHsf genes had G-Box and MBS elements, while most genes lacked motif IIb, ERE, and TCA-element 2, even all TATC-box and CE I (Table S5), suggesting that BdHsf genes may be mainly induced by abiotic stresses, not hormones. Likewise, only one BdHsp20 gene member contained one cis-element including TATC-box, TCA-element 2, CE I, and motif IIb which associated with hormone response, while most BdHsp20 gene members contained ARE and MBS elements (Table S5). Most importantly, seven BdHsp20 genes (BdHsp16.8-CIV, BdHsp15.8-PX, BdHsp17.3-CI, BdHsp24.2-ER, BdHsp18.3-CIII, BdHsp26.4-CP, and BdHsp21.0-CV) contained HSE elements (Table S5).
Figure 5. Phylogenetic relationships and cis-element analysis with expression profiles of BdHsf and BdHsp20 genes under different abiotic stresses.
(A–B) The phylogenetic tree was constructed from the CDS sequences using the ML program from MEGA 5, and represents relationships between BdHsf and BdHsp20 genes. (C–D) Promoter analysis of BdHsf and BdHsp20 genes. All promoter sequences (−2000 bp) were analyzed. The scale bar at the top indicates the length of the promoter sequence. (E–F) Expression patterns of BdHsf and BdHsp20 genes under different abiotic stresses including dark, heat, ABA, and UV-B treatments.
We then investigated the transcript levels of BdHsp20s and BdHsfs under multiple abiotic stresses and ABA treatment. The results showed that most BdHsp20s and BdHsfs, such as BdHsfA3a, were strongly induced by heat stress, were moderately regulated by dark treatment, and were almost not affected by ABA and UV-B treatments (Fig. 5E and 5F). However, there were some differences in the expression profiles of some genes. For example, the expression levels of BdHsfA6a and BdHsfA5b significantly increased after ABA and UV-B treatments, while BdHsfA2b increased in the ABA condition (Fig. 5E), which was consistent with their corresponding cis-element distributions. The expression levels of BdHsfA6a were up-regulated after heat stress, peaking at 6 h, and then recovered to relatively low levels (Fig. 5E and Table S6). Interestingly, many BdHsp20 genes (including BdHsp23.2-M and BdHsp26.4-CP) showed growth under early dark treatment, peaking at 3 h, and then dropping to very low levels (Fig. 5F). Heat stress had no obvious influence on the expression of BdHsp18.7-CI, BdHsp20.5-ER, and BdHsp16.8-CIV, which may be due to their missing HSE element (Fig. 5F and Table S6). These results indicated that BdHsp20s and BdHsfs were all strongly regulated by heat stress and regulated the expressions of downstream genes.
To further survey the potential factor of gene expression, we calculated the correlation coefficient between 3′UTR length and BdHsp20 gene expression under heat stress. The result revealed that all of the BdHsp20 genes had a significant negative correlation with their 3′UTR length (Fig. S2). The shorter 3′UTR length, the higher expression of BdHsp20 genes under heat stress, which may be because of the escape from the inhibition of some miRNAs (Zhang et al., 2019). However, under heat stress, the 5′UTR length had no significant effect on the expression of BdHsp20 genes, which fully conflicted with the correlation of 3′UTR (Fig. S3). Even so, we found that the 5′UTR lengths showed a negative correlation with gene expression levels of BdHsp20 genes under heat stress, which was in accordance with a previous study (Rao et al., 2013).
BdHsp20 and BdHsf gene regulatory network
Together, Hsfs and Hsp20s usually formed signaling modules in response to environmental stresses, especially heat shock stimuli (Lin et al., 2011). Hsp20 expression can be strongly induced by heat and other abiotic stresses, while Hsfs can initially perceive the stimuli and facilitate the expression of Hsps by binding to the HSE of the Hsp promoter region (Schoffl, Prandl & Reindl, 1998). To explore the potential regulatory network between BdHsfs and the downstream protein BdHsp20s under heat stress, we calculated the correlation coefficient between their expression results and constructed the correlation expression network (consisting of 111 nodes and 402 interactions) using Cytoscape software (Fig. 6). We selected the molecular modules if the values of their correlation coefficients were greater than 0.7. The results showed that eight BdHsp20 genes (BdHsp18-CII, BdHsp18.3-CIII, BdHsp15.8-PX, BdHsp17.3-CI, BdHsp40.0-M, BdHsp16.4-CI, BdHsp18.7-CI, and BdHsp16.8-CIV) were regulated by almost all BdHsf genes (Fig. 6). For instance, the BdHsp18-CII gene could be regulated by 12 BdHsf genes, while BdHsp15.8-PX and BdHsp18.3-CIII could be regulated by 11 BdHsf genes (Fig. 6). Other BdHsp20 genes might be regulated by other BdHsf genes under other abiotic stresses. Moreover, all BdHsf genes, except BdHsfC2b, could regulate more than one BdHsp20 gene. For example, BdHsfB4a could regulate eleven BdHsp20 genes (Fig. 6). Furthermore, it was notable that BdHsfA5b and BdHsfA7b expression showed significant negative correlation with BdHsp20s, suggesting that they may be transcriptional repressors. These results indicated a complex signaling regulatory network between BdHsp20s and BdHsfs, which may exhibit the same functions or interactions in response to heat stress treatment.
Figure 6. Visualization for the predicted results of the expression network between BdHsfs and BdHsp20s, constructed using the Cytoscape tool.
Each node represents a protein, and each line represents an interaction. The orange and blue node color indicates the BdHsp20 and BdHsf gene families, respectively. Line thickness reflects the correlation strength of the expression. Font size and circle color indicate the correlated expression number. Our results showed that there were 143 pairs of potential expression pairs. The network was generated using the Cytoscape tool.
BdHsp20 gene overexpression improved plant seed germination under heat stress
To investigate the effect of high-temperature stress on BdHsp20 seed germination, we examined the seed germination rate and trypan blue staining assays of heterologous expression in A. thaliana across five BdHsp20 genes. We found that the germination rate of BdHsp20 genes in transgenic plants was higher than in WT seedlings (Fig. 7 and Table S7). For example, the germination rate of BdHsp17.2B-CI transgenic plants was 69.5 after treatment, while WT seedlings showed a germination rate of 37 (Fig. 7 and Table S7). This finding indicated that BdHsp20 transgenic plants had a higher survival rate under high-temperature stress. To further explore the details of cell survival under high-temperatures, the trypan blue staining of the leaf blade was executed. The results showed that the WT leaf was a deeper blue when compared to BdHsp20 transgenic leaves under high-temperature stress (Fig. 7), indicating that there was more cell death in WT plants. Therefore, we concluded that these BdHsp20 genes can enhance resistance to heat stress.
Figure 7. The effect of BdHsp20 overexpression on seed germination and leaf cell death after heat treatment.
Discussion
Phylogeny, evolution, and duplication analyses of BdsHsp20 and BdHsf gene families
In this study, we investigated the evolution of BdsHsp20 and BdHsf gene families. We first retrieved 18 sHsp20s and 24 Hsfs in B. distachyon and then reconstructed their ML phylogenetic trees (Fig. 1). B. distachyon BdHsfs and BdsHsp20s were divided into three and ten subfamilies, respectively (Fig. 1), similar to A. thaliana and rice (Scharf et al., 2012). Further analysis showed that the pattern of BdsHsp20 and BdHsf gene families’ intron-exon organization was closely related to their phylogenetic branch (Fig. 1). In addition, BdHsp20s and BdHsfs were all located in the low-density region of CpG islands, and were distributed tendentiously on all five chromosomes (Fig. 2), which was in accordance with previous research (Jiang et al., 2015). We also found that four pairs of duplicates showed tandem duplication in subgroup CI BdsHsp20s (Fig. 2), suggesting that tandem duplication contributed to the expansion of these genes, similar to the patterns of the MKK gene family (Jiang & Chu, 2018; Jiang, Li & Wang, 2021). Moreover, domain and 3D structure analyses showed that BdHsp20s and BdHsfs were highly conserved (Fig. 3). Most importantly, these results indicated that the BdsHsp20 and BdHsf gene families were highly conserved.
Correlation regulatory networks across BdHsp20 and BdHsf genes
Numerous Hsp and Hsf genes have been reported in higher plants such as Prunus mume (Wan et al., 2019) and Populus trichocarpa (Zhang et al., 2015). It is well-known that tissue-specific expression patterns may enable Hsp and Hsf proteins to have distinct functions in different signaling pathways, although Hsp and Hsf proteins usually show low abundance under normal conditions. For example, PtHsf-A2 showed high transcript levels in internode 9, PtHsf-B2a showed high transcript levels in internode 5, and PtHsf-A6a showed high levels in female catkins, which was in accordance with a previous study that reported that these genes are key regulators of downstream Hsps (Nishizawa et al., 2006). Therefore, we examined the expression profiles of different tissues in B. distachyon. BdHsfB2a was highly expressed in four tissues, and BdHsfA6a in root, sheet, and caryopsis (Fig. 4E), implying that they might have a unique signaling pathway. Moreover, almost all BdHsp20s and BdHsfs were significantly up-regulated under heat stress (Fig. 5E), indicating that these genes play a significant role in heat stress response. Furthermore, correlation networks between PtHsfs and PtHsps under various stresses (Zhang et al., 2015) and HSF/HSP with switchgrass Cd tolerance (Song et al., 2018) have been constructed. In this study, BdHsfB4a, BdHsfA5b and BdHsfA7b revealed high correlation levels with BdsHsps (Fig. 6), suggesting that these BdHsfs may be the key regulators for many BdsHsps. On the other hand, several BdHsp20s, including BdHsp18-CII, BdHsp18.3-CIII, and BdHsp15.8-PX, displayed co-expression with BdHsfs (Fig. 6), indicating that these genes might play a crucial role in the heat stress response. However, the regulatory mechanisms of herbaceous Hsf and sHsp plant growth, development, and stress responses require further study.
Functional analysis of BdHsp20s in B. distachyon under high temperatures
Previous studies suggested that VcHSP17.7 was associated with the chilling injury resistance of blueberry (Vaccinium corymbosum) fruit induced by heat shock treatment (Shi et al., 2017). Rosa chinensis RcHsp17.8 confers stress tolerance to heat, cold, salt, drought, and osmotic and oxidative conditions (Jiang et al., 2009), while OsHSP18.6 overexpression improved thermotolerance (Wang et al., 2015). In this study, BdHsp17.2B-CI expression was strongly up-regulated under heat conditions (Fig. 5), whereas overexpression in A. thaliana plants enhanced their tolerance to heat stress by decreasing cell death (Fig. 7), indicating that BdHsp17.2B-CI conferred resistance to heat stress. Similarly, AtHSP17.4 and CsHSP17.2 were also more actively expressed in transgenic overexpressed plants under heat shock (Wahid et al., 2007; Wang et al., 2017). The rest of the four overexpressed BdHsp20s in A. thaliana also showed improved tolerance to heat stress (Fig. 7), which supported previous transgenic research (Guo et al., 2020). The heterologous expression of LimHSP16.45 in Arabidopsis enhanced their tolerance to various stresses by forming heat shock particles (HSGs) (Mu et al., 2013). Although it is well-known that HSP20 genes can be induced by heat stress, some heat-regulated genes were almost expressed or downregulated under UA-B and/or dark conditions (Fig. 5F), suggesting that HSP20 genes may have a negative or only slight effect under UA-B and/or dark stress. These results suggest that the signaling pathways may be different in a plant’s response to heat and light stress. Collectively, these results provide a basis for further functional research on the roles of these families in herbaceous plant stress responses.
Conclusion
In this study, we retrieved 24 Hsf and 18 sHsp genes in B. distachyon using bioinformatics approaches. Based on the phylogenetic tree, subcellular localization, and domain analyses, the BdHsfs were categorized into three classes, and the BdHsp20s were divided into ten subfamilies. Notably, we found that 3′UTR length might negatively affect BdHsp20 gene expression. Expression analysis showed that most BdHsp20s and BdHsfs were strongly and rapidly regulated by heat stimuli, moderately induced by dark treatment, and almost not affected at all by ABA and UV-B treatments. More importantly, the correlation regulatory network between BdHsp20s and BdHsfs under heat stress treatments showed a complex signaling regulatory network. Additionally, seed germination and trypan blue staining analyses revealed that BdHsp20s enhanced high-temperature tolerance. These results provide useful information for future research on the regulatory networks of BdHsp20s’ and BdHsfs’ response to environmental stresses, and improved our understanding of the functions of Hsf and sHsp genes in herbaceous plants.
Supplemental Information
Logos were generated using the Weblogo3 application (http://weblogo.threeplusone.com/).
Funding Statement
This work was funded by a grant from the Shanghai Sailing Program (19YF1414800). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Na Li conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Min Jiang conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Peng Li and Xiwen Li analyzed the data, prepared figures and/or tables, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The data of RT-qPCR experiments are available in the Supplemental Table.
References
- Bailey et al. (2015).Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Research. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird (1986).Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bondino, Valle & Have (2012).Bondino HG, Valle EM, Ten Have A. Evolution and functional diversification of the small heat shock protein/alpha-crystallin family in higher plants. Planta. 2012;235:1299–1313. doi: 10.1007/s00425-011-1575-9. [DOI] [PubMed] [Google Scholar]
- Cannon et al. (2004).Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biology. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan et al. (2014).Catalan P, Chalhoub B, Chochois V, Garvin DF, Hasterok R, Manzaneda AJ, Mur LA, Pecchioni N, Rasmussen SK, Vogel JP, Voxeur A. Update on the genomics and basic biology of Brachypodium: International Brachypodium Initiative (IBI) Trends in Plant Science. 2014;19:414–418. doi: 10.1016/j.tplants.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Cui et al. (2018).Cui J, Jiang N, Zhou X, Hou X, Yang G, Meng J, Luan Y. Tomato MYB49 enhances resistance to Phytophthora infestans and tolerance to water deficit and salt stress. Planta. 2018;248:1487–1503. doi: 10.1007/s00425-018-2987-6. [DOI] [PubMed] [Google Scholar]
- Garvin et al. (2008).Garvin DF, Gu YQ, Hasterok R, Hazen SP, Jenkins G, Mockler TC, Mur LAJ, Vogel JP. Development of genetic and genomic research resources for Brachypodium distachyon. A new model system for grass crop research. Crop Science. 2008;48:S69–S84. doi: 10.2135/cropsci2007.04.0208. [DOI] [Google Scholar]
- Giorno et al. (2010).Giorno F, Wolters-Arts M, Grillo S, Scharf KD, Vriezen WH, Mariani C. Developmental and heat stress-regulated expression of HsfA2 and small heat shock proteins in tomato anthers. Journal of Experimental Botany. 2010;61:453–462. doi: 10.1093/jxb/erp316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo et al. (2020).Guo LM, Li J, He J, Liu H, Zhang HM. A class I cytosolic HSP20 of rice enhances heat and salt tolerance in different organisms. Scientific Reports. 2020;10:1383. doi: 10.1038/s41598-020-58395-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo et al. (2008).Guo J, Wu J, Ji Q, Wang C, Luo L, Yuan Y, Wang Y, Wang J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. Journal of Genetics and Genomics. 2008;35:105–118. doi: 10.1016/S1673-8527(08)60016-8. [DOI] [PubMed] [Google Scholar]
- Hahn et al. (2011).Hahn MA, Wu X, Li AX, Hahn T, Pfeifer GP. Relationship between gene body DNA methylation and intragenic H3K9me3 and H3K36me3 chromatin marks. PLOS ONE. 2011;6:e18844. doi: 10.1371/journal.pone.0018844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, Bracher & Hayer-Hartl (2011).Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hu et al. (2015).Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang et al. (2014).Hwang SM, Kim DW, Woo MS, Jeong HS, Son YS, Akhter S, Choi GJ, Bahk JD. Functional characterization of Arabidopsis HsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions. Plant Cell and Environment. 2014;37:1202–1222. doi: 10.1111/pce.12228. [DOI] [PubMed] [Google Scholar]
- International Brachypodium (2010).International Brachypodium I. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Jiang & Chu (2018).Jiang M, Chu ZQ. Comparative analysis of plant MKK gene family reveals novel expansion mechanism of the members and sheds new light on functional conservation. BMC Genomics. 2018;19:407. doi: 10.1186/s12864-018-4793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Li & Wang (2021).Jiang M, Li P, Wang W. Comparative analysis of MAPK and MKK gene families reveals differential evolutionary patterns in Brachypodium distachyon inbred lines. PeerJ. 2021;9:e11238. doi: 10.7717/peerj.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang et al. (2015).Jiang M, Wen F, Cao J, Li P, She J, Chu Z. Genome-wide exploration of the molecular evolution and regulatory network of mitogen-activated protein kinase cascades upon multiple stresses in Brachypodium distachyon. BMC Genomics. 2015;16:228. doi: 10.1186/s12864-015-1452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang et al. (2009).Jiang CH, Xu JY, Zhang H, Zhang X, Shi JL, Li M, Ming F. A cytosolic class I small heat shock protein, RcHSP17.8, of Rosa chinensis confers resistance to a variety of stresses to Escherichia coli, yeast and Arabidopsis thaliana. Plant Cell and Environment. 2009;32:1046–1059. doi: 10.1111/j.1365-3040.2009.01987.x. [DOI] [PubMed] [Google Scholar]
- Kaur et al. (2015).Kaur H, Petla BP, Kamble NU, Singh A, Rao V, Salvi P, Ghosh S, Majee M. Differentially expressed seed aging responsive heat shock protein OsHSP18.2 implicates in seed vigor, longevity and improves germination and seedling establishment under abiotic stress. Frontiers in Plant Science. 2015;6:713. doi: 10.3389/fpls.2015.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak et al. (2007).Kotak S, Larkindale J, Lee U, Koskull-Doring Pvon, Vierling E, Scharf KD. Complexity of the heat stress response in plants. Current Opinion in Plant Biology. 2007;10:310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Kriehuber et al. (2010).Kriehuber T, Rattei T, Weinmaier T, Bepperling A, Haslbeck M, Buchner J. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB Journal. 2010;24:3633–3642. doi: 10.1096/fj.10-156992. [DOI] [PubMed] [Google Scholar]
- Krishna et al. (1995).Krishna P, Sacco M, Cherutti JF, Hill S. Cold-induced accumulation of hsp90 transcripts in Brassica napus. Plant Physiology. 1995;107:915–923. doi: 10.1104/pp.107.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch et al. (2012).Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, Karthikeyan AS, Lee CH, Nelson WD, Ploetz L, Singh S, Wensel A, Huala E. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Research. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot et al. (2002).Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, VandePeer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin et al. (2011).Lin YX, Jiang HY, Chu ZX, Tang XL, Zhu SW, Cheng BJ. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics. 2011;12:76. doi: 10.1186/1471-2164-12-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist & Craig (1988).Lindquist S, Craig EA. The heat-shock proteins. Annual Review of Genetics. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Luan et al. (2018).Luan Y, Cui J, Li J, Jiang N, Liu P, Meng J. Effective enhancement of resistance to Phytophthora infestans by overexpression of miR172a and b in Solanum lycopersicum. Planta. 2018;247:127–138. doi: 10.1007/s00425-017-2773-x. [DOI] [PubMed] [Google Scholar]
- Marrs et al. (1993).Marrs KA, Casey ES, Capitant SA, Bouchard RA, Dietrich PS, Mettler IJ, Sinibaldi RM. Characterization of two maize HSP90 heat shock protein genes: expression during heat shock, embryogenesis, and pollen development. Developmental Genetics. 1993;14:27–41. doi: 10.1002/dvg.1020140105. [DOI] [PubMed] [Google Scholar]
- Mayer & Bukau (2005).Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cellular and Molecular Life Sciences. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell et al. (2019).Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, Gough J, Haft DR, Huang H, Letunic I, Lopez R, Luciani A, Madeira F, Marchler-Bauer A, Mi H, Natale DA, Necci M, Nuka G, Orengo C, Pandurangan AP, Paysan-Lafosse T, Pesseat S, Potter SC, Qureshi MA, Rawlings ND, Redaschi N, Richardson LJ, Rivoire C, Salazar GA, Sangrador-Vegas A, Sigrist CJA, Sillitoe I, Sutton GG, Thanki N, Thomas PD, Tosatto SCE, Yong SY, Finn RD. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Research. 2019;47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler (2006).Mittler R. Abiotic stress, the field environment and stress combination. Trends in Plant Science. 2006;11:15–19. doi: 10.1016/j.tplants.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Mu et al. (2011).Mu C, Wang S, Zhang S, Pan J, Chen N, Li X, Wang Z, Liu H. Small heat shock protein LimHSP16.45 protects pollen mother cells and tapetal cells against extreme temperatures during late zygotene to pachytene stages of meiotic prophase I in David Lily. Plant Cell Reports. 2011;30:1981–1989. doi: 10.1007/s00299-011-1106-y. [DOI] [PubMed] [Google Scholar]
- Mu et al. (2013).Mu C, Zhang S, Yu G, Chen N, Li X, Liu H. Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses. PLOS ONE. 2013;8:e82264. doi: 10.1371/journal.pone.0082264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa et al. (2006).Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant Journal. 2006;48:535–547. doi: 10.1111/j.1365-313X.2006.02889.x. [DOI] [PubMed] [Google Scholar]
- Nover et al. (2001).Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf KD. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones. 2001;6:177–189. doi: 10.1379/1466-1268(2001)006<0177:AATHST>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama et al. (2016).Ohama N, Kusakabe K, Mizoi J, Zhao H, Kidokoro S, Koizumi S, Takahashi F, Ishida T, Yanagisawa S, Shinozaki K, Yamaguchi-Shinozaki K. The transcriptional cascade in the heat stress response of Arabidopsis is strictly regulated at the level of transcription factor expression. The Plant Cell. 2016;28:181–201. doi: 10.1105/tpc.15.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opanowicz et al. (2008).Opanowicz M, Vain P, Draper J, Parker D, Doonan JH. Brachypodium distachyon: making hay with a wild grass. Trends in Plant Science. 2008;13:172–177. doi: 10.1016/j.tplants.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Ouyang et al. (2009).Ouyang Y, Chen J, Xie W, Wang L, Zhang Q. Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Mol Biol. 2009;70:341–357. doi: 10.1007/s11103-009-9477-y. [DOI] [PubMed] [Google Scholar]
- Ouyang et al. (2007).Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Research. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain, Gelly & Flatters (2010).Poulain P, Gelly JC, Flatters D. Detection and architecture of small heat shock protein monomers. PLOS ONE. 2010;5:e9990. doi: 10.1371/journal.pone.0009990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao et al. (2019).Qiao X, Li Q, Yin H, Qi K, Li L, Wang R, Zhang S, Paterson AH. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biology. 2019;20:38. doi: 10.1186/s13059-019-1650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao et al. (2013).Rao YS, Wang ZF, Chai XW, Nie QH, Zhang XQ. Relationship between 5 ’ UTR length and gene expression pattern in chicken. Genetica. 2013;141:311–318. doi: 10.1007/s10709-013-9730-9. [DOI] [PubMed] [Google Scholar]
- Sarkar, Kim & Grover (2009).Sarkar NK, Kim YK, Grover A. Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics. 2009;10:393. doi: 10.1186/1471-2164-10-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf et al. (2012).Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochimica Et Biophysica Acta/General Subjects. 2012;1819:104–119. doi: 10.1016/j.bbagrm.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Scharf et al. (1990).Scharf KD, Rose S, Zott W, Schoffl F, Nover L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. The EMBO Journal. 1990;9:4495–4501. doi: 10.1002/j.1460-2075.1990.tb07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf, Siddique & Vierling (2001).Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins) Cell Stress Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:TEFOAT>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer, Lindquist & Vierling (1994).Schirmer EC, Lindquist S, Vierling E. An Arabidopsis heat shock protein complements a thermotolerance defect in yeast. The Plant Cell. 1994;6:1899–1909. doi: 10.1105/tpc.6.12.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoffl, Prandl & Reindl (1998).Schoffl F, Prandl R, Reindl A. Regulation of the heat-shock response. Plant Physiology. 1998;117:1135–1141. doi: 10.1104/pp.117.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon et al. (2003).Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi et al. (2017).Shi WJ, Su SC, Li BB, Yang JF, Gong ZZ, Hou ZX. Molecular characterisation and expression analysis of a sHSP gene involved in heat shock treatment-induced chilling tolerance in highbush blueberry fruit. Journal of Horticultural Science & Biotechnology. 2017;92:455–464. doi: 10.1080/14620316.2017.1304167. [DOI] [Google Scholar]
- Siddique et al. (2008).Siddique M, Gernhard S, Koskull-Doring Pvon, Vierling E, Scharf KD. The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones. 2008;13:183–197. doi: 10.1007/s12192-008-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song et al. (2018).Song G, Yuan SX, Wen XH, Xie ZI, Lou LQ, Hu BY, Cai QS, Xu B. Transcriptome analysis of Cd-treated switchgrass root revealed novel transcripts and the importance of HSF/HSP network in switchgrass Cd tolerance. Plant Cell Reports. 2018;37:1485–1497. doi: 10.1007/s00299-018-2318-1. [DOI] [PubMed] [Google Scholar]
- Srivastava et al. (2018).Srivastava AK, Lu Y, Zinta G, Lang Z, Zhu JK. UTR-dependent control of gene expression in plants. Trends in Plant Science. 2018;23:248–259. doi: 10.1016/j.tplants.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun & Macrae (2005).Sun Y, Macrae TH. Small heat shock proteins: molecular structure and chaperone function. Cellular & Molecular Life Sciences. 2005;62:2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura et al. (2011).Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel et al. (2018).Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, VandePeer Y, Coppens F, Vandepoele K. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Research. 2018;46:D1190–D1196. doi: 10.1093/nar/gkx1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid et al. (2007).Wahid A, Gelani S, Ashraf M, Foolad MR. Heat tolerance in plants: an overview. Environmental and Experimental Botany. 2007;61:199–223. doi: 10.1016/j.envexpbot.2007.05.011. [DOI] [Google Scholar]
- Wan et al. (2019).Wan X, Yang J, Guo C, Bao M, Zhang J. Genome-wide identification and classification of the Hsf and sHsp gene families in Prunus mume, and transcriptional analysis under heat stress. PeerJ. 2019;7:e7312. doi: 10.7717/peerj.7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2020).Wang N, Liu W, Yu L, Guo Z, Chen Z, Jiang S, Xu H, Fang H, Wang Y, Zhang Z, Chen X. HEAT SHOCK FACTOR A8a modulates flavonoid synthesis and drought tolerance. Plant Physiology. 2020;184:1273–1290. doi: 10.1104/pp.20.01106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Vinocur & Altman (2003).Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2004).Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends in Plant Science. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2015).Wang AQ, Yu XH, Mao Y, Liu Y, Liu GQ, Liu YS, Niu XL. Overexpression of a small heat-shock-protein gene enhances tolerance to abiotic stresses in rice. Plant Breeding. 2015;134:384–393. doi: 10.1111/pbr.12289. [DOI] [Google Scholar]
- Wang et al. (2017).Wang M, Zou Z, Li Q, Sun K, Chen X, Li X. The CsHSP17.2 molecular chaperone is essential for thermotolerance in Camellia sinensis. Scientific Reports. 2017;7:1237. doi: 10.1038/s41598-017-01407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters (2013).Waters ER. The evolution, function, structure, and expression of the plant sHSPs. Journal of Experimental Botany. 2013;64:391–403. doi: 10.1093/jxb/ers355. [DOI] [PubMed] [Google Scholar]
- Waters, Aevermann & Sanders-Reed (2008).Waters ER, Aevermann BD, Sanders-Reed Z. Comparative analysis of the small heat shock proteins in three angiosperm genomes identifies new subfamilies and reveals diverse evolutionary patterns. Cell Stress Chaperones. 2008;13:127–142. doi: 10.1007/s12192-008-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2014).Yang Z, Wang Y, Gao Y, Zhou Y, Zhang E, Hu Y, Yuan Y, Liang G, Xu C. Adaptive evolution and divergent expression of heat stress transcription factors in grasses. BMC Evolutionary Biology. 2014;14:147. doi: 10.1186/1471-2148-14-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng et al. (2016).Zeng JK, Li X, Zhang J, Ge H, Yin XR, Chen KS. Regulation of loquat fruit low temperature response and lignification involves interaction of heat shock factors and genes associated with lignin biosynthesis. Plant Cell and Environment. 2016;39:1780–1789. doi: 10.1111/pce.12741. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2015).Zhang J, Liu B, Li J, Zhang L, Wang Y, Zheng H, Lu M, Chen J. Hsf and Hsp gene families in Populus: genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genomics. 2015;16:181. doi: 10.1186/s12864-015-1398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2019).Zhang R, Pan B, Li Y, Li X. SNP rs4937333 in the miRNA-5003-binding site of the ETS1 3′-UTR decreases ETS1 expression. Frontiers in Genetics. 2019;10:581. doi: 10.3389/fgene.2019.00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Logos were generated using the Weblogo3 application (http://weblogo.threeplusone.com/).
Data Availability Statement
The following information was supplied regarding data availability:
The data of RT-qPCR experiments are available in the Supplemental Table.