Abstract
Purpose
To develop a proof-of-concept convolutional neural network (CNN) to synthesize T2 maps in right lateral femoral condyle articular cartilage from anatomic MR images by using a conditional generative adversarial network (cGAN).
Materials and Methods
In this retrospective study, anatomic images (from turbo spin-echo and double-echo in steady-state scans) of the right knee of 4621 patients included in the 2004–2006 Osteoarthritis Initiative were used as input to a cGAN-based CNN, and a predicted CNN T2 was generated as output. These patients included men and women of all ethnicities, aged 45–79 years, with or at high risk for knee osteoarthritis incidence or progression who were recruited at four separate centers in the United States. These data were split into 3703 (80%) for training, 462 (10%) for validation, and 456 (10%) for testing. Linear regression analysis was performed between the multiecho spin-echo (MESE) and CNN T2 in the test dataset. A more detailed analysis was performed in 30 randomly selected patients by means of evaluation by two musculoskeletal radiologists and quantification of cartilage subregions. Radiologist assessments were compared by using two-sided t tests.
Results
The readers were moderately accurate in distinguishing CNN T2 from MESE T2, with one reader having random-chance categorization. CNN T2 values were correlated to the MESE values in the subregions of 30 patients and in the bulk analysis of all patients, with best-fit line slopes between 0.55 and 0.83.
Conclusion
With use of a neural network–based cGAN approach, it is feasible to synthesize T2 maps in femoral cartilage from anatomic MRI sequences, giving good agreement with MESE scans.
See also commentary by Yi and Fritz in this issue.
Keywords: Cartilage Imaging, Knee, Experimental Investigations, Quantification, Vision, Application Domain, Convolutional Neural Network (CNN), Deep Learning Algorithms, Machine Learning Algorithms
© RSNA, 2021
Keywords: Cartilage Imaging, Knee, Experimental Investigations, Quantification, Vision, Application Domain, Convolutional Neural Network (CNN), Deep Learning Algorithms, Machine Learning Algorithms
Summary
A neural network was developed to produce synthetic T2 maps of cartilage based on anatomic MRI scans that were not designed for T2 mapping.
Key Points
■ Two readers assessed the synthetic T2 maps and found them to be comparable to multiecho spin-echo T2 maps.
■ Linear regression analysis of synthesized and baseline T2 values revealed a significant correlation, both in the whole cartilage region in a section as well as in smaller subregions of the cartilage.
■ The synthetic T2 maps produced by the model were free of image artifacts caused by pulsation on the baseline T2 maps.
Introduction
MRI often involves applying several acquisition sequences that together comprise an MRI protocol. Each sequence is chosen for specific tissue contrast, such as T1- or T2-weighted, to add information provided by the examination. A purely quantitative T2 contrast, showing the absolute value of T2 relaxation at each pixel, has been found to have clinical value in knee imaging. A T2 measurement can help assess cartilage matrix integrity, particularly that of collagen, potentially indicating early tissue degradation in osteoarthritis (OA) (1–4). Articular cartilage contains type II collagen fibrils that have different orientations at different cartilage depths (5). Previous studies suggest that cartilage T2 is affected by changes in the orientation and anisotropy of the collagen matrix and in the water content from tissue degradation (6–8). As the MRI signal decays exponentially with the inverse of T2 in a spin-echo experiment, a “T2 map” can be estimated by fitting signals from a multiecho spin-echo (MESE) scan at several echo times to an exponential curve (2).
MESE MRI examinations can be time-consuming and are not performed routinely in knee MRI. In the Osteoarthritis Initiative (OAI), however, in which MRI scans were obtained in both knees of 4796 patients (across four imaging centers and at five annual time points), MESE scans were acquired (9), and, thus, this dataset is of value for machine learning applications. In the OAI, several anatomic (nonquantitative) MRI examinations, including double-echo in steady-state (DESS) and turbo spin-echo (TSE, with and without fat suppression) sequences, were performed in both knees. However, because MESE MRI involved acquiring seven echoes and took almost 11 minutes (more than twice as long as some anatomic sequences), it was performed in only one knee (usually the right) (10). As such, there would be a clear benefit in obtaining T2 maps without an MESE acquisition, potentially by estimating them from anatomic sequences such as DESS and sagittal and coronal TSE sequences. Such anatomic T2 estimation could potentially enable its more widespread use in clinical practice, providing imaging features to discriminate between healthy individuals and those with degenerative joint disease (11). The anatomic contrast dependency on T2 is more complicated than that of MESE; therefore, estimating T2 analytically from these sequences would be nontrivial.
Recently, neural networks have emerged as an efficient means of obtaining models that describe various probabilistically distributed data. By iteratively adjusting weights in a large neural network through back-propagation, the neural network can be trained as a nonlinear regression model to produce desired output data from given input data. This approach has been used in medical imaging for image reconstruction (12), classification (13), and segmentation (14), and for achieving super-resolution (15). Furthermore, neural networks have shown that they are capable of generating realistic images by using the generative adversarial network (GAN) approach (16), in which two networks, the “generator” and the “discriminator,” compete in synthesizing data and determining whether data are synthesized or real. In a refinement of this methodology, conditional GANs (cGANs) can be trained to perform image-to-image translation by conditioning on an input image to generate a corresponding output image (17). Such cGANs have previously been used for image segmentation (14) and for generating contrast in the spine (18). To our knowledge, synthesis of quantitative T2 maps as described in this study has not yet been performed.
In this proof-of-concept study, we describe the use of a convolutional neural network (CNN) to learn the relationship between the pixel intensities from anatomic OAI scans and their corresponding values on a T2 map, producing what is referred to in this work as “CNN T2 maps,” in right lateral femoral condyle cartilage. We compare the CNN T2 maps with those produced with a ground truth MESE T2-mapping sequence.
Materials and Methods
The images for this retrospective study were collected between 2004 and 2006 as part of the OAI study. This dataset has been used in over 400 publications. The applications of OAI in machine learning described in recent publications (19–21) include automatic classification of OA severity from radiographs, prediction of knee replacements, and diagnosing OA from analytical T2 maps. The current study is focused on the feasibility of T2 map generation in the lateral femoral condyles with the aid of cartilage masks. Participants provided informed consent, as required by the institutional review boards of the respective OAI imaging centers. The study was in compliance with Health Insurance Portability and Accountability Act (HIPAA) regulations, with language pertaining to the acquisition, use, and disclosure of protected health information incorporated into consent forms and participants signing all necessary HIPAA authorizations prior to the public release of their data. We obtained institutional approval for use of these data.
Study Cohort
The OAI was an observational cohort study sponsored jointly by the National Institutes of Health and the pharmaceutical industry to identify imaging and biochemical biomarkers of knee OA. A total of 4796 men and women participants between the ages of 45 and 79 years who had or were at risk for knee OA were enrolled and underwent imaging with Siemens Trio 3T MRI scanners (Siemens Healthineers). In addition, clinical and radiologic information was collected and included data from questionnaires assessing pain and disability. These imaging examinations and data collections were performed at baseline and at four subsequent annual time points at four centers in the United States (The Ohio State University, Columbus; University of Maryland School of Medicine, Baltimore; University of Pittsburgh School of Medicine; Brown University and Memorial Hospital of Rhode Island, Pawtucket [closed in 2018]), with data coordinating centered at the University of California, San Francisco, School of Medicine and the results made publicly available (9). Patients were recruited in four stages: (a) initial contact through mailing, advertisements, public presentations, and websites; (b) eligibility telephone interview; (c) screening visit; and (d) baseline visit for data collection (22).
Anonymized data were extracted from the baseline OAI data point on a per-patient basis to produce CNN T2 maps (synthesized maps) of the right lateral femoral condyles. We chose to focus on the right knees, as this would include the majority of the T2 maps and because expanding the preprocessing and analysis to include the few left knees for which T2 maps were available would only marginally enlarge the dataset. Patients were referred to by their unique identification number that had been assigned during initial screening. The mapping of identifiable patient data to these numbers is not possible with use of publicly available information. Information that could unmask a participant’s identity, such as clinic location, rare medical condition(s), or uncommon combinations of demographic characteristics, was not retrievable from the data (22).
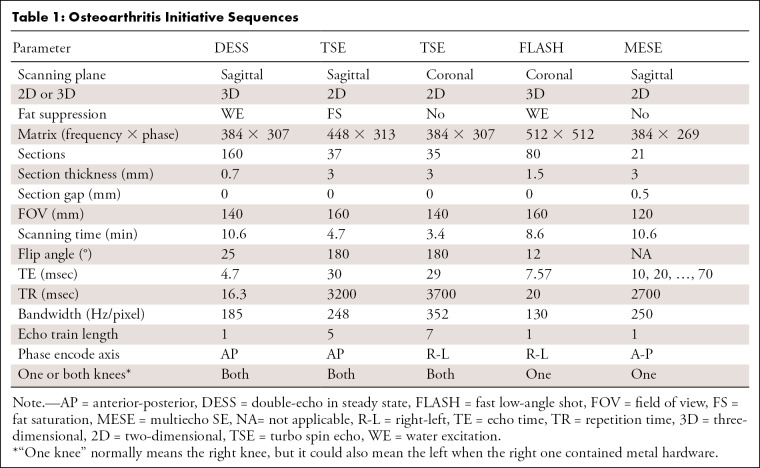
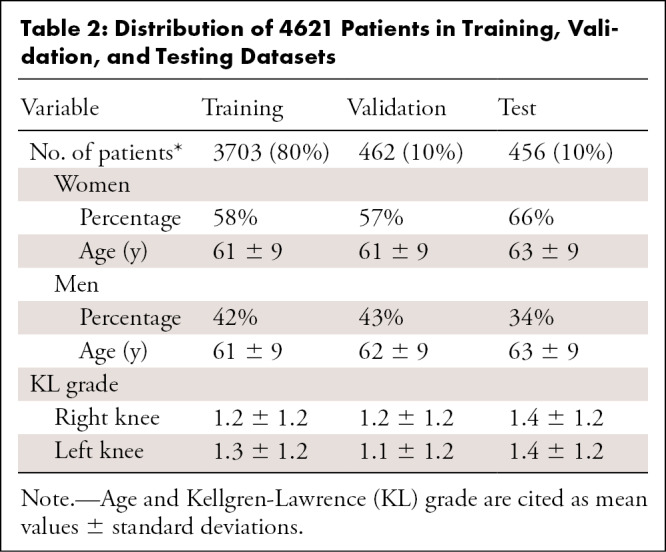
In our preprocessing, there was only one exclusion criterion: If our preprocessing program (described in the Data Preprocessing section) could not recognize the sequences from the Digital Imaging and Communications in Medicine (DICOM) image headers, as described in Table 1 (TSE, DESS, fast low-angle shot [FLASH]) or the full MESE scans for the right knee were not found on the basis of sequence names (eg, the sequence name would be checked for “DESS” and “RIGHT” to see if the dataset contained a DESS scan for the right knee), then the dataset was omitted. This could be for various reasons, including errors in the sequence name or the MESE sequence being run in the left knee instead of the right. This exclusion resulted in a total of 4621 patient datasets from the initial 4796 patients, thus excluding 175 patients or about 3.6% of the total. Inspection of two randomly selected patients who were omitted revealed that for one patient only four MESE echo times were collected instead of the normal seven; we considered this an incomplete acquisition. The other patient had metal in the right knee, so MESE and FLASH sequences were run on the left side. The data were ordered by patient identification numbers, and the first 3703 patients were used for training; the next 462 patients, for validation; and the final 456 patients, for testing. As a result, the split between training, validation, and testing datasets shown in Table 2 had a similar distribution between the sexes, their respective age distributions, and mean Kellgren-Lawrence (KL) grades of OA severity.
Table 1:
Osteoarthritis Initiative Sequences
Table 2:
Distribution of 4621 Patients in Training, Validation, and Testing Datasets
Data Preprocessing
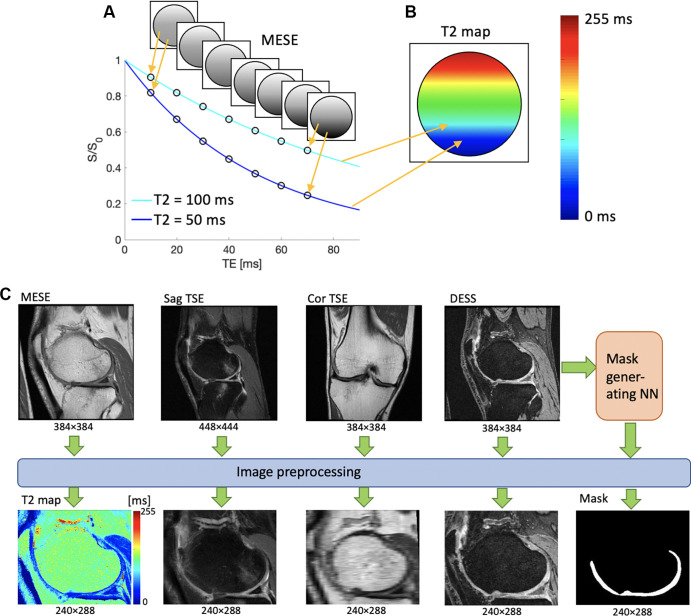
The data were preprocessed as shown in Figure 1. First, a T2 map was generated from the MESE data by means of simple exponential fitting of the images that corresponded to the seven MESE echo times. Such maps can provide information about the cartilage microstructure (Fig 1). Next, the anatomic images (DESS, sagittal TSE, coronal TSE) were reformatted and resampled to align with the voxel locations of the MESE T2 map by using location information in the DICOM headers so that all of the resulting images had the same scanning plane, spatial resolution, and field of view. As shown in Figure 1C, the reformatted coronal TSE data appeared to be more pixelated than the other sequence data owing to its different scanning plane; however, it was included because it could add information that was not available from the other scans. Next, a mask of the femoral cartilage was generated from the DESS data by using a previously developed automatic segmentation neural network (also trained on DESS data) (23). Finally, the images were cropped to focus on the femoral condyles, without changing the pixel size. This was done to eliminate the edge artifacts observed in some OAI data, as well as to prevent the network from focusing on noisy regions outside the tissue and reduce the data size. By using an iterative process, starting with a small cropping area and increasing that area until it captured the features of about 30 datasets well, it was determined that a cropped image size of 240 × 288 pixels would accomplish this goal. This resulted in a quintuple dataset for each T2 section obtained from the MESE sequence and was used as the resampling template for other sequences, as shown in Figure 1C. This process was applied to all MESE T2 sections for every patient in the sample, yielding about 123 000 datasets with five images each, or about 26 datasets on average per patient, with about 99 000 datasets for training and 12 000 datasets for both validation and testing.
Figure 1:
(A, B) Schematic illustrations demonstrate the meaning of the pixel values of a T2 map. (A) Several signals are acquired with different echo times (TEs) by using a multiecho spin-echo (MESE) MRI sequence. When plotted as a function of TE, the intensities of each pixel form an exponential decay curve. The shorter the T2, the steeper the curve. (B) An exponential fit of the form S(t) = Soexp (−t/T2), where S(t) is the measured signal at time t, So is the signal before T2 decay, exp is the natural exponential function, and (−t/T2) is the negative ratio of the measurement time t and the T2 relaxation rate, is performed on the data points, resulting in a T2 value for each point, here drawn on a colored map. The color mapping of two sample points is shown for illustrative purposes. (C) Preprocessing workflow of image data. T2 maps are generated from exponential fitting of the time series MESE data, and the anatomic images are reformatted so that they all have the same spatial resolution, field of view, and image plane. Cor = coronal, DESS = double-echo in steady-state, NN = neural network, Sag = sagittal, S/So = signal with/before T2 decay, TSE = turbo spin echo.
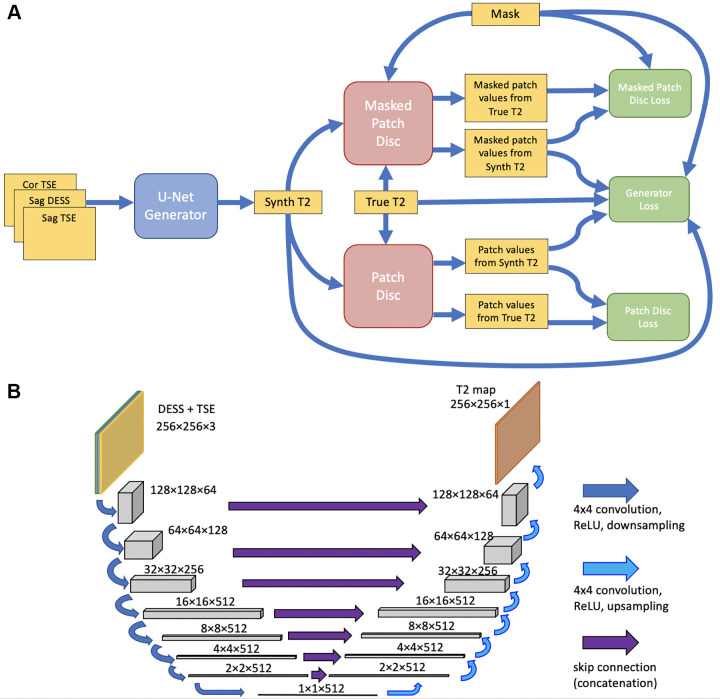
Network Implementation and Training
Figure 2A shows the network structure, based on the pix2pix approach (24), in which a cGAN is used for image-to-image translation. As in pix2pix, a U-Net (24,25) (Fig 2B) generator created CNN T2 maps from anatomic data, and a patch-based encoder discriminator determined whether a T2 map was synthetic or real.
Figure 2:
(A) Schematic architecture of the neural network. A U-Net–based generator (blue) synthesizes a T2 map estimate from three anatomic scans. Patch-based discriminators (red) compare the synthesized (Synth) map to a true T2 map corresponding to the same anatomy. Loss functions (green), consisting of a combination of sigmoid cross-entropy and L1 loss, were computed and used to update the generator and the two discriminators. (B) Schematic illustration of the U-Net, taking the double-echo in steady-state (DESS) and turbo spin-echo (TSE) scans as input and producing an estimated T2 map as output. Cor = coronal, ReLU = rectified linear unit, Sag = sagittal.
Generator.— Gray-scale 8-bit anatomic images were input into a 16-layer U-Net generator in three separate image channels, scaled to a range between −1 and 1. Because the dataset was large, data augmentation was deemed unnecessary. As in pix2pix, the U-Net consisted of a series of 4 × 4 two-stride convolutional downsampling layers followed by 4 × 4 two-stride deconvolutional upsampling layers. The first three downsampling layers and the last three upsampling layers had 64, 128, and 256 filters, while all other layers had 512 filters. The first three upsampling layers used dropout with a rate of 0.5. The network used a skip connection between the input and the U-Net T2 estimate. The concatenated data were then passed through a simple convolutional layer with a 4 × 4 kernel. The generator loss function was computed as follows:
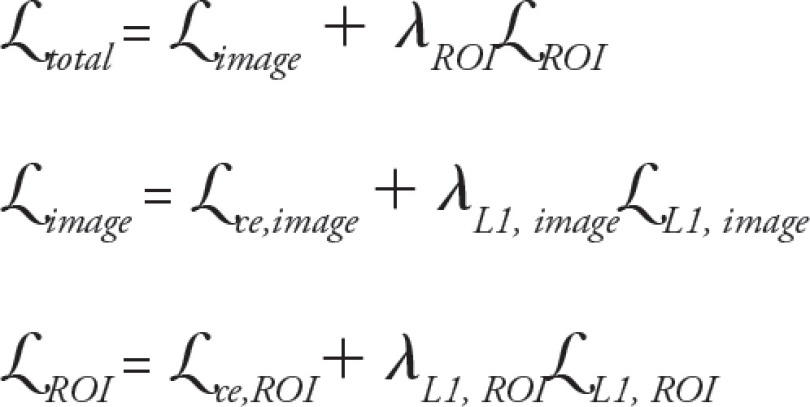 |
Here, image refers to the loss function computed over the whole image, while ROI refers to the region of interest determined by the cartilage mask and ce, to sigmoid cross-entropy loss. The weighting parameters were λL1,image = λL1,ROI = 25, and λROI = 3.
Discriminators.— Two encoder discriminators were used, one for the whole image and one for the cartilage region, by employing the cartilage mask (Fig 1C). The whole-image discriminator had three downsampling layers with 64, 128, and 256 filters, using 4 × 4 convolution kernels, and processed 70 × 70 image patches, while the discriminator focusing on the cartilage was simpler, with one 64-filter downsampling layer with a 3 × 3 kernel, and it processed smaller 11 × 11 patches. Both the generator and the discriminators used modules of the form convolution-batch normalization-rectified linear unit (26).
Training.— The network used a batch size of 16. The generator and the discriminators used the Adam optimizer with a learning rate of 0.0002 and β1 equal to 0.5. As in the original pix2pix work, the only noise provided was that in the form of generator dropout. Training was performed over 20 epochs, with initially random weights. One hundred twenty models were trained, with model performance deemed to be of sufficiently high quality for further analysis based quantitatively on the average L1 error over the cartilage mask not exceeding 5 msec per section and 3 msec per patient over the 12 000 validation sections, and based qualitatively on the image quality of randomly selected validation sections, defined as the (nonradiologist) developer not observing any unrealistic features compared with the MESE T2 maps.
Software and Hardware
The network was implemented by using TensorFlow, version 1.10.0 (27,28), and Keras, version 2.2.2 (29), software with use of eager execution. Development took place on a workstation with two NVIDIA Pascal-architecture GeForce GTX 1080 Ti graphics processing units (Nvidia). Coding was performed in Python, version 3.5.2 (https://www.python.org/). DICOMs were read in preprocessing by using Pydicom (https://pydicom.github.io/), version 1.2.2, and the preprocessed data were stored as JPEGs by using Pillow, version 5.2.0 (https://pypi.org/project/Pillow/5.2.0/). These JPEGs were then read into the network by using the read_file functionality of TensorFlow and saved as PNGs, also using Pillow. To be input into the mask-generating network, DESS images were converted to h5 files by using h5py, version 2.8.0 (https://pypi.org/project/h5py/2.8.0/). Our training code, which is based on and builds on the original pix2pix code and its implementation on the Google TensorFlow website (24,28), and our image preprocessing code are available at https://github.com/bsvmgh/CNNT2.
Image Evaluation
Detailed evaluation of limited test dataset.— Of the 456 patients in the test dataset, 30 were randomly selected for manual evaluation. This was considered to be the highest number of datasets that is feasible for thorough manual processing while ensuring a manageable evaluation workload. The purpose of analyzing this smaller dataset was twofold: (a) to perform more detailed laminar evaluation of the cartilage, which was not considered feasible with use of the automatically generated cartilage mask, and (b) for board-certified radiologists’ evaluation to determine whether the CNN T2 maps were realistic or their quality differed from that of MESE T2 maps.
For these 30 patients, a section prominently displaying the lateral condyle was selected; this section was defined as being visually estimated to have the largest condyle cartilage angular span. This section was examined to keep the evaluation workload manageable while allowing laminar T2 map analysis, by dividing the femoral cartilage of that section into six regions: deep or superficial in the anterior, central, or posterior locations. These regions were defined as anterior, covering the cartilage anterior to the tibial plateau; posterior, similarly covering the cartilage posterior to the tibial plateau; and central, covering the region between them, with deep and superficial covering the superior and inferior halves of the cartilage thickness, respectively. The mean T2 was computed for each region in all 30 patients. This was done for both the MESE and the CNN T2 maps, and the two estimates were compared by using linear least-squares regression. The best-fit line through the points, as well as R2 and P values, were computed to assess agreement. The data were tested for normality by using the Kolmogorov-Smirnov test, with α equal to .05. The root-mean-square difference and average absolute difference between the methods, as well as the coefficient of variation of each method, were computed in each region.
Two board-certified musculoskeletal radiologists (reader 1 [M.T.] and reader 2 [G.E.G.], with 23 and 27 years of experience in reading musculoskeletal MR images, respectively) assessed the resulting maps. The readers were provided with the MESE and CNN T2 maps from each of the 30 patients but were blinded as to which were MESE T2 maps and which were CNN T2 maps. These readers were not involved with any other aspect of this study, such as the design of the network. They assessed which image was a CNN T2 map and which was MESE T2 map, with correct and incorrect assessments assigned scores of 1 and −1, respectively. The readers also reported a confidence level of 1 (least confident) to 5 (most confident) to produce a weighted assessment—for example, a correct assessment with a confidence level of 3 yielded an unweighted score of 1 but a weighted score of 0.6.
After scoring all 30 image sets in this way, the readers were made aware of which image was the CNN T2 map and compared it with the MESE T2 map, evaluating the signal-to-noise ratio (SNR), sharpness, artifacts, and overall quality on a five-point quality scale, with 1 indicating much worse than the MESE and 5 being much better. Two-sided t tests with α equal to .05 were performed to assess whether the readers distinguished the MESE maps from the CNN maps significantly differently than what would be expected from random assessment (mean significantly different from 0) and whether the CNN T2 map quality was significantly different from the MESE T2 map quality (mean significantly different from 3). These metrics of similarity and quality indicated whether the CNN T2 maps had benefits that were comparable to those of the MESE T2 maps for radiologic assessment.
Last, a failure analysis was performed wherein the set of 30 patients was split into a “success” set, in which the CNN map convinced at least one reader that it was the MESE T2 map, and a “failure” set, in which it failed to do so. The mean T2 in the six subregions, as well as the ratio of female to male patients, mean age, and mean KL grade, were calculated for both subsets, and these values were compared by using two-sided t tests, with α equal to .05.
Evaluation of full test dataset.— In each of the 456 patients in the test dataset, an ROI covering the whole femoral cartilage was drawn in a section in the lateral condyle of the MESE and CNN T2 maps. This ROI was selected in the same manner as in the 30 patients who were randomly selected for manual evaluation. The ROI was drawn by a technical MRI researcher with 9 years of experience in drawing and analyzing ROIs in knee cartilage (B.S). The mean T2 over the ROI was recorded. As for the laminar analysis of the limited test dataset, agreement was estimated with linear least-squares regression analysis. Data were again tested for normality by using the Kolmogorov-Smirnov test, with α equal to .05. The root-mean-square difference, average absolute difference, and coefficient of variation were computed as they were for the 30-patient subregional analysis.
Results
Model and Map Overview
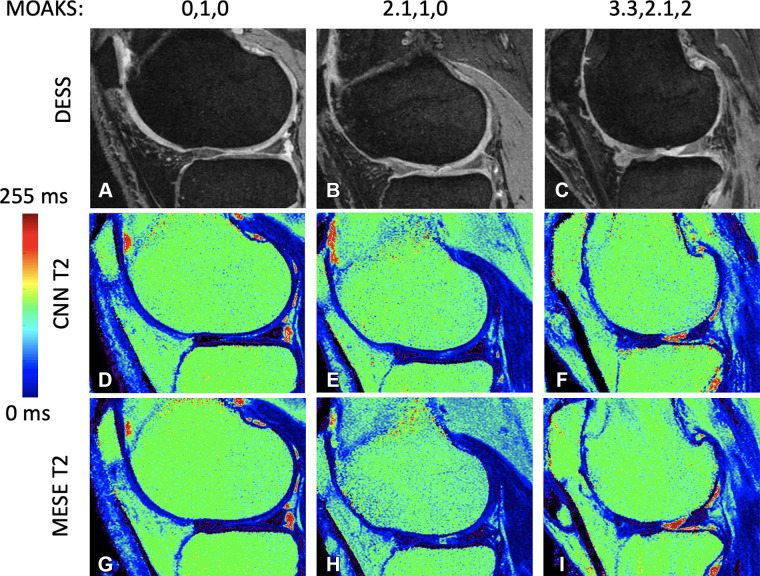
Each training epoch took about 1.5 hours, totaling 30 hours of training for the 20 epochs. Figure 3 shows sample results in three patients with progressively more cartilage loss, which was measured by using the MRI OA knee score (MOAKS) (30). Reference DESS images, CNN T2 maps, and MESE T2 maps are shown in Figure 3A–3C, 3D–3F, and 3G–3I, respectively. In some instances, the network was observed to either remove artifacts that were present on the MESE images or introduce artifacts on the CNN maps, as shown in Figure 4.
Figure 3:
(A–C) Double-echo in steady-state (DESS) images for progressive cartilage degeneration, measured on the lateral femur by using the MRI osteoarthritis knee score (MOAKS). (D–F) Convolutional neural network (CNN) T2 maps corresponding to images A, B, and C, respectively. (G–I) Multiecho spin-echo (MESE) T2 maps corresponding to images A, B, and C, respectively. The MOAKS values listed above the top row of images are defined as the individual anterior, center, and posterior scores, respectively. The scores show any cartilage loss in the respective region; if the patient had full-depth cartilage loss in the region as well, this is added as a separate score after a period. Both scores (the one before the period representing any cartilage loss and the one after the period representing full-depth loss) range from 0 to 3, with 0 representing no loss, 1 representing loss of 1%–10% of the area, 2 representing loss of 10%–75% of the area, and 3 representing loss of more than 75% of the area.
Figure 4:
Sample images show artifacts on multiecho spin-echo (MESE) or convolutional neural network (CNN) T2 maps. In patient 1, the CNN T2 map has removed pulsation artifacts that were apparent on the MESE T2 map. In patients 2 and 3, the CNN T2 map has visible artifacts. For patient 2, patient movement is seen along the superior-inferior axis on the double-echo in steady-state (DESS) scan, likely causing the artifact, as the discrepancy between the anatomic images confuses the CNN when predicting the T2 map. The source of the artifact for patient 3 is less clear, but it could be due to the network picking up incomplete fat suppression in the bone marrow and interpreting it as T2 variation. Such artifacts in the bone marrow do not affect the analysis of the femoral condyle cartilage. Cor = coronal, Sag = sagittal, TSE = turbo spin echo.
Detailed Evaluation of Limited Test Dataset
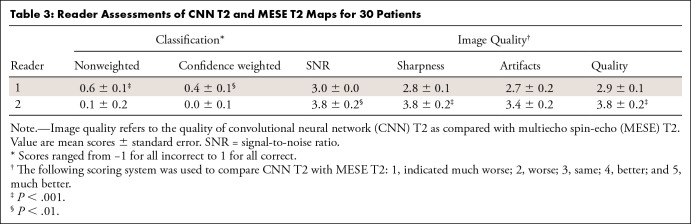
Reader assessments are tabulated in Table 3, which shows results for the 30 patients randomly chosen for the detailed evaluation. One reader could determine which image was created by the CNN (mean confidence-weighted score, 0.4 ± 0.1 [standard error]; P < .01). The same reader did not see a significant difference in image quality metrics. The other reader could not significantly discern which map was created by the CNN but found the CNN maps to have a significantly better SNR, sharpness, and overall quality. The Cohen κ for the nonweighted classification of the CNN and MESE maps between the two readers was 0.03, indicating slight agreement.
Table 3:
Reader Assessments of CNN T2 and MESE T2 Maps for 30 Patients
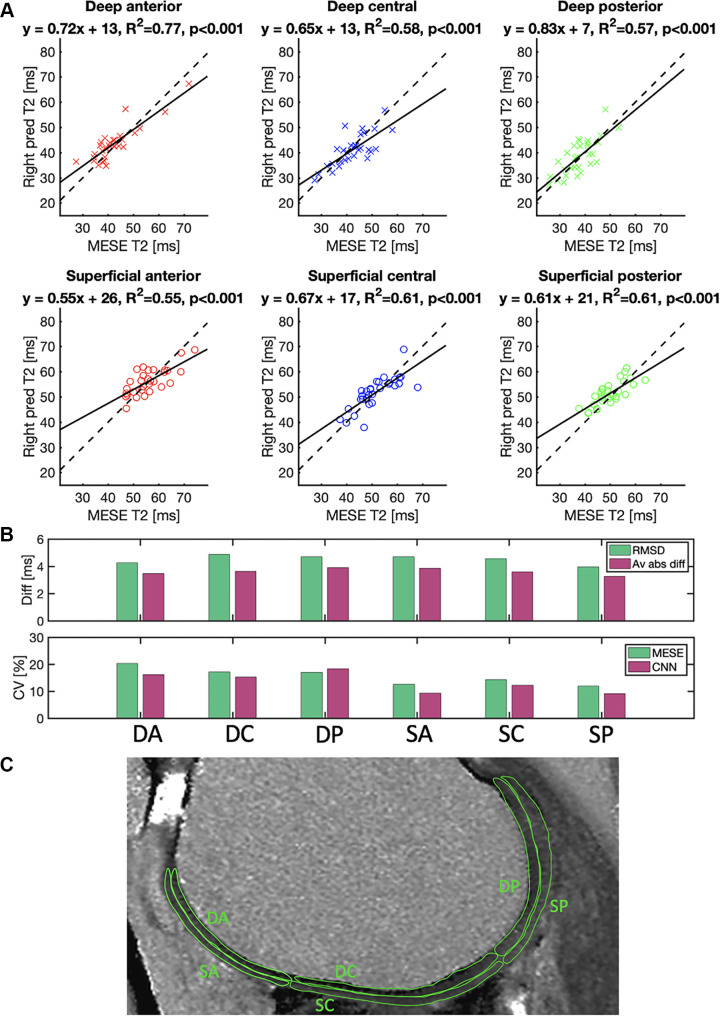
Figure 5 shows the subregional analysis in the 30 patients. The best-fit line slope was between 0.55 and 0.83, with R2 values between 0.55 and 0.77 (P < .001 for all). In all subregions, the Kolmogorov-Smirnov test did not reject the null hypothesis of the MESE data or CNN data being normal (P > .25).
Figure 5:
(A) Mean T2 in a region of interest drawn in the deep or superficial anterior (red), central (blue), or posterior (green) position on the femoral cartilage on images from 30 patients. Each point represents a value from a region of interest in a section of the right knee of one patient. The plots compare the reference multiecho spin-echo (MESE) T2 estimates of the right knee (x-axis) with the T2 predicted (pred) by the neural network (y-axis) of the same knee. Solid lines represent best-fit lines, and dashed lines represent y equal to x. (B) Root-mean-square differences (RMSD), average absolute differences (Av abs diff) (top), and coefficients of variation (CV) (bottom) in the cartilage regions. (C) Typical example of the different cartilage regions. CNN = convolutional neural network, DA = deep anterior, DC = deep central, diff = difference, DP = deep posterior, SA = superficial anterior, SC = superficial central, SP = superficial posterior.
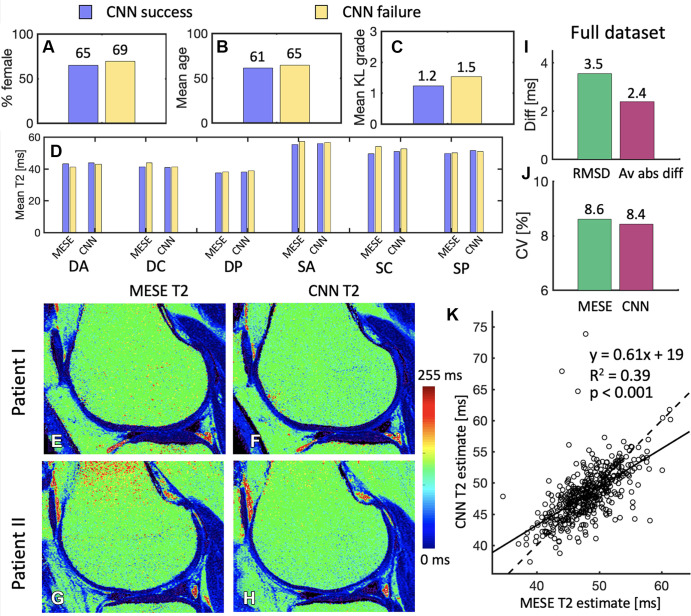
Figure 6A–6D show the failure analysis, comparing the “success” and “failure” subsets. These contained 17 and 13 patients, respectively. The failure set had a slightly higher proportion of women, higher mean age, and higher KL grade, but these differences were not significant. Similarly, no clear pattern or significance of T2 differences was observed, either for the MESE T2 or the CNN T2 values.
Figure 6:
(A–C) Comparison of the subsets of the randomly selected 30 patients evaluated by the radiologists, whereby the convolutional neural network (CNN) succeeded in convincing at least one radiologist (17 patients) that it was the multiecho spin-echo (MESE) map and where it did not (13 patients). In B, age is presented in years. (D) Comparison between the success and failure sets of multiecho spin-echo (MESE) T2 and CNN T2 in the six different cartilage subregions. (E, F) Sample (E) MESE T2 map and (F) CNN T2 map in a patient for which both readers failed to recognize which map was the CNN T2 map. (G, H) Sample (G) MESE T2 map and (H) CNN T2 map on which both readers accurately recognized which image was the CNN T2 map but one or both deemed the CNN T2 map to be better in terms of signal-to-noise ratio, sharpness, artifacts, and overall quality. (I) Root-mean-square difference (RMSD) and average absolute difference (Av abs diff) between the MESE T2 and CNN T2 maps. (J) Coefficients of variation (CV) for the MESE and CNN maps. (K) Comparison of average values from the MESE and CNN T2 maps. Plot shows the results for all 456 patients in the test dataset. One point represents one patient. The solid line represents the least-squares fit line through the points, while the dashed line represents the y equal to x line. DA = deep anterior, DC = deep central, diff = difference, DP = deep posterior, KL = Kellgren-Lawrence, SA = superficial anterior, SC = superficial central, SP = superficial posterior.
Figure 6E and 6F show a patient image on which both readers failed to determine which image was the CNN T2 map. Figure 6G and 6H show a patient image on which both radiologists accurately determined which image was the CNN T2 map but deemed it to have higher quality than the MESE T2 map. By using the metrics in Table 3, reader 1 assigned the scores 3, 4, 4, and 4 for SNR, sharpness, artifacts, and overall quality of this CNN T2 map, respectively, as compared with these parameters of the MESE T2 map, while reader 2 assigned the scores 5, 5, 5, and 5, respectively, for these parameters.
Evaluation of Full Test Dataset
The comparison of the MESE T2 and CNN T2 maps for the 456 test dataset patients is shown in Figure 6I–6K. Regression analysis of the MESE versus CNN estimates showed a significant correlation (P < .001) between the two approaches, resulting in a line with a slope of 0.61 and an R2 of 0.39. As in the subregional analysis, the Kolmogorov-Smirnov test did not reject the null hypothesis of the MESE or CNN data being normally distributed (P = .62 and P = .22 for MESE data and CNN data, respectively).
Discussion
In this study, we investigated whether synthetic T2 maps, of comparable utility to T2 maps conventionally acquired with MESE, could be estimated in the femoral cartilage of the lateral condyles in the right knee from anatomic DESS and TSE sequences by using a CNN. Our motivation was that such an approach could potentially eliminate the need for a specific T2 acquisition in a scan protocol. A more detailed comparison with MESE was performed for 30 patients. This involved a reader assessment, which showed the CNN T2 maps to be similar to or better than the MESE maps, and a cartilage subregional analysis, which showed a large correlation between the two methods. A larger quantitative comparison to the MESE T2 in the whole cartilage of an examined section was performed in 456 patients in the test datasets (approximately 10% of the included patients), and it likewise showed a large correlation but incomplete absolute numeric agreement between the two methods. While the results showed the CNN T2 maps to be visually similar and correlated to baseline T2 maps from MESE measurements, there is still room for improvement in the quantitative values in terms of absolute agreement.
The quality assessment performed by one reader indicated a similar or better SNR, sharpness, and overall quality of the CNN maps as compared with MESE, while another reader found no significant difference in quality. A failure analysis in which images on which the network succeeded in convincing at least one radiologist of having generated MESE T2 maps were compared with images on which the network failed to do so did not yield significant differences in cartilage region T2 values, sex, age, or disease severity measured with KL grades. However, it is interesting to note that the largest relative difference in these metrics between the two sets was in the KL grade (25%). This could indicate a worse performance for patients with more severe OA. However, a larger sample would be required to investigate this and potentially reach statistical significance, and this could be pursued in future studies.
While complete numeric agreement between any two estimation methods is always ideal, even imperfect agreement can be valuable if the two sets of results are well correlated. For example, if both methods show an increase in measurements with disease severity, either method could be used for such estimation if it was used consistently throughout the study. Our measurement correlations, both for whole-cartilage areas and subregions, may indicate that CNN T2 maps could be used for such internal assessment. Furthermore, it should be noted that estimation methods for MRI parameters such as T2 do not always agree, even under the best circumstances. Our detailed evaluation demonstrated agreement similar to what was previously reported between different T2 estimation methods in cartilage, where similar regression analysis comparing different methodologies to spin-echo measurements yielded slopes, intercepts, and R2 values in the range of 0.41 to 1.31, −2 to 19, and 0.25 to 0.64, respectively, for data representing both deep and superficial cartilage (31). The T2 in deep cartilage was generally lower than that in superficial cartilage, in agreement with data in the literature (1).
The ultimate goal of using deep learning for T2 mapping is to replace the conventional T2 mapping sequence in a clinical scanning protocol. This requires the CNN T2 map to be quantitatively accurate, as well as retain or improve on the image quality of conventional maps. To assess the quantitative accuracy of CNN T2 maps, we performed a quantitative ROI-based comparison of CNN T2 maps and MESE T2 maps, and to assess the capability of CNN T2 maps to retain or improve on the image quality of conventional maps, experienced radiologists performed a quality assessment. Figure 6I demonstrates an average root-mean-square difference of 3.5 msec and an average absolute difference of 2.4 msec between the two methods in an ROI covering the whole femoral cartilage in a lateral condyle section. Larger average differences were observed in the subregions, as shown in Figure 5B. Prior studies suggest an average difference of 4–5 msec in lateral femur T2 between healthy individuals and those with mild or severe OA (32). Other studies suggest a larger difference of 12 msec in the lateral condyle between healthy controls and those having cartilage injury damage (33). These data indicate that an average CNN T2 map can be accurate enough to have radiologic value. However, Figures 5A and 6K demonstrate substantial variability when looking at individual images.
While large correlation effects can have value in research studies, as previously noted, this is not the case for individual clinical assessments, in which such false-negative and false-positive results can lead to inappropriate treatments. Therefore, even stronger linearity is desirable to reduce the odds of false-negative and false-positive assessments. Future work aimed at obtaining such strong linearity could also be used to investigate whether internal relationships between cartilage regions are conserved on the CNN T2 maps, as compared with the MESE T2 maps, within a scaling factor. For example, if the difference between a patient’s deep anterior and deep posterior measurements is 10% with both the CNN and the MESE approaches, this could have clinical value, even if the CNN approach yielded consistently higher values. The ratio of the relative differences for the two approaches should then be 1 for all cartilage regions. In terms of our results, computing this ratio over all patients (using the deep anterior region as the baseline) and then computing the average value for each patient resulted in a distribution of averages with a mean of 0.98 and standard deviation of 0.08.
A study to further investigate the clinical value of our approach would be an important expansion of this work. In such a study, the reader assessment could involve estimating disease severity on the basis of the MESE and CNN T2 maps to learn which estimate is closer to the baseline estimate included with the OAI data. This should take into account the fact that an assessment of cartilage damage is not based solely on a T2 map; rather, it includes multiple contrasts, including that on proton-density–weighted TSE images. In addition, the study could involve initial assessment of data from proton-density–weighted TSE MRI, and the T2 map could be used to confirm elevated areas of T2, indicating lesions. This could be accomplished by using a modified Outerbridge classification (34) with multiple observers and a consistent scoring system.
In some cases, the CNN maps outperformed the MESE maps by removing pulsation artifacts, as shown in patient 1 (Fig 4). Therefore, the CNN could be used to reduce pulsation ghosting artifacts at T2 mapping. However, image artifacts that were not present on the MESE T2 maps were sometimes observed on the CNN T2 maps. Examples of such artifacts are shown in patients 2 and 3 (Fig 4). In patient 2, the DESS scan shows substantial motion along the superior-inferior axis. This prevents the network from effectively comparing pixel intensities, leading to CNN T2 artifacts. In patient 3, the source of a smaller artifact in the bone marrow is less clear, but it could be the result of the network interpreting incomplete fat suppression in the bone marrow as T2 differences, although pure machine learning sources such as hallucinations cannot be ruled out. Such bone marrow artifacts would not interfere with the study goal of estimating cartilage T2. Recent developments allow the computation of quantitative maps in addition to anatomic images directly by using DESS (35–38), which has the potential to mitigate pulsation artifacts and intersequence motion artifacts; however, this requires a modified DESS sequence that is not used in the OAI.
This work had certain limitations: As mentioned, the reformatted coronal TSE MR images had a pixelated appearance owing to lower spatial resolution along the anterior-posterior axis. While we considered it reasonable to assume that the network would set its weights for this data to contribute to the image quality and not negatively affect it, we cannot rule out the possibility that this might have had a detrimental effect on the output. While such effects would apply systematically to the whole dataset, higher–spatial-resolution or isotropic data could improve the results. Also, although data randomization and augmentation were not deemed to be necessary (owing to the anonymization process and the size of the dataset, respectively), the possible benefits of performing such steps could be investigated further. Data augmentation in the form used in the original pix2pix approach (24,28) was tried in the very early design stages of the network and was not observed to yield an improvement, so it was skipped in the subsequent design on the basis of that preliminary testing. Augmentation was again tried on the final design of the network, but it led to increases in average L1 over the cartilage mask of 2.2 msec and 2.5 msec per section and per patient, respectively, in the validation set. This could have occurred because the data were so standardized that augmentation (altering the field of view and orientation) became unrepresentative. Nonetheless, different types of augmentation applied throughout training could possibly yield performance improvements; this application could be investigated in future studies.
It should also be noted that this work was focused on the right femoral condyles only and no other cartilage regions. This was partly owing to our intention to show proof of concept in a commonly examined region and partly because the network used to generate the cartilage mask had been validated mainly for this cartilage type. In addition, the network is trained to produce CNN T2 maps on the basis of sequences and imaging parameters in the OAI dataset, and using this approach for other datasets would require retraining the network. Furthermore, the data used for training, validation, and testing in this work were not split according to patient characteristics such as age, sex, or disease stage. Grouping patients on the basis of such features potentially could yield more specialized networks. It is important to note that the analysis was not substantially focused on disease severity, apart from what is reported in Figure 6C. Analyzing performance in terms of both the quantitative agreement and the reader analysis for different stages of disease (based on scales such as KL and MOAKS scales) could yield important information about whether the network performs better at certain stages of disease and how it performs in diagnosing disease. While such analysis would require a larger number of patients than the 30 patients examined in this work for detailed evaluation, this would be an interesting direction of future research.
In conclusion, the results of this proof-of-concept study show that a neural network can be trained to produce quantitative CNN T2 maps (from anatomic OAI scans) that are comparable to MESE T2 maps. This approach represents an important innovation that demonstrates the potential of neural networks to derive quantitative data from anatomic images and is a step forward in the direction of research on deep learning in medical imaging.
Acknowledgments
Acknowledgments
The authors thank Will Lamond for helpful advice concerning neural network theory. Data used in the preparation of this manuscript were obtained and analyzed from the controlled access datasets distributed from the OAI, which is a collaborative informatics system created by the National Institute of Mental Health and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) to provide a worldwide resource to quicken the pace of biomarker identification, scientific investigation, and OA drug development.
Supported by DARPA (grant 2016D006054) and the National Institutes of Health (grant K99AG066815).
Disclosures of Conflicts of Interest: B.S. Activities related to the present article: institution/author receives funding from the NIH (K99AG066815, PI B.S.); institution received grant from Defense Advanced Research Projects Agency (DARPA 2016D006054, PI M.S.R.). Activities not related to the present article: author is listed as inventor of US patents 9,389,294 and 10,775,463; royalties from these patents are paid to Stanford University, the patent owner, and Stanford University in turn forwards a part of the royalties to the author; the methods described in these patents are not used in the work presented; the author’s laboratory is engaged with GE Healthcare in a research project to reduce noise in medical images with the help of AI-based image reconstruction and the laboratory receives funds from GE Healthcare as a part of this (author is not involved with this project). Other relationships: disclosed no relevant relationships. A.S.C. Activities related to the present article: institution received grants from NIH, GE Healthcare, and Philips. Activities not related to the present article: author serves on the scientific advisory boards of BrainKey and Chondrometrics; author is a paid consultant for Skope MR, Subtle Medical, Chrondrometrics, Image Analysis Group, Edge Analytics, ICM, and Culvert Engineering; institution receives grants from GE Healthcare and Philips; author has stock or stock options in LVIS, BrainKey, and Subtle Medical. Other relationships: disclosed no relevant relationships. B.Z. disclosed no relevant relationships. N.K. Activities related to the present article: institution receives grants from DARPA and GE Healthcare; institution receives support for travel from DARPA and GE Healthcare. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. M.T. disclosed no relevant relationships. G.E.G. disclosed no relevant relationships. M.S.R. Activities related to the present article: institution receives grant from DARPA (PI: M.S.R.) and GE Healthcare (PI: M.S.R). Activities not related to the present article: author is a paid consultant to Synex (unrelated to the work in the manuscript). Other relationships: M.S.R. is a co-founder and equity holder of Hyperfine Research, Vizma Life Sciences, Intact Data Services, Q4ML, and BlinkAI. None of these companies are related to the present manuscript.
Abbreviations:
- cGAN
- conditional GAN
- CNN
- convolutional neural network
- DICOM
- Digital Imaging and Communications in Medicine
- DESS
- double-echo in steady state
- GAN
- generative adversarial network
- KL
- Kellgren-Lawrence
- MOAKS
- MRI OA knee score
- MESE
- multiecho spin echo
- OA
- osteoarthritis
- OAI
- Osteoarthritis Initiative
- ROI
- region of interest
- SNR
- signal-to-noise ratio
- TSE
- turbo spin echo
References
- 1. Dardzinski BJ , Mosher TJ , Li S , Van Slyke MA , Smith MB . Spatial variation of T2 in human articular cartilage . Radiology 1997. ; 205 ( 2 ): 546 – 550 . [DOI] [PubMed] [Google Scholar]
- 2. Gold GE , Burstein D , Dardzinski B , Lang P , Boada F , Mosher T . MRI of articular cartilage in OA: novel pulse sequences and compositional/functional markers . Osteoarthritis Cartilage 2006. ; 14 ( Suppl A ): A76 – A86 . [DOI] [PubMed] [Google Scholar]
- 3. Mosher TJ , Dardzinski BJ . Cartilage MRI T2 relaxation time mapping: overview and applications . Semin Musculoskelet Radiol 2004. ; 8 ( 4 ): 355 – 368 . [DOI] [PubMed] [Google Scholar]
- 4. Chaudhari AS , Kogan F , Pedoia V , Majumdar S , Gold GE , Hargreaves BA . Rapid Knee MRI Acquisition and Analysis Techniques for Imaging Osteoarthritis . J Magn Reson Imaging 2020. ; 52 ( 5 ): 1321 – 1339 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buckwalter J , Mankin HJ . Instructional Course Lectures, The American Academy of Orthopaedic Surgeons - Articular Cartilage. I. Tissue Design and Chondrocyte-Matrix Interactions . J Bone Joint Surg 1997. ; 79 ( 4 ): 600 – 611 . [Google Scholar]
- 6. Henkelman RM , Stanisz GJ , Kim JK , Bronskill MJ . Anisotropy of NMR properties of tissues . Magn Reson Med 1994. ; 32 ( 5 ): 592 – 601 . [DOI] [PubMed] [Google Scholar]
- 7. Rubenstein JD , Kim JK , Morova-Protzner I , Stanchev PL , Henkelman RM . Effects of collagen orientation on MR imaging characteristics of bovine articular cartilage . Radiology 1993. ; 188 ( 1 ): 219 – 226 . [DOI] [PubMed] [Google Scholar]
- 8. Lüsse S , Claassen H , Gehrke T , et al . Evaluation of water content by spatially resolved transverse relaxation times of human articular cartilage . Magn Reson Imaging 2000. ; 18 ( 4 ): 423 – 430 . [DOI] [PubMed] [Google Scholar]
- 9. Eckstein F , Wirth W , Nevitt MC . Recent advances in osteoarthritis imaging: the osteoarthritis initiative . Nat Rev Rheumatol 2012. ; 8 ( 10 ): 622 – 630 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peterfy CG , Schneider E , Nevitt M . The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee . Osteoarthritis Cartilage 2008. ; 16 ( 12 ): 1433 – 1441 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Link TM , Li X . Establishing compositional MRI of cartilage as a biomarker for clinical practice . Osteoarthritis Cartilage 2018. ; 26 ( 9 ): 1137 – 1139 . [DOI] [PubMed] [Google Scholar]
- 12. Zhu B , Liu JZ , Cauley SF , Rosen BR , Rosen MS . Image reconstruction by domain-transform manifold learning . Nature 2018. ; 555 ( 7697 ): 487 – 492 . [DOI] [PubMed] [Google Scholar]
- 13. Hosny A , Parmar C , Quackenbush J , Schwartz LH , Aerts HJWL . Artificial intelligence in radiology . Nat Rev Cancer 2018. ; 18 ( 8 ): 500 – 510 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaj S , Yang M , Nakamura K , Li X . Automated cartilage and meniscus segmentation of knee MRI with conditional generative adversarial networks . Magn Reson Med 2020. ; 84 ( 1 ): 437 – 449 . [DOI] [PubMed] [Google Scholar]
- 15. Chaudhari AS , Fang Z , Kogan F , et al . Super-resolution musculoskeletal MRI using deep learning . Magn Reson Med 2018. ; 80 ( 5 ): 2139 – 2154 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodfellow IJ , Pouget-Abadie J , Mirza M , et al . Generative Adversarial Nets . In: Proceedings of the 27th International Conference on Neural Information Processing Systems, 2, 2672–2680 . 2014. ; 2 : 2672 – 2680 . https://dl.acm.org/doi/10.5555/2969033.2969125 . [Google Scholar]
- 17. Mirza M , Osindero S . Conditional Generative Adversarial Nets . arXiv 1411.1784 [preprint] http://arxiv.org/abs/1411.1784. Posted November 6 , 2014. . Accessed November 2018. [Google Scholar]
- 18. Galbusera F , Bassani T , Casaroli G , et al . Generative models: an upcoming innovation in musculoskeletal radiology? A preliminary test in spine imaging . Eur Radiol Exp 2018. ; 2 ( 1 ): 29 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thomas KA , Kidziński Ł , Halilaj E , et al . Automated Classification of Radiographic Knee Osteoarthritis Severity Using Deep Neural Networks . Radiol Artif Intell 2020. ; 2 ( 2 ): e190065 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leung K , Zhang B , Tan J , et al . Prediction of total knee replacement and diagnosis of osteoarthritis by using deep learning on knee radiographs: data from the osteoarthritis initiative . Radiology 2020. ; 296 ( 3 ): 584 – 593 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pedoia V , Lee J , Norman B , Link TM , Majumdar S . Diagnosing osteoarthritis from T2 maps using deep learning: an analysis of the entire Osteoarthritis Initiative baseline cohort . Osteoarthritis Cartilage 2019. ; 27 ( 7 ): 1002 – 1010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nevitt M , Felson D , Lester G . The Osteoarthritis Initiative: Protocol for the Cohort Study . https://nda.nih.gov/oai. Published 2006. Accessed May 2020 .
- 23. Desai AD , Gold GE , Hargreaves BA , Chaudhari AS . Technical Considerations for Semantic Segmentation in MRI using Convolutional Neural Networks . arXiv 1902.01977 [preprint]http://arxiv.org/abs/1902.01977. Posted February 5, 2019. Accessed May 2020.
- 24. Isola P , Zhu JY , Zhou T , Efros AA . Image-to-image translation with conditional adversarial networks . In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) , Honolulu, HI , July 21–26, 2017 . Piscataway, NJ: : IEEE; , 2017. ; 5967 – 5976 . [Google Scholar]
- 25.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, eds. Medical Image Computing and Computer-Assisted Intervention: MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Cham, Switzerland: Springer, 2015; 234–241. [Google Scholar]
- 26. Long J , Shelhamer E , Darrel T . Fully Convolutional Networks for Semantic Segmentation . In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) , Boston, Mass , June 7–12, 2015 . Piscataway, NJ; : IEEE; , 2015. ; 3431 – 3440 . [Google Scholar]
- 27. Abadi M , Barham P , Chen J , et al . Tensorflow: A system for large-scale machine learning . In: Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation , 2016. ; 265 – 283 . [Google Scholar]
- 28. Pix2Pix: Image-to-image translation with a conditional GAN . https://www.tensorflow.org/tutorials/generative/pix2pix. Accessed May 2020 .
- 29. Chollet F. Keras . GitHub . https://github.com/fchollet/keras. Published 2015. Accessed May 2020 .
- 30. Hunter DJ , Guermazi A , Lo GH , et al . Evolution of semi-quantitative whole joint assessment of knee OA: MOAKS (MRI Osteoarthritis Knee Score) . Osteoarthritis Cartilage 2011. ; 19 ( 8 ): 990 – 100 . [Published correction appears in Osteoarthritis Cartilage 2011; 19 (9):1168.] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matzat SJ , McWalter EJ , Kogan F , Chen W , Gold GE . T2 Relaxation time quantitation differs between pulse sequences in articular cartilage . J Magn Reson Imaging 2015. ; 42 ( 1 ): 105 – 113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dunn TC , Lu Y , Jin H , Ries MD , Majumdar S . T2 relaxation time of cartilage at MR imaging: comparison with severity of knee osteoarthritis . Radiology 2004. ; 232 ( 2 ): 592 – 598 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu J , Xie G , Di Y , Bai M , Zhao X . Value of T2-mapping and DWI in the diagnosis of early knee cartilage injury . J Radiol Case Rep 2011. ; 5 ( 2 ): 13 – 18 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Outerbridge RE . The etiology of chondromalacia patellae . J Bone Joint Surg Br 1961. ; 43-B ( 4 ): 752 – 757 . [DOI] [PubMed] [Google Scholar]
- 35. Sveinsson B , Chaudhari AS , Gold GE , Hargreaves BA . A simple analytic method for estimating T2 in the knee from DESS . Magn Reson Imaging 2017. ; 38 ( 63 ): 70 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sveinsson B , Gold GE , Hargreaves BA , Yoon D . SNR-weighted regularization of ADC estimates from double-echo in steady-state (DESS) . Magn Reson Med 2019. ; 81 ( 1 ): 711 – 718 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chaudhari AS , Black MS , Eijgenraam S , et al . Five-minute knee MRI for simultaneous morphometry and T2 relaxometry of cartilage and meniscus and for semiquantitative radiological assessment using double-echo in steady-state at 3T . J Magn Reson Imaging 2018. ; 47 ( 5 ): 1328 – 1341 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chaudhari AS , Stevens KJ , Sveinsson B , et al . Combined 5-minute double-echo in steady-state with separated echoes and 2-minute proton-density-weighted 2D FSE sequence for comprehensive whole-joint knee MRI assessment . J Magn Reson Imaging 2019. ; 49 ( 7 ): e183 – e194 . [DOI] [PMC free article] [PubMed] [Google Scholar]