Abstract
Purpose
To evaluate the performance of a deep learning–based algorithm for automatic detection and labeling of rib fractures from multicenter chest CT images.
Materials and Methods
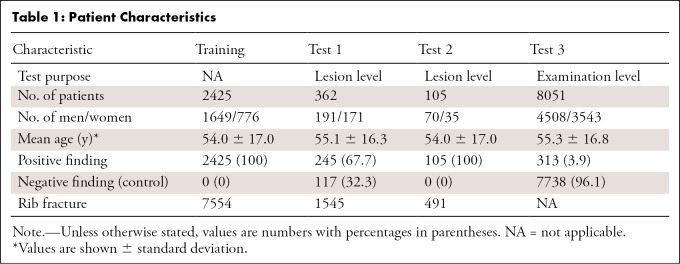
This retrospective study included 10 943 patients (mean age, 55 years; 6418 men) from six hospitals (January 1, 2017 to December 30, 2019), which consisted of patients with and without rib fractures who underwent CT. The patients were separated into one training set (n = 2425), two lesion-level test sets (n = 362 and 105), and one examination-level test set (n = 8051). Free-response receiver operating characteristic (FROC) score (mean sensitivity of seven different false-positive rates), precision, sensitivity, and F1 score were used as metrics to assess rib fracture detection performance. Area under the receiver operating characteristic curve (AUC), sensitivity, and specificity were employed to evaluate the classification accuracy. The mean Dice coefficient and accuracy were used to assess the performance of rib labeling.
Results
In the detection of rib fractures, the model showed an FROC score of 84.3% on test set 1. For test set 2, the algorithm achieved a detection performance (precision, 82.2%; sensitivity, 84.9%; F1 score, 83.3%) comparable to three radiologists (precision, 81.7%, 98.0%, 92.0%; sensitivity, 91.2%, 78.6%, 69.2%; F1 score, 86.1%, 87.2%, 78.9%). When the radiologists used the algorithm, the mean sensitivity of the three radiologists showed an improvement (from 79.7% to 89.2%), with precision achieving similar performance (from 90.6% to 88.4%). Furthermore, the model achieved an AUC of 0.93 (95% CI: 0.91, 0.94), sensitivity of 87.9% (95% CI: 83.7%, 91.4%), and specificity of 85.3% (95% CI: 74.6%, 89.8%) on test set 3. On a subset of test set 1, the model achieved a Dice score of 0.827 with an accuracy of 96.0% for rib segmentation.
Conclusion
The developed deep learning algorithm was capable of detecting rib fractures, as well as corresponding anatomic locations on CT images.
Keywords CT, Ribs
© RSNA, 2021
Keywords: CT, Ribs
Summary
A deep learning algorithm was developed to detect rib fractures and anatomic location of each fracture on chest CT images.
Key Points
■ The proposed deep learning–based method was able to detect rib fractures on chest CT scans and also indicate the anatomic location of each rib fracture.
■ For lesion-level detection, the model achieved a free-response operating characteristic score of 84.3% and a detection performance (precision, 82.2%; sensitivity, 84.9%) comparable to that of three radiologists (precision, 81.7%, 98.0%, 92.0%; sensitivity, 91.2%, 78.6%, 69.2%).
■ For examination-level detection, a large clinical dataset was evaluated with the developed model and attained an area under the receiver operating characteristic curve of 93% (95% CI: 91%, 94%), sensitivity of 87.9% (95% CI: 83.7%, 91.4%), and specificity of 85.3% (95% CI: 74.6%, 89.8%).
■ With the assistance of the deep learning model, the mean sensitivity of three radiologists increased (from 79.7% to 89.2%), with precision achieving similar performance (from 90.6% to 88.4%).
Introduction
Rib fractures are the most common complication in chest trauma, which is mainly caused by external forceful impact on the patient’s chest (1). Although only conservative treatment is required for most rib fractures, more than 90% of rib fractures have one or more associated injuries, such as the laceration of abdominal organs (the spleen, liver, or kidneys) and the damage of the brachial plexus and subclavian vessels (2). Therefore, the location, alignment, and quantity of rib fractures could potentially affect the clinical treatment of patients. Radiography and CT are two common diagnostic modalities for radiologists to investigate rib fractures. Compared with radiography, CT has the advantage of higher contrast resolution, which can display lesions in all directions and usually has a higher detection rate of rib fractures (3). However, radiologists need to investigate the rib fractures from hundreds of CT images on a section-by-section basis, which is a time-consuming process with the potential for missed fraction detection. Therefore, automatic rib fracture detection and labeling methods could reduce the number of missed fractures and improve the efficiency of clinical diagnosis.
Deep learning–based algorithms have been applied to the field of medical image processing, such as image registration (4,5), detection (6–9), segmentation (10–12), and disease prognosis (13,14). However, to the best of our knowledge, there are limited studies available on the detection of rib fractures on CT images using deep learning methods. Zhou et al (15) proposed a deep learning–based method to automatically detect the rib fractures from thoracic CT images and classify the rib fractures into three categories (fresh fractures, healing fractures, and old fractures). Weikert et al (16) presented rib fracture detection performance of a deep learning algorithm on a per-examination level and a per-finding level, as well as an analysis on the false-positive findings detected from the model. However, both of these studies used relatively small test datasets to evaluate the performance of the respective models, and the anatomic location of the rib fractures was also not assessed.
Therefore, this study aimed to evaluate the performance of a deep learning–based algorithm for automatic detection and anatomic location of the rib fractures from multicenter chest CT images.
Materials and Methods
Dataset
This retrospective study was approved by our medical ethical committee (approval no. YB-2020–445); the requirement to obtain informed consent was waived. A total of 11 229 consecutive examinations were collected from sites in different centers (A, B, C, D, E, and F) in China between January 2017 and December 2019. Scans of patients (n = 286; average age, 50 years; 210 men) with a history of fracture surgery, bone tumor, and notable artifacts were excluded, leaving a final dataset of 10 943 examinations with no further exclusions.
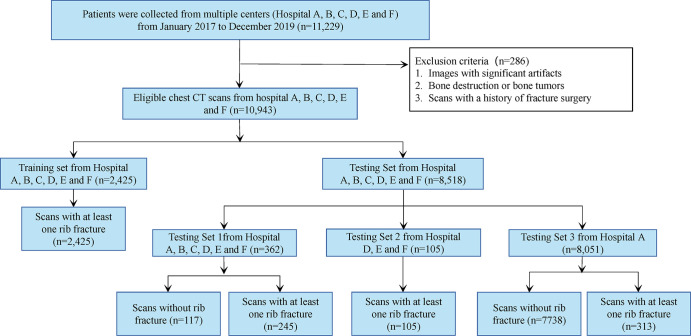
Of the 10 943 scans, we randomly chose 2425 that were positive for rib fracture as our training data, and the remaining 8518 scans were divided into three subtest sets (test set 1, 2, and 3), in which test set 1 and test set 2 were used for lesion-level assessment and test set 3 was used for examination-level assessment. Test set 1 consisted of 245 examinations with positive findings for rib fractures and 117 control examinations (without rib fractures) from six centers (A, B, C, D, E, and F), while test set 2 contained 105 examinations with positive findings for rib fractures from three centers (D [n = 31], E [n = 42], and F [n = 32]). Test set 3 was drawn from electronic clinical reports of patients who underwent chest CT examinations in hospital A between January 2017 and December 2019, which included 313 examinations with positive findings for rib fractures and 7738 control examinations. In addition, we randomly selected a subset from test set 1 (n = 74) to evaluate the performance of the rib labeling algorithm. Figure 1 summarizes the data distribution. Table 1 details patient characteristics.
Figure 1:
Flowchart of patient inclusion.
Table 1:
Patient Characteristics
CT Acquisition
CT examinations were performed using six different CT scanners (Philips Brilliance 16, Philips Medical Systems; SOMATOM Definition AS+, SOMATOM Definition Flash, Siemens Healthineers; GE Revolution CT, and GE LightSpeed VCT, GE Healthcare). The tube voltage was 120 kV, and the tube current was performed with an automatic modulation. Images have high variations with both section spacing (0.44 to 5 mm) and thickness (0.625 to 5 mm).
Ground Truth Annotation
The rectangular bounding box and polygon annotation were drawn on each CT section for rib fractures and ribs, respectively. For fracture detection, a total of 9590 rib fractures were annotated by radiologists, of which 7554 were in the training dataset. For rib segmentation, a total of 160 scans and 3792 ribs were annotated, of which 86 scans and 2046 ribs were in the training dataset. ITK-SNAP (version:3.6.0, http://www.itksnap.org) (17) was used as the labeling tool, and all types of rib fractures were included (fresh fracture, healing fracture, and healed fracture). The training set and test set 1 were annotated by a radiologist (10 years of experience). Test set 2 was annotated by three radiologists (6, 10, and 14 years of experience) and checked by one senior radiologist (18 years of experience). Test set 3 was collected from the picture archiving and communication system using keyword searching in hospital A from January 2017 to December 2019.
Model Overview
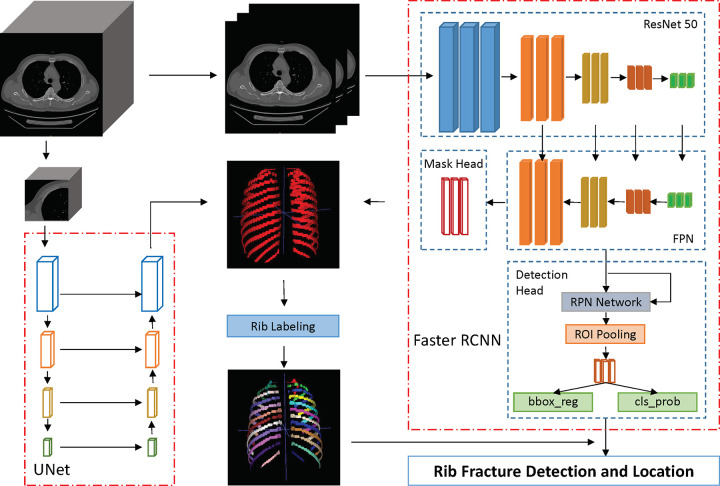
The pipeline of the rib fracture detection and location model is illustrated in Figure 2. In this study, we first used a two-dimensional (2D) detection network to scan the rib fractures and segment the ribs section by section. Then a three-dimensional (3D) network was selected to improve the accuracy of rib segmentation. The ensemble segmentation result of the 2D network and the 3D network was used to obtain the rib labeling results through a postprocessing algorithm. Finally, for each rib fracture output from the detection model, we determined the anatomic position from the rib labeling mask.
Figure 2:
Model development. The framework consists of a two-dimensional detection model (Faster R-CNN) to detect the rib fractures from the whole CT images and a three-dimensional segmentation model (U-Net) to segment the ribs. bbox_reg = bounding box regression, cls_prob = classification probability, FPN = feature pyramid network, ROI = region of interest, RPN = region proposal network.
Rib Fracture Detection
The algorithms used for object detection on the basis of deep learning can generally be grouped into two categories: two-stage detection and one-stage detection. The two-stage methods (18–20) first use a region proposal network to generate potential bounding boxes in an image, and then a classifier is run to obtain the class probabilities of these proposed boxes, while the one-stage methods (21–23) use object detection as a regression problem and obtain the bounding box coordinates and class probabilities from the image pixels directly.
We used the Faster R-CNN (20) model, a two-stage detection network, to detect the rib fractures on the chest CT scans. Specifically, the ResNet50 (24) network pretrained on the ImageNet competition dataset (25) was selected as the backbone to speed up training and improve the stability of the model. We added the Feature Pyramid Network (26) structure into the basic Faster R-CNN (20) detector, which consists of top-down architecture with lateral connections to fuse multiscale features and substantially improve the performance of the detector. The feature maps generated from the Feature Pyramid Network structure were then input to two different heads: a detection head for fracture detection and a mask head for rib segmentation. For the detection head, the region proposal network (20) was used to generate proposal regions, and the region of interest pooling layer was used here to convert the features of different sizes into a series of feature maps with a fixed size. Finally, joint training of classification probabilities and border regressions was implemented using softmax cross-entropy loss and smooth L1 loss. For the mask head, the feature maps from the last layer of the Feature Pyramid Network (26) were refined by several 3 × 3 convolutions and obtained the rib segmentation result. Formally, an anchor was considered as a positive label if it had an intersection over union (IoU) greater than 0.5 with any ground truth box or the highest IoU for a given ground truth box, and the anchor that had an IoU less than 0.3 for all ground truth boxes was defined as a negative label. The sizes of anchors were set as (16 × 16, 32 × 32, 64 × 64, 128 × 128, 256 × 256) with the multi-aspect ratios of (1:2, 1:1, 2:1) at each level.
For data preprocessing, we first clipped the intensities of all scans to the range of (−600,1200) HU and normalized them to the range of (0,1). Then we conducted random horizontal flip augmentation to make models learn invariant features to geometric perturbations.
For model training, each input section image was scaled to 1024 × 1024. We trained the network with TensorFlow (27) using four NVIDIA GeForce GTX 1080 GPUs, and the batch size was set as 4. Stochastic gradient descent with momentum (β = .9) was used to update the weights of the network. The initial learning rate was set to 0.001 and multiplied by 0.1 after each epoch.
Rib Segmentation and Labeling
The 2D networks with deep convolutions ignore the spatial information along the z dimension and therefore lose the ability to capture the 3D features of data. In this study, we used a 3D deep neural network, U-Net (28), which consists of a series of 3D convolutions to efficiently capture stereoscopic features and a decoder to recover the spatial information, to improve the accuracy of rib segmentation. Specifically, we divided the image into a series of 3D patches by sliding windows with a size of 256 × 256 × 64 and a stride of 32 × 32 × 16 from the whole volume data, then the patches were input into the neural network to obtain the rib masks. The final binary rib mask was the ensemble result of the masks from the Faster R-CNN (20) mask head and the 3D U-Net (28) model, and the ensemble weight was set as 0.5. For rib labeling, we first used the connected domain algorithm, which was implemented via the Scikit-Image (29) library to distinguish the ribs, then the ribs were anatomically labeled (left to right, and superior to inferior) according to their centroid coordinates.
For data preprocessing, we resampled the volume data to 3 mm on the z direction. Then the intensities of all scans were truncated into the interval (−361, 797) HU, and the min-max normalization method was used to normalize the data to (0,1). In the training stage, we adopted random rotation (90, 180, 270), scale (0.8–1.2), and flipping to alleviate the overfitting problem.
For model training, we adopted Keras (30) to train the model using one NVIDIA GeForce GTX 1080 GPU, and the batch size was set as 2. The Adam optimizer with a learning rate initialized as 0.001 was used to update the parameter of the model.
Assessment of the Algorithms
To evaluate the performance of our detection model, both the lesion-level and examination-level assessments were considered. For lesion-level assessment, we defined a predicted fracture as a true-positive finding if it had an IoU higher than 0.1 on the transverse plane and an intersection on z direction for any given ground truth. The reason we chose an IoU of higher than 0.1 as the hit detection criterion was because IoU tends to be varied for elongated objects. We used the free-response receiver operating characteristic (FROC), which measures both the detection rate and average false-positive rate per scan on test set 1. Seven false-positive rates (0.125, 0.25, 0.5, 1, 2, 4, 8 false-positive findings per scan) were included in this evaluation scheme. In addition, we compared the detection performance of our detection model with three experienced radiologists using sensitivity, precision, and F1 score on test set 2. Specifically, the radiologist’s results were annotated by themselves. For the results of the radiologist with the artificial intelligence (AI) model, we asked the radiologists to annotate on the CT sections that were drawn with the AI results (a series of bounding boxes). To ensure the fairness of the experiment, the AI-aided radiologist portion of the study was conducted 3 months after that of the radiologist annotating CT images without AI assistance. For examination-level assessment, we employed the area under the receiver operating characteristic curve (AUC), sensitivity, and specificity to evaluate the classification performance on the test set 3.
We defined a true-positive finding as when the algorithm detected a fracture on a positive scan that had at least one rib fracture and a false-positive finding as a detection label placed on a scan that was negative for a rib fracture. We defined a true-negative finding as when there was no model detection result on a negative image and a false-negative finding if the model detection label on a positive image was absent.
For the rib segmentation and labeling, we first used the Dice coefficient to evaluate the segmentation performance of different models, then mean Dice score and mean accuracy were selected to evaluate the performance of rib labeling. In this study, a rib is considered as labeled correctly if it has a Dice coefficient higher than 0.5 for its corresponding rib mask in the ground truth.
Statistical Analysis
We used software (R version 3.5.1; R Foundation for Statistical Computing) for statistical analyses. The nonparametric method proposed by DeLong et al (31) was used to calculate the AUC and its 95% CIs. The McNemar test was used to compare the sensitivity and precision of radiologists with and without AI assistance.
Results
Performance of Deep Learning Model on Rib Fracture Detection from Multicenter Evaluation
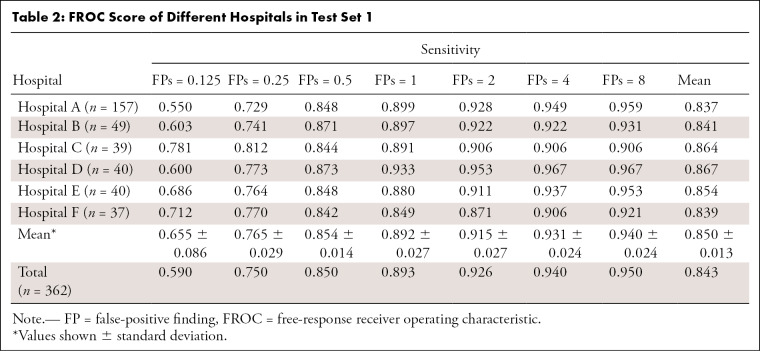
Table 2 lists the detection sensitivities at different false-positive rates on the six subsets of test set 1. The sensitivity for the six subsets of the model on test dataset 1 were all greater than 84%, with a mean sensitivity of 85.4% and a false-positive rate of 0.5. The mean sensitivity of the seven false-positive rates (0.125, 0.25, 0.5, 1, 2, 4, and 8 per scan) for the different subtest sets were all greater than 83%.
Table 2:
FROC Score of Different Hospitals in Test Set 1
Comparison of Detection Performance between Deep Learning Model and Radiologists
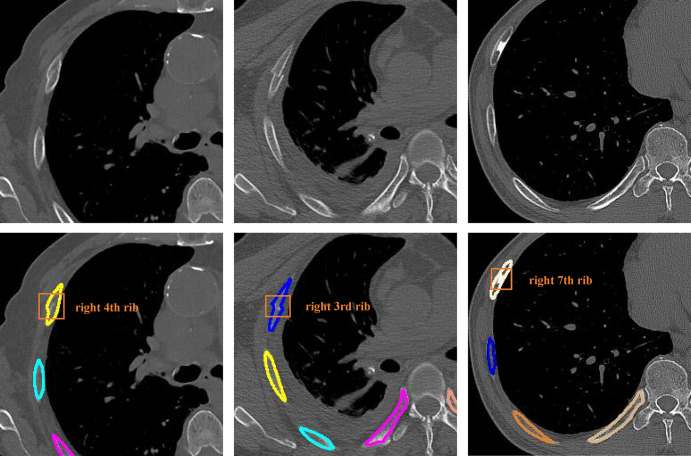
We evaluated the performance of radiologists for detecting fractures with and without the use of the AI model. As shown in Figure 3, the points of radiologist 1 and 2 without the assistance of AI were both above the FROC curve on test set 2, while radiologist 3 had a lower sensitivity than the model at the same false-positive rate. Radiologist 1 achieved a high sensitivity (91.7% [198 of 216], 91.0% [151 of 166], and 90.8% [99 of 109]) for the three different test subsets of test set 2 and a modest precision (81.0% [209 of 258], 83.3% [155 of 186], and 80.7% [96 of 119]), while radiologist 2 had a high precision (97.8% [179 of 183], 97.6% [120 of 123], and 98.9% [92 of 93]) and modest sensitivity (80.1% [173 of 216], 72.3% [120 of 166], and 83.5% [91 of 109]). Radiologist 3 had a higher precision (95.1% [154 of 162], 88.9% [104 of 117], and 91.9% [79 of 86]) than radiologist 1 but a much lower sensitivity (69.9% [151 of 216], 63.3% [105 of 166], and 74.3% [81 of 109]; Table 3).
Figure 3:
Free-response receiver operating characteristic curve for test set 2. The performance of the three experienced radiologists (red circles) and radiologists with the artificial intelligence (AI)–aided annotation (green circles) are shown. FP = false-positive finding.
Table 3:
Lesion-level Detection Performance on Test Set 2
The sensitivities of the three radiologists on three subsets of test set 2 were all higher with the assistance of the deep learning model. Radiologist 1 achieved a mean sensitivity of 92.5% (from 91.2%, P = .36) on the three subsets, while radiologist 2 and radiologist 3 attained a mean sensitivity of 88.3% (from 78.6%, P < .001) and 86.8% (from 69.2%, P < .001), respectively. We found no difference in the mean precision of radiologist 1 (81.7%–80.6%; P = .69); however, the precision of radiologist 2 was lower with the model (98.1%–91.2%; P < .001). Radiologist 3 attained a slightly higher mean precision from 92.0% to 93.4% (P < .001) on the three subsets of test set 2. The mean sensitivity and F1 score of the three radiologists on test set 2 was higher with AI assistance (mean sensitivity, 79.7%–89.2%; mean F1 score, 84.1%–88.6%), while the mean precision was lower with the use of the AI model (from 90.6% to 88.4%).
Moreover, we compared the annotation time between the radiologists without AI assistance and the radiologists with AI assistance. The average time for radiologists without AI assistance was about 7 minutes per case, while the time for radiologists with AI assistance was about 3 minutes per case, which demonstrated the great potential of the deep learning method in assistance of rib fracture detection.
Performance of Deep Learning Model on a Per-Examination Level
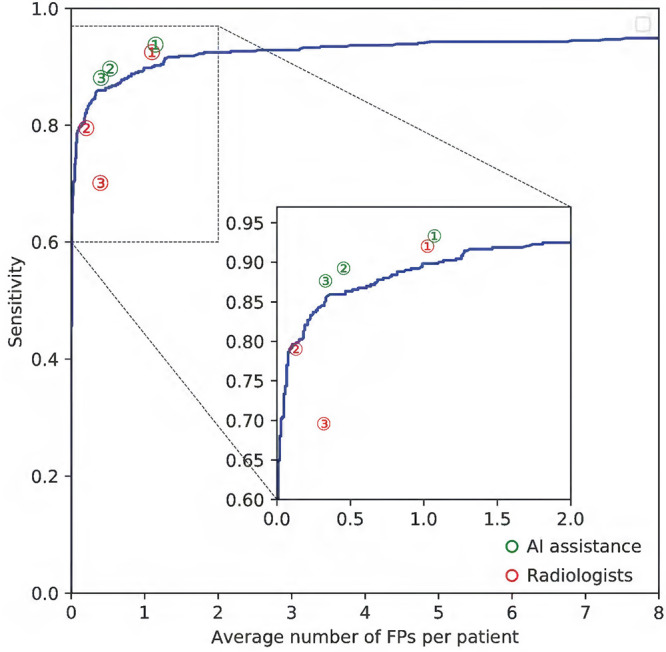
The model produced a total of 275 true-positive findings, 1138 false-positive findings, 6600 true-negative findings, and 38 false-negative findings on a per-examination level on test set 3, corresponding to a sensitivity of 87.9% (275 of 313 with positive findings for rib fractures; 95% CI: 83.7%, 91.4%) and specificity of 85.3% (6600 of 7738 control scans; 95% CI: 74.6%, 89.8%). As shown in Figure 4, the algorithm attained an AUC of 0.93 (95% CI: 0.91, 0.94) for lesion detection. However, the model had a low positive predictive value of 19.5% (275 of 1413) and a high negative predictive value of 99.4% (6600 of 6638) with the threshold of 0.5 on test set 3, which was due to the low prevalence (3.9%, 313 of 8051) of rib fracture in the clinical setting.
Figure 4:
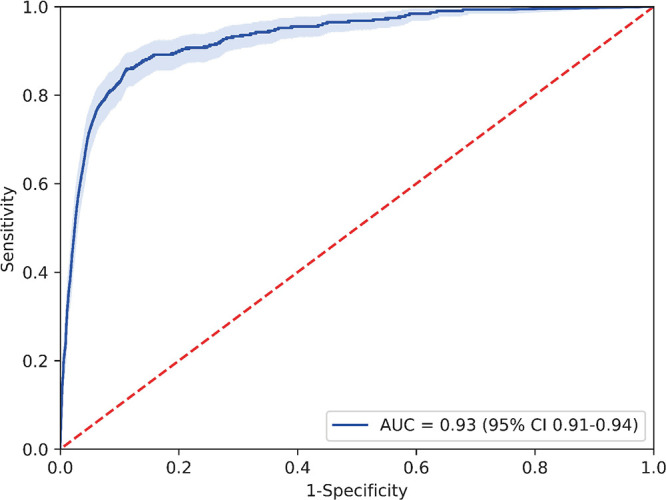
Receiver operating characteristic curve for test set 3. AUC = area under the receiver operating characteristic curve.
Performance of Rib Segmentation and Labeling
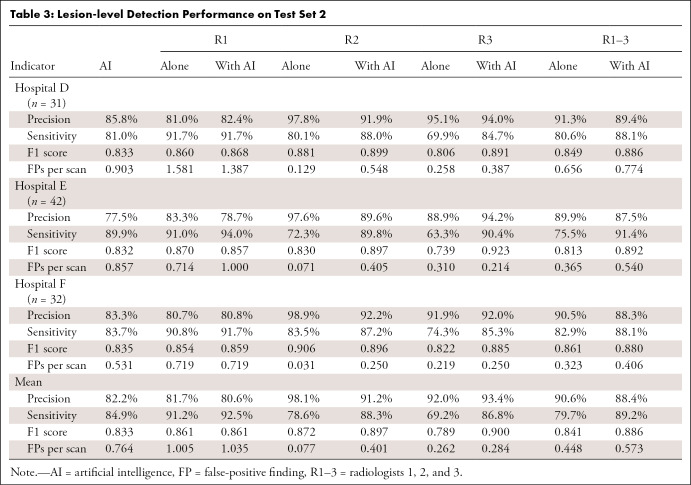
The segmentation performance of the 2D and 3D models was assessed next. The 2D Fast D-CNN achieved Dice score (0.850 ± 0.028) and IoU (0.740 ± 0.042) comparable to the 3D U-Net (0.867 ± 0.024 and 0.767 ± 0.037, respectively). The ensemble model, defined as the weighted average of probabilities predicted by the 2D Faster R-CNN network and the 3D U-Net network, achieved the highest Dice score (0.872 ± 0.024) and IoU (0.775 ± 0.038). We tested the ensemble rib mask model on a subset (n = 74) of test dataset 1 and obtained a mean Dice score of 0.827 ± 0.084 and a mean accuracy of 0.960 ± 0.094. Figure 5 shows an example of detection and labeling results from the model on CT images (two true-positive findings and one false-positive finding).
Figure 5:
Detection and segmentation examples shown on CT images. The first row images are sections cropped from the raw CT images, and the second row images are the corresponding results from the deep learning model, with colored outlines indicating the different ribs. The first two columns successfully detected rib fractures, while the last column is a false-positive finding.
Discussion
We developed a deep learning–based method for the automatic detection and location of rib fractures by using deep learning from a set of 2425 patients to develop our model, and the performance of the algorithm was assessed on a large set of 8518 patients. The results demonstrated that our model achieved a good performance in the detection of rib fractures on both lesion level and examination level, as verified by multicenter test sets. In addition, we compared the diagnosis accuracy between our model and three experienced radiologists, and the model had a comparable performance with radiologists.
Despite recent efforts to apply deep learning technology to diagnostic imaging, large datasets are still a necessity for models to achieve expert-level performance. Our training dataset consisted of 2545 chest CT data from six different hospitals, and all fractures were annotated by a radiologist with 10 years of diagnostic experience. To validate the performance and generalization of the model, we collected 8517 chest CT scans for testing, in which 467 cases were used for lesion-level assessment, and the other 8051 cases were for examination-level assessment. This large-scale training set and test set, combined with our preprocessing and network optimization technology, enabled our deep learning algorithm to be successfully developed. We compared the performance of the algorithm with three experienced radiologists and found that there were certain differences in the diagnostic performance among different radiologists. Radiologist 1 had higher sensitivity but lower specificity, whereas radiologists 2 and 3 achieved higher specificity but lower sensitivity. The sensitivities of three radiologists were all consistently higher with the use of AI assistance, which demonstrated that the algorithm could assist radiologists in the diagnosis of rib fracture and improve the diagnosis efficiency.
To our best knowledge, limited studies have been proposed for the detection of rib fractures on CT scans. Zhou et al (15) used the original Faster R-CNN model to detect the rib fractures and classify the fracture types. Weikert et al (16) first used a 3D convolutional deep neural network to provide proposals for suspected rib fractures, and then a Fast Region-based CNN was used to reduce the false-positive findings. Although the test data in the previously mentioned studies were from different centers, the datasets were relatively small to validate the performance and generalizability of the model. In addition, neither of the previously mentioned methods outputs the anatomic position of rib fractures, which also plays an important role in clinical diagnosis and treatment. In this study, we first designed a semantic mask head for the Faster R-CNN model so that the model can segment the ribs from the CT scans while detecting rib fractures. Then, we used a 3D U-Net model to further improve the performance of rib segmentation.
Our research study had several limitations. First of all, although our model can detect the rib fractures on chest CT scans and identify the anatomic location of each rib fracture, the types of rib fractures (fresh fractures, old fractures, and healing fractures) have not been specified. In the future, a 3D convolutional neural network can be used as a classification model to further classify the detected rib fracture types from the current model. In addition, the performance of the rib segmentation and labeling algorithm requires further improvement, especially for the scans in which the ribs are seriously misaligned due to rib fractures. We plan to increase the data annotations of such cases to help the model better segment and label the ribs. Finally, although the performances of the three radiologists with the assistance of AI were all improved, whether their diagnostic efficiency has been improved needs to be further investigated in the clinical environment.
In conclusion, we developed a deep learning algorithm to detect rib fractures on chest CT images, as well as to identify the location of each rib fracture. The model had good performance on the large-scale test datasets collected from multiple centers at both the lesion and examination level, which demonstrates generalization of the model. We found that radiologist sensitivity was higher with the use of the AI model, and the examination time was reduced. Together, these results warrant further investigation into the use of AI assistance for rib fracture detection.
M.W. and Z.C. contributed equally to this work.
Supported by the Shenzhen Science and Technology Program (grant JCYJ20180507182410327) and National Natural Science Foundation of China (grant 62072452).
Disclosures of Conflicts of Interest: M.W. disclosed no relevant relationships. Z.C. disclosed that author’s institution received grant from Shenzhen Science and Technology Program (No. JCYJ20180507182410327). G.Q. disclosed that author’s institution received grant from the National Natural Science Foundation of China (Project no. 62072452); this submission is a part of work under the grant. H.L. disclosed that author’s institution received grant from Key-Area Research and Development Program of Guangdong Province, China (2020B010165004), Shenzhen Science and Technology Program (No. JCYJ20180507182410327), and the National Natural Science Foundation of China (project no. 62072452). Q.W. disclosed that author’s institution received grant from NSFC. L.W. received consulting fee or honorarium from Xiamen University. H.C. disclosed no relevant relationships.
Abbreviations:
- AI
- artificial intelligence
- AUC
- area under the receiver operating characteristic curve
- FROC
- free-response receiver operating characteristic
- IoU
- intersection over union
- 3D
- three dimensional
- 2D
- two dimensional
References
- 1. Miller LA . Chest wall, lung, and pleural space trauma . Radiol Clin North Am 2006. ; 44 ( 2 ): 213 – 224, viii . [DOI] [PubMed] [Google Scholar]
- 2. Sirmali M , Türüt H , Topçu S , et al . A comprehensive analysis of traumatic rib fractures: morbidity, mortality and management . Eur J Cardiothorac Surg 2003. ; 24 ( 1 ): 133 – 138 . [DOI] [PubMed] [Google Scholar]
- 3. Cho SH , Sung YM , Kim MS . Missed rib fractures on evaluation of initial chest CT for trauma patients: pattern analysis and diagnostic value of coronal multiplanar reconstruction images with multidetector row CT . Br J Radiol 2012. ; 85 ( 1018 ): e845 – e850 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu G , Kim M , Wang Q , Munsell BC , Shen D . Scalable High-Performance Image Registration Framework by Unsupervised Deep Feature Representations Learning . IEEE Trans Biomed Eng 2016. ; 63 ( 7 ): 1505 – 1516 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nazib A , Fookes C , Perrin D . Towards Extreme-Resolution Image Registration with Deep Learning . In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) . IEEE; , 2019. : 512 – 516 . [Google Scholar]
- 6. Jiang H , Ma H , Qian W , et al . An automatic detection system of lung nodule based on multi-group patch-based deep learning network . IEEE J Biomed Health Inform 2018. ; 22 ( 4 ): 1227 – 1237 . [DOI] [PubMed] [Google Scholar]
- 7. Gu Y , Lu X , Yang L , et al . Automatic lung nodule detection using a 3D deep convolutional neural network combined with a multi-scale prediction strategy in chest CTs . Comput Biol Med 2018. ; 103 ( 220 ): 231 . [DOI] [PubMed] [Google Scholar]
- 8. Kuo W , Hӓne C , Mukherjee P , Malik J , Yuh EL . Expert-level detection of acute intracranial hemorrhage on head computed tomography using deep learning . Proc Natl Acad Sci U S A 2019. ; 116 ( 45 ): 22737 – 22745 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu F , Zhou Z , Samsonov A , et al . Deep learning approach for evaluating knee MR images: Achieving high diagnostic performance for cartilage lesion detection . Radiology 2018. ; 289 ( 1 ): 160 – 169 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang W , Li R , Deng H , et al . Deep convolutional neural networks for multi-modality isointense infant brain image segmentation . Neuroimage 2015. ; 108 ( 214 ): 224 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Norman B , Pedoia V , Majumdar S . Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry . Radiology 2018. ; 288 ( 1 ): 177 – 185 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Monteiro M , Newcombe VFJ , Mathieu F , et al . Multiclass semantic segmentation and quantification of traumatic brain injury lesions on head CT using deep learning: an algorithm development and multicentre validation study . Lancet Digit Health 2020. ; 2 ( 6 ): e314 – e322 . [DOI] [PubMed] [Google Scholar]
- 13. Suk HI , Wee CY , Lee SW , Shen D . State-space model with deep learning for functional dynamics estimation in resting-state fMRI . Neuroimage 2016. ; 129 ( 292 ): 307 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huseyn E . Deep learning method for early prognosis of Parkinson’s disease acuteness . Nat Sci 2020. ; 02 ( 03 ): 7 – 12 . [Google Scholar]
- 15. Zhou QQ , Wang J , Tang W , et al . Automatic detection and classification of rib fractures on thoracic CT using convolutional neural network: Accuracy and Feasibility . Korean J Radiol 2020. ; 21 ( 7 ): 869 – 879 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weikert T , Noordtzij LA , Bremerich J , et al . Assessment of a deep learning algorithm for the detection of rib fractures on whole-body trauma computed tomography . Korean J Radiol 2020. ; 21 ( 7 ): 891 – 899 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yushkevich PA , Piven J , Hazlett HC , et al . User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability . Neuroimage 2006. ; 31 ( 3 ): 1116 – 1128 . [DOI] [PubMed] [Google Scholar]
- 18. Girshick R , Donahue J , Darrell T , Malik J . Rich feature hierarchies for accurate object detection and semantic segmentation . In: 2014 IEEE conference on computer vision and pattern recognition, Columbus, OH, June 23–28, 2014 . Piscataway, NJ : IEEE , 2014. ; 580 – 587 . [Google Scholar]
- 19. Girshick R. Fast R-CNN . In: 2015 IEEE international conference on computer vision (ICCV), Santiago, Chile, December 7–13, 2015 . Piscataway, NJ : IEEE , 2015. ; 1440 – 1448 . [Google Scholar]
- 20. Ren S , He K , Girshick R , Sun J. Faster R-CNN: . Towards real-time object detection with region proposal networks . IEEE Trans Pattern Anal Mach Intell 2017. ; 39 ( 6 ): 1137 – 1149 . [DOI] [PubMed] [Google Scholar]
- 21. Redmon J , Divvala S , Girshick R , et al . You only look once: Unified, real-time object detection . In: Proceedings of the IEEE conference on computer vision and pattern recognition . IEEE; , 2016. ; 779 – 788 . [Google Scholar]
- 22. Redmon J , Farhadi A . YOLO9000: Better, Faster, Stronger . In: Proceedings of the IEEE conference on computer vision & pattern recognition, Conference . IEEE; , 2017. ; 7263 – 7271 . [Google Scholar]
- 23. Redmon J , Farhadi A . YOLOv3: An Incremental Improvement . arXiv:1804.02767 [preprint] https://arxiv.org/abs/1804.02767. Posted 2018. Accessed April 2019. [Google Scholar]
- 24. He K , Zhang X , Ren S , et al . Deep residual learning for image recognition . In: Proceedings of the IEEE conference on computer vision and pattern recognition . IEEE; , 2016. ; 770 – 778 . [Google Scholar]
- 25. Deng J , Dong W , Socher R , et al . ImageNet: A large-scale hierarchical image database . In: 2009 IEEE conference on computer vision and pattern recognition . IEEE; , 2009. ; 248 – 255 . [Google Scholar]
- 26. Lin TY , Dollár P , Girshick R , et al . Feature Pyramid Networks for Object Detection . 2016. . [Google Scholar]
- 27. Abadi M , Barham P , Chen J , et al . TensorFlow: A system for large-scale machine . 12th USENIX symposium on operating systems design and implementation (OSDI) 16 . TensorFlow: , 2016. ; 265 – 283 . [Google Scholar]
- 28. Ronneberger O , Fischer P , Brox T . U-net: Convolutional networks for biomedical image segmentation . In: International conference on medical image computing and computer-assisted intervention . Cham, Switzerland: : Springer; , 2015. ; 234 – 241 . [Google Scholar]
- 29. van der Walt S , Schönberger JL , Nunez-Iglesias J , et al . scikit-image: image processing in Python . PeerJ 2014. ; 2 e453 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chollet F. Keras . https://github.com/fchollet/keras Published 2015. Accessed December 2017.
- 31. DeLong ER , DeLong DM , Clarke-Pearson DL . Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach . Biometrics 1988. ; 44 ( 3 ): 837 – 845 . [PubMed] [Google Scholar]