Abstract
To investigate the mechanisms of transcriptional enhancement by the p300 coactivator, we analyzed wild-type and mutant versions of p300 with a chromatin transcription system in vitro. Estrogen receptor, NF-κB p65 plus Sp1, and Gal4-VP16 were used as different sequence-specific activators. The CH3 domain (or E1A-binding region) was found to be essential for the function of each of the activators tested. The bromodomain was also observed to be generally important for p300 coactivator activity, though to a lesser extent than the CH3 domain/E1A-binding region. The acetyltransferase activity and the C-terminal region (containing the steroid receptor coactivator/p160-binding region and the glutamine-rich region) were each found to be important for activation by estrogen receptor but not for that by Gal4-VP16. The N-terminal region of p300, which had been previously found to interact with nuclear hormone receptors, was not seen to be required for any of the activators, including estrogen receptor. Single-round transcription experiments revealed that the functionally important subregions of p300 contribute to its ability to promote the assembly of transcription initiation complexes. In addition, the acetyltransferase activity of p300 was observed to be distinct from the broadly essential activation function of the CH3 domain/E1A-binding region. These results indicate that specific regions of p300 possess distinct activation functions that are differentially required to enhance the assembly of transcription initiation complexes. Interestingly, with the estrogen receptor, four distinct regions of p300 each have an essential role in the transcription activation process. These data exemplify a situation in which a network of multiple activation functions is required to achieve gene transcription.
Transcription by RNA polymerase II requires the action of sequence-specific DNA-binding factors, coactivators, chromatin remodeling and modifying factors, and the basal transcriptional machinery. The packaging of DNA into chromatin causes a general repression of gene activity, and transcription factors function with chromatin remodeling and modifying factors to relieve this chromatin-mediated repression (for reviews, see references 7, 11, 20, 23, 26, 33, 37, 58, 59, 61, 70, 72, 75–77).
In this study, we focus upon the function of the coactivator, p300, which is required for transcriptional activation by many sequence-specific DNA-binding factors that include nuclear hormone receptors, NF-κB, cyclic AMP response element binding protein (CREB), p53, STAT-1, and others. p300 is closely related to CREB-binding protein (CBP), and the two proteins are often referred to as p300/CBP. p300 was identified as an adenovirus E1A oncoprotein-associated factor (19), whereas CBP was discovered on the basis of its binding to CREB (16, 43).
p300 and CBP have key roles in cellular differentiation, growth control, and homeostasis. The importance of p300/CBP for normal cellular functioning was revealed by results from gene knockout studies and various disease states involving the factors (21, 22). Gene knockouts in mice indicated that p300 and CBP are required for normal embryonic development and viability (68, 80). Mutations in the human CBP gene are associated with Rubinstein-Taybi syndrome, a haploinsufficiency disorder resulting in mental retardation and various other developmental abnormalities (60). Chromosomal translocations resulting in the fusion of CBP with monocytic leukemia zinc finger protein or mixed lineage leukemia protein have been found in subtypes of acute myeloid leukemia (4, 66). Finally, p300 gene alterations have been detected in colorectal and gastric carcinomas (50).
p300 and CBP are large proteins with multiple functional domains (Fig. 1A). The conserved motifs in p300/CBP include a bromodomain, a glutamine-rich (Q-rich) region, and three cysteine-histidine (CH)-rich regions (CH1, CH2, and CH3). The bromodomain is found in many chromatin- and transcription-related factors and is thought to play a role in protein-protein interactions and the association of factors with chromatin (17, 28, 31, 56). The Q-rich region, which is located at the C terminus of the protein, has features similar to those of the glutamine-rich transcriptional activation domains found in a number of transcriptional activators (1, 16, 19). Importantly, within the Q-rich region, there is a site for the binding of the steroid receptor coactivator (SRC)/p160 family of coactivator proteins (36). The CH3 region is the site of interaction with many different factors, which include the adenovirus E1A protein (1, 54, 78), the coactivator PCAF (78), RNA polymerase-containing complexes (51, 52), and TFIIB (43, 54). In addition, other regions in p300 serve as binding sites for a variety of DNA-binding transcriptional activator proteins.
FIG. 1.
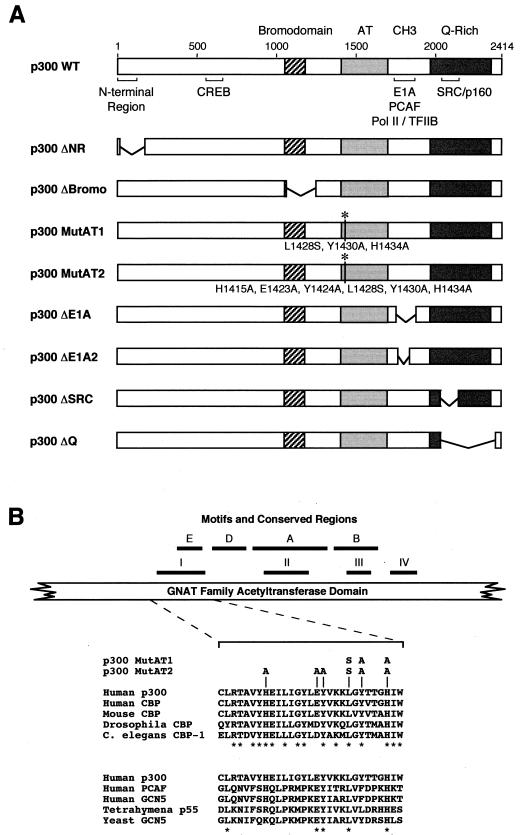
Wild-type and mutant human p300 proteins. (A) Schematic diagram of wild-type (WT) and mutant variants of p300. Specific regions of p300 are indicated: N-terminal region (NR), CREB-binding region, bromodomain, AT domain, CH3 domain/E1A-binding region, Q-rich region, and SRC/p160-binding region. The deleted regions of p300 are as follows: ΔNR lacks residues 3 to 173, ΔBromo lacks residues 1071 to 1241, ΔE1A lacks residues 1739 to 1871; ΔE1A2 lacks residues 1751 to 1813, ΔSRC lacks residues 2042 to 2157, and ΔQ lacks residues 2042 to 2375. The amino acid substitutions in the MutAT1 and MutAT2 proteins are shown. (B) Alignment of p300/CBP family members with the conserved region I/motif E of GNAT family member AT domains. The schematic diagram of the GNAT family AT domain depicts the locations of motifs E, D, A, and B as well as conserved regions I to IV, as previously defined (8, 12, 47, 53). In the alignments of conserved region I/motif E, the asterisks denote amino acid residues that are identical in each group of proteins. In addition, the specific amino acid substitutions in p300MutAT1 and p300MutAT2 are shown.
p300 has acetyltransferase (AT) activity and possesses a region of similarity to members of the GCN5-related N-AT (GNAT) family of proteins (Fig. 1B) (47, 53). The AT activity of p300 is capable of acetylating histones (2, 55), nonhistone chromatin-associated proteins (49), components of the basal transcriptional machinery (30), and transcriptional activator proteins (5, 24, 29, 45).
In this work, we examined the roles of specific subregions of p300 in the transcription activation process. A key feature of these experiments is the use of a biochemical approach, which includes the use of chromatin templates and purified p300 proteins, that has enabled the separate analysis of distinct p300 activities. In this manner, the results provide a different perspective on p300 function than those of previous mutational analyses of transcriptional enhancement by p300/CBP that were performed in cultured cells by the use of transient-transfection or microinjection assays (38, 42, 47, 62).
Thus, by using purified wild-type or mutant versions of p300, we investigated the relation between p300 AT activities, transcriptional coactivator activities of p300 (in a single round or in multiple rounds of transcription) with different promoter-binding factors, and the interaction of p300 with E1A. The data indicate that multiple, distinct biochemical functions of p300 can each have essential roles in the transcription process.
MATERIALS AND METHODS
Synthesis and purification of recombinant proteins.
FLAG-tagged human estrogen receptor (ER) was synthesized in Sf9 cells by using a baculovirus expression system and was purified as previously described (39, 40). His6-tagged NF-κB p65 subunit (46) was synthesized in Sf9 cells by using a recombinant baculovirus kindly provided by J. Hiscott (Lady Davis Institute, Montreal, Canada). The p65 protein was synthesized and purified essentially as described for His6-tagged p300 (40). Purified Sp1 was obtained from Promega (Madison, Wis.). Gal4-VP16 was synthesized in Escherichia coli and purified as previously described (15). His6-tagged human p300 was synthesized in Sf9 cells by using a baculovirus expression system and purified as previously described (39, 40). The panel of p300 mutants was generated by inserting PCR-amplified fragments into the appropriate restriction sites in the full-length His6-tagged p300 cDNA. The integrity of all PCR-amplified DNA fragments was analyzed by DNA sequencing. The specific mutations are described in the legend to Fig. 1A. The mutant p300 cDNAs were used to generate recombinant baculoviruses, and the mutant p300 proteins were synthesized and purified as described for wild-type p300. Glutathione S-transferase (GST), GST-E1A, and GST-E1AΔ were synthesized in E. coli and purified as previously described (52).
Protein acetylation assays.
Purified native Drosophila chromatin (average DNA fragment length of approximately 1 kbp) and purified Drosophila core histones were prepared as previously described (9). To assay the histone AT (HAT) and autoacetylase activities of the wild-type and mutant p300 proteins, 400 ng of purified wild-type or mutant p300 was incubated for 30 min at 27°C in a 30-μl reaction mixture containing 4 μg of core histones (either as purified core histones or as native chromatin) and 5 mM [3H]acetyl coenzyme A ([3H]acetyl-CoA) (New England Nuclear). After this incubation, the samples were divided into two tubes and run on two separate sodium dodecyl sulfate (SDS)–15% polyacrylamide gels. One gel was subjected to fluorography to visualize the 3H-labeled proteins. The other gel was either stained with Coomassie brilliant blue R-250 to visualize the histones or transferred to nitrocellulose for immunoblotting with anti-p300 antibodies (Upstate Biotechnology or Santa Cruz Biotechnology). To quantify the amounts of [3H]p300 and [3H]histone proteins in the gels, the corresponding bands were excised and subjected to liquid scintillation counting. The HAT and autoacetylase activities of the mutant p300 proteins were each expressed as a percentage of the wild-type p300 activity. The amount of acetylation of the histones in chromatin as well as the autoacetylation of p300 occurred with approximately 2% of the efficiency of acetylation of free core histones in these assays. Similar HAT assay results were observed with free core histone substrates when the reactions were analyzed by trichloroacetic acid precipitation and filter binding techniques (data not shown).
To assess the effects of the GST-E1A fusion proteins on p300 HAT activity, reactions were performed as described above with purified core histones in the presence of the purified GST fusion proteins. The HAT assay reaction mixtures contained the same concentrations of p300 and GST fusion proteins as those used in the corresponding transcription assays. The reactions were analyzed by trichloroacetic acid precipitation and filter binding with subsequent liquid scintillation counting. Each AT experiment was performed a minimum of two times, but more typically three or four times, to ensure reproducibility.
DNA templates.
2ERE-AdE4 contains two tandem copies of the Xenopus vitellogenin A2 gene ER element (ERE) (73) located upstream of the adenovirus E4 core promoter in pIE0 (57). 2ERE-pS2 (48) contains two tandem copies of the Xenopus vitellogenin A2 gene ERE located upstream of the human pS2 gene core promoter (32). The wild-type human interferon regulatory factor type 1 (IRF-1) template contains 1.3 kb of the native gene, which includes the core promoter and the upstream regulatory region containing NF-κB and Sp1 binding sites (10, 64). 2Gal4-pS2 and 2Gal4-IRF contain two tandem Gal4 binding sites located upstream of the human pS2 gene core promoter and the human IRF-1 gene core promoter, respectively.
Chromatin assembly and in vitro transcription reactions.
Chromatin assembly reactions were performed with a chromatin assembly extract derived from Drosophila embryos (9, 35) as previously described (39, 40). All transcriptional activator proteins (ER plus E2, NF-κB p65 plus Sp1, and Gal4-VP16) were added during the chromatin assembly reactions, whereas the p300 or GST fusion proteins (such as GST-E1A) were added after the chromatin assembly reactions were complete. The reaction mixtures were incubated for an additional 30 min at 27°C after the p300 or GST fusion proteins were added to allow interaction of these proteins with the chromatin templates.
In vitro transcription reactions were performed with HeLa cell nuclear extracts that were prepared essentially by the method of Dignam et al. (18) with 0.42 M KCl for the extraction of nuclei instead of 0.42 M NaCl. Multiple-round transcription reactions (used for all experiments except those shown in Fig. 5) were set up under conditions described previously (39, 40). The single-round transcription experiments (see Fig. 5) were performed as described previously (40), except that transcription preinitiation complexes were formed for 45 min at 27°C after the addition of the HeLa cell nuclear extract. The RNA products from the in vitro transcription reactions were analyzed by primer extension analysis (40). All reactions were performed in duplicate, and each experiment was performed a minimum of two separate times to ensure reproducibility. The data were analyzed and quantified with a PhosphorImager (Molecular Dynamics).
FIG. 5.
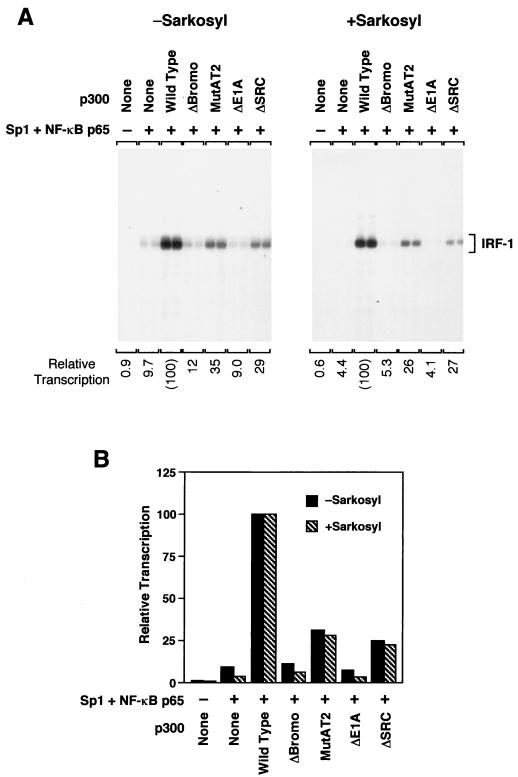
p300 mutant proteins exhibit impaired phenotypes in a single round of transcription. (A) Comparison of the transcriptional activities of p300 proteins in multiple (−Sarkosyl) versus single (+Sarkosyl) rounds of transcription. The coactivator activities of the p300 mutants were examined in transcription experiments in the presence or the absence of 0.2% Sarkosyl with purified human NF-κB p65 plus Sp1 and the human IRF-1 promoter, as described in the legend to Fig. 4A. The production of RNA transcripts was analyzed and quantified by primer extension analysis. The final concentration of purified exogenous p300 protein in each of the transcription reaction mixtures was 15 nM. (B) Summary and comparison of results from experiments such as those represented in panel A. The data are expressed as a percentage of wild-type p300 activity and are the means from two independent determinations.
p300-E1A interaction assays.
Assays to determine interactions between wild-type (full-length) or mutant p300 proteins and full-length adenovirus 12S E1A proteins were performed with baculovirus-infected Sf9 cells. Cells (one 80%-confluent 10-cm-diameter dish per sample) were coinfected with recombinant viruses for wild-type or mutant p300 and FLAG-tagged E1A (78). The amount of virus used to yield approximately equal expression of the p300 proteins was determined empirically. The cells were incubated with the virus for 3 days at 26°C under standard growth conditions. After the incubations, the cells were collected, washed in ice-cold phosphate-buffered saline, and homogenized on ice with a mini-Dounce in 1.2 ml of lysis buffer (10 mM Tris-HCl [pH 7.5], 300 mM NaCl, 0.1 mM EDTA, 10% [wt/vol] glycerol, 0.1% [wt/vol] NP-40, 2 mM 2-mercaptoethanol, 2 mM phenylmethylsulfonyl fluoride, 20 μg of leupeptin per ml, 20 μg of aprotinin per ml). Cell debris was removed from the cell lysates by centrifugation in a microcentrifuge for 10 min at 4°C. The supernatants were transferred to new 1.5-ml tubes, and 10 μl of anti-FLAG M2 affinity resin (Sigma) which had been previously equilibrated in lysis buffer was added to each tube. The samples were incubated at 4°C with gentle mixing for 2 h. After the incubation, the resin was washed four times with 1 ml of ice-cold wash buffer (10 mM Tris-HCl [pH 7.5], 300 mM NaCl, 10% [wt/vol] glycerol, 0.02% [wt/vol] SDS, 2 mM 2-mercaptoethanol, 2 mM phenylmethylsulfonyl fluoride, and the proteins were eluted in 40 μl of wash buffer containing 0.3 mg of FLAG peptide per ml. The samples were subjected to SDS–8% polyacrylamide gel electrophoresis and immunoblotting with anti-p300 and anti-FLAG antibodies followed by detection with 125I-protein A. Each experiment was performed a minimum of two separate times to ensure reproducibility. The data were analyzed and quantified with a PhosphorImager (Molecular Dynamics).
RESULTS
Synthesis and purification of mutant versions of human p300.
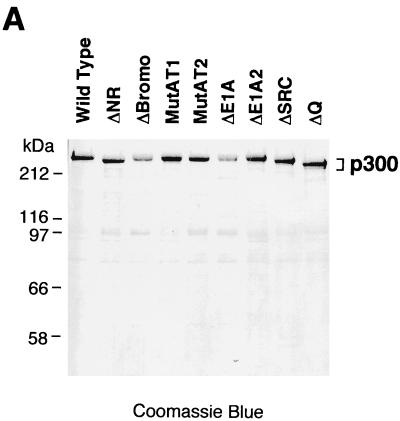
To investigate the mechanism of p300/CBP function in vitro, we generated a series of mutant versions of human p300. As depicted in Fig. 1A, the mutations are focused upon conserved protein motifs and regions of p300 that were found to be important for interactions with other transcription factors. The wild-type and mutant p300 proteins were synthesized with a baculovirus expression system and purified to near homogeneity (Fig. 2A).
FIG. 2.
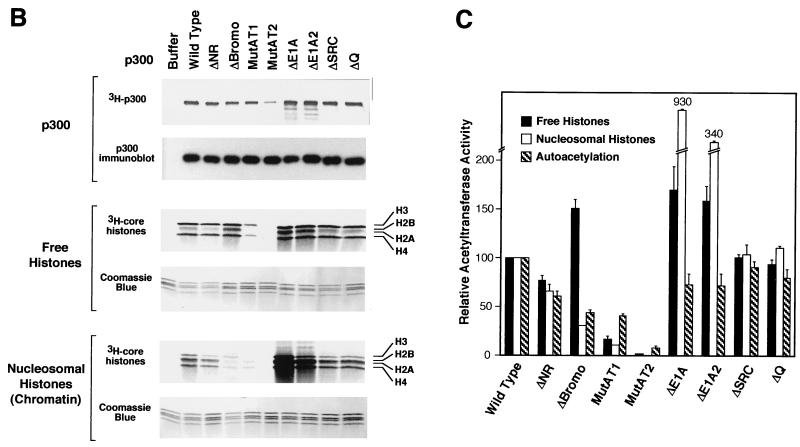
HAT and autoacetylase activities of wild-type and mutant p300 proteins. (A) SDS-polyacrylamide gel analysis of purified wild-type and mutant p300 proteins. His6-tagged wild-type and mutant p300 proteins were synthesized in Sf9 cells by using a baculovirus expression system and purified by nickel chelate chromatography. The proteins were subjected to SDS–8% polyacrylamide gel electrophoresis and then visualized by staining with Coomassie brilliant blue R-250. (B) Analysis of HAT and autoacetylase activities of wild-type and mutant p300 proteins. Purified wild-type or mutant p300 proteins were incubated in reaction mixtures containing either purified free histones or nucleosomal histones (in purified native chromatin) along with [3H]acetyl-CoA. The reactions were then subjected to SDS–15% polyacrylamide gel electrophoresis and fluorography. In parallel, aliquots from the same reaction mixtures were run on similar gels for immunoblotting with anti-p300 antibodies or for staining with Coomassie blue R-250. The core histones (H2A, H2B, H3, and H4) are indicated. (C) HAT and autoacetylase activities of wild-type and mutant p300 proteins. The bands from the gels shown in panel B were excised and quantified by liquid scintillation counting. The data are expressed as a percentage of wild-type activity and represent the mean + the standard error of the mean from two or more independent determinations.
The specific alterations in the mutant p300 proteins are as follows. In p300ΔNR, there is a deletion of the N-terminal region of p300, which has been found to interact with nuclear hormone receptors (13, 36). p300ΔBromo lacks the bromodomain. The p300MutAT1 and p300MutAT2 proteins contain triple and sextuple amino acid substitutions in the conserved region I/motif E of the p300 AT region (Fig. 1B). Notably, various mutations in the analogous regions of yeast GCN5 and mouse CBP have been found to reduce HAT activity (41, 47, 74). The p300ΔE1A and p300ΔE1A2 proteins lack different amounts of the CH3 domain, which interacts with E1A, PCAF, RNA polymerase II-containing complexes, and TFIIB. Lastly, we constructed and purified p300ΔSRC and p300ΔQ. In p300ΔSRC, there is a deletion of the C-terminal segment of p300 that binds to the SRC/p160 family of coactivator proteins. The p300ΔQ protein lacks the majority of the C-terminal Q-rich segment of p300, which includes the SRC/p160-binding region.
HAT and autoacetylase activities of wild-type and mutant p300 proteins.
To assess the AT activities of the mutant p300 proteins, we used purified recombinant p300 proteins with either free core histones or nucleosomal histones (in native chromatin) as substrates in the presence of [3H]acetyl-CoA. The reaction products were resolved on SDS-polyacrylamide gels, and the acetylated polypeptides were identified by fluorography. As seen previously (2, 55), wild-type p300 exhibits a preference for the acetylation of H3-H4 relative to that of H2A-H2B as free histones but not as nucleosomal histones (Fig. 2B). In addition, we observed that p300 was autoacetylated under the same conditions (Fig. 2B).
Next, we assayed the panel of p300 mutant proteins for HAT and autoacetylase activities (Fig. 2B and C). The p300MutAT1 and p300MutAT2 proteins, which have triple and sextuple amino acid substitutions in the AT region, exhibited ∼10% and <1% of the nucleosomal HAT activity of wild-type p300, respectively. The AT activities of the p300ΔNR, p300ΔSRC, and p300ΔQ proteins were similar to those of the wild-type protein. p300ΔNR did, however, consistently exhibit less acetylation of H3 relative to that of H4 with nucleosomal histones but not with free histones.
Interestingly, p300ΔBromo was impaired in its ability to acetylate nucleosomal histones (in native chromatin) relative to free histones. These results suggest that the bromodomain contributes to the ability of HATs to acetylate nucleosomal histones and are similar to data obtained with yeast Gcn5p in the SAGA complex (67). In contrast, deletions in the CH3 region (p300ΔE1A and p300ΔE1A2) led to significantly increased nucleosomal HAT activity. Moreover, when compared with the wild-type protein, p300ΔBromo, p300ΔE1A, and p300ΔE1A2 exhibited enhanced acetylation of H2A-H2B relative to that of H3-H4 with free histones. These results indicate that the conserved AT region is important for the AT activity of p300 and further reveal that segments of p300 that are outside of the core AT region can significantly affect the HAT activity of the protein.
Transcriptional activity of wild-type and mutant p300 proteins with human ERα.
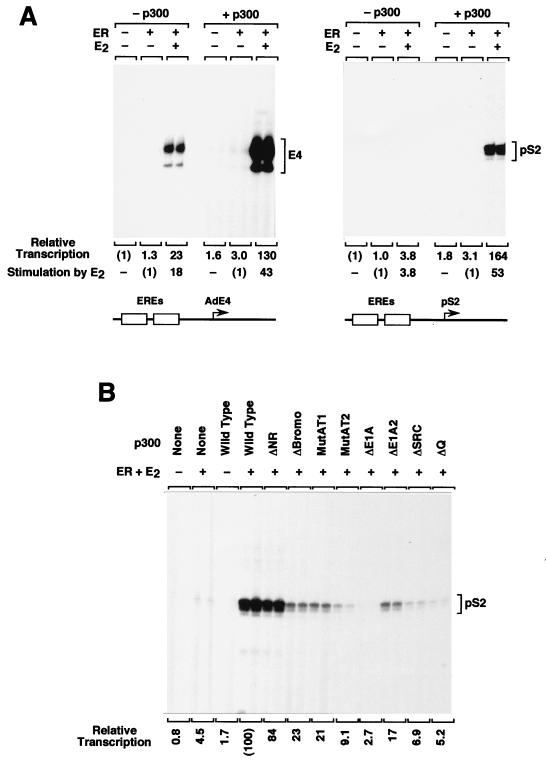
To test the transcriptional activity of the mutant p300 proteins, we carried out in vitro transcription reactions with the human ERα in conjunction with chromatin templates. Members of our group have previously shown that ligand-activated ER and p300 function synergistically in the activation of transcription in vitro with chromatin templates (39). In that earlier study, a reporter promoter construction that contains two EREs upstream of the adenovirus E4 core promoter was used. More recently, we have been using a reporter gene that contains two EREs upstream of the core promoter from the estrogen-responsive pS2 gene (3, 6). With the pS2-based promoter construction, we observed more potent transcriptional synergy between ER and p300 than with the adenovirus E4 reporter (Fig. 3A).
FIG. 3.
Transcriptional enhancement by wild-type and mutant p300 proteins with human ERα. (A) Wild-type p300 significantly enhances transcriptional activation by ER with chromatin templates containing the pS2 core promoter. Templates containing two ER binding sites upstream of the adenovirus E4 core promoter (2ERE-AdE4) or the human pS2 core promoter (2ERE-pS2) were assembled into chromatin in the presence of purified ER and 17β-estradiol (E2), as noted. The chromatin samples were then subjected to in vitro transcription analysis in the presence or absence of purified wild-type human p300. The production of RNA transcripts was analyzed and quantified by primer extension analysis. The final concentrations of ER, E2, and p300 in the transcription reactions were 4.5, 30, and 15 nM, respectively. (B) Transcriptional coactivator activity of mutant p300 proteins with liganded ER. The 2ERE-pS2 template was assembled into chromatin in the presence of purified human ER and E2, where noted, and the samples were subjected to in vitro transcription analysis in the presence or absence of purified wild-type or mutant human p300 proteins.
By using the chromatin transcription system with ligand-activated ER and the pS2 reporter construction, the coactivator activity of each of the mutant p300 proteins was tested (Fig. 3B). Deletion or mutation of the bromodomain, the AT region, the CH3 domain/E1A-binding region, or the SRC/p160-binding region led to substantial reduction in p300 coactivator activity. In contrast, deletion of the N-terminal region had little or no effect on p300 activity with ER. The strongest effects were seen with the large deletion of the CH3 domain/E1A-binding region (p300ΔE1A), the six-amino-acid substitution in the AT region (p300MutAT2), and the two C-terminal deletions (p300ΔSRC and p300ΔQ). Notably, the p300MutAT2 sextuple-substitution mutant exhibited lower AT and transcriptional activities than the p300MutAT1 triple-substitution mutant, which suggests that the decrease in the AT activity of p300 causes a decrease in its coactivator activity with ER. As a control, an immunoblot analysis of aliquots from the transcription reactions before the initiation of transcription or after the completion of the reactions indicated that the wild-type and mutant p300 proteins remained intact throughout the transcription process (data not shown). These results reveal that multiple regions of p300 are required for the synergistic enhancement of transcription with ligand-activated ER.
Transcriptional activity of wild-type and mutant p300 proteins with different activators and promoters.
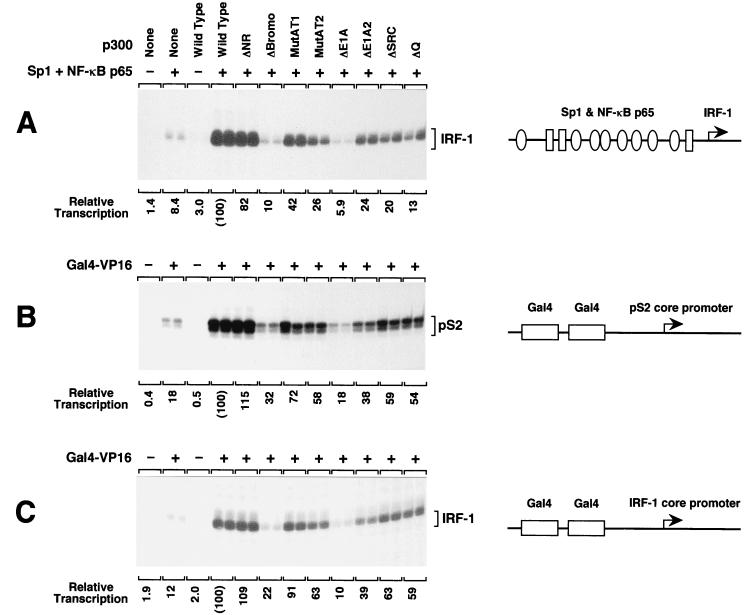
To investigate further the role of specific regions of p300 in transcriptional activation, we tested the properties of the mutant p300 proteins with other promoter-binding activators. We used the NF-κB p65 subunit (RelA) and Sp1 with the human IRF-1 gene promoter (from −1312 to +38, relative to the transcription start site) (10, 64). We also used the Gal4-VP16 activator with promoter constructions containing two Gal4 binding sites upstream of either the TATA box-containing core promoter from the pS2 gene (32) or the TATA-less core promoter from the IRF-1 gene.
With p65 plus Sp1 and the native human IRF-1 promoter, deletion of the bromodomain or the CH3 domain/E1A-binding region (p300ΔBromo and p300ΔE1A, respectively) led to the strongest reduction in p300 transcriptional activity (Fig. 4A). A moderate loss of p300 coactivator activity was seen with the p300MutAT2, p300ΔE1A2, p300ΔSRC, and p300ΔQ proteins. As with the ER, deletion of the N-terminal region did not cause a significant decrease in p300 transcriptional activity with p65 and Sp1. Relative to the ER, p300 coactivator function with NF-κB p65 and Sp1 appeared to be less dependent on the AT, SRC/p160, and Q-rich regions and more dependent on the bromodomain.
FIG. 4.
Analysis of the transcriptional activity of wild-type and mutant p300 proteins with different activators and promoters. DNA templates, illustrated schematically on the right, were assembled into chromatin in the presence of purified transcriptional activators, as noted. The samples were then subjected to in vitro transcription analysis in the presence or absence of purified wild-type or mutant human p300 proteins. The production of RNA transcripts was analyzed and quantified by primer extension analysis. The final concentration of purified exogenous p300 protein in each of the transcription reaction mixtures was 15 nM. (A) Transcriptional activity of p300 proteins with purified human NF-κB p65 and Sp1 in the context of the human IRF-1 promoter and upstream regulatory region. The ovals and rectangles represent binding sites for Sp1 and NF-κB, respectively (64), and are not drawn to scale. The final concentrations of p65 and Sp1 in the transcription reaction mixtures were 60 and 9 nM, respectively. (B) Transcriptional activity of p300 proteins with purified Gal4-VP16 in conjunction with a DNA template containing two Gal4 binding sites upstream of the human pS2 gene core promoter. The final concentration of Gal4-VP16 in the transcription reaction mixture was 7.5 nM. (C) Transcriptional activity of p300 proteins with purified Gal4-VP16 in conjunction with a DNA template containing two Gal4 binding sites upstream of the human IRF-1 core promoter. The final concentration of Gal4-VP16 in the transcription reaction mixture was 7.5 nM.
With Gal4-VP16 and either the pS2 or the IRF-1 core promoters, the strongest effects were seen upon deletion of the CH3 domain/E1A-binding region or the bromodomain (Fig. 4B and C). Gal4-VP16 was not significantly affected (less than twofold) by mutations in the N-terminal region, the AT region, and the C terminus. These findings suggest that the synthetic Gal4-VP16 protein has broader activation functions than the naturally occurring Sp1, p65, and ER proteins.
Transcriptionally impaired, mutant p300 proteins exhibit a reduced ability to enhance transcription initiation.
To determine the step in the transcription process that is affected by mutations in p300, we carried out single-round transcription experiments to compare the abilities of wild-type versus mutant p300 proteins to promote the assembly of productive initiation complexes. To this end, we assembled transcription complexes on chromatin templates (as in Fig. 3 and 4), initiated transcription by addition of the four ribonucleoside 5′-triphosphates, and then added the anionic detergent Sarkosyl at a concentration that allows elongation of transcriptionally engaged polymerases but inhibits reassembly of transcription initiation complexes. In these experiments, we used NF-κB p65 plus Sp1 as the sequence-specific activators instead of ER because members of our group had previously found that ER functions mainly to promote transcription reinitiation and thus yields only low levels of transcription in a single round (39).
As seen in Fig. 5, the transcriptional defects in the mutant p300 proteins (relative to the wild-type p300 as a reference) in a single round of transcription were similar to those seen when transcription was not limited to a single round, in which approximately three rounds of transcription were observed. These results thus indicate that the subregions of p300 contribute to the assembly of productive transcription initiation complexes.
An N-terminal fragment of E1A inhibits the transcriptional activity of p300, but not its HAT activity.
Because the integrity of the CH3 domain/E1A-binding domain was observed to be particularly important for p300 coactivator activity, we explored the function of this region in greater detail. Previous studies identified interactions between the CH3 domain and various factors, which include E1A (an inhibitor of p300 activity) (1, 54, 78), PCAF (a coactivator with AT activity) (78), and components of the basal transcriptional machinery (RNA polymerase II-containing complexes and TFIIB) (43, 51, 52, 54). Moreover, interactions between CBP and RNA polymerase II-containing complexes can be blocked by an N-terminal fragment of E1A that binds to the CH3 region (52). It was also observed that E1A can block the binding of PCAF to the CH3 domain of p300 (78).
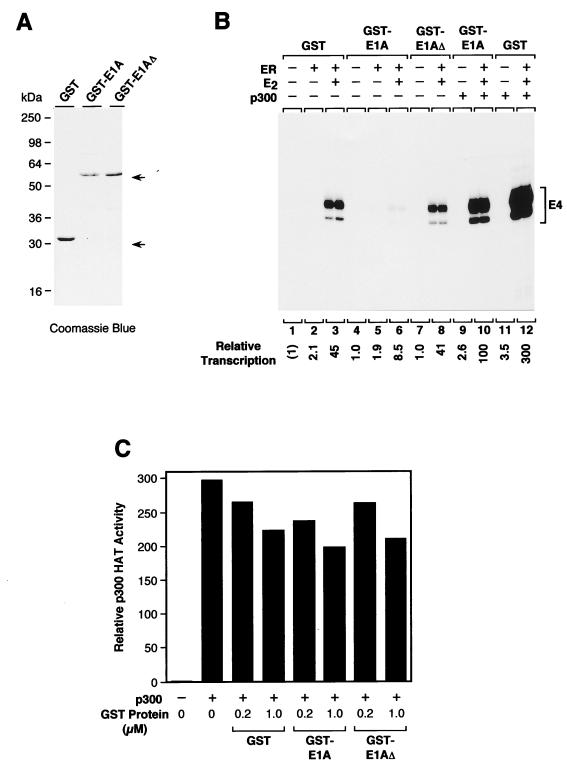
Based on these findings, we tested whether an N-terminal fragment of E1A could inhibit the transcriptional activity of p300. Specifically, we used a fusion of GST with an N-terminal fragment of Ad5 E1A (residues 1 to 139) that had been previously shown to bind to CBP and to inhibit transcriptional activation by phosphorylated (Ser-133) CREB (52). In addition, as controls, we used GST alone as well as GST-E1AΔ, which is identical to GST-E1A except that it lacks amino acid residues 4 to 25 of E1A and does not bind to p300/CBP (52).
The fusion proteins were purified (Fig. 6A) and then tested for their ability to affect the transcription of the AdE4 core promoter construction by ER and p300 (Fig. 6B). These experiments revealed that GST-E1A, but not GST-E1AΔ, caused a fivefold inhibition of transcriptional activation by liganded ER in the absence of exogenously added p300 (Fig. 6B, lanes 3, 6, and 8). This inhibition by GST-E1A most likely reflects the inactivation of the endogenous p300/CBP in the HeLa cell nuclear extract that was used in the reactions because exogenously added p300 was able to relieve the transcriptional inhibition by GST-E1A (Fig. 6B, lanes 9 and 10). Moreover, we observed similar effects when we examined the ability of GST-E1A to inhibit transcriptional activation by liganded ER in the presence of exogenously added p300 with the pS2 promoter (data not shown).
FIG. 6.
Binding of an N-terminal fragment of E1A to p300 inhibits transcriptional activity but not HAT activity. (A) Purification of GST-E1A fusion proteins. GST-E1A is a fusion of GST with amino acid residues 1 to 139 of Ad5 E1A protein. GST-E1AΔ is identical to GST-E1A except for the deletion of E1A amino acid residues 4 to 25, which are required for the binding of E1A to p300. These proteins, along with a GST control, were synthesized in E. coli, purified as described previously (52), subjected to SDS–12% polyacrylamide gel electrophoresis, and visualized by staining with Coomassie brilliant blue R-250. (B) GST-E1A, but not GST-E1AΔ, inhibits ER-dependent transcription in vitro with HeLa transcription extracts containing endogenous p300/CBP. Chromatin assembly and in vitro transcription reactions were performed with the 2ERE-AdE4 template, as in Fig. 3A, with the indicated components. Where noted, purified GST, GST-E1A, or GST-E1AΔ in the presence or absence of exogenous purified p300 was added to the templates after the completion of chromatin assembly. The chromatin samples were then incubated for an additional 30 min at 27°C prior to the transcriptional analysis. The final concentrations of ER, E2, p300, and GST proteins (GST, GST-E1A, or GST-E1AΔ) in the transcription reaction mixtures were 4.5, 30, 15, and 300 nM, respectively. (C) GST-E1A does not inhibit p300 HAT activity. HAT assays were performed with purified recombinant p300, purified core histones, and [3H]acetyl-CoA in the presence or absence of GST, GST-E1A, or GST-E1AΔ. HAT activity was measured by the incorporation of 3H into the core histones. The amount of p300 in the AT reaction mixtures was identical to that used in the chromatin transcription experiments (B). The GST proteins were used at concentrations that correspond to a fourfold or 20-fold molar excess relative to the concentration of p300. The data shown are from a representative experiment.
Recent studies suggest that E1A can inhibit p300 HAT activity (14, 25) and that these effects are mediated primarily through the C-terminal region of E1A (14). Thus, as a control, we tested whether the GST-E1A fusion protein (containing an N-terminal fragment of E1A) that we had used in the transcription studies (Fig. 6B) also affected p300 HAT activity. As shown in Fig. 6C, purified p300 exhibits approximately the same HAT activity in the presence of GST-E1A, GST-E1AΔ, or GST alone when tested under conditions similar to those used in the transcription assays. Therefore, GST-E1A inhibits the coactivator activity of p300 but not its HAT activity. (Because GST-E1A contains only the N-terminal portion of E1A, these results are in accord with the finding that the C terminus of E1A is primarily responsible for the inhibition of p300 HAT activity by E1A [14]). Hence, consistent with the finding that the p300ΔE1A mutant protein is defective for transcription but not HAT activity, these results indicate that E1A can inhibit p300-mediated transcriptional enhancement via a mechanism that is independent of the p300 HAT activity.
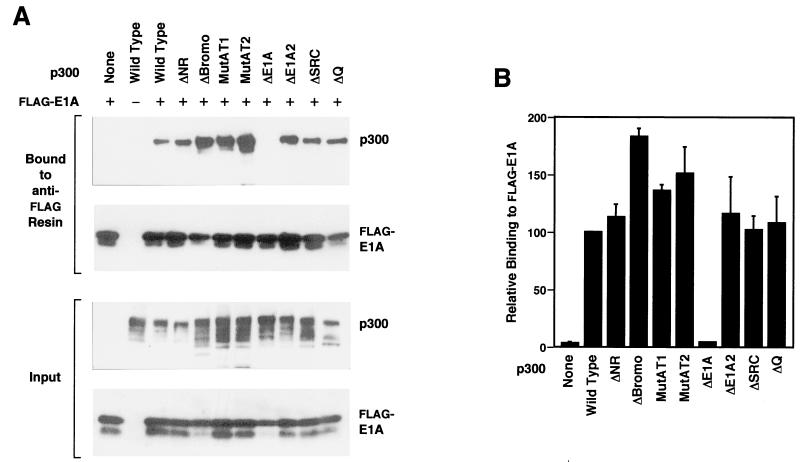
To investigate whether the binding of E1A to p300 correlates with a loss of p300 transcriptional activity, we examined the binding of full-length E1A to the series of mutant p300 proteins. In these experiments, recombinant baculoviruses encoding full-length, FLAG-tagged E1A and wild-type or mutant p300 were used to coinfect Sf9 cells, and then FLAG-E1A and associated p300 proteins were copurified with an anti-FLAG affinity resin. As shown in Fig. 7, all of the mutant p300 proteins, except for p300ΔE1A, were observed to bind efficiently to E1A. There was no detectable binding of p300ΔE1A to full-length FLAG-E1A. Thus, the ΔE1A deletion of the CH3 region, which causes a loss of coactivator activity, also results in the elimination of E1A binding to p300. Yet, it is also notable that we observed efficient binding of E1A to p300ΔE1A2 (which contains a smaller deletion in the CH3 region than the p300ΔE1A protein does), which is partially defective for transcription (Fig. 3 and 4). Thus, these data collectively suggest that the portion of the CH3 domain that is important for p300 coactivator activity overlaps, but does not coincide precisely with, the high-affinity E1A-binding site. In addition, these results suggest that the high-affinity binding of E1A to p300 does not require an intact N-terminal region, bromodomain, or C-terminal region.
FIG. 7.
Deletion of the CH3 region of p300 that is required for coactivator activity also results in the loss of high-affinity binding of E1A to p300. (A) Interactions between the panel of mutant p300 proteins and full-length, FLAG-tagged adenovirus E1A protein. Sf9 cells were coinfected with either wild-type or mutant p300 baculovirus and a FLAG-tagged E1A baculovirus, as indicated. Whole-cell extracts were prepared and incubated with anti-FLAG M2 affinity resin. After a washing, the bound proteins were eluted with FLAG peptide. The eluted proteins and samples of the whole-cell extracts were subjected to SDS–8% polyacrylamide gel electrophoresis and Western blotting for p300 or E1A. (B) Quantification of the p300-E1A interactions. Data from experiments such as those represented in panel A are reported as relative to the binding of FLAG-E1A to wild-type p300 and are the means + the standard errors of the mean from two or more separate determinations.
DISCUSSION
In this study, we performed a biochemical analysis of different activities of the p300 transcriptional coactivator by using wild-type and mutant versions of the protein. The results are summarized in Fig. 8. Specific results and conclusions are discussed below.
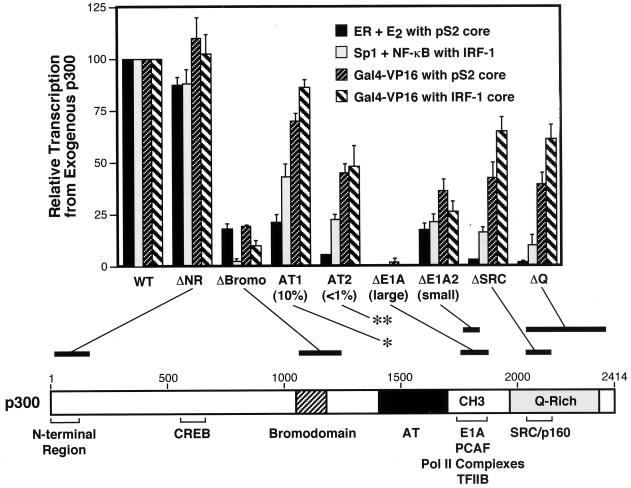
FIG. 8.
Distinct activation functions in p300 are differentially required for transcriptional activation. Summary of in vitro transcription data with wild-type or mutant p300 proteins with different activators and promoters. The transcriptional enhancement derived from the exogenously added p300 protein is obtained by subtraction of the background due to the endogenous p300/CBP. The data are expressed as a percentage of wild-type activity and are the means + the standard errors of the mean from two or more separate determinations. The schematic diagram indicates the locations of the mutations in p300.
The CH3 domain/E1A-binding region.
The CH3 domain/E1A-binding region appears to be a critical element for p300 coactivator activity. The larger deletion of this motif led to a nearly complete loss of p300 transcriptional activity with all activators tested (Fig. 3 to 5), and the addition of a GST-E1A fragment that binds to this region of p300/CBP (but not a control GST-E1AΔ fragment) caused a reduction in p300 activity (Fig. 6). Furthermore, AT activity appears to be distinct from the function of the CH3 domain/E1A-binding region. The GST-E1A fragment that inhibits CBP/p300-mediated activation does not affect the HAT activity of p300 (Fig. 6C). Furthermore, deletion of the CH3 domain/E1A-binding motif in p300 leads to an increase in AT activity (Fig. 2). Thus, these data support a model for the function of the CH3 domain/E1A-binding region as a site for interaction with other cellular factors, such as RNA polymerase II complex, TFIIB, or PCAF (43, 51, 52, 54, 78). The generality of the role of the CH3 region in transcriptional activation remains to be assessed. For instance, in transient-cotransfection assays, it was observed that the CH3 domain is not required for activation by the retinoic acid receptor (42). In addition, E1A has been shown to interact with non-CH3 regions of CBP (42). In this study, we did not observe the binding of E1A to regions outside of the CH3 region of p300 (Fig. 7), probably due to the high stringency of the wash conditions (including 300 mM NaCl and 0.02% SDS) that we had used to detect high-affinity interactions.
The bromodomain.
The bromodomain was originally identified as a conserved sequence that is present in many chromatin-related proteins (28, 31). In our studies, we found that the bromodomain is broadly required for full p300 coactivator function and is particularly important for transcriptional activation by NF-κB p65 and Sp1 (Fig. 4 and 5). Analysis of the HAT activity of the p300ΔBromo protein revealed that it is impaired in its ability to acetylate nucleosomal histones (in native chromatin) but not free histones (Fig. 2). A similar requirement for the bromodomain in the acetylation of nucleosomal histones was observed with yeast Gcn5p in the SAGA complex (67). In addition, these results are consistent with recent experiments suggesting that the bromodomain is involved in protein-protein interactions with the N-terminal tails of histones (17, 56).
AT activity.
We constructed two mutant p300 proteins for the analysis of its AT activity, p300MutAT1 and p300MutAT2, which contain triple and sextuple amino acid substitutions in the AT region, respectively (Fig. 1 and 2). With nucleosomal histone substrate, p300MutAT1 and p300MutAT2 exhibited ∼10% and <1% HAT activity relative to that of wild-type p300 (Fig. 2). Transcriptional studies revealed that the AT mutations cause a significant reduction in the activity of the ER but have only a minor effect on the function of Gal4-VP16 (Fig. 3 and 4). This sort of selective requirement for AT activities was previously observed with other activators, including Myo-D, Stat-1, CREB, and retinoic acid receptor (38, 62).
Interestingly, there was a general correlation between nucleosomal HAT activity and p300 coactivator function with ER. That is, the triple mutant p300MutAT1 has partial nucleosomal HAT activity (10%) and coactivator activity (∼17%), and the sextuple mutant p300MutAT2 has more significantly reduced nucleosomal HAT activity (<1%) and coactivator activity (∼5%). These results suggest that the decrease in the AT activity of p300 contributes to the decrease in the coactivator activity of p300 with ER.
It is important to note, however, that the key acetylation substrate(s) that is responsible for the transcriptional effects has not yet been identified. p300/CBP has been found to acetylate components of the basal transcriptional machinery (e.g., TFIIEβ and TFIIF) (30), sequence-specific transcriptional activator proteins (e.g., p53, NF-Y, and GATA-1) (5, 24, 29, 45), nonhistone chromatin proteins (e.g., HMG-I/Y) (49), and p300 itself (Fig. 2). It remains to be determined whether any of these factors and/or other proteins are functionally important downstream targets of acetylation by p300/CBP in vivo.
Unexpectedly, deletion of the CH3 domain/E1A-binding region in the p300ΔE1A protein led to a ninefold increase in the ability of p300 to acetylate nucleosomal histones (Fig. 2). In the absence of an intact CH3 domain/E1A-binding region, the enhanced AT activity alone is not sufficient for transcriptional activation because the p300ΔE1A protein is almost completely inactive as a coactivator (Fig. 3 to 5). (In addition, the GST-E1A fragment that binds to the CH3 domain inhibits p300 transcriptional activity but does not affect [increase or decrease] its HAT activity [Fig. 6].) It is possible, however, that there is a self-regulatory motif in p300 that limits its AT activity, and some studies suggest that such regulation of p300 AT activity can be mediated by factors that interact with p300 (14, 25).
In addition, it was generally observed that p300 autoacetylation was less affected by the various mutations than p300-mediated acetylation of free or nucleosomal histones. For instance, with the p300MutAT1 protein, there is a significant decrease in the acetylation of free or nucleosomal histones but only a twofold decrease in the autoacetylation activity. It is only possible to speculate on the basis for these results, though it may be relevant that p300 autoacetylation could be an intramolecular event, whereas p300-mediated histone acetylation is an intermolecular reaction.
The SRC/p160-binding region.
The C-terminal portion of p300 contains an extended Q-rich stretch that encompasses a smaller SRC/p160-binding region. The deletion of these motifs (p300ΔQ and p300ΔSRC) had no apparent effect on the AT activities of p300 (Fig. 2). In contrast, the transcriptional studies revealed that the C-terminal region of p300 was important for transcriptional enhancement by ligand-activated ER as well as by NF-κB p65 and Sp1 but was less essential for activation by Gal4-VP16 (Fig. 3 to 5). These results indicate that the function of the SRC/p160-binding region of p300 is distinct from its AT activity, and they are consistent with the model in which the binding of SRC/p160 proteins to the C-terminal region of p300 is important for transcriptional activation by a subset of activators that include ER and NF-κB p65 (27, 34, 36, 44, 63, 65, 69, 71, 79).
Summary and conclusions.
In this study, we carried out a biochemical analysis of distinct activation functions in p300: (i) the CH3 domain/E1A-binding region, which is thought to recruit transcription factors, such as RNA polymerase II complex, TFIIB, or PCAF; (ii) the bromodomain, which may be important for the association of p300 with chromatin; (iii) the AT region, which catalyzes the acetylation of histones and other factors; and (iv) the SRC/p160-binding region at the C terminus. We observed that the CH3 domain/E1A-binding region and the bromodomain are broadly required for p300-mediated transcriptional enhancement, whereas the AT activity and C-terminal region (containing the SRC/p160-binding region and the Q-rich region) are important for p300 coactivator function with some sequence-specific activators but not with others (Fig. 8). In addition, the bromodomain, the CH3 domain/E1A-binding region, and the SRC/p160-binding region each appeared to act independently of the ability of p300 to acetylate free histones (Fig. 2). Significantly, these regions of p300 were found to be important for its ability to enhance the assembly of productive initiation complexes (Fig. 5). Thus, in conclusion, these results support a model in which specific regions of p300 possess distinct activation functions that are differentially required for the assembly of transcription initiation complexes. Interestingly, with ER, there is a strong requirement for the bromodomain, the CH3 domain/E1A-binding region, and the SRC/p160-binding region. In this instance, four distinct regions of p300 each have an essential role in the transcription activation process. These data exemplify a situation in which a network of multiple activation functions is required to achieve gene transcription.
ACKNOWLEDGMENTS
We thank John Lis, Patricia Willy, Steve Nordeen, Dmitry Fyodorov, Mark Levenstein, and Paul Mason for critical reading of the manuscript. We are grateful to John Hiscott for the p65 baculovirus, Marc Montminy for the GST-E1A fusions, Jessica Tyler for advice on the AT assays, Yoshihiro Nakatani for the FLAG-E1A baculovirus, Benita Katzenellenbogen for the 2ERE-pS2-CAT construct, and Y. Cha Henderson, A. Deisseroth, and T. Burke for the hIRF-1 constructs.
This work was supported by a grant from the National Institutes of Health (GM46995) to J.T.K. W.L.K. was supported by a postdoctoral fellowship from the American Cancer Society, California Division, and by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
REFERENCES
- 1.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 2.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 3.Berry M, Nunez A M, Chambon P. Estrogen-responsive element of the human pS2 gene is an imperfectly palindromic sequence. Proc Natl Acad Sci USA. 1989;86:1218–1222. doi: 10.1073/pnas.86.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrow J, Stanton V P, Jr, Andresen J M, Becher R, Behm F G, Chaganti R S, Civin C I, Disteche C, Dube I, Frischauf A M, Horsman D, Mitelman F, Volinia S, Watmore A E, Housman D E. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 5.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 6.Brown A M, Jeltsch J M, Roberts M, Chambon P. Activation of pS2 gene transcription is a primary response to estrogen in the human breast cancer cell line MCF-7. Proc Natl Acad Sci USA. 1984;81:6344–6348. doi: 10.1073/pnas.81.20.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brownell J E, Allis C D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 8.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 9.Bulger M, Kadonaga J T. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol Genet. 1994;5:241–262. [Google Scholar]
- 10.Burke T W, Kadonaga J T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 12.Candau R, Zhou J X, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 14.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 15.Chasman D I, Leatherwood J, Carey M, Ptashne M, Kornberg R D. Activation of yeast polymerase II transcription by herpesvirus VP16 and GAL4 derivatives in vitro. Mol Cell Biol. 1989;9:4746–4749. doi: 10.1128/mcb.9.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 17.Dhalluin C, Carlson J E, Zeng L, He C, Aggarwal A K, Zhou M-M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 18.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 20.Freedman L P. Increasing the complexity of coactivation in nuclear receptor signaling. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 21.Giles R H. Update CBP/p300 transgenic mice. Trends Genet. 1998;14:214. doi: 10.1016/s0168-9525(98)01500-5. [DOI] [PubMed] [Google Scholar]
- 22.Giles R H, Peters D J, Breuning M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 23.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 24.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 25.Hamamori Y, Sartorelli V, Ogryzko V, Puri P L, Wu H Y, Wang J Y, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 26.Hampsey M, Reinberg D. RNA polymerase II as a control panel for multiple coactivator complexes. Curr Opin Genet Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- 27.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haynes S R, Dollard C, Winston F, Beck S, Trowsdale J, Dawid I B. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung H-L, Lau J, Kim A Y, Weiss M J, Blobel G A. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol. 1999;19:3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 31.Jeanmougin F, Wurtz J M, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 32.Jeltsch J M, Roberts M, Schatz C, Garnier J M, Brown A M, Chambon P. Structure of the human oestrogen-responsive gene pS2. Nucleic Acids Res. 1987;15:1401–1414. doi: 10.1093/nar/15.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 34.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamakaka R T, Bulger M, Kadonaga J T. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 36.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 37.Kornberg R D, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 38.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 39.Kraus W L, Kadonaga J T. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kraus W L, Kadonaga J T. Ligand- and cofactor-regulated transcription with chromatin templates. In: Picard D, editor. Nuclear receptors: a practical approach. Oxford, England: Oxford University Press; 1999. pp. 167–189. [Google Scholar]
- 41.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurokawa R, Kalafus D, Ogliastro M-H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 43.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 44.Li H, Chen J D. The receptor-associated coactivator 3 activates transcription through CREB-binding protein recruitment and autoregulation. J Biol Chem. 1998;273:5948–5954. doi: 10.1074/jbc.273.10.5948. [DOI] [PubMed] [Google Scholar]
- 45.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko V V, Nakatani Y, Wolffe A P. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin R, Gewert D, Hiscott J. Differential transcriptional activation in vitro by NF-κB/Rel proteins. J Biol Chem. 1995;270:3123–3131. doi: 10.1074/jbc.270.7.3123. [DOI] [PubMed] [Google Scholar]
- 47.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montano M M, Kraus W L, Katzenellenbogen B S. Identification of a novel transferable cis element in the promoter of an estrogen-responsive gene that modulates sensitivity to hormone and antihormone. Mol Endocrinol. 1997;11:330–341. doi: 10.1210/mend.11.3.9899. [DOI] [PubMed] [Google Scholar]
- 49.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 50.Muraoka M, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Shitara N, Chong J M, Iwama T, Miyaki M. p300 gene alterations in colorectal and gastric carcinomas. Oncogene. 1996;12:1565–1569. [PubMed] [Google Scholar]
- 51.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 52.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 53.Neuwald A F, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 54.O’Connor M J, Zimmermann H, Nielsen S, Bernard H-U, Kouzarides T. Characterization of an E1A-CBP interaction defines a novel transcriptional adapter motif (TRAM) in CBP/p300. J Virol. 1999;73:3574–3581. doi: 10.1128/jvi.73.5.3574-3581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 56.Ornaghi P, Ballario P, Lena A M, Gonzàlez A, Filetici P. The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J Mol Biol. 1999;287:1–7. doi: 10.1006/jmbi.1999.2577. [DOI] [PubMed] [Google Scholar]
- 57.Pazin M J, Kamakaka R T, Kadonaga J T. ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science. 1994;266:2007–2011. doi: 10.1126/science.7801129. [DOI] [PubMed] [Google Scholar]
- 58.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 59.Perlmann T, Evans R M. Nuclear receptors in Sicily: all in the famiglia. Cell. 1997;90:391–397. doi: 10.1016/s0092-8674(00)80498-5. [DOI] [PubMed] [Google Scholar]
- 60.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C, Masuno M, Tommerup N, van Ommen G J, Goodman R H, Peters D J, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional coactivator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 61.Pollard K J, Peterson C L. Chromatin remodeling: a marriage between two families? Bioessays. 1998;20:771–780. doi: 10.1002/(SICI)1521-1878(199809)20:9<771::AID-BIES10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 62.Puri P L, Sartorelli V, Yang X J, Hamamori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 63.Sheppard K A, Phelps K M, Williams A J, Thanos D, Glass C K, Rosenfeld M G, Gerritsen M E, Collins T. Nuclear integration of glucocorticoid receptor and nuclear factor-kappaB signaling by CREB-binding protein and steroid receptor coactivator-1. J Biol Chem. 1998;273:29291–29294. doi: 10.1074/jbc.273.45.29291. [DOI] [PubMed] [Google Scholar]
- 64.Sims S H, Cha Y, Romine M F, Gao P-Q, Gottlieb K, Deisseroth A B. A novel interferon-inducible domain: structural and functional analysis of the human interferon regulatory factor 1 gene promoter. Mol Cell Biol. 1993;13:690–702. doi: 10.1128/mcb.13.1.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith C L, Onate S A, Tsai M J, O’Malley B W. CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sobulo O M, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, Housman D, Doggett N A, Rowley J D, Zeleznik-Le N J. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3) Proc Natl Acad Sci USA. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sterner D E, Grant P A, Roberts S M, Duggan L J, Belotserkovskaya R, Pacella L A, Winston F, Workman J L, Berger S L. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci USA. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 70.Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 71.Voegel J J, Heine M J, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wade P A, Pruss D, Wolffe A P. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 73.Walker P, Germond J E, Brown-Luedi M, Givel F, Wahli W. Sequence homologies in the region preceding the transcription initiation site of the liver estrogen-responsive vitellogenin and apo-VLDLII genes. Nucleic Acids Res. 1984;12:8611–8626. doi: 10.1093/nar/12.22.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang L, Liu L, Berger S L. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 76.Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 77.Xu L, Glass C K, Rosenfeld M G. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 78.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 79.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch’ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]