Abstract
Purpose
To compare the inter- and intraobserver agreement and reading times achieved when assigning Lung Imaging Reporting and Data System (Lung-RADS) categories to baseline and follow-up lung cancer screening studies by using a dedicated CT lung screening viewer with integrated nodule detection and volumetric support with those achieved by using a standard picture archiving and communication system (PACS)-like viewer.
Materials and Methods
Data were obtained from the National Lung Screening Trial (NLST). By using data recorded by NLST radiologists, scans were assigned to Lung-RADS categories. For each Lung-RADS category (1 or 2, 3, 4A, and 4B), 40 CT scans (20 baseline scans and 20 follow-up scans) were randomly selected for 160 participants (median age, 61 years; interquartile range, 58–66 years; 61 women) in total. Seven blinded observers independently read all CT scans twice in a randomized order with a 2-week washout period: once by using the standard PACS-like viewer and once by using the dedicated viewer. Observers were asked to assign a Lung-RADS category to each scan and indicate the risk-dominant nodule. Inter- and intraobserver agreement was analyzed by using Fleiss κ values and Cohen weighted κ values, respectively. Reading times were compared by using a Wilcoxon signed rank test.
Results
The interobserver agreement was moderate for the standard viewer and substantial for the dedicated viewer, with Fleiss κ values of 0.58 (95% CI: 0.55, 0.60) and 0.66 (95% CI: 0.64, 0.68), respectively. The intraobserver agreement was substantial, with a mean Cohen weighted κ value of 0.67. The median reading time was significantly reduced from 160 seconds with the standard viewer to 86 seconds with the dedicated viewer (P < .001).
Conclusion
Lung-RADS interobserver agreement increased from moderate to substantial when using the dedicated CT lung screening viewer. The median reading time was substantially reduced when scans were read by using the dedicated CT lung screening viewer.
Keywords: CT, Thorax, Lung, Computer Applications-Detection/Diagnosis, Observer Performance, Technology Assessment
Supplemental material is available for this article.
© RSNA, 2021
Keywords: CT, Thorax, Lung, Computer Applications-Detection/Diagnosis, Observer Performance, Technology Assessment
Summary
An observer study with seven radiologists showed that using a dedicated CT lung screening viewer (with computer-aided detection, a segmentation algorithm, and automatic Lung Imaging Reporting and Data System [Lung-RADS] categorization) improved interobserver agreement for Lung-RADS categorization while substantially reducing the reading time.
Key Points
■ Interobserver agreement for the Lung Imaging Reporting and Data System categorization of low-dose chest CT scans was moderate when using a standard picture archiving and communication system (PACS) viewer and increased to substantial agreement when using a dedicated CT lung screening viewer (Fleiss κ values of 0.58 [95% CI: 0.55, 0.60] vs 0.66 [95% CI: 0.64, 0.68], respectively).
■ The median reading time per study was significantly reduced from 160 seconds when using a standard PACS viewer to 86 seconds when using the dedicated viewer for lung cancer screening CT scans (P < .001).
Introduction
Lung cancer is the primary cause of cancer-related deaths worldwide (1), which is largely attributable to the fact that most cases are diagnosed at an advanced stage. In the last decade, various trials have found that screening individuals at high risk by using CT led to a significant reduction in lung cancer mortality in the study group compared with a control group (2–6). As a result, screening is being implemented in several countries, and elsewhere, various lung cancer screening pilots and feasibility studies are underway (7–10).
The implementation of lung cancer screening will require a substantial reading effort from radiologists. Expert readers need to make the distinction between high- and low-risk nodules and assign an appropriate diagnostic workup. A widely used lung cancer screening guideline has been provided by the American College of Radiology—the Lung Imaging Reporting and Data System (Lung-RADS) assessment categories—for standardized reporting and nodule management recommendations (11). As shown in a previous study, there is a fair amount of interreader variability in determining the Lung-RADS category for a screening examination: 8% of the patients would have received different management recommendations from different radiologists (12).
The use of dedicated software may increase efficiency and agreement among radiologists. Computer-aided detection (CAD) algorithms in particular would likely make lung cancer screening more efficient and less expensive (13–18). Double reading by combining CAD and human readers has consistently been shown to enable a higher nodule detection sensitivity than other reading strategies (19–24). Although there are indications that algorithms can also help to reduce the average reading time (19), to the best of our knowledge, no study has yet compared the workflow in a dedicated viewer with integrated CAD support with that in a typical picture archiving and communication system (PACS) viewer without dedicated computerized tools.
The purpose of this study was to test the hypothesis that using a dedicated CT lung screening viewer would increase interreader agreement and reduce the reading time compared with using a standard PACS viewer.
Materials and Methods
Study Data
Scans were obtained from the National Lung Screening Trial (NLST), which took place from August 2002 to August 2004 (3). Participants were randomized to three annual rounds of CT or chest radiographic screening (control group); the median follow-up time was 6.5 years. The NLST was approved by the institutional review boards of participating centers, and all participants provided informed consent.
We received all CT scans (annual rounds T0, T1, and T2) from a random sample of 4512 participants in the NLST (project identifier: NLST-187; https://cdas.cancer.gov/approved-projects/982/). We included T0 and T1 scans in our study to be able to investigate the Lung-RADS categorization of baseline and follow-up scans.
The NLST database provided information regarding nodules detected on each scan, including the type (solid, part solid, pure ground glass, and calcified), size, and lobe location. Each CT scan was assigned a Lung-RADS category on the basis of these annotations. Lung-RADS version 1.0 was used in this study. The size of the solid component in part-solid nodules is used in Lung-RADS but was not recorded in the NLST annotations. A medical student (in their 4th year of their medical degree program) who was trained to read CT scans was tasked with identifying the part-solid nodules and semiautomatically segmented the solid component to obtain diameter measurements of the solid component for subsequent Lung-RADS categorization.
Because we were unsure of the effect size prior to this study, we did not perform power calculations but determined the number of cases on the basis of a trade-off between the sample size and the reading effort. An enriched group of scans was compiled by selecting 20 scans at random for each Lung-RADS category (1 or 2, 3, 4A, and 4B) and for each time point (T0 and T1). For the T1 scans, the corresponding T0 scan of the same participant was also selected. For verification, all scans were checked by a researcher (S.J.v.R., PhD candidate, MSc in Medicine) who was trained in annotating pulmonary nodules in consensus with an experienced radiologist (C.S.P., with >10 years of experience as a thoracic radiologist) to confirm the Lung-RADS categories of the included scans. The final data set consisted of 80 T0 CT scans and 80 T1 CT scans as well as the corresponding T0 CT scans from 160 distinct participants. Whether lung cancer was diagnosed and whether death from lung cancer or other causes occurred were known.
All CT scans were acquired by using a multidetector CT scanner with a minimum of four detectors, a section thickness ranging from 1.0 to 3.2 mm, and a low-dose protocol with an average effective dose of 1.5 mSv (3). If multiple reconstructions were available for a CT study, a random selection was made, and only this image was presented to the readers. Table E1 (supplement) includes the CT characteristics for the included participants.
Study End Points
The main aim of this study was to investigate the impact of using a dedicated viewer for CT lung cancer screening on the level of interobserver agreement for Lung-RADS categorization. Therefore, the primary end point of this study was the difference between the level of interobserver agreement achieved by using the standard viewer and the level of interobserver agreement achieved by using the dedicated CT lung screening viewer. The second aim of this study was to assess the reading time of all readers for both viewers; therefore, the secondary end point was the difference in reading times between the two viewers. Finally, we aimed to investigate which factors played an important role in interobserver agreement for Lung-RADS categorization and how these varied when the readers used the two different viewers. The categorization of a CT scan into a Lung-RADS category is determined by identifying the risk-dominant nodule and by subsequently characterizing that nodule in terms of the nodule type and nodule size. We aimed to analyze whether readers agreed more for each of these factors that play a role in Lung-RADS categorization. These were the exploratory end points of our study.
CT Image Viewers
Two viewers were used for the observer study: a dedicated viewer developed to optimize the workflow of reading lung cancer screening chest CT scans (CIRRUS Lung Screening, Diagnostic Image Analysis Group, Radboud University Medical Center), and a standard PACS-like viewer without dedicated computerized tools for reading CT screening studies (CIRRUS Essentials, Diagnostic Image Analysis Group, Radboud University Medical Center).
In the dedicated CT lung screening viewer, a commercially available CAD algorithm for nodule detection with regulatory approval for clinical use (Veolity Lung CAD, version 1.5, MeVis Medical Solutions) was integrated. Additionally, nodules annotated on prior (T0) scans were propagated as marks on T1 scans in the dedicated CT lung screening viewer. Both CAD and propagated nodule prompts were displayed as colored squares, visible from any orthogonal direction, and could be accepted or rejected by the observer. New nodule annotations could be created by double-clicking at the location of the nodule on the scan. Accepted marks and new nodule annotations were automatically segmented by a volumetric segmentation algorithm. The observers defined nodules as solid, part solid, nonsolid, or calcified, which would affect the segmentation of the nodule. The segmentation algorithm distinguished between solid and ground-glass components, including in part-solid nodules. Two parameters, the attenuation threshold value and the roundness versus irregularity, could be adjusted by the user to optimize the segmentation if this was felt to be necessary. The segmentation algorithm uses the volumetric segmentation to compute the longest and perpendicular diameter on any axial section and presents that to the user. If no satisfactory segmentation could be achieved by tuning of the parameters, the reader could use a manual diameter measurement of the nodule. Finally, the Lung-RADS category was automatically determined on the basis of this information and was presented in a dedicated report. When a prior scan was available, the software used a nonrigid registration to allow the user to scroll through both scans simultaneously (linked scrolling).
In the standard PACS-like viewer, nodule segmentation, CAD marks, linked scrolling, and automatic calculation of the Lung-RADS category were not available. Observers manually searched the scan, annotated the risk-dominant nodule by double-clicking the location of the nodule on the scan, selected the corresponding nodule type, and measured the longest and perpendicular diameters on axial sections by using electronic calipers.
All scans were read in both viewers by all observers. For follow-up cases, the T0 and T1 scans were shown side by side on two separate monitors. The T0 scans of the follow-up cases were preread by a researcher (S.J.v.R., PhD candidate, MSc in Medicine), and these readings were subsequently checked by an expert radiologist (C.S.P., with >10 years of experience as a thoracic radiologist) who was not involved in this observer study. Their annotations and Lung-RADS categories were available to the observers when categorizing the T1 scan. Both viewers included all standard radiologic viewing tools, such as window-level adjustment and magnification.
Observer Study
Three radiologists (E.T.S., B.d.H., and B.G., with >10, >5, and >5 years of experience in reading chest CT scans, respectively) and four radiology residents (R.W., M.M.W.W., R.S., and O.M.M., all 5th-year radiology residents) from five different institutions in the Netherlands and Denmark participated as observers. By inviting observers from different institutions and countries, and with different expertise levels, we aimed to increase the applicability of our results in lung cancer screening practice. Experience for the radiologists in reading chest CT scans ranged from 4 to 30 years. The residents were all in the 5th year of their residency. One radiologist had experience in reading screening CT studies. The observers were instructed to read the complete CT scan for the presence of lung nodules, annotate the risk-dominant nodule(s), and determine the correct Lung-RADS category. In cases that were assigned a Lung-RADS category of 1 or 2, annotating a risk-dominant nodule was not needed.
All observers read all cases twice: once in the dedicated CT lung screening viewer and once in the standard viewer. The reading process was split up into two separate reading sessions with at least 2 weeks in between to minimize any effect of memory bias. In each reading session, half of the cases were read with the dedicated CT lung screening viewer, and the other half of the cases were read with the standard viewer. The order of cases and the order of viewers were randomized for all readers. For all observers, the time between opening and signing off on each scan was recorded by the viewers automatically. The time duration therefore included the search for and measurement of pulmonary nodules and the assignment of the Lung-RADS category on the dedicated report as generated by the software. A review of the soft-tissue and bone windows for additional findings was not required. To get acquainted with the viewers, observers annotated 24 example cases.
Statistical Analysis
Because the NLST did not assign Lung-RADS categories to CT scans, there was no reference standard. For comparing the overall agreement between the two viewers, the Fleiss κ value for multiple raters (25) was calculated. For each observer, pairwise interobserver agreement on Lung-RADS categories with the remaining six observers was calculated by using Cohen weighted κ values with linear weights for each viewer separately (26). To assess whether observers rated the same scans differently between viewers, intraobserver agreement was also measured by using Cohen weighted κ values. κ values were interpreted by using the Landis and Koch guidelines (27).
Differences in reading times between the two viewers were analyzed by using a Wilcoxon signed ranked test (a Kolmogorov-Smirnov test found data not normally distributed). Bonferroni correction for multiple testing was applied for the individual tests of each observer; a P value less than .0071 (.05 divided by 7) was considered to indicate statistical significance. For the pooled results, Bonferroni correction was not applied, and the threshold for statistical significance was thus set to a P value less than .05. Analyses were performed in R version 3.6.3 (R Foundation for Statistical Computing) (28).
Results
Participant and Lung-RADS Overview
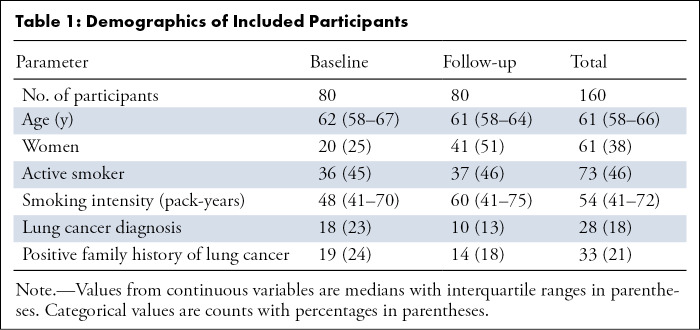
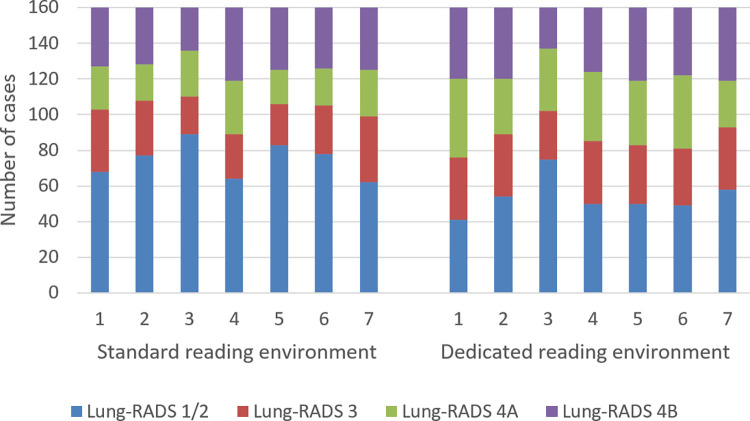
A total of 160 participants (median age, 61 years; interquartile range [IQR], 58–66 years; 61 women) were included. Demographics of the included participants are summarized in Table 1. The distribution of Lung-RADS classifications for each observer is shown in Figure 1. A total of 1120 reads were performed (seven readers for the 160 participants). The values for the percentage of scans per Lung-RADS category for the standard viewer and the dedicated lung screening viewer, respectively, were as follows: Lung-RADS category 1 or 2, 47% (521 of 1120) and 34% (377 of 1120); Lung-RADS category 3, 18% (199 of 1120) and 21% (232 of 1120); Lung-RADS category 4A, 15% (166 of 1120) and 23% (252 of 1120); and Lung-RADS category 4B, 21% (234 of 1120) and 23% (259 of 1120). These proportions show that the number of scans with a Lung-RADS category of 1 or 2 was substantially reduced when using the dedicated CT lung screening viewer. The total number of positive screening results (Lung-RADS category 3, 4A, or 4B) increased from 54% to 67% when using the dedicated CT lung screening viewer.
Table 1:
Demographics of Included Participants
Figure 1:
Distribution of Lung Imaging Reporting and Data System (Lung-RADS) classifications of all cases (n = 160) among seven observers when using the standard CT lung screening viewer and when using the dedicated CT lung screening viewer.
Inter- and Intraobserver Agreement
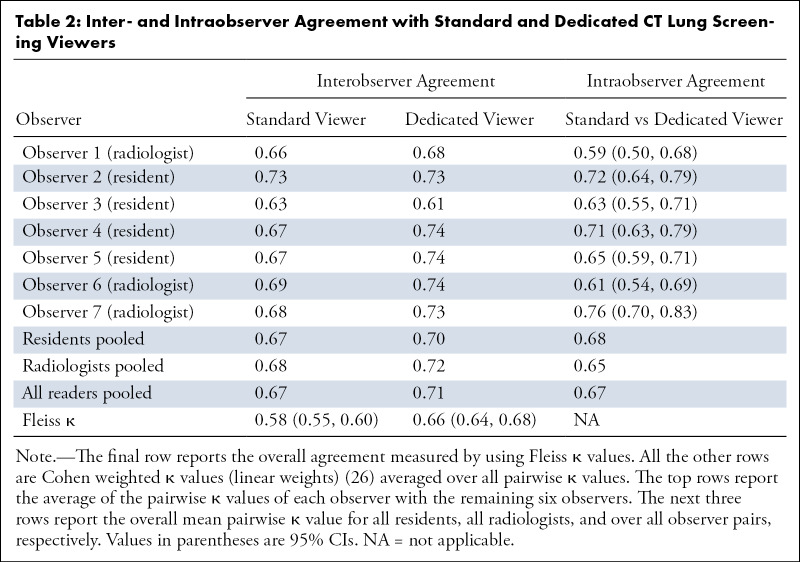
When using the standard PACS-like viewer, the interobserver agreement was moderate, with a Fleiss κ value of 0.58 (95% CI: 0.55, 0.60) (Table 2). The Fleiss κ value from readings conducted in the dedicated CT lung screening viewer was substantial at 0.66 (95% CI: 0.64, 0.68).
Table 2:
Inter- and Intraobserver Agreement with Standard and Dedicated CT Lung Screening Viewers
The mean pairwise Cohen weighted κ values of each observer with the remaining six observers ranged from 0.63 to 0.73 for the standard viewer and from 0.61 to 0.74 for the dedicated CT lung screening viewer (Table 2). For both viewers, no differences were seen between the subgroup of radiologists and the subgroup of residents (Table 2).
The intraobserver agreement, across the viewers, was substantial, with a mean Cohen weighted κ value of 0.67. Note that this was similar to the pairwise interobserver agreement. Weighted κ values ranged from 0.59 (95% CI: 0.50, 0.68) to 0.76 (95% CI: 0.70, 0.83) for each observer (Table 2).
Causes of Lung-RADS Disagreement
Next, disagreements were assessed among the readers and between the two viewers. There were a total of 3360 possible observer pairs for comparison (seven readers across 160 participants). When using the standard and dedicated CT lung screening viewers, disagreement regarding Lung-RADS categories occurred in 29% (971 of 3360) and 25% (853 of 3360) of the readings, respectively (Table 3). Thus, we found that there were 12% (118 of 971) fewer disagreements between observer pairs when using the dedicated CT lung screening viewer than there were when using the standard PACS-like viewer. Most disagreements were attributed to the annotation of a different risk-dominant nodule (74% [721 of 971] for the standard viewer and 87% [744 of 853] for the dedicated viewer). Among these disagreement pairs, it was often the case that one of the readers annotated a risk-dominant nodule (Lung-RADS category >2) but that the other reader did not (65% [468 of 721] for the standard viewer and 75% [556 of 744] for the dedicated viewer). The remainder of the disagreements were due to differences in the type or size of the same risk-dominant nodules (26% [250 of 971] for the standard viewer and 13% [109 of 853] for the dedicated viewer). We found that there were 67% (207 vs 68, see Table 3) fewer disagreement pairs that were due to different nodule diameter measurements when using the dedicated CT lung screening viewer.
Table 3:
Factors of Disagreement for Standard and Dedicated CT Lung Screening Viewers for All Pairwise Lung-RADS Comparisons
For the 971 disagreements (see Table 3) that were found for the standard PACS-like viewer, there were 480 (49%) disagreements about the baseline scans, and there were 491 (51%) disagreements about the follow-up scans. For the 853 disagreements (see Table 3) that were found when using the dedicated CT lung screening viewer, there were 403 (47%) disagreements about the initial CT scans and 450 (53%) disagreements about the annual repeat CT scans.
Examples of cases in which disagreements occurred are depicted in Figures 2 and 3.
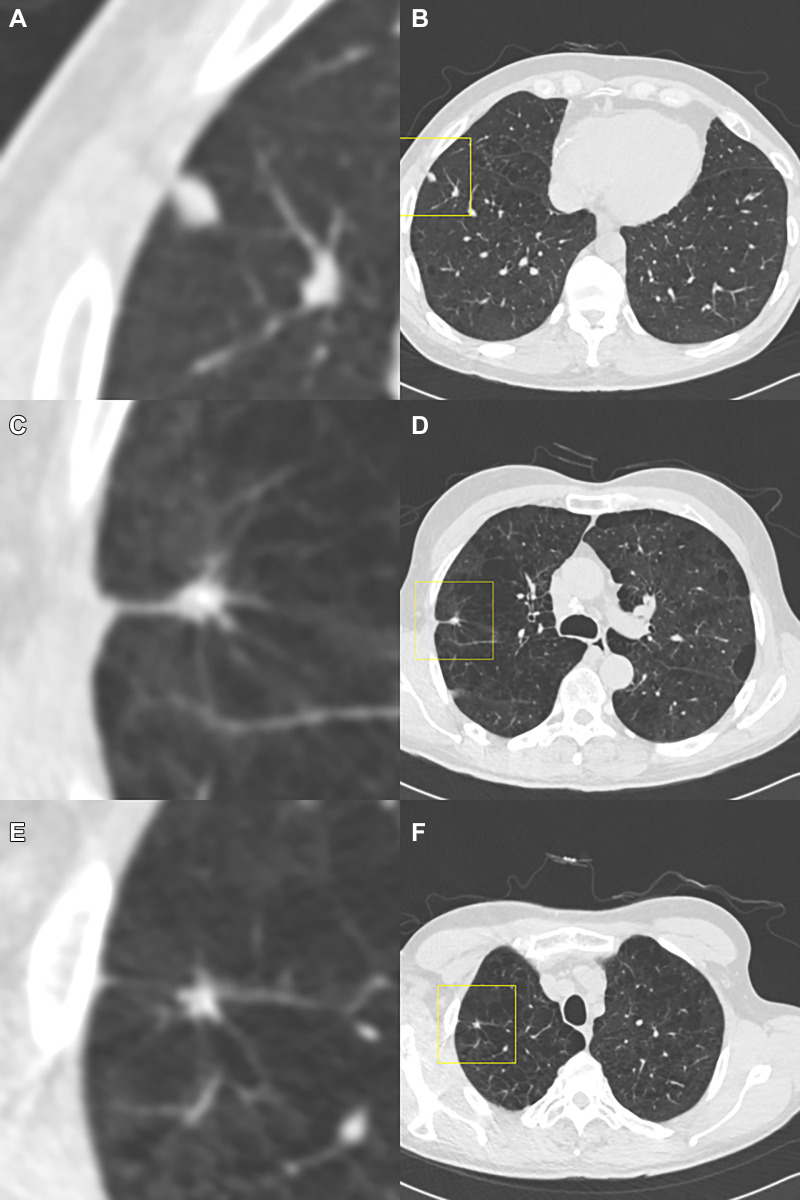
Figure 2:
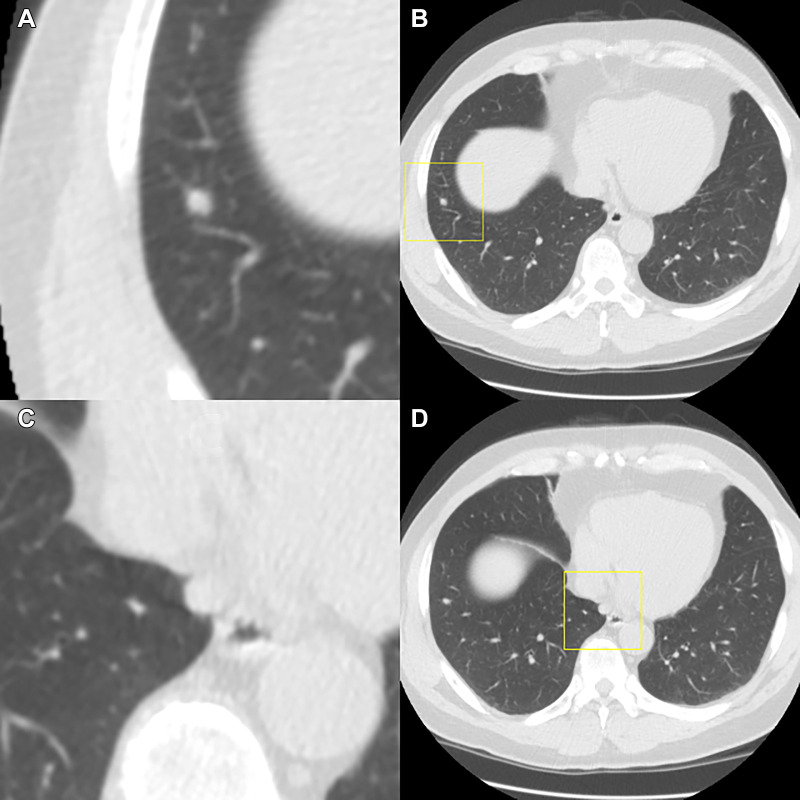
Example of a case in which the observers disagree on the Lung Imaging Reporting and Data System (Lung-RADS) category when reading using the dedicated CT lung screening viewer. This is a case for which the baseline scan was read by all observers. Each image pair (A and B, C and D, and E and F) shows one focal abnormality in a magnified view in the center (axial, field of view of 60 × 60 mm) and in a normal view with a square indicating the location. The three presented focal abnormalities are all detected by the computed-aided detection program. The (A, B) top and (E, F) bottom row show focal abnormalities that were accepted as a nodule by all observers and were measured as being 7 mm and 5 mm in diameter, respectively, by the volumetric software, which would result in a Lung-RADS 3 categorization. (C, D) The middle focal abnormality was only accepted by three of the seven observers as a nodule, was measured as being 8 mm in diameter by the volumetric software, and was determined to not be a nodule (apical fibrosis) by the remaining four observers. As a result, the three observers who accepted the middle focal abnormality as a nodule gave this case a Lung-RADS 4A score, whereas the remaining four observers gave this case a Lung-RADS 3 score.
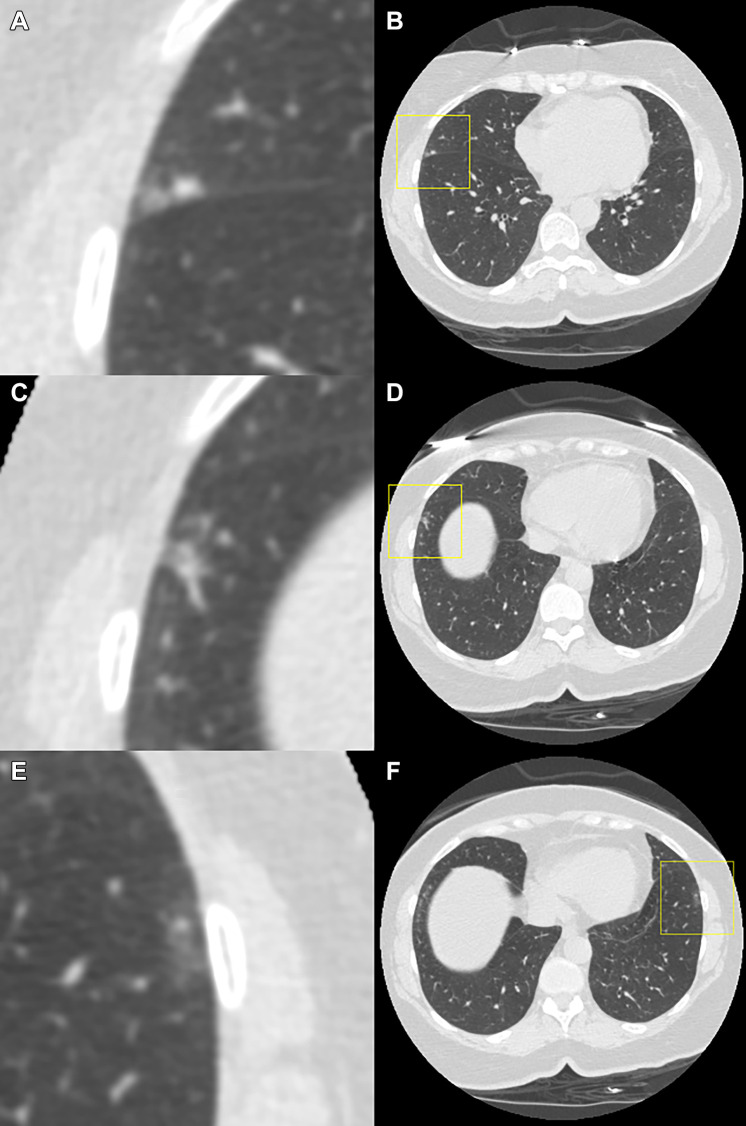
Figure 3:
Example of a case in which the observers disagree on the Lung Imaging Reporting and Data System (Lung-RADS) category when reading using the dedicated CT lung screening viewer. This is a case for which the follow-up scan was read by the observers. Each image pair (A and B, C and D, and E and F) shows one focal abnormality in a magnified view in the center (axial, field of view of 60 × 60 mm) and in a normal view with a square indicating the location. All of the three presented focal abnormalities were not visible on the baseline scan, and none of them were detected by the computed-aided detection program. The focal abnormality shown at the (A, B) top was scored by six out of seven observers as a solid nodule and was measured as 4 mm in diameter by the volumetric software. (C, D) The middle focal abnormality was scored as a solid nodule by two out of the seven observers and was measured as 7 and 5 mm in diameter, respectively. (E, F) The bottom focal abnormality was a ground-glass lesion that was only annotated by one observer and was measured as 8 mm in diameter. As a result, the Lung-RADS categories for this case varied between Lung-RADS 1 (one observer did not annotate any of the presented abnormalities), Lung-RADS 2, Lung-RADS 3, and Lung-RADS 4A (one observer measured the middle abnormality as a new solid nodule with a 7-mm diameter).
CAD Marks and Automatic Segmentation
It was only in the dedicated CT lung screening viewer that CAD marks were displayed to the users. Among the baseline (T0 scans only) cases, 359 CAD marks were shown (4.49 per scan on average). On average, 177 CAD marks (49%) were accepted on the baseline scans (range of 106–229 across observers). On the follow-up scans, 236 propagated marks and 179 CAD marks were displayed to the users. For the 179 CAD marks, 77 CAD marks (43%) were accepted on average on the follow-up scans (range, 56–105). For the 236 propagated nodule marks on the follow-up scans, an average of 187 marks (79%) were accepted by the observers (range, 151–217). Among the baseline cases, 48 nodules on average (range, 23–88) were manually added; 30 (range, 8–94) were added among the follow-up cases. No conclusions about CAD sensitivity or the false-positive rate can be drawn for this study because observers were not instructed to annotate and segment all visible nodules; the only requirement was to annotate the risk-dominant nodule in cases with a Lung-RADS category of 3 or higher.
A satisfactory nodule segmentation was achieved for almost all nodules shown in the dedicated CT lung screening viewer. Manual tuning of the segmentation parameters was performed by the observers in 28% of the nodule segmentations. Three out of seven observers deemed a manual diameter measurement necessary for three (n = 1 observer) or two (n = 2 observers) nodules.
Reading Time
Table 4 reports the observers’ reading times for each viewer. Pooling all results, the median reading time of 86 seconds (IQR, 51–141 seconds) when using the dedicated viewer was lower than the median reading time of 160 seconds (IQR, 96–245 seconds; P < .001) when using the standard viewer. For four of the seven observers, using the dedicated CT lung screening viewer reduced the reading time by more than half. For only a single observer (observer 4, O.M.M., 5th year resident), we found no evidence of a difference between the reading time when using the standard screening viewer and that when using the dedicated CT lung screening viewer (104 vs 82 seconds; P = .009 using Bonferroni correction). For the baseline (n = 80) CT scans, the median reading times were 129 seconds (IQR, 81–196 seconds) and 79 seconds (IQR, 46–123 seconds) when using the standard and dedicated CT lung screening viewers, respectively. For the annual repeat CT scans (n = 80), the median reading times were 196 seconds (IQR, 122–282 seconds) and 100 seconds (IQR, 59–158 seconds) when using the standard and dedicated CT lung screening viewers, respectively.
Table 4:
Reading Time in Standard and Dedicated CT Lung Screening Viewers per Observer
Lung Cancer Cases
In 13 of the 28 participants with a lung cancer diagnosis (Table 1), lung cancer was diagnosed within 1 year of the screening CT included in this study. For all 91 (13 lesions across seven readers) readings of the cases, 96% (87 of 91) and 91% (83 of 91) were assigned a Lung-RADS category of 4A or 4B in the standard and dedicated viewers, respectively. One case was assigned a Lung-RADS category of 1, 2, or 3 by four out of seven observers in the standard viewer and by all seven observers in the dedicated CT lung screening viewer (see Fig 4). For this participant, one small new nodule was visible on the T1 scan in the periphery of the right lower lobe. This nodule was detected by the CAD system and was measured by the segmentation software as having a mean diameter of 5.2 mm, and, as a result, all observers scored this nodule as Lung-RADS category 3 in the dedicated CT lung screening viewer. In the standard viewer, one observer measured this nodule as having a diameter of 6 mm, leading to a Lung-RADS 4A score. The other two observers who assigned a Lung-RADS 4A score to this case did measure this lesion as having a diameter of 5 mm but found another subpleural lesion close to the heart that measured 6 mm in diameter. The NLST records indicate that the cancer was found in the right hilum after a positive T1 screening with reason of “other,” suggesting that the subpleural lesion close to the heart was malignant.
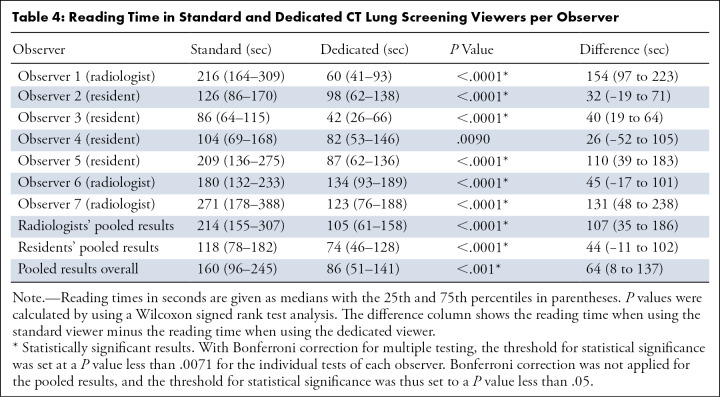
Figure 4:
Example of a participant with a lung cancer diagnosis in the same year. The T1 scan of this participant was included in the observer study; the T0 scan showed only a 2.5-mm solid nodule in the left upper lobe. The T1 scan shows two nodules, which are depicted in this figure. Each image pair (A and B and C and D) shows one nodule displayed in a magnified view in the center (left column, field of view of 60 × 60 mm) and in a normal view (right column) on the T1 scan. All observers categorized the scan as Lung Imaging Reporting and Data System (Lung-RADS) 3 when using the dedicated CT lung screening viewer, whereas three observers categorized the scan as Lung-RADS 4A when using the standard viewer. (A, B) The top images show a new, small solid nodule in the right lower lobe, which was detected by the computer-aided detection (CAD) program and was measured as 5.2 mm in diameter by the segmentation software. As a result, all observers gave this nodule a Lung-RADS 3 categorization when using the dedicated CT lung screening viewer. (C, D) The bottom images show a new subpleural nodule close to the heart that was not detected by the CAD program. This nodule was only detected by two of seven readers when using the standard viewer and was detected by none of the readers in the dedicated CT lung screening viewer. One reader measured the nodule in the right lower lobe as having a diameter of 6 mm when using the standard viewer, also leading to a 4A categorization of the scan, while still missing the nodule close to the heart.
Discussion
We have compared the interobserver agreement for Lung-RADS categorization and the reading times between a dedicated CT lung screening viewer with several supporting computerized tools and a standard PACS-like viewer without supporting tools. We hypothesized that the interobserver agreement would increase when using the dedicated CT lung screening viewer because of the availability of CAD marks and volumetric measurements, that, when relied on, should have increased reading consistency across observers. We found in this study that there were 12% (118 of 971) fewer disagreements between observer pairs when the dedicated CT lung screening viewer was used compared with when the standard PACS-like viewer was used (Table 3). The Fleiss κ value showed a significant increase in agreement when the dedicated CT lung screening viewer was used.
We expected that the use of computer support would reduce measurement differences and reduce the number of missed nodules (29,30). Indeed, our study suggests that automatic nodule segmentation allows a more precise measurement of nodule size, especially when a prior scan is available. This is reflected in the finding that there were 67% (207 vs 68, see Table 3) fewer disagreement pairs that were due to different nodule diameter measurements when the dedicated CT lung screening viewer was used.
An important finding of our study is that most disagreements were related to determining the risk-dominant nodule, a task that was not automated but was performed by the individual observers. Thus, although our observers were prompted by CAD, there was still substantial disagreement among readers as to what constituted a nodule. A previous study investigating the variability in the number of reported noncalcified nodules among the 112 radiologists in 32 screening centers in the NLST reported a substantial variability and referred to lesion detection, classification, and measurement (ie, whether the nodule was ≥4 mm) as possible reasons (31). This study suggests that a large proportion of the variability is caused by classification differences regarding whether a finding is a nodule or not. Note that this prior study included only one reader with experience in reading lung cancer screening CT scans; this may have affected the variability related to the designation of the risk-dominant nodule.
Regarding our second objective of determining whether the dedicated viewer could reduce reading times, the reading times in the dedicated viewer were nearly half those reported in the standard viewer (160 vs 86 seconds). Regardless of each observer’s reading speed in a standard viewer, there was a reduction in the reading times in the dedicated CT lung screening viewer (Table 4). Our hypothesis that there would be a shortened reading time in the dedicated viewer includes three main factors: less time needed to measure nodules (automatic volumetry vs manual measurement of the longest and perpendicular diameter on axial planes), no time loss caused by manual synchronization of the baseline and follow-up scan sections (only applicable to the follow-up scans in our data set), and less time needed to record information (automatic recording of the nodule size and Lung-RADS category vs manual reporting of the diameters and Lung-RADS category). However, because the data of this study do not allow for a precise time analysis of these events, we have no data to test our hypotheses. Note that a complete review of lung cancer screening CT scans for potential lung cancer findings other than pulmonary nodules (like endobronchial lesions or lymphadenopathy) and incidental findings unrelated to lung cancer would add to the overall reading times measured in this study.
In this study, we found that use of the dedicated CT lung screening viewer led to a higher proportion of positive screening results (Lung-RADS categories 3, 4A, or 4B) than did use of the standard viewer (67% vs 54%). This is an important finding for future research. On the basis of the data from this study, it is difficult to draw conclusions about the exact cause of this increase. We found that a higher number of risk-dominant nodules were annotated when the dedicated viewer was used. However, because our readers only had to annotate the risk-dominant nodule, we cannot make claims about whether readers missed more nodules when using the standard viewer without CAD support or whether they characterized the nodules differently without a dedicated viewer automatically highlighting and volumetrically measuring them. It is important to note that the division of Lung-RADS results for the dedicated CT lung screening viewer are more in line with how we selected the cases from the NLST database (25% in each category).
Our study adds to existing research by analyzing the effect of computer support on interobserver agreement for the categorization of screening CT scans into Lung-RADS categories. Analyzing sources of variability in studies like the one presented here will help to further standardize CT reporting of lung screening CT scans in the future. A recent study by Gierada et al (32) focused on the task of nodule measurement and showed that semiautomated nodule volumetry led to an increased interobserver agreement for Lung-RADS categories. This is in line with the results presented in this study.
The benefit of assisted reading workflows will depend on successful integration of computer support into clinical workflows. Integration of computer support into clinical workflows can be accomplished in different ways, ranging from loose integration into PACS systems by sending Digital Imaging and Communications in Medicine objects with CAD findings to use of a separate dedicated CT lung screening viewer that would allow interactive handling of CAD findings. The latter will most likely lead to the most efficient reading workflow but may only be acceptable if radiologists need to review larger numbers of screening scans consecutively.
We acknowledge that a more recent version of the Lung-RADS guidelines (version 1.1) has been released since this study was performed (33). Two main changes are the definition and categorization of perifissural nodules and the introduction of volumetric size thresholds. However, if we were to have used the new version, our results should not have been influenced by the inclusion of volumetric size thresholds because the diameter thresholds in the original Lung-RADS guidelines (version 1.0) were defined as the average diameters, which are ideally derived from volumetric measurements. A recent study showed that there is little benefit to using the volume instead of the mean diameter as a predictor for lung cancer risk in a logistic regression model (34).
Our study had some limitations that should be taken into consideration. First, we used a relatively small sample size of scans. However, it was an enriched cohort consisting of 20 cases from each Lung-RADS category (1 or 2, 3, 4A, and 4B). We chose this approach to compensate for the disproportionately large number of Lung-RADS category 1 or 2 cases in a screening population to make the best use of observer time for our study. Additionally, the results from seven observers could be pooled together. Second, the κ values reported in this study are based on an enriched set and are not representative of a random set of screening CT scans. In screening practice, the large majority of CT scans will show Lung-RADS category 1 or 2 findings, and the presented results on the enriched set of cases therefore have to be interpreted with this limitation in mind.
Third, we note that Lung-RADS category 4X was omitted from our study. This category provides radiologists the opportunity to upgrade a Lung-RADS 3 or 4A nodule on the basis of an increased level of malignancy suspicion so that it may undergo a Lung-RADS 4B workup. We feel that the addition of the 4X category might have introduced extra subjectivity and disagreement, thereby confounding the results. Moreover, our study’s purpose was not to assess the accuracy of malignancy identification when using different viewers. This is nevertheless an interesting research question that future studies should focus on to study the effect of using dedicated CT lung screening viewers on diagnostic accuracy.
Fourth, the majority of the CT data used in this study were acquired with a 2.0- or 2.5-mm section thickness. At present, the guidelines recommend the acquisition of CT scans with 1-mm section thickness, and it is unclear what effect this would have on our results. Potentially, a better resolution would further decrease the variability in nodule measurement, but this will need further investigation.
In summary, it is possible to reduce the median reading times of lung cancer CT scans by almost half with the support of CAD systems and nodule volume measurements that are integrated into an optimized CT lung screening viewer. Our findings suggest that use of the dedicated CT lung screening viewer led to a significant improvement in the interobserver agreement during follow-up management among radiologists compared with use of the standard PACS-like viewer, while significantly reducing the reading time.
Acknowledgments
The authors thank the National Cancer Institute for access to the data collected by the NLST. The statements contained herein are solely those of the authors and do not represent or imply concurrence or endorsement by the National Cancer Institute.
Supported by a research grant from MeVis Medical Solutions, Bremen, Germany.
Disclosures of Conflicts of Interest: C.J. MeVis Medical Solutions (Bremen, Germany) supplied a research grant to conduct this study and allowed the use of their commercial CAD system; research grant from Thirona (Nijmegen, the Netherlands); institution received royalties from MeVis Medical Solutions for the development of Veolity, a reading platform for lung cancer screening. A.S. disclosed no relevant relationships. S.J.v.R. disclosed no relevant relationships. E.T.S. disclosed no relevant relationships. R.W. disclosed no relevant relationships. M.M.W.W. disclosed no relevant relationships. B.d.H. disclosed no relevant relationships. R.S. disclosed no relevant relationships. O.M.M. disclosed no relevant relationships. B.G. author’s institution receives consulting fee from Siemens Healthineers; stock/stock options in Clovis Oncology, Infinity Pharmaceuticals, CRISPR Therapeutics, Nanostring Technologies, and Bionano Genomics. M.P. disclosed no relevant relationships. C.S.P. royalties from Springer, Thieme, and Elsevier; payment or honoraria from Boehringer. B.v.G. grants from Thirona and MeVis Medical Solutions, Delft Imaging, Canon, Dutch Research Council, and European Union Funds; royalties from Thirona, MeVis Medical Solutions, and Delft Imaging; leadership or fiduciary role with Fleischner Society; stock/stock options in Thirona.
Abbreviations:
- CAD
- computer-aided detection
- IQR
- interquartile range
- Lung-RADS
- Lung Imaging Reporting and Data System
- NLST
- National Lung Screening Trial
- PACS
- picture archiving and communication system
References
- 1. Global Health Observatory: tobacco control . World Health Organization Web site . https://www.who.int/gho/tobacco/use/en/. Published 2016. Accessed June 2020 .
- 2. Becker N , Motsch E , Trotter A , et al . Lung cancer mortality reduction by LDCT screening: results from the randomized German LUSI trial . Int J Cancer 2020. ; 146 ( 6 ): 1503 – 1513 . [DOI] [PubMed] [Google Scholar]
- 3. Aberle DR , Adams AM , et al. ; National Lung Screening Trial Research Team . Reduced lung-cancer mortality with low-dose computed tomographic screening . N Engl J Med 2011. ; 365 ( 5 ): 395 – 409 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nawa T , Fukui K , Nakayama T , et al . A population-based cohort study to evaluate the effectiveness of lung cancer screening using low-dose CT in Hitachi city, Japan . Jpn J Clin Oncol 2019. ; 49 ( 2 ): 130 – 136 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pastorino U , Silva M , Sestini S , et al . Prolonged lung cancer screening reduced 10-year mortality in the MILD trial: new confirmation of lung cancer screening efficacy . Ann Oncol 2019. ; 30 ( 10 ): 1672 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Koning HJ , van der Aalst CM , de Jong PA , et al . Reduced lung-cancer mortality with volume CT screening in a randomized trial . N Engl J Med 2020. ; 382 ( 6 ): 503 – 513 . [DOI] [PubMed] [Google Scholar]
- 7. Crosbie PA , Balata H , Evison M , et al . Implementing lung cancer screening: baseline results from a community-based ‘Lung Health Check’ pilot in deprived areas of Manchester . Thorax 2019. ; 74 ( 4 ): 405 – 409 . [DOI] [PubMed] [Google Scholar]
- 8. Delva F , Laurent F , Paris C , et al . LUCSO-1: French pilot study of LUng Cancer Screening with low-dose computed tomography in a smokers population exposed to Occupational lung carcinogens: study protocol . BMJ Open 2019. ; 9 ( 3 ): e025026 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Field JK , Duffy SW , Baldwin DR , et al . The UK Lung Cancer Screening Trial: a pilot randomised controlled trial of low-dose computed tomography screening for the early detection of lung cancer . Health Technol Assess 2016. ; 20 ( 40 ): 1 – 146 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee J , Lim J , Kim Y , et al . Development of Protocol for Korean Lung Cancer Screening Project (K-LUCAS) to evaluate effectiveness and feasibility to implement national cancer screening program . Cancer Res Treat 2019. ; 51 ( 4 ): 1285 – 1294 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lung CT Screening Reporting & Data System (Lung-RADS) v1.0 . American College of Radiology Web site . https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/Lung-Rads. Published 2014. Accessed June 2020 .
- 12. van Riel SJ , Jacobs C , Scholten ET , et al . Observer variability for Lung-RADS categorisation of lung cancer screening CTs: impact on patient management . Eur Radiol 2019. ; 29 ( 2 ): 924 – 931 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murphy A , Skalski M , Gaillard F . The utilisation of convolutional neural networks in detecting pulmonary nodules: a review . Br J Radiol 2018. ; 91 ( 1090 ): 20180028 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Benzaquen J , Boutros J , Marquette C , Delingette H , Hofman P . Lung cancer screening, towards a multidimensional approach: why and how? Cancers (Basel) 2019. ; 11 ( 2 ): E212 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simon AF , Holmes JH , Schwartz ES . Decreasing radiologist burnout through informatics-based solutions . Clin Imaging 2020. ; 59 ( 2 ): 167 – 171 . [DOI] [PubMed] [Google Scholar]
- 16. Lee SM , Park CM . Application of artificial intelligence in lung cancer screening . Korean J Radiol 2019. ; 80 ( 5 ): 872 . [Google Scholar]
- 17. Seijo LM , Peled N , Ajona D , et al . Biomarkers in lung cancer screening: achievements, promises, and challenges . J Thorac Oncol 2019. ; 14 ( 3 ): 343 – 357 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu B , Chi W , Li X , et al . Evolving the pulmonary nodules diagnosis from classical approaches to deep learning-aided decision support: three decades’ development course and future prospect . J Cancer Res Clin Oncol 2020. ; 146 ( 1 ): 153 – 185 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ritchie AJ , Sanghera C , Jacobs C , et al . Computer vision tool and technician as first reader of lung cancer screening CT scans . J Thorac Oncol 2016. ; 11 ( 5 ): 709 – 717 . [DOI] [PubMed] [Google Scholar]
- 20. Christe A , Leidolt L , Huber A , et al . Lung cancer screening with CT: evaluation of radiologists and different computer assisted detection software (CAD) as first and second readers for lung nodule detection at different dose levels . Eur J Radiol 2013. ; 82 ( 12 ): e873 – e878 . [DOI] [PubMed] [Google Scholar]
- 21. White CS , Pugatch R , Koonce T , Rust SW , Dharaiya E . Lung nodule CAD software as a second reader: a multicenter study . Acad Radiol 2008. ; 15 ( 3 ): 326 – 333 . [DOI] [PubMed] [Google Scholar]
- 22. Rubin GD , Lyo JK , Paik DS , et al . Pulmonary nodules on multi-detector row CT scans: performance comparison of radiologists and computer-aided detection . Radiology 2005. ; 234 ( 1 ): 274 – 283 . [DOI] [PubMed] [Google Scholar]
- 23. Wormanns D , Beyer F , Diederich S , Ludwig K , Heindel W . Diagnostic performance of a commercially available computer-aided diagnosis system for automatic detection of pulmonary nodules: comparison with single and double reading . Rofo 2004. ; 176 ( 7 ): 953 – 958 . [DOI] [PubMed] [Google Scholar]
- 24. Godoy MCB , Kim TJ , White CS , et al . Benefit of computer-aided detection analysis for the detection of subsolid and solid lung nodules on thin- and thick-section CT . AJR Am J Roentgenol 2013. ; 200 ( 1 ): 74 – 83 . [DOI] [PubMed] [Google Scholar]
- 25. Fleiss JL . Measuring nominal scale agreement among many raters . Psychol Bull 1971. ; 76 ( 5 ): 378 – 382 . [Google Scholar]
- 26. Cohen J . Weighted kappa: nominal scale agreement with provision for scaled disagreement or partial credit . Psychol Bull 1968. ; 70 ( 4 ): 213 – 220 . [DOI] [PubMed] [Google Scholar]
- 27. Landis JR , Koch GG . The measurement of observer agreement for categorical data . Biometrics 1977. ; 33 ( 1 ): 159 – 174 . [PubMed] [Google Scholar]
- 28. R Core Team . R: a language and environment for statistical computing . R Foundation for Statistical Computing Web site . https://www.R-project.org/. Accessed June 2020 .
- 29. Roos JE , Paik D , Olsen D , et al . Computer-aided detection (CAD) of lung nodules in CT scans: radiologist performance and reading time with incremental CAD assistance . Eur Radiol 2010. ; 20 ( 3 ): 549 – 557 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liang M , Tang W , Xu DM , et al . Low-dose CT screening for lung cancer: computer-aided detection of missed lung cancers . Radiology 2016. ; 281 ( 1 ): 279 – 288 . [DOI] [PubMed] [Google Scholar]
- 31. Pinsky PF , Gierada DS , Nath PH , Kazerooni E , Amorosa J . National lung screening trial: variability in nodule detection rates in chest CT studies . Radiology 2013. ; 268 ( 3 ): 865 – 873 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gierada DS , Rydzak CE , Zei M , Rhea L . Improved interobserver agreement on Lung-RADS classification of solid nodules using semiautomated CT volumetry . Radiology 2020. ; 297 ( 3 ): 675 – 684 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lung CT Screening Reporting & Data System (Lung-RADS) v1.1 . American College of Radiology Web site . https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/Lung-Rads. Published 2019. June 2020 .
- 34. Tammemagi M , Ritchie AJ , Atkar-Khattra S , et al . Predicting malignancy risk of screen-detected lung nodules-mean diameter or volume . J Thorac Oncol 2019. ; 14 ( 2 ): 203 – 211 . [DOI] [PubMed] [Google Scholar]