Abstract
Filamentous fungi are the organisms of choice for most industrial biotechnology. Some species can produce a variety of secondary metabolites and enzymes of commercial interest, and the production of valuable molecules has been enhanced through different molecular tools. Methods for genetic manipulation and transformation have been essential for the optimization of these organisms. The genus Simplicillium has attracted increased attention given several potential biotechnological applications. The Simplicillium genus harbors several entomopathogenic species and some isolates have been explored for bioremediation of heavy metal contaminants. Furthermore, the myriad of secondary metabolites isolated from Simplicillium spp. render these organisms as ideal targets for deep exploration and further biotechnological mining possibilities. However, the lack of molecular tools hampered the exploration of this genus. Thus, an Agrobacterium tumefaciens-mediated transformation method was established for Simplicillium subtropicum, employing the far-red fluorescent protein TURBOFP635/Katushka, as a visual marker, and the selection marker SUR gene, that confers resistance to chlorimuron ethyl. Notably, one round of transformation using the established method yielded almost 400 chlorimuron resistant isolates. Furthermore, these transformants displayed mitotic stability for, at least, five generations. We anticipate that this method can be useful for deep molecular exploration and improvement of strains in the Simplicillium genus.
Keywords: Simplicillium, Simplicillium subtropicum, Agrobacterium tumefaciens, Katushka, Agrobacterium tumefaciens-mediated transformation
Introduction
The family Cordycipitaceae harbors filamentous fungi of commercial and scientific interest. The most recognizable member of this family, the Beauveria genus, embraces entomopathogenic species (Zimmermann, 2007). Several commercial formulations of Beauveria bassiana are available worldwide for agricultural pests biological control (Faria and Wraight, 2007). Similarly, species from the Cordyceps genus are retailed, given several potential health benefits (Tuli et al., 2014). Furthermore, some species from this family can be mycoparasitic (Kepler et al., 2017). For instance, the dry bubble disease caused by Lecanicillium fungicola is a persistent problem in the cultivation of Agaricus bisporus (Berendsen et al., 2010), while Akanthomyces psalliotae is an entomopathogenic, mycoparasitic, and nematophagous fungus (Harm et al., 2018).
The earliest diverging lineage in the family Cordycipitaceae is the Simplicillium genus (Kepler et al., 2017). This genus has attracted interest for several potential applications, ranging from biological control to remediation by toxic metal accumulation. Some mycoparasitic Simplicillium species can be employed for biological control of oomycetes and fungi, as Simplicillium lamellicola and Simplicillium lanosoniveum, while the latter species can also be a plant pathogen (Chen et al., 2008; Ward et al., 2012; Shin et al., 2017). Simplicillium chinense has been explored for biosorption of cadmium and lead (Jin et al., 2019; Jin et al., 2020). Additionally, this species can ameliorate the phytoremediation performance of the water-reed Phragmites communis (Jin et al., 2019).
Several secondary metabolites have been isolated from Simplicillium spp. Simplicilliumtides, from Simplicillium obclavatum, possess a broad range of biological activities, including antibacterial, antifungal, antiviral, antifouling, cytotoxic, as well as acetylcholinesterase inhibitory activity (Youssef et al., 2019). Aogacillins A and B, produced by Simplicillium sp. FKI-5985, circumvent arbekacin resistance in methicillin-resistant Staphylococcus aureus (Takata et al., 2013). Verlamelins A and B and sinulariapeptide A, isolated from a soft coral-associated Simplicillium sp., showed antifungal activity against Pyricularia oryzae and Colletotrichum asianum, respectively (Dai et al., 2018). Simpotentin, isolated from the culture broth of Simplicillium minatense, has been described as a potentiator of amphotericin B activity against Candida albicans and Cryptococcus neoformans (Uchida et al., 2019). Furthermore, several compounds isolated from Simplicillium lanosoniveum have shown antibacterial, antifungal and phosphodiesterase 5 inhibitory activity (Rukachaisirikul et al., 2019).
Contrasting with the biotechnological potential of the Simplicillium genus, there is no method for genetic modification of these species. Thus, to address this obstacle, a highly efficient Agrobacterium tumefaciens-mediated transformation (ATMT) method was standardized for Simplicillium subtropicum, employing, as an in vivo tag, the reporter far-red fluorescent protein TURBOFP635/ Katushka (Kat) (Shcherbo et al., 2007), and, as a selection marker, the SUR gene.
Material and Methods
Strains and culture media
S. subtropicum strain IBCB 79 was originally isolated from a dead Leptopharsa heveae collected in Mato Grosso, Brazil. This strain is deposited in the fungal collection “Coleção de Culturas de Entomopatógenos Oldemar Cardim Abreu” located in São Paulo, Brazil. Before subsequent experiments, this strain was grown at 28 °C in solid Cove’s Complete Medium (MCc), as previously described (Sbaraini et al., 2019). Maintenance of transformants were in solid Cove’s Medium (MC), as described by Sbaraini and coworkers (2019), and 10 μg/mL of Chlorimuron Ethyl (CE) was employed. Escherichia coli TG2 was employed in routine cloning, and Agrobacterium tumefaciens strains EHA105 and LBA1100 were employed to perform the ATMT of S. subtropicum strain IBCB 79. Bacteria were obtained from the laboratory’s collection and maintained in Luria-Bertani (LB) medium with the appropriate antibiotics (Green and Sambrook, 2012).
Species identification and phylogeny
To ensure that strain IBCB 79 is a member of the Simplicillium genus, the internal transcribed spacer (ITS) region was sequenced and analyzed. DNA was extracted employing the standard phenol/chloroform method (Green and Sambrook, 2012). PCR reaction was performed following the standard protocols. DNA sequencing was performed by ACTGene Análises Moleculares (Brazil, RS) employing Sanger sequencing (Applied Biosystems, AB3500). The sequence (MT822178) was amended together with other sequences employed in the proposed Simplicillium genus tree described previously (Crous et al., 2018). The DNA barcode sequences were subjected to alignment reliability analyses using GUIDANCE 2.0 (alignment is shown in Supplementary Data S1), using the PRANK algorithm for sequence alignment with 100 bootstrap replicates and variable gap penalties (Loytynoja and Goldman, 2010; Sela et al., 2015). Additionally, a GUIDANCE 2.0 score cutoff of 0.93 for site removal was employed (Sela et al., 2015). The phylogenetic reconstruction was conducted with PhyML 3.1 (Maximum Likelihood) with aLRT SH-like (approximate likelihood ratio test Shimodaira-Hasegawa) branch support estimation (Guindon et al., 2010), employing GTR+I+G as the evolutionary model.
Katushka reporter plasmid construction
For expression of the KAT gene, first, the Magnaporthe grisea acetolactate synthase enconding gene (SUR), which confers resistance to CE (Lin et al., 2011), was PCR-amplified with primers pPZP_EcoRV_SUR_F and pPZP_EcoRV_SUR_R (Table S1) and introduced in the EcoRV site of the binary vector pPZP201BK (Walton et al., 2005), to generate the plasmid pPZP201BK::SUR. The isolation of the SUR gene (sulfonylurea resistance allele M. grisea ILV1) has been described previously (Sweigard et al., 1997). The SUR gene, along with its native promoter and terminator, was obtained from plasmid pCB1532 (kindly provided by Aline S. Romão-Dumaresq and Nicholas Talbot). The Kat coding sequence was PCR amplified with primers gpdA_CDSKat_F and TtrpC_CDSKat_Rfrom the plasmid pJAF15::H3P::Kat::H3T (i. e., the gene is under control of the Histone 3 promoter and terminator of Cryptococcus neoformans; the plasmid was kindly provided by Marilene Henning Vainstein) and cloned in the NcoI and BamHI sites of the plasmid pAN::gpdA::BAR::TrpC, to generate the plasmid pAN::gpdA::Kat::TrpC (i. e., NcoI and BamHI digestion released the BAR gene) (Haleva et al., 2020). The Kat gene expression cassette, gpdA::Kat::TrpC was PCR amplified with primers pPZP_HindIII_gpdA_Kat_F and pPZP_HindIII_trpC_Kat_R, and cloned in the EcoRV site of the plasmid pPZP201BK::SUR to generate the plasmid pPZP201BK::SUR::gpdA::Kat::TrpC (primers, plasmid map, and plasmid sequence were included as Table S1, Figure S1, and Supplementary Data S2, respectively). All cloning steps were performed employing the Hot fusion protocol (Fu et al., 2015).
Agrobacterium tumefaciens-mediated transformation
The protocol used for the transformation of S. subtropicum was similar to the ATMT method used for Aspergillus awamori (Michielse et al., 2008), and Paracoccidioides brasiliensis (Almeida et al., 2007; Menino et al., 2012). Furthermore, these employed protocols were based on the work of Bundock et al. (1995), and de Groot et al. (1998). In brief, cultures of A. tumefaciens EHA105 or LBA1100 (carrying the Katushka plasmid), which were overnight grown in LB broth supplemented with antibiotics, were inoculated in 10 mL of freshly prepared induction medium (IM) supplemented with 400 μM acetosyringone (AS) and antibiotics. The cells were grown in IM at 28 °C and 180 rpm until reaching OD600 nm of 0.8-0.9. Concomitantly, five days old MCc plates presenting S. subtropicum growth were used to prepare a fresh spore suspension. The plates were washed with Tween 80 0.01 % (w/v) solution, the spores recovered and washed two times with liquid IM without antibiotics or AS. Spore counting was performed employing a hemocytometer and spore concentration was adjusted to the desired concentration (1 x 106, 1 x 107, or 1 x 108 spores/mL) with liquid IM without antibiotics or AS. Subsequently, one hundred μL of the A. tumefaciens resulting growth was mixed with 50 μL of the S. subtropicum spore suspension. The A.tumefaciens-S. subtropicum mixture was kindly homogenized and pipetted over 0.45 μm Hybond N+ filter membranes disposed over solid IM supplemented with 400 μM AS and antibiotics. The mixture was left evaporating for 30 min in the dark before co-cultivation at 24 °C for 24, 48, and 72 h. After co-cultivation, the resulting growth (over the filter membranes), was scrapped in liquid MC supplemented with 200 µg/mL of cefotaxime and 10 µg/mL of CE and spread over MC plates supplemented with 200 µg/mL of cefotaxime and 10 µg/mL of CE. The plates were incubated at 28 °C until the transformants emergence. Potential transformants usually start to appear after 2 days. All transformations were performed in triplicates in two independent experiments. Emerging transformants were transferred to a new MC plate supplemented with 10 µg/mL CE for additional selection. Subsequently, selected transformants were cultured in MCc medium without the selection agent for five generations to evaluate mitotic stability.
Screening of the transformants by fluorescent imaging
The first screening for Kat expression was carried out by qualitative detection of the far-red fluorescence using the Living Image 3.1 software in IVIS Lumina II (PerkinElmer). The parameters were set to 60 second exposure time, excitation at 535 nm and 465 nm (for background removal), using the dsRed emission filter. Not only transformant fungal colonies were inspected, as we also checked A. tumefaciens EHA105 harboring the plasmid pPZP201BK::SUR::gpdA::Kat::TrpC (i.e., the observed fluorescence could come from a bacterial contaminant rather than the selected mutants). Furthermore, intracellular expression of Kat was evaluated using the FLoid Cell Imaging Station (Thermo Fisher Scientific) with the red filter parameters (excitation: 586 nm; emission: 646 nm). Moreover, for detailed microscopic visualization of the Kat-fluorescence, the FLoid Cell Imaging Station was also employed. In this assay, wild-type and mutant strains were microcultured in MCc (28 °C, 4 days) and posteriorly inspected.
Genomic DNA of potential transformants was extracted employing the phenol: chloroform method (Green and Sambrook, 2012). PCR to amplify SUR and KAT were also performed to evaluate the mutants (Primer sequences are described in Table S1). Subsequently, three selected mutants and the wild-type strain were evaluated by Southern blotting (Alkphos Direct Labeling and Detection System, GE Healthcare), to confirm the insertion of the Kat expression cassette and potentially evaluate the number of insertions in the genome. Genomic DNA was digested with StuI and further hybridized with a KAT CDS probe.
Statistics
Statistical analyses were conducted with GraphPad Prism 6 (GraphPad software). Transformation efficiencies were plotted in number of isolated transformants. Effective transformation regimes were determined by one-way ANOVA with posthoc Tukey’s test (p < 0.01). The letters above bars indicate the statistical difference between transformation regimes.
Results
S. subtropicum phylogeny
To characterize the genetic identity of strain IBCB 79, ITS sequencing and phylogeny were employed. This step was performed due to the inherent difficulty to distinguish Simplicillium isolates from Sporothrix insectorum isolates. The ITS sequence was compared with other Simplicillium sequences used to describe Simplicillium filiforme, a new endophytic species, recently described (Crous et al., 2018). The inferred phylogenetic tree supports the inclusion of the strain IBCB 79 as S. subtropicum (Figure 1). Originally isolated from soil in Japan (Nonaka et al., 2013), S. subtropicum has been explored for bioremediation of copper polluted areas, displaying the best results among few selected species (Ong et al., 2017).
Figure 1 -. Simplicillium subtropicum species phylogenetic tree. A species tree based on ITS sequences was constructed to establish the relationships between the Simplicillium species with available ITS sequences and strain IBCB 79. Bionectria vesiculosa, Trichoderma atroviride, and Verticillium chlamydosporium entries were employed as outgroup and the tree was rooted in these species. Phylogenetic analysis was performed using Maximum-likelihood. Branch support values (aLRT SH-like supports) are associated with nodes. Polytomies were included when branch support values were lower than 0.80.
Agrobacterium tumefaciens-mediated transformation of S. subtropicum
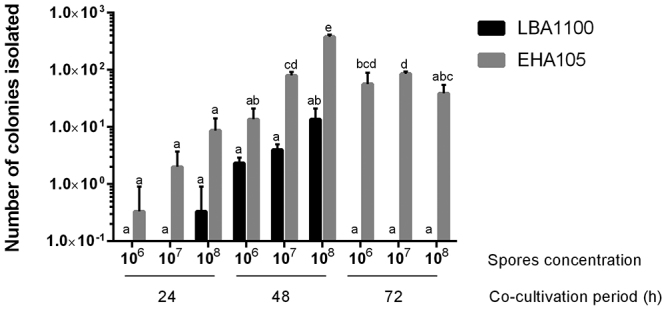
For several filamentous fungi (e.g., A. awamori and Paracoccidioides spp.) the strain LBA1100 of A. tumefaciens is the most used for ATMT (Michielse et al., 2008; Fernandes et al., 2017), while, for species from Hypocreales order, the strains EHA105 or AGL1 are usually employed (Staats et al., 2007; Padilla-Guerrero and Bidochka, 2017). Thus, the A. tumefaciens strains EHA105 and LBA1100 were evaluated for S. subtropicum transformation. Furthermore, the effect of different spore concentrations, during co-cultivation, was also assessed. Finally, different co-cultivation periods (24, 48, and 72 h) were also evaluated. The best results were obtained employing A. tumefaciens strain EHA105 and 1 x 108 spores/mL of S. subtropicum during 48 h of co-cultivation (Figure 2). Almost 400 transformants were obtained employing 1 x 108 spores/mL, although, it is important to notice, that 1 x 107 spores/mL can be more suitable for routine experiments since fewer spores are needed and transformants are easily isolated. A similar number of transformants were obtained employing 1 x 107 spores/mL of S. subtropicum with 48 and 72 h of co-cultivation (Figure 2) Furthermore, the results discourage the use of A. tumefaciens strain LBA1100 for the transformation of S. subtropicum. Notably, transformants were not obtained when A. tumefaciens strain LBA1100 was co-cultured with S. subtropicum for 72 h (Figure 2).
Figure 2 -. Transformation efficiency. To determine the most suitable transformation protocol for S. subtropicum, different strains of A. tumefaciens were evaluated (EHA105 and LBA1100), as well as different spore concentrations (1 x 106, 1 x 107, or 1 x 108 spores/mL) and co-cultivation times (24, 48, and 72 h). The different letters above bars indicate statistical differences between transformation regimes according to one-way ANOVA analysis followed by posthoc Tukey’s test (p < 0.01).
Kat-fluorescence detection and mitotic stability analysis
Transformants from the ATMT experiments, selected based on CE resistance, were first evaluated for far-red Kat-fluorescence using the Living Image 3.1 software implemented in the IVIS Lumina II equipment followed by intracellular analysis of Kat expression with the FLoid Cell Imaging Station. As expected, given the potential scattered insertion of the Kat expression cassette in the genome of the recipients, different mutants displayed different levels of fluorescence. Of 69 mutants evaluated for far-red Kat-fluorescence, 58 exhibited high levels of positive fluorescence (Figure 3).
Figure 3 -. Macroscopic evaluation of far-red Kat-fluorescence. Fifty-eight of sixty-nine mutants evaluated displayed the far-red Kat-fluorescence (84 %). The parameters on Living Image 3.1 (implemented in the IVIS Lumina II equipment) were set to 60 second exposure time, excitation at 535 nm and 465 nm (for background removal), using the dsRed emission filter. Left Panel, White Light. Right Panel, dsRed emission filter.
To rule out that the observed fluorescence could be due to the presence of residual A. tumefaciens cells, we also evaluated the generated mutants by fluorescence microscopy analysis. As expected, due to the use of a strong and constitutive promoter, the Kat fluorescence was detected in both mycelia and spores of a representative transformant (Figure 4). Noteworthy, while we could detect fluorescence in fungal cells, the bacterial strain harboring the plasmid pPZP201BK::SUR::gpdA::Kat::TrpC has no clear phenotype (Figure S2). Therefore, besides showing CE resistance, several transformants also carry the active Kat expression cassette, reiterating the success of the developed ATMT method.
Figure 4 -. Microscopic evaluation of far-red Kat-fluorescence. Isolated mutants were evaluated for Kat-fluorescence employing the FLoid Cell Imaging Station. A) S. subtropicum wild-type strain relief phase. B) Wild-type red fluorescence. C) S. subtropicum mutant strain, gpdA-Kat-Sur (+), relief phase. D) gpdA-Kat-Sur (+) red fluorescence. Scale bar, 100 μm, for all images.

Six mutants, which displayed the brightest far-red Kat-fluorescence, were chosen to assess mitotic stability. These mutants were cultivated in MCc medium without selection of CE during five generations. All mutants maintained the fluorescence even in MCc (data not shown). Moreover, the DNA of these mutants was extracted, and PCR was employed to evaluate the presence of the SUR gene and the Kat expression cassette (Figure 5A). All mutants presented amplification for the SUR gene (~ 2800 bp; Figure 5B). Additionally, three selected mutants were also PCR-assayed for the KAT CDS (Figure 5C), and the same mutants were inspected by Southern blotting (Figure 5D). Therefore, even with the brightest far-red Kat-fluorescence (i. e., pointing for putative stronger expression/ higher protein content) the insertions in the S. subtropicum genome were stable for at least five generations. Notably, the mutants that displayed the brightest far-red Kat-fluorescence presented at least two insertions of the Kat expression cassette in the genome (Figure 5D). The stability of the construct, together with the usefulness of the macro- and micro-visualization of the Kat fluorescence (Figures 3 and 4, respectively) can be valuable for genetic studies in Simplicillium.
Figure 5 -. Evaluation of the SUR gene and Kat expression cassette stability in the S. subtropicum mutants. Selected S. subtropicum mutants with the brightest far-red Kat-fluorescence were cultivated for five generations in M Cc without CE to evaluate the mitotic stability. A) The Kat gene expression cassette (gpdA::Kat::TrpC) was cloned next to the SUR expression cassette (as displayed) in the plasmid pPZP201BK::SUR, to generate the plasmid pPZP201BK::SUR::gpdA::Kat::TrpC. B) The PCR results amplifying the SUR gene (~2800 bp). C) The PCR results amplifying the KAT CDS (~700 bp). D) The Southern blotting results employing the KAT CDS as a probe. M - DNA Ladder; N - Negative control (without DNA); + - pPZP201BK::SUR::gpdA::Kat::TrpC plasmid; WT - S. subtropicum wild-type strain; gpdA-Kat-Sur (+) - S. subtropicum mutant strains harboring the Kat gene expression cassette and the SUR gene integrated into the genome.

Discussion
Methods for the transformation of fungi are basic to understand molecular aspects of these species. In addition, the impact of genetic manipulation has revolutionized modern biotechnology, and filamentous fungi are a well-established and important source of enzymes and bioactive molecules (Idnurm and Meyer, 2014; Khan et al., 2016). The implementation of A. tumefaciens as a tool remodeled several approaches to discover and understand gene function in many fungal species (Idnurm et al., 2017). The Simplicillium genus has drawn increase scientific interest and a method for genetic transformation was still absent.
Different strategies have been used for the transformation of filamentous fungi (Ruiz-Díez, 2002; Li et al., 2017). These methods range from shock-wave-mediated transformation to protoplast-mediated transformation and ATMT (Li et al., 2017). Although laborious, the ATMT method has been explored in several organisms, being, usually, the first method standardized for fungi that lack established genetic transformation strategies (Idnurm et al., 2017). Although there are several ATMT protocols established, the co-cultivation of A. tumefaciens-fungus is a central step. Small variations in A. tumefaciens strains, membranes employed/solid support, as well as mutant plating and selection, are crucial for transformation success/failure and efficiency. The robust protocol developed for ATMT of A. awamori has been previously adapted for hard-to-transform fungi (Michielse et al., 2008), as Paracoccidioides spp. (Almeida et al., 2007; Menino et al., 2012; Bailão et al., 2014; Fernandes et al., 2017; Nora et al., 2019; Silva et al., 2020), and, for that reason, this method was chosen.
As entomopathogenic species, future studies in Simplicillium spp. can focus on the heterologous expression of toxins and virulence determinants. This approach has been successfully implemented in Metarhizium spp. (Wang and St Leger, 2007; Bilgo et al., 2017). Recently, a semifield trial of a transgenic Metarhizium pingshaense expression insect-specific toxins has shown high efficiency (Lovett et al., 2019). Similarly, genetic engineering can improve the biosorption capacity of Simplicillium spp. The expression of cell wall metal-binding chimeric ligands increased Cd2+ and Zn2+ recovery in Saccharomyces cerevisiae (Vinopal et al., 2007). Furthermore, T-DNA libraries can be useful for the characterization of S. subtropicum genes of overall importance, enrolled in heavy metal tolerance/assimilation and virulence determinants (Zhao et al., 2014).
A diverse array of secondary metabolites has been isolated from Simplicillium species. In recent years, genome mining of secondary metabolite biosynthetic gene clusters (BGCs), coupled with knockout strains, overexpression of transcription factors, and heterologous expression of BGCs have been largely employed for the discovery of biosynthetic pathways (Gilchrist et al., 2018). Although genomes from Simplicillium spp. are not yet available, that should be one of the main goals going forward. Besides, secondary metabolites, the genome sequencing of these species, coupled with the standardized ATMT method, can reveal important aspects of the life and infection cycle of fungi from the Simplicillium genus.
Acknowledgements
This study is part of the Advanced Network of Computational Biology (RABICÓ). The manuscript has been read and approved by all named authors. The authors would like to thank to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for financial and scholarship support.
Supplementary material
References
- Almeida AJ, Carmona JA, Cunha C, Carvalho A, Rappleye CA, Goldman WE, Hooykaas PJ, Leão C, Ludovico P, Rodrigues F. Towards a molecular genetic system for the pathogenic fungus Paracoccidioides brasiliensis. Fungal Genet Biol. 2007;44:1387–1398. doi: 10.1016/j.fgb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Bailão EFLC, Parente JA, Pigosso LL, de Castro KP, Fonseca FL, Silva-Bailão MG, Báo SN, Bailão AM, Rodrigues ML, Hernandez O, et al. Hemoglobin uptake by Paracoccidioides spp. is receptor-mediated. PLoS Negl Trop Dis. 2014;8:e2856. doi: 10.1371/journal.pntd.0002856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen RL, Baars JJP, Kalkhove SIC, Lugones LG, Wosten HAB, Bakker PAHM. Lecanicillium fungicola: causal agent of dry bubble disease in white-button mushroom. Mol Plant Pathol. 2010;11:585–595. doi: 10.1111/j.1364-3703.2010.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgo E, Lovett B, Fang W, Bende N, King GF, Diabate A, St. Leger RJ. Improved efficacy of an arthropod toxin expressing fungus against insecticide-resistant malaria-vector mosquitoes. Sci Rep. 2017;7:3433. doi: 10.1038/s41598-017-03399-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas PJ. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995;14:3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R-S, Huang C-C, Li J-C, Tsay J-G. First report of Simplicillium lanosoniveum causing brown spot on Salvinia auriculata and S. molesta in Taiwan. Plant Dis. 2008;92:1589. doi: 10.1094/PDIS-92-11-1589C. [DOI] [PubMed] [Google Scholar]
- Crous PW, Luangsa-Ard JJ, Wingfield MJ, Carnegie AJ, Hernández-Restrepo M, Lombard L, Roux J, Barreto RW, Baseia IG, Cano-Lira JF, et al. Fungal Planet description sheets: 785-867. Persoonia. 2018;41:238–417. doi: 10.3767/persoonia.2018.41.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Lin Y, Pang X, Luo X, Salendra L, Wang J, Zhou X, Lu Y, Yang B, Liu Y. Peptides from the Soft Coral-associated fungus Simplicillium sp. SCSIO41209. Phytochemistry. 2018;154:56–62. doi: 10.1016/j.phytochem.2018.06.014. [DOI] [PubMed] [Google Scholar]
- de Groot MJ, Bundock P, Hooykaas PJ, Beijersbergen AG. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16:839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- Faria MR de, Wraight SP. Mycoinsecticides and Mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biol Control. 2007;43:237–256. [Google Scholar]
- Fernandes FF, Oliveira AF, Landgraf TN, Cunha C, Carvalho A, Vendruscolo PE, Gonçales RA, Almeida F, da Silva TA, Rodrigues F, et al. Impact of Paracoccin gene silencing on Paracoccidioides brasiliensis virulence. MBio. 2017;8:e00537-17. doi: 10.1128/mBio.00537-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Donovan WP, Shikapwashya-Hasser O, Ye X, Cole RH. Hot fusion: An efficient method to clone multiple DNA fragments as well as inverted repeats without ligase. PLoS One. 2015;9:e115318. doi: 10.1371/journal.pone.0115318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist CLM, Li H, Chooi Y-H. Panning for gold in mould: can we increase the odds for fungal genome mining? Org Biomol Chem. 2018;16:1620–1626. doi: 10.1039/c7ob03127k. [DOI] [PubMed] [Google Scholar]
- Green M, Sambrook J. Molecular Cloning: A Laboratory Manual. 4. Cold Spring Harbour, Spring Harbor Laboratory Press; New York: 2012. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Haleva L, Lopes W, Barcellos VA, Schrank A, Vainstein MH. The contest of microbial pigeon neighbors: Interspecies competition between Serratia marcescens and the human pathogen Cryptococcus neoformans. Fungal Biol. 2020;124:629–638. doi: 10.1016/j.funbio.2020.03.004. [DOI] [PubMed] [Google Scholar]
- Harm GFS, Papanicolaou A, Cuddy WS, Park RF, Moffitt MC. Draft genome sequence of the fungus Lecanicillium psalliotae strain HWLR35, isolated from a wheat leaf infected with leaf rust (Caused by Puccinia triticina) Genome Announc. 2018;6:e01442-17. doi: 10.1128/genomeA.01442-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Bailey AM, Cairns TC, Elliott CE, Foster GD, Ianiri G, Jeon J. A silver bullet in a golden age of functional genomics: the impact of Agrobacterium-mediated transformation of fungi. Fungal Biol Biotechnol. 2017;4:6. doi: 10.1186/s40694-017-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Meyer V. Welcome to fungal biology and biotechnology. Fungal Biol Biotechnol. 2014;1:8. doi: 10.1186/s40694-014-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Deng S, Wen Y, Jin Y, Pan L, Zhang Y, Black T, Jones KC, Zhang H, Zhang D. Application of Simplicillium chinense for Cd and Pb biosorption and enhancing heavy metal phytoremediation of soils. Sci Total Environ. 2019;697:134148. doi: 10.1016/j.scitotenv.2019.134148. [DOI] [PubMed] [Google Scholar]
- Jin Z, Xie L, Zhang T, Liu L, Black T, Jones KC, Zhang H, Wang X, Jin N, Zhang D. Interrogating cadmium and lead biosorption mechanisms by Simplicillium chinense via infrared spectroscopy. Environ Pollut. 2020;263:114419. doi: 10.1016/j.envpol.2020.114419. [DOI] [PubMed] [Google Scholar]
- Kepler RM, Luangsa-Ard JJ, Hywel-Jones NL, Quandt CA, Sung G-H, Rehner SA, Aime MC, Henkel TW, Sanjuan T, Zare R, et al. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales) IMA Fungus. 2017;8:335–353. doi: 10.5598/imafungus.2017.08.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Ullah MW, Siddique R, Nabi G, Manan S, Yousaf M, Hou H. Role of recombinant DNA technology to improve life. Int J Genomics. 2016;2016:2405954. doi: 10.1155/2016/2405954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Tang Y, Lin J, Cai W. Methods for genetic transformation of filamentous fungi. Microb Cell Fact. 2017;16:168. doi: 10.1186/s12934-017-0785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Wang F, Wei D. Chlorimuron ethyl as a new selectable marker for disrupting genes in the insect-pathogenic fungus Metarhizium robertsii. J Microbiol Methods. 2011;87:241–243. doi: 10.1016/j.mimet.2011.07.018. [DOI] [PubMed] [Google Scholar]
- Lovett B, Bilgo E, Millogo SA, Ouattarra AK, Sare I, Gnambani EJ, Dabire RK, Diabate A, St. Leger RJ. Transgenic Metarhizium rapidly kills mosquitoes in a malaria-endemic region of Burkina Faso. Science. 2019;364:894–897. doi: 10.1126/science.aaw8737. [DOI] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics. 2010;11:6. doi: 10.1186/1471-2105-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menino JF, Almeida AJ, Rodrigues F. Gene knockdown in Paracoccidioides brasiliensis using antisense RNA. Methods Mol Biol. 2012;845:187–198. doi: 10.1007/978-1-61779-539-8_12. [DOI] [PubMed] [Google Scholar]
- Michielse CB, Hooykaas PJJ, van den Hondel CAMJJ, Ram AFJ. Agrobacterium-mediated transformation of the filamentous fungus Aspergillus awamori. Nat Protoc. 2008;3:1671–1678. doi: 10.1038/nprot.2008.154. [DOI] [PubMed] [Google Scholar]
- Nonaka K, Kaifuchi S, Ōmura S, Masuma R. Five new Simplicillium species (Cordycipitaceae) from soils in Tokyo, Japan. Mycoscience. 2013;54:42–53. [Google Scholar]
- Nora LC, Gonçales RA, Martins-Santana L, Ferreira BH, Rodrigues F, Silva-Rocha R. Synthetic and minimalist vectors for Agrobacterium tumefaciens-mediated transformation of fungi. Genet Mol Biol. 2019;42:395–398. doi: 10.1590/1678-4685-GMB-2018-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong GH, Ho XH, Shamkeeva S, Manasha Savithri Fernando AS, Wong LS. Biosorption study of potential fungi for copper remediation from Peninsular Malaysia. Remediat J. 2017;27:59–63. [Google Scholar]
- Padilla-Guerrero IE, Bidochka MJ. Agrobacterium-Mediated Co-transformation of multiple genes in Metarhizium robertsii. Mycobiology. 2017;45:84–89. doi: 10.5941/MYCO.2017.45.2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Díez B. Strategies for the transformation of filamentous fungi. J Appl Microbiol. 2002;92:189–195. doi: 10.1046/j.1365-2672.2002.01516.x. [DOI] [PubMed] [Google Scholar]
- Rukachaisirikul V, Chinpha S, Saetang P, Phongpaichit S, Jungsuttiwong S, Hadsadee S, Sakayaroj J, Preedanon S, Temkitthawon P, Ingkaninan K. Depsidones and a dihydroxanthenone from the endophytic fungi Simplicillium lanosoniveum (J.F.H. Beyma) Zare & W. Gams PSU-H168 and PSU-H261. Fitoterapia. 2019;138:104286. doi: 10.1016/j.fitote.2019.104286. [DOI] [PubMed] [Google Scholar]
- Sbaraini N, Bellini R, Penteriche AB, Guedes RLM, Garcia AWA, Gerber AL, Vainstein MH, de Vasconcelos ATR, Schrank A, Staats CC. Genome-wide DNA methylation analysis of Metarhizium anisopliae during tick mimicked infection condition. BMC Genomics. 2019;20:836. doi: 10.1186/s12864-019-6220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 2015;43:w7–w14. doi: 10.1093/nar/gkv318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, et al. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- Shin TS, Yu NH, Lee J, Choi GJ, Kim J-C, Shin CS. Development of a biofungicide using a mycoparasitic fungus Simplicillium lamellicola BCP and its control efficacy against gray mold diseases of tomato and ginseng. Plant Pathol J. 2017;33:337–344. doi: 10.5423/PPJ.FT.04.2017.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MG, de Curcio JS, Silva-Bailão MG, Lima RM, Tomazett MV, de Souza AF, Cruz-Leite VRM, Sbaraini N, Bailão AM, Rodrigues F, et al. Molecular characterization of siderophore biosynthesis in Paracoccidioides brasiliensis. IMA Fungus. 2020;11:11. doi: 10.1186/s43008-020-00035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staats CC, Junges A, Fitarelli M, Furlaneto MC, Vainstein MH, Schrank A. Gene inactivation mediated by Agrobacterium tumefaciens in the filamentous fungi Metarhizium anisopliae. Appl Microbiol Biotechnol. 2007;76:945–950. doi: 10.1007/s00253-007-1043-4. [DOI] [PubMed] [Google Scholar]
- Sweigard JA, Chumley F, Carroll A, Farrall L, Valent B. A series of vectors for fungal transformation. Fungal Genet Rep. 1997;44:52–53. [Google Scholar]
- Takata K, Iwatsuki M, Yamamoto T, Shirahata T, Nonaka K, Masuma R, Hayakawa Y, Hanaki H, Kobayashi Y, Petersson GA, et al. Aogacillins A and B produced by Simplicillium sp. FKI-5985: new circumventors of arbekacin resistance in MRSA. Org Lett. 2013;15:4678–4681. doi: 10.1021/ol401975z. [DOI] [PubMed] [Google Scholar]
- Tuli HS, Sandhu SS, Sharma AK. Pharmacological and therapeutic potential of Cordyceps with special reference to Cordycepin. 3 Biotech. 2014;4:1–12. doi: 10.1007/s13205-013-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida R, Kondo A, Yagi A, Nonaka K, Masuma R, Kobayashi K, Tomoda H. Simpotentin, a new potentiator of amphotericin B activity against Candida albicans, produced by Simplicillium minatense FKI-4981. J Antibiot (Tokyo) 2019;72:134–140. doi: 10.1038/s41429-018-0128-x. [DOI] [PubMed] [Google Scholar]
- Vinopal S, Ruml T, Kotrba P. Biosorption of Cd2+ and Zn2+ by cell surface-engineered Saccharomyces cerevisiae. Int Biodeterior Biodegradation. 2007;60:96–102. [Google Scholar]
- Walton FJ, Idnurm A, Heitman J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol. 2005;57:1381–1396. doi: 10.1111/j.1365-2958.2005.04779.x. [DOI] [PubMed] [Google Scholar]
- Wang CS, St Leger RJ. A scorpion neurotoxin increases the potency of a fungal insecticide. Nat Biotechnol. 2007;25:1455–1456. doi: 10.1038/nbt1357. [DOI] [PubMed] [Google Scholar]
- Ward NA, Robertson CL, Chanda AK, Schneider RW. Effects of Simplicillium lanosoniveum on Phakopsora pachyrhizi, the soybean rust pathogen, and its use as a biological control agent. Phytopathology. 2012;102:749–760. doi: 10.1094/PHYTO-01-11-0031. [DOI] [PubMed] [Google Scholar]
- Youssef FS, Ashour ML, Singab ANB, Wink M. A comprehensive review of bioactive peptides from marine fungi and their biological significance. Mar Drugs. 2019;17(10):559. doi: 10.3390/md17100559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Xu C, Lu H-L, Chen X, St. Leger RJ, Fang W. Host-to-Pathogen gene transfer facilitated infection of insects by a pathogenic fungus. PLoS Pathog. 2014;10:e1004009. doi: 10.1371/journal.ppat.1004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G. Review on safety of the entomopathogenic fungi Beauveria bassiana and Beauveria brongniartii. Biocontrol Sci Technol. 2007;17:553–596. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




