ABSTRACT
A new paradigm has emerged proposing that the crosstalk between nuclear transcription and cytoplasmic mRNA stability keeps robust mRNA levels in cells under steady-state conditions. A key piece in this crosstalk is the highly conserved 5′–3′ RNA exonuclease Xrn1, which degrades most cytoplasmic mRNAs but also associates with nuclear chromatin to activate transcription by not well-understood mechanisms. Here, we investigated the role of Xrn1 in the transcriptional response of Saccharomyces cerevisiae cells to osmotic stress. We show that a lack of Xrn1 results in much lower transcriptional induction of the upregulated genes but in similar high levels of their transcripts because of parallel mRNA stabilization. Unexpectedly, lower transcription in xrn1 occurs with a higher accumulation of RNA polymerase II (RNAPII) at stress-inducible genes, suggesting that this polymerase remains inactive backtracked. Xrn1 seems to be directly implicated in the formation of a competent elongation complex because Xrn1 is recruited to the osmotic stress-upregulated genes in parallel with the RNAPII complex, and both are dependent on the mitogen-activated protein kinase Hog1. Our findings extend the role of Xrn1 in preventing the accumulation of inactive RNAPII at highly induced genes to other situations of rapid and strong transcriptional upregulation.
KEYWORDS: Xrn1, RNA polymerase II, osmotic stress, backtracking, mRNA stability, transcription, Hog1
Introduction
Gene expression is a highly regulated process that adapts cellular content in RNAs and proteins to the cellular context. Under steady-state conditions, cellular mRNA homoeostasis is robustly maintained [1]; however, changes in the environment occur frequently and force cells to quickly adjust the composition of their transcriptome and proteome in order to survive and resume growth. For this, gene expression is reprogrammed using diverse transcriptional and post-transcriptional mechanisms to fine-tune the levels of each RNA and protein in the cell. One of the best-known models to study the molecular mechanisms underlying stress responses in eukaryotic cells is the response of the budding yeast Saccharomyces cerevisiae to high osmolarity [2–5].
Osmotic stress induces profound changes in yeast physiology and triggers a deep reprogramming of gene expression. Several hundreds of yeast genes change their expression, including both genes involved in stress protection and adaptation that undergo strong and transitory activation, and housekeeping and proliferation genes that are quickly repressed and return later to their initial levels [6–9]. Although it is probable that changes in gene expression under osmotic stress are mostly the result of the regulation of the different phases of transcription (reviewed in [2,4,5]), post-transcriptional regulation of mRNA stability [8–11], mRNA export [12], and translation [13–17] also influence variations in the cellular transcriptome and proteome along the stress gradient.
Most changes in gene expression upon osmotic stress are regulated by the mammalian p38-homologous mitogen-activated protein kinase (MAPK) Hog1, which is essential for the survival and adaptation of yeast cells to osmotic stress [3]. The response to a hyperosmotic shock starts with the activation of sensors at the plasma membrane where the high osmolarity glycerol (HOG) MAPK pathway components assemble to activate Hog1 [18–21]. At the nucleus, the increased ion concentration provokes an immediate dissociation of most proteins from chromatin and global transcription drops [8,22]. However, activated Hog1 rapidly localizes in the nucleus, where it binds to chromatin through its interaction with specific osmotic stress transcription factors; it then redirects RNA polymerase II (RNAPII) to positively responsive genes to promote their transcription [8,23–25]. Hog1 affects the recruitment of different factors acting during the transcription initiation process, such as chromatin remodellers, and modifying activities (reviewed in [2,4,5]) to modify chromatin at osmotic-stress genes [26,27]. Transcription elongation also seems to be regulated by Hog1, given that the kinase accompanies the RNAPII through the elongation process and regulates several factors that facilitate RNAPII elongation, including the remodelling the structure of chromatin (RSC) complex, the ubiquitin protease Ubp3, and the elongation factor Spt4 [28–31].
Transcription elongation by RNAPII must be efficient during stress responses to produce huge increases in mRNA levels of activated genes. Backtracking of RNAPII during elongation diminishes its efficacy and is suggested to be a major effector for rapid activation of stress-inducible genes in human cells [32]; it also seems to be important for regulating gene expression in yeast [33]. RNAPII may backtrack when it incorporates a wrong base or collides with an elongation barrier like a nucleosome or DNA-binding factor. The backwards movement of the polymerase results in the nascent RNA chain disengaging from the catalytic site. This backtracking is resolved by the polymerase cleaving the RNA 3′-end with the help of the elongation factor TFIIS [34–36]. How the regulation of the RNAPII backtracking contributes to the burst upregulation of stress-responsive genes in yeast has not thus far been determined.
The kinetics of transcript levels is controlled not only through transcription but also by the modification of mRNA decay rates during stress. Variation in the stability of an mRNA molecule serves to regulate the speed of the response and can be used to narrow the peak of the mRNA induction in upregulated genes and to accelerate the decrease in mRNA levels in downregulated ones [8–10,37-41]. mRNA stability depends on the action of general decay machineries that have transcriptome-wide effects. After an initial shortening of the polyA tail by the Ccr4–Not complex [42], degradation of eukaryotic mRNAs follows one of two main cytoplasmic decay pathways: the exosome 3′-5′ exonuclease pathway; and the 5′–3′ pathway that starts with a decapping complex and its activating factors, followed by the Xrn1 exonuclease [42,43].
In recent years a new paradigm in gene expression has been established, of transcript buffering by the coordinated regulation of the synthesis (transcription) and degradation (stability) of the mRNAs [44]. This has led to the hypothesis of crosstalk between the two processes [45]. Several decay factors, such as Xrn1, Dhh1, Lsm1, and Ccr4, along with components of the RNA polymerase complex, such as Rpb4/7, may function in this crosstalk by regulating both RNA synthesis and degradation. They have therefore been named ‘synthegradase’ factors (reviewed in [46–49]). Together with components of the RNAPII complex (Rpb4/7) and the decay factor Ccr4, Xrn1 has a prominent role in the crosstalk hypothesis [50–53]. Under steady-state conditions, nuclear Xrn1 is detected associated with the chromatin, at positions immediately before the transcription start site (TSS) [45]. It has also been detected on the GAL1 gene body upon activation [54]. In these conditions, the xrn1 mutant shows a reduction in mRNA synthesis in yeast and mammalian cells [45,55], and a general increase in mRNAs half-life (HL) [45]. Both effects are stronger for genes with, respectively, a higher decay constant and higher synthesis rates [54]. However, the precise role of Xrn1 in transcription by RNAPII has yet to be elucidated. Although there is clear agreement on the role of Xrn1 in controlling the homoeostasis of mRNA concentration in the cell, Xrn1 has been proposed to act both as an activator of transcription initiation and elongation [45,56] and as a transcriptional repressor [44]. The connection between Xrn1 and transcription could be even more convoluted: a recent study found that higher cytoplasmic activity of mammalian Xrn1 had a negative impact on transcription due to the increased nuclear localization of cytoplasmic poly(A)-binding protein, which acts as a transcriptional repressor [57].
In the present work we have addressed the role of Xrn1 in the regulation of transcription under osmotic stress. Our results show that Xrn1 is necessary to maintain proper kinetics of up- and downregulated genes, with more widespread initial effects on transcription throughout the genome than those caused by the lack of Hog1 MAPK. For osmotic stress-upregulated genes, transcription is strongly reduced in the xrn1Δ mutant whereas their corresponding mRNAs are stabilized. The lack of Xrn1 also strongly affects the recovery of transcription of downregulated genes. Our data suggest that Xrn1 acts as a transcription elongation factor by binding to upregulated genes in a Hog1-dependent manner to prevent the accumulation of inactive backtracked RNAPII. Finally, we show that the function of Xrn1 as a regulator of RNAPII elongation is not exclusive to the osmotic response and can be extended to other environmental conditions in which a competent elongating RNAPII is necessary for rapid and strong gene induction.
Materials and methods
Strains and growth conditions
S. cerevisiae strains used in this study are listed in Supplementary Table S1. All strains used were generated in the S. cerevisiae haploid wild-type strain BY4741. Yeast cells were grown at 30°C to mid-log phase in liquid YPD medium (1% yeast extract, 2% peptone, 2% glucose). Osmotic stress was applied by adding solid KCl to reach 0.6 M and incubating at 30°C for the indicated time. For the induction of GAL1, yeast cells from an exponentially growing culture in YPRaf (1% yeast extract, 2% peptone, 2% raffinose) were collected, washed, and resuspended in YPGal medium (2% galactose). Cells were then collected at the indicated times.
Stress tolerance and viability experiments
Phenotypic analyses were performed by plating serial dilutions of the indicated strains at mid-log phase on YPD plates containing the corresponding KCl osmotic stress and then incubation at 30°C. Growth curves were constructed for the wild type and xrn1Δ mutant by measuring the optical density at 600 nm (OD600) every 30 min for a total of nine time points. The doubling time was estimated from this growth curve and statistics were calculated from four independent experiments.
For viability experiments, yeast cells were grown until the mid-log phase in YPD and were subjected to osmotic stress by adding 0.6 M KCl; for the control condition they were maintained in YPD. After stress incubation, 100 µL of the culture was harvested by centrifugation at 13,000 rpm for 30 s and cells were resuspended in 400 mL of PBS buffer (140 mM NaCl, 40 mM Na2HPO4, 2.7 mM KCl, 1.8 mM KH2PO4). To identify dead cells, propidium iodide (PI) 15 mM dye was added and the cells incubated at room temperature for 5 min. Fluorescence was then measured in a flow cytometer (EPICS XL-MCL, Beckman Coulter). The yeast cells kept in YPD were used as a 100% viability control. The fluorescence of the cells under the control conditions without PI was used as a baseline.
Genomic run-on, normalizations, and bioinformatics analysis
To determine the transcription rate (TR) in the strains BY4741 (wild type) and xrn1Δ mutant, a genomic run-on (GRO) assay was performed as previously described in [58] with modifications as indicated in [8] and [59]. Yeast samples were grown to mid-log phase and osmotic stress was applied by treatment with 0.6 M KCl for 0, 8, 15, 30 and 45 min as described previously. Three independent experiments were performed with the same number of cells in each sample. The scanned macroarray images were quantified using Array Vision software (Imaging Research Inc., St Catharines, ON, Canada). Values that were at least 1.2 times higher than the local background were taken as valid measurements. An average data set for each time point was created using median absolute deviation normalization by ArrayStat software (Imaging Research Inc.). To determine the global mRNA amount (RA), we used a dot-blot procedure to determine the total poly(A) mRNA per cell as previously described [58] and normalized by the cell volume to obtain total concentration (RA) in relative values for each sample. The median values of cell volumes for each strain were calculated using a Coulter-Counter Z series device (Coulter Inc., Middlebury, IN, USA).
Yeast genes upregulated and downregulated by osmotic stress were obtained according to previous GRO data [8] or according to the conventionally defined groups of genes of the general environmental stress response (ESR) [60]. The classification of yeast genes in five distinct regulons based on their transcriptional dependency on Hot1, Msn2, Msn4, Sko1, and Smp1 was obtained from the YEASTRACT database [61]. These regulons were then cross-referenced with our GRO data to select the genes upregulated during osmotic stress treatment. The groups of ribosomal proteins (RPs) were obtained from the Saccharomyces Genome Database (SGD) and the group of ribosome biogenesis (RiBi) proteins were defined according to [62].
Raw and processed data are stored in the Gene Expression Omnibus (GEO) database, with the accession number GSE151736.
RNA extraction, retrotranscription to cDNA and RT-quantitative PCR
Yeast cells were grown at 30°C in YPD media until the mid-log phase and then subjected to osmotic stress by adding 0.6 M KCl as previously described. Next, 20 mL of culture cells were centrifuged and the pellet was frozen. Total RNA extraction, cDNA synthesis by retrotranscription, and quantitative PCR (RT-qPCR) were performed as previously described [59]. The mRNA levels of the indicated genes were quantified using the primers listed in Supplementary Table S2 and were normalized with the ACT1 mRNA level in the same sample.
Determination of mRNA stability
For the mRNA stability analysis, we used a transcription shut-off assay as described in [63]. For this, yeast cells were grown to mid-log phase in YPD and then treated with 0.6 M KCl for 30 min. Samples were collected at 5, 10, 20, 30, 40, 50 and 60 min after the addition of thiolutin (5 µg/mL). STL1 and GRE3 mRNA levels were determined with RT-qPCR using specific primers and normalized against the 18S rRNA (Supplementary Table S2). mRNA stability was estimated using the percentage of the remaining mRNA with respect to the initial amount (100%) along the treatment time.
Chromatin immunoprecipitation
Yeast cells were grown at 30°C in YPD media until the mid-log phase and were subjected to 0.6 M KCl osmotic stress for the indicated time. For the cross-linking reaction, formaldehyde was added (1% final concentration) to a volume of 45 mL of culture cells for 15 min at room temperature with occasional inversion. The chromatin immunoprecipitation (ChIP) experiments were performed as previously described [59] with the following modifications: after reversing formaldehyde-mediated cross-linking, samples were treated with proteinase K, and DNA was purified using the GeneJET PCR Purification Kit (Fermentas cat. no. K0702) according to the manufacturer’s instructions. To determine the enrichment of the DNA regions bound by the protein of interest, qPCR was run as described [11] using the primers listed in Supplementary Table S2. The qPCR amplification data were normalized with the total input DNA value in the corresponding sample and it was checked that no enrichment over background signal was observed in samples in which the corresponding antibody was not added. Immunoprecipitations of RNAPII were made with anti-RPB3 antibody [1Y26] (Abcam, cat. no. ab202893) with Dynabeads Pan Mouse IgG (Invitrogen, cat. no. 110.41) and anti-RPB1 Ser2-P antibody (Abcam, cat. no. ab5095) with Dynabeads Protein A (Invitrogen, cat. no. 100.02D). Immunoprecipitations of Xrn1-FLAG were made with anti-FLAG antibody (M2, Sigma-Aldrich, cat. no. F1804) with Dynabeads Pan Mouse IgG (Invitrogen, cat. no. 110.41). Immunoprecipitations of HA-TFIIS and Spt5-myc were made with anti-HA antibody (3F10; Roche, cat. no. 11,867,423,001) with Dynabeads anti-Rat IgG (Invitrogen, cat. no. 11035) and anti-C-Myc antibody (9E10; Santa Cruz Biotechnology, Inc., cat. no. sc-40) with Dynabeads Pan Mouse IgG (Invitrogen, cat. no. 110.41), respectively. For the ChIPs in which the association of two proteins was compared (TFIIS or Spt5 vs. RNAPII) or those for checking phosphorylation at Rpb1-CTDSer2 respect total RNAPII, a whole cell extract was prepared from a volume of 90 mL of culture cells and then was divided in two aliquots, one for each antibody used.
Results
Transcription by RNAPII during osmotic stress is strongly reduced in xrn1∆ mutants
It has been proposed that Xrn1 has a role in regulating transcription, although its specific and mechanistic modes of action are still controversial [44,45]. We took advantage of the rapid transcriptional response of yeast cells to a sudden change in media osmolarity to study the effect of a lack of Xrn1 on the changes in transcription that are mainly governed by the signalling MAPK Hog1 [5].
We performed GRO experiments that allow the measurement of TRs during a rapid kinetic change under osmotic stress (5063 yeast genes analysed) [8,58]. As described previously [8,59] and shown in Fig. 1A, global RNAPII TR drops in wild-type cells during the first minutes of stress and recovers soon after, showing a certain transitory increase. Under non-stress conditions (time 0) and as reported previously [45,54], global transcription is reduced in the xrn1Δ mutant (Fig. 1B). Under osmotic stress caused by 0.6 M KCl, global TR dropped rapidly (8 min) in xrn1Δ similarly to wild type, but transcription did not then follow the transient over-activation observed in the wild type (Fig. 1A).
Figure 1.
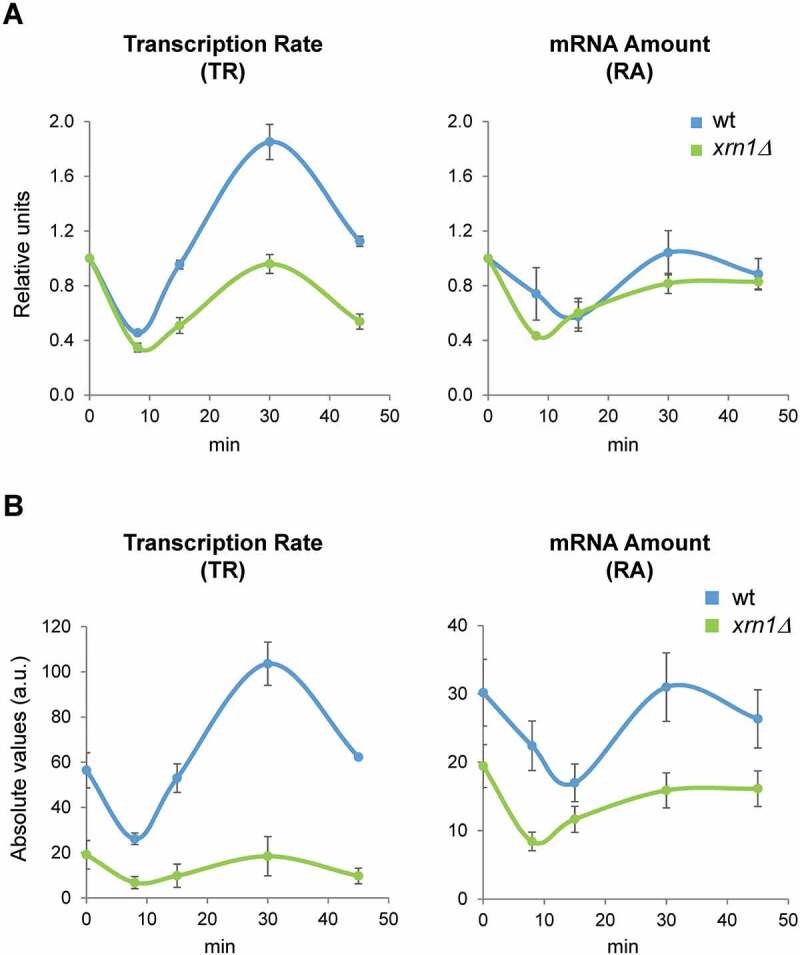
Deletion of Xrn1 causes changes in global RNAPII transcription rate (TR) and mRNA amount (RA) during the response to osmotic stress. Exponentially growing cultures of wild type (wt, BY4741) and xrn1Δ mutant yeast strains were grown in YPD medium and treated with 0.6 M KCl for 0, 8, 15, 30 and 45 min. Samples were obtained as indicated in the Materials and Methods section. Genomic data represent the median of three independent genomic run-on (GRO) experiments in wt (blue) and xrn1Δ (green) cells. Global data for TR were normalized to an arbitrary value of 1 at t = 0 (A) or represented in absolute arbitrary units (B). Global RA was determined by quantitative RNA extraction and poly A determination as described in Materials and Methods. Error bars represent the standard deviation of three independent experiments
We also analysed global mRNA cellular concentration and saw a drop in both strains, the xrn1Δ mutant and wild type, although the global RA levels fell a little more and quicker in the mutant (Fig. 1A, B). In this case, the recovery in global RA level was similar between the wild type and xrn1Δ mutant, suggesting that the global mRNA stabilization characteristic of this mutant [45] has a compensatory effect.
Lack of Xrn1 impairs rapid recovery of growth during osmotic stress
Results shown above indicate that the xrn1Δ mutant has an important defect in RNAPII transcription during the response to osmotic stress; however, global RA kinetics are not affected owing to the compensatory effect of higher mRNA stabilization in xrn1Δ. It was, therefore, difficult to predict whether a phenotype would be associated with the lack of Xrn1. To address this, we performed different experiments to compare the behaviour of wild-type and xrn1Δ cells under osmotic stress conditions.
First, we checked growth on plates with media containing 0.6 M KCl or 0.4 M NaCl and observed that the xrn1∆ mutant grew slower than wild type but that the difference was similar to that observed in control media without salt (Fig. 2A), conditions in which it is known that the xrn1Δ mutant grows slower than wild type [45]. Changes in viability gave similar results in both strains under mild osmotic stress for longer periods of exposure (Figs. 2B, 120 and 240 min of 0.6 M KCl), although the percentage of dead cells was significantly higher in the mutant strain compare to wild type under non-stress and stress conditions (Fig. 2B). Interestingly, the value of relative dead cells was higher for wild type than for the mutant at 30 min of stress, suggesting that lack of Xrn1 allows for higher survival of yeast cells when exposed for a short time to osmotic stress. However, when measuring the resumption of growth after the first minutes of mild osmotic stress, the xrn1∆ strain showed an increase in the ratio of relative duplication time compared to wild type (Fig. 2C). Mutants grew slower under non-stress (ratio of duplication time of 1.4 for the xrn1Δ mutant vs wild type), but the difference was even higher under osmotic stress (ratio of 1.9). These results suggest that defects in the transcriptional response due to lack of Xrn1 negatively affect the rapid recovery of growth after the initial stress.
Figure 2.
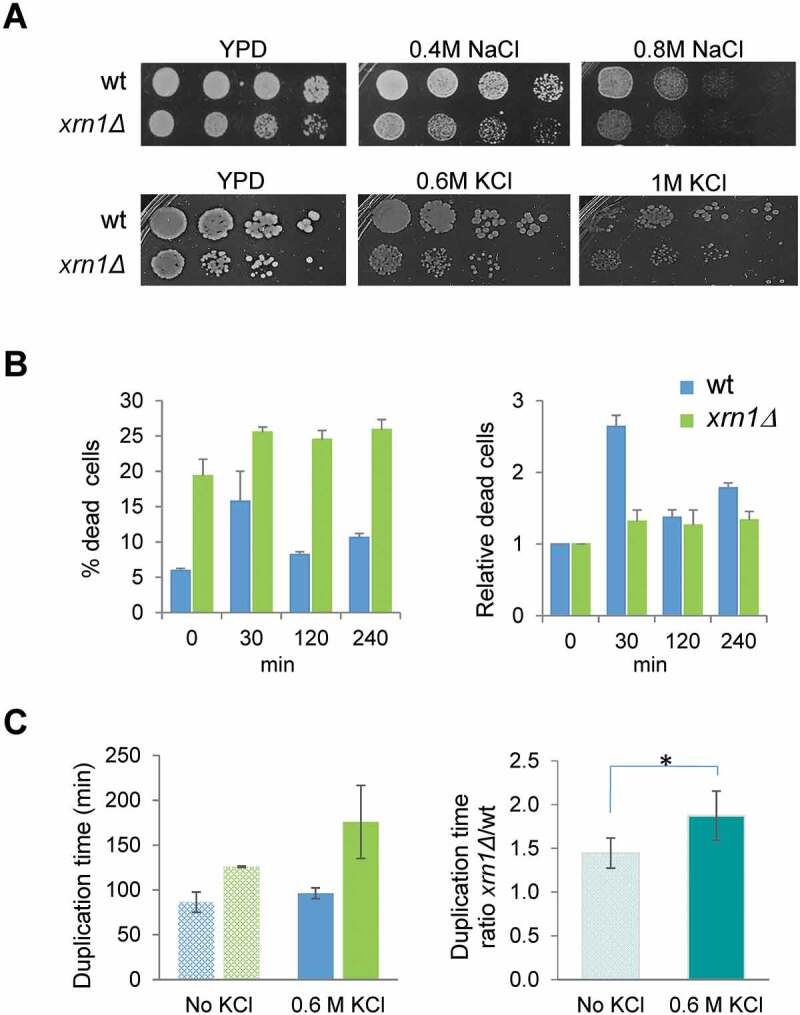
Sensitivity to different osmotic stress conditions of the xrn1Δ mutant cells. (A) Exponentially growing cells were spotted on YPD plates supplemented, or not, with NaCl or KCl at the indicated concentrations. Plates were incubated at 30°C for 2 days. (B) Viability analysis of the xrn1Δ mutant during osmotic stress. Exponentially growing wild type (wt) and mutant cells in YPD were subjected to 0.6 M KCl. The left panel shows the percentage of dead cells. The right panel shows these data normalized to an arbitrary value of 1 at t = 0. (C) The left panel shows the duplication time measurements in growing cells on YPD medium (no KCl) and YPD supplemented with 0.6 M KCl for wt and xrn1Δ. The right panel shows the ratio between the duplication time for the xrn1Δ mutant with respect to wt in the indicated conditions. The mean of at least three independent experiments is shown; *p < 0.05
Xrn1 is needed for the transcriptional induction of upregulated genes and the recovery of transcription of downregulated genes under osmotic stress
Given that the transcriptional response to osmotic stress seemed to be affected in the xrn1Δ mutant, we went on to study genes known to be responsive to osmotic stress [8]. This allowed us to extract two sets from the analysed genes: 1) genes that increase their mRNA levels mostly due to increases in transcription rates (upregulated genes, n = 245); and 2) genes showing downregulation of their mRNA levels, caused primarily by a drop in TR (downregulated genes, n = 355). Upregulated genes showed a rapid and transient increase in their TRs that was much lower in the xrn1Δ mutant, reaching only 42.5% of the maximum increase observed in the wild type (Fig. 3A).
Figure 3.
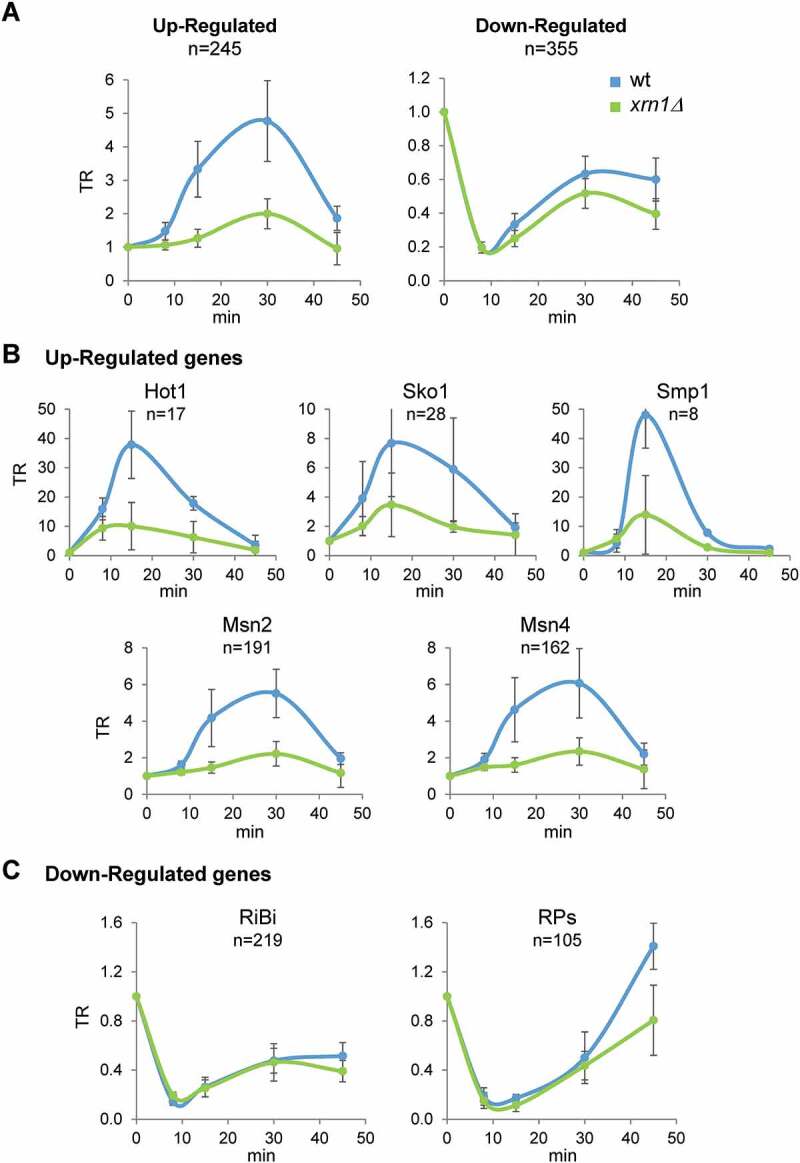
Absence of Xrn1 reduces the transcriptional response of genes up- and downregulated during osmotic stress. Genome wide data were obtained for wild type and xrn1Δ treated with 0.6 M KCl for 0, 8, 15, 30 and 45 min as indicated in Fig. 1. (A) Transcription rate (TR) medians for 245 upregulated and 355 downregulated genes in response to osmotic stress. (B) TR medians of upregulated genes divided into five distinct regulons based on their transcriptional dependency on Hot1, Sko1, Smp1, Msn2, and Msn4. (C) TR medians of downregulated genes belonging to the ribosome biogenesis (RiBi) and ribosomal proteins (RP) genes. All data were normalized to an arbitrary value of 1 at t = 0. It is indicated the number of genes analized in each regulon (N). Error bars represent the standard deviation of three independent experiments
The vast majority of the upregulated genes are controlled by the MAPK Hog1; however, different transcription factors (e.g. Hot1, Msn2/4, Sko1, Smp1), acting downstream of Hog1, control subgroups of the induced genes [2]. We examined whether the role of Xrn1 in promoting gene induction was specifically linked to any of these subgroups. However, as seen in Fig. 3B, transcription was similarly reduced in the xrn1Δ mutant for all sets of upregulated genes with respect to wild type during osmotic stress.
For downregulated genes, we observed the same timing in the drop and recovery of transcription between wild type and the xrn1Δ mutant; however, the xrn1Δ mutant showed slightly lower recovery of TRs following the initial drop in transcription (Fig. 3A). We also examined the different functional groups of osmotic stress-downregulated genes on the basis that different functional groups usually show co-regulation under different stress conditions [9]. At 45 min of stress, the RP group showed lower levels of TR recovery in the xrn1Δ mutant than in the wild type, whereas the RiBi group did not show any important differences in TR kinetics between the wild type and xrn1Δ mutant (Fig. 3C). This behaviour could be related to the known preference of Xrn1 for activating genes with higher TRs, such as RP genes [54].
Altogether, our results indicate that a lack of the exonuclease Xrn1 yields specific and important defects in transcriptional induction for most genes upregulated by osmotic stress, and only slight defects in the transcriptional recovery of downregulated genes, specifically RP genes. We also confirmed that these defects are not constrained by the definition of osmotic stress-responsive genes [8], because we obtained similar results for the TR kinetics analysis made with conventionally defined groups of genes of the general ESR (Supplementary Figure S1) [60].
As described previously, the transcriptional response to osmotic stress is governed by the MAPK Hog1, which controls most of the upregulation but only some of the downregulation [8,27]. To determine if the effect of osmotic stress on transcription in the xrn1Δ mutant resembles that in the hog1 mutant, we reviewed genomic TR data obtained with hog1Δ [8] and xrn1Δ (this work) under mild osmotic stress. For upregulated genes, the TR response to osmotic stress at 15 min is defective in both mutants. Loss of Hog1 has a stronger effect than loss of Xrn1, as shown by a lower correlation with the wild-type stress response (Supplementary Figure S2A). For downregulated genes, loss of Hog1 and Xrn1 has more similar effects, although they were a bit stronger for the xrn1Δ mutant at the short time stress (8 min) (Supplementary Figure S2B). To summarize, loss of Xrn1 does not exactly reflect loss of Hog1; and since the effects of Xrn1 loss seem to be a bit stronger for the rapid down-regulation of genes upon stress, this could be indicative of, at least in part, a Hog1-independent function of Xrn1 during stress.
Kinetics of osmotic stress-induced mRNAs in the xrn1∆ mutant result from defects in transcription and increased mRNA stabilization
To validate our genome-wide data, we measured the changes in RA of typical upregulated genes, such as STL1, which encodes for a glycerol transporter strongly induced upon osmotic stress in a Hog1-dependent way through the osmotic stress transcription factor Hot1 (Fig. 4A) [6,8,23]. The quantification of STL1 mRNA levels (relative to t0) by qPCR from cells treated with 0.6 M KCl showed a much reduced increase in xrn1Δ mutant cells with respect to the wild type (Fig. 4B, upper left panel), which correlates with the defective increase in STL1 TR observed in the xrn1Δ mutant (Fig. 4A, left panel). This result is qualitatively similar to the average of upregulated genes shown in Fig. 3A, although STL1 is much more highly expressed and activated than the average of this group. A similar result, although with lower activation levels, is seen for GRE3, which encodes an aldose reductase induced by osmotic stress (and other stressors) under the control of Hog1 (Fig. 4B, upper right panel, and Fig. 4A, right panel) [8,64].
Figure 4.
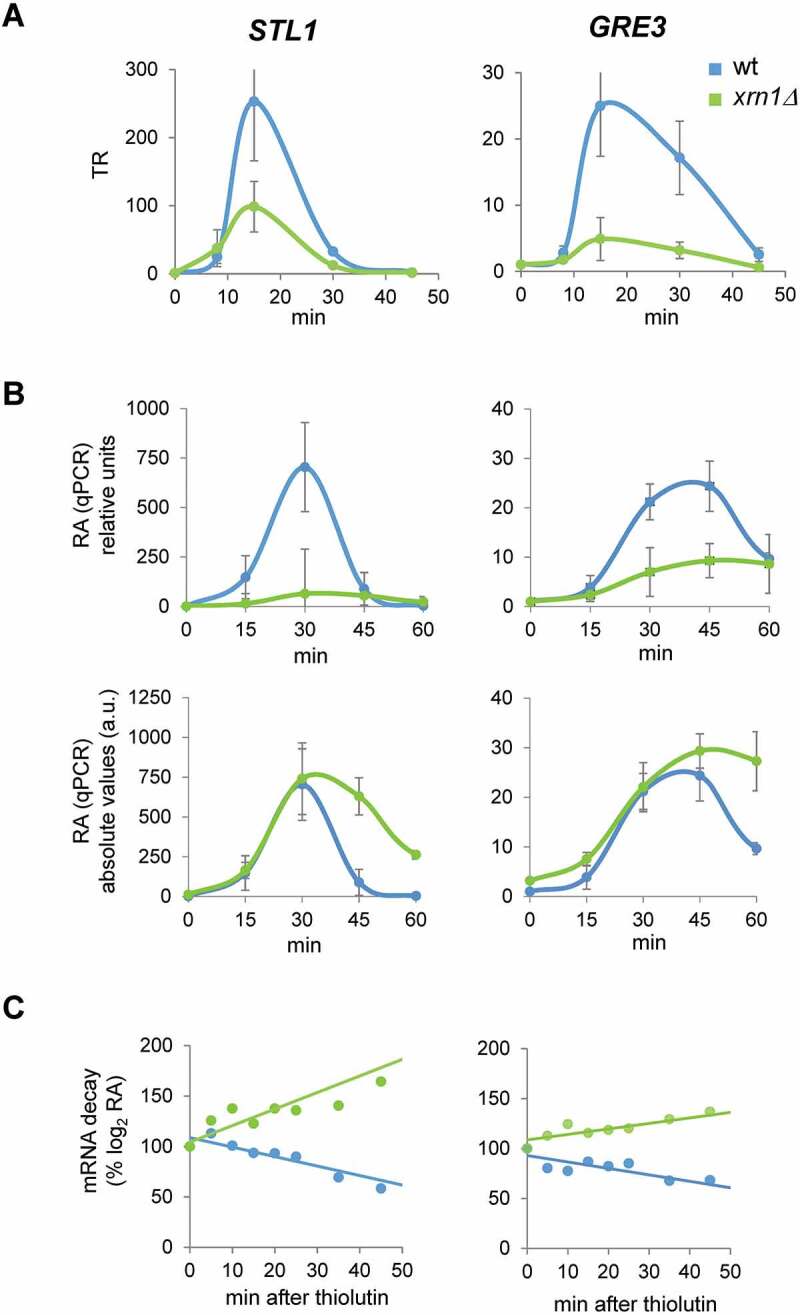
Lack of Xrn1 causes low transcription and high mRNA stabilization of typical osmotic stress-upregulated genes. (A) Transcription rates (TR) of the osmotic stress-upregulated genes STL1 and GRE3 obtained by genomic run-on (GRO) as detailed in Fig. 1. (B) RT-qPCR analyses of STL1 and GRE3 mRNA expression at the same conditions for the GRO experiment and for the indicated times of osmotic stress by treatment with 0.6 M KCl. mRNA expression was normalized against the reference gene ACT1. Data are presented as relative to an arbitrary value of 1 for each strain at t = 0 (upper panels) or in absolute arbitrary units as fold-change relative to wild type (wt) non-stress of each experiment (lower panels). Averages and standard deviations of three independent experiments are presented. (C) STL1 and GRE3 mRNA stability determination for wt and xrn1Δ strains. Cells were treated with 0.6 M KCl for 30 min and then 5 µg/mL of thiolutin was added to stop transcription. Samples were taken at different times, analysed by RT-qPCR using specific primers for STL1 and GRE3 (Supplementary Table S2) and normalized using 18S RNA. Data are presented as the percentage of the remaining mRNA with respect to the initial (100%) over time. One representative experiment is shown (a second experiment is shown in Supplementary Figure S3)
However, examining absolute mRNA levels (Fig. 4B, lower panels) shows that mRNA levels in the xrn1Δ mutant reached similar peak values as in the wild type, despite the lower activation. Additionally, the RA kinetics followed different patterns for wild type and xrn1Δ: in wild-type STL1 mRNA level dropped quickly and returned to basal level after 45 min and GRE3 mRNA dropped at 60 min to less than 50% of the maximum reached level. However, in xrn1Δ, mRNA levels dropped slowly for both mRNAs and were still high at 60 min (Fig. 4B, lower panels). The high STL1 and GRE3 mRNA levels observed in xrn1Δ mutant, in which transcription of these genes is much reduced, could be explained by an increase in mRNA stability [8,10,37]. To check this, we measured the decay of the upregulated STL1 and GRE3 mRNAs after shutting down RNAPII transcription with thiolutin at 30 min of treatment with 0.6 M KCl (Fig. 4C and Supplementary Figure S3). It has been shown that transcriptional stop with thiolutin does not occur immediately, so under osmotic stress treatment, upregulated genes may still show increases in their mRNA levels during the first minutes of thiolutin addition [8,17]. Our results showed that, upon thiolutin addition there was a rapid drop in STL1 and GRE3 mRNA in the wild type cells, indicating destabilization. However, the mRNA levels in xrn1Δ cells did not drop, instead showing an increase, suggesting a strong mRNA stabilization of these upregulated genes in the mutant.
Together, these results indicate that in wild-type cells Xrn1 boosts the induction of transcription of upregulated genes but also promotes destabilization of their mRNAs, probably to configure the rapid and transitory induction of genes involved in the survival of and adaptation to osmotic stress.
Transcriptionally inactive RNAPII accumulates at osmotic stress-responsive genes in the absence of Xrn1
As we reported above, one of the main defects in the response to osmotic stress caused by the lack of Xrn1 is an important reduction in the global transcription measured by GRO, and, specifically, a much more reduced transcriptional induction of osmotic stress-upregulated genes. To corroborate this, we investigated occupancy of the RNAPII to stress-responsive genes in the xrn1Δ mutant. We performed ChIP experiments using antibodies against the Rpb3 subunit of RNAPII. In the upregulated STL1, RNAPII was recruited to similar levels in xrn1Δ and wild-type cells at 15 min of osmotic stress (Fig. 5, left panels). A higher association of the polymerase to STL1 and GRE3 was then observed in xrn1Δ at 30 min. Finally, the polymerase was almost absent from the genes at 45 min in both strains (Fig. 5A). Therefore, and unexpectedly, not less but even more polymerase was present in stress-upregulated genes in the absence of Xrn1, although the genes had much lower transcription (Figs. 4 and 5A). Phosphorylation of Ser2 at the carboxy-terminal repeat domain (CTD) of the catalytic Rpb1 subunit of the RNAPII has been described as a mark of an active elongating polymerase [65], so we investigated if polymerase bound to upregulated genes was phosphorylated at this position in xrn1Δ cells. As shown in Fig. 5B, more CTD Ser2 phosphorylation was detected in the xrn1Δ mutant at shorter times, 15 min, showing higher ratios of Ser2-P with respect to Rpb3 (Fig. 5C); therefore, RNAPII was initially Ser2-hyperphosphorylated in xrn1Δ. However, at 30 min of stress, a lower Ser2-P/Rpb3 ratio in xrn1Δ respect to wild-type was detected.
Figure 5.
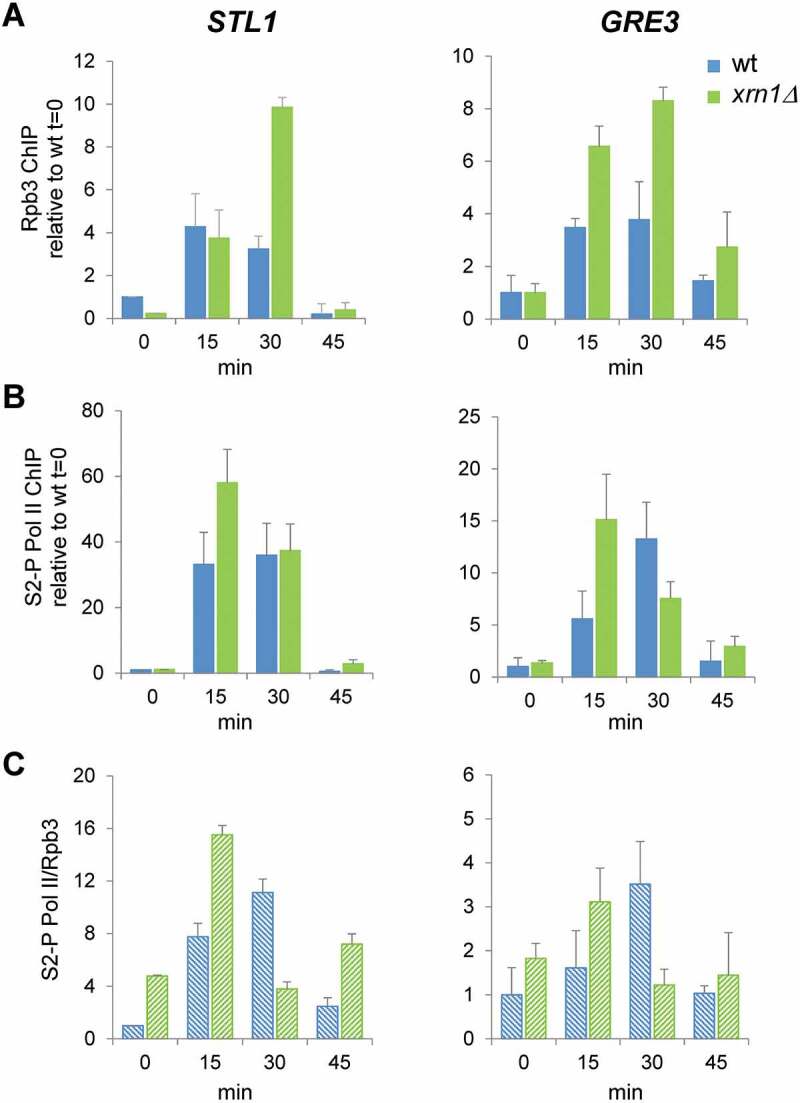
Lack of Xrn1 provokes an increased association of RNAPII with osmotic stress-upregulated genes and affects the dynamics of Ser2 phosphorylation at the Rpb1-carboxy-terminal repeat domain (CTD). Chromatin immunoprecipitation (ChIP) analysis of RNAPII recruitment in wild type (wt) and xrn1Δ under 0.6 M KCl for the indicated times. ChIP of RNAPII was performed using the antibodies anti-Rpb3 (A) and anti-Rpb1-CTDSer2-P (B) in two aliquots of the same whole cell extract sample. The ChIP DNA was used to quantify STL1 (left panels) and GRE3 (right panels) open reading frames (ORFs) by qPCR. The percentage of the signal obtained in each ChIP sample with respect to the signal obtained with the DNA from the corresponding whole cell extract was calculated. Data are normalized against the percentage ChIP of wt at time 0. (C) The proportion of RNAPII phosphorylated at Rpb1-CTDSer2-P with respect to the total RNAPII bound to STL1 and GRE3 (ratio S2-P PolI/Rpb3) is presented. Mean and standard deviation from three independent experiments are shown
To summarize, the run-on data indicate a decrease in the RNAPII elongating complex, while the ChIP data show high RNAPII recruitment to the same genes, even with higher phosphorylation at Ser2-P at shorter times. This unexpected result was not found for other mutants with a negative impact on RNAPII transcription during osmotic stress, e.g. hog1 and cbc1, that were previously analysed using the same techniques [8,24,59]. It has previously been reported that run-on assays are sensitive to backtracking, because in the backtracked polymerase the 3′ end of the nascent RNA is displaced from the active site and transcription elongation cannot proceed during the in vitro assay [66]. Therefore, all our above results strongly suggest that the high RNAPII ChIP signals but low GRO signals in xrn1Δ during osmotic stress are due to an accumulation of inactive, backtracked RNAPII molecules, unable to elongate at upregulated genes. Moreover, after the initial accumulation of Ser2-hyperphosphorylated RNAPII, the following Ser2-hypophosphorylation suggests that blocked polymerases accumulate and are not reactivated in xrn1Δ mutant.
Coordinated recruitment of Xrn1 and RNAPII at osmotic stress-induced genes is mediated by the MAPK Hog1
It has been reported that under non-stress conditions Xrn1 is associated with promoters and gene bodies to stimulate transcription [45]. We wondered if Xrn1 is acting directly through its binding to promote the activity of RNAPII in osmotic stress-upregulated genes. We performed ChIP experiments using a functional Xrn1-FLAG tagged version (Supplementary Figure S4) to check binding to chromatin along a mild osmotic stress. As shown in Fig. 6, Xrn1 was recruited to STL1 and GRE3 promoters and open reading frame (ORF) regions after a few minutes of stress, reaching a peak at 20 min of stress. On the contrary, binding to RPL30, which is highly expressed under non-stress conditions and downregulated under stress, lowered much upon 0.6 M KCl treatment. Then, binding of Xrn1 to RPL30 was slowly increasing and a high binding signal was observed at the RPL30 promoter and lower at the ORF at 30 min of stress, suggesting that transcription is recovering at this RP gene (Fig. 6C).
Figure 6.
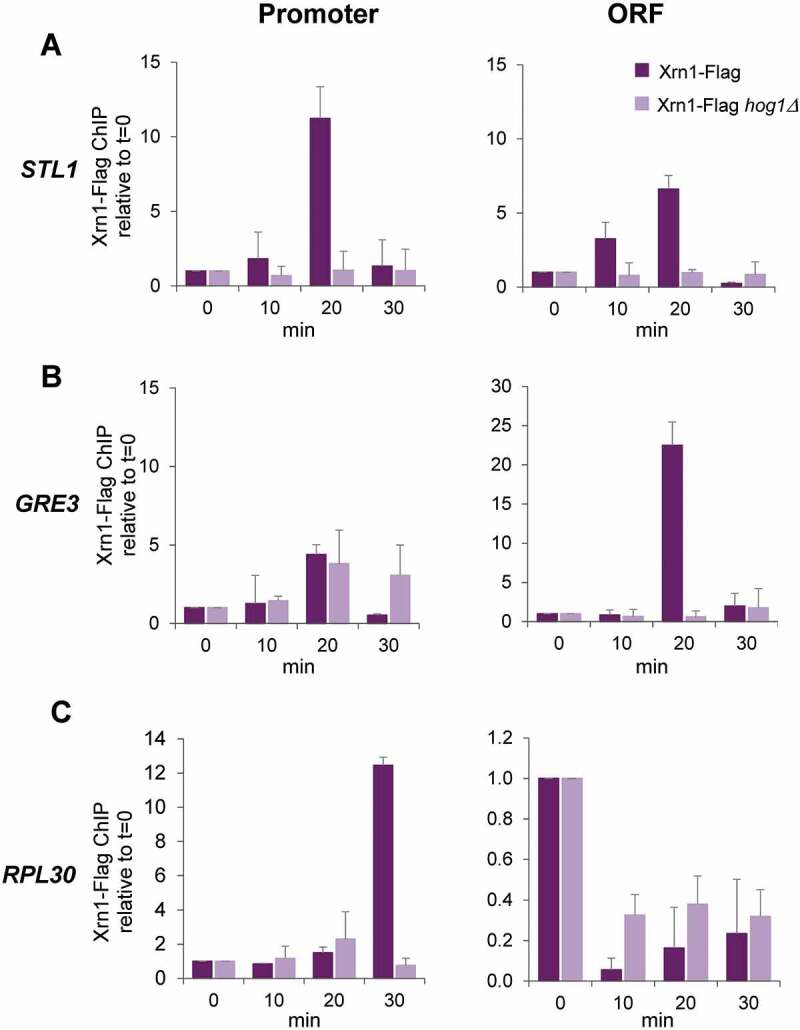
Xrn1 binds to osmotic stress-induced genes upon activation in a Hog1-dependent manner. Chromatin immunoprecipitation (ChIP) analysis of Xrn1 binding to the osmotic stress-induced genes STL1 (A) and GRE3 (B) and to the osmotic stress-downregulated gene RPL30 (C) in cells treated with 0.6 M KCl for the indicated time. Wild type and hog1 mutant strains carrying functional Xrn1-Flag (Supplementary Figure S4) were used. Immunoprecipitated samples were analysed for the binding of Xrn1-Flag to the promoter (left panels) and open reading frame (ORF) (right panels) regions of the regulated genes. Data were obtained as described in Figure 5 and normalized respect to the signal at t = 0. Mean and standard deviation from three independent experiments are shown
Next, we interrogated the role of Hog1 MAPK in Xrn1 recruitment to genes under osmotic stress. Binding of Xrn1 to Hog1-regulated STL1 and GRE3 was almost completely abolished in a hog1 mutant (Fig. 6), as occurs with binding of RNAPII [59]. At the RPL30 gene (Fig. 6C), loss of Xrn1 binding was slightly lower in the hog1 mutant than in the wild type, correlating with the lower downregulation of housekeeping genes observed in the MAPK mutant [8]. However, recovery of Xrn1 binding at longer periods was dependent on Hog1 (Fig. 6C), also correlating with the need of functional Hog1 to return to growth [3].
These results indicate Xrn1 responds to osmotic stress to specifically bind upregulated genes and dissociate from downregulated ones. Additionally, Xrn1 follows recruitment kinetics to upregulated genes (Fig. 6) similar to those of RNAPII (see Fig. 5, figures in next sections of Results and [24,59]) and dependent on the signalling kinase Hog1. In response to osmotic stress, the MAPK binds to upregulated genes and promotes the recruitment of transcription factors and the RNAPII complex [23,24]; thus, in the hog1 mutant the lack of RNAPII at upregulated promoters may cause the lack of recruitment of Xrn1. Our results above and here suggest that the coordinated binding of Xrn1 protein and RNAPII at the chromatin of upregulated genes upon stress is a requisite for reaching high efficient elongation.
Xrn1 binds to GAL1 in response to galactose to prevent the accumulation of inactive RNAPII
Our results suggest a direct role of Xrn1 in the formation of an elongation-competent RNAPII complex to prevent its accumulation in an inactive state at genes that are induced under osmotic stress. We wondered whether this role was specific for osmotic stress or whether it could be extended to different conditions in which a rapid and strong induction of transcription is needed. Recent studies have shown that rescue of RNAPII from backtracking is a major stimulus of rapid transcriptional elongation in stress-inducible genes [32]. To address this, we studied the induction of GAL1 in yeast cells in response to a sudden change to a media with galactose as the only carbon source (Fig. 7A) [67]. A previous study comparing mRNA levels and stability suggested a defect in the transcriptional induction of GAL genes in the xrn1Δ mutant [45] and it was also shown that Xrn1 is associated to the GAL1 ORF when cells were grown in galactose media [54]. Recently, it has been shown that depletion of Xrn1 in cells growing exponentially in galactose medium reduces the RNAPII run-on signal of GAL1 without changing the amount of polymerase bound to the gene [56]. Here, we studied the binding kinetics of both Xrn1-FLAG and RNAPII to GAL1 upon sudden activation conditions (Fig. 7A). For the latter, we studied RNAPII binding with Rpb3 and Rpb1-CTD Ser2-P antibodies both in wild type and xrn1Δ strains. As shown in Fig. 7B, the transfer of wild-type cells to galactose media induced the binding of Xrn1 protein to the responsive GAL1 gene. RNAPII binding also increased within minutes of induction by galactose and higher levels of total polymerase (Rpb3) and Rpb1-CTDSer2-P were observed in wild-type cells during the first 60 min of induction (Fig. 7C). RNAPII binding, similar to the results with upregulated genes under osmotic stress (Fig. 5), was much stronger in the xrn1Δ mutant that in wild type. The accumulated RNAPII at GAL1 seems to be initially Ser2-hyperphosphorylated and later hypophosphorylated (Fig. 7C). This is again similar to that seen in osmotic stress-induced genes (Fig. 5).
Figure 7.
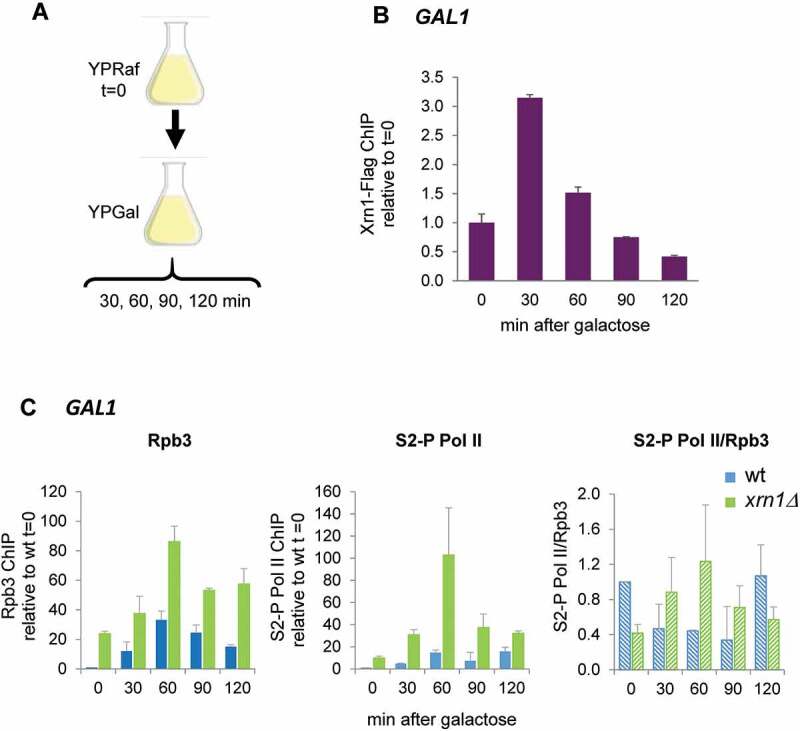
Xrn1 binds to GAL1 upon activation and prevents the accumulation of RNAPII. (A) Scheme of the assay. Exponentially growing cells in raffinose (YPRaf) medium were washed and transferred to galactose medium (YPGal), and samples were collected at the indicated times. (B) Chromatin immunoprecipitation (ChIP) analysis of Xrn1-Flag was made using anti-Flag antibodies and binding to GAL1 ORF was determined. (C) ChIP analysis of RNAPII binding at GAL1 ORF in wild type (wt) and xrn1Δ cells. RNAPII was pulled down with anti-Rpb3p (left panel) and anti-Rpb1-CTDSer2-P (middle panel) antibodies in two aliquots of the same whole cell extract sample. Ratio between Rpb1-CTDSer2-P (S2-P Pol II) and Rpb3 ChIP is shown (right panel). ChIP data were calculated and normalized as described in Figure 5. Mean and standard deviation from three independent experiments are shown
These results suggest that Xrn1 binds to rapidly induced genes, not only upon osmotic stress but also under other circumstances of rapid activation and, therefore independently of the activation signal, to facilitate RNAPII elongation by preventing backtracking of the polymerase.
High levels of TFIIS and Spt5 are found in parallel to the accumulation of RNAPII at osmotic stress upregulated genes in xrn1Δ mutant cells
Rescue from backtracking involves cleaving the 3′ end of the nascent RNA to reactivate stalled RNAPII, a process that is promoted by TFIIS [34,35]. Resolving backtracking by TFIIS thus accelerates transcription through the body of the genes containing histone-barriers or other pausing features [32,68]. Other elongation factors necessary for maintaining the RNAPII elongation rate are Spt4 and Spt5, which form a complex involved in coupling chromatin modification states to elongation [69–71]. Recently, it has been shown that processivity of RNAPII at osmotic stress-induced genes is stimulated by the Spt4–Spt5 complex through the phosphorylation of Spt4 by Hog1 [31]. To further understand the function of Xrn1 in preventing/rescuing RNAPII backtracking, we measured the recruitment of TFIIS and the Spt4–Spt5 complex in relation to the amount of RNAPII.
We observed higher levels of TFIIS and Spt5 in the mutant than in the wild type at osmotic stress-upregulated genes (Fig. 8A and B, respectively). The higher binding of the elongation factors in xrn1Δ was in parallel to the higher recruitment of RNAPII. Then, we obtained the TFIIS/RNAPII and Spt5/RNAPII ratios only at the times of gene induction (10 and 20 min) to avoid putative artefacts in the ratios due to the low signals at other times of stress treatment. We observed similar TFIIS/RNAPII ratios in the xrn1Δ mutant than in wild type at STL1 at 10 min and 20 min, and at GRE3 at 10 min, but the ratio was lower at GRE3 at 20 min of osmotic stress (Fig. 8A). No significant differences in the Spt5/RNAPII ratios were found between cells with or without Xrn1 (Fig. 8B). These results indicate that accumulation of RNAPII at osmotic stress-upregulated genes runs in parallel to a rise in the binding of TFIIS and Spt5 elongation factors that seems to be quantitatively similar at most times of stress. Therefore, the accumulation of backtracked RNAPII does not seem to be due to the lack of recruitment of these two elongation factors, despite the fact that the lower TFIIS/RNAPII ratio found in one gene and one time (GRE3 at 20 min) does not allow us to completely rule out the possibility of a negative effect on the recruitment of this elongation factor by the absence of Xrn1.
Figure 8.
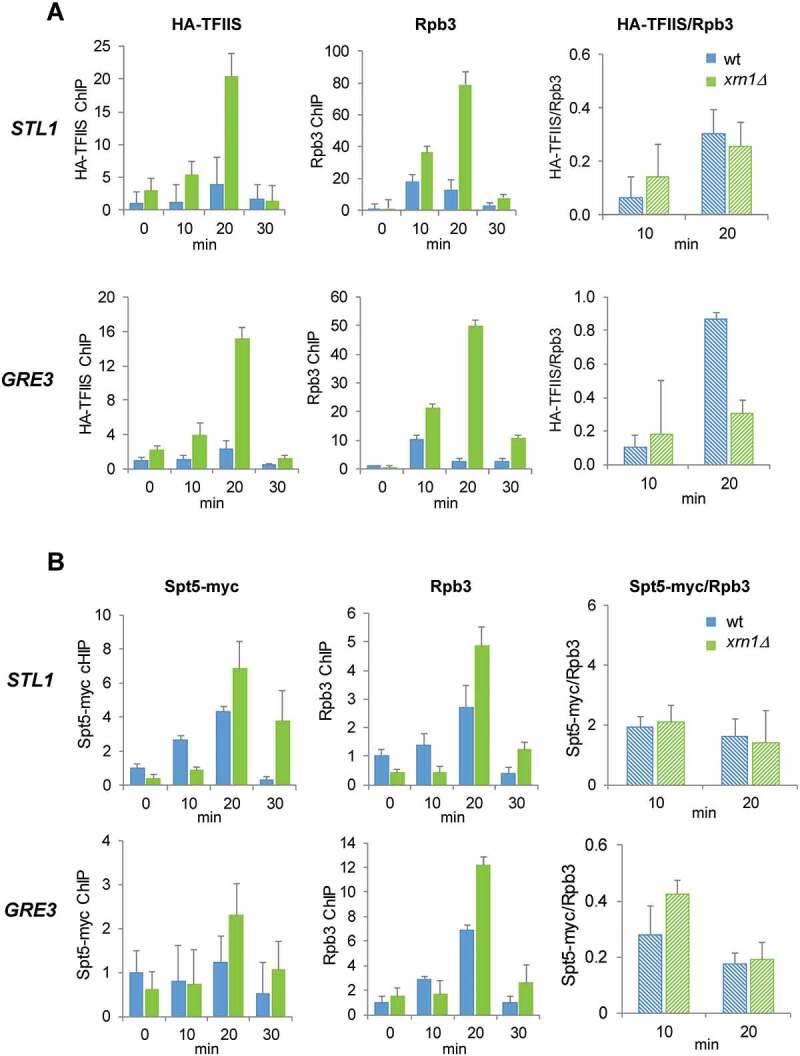
Deletion of XRN1 causes high levels of TFIIS and Spt5 recruitment to elongating RNAPII in genes upregulated by osmotic stress. (A) Parallel increase in TFIIS and Rpb3 recruitment at osmotic stress-upregulated STL1 and GRE3 in an xrn1Δ mutant. (B) The amount of Spt5/Rpb3 found at osmotic stress-upregulated STL1 and GRE3 is not changed in an xrn1Δ mutant. (A, B) Binding of HA-TFIIS or Spt5-myc (left panels) and Rpb3 (middle panels) and HA-TFIIS (or Spt5-myc)/Rpb3 ratio (right panels) on STL1 (upper panels) and GRE3 (lower panels) ORFs were analysed by chromatin immunoprecipitation (ChIP) in wild-type (wt) cells and xrn1Δ mutant cells. Cells were treated with 0.6 M KCl for the indicated time. ChIP data were calculated and normalized as described in Figure 5. Mean and standard deviation from three independent experiments are shown
Discussion
Xrn1, the evolutionarily conserved 5′ to 3′ mRNA exonuclease, seems to have a central role in the crosstalk between transcription and mRNA degradation in steady-state conditions [44,45,55]. The precise role of Xrn1 in transcription by RNAPII has yet to be elucidated and the involvement of Xrn1 during fast transcriptional responses was unknown. In this work, we have addressed the role of Xrn1 in the rapid and strong changes in TRs and mRNA concentrations that take place when cells respond to stress. Using the yeast S. cerevisiae and osmotic stress as a model, we discovered that i) Xrn1 is specifically recruited to genes strongly upregulated in a Hog1-dependent manner and in parallel to RNAPII recruitment; ii) acts as a transcription factor that promotes elongation by RNAPII by preventing its accumulation at genes in a backtracked state; iii) the activity of Xrn1 is necessary to change the stability of mRNAs during osmotic stress to shape their rapid and transitory kinetics of expression; and iv) Xrn1 binds and prevents backtracked RNAPII accumulation in other genes (e.g. GAL genes) that need a burst of transcription upon activation conditions.
Our results here and from three recent papers provide new evidence on the mechanism by which Xrn1 regulates transcription, all of which points to Xrn1 having a specific role in regulating transcription elongation. Begley et al. showed by run-on and ChIP analysis that fast depletion of Xrn1 in cells growing in galactose medium caused a strong decrease in elongating RNAPII on GAL1, without a change in the total RNAPII occupancy, suggesting RNAPII backtracking [56]. This study has then been extended genome-wide using CRAC and BioGRO-seq to show that the lack of Xrn1 affects the RNAPII profiles across the genome [72]. Fisher et al., using NET-seq and BioGRO-seq data, observed a high amount of chromatin-bound RNAPII compared to the levels of productive elongating RNAPII, again suggesting that RNAPII enters a state which is incompatible with elongation, such as a backtracked configuration. Moreover, they found that disruption of XRN1 causes a reduction of RNAPII occupancy downstream of TSSs but also an accumulation at 3′ regions proximal to the polyadenylation site [73]. In our study, we report a dramatic reduction of active elongating RNAPII in genes strongly upregulated by osmotic stress in parallel with an accumulation of inactive bound polymerase. Therefore, the four studies support the role of Xrn1 in maintaining a functional RNAPII complex along the transcriptional elongation process.
Our experimental model of transcriptional induction upon stress (either osmotic or by a sudden change in the carbon source) further extends our comprehension of how Xrn1 intervenes in the control of transcription elongation. We observed that Xrn1 binds to the promoters and ORFs of induced genes in a concomitant way to RNAPII binding. For both RNAPII and Xrn1, recruitment to osmotic stress-upregulated genes depends on the activity and, probably, binding of the MAPK Hog1, which activates in response to stress and interacts with the polymerase to allow transcription [23,24]. On the other hand, lack of Xrn1 seems to affect downregulated genes to a greater extent than lack of Hog1. Osmotic stress downregulated genes are regulated in a mostly Hog1-independent way [6,8,9]. So, Xrn1 function may be linked not to Hog1 but to its association with the RNAPII elongation complex. Supporting this, it has been reported that Xrn1 interacts physically [74] and functionally [75] with RNAPII and other factors involved in transcription [76–78]. We previously found that Xrn1 affects the TR of most yeast genes to an extent directly dependent on the TR value [54]. All these results support the idea that Xrn1 acts by helping RNAPII mainly in transcription-demanding contexts.
During the response to stress, lack of Xrn1 produced an accumulation of RNAPII at induced genes, which was then rapidly phosphorylated at Ser2 of its Rpb1 CTD. Although Ser2 phosphorylation is the RNAPII mark most clearly associated with the elongation step of transcription and is linked to the recruitment of 3′ pre-mRNA cleavage factors [79], our results in the xrn1Δ mutant indicate that the Ser2-P polymerase remains in a backtracked unproductive conformation (GRO vs RNAPII ChIP results). A similar result with over-phosphorylation at Ser2 of RNAPII is also observed in other genes highly transcribed under normal conditions in cells lacking Xrn1 [72]. These results suggest that in some circumstances, e.g. lack of Xrn1, Ser2 phosphorylation of RNAPII is not necessarily associated with active polymerase.
Release of paused polymerases and specifically backtracked RNAPII has been identified as essential for rapid activation of mammalian genes upon biological perturbations [32,80]. To reactivate stalled RNAPII, the conserved factor TFIIS enhances cleavage of RNAs in backtracked transcription complexes and dramatically accelerates transcription; meanwhile, the Spt4–Spt5 elongation complex stimulates polymerase processivity during elongation [68]. In our study, we found high levels of TFIIS and Spt5 at osmotic stress upregulated genes in xrn1Δ cells, in parallel to high levels of RNAPII at the same times of stress. Therefore, these results do not explain the accumulation of backtracked RNAPII molecules at stress-induced genes. In steady-state conditions (cells grown in galactose media), it has been found higher ratios of TFIIS/RNAPII at GAL1 in xrn1Δ cells and interpreted as a mark of excess backtracked RNAPII. However, the higher proportion of TFIIS with respect to RNAPII was not found in all genes, and RP genes, which are highly expressed and whose expression is highly negatively impacted by Xrn1 loss, did not show this accumulation of TFIIS [56]. More work should be done to determine the functional connection between Xrn1 and TFIIS and the precise role of Xrn1 in keeping an active elongating RNAPII along the transcribed genes.
Cytoplasmic Xrn1 has long been known to act as an exonuclease that digests 5′ to 3′ decapped mRNAs in one of the major routes of cytoplasmic mRNA degradation in eukaryotes [43,81]. Mutation of Xrn1 has wider effects on mRNA stability [44,45], although it seems to have certain specificity for unstable mRNAs [54,82]. Additionally, Xrn1 participates in specialized pathways that control the quality of mRNAs, e.g. nonsense-mediated and non-stop decay pathways, and in the digestion of non-coding RNAs [83,84]. As mentioned above, studies in yeast and recently in mammalian cells using mutants in XRN1 and other decay factors showed that alterations in mRNA half-life were inversely associated with variations in TRs, and mostly did not result in changes in transcript abundance. However, these studies were done with the knockdown mutants grown under steady-state conditions, in which mRNA cellular metabolism must have adapted to the growth. In our work, however, we studied the changes in RNA levels under stress situations in which rapid but transitory changes in mRNA levels occur that require variations in the rate of synthesis and of mRNA degradation [37] thus, if both increase, the kinetics are faster [37,41]. Our results show that Xrn1 is necessary for maintaining the kinetics of up- and downregulation in response to stress. For upregulated genes, lack of Xrn1 decrease TR and decay rate. mRNA stabilization compensates for the defects in transcription, so similar mRNA levels are reached in the xrn1Δ mutant. However, the quick drop in mRNA levels observed in upregulated genes in wild type is delayed in xrn1Δ. Conversely, during stress at times when mRNA levels are similar to initial levels in wild type, downregulated genes do not recover in xrn1Δ, so mRNA stabilization does not compensate for transcription defects in this gene group. Therefore, although in xrn1Δ there is homoeostatic compensation owing to its dual action in mRNA synthesis and degradation, this compensation cannot configure the proper kinetics profiles of the mRNA levels during stress.
Differences in the quantitative effect of Xrn1 in the metabolism of up- and downregulated transcripts may be due to different specificity, as it has been reported under steady-state conditions [54,56,73]. Additionally, we cannot rule out that stabilization experienced by the stress-responsive genes could be due to an alteration in their specific mRNA nuclear imprinting in the xrn1Δ mutant. Thus, we can generally conclude that Xrn1 is responsible for the fine-tunning necessary for quick changes in mRNAs during a rapid response to stress. One of the consequences of this lack of fine-tuned regulation in xrn1Δ could be the specific growth defect under osmotic stress observed in the mutant.
Supplementary Material
Acknowledgments
We thank Mordechai Choder (Technion-Israel Institute of Technology) and Sebastian Chavez (IBiS and Universidad de Sevilla) for yeast strains and valuable comments. We also thank all members of the GFL laboratory (Universitat de València) for helpful discussion.
Funding Statement
BFU2016–77728-C3–3-P to J.E.P.-O. and P.A., BFU2015–71978-REDT to J.E.P.-O. and the Regional Valencian Government [AICO2020/086 to P.A. and AICO2019/088 to J.E.P-O.
Data availability
The GRO data are stored in the GEO repository (accession number GSE151736).
Disclosure statement
No potential conflict of interest was reported by the authors.
Disclosure of interest
The authors report no conflict of interest.
Supplementary Material
Supplemental data for this article can be accessed here.
References
- [1].Perez-Ortin JE, Tordera V, Chavez S.. Homeostasis in the central dogma of molecular biology: the importance of mRNA instability. RNA Biol. 2019;16:1659–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].de Nadal E, Alepuz PM, Posas F. Dealing with osmostress through MAP kinase activation. EMBO Rep. 2002;3:735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hohmann S, Krantz M, Nordlander B. Yeast osmoregulation. Methods Enzymol. 2007;428:29–45. [DOI] [PubMed] [Google Scholar]
- [4].de Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nat Rev Genet. 2011;12:833–845. [DOI] [PubMed] [Google Scholar]
- [5].de Nadal E, Posas F. Osmostress-induced gene expression–a model to understand how stress-activated protein kinases (SAPKs) regulate transcription. Febs J. 2015;282:3275–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Posas F, Chambers JR, Heyman JA, et al. The transcriptional response of yeast to saline stress. J Biol Chem. 2000;275:17249–17255. [DOI] [PubMed] [Google Scholar]
- [7].Martinez-Montanes F, Pascual-Ahuir A, Proft M. Toward a genomic view of the gene expression program regulated by osmostress in yeast. Omics. 2010;14:619–627. [DOI] [PubMed] [Google Scholar]
- [8].Romero-Santacreu L, Moreno J, Perez-Ortin JE, et al. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA (New York, N.Y. 2009;15:1110–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Canadell D, Garcia-Martinez J, Alepuz P, et al. Impact of high pH stress on yeast gene expression: A comprehensive analysis of mRNA turnover during stress responses. Biochim Biophys Acta. 2015;1849:653–664. [DOI] [PubMed] [Google Scholar]
- [10].Molin C, Jauhiainen A, Warringer J, et al. mRNA stability changes precede changes in steady-state mRNA amounts during hyperosmotic stress. RNA (New York, N.Y). 2009;15:600–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Garre E, Romero-Santacreu L, Barneo-Munoz M, et al. Nonsense-mediated mRNA decay controls the changes in yeast ribosomal protein pre-mRNAs levels upon osmotic stress. PLoS One. 2013;8:e61240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Regot S, de Nadal E, Rodriguez-Navarro S, et al. The Hog1 stress-activated protein kinase targets nucleoporins to control mRNA export upon stress. J Biol Chem. 2013;288:17384–17398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Melamed D, Pnueli L, Arava Y. Yeast translational response to high salinity: global analysis reveals regulation at multiple levels. RNA (New York, N.Y. 2008;14:1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Warringer J, Hult M, Regot S, et al. The HOG pathway dictates the short-term translational response after hyperosmotic shock. Mol Biol Cell. 2010;21:3080–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Garre E, Romero-Santacreu L, De Clercq N, et al. Yeast mRNA cap-binding protein Cbc1/Sto1 is necessary for the rapid reprogramming of translation after hyperosmotic shock. Mol Biol Cell. 2012;23:137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wu CC, Zinshteyn B, Wehner KA, et al. High-resolution ribosome profiling defines discrete ribosome elongation states and translational regulation during cellular stress. Mol Cell. 2019;73:959–970 e955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Garre E, Pelechano V, Del Pino S, et al. The Lsm1-7/Pat1 complex binds to stress-activated mRNAs and modulates the response to hyperosmotic shock. PLoS Genet. 2018;14:e1007563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Saito H, Posas F. Response to hyperosmotic stress. Genetics. 2012;192:289–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tatebayashi K, Yamamoto K, Nagoya M, et al. Osmosensing and scaffolding functions of the oligomeric four-transmembrane domain osmosensor Sho1. Nat Commun. 2015;6:6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zuzuarregui A, Kupka T, Bhatt B, et al. M-Track: detecting short-lived protein-protein interactions in vivo. Nat Methods. 2012;9:594–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zuzuarregui A, Li T, Friedmann C, et al. Msb2 is a Ste11 membrane concentrator required for full activation of the HOG pathway. Biochim Biophys Acta. 2015;1849:722–730. [DOI] [PubMed] [Google Scholar]
- [22].Proft M, Struhl K. MAP kinase-mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell. 2004;118:351–361. [DOI] [PubMed] [Google Scholar]
- [23].Alepuz PM, Jovanovic A, Reiser V, et al. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell. 2001;7:767–777. [DOI] [PubMed] [Google Scholar]
- [24].Alepuz PM, de Nadal E, Zapater M, et al. Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. Embo J. 2003;22:2433–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nadal-Ribelles M, Conde N, Flores O, et al. Hog1 bypasses stress-mediated down-regulation of transcription by RNA polymerase II redistribution and chromatin remodeling. Genome Biol. 2012;13:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nadal-Ribelles M, Mas G, Millan-Zambrano G, et al. H3K4 monomethylation dictates nucleosome dynamics and chromatin remodeling at stress-responsive genes. Nucleic Acids Res. 2015;43:4937–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Perez-Martinez ME, Benet M, Alepuz P, et al. Nut1/Hos1 and Sas2/Rpd3 control the H3 acetylation of two different sets of osmotic stress-induced genes. Epigenetics. 2020;15:251–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Proft M, Mas G, de Nadal E, et al. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell. 2006;23:241–250. [DOI] [PubMed] [Google Scholar]
- [29].Mas G, de Nadal E, Dechant R, et al. Recruitment of a chromatin remodelling complex by the Hog1 MAP kinase to stress genes. Embo J. 2009;28:326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sole C, Nadal-Ribelles M, Kraft C, et al. Control of Ubp3 ubiquitin protease activity by the Hog1 SAPK modulates transcription upon osmostress. Embo J. 2011;30:3274–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Silva A, Cavero S, Begley V, et al. Regulation of transcription elongation in response to osmostress. PLoS Genet. 2017;13:e1007090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sheridan RM, Fong N, D’Alessandro A, et al. Widespread backtracking by RNA Pol II is a major effector of gene activation, 5ʹ pause release, termination, and transcription elongation rate. Mol Cell. 2019;73:107–118 e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gomez-Herreros F, de Miguel-jimenez L, Morillo-Huesca M, et al. TFIIS is required for the balanced expression of the genes encoding ribosomal components under transcriptional stress. Nucleic Acids Res. 2012;40:6508–6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lisica A, Engel C, Jahnel M, et al. Mechanisms of backtrack recovery by RNA polymerases I and II. Proc Natl Acad Sci U S A. 2016;113:2946–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zatreanu D, Han Z, Mitter R, et al. Elongation factor TFIIS prevents transcription stress and r-loop accumulation to maintain genome stability. Mol Cell. 2019;76:57–69 e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Perez-Ortin JE, Alepuz PM, Moreno J. Genomics and gene transcription kinetics in yeast. Trends Genet. 2007;23:250–257. [DOI] [PubMed] [Google Scholar]
- [38].Rabani M, Levin JZ, Fan L, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;29:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yang E, van Nimwegen E, Zavolan M, et al. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tani H, Mizutani R, Salam KA, et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shalem O, Dahan O, Levo M, et al. Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol. 2008;4:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191:671–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Perez-Ortin JE, Alepuz P, Chavez S, et al. Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J Mol Biol. 2013;425:3750–3775. [DOI] [PubMed] [Google Scholar]
- [44].Sun M, Schwalb B, Pirkl N, et al. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol Cell. 2013;52:52–62. [DOI] [PubMed] [Google Scholar]
- [45].Haimovich G, Medina DA, Causse SZ, et al. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. [DOI] [PubMed] [Google Scholar]
- [46].Braun KA, Young ET. Coupling mRNA synthesis and decay. Mol Cell Biol. 2014;34:4078–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Das S, Sarkar D, Das B. The interplay between transcription and mRNA degradation in Saccharomyces cerevisiae. Microb Cell. 2017;4:212–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Timmers HTM, Tora L. Transcript buffering: a balancing act between mRNA synthesis and mRNA degradation. Mol Cell. 2018;72:10–17. [DOI] [PubMed] [Google Scholar]
- [49].Hartenian E, Glaunsinger BA. Feedback to the central dogma: cytoplasmic mRNA decay and transcription are interdependent processes. Crit Rev Biochem Mol Biol. 2019;54:385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Harel-Sharvit L, Eldad N, Haimovich G, et al. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell. 2010;143:552–563. [DOI] [PubMed] [Google Scholar]
- [51].Haimovich G, Choder M, Singer RH, et al. The fate of the messenger is pre-determined: a new model for regulation of gene expression. Biochim Biophys Acta. 2013;1829:643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Miller JE, Reese JC. Ccr4-Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol. 2012;47:315–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Collart MA. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip Rev RNA. 2016;7:438–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Medina DA, Jordan-Pla A, Millan-Zambrano G, et al. Cytoplasmic 5ʹ-3ʹ exonuclease Xrn1p is also a genome-wide transcription factor in yeast. Front Genet. 2014;5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Singh P, James RS, Mee CJ, et al. mRNA levels are buffered upon knockdown of RNA decay and translation factors via adjustment of transcription rates in human HepG2 cells. RNA Biol. 2019;16(9): 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Begley V, Corzo D, Jordan-Pla A, et al. The mRNA degradation factor Xrn1 regulates transcription elongation in parallel to Ccr4. Nucleic Acids Res. 2019;47:9524–9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gilbertson S, Federspiel JD, Hartenian E, et al. Changes in mRNA abundance drive shuttling of RNA binding proteins, linking cytoplasmic RNA degradation to transcription. Elife. 2018;7:e37663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Garcia-Martinez J, Aranda A, Perez-Ortin JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell. 2004;15:303–313. [DOI] [PubMed] [Google Scholar]
- [59].Li T, De Clercq N, Medina DA, et al. The mRNA cap-binding protein Cbc1 is required for high and timely expression of genes by promoting the accumulation of gene-specific activators at promoters. Biochim Biophys Acta. 2016;1859:405–419. [DOI] [PubMed] [Google Scholar]
- [60].Gasch AP, Spellman PT, Kao CM, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Teixeira MC, Monteiro PT, Guerreiro JF, et al. The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:D161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jorgensen P, Rupes I, Sharom JR, et al. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Garrido-Godino AI, Garcia-Lopez MC, Garcia-Martinez J, et al. Rpb1 foot mutations demonstrate a major role of Rpb4 in mRNA stability during stress situations in yeast. Biochim Biophys Acta. 2016;1859:731–743. [DOI] [PubMed] [Google Scholar]
- [64].Aguilera J, Prieto JA. The Saccharomyces cerevisiae aldose reductase is implied in the metabolism of methylglyoxal in response to stress conditions. Curr Genet. 2001;39:273–283. [DOI] [PubMed] [Google Scholar]
- [65].Joo YJ, Ficarro SB, Chun Y, et al. In vitro analysis of RNA polymerase II elongation complex dynamics. Genes Dev. 2019;33:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hirayoshi K, Lis JT. Nuclear run-on assays: assessing transcription by measuring density of engaged RNA polymerases. Methods Enzymol. 1999;304:351–362. [DOI] [PubMed] [Google Scholar]
- [67].Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987;51:458–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sanders TJ, Lammers M, Marshall CJ, et al. TFS and Spt4/5 accelerate transcription through archaeal histone-based chromatin. Mol Microbiol. 2019;111:784–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Schulz S, Gietl A, Smollett K, et al. TFE and Spt4/5 open and close the RNA polymerase clamp during the transcription cycle. Proc Natl Acad Sci U S A. 2016;113:E1816–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Crickard JB, Lee J, Lee TH, et al. The elongation factor Spt4/5 regulates RNA polymerase II transcription through the nucleosome. Nucleic Acids Res. 2017;45:6362–6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hartzog GA, Fu J. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta. 2013;1829:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Victoria Begley AJ-P, Peñate X, Garrido-Godino AI, et al. Xrn1 influences RNA pol II-dependent transcription elongation rates across the yeast genome and this control is particularly relevant for late elongation of regulatory genes. RNA Biol. 2020. Dec 1;1-14. DOI:10.1080/15476286.2020.1845504. Online ahead of print. [Google Scholar]
- [73].Fischer J, Song YS, Yosef N, et al. The yeast exoribonuclease Xrn1 and associated factors modulate RNA polymerase II processivity in 5ʹ and 3ʹ gene regions. J Biol Chem. 2020. DOI: 10.1074/jbc.RA120.013426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Harlen KM, Churchman LS. Subgenic Pol II interactomes identify region-specific transcription elongation regulators. Mol Syst Biol. 2017;13:900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Srivas R, Shen JP, Yang CC, et al. A network of conserved synthetic lethal interactions for exploration of precision cancer therapy. Mol Cell. 2016;63:514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Costanzo M, VanderSluis B, Koch EN, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science (New York, N.Y. 2016;353. DOI: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Collins SR, Miller KM, Maas NL, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. [DOI] [PubMed] [Google Scholar]
- [78].Krogan NJ, Cagney G, Yu H, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. [DOI] [PubMed] [Google Scholar]
- [79].Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Bartman CR, Hamagami N, Keller CA, et al. Transcriptional burst initiation and polymerase pause release are key control points of transcriptional regulation. Mol Cell. 2019;73:519–532 e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Geisler S, Coller J. XRN1: A major 5ʹ to 3ʹ exoribonuclease in eukaryotic cells. Enzymes. 2012;31:97–114. [DOI] [PubMed] [Google Scholar]
- [82].Jones CI, Zabolotskaya MV, Newbury SF. The 5ʹ –> 3ʹ exoribonuclease XRN1/Pacman and its functions in cellular processes and development. Wiley Interdiscip Rev RNA. 2012;3:455–468. [DOI] [PubMed] [Google Scholar]
- [83].Wolin SL, Maquat LE. Cellular RNA surveillance in health and disease. Science (New York, N.Y. 2019;366:822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Labno A, Tomecki R, Dziembowski A. Cytoplasmic RNA decay pathways - Enzymes and mechanisms. Biochim Biophys Acta. 2016;1863:3125–3147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GRO data are stored in the GEO repository (accession number GSE151736).


