Abstract
The mammalian Y chromosome is critical for male sex determination and spermatogenesis. However, linking each Y gene to specific aspects of male reproduction has been challenging. As the Y chromosome is notoriously hard to sequence and target, functional studies have mostly relied on transgene-rescue approaches using mouse models with large multi-gene deletions. These experimental limitations have oriented the field toward the search for a minimum set of Y genes necessary for male reproduction. Here, considering Y-chromosome evolutionary history and decades of discoveries, we review the current state of research on its function in spermatogenesis and reassess the view that many Y genes are disposable for male reproduction.
Introduction
In therian mammals, with some exceptions, whether an embryo will develop into a male or a female is defined by the presence or absence of a Y chromosome (Capel, 2017). Males bear an X and a Y, whereas females carry two X chromosomes. This is the most fundamental genetic difference between the two sexes and has been the subject of numerous studies.
Historically, the Y chromosome’s biological function has been misunderstood. From the 1950s, it was considered to be a genetic wasteland as studies of human pedigrees were only uncovering traits with autosomal or X-linked inheritance (Stern, 1957). In 1959, it was shown that the male-determining gene is Y-linked, but this was considered an exception on an otherwise functionally inert chromosome (Ford et al., 1959; Jacobs and Strong, 1959). When transcription units were first discovered on the Y chromosome (Agulnik et al., 1994; Arnemann et al., 1991; Page et al., 1987; Reijo et al., 1995; Salido et al., 1992; Sinclair et al., 1990), they were thought to be inactive vestiges of their former autosomal ancestors (Marshall Graves, 1995). More recently, the ‘impending demise’ theory hypothesised a continuous loss of Y-protein-coding genes predicting the eventual loss of the Y chromosome (Aitken and Marshall Graves, 2002; Marshall Graves, 2004). We now know that the view of the Y as a disappearing genetic desert is incorrect. Decades of research have proved that besides controlling male gonadal sex determination, the Y chromosome is crucial for initialisation, maintenance, and completion of spermatogenesis.
In this review, we first describe the evolutionary history of the X-Y chromosome pair, then use it as a paradigm with which to understand how the Y chromosome has become functionally specialised across mammals. Focusing on humans and mice, we discuss the early evidence that the Y chromosome is more than just a sex switch, and the subsequent efforts to discover the Y genes involved in spermatogenesis. We then highlight how experimental limitations have impacted progress in the field and propose approaches to enrich our understanding of Y-chromosome function.
Evolution of the mammalian sex chromosomes
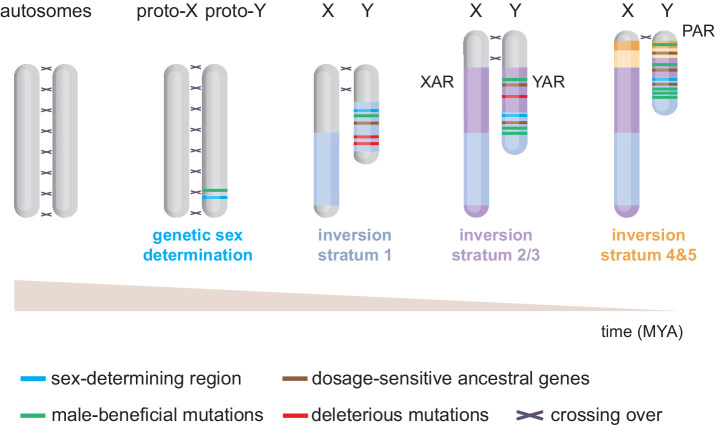
The mammalian heteromorphic X and Y chromosomes have evolved from an ancestral pair of autosomes (Figure 1). This seminal concept, like many principles governing sex chromosome evolution, was hypothesised by Ohno, 1967. Henceforth, breakthroughs in Y-chromosome sequencing and assembly, which were long hindered by the Y’s repeat-rich nature and homology to the X chromosome, have been crucial to understand sex-chromosome evolution (Bellott et al., 2014; Hughes et al., 2012; Hughes et al., 2010; Li et al., 2013; Skaletsky et al., 2003; Soh et al., 2014). We now know that divergence into a ‘proto-Y’ started following mutations of Sox3, creating the sex-determining region Sry gene (Foster and Graves, 1994; Marshall Graves, 1998). The evolution of Sry-specific sex determination is thought to have happened around 160–166 million years ago after the split between therians and monotremes (egg-laying mammals) (Potrzebowski et al., 2008; Veyrunes et al., 2008), and prior to the split between marsupials (metatherians) and eutherians (Luo et al., 2011).
Figure 1. Evolution of the eutherian sex chromosomes.
The X and Y chromosomes evolved from a pair of autosomes. First, the testis-determining gene Sry evolved on the proto-Y. Then, successive stratification events occurred, wherein X-Y meiotic recombination arrests along specific regions of the Y, likely due to inversions. The first stratum (i.e. discrete non-recombining region) includes Sry and originated in the last common therian ancestor ~166 million years ago (MYA). After the split from marsupials, the X/Y-added regions (XAR/YAR) fused from an autosome to the sex chromosomes in the eutherian ancestor. A second stratification event occurred independently in both marsupial and eutherian ancestors ~ 97–117 MYA. Concomitantly, in eutherians, a third stratum was formed encompassing the YAR. A fourth and fifth stratum evolved in the ancestor of old-world monkeys ~ 25–44 MYA. Recombination now only occurs within the pseudoautosomal region (PAR).
The male-specific inheritance of the proto-Y exposed it to unique evolutionary forces (Larson et al., 2018). Notably, it facilitated selection and accumulation of male-beneficial mutations in the vicinity of Sry (Figure 1). The linkage between Sry and male-beneficial loci in turn created selective pressure to supress X-Y crossing over. Eventually, multiple discrete inversions on the Y supported and amplified meiotic-recombination arrest in specific regions, or strata (Figure 1; Bellott et al., 2014; Cortez et al., 2014; Lahn and Page, 1999). Stratification expanded the non-recombining male-specific region at the expense of the recombining portion of the chromosome known as the pseudoautosomal region (PAR) (Figure 1). In addition to these strata, (retro)transpositions/translocations from autosomes have occurred more recently in various mammalian lineages (Bellott et al., 2014; Cortez et al., 2014; Hughes et al., 2012).
Importantly, the loss of recombination associated with stratification allowed accumulation of Y-linked loss-of-function mutations, triggering chromosome-wide gene decay and amplification of repetitive DNA (Bachtrog, 2013). As a mechanism to maintain male fitness, large deletions of deleterious and non-functional DNA regions occurred within each non-recombining stratum (Bachtrog, 2013; Hughes and Page, 2015). Although 98% of the ancestral human X-genes survived (Mueller et al., 2013), only 3% persisted on the human Y (Bellott et al., 2014; Skaletsky et al., 2003). However, although the Y experienced rapid decay early in evolution, degeneration then dramatically slowed down, with genes having survived this ‘purge’ showing stability (Bellott et al., 2014; Cortez et al., 2014; Hughes et al., 2012; Hughes et al., 2010; Hughes et al., 2005). For instance, of the 18 ancestral genes present in the ~97 million year old eutherian ancestor (Bellott et al., 2014), humans retain 14, bull 13, chimp 11, and mouse 9. In humans, no Y genes appear to have been lost in the last 44 million years.
In addition, the longevity of certain X-Y gene pairs has been convergent between eutherians and marsupials. Indeed, one stratum independently evolved in both therian lineages (Figure 1). Two genes on this stratum have convergently survived, suggesting that Y gene decay is not stochastic (Bellott et al., 2014). Altogether, it is likely that X-Y gene-pair retention did not occur at random. Generally, it is thought that two evolutionary strategies have protected a specific set of Y genes: maintenance of ancestral X-Y gene pairs which are sensitive to transcriptional dosage; or retention and amplification of testis-biased gene families.
Evolutionary forces and functional specialisation of the Y chromosome
Whereas X-chromosome content has remained mostly unchanged over evolutionary time, the Y chromosome has experienced extensive genetic decay. As a result, females carry two copies of the numerous X-linked genes, whereas males carry a single copy. This creates an imbalance between X and autosome transcriptional dosage (X:A ratio) in males as well as between sexes (Table 1). This led to the evolution of dosage-compensation mechanisms, as hypothesised by Ohno, 1967. Firstly, twofold X-chromosome upregulation in both males and females re-established X:A equilibrium in males (Table 1; Deng et al., 2014). However, without other mechanisms, the X:A ratio in females would be two. This was prevented by the evolution of X-chromosome inactivation, a process by which one X is transcriptionally silenced in females, bringing the overall X-output level to that of the autosomes (Table 1; Brockdorff et al., 2020; Loda and Heard, 2019; Lyon, 1961).
Table 1. Dosage compensation of the sex chromosomes.
Without dosage compensation, the transcriptional output of each cell is 0.5 for individual chromosomes, and one for chromosome pairs. X-chromosome upregulation (XCU) doubles each X-chromosome’s output (shown in red). X-chromosome inactivation (XCI) silences output of one X in females (shown in blue).
| No compensation | XCU | XCI | |
|---|---|---|---|
| female | AA = 1XX = 1 | AA = 1 XX = 2 |
AA = 1XX = 1 |
| male | AA = 1XY = 0.5 | AA = 1XY = 1 | AA = 1XY = 1 |
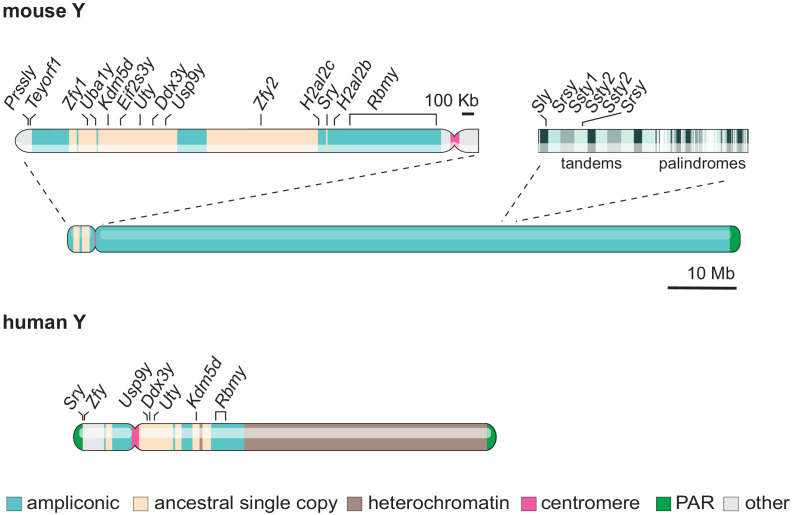
X-upregulation and X-inactivation can ultimately compensate for genes lost on the Y. However, as an X-Y gene pair loses its Y-copy due to decay, it goes through a transition period during which there will be gene-dosage imbalance. During that intermediate state, the pair has a non-functional Y gene and a non-compensated X gene. For haploinsufficient genes, with function sensitive to their expression dosage, this transition state would be disadvantageous and selected against. As such, dosage-sensitive Y genes have been under strong selective pressure to survive. For instance, studies indicate that Y ancestral genes that share ubiquitous housekeeping regulatory functions with their X homologue, which would be very sensitive to change in expression levels, have preferentially survived on the Y (Bellott et al., 2014; Cortez et al., 2014; Jegalian and Page, 1998; Lahn and Page, 1997). Indeed, the annotations of remaining X-Y pairs link them to the sensitive regulation of histone lysine demethylation, stem-cell self-renewal, splicing, translation initiation and deubiquitylation (Bellott et al., 2014; Lahn and Page, 1997). A key prediction is that if an X-Y pair survives and maintains ancestral gene expression and function, the X copy should escape X-inactivation in females to avoid dosage imbalance. Indeed, the study of allele-specific expression in human, mouse and opossum has revealed that a higher proportion of X-linked genes with a surviving Y homologue escape X-inactivation, compared to genes with no Y homologues or with those showing signs of functional differentiation (Bellott et al., 2014; Cortez et al., 2014). The strict dosage requirement of sex-linked genes can be further inferred from individuals with Turner syndrome (XO), who only have a single full X chromosome (Gravholt et al., 2019). Indeed, human X monosomy is associated with poor embryonic viability and individuals surviving into adulthood tend to be mosaic for at least part of the second sex chromosome. This illustrates the severity of X-X or X-Y dosage imbalance. Interestingly, Turner syndrome is less severe in mice, even in fully XO animals (Burgoyne et al., 1983b; Burgoyne et al., 1983a; Burgoyne and Baker, 1981). This could be in part because mice have fewer surviving Y-linked ancestral genes (9 vs 17 in humans) (Figure 2).
Figure 2. Gene content and structure of the mouse and human Y chromosome.
The short arm of the mouse Y chromosome (Yp) has retained ancestral non-ampliconic genes. The mouse Y chromosome long arm (Yq) contains alternating tandem and palindromic repeats of ampliconic core blocks throughout. Ampliconic core blocks encompass rodent-specific gene clusters. The human Y shares seven ancestral protein-coding genes with the murine Y (labelled). Whereas the human Y chromosome is roughly 60 % heterochromatic, the mouse Y chromosome is 99.9% euchromatic (Skaletsky et al., 2003; Soh et al., 2014). The 10 Mb scale bar applies to both mouse and human Y chromosomes.
Another strategy thought to have protected Y-genes from regulatory and structural decay is the specialisation towards functions in male fertility. Indeed, ancestral, single-copy genes have sometimes diverged towards male-specific reproductive functions. The expression of some Y genes became almost exclusively limited to the testis, gradually losing expression in other organs to become testis-biased (Jegalian and Page, 1998; Mitchell, 2000). In parallel, the Y chromosome has gained additional testis-biased genes through (retro)transposition and translocation events from autosomes (Burgoyne, 1998). Importantly, some recently acquired testis-biased X/Y gene homologues have been massively amplified, forming tandem and palindromic repeats (Hughes et al., 2020; Rozen et al., 2003; Skaletsky et al., 2003; Soh et al., 2014; Trombetta and Cruciani, 2017; Figure 2). Following amplification, repeats have been maintained and homogenised through intra-chromosomal recombination (Trombetta and Cruciani, 2017). This recombination within the Y can either remove or fix mutations, beneficial or deleterious, and male-advantageous mutations would have then been selected for over time. Overall, amplification seems to have contributed to the longevity of some testis-biased Y genes and it has occurred independently in many lineages. In mice, bull and likely other mammals, amplification of sex chromosomes is thought to have been driven by the presence of antagonistic meiotic drivers (selfish genes) on the X and Y, each competing for transmission to the next generation (Cocquet et al., 2012; Ellis et al., 2011; Hughes et al., 2020; Soh et al., 2014).
Altogether, evolutionary insights have shown that surviving mammalian Y genes tend to fall into two categories: testis-specialised fertility genes or dosage-sensitive ubiquitous housekeepers (Lahn and Page, 1997). This is unlike mammalian autosomes which harbour genes with highly variable biological functions and expression patterns. However, despite the apparent specialisation of the Y chromosome, its exact somatic and fertility functions remain to be fully dissected. Particularly interesting are the functions of the Y chromosome in the context of spermatogenesis, which will be discussed in the rest of this review.
Mammalian spermatogenesis
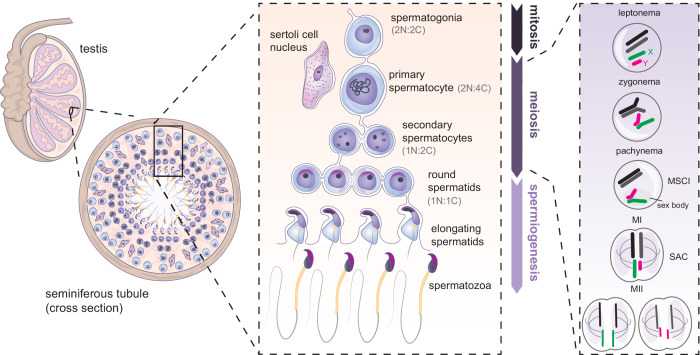
Spermatogenesis allows continuous formation of gametes from spermatogonial stem cells (Figure 3). It occurs in cycles and can broadly be divided in three phases: mitosis, meiosis and spermiogenesis (Figure 3). All stages happen in the epithelium of the seminiferous tubules of the testis. As cells progress through development, they migrate away from the basal seminiferous membrane towards the lumen (Figure 3). The spermatogenic cycle has been mostly studied in rodents but the mechanisms are believed to be well-conserved across mammals. A major difference seems to be in developmental tempo. In mouse, one cycle can be dissected into 12 stages occurring over 8.6 days, and four cycles (35 days) are required to complete spermatogenesis, from spermatogonial stem cells to mature spermatozoa (de Rooij, 2017; Oakberg, 1956a; Oakberg, 1956b). In humans, one spermatogenic cycle is completed every 16 days, and ~72 days are needed for full differentiation of spermatogonial stem cells into spermatozoa (Muciaccia et al., 2013).
Figure 3. Mammalian spermatogenesis in seminiferous tubules of the testis.
Spermatogenesis occurs in three main phases: mitosis, meiosis and spermiogenesis. First, undifferentiated spermatogonial cells undergo multiple mitotic divisions. Subsequently, germ cells commit to meiosis, undergoing meiotic DNA replication (becoming 2 N:4 C) synapsis and recombination between homologues (shown in black and dark gray), and silencing of the X (green) and Y (magenta) chromosomes. Spermatocytes then undergo two rounds of cell division (becoming 1N:1C). The resulting round spermatids later undergo spermiogenesis, further specialising into mature spermatozoa. N = number of chromosomes, C = number of chromatids. SAC: spindle assembly checkpoint.
Early in embryogenesis, at embryonic day (E) 6.25 in mouse and E12 in humans, the primordial germ cells are specified and subsequently migrate to the genital ridge (Kurimoto and Saitou, 2019; Sybirna et al., 2019; Tang et al., 2016). The early bipotent gonad is formed at E10.5–11.5 in mouse and week six in humans. Sry then triggers a signalling cascade resulting in the supporting cell lineage differentiating into Sertoli cells (Capel, 2017; Kurimoto and Saitou, 2019). Sertoli cells then direct adoption of the testicular state within the gonad. Germ cell proliferation will then occur until developmental arrest at E13.5 in mouse and after week 20 in human (Kurimoto and Saitou, 2019; Tam and Snow, 1981).
After birth, at around postnatal day (P) three in mouse and 2 months in humans, germ cells resume cell division, giving rise to undifferentiated spermatogonial cells that are found at the basal compartment of seminiferous tubules. Amongst this cell population lie spermatogonial stem cells that can both self-renew and multiply through mitotic divisions. The spermatogonial differentiation and proliferation through mitosis marks the starting point of spermatogenesis. It is a specific subset of spermatogonial cells, referred as type-B, that are now ready to undergo meiotic DNA replication and differentiate into pre-leptotene spermatocytes (de Rooij, 2017; Wu et al., 2009).
Male meiosis is initiated in a synchronous manner during puberty, at P10 in mouse and ~13 years in humans. Meiosis occurs through two rounds of cell division. During prophase I, which takes 2 weeks in mouse, diploid primary spermatocytes (2 N:4 C) enter leptonema, zygonema, pachynema, and diplonema successively (Figure 3; Baudat et al., 2013; Oakberg, 1956b). At leptonema, meiotic DNA double-strand breaks are formed. At zygonema, homologous chromosomes synapse and form bivalents (Figure 3). This allows crossing over via reciprocal recombination between homologues. Unlike autosomes, the X and Y are transcriptionally silenced at pachynema through the process of meiotic sex chromosome inactivation (MSCI) (Turner, 2015) and compartmentalised into a specialised chromatin domain called the sex body (Figure 3; McKee and Handel, 1993; Solari, 1974). MSCI failure leads to pachytene arrest and apoptosis. MSCI persists throughout diplonema, when homologous chromosomes start to separate (Burgoyne et al., 2009). The molecular mechanisms and roles of MSCI in fertility have been reviewed previously (Turner, 2015).
After prophase I, metaphase I takes place and chromosome homologues are lined up along the metaphase plate. At this stage, the spindle assembly checkpoint (SAC) ensures proper chromosome alignment and spindle attachment (Figure 3; Lane and Kauppi, 2019). Upon satisfaction of the SAC, homologues finally separate to opposite poles of the meiotic cells and the first division occurs with cytokinesis, giving rise to haploid secondary spermatocytes (1 N:2 C). The second meiotic division separates sister chromatids, leading to the formation of round spermatid cells (1 N:1 C) (Figure 3). These haploid cells then undergo a complex post-meiotic differentiation programme called spermiogenesis (Figure 3).
Spermiogenesis is characterised by the emergence of unique sperm-specific morphological features and by dramatic epigenetic reprogramming (Bao and Bedford, 2016; O’Donnell, 2014). Importantly, DNA of differentiating spermatids undergo increasing compaction in the nucleus (O’Donnell, 2014). This is achieved notably through near genome-wide replacement of histones with protamines. Protamines permit an up to 20-fold increase in chromatin condensation (Balhorn, 2007). Structurally, the acrosome starts to form over the anterior part of the spermatid nucleus and the flagellum, the propeller of the sperm, elongates on the posterior side (Figure 3). Additionally, the cytoplasm is transported along the spermatid tail and finally disposed (O’Donnell, 2014). Interestingly, most X/Y-linked genes remain repressed throughout spermiogenesis (Greaves et al., 2006; Namekawa et al., 2006; Sin et al., 2012; Turner et al., 2006), but some are re-activated (Hendriksen, 1999; Hendriksen et al., 1995; Mueller et al., 2013; Mueller et al., 2008; Wang et al., 2005). By the end of this post-meiotic stage, sperm are freed into the lumen of the tubule.
The Y chromosome: more than just a sex switch
The first evidence of the Y chromosome having any biological function arose from studies of mice (Welshons and Russell, 1959) and humans with Turner (XO) or Klinefelter (XXY) syndrome (Ford et al., 1959; Jacobs and Strong, 1959). These reports showed that in eutherian mammals, sex was determined by the presence/absence of the Y, independent of the number of X chromosomes. Later research on sex-reversed men; rare individuals which are phenotypically males but have an XX karyotype, narrowed down the search for the male-determining region to the short arm of the Y chromosome. For instance, using DNA hybridisation with several Y-DNA probes, it was showed that these XX males carried translocated DNA from the Y chromosome on one of their X chromosome (Guellaen et al., 1984; Page, 1986; Page et al., 1987; Page et al., 1985). As additional XX males were studied and the number of markers increased, the candidate region was narrowed to 35 kb containing SRY (Sinclair et al., 1990). In the span of 2 years, the evidence for SRY being the testis-determining factor became undeniable. The mouse orthologue was cloned and found to be expressed in specific somatic cells of the embryonic gonad at the time of testis differentiation (Gubbay et al., 1990; Koopman et al., 1990). It was also shown that both sex-reversed XY mice and women had mutations in Sry (Berta et al., 1990; Jäger et al., 1990; Lovell-Badge and Robertson, 1990). Finally, it was shown that, in mouse, Sry alone is sufficient to induce testicular development (Koopman et al., 1991), as XX mice transgenic for Sry developed as males.
Sry is a transcription factor of the SOX family that encodes a protein with an high mobility group (HMG)-box DNA binding domain (Table 2; Kashimada and Koopman, 2010; Figure 2). Much of what is known about the function of Sry in testis determination result from mouse studies. During murine embryogenesis, Sry upregulates Sox9 in Sertoli cell precursors between E11.5 and E12.5 (Kashimada and Koopman, 2010). SOX9 then activates fibroblast growth factor 9 (Fgf9) in a feedforward loop, which represses Wnt4 and the female pathway that would otherwise trigger ovary development (Capel, 2017; Kashimada and Koopman, 2010). Importantly, it has been shown that precise regulation of levels and onset of Sry expression is key for Sertoli cell differentiation. Indeed, minor alteration in amount and timing of Sry expression leads to sex-reversal phenotypes (Kashimada and Koopman, 2010).
Table 2. Summary of mouse Y chromosome genes and their known functions.
A gene is classified as ‘ancestral’ if it is predicted to have been present in the last common eutherian ancestor. The list of animals where each gene is conserved only includes organisms where a high-quality Y chromosome sequence is available, and the presented list is not exhaustive. *Rbmy was initially thought to be important in spermatid morphogenesis, but this was later questioned (Szot et al., 2003).
| Y gene | Sequence class | X-homologue | Y copy number | Conserved in | Reported functions in the testis | Key gene ontology process | Expression pattern |
|---|---|---|---|---|---|---|---|
| Sry | Ancestral | Sox3 | 1 | Opossum, bull, rat, mouse, marmoset, rhesus, chimp, human |
Testis determination | DNA-binding transcription factor activity | Somatic cells in genital ridge, germ cell specific in adult |
| Rbmy array | Ancestral | Rbmx | ~30 | Opossum, bull, rat, mouse, marmoset, rhesus, chimp, human | Unknown * | RNA splicing | Testis biased |
| Zfy1, Zfy2 | Ancestral | Zfx | 2 | Bull, rat, mouse, marmoset, rhesus, chimp, human |
Apoptotic elimination of univalent spermatocytes at metaphase I, meiosis II completion, MSCI initiation and maintenance, spermatid head and tail morphogenesis | Transcription activator | Spermatogenic cells specific |
| Kdm5d | Ancestral | Kdm5c | 1 | Opossum, rat, mouse, marmoset, rhesus, chimp, human |
Unknown | Histone demethylase that specifically demethylates 'Lys-4' of histone H3 | Ubiquitous |
| Uty | Ancestral | Utx | 1 | Bull, rat, mouse, marmoset, rhesus, chimp, human | Unknown | Histone demethylase activity (H3-K27 specific) | Ubiquitous |
| Ddx3y | ancestral | Ddx3x | 1 | Bull, rat, mouse, marmoset, rhesus, chimp, human |
Unknown | ATP-dependent RNA helicase | Ubiquitous |
| Usp9y | Ancestral | Usp9x | 1 | Bull, rat, mouse, marmoset, rhesus, chimp, human | Unknown | Ubiquitination regulator, peptidase C19 | Testis biased |
| Uba1y | Ancestral | Uba1x | 1 | Opossum, bull, rat, mouse | Unknown | U1 ubiquitin activator | Testis biased, mostly spermatogonia and round spermatid |
| Eif2s3y | Ancestral | Eif2s3x | 1 | Bull, rat, mouse | Spermatogonial proliferation and differentiation | Translation initiation | Ubiquitous |
| Teyorf | Acquired | – | 1 | Mouse | Unknown | Claudin transmembrane | Testis specific |
| Prslly | Acquired | – | 1 | Mouse | Unknown | Serine-type endopeptidase activity | Testis specific |
| H2al2b, H2al2c | Acquired | H2al1 | 2 | Mouse | Unknown | DNA packaging, pericentric heterochromatin regulation | Testis specific, expressed from round spermatid stage |
| Sly | Acquired | Slx, Slxl1 | 126 | Mouse | Interacts with SSTY to recruit SMRT/N-Cor in turn mediating spermatid-specific gene expression | Chromatin remodelling | Testis biased |
| Srsy | Acquired | Srsx | 197 | Mouse | Unknown | Unknown | Testis biased |
| Ssty1&2 | Acquired | Sstx | 85&221 | Mouse | H3K4me3-reader at the promoter of spermatid-specific genes, recruits SLY and SLX/SLX1 | Methylated histone binding | Testis biased |
| Rbm31y | Acquired | Rbm31x | 2 | Mouse | Unknown | RNA binding | Testis biased |
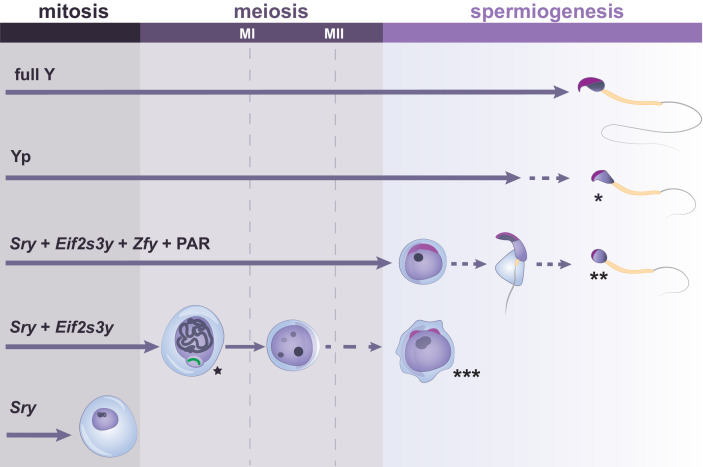
The race to discover the sex-determining region brought spotlight to the Y chromosome. However, the Y chromosome is far from being just a sex switch. It has a clear role in spermatogenesis. Mice with one X chromosome and transgenic expression of Sry (XOSry) develop testis populated with pro-spermatogonia that fail to proliferate, leading to absence of meiotic and post-meiotic cells (Figure 4; Mazeyrat et al., 2001). A similar phenotype can be observed in XO germ cells within XO/XY/XYY male mosaic mice (Levy and Burgoyne, 1986).
Figure 4. Y-linked gene complements and developmental progression through spermatogenesis.
Specific Y genes control the developmental progress of germ cells in spermatogenesis. The presence of the Y short arm (Yp) allows formation of sperm which have abnormal heads, exhibiting reduced curvature and poor chromatin compaction (indicated by *). The presence of Sry, Eif2s3y, Zfy genes and a PAR allows progression to the round spermatid stage, but spermatid elongation is abnormal and delayed, with rare production of sperm with poorly structured heads (indicated by **). Germ cells expressing only Sry and Eif2s3y have incomplete MSCI (indicated by the star next to the X chromosome shown in green). Moreover, most germ cells of this model arrest before the second meiotic division (MII), with occasional progression to form mostly diploid and abnormally shaped round spermatids (indicated by ***). When only Sry is expressed, spermatogonial cells fail to proliferate and to enter meiosis. Arrowed lines show germ cell progression through spermatogenesis, with dotted lines indicating abnormal cell differentiation at a low frequency.
Furthermore, it seems that additional Y genes are necessary not only in germ cells, but also in testicular somatic cells that are key to support successful spermatogenesis. Indeed, XY spermatogonia transplanted into XXSry testis cannot complete spermatogenesis, whereas segments of immature XY seminiferous tubule transplanted into XXSry testis display normal spermatogenic development of XY germ cells (Ishii et al., 2007). Altogether this highlights the importance of other Y genes besides Sry for both somatic and germ cell functions in the mouse testis.
In humans, the first sign that the Y chromosome may be involved in gamete production after testis determination came from the study of sub/infertile men with de novo deletions on the Y (Tiepolo and Zuffardi, 1976). The authors proposed the existence of at least one spermatogenesis factor or ‘azoospermia factor’ (AZF) on the Y. With an increasing number of studies finding sub-fertile males with Y deletions, and with the advances of physical and molecular mapping, three recurrently deleted regions of the human Y long arm were encountered: AZFa, AZFb, and AZFc (Vogt et al., 1996).
By the end of the twentieth century, there was undeniable evidence that both human and mouse Y chromosomes are crucial for spermatogenesis. However, the nature and number of Y-linked genes involved was still completely unknown. The importance of identifying such spermatogenesis factors became clear, especially given their clinical relevance for human fertility.
The search for spermatogenesis factors
Historically, functional studies of the human Y chromosome have been very challenging. They have relied on genetic analyses of individuals exhibiting vastly different genotypes and fertility phenotypes. A main aim of these heterogeneous studies has been to identify deleted/mutated genes within AZFa, b, and c. However, It has been difficult to draw clear associations between individual genes and specific reproduction phenotypes, especially given the historical lack of good Y assemblies (Colaco and Modi, 2018). Much remains unknown about the role of both ubiquitous and testis-specific genes for human reproduction (Colaco and Modi, 2018). These difficulties have highlighted the need for a genetically tractable model organism to study individual Y-gene functions. For decades, the mouse has been the mammal of choice to study Y-chromosome biology and spermatogenesis. As a result, this review focuses on the biology of the mouse Y chromosome. However, mammalian Ys can be highly divergent, and it is still unclear to what extent findings in the murine model will extend to other mammals.
The realisation that the mouse Y contained key factors for spermatogenesis resulted in attempts to perform loss of function studies using gene knockout. However, classic gene targeting strategies based on homologous recombination in mouse embryonic stem cells, which had worked to study autosomal and X gene functions, proved to be unsuccessful for the Y chromosome. This has been attributed to several factors. Firstly, sequencing data for the Y was sparse, with genomic characterisation relying on incomplete transcription-mapping approaches (Lahn and Page, 1997; Mazeyrat et al., 1998; Soh et al., 2014). The highly repetitive nature of the Y, besides making sequencing difficult using classical approaches, also hindered gene targeting attempts. This limited the field to the use of a few mouse models, which have different deletion variants spanning multiple Y genes, referred as Y-deletants hereafter. By adding transgenes of individual Y genes to the different Y-deletants, researchers investigated the rescue of previously identified spermatogenic failures. Transgenes have also been used in knockdown experiments to express interfering RNAs against ampliconic long arm Y gene (see: Y genes for meiotic progression and spermiogenesis). Overall, transgene rescue strategies, despite some caveats (see: the other indisposable Y genes), have been key to shape our current knowledge of Y-chromosome biology.
Are only two Y genes essential for male fertility?
In 2001, Mazeyrat et al., 2001 used BAC or cosmid clones of Y genes to generate individual transgenic mouse lines for three single-copy Y genes. These were then bred into two mouse models that express Sry but lack most or all of the remaining Y. Whereas Y-deletants experience arrest in spermatogonial proliferation, a clear amelioration was seen for mice expressing an Eif2s3y transgene. This ancestral gene has been lost in humans but is ubiquitously expressed in mouse and encodes a subunit of the translation initiation factor Eif2 (Figure 2 and Table 2).
Testes of these ‘rescued’ mice were larger than XOSry controls. Furthermore, the proportion of spermatogonia in rescued mice was similar to that of wild types. Importantly, germ cells progressed to the secondary spermatocyte stage, only arresting before the second meiotic division (Figure 4). Occasionally, spermatid-like cells were observed in these mouse models, although most of them were abnormally shaped and diploid (Figure 4; Mazeyrat et al., 2001; Vernet et al., 2011). Altogether, the authors reported having found the sought-after spermatogonial proliferation factor.
This study would become very influential for later investigations. For instance, Yamauchi and colleagues sought to generate offspring from the sterile mouse models described 13 years prior (Yamauchi et al., 2014). Using assisted reproduction, they were able to fertilise donor oocytes by injecting the rare round spermatids found in the testis of Sry and Eif2s3y-complemented mouse models. These fertilised oocytes were further transferred into recipient females, leading to the birth of a few offspring. The authors speculated that it may be possible to dispose of the mouse Y chromosome if replacements were made for Sry and Eif2s3y.
However, the many spermatogenic abnormalities seen when most of the Y chromosome is missing highlight that other Y genes are required for fertility. For instance, Yamauchi and colleagues showed that in mice expressing an additional fragment of the Y chromosome short arm, the abundance of later stage spermatids and the success of assisted reproduction was increased. This stresses the importance of additional Y short-arm genes to complete meiosis and differentiation from spermatid to spermatozoa (Yamauchi et al., 2014). In particular, a family of duplicated paralogues, the zinc-finger Y-linked Zfy1 and Zfy2 (Figure 2 and Table 2), has received much attention in the last decade and has been described as a key regulator of multiple spermatogenesis events.
Y genes for meiotic progression and spermiogenesis
The Zfy transcription factors are conserved across eutherians (Table 2). In humans, the single ZFY has two splice variants. Contrary to the short variant, the full-length variant has the ability to control transcription via transactivation (Decarpentrie et al., 2012). This seems to be reflected in the mouse, as Zfy2 expresses the long isoform with higher transactivation activity compared to Zfy1 (Decarpentrie et al., 2012).
Zfy genes are thought to play multiple roles in spermatogenesis. It has been suggested that Zfy genes are needed during pachynema for initiation and maintenance of MSCI, as well as apoptotic elimination of cells in which this process is defective (Figures 3 and 4; Vernet et al., 2016a). Later, during the first meiotic metaphase, Zfy2 regulates the SAC and is required to eliminate cells with unpaired sex chromosomes (Vernet et al., 2011). In Y-deletant models lacking a PAR, sex chromosomes cannot pair, causing spermatocyte univalence. In these models, the checkpoint-mediated apoptosis of univalent spermatocytes fails to take place and aneuploid round spermatids are produced. The addition of a Zfy2 transgene rescues this phenotype and ensures elimination of univalent spermatocytes (Figure 4; Vernet et al., 2014; Vernet et al., 2011). Zfy2, and to a lesser extent Zfy1, is also required to promote completion of meiosis II, rescuing the meiotic block seen in Y-deletant models (Figure 4; Vernet et al., 2014). Zfy2 is also involved in spermatid morphogenesis, as transgenic integration of Zfy2 in Y-deletants improves spermatid morphology and increases the success rate of assisted reproduction (Figure 4; Yamauchi et al., 2015). Indeed, addition of Zfy2 contributes to the restructuring of the sperm head and development of the sperm tail (Figure 4; Vernet et al., 2016b). Specifically, these morphogenic changes are thought to be regulated by Zfy2 through strong expression of the ZFY2 transactivation domain from an alternative, spermatid-specific Cypt-derived promoter. However, the molecular mechanisms underlying Zfy genes’ functions remain unclear.
In the mouse, the ancestral genes that are mostly conserved across mammals are within the Y chromosome short arm (Figure 2). This partly explains why studies looking for ‘essential’ spermatogenesis factors have mostly focused on this region. However, the Y short arm only represents ~ 2.5% of the full mouse Y length (Figure 2). The rest of the mouse Y euchromatin is encompassed within the long arm, which until 2014 remained mostly un-sequenced (Soh et al., 2014). The mouse Y long arm contains highly repetitive sequences and testis and murine-specific gene clusters consisting of three main gene families: Sycp3-like Y-linked (Sly), Spermiogenesis-specific transcript on the Y (Ssty) and Serine-rich Secreted Y-linked (Srsy) (Figure 2 and Table 2). Since the many copies of these gene families are intermingled (Figure 2), it is very challenging to functionally study Sly, Ssty, and Srsy independently. Indeed, any deletion of one or more amplicon on the Y long arm will encompass all the three main gene families. Y deletant models, which have depletions of all three gene families, show infertility and severe sperm abnormalities (Figure 4), associated with upregulation of spermatid-expressed sex chromosome genes (Conway et al., 1994; Ellis et al., 2005; Styrna et al., 1991; Touré et al., 2004; Yamauchi et al., 2010).
Knockdown of the Sly family using transgenic expression of interfering RNAs has shown that SLY depletion is at least partly responsible for the phenotypes seen in long-arm Y deletant mice. Indeed, knockdown of Sly causes subfertility and upregulation of spermatid genes (Cocquet et al., 2009; Riel et al., 2013). When knockdown males were able to breed, SLY depletion was also linked to female-biased litters. Conversely, knockdown of Slx/Slxl1, the X homologues to Sly, causes spermatid-specific sex-chromosome gene downregulation, as well as male-biased litters (Cocquet et al., 2012; Cocquet et al., 2010; Kruger et al., 2019). These sex-ratio skews likely result from differential fertilising abilities between X- and Y-bearing sperms in Sly and Slx/Slxl1 depleted models (Kruger et al., 2019; Rathje et al., 2019). Moreover, knockdown of both X and Y families re-establishes correct sex ratio and transcriptional output.
Mechanistically, SLY and SLX/SLXL1 compete for chromatin occupancy at the loci of spermatid-expressed genes, especially on the sex chromosomes (Moretti et al., 2020). This is where SLY is thought to interact with the histone-reader SSTY to recruit repressive transcriptional complexes, therefore modulating global spermatid-gene expression (Comptour et al., 2014; Moretti et al., 2020).
Altogether, the pressure to keep a balance in postmeiotic sex-chromosome expression and in sex ratio has created intragenomic conflict between sex chromosomes, with Sly and Slx/Slxl1 acting as antagonistic meiotic drivers (see: Evolutionary forces and functional specialisation of the Y chromosome) (Cocquet et al., 2012; Ellis et al., 2011; Larson et al., 2018). This ‘arms race’ has promoted the amplification of long-arm Y genes and their X homologues shaping the functional and evolutionary dynamic of these loci.
Altogether, the advent of the 21th century has seen an increasing interest in studying Y-linked genes. These efforts have provided valuable information on how some Y genes may participate in the developmental control of spermatogenesis (Figure 4). However, many questions remain. Firstly, even for well-studied genes such as Eif2s3y and Zfy2, the mechanism of action and molecular interactions of these spermatogenic factors are mostly unknown. Secondly, the majority of Y-linked genes have not yet been assigned to specific spermatogenic function (Table 2). Are they therefore disposable for male fertility?
The other indisposable Y genes
There is undeniable evidence that additional Y-linked genes are necessary for non-assisted reproduction. Although mice carrying only Sry, Eif2s3y, and Zfy can produce offspring through assisted reproduction, they are infertile. This is in part due to the lack of the Y chromosome long arm. However, there are other genes on the Y short arm that contribute to spermatogenesis. For instance, mice retaining most of the short arm, but no long arm, develop sperm with better morphology and have increased assisted-reproduction success compared to individuals expressing only Sry, Eif2s3y, and Zfy complements (Figure 4; Yamauchi et al., 2015).
How should research proceed to characterise the function of these indisposable genes on both the short and long arm of the Y chromosome? Can previously described multi-gene deletion mouse models and transgene strategies still be informative, or has their utility reached a plateau? These time-consuming approaches have been successful but also have inherent limitations to study genes individually. Firstly, transgenes usually integrate at random genomic locations (Yan et al., 2013), which can disturb expression of neighbouring genes. Furthermore, transgenes often integrate in multiple copies and therefore have varying expression levels (Mazeyrat et al., 2001; Royo et al., 2010). These caveats can complicate interpretation of transgene rescue experiments. Even if transgenes are integrated at defined genomic locations, their expression levels can differ from wild-type (Vernet et al., 2016b; Vernet et al., 2014). This is limiting because dosage of ancestral Y genes is thought to be crucial and sensitive for sex-chromosome function (see: Evolutionary forces and functional specialisation of the Y chromosome).
With the inability to perform gene knockouts on the Y, scientists had to make do with transgene-complementation approaches. However, new technologies for gene targeting have been developed, and better-quality Y-chromosome sequence information is now available. Indeed, the rise of gene-editing strategies such as transcription activator-like effector nuclease (TALEN) and CRISPR/Cas9 has opened new avenues for unbiased dissection of Y-gene functions. Targeting of the two most studied Y genes, Sry and Eif2s3y, recapitulated the expected sex-reversed and spermatogonial-arrest phenotypes, respectively (Kato et al., 2013; Matsubara et al., 2015; Song et al., 2017; Wang et al., 2013; Zuo et al., 2017). These studies provided proof of principles that genome editing was possible on the Y chromosome.
Additionally, other Y chromosome genes have been targeted in the mouse. In some instances, potential spermatogenic functions of targeted Y genes were not investigated in detail. This has been the case for the Ubiquitously Transcribed Y Chromosome Tetratricopeptide Repeat (Uty) gene. It was shown that this deeply conserved ancestral gene has partially-redundant functions with its X homologue Utx during embryonic development (Shpargel et al., 2012; Wang et al., 2013). However, possible spermatogenic abnormalities and subfertility were not examined.
When gene-editing studies of short-arm mouse Y genes have looked at spermatogenic functions, they have faced a major difficulty. Indeed, it has been difficult to ensure complete Y protein abrogation. To date, Y genes have either been targeted at the vicinity of the start codon, e.g. for Zfy1 and Zfy2 (Nakasuji et al., 2017); or at an exon not thought to encode a key domain, e.g. DEAD-Box Helicase 3 Y-Linked (Ddx3y), Ubiquitin Specific Peptidase 9 Y-Linked (Usp9y), Ubiquitin-like modifier-activating enzyme 1 Y (Uba1y) and Lysine Demethylase 5D (Kdm5d) (Matsumura et al., 2019; Zuo et al., 2017). However, we now know that following CRISPR/Cas9 editing, transcription through alternative start codons or skipping of mutated/deleted exons can allow functional protein formation (Lindeboom et al., 2016; Lindeboom et al., 2019 Sharpe and Cooper, 2017; Smits et al., 2019; Wiles, 2020). Because effective antibodies are not currently available for most Y-encoded proteins, it is unclear whether in these deletion models the Y-proteins in question had been completely abolished. As a result, although using nuclease-mediated gene editing is a step in the right direction, only carefully designed strategies ensuring complete protein ablation will help dissect the functions of Y-linked genes.
Although most Y-genes do not yet have ascribed functions, many could be important for spermatogenesis. Non-conserved Y genes that are acquired within mammalian lineages tend to show testis-biased expression, and are therefore predicted to be linked to male fertility (Bellott et al., 2014; Godfrey et al., 2020; Mitchell, 2000; Soh et al., 2014). In the case of the mouse, Testis expressed, Chromosome Y open reading frame 1 (Teyorf1), Protease, serine-like, Y (Prssly) and H2A histone family member L2 (H2al2) (Soh et al., 2014; Figure 2 and Table 2) were discovered only recently, and their functions in fertility have therefore not been investigated.
Ancestral genes are also interesting, because they have been retained on the Y chromosome during evolution and are implicated in critical housekeeping functions. Examples include the ubiquitination proteins Uba1y and Usp9y, splicing factor Rbmy, RNA helicase Ddx3y and chromatin modifiers Uty and Kdm5d (Table 2). Interestingly, some of these ancestral genes seem to have functionally diverged from their X-homologues (Bellott et al., 2014; Snell and Turner, 2018) and evolved testis-biased expression, suggesting a specialised role in spermatogenesis. Preferential testis-expression is observed for Ddx3y in primates, Uba1y in mice, rats, bulls, and opossums, Usp9y in mice and rats and Rbmy in bull, rodents and primates (Table 2; Bellott et al., 2014). Furthermore, deletions encompassing DDX3Y and USP9Y have been recorded in sub-fertile men (Sun et al., 1999; Vogt et al., 2008).
In addition to protein-coding loci, the potential spermatogenic roles of non-coding regulatory regions on the Y chromosome must not be underestimated. Divergence between Y and X chromosome regulatory regions can lead to expression, and potentially phenotypic differences between XY and XX individuals (Godfrey et al., 2020; Naqvi et al., 2019; Naqvi et al., 2018). One could imagine that, just like Y protein-coding genes, Y regulatory regions have experienced pressure to specialise in regulating male fertility, especially through dosage control of nearby Y protein-coding genes.
Outlook
The Y chromosome has historically been misunderstood. Evolutionary insights and discoveries of spermatogenic factors have shown that the Y chromosome is far from being an ever-degenerating functional wasteland. For a long time, however, technical limitations have restricted the investigation of individual Y gene functions. In an era dominated by fast-evolving gene-editing techniques and omics technologies, the stage is now set for systematic Y-gene deletions and functional characterisation of resulting mutants. Altogether, fully dissecting the link between Y genes and key reproductive processes in the mouse will be paramount to deepen our understanding of genetic components influencing human male fertility and inform us on the functional evolution of the Y chromosome in mammals.
Acknowledgements
The authors would like to thank members of the Turner lab for providing feedback on the manuscript. This review is dedicated to the memory of Paul Burgoyne.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
James Turner, Email: james.turner@crick.ac.uk.
Blanche Capel, Duke University, United States.
Kathryn Song Eng Cheah, The University of Hong Kong, Hong Kong.
Funding Information
This paper was supported by the following grants:
Cancer Research UK FC001193 to Jeremie Subrini, James Turner.
Medical Research Council FC001193 to Jeremie Subrini, James Turner.
Wellcome Trust FC001193 to Jeremie Subrini, James Turner.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Writing - original draft, Writing - review and editing.
Conceptualization, Supervision, Writing - review and editing.
References
- Agulnik AI, Mitchell MJ, Lerner JL, Woods DR, Bishop CE. A mouse y chromosome gene encoded by a region essential for spermatogenesis and expression of male-specific minor histocompatibility antigens. Human Molecular Genetics. 1994;3:873–878. doi: 10.1093/hmg/3.6.873. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, Marshall Graves JA. The future of sex. Nature. 2002;415:963. doi: 10.1038/415963a. [DOI] [PubMed] [Google Scholar]
- Arnemann J, Jakubiczka S, Thüring S, Schmidtke J. Cloning and sequence analysis of a human y-chromosome-derived, testicular CDNA, TSPY. Genomics. 1991;11:108–114. doi: 10.1016/0888-7543(91)90107-P. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. Y-chromosome evolution: Emerging insights into processes of y-chromosome degeneration. Nature Reviews. Genetics. 2013;14:113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R. The protamine family of sperm nuclear proteins. Genome Biology. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Bedford MT. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction. 2016;151:R55–R70. doi: 10.1530/REP-15-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: Localization and regulation. Nature Reviews. Genetics. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho T-J, Koutseva N, Zaghlul S, Graves T, Rock S, Kremitzki C, Fulton RS, Dugan S, Ding Y, Morton D, Khan Z, Lewis L, Buhay C, Wang Q, Watt J, Holder M, Lee S, Nazareth L, Alföldi J, Rozen S, Muzny DM, Warren WC, Gibbs RA, Wilson RK, Page DC. Mammalian y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta P, Hawkins JR, Sinclair AH, Taylor A, Griffiths BL, Goodfellow PN, Fellous M. Genetic evidence equating SRY and the testis-determining factor. Nature. 1990;348:448–450. doi: 10.1038/348448a0. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Bowness JS, Wei G. Progress toward understanding chromosome silencing by xist RNA. Genes & Development. 2020;34:733–744. doi: 10.1101/gad.337196.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne PS, Baker TG. Oocyte depletion in XO mice and their XX sibs from 12 to 200 days post partum. Journal of Reproduction and Fertility. 1981;61:207–212. doi: 10.1530/jrf.0.0610207. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Evans EP, Holland K. XO monosomy is associated with reduced birthweight and lowered weight gain in the mouse. Journal of Reproduction and Fertility. 1983a;68:381–385. doi: 10.1530/jrf.0.0680381. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Tam PP, Evans EP. Retarded development of XO conceptuses during early pregnancy in the mouse. Journal of Reproduction and Fertility. 1983b;68:387–393. doi: 10.1530/jrf.0.0680387. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS. The mammalian y chromosome: A new perspective. BioEssays. 1998;20:363–366. doi: 10.1002/(SICI)1521-1878(199805)20:5<363::AID-BIES2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Mahadevaiah SK, Turner JMA. The consequences of asynapsis for mammalian meiosis. Nature Reviews. Genetics. 2009;10:207–216. doi: 10.1038/nrg2505. [DOI] [PubMed] [Google Scholar]
- Capel B. Vertebrate sex determination: Evolutionary plasticity of a fundamental switch. Nature Reviews. Genetics. 2017;18:675–689. doi: 10.1038/nrg.2017.60. [DOI] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJI, Yamauchi Y, Mahadevaiah SK, Affara NA, Ward MA, Burgoyne PS. The multicopy gene sly represses the sex chromosomes in the male mouse germline after meiosis. PLOS Biology. 2009;7:e1000244. doi: 10.1371/journal.pbio.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJI, Yamauchi Y, Riel JM, Karacs TPS, Rattigan A, Ojarikre OA, Affara NA, Ward MA, Burgoyne PS. Deficiency in the multicopy sycp3-like x-linked genes Slx and slxl1 causes major defects in spermatid differentiation. Molecular Biology of the Cell. 2010;21:3497–3505. doi: 10.1091/mbc.E10-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJI, Mahadevaiah SK, Affara NA, Vaiman D, Burgoyne PS. A genetic basis for a postmeiotic x versus y chromosome intragenomic conflict in the mouse. PLOS Genetics. 2012;8:e1002900. doi: 10.1371/journal.pgen.1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaco S, Modi D. Genetics of the human Y chromosome and its association with male infertility. Reproductive Biology and Endocrinology. 2018;16:14. doi: 10.1186/s12958-018-0330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comptour A, Moretti C, Serrentino ME, Auer J, Ialy-Radio C, Ward MA, Touré A, Vaiman D, Cocquet J. SSTY proteins co-localize with the post-meiotic sex chromatin and interact with regulators of its expression. The FEBS Journal. 2014;281:1571–1584. doi: 10.1111/febs.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway SJ, Mahadevaiah SK, Darling SM, Capel B, Rattigan AM, Burgoyne PS. Y353/B: A candidate multiple-copy spermiogenesis gene on the mouse Y chromosome. Mammalian Genome. 1994;5:203–210. doi: 10.1007/BF00360546. [DOI] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grützner F, Kaessmann H. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508:488–493. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- de Rooij DG. The nature and dynamics of spermatogonial stem cells. Development. 2017;144:3022–3030. doi: 10.1242/dev.146571. [DOI] [PubMed] [Google Scholar]
- Decarpentrie F, Vernet N, Mahadevaiah SK, Longepied G, Streichemberger E, Aknin-Seifer I, Ojarikre OA, Burgoyne PS, Metzler-Guillemain C, Mitchell MJ. Human and mouse ZFY genes produce a conserved testis-specific transcript encoding a zinc finger protein with a short acidic domain and modified transactivation potential. Human Molecular Genetics. 2012;21:2631–2645. doi: 10.1093/hmg/dds088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Berletch JB, Nguyen DK, Disteche CM. X chromosome regulation: Diverse patterns in development, tissues and disease. Nature Reviews. Genetics. 2014;15:367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis PJI, Clemente EJ, Ball P, Touré A, Ferguson L, Turner JMA, Loveland KL, Affara NA, Burgoyne PS. Deletions on mouse YQ lead to upregulation of multiple x- and y-linked transcripts in spermatids. Human Molecular Genetics. 2005;14:2705–2715. doi: 10.1093/hmg/ddi304. [DOI] [PubMed] [Google Scholar]
- Ellis PJI, Bacon J, Affara NA. Association of Sly with sex-linked gene amplification during mouse evolution: a side effect of genomic conflict in spermatids? Human Molecular Genetics. 2011;20:3010–3021. doi: 10.1093/hmg/ddr204. [DOI] [PubMed] [Google Scholar]
- Ford CE, Jones KW, Polani PE, De Almedia JC, Briggs JH. A sex-chromosome anomaly in a case of gonadal dysgenesis (Turner’s syndrome. Lancet. 1959;1:711–713. doi: 10.1016/s0140-6736(59)91893-8. [DOI] [PubMed] [Google Scholar]
- Foster JW, Graves JA. An sry-related sequence on the marsupial x chromosome: Implications for the evolution of the mammalian testis-determining gene. PNAS. 1994;91:1927–1931. doi: 10.1073/pnas.91.5.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey AK, Naqvi S, Chmátal L, Chick JM, Mitchell RN, Gygi SP, Skaletsky H, Page DC. Quantitative analysis of y-chromosome gene expression across 36 human tissues. Genome Research. 2020;30:860–873. doi: 10.1101/gr.261248.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravholt CH, Viuff MH, Brun S, Stochholm K, Andersen NH. Turner syndrome: mechanisms and management. Nature Reviews. Endocrinology. 2019;15:601–614. doi: 10.1038/s41574-019-0224-4. [DOI] [PubMed] [Google Scholar]
- Greaves IK, Rangasamy D, Devoy M, Marshall Graves JA, Tremethick DJ. The X and Y chromosomes assemble into H2A.Z-containing [corrected] facultative heterochromatin [corrected] following meiosis. Molecular and Cellular Biology. 2006;26:5394–5405. doi: 10.1128/mcb.00519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Münsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- Guellaen G, Casanovat M, Geldwerth D, Andre G, Felloust M, Weissenbach J. Human XX males with Y single-copy DNA fragments. Nature. 1984;307:30–31. doi: 10.1038/307172a0. [DOI] [PubMed] [Google Scholar]
- Hendriksen PJM, Hoogerbrugge JW, Themmen APN, Koken MHM, Hoeijmakers JHJ, Oostra BA, van der Lende T, Grootegoed JA. Postmeiotic transcription of X and Y chromosomal genes during spermatogenesis in the mouse. Developmental Biology. 1995;170:730–733. doi: 10.1006/dbio.1995.1252. [DOI] [PubMed] [Google Scholar]
- Hendriksen PJM. Do X and Y spermatozoa differ in proteins? Theriogenology. 1999;52:1295–1307. doi: 10.1016/S0093-691X(99)00218-6. [DOI] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, Minx PJ, Graves T, Rozen S, Wilson RK, Page DC. Conservation of y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature. 2005;437:100–103. doi: 10.1038/nature04101. [DOI] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SKM, Minx PJ, Fulton RS, McGrath SD, Locke DP, Friedman C, Trask BJ, Mardis ER, Warren WC, Repping S, Rozen S, Wilson RK, Page DC. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463:536–539. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Graves T, Fulton RS, Dugan S, Ding Y, Buhay CJ, Kremitzki C, Wang Q, Shen H, Holder M, Villasana D, Nazareth LV, Cree A, Courtney L, Veizer J, Kotkiewicz H, Cho T-J, Koutseva N, Rozen S, Muzny DM, Warren WC, Gibbs RA, Wilson RK, Page DC. Strict evolutionary conservation followed rapid gene loss on human and rhesus y chromosomes. Nature. 2012;483:82–86. doi: 10.1038/nature10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Page DC. The biology and evolution of mammalian Y chromosomes. Annual Review of Genetics. 2015;49:507–527. doi: 10.1146/annurev-genet-112414-055311. [DOI] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, Koutseva N, Raudsepp T, Brown LG, Bellott DW, Cho TJ, Dugan-Rocha S, Khan Z, Kremitzki C, Fronick C, Graves-Lindsay TA, Fulton L, Warren WC, Wilson RK, Owens E, Womack JE, Murphy WJ, Muzny DM, Worley KC, Chowdhary BP, Gibbs RA, Page DC. Sequence analysis in Bos taurus reveals pervasiveness of X-Y arms races in mammalian lineages. Genome Research. 2020;30:1716–1726. doi: 10.1101/gr.269902.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Tachiwana T, Hoshino A, Tsunekawa N, Hiramatsu R, Matoba S, Kanai-Azuma M, Kawakami H, Kurohmaru M, Kanai Y. Potency of testicular somatic environment to support spermatogenesis in XX/SRY transgenic male mice. Development. 2007;134:449–454. doi: 10.1242/dev.02751. [DOI] [PubMed] [Google Scholar]
- Jacobs BPA, Strong DJA. A case of human intersexuality having possible XXY sex-determining mechanism. Nature. 1959;1:7–8. doi: 10.1038/183302a0. [DOI] [PubMed] [Google Scholar]
- Jäger RJ, Anvret M, Hall K, Scherer G. A human XY female with a frame shift mutation in the candidate testis-determining gene SRY. Nature. 1990;348:452–454. doi: 10.1038/348452a0. [DOI] [PubMed] [Google Scholar]
- Jegalian K, Page DC. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become x inactivated. Nature. 1998;394:776–780. doi: 10.1038/29522. [DOI] [PubMed] [Google Scholar]
- Kashimada K, Koopman P. SRY: The master switch in mammalian sex determination. Development. 2010;137:3921–3930. doi: 10.1242/dev.048983. [DOI] [PubMed] [Google Scholar]
- Kato T, Miyata K, Sonobe M, Yamashita S, Tamano M, Miura K, Kanai Y, Miyamoto S, Sakuma T, Yamamoto T, Inui M, Kikusui T, Asahara H, Takada S. Production of sry knockout mouse using TALEN via oocyte injection. Scientific Reports. 2013;3:3136. doi: 10.1038/srep03136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman P, Münsterberg A, Capel B, Vivian N, Lovell-Badge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990;348:450–452. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- Kruger AN, Brogley MA, Huizinga JL, Kidd JM, de Rooij DG, Hu YC, Mueller JL. A neofunctionalized x-linked ampliconic gene family is essential for male fertility and equal sex ratio in mice. Current Biology. 2019;29:3699–3706. doi: 10.1016/j.cub.2019.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Saitou M. Germ cell reprogramming. Current Topics in Developmental Biology. 2019;135:91–125. doi: 10.1016/bs.ctdb.2019.04.005. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Lane S, Kauppi L. Meiotic spindle assembly checkpoint and aneuploidy in males versus females. Cellular and Molecular Life Sciences. 2019;76:1135–1150. doi: 10.1007/s00018-018-2986-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EL, Kopania EEK, Good JM. Spermatogenesis and the evolution of mammalian sex chromosomes. Trends in Genetics. 2018;34:722–732. doi: 10.1016/j.tig.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy ER, Burgoyne PS. The fate of XO germ cells in the testes of xo/xy and xo/xy/xyy mouse mosaics: Evidence for a spermatogenesis gene on the mouse Y chromosome. Cytogenetic and Genome Research. 1986;42:208–213. doi: 10.1159/000132280. [DOI] [PubMed] [Google Scholar]
- Li G, Davis BW, Raudsepp T, Pearks Wilkerson AJ, Mason VC, Ferguson-Smith M, O’Brien PC, Waters PD, Murphy WJ. Comparative analysis of mammalian Y chromosomes illuminates ancestral structure and lineage-specific evolution. Genome Research. 2013;23:1486–1495. doi: 10.1101/gr.154286.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom RGH, Supek F, Lehner B. The rules and impact of nonsense-mediated mrna decay in human cancers. Nature Genetics. 2016;48:1112–1118. doi: 10.1038/ng.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom RGH, Vermeulen M, Lehner B, Supek F. The impact of nonsense-mediated mrna decay on genetic disease, gene editing and cancer immunotherapy. Nature Genetics. 2019;51:1645–1651. doi: 10.1038/s41588-019-0517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loda A, Heard E. Xist RNA in action: Past, present, and future. PLOS Genetics. 2019;15:e1008333. doi: 10.1371/journal.pgen.1008333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell-Badge R, Robertson E. XY female mice resulting from a heritable mutation in the primary testis-determining gene, Tdy. Development. 1990;109:635–646. doi: 10.1242/dev.109.3.635. [DOI] [PubMed] [Google Scholar]
- Luo ZX, Yuan CX, Meng QJ, Ji Q. A jurassic eutherian mammal and divergence of marsupials and placentals. Nature. 2011;476:442–445. doi: 10.1038/nature10291. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L. Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Marshall Graves JA. The origin and function of the mammalian y chromosome and y‐borne genes – an evolving understanding. BioEssays. 1995;17:950170407. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Marshall Graves JA. Evolution of the mammalian Y chromosome and sex-determining genes. The Journal of Experimental Zoology. 1998;1:2-B. doi: 10.1002/(SICI)1097-010X(19980801)281:53.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Marshall Graves JA. the degenerate y chromosome - can conversion save it? Reproduction, Fertility, and Development. 2004;1:RD03096. doi: 10.1071/RD03096. [DOI] [PubMed] [Google Scholar]
- Matsubara Y, Kato T, Kashimada K, Tanaka H, Zhi Z, Ichinose S, Mizutani S, Morio T, Chiba T, Ito Y, Saga Y, Takada S, Asahara H. Talen-mediated gene disruption on Y chromosome reveals critical role of EIF2S3Y in mouse spermatogenesis. Stem Cells and Development. 2015;24:1164–1170. doi: 10.1089/scd.2014.0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura T, Endo T, Isotani A, Ogawa M, Ikawa M. An azoospermic factor gene, DDX3Y and its paralog, DDX3X are dispensable in germ cells for male fertility. The Journal of Reproduction and Development. 2019;65:121–128. doi: 10.1262/jrd.2018-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazeyrat S, Saut N, Sargent CA, Grimmond S, Longepied G, Ehrmann IE, Ellis PS, Greenfield A, Affara NA, Mitchell MJ. The mouse y chromosome interval necessary for spermatogonial proliferation is gene dense with syntenic homology to the human AZFA region. Human Molecular Genetics. 1998;7:1713–1724. doi: 10.1093/hmg/7.11.1713. [DOI] [PubMed] [Google Scholar]
- Mazeyrat S, Saut N, Grigoriev V, Mahadevaiah SK, Ojarikre OA, Bishop C, Eicher EM, Mitchell MJ, Burgoyne PS. A y-encoded subunit of the translation initiation factor eif2 is essential for mouse spermatogenesis. Nature Genetics. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- McKee BD, Handel MA. Sex chromosomes, recombination, and chromatin conformation. Chromosoma. 1993;102:71–80. doi: 10.1007/BF00356023. [DOI] [PubMed] [Google Scholar]
- Mitchell MJ. Spermatogenesis and the mouse y chromosome: Specialisation out of decay. Results and Problems in Cell Differentiation. 2000;28:233–270. doi: 10.1007/978-3-540-48461-5_10. [DOI] [PubMed] [Google Scholar]
- Moretti C, Blanco M, Ialy-Radio C, Serrentino M-E, Gobé C, Friedman R, Battail C, Leduc M, Ward MA, Vaiman D, Tores F, Cocquet J. Battle of the sex chromosomes: Competition between x and Y chromosome-encoded proteins for partner interaction and chromatin occupancy drives multicopy gene expression and evolution in muroid rodents. Molecular Biology and Evolution. 2020;37:3453–3468. doi: 10.1093/molbev/msaa175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muciaccia B, Boitani C, Berloco BP, Nudo F, Spadetta G, Stefanini M, de Rooij DG, Vicini E. Novel stage classification of human spermatogenesis based on acrosome development. Biology of Reproduction. 2013;89:60. doi: 10.1095/biolreprod.113.111682. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JMA. The mouse x chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nature Genetics. 2008;40:794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JL, Skaletsky H, Brown LG, Zaghlul S, Rock S, Graves T, Auger K, Warren WC, Wilson RK, Page DC. Independent specialization of the human and mouse X chromosomes for the male germ line. Nature Genetics. 2013;45:1083–1087. doi: 10.1038/ng.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasuji T, Ogonuki N, Chiba T, Kato T, Shiozawa K, Yamatoya K, Tanaka H, Kondo T, Miyado K, Miyasaka N, Kubota T, Ogura A, Asahara H. Complementary critical functions of zfy1 and Zfy2 in mouse spermatogenesis and reproduction. PLOS Genetics. 2017;13:e1006578. doi: 10.1371/journal.pgen.1006578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang L-F, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Current Biology. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- Naqvi S, Bellott DW, Lin KS, Page DC. Conserved microrna targeting reveals preexisting gene dosage sensitivities that shaped amniote sex chromosome evolution. Genome Research. 2018;28:474–483. doi: 10.1101/gr.230433.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi S, Godfrey AK, Hughes JF, Goodheart ML, Mitchell RN, Page DC. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science. 2019;365:eaaw7317. doi: 10.1126/science.aaw7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakberg EF. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. The American Journal of Anatomy. 1956a;99:391–413. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- Oakberg EF. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. The American Journal of Anatomy. 1956b;99:507–516. doi: 10.1002/aja.1000990307. [DOI] [PubMed] [Google Scholar]
- Ohno S. Sex Chromosomes and Sex-Linked Genes., Monographs on Endocrinology. Berlin, Heidelberg, New York: Springer Verlag; 1967. [DOI] [Google Scholar]
- O’Donnell L. Mechanisms of spermiogenesis and spermiation and how they are disturbed. Spermatogenesis. 2014;4:e979623. doi: 10.4161/21565562.2014.979623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page DC, de la Chapelle A, Weissenbach J. Chromosome y-specific DNA in related human xx males. Nature. 1985;315:224–226. doi: 10.1038/315224a0. [DOI] [PubMed] [Google Scholar]
- Page DC. Cold Spring Harbor Symposia on Quantitative Biology. Sex reversal: Deletion mapping the male-determining function of the human Y chromosome; 1986. pp. 229–235. [DOI] [PubMed] [Google Scholar]
- Page DC, Mosher R, Simpson EM, Fisher EMC, Mardon G, Pollack J, McGillivray B, de la Chapelle A, Brown LG. The sex-determining region of the human Y chromosome encodes a finger protein. Cell. 1987;51:1091–1104. doi: 10.1016/0092-8674(87)90595-2. [DOI] [PubMed] [Google Scholar]
- Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jégou B, Kaessmann H. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLOS Biology. 2008;6:e80. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathje CC, Johnson EEP, Drage D, Patinioti C, Silvestri G, Affara NA, Ialy-Radio C, Cocquet J, Skinner BM, Ellis PJI. Differential sperm motility mediates the sex ratio drive shaping mouse sex chromosome evolution. Current Biology. 2019;29:3692–3698. doi: 10.1016/j.cub.2019.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, de la Chapelle A, Silber S, Page DC. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nature Genetics. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- Riel JM, Yamauchi Y, Sugawara A, Li HYJ, Ruthig V, Stoytcheva Z, Ellis PJI, Cocquet J, Ward MA. Deficiency of the multi-copy mouse y gene sly causes sperm dna damage and abnormal chromatin packaging. Journal of Cell Science. 2013;126:803–813. doi: 10.1242/jcs.114488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo H, Polikiewicz G, Mahadevaiah SK, Prosser H, Mitchell M, Bradley A, de Rooij DG, Burgoyne PS, Turner JMA. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Current Biology. 2010;20:2117–2123. doi: 10.1016/j.cub.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC. Abundant gene conversion between arms of palindromes in human and APE y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- Salido EC, Yen PH, Koprivnikar K, Yu LC, Shapiro LJ. The human enamel protein gene amelogenin is expressed from both the x and the y chromosomes. American Journal of Human Genetics. 1992;50:303–316. [PMC free article] [PubMed] [Google Scholar]
- Sharpe JJ, Cooper TA. Unexpected consequences: Exon skipping caused by crispr-generated mutations. Genome Biology. 2017;18:1240–1243. doi: 10.1186/s13059-017-1240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLOS Genetics. 2012;8:e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin HS, Ichijima Y, Koh E, Namiki M, Namekawa SH. Human postmeiotic sex chromatin and its impact on sex chromosome evolution. Genome Research. 2012;22:827–836. doi: 10.1101/gr.135046.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved dna-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou SF, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang SP, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Smits AH, Ziebell F, Joberty G, Zinn N, Mueller WF, Clauder-Münster S, Eberhard D, Savitski MF, Grandi P, Jakob P, Michon AM, Sun H, Tessmer K, Bürckstümmer T, Bantscheff M, Steinmetz LM, Drewes G, Huber W. Biological Plasticity Rescues Target Activity in CRISPR Knockouts. bioRxiv. 2019 doi: 10.1101/716019. [DOI] [PubMed]
- Snell DM, Turner JMA. Sex Chromosome Effects on Male-Female Differences in Mammals. Current Biology. 2018;28:R1313–R1324. doi: 10.1016/j.cub.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh YQS, Alföldi J, Pyntikova T, Brown LG, Graves T, Minx PJ, Fulton RS, Kremitzki C, Koutseva N, Mueller JL, Rozen S, Hughes JF, Owens E, Womack JE, Murphy WJ, Cao Q, de Jong P, Warren WC, Wilson RK, Skaletsky H, Page DC. Sequencing the mouse y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell. 2014;159:800–813. doi: 10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari AJ. The behavior of the XY pair in mammals. International Review of Cytology. 1974;38:273–317. doi: 10.1016/S0074-7696(08)60928-6. [DOI] [PubMed] [Google Scholar]
- Song Y, Liu T, Wang Y, Deng J, Chen M, Yuan L, Lu Y, Xu Y, Yao H, Li Z, Lai L. Mutation of the Sp1 binding site in the 5’ flanking region of SRY causes sex reversal in rabbits. Oncotarget. 2017;8:38176–38183. doi: 10.18632/oncotarget.16979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern C. The problem of complete y-linkage in man. American Journal of Human Genetics. 1957;9:147–166. [PMC free article] [PubMed] [Google Scholar]
- Styrna J, Imai HT, Moriwaki K. An increased level of sperm abnormalities in mice with a partial deletion of the y chromosome. Genetical Research. 1991;57:195–199. doi: 10.1017/S0016672300029268. [DOI] [PubMed] [Google Scholar]
- Sun C, Skaletsky H, Birren B, Devon K, Tang Z, Silber S, Oates R, Page DC. An azoospermic man with a de novo point mutation in the y-chromosomal gene USP9Y. Nature Genetics. 1999;23:429–432. doi: 10.1038/70539. [DOI] [PubMed] [Google Scholar]
- Sybirna A, Wong FCK, Surani MA. Genetic basis for primordial germ cells specification in mouse and human: Conserved and divergent roles of PRDM and SOX transcription factors. Current Topics in Developmental Biology. 2019;135:35–89. doi: 10.1016/bs.ctdb.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Szot M, Grigoriev V, Mahadevaiah SK, Ojarikre OA, Touré A, von Glasenapp E, Rattigan A, Turner JMA, Elliott DJ, Burgoyne PS. Does Rbmy have a role in sperm development in mice? Cytogenetic and Genome Research. 2003;103:330–336. doi: 10.1159/000076821. [DOI] [PubMed] [Google Scholar]
- Tam PP, Snow MHL. Proliferation and migration of primordial germ cells during compensatory growth in mouse embryos. Journal of Embryology and Experimental Morphology. 1981;64:133–147. [PubMed] [Google Scholar]
- Tang WWC, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nature Reviews. Genetics. 2016;17:585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Human Genetics. 1976;34:119–124. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- Touré A, Szot M, Mahadevaiah SK, Rattigan A, Ojarikre OA, Burgoyne PS. A new deletion of the mouse y chromosome long arm associated with the loss of SSTY expression, abnormal sperm development and sterility. Genetics. 2004;166:901–912. doi: 10.1534/genetics.166.2.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta B, Cruciani F. Y chromosome palindromes and gene conversion. Human Genetics. 2017;136:605–619. doi: 10.1007/S00439-017-1777-8. [DOI] [PubMed] [Google Scholar]
- Turner JMA, Mahadevaiah SK, Ellis PJI, Mitchell MJ, Burgoyne PS. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Developmental Cell. 2006;10:521–529. doi: 10.1016/j.devcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Turner JMA. Meiotic silencing in mammals. Annual Review of Genetics. 2015;49:395–412. doi: 10.1146/annurev-genet-112414-055145. [DOI] [PubMed] [Google Scholar]
- Vernet N, Mahadevaiah SK, Ojarikre OA, Longepied G, Prosser HM, Bradley A, Mitchell MJ, Burgoyne PS. The y-encoded gene zfy2 acts to remove cells with unpaired chromosomes at the first meiotic metaphase in male mice. Current Biology. 2011;21:787–793. doi: 10.1016/j.cub.2011.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet N, Mahadevaiah SK, Yamauchi Y, Decarpentrie F, Mitchell MJ, Ward MA, Burgoyne PS. Mouse y-linked zfy1 and zfy2 are expressed during the male-specific interphase between meiosis I and meiosis ii and promote the 2nd meiotic division. PLOS Genetics. 2014;10:e1004444. doi: 10.1371/journal.pgen.1004444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet N, Mahadevaiah SK, de Rooij DG, Burgoyne PS, Ellis PJI. Zfy genes are required for efficient meiotic sex chromosome inactivation (MSCI) in spermatocytes. Human Molecular Genetics. 2016a;25:5300–5310. doi: 10.1093/hmg/ddw344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet N, Mahadevaiah SK, Decarpentrie F, Longepied G, de Rooij DG, Burgoyne PS, Mitchell MJ. Mouse y-encoded transcription factor Zfy2 is essential for sperm head remodelling and sperm tail development. PLOS ONE. 2016b;11:e0145398. doi: 10.1371/journal.pone.0145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, Grützner F, Deakin JE, Whittington CM, Schatzkamer K, Kremitzki CL, Graves T, Ferguson-Smith MA, Warren W, Marshall Graves JA. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Research. 2008;18:965–973. doi: 10.1101/gr.7101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Köhn FM, Schill WB, Farah S, Ramos C, Hartmann M, Hartschuh W, Meschede D, Behre HM, Castel A, Nieschlag E, Weidner W, Gröne HJ, Jung A, Engel W, Haidl G. Human y chromosome azoospermia factors (AZF) mapped to different subregions in yq11. Human Molecular Genetics. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- Vogt PH, Falcao CL, Hanstein R, Zimmer J. The AZF proteins. International Journal of Andrology. 2008;31:383–394. doi: 10.1111/j.1365-2605.2008.00890.x. [DOI] [PubMed] [Google Scholar]
- Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Human Molecular Genetics. 2005;14:2911–2918. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu YC, Markoulaki S, Welstead GG, Cheng AW, Shivalila CS, Pyntikova T, Dadon DB, Voytas DF, Bogdanove AJ, Page DC, Jaenisch R. Talen-mediated editing of the mouse Y chromosome. Nature Biotechnology. 2013;31:530–532. doi: 10.1038/nbt.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshons WJ, Russell LB. The y-chromosome as the bearer of male determining factors in the mouse. PNAS. 1959;45:560–566. doi: 10.1073/pnas.45.4.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles V. Genes adapt to outsmart gene targeting strategies in mutant mouse strains by skipping exons to reinitiate transcription and translation. Genome Biology. 2020;168:1–26. doi: 10.1186/s13059-020-02086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Schmidt JA, Avarbock MR, Tobias JW, Carlson CA, Kolon TF, Ginsberg JP, Brinster RL. Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. PNAS. 2009;106:21672–21677. doi: 10.1073/pnas.0912432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Riel JM, Stoytcheva Z, Burgoyne PS, Ward MA. Deficiency in mouse y chromosome long arm gene complement is associated with sperm dna damage. Genome Biology. 2010;11:R66. doi: 10.1186/gb-2010-11-6-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Riel JM, Stoytcheva Z, Ward MA. Two Y genes can replace the entire y chromosome for assisted reproduction in the mouse. Science. 2014;343:69–72. doi: 10.1126/science.1242544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Riel JM, Ruthig V, Ward MA. Mouse y-encoded transcription factor ZFY2 is essential for sperm formation and function in assisted fertilization. PLOS Genetics. 2015;11:e1005476. doi: 10.1371/journal.pgen.1005476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan BW, Zhao YF, Cao WG, Li N, Gou KM. Mechanism of random integration of foreign DNA in transgenic mice. Transgenic Research. 2013;22:983–992. doi: 10.1007/s11248-013-9701-z. [DOI] [PubMed] [Google Scholar]
- Zuo E, Cai Y-J, Li K, Wei Y, Wang B-A, Sun Y, Liu Z, Liu J, Hu X, Wei W, Huo X, Shi L, Tang C, Liang D, Wang Y, Nie Y-H, Zhang C-C, Yao X, Wang X, Zhou C, Ying W, Wang Q, Chen R-C, Shen Q, Xu G-L, Li J, Sun Q, Xiong Z-Q, Yang H. One-step generation of complete gene knockout mice and monkeys by CRISPR/Cas9-mediated gene editing with multiple sgRNAs. Cell Research. 2017;27:933–945. doi: 10.1038/cr.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]