ABSTRACT
Circular RNA, typically generated from backsplicing reaction, is a class of single-stranded and covalently linked RNA. Although most circular RNAs are lowly expressed, some of them are able to accumulate to high levels and even exceed their cognate mRNAs due to their longer half-lives. Once produced in the nucleus, the majority of circular RNAs are exported to the cytoplasm for their proper functions or degradation. In this review, we will summarize the biogenesis and classification of circular RNAs and highlight the recent advances in our understanding of circular RNA nuclear export and degradation.
KEYWORDS: Pre-mRNA splicing, backsplicing, RNA-binding protein, circular RNA, nuclear export, degradation
Introduction
Most eukaryotic genes were split by introns which must be removed from precursor message RNA (pre-mRNA) through joining an upstream 5ʹ splicing site (5ʹ ss) to a downstream 3ʹ splicing site (3ʹ ss) by the splicing machinery in linear order [1]. It has been well-established that the standard splicing process is mostly executed in a co-transcriptional manner and highly regulated to yield a mature mRNA that will be exported subsequently to the cytoplasm fraction for translation [2–4]. However, only recently has it been appreciated that the splicing event can also happen in an alternative or noncanonical way to allow the backsplicing reaction by joining a downstream 5ʹ ss to an upstream 3ʹ ss, thereby generating a circular RNA with covalently linked structure [5–11].
With the development of high-throughput RNA sequencing (RNA-seq) technique, thousands of circular RNAs have been detected or identified across different cell lines, tissues, and species [12–17]. Although the majority of circular RNAs are rarely produced comparing with their cognate mRNAs, they, at least for some of them, could still accumulate to high levels as they naturally lack features utilized by diverse exonucleases [18]. For instance, a circular RNA generated from the laccase 2 gene is 10-fold more abundant than its linear cognate mRNA in Drosophila DL1 cells [19]. Also, circular RNAs have been found as the major products from roughly 50 human genes in different cell lines [13]. These studies suggest that at least some of circular RNAs may have important functional roles.
The biogenesis of circular RNAs
The majority of circular RNAs identified so far are generated from exon(s) of protein-coding genes (Fig. 1). We thus call them exonic circular RNAs (EcircRNAs). Many EcircRNAs contain multiple exons, usually two or three, but it is important to note that there is a minimal length of a single exon that allows circularization [18–20]. In human cells, a median length of 353 nt of a circularized exon is required for single-exon backsplicing, whereas only 112–130 nt per exon for multiple-exon backsplicing is required [21]. Moreover, a series of plasmid experiments have successfully demonstrated that only exons > 300 nt could be circularized efficiently in Drosophila system [19]. Besides the role of exonic sequences play in circular RNA formation, a previous study on mouse Sry revealed that the reverse complementary sequence flanking the circularized exon is important for the generation of a circular RNA [8]. Indeed, many sequence feature analyses have proved that the flanking introns are longer than average and are more likely to have reverse complementary sequences, such as Alu repeats in human [18,21]. Base pairing between the flanking introns brings together the 5ʹ ss and 3ʹ ss in close intervene, which facilitates backsplicing reactions (Fig. 1) [18,21]. In some cases, short reverse complementary sequences, even as short as 30–40 nt, are sufficient to promote backsplicing [18]. However, it should be noted that all circularized exons lack noticeable flanking intronic sequences in yeast, indicating that the reverse repeats-mediated mechanism may not account for circularization in lower eukaryotes [20,22]. In fact, the formation of a circular RNA can proceed through an exon skipping-mediated mechanism (also called indirect backsplicing reaction) by re-splicing of an exon-containing lariat precursor in Schizosaccharomyces pombe (Fig. 1) [22]. There is another category of exon-containing circular RNAs, named as exon-intron circular RNAs (EIciRNAs) that consist of both exon(s) and intron(s) [23]. In this case, the intron between the circularized exons is retained during splicing. But the backsplicing process of EIciRNAs is similar to EcircRNAs.
Figure 1.
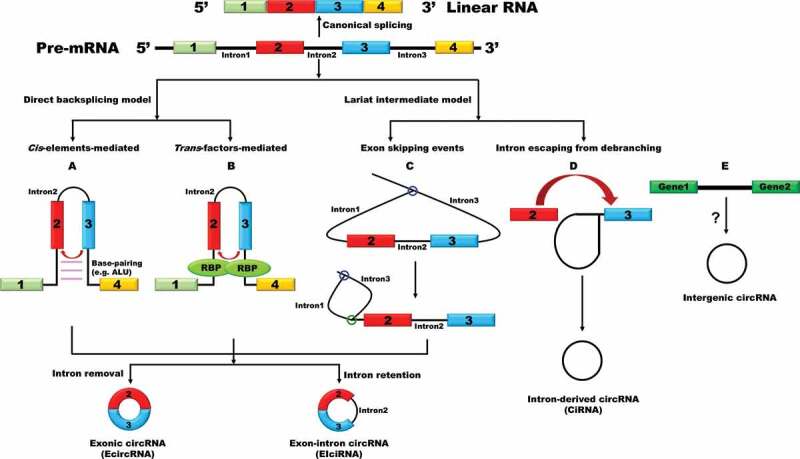
Circular RNAs biogenesis
A model shows a pre-mRNA with 4 exons and how different types of circular RNAs are generated. (A) Base-pairing between the two flanking introns, which contain reverse complementary sequences, brings together a downstream 5ʹ ss and an upstream 3ʹ ss and facilitates backsplicing reactions to produce circular RNAs. (B) RBPs binding to the flanking introns can act analogously as flanking complementary sequences to promote backsplicing of circularized exons. (C) Exon skipping generates a mature mRNA and a lariat RNA. Instead of debranching and degradation, the lariat RNA goes for another round of splicing to generate a circular RNA and a double lariat RNA. (D) During canonical splicing, lariat RNAs fail to be debranched and accumulate as circular intronic RNAs (ciRNAs). (E) Some circular RNAs might be products of the intergenic genome region.
Besides exon-containing circular RNAs, circular intronic RNA (ciRNA) is a type of lariat RNAs that fails to be debranched during the process of splicing and accumulates in the nucleus instead (Fig. 1) [24–26]. ciRNAs were identified in different species, and most of them are 100–500 nt in length [24–26]. Interestingly, a large proportion of them tend to contain an unusual cytosine at the branchpoint rather than the more common adenine [25]. These cytosine branchpoints may account for the resistance of the stable lariat intronic RNAs (ciRNAs) to debranching enzyme [25]. A study on a ciRNA derived from the second intron of ANKRD52 revealed that a consensus motif containing a 7-nt GU-rich element near the 5ʹ ss and an 11-nt C-rich element near the branchpoint are able to stabilize ciRNAs, but the detailed molecular mechanisms are still unclear [24]. In addition, a few of bioinformatics studies suggested that some circular RNAs are products of the intergenic genome region [27–29]. Nevertheless, the accuracy of these intergenic circular RNAs needs to be further validated in the future through specialized techniques such as RNase R-treated assays and northern blotting experiments.
Notably, neither reverse repeats nor lariat production is sufficient to give rise to circular RNAs, indicating an additional parameter regulating the generation of circular RNAs. Indeed, beyond cis-elements, a number of RNA-binding proteins (RBPs) were found to actively modulate circular RNA biogenesis (Fig. 1 and Table 1) [19,30–39]. Adenosine deaminase 1 acting on RNA (ADAR1), a double-stranded RNA-binding protein, is able to inhibit exon circularization through converting A to I which disrupts the base pairing between the two flanking introns [30]. The alternative splicing factor Quaking (QKI) increases the production of a subset of highly expressed circular RNAs during human epithelial-mesnchymal transition (EMT) [32]. QKI is able to recognize the QKI binding sites in the flanking introns of circularized exons, thereby acting analogously as reverse complementary sequences to facilitate circularization. Inserting of QKI motifs to the flanking introns is sufficient to generate certain circular RNAs from exons that do not produce circular RNAs normally in cells. In Drosophila, the splicing factor Muscleblind (Mbl) promotes the production of circular RNAs through binding to the flanking introns of circularized exon when there is an excess amount of the protein, thereby fine-tuning its host gene expression [33]. Afterwards, the RBP fused in sarcoma (FUS), a well-characterized splicing factor that functions in amyotrophic lateral sclerosis (ALS), was also described as a potent regulator of circular RNA biogenesis in mouse motor neurons [39]. In addition to the RBPs regulating the production of circular RNAs through modulating the base pairing between flanking introns, depletion of spliceosome components or inhibiting splicing activity pharmacologically resulted in shifting the outputs of at least a subset of genes towards the production of circular RNAs while their linear cognate mRNA levels were damped in both fly and human cells [37]. In line with this, similar expression pattern of circular RNAs and their cognate mRNAs was observed in rat primary neurons treated with splicing inhibitor [38].
Table 1.
RBPs involved in circular RNA biogenesis
| Species | Trans factors | REFs |
|---|---|---|
| Homo sapiens | QKI (Quaking) | 32 |
| Homo sapiens | ADAR1 (adenosine deaminase 1 acting on RNA) | 30 |
| Homo sapiens | RBM20 (RNA-binding motif protein 20) | 35 |
| Homo sapiens | NF90/NF110 (nuclear factor 90 and its 110 isoform NF110) | 34 |
|
Homo sapiens Drosophila melanogaster |
HnRNP (heterogeneous nuclear ribonucleoprotein) | 19, 36 |
| Homo sapiens | DHX9 (DExH-box helicase 9) | 31 |
| Drosophila melanogaster | Mbl (muscleblind) | 33 |
| Drosophila melanogaster | SR (serine-arginine) proteins | 19 |
| Mus musculus | FUS (fused in sarcoma) | 39 |
|
Drosophila melanogaster Rattus norvegicus |
Spliceosome | 37, 38 |
The nuclear export of circular RNAs
Different types of circular RNAs have distinct localizations, which might be regulated by nuclear localization sequences or nuclear export sequences positioned in the circularized exons or retained introns. In fact, ciRNAs and EIciRNAs were found to be mainly retained in the nucleus [23,24], whereas the vast majority of EcircRNAs were enriched in the cytoplasmic fraction. Nucleus-localized circular RNAs were proposed to regulate gene expression through modulating either transcription or alternative splicing pattern [23,40]. Cytoplasmic circular RNAs also have diverse functions such as acting as microRNA sponges and translation templates [41]. Considering that a large amount of circular RNAs are detected in non-dividing cells, such as sperm and neuronal cells, the changes in circular RNA cellular distribution may not simply rely on nuclear envelope breakdown during the cycle of cell division or chromosome segregation. Therefore, it is more likely that active transport processes control the nuclear export of cytoplasm-localized circular RNAs. Indeed, a comprehensive circular RNA profiling in HepG2 cells revealed that cytoplasm-enriched circular RNAs consist of motifs recognized by nuclear export RBPs through analysing the binding motif distribution of 19 nuclear export RBPs [42]. This study suggests that RBP-mediated selective transportation maybe responsible for cells to distinguish between linear RNAs and circular RNAs for their nuclear export.
Using a focused RNAi screening, Huang et al. found that depletion of Drosophila DExH/D-box helicase Hel25E resulted in a robust accumulation of long (> 800 nt) but not short circular RNAs in the nucleus [43](Fig. 2). Interestingly, the two human homologs of Hel25E (UAP56 and URH49) are also involved in the localization of circular RNAs but with distinct characteristics. UAP56 is responsible for exporting of long circular RNAs (>1300 nt), while URH49 is required for short circular RNAs (< 400 nt) nuclear export. Moreover, through generating chimeric proteins and rescue experiments, a four-amino-acid motif was found to be necessary and sufficient to dictate circular RNA export preferences [43–45]. However, the detailed mechanism whereby the length of circular RNAs is measured by the two homologs remains unclear.
Figure 2.
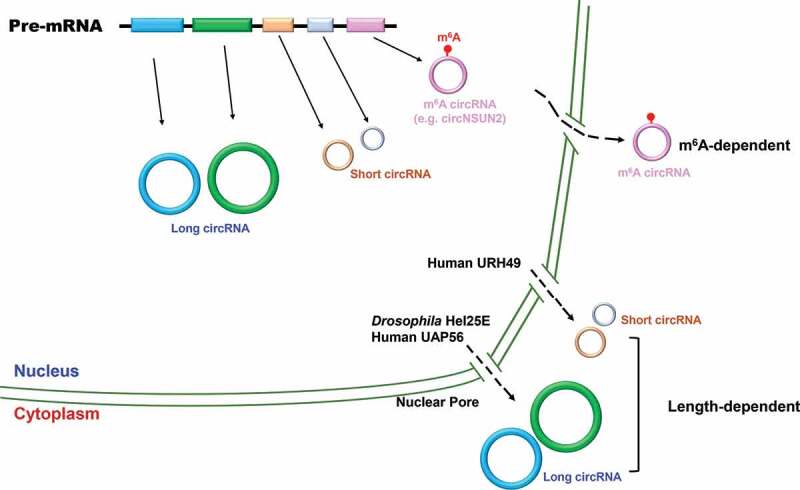
Circular RNA nuclear export
Once generated, circular RNAs can be fed into different export pathways. Drosophila Hel25E and human UAP56 regulate the nuclear export of long circular RNAs. Human URH49 controls short circular RNA export. YTHDC1 promotes the export of m6A-modified circNSUN2 in human cells.
N6-methyladenosine is the most abundant internal modification on RNAs [46,47], and it plays diverse roles in gene expression at the transcriptional, post-transcriptional, and/or translational levels [48]. Intriguingly, m6A circular RNAs are frequently derived from exon(s) that are not methylated in linear mRNAs [49]. m6A modification has been proposed to participate in circular RNA nuclear export [50]. For example, a circular RNA (circNSUN2) derived from 4th and 5th exon of the NSUN2 gene was found to interact with YTH domain-containing protein 1 (YTHDC1), an m6A reader, via an m6A motif (GAACU) formed in the backsplicing junction site [50]. Knocking down YTHDC1 or m6A deposit methyltransferase METTL3 led to a significant accumulation of circNSUN2 in the nucleus, and this phenomenon could be rescued by re-expression of wild type proteins [50]. But m6A-mediated circular RNA nuclear export appears to be unique to a few cases as only a small number of circular RNAs have m6A modification [51]. A case in point is that the subcellular localization of circZNF609 was not altered upon YTHDC1 depletion [52]. Nevertheless, our current understanding on how circular RNAs are exported to the cytoplasm and/or which factor determines their localization is still very limited. With the development of single molecule tracking system and CRISPR-based genome engineering or nucleic acid detection [53–58], we believe future researches will provide more insights into orchestrated circular RNA transportation pathways.
The degradation of circular RNAs
Naturally closed characteristics of circular RNAs theoretically prevent them from degradation by exonucleases that typically degrade linear RNAs from either 5ʹ or 3ʹ end. Therefore, circular RNAs have much longer half-lives than their linear cognate RNAs generally [59]. Although it is not fully understood how circular RNAs are degraded in steady-state condition, there have been exciting studies beginning to shed light on the clearance of circular RNAs through multiple pathways.
(1) MicroRNA-mediated circular RNA degradation (Fig. 3A)
Figure 3.
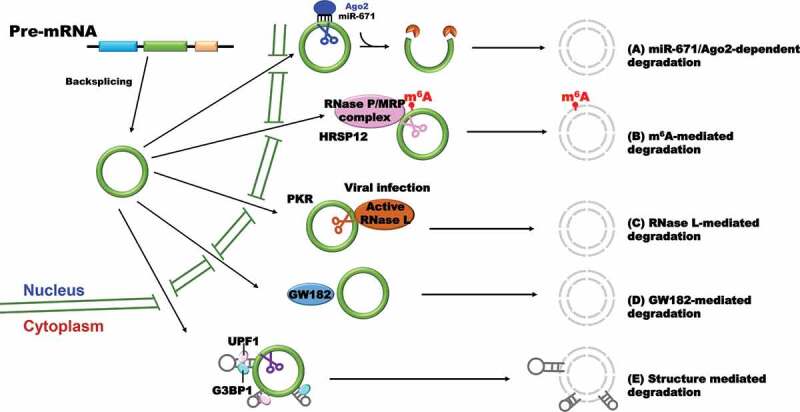
Circular RNA degradation
A model shows how different types of circular RNAs are degraded. (A) microRNA-mediated circular RNA degradation; (B) m6A-mediated circular RNA degradation; (C) RNase L-mediated circular RNA degradation; (D) GW182-mediated circular RNA degradation; (E) structure-mediated circular RNA degradation.
Circular RNA CDR1as/ciRS-7 has a near perfect target site for miR-671 [16]. The binding of miR-671 to CDR1as leads to direct RNA cleavage by endonuclease Argonaute-2 (Ago2), thereby generating a 5ʹ and 3ʹ free end. The cleaved RNA is subjected subsequently to fully degradation by different exonucleases on both ends. However, whether other circular RNAs can be degraded though microRNA-mediated endonucleolytic cleavage is still unclear, because only few circular RNAs exhibit properties expected of microRNA sponges for their lack of potential microRNA binding sites [15]. Based on the proof-of-concept experiments in hand, the levels of many circular RNAs (e.g. Drosophila circdati and circlaccase2) were unaffected by knocking down of microRNA silencing factors [60].
(2) m6A-mediated circular RNA degradation (Fig. 3B).
In a recent study, Park and colleagues found that the m6A reader protein YTHDF2 is able to recognize m6A modification deposited on circular RNAs and interacts with the adaptor protein HRSP12 which further recruit RNase P/MRP complex to specifically degrade circular RNAs [61]. Although the finding first demonstrates the molecular mechanisms of circular RNA degradation mediated by m6A modification, it is not likely a general mechanism for the regulation of circular RNA metabolism based on the reported characteristics that most of circular RNAs lack m6A modification.
(3) RNase L-mediated circular RNA degradation (Fig. 3C)
Upon poly(I:C) treatment or encephalomyocarditis (EMCV) RNA virus infection, endogenous circular RNA level falls rapidly due to the activation of endonuclease RNase L [62]. In this case, many circular RNAs are able to interact with protein kinase (PKR) though their intra-molecularly imperfect 16–26 bp duplex, a special topological structure, to suppress the activity of PKR in steady state. Once infected by EMCV, rapid circular RNA degradation by RNase L is likely to release the circular RNA-associated PKR which is further activated and engaged in early cellular immune response. Furthermore, elevated PKR phosphorylation level and global circular RNA reduction were detected in cells from patients with autoimmune disease systemic lupus erythematosus, indicating that the regulation of circular RNA stability is an important therapeutic strategy for certain diseases. Nevertheless, the question whether other virus infection also induces the activation of RNase L which leads to cleavage of RNAs is still waiting for further investigation.
(4) GW182 (gawky)-mediated circular RNA degradation (Fig. 3D).
Through a focused RNAi screening, Jia et al. found that circular RNA levels increased upon GW182 depletion in Drosophila DL1/S2 cells, and the changes can only be observed in pre-existing circular RNAs but not newly-synthesized transcripts, indicating GW182 is specifically responsible for circular RNA degradation [60]. As an evolutionarily conserved protein, GW182 is usually thought as the key factor involved in microRNA-mediated gene silencing and localizes to the processing body (P-body) which contains multiple factors for mRNA decay and translation repression [63,64]. Moreover, three human homologs of GW182 (TNRC6A, TNRC6B, and TNRC6C) showed similar roles in regulating endogenous circular RNA degradation in HeLa cells. Extensive mutagenesis of GW182 further revealed that the Mid domain itself may function as a scaffold to mediate the degradation of circular RNAs by ribonucleases, and thus the process is likely independent of P-body.
(5) Structure-mediated circular RNA degradation (Fig. 3E).
A recent work found that the RBP UPF1 and its associated factor G3BP1 cooperatively control the degradation of linear RNAs in a structure-dependent manner [65]. In this pathway, target mRNAs typically consist of highly structured 3ʹ UTRs which are recognized and bound by UPF-1 and G3BP1. Concerning circular RNAs, highly structured circular RNAs were also upregulated when UPF1 or G3BP1 was depleted from multiple cell lines. G3BP1 was found to be specifically bound to highly structured circular RNAs, while UPF1 lacks specificity for the degree of structure of circular RNAs, suggesting that G3BP1 plays a critical role in recognizing highly structured circular RNAs in structure-mediated degradation pathway [65].
The functional roles of circular RNAs
The functions of most circular RNAs remain largely unexplored, but emerging evidence is starting to reveal that circular RNAs play important roles in many biological processes.
(1) Sponging microRNA and modulating expression of its target genes
One of the typical and most studied circular RNAs is CDR1as/ciRS-7 which contains over 60 evolutionarily conserved miR-7 binding sites and one miR-671 binding site [16]. It was originally thought that CDRas circular RNA sequesters many miR-7 in the cytoplasm and ultimately leads to the increased level of miR-7 target mRNAs [16]. However, this hypothesis was challenged by a follow up observation that miR-7 level increased and its target mRNA level decreased when CDR1as was knocked out through removing the whole gene locus in mouse model [66]. Besides this classical circular RNA, some other circular RNAs were found to be able to sponge microRNAs, although most of them only contain a limited number of microRNA binding sites [41,67,68]. As most circular RNAs are lowly expressed, further studies are needed to determine in what extend circular RNAs could sponge microRNAs in different context.
(2) Affecting the splicing of their host pre-mRNAs.
In Arabidopsis, circSEP3, which is derived from exon 6 of SEPALLATA3 (SEP3), tightly binds to the single-strand DNA of its cognate gene locus and forms an R-loop (RNA:DNA), while its linear cognate mRNAs cannot form stable interaction with DNA [40]. The R-loop structure inhibits RNA Polymerase II (Pol II) elongation and ultimately leads to the generation of its host transcript without exon 6 [40].
(3) Promoting RNA Pol II mediated transcription.
A considerable number of EIciRNAs were found to be enriched in the nucleus and associated with Pol II [23]. As one of the example, EIciRNA EIF3J interacts with U1 snRNP via specific RNA-RNA interaction to promote transcription of its host gene through forming a larger complex with Pol II in the promoter region. Knocking down EIciRNA EIF3J by the antisense oligonucleotide (ASO) or siRNA specifically targeting junction region resulted in decreased expression of its cognate mRNA. In addition, the second intron of ANKRD52 derived ciRNA is also predominantly enriched in the nucleus, and it was found to positively regulate its host gene expression [24]. However, whether the effect of these nuclear-retained circular RNAs on their host gene represents a general phenomenon is still unknown.
(4) Acting as protein decoys to modulate protein function.
Circular RNAs were also found to have diverse functions via direct interaction with different proteins [69–71]. CircPABPN1 was reported to suppress translation by sequestering HuR from binding to its host PABPN1 mRNA [69]. Both circFoxo3 and circANRIL are able to regulate cell proliferation by interacting with specific proteins. CircFoxo3 inhibits cell division protein kinase2 (CDK2) activity and blocks cell proliferation by forming ternary complex with CDK2 and cell cycle-dependent kinase inhibitor 1 (p21) [70]. INK4 antisense derived circANRIL is able to induce nucleolar stress and p53 activation. CircANRIL induces apoptosis and inhibits proliferation by binding to pescadillo homologue (PES1) which is important for ribosome biogenesis in vascular smooth muscle cells and macrophages [71]. Moreover, some circular RNAs could also bind to NF90/NF110, PKR, or cGAS and are involved in innate/autoimmune response [34,62,72].
(5) Translating to proteins
Generation of protein isoforms via circularization of selected exon(s) from pre-mRNA transcripts is an intriguing field that raises many studies. Several lines of evidence revealed that a few circular RNAs could be translated into proteins through internal ribosome entry site (IRES) in a cap-independent manner [73,74]. For example, circZNF609, derived from the second exon of the ZNF609 gene, is translatable during myoblast proliferation. It contains a complete ORF that shares the same start codon with its cognate gene and has a stop codon 3 nt right after the splice junction [74]. However, the function of circZNF609-derived protein remains elusive. Another nice work by Weigelt et al. demonstrated that insulin-sensitive circSf1 is able to translated into a peptide which shares the N terminal with the full-length Sfl protein encoded by the sulfateless gene [75]. Interestingly, circSf1-derived peptide is sufficient to extend lifespan of fruit flies [75]. Besides employing IRES, circular RNAs with ORF were proposed to be translated when the m6A modification site exists, and the translation efficiency is positively correlated with the number of m6A sites [76]. m6A recruits translation initiation factors through YTHDF3, a reader protein of m6A, and the translation efficiency can be reversibly regulated by both METTL3/14 methyltransferases and fat mass and obesity-associated (FTO) protein (demethylase) [76]. However, a recent comprehensive study on circZNF609 produced from plasmid revealed that its translation is independent of IRES or METTL3/METTL4-mediated m6A modification, but from trans-splicing by-products of plasmid, suggesting that future works on the translation of circular RNAs should be evaluated carefully, especially when a plasmid system is used [77].
(6) Regulating mitochondrial entry of nuclear-encoded proteins and mitochondrial permeability transition pore
Eukaryotic cells contain many copies of mitochondrial DNA [78–81]. Although it has long been thought that there is no splicing during the transcription cycle of mitochondria genome-encoded genes, hundreds of backsplicing junction reads supported the existence of mitochondria-encoded circular RNAs (MecciRNAs) in different vertebrate species [82]. Detailed studies on MecciND1 and MecciDN5 further suggested that they could interact with nuclear-encoded proteins and promote their importing into mitochondria. Another interesting work by Zhao et al. demonstrated that mitochondria-located circular RNA SCAR is dysregulated in non-alcoholic fatty liver disease (NAFLD) patients’ liver fibroblasts and can turn off mitochondrial permeability transition pore (mPTP) via direct interaction with ATP5B, a subunit of mitochondrial ATP synthase [83]. With further in vivo experiments, the authors proved that the human circular RNA SCAR can interact with mouse ATP5B and is able to alleviate high fat diet-induced cirrhosis in mice, providing a potential therapeutic strategy.
Perspectives and moving forward
Localization of circular RNAs to specific location within cells is an important aspect of the control of gene expression during differentiation, development, ageing, and multiple diseases. EIciRNAs and ciRNAs are retained in the nucleus to regulate their host gene transcription [23,24], while the EcircRNAs are mostly enriched in the cytoplasmic fraction to modulate gene expression post-transcriptionally through sponging microRNAs or decoy functional proteins. Our current understanding is only limited to Hel25E and its human homologs (UAP56 and URH49) which mediate circular RNA nuclear export in a length-dependent manner [43]. In the future, it will be important to figure out the detailed mechanisms whereby circular RNAs are exported from the nucleus and the question whether there are other factors that are able to specifically recognize and facilitate the exporting process. Furthermore, it is not intuitive why intron-containing circular RNAs are retained in the nucleus and how cells may distinguish them from EcircRNAs. Our group is working on the question currently.
On the other hand, cells under stress conditions not only undergo a huge change in gene expression, but also in dyslocalization of RNAs and proteins, such as the components of nuclear pore complex (NPC) and different RBPs. Therefore, it would also be intriguing to evaluate the localization of circular RNAs in response to different stresses and their potential roles in defence against stresses.
In addition to circular RNA intracellular transportation, circular RNAs could be sorted and packed into exosomes which are nanoscale (30–150 nm) extracellular vesicles of endocytic origin released by many cells and exported out of cells [84–86]. However, whether circular RNAs are randomly wrapped or selective sorted into exosomes is unclear. Furthermore, some parasitic plants, such as Cuscuta campestris, can facilitate bidirectional movement of proteins and mRNAs between parasite and host during parasitism. In fact, some microRNAs of Cuscuta campestris were found to act as trans-species regulators of host gene expression in Arabidopsis thaliana [87]. This raises an interesting question whether circular RNAs could also be exchanged between cells or even species. If so, what are the roles of these circular RNAs?
Multiple pathways have been proposed to regulate the degradation of circular RNAs either in normal cells or under virus infection, but the detailed molecular mechanisms are still obscure and many attractive questions are raised. For instance, (1) does the degradation of circular RNA represent a general phenomenon upon virus infection? (2) What are the responses and functions of circular RNAs when cells are challenged with different viruses? (3) How do the distinct structures of circular RNAs determine their functions and stability? (4) Is there a specific endoribonuclease which is solely responsible for circular RNA turnover in normal cells?
Considering that the dysregulation of circular RNA transportation or degradation may result in various pathological conditions, such as cancers and brain disorders [66,88–92], future exploration on the transportation and degradation of different types of circular RNAs will shed more insights on the functions of circular RNAs and provide novel diagnostic or therapeutic strategies for human diseases.
Acknowledgement
We thank the members of the Shan lab (University of Science and Technology of China) for discussions.
Funding Statement
This work was sponsored by the National Natural Science Foundation of China [32070633], the Natural Science Foundation of Chongqing, China [cstc2019jcyjmsxmX0085], the Innovation Support Program for Overseas Returned Scholars of Chongqing, China [cx2019142], the Fundamental Research Funds for the Central Universities of China [2020CDJQY-A076], and the 100 Talent Program of Chongqing University [0304001104433].
Author contributions
MZ, MX, ZL, and CH wrote the paper. MZ contributed to the figures in the paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Wahl MC, Will CL, Luhrmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. [DOI] [PubMed] [Google Scholar]
- [2].Herzel L, Ottoz DSM, Alpert T, et al. Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nat Rev Mol Cell Biol. 2017;18:637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Merkhofer EC, Hu P, Johnson TL. Introduction to cotranscriptional RNA splicing. Methods Mol Biol. 2014;1126:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chen L, Huang C, Wang XL, et al. Circular RNAs in eukaryotic cells. Curr Genomics. 2015;16:312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell. 1991;64:607–613. [DOI] [PubMed] [Google Scholar]
- [7].Cocquerelle C, Mascrez B, Hetuin D, et al. Mis-splicing yields circular RNA molecules. Faseb J. 1993;7:155–160. [DOI] [PubMed] [Google Scholar]
- [8].Capel B, Swain A, Nicolis S, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. [DOI] [PubMed] [Google Scholar]
- [9].Huang C, Shan G. What happens at or after transcription: insights into circRNA biogenesis and function. Transcription. 2015;6:61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–490. [DOI] [PubMed] [Google Scholar]
- [11].Wilusz JE. Circular RNAs: unexpected outputs of many protein-coding genes. RNA Biol. 2017;14:1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Westholm JO, Miura P, Olson S, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- [17].Xiao MS, Wilusz JE. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3ʹ ends. Nucleic Acids Res. 2019;47:8755–8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kramer MC, Liang D, Tatomer DC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li X, Liu S, Zhang L, et al. A unified mechanism for intron and exon definition and back-splicing. Nature. 2019;573:375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. [DOI] [PubMed] [Google Scholar]
- [22].Barrett SP, Wang PL, Circular SJ. RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. [DOI] [PubMed] [Google Scholar]
- [24].Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. [DOI] [PubMed] [Google Scholar]
- [25].Talhouarne GJS, Gall JG. Lariat intronic RNAs in the cytoplasm of vertebrate cells. Proc Natl Acad Sci U S A. 2018;115:E7970–E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cheng J, Zhang Y, Li Z, et al. A lariat-derived circular RNA is required for plant development in Arabidopsis. Sci China Life Sci. 2018;61:204–213. [DOI] [PubMed] [Google Scholar]
- [27].Zhao X, Duan X, Fu J, et al. Genome-wide identification of circular RNAs revealed the dominant intergenic region circularization model in apostichopus japonicus. Front Genet. 2019;10:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gaffo E, Boldrin E, Dal Molin A, et al. Circular RNA differential expression in blood cell populations and exploration of circRNA deregulation in pediatric acute lymphoblastic leukemia. Sci Rep. 2019;9:14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang J, Chen S, Yang J, et al. Accurate quantification of circular RNAs identifies extensive circular isoform switching events. Nat Commun. 2020;11:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. [DOI] [PubMed] [Google Scholar]
- [31].Aktas T, Avsar Ilik I, Maticzka D, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. [DOI] [PubMed] [Google Scholar]
- [32].Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. [DOI] [PubMed] [Google Scholar]
- [33].Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. [DOI] [PubMed] [Google Scholar]
- [34].Li X, Liu CX, Xue W, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67:214–27 e7. [DOI] [PubMed] [Google Scholar]
- [35].Khan MA, Reckman YJ, Aufiero S, et al. RBM20 regulates circular RNA production from the titin gene. Circ Res. 2016;119:996–1003. [DOI] [PubMed] [Google Scholar]
- [36].Fei T, Chen Y, Xiao T, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci U S A. 2017;114:E5207–E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liang D, Tatomer DC, Luo Z, et al. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol Cell. 2017;68:940–54 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang M, Hou J, Muller-McNicoll M, et al. Long and repeat-rich intronic sequences favor circular RNA formation under conditions of reduced spliceosome activity. iScience. 2019;20:237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Errichelli L, Dini Modigliani S, Laneve P, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Conn VM, Hugouvieux V, Nayak A, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3:17053. [DOI] [PubMed] [Google Scholar]
- [41].Xiao MS, Ai Y, Wilusz JE. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. 2020;30:226–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang J, Zhang X, Li C, et al. Circular RNA profiling provides insights into their subcellular distribution and molecular characteristics in HepG2 cells. RNA Biol. 2019;16:220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Huang C, Liang D, Tatomer DC, et al. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Li Z, Kearse MG, Huang C. The nuclear export of circular RNAs is primarily defined by their length. RNA Biol. 2019;16:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wan Y, Hopper AK. Size matters: conserved proteins function in length-dependent nuclear export of circular RNAs. Genes Dev. 2018;32:600–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liu M, Wang Q, Shen J, et al. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019;16:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chen J, Fang X, Zhong P, et al. N6-methyladenosine modifications: interactions with novel RNA-binding proteins and roles in signal transduction. RNA Biol. 2019;16:991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608–624. [DOI] [PubMed] [Google Scholar]
- [49].Zhou C, Molinie B, Daneshvar K, et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20:2262–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chen RX, Chen X, Xia LP, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Paramasivam A, Vijayashree Priyadharsini J. Novel insights into m6A modification in circular RNA and implications for immunity. Cell Mol Immunol. 2020;17:668–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Di Timoteo G, Dattilo D, Centron-Broco A, et al. Modulation of circRNA metabolism by m(6)A modification. Cell Rep. 2020;31:107641. [DOI] [PubMed] [Google Scholar]
- [53].Teng F, Cui T, Feng G, et al. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov. 2018;4:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Li SY, Cheng QX, Wang JM, et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Catarino RR, Stark A. Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev. 2018;32:202–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Teng F, Cui T, Gao Q, et al. Artificial sgRNAs engineered for genome editing with new Cas12b orthologs. Cell Discov. 2019;5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kocks C, Boltengagen A, Piwecka M, et al. In situ hybridization (FISH) of circular RNA CDR1as. Methods Mol Biol. 2018;1724:77–96. [DOI] [PubMed] [Google Scholar]
- [59].Enuka Y, Lauriola M, Feldman ME, et al. RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jia R, Xiao MS, Li Z, et al. Defining an evolutionarily conserved role of GW182 in circular RNA degradation. Cell Discov. 2019;5:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Park OH, Ha H, Lee Y, et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol Cell. 2019;74:494–507 e8. [DOI] [PubMed] [Google Scholar]
- [62].Liu CX, Li X, Nan F, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–80 e21. [DOI] [PubMed] [Google Scholar]
- [63].Ding L, Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17:411–416. [DOI] [PubMed] [Google Scholar]
- [64].Liu J, Rivas FV, Wohlschlegel J, et al. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fischer JW, Busa VF, Shao Y, et al. Structure-mediated RNA decay by UPF1 and G3BP1. Mol Cell. 2020;78:70–84 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Piwecka M, Glazar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. In: Science. 2017. p. 357:eaam8526. [DOI] [PubMed] [Google Scholar]
- [67].Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. [DOI] [PubMed] [Google Scholar]
- [68].Zhang X, Yan Y, Lin W, et al. Circular RNA Vav3 sponges gga-miR-375 to promote epithelial-mesenchymal transition. RNA Biol. 2019;16:118–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Holdt LM, Stahringer A, Sass K, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Xia P, Wang S, Ye B, et al. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity. 2018;48:688–701 e7. [DOI] [PubMed] [Google Scholar]
- [73].Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Weigelt CM, Sehgal R, Tain LS, et al. An insulin-sensitive circular RNA that regulates lifespan in drosophila. Mol Cell. 2020;79:268–79 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ho-Xuan H, Glazar P, Latini C, et al. Comprehensive analysis of translation from overexpressed circular RNAs reveals pervasive translation from linear transcripts. Nucleic Acids Res. 2020;48:10368–10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Priesnitz C, Becker T. Pathways to balance mitochondrial translation and protein import. Genes Dev. 2018;32:1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Jin X, Cheng Z, Wang B, et al. Precise annotation of human, chimpanzee, rhesus macaque and mouse mitochondrial genomes leads to insight into mitochondrial transcription in mammals. RNA Biol. 2020;17:395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Caudron-Herger M, Diederichs S. Mitochondrial mutations in human cancer: curation of translation. RNA Biol. 2018;15:62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ji H, Xu X, Jin X, et al. Using high-resolution annotation of insect mitochondrial DNA to decipher tandem repeats in the control region. RNA Biol. 2019;16:830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Liu X, Wang X, Li J, et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci China Life Sci. 2020;63:1429–1449. [DOI] [PubMed] [Google Scholar]
- [83].Zhao Q, Liu J, Deng H, et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell. 2020;183:76–93 e22. [DOI] [PubMed] [Google Scholar]
- [84].Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Fanale D, Taverna S, Russo A, et al. Circular RNA in exosomes. Adv Exp Med Biol. 2018;1087:109–117. [DOI] [PubMed] [Google Scholar]
- [86].Lasda E, Circular PR. RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS One. 2016;11:e0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Shahid S, Kim G, Johnson NR, et al. MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature. 2018;553:82–85. [DOI] [PubMed] [Google Scholar]
- [88].Lukiw WJ. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet. 2013;4:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Patop IL, Kadener S. circRNAs in Cancer. Curr Opin Genet Dev. 2018;48:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Xu K, Chen D, Wang Z, et al. Annotation and functional clustering of circRNA expression in rhesus macaque brain during aging. Cell Discov. 2018;4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15:995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Tan J, Gu S, Zheng Y, et al. Expression profile of circular RNAs in myocardial ischemia/reperfusion with and without intermittent hypobaric hypoxia preconditioning. Sci China Life Sci. 2019;62:1104–1106. [DOI] [PubMed] [Google Scholar]


