Abstract
To investigate the transcriptional apparatus in wheat mitochondria, mitochondrial extracts were subjected to column chromatography and protein fractions were analyzed by in vitro transcription and mobility shift assays. Fractions eluting from DEAE-Sephacel between 0.2 and 0.3 M KCl displayed DNA-binding activity and supported specific transcription initiated from a wheat cox2 promoter. The active DEAE-Sephacel pool was further resolved by chromatography on phosphocellulose. Fractions that exhibited DNA-binding activity and that stimulated both specific and nonspecific transcription in vitro were highly enriched in a 63-kDa protein (p63). From peptide sequence obtained from purified p63, a cDNA encoding the protein was assembled. The predicted amino acid sequence (612 amino acid residues, 69 kDa) contains a basic N-terminal targeting sequence expected to direct transport of the protein into mitochondria. The p63 sequence also features an acidic domain characteristic of transcriptional activation factors, as well as sequence blocks displaying limited similarity to positionally equivalent regions in sigma factors from eubacteria related to mitochondria. Recombinant p63 possesses DNA-binding activity, exhibiting an affinity for the core cox2 promoter element and upstream regions in gel shift assays and having the ability to enhance specific transcription in vitro. Transcripts encoding p63 are expressed at an early stage in the germination of isolated wheat embryos, in a temporal pattern parallelling that of newly synthesized precursors of cox2, a mitochondrial gene. Taken together, these data suggest a role for p63 in transcription in wheat mitochondria.
Mitochondrial DNA (mtDNA) is extraordinarily diverse in size, gene content, and genome organization (24). In particular, the mitochondrial genomes of flowering plants (angiosperms) are the largest and structurally most complex mtDNAs known (30, 42, 60). Although the collection of completely sequenced mitochondrial genomes is expanding rapidly among the broad range of eukaryotes, including unicellular protists (26) and land plants (58), studies of the mode of mtDNA transcription have so far been limited to only a few species.
In the yeast Saccharomyces cerevisiae, transcription of the 80- to 85-kbp mtDNA initiates from at least 13 different promoters that comprise a highly conserved 9-bp motif (54). In this system, all transcripts appear to be composed of two or more coding sequences (57). Initiation of promoter-specific transcription requires two components: a single-polypeptide, T3/T7 bacteriophage-like RNA polymerase (45) and a 43-kDa specificity factor (mtTFB) sharing limited sequence similarity with eubacterial sigma factors (36). Both of these mitochondrial transcription components are encoded in the nuclear genome.
In humans, the highly compact 16-kbp mitochondrial genome is symmetrically transcribed from one heavy-strand and one light-strand promoter in the displacement (D) loop region (14); a protein factor (mtTFA) distinct from mtTFB and containing domains characteristic of high-mobility-group proteins is required for maximal levels of specific transcription (17, 18, 49). In Xenopus, mitochondrial genome organization and promoter placement are almost identical to the situation in human mitochondria (6); in this system, mtTFA is not required for basal transcription, but a 40-kDa protein is essential for initiation of specific transcription (2, 7). The gene encoding a human mitochondrial RNA polymerase has recently been cloned (56), but a human homolog of yeast specificity factor mtTFB remains to be identified.
In plant mitochondria, both core (YRTAT in monocots and CRTAAGAGA in dicots) and upstream sequences are required for optimal promoter-specific transcription (4, 5, 9, 21, 25, 50). The RNA polymerase interacting with these transcription elements has not yet been directly identified; however, the complete sequences of nucleus-encoded phage-like RNA polymerase genes (presumed to encode the mitochondrial RNA polymerase) have been determined for two dicot plants, Chenopodium album (59) and Arabidopsis thaliana (32), and two monocots, maize (13) and wheat (35). In the case of Arabidopsis, three different but related phage-type RNA polymerase genes have been reported (32, 52); in vitro import studies suggest that two of these genes encode a mitochondrial RNA polymerase whereas the third specifies a chloroplast RNA polymerase (8, 34). Similarly, one of the two maize gene products has been shown to be targeted to and imported into chloroplasts, whereas the other phage-like RNA polymerase is specifically localized to mitochondria (13).
Detailed biochemical studies of these RNA polymerases and of any specificity factor(s) necessary for promoter recognition in plant mitochondria have not yet been reported. Elsewhere we describe the characterization of two nucleus-encoded, phage-like RNA polymerase sequences from wheat and the identification of one of these sequences with an RNA polymerase isolated from wheat mitochondria (35). Here we report the isolation and functional analysis of a wheat mitochondrial DNA-binding protein. This protein (p63) displays affinity for a wheat mitochondrial cox2 promoter region and enhances specific transcription from this promoter in vitro, observations suggesting that p63 plays a role in transcription in wheat mitochondria.
MATERIALS AND METHODS
Purification of a DNA-binding protein from extracts of wheat mitochondria.
Preparation of a transcriptionally active protein fraction from wheat mitochondria was carried out as described previously (29). Mitochondria were isolated from germinating wheat embryos (24 h posthydration), purified on sucrose gradients, and lysed in the presence of 0.5% Triton X-100 and 1.05 M KCl. The supernatant from a 100,000 × g centrifugation was subjected to fractional precipitation with (NH4)2SO4, and proteins in the 20 to 50% (wt/vol) (NH4)2SO4 fraction were further resolved by a series of chromatographies on DEAE-Sephacel (DS) and phosphocellulose (PC). Mitochondrial proteins were first resolved by stepwise elution with KCl from DS. The active DS pool (0.2 M and 0.3 M KCl fractions) was then diluted with column buffer (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, 7.5% glycerol) to reduce the KCl concentration to ∼0.1 M and was then loaded onto a 5-ml column of PC that was equilibrated with column buffer containing 0.1 M KCl. After extensive washing of the column, proteins were eluted with a linear gradient of KCl (0.1 to 1.0 M) in 50 ml of column buffer at a flow rate at 15 ml/h.
For analysis of protein composition, protein fractions were precipitated with 80% acetone, dissolved in sample loading buffer (50 mM Tris-HCl [pH 6.8], 0.1 M DTT, 5% sodium dodecyl sulfate [SDS]), and heated at 65°C for 5 min. Samples were separated by electrophoresis in an SDS–12% polyacrylamide gel (38) and visualized by Silver Stain Plus (Bio-Rad). Transcription assays were carried out as described previously (29) in 20 μl of reaction mixture containing 10 mM Tris-HCl (pH 8), 10 mM MgCl2, 6.25 mM KCl, 1 mM DTT, 100 μg of bovine serum albumin per ml, 150 μM each CTP and GTP, 400 μM ATP, 5 μM unlabeled UTP, and 2.5 μCi of [α-32P]UTP (specific activity, 800 Ci/mmol). The transcription template was an EcoRI-restricted plasmid (pHJ2-15-9) containing a portion of the region upstream of the wheat mitochondrial cox2 gene (29).
Gel mobility shift assay.
DNA-binding activity in column-fractionated wheat mitochondrial extracts was detected in a gel mobility shift assay in which a ScaI-EcoRI fragment containing the cox2 core promoter sequence was 5′-end labeled with [α-32P]dATP, using Klenow fragment Escherichia coli DNA polymerase I. A competition assay employed DNA fragments prepared by PCR using Taq DNA polymerase with plasmids pHJ2-15-9 or pHJ2-15-9/M-10 as template, the latter containing an 8-bp deletion spanning the core promoter sequence. Fragments A and B encompassing the upstream region of wheat cox2 (positions −228 to −115 from the translation start site) were amplified with primer pair 5 plus 3 (Table 1), using pHJ2-15-9 and pHJ2-15-9/M-10 as templates, respectively. Fragment C (upstream of the core promoter sequence) was amplified with primer pair 5.1 plus 3.1 (Table 1), whereas fragment D (downstream of the core promoter) was amplified with primer pair 5.2 plus 3.2 (Table 1). PCR was performed in a buffer containing 16 mM (NH4)2SO4, 2 mM MgCl2, 25 mM Tricine (pH 8.5), 0.4 mM deoxynucleoside triphosphates, 1 U of Taq DNA polymerase (Gibco BRL), 10 pmol of each primer, and 5 ng of each template, using 30 cycles of denaturation (94°C, 30 s), annealing (55°C, 30 s), and extension (72°C, 20 s). PCR products were purified by polyacrylamide gel electrophoresis (PAGE), and fragment A was labeled with [γ-32P]ATP, using T4 polynucleotide kinase. For the mobility shift experiments, protein fractions were incubated in a reaction mix containing 10 mM Tris-HCl (pH 8.0), 60 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.1 μg of bovine serum albumin per ml, 12% glycerol, and 0.1 μg of poly(dI-dC). After 10 min of preincubation, approximately 10,000 cpm of labeled DNA was added to the mixture, which was then incubated at 25°C for 30 min. Samples were loaded directly onto a 4% polyacrylamide gel (prerun for 1.5 h at 150 V) and electrophoresed for 2 h at 200 V in 50 mM Tris-borate (pH 8.3)–2 mM EDTA. Gels were frozen at −75°C and subjected to autoradiography.
TABLE 1.
Oligonucleotide primers used in PCR amplification experiments
| Primer | Sequence (5′ to 3′) | Corresponding amino acid sequence |
|---|---|---|
| 1-1 | TCIaACICC(T/C)TCIGC(T/C)TGCAT | MQAEGVE |
| 1-2 | CA(G/A)GCIGA(G/A)GGIGTIGAACC | QAEGVEPD |
| 1-3 | A(G/A)(G/A)TCIGG(T/C)TCIACICC(T/C)TC | EGVEPDL |
| 1-4 | AAGCTCCTTGTGGACACCAA | |
| 1-5 | AGGGCAGCAGCTCGATATCT | |
| 1-9 | ATTGAAGACTTCCTCTGACT | |
| 1-11 | GTTCTGTATACAATCTCACC | |
| 1-13 | GGAGCTTCCAACATCACCT | |
| 1-14 | CGGGGATCCGCAAGCCTTGACGTA | |
| 1-15 | CTACTCGAGCATGTCAGATA | |
| 3 | ATTCTAGATGCCTCGTAACGC | |
| 3.1 | TTTTCTGAAGAGCAACAACG | |
| 3.2 | TTGACGTCGGCGTTGGTTTT | |
| 5 | ATGGTATCCAGGTTCACCTAT | |
| 5.1 | CCGACTAAGTAGTACTCACG | |
| 5.2 | GTAAGTAGTCTTCGTCGATG | |
| gt11F2 | ACTCCTGGAGCCCGTCAGTA | |
| gt11R | CAGACCAACTGGTAATGGTAGCG | |
| Anchor | ACTCCTGGAGCCCGTCAGTAGGGIIGGGIIGGGIIG |
I, inosine.
Assembly and cloning of a cDNA sequence encoding p63.
PC fractions containing p63 were resolved by SDS-PAGE, and the band corresponding to p63 was blotted onto a polyvinylidene difluoride membrane. The transferred protein was digested, internal peptides were isolated, and their amino acid sequences were determined by the Edman degradation method at the Harvard Microchemistry Facility. From the peptide sequences obtained by this procedure (VVESMQAEGVEPDLLFQATIAK and TLDGGNTFDRSDIFYVIMNLTK), degenerate primers were designed to obtain a cDNA encoding p63. All PCR products were cloned into pT7Blue T-vector (Novagen) and sequenced by means of Sequenase version 2 (Amersham).
Expression of recombinant p63.
To generate recombinant p63, a first-strand cDNA was synthesized by using primer 1-5, followed by PCR with primers 1-14 and 1-15 (Table 1). This strategy amplified a region extending from Ala at position 25 to the C-terminal end of the corresponding amino acid sequence, thereby excluding a putative mitochondrial targeting sequence. PCR was carried out with a mixture of Taq DNA polymerase (1 U) and Pfu DNA polymerase (0.01 U) to ensure accurate amplification (3). The cloned cDNA fragment was inserted into the pTrcHis vector (Invitrogen) and expressed with a histidine tag in E. coli DH5α. The expressed protein was denatured in a buffer containing 6 M guanidinium-HCl and was recovered from a nickel affinity column (TALON; Stratagene) with a buffer containing 8 M urea according to a protocol supplied by the manufacturer. Purified protein fractions were refolded by stepwise dialysis against a buffer containing 10 mM Tris-HCl, 1 mM EDTA (pH 8.0), and decreasing concentrations of urea. Renatured p63 was used for gel mobility shift and in vitro transcription assays.
Southern blot analysis.
Samples of wheat seedling total DNA (8 μg) were separately hydrolyzed with BamHI, DraI, EcoRI, or HindIII. Products were electrophoresed in a 0.8% agarose slab gel cast in TAE buffer (51), and DNA fragments were transferred to a nylon membrane [BIOTRANS(+); ICN] in 0.4 M NaOH. A p63 gene-specific sequence was amplified with primers 1-14 and 1-15 (Table 1), and amplification products were labeled by random hexamer-primed synthesis in the presence of [α-32P]dATP. Hybridization was carried out in a buffer containing 5× SSPE (0.8 M NaCl, 50 mM Na2HPO4, 5 mM EDTA [pH 8.3]), 2× Denhardt’s reagent (15), 0.1% SDS, 100 μg of denatured salmon sperm DNA per ml, and 50% formamide at 42°C (16). The membrane was rinsed with 2× SSPE–0.1% SDS for 30 min at room temperature and then washed with 0.1× SSPE–0.1% SDS for 30 min at 65°C.
Northern blot analysis.
Salt-insoluble RNA fractions (25 μg) isolated from embryos before (0 h) and after (1, 5, 15, and 25 h) imbibition were kindly provided by B. G. Lane (University of Toronto). These RNA samples were electrophoresed in a 1% agarose gel in 1× MOPS (morpholine-propanesulfonic acid) buffer containing 0.66 M formaldehyde and blotted onto a nylon membrane [BIOTRANS(+); ICN]. Amplification and labeling of a p63 probe and hybridization were carried out as above for Southern blot analysis. The membrane was rinsed with 2× SSPE–0.1% SDS for 20 min at room temperature and then washed with 0.2× SSPE–0.1% SDS for 30 min at 37°C and for 30 min at 42°C.
RESULTS
Purification of p63 from wheat mitochondria.
To identify a protein factor(s) that might be involved in promoter-specific transcription in wheat, we used a gel mobility shift assay to screen for DNA-binding activity in a fractionated wheat mitochondrial extract (Fig. 1). Such activity was confined to transcriptionally active fractions eluting from DS between 0.2 and 0.3 M KCl (data not shown). Distinct band shifts (1 and 2 in Fig. 2B and C) were produced with these DS fractions by using a labeled DNA fragment containing a cox2 promoter sequence (dashed line in Fig. 2A). Some variability in the fractionation procedure was observed, with specific transcription and DNA-binding activities mostly concentrated in the D3 fraction in some experiments but in D4 in others.
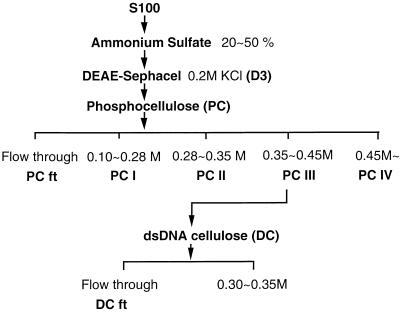
FIG. 1.
Flow chart depicting the fractionation of a wheat mitochondrial S100 extract (100,000 × g supernatant of a Triton X-100 lysate) by ammonium sulfate precipitation followed by successive chromatographies on DS, PC, and double-stranded DNA (dsDNA) cellulose. Details of the fractionation procedure are given in Materials and Methods.
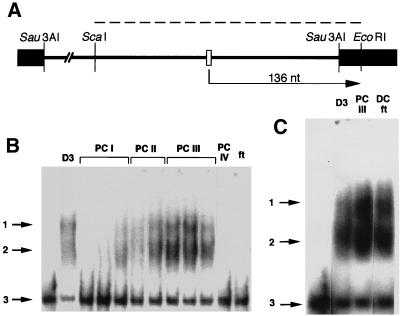
FIG. 2.
(A) Restriction map of plasmid template (pHJ2-15-9) spanning a promoter-containing region upstream of the wheat cox2 coding sequence (29). Plasmid was linearized by digestion with EcoRI and used for assay of runoff transcription in vitro (the expected 136-nucleotide [nt] runoff transcript is depicted, with the arrow indicating direction of transcription). The core promoter sequence of the wheat cox2 gene and portions of the pUC19 vector are denoted by open and solid rectangles, respectively. The dashed line indicates a DNA fragment generated by digestion with ScaI and EcoRI and used for the gel mobility shift assay. (B) DNA-binding activity exhibited by protein fractions isolated by chromatography on DS (D3, eluted with 0.2 M KCl) and PC. Salt concentrations at which the various PC fractions were eluted are given in Fig. 1. Arrows indicate putative dimer (band 1) and monomer (band 2) forms of the DNA-binding activity (see text) and the free radiolabeled ∼240-bp ScaI-EcoRI restriction fragment (band 3) used in the gel shift assay. (C) DNA-binding activity in pooled fractions isolated during chromatography on DS (D3), PC (PC III), and DNA cellulose (DC ft) (Fig. 1). Numbered arrows are as in panel B.
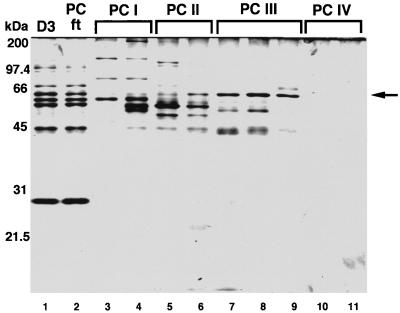
To isolate the DNA-binding protein(s), the D3 fraction was further resolved by chromatography on PC (Fig. 1). Analysis of material recovered after PC chromatography indicated that DNA-binding activity was mainly concentrated in fractions eluting between 0.35 and ∼0.45 M KCl (PC III in Fig. 2B). The most prominent protein in these PC fractions, with an estimated size of 63 kDa (Fig. 3), was designated p63. PC fractions enriched in p63 (PC II and PC III [Fig. 3, lanes 6 to 9]) stimulated both specific and nonspecific transcription in vitro when added to a transcriptionally active DS fraction (D4 in this case [Fig. 4, lanes 3 and 4]).
FIG. 3.
Profiles of wheat mitochondrial protein fractions isolated by chromatography on DS (D3) and PC and separated by SDS-PAGE in a 12% gel. Proteins were visualized by silver staining. Descriptions of protein fractions are given in the legends to Fig. 1 and 2. The position of p63 is indicated by the arrow.
FIG. 4.
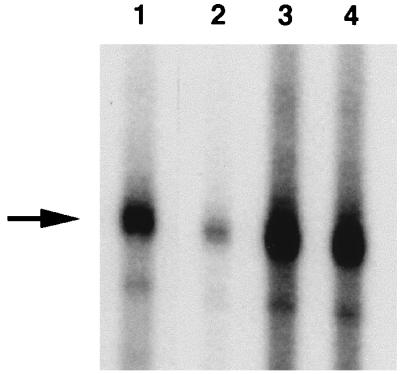
Effect of various protein fractions on in vitro transcription supported by DS fraction D3; 5 μl of each fraction was used. Lane 1, D3; lane 2, D3 plus PC I; lane 3, D3 plus PC II; lane 4, D3 plus PC III. Descriptions of protein fractions are provided in the legends to Fig. 1 and 2. The arrow indicates the position of the 136-nucleotide cox2 runoff transcript (Fig. 2A).
As is evident in Fig. 2, shifted bands were quite broad and appeared as doublets (1 and 2 in Fig. 2B and C) in these initial experiments. We attribute this pattern to a high ratio of extract to reaction mix: when a lesser amount of extract was used, only the more rapidly migrating band (band 2) appeared. The more slowly migrating band (band 3) is likely due to dimerization of the DNA-binding activity. Single, sharp bands were obtained in later mobility shift experiments using D3 and D4 (data not shown).
We attempted further resolution of the PC III fraction by chromatography on DNA cellulose; however, for reasons that are unclear, p63 appeared both in the flowthrough (DC ft [Fig. 1]) and in fractions eluting between 0.3 and ∼0.35 M KCl. The DC ft displayed DNA-binding activity that gave the same mobility shift as that observed with both D3 and PC III fractions (Fig. 2C); moreover, the DC ft stimulated both specific and nonspecific transcription in vitro when added to a transcriptionally competent DS fraction (data not shown). Because p63 was also the only protein detectable in the DC ft by silver staining (data not shown), we were encouraged to continue its characterization.
Cloning of a cDNA sequence encoding p63.
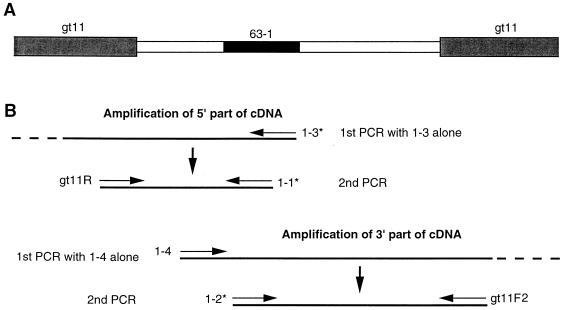
Initially, we were unable to amplify any p63 cDNA fragments from a cDNA library constructed by using mRNA isolated from germinating wheat embryos, suggesting that this library did not contain a sequence encompassing both of the peptide sequences on which PCR primer design was based. Therefore, we resorted to a heminested PCR approach, as depicted in Fig. 5. First, to amplify the 5′ portion of the cDNA from a wheat cDNA library, PCR was carried out with a single degenerate primer, 1-3 (Table 1), complementary to the predicted DNA sequence specifying the amino acid sequence EGVEPDL. A second round of PCR was carried out with degenerate primer 1-1 (Table 1), complementary to the predicted DNA sequence encoding the sequence MQAEGVE, plus primer gt11R (Table 1), which is specific for the 5′-flanking sequence at the cloning site of λgt11. The target site of primer 1-1 overlapped and was slightly N terminal to that of the first primer, 1-3. Using the sequence information obtained in this way, a further round of heminested PCR starting with a specific primer 1-4 (Table 1) was carried out to clone the 3′ end of the cDNA. A subsequent PCR used degenerate primer 1-2 (Table 1), the sequence of which is based on the predicted DNA sequence for the amino acid sequence QAEGVEPD, plus primer gt11F2 (Table 1), which is specific for 3′-flanking sequence at the cloning site of λgt11. With this method, we obtained about 1.4 kbp of cDNA sequence that contained a poly(A) tract at the 3′ end.
FIG. 5.
Schematic diagram illustrating the heminested PCR approach used to assemble a cDNA sequence encoding wheat mitochondrial p63. (A) Portions of the λgt11 vector (hatched boxes), cDNA insert (open boxes), and a region encoding peptide sequence 63-1 (filled box [Fig. 6]) are shown. (B) Construction of degenerate primers 1-1, 1-2, and 1-3 (indicated by ∗) was based on peptide sequence 63-1 (Fig. 6). λgt11-specific primers (gt11F2 and gt11R) were used for the second PCRs. A perfect-match primer (1-4) was constructed based on the sequence of the PCR product generated with primers 1-1 and gt11R. Additional details are given in the text; primer sequences are listed in Table 1.
To obtain the 5′ end of the cDNA, the 5′ RACE (rapid amplification of 5′ cDNA ends) method (20) was applied according to a Gibco BRL protocol. Total RNA from germinating embryos (24 h posthydration) was prepared as described by Goodall et al. (23), and poly(A)+ RNA was selected by using the PolyATtract mRNA isolation system (Promega). First-strand cDNA was synthesized by using primer 1-3 (Table 1) together with SuperScript II reverse transcriptase (Gibco BRL) at 46°C for 1 h. Homopolymeric dC tailing used terminal deoxynucleotidyltransferase at 37°C for 10 min. The first PCR used an anchor primer (Table 1), containing the gt11F2 sequence within its 5′ half, and primer 1-9 (Table 1). To obtain specific products, nested PCR was carried out with primer gt11F2 and primer 1-11 or 1-13 (Table 1).
Melding the 5′ (N-terminal) and 3′ (C-terminal) portions of the p63 coding sequence yielded a 2,063-bp cDNA that included 5′ and 3′ untranslated regions of 19 and 208 bp, respectively. The 3′ untranslated region contains a poly(A) tract (data not shown). The assembled open reading frame predicts a protein of 612 amino acids with an estimated molecular size of 69 kDa, somewhat larger than the size estimate (63 kDa) obtained by SDS-PAGE. At least part of this difference reflects the presence in the inferred protein sequence of a mitochondrial targeting sequence (see below) that would presumably be removed during import of the protein into mitochondria.
Characteristics of the predicted amino acid sequence.
The N-terminal end of the predicted amino acid sequence contains a domain (underlined in Fig. 6) that forms a basic amphipathic α helix typical of a targeting sequence required for transport of the protein into mitochondria (47). Also present is a domain (double underlined in Fig. 6) having the potential to form an acidic amphipathic α helix characteristic of transcriptional activation factors (22). Although the sequence downstream of this domain is also rich in acidic amino acids, this downstream region does not have the potential to form an amphipathic α helix.
FIG. 6.
Amino acid sequence of p63, deduced from the assembled cDNA sequence. The N-terminal region expected to form a basic amphipathic helix is underlined; a region with the potential to form an acidic amphipathic helix is double underlined; internal peptide sequences determined by direct protein microsequencing (63-1 and 63-2) are boxed. Acidic (−) and basic (+) amino acids are indicated. Sequencing of individual full-length PCR clones revealed polymorphic sites at amino acid positions 23 (S or G), 35 (A or T), 40 (Q or R), 42 (C or S), 61 (R or Q), 70 (T or M), 80 (G or S), 127 (L or S), 292 (G or R), 337 (V or I), 437 (E or K), 468 (E or K), and 590 (N or S) and between 603 and 604 (− or K) (34a).
Database searches using the BLAST program (1) revealed a large family of related plant proteins, most of which are between 400 and 700 amino acids long, displaying a highly significant level of sequence similarity to wheat mitochondrial p63. Table 2 lists those National Center for Biotechnology Information (NCBI) entries having a BLAST E value of less than 10−12. Most of these hits are to putative Arabidopsis protein sequences identified as a result of cDNA or genomic sequencing; in these cases, the function of the putative protein is unknown, although similarity to (membrane-associated) salt-inducible proteins is frequently noted. Based on PSORT analysis (47), most of the putative p63 homologs (like p63 itself) appear to be targeted to mitochondria, including the six NCBI entries displaying highest BLAST similarity to p63. Individual alignments of p63 with these plant protein sequences did not reveal any long stretches of identity or similarity; rather, shared amino acids or conservative substitutions are scattered uniformly throughout the sequences, with overall identities ranging for the most part from 20 to 25% over the compared regions. A notable exception is A. thaliana hypothetical protein gi:5103846, which shares 47% identity (including several blocks >10 amino acids long) and 66% similarity with p63 over a continuous stretch of 525 positions. The database search did not reveal similarity to any proteins involved in gene expression, with the exception of the product of the maize crp1 gene, a nuclear gene that has been reported to activate the translation and processing of specific chloroplast mRNAs (19). This gene is related to nuclear genes in fungi that play an analogous role in mitochondrial gene expression.
TABLE 2.
Amino acid similarity between wheat mitochondrial p63 and apparent homologs in public-domain databases
| Accession no. (GenBank identifier) | Organism | Protein description | Scorea | E valuea | Size (amino acids) | Localizationb |
|---|---|---|---|---|---|---|
| 5103846 | Arabidopsis thaliana | Hypothetical | 515 | e-145 | 623 | Mitochondria |
| 2462744 | A. thaliana | Hypothetical | 180 | 2e-44 | 491 | Mitochondria |
| 4454477 | A. thaliana | Hypothetical | 173 | 4e-42 | 490 | Mitochondria |
| 4263527 | A. thaliana | Hypothetical | 164 | 2e-39 | 532 | Mitochondria |
| 2317908 | A. thaliana | Unknown | 149 | 4e-35 | 524 | Mitochondria |
| 4558568 | A. thaliana | Hypothetical | 130 | 2e-29 | 501 | Mitochondria |
| 3107905 | Ipomoea nil | Leaf | 94 | 3e-18 | 665 | Cytoplasm |
| 3004555 | A. thaliana | Similar to salt inducible | 88 | 2e-16 | 822 | Cytoplasm |
| 3298551 | A. thaliana | Putative salt inducible | 87 | 4e-16 | 624 | Mitochondria |
| 3668091 | A. thaliana | Hypothetical | 86 | 7e-16 | 591 | Mitochondria |
| 3289002 | Zea mays | crp1 gene product | 83 | 4e-15 | 668 | Cytoplasmc |
| 4006835 | A. thaliana | Hypothetical | 80 | 4e-14 | 536 | Microbody |
| 2809247 | A. thaliana | F21B7.16 gene product | 80 | 6e-14 | 660 | Mitochondria |
| 2335106 | A. thaliana | Salt-inducible protein-like | 79 | 1e-13 | 810 | Chloroplast |
| 4539460 | A. thaliana | Putative | 78 | 1e-13 | 566 | Outside |
| 4584524 | A. thaliana | Putative | 78 | 2e-13 | 1,112 | Endoplasmic reticulum membrane |
| 4220445 | A. thaliana | Similar to salt inducible | 76 | 7e-13 | 650 | Mitochondria |
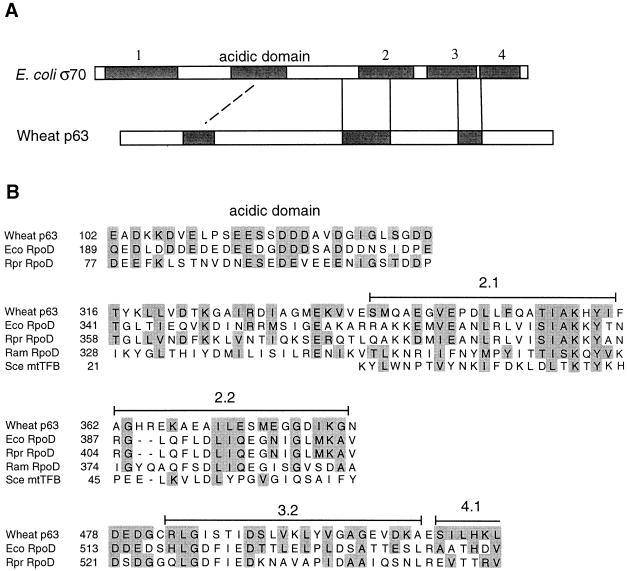
Because the biochemical data seemed to implicate p63 in transcription in wheat mitochondria, we searched for more limited amino acid sequence similarities to eubacterial sigma factors, using a local sequence alignment program, MACAW (multiple alignment construction and analysis workbench) (53). With the major sigma factor (RpoD) sequence of Rickettsia prowazekii, which belongs to the group of α-subdivision proteobacteria most closely related to the ancestral eubacterium-like endosymbiont that gave rise to mitochondria (24), and RpoD of E. coli, a member of the γ subdivision of proteobacteria, we could discern several sequence blocks sharing similarity with the p63 sequence (Fig. 7). One block contains regions 2.1 and 2.2, which are highly conserved among sigma factors. Region 2.1 may be involved in catalyzing DNA melting during open promoter complex formation (33, 43), and both regions 2.1 and 2.2 are important for interaction with the core polymerase (37, 41). In addition, the p63 sequence displays extended similarity to that of R. prowazekii RpoD upstream of region 2.1 (Fig. 7). Another block of similarity spans region 3.2, which is rich in acidic residues and is involved in core polymerase binding (61). We also detected similarity between RpoD proteins and p63 within the highly acidic region near the N terminus (Fig. 7). This region is not conserved among other sigma factors and may play a role in inhibiting interaction of sigma factors with the promoter sequence in the absence of core RNA polymerase (44). The sequence upstream of this acidic region in p63 has the potential to form an acidic amphipathic α helix, which could conceivably mediate interactions with protein components of the transcriptional machinery.
FIG. 7.
Amino acid sequence similarity between wheat mitochondrial p63 and various eubacterial and mitochondrial sigma factors (RpoD) and a mitochondrial transcription factor (mtTFB). (A) Schematic diagram showing the locations of blocks of sequence similarity (shaded) within E. coli ς70 (a eubacterial RpoD) and wheat p63. (B) Amino acid sequence alignments of similarity blocks that correspond positionally within wheat p63 and eubacterial (E. coli [Eco] and R. prowazekii [Rpr]) and mtDNA-encoded (R. americana [Ram]) RpoD proteins. Also included in the alignment is the yeast (S. cerevisiae [Sce]) mitochondrial transcription factor mtTFB. Similar amino acids (shaded) are defined as members of the following groups: I, L, M, V; H, K, R; D, E, N, Q; A, G; F, Y, W; S, T; P; and C. NCBI accession numbers are P07336 (E. coli RpoD), P33451 (R. prowazekii RpoD), AAD11909 (R. americana mitochondrial RpoD), P14908 (yeast mtTFB), and F091837 (wheat p63).
Characterization of a recombinant p63.
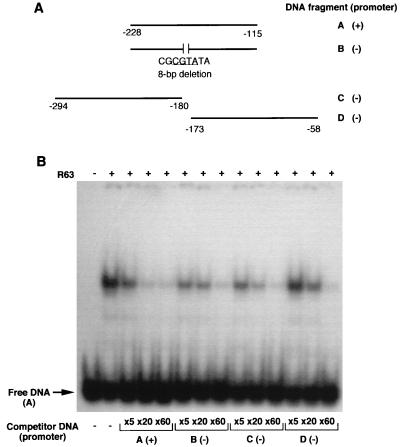
Recombinant p63 was expressed in E. coli from a portion of the complete cDNA extending from amino acid position 25 to the C-terminal end of the protein. The expressed protein formed insoluble inclusion bodies in E. coli, which were dissolved in 6 M guanidinium-HCl. Recombinant p63 was then refolded in the presence of decreasing concentrations of urea, and the renatured protein was subjected to gel mobility shift assay. In competition with labeled fragment A, unlabeled fragment A competed much more effectively than did other fragments that lack the promoter sequence (Fig. 8B). In the experiment shown here, a 20-fold molar excess of fragment A almost completely eliminated the band shift. On the other hand, a 60-fold molar excess of fragment B, which spans the same region but lacks the core promoter sequence, was required to achieve a comparable reduction in the intensity of the band shift. With fragment C, which encompasses a region upstream of the core promoter sequence, a 20-fold molar excess was sufficient to diminish the band shift significantly. The weakest competitor was fragment D: even a 20-fold excess of this fragment had a relatively small effect on the intensity of the shifted band (Fig. 8B). In a competition assay with transcriptionally active DS fractions, fragment D also did not compete as effectively as fragments A, B, and C (data not shown). This analysis suggests that recombinant p63 has higher binding affinity for DNA fragments containing the core promoter sequence and regions upstream of it than to fragments containing the downstream region.
FIG. 8.
Gel mobility shift competition assay with recombinant p63. (A) Diagram showing the scheme for generation of DNA fragments A to D. Numbers indicate positions upstream from the start of the cox2 coding sequence. The presence (+) or absence (−) of the core promoter sequence is indicated. The sequence of an 8-bp deletion is shown below fragment B, with the core promoter sequence underlined. (B) Autoradiogram showing gel shifts obtained with various DNA fragments in the presence of recombinant p63. Fragment A containing the core promoter sequence was labeled by using T4 polynucleotide kinase in the presence of [γ-32P]ATP; recombinant p63 (R63) was included in the reaction mixtures. After a 5-min preincubation, competitor DNAs were added at three different molar excesses (5-, 20-, and 60-fold [×5, ×20, and ×60]) and incubated for 30 min at room temperature. Radiolabeled protein-DNA complexes were separated from uncomplexed DNA fragments by electrophoresis in a 4% native polyacrylamide gel.
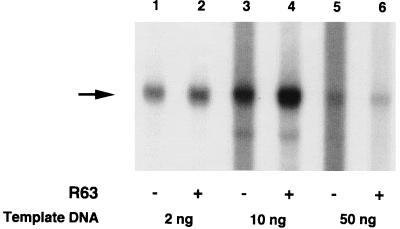
To study the effect of the recombinant p63 in an in vitro transcription assay, the protein was mixed with a transcriptionally active DS fraction (D4). In contrast to the effect of crude, native p63 fractions, the recombinant protein not only enhanced specific transcription but also suppressed nonspecific transcription in vitro (Fig. 9; compare lanes 1 and 2 and lanes 3 and 4). This enhancement (approximately fourfold, based on measurement of unsaturated autoradiograph signals) was observed with amounts of the plasmid template ranging from 2 to 10 ng in the reaction mixture. With excess template (50 ng), the level of specific transcription was substantially reduced (Fig. 9, lanes 5 and 6).
FIG. 9.
Enhancement of in vitro transcription by recombinant p63, using a transcriptionally active DS fraction. About 0.05 μg (in 5 μl) recombinant p63 (R63) was added (+) or not added (−) to 5 μl of D4 (the fraction eluting at 0.3 M KCl during DS chromatography). Three different amounts (2, 10, and 50 ng) of template DNA (EcoRI digest of pHJ2-15-9) were used.
Genomic distribution of the p63 coding sequence.
Southern blot analysis suggests that the wheat p63 gene is present in at least three distinct copies in wheat nuclear DNA, judging from the three equally intense bands seen in EcoRI, DraI, and particularly HindIII digests of wheat DNA (data supplied to reviewers but not shown). A single, very intense band seen in the EcoRI digest suggests that these three copies share an identical EcoRI fragment of about 2.2 kbp that encompasses the internal region represented in the p63 gene probe. Sequence analysis of individual full-length clones obtained by PCR has provided evidence of at least two distinct p63 sequences, based on covariation of polymorphic amino acid positions (34a) (Fig. 6).
Expression of the p63 gene in wheat embryos during germination.
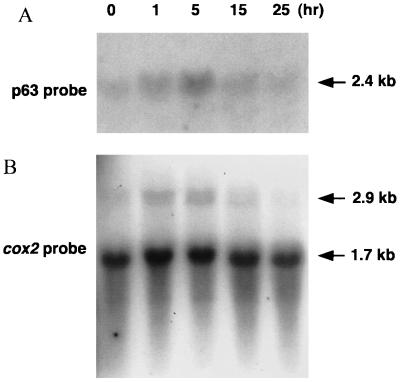
To assess expression of the gene encoding p63 in the course of wheat embryo germination, Northern blot analysis was carried out with RNA (a 2.5 M NaCl-insoluble fraction of total RNA [28]) isolated from embryos before (0 h) and after (1, 5, 15 and 25 h) the start of imbibition. Although the amount of p63 transcript is very low in dry embryos, transcript levels increase after imbibition, showing an apparent peak at around 5 h posthydration (Fig. 10A). Parallel experiments carried out with a mtDNA-specific gene probe (cox2) showed a corresponding temporal accumulation of cox2 intron-containing transcripts (i.e., newly synthesized, unspliced cox2 mRNAs) during the same time period (Fig. 10B). Because some RNA degradation is evident in the 15- and 25-h samples upon ethidium bromide staining, and because the level of mature cox2 transcript (which serves as an internal control) is slightly lower in these two samples than in the 0-, 1-, and 5-h preparations, the observed peak at 5 h may be less pronounced than it appears. Nevertheless, it is clear that p63 gene expression is activated shortly after imbibition of wheat embryos, in concert with renewed mitochondrial transcription.
FIG. 10.
Northern blot analysis of nucleus-encoded p63 (A) and mitochondrially encoded cox2 (B) transcripts. Salt-insoluble (2.5 M NaCl) RNA fractions (25 μg) isolated from embryos before (0 h) and after (1, 5, 15, and 25 h) imbibition were electrophoresed in a 1% agarose gel in 1× MOPS buffer containing 0.66 M formaldehyde; separated RNAs were blotted onto nylon membrane. As probe for p63 transcripts, a portion of the 63-kDa cDNA was amplified with primers 1-14 and 1-15 (Table 1), and the product was labeled with [α-32P]dATP in the presence of random hexamers. The cox2 probe contained the first exon of the wheat cox2 gene plus flanking 5′ untranslated and 3′ intron sequences. Transcript sizes in the lower panel indicate intron-containing (unspliced) cox2 pre-mRNA (2.9 kb) and mature cox2 mRNA (1.7 kb).
DISCUSSION
We isolated a 63-kDa protein (p63) from wheat mitochondria on the basis of its ability to bind DNA and enhance transcription in an in vitro assay. In combination with 5′ RACE and heminested PCR, we assembled about 2.1 kbp of cDNA sequence encoding p63. Northern blot analysis revealed a transcript of about 2.4 kb (Fig. 10A), and so we expect that the 5′ untranslated region extends beyond the sequence that we obtained. Southern blot analysis suggests that the p63 gene is present in at least three copies in the wheat nuclear genome, whereas analysis of individual full-length PCR clones provided evidence of at least two major p63 sequence variants (data not shown). Considering the hexaploid nature of the wheat genome, the p63 coding sequence could be present as a single-copy gene in each of its constituent genomes (A, B, and D). The predicted amino acid sequence of p63 contains a typical N-terminal signal peptide required for transport of the protein into mitochondria; in fact, a computer-assisted analysis (47) of this sequence clearly predicts that p63 should be targeted to mitochondria. Recombinant p63 expressed from the cDNA clone possesses DNA-binding activity, displaying weak affinity for the core promoter of the wheat cox2 gene and upstream regions (in maize, a close relative of wheat, a core promoter sequence and an upstream region both contribute to optimal transcriptional activity [9, 50]). These results suggest that recombinant p63 binds to a region of DNA directing specific transcription. However, although we conducted footprinting (DNase I protection) and exonuclease III protection studies to define a precise protein-binding DNA sequence in the vicinity of the cox2 promoter, we were unable to detect any specific region protected either by active DS fractions or by recombinant p63.
We did find a difference between native and recombinant p63 proteins in their effect on in vitro transcription: whereas fractions containing native p63 stimulated both nonspecific and specific transcription, recombinant p63 appeared to enhance specific transcription and suppress nonspecific transcription. Fractions containing partially purified, native p63 may well contain other factors involved in or influencing transcription in the in vitro assay. Because all of the transcriptionally active DS fractions that we tested supported specific as well as nonspecific transcription in vitro, it is not clear at this point whether p63 functions directly as a specificity factor that is essential for basal transcription or as an activator that enhances transcription but is not itself required for basal function. Preparation of a core RNA polymerase fraction devoid of transcription factors will be necessary before we can distinguish between these two possibilities.
Transcripts encoding p63 are present at a very low level in dry wheat embryos but accumulate to readily detectable levels by 5 h after the start of imbibition (Fig. 10A). Unprocessed cox2 transcripts (synthesized de novo) also appear in detectable amounts between 1 to 5 h following embryo imbibition (Fig. 10B); thus, the expression of nucleus-encoded p63 correlates temporally with de novo transcription of a mitochondrial gene during germination. This lends credence to the idea that p63 may be involved in the regulation of transcription of mitochondrial genes, as is mtTFA in vertebrate mitochondria (40, 46). Because we used isolated embryos lacking endosperm and seed coat for this analysis, the apparent decrease in transcript levels after 5 h of imbibition may not represent the actual situation in intact wheat seeds during germination. Further analysis will be necessary to clarify the relationship between the expression of p63 and wheat mitochondrial biogenesis.
In searches of public-domain protein databases, we could not detect any similarities between p63 and proteins known to be involved in transcription in mitochondria, including yeast mtTFB and human mtTFA. In view of evidence supporting the eubacterial origin of mitochondria (24) and reports of a limited similarity of yeast mtTFB to eubacterial sigma factors (36), we considered the possibility that p63, characterized here as a candidate transcription factor, might be distantly related to eubacterial sigma factors. Indeed, the local sequence similarities deduced between the wheat mitochondrial p63 sequence and eubacterial RpoD sequences (Fig. 7), coupled with the positional correspondence of these regions within the compared proteins, support this possibility. On the other hand, we could not discern clear similarities in other regions conserved among different sigma factors, including the helix-turn-helix DNA-binding motif in regions 3 and 4.2 (33).
Recently, Lang et al. (39) reported that in the protist Reclinomonas americana, the mitochondrial genome carries a set of genes (rpoA to rpoD) that encode the subunits that comprise a eubacterium-type RNA polymerase (α2ββ′ς). Because the R. americana mitochondrial genome appears to be an ancestral type of mtDNA, representative of an early stage in the evolution of this organellar genome (26), one might expect that the mtDNA-encoded sigma factor would have been recruited as a transcription factor for mitochondrial gene expression, even after the evolutionary replacement of the mtDNA-encoded, eubacterium-like RNA polymerase by the nucleus-encoded, phage-type enzyme that now functions as the mitochondrial RNA polymerase in virtually all eukaryotes (11, 12, 27). However, in our alignment, the p63 sequence does not show appreciably higher similarity to the R. americana mitochondrial RpoD, which lacks the acidic domain and region 3.1, than to the eubacterial RpoD sequences (Fig. 7). More detailed biochemical analysis will be required to explore the functional relevance of these similarities. Given the overall low level of similarity, it may be significant that a mutagenesis study of the yeast mtTFB and amino acid sequence comparison with a mtTFB homolog from closely related species both suggest that the mechanism of promoter recognition by the fungal mtTFB is different from that used by eubacterial sigma factors (10, 55).
Two promoter-specific DNA-binding proteins (34 and 44 kDa) have been identified in the mitochondria of pea, a dicotyledonous plant (31). As noted above, our sequence similarity searches have identified Arabidopsis protein sequences bearing significant similarity to the wheat p63 sequence. Whether either of the pea proteins is homologous to the wheat p63 and Arabidopsis proteins is not known at this time. Because the core promoter sequence differs substantially between dicotyledonous and monocotyledonous plants, it would not be surprising if the corresponding RNA polymerases required different transcription factors, although similarities in the basal transcription machinery might be expected. In maize, a nonconsensus promoter has been found in lines having Zea perennis mitochondria (48); whether this alternative promoter interacts with a different transcription factor(s) than does the standard promoter remains to be ascertained. Further analysis will be necessary to definitively identify the minimal protein components essential for specific transcription in plant mitochondria and to better define differences in this process in monocotyledonous and dicotyledonous plants.
ACKNOWLEDGMENTS
We are indebted to B. G. Lane (University of Toronto) for the kind gift of a wheat cDNA library and for samples of wheat RNA isolated from different stages during imbibition of isolated wheat embryos. We thank members of the Gray lab for valuable comments on this work.
The work reported here was supported by grant MT-4124 to M.W.G. from the Medical Research Council of Canada, as well as by a grant to T.M.I. from the Research Development Fund in the Sciences, Dalhousie University. Salary support to M.W.G. in the form of a fellowship from the Canadian Institute for Advanced Research (Program in Evolutionary Biology) is also gratefully acknowledged.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoshechkin I, Bogenhagen D F. Distinct roles for two purified factors in transcription of Xenopus mitochondrial DNA. Mol Cell Biol. 1995;15:7032–7042. doi: 10.1128/mcb.15.12.7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes W M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from λ bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binder S, Brennicke A. Transcription initiation sites in mitochondria of Oenothera berteriana. J Biol Chem. 1993;268:7849–7855. [PubMed] [Google Scholar]
- 5.Binder S, Hatzack F, Brennicke A. A novel pea mitochondrial in vitro transcription system recognizes homologous and heterologous mRNA and tRNA promoters. J Biol Chem. 1995;270:22182–22189. doi: 10.1074/jbc.270.38.22182. [DOI] [PubMed] [Google Scholar]
- 6.Bogenhagen D F, Yoza B K, Cairns S S. Identification of initiation sites for transcription of Xenopus laevis mitochondrial DNA. J Biol Chem. 1986;261:8488–8494. [PubMed] [Google Scholar]
- 7.Bogenhagen D F. Interaction of mtTFB and mtRNA polymerase at core promoters for transcription of Xenopus laevis mtDNA. J Biol Chem. 1996;271:12036–12041. [PubMed] [Google Scholar]
- 8.Börner T, Hedtke B, Hess W R, Legen J, Herrmann R G, Weihe A. Phage-type RNA polymerases in higher plants. In: Argyroudi-Akoyunoglou J, editor. The chloroplast: from molecular biology to biotechnology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 73–78. [Google Scholar]
- 9.Caoile A G F S, Stern D B. A conserved core element is functionally important for maize mitochondrial promoter activity in vitro. Nucleic Acids Res. 1997;25:4055–4060. doi: 10.1093/nar/25.20.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrodeguas J A, Yun S, Shadel G S, Clayton D A, Bogenhagen D F. Functional conservation of yeast mtTFB despite extensive sequence divergence. Gene Expr. 1996;6:219–230. [PMC free article] [PubMed] [Google Scholar]
- 11.Cermakian N, Ikeda T M, Cedergren R, Gray M W. Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucleic Acids Res. 1996;24:648–654. doi: 10.1093/nar/24.4.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cermakian N, Ikeda T M, Miramontes P, Lang B F, Gray M W, Cedergren R. On the evolution of the single-subunit RNA polymerases. J Mol Evol. 1997;45:671–681. doi: 10.1007/pl00006271. [DOI] [PubMed] [Google Scholar]
- 13.Chang C-C, Sheen J, Bligny M, Niwa Y, Lerbs-Mache S, Stern D B. Functional analysis of two maize cDNAs encoding T7-like RNA polymerases. Plant Cell. 1999;11:911–926. doi: 10.1105/tpc.11.5.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clayton D A. Transcription and replication of animal mitochondrial DNAs. Int Rev Cytol. 1992;141:217–232. doi: 10.1016/s0074-7696(08)62067-7. [DOI] [PubMed] [Google Scholar]
- 15.Denhardt D T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966;23:641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- 16.Farrell R E., Jr . RNA methodologies. A laboratory guide for isolation and characterization. San Diego, Calif: Academic Press; 1993. [Google Scholar]
- 17.Fisher R P, Clayton D A. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J Biol Chem. 1985;260:11330–11338. [PubMed] [Google Scholar]
- 18.Fisher R P, Clayton D A. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisk D G, Walker M B, Barkan A. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999;18:2621–2630. doi: 10.1093/emboj/18.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giese A, Thalheim C, Brennicke A, Binder S. Correlation of nonanucleotide motifs with transcript initiation of 18S rRNA genes in mitochondria of pea, potato and Arabidopsis. Mol Gen Genet. 1996;252:429–436. doi: 10.1007/BF02173008. [DOI] [PubMed] [Google Scholar]
- 22.Giniger E, Ptashne M. Transcription in yeast activated by a putative amphipathic α helix linked to a DNA binding unit. Nature. 1987;330:670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- 23.Goodall G J, Wiebauer K, Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- 24.Gray M W. The endosymbiont hypothesis revisited. Int Rev Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- 25.Gray M W, Hanic-Joyce P J, Covello P S. Transcription, processing and editing in plant mitochondria. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:145–175. [Google Scholar]
- 26.Gray M W, Lang B F, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn T G, Plante I, Rioux P, Saint-Louis D, Zhu Y, Burger G. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray M W, Lang B F. Transcription in chloroplasts and mitochondria: a tale of two polymerases. Trends Microbiol. 1998;6:1–3. doi: 10.1016/S0966-842X(97)01182-7. [DOI] [PubMed] [Google Scholar]
- 28.Haffner M H, Chin M B, Lane B G. Wheat embryo ribonucleates. XII. Formal characterization of terminal and penultimate nucleoside residues at the 5′-ends of ‘capped’ RNA from imbibing wheat embryos. Can J Biochem. 1978;56:729–733. doi: 10.1139/o78-109. [DOI] [PubMed] [Google Scholar]
- 29.Hanic-Joyce P J, Gray M W. Accurate transcription of a plant mitochondrial gene in vitro. Mol Cell Biol. 1991;11:2035–2039. doi: 10.1128/mcb.11.4.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson M R, Folkerts O. Structure and function of the higher plant mitochondrial genome. Int Rev Cytol. 1995;141:129–172. [Google Scholar]
- 31.Hatzack F, Dombrowski S, Brennicke A, Binder S. Characterization of DNA-binding proteins from pea mitochondria. Plant Physiol. 1998;116:519–527. doi: 10.1104/pp.116.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hedtke B, Börner T, Weihe A. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science. 1997;277:809–811. doi: 10.1126/science.277.5327.809. [DOI] [PubMed] [Google Scholar]
- 33.Helmann J D, Chamberlin M J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 34.Hess W R, Börner T. Organellar RNA polymerases of higher plants. Int Rev Cytol. 1999;190:1–59. doi: 10.1016/s0074-7696(08)62145-2. [DOI] [PubMed] [Google Scholar]
- 34a.Ikeda, T. M. Unpublished results.
- 35.Ikeda T M, Gray M W. Identification and characterization of T3/T7 bacteriophage-like RNA polymerase sequences in wheat. Plant Mol Biol. 1999;40:567–578. doi: 10.1023/a:1006203928189. [DOI] [PubMed] [Google Scholar]
- 36.Jang S H, Jaehning J A. The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial ς factors. J Biol Chem. 1991;266:22671–22677. [PubMed] [Google Scholar]
- 37.Joo D M, Ng N, Calendar R. A ς32 mutant with a single amino acid change in the highly conserved region 2.2 exhibits reduced core RNA polymerase affinity. Proc Natl Acad Sci USA. 1997;94:4907–4912. doi: 10.1073/pnas.94.10.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Lang B F, Burger G, O’Kelly C J, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Gray M W. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 40.Larsson N-G, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh G S, Clayton D A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 41.Léonetti J-P, Wong K, Geiduschek E P. Core-sigma interaction: probing the interaction of the bacteriophage T4 gene 55 promoter recognition protein with E.coli RNA polymerase core. EMBO J. 1998;17:1467–1475. doi: 10.1093/emboj/17.5.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levings C S, III, Brown G G. Molecular biology of plant mitochondria. Cell. 1989;56:171–179. doi: 10.1016/0092-8674(89)90890-8. [DOI] [PubMed] [Google Scholar]
- 43.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malhotra A, Severinova E, Darst S A. Crystal structure of a ς70 subunit fragment from E. coli RNA polymerase. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 45.Masters B S, Stohl L L, Clayton D A. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 46.Montoya J, Perez-Martos A, Garstka H L, Wiesner R J. Regulation of mitochondrial transcription by mitochondrial transcription factor A. Mol Cell Biochem. 1997;174:227–230. [PubMed] [Google Scholar]
- 47.Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newton K J, Winberg B, Yamato K, Lupold S, Stern D B. Evidence for a novel mitochondrial promoter preceding the cox2 gene of perennial teosintes. EMBO J. 1995;14:585–593. doi: 10.1002/j.1460-2075.1995.tb07034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parisi M A, Clayton D A. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 50.Rapp W D, Lupold D S, Mack S, Stern D B. Architecture of the maize mitochondrial atp1 promoter as determined by linker-scanning and point mutagenesis. Mol Cell Biol. 1993;13:7232–7238. doi: 10.1128/mcb.13.12.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Sánchez, H., and W. Schuster. 1997. GenBank accession no. AJ001037.
- 53.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 54.Shadel G S, Clayton D A. Mitochondrial transcription initiation. Variation and conservation. J Biol Chem. 1993;268:16083–16086. [PubMed] [Google Scholar]
- 55.Shadel G S, Clayton D A. A Saccharomyces cerevisiae mitochondrial transcription factor, sc-mtTFB, shares features with sigma factors but is functionally distinct. Mol Cell Biol. 1995;15:2101–2108. doi: 10.1128/mcb.15.4.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tiranti V, Savoia A, Forti F, D’Apolito M-F, Centra M, Rocchi M, Zeviani M. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum Mol Genet. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- 57.Tzagoloff A, Myers A M. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- 58.Unseld M, Marienfeld J R, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 59.Weihe A, Hedtke B, Börner T. Cloning and characterization of a cDNA encoding a bacteriophage-type RNA polymerase from the higher plant Chenopodium album. Nucleic Acids Res. 1997;25:2319–2325. doi: 10.1093/nar/25.12.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolstenholme D R, Fauron C M-R. Mitochondrial genome organization. In: Levings III C S, Vasil I K, editors. The molecular biology of plant mitochondria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–59. [Google Scholar]
- 61.Zhou Y N, Walter W A, Gross C A. A mutant ς32 with a small deletion in conserved region 3 of ς has reduced affinity for core RNA polymerase. J Bacteriol. 1992;174:5005–5012. doi: 10.1128/jb.174.15.5005-5012.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]