Abstract
We utilized the mouse mammary tumor virus (MMTV) long terminal repeat (LTR) in vivo to understand how the interaction of the glucocorticoid receptor (GR) with a nucleosome-assembled promoter allows access of factors required for the transition from a repressed promoter to a derepressed, transcriptionally competent promoter. A mutation (C644G) in the ligand binding domain (LBD) of the mouse GR has provided information regarding the steps required in the derepression/activation process and in the functional significance of the two major transcriptional activation domains, AF1 and AF2. The mutant GR activates transcription from a transiently transfected promoter that has a disordered nucleosomal structure, though significantly less well than the wild-type GR. With an integrated, replicated promoter, which is assembled in an ordered nucleosomal array, the mutant GR does not activate transcription, and it fails to induce chromatin remodeling of the MMTV LTR promoter, as indicated by nuclease accessibility assays. Together, these findings support a two-step model for the transition of a nucleosome-assembled, repressed promoter to its transcriptionally active, derepressed form. In addition, we find that the C-terminal GR mutation is dominant over the transcription activation function of the N-terminal GR activation domain. These findings suggest that the primary activation function of the C-terminal activation domain is different from the function of the N-terminal activation domain and that it is required for derepression of the chromatin-repressed MMTV promoter.
Two questions are central to understanding how the initiation of transcription is regulated. The first is how proteins associated with the transcription initiation complex gain access to promoters assembled in complex nucleoprotein structures termed chromatin. The second is, what interactions among those proteins and promoter-specific transcription factors are necessary to coordinate the multiple functions needed to initiate gene activation. The regulatory role of chromatin remodeling in gene regulation is clear, but the mechanisms that are relevant in vivo are not (40, 58, 67).
Chromatin derepression and subsequent transcription initiation can be functionally separated, but how a transcriptional activator, such as the glucocorticoid receptor (GR), mediates these activities remains an open question. For the GR to activate transcription in vivo from the mouse mammary tumor virus (MMTV) long terminal repeat (LTR), the repressive effects of the chromatin, in which glucocorticoid response elements (GREs) are found, must be overcome (3, 61, 62, 80). The GR has thus provided a useful model to uncover mechanisms underlying chromatin-regulated gene expression. On the MMTV LTR, GR binding induces an open chromatin conformation that allows access of proteins to the promoter (13, 43). Direct or indirect protein interactions must then be made to bring the GR activation domain(s) and associated proteins into proximity with the basal transcription complex to effect transcription initiation.
A bimodal, or two-step, mechanism for transcriptional activation has been described for chromatin-assembled promoters including the MMTV LTR (5), the Pho5 activator (68), Cha1 in yeast (51), and the myogenin promoter (27), among others. In this model, the first step is chromatin remodeling, which allows subsequent protein-protein interactions necessary to bring enhancer-bound factors into contact with the basal transcription complex at the TATA box. Archer et al. (5) showed that GR-mediated transactivation on the MMTV LTR is bimodal, when they found that the transcription factor NF-1 is unable to bind to a closed or chromatin-assembled MMTV LTR, but that it binds constitutively to an MMTV LTR in an open chromatin conformation. Additionally, they found that even when NF-1 is constitutively bound to the open MMTV LTR, transcription is initiated only after the GR is activated by the addition of hormone. Thus, when the MMTV LTR is found in a closed (repressed) chromatin conformation, the first step in activation is hormone-dependent chromatin remodeling, mediated by the GR. After the chromatin is opened, or derepressed, and accessible to factors such as NF-1, there must be a second step in which contacts are presumably made between GR and/or GR-associated proteins and the basal transcription machinery to allow transcriptional initiation. This indicates that two functionally different activities are required for an activated initiation complex. How a transcriptional activator mediates these two mechanistically different functions has not been elucidated.
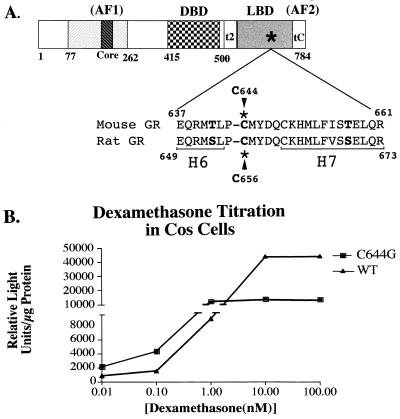
The GR is a member of the steroid nuclear receptor family that includes the progesterone, androgen, mineralocorticoid, and estrogen receptors, which in turn are part of the superfamily of nuclear receptors with the thyroid hormone receptor, the retinoic acid receptor, the vitamin D receptor, and orphan receptors (70). Nuclear receptors share homologies in their domain structure that include two transcription activation domains (AF1 and AF2), a DNA binding domain, and a ligand binding domain (LBD) (Fig. 1A) (17, 28, 36, 45). Steroid receptors have a third activation domain designated tau2 (GR) or AF2a (estrogen receptor) at the N-terminal end of the LBD (49, 53).
FIG. 1.
(A) Domain structure of steroid nuclear receptors. The hatched box represents the amino-terminal activation domain (AF1), and the darker hatched box represents the core activation domain. The checked box indicates the DNA binding domain (DBD), and the shaded box represents the carboxy-terminal LBD. The asterisk indicates the approximate location of the C644G mutation. The two regions in the C-terminal domain that have transcription activation potential are indicated by t2 and tC or AF2. Numbers indicate amino acid numbers in the mouse GR. The amino acid sequence surrounding the C644G mutation is also shown in comparison with the homologous sequence in the rat GR. Some of the residues that differ in the two sequences are in boldface. H6 and H7 represent the predicted boundaries of those α helices in the LBD based on crystal structures of other members of the nuclear receptor family. (B) Hormone titration curve of C644G versus wtGR. Cos-7 cells were transfected with 5 μg of either pCI-nH6HA-C644G or pCI-nH6HA-wtGR. Approximately 16 h following transfection, the cells were treated with the indicated amount of Dex for 6 h. Cells were harvested and assayed for luciferase activity. The number of receptors per cell was calculated by whole-cell binding of [3H]TA (see Materials and Methods). There were threefold more C644G receptors per cell than wtGRs. The data have been normalized for this difference. Note the break in the vertical scale where lower values are expanded relative to upper values.
The AF1 transactivation domain is located in the N-terminal region of GR and has been shown by mutational analysis to contain critical residues for GR-mediated transcriptional activation (reference 1 and references therein; 35, 48). The AF2 domain is located in the C terminus of GR and is contained within the LBD. In contrast to the ligand-independent transactivation mediated by the AF1 domain, transactivation by the AF2 domain is ligand dependent (31, 74). The liganded and unliganded crystal structures of the LBD of a number of other members of the nuclear receptor superfamily, but not GR, have been resolved, and the structure among the different receptors is highly conserved (for reviews, see references 50 and 78). There is a correlation between residues in the LBD/AF2 domain that disrupt transactivation when mutagenized and those that undergo a conformational change upon ligand binding. These data support the idea that conformational changes induced by ligand binding are involved in the generation of a transcriptionally active conformation of the AF2 domain (8, 60, 63, 69, 73, 75).
We show that the mouse GR, with a point mutation at cysteine 644 in the LBD, can activate transcription in vivo from a transiently transfected MMTV-luciferase reporter that does not have an ordered nucleosomal array, but not as effectively as the wild-type GR (wtGR). The mutant GR cannot activate transcription from a stably replicating reporter assembled in an ordered nucleosomal array and exhibits dominant negative repression on transcriptional activation mediated by the wtGR. The repression is the result of a closed (repressed) chromatin conformation, as indicated by a decrease in accessibility of a DNA hypersensitive site on the inactive promoter. Because this GR, with a single mutation, is functionally impaired in both chromatin remodeling and transcriptional initiation, the data provide a functional link between these two mechanistically separate events. We also find that the C-terminal GR mutation in the LBD is dominant over the transcription activation function of the N-terminal GR activation domain. This result suggests that the primary activation function of the C-terminal activation domain is different from the function of the N-terminal activation domain.
MATERIALS AND METHODS
Cell lines and plasmids.
The two cell lines used are designated 1471.1 and 3134. 1471.1 cells have been described previously (2, 10) and contain multiple copies of the stably replicating MMTV-chloramphenicol acetyl transferase (CAT) reporter. The 3134 cell line contains multiple copies of stably replicating MMTV-Ras reporter. Both cell lines were derived from the mouse adenocarcinoma line C127. Cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% fetal calf serum. The medium was replaced with DMEM with 10% 2× charcoal-stripped calf serum 18 to 24 h prior to transfection.
The mutant C644G was made by site-directed mutagenesis (41) of the mouse GR in plasmid pSV2wRec (17) as specified by the manufacturer (TaKaRa Biochemical). The oligonucleotide used for mutagenesis was 1916GAATGACTCTACCCGGCATGTATGACCAA1944. The mutagenized base is in boldface, and numbers indicate base numbers. The mutant GR, C644G, was amplified by PCR with a 5′ KpnI restriction site and a 3′ SmaI site and subcloned into the KpnI/SmaI-cut vector, pCI-nH6HA. pCI-nH6HA was derived from the Promega pCI plasmid as described previously (64). Plasmid pCI-nH6HA-C644G was fully sequenced from the 5′ to 3′ junctions of the GR and the vector plasmid. pCI-nH6HA-Mouse wtGR was made by replacing the EheI/NsiI fragment that contains the C644G mutation with the wild-type fragment. Other plasmids used have been described previously; they are pLTRluc (full-length MMTV LTR driving the luciferase gene) (46), pCMVIL2R (cytomegalovirus promoter driving the interleukin-2 receptor [IL-2R] gene) (29), and pCI-nH6HA-C656G (the rat GR with the homologous mutation to C644G at C656G) (64) (Fig. 1A).
The mouse and rat GR chimeric plasmids were made by cutting pCI-nH6HA-C644G with KpnI, ApaI, and NotI and pCI-nH6HA-C656G with PvuII, ApaI, and NotI. The common ApaI site in proline 319 in the rat GR and proline 307 in the mouse GR was used to ligate the N-terminal and C-terminal fragments. The mouse-rat chimera was made with the pCI-nH6HA-C644G vector cut at KpnI/NotI and ligated to the mouse KpnI/ApaI and rat ApaI/NotI fragments. The rat-mouse chimera was made with the pCI-nH6HA-C656G vector cut with PvuII/NotI ligated to the PvuII/ApaI rat fragment and the ApaI/NotI mouse fragment.
Transfections and MACS.
Cells were transfected with 5 μg of pCMVIL2 receptor plasmid, 10 μg of pLTRluc, and from 0 to 15 μg of pCI-nH6HA-C644G, pCI-nH6HA-C656G, or pCI-nH6HA Mouse wtGR plasmid DNA by electroporation with a BTX600 electroporator (BTX, Inc., San Diego, Calif.). Briefly, cells were trypsinized from flasks, rinsed in phosphate-buffered saline (PBS), resuspended at 2 × 107 cells in 300 μl of Dulbecco’s PBS, and chilled for 5 to 15 min on ice. Electroporation was at 960-μF capacitance, 129-ohm resistance, and 240 to 280 V in chilled cuvettes. After electroporation, cells were plated in DMEM with 1 to 10% 2× charcoal-stripped serum. After 15 to 20 h of recovery, cells were treated and harvested. Magnetic affinity cell sorting (MACS) (29) was used to separate transfected from nontransfected cells. Goat anti-mouse immunoglobulin G-coated magnetic beads (Dynal Inc., Lake Success, N.Y.) were incubated overnight with IL-2R monoclonal antibody (Upstate Biotechnology, Lake Placid, N.Y.). Beads were rinsed and diluted in medium S (4 mM EGTA, chondroitin sulfate [100 μg/ml], 0.1% gelatin, 10 mM HEPES [pH 8.0], 1 mM MgCl2, 1 mM MgSO4, nonfat dry milk [8 mg/ml], bovine serum albumin and [100 μg/ml] in PBS without Ca2+ or Mg2+ at a ratio of 15 μl of beads at 50 mg/ml to 1 ml of medium S. Cells were incubated for 15 min at 37°C with the IL-2R-coated beads in medium S and then placed between magnetic plates to separate those expressing IL-2R from those that were not. Sorted cells were resuspended in 0.25 M Tris (pH 7.5), lysed by three cycles of freeze-thaw, and assayed for CAT or luciferase activity.
CAT, luciferase, and protein analysis.
Cell lysates were analyzed for CAT activity by the thin-layer chromatography method (32). Five micrograms of total protein was assayed for 30 min at 37°C with [14C]chloramphenicol. Visualization of the acetylated products was done with a Molecular Dynamics PhosphorImager and the ImageQuant analysis program. Luciferase assays were carried out as previously described (46), using a Berthold Microlumat LB 96V luminometer. The reaction mix was 20 mM glycyl glycine (pH 7.5)–12 mM MgSO4–4 mM ATP–0.2 mM luciferin. Protein analysis was done by the Bradford method with reagent from Bio-Rad.
Nuclease hypersensitivity assays.
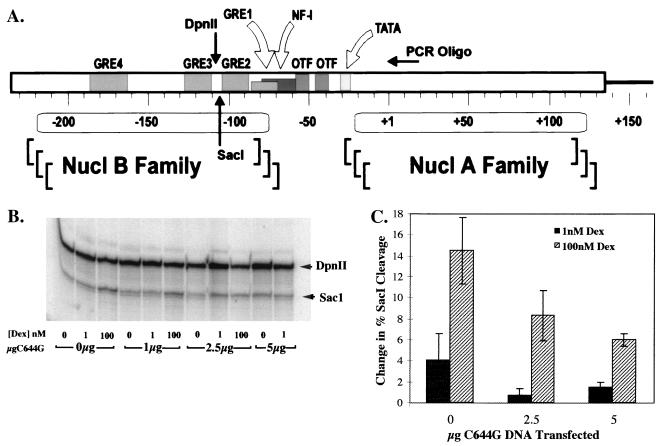
Nuclei were isolated from magnetically sorted cells, and 100 μg of nuclei was subjected to nuclease digestion with SacI (10 U/μg) in 50 mM NaCl–50 mM Tris (pH 8.0)–1 mM MgCl2–1 mM β-mercaptoethanol–2.5% glycerol for 15 min at 30°C. The reaction was terminated, and genomic DNA was extracted in 5 volumes of 10 mM Tris (pH 7.5)–10 mM EDTA–0.5% sodium dodecyl sulfate (SDS)–proteinase K (100 μg/ml) overnight at 37°C. DNA was purified by phenol-chloroform-isoamyl alcohol extraction and digested to completion with DpnII at 5 U of DNA/μg. Linear amplification of the digested fragments was carried out by PCR primer extension with a 32P-end-labeled primer to MMTV bases +1 to +27 to allow visualization of the SacI and DpnII fragments (see Fig. 5A) that were electrophoretically separated on 8% denaturing polyacrylamide gels. Hormone-induced SacI cleavage was calculated as the amount of cleavage in the SacI band divided by the total cleavage in both DpnII and SacI bands. PhosphorImager analysis was quantified by ImageQuant (Molecular Dynamics).
FIG. 5.
(A) Representation of the B nucleosome region of the MMTV LTR. The oligonucleotide used for PCR primer extension is indicated and extends from bases +1 to +27 of the MMTV coding region. The SacI and DpnII sites are indicated at −105 (SacI) and −113 (DpnII), as are the four GREs and the NF-1 and OTF binding sites. Numbers indicate base pairs of DNA with +1 as the transcription start site. (B) Nuclease hypersensitivity assays to evaluate whether chromatin was accessible with increasing amounts of C644G transfected into the cells. SacI digests were done on nuclei treated for 1 h with Dex and isolated from 3134 cells transfected with the indicated amount of C644G DNA and 5 μg of pCMV-IL2R DNA to allow magnetic sorting. Aliquots of nuclei containing 100 μg of DNA were subjected to digestion with the restriction enzyme SacI (10 U/μg) for 15 min at 30°C. Genomic DNA was isolated and digested to completion with 5 U of DpnII/μg of DNA. PCR primer extension reactions were run with the radiolabeled primer described above. Reaction products were resolved on 8% denaturing polyacrylamide gels with the SacI band migrating at 132 bp and the DpnII band migrating at 140 bp. (C) Gel analysis with the PhosphorImager program ImageQuant (Molecular Dynamics). Fractional SacI cleavage was calculated as the ratio of the amount of SacI digestion product to the total amount of digestion products (SacI plus DpnII). The change in percent SacI cleavage was calculated by subtracting the fractional cleavage observed in the absence of Dex from that in the presence of Dex. The standard errors of the means represent data from three separate experiments per treatment.
Electrophoretic mobility shift assays (EMSAs).
1471.1 cells were transfected with 5 μg of C644G DNA and 5 μg of IL-2R DNA as described above. Following recovery in DMEM–1% 2× charcoal-stripped calf serum, cells were treated with 1 nM dexamethasone (Dex) for 30 to 40 min and then sorted by MACS into transfected and nontransfected populations. Nuclear extracts were prepared from the transfected and the nontransfected cells. Miniextracts were made by the method of Lee et al. (44), in which cells are lysed in buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT]). Nuclei were isolated and extracted in buffer C (20 mM HEPES [pH 7.9], 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride 0.5 mM DTT). Dialysis was against buffer D (20 mM HEPES [pH 7.9], 20% glycerol, 0.1 M KCl, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM DTT, 5 mM MgCl2), and extracts were flash frozen. Protein was measured by Bradford assay (Bio-Rad).
The EMSA reaction mixture included 10 μg of nuclear extract incubated in buffer D with 30 to 60 fmol of 32P-end-labeled probe, either wtGRE (5′-GATCCGGTacaATCtgtTCTA-3′) (20) or mutant (5′-GATCCGGTcacATCgtgTCTA-3′) (lowercase letters indicate mutagenized bases), and 0.2 μg of poly(dI-dC) · poly(dI-dC) as nonspecific competitor in a 20-μl total reaction. A 30-min incubation at room temperature was followed by separation of shifted bands on a 5% acrylamide–0.5× Tris-borate-EDTA gel at 4°C. Gels were dried and analyzed by PhosphorImager analysis (Molecular Dynamics).
S1 nuclease analysis.
3134 cells were transfected by electroporation and sorted as described above. Following treatment with hormone, cells were lysed in 0.2 M Tris-HCl (pH 8.0)–140 mM NaCl–2 mM MgCl2–0.5% NP-40, and nuclei were pelleted. Cellular supernatant was extracted in STE (0.2% SDS, 5 mM Tris [pH 8.5], 2 mM EDTA) and phenol-chloroform-isoamyl alcohol. Radiolabeled probe was prepared by PCR primer extension from linearized pMTVβglobin or pT7-Actin-F as described by Smith et al. (64). Ten micrograms of total RNA plus labeled probe was hybridized at 42°C overnight. After treatment with 100 U of S1 nuclease (Gibco/BRL) for 1 h at room temperature, products were separated on an 8% denaturing polyacrylamide gel and visualized by PhosphorImager analysis. Quantitation was by ImageQuant (Molecular Dynamics).
Whole-cell hormone binding assay.
The average number of receptors per cell was determined by using a whole-cell hormone binding assay (14). Transfected cells were harvested and sorted by MACS, and a portion was counted; 1 × 104 to 4 × 104 cells were used in each assay. Cells were incubated with 50 nM [3H]triamcinalone acetonide (TA) (specific activity, 44.3 Ci/mmol) alone or with 5 μM unlabeled TA in a volume of 275 μl of 25 mM HEPES (pH 7.15)–PBS for 30 min at 37°C. Cells were rinsed twice in PBS, lysed with 2% SDS, and counted in Hydrofluor (National Diagnostics, Atlanta, Ga.). The average number of receptors per cell was determined by calculating the number of molecules of [3H]TA incorporated and dividing by the number of cells assayed. To calculate the number of mutant receptors per cell, the number of receptors in untransfected cells (endogenous receptors) was subtracted from the number of receptors in the pCI-nH6HA-C644G-transfected cells.
Experiments for receptor-hormone binding data for Scatchard analysis and nonlinear regression analysis were done essentially as described above (14) except that cells were incubated with [3H]TA for 1 h at 37°C in 10% 2× stripped serum–DMEM plus 25 mM HEPES (pH 7.15). Nonsaturable bound hormone was measured after a parallel incubation with a saturating concentration of unlabeled TA plus labeled TA. The nonsaturable value was subtracted from the amount of bound TA at each concentration of [3H]TA measured. A range of concentrations of hormone from 0.01, to 100 nM [3H]TA was used in each assay, and the data were analyzed by a two-binding-site analysis because all cells used in the analyses except EDR3 liver cells have some endogenous wild-type receptor (12). Cells used for analysis were Cos-7, EDR3, and 1471.1 cells, all transfected with either C644G DNA or wtGR in the same vector as the C644G mutant DNA. Following incubation and initial spin of cells, 20 μl of supernatant was saved for measurement of unbound hormone. All washes of cells were done immediately following the 1-h incubation with room temperature PBS. After two successive washes, the cells were lysed with 2% SDS and counted as described above. Binding data were analyzed to determine Kd by the GraphPad Prism program (San Diego, Calif.) using nonlinear regression analysis.
RESULTS
C644G binds hormone with higher affinity than the wtGR.
A point mutation was made in the LBD of the mouse GR that is homologous to a mutation in the rat GR designated C656G, described by Chakraborti et al. (9). (Fig. 1A). The mouse mutation is at cysteine 644, and the change is to glycine (C644G). The rat mutation increases steroid binding affinity to the extent that the receptor can be activated by a >6-fold-lower concentration of Dex than is required to activate the wtGR. This allows transcriptional activation by the mutant to be distinguished from activation by an endogenous wtGR when introduced into cells in culture. To compare induction of C644G to that of wtGR, we transfected either pCI-nH6HA-C644G or pCI-nH6HA-Mouse wtGR into Cos-7 cells that have a nonfunctional GR, along with the pLTRluc reporter plasmid, a fusion of the MMTV LTR and the structural gene for luciferase. We found that at 1 nM Dex, activation of C644G receptors was maximal (Fig. 1B). In contrast, at 1 nM Dex, activation of the mouse wtGR was only 15 to 20% of the maximum reached at 100 nM Dex. This confirmed that with 1 nM Dex, the mutant mouse GR can be maximally activated whereas the wtGR is activated only to 15 to 20% of maximum.
To determine hormone binding affinity of C644G versus wtGR, we did whole-cell hormone binding assays and determined Kds by nonlinear regression analysis on transfected Cos-7, EDR3, and 1471.1 cells. Analysis of wtGR was also done on untransfected 1471.1 cells. Cells were incubated with [3H]TA for 1 h at 37°C. The calculated Kd for C644G is 0.68 nM ± 0.32 (standard deviation) (n = 5), and that for endogenous wild-type receptor is 2.7 nM ± 0.61 (standard deviation) (n = 7). The independent experiments (n = 5 and 7) included data from analyses in all three cell lines. The fourfold difference in these values is in reasonable agreement with results of Chakraborti et al. (9), taking into account that those analyses were done with [3H]Dex rather than [3H]TA and were done by cytosol assay rather than by whole-cell assay. They reported a Kd of 0.55 nM ± 0.16 for the C656G rat mutant and 4.73 nM ± 2.04 for the rat wtGR.
Transactivation from a nonphased nucleosomal promoter by C644G.
To compare transcriptional activation by C644G from a promoter assembled into a regularly phased nucleosomal array to one with a disordered nucleosomal array, we used the mouse adenocarcinoma-derived cell line 1471.1. The cells have endogenous wtGR and stably maintain multiple copies of the reporter MMTV-CAT, a fusion of the MMTV LTR and the CAT gene. In these cells, the MMTV LTR driving the CAT reporter is assembled in an ordered array of nonrandomly positioned nucleosomes and is repressed, or closed, in the absence of hormone (24, 61). When the GR is activated by hormone, it binds to the GREs found in the B nucleosome of the MMTV LTR (see Fig. 5A). A structural transition in the chromatin allows access of Oct-1, NF-1, and basal transcription factors to the promoter (13, 25, 43, 59). In contrast, the transiently transfected template, pLTRluc, is nonreplicating, lacks an ordered nucleosomal array, and has a constitutively open conformation, as was shown by the binding of NF-1 and Oct-1 to their sites in the absence of hormone (5, 43). When the hormone-activated GR binds to the pLTRluc reporter, no obvious structural transition in the template occurs, but basal transcription factors are recruited to the promoter, which results in transcriptional activation (5). Thus, activation of the two reporters can be compared in the same cell, to distinguish between requirements for transcription that are part of the chromatin remodeling process from those that are not.
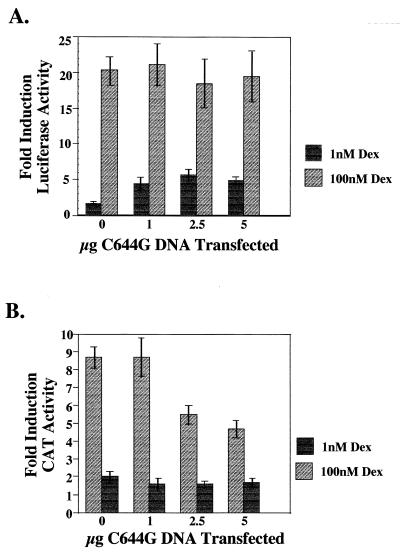
The 1471.1 cells were transfected with pCI-nH6HA-C644G, the pLTRluc reporter, and an IL-2R expression plasmid. IL-2R is expressed on the surface of transfected cells, allowing MACS of the cells with beads coated with anti-IL-2R antibody (29). This method enabled us to isolate an enriched population of cells expressing C644G and the transiently expressed pLTRluc reporter. At 12 to 16 h following transfection, the cells were induced with 0, 1, or 100 nM Dex for 4 h. Cells were then sorted, and the transfected cells were lysed and assayed for luciferase and CAT activity. As shown in Fig. 2A, there was an increase in reporter activity from the pLTRluc reporter with both 1 nM (about 5-fold) and 100 nM (about 20-fold) Dex. At 1 nM Dex, C644G receptors are maximally activated and endogenous wtGR is activated to 15 to 20% of maximum (Fig. 1B). This results in a four- to fivefold induction over Luc activity when no mutant receptor is present (Fig. 2A; compare 1 nM at 0 μg of C644G with 2.5 μg of C644G). At 100 nM Dex, both the wild-type and mutant receptors are maximally activated (Fig. 1B). C644G is able to activate transcription from the disordered nucleosomal reporter, pLTR Luc, but less well than the endogenous wtGR (Fig. 2A; compare 1 and 100 nM Dex at any amount of C644G transfected).
FIG. 2.
(A) Titration of the C644G receptor. 1471.1 cells were transfected with the indicated amount of C644G plasmid DNA along with 10 μg of pLTRluc and 5 μg of pCMV-IL2R DNA. Twelve to sixteen hours posttransfection, the cells were treated with 0, 1, or 100 nM Dex for 4 h. Cells were magnetically sorted, and lysates were analyzed for luciferase and CAT activity. Data are expressed as fold induction over basal activity at 0 nM Dex. At 1 nM Dex, only C644G is more active than wtGR; at 100 nM Dex, both C644G and the endogenous GR are activated. Fold induction was calculated by dividing activity at either 1 or 100 nM Dex by the activity in cells treated with vehicle (ethanol) only (0 nM Dex). Error bars represent standard error of the mean with n = 12 to 16. (B) The lysates used for panel A were assayed for CAT activity; 5 μg of total protein was used in the CAT assays, and reaction mixtures were incubated for 30 min at 37°C. Thin-layer chromatography was used to separate the acetylated forms of chloramphenicol, and PhosphorImager analysis was used for quantitation (ImageQuant; Molecular Dynamics). Fold induction is activity over basal induction from cells treated with vehicle (ethanol) only.
C644G cannot activate transcription and is a dominant negative repressor of transcription by wtGR on a phased nucleosomal promoter.
In striking contrast to the results for the luciferase reporter, there was very little activation with 1 nM Dex (Fig. 2B) with or without C644G DNA when we assayed for CAT activity. The small amount of activation (less than twofold) is probably due to the low activation of the endogenous wtGR in these cells (Fig. 1B and 5B). In addition, at 100 nM Dex there was an 8- to 10-fold activation of CAT activity when no C644G was present. As increasing amounts of C644G DNA were added to the cells, we observed a decrease in activation by the endogenous GR that had not been seen with the luciferase reporter at 2.5 and 5 μg of transfected DNA (compare Fig. 2A and B at 100 nM Dex). We do not detect the dominant negative effect on CAT activity at 1 nM Dex. We attribute this to the low levels of CAT activity from the 15 to 20% of maximum wtGR activity at 1 nM Dex and an insensitivity of the assay at this low level of expression. We believe that if the assay were more sensitive, we would be able to see a dominant negative effect on the wtGR by C644G at 1 nM Dex as we can in S1 analysis of reporter mRNA (Fig. 3) and in nuclease hypersensitivity assays to be described (see Fig. 5B and C).
FIG. 3.
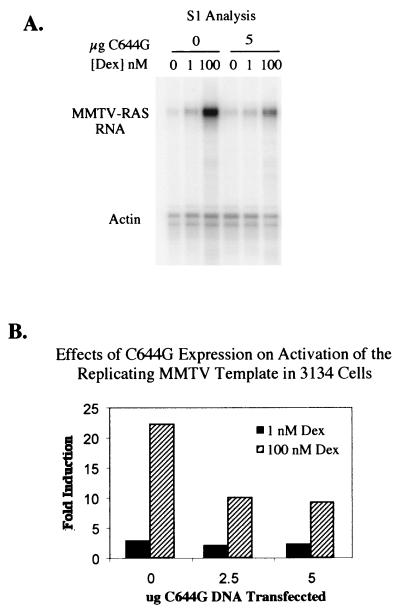
(A) S1 analysis of 3134 cells. An 8% denaturing polyacrylamide gel was used to analyze mRNA in 3134 cells following treatment with Dex. Cells were transfected with either 0 (vehicle), 2.5 (not shown), or 5 μg of C644G DNA as indicated and treated with 0, 1, or 100 nM Dex as for Fig. 2. Two radiolabeled probes were used for mRNA protection, MMTV-Ras (stably integrated reporter) and actin F (control). (B) Quantitation of the gel in panel A by PhosphorImager analysis and ImageQuant. The zero Dex value was used as the baseline for each amount of C644G transfected. All values were normalized for loading differences based on the actin band.
S1 nuclease analysis of RNA from the stably integrated MMTV-Ras reporter in 3134 cells.
S1 nuclease analysis was done on a stably integrated MMTV-Ras reporter gene in 3134 cells. Cells were transfected with 0, 2.5, or 5 μg of C644G and treated with 0, 1, or 100 nM Dex for 4 h. There is very little Ras RNA present at either 0 or 1 nM Dex and about a 50% decrease in RNA when C644G is expressed in the cells treated with 100 nM Dex (compare 0 and 5 μg of C644G at 100 nM Dex in Fig. 3A and B). These data demonstrate the same dominant negative effect observed in 1471.1 cells (Fig. 2B).
Together, these data demonstrate the inability of C644G to activate transcription from a chromatin-assembled promoter; furthermore, when coexpressed with the endogenous wtGR, it acts as a dominant negative repressor of transactivation from the endogenous wtGR.
wtGR can complement the dominant negative effect of C644G.
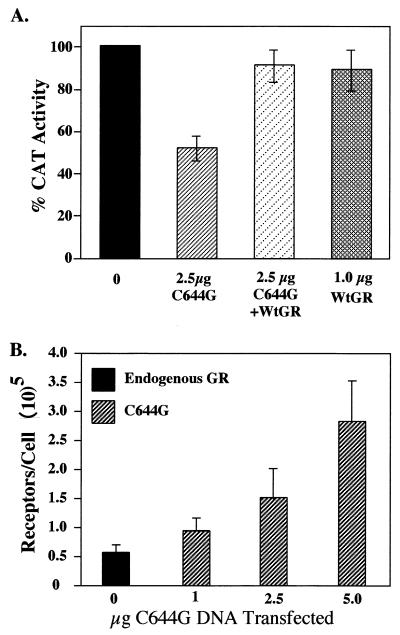
Complementation of the dominant negative phenotype of C644G with the addition of wtGR demonstrates that the phenotype is not due to squelching of basal transcription factors that would result in a general decrease in transcription (Fig. 4A). 1471.1 cells were transfected with C644G alone, the mouse wtGR alone, or C644G and wtGR together. Cells were treated with 0 or 100 nM Dex for 4 h and harvested, and CAT activity was assayed. The dominant negative effect of C644G inhibits CAT activity to about 50% of that seen when no mutant is present (compare 0 and 2.5 μg of C644G). When 2.5 μg of C644G and 0.5 μg of wtGR DNA are cotransfected into the cells, transcription of the CAT reporter gene is restored to almost the same level as when only the endogenous GR or the wtGR is present. In this experiment, the wtGR was introduced into the cells on a plasmid as was C644G. The number of receptors expressed when 0.5 μg of wtGR DNA was transfected into the cells is approximately equal to the number of endogenous receptors (Fig. 4B).
FIG. 4.
(A) Complementation of C644G by wtGR. 1471.1 cells were not transfected (0) or transfected with 2.5 μg of pCI-nH6HA-C644G DNA (C644G) alone or with 2.5 μg of C644G plus 0.5 μg of pCI-nH6HA-wtGR (WtGR) or 1 μg of wtGR alone, as indicated, as well as 5 μg pCMV-IL2R DNA. Cells were treated with vehicle (ethanol) or 100 nM Dex for 4 h, magnetically sorted, lysed, and assayed for CAT activity. Data are expressed as percent CAT activity above or below the sample with endogenous GR alone (no transfected DNA) at 100 nM Dex (solid bar). Error bars represent standard errors of the means with three to six points per treatment. (B) Whole-cell binding assays to determine the number of receptors per cell. 1471.1 cells were transfected with the indicated amount of pCI-nH6HA-C644G DNA and 5 μg of pCMV-IL2R DNA and allowed to recover 16 to 20 h. Cells were harvested, sorted, and assayed as described in Methods and Materials. In each whole-cell binding assay, the number of receptors in untransfected cells (endogenous receptors) was subtracted from the number of receptors in the C644G-transfected cells. Error bars represent standard errors of the means (n = 5 to 8).
To determine how many receptors were expressed when pCI-nH6HA-C644G DNA was transfected into 1471.1 cells, cells were transfected and magnetically sorted, and whole-cell hormone binding assays were done. Endogenous receptors were measured in the unsorted cells (cells not bound to magnetic beads) in each experiment. As shown in Fig. 4B, the number of endogenous receptors in the 1471.1 cells is approximately 5 × 104/cell. This value was measured in all C644G transfections shown. Receptor number shown is the number of C644G receptors at each concentration of C644G DNA transfected minus the endogenous receptor number. At 1 μg of C644G DNA transfected, the average number of mutant receptors per cell is 105, at 2.5 μg the average number is about 1.5 × 105, and at 5 μg the average number is about 3 × 105. Therefore, at 1 μg of C644G DNA transfected, there is a ratio of 1:1.5 of endogenous to mutant receptor, at 2.5 μg there is a 1:3 ratio, and at 5 μg there is a 1:6 ratio. Parallel determinations were done in 3134 cells, and the receptor expression levels were the same as in 1471.1 cells. These data confirm that C644G was being expressed in the cells and at levels greater than endogenous GR. This suggests that there should be enough mutant receptor available to activate the reporters MMTV-Luc, MMTV-CAT, and MMTV-Ras.
C644G is unable to remodel chromatin.
Our observation that the C644G receptor can activate transcription from the transiently expressed (pLTRluc) but not the nucleosome-assembled (MMTV-CAT) template led us to investigate whether C644G is defective in mediating chromatin access. An endonuclease (SacI) cleavage site is present in the stably replicating MMTV-CAT reporter within the cluster of the three GREs in the B nucleosome region of the promoter (Fig. 5A). Access of SacI to this cleavage site increases 10 to 20% upon addition of Dex and is therefore an indicator of a GR-induced structural transition in chromatin (25) at the MMTV LTR.
The cell line 3134 was transfected with increasing amounts of C644G DNA and induced for 1 h with 0, 1, or 100 nM Dex. One hour was chosen because in a time course of SacI cleavage in Dex-induced cells, the amount of cleavage was optimal at 1 h and decreased after that, even in the continued presence of hormone (4). Like 1471.1 cells, 3134 cells are derived from C127 mouse mammary adenocarcinoma cells but carry a stably integrated MMTV-Ras reporter instead of MMTV-CAT. This cell line was used because the magnitude of the change in SacI cleavage is greater than in 1471.1 cells upon activation of wtGR, and thus any modulations caused by the expression of C644G would be more apparent (our data and reference 25). SacI experiments were also done with 1471.1 cells, and the results paralleled the results from the 3134 cells (data not shown).
Maximum change in percent SacI cleavage between 0 and 100 nM Dex by the endogenous wtGR, when no C644G DNA was present in the cells, was about 15% (Fig. 5B and C). The 15% value represents the increase in cleavage at 100 nM Dex over basal cleavage at 0 nM Dex. This amount of cleavage by the endogenous GR is consistent with results reported by Fragoso et al. (25), in which cell line 3134 was extensively characterized for cleavage by various restriction enzymes with sites in the hypersensitive region of MMTV-LTR. The change in percent cleavage of the chromatin templates decreased at 100 nM Dex as increasing amounts of C644G DNA were expressed in the cells to about 9 and 7% when cells were transfected with 2.5 and 5 μg of C644G DNA (Fig. 5C). The changes in SacI cleavage when C644G is expressed represent a 50% drop in template accessibility compared to the amount of cleavage when no C644G is expressed. This result is comparable to the decrease in CAT activity in 1471.1 cells (Fig. 2B) and the decrease in Ras RNA levels in 3134 cells when increasing amounts of C644G are added to cells (Fig. 3). Thus, the dominant negative effect of C644G can be accounted for by decreased accessibility to the chromatin-assembled promoter. A decrease in accessibility can also be detected in this assay at 1 nM Dex upon addition of C644G (Fig. 5B and C), indicating that the cleavage at 1 nM Dex, when no C644G is transfected, is probably due to the 15 to 20% of endogenous GR activity at 1 nM Dex (Fig. 1B).
We also examined whether C644G could mediate a structural change in the chromatin at 1 nM Dex. Cells transfected with C644G were treated for 1 h with 1 nM Dex. Data in Fig. 5C show that the change in percent SacI cleavage at 1 nM Dex when no C644G DNA was added to the cells was about 4%, which is three- to fourfold-lower than that seen with full induction at 100 nM Dex with no C644G present. At 1 nM Dex, when the cells were transfected with 2.5 or 5 μg of pCI-nH6HA-C644G, there was no increase in SacI cleavage but rather a decrease reflecting the dominant negative effect observed at 100 nM Dex (Fig. 5B, 3, 2B, and 5C). We therefore conclude that C644G alone is unable to induce a structural transition when the GRE is in a repressed chromatin-assembled context and that is the reason for the lack of transcriptional activity (CAT activity and Ras mRNA) seen at 1 nM Dex (Fig. 2B and 3).
C644G is able to bind to the GRE in EMSAs.
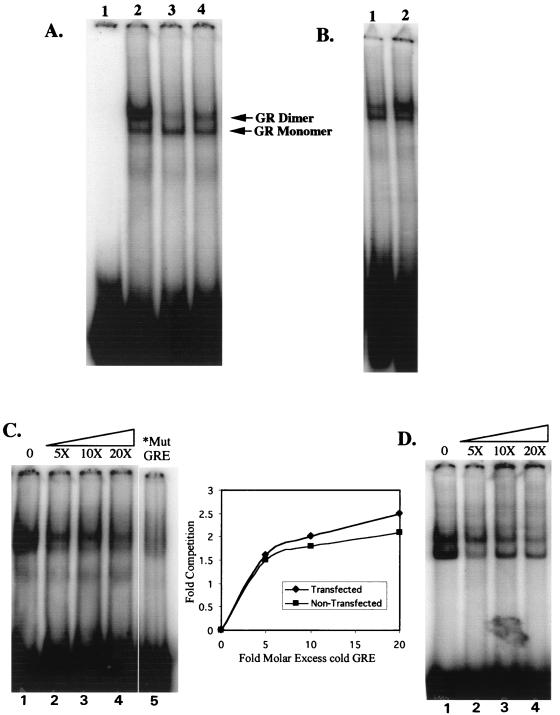
The GR binds to a GRE as a monomer and a dimer that can be detected in gel shift assays (15, 19, 71). To ascertain that C644G can bind to a GRE, we did EMSAs with nuclear extracts prepared from 1471.1 cells. Cells were transfected with 5 μg of C644G and 5 μg of IL-2R and allowed to recover overnight. Cells were incubated with 1 nM Dex for 40 min and then sorted as described above. Mini-nuclear extracts were made (44) from magnetically selected (transfected) cells and from the nonselected (nontransfected) cells. The cytosolic fraction from each extraction was also saved and dialyzed against buffer D as was the nuclear extract.
EMSAs were performed on the extracts as shown in Fig. 6. We treated transfected 1471.1 cells with 1 nM Dex prior to preparing nuclear extracts to specifically move C644G into the nucleus and minimize the amount of wtGR in the nucleus. Shifts were done with a radiolabeled consensus GRE (20), and 10 μg of nuclear extract was used in all reactions. Lane 3 in Fig. 6A shows a dimer/monomer pattern in nontransfected cell nuclear extracts. Lane 2 shows a large increase in the dimer band when nuclear extracts are made from cells transfected with C644G. The increase in the dimer band is the result of the increased number of transfected C644G GRs in the cells which shifts the binding equilibrium to the dimer complex. It has been shown previously that at low concentrations of GR, the monomeric band predominates in shift assays, but as the concentration of receptor increases, the dimer form also increases (15, 19, 71). We show this in Fig. 6B, in which we doubled the amount of GR in the shift by doubling the amount of lysate added to the shift (compare lanes 1 and 2, with 10 and 20 μg of nuclear extract). Quantitation of the shifted dimer form, as a percentage of the total amount of probe loaded, was twofold higher with 20 μg of extract than with 10 μg of extract. One would expect less GR in the nucleus and therefore little of the dimer form in untransfected cells treated with 1 nM Dex. At 1 nM Dex, very little of the endogenous GR would be activated, and so it should remain in the cytoplasmic compartment of the cell. This is what is seen when lanes 2 and 3 in Fig. 6A are compared. In lane 4, the shift is with 10 μg of cytosolic extract from the transfected cells treated with 1 nM Dex. More of the dimer form is seen than in the untransfected cells, presumably because both endogenous GR and some C644G are left in the cytoplasm in these cells.
FIG. 6.
(A) C644G can bind to a GRE by EMSA. Lane 1, free GRE (probe) (described in Methods and Materials) end labeled with 32P; lane 2, 10 μg of nuclear extract from C644G-transfected cells treated with 1 nM Dex; lane 3, 10 μg of nuclear extract from nontransfected cells treated with 1 nM Dex; lane 4, 10 μg of cytosolic extract from transfected cells treated with 1 nM Dex. Arrows indicate shifted bands corresponding to the GR monomer (lower) and dimer (upper). (B) As more receptor is added to shifts, the dimerized form of the GR becomes the predominant band. In lane 1, 10 μg of nuclear extract from nontransfected 1471.1 cells treated with 1 nM Dex was incubated with the radiolabeled GRE; in lane 2, 20 μg of the same nuclear extract was incubated with the labeled GRE. (C) Competition for binding with unlabeled GRE with nuclear extracts from C644G-transfected 1471.1 cells. In lanes 1 to 4, no competitor to 20-fold excess unlabeled GRE was incubated with 10 μg nuclear extract from C644G-transfected cells treated with 1 nM Dex and 32P-labeled GRE; lane 5 shows a shift with radiolabeled mutant (Mut) GRE and the same nuclear extract used in the competitions. The inset shows the amount of competition at the indicated molar excess of unlabeled GRE in each lane of the gels shown in panels C and D. (D) Binding competition as for panel C except that nuclear extracts were prepared from untransfected 1471.1 cells. Quantitation in the untransfected cells was of the dimerized band.
From these data, we can conclude that in 10 μg of transfected (C644G) nuclear extract, there are more receptors in the extract that can bind to the GRE, and so we see more of the dimer form shifted than with 10 μg of nuclear extract from untransfected cells. Extracts in Fig. 6B were prepared on a different day than extracts in Fig. 6A, and there is slightly more of the dimer form in 10 μg of extract in Fig. 6B. At 1 nM Dex there is enough endogenous GR in the nucleus to activate the luciferase and CAT reporters at very low levels (Fig. 2), and there is some variation from experiment to experiment in the amount of endogenous GR that is found in the nucleus.
To demonstrate that wtGR and C644G have similar interactions with the GRE, we did EMSA competitions using 5- to 20-fold molar excess unlabeled GRE to compare nuclear extracts from 1471.1 cells transfected with C644G (Fig. 6C) and untransfected 1471.1 cells (Fig. 6D). The untransfected cells have endogenous GR alone. Extracts were prepared from transfected 1471.1 cells as described for Fig. 6A except that transfections were done with 10 μg of C644G DNA rather than 5 μg. Each shift reaction had 58 fmol of 32P-labeled GRE and the indicated amount of unlabeled competitor GRE added as well as 10 μg of nuclear extract. As shown in Fig. 6C and D, a 5-fold molar excess of unlabeled GRE produces a similar and significant competition of both the endogenous wtGR and the C644G GR (compare lanes 1 and 2). At 10- and 20-fold molar excess GRE, there is almost complete competition for binding with the labeled probe. The monomer and dimer forms of the GR-GRE complex are distinct in the untransfected cells but not in the transfected cells. We attribute this to the high levels of GR in the transfected cells in this experiment, consistent with data in Fig. 6A, lane 2. DNA binding characteristics of wtGR and C644G GR for the GRE were essentially the same (graph in Fig. 6) in the EMSA competitions.
We also show that both C644G GR and endogenous GR bind specifically to the GRE. There is no shift when 32P-labeled mutant GRE is used to shift the same nuclear extract used in the transfected cell competitions (Fig. 6C; compare lane 5 with lane 1). The EMSA data provide evidence that the chromatin remodeling defect of C644G is not due to a defect in DNA binding.
The LBD mutation is dominant over the N-terminal transactivation domain.
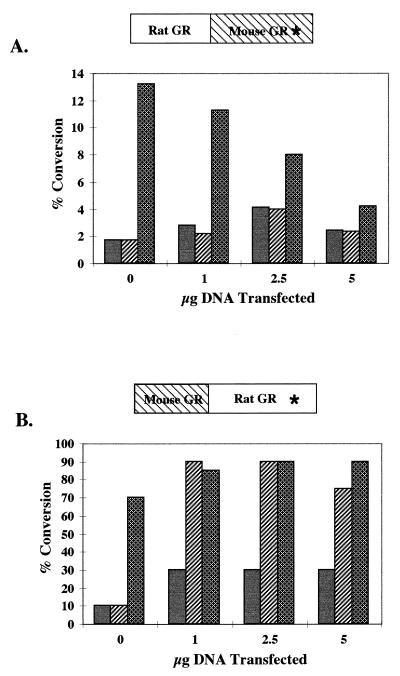
We were surprised by the inability of C644G to remodel chromatin and activate transcription, because the homologous mutation in the rat GR, C656G, changes the receptor’s affinity for steroid but does not affect its ability to activate transcription and remodel chromatin (64). The two receptors are 95% homologous, and we wondered whether the mutation in the mouse LBD would have the same phenotype in the context of the N-terminal portion of the rat GR. We therefore made two chimeras, one a rat-mouse GR, in which the entire carboxy-terminal portion of the rat GR, including the DNA binding domain to the stop codon, was replaced with the mouse C644G-containing carboxy terminus (Fig. 7A). We also made a mouse-rat GR, in which the entire carboxy-terminal portion of the mouse GR was replaced with the rat C656G-containing carboxy terminus (Fig. 7B). We transfected 1471.1 cells with increasing amounts of chimeric DNA, as was previously done with C644G DNA, and assayed for luciferase and CAT activity. The results from a representative CAT assay (Fig. 7A) demonstrate that the mouse mutation, in the context of the rat N terminus, confers the dominant negative phenotype of C644G on the receptor at 100 nM Dex, and no CAT activity was seen with 1 nM Dex. The rat mutation, C656G, in the context of the mouse N terminus confers the phenotype of C656G, in which there is activation of transcription with both 1 and 100 nM Dex (Fig. 7B and reference 64). Both chimeras activated the pLTRluc reporter at 1 and 100 nM Dex, consistent with previous results with C644G and C656G (Fig. 2A reference 64), in which there was activation of the reporter at both 1 and 100 nM Dex with both chimeras (data not shown). Background CAT activity levels vary from experiment to experiment and increase as more DNA is transfected. The main observation, that there is as much activity at 1 nM Dex as at 100 nM Dex with the mouse-rat chimera but essentially no activity above background with the rat-mouse chimera at 1 nM Dex, is very consistent. There is also no indication of a dominant negative effect at 100 nM with the mouse-rat chimera as there is with the rat-mouse chimera.
FIG. 7.
(A) 1471.1 cells were transfected with 0, 1, 2.5, or 5 μg of the rat-mouse chimera that carries the C-terminal mutation C644G indicated by an asterisk. The cells were induced with 0, 1, or 100 nM Dex for 4 h, sorted, and lysed; 5 μg of total protein was used to measure CAT activity. Assay mixtures were incubated for 30 min at 37°C. Thin-layer chromatography was used to separate the acetylated forms of chloramphenicol, and PhosphorImager analysis (ImageQuant; Molecular Dynamics) was used for quantitation. Solid bars, cells treated with vehicle only; hatched bars, cells treated with 1 nM Dex; cross-hatched bars, cells treated with 100 nM Dex. (B) 1471.1 cells were transfected with the mouse-rat chimera that carries the C656G mutation in the rat C-terminal domain, treated, and assayed as described above.
These results show that the mutation in the carboxy-terminal portion of the mouse GR is responsible for the transcriptional defect seen in the intact C644G receptor. Furthermore, they suggest that AF1 and AF2 provide two separable functions for transcriptional activation and that AF2 function is essential for chromatin derepression and may be necessary for AF1 to have functional activity on a chromatin-assembled promoter.
DISCUSSION
C644G is defective in two distinct events necessary for transactivation.
In the bimodal model for transcriptional activation from a chromatin-assembled promoter, the repressive effects of chromatin must first be overcome by chromatin remodeling. After chromatin is remodeled, contacts between the basal transcription complex at the TATA box and the promoter-specific activator must be made. At the MMTV LTR both of these steps are hormone dependent (5), which indicates that an activated hormone receptor is essential for both steps. Our data support this model and furthermore provide in vivo evidence that the two steps are functionally linked either through a single protein-protein interaction or through multiple protein-protein interactions that are mediated by and dependent on the GR.
C644G is defective in two mechanistically distinct events leading to transactivation. The first is in chromatin remodeling, as shown by the failure of C644G to induce a SacI cleavage at 1 nM Dex (Fig. 5B and C) and by the decrease in accessibility to the MMTV LTR as increasing amounts of C644G DNA are transfected into cells treated with 100 nM Dex (dominant negative effect) (Fig. 5B and C). As more C644G is expressed in the cells, fewer promoters are remodeled, which suggests that as more of the dimerized mutant is bound to the GRE, the endogenous wtGR is outcompeted for binding and remodeling. The data also indicate that C644G is a less effective transcriptional activator than is the endogenous wtGR on a promoter that is not in an ordered nucleosomal array. Figure 2A shows that there is a 15- to 20-fold increase in transactivation when the endogenous GR and C644G are both activated by 100 nM Dex, but at 1 nM Dex, when C644G is maximally activated, there is only a 4- to 5-fold increase in transactivation over the 1.5-fold induction by wtGR at 1 nM Dex. Thus, C644G has two defects: (i) an inability to remodel chromatin and (ii) an impaired ability to act as an effective transcriptional activator on an open promoter and provide a functional link between chromatin remodeling and transcription initiation.
What interactions might account for the C644G phenotype?
The obvious link between chromatin remodeling and the activation of transcription is through the contacts between the large chromatin remodeling complexes and the RNA polymerase II holoenzyme positioned at the TATA box (76). Transcription factors, such as ligand-activated nuclear receptors, target specific promoters for activation and provide a nucleation site for protein interactions with coactivator proteins that are often found in large chromatin remodeling complexes. Transcription factors also provide sites for interaction with the basal transcription machinery through multiprotein complexes (for a review, see reference 6). It is likely that multiple protein-protein interactions are required to bring the GR and its associated proteins into proximity with the basal transcription complex. If C644G interacts ineffectively with one protein, it could affect subsequent protein-protein interactions. There are many possible interactions between transcription factors, transcription factor-associated chromatin remodeling factors, and RNA polymerase II-associated factors, but it remains to be determined which of those interactions are functionally relevant in vivo (for reviews see references 39 and 67).
The inability of C644G to remodel chromatin, in addition to its weak transactivation potential, suggests that it may not interact well with one or more of the coactivators or with the basal transcription machinery. The GR can promote the establishment of an open chromatin conformation in vivo, and it associates with large protein complexes that contain coactivators such as p300/CBP, SRC-1, and GRIP-1, in addition to SWI/SNF/BRG1 (references 27 and 57 and references therein). Histone acetylase/deacetylase activity is associated with coactivators (38, 54, 66), chromatin remodeling, and subsequent gene activation 7, 42, 72; reviewed in references 33, 39, and 77). The coactivators themselves may interact directly with the basal complex and/or they may make an essential contact with other proteins that can then interact with the basal machinery, thus bridging the gap between the promoter-specific transcriptional activator and the basal transcription complex (52). Whether C644G is able to interact with known coactivators or as yet unidentified factors is now being investigated.
Interactions of coactivators with the GR and with other nuclear receptors are largely mediated by the LBD in which the AF2 domain resides and the interactions are ligand dependent (22, 30). From the resolved crystal structures of other nuclear receptor family members, the LBD consists of 12 α helices as well as four β strands (73, 78). The AF2 activation domain core is found in helix 12 (78) and is essential for transactivation (16). Specific residues in the LBD in helices 3, 4, 5, and 12 (11, 22) have been identified as sites for interactions with coactivators such as SRC-1 (55) and GRIP-1 (37) among others, which indicates that one role of the LBD is the recruitment of factors that are involved in chromatin remodeling. The mutation C644G or the homologous residue in other members of the family falls between helices 6 and 7 (Fig. 1A) in all of the LBDs crystallized to date. This region of the LBD has not been defined by any other mutations as an important site for interactions with any of the known coactivators, but it is deep in the LBD pocket where direct contact is made with ligand (9, 65). An amino acid change at this site could have structural consequences that affect protein interactions with the LBD. It is interesting that in the ligand-bound state, helix 3 lies under helix 6 (30) and helices 3, 4, 5, and 12 make a hydrophobic groove in the thyroid hormone receptor β LBD through which contact with coactivators is made (18). Although the GR LBD has not been crystallized, based on the high degree of structural homology in the LBDs found among the various receptors and the demonstrated association of coactivators with the LBD of GR, it is likely that the predicted structures apply to the GR as well as to other members of the nuclear receptor family.
Different phenotypes of the mutation in the mouse GR versus the rat GR.
It is curious that unlike C644G, the analogous mutation in the highly homologous rat GR can remodel chromatin and activate transcription. The primary differences in the immediate area of the C644G and C656G mutations are in two residues that flank the mutant site (Fig. 1A). In the rat GR they are serines 653 and 669; at the homologous positions in the mouse GR, they are threonines 641 and 657. Either serine or threonine is conserved at the homologous position to mouse T641 in GRs from all species sequenced thus far (except Xenopus), but at the position homologous to T657 in the mouse GR there is a serine in the GR in other species. We do not know whether these residues contribute to the very different phenotypes that we observe between the rat and the mouse GR LBD mutants. Serine and threonine both have aliphatic hydroxyl side groups, but threonine has a methyl group that serine does not have, which could affect local hydrogen bonding or van der Waals interactions with associated proteins. Crystal structures of the LBDs of the estrogen and progesterone receptors with agonist and antagonists bound demonstrate that conformational changes have profound effects on the potential interactions between amino acids in the LBD with associated proteins (50, 63, 69). The basis for the differences that we see between the mouse and rat GR is now being investigated.
Different roles for the C-terminal and N-terminal activation domains of GR?
The AF1 and AF2 domains in the GR are functionally different since AF1 is constitutively active and the activity of the AF2 domain is ligand dependent (31 and references therein). This raises the question as to whether the roles that they play in transcriptional activation in vivo are also different. MyoD, a transcriptional factor in the myogenic family of basic helix-loop-helix transcription factors, has a transcription activation domain that is independent of another domain that mediates chromatin remodeling (27). Pho4, a transcriptional activator that is activated in response to high phosphate levels in yeast, also requires a chromatin transition to activate transcription, but in this case both remodeling and activation are mediated by a single activation domain (21, 47). Together the data suggest that activation domains can carry out multiple functions, and when more than one activation domain is present in a transcription factor, the role of each may be different.
Our experiments with GR chimeras suggest that the AF1 and AF2 domains of GR have different functional roles in transcriptional activation at the MMTV LTR. The C644G mutation in all likelihood affects the local conformation of the LBD within which the AF2 domain is found, leaving the AF1 domain functionally intact. We know that many coactivator interactions with steroid receptors are mediated through the LBD and/or AF2 domain and that the coactivators have functional activity that is associated with chromatin remodeling. Our data suggests that in vivo, a functional role of AF2 is the recruitment of chromatin remodeling factors that may also act as bridging and/or stabilizing factors with the basal transcription complex positioned at the TATA box. C644G may be unable to recruit or make functionally effective contacts with proteins that then make contact with the basal complex or that stabilize interactions between the GR and the basal complex. It is also possible that C644G is unable to maintain an effectively open chromatin conformation to allow factors such as NF1 to load onto the promoter.
Several studies have shown that various cofactors including Ada2 (1) (34), the SWI-SNF complex (79), and the coactivator SRC-1 (56) can interact with either or both AF1 and AF2 in vitro. We also know that AF1 makes contacts with the basal complex through TFIID and/or TBP in vitro (23) that may stabilize interactions between proteins at the GRE with those at the TATA box. Which of these interactions occur or have a functional role at promoters in repressed chromatin in vivo is not known. It is possible that there is functional redundancy between AF1 and AF2, or that both are required for stable and functional interactions with coactivators or chromatin remodeling complexes. Our expectation is that C644G will be a useful tool in elucidating and defining some of the required interactions of proteins for chromatin derepression and subsequent transcriptional activation in vivo from the MMTV LTR in mammalian cells.
ACKNOWLEDGMENTS
We thank members of the Hager group at the NCI, particularly Barbour Warren for advice on EMSAs, and Jan Richardson and Aniko and Geza Fejes-Toth at Dartmouth Medical School for helpful discussions.
This work was supported by NIH grants DK03535 (A.U.M.) and DK45337 (J.E.B.).
REFERENCES
- 1.Almhöf T, Wallberg A E, Gustafsson J-Å, Wright A P H. Role of important hydrophobic amino acids in the interaction between the glucocorticoid receptor t1-core activation domain and target factors. Biochemistry. 1998;37:9586–9594. doi: 10.1021/bi973029x. [DOI] [PubMed] [Google Scholar]
- 2.Archer T K, Cordingley M G, Marsaud V, Richard-Foy H, Hager G L. Proceedings: Second International CBT Symposium on the Steroid/Thyroid Receptor Family and Gene Regulation. Berlin: Birkhauser Verlag AG; 1989. pp. 221–238. [Google Scholar]
- 3.Archer T K, Cordingley M G, Wolford R G, Hager G L. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991;11:688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archer T K, Lee H-L, Cordingley M G, Mymryk J S, Fragoso G, Berard D S, Hager G L. Differential steroid hormone induction of transcription from the mouse mammary tumor virus promoter. Mol Endocrinol. 1994;8:568–576. doi: 10.1210/mend.8.5.8058066. [DOI] [PubMed] [Google Scholar]
- 5.Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 6.Blackwood E M, Kadonaga J T. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 7.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 8.Brzozowski A M, Pike A C W, Zbigniew D, Hubbard R E, Bonn T, Engström O, Öhman L, Greene G L, Gustafsson J-Å, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 9.Chakraborti P K, Garabedian M J, Yamamoto K R, Simons S S., Jr Creation of “super” glucocorticoid receptors by point mutations in the steroid binding domain. J Biol Chem. 1991;266:22075–22078. [PubMed] [Google Scholar]
- 10.Charron J, Richard-Foy H, Berard D S, Hager G L, Drouin J. Independent glucocorticoid induction and repression of two contiguous responsive genes. Mol Cell Biol. 1989;9:3127–3131. doi: 10.1128/mcb.9.7.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collingwood T N, Wagner R, Matthews C H, Clifton-Bligh R J, Gurnell M, Rajanayagam O, Agostini M, Fletterick R J, Beck-Peccoz P, Reinhardt W, Binder G, Ranke M B, Hermus A, Hesch R D, Lazarus J, Newrick P, Parfitt V, Raggatt P, de Zegher F, Chatterjee K V. A role for helix 3 of the TRβ ligand-binding domain in coactivator recruitment identified by characterization of a third cluster of mutations in resistance to thyroid hormone. EMBO J. 1998;17:4760–4770. doi: 10.1093/emboj/17.16.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook P W, Swanson K T, Edwards C P, Firestone G L. Glucocorticoid receptor-dependent inhibition of cellular proliferation in dexamethasone-resistant and hypersensitive rat hepatoma cell variants. Mol Cell Biol. 1988;8:1449–1459. doi: 10.1128/mcb.8.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordingley M G, Riegel A T, Hager G L. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987;48:261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- 14.Crabtree G R, Smith K A, Munck A. Glucocorticoid Receptors. In: Catovsky D, editor. The leukemic cell. Vol. 2. Edinburgh, Scotland: Churchill Livingstone, Longman Group Limited; 1981. pp. 252–269. [Google Scholar]
- 15.Dahlman-Wright K, Wright A, Gustafsson J-Å, Carlstedt-Duke J. Interaction of the glucocorticoid receptor DNA-binding domain with DNA as a dimer is mediated by a short segment of five amino acids. J Biol Chem. 1991;266:3107–3112. [PubMed] [Google Scholar]
- 16.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danielsen M, Northrop J P, Ringold G M. The mouse glucocorticoid receptor: mapping of functional domains by cloning, sequencing and expression of wild-type and mutant receptor proteins. EMBO J. 1986;5:2513–2522. doi: 10.1002/j.1460-2075.1986.tb04529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drouin J, Sun Y L, Tremblay S, Lavender P, Schmidt T J, de Lean A, Nemer M. Homodimer formation is rate-limiting for high affinity DNA binding by glucocorticoid receptor. Mol Endocrinol. 1992;6:1299–1309. doi: 10.1210/mend.6.8.1406707. [DOI] [PubMed] [Google Scholar]
- 20.Evans R M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fascher K-D, Schmitz J, Hörz W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 1990;9:2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng W, Ribeiro R C J, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 23.Ford J, McEwan I J, Wright A P H, Gustafsson J-Å. Involvement of the transcription factor IID protein complex in gene activation by the N-terminal transactivation domain of the glucocorticoid receptor in vitro. Mol Endocrinol. 1997;11:1467–1475. doi: 10.1210/mend.11.10.9995. [DOI] [PubMed] [Google Scholar]
- 24.Fragoso G, John S, Roberts M S, Hager G L. Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev. 1995;9:1933–1947. doi: 10.1101/gad.9.15.1933. [DOI] [PubMed] [Google Scholar]
- 25.Fragoso G, Pennie W D, John S, Hager G L. The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames. Mol Cell Biol. 1998;18:3633–3644. doi: 10.1128/mcb.18.6.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 27.Gerber A N, Klesert T R, Bergstrom D A, Tapscott S J. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 28.Giguère V, Hollenberg S M, Rosenfeld M G, Evans R M. Functional domains of the human glucocorticoid receptor. Cell. 1986;46:645–652. doi: 10.1016/0092-8674(86)90339-9. [DOI] [PubMed] [Google Scholar]
- 29.Giordano T, Howard T H, Coleman J, Sakamoto K, Howard B H. Isolation of a population of transiently transfected quiescent and senescent cells by magnetic affinity cell sorting. Exp Cell Res. 1991;192:193–197. doi: 10.1016/0014-4827(91)90175-t. [DOI] [PubMed] [Google Scholar]
- 30.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 31.Godowski P J, Picard D, Yamamoto K R. Signal transduction and transcriptional regulation by glucocorticoid receptor-LexA fusion proteins. Science. 1988;241:812–816. doi: 10.1126/science.3043662. [DOI] [PubMed] [Google Scholar]
- 32.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 34.Henriksson A, Almhöf T, Ford J, McEwan I J, Gustafsson J-Å, Wright A P H. Role of the Ada adaptor complex in gene activation by the glucocorticoid receptor. Mol Cell Biol. 1997;17:3065–3073. doi: 10.1128/mcb.17.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollenberg S M, Evans R M. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell. 1988;55:899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- 36.Hollenberg S M, Weinberger C, Ong E S, Cerelli G, Oro A, Lebo R, Thompson E B, Rosenfeld M G, Evans R M. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature. 1985;318:19–26. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong H, Kohli K, Garabedian M J, Stallcup M R. Grip1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP-1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 40.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 41.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 42.Kuo M-H, Zhou J, Jambeck P, Churchill M E A, Allis C D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee H-L, Archer T K. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vivo. Mol Cell Biol. 1994;14:32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee K A, Bindereif A, Green M R. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 45.Lees J A, Fawell S E, Parker M G. Identification of two transactivation domains in the mouse oestrogen receptor. Nucleic Acids Res. 1989;17:5477–5489. doi: 10.1093/nar/17.14.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lefebvre P, Berard D S, Cordingley M G, Hager G L. Two regions of the mouse mammary tumor virus long terminal repeat regulate the activity of its promoter in mammary cell lines. Mol Cell Biol. 1991;11:2529–2537. doi: 10.1128/mcb.11.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McAndrew P C, Svaren J, Martin S R, Hörz W, Goding C R. Requirements for chromatin modulation and transcription activation by the Pho4 acidic activation domain. Mol Cell Biol. 1998;18:5818–5827. doi: 10.1128/mcb.18.10.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milhon J, Kohli K, Stallcup M R. Genetic analysis of the N-terminal end of glucocorticoid receptor hormone binding domain. J Steroid Biochem Mol Biol. 1994;51:11–19. doi: 10.1016/0960-0760(94)90110-4. [DOI] [PubMed] [Google Scholar]
- 49.Milhon J, Lee S, Kohli K, Chen D, Hong H, Stallcup M R. Identification of amino acids in the t-2 region of the mouse glucocorticoid receptor that contribute to hormone binding and transcriptional activation. Mol Endocrinol. 1997;11:1795–1805. doi: 10.1210/mend.11.12.0018. [DOI] [PubMed] [Google Scholar]
- 50.Moras D, Gronemeyer H. The nuclear receptor ligand binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 51.Moreira J M A, Holmberg S. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA polII mutants defective in vivo in response to acidic activators. EMBO J. 1998;17:6028–6038. doi: 10.1093/emboj/17.20.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakajima T U, Anderson C S, Parvin J, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 53.Norris J D, Fan D, Kerner S A, McDonnell D P. Identification of a third autonomous activation domain within the human estrogen receptor. Mol Endocrinol. 1997;11:747–754. doi: 10.1210/mend.11.6.0008. [DOI] [PubMed] [Google Scholar]
- 54.Ogryzko V, Schlitz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 55.Oñate S, Tsai S Y, Tsai M-J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 56.Oñate S A, Boonyaratanakornkit V, Spencer T E, Tsai S Y, Tsai M-J, Edwards D P, O’Malley B W. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 57.Östlund Farrants A-K, Blomquist P, Kwon H, Wrange Ö. Glucorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol Cell Biol. 1997;17:895–905. doi: 10.1128/mcb.17.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Owens-Hughes T, Workman J L. Experimental analysis of chromatin function in transcriptional control. Crit Rev Eukaryotic Gene Expr. 1994;4:403–441. [PubMed] [Google Scholar]
- 59.Pennie W D, Hager G L, Smith C L. Nucleoprotein structure influences the response of the mouse mammary tumor virus promoter to activation of the cyclic AMP signalling pathway. Mol Cell Biol. 1995;15:2125–2134. doi: 10.1128/mcb.15.4.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Renaud J-P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-y ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 61.Richard-Foy H, Hager G. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rigaud G, Roux J, Pictet R, Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991;67:977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- 63.Shiau A K, Barstad D, Loria P, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 64.Smith C L, Htun H, Wolford R G, Hager G L. Differential activity of progesterone and glucocorticoid receptors on mouse mammary tumor virus templates differing in chromatin structure. J Biol Chem. 1997;272:14227–14235. doi: 10.1074/jbc.272.22.14227. [DOI] [PubMed] [Google Scholar]
- 65.Smith L I, Bodwell J E, Mendel D B, Ciardelli T, North W G, Munck A. Identification of cysteine-644 as the covalent site of attachment of dexamethasone 21-mesylate to murine glucocorticoid receptors in WEHI-7 cells. Biochemistry. 1988;27:3747–3753. doi: 10.1021/bi00410a034. [DOI] [PubMed] [Google Scholar]
- 66.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Oñate S A, Tsai S Y, Tsai M-J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 67.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 68.Svaren J, Hörz W. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci. 1997;22:93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 69.Tanenbaum D M, Wang Y, Williams S P, Sigler P B. Crystallographic comparison of the estrogen and progesterone receptor’s ligand binding domains. Proc Natl Acad Sci USA. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsai M-J, O’Malley B W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 71.Tsai S Y, Carlstedt-Duke J, Weigel N L, Tsai M-J, O’Malley B W. Molecular interactions of steroid hormone receptor with its enhancer element: evidence for receptor dimer formation. Cell. 1988;55:361–369. doi: 10.1016/0092-8674(88)90059-1. [DOI] [PubMed] [Google Scholar]
- 72.Utley R T, Ikeda K, Grant P A, Côté J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 73.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 74.Webster N J G, Green S, Jin J R, Chambon P. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell. 1988;54:199–207. doi: 10.1016/0092-8674(88)90552-1. [DOI] [PubMed] [Google Scholar]
- 75.Williams S P, Sigler P B. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 76.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 77.Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 78.Wurtz J-M, Bourguet W, Renaud J-P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 79.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 80.Zaret K S, Yamamoto K R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984;38:29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]