Abstract
Propagation of the yeast protein-based non-Mendelian element [PSI], a prion-like form of the release factor Sup35, was shown to be regulated by the interplay between chaperone proteins Hsp104 and Hsp70. While overproduction of Hsp104 protein cures cells of [PSI], overproduction of the Ssa1 protein of the Hsp70 family protects [PSI] from the curing effect of Hsp104. Here we demonstrate that another protein of the Hsp70 family, Ssb, previously implicated in nascent polypeptide folding and protein turnover, exhibits effects on [PSI] which are opposite those of Ssa. Ssb overproduction increases, while Ssb depletion decreases, [PSI] curing by the overproduced Hsp104. Both spontaneous [PSI] formation and [PSI] induction by overproduction of the homologous or heterologous Sup35 protein are increased significantly in the strain lacking Ssb. This is the first example when inactivation of an unrelated cellular protein facilitates prion formation. Ssb is therefore playing a role in protein-based inheritance, which is analogous to the role played by the products of mutator genes in nucleic acid-based inheritance. Ssb depletion also decreases toxicity of the overproduced Sup35 and causes extreme sensitivity to the [PSI]-curing chemical agent guanidine hydrochloride. Our data demonstrate that various members of the yeast Hsp70 family have diverged from each other in regard to their roles in prion propagation and suggest that Ssb could serve as a proofreading component of the enzymatic system, which prevents formation of prion aggregates.
Prions are infectious proteins which are believed to reproduce by turning the normal protein into prion form. The PrPSc protein, a prion isoform of the mammalian prion protein PrP, is associated with neurodegenerative diseases such as sheep scrapie, bovine spongiform encephalopathy (mad cow disease), and human Creutzfeldt-Jacob disease (see reference 48 for a review). Characteristics of brain damage, observed in prion-infected patients, resemble those observed in patients with other types of neural inclusion diseases, such as Alzheimer’s and Huntington diseases, indicating similarities in the mechanisms of cellular toxicity between these disorders.
In yeast and fungi, prions manifest themselves as non-Mendelian elements inherited via the cytoplasm (58). Therefore, prion proteins serve as genetic material transmitting information about inherited traits (60). Examples of yeast and fungal prion-based genetic elements characterized to date include Saccharomyces cerevisiae [URE3] (1, 58) and [PSI] (15, 17, 58) and Podospora anserina [Het-S] (14). Among those, [PSI] element is of specific interest.
Yeast [PSI] is a prion-like polymerized derivative of the ribosome release factor Sup35 (eRF3) (see references 34 and 59 for reviews). The polymerized Sup35 protein (Sup35PSI) is defective in termination of translation, resulting in translational readthrough of termination codons (nonsense suppression). Properties of the Sup35PSI prion aggregates, observed in vivo (44, 45) and in vitro (27, 31, 46), resemble those of PrPSc. The N-terminal portion of the Sup35 protein (Sup35N), which is responsible for in vivo [PSI] formation (21) and propagation (23, 56) and for in vitro Sup35 polymerization (27), possesses similarities to both PrP (17, 33) and huntingtin (20). Considering that Sup35 is an evolutionarily conserved eukaryotic protein which is essential for such an important process as translation, it seems evident that understanding the specifics of [PSI] formation and propagation will shed light on the general mechanisms of protein-based infectivity and inheritance.
Considering prions as protein mutants, one could suggest the existence of cellular protein repair systems, which would normally prevent the appearance and propagation of such mutants. Chaperones, which are involved in protein folding and assembly/disassembly of the multiprotein complexes, are likely participants in such protein repair pathways. Thus far, the yeast Sup35/[PSI] system is the only experimental model providing direct evidence for the chaperone role in prion propagation. We have previously shown that [PSI] can be cured by either overproduction or inactivation of the chaperone protein Hsp104 (11). Hsp104 is implicated in disaggregating the heat-damaged proteins in vivo (43) and in vitro (26). Biochemical evidence confirms that excess Hsp104 leads to the solubilization of the Sup35PSI aggregates in vivo (44, 45). However, high temperature and some other stresses, which induce Hsp104 and other heat shock proteins, do not efficiently cure cells of [PSI] (18, 54). Further investigating this phenomenon, we have found out that overproduction of the Ssa1 protein, a stress-inducible member of the Hsp70 family, protects [PSI] from curing by excess Hsp104 and increases translational readthrough by [PSI] (39). This suggests that prions can use cellular stress defense systems for their own advantage. As in the case of the DNA repair machinery, the protein repair machinery can be error prone.
The yeast Hsp70 family includes several subfamilies, of which the Ssa and Ssb subfamilies are the most extensively investigated. The Ssb subfamily includes two essentially identical proteins, Ssb1 and Ssb2 (38), collectively designated Ssb protein. While the Ssb subfamily was initially observed only in yeast (3), recent data have uncovered the existence of Ssb homologs in genomes of other eukaryotes, such as the nematode Caenorhabditis elegans (13). In contrast to Ssa, Ssb protein is not required for viability and is not involved in response to high-temperature stress in yeast (38). Genetic and biochemical data suggest that Ssb is involved in cotranslational folding of the nascent polypeptide (38, 47) and in protein turnover (41). Ssa and Ssb proteins are not functionally interchangeable, although the molecular basis of their divergence remains unclear (29).
In this study, we demonstrate that in strong contrast to Ssa, the Ssb chaperone exhibits antagonistic effects on [PSI]. Formally speaking, the Ssb− derivatives manifest themselves as mutators in regard to the protein-based hereditary system. Therefore, two major subfamilies of the Hsp70 family, Ssa and Ssb, have functionally diverged from each other in regard to their roles in prion formation and propagation. Our data are in agreement with the existence of the proofreading system, which involves Ssb protein and is aimed at preventing formation of the prion aggregates.
MATERIALS AND METHODS
Yeast strains.
The S. cerevisiae strains used in this study are described in Table 1. The haploid [PSI+] strains OT55 and OT56, also called [PSI+]1-1-74-D694 and [PSI+]7-74-D694, respectively (21, 22, 39), and isogenic [psi− PIN+] strain OT60, also called [psi−]-74-D694 (11, 22), were described earlier. The haploid [PSI+] strains GT81-1C and GT81-1D were obtained by A. Galkin as a result of sporulating and dissecting the diploid strain GT81, constructed via HO-mediated self-homozygotization of the haploid strain GT56-34D (12). The [PSI+] strains GT128, containing the ssb1Δ::HIS3 disruption, and GT127, containing the double ssb1Δ::HIS3 ssb2Δ::URA3 disruption (ssb1/2Δ), are derivatives of strain GT81-1C, constructed as described below. Strain GT146 is a spontaneous Ura− derivative of strain GT127, selected on medium containing 5-fluoroorotic acid (30). The [psi− PIN+] derivatives of strains GT81-1C and GT146 were obtained by transforming these strains with plasmid pYS-GAL104 (see below), which bears the HSP104 gene under control of the GAL promoter, and curing [PSI] as a result of galactose-induced overproduction of Hsp104 as described earlier (11, 22). The resulting [psi−] derivatives were subsequently cured of plasmid pYS-GAL104. The [psi− pin−] derivatives of strains GT81-1C, GT127, and GT146 were obtained by curing cells of [PSI] and [PIN] after growth on YPD medium containing 5 mM guanidine hydrochloride (GuHCl) (22). The haploid strain GT84-5A is isogenic to GT81-1C except that it contains the hsp104Δ::URA3 disruption, which eliminates [PSI]. This strain was constructed by sporulating and dissecting diploid strain GT84, which is a GT81 derivative with one of the HSP104 copies disrupted via direct transplacement with the hsp104Δ::URA3 allele as described above (11). All strains contain the [PSI]-suppressible UGA mutation ade1-14, so that [PSI+] strains are white and Ade+, while [psi−] strains are red and Ade−, as described previously (11).
TABLE 1.
S. cerevisiae strains used
| Strain(s) | Genotype or description | Reference(s) |
|---|---|---|
| OT60 ([psi−]-74-D694) | MATa ade1-14 (UGA) his3 leu2 trp1-289 (UAG) ura3 [psi− PIN+] | 11, 22 |
| OT56 and OT55 | [PSI+] derivatives of OT60 | 21, 22, 39 |
| GT81-1C | MATa ade1-14 (UGA) his3 leu2 lys2 trp1Δ ura3 [PSI+] | 12 |
| GT81-1D | MATα [PSI+] strain isogenic to GT81-1C | 12 |
| GT128 | ssb1Δ::HIS3 derivative of GT81-1C | This study |
| GT127 | ssb2Δ::URA3 derivative of GT128 | This study |
| GT146 | Spontaneous Ura− derivative of GT127 | This study |
| GT159 | [psi− PIN+] derivative of GT81-1C | This study |
| GT157 | [psi− PIN+] derivative of GT146 | This study |
| GT117 | [psi− pin−] derivative of GT81-1C | |
| GT174 | [psi− pin−] derivatives of GT81-1C | This study |
| GT175 | [psi− pin−] derivative of GT146 | This study |
| GT139 | [psi− pin−] derivative of GT127 | This study |
| GT84-5A | hsp104Δ::URA3 strain isogenic to GT81-1D | This study |
Plasmids.
The centromeric URA3 plasmid pRS316 (53) and 2μm DNA-based multicopy plasmids YEp13, bearing the LEU2 marker (4), and pEMBL-yex, bearing the URA3 and LEU2-d markers (5), were described previously. Plasmid pRS316GAL is a pRS316 derivative bearing the galactose-inducible (GAL) promoter GAL1,10 (37). The centromeric plasmids pLA1, which contains the HIS3 marker and GAL promoter, and pH28, which is a pLA1 derivative bearing the HSP104 gene control of under the GAL promoter, were described previously (39). The centromeric plasmid pYS-GAL104 (11, 35) contains the HSP104 gene under control of the GAL promoter and the URA3 marker. The centromeric LEU2 plasmid pLH105, kindly provided by S. Lindquist, contains the HSP104 gene under control of the constitutively expressed GPD promoter. The previously described plasmids pEMBL-SUP35 (also called pEMBL-SUP2) and pEMBL-SUP35-ΔBal (also called pEMBL-SUP2-ΔBal) are pEMBL-yex derivatives which bear the complete S. cerevisiae SUP35 gene and the N-terminal 154 codons of the SUP35 gene, respectively, under control of the endogenous SUP35 promoter (55). The centromeric URA3 plasmid CEN-GAL-SUP35 (also called pVK71) contains the complete S. cerevisiae SUP35 gene under control of the GAL promoter (21). Plasmid pRS316GAL-SUP35Pm (12) is a pRS316GAL derivative which contains the Pichia methanolica SUP35 open reading frame (ORF) (32) placed under the control of the GAL promoter. The centromeric URA3 plasmid YCp-SSB1, which is a pRS316 derivative bearing the wild-type SSB1 gene under control of its endogenous promoter, was kindly provided by M. Ohba (41). Plasmids pRS316GAL-SSB1#1 and pRS316GAL-SSB1#2 were constructed by PCR amplifying the SSB1 ORF from the genome of S. cerevisiae S288C and placing it into pRS316GAL under control of the GAL promoter. Primers SSB1.EXT5 (5′-TACAGGATCCGTCCCAAGATCATTAC-AGTATT) and SSB1.EXT3NEW (5′-TACAGCGGCCGCCATATATATGTGATGAATGCAG), which are complementary to the 5′ flanking region (positions −35 to −14) and 3′ flanking region (positions +1845 to +1866) of the S. cerevisiae SSB1 ORF, respectively, were used in PCR with Pfu polymerase. These primers contain extensions bearing the restriction sites for BamHI and NotI, correspondingly. The PCR products were cut with BamHI and NotI and ligated into the BamHI-NotI cut pRS316GAL. To minimize the possible effect of PCR-generated errors, two independent PCRs were performed, each leading to one plasmid construct (1 or 2). Resulting plasmids pRS316GAL-SSB#1 and pRS316GAL-SSB#2 produced identical results in all experiments described below and are further designated pRS316GAL-SSB1. On galactose medium, either plasmid compensated for the ssb1/2Δ-associated phenotypes, such as paromomycin sensitivity (Pars) (38) and GuHCl sensitivity (Ghcs) (see below). Western blot results confirm that yeast strains bearing any of these plasmids exhibit increased levels of the Ssb protein on galactose medium (40). Plasmids pBS-HIS3-I and pBS-URA3-I, used in gene disruption experiments (see below), were constructed by inserting the 1.7-kb fragment bearing the complete HIS3 gene and the 1.1-kb fragment bearing the complete URA3 gene, respectively, into pBluescript KS II(+) polylinker. The URA3 centromeric plasmids pUKC815 and pUKC819, which were used for the quantitative measurement of UGA suppression, were kindly provided by M. F. Tuite. These plasmids contain the in-frame fusions PGK-lacZ and PGK-UGA-lacZ, respectively (25).
Gene disruptions.
The direct PCR-mediated transplacement protocol (2) was used to disrupt SSB1 and SSB2 genes. Primers SSB1-BLUE.PRO (5′-GATGT CCCAAGATCATTACAGTATTTTAATTGAACCTCACTATAGGGCGAA) and SSB1-BLUE.TER (5′-TAAGTAATATTCATATATATGTGATGAATGCAGTCCCTCACTAAAGGGAACAA) were designed in such a way that their 5′ flanking portions were complementary to the 5′ and 3′ flanks of the chromosomal SSB1 gene, respectively. The 3′ portions of the same primers were complementary to sequences of the pBluescript KS II(+) polylinker, located on the 5′ and 3′ sides, respectively, of the HIS3 insertion in plasmid pBS-HIS3-I. These primers were used to PCR amplify the HIS3 fragment from pBS-HIS3-I in a Taq polymerase-mediated reaction. The resulting HIS3 fragment, which contains the complete HIS3 gene flanked by extensions homologous to the flanks of the chromosomal copy of SSB1, was gel purified and transformed into strain GT81-1C. Transformants were selected on His-deficient (−His) medium. The ssb1Δ::HIS3 disruptants, in which the complete SSB1 ORF is deleted and replaced by the HIS3 gene in the same orientation, were verified by Southern hybridization as described below. One of these disruptants, GT128, was used in further transplacement experiments employing primers SSB2-BLUE.PRO (5′-TTTCAAGA AACCAAGAACCAATATCCTCATTAACACTCACTATAGGGCGAATT) and SSB2-BLUE.TER (5′-ATATATATGTGTATAACCTTAACCAGAATGACATCCCTCACTAAAGGGAACAA) to amplify the URA3 gene from plasmid pBS-URA3-I. The resulting URA3 fragment, flanked by extensions homologous to the 5′ and 3′ flanks of the chromosomal SSB2 gene in the same orientation, was gel purified and transformed into strain GT128. Transformants were selected on −Ura medium. One, containing a double ssb1Δ::HIS3 ssb2Δ::URA3 (ssb1/2Δ) disruption, was verified by Southern hybridization as described below. This ssb1/2Δ strain and its derivatives exhibited slow growth at low temperature (20 to 25°C) and sensitivity to the aminoglycoside antibiotics paromomycin and hygromycin, as described previously (38).
Media and growth conditions.
Standard yeast media, cultivation conditions, and standard procedures for yeast growth, sporulation, micromanipulation, and tetrad analysis were used (30). Gal medium contained 2% galactose instead of glucose. Gal+Raf medium contained 2% galactose and 2% raffinose instead of glucose. Transformation was performed according to a modified Li+ protocol (30). Yeast cultures were grown at 30°C unless specified otherwise. Liquid cultures were grown on a shaker, normally at 200 to 250 rpm, with a liquid/flask volume ratio of 1:5 or more.
[PSI] curing assays.
To assay for [PSI] curing by the overproduced Hsp104, yeast cultures bearing the GAL-HSP104 construct or matching controls were grown to 2 × 106 to 1 × 107 cells/ml in liquid synthetic glucose medium selective for the plasmid(s), washed twice with H2O, and inoculated into the corresponding synthetic Gal+Raf medium at the starting concentration of 2.5 × 105 to 5 × 105 cells/ml. Aliquots were taken before and after incubation (usually 22 to 24 h) and plated onto the solid synthetic glucose medium selective for the plasmid(s). The colonies grown after 4 to 5 days were velveteen replica plated onto YPD and −Ade medium. [PSI+] colonies were identified by white color on YPD and growth on −Ade medium after 4 to 6 days of incubation.
[PSI] induction assays.
The plate assays for [PSI] induction were performed as described previously (21). Yeast transformants bearing plasmid CEN-GAL-SUP35 or pRS316GAL-SUP35Pm, or matching control plasmid pRS316GAL, were grown on glucose −Ura medium and velveteen replica plated onto −Ura and −Ura/Gal media. After 3 to 4 days of incubation, each plate was velveteen replica plated onto glucose −Ade medium. [PSI] induction was detected as heterogeneous growth on −Ade medium after 7 to 10 days of incubation. To assay [PSI] induction quantitatively, transformants bearing the CEN-GAL-SUP35 plasmid were grown in the synthetic glucose −Ura medium, washed twice with H2O, and inoculated into the −Ura/Gal+Raf medium at the starting concentration 105 cells/ml. The aliquots were taken before incubation and after various periods of incubation and plated onto the glucose −Ura medium. The colonies grown after 4 to 5 days were velveteen replica plated onto the YPD and −Ade media. [PSI+] colonies were identified by white color on YPD medium and growth on −Ade medium after 6 to 8 days of incubation.
Quantitation of the spontaneous [PSI] formation.
To measure the frequencies of the spontaneous [PSI] formation, yeast strains were streaked out on YPD medium at 30°C to obtain single colonies originating from individual cells; 11 to 12 colonies were analyzed for each strain. The whole colony was resuspended in 200 μl of H2O; 10 μl of each solution was taken to perform serial dilutions in H2O, and the rest of each solution was plated onto −Ade medium. A 1/10 aliquot of the first (1/10) dilution was taken for further serial dilutions, while the rest of the 1/10 dilution was also plated on −Ade medium. Further serial dilutions were plated on YPD medium in order to determine the concentrations of viable cells (CFU). Ade+ colonies were counted on −Ade plates following 10 days of incubation at 30°C. Numbers of Ade+ colonies in undiluted cultures and 1/10 dilutions were roughly proportional to the degree of dilution, suggesting that residual growth on −Ade medium is minimal and has no significant effect on the frequency of Ade+ derivatives. For each sample, the numbers of Ade+ colonies detected in the undiluted culture and in 1/10 dilution were summed to determine the total number of Ade+ derivatives in the sample. Median frequencies of the Ade+ colonies were calculated for each strain and shown in Table 2. [PSI] formation rates were calculated according to the formula R = f/ln(NR), where R is rate of [PSI] formation, f is the observed frequency of [PSI+] colonies, and N is number of cells in the culture. This approach is based on the formula used previously for measuring spontaneous mutation rates (24).
TABLE 2.
Effect of Ssb on the rates of spontaneous [PSI] formation in [psi− PIN+] background
| Strain | Frequency of Ade+ colonies
|
Rate of Ade+ appearance
|
||
|---|---|---|---|---|
| Median | 95% confidence limit | Median | 95% confidence limit | |
| GT159 (Ssb+) | 1.7 × 10−6 | (1.0–3.5) × 10−6 | 7.3 × 10−7 | (4.0–11.0) × 10−7 |
| GT157 (Ssb−) | 3.1 × 10−5 | (1.5–3.5) × 10−5 | 6.4 × 10−6 | (3.3–7.2) × 10−6 |
DNA and protein analysis.
Standard protocols were used for DNA isolation, restriction digestion, ligation, and Escherichia coli transformation (49). Oligonucleotides were purchased from Gibco BRL. Restriction endonucleases were purchased from New England Biolabs and Gibco BRL. Polymerases were purchased from Gibco BRL, Promega, and Stratagene. The thermal cycler was from Ericomp. Southern hybridization was with a chemiluminescent probe as described in the Amersham protocol. The 1.9-kb BamHI-SacII fragment of plasmid pRS316GAL-SSB1, labeled by the random priming procedure, was used as a probe. It hybridizes to both SSB1 and SSB2 genes due to high homology between their ORFs. The chromosome DNA was cut with SacII to produce a 2.8-kb fragment corresponding to the SSB1 gene and large (>20-kb) fragment corresponding to the SSB2 gene. These bands were both absent in the double ssb1Δ::HIS3 ssb2Δ::URA3 disruptant. Additional digests with XbaI and with XbaI plus SacII were also analyzed to confirm disruptions. The blot was also probed with the labeled HSP104 gene used as a DNA loading control. Proteins were isolated as described previously (39) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Antibodies specific to the Hsp104, Hsp26, Hsp82, and Sup35 proteins were kindly provided by S. Lindquist. Antibodies specific to Ssa protein were kindly provided by E. Craig. Reactions with antibodies specific to Ssb protein were performed by M. Patino in S. Lindquist’s lab. Reactions with antibodies were detected by using the ECL (enhanced chemiluminescence) detection system according to the Amersham protocol. Relative amounts of proteins were determined by densitometry where necessary by using the Image Tool program as described previously (39). The amounts of proteins and times of exposure were kept within the linear range of the ECL detection system, determined on the basis of our previous data.
β-Galactosidase activity assays.
β-Galactosidase activity was measured by using the chemiluminescence assay as described previously (39). Expression of lacZ in the PGK-UGA-lacZ construct is possible only if the UGA nonsense codon is read through. Efficiency of nonsense suppression was determined as the ratio between β-galactosidase activity in the strain bearing the PGK-UGA-lacZ construct (pUKC819) and β-galactosidase activity in the isogenic strain bearing the in frame PGK-lacZ construct (pUKC815), which does not contain a nonsense codon between the PGK and lacZ genes. At least four independent transformants were measured for each strain-plasmid combination.
RESULTS
Ssb overproduction increases [PSI] curing by overproduced Hsp104.
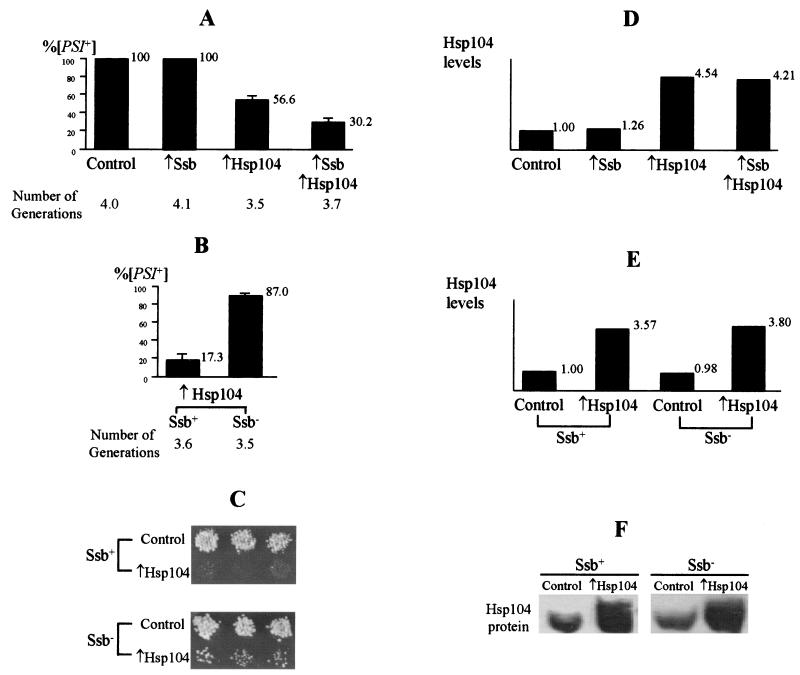
We have previously shown that transient overproduction of the Hsp104 chaperone causes loss of [PSI] (11), while simultaneous overproduction of the Ssa1 protein of the Hsp70 family protects [PSI] from the curing effect of the overproduced Hsp104 (39). To check whether Ssb1 protein, which also belongs to the Hsp70 family, affects [PSI], the [PSI+] yeast strains were transformed with plasmids bearing GAL-SSB1 and GAL-HSP104 constructs, or with matching control plasmids, in all possible combinations. Resulting transformants were incubated in Gal+Raf medium, selective for the plasmids, to induce the GAL-SSB1 and GAL-HSP104 constructs. Western blotting followed by densitometry assays (not shown) confirmed that in Gal+Raf medium, transformants bearing the GAL-SSB1 constructs contained 3.8-fold more Ssb protein than the isogenic transformants bearing the matching control plasmid. [PSI] curing was assayed as described in Materials and Methods. We have observed that overproduced Ssb1 protein does not affect [PSI] propagation by itself but increases the [PSI]-curing effect of overproduced Hsp104. This increase was detected in six repeats of the experiment with three different [PSI+] strains and was therefore statistically significant. Results of the representative experiment are shown in Fig. 1A. Neither growth parameters in Gal+Raf medium (Fig. 1A) nor levels of the Hsp104 protein (Fig. 1D) were affected by Ssb1 overproduction. Therefore, the effect of overproduced Ssb1 protein on [PSI] curing by Hsp104 is opposite that of overproduced Ssa1 protein.
FIG. 1.
Effects of Ssb protein on [PSI] curing by overproduced Hsp104. (A) Ssb overproduction increases [PSI] curing by overproduced Hsp104 in yeast strain OT56. Plasmid combinations: Control, pRS316GAL plus pLA1; ↑Ssb, pRS316GAL-SSB1 plus pLA1; ↑Hsp104, pRS316GAL plus pH28; ↑Ssb ↑Hsp104, pRS316GAL-SSB1 plus pH28. The isogenic weak [PSI+] strain OT55 (39) also was not cured of [PSI] by Ssb overproduction alone (not shown). [PSI] curing was measured after 22.5 h of induction in Gal+Raf liquid medium at 25°C as described in Material and Methods. Number of generations (G) was calculated according to the formula G = log2(C2/C1), where C2 and C1 are concentrations of CFU at the end and start of incubation in Gal+Raf medium, respectively. Only plasmid-containing colonies were counted. (B) Ssb depletion inhibits [PSI] curing by galactose-induced Hsp104. Strains were GT81-1C (Ssb+) and GT146 (Ssb−). The Hsp104-overproducing plasmid was pYS-GAL104. [PSI] curing was measured after 24 h of induction in Gal+Raf liquid medium at 30°C. Numbers of generations were calculated as described above. Differences between the samples, which express normal levels of Ssb and elevated levels of Hsp104, in panels A and B are apparently due to use of different [PSI+] strains. (C) Ssb depletion decreases [PSI] curing by constitutively overproduced Hsp104. Strains were GT81-1C (Ssb+) and GT127 (Ssb−). The Hsp104-overproducing plasmid was pLH105; the matching control plasmid was YEp13. Transformants were selected on −Leu medium and velveteen replica plated onto −Ade medium, which is not selective for the plasmid. Growth is indicative of [PSI+]. (D) Relative Hsp104 levels determined by Western blotting and densitometry after 22.5 h of induction in Gal+Raf medium at 25°C. Yeast cultures and strain and plasmid designations are the same as for panel A. Amounts of total protein loaded were normalized according to the total Sup35 protein levels (not shown). (E and F) Relative Hsp104 levels determined by densitometry (E) and photographs of the Western blots (F) after 24 h of induction in Gal+Raf medium at 30°C. Strains were GT81-1C (Ssb+) and GT146 (Ssb−). Plasmids: Control, pRS316GAL; ↑Hsp104, pYS-GAL104. Equal amounts of total protein were loaded, according to Coomassie blue staining (not shown).
[PSI] curing by overproduced Hsp104 is decreased in the Ssb− background.
To check whether the absence of Ssb protein would affect [PSI], we disrupted SSB1 and SSB2 genes in the [PSI+] strain GT81-1C as described in Materials and Methods. Resulting Ssb− disruptants remained [PSI+], and [PSI] remained stable in mitotic divisions. Moreover, [PSI] curing by both the GAL-HSP104 construct, induced in the Gal+Raf medium (Fig. 1B), and the constitutively expressed GPD-HSP104 construct (Fig. 1C) were less efficient in the Ssb− strain than in the isogenic Ssb+ strain. Neither parameters of growth in Gal+Raf medium (Fig. 1B) nor levels of the Hsp104 protein (Fig. 1E and F) were significantly affected, suggesting that the absence of Ssb influences [PSI] curing by Hsp104 rather than the number of generations or Hsp104 expression. This result confirms that Ssb protein assists in [PSI] curing by excess Hsp104 protein.
[PSI] curing by inactivated Hsp104 is not affected in the Ssb− background.
Inactivation of the Hsp104 protein has been shown to cure yeast cells of [PSI] (11). We checked whether [PSI] loss, caused by Hsp104 inactivation, requires Ssb protein. The [PSI+] ssb1/2Δ strain was crossed to the isogenic hsp104Δ strain GT84-5A. The resulting diploid was sporulated and dissected. Among 19 complete tetrads analyzed, we identified 2 which contained only two Ura+ spores each. In such tetrads, the ssb2Δ::URA3 and hsp104Δ::URA3 disruptions had to segregate together. The ssb1Δ::HIS3 disruption was monitored by the His+ phenotype, and the double ssb1/2Δ disruptants were also verified by the Pars phenotype. Using these approaches, we determined that three of four Ura+ spores recovered from these two tetrads contained all three disruptions simultaneously (ssb1Δ::HIS3 ssb2Δ::URA3 hsp104Δ::URA3). One of these spore clones was also tested by Southern hybridization to confirm the presence of all three disruptions. All three triple disruptant spores were [psi−], confirming that [PSI] curing by Hsp104 inactivation does not require the presence of Ssb protein.
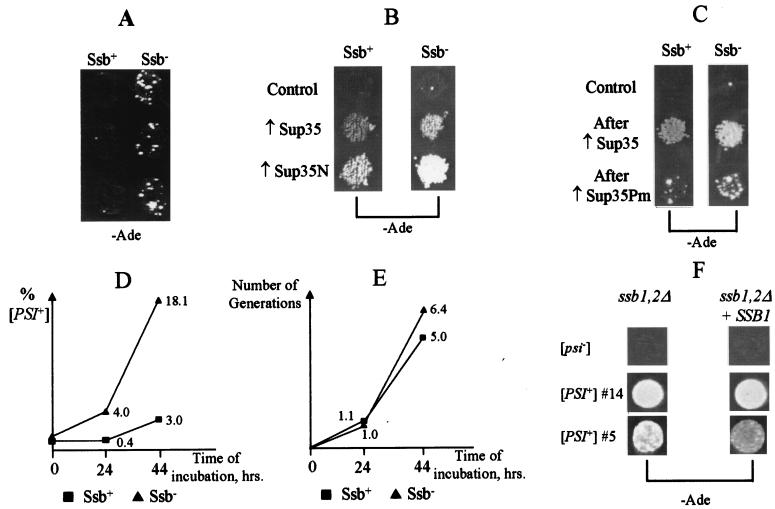
Spontaneous appearance of [PSI] is increased in the Ssb− background.
Spontaneous appearance of [PSI] is easier to detect in strains containing the non-Mendelian determinant [PIN], which increases the frequency of [PSI] formation (22). We observed that the [psi− PIN+] Ssb− strain exhibited 1-order-of-magnitude-higher frequency and rate of spontaneous Ade+ revertants compared to the isogenic [psi− PIN+] Ssb+ strain (Fig. 2A and Table 2). In all 12 independently arisen spontaneous Ssb− Ade+ colonies tested, the Ade+ phenotype was cured by growth in the presence of GuHCl, as described previously for the [PSI] element (57). This finding suggests that the increased frequency of the Ade+ revertants in the Ssb− strain is due to increased spontaneous appearance of the [PSI] prion.
FIG. 2.
Effects of Ssb depletion on [PSI] formation. All experiments were performed in the [psi− PIN+] yeast strains GT159 (Ssb+) and GT157 (Ssb−). (A) Double ssb1/2Δ deletion increases frequency of the spontaneous appearance of [PSI+] (Ade+) colonies. Several independent colonies of each strain were patched on YPD medium, grown, and velveteen replica plated onto −Ade medium. The −Ade plate was photographed after 10 days of incubation. (B) Double ssb1/2Δ deletion increases suppression of ade1-14 (UGA) by the overproduced Sup35 or Sup35N. Plasmids: Control, pEMBL-yex; ↑Sup35, pEMBL-SUP35; ↑Sup35N, pEMBL-SUP35-ΔBal. The −Ade plates were photographed after 6 days of incubation. (C) Double ssb1/2Δ deletion increases efficiency of [PSI] induction by the overproduced S. cerevisiae Sup35 protein or P. methanolica Sup35 (Sup35Pm) protein (qualitative assay). Plasmids: Control, pRS316; ↑Sup35, CEN-GAL-SUP35; ↑Sup35Pm, pRS316GAL-SUP35Pm. Transformants were incubated on −Ura/Gal medium and velveteen replica plated onto glucose −Ade medium. Plates were photographed after 8 days of incubation. Eleven independent Ade+ derivatives, induced by CEN-GAL-SUP35, were checked further. All maintained the Ade+ phenotype after the loss of plasmid and were GuHCl curable, which confirms the presence of [PSI]. (D) Double ssb1/2Δ deletion increases efficiency of [PSI] induction by the overproduced S. cerevisiae Sup35 protein (quantitative assay). Transformants bearing the CEN-GAL-SUP35 plasmid were grown in the −Ura/Gal+Raf medium, where expression of the GAL-SUP35 construct is induced. Frequencies of the [PSI+] cells in each culture were determined after various periods of time as described in Materials and Methods. The experiments were repeated with two independent transformants per strain, with similar results. The average numbers are shown. (E) Double ssb1/2Δ deletion has only slight effect on growth rate of the [psi− PIN+] strain, which overexpresses the Sup35 protein. Strains and plasmids are the same as for panel D. Numbers of generations in Gal+Raf medium were determined as described for Fig. 1A. Results for one representative transformant are shown. (F) Effects of Ssb protein on the ade1-14 (UGA) suppression mediated by [PSI]. The [PSI+] derivatives, obtained in the ssb1/2Δ [psi− PIN+] strain GT157, were transformed with either plasmid YCp-SSB1 (ssb1/2Δ + SSB1) or control matching plasmid pRS316 (ssb1/2Δ). The resulting transformants were patched on −Ura medium and velveteen replica plated onto −Ura-Ade medium. Plates were photographed after 5 days of incubation. A total of 14 independent [PSI+] derivatives were tested, and at least eight transformants were tested for each strain-plasmid combination. The representative examples are shown. See also comments in the text.
Nonsense suppression and [PSI] induction by the overproduced Sup35 protein are increased in the Ssb− background.
Overproduction of the Sup35 protein causes translational readthrough, or nonsense suppression (9), and induces formation of [PSI] prion (10) in [psi− PIN+] strains (22) of S. cerevisiae. Overproduction of the Sup35N prion-forming domain alone also induces nonsense suppression (55) and [PSI] formation (21) in both [psi− PIN+] and [psi− pin−] strains (22). We have observed that nonsense suppression by overproduced Sup35 or Sup35N in the [psi− PIN+] background (Fig. 2B) is greater in the Ssb− strain than in the isogenic Ssb+ strain. Nonsense suppression by overproduced Sup35N in the [psi− pin−] background was also increased in the Ssb− strain (not shown), indicating that the Ssb effect is not [PIN] specific. The frequency of [PSI] formation, induced by the transient Sup35 overproduction in the [psi− PIN+] background, was also increased 6- to 10-fold in the strain lacking Ssb protein (Fig. 2C and D). The growth rate of the [psi− PIN+] Sup35 overproducer was only slightly influenced by ssb1/2Δ (Fig. 2E), which was not sufficient to explain a difference in [PSI] induction rates.
[PSI] induction by the heterologous Sup35 protein is increased in the Ssb− background.
The sequence variation between the prion proteins of different origins decreases the efficiency of prion conversion in the heterologous system, which is recognized as a species barrier in prion transmission (for a review, see reference 48). We have shown that the highly divergent Sup35NM domain, originated from the distantly related yeast species Pichia methanolica (32), can form a prion in the S. cerevisiae cellular environment (12). Moreover, overproduction of Pichia Sup35 induces formation of the S. cerevisiae [PSI] prion; however, the efficiency of such heterologous prion induction is much lower than that of the homologous induction by overproduction of the endogenous S. cerevisiae Sup35 (12). Our data (Fig. 2C) indicate that the heterologous [PSI] induction by P. methanolica Sup35 is increased in the Ssb− background. Therefore, Ssb depletion makes Sup35 more sensitive to the heterologous prion-inducing agents and helps to overcome the species barrier.
Effects of the Ssb protein on the suppressor efficiency of [PSI].
One possible explanation for the increased [PSI] appearance in the Ssb− background could be that Ssb, which is a ribosome-associated protein (38, 47), antagonizes the translational readthrough, caused by [PSI], or prevents folding and/or facilitates destruction of the readthrough products, which likely contain the wrong amino acids inserted. In this case, the efficiency of nonsense suppression would be increased in the absence of Ssb. Therefore, some weak [PSI+] derivatives, which are not detectable in an Ssb+ background, could become detectable in an Ssb− background. Indeed, in some experiments the Ssb− derivative of the [PSI+] strain GT81-1C grew a little better than the original Ssb+ strain on −Ade medium, although this difference was difficult to reproduce (not shown). To check it quantitatively, we measured nonsense suppressor efficiencies by using the PGK-UGA-lacZ construct. Our results show that while the effect of ssb1/2Δ on UGA suppression in the [psi−] strain was insignificant, the level of UGA suppression by [PSI] was increased about fivefold in the Ssb− background (Table 3). To check whether such an effect could compromise the [PSI] formation assays, we transformed three independent spontaneous [PSI+] derivatives of the [psi− PIN+] ssb1/2Δ strain and 11 independent [PSI+] derivatives induced by Sup35 overproduction in the same strain with plasmid YCp-SSB1, bearing the wild-type SSB1 gene. Our results (e.g., Fig. 2F) confirm that all [PSI+] derivatives obtained in the absence of Ssb remain able to grow on −Ade medium in the presence of Ssb. Therefore, while Ssb protein appears to affect nonsense suppression by [PSI], results of the [PSI] formation assays cannot be explained by the Ssb effect on nonsense suppression. Moreover, reintroduction of SSB1 caused a detectable decrease in growth on −Ade medium only in 3 of 14 independent [PSI+] derivatives tested. We previously reported that various strains of the [PSI] prion could be recovered in the same genotypic background, and so differences between these strains are apparently controlled by the prion protein itself (21). Thus, the Ssb protein appears to have differential effects on the different prion strains.
TABLE 3.
Readthrough of the PGK-UGA-lacZ construct in the Ssb+ and Ssb− backgrounds
| Strain | β-Galactosidase (ng)/mg of total protein
|
Suppression (%) | |
|---|---|---|---|
| UGA (pUKC819) | Control (pUKC815) | ||
| GT174 ([psi−] Ssb+) | 0.5 | 448 | 0.11 |
| GT175 ([psi−] Ssb−) | 0.5 | 332 | 0.15 |
| GT81-1C ([PSI+] Ssb+) | 7.5 | 713 | 1.05 |
| GT146 ([PSI+] Ssb−) | 31.0 | 552 | 5.62 |
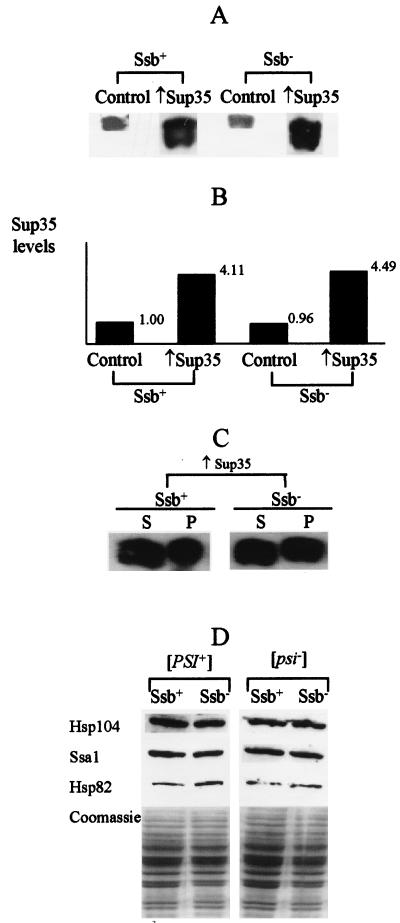
The Ssb depletion does not affect Sup35 or chaperone protein levels.
Effects of Ssb depletion on suppression and [PSI] formation are not readily explained by alterations of the Sup35 protein levels or its distribution between soluble and precipitated (that is, aggregated) fractions, since neither of these parameters appears to be affected in the [psi− PIN+] cells lacking Ssb (Fig. 3A to C). To determine whether ssb1/2Δ acts on [PSI] indirectly by altering other chaperone levels, we compared levels of several chaperone proteins in the isogenic Ssb+ and Ssb− strains. Our results confirm that levels of Hsp104 protein on glucose medium are unaffected by ssb1/2Δ (Fig. 3D), as also shown above for Gal+Raf medium (Fig. 1E and F). Levels of Ssa protein were also unaffected (Fig. 3D). There appeared to be a slight increase of Hsp82 levels in the Ssb− background (Fig. 3D); however, it has previously been shown that [PSI] is not affected by Hsp82 levels (39). Neither Ssb+ nor Ssb− cells produced Hsp26 protein in the exponential phase, independently of whether they were [PSI+] or [psi−] (not shown). These data confirm that Ssb depletion does not lead to a significant stress response, making indirect effects of ssb1/2Δ on [PSI] unlikely.
FIG. 3.
Levels of the Sup35 and heat shock proteins in the Ssb+ and Ssb− strains. (A and B) Double ssb1/2Δ deletion does not affect levels of the overproduced Sup35 protein. Plasmids: Control, pRS316GAL; ↑Sup35, CEN-GAL-SUP35. Total protein lysates were prepared from the cells grown for 44 h in −Ura/Gal+Raf medium, separated by SDS-PAGE, and Western blotted with the Sup35-specific antibodies. Western blot photograph (A) and relative Sup35 protein levels as determined by densitometry (B) are shown. Equal amounts of total protein were loaded, according to Coomassie blue staining (not shown). (C) Double ssb1/2Δ deletion does not affect distribution of the overproduced Sup35 protein between the soluble (supernatant [S]) and insoluble (pellet [P]) fractions. The protein extracts prepared from the CEN-GAL-SUP35 transformants after 44 h of incubation in −Ura/Gal+Raf medium, were fractionated by centrifugation as described previously (39). The S and P fractions were separated by SDS-PAGE and Western blotted with Sup35-specific antibodies. (D) Levels of heat shock proteins in Ssb+ and Ssb− strains. Strains [PSI+] Ssb+, GT81-1C; [PSI+] Ssb−, GT146; [psi−] Ssb+, GT174; [psi−] Ssb−, GT175. The protein extracts were subjected to SDS-PAGE and Western blotted with the antibodies specific to Hsp104, Ssa1, or Hsp82. Equal amounts of each sample, run on a gel stained with Coomassie blue, are shown for comparison.
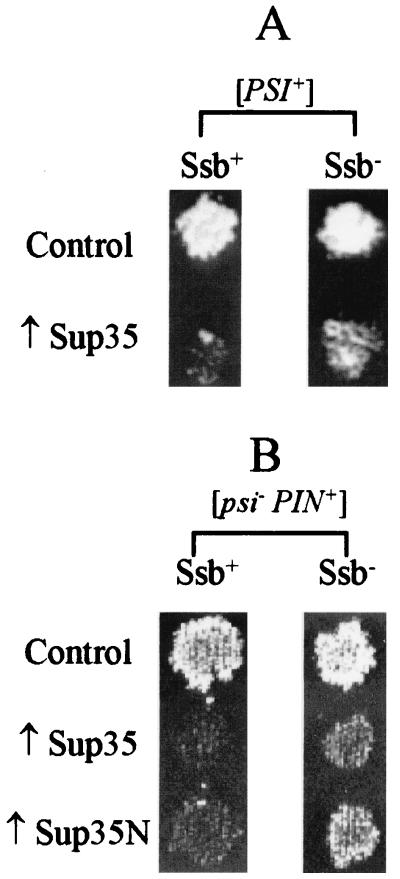
Ssb influences toxicity of the overproduced Sup35 protein.
Overproduced Sup35 protein (8, 9, 19) or Sup35N domain (55) inhibits growth of the [PSI+] strains, apparently due to increased accumulation of prion aggregates. Moreover, high-copy-number plasmids, coding for Sup35 or Sup35N, are also toxic for the [psi− PIN+] strains, and this toxicity requires the same sequence elements as are required for [PSI] induction, suggesting that toxicity could be related to prion formation (21, 22). We have checked the effect of the ssb1/2Δ on toxicity of overproduced Sup35 and Sup35N. Surprisingly, growth-inhibitory effects of the overproduced Sup35 in [PSI+] strains (Fig. 4A), as well as those of the hyperamplified SUP35 and SUP35N plasmids in the [psi− PIN+] strains (Fig. 4B), were both decreased rather than increased in an Ssb− background. Thus, Ssb effects on [PSI] formation and toxicity are opposite each other. One could suggest that the decrease in toxicity of the overproduced Sup35 in an Ssb− background contributes to the increase in [PSI] induction that we have observed. However, ssb1/2Δ influenced the growth of moderate Sup35 overproducers, used in [PSI] induction experiments, only slightly (Fig. 2E). Moreover, a 1-order-of-magnitude difference in percentage of [PSI+] cells was observed between the Ssb+ and Ssb− cultures that had undergone equal numbers of cell divisions (Fig. 2D and E). Besides, spontaneous [PSI] formation in the absence of Sup35 overproduction was also greater in the Ssb− strain than in the Ssb+ strain (Fig. 2A and Table 2), while growth rates of the [PSI+] strains which did not overproduce the Sup35 protein were not increased by ssb1/2Δ (Fig. 5B). This finding suggests that differences in toxicity of the overproduced Sup35 or Sup35N between Ssb+ and Ssb− strains are not sufficient to explain the difference in frequency of [PSI] formation.
FIG. 4.
Toxicity of overproduced Sup35 protein in Ssb+ and Ssb− strains. (A) The ssb1/2Δ deletion decreases toxicity of the overproduced Sup35 protein in the [PSI+] strain. Yeast strains: Ssb+, GT81-1C; Ssb−, GT146. Plasmids, Control, pRS316GAL; ↑Sup35, CEN-GAL-SUP35. The −Ura/Gal plates, on which the GAL-SUP35 construct is activated, were photographed after 7 days of incubation. At least eight independent transformants were tested for each strain-plasmid combination; representative examples are shown. No differences between the strains and plasmids were observed on glucose −Ura medium, where GAL-SUP35 is inactive. (B) The ssb1/2Δ deletion decreases toxicity of multicopy SUP35 and SUP35N plasmids in the [psi− PIN+] strain. Yeast strains: Ssb+, GT159; Ssb−, GT157. Plasmids: Control, pEMBL-yex; ↑Sup35, pEMBL-SUP35; ↑Sup35N, pEMBL-SUP35-ΔBal. Transformants were selected on −Ura medium and velveteen replica plated onto −Leu medium. Growth on −Leu requires plasmid amplification up to about 100 copies per cell (21). The −Leu plates were photographed after 2 days of incubation.
FIG. 5.
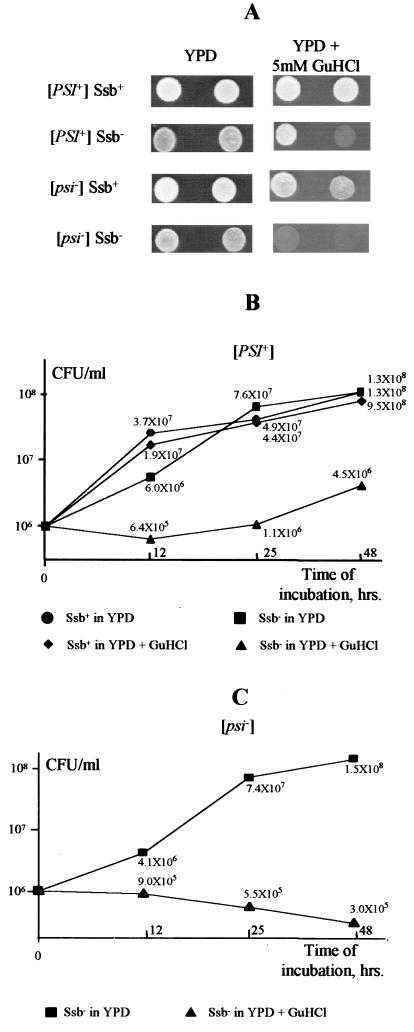
Sensitivity of Ssb− strains to GuHCl. (A) Qualitative assay. Yeast strains: [PSI+] Ssb+, GT81-1C; [PSI+] Ssb−, GT127, [psi−] Ssb+, GT117; [psi−] Ssb−, GT139. The [psi−] strains shown are [pin−]. However, similar results were obtained for the [psi− PIN+] strains GT159 and GT157 (not shown), indicating that levels of resistance to GuHCl are not affected by the presence or absence of the [PIN] factor. Yeast cells grown on YPD plates were diluted in sterile H2O at approximately 107 cells/ml. The original solution and the 10-fold dilution (from left to right) were plated onto YPD and YPD–5 mM GuHCl media. Plates were photographed after 1 day of incubation at 30°C. Differences remained after longer periods of incubation. (B and C) Quantitative assays. Yeast [PSI+] strains (B) were GT81-1C (Ssb+) and GT127 (Ssb−); the yeast [psi−] Ssb− strain (C) was GT139. Yeast cultures were grown in the liquid YPD medium with or without 5 mM GuHCl with agitation. After certain periods of time, aliquots of the cultures were taken, plated onto YPD medium, and grown for 4 to 5 days. Concentrations of CFU, which correspond to the viable cells, were determined.
Ssb depletion causes sensitivity to GuHCl.
Since Ssb protein assists in [PSI] curing by overproduced Hsp104, we checked whether Ssb would effect another [PSI]-curing agent, GuHCl. Surprisingly, we found that growth of Ssb− strains is inhibited by the millimolar concentrations of GuHCl that are normally used to cure [PSI] (Fig. 5). While GuHCl remained capable of curing the Ssb− strains of [PSI], it was not possible to compare efficiencies of curing in Ssb+ and Ssb− backgrounds due to differences in the growth rates. Interestingly, the Ssb− [psi−] strain was slightly more sensitive to GuHCl than the isogenic Ssb− [PSI+] strain (Fig. 5). Moreover, the concentration of the viable cells in Ssb− [psi−] culture was decreasing upon incubation in medium containing GuHCl (Fig. 5C), indicating that GuHCl does not simply inhibit growth but actually kills the Ssb− [psi−] cells. The Ghcs phenotype was complemented by the plasmid bearing the SSB1 gene (not shown) and cosegregated with Pars, a phenotypic marker of the double ssb1/2Δ disruption, in tetrad analysis (Table 4). ssb1/2Δ is the first mutation shown to cause sensitivity to millimolar concentrations of GuHCl. While we have observed that hsp104Δ also causes slight sensitivity to GuHCl in the GT81-1C genotypic background (6), the effect of ssb1/2Δ was much stronger than that of hsp104Δ.
TABLE 4.
Cosegregation of Pars and Ghcs in meiotic progeny of the diploid heterozygous by ssb1Δ and ssb2Δa
| Phenotype | No. of spores |
|---|---|
| Parr Ghcr (wild type) | 40 |
| Parr Ghcs | 0 |
| Pars Ghcr | 0 |
| Pars Ghcs | 14 |
| Total | 54 |
Two independent [PSI+] derivatives of GT146 were crossed to GT81-1D. Resulting diploids were sporulated and dissected. Cumulative values are shown.
DISCUSSION
Despite significant progress in understanding propagation of preexisting prions, the mechanism of initial prion formation remains a mystery. In vitro experiments suggest that prion-forming domains tend to aggregate spontaneously (27, 31). However, this aggregation potential is apparently suppressed in vivo, since in the absence of a prion seed, the potential prion-forming proteins remain soluble and frequency of spontaneous aggregate formation is low. This suggests that spontaneous prion formation could be caused by dysfunction or imbalance of proteins, which influence the normal process of folding and suppress the aggregation potential of the prion-forming domains. Here, we demonstrate for the first time that inactivation of the chaperone protein (Ssb) may increase the frequency of spontaneous prionization of another protein (Sup35). If prion conformers are considered analogous to the mutants, this is equivalent to discovery of the mutator genes.
Quite remarkably, representatives of two major cytosolic Hsp70 subfamilies, Ssa and Ssb, exhibit opposite effects on [PSI] prion. While Ssa protein assists [PSI] and protects it from curing by Hsp104, Ssb protein antagonizes [PSI] formation and assists Hsp104 in [PSI] curing. Functional divergence between Ssa and Ssb has been noted previously in other assays (29, 38). While the Ssa subfamily is essential for vegetative growth, inducible by high temperature and some other stresses, and involved in stress defense, Ssb protein is dispensable for viability and stress response and does not appear to be heat inducible. Genetic and biochemical data demonstrate an Ssb association with translating ribosomes (38), in particular with nascent polypeptide (47). It has been suggested that Ssb is involved in cotranslational protein folding (38). However, Ssb is certainly not required for this process, since its inactivation causes only slight growth deficiencies. Phenotypes associated with the double ssb1/2 deletion, i.e., cold sensitivity and sensitivity to translational inhibitors, do not contradict the role of Ssb in cotranslational folding but cannot be easily explained as a simple consequence of the protein folding defects.
Another cellular function attributed to the Ssb protein is stimulation of proteasome-dependent protein turnover (41). The prokaryotic Hsp70 homolog, DnaK, is also involved in protein degradation mediated by proteases La and ClpA/B (52). It is worth noting that ClpB, a subunit of the protease complex which converts multiprotein substrates to a form accessible for protease, is a prokaryotic homolog of the yeast Hsp104 protein (51). It has been suggested that yeast Ssb protein is responsible for proofreading of the newly synthesized and folded polypeptides and for targeting the incorrectly folded products for proteasome-mediated degradation (41). This does not rule out the possibility of Ssb being involved in the actual process of folding, since coupling of the processes of synthesis and breakdown is frequently observed in biological systems. The proofreading role for Ssb protein would be consistent with our data.
We propose that Ssb either prevents formation of the misfolded intermediates which serve as the raw material for the initial formation of prion aggregates, stimulates degradation of such intermediates, or both. This is why spontaneous [PSI] formation, and especially [PSI] formation induced by Sup35 (or Sup35N) overproduction is increased in Ssb− cells. Ssb could also assist Hsp104 in curing yeast cells of [PSI] by two mechanisms: (i) decreasing the amount of the misfolded newly synthesized Sup35 protein and therefore eliminating the substrate for generation of the new prion polymers and (ii) refolding the misfolded Sup35 molecules, generated due to Hsp104-mediated breakdown of preexisting prion polymers, or targeting these molecules for proteasome-mediated degradation and therefore preventing them from reverting to prion form. Since we detected no significant effect of Ssb levels on prion propagation under the normal circumstances (that is, when Hsp104 levels are low), the latter mechanism seems more likely. This would mean that the proofreading function of Ssb is not restricted to the newly synthesized proteins. However, one has to remember that Sup35 is a ribosome-binding protein, and thus at least a fraction of the Ssb and Sup35 molecules are located close to each other. This might enable Ssb to monitor conformation of some preexisting Sup35 molecules.
Other phenotypes of the double ssb1/2Δ deletion are also consistent with the proofreading function of the Ssb protein. Both translational inhibitors, such as paromomycin and hygromycin (42), and growth at low temperature (28) increase translational misreading, resulting in production of the erroneous and potentially misfolded proteins. If such proteins are not corrected or removed by the Ssb-mediated proofreading system, they may cause deleterious effects. Interestingly, we have observed for the first time that Ssb− strains are extremely sensitive to the protein-denaturing agent GuHCl. Growth in the presence of 1 to 5 mM GuHCl has been shown to efficiently cure yeast cells of prions [PSI] (18, 57) and [URE3] (58) and of the non-Mendelian element [PIN] (22), even though such low concentrations of GuHCl are not sufficient to cause significant protein denaturing and solubilize protein aggregates in vitro. GuHCl was shown to induce expression of some heat shock proteins including Hsp104 (36), leading to the hypothesis that the GuHCl effect is mediated by Hsp104 induction (11, 36). However, Hsp104 overproduction does not appear to cure yeast cells of [URE3] (7, 59) and [PIN] (22). Extreme sensitivity of the Ssb− strains to the millimolar concentrations of GuHCl indicates that low concentrations of GuHCl can specifically target cellular processes involving Ssb, in particular, cotranslational protein folding. It is possible that growth in the presence of GuHCl results in synthesis of the amorphous unfolded polypeptide products, which are unable both to perform their normal function and convert to the prion form. If these are not corrected or targeted for degradation due to Ssb action, they are toxic for the yeast cells. Dynamics of [PSI] loss in the presence of GuHCl confirm that GuHCl prevents formation of new prions rather than acts on preexisting ones (16).
Interestingly, toxicity of the overproduced Sup35 appears to decrease in the Ssb− background, even though translational readthrough and [PSI] induction increase. This finding suggests that growth defects caused by the overproduced Sup35 protein may not necessarily result directly from translational readthrough and aggregate formation. One possibility is that prion protein incorporated in the huge aggregates is harmless, so that aggregate formation might in fact protect cells by localizing and compartmentalizing prions. Proteins like Ssb, which interfere with prion formation and/or promote interactions between prion conformer and cellular metabolic systems (e.g., proteolysis machinery), may inadvertently increase toxicity, since wrongly shaped prion conformers or misfolded intermediates inhibit processes in which they become involved. Recent data indicate that in some human neural inclusion diseases, such as Huntington disease, aggregate formation may indeed protect cells by localizing and inactivating the misfolded protein rather than contribute to toxicity (50). This may also explain why the presence of [PSI] prion slightly increases resistance of the Ssb− strains to GuHCl (Fig. 5). This raises the possibility of yeast prions being by-products of the cellular processes aimed at protecting cells from the toxic effects of misfolded and mislocalized proteins. By analogy with mutagenic DNA repair systems, protein repair pathways, which are supposed to correct and/or remove damaged proteins, can generate protein “mutations” becoming reproducible in a prion-like fashion. Further experiments to test this hypothesis are under way.
Protein-based transmission of the phenotypic traits mediated by prion-like elements constitutes a new mechanism of inheritance. The very fact that several proteins of different functions and origins exhibit this phenomenon suggests a wide distribution of the protein-based systems of structural (as opposed to sequential) coding in nature. Our research uncovers the enzymatic machinery for protein-based inheritance, playing a role comparable to that of the DNA repair machinery in DNA-based inheritance. This machinery is composed of evolutionarily conserved proteins of the Hsp100 and Hsp70 families, suggesting that we are dealing with ancient phenomenon, probably having implications for organisms other than yeast.
ACKNOWLEDGMENTS
We thank A. Galkin for help with strain constructions, M. Patino for help with determining levels of the Ssb protein in the overproducer strains, R. Wegrzyn for help with some [PSI] curing assays, D. Gordenin for valuable comments on spontaneous rate measurements, and E. Craig, S. Lindquist, M. Ohba, and M. Tuite for plasmids and antibodies.
This work was supported in parts by grant R01GM58763 from the National Institute of General Medical Sciences and by grants from the Amyotrophic Lateral Sclerosis Association and the Emory-Georgia Tech Biomedical Technology Research Foundation to Y.O.C.
REFERENCES
- 1.Aigle M, Lacroute F. Genetical aspects of [URE3], a non-mitochondrial cytoplasmically inherited mutation in yeast. Mol Gen Genet. 1975;136:327–335. doi: 10.1007/BF00341717. [DOI] [PubMed] [Google Scholar]
- 2.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boorstein W R, Ziegelhoffer T, Craig E A. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- 4.Broach J R, Strathern J N, Hicks J B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979;8:121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- 5.Cesareni G, Murray A H. Plasmid vectors carrying the replication origin of filamentous single-stranded phages. In: Setlow J K, editor. Genetic engineering: principles and methods. Vol. 4. New York, N.Y: Plenum Press; 1987. pp. 13–154. [Google Scholar]
- 6.Chernoff, Y. O., and J. Kumar. Unpublished data.
- 7.Chernoff, Y. O., and S. W. Liebman. Unpublished data.
- 8.Chernoff Y O, Derkach I L, Tikhomirova V L, Dagkesamanskaya A R, Ter-Avanesyan M D, Inge-Vechtomov S G. Nonsense-suppression by amplification of translation protein factor gene. Dokl Akad Nauk. 1988;301:1227–1229. . (In Russian.) [PubMed] [Google Scholar]
- 9.Chernoff Y O, Inge-Vechtomov S G, Derkach I L, Ptyushkina M V, Tarunina O V, Dagkesamanskaya A R, Ter-Avanesyan M D. Dosage-dependent translational suppression in yeast Saccharomyces cerevisiae. Yeast. 1992;8:489–499. doi: 10.1002/yea.320080702. [DOI] [PubMed] [Google Scholar]
- 10.Chernoff Y O, Derkach I L, Inge-Vechtomov S G. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 11.Chernoff Y O, Lindquist S L, Ono B, Inge-Vechtomov S G, Liebman S W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 12.Chernoff, Y. O., A. P. Galkin, E. Lewitin, T. A. Chernova, G. P. Newnam, and S. M. Belenkiy. Submitted for publication. [DOI] [PubMed]
- 13.Chervitz S A, Aravind L, Sherlock G, Ball C A, Koonin E V, Dwight S S, Harris M A, Dolinski K, Mohr S, Smith T, Weng S, Cherry J M, Botstein D. Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science. 1998;282:2022–2028. doi: 10.1126/science.282.5396.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox B S. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- 16.Cox B S. Psi phenomena in yeast. In: Hall M N, Linder P, editors. The early days of yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 219–239. [Google Scholar]
- 17.Cox B S. Prion-like factors in yeast. Curr Biol. 1994;4:744–748. doi: 10.1016/s0960-9822(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 18.Cox B S, Tuite M F, McLaughlin C S. The PSI factor of yeast: a problem in inheritance. Yeast. 1988;4:159–178. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- 19.Dagkesamanskaya A R, Ter-Avanesyan M D. Interactions of the yeast omnipotent suppressors SUP1 (SUP45) and SUP2 (SUP35) with non-Mendelian factors. Genetics. 1991;128:513–520. doi: 10.1093/genetics/128.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Pace A H, Santoso A, Hillner P, Weissman J S. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 21.Derkatch I L, Chernoff Y O, Kushnirov V V, Inge-Vechtomov S G, Liebman S W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derkatch I L, Bradley M, Zhou P, Chernoff Y O, Liebman S W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doel S M, McCready S J, Nierras C R, Cox B S. The dominant PNM2-mutation which eliminates the PSI factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics. 1994;137:659–670. doi: 10.1093/genetics/137.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drake J W. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Firoozan M, Grant C, Duarte J A B, Tuite M F. Quantitation of readthrough of termination codons in yeast using a novel gene fusion assay. Yeast. 1991;7:173–183. doi: 10.1002/yea.320070211. [DOI] [PubMed] [Google Scholar]
- 26.Glover J R, Lindquist S. Hsp104, Hsp70 and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:1–20. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 27.Glover J R, Kowal A S, Schirmer E C, Patino M M, Liu J J, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of Saccharomyces cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 28.Inge-Vechtomov S G, Tikhodeev O N, Tikhomirova V L. Nonsense suppression in yeasts upon substitution of carbon sources and a decrease in temperature, mediated by nonchromosomal genetic determinants. Genetika. 1988;24:1159–1165. . (In Russian.) [Google Scholar]
- 29.James P, Pfund C, Craig E A. Functional specificity among Hsp 70 molecular chaperones. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 31.King C Y, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kushnirov V V, Ter-Avanesyan M D, Didichenko S A, Smirnov V N, Chernoff Y O, Derkach I L, Novikova O N, Inge-Vechtomov S G, Neistat M A, Tolstorukov I I. Divergence and conservation of SUP2 (SUP35) gene of yeasts Pichia pinus and Saccharomyces cerevisiae. Yeast. 1990;6:461–472. doi: 10.1002/yea.320060603. [DOI] [PubMed] [Google Scholar]
- 33.Kushnirov V V, Ter-Avanesyan M D, Smirnov V N. Structural and functional similarity of Sup35p and Ure2p yeast proteins and mammalian prions. Mol Biol. 1995;29:427–430. . (In Russian.) [PubMed] [Google Scholar]
- 34.Liebman S W, Derkatch I L. The yeast [PSI+] prion: making sense of nonsense. J Biol Chem. 1999;274:1181–1184. doi: 10.1074/jbc.274.3.1181. [DOI] [PubMed] [Google Scholar]
- 35.Lindquist S, Kim G. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc Natl Acad Sci USA. 1996;93:5301–5306. doi: 10.1073/pnas.93.11.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindquist S, Patino M M, Chernoff Y O, Kowal A S, Singer M A, Lee K-H, Blake T, Liebman S W. The role of Hsp104 in stress tolerance and [PSI+] propagation in Saccharomyces cerevisiae. Cold Spring Harbor Symp Quant Biol. 1995;60:451–460. doi: 10.1101/sqb.1995.060.01.050. [DOI] [PubMed] [Google Scholar]
- 37.Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson R J, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig E A. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 39.Newnam G P, Wegrzyn R D, Lindquist S L, Chernoff Y O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newnam, G. P., Y. O. Chernoff, and M. M. Patino. Unpublished data.
- 41.Ohba M. Modulation of intracellular protein degradation by SSB1-SIS1 chaperon system in yeast Saccharomyces cerevisiae. FEBS Lett. 1997;409:307–311. doi: 10.1016/s0014-5793(97)00535-8. [DOI] [PubMed] [Google Scholar]
- 42.Palmer E, Wilhem J, Sherman F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- 43.Parsell D A, Kowal A S, Singer M A, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 44.Patino M M, Liu J J, Glover J R, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 45.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 46.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. In vitro propagation of the prion-like state of yeast Sup35 protein. Science. 1997;277:381–383. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- 47.Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilike B A, Lopez-Buesa P, Walter W A, Wiedmann M, Craig E A. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component off the ribosome-nascent chain complex. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prusiner S B, Scott M R, De Armond S J, Cohen F E. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Saudou F, Finkbeiner S, Devys D, Greenberg M E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 51.Schirmer E C, Glover J R, Singer M A, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 52.Sherman M Y, Goldberg A L. Involvement of the chaperonine dnaK in the rapid degradation of a mutant protein in Escherichia coli. EMBO J. 1992;11:71–77. doi: 10.1002/j.1460-2075.1992.tb05029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh A, Helms C, Sherman F. Mutation of the non-mendelian suppressor, psi+, in yeast by hypertonic media. Proc Natl Acad Sci USA. 1979;76:1952–1956. doi: 10.1073/pnas.76.4.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ter-Avanesyan M D, Kushnirov V V, Dagkesamanskaya A R, Didichenko S A, Chernoff Y O, Inge-Vechtomov S G, Smirnov V N. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 56.Ter-Avanesyan M D, Dagkesamanskaya A R, Kushnirov V V, Smirnov V N. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuite M F, Mundy C R, Cox B S. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wickner R B. [URE3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 59.Wickner R B, Chernoff Y O. Prions of fungi: [URE3], [PSI] and [Het-s] discovered as heritable traits. In: Prusiner S B, editor. Prion biology and diseases. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 229–272. [Google Scholar]
- 60.Wickner R B, Edskes H K, Maddelein M L, Taylor K L, Moriyama H. Prions of yeast and fungi: proteins as genetic material. J Biol Chem. 1999;274:555–558. doi: 10.1074/jbc.274.2.555. [DOI] [PubMed] [Google Scholar]