ABSTRACT
Long non-coding RNA HOTAIR has been reported to play a key role in regulating various biological processes in various cancers. However, the roles and mechanisms of HOTAIR in acute myeloid leukaemia (AML) are still unclear and need to be investigated. In this study, we induced differentiation of four AML cell lines by all-trans retinoic acid (ATRA) and found HOTAIR was significantly upregulated in the process. Chromatin immunoprecipitation (ChIP) assays indicated that C/EBPβ upregulated HOTAIR during ATRA induced differentiation in HL-60 cells. By gain- and loss-of-function analysis, we then observed that HOTAIR expression was positively correlated with ATRA-induced differentiation and negatively regulated G1 phase arrest in HL-60 cells. In addition, we found that HOTAIR promoted ATRA-induced differentiation via the regulation of the cell cycle regulator p21 via miR-17-5p. Moreover, we detected the expression of HOTAIR in 84 de novo AML patients, HOTAIR was found significantly downregulated in the AML patients compared to the iron deficiency anaemia (IDA) control group, negatively correlated with the platelet level in M2 patients. In all, our data suggest that HOTAIR may be subtype-specific in AML-M2 patients, also HOTAIR regulates AML differentiation by C/EBPBβ/HOTAIR/miR-17-5p/p21 pathway. The findings of the present study provide a novel insight into the mechanism of lncRNA-mediated differentiation and indicate that HOTAIR may be a promising therapeutic target for leukaemia, especially for AML with M2 type.
Abbreviation: AML: acute myeloid leukaemia; APL: acute promyelocytic leukaemia; ATRA: all-trans retinoic acid; CCK8: cell Counting Kit-8; CDKs: cyclin-dependent kinases ; CeRNA: competing endogenous RNAs; ChIP: chromatin immunoprecipitation; CHX: cycloheximide; FAB: French–American–British; FCM: flow cytometry; HOTAIR: HOX transcript antisense RNA; IDA: iron-deficiency anemia; lncRNA: long non-coding RNA; 3′UTR: 3′untranslated region; MT: Mutation type; WT: Wild type; qRT-PCR: Quantitative real-time PCR
KEYWORDS: HOTAIR, acute myeloid leukaemia, differentiation, ATRA, p21, miR-17-5p
Introduction
Acute myeloid leukaemia (AML) is a kind of malignant clonal disease that originates from stem cells of the myeloid system. According to the French–American–British (FAB) criteria, AML is classified into eight subtypes, M0-M7 [1]. The incidence of AML is age-related and is commonly seen in the elderly (>60 years old), with significant heterogeneity [2]. Induction chemotherapy and haematopoietic stem cell transplantation are commonly used in the clinical treatment of AML. However, the long-term prognosis of AML patients is still unsatisfactory [3]. AML cells fail to exit the cell cycle and are blocked at a certain stage of the haematopoietic differentiation [4]. All-trans retinoic acid (ATRA) could induce myeloid leukaemia cells to differentiate to promote myeloid differentiation by inducing cell cycle arrest at the G1 phase [5]. Thus, the application of ATRA has made great progress in the treatment and prognosis of the most dangerous AML subtype, acute promyelocytic leukaemia (APL), also known as AML-M3 [6]. However, the treatment and prognosis of the other subtypes of AML are still not satisfying, further pathogenetic and therapeutic approaches need to be explored [3,7].
Recent studies have identified that long non-coding RNA (lncRNA), a kind of non-protein-coding transcripts longer than 200 nucleotides, plays a vital role in numerous biological processes including epigenetic regulation [8], differentiation [9–12], and tumour progression [13]. HOTAIR is a well-documented lncRNA, and it is highly expressed in many tumours [14–16], and known to inhibit its target genes via direct interaction with histone modification complexes [17–19]. Evidence suggests it acts as a competing endogenous RNAs (ceRNA) to regulate miRNA [15,20]; and is considered as a negative biomarker for tumour metastasis and prognosis [21,22]. However, the link between HOTAIR and AML differentiation has not yet been extensively studied.
In this study, we investigated the expression, function, and the potential mechanism of HOTAIR in AML differentiation. Our data showed that HOTAIR was upregulated in AML cell line HL-60 after ATRA treatment and revealed that overexpressed HOTAIR promoted HL-60 cell differentiation through p21/cyclin D1/CDK4 pathway via miR-17-5p. In addition, our data indicated that ATRA upregulated HOTAIR expression via C/EBPβ. Moreover, HOTAIR was found significantly downregulated in the AML patients, negatively correlated with the platelet level in M2 patients. Therefore, this study will shed some light on the role of HOTAIR in the differentiation of AML; and helping determine if HOTAIR could be used as a potential target to treat AML (non-APL).
Result
ATRA treatment upregulated HOTAIR expression in AML cells
A lot of evidence suggests that HOTAIR plays a critical role in tumorigenesis and tumour progression. However, the role of HOTAIR in myeloid differentiation remains unknown. To this aim, we firstly induced myeloid differentiation of acute myeloid leukaemia cell lines (HL-60, NB4, U937, and THP-1) by ATRA, and CD11b was used to measure the differentiation by flow cytometry (FCM). As shown in Fig. 1, CD11b was highly expressed in the four ATRA-treated cell lines. Additionally, all four cell lines exhibited a higher expression level of HOTAIR after the ATRA treatment and the level was markedly upregulated in HL-60 (Fig. 1). This result suggests HOTAIR may be essential for myeloid cell differentiation.
Figure 1.
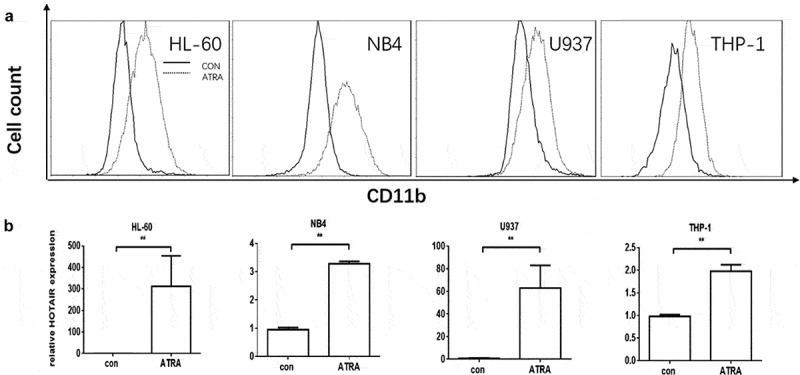
CD11b was highly expressed in four AML cell lines. CD11b was detected by flow cytometry (FCM); the full lines represent negative control, and the dotted lines represent ATRA-treatment (A). The relative expression of HOTAIR: HOTAIR was highly expressed in the four AML cell lines after the ATRA-treatment (B). ** represents P < 0.05, con represents control, ATRA represents all-trans retinoic acid
HOTAIR regulated cell differentiation in HL-60 cells
To further investigate whether HOTAIR is required for myeloid differentiation, specific small interfering RNA (siRNA) target HOTAIR (si-HOTAIR), and negative control siRNA (si-NC) were used to transfect the AML cell lines (Fig. 2), and the corresponding knockdown efficiency of the siRNA were shown in Fig. S1. Myeloid differentiation was induced by ATRA for 48 hours after the siRNA transfection, CD11b, the biomarker of myeloid differentiation was then tested by FCM. The results showed a significant reduction of CD11b expression after HOTAIR knockdown in HL-60 cells (Fig. 2A); however, no significant changes of CD11b expression were observed in NB4, U937, and THP-1 cell lines throughout the myeloid differentiation (Fig. 2).
Figure 2.
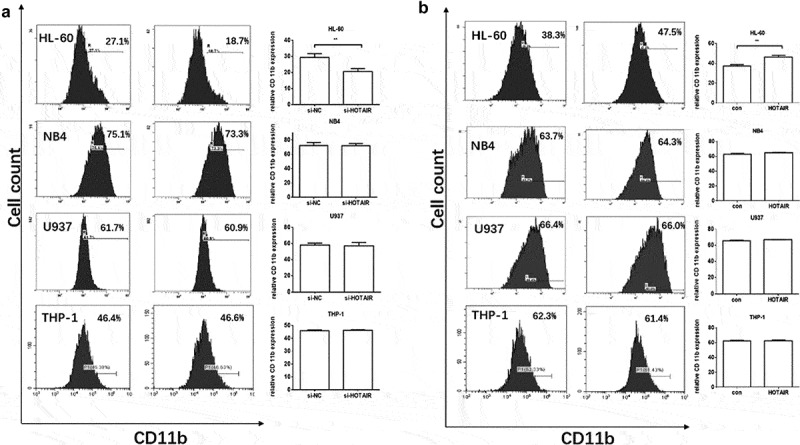
Effect of HOTAIR on the expression of CD11b. (A) Knockdown of HOTAIR by siRNA decreased the expression of CD11b in HL-60 cell, no expression difference observed in NB4, U937, and THP-1 cells compared with their si-negative control (si-NC) group. (B) overexpression of HOTAIR by plasmid increased the expression of CD11b in HL-60 cell, no expression difference in NB4, U937, and THP-1 cells compared with controls (con). ** represents P < 0.05
To verify the above observation, HOTAIR plasmid was conducted, as shown in Fig. 2B, overexpression of HOTAIR significantly increased CD11b expression in HL-60 cells, indicating an enhancement of myeloid differentiation, but not in NB4, U937, and THP-1 cell lines. Interestingly, we overexpressed HOTAIR in HL-60 cells but without ATRA treatment and the CD11b was higher in the HOTAIR overexpression group (Fig. S2). Therefore, HOTAIR seems to be cell type-specific, hence HL-60 cell was chosen to explore the mechanisms of HOTAIR.
C/EBPβ upregulated HOTAIR during ATRA-induced differentiation
HOTAIR may play an essential role in HL-60 cell differentiation, but how ATRA induces HOTAIR expression was not clear. To address this, we used a translation inhibitor, cycloheximide (CHX), to treat the HL-60 cells prior to the addition of ATRA. As shown in Fig. 3A, CHX significantly impaired the upregulated expression of HOTAIR, indicating the induction of HOTAIR requires newly synthesized intermediate protein.
Figure 3.
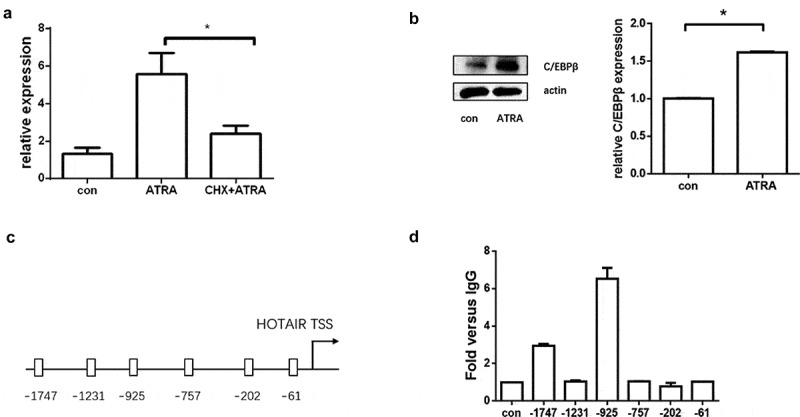
C/EBPβ binding to the promoter region of HOTAIR during differentiation. (A) Cycloheximide (CHX) significantly impaired the up-regulated expression of HOTAIR by ATRA. (B) C/EBPβ was upregulated after ATRA-treatment. (C) Schematic representation of predicted C/EBPβ binding sites around the promoter region of HOTAIR. (D) The predicted binding site of C/EBPβ in HOTAIR promoter (−1747 and −925) was significantly increased after ATRA treatment
Then, the online database JASPAR and PROMO were used to predict that the HOTAIR promoter region possesses transcription factors binding sites. Among the predicted transcription factors, C/EBPβ has been documented to play an important role during myeloid differentiation [23], and our data showed that C/EBPβ was significantly upregulated during myeloid differentiation by ATRA treatment (Fig. 3B). To probe further, we validated the binding capacity of C/EBPβ to HOTAIR by ChIP and ChIP-PCR experiments; specific primers were designed and synthesized for each predicted binding site (Fig. 3C and Table S1). The results of the ChIP-PCR assays showed that the binding site of C/EBPβ in HOTAIR promoter (located in −1747 and −925) was significantly amplified after the ATRA treatment in HL-60 cells (Fig. 3D).
HOTAIR regulated HL-60 cell differentiation via modulating cell cycle
It is well known that ATRA promotes myeloid differentiation by inducing cell cycle arrest at the G0/G1 phase [5], therefore; we wondered whether the upregulation of HOTAIR may affect the cell cycle. To address the hypothesis, we investigated the cell cycle phase distribution of HL-60 cells by FCM. As shown in Fig. 4A and Fig. 4B, the cell proliferation rate was increased, and the G0/G1 phase was significantly lower in the ATRA + si -HOTAIR group than the ATRA + si-NC group. Additionally, the cell proliferation rate was decreased, and the G0/G1 phase was significantly higher in the ATRA + HOTAIR plasmid group than the ATRA+NC plasmid group (Fig. 4C, D).
Figure 4.
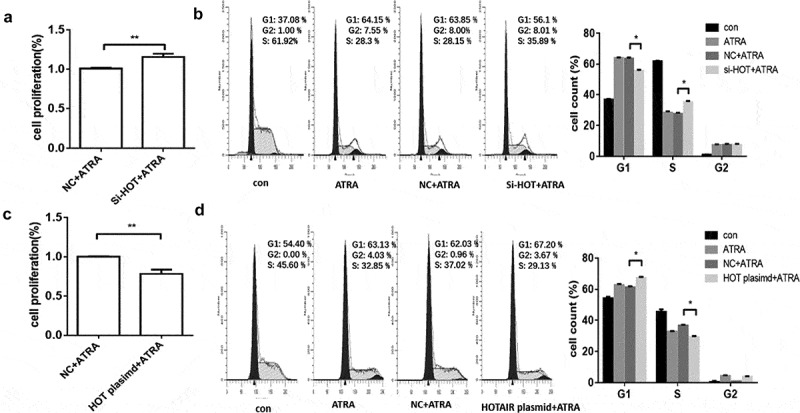
Effect of HOTAIR on the cell cycle. (A) The proliferation rate of HL-60 cells of si-HOTAIR and si-NC. (B) Flow cytometry was performed to analyse the cell cycle of si-HOTAIR and si-NC, and representative histograms are presented. (C) The proliferation rate of HL-60 cells of HOTAIR plasmid and NC plasmid. (D) Flow cytometry was performed to analyse the cell cycle of HOTAIR plasmid and NC plasmid, and representative histograms are presented. * represent P < 0.05, NC represents negative control
HOTAIR modulated cell cycle through p21/cyclin D1/CDK4 pathway
A group of proteins named cyclin-dependent kinases (CDKs) controls the cell cycle progression in mammals. CDK4 has been identified to regulate cell cycle progression from the G1 phase to the S phase by binding to cyclin D1. Whereas p21 inhibits the cyclin D1/CDK4 complex, the CIP/KIP family members regulate the induction of differentiation [24]. Our data showed increased cyclin D1, and CDK4 expression levels, but decreased p21 protein, in the HOTAIR siRNA group than in the NC siRNA group. On the contrary, we observed decreased cyclin D1, CDK4 expression levels but increased p21 protein level in the HOTAIR plasmid group than in the NC plasmid group (Fig. 5).
Figure 5.
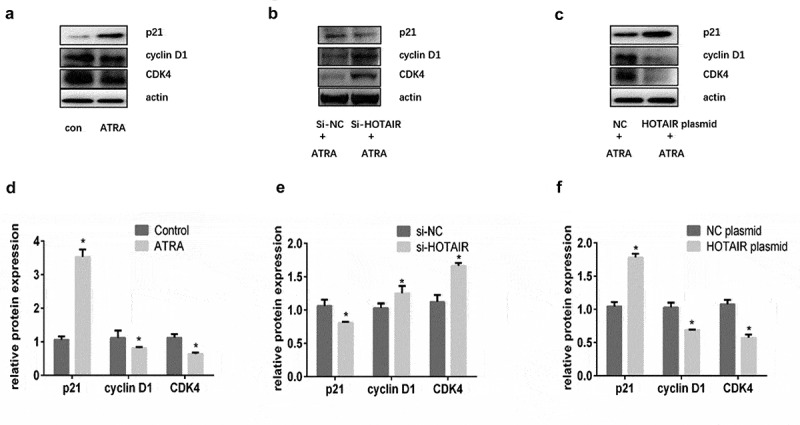
Effects of HOTAIR on p21, cyclin D1 and CDK4. (A) Western blot analysis showing the protein levels of p21, cyclin D1 and CDK4 in HL-60 cells treated with PBS and ATRA. β-actin served as a loading control, and (D) relative protein levels (means ± SD, n = 3) of p21, cyclin D1 and CDK4 were measured. (B) Western blot analysis showing the protein levels of p21, cyclin D1 and CDK4 in HL-60 cells transfected with si-HOTAIR and si-NC. β-actin served as a loading control, (E) relative protein levels (means ± SD, n = 3) of p21, cyclin D1 and CDK4 were measured. (C) Western blot analysis showing the protein levels of p21, cyclin D1 and CDK4 in HL-60 cells transfected with HOTAIR plasmid and NC plasmid. β-actin served as a loading control, and (F) relative protein levels (means ± SD, n = 3) of p21, cyclin D1 and CDK4 were measured. * represent P < 0.05
HOTAIR acted as a ceRNA to regulate p21 via miR-17-5p
HOTAIR acts as a ceRNA in numerous physiological and pathological processes [15,20]. To investigate the potential mechanistic role of HOTAIR, we used the online database Starbase to predict miRNAs that might interact with HOTAIR and p21. We found miR-17-5p, one of the predicted miRNAs (miR-20a-5p, miR-20b-5p, miR-106a-5p, miR-106b-5p, miR-93-5p, miR-519d-3p), as the most downregulated in HL-60 differentiation (Fig. 6A). Then, we constructed luciferase reporter vectors containing the predicted wild type (WT) and mutated type (MT) binding site of miR-17-5p for HOTAIR and p21 (Fig. 6B, C). We co-transfected HEK293T cells with these vectors and miR-17-5p mimics or negative control (miR-NC), and then examined them for luciferase activity. As shown in Fig. 6D, E, the luciferase activity was significantly decreased in HOTAIR WT and p21-3ʹUTR WT vectors than HOTAIR MT and p21-3ʹUTR MT. Besides, miR-17-5p restored the si-HOTAIR induced downregulation of p21 protein (Fig. 6F).
Figure 6.
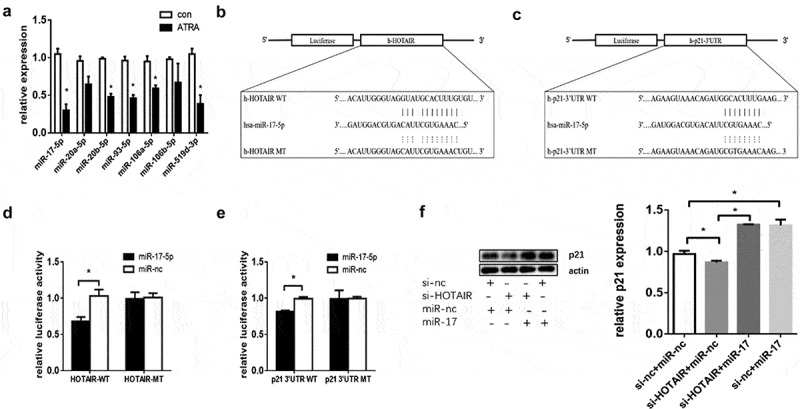
HOTAIR regulated p21 via miR-17-5p. (A) miR-17-5p was downregulated after ATRA treatment. (B) HOTAIR and corresponding fragments were inserted into the region immediately downstream of the luciferase gene in the psiCHECK2 vector. The sequences of the predicted miR-17-5p binding sites within the HOTAIR or p21, including the wild-type (WT) or mutant (MT) segments (dotted lines), are shown. (C) p21-3ʹUTR and corresponding fragments were inserted into the region immediately downstream of the luciferase gene in the psiCHECK2 vector. The sequences of the predicted miR-17-5p binding sites within the p21-3ʹUTR, including the wild-type 3ʹUTR or mutant 3ʹUTR segments (dotted lines), are shown. (D) The predicted binding sites of miR-17-5p to HOTAIR were confirmed by dual-luciferase reporter gene assay. (E) The predicted binding sites of miR-17-5p to p21-3ʹUTR confirmed by dual-luciferase reporter gene assay. (F) Western blot analysis showing the protein levels of p21 in HL-60 cells treated with ATRA after co-transfection with HOTAIR siRNA and or miR-17-5p inhibitor
Above all, our data indicated C/EBPβ upregulated HOTAIR and modulated cell cycle through the p21/cyclin D1/CDK4 pathway via miR-17-5p in the myeloid differentiation (Fig. 7).
Figure 7.
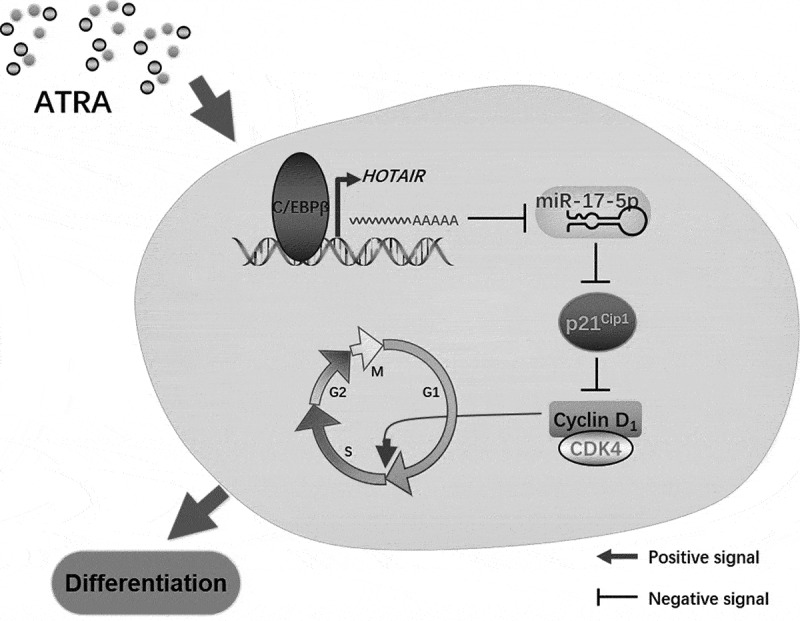
The hypothetical mechanism of HOTAIR in AML differentiation
The expression of HOTAIR in AML patients
The expression of HOTAIR in AML patients remains largely undetermined; to this aim, we analysed HOTAIR expression in 84 de novo AML patients and 20 IDA patients’ bone marrow cells by PCR (Fig. 8A). Our data revealed a lower expression level of HOTAIR in AML patients than IDA patients (Fig. 8B); and FAB subtype, gender, and age did not influence the expression level (Fig. 8C and Fig. S3).
Figure 8.
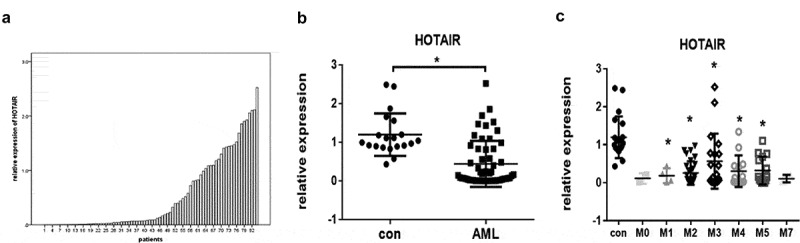
The expression of HOTAIR in AML patients. (A) The distribution of HOTAIR in AML patients. (B) Relative expression of HOTAIR in AML compared to IDA patients. (C) Relative expression of HOTAIR in AML according to FAB classification. * represents P < 0.05
Furthermore, we explored the relationship between HOTAIR and AML patients’ clinical data; the characteristics of the patients are listed in Table 1. There were no correlations between the HOTAIR expression and all the AML patients’ clinical parameters in this cohort (data not shown). Then, we re-analysed the relationship between HOTAIR and AML patients’ clinical parameters according to FAB-subtypes. We found HOTAIR negatively correlated with platelet levels in AML-M2 patients (Table 2); however, we did not find a correlation in the other subtypes (data not shown), respectively.
Table 1.
Characteristics of the newly diagnosed AML patients
| Characteristics | Values |
|---|---|
| Age (Median, range) | 45, years (0–80) |
| Male (no. (%)) | 42 (50%) |
| WBC (Median, range) | 8.31, (0.09–342.24) |
| Haemoglobin (Median, range) | 72, g/L (31–121) |
| PLT (Median, range) | 34.5, (6–523) |
| FAB (no. (%)) | |
| M0 | 2 (2.4%) |
| M1 | 4 (4.8%) |
| M2 | 33 (39.3%) |
| M3 | 15(17.9) |
| M4 | 13 (15.5%) |
| M5 | 15 (17.9%) |
| M7 | 2 (2.4%) |
Table 2.
The correlation between HOTAIR and clinical parameters in AML-M2 patients
| HOTAIR |
||
|---|---|---|
| R2 | P | |
| WBC | 0.02 | 0.993 |
| Haemoglobin | 0.169 | 0.348 |
| Platelets | −0.433 | 0.012* |
| Albumin | −0.25 | 0.891 |
| Creatinine | −0.335 | 0.057 |
| Lactate dehydrogenase | 0.176 | 0.326 |
* represent P < 0.05
Discussion
ATRA improves the survival and prognosis of APL, but the other AML subtypes mainly remains an incurable disease, there is a continued need for novel therapies [25]. LncRNA has been proven to play an essential role in biological process and cancer development by targeting multiple signalling pathways [14,15,26]. In this study, we found that HOTAIR regulates the differentiation via miR-17-5p/p21 pathway in AML cell; also, ATRA upregulated HOTAIR expression via C/EBPβ.
Recent studies suggest that lncRNA plays a vital role in leukaemia cell differentiation. For instance, NEAT1 is downregulated in APL patients compared with healthy donors and elevated during ATRA induced NB4 cell differentiation, in addition, si-NEAT1 blocks ATRA-induced differentiation [9]. PVT1 was highly expressed in APL patients compared with healthy donors and its expression decreased during ATRA-induced differentiation [10]. HOTAIRM1 was upregulated in myeloid cell lines after ATRA treatment, and HOTAIRM1 knockdown decreased granulocytic maturation in NB4 cells [11,27,28]. Besides, CCAT1 was upregulated in AML patients compared with the normal controls, and its expression was decreased during phorbol myristate acetate induced monocytic differentiation of HL-60 cells, moreover, CCAT1 knockdown resulted in more differentiation of HL-60 cells [12]. In this study, we found that HOTAIR was upregulated in AML cell lines during ATRA-induced differentiation, and decreased expression of HOTAIR results in less ATRA-induced differentiation in HL-60 cells. Furthermore, CD11b was also upregulated in the HOTAIR overexpressed group, with or without ATRA treatment in HL-60 cells. These results demonstrate that HOTAIR mediates leukaemia differentiation and may be a major downstream trigger of differentiation. However, our data suggests the function of HOTAIR may be specific to HL-60 cells. We speculated on the following probable reasons: firstly, differentiation blocks do not have a uniform underlying mechanism [29]; secondly, the disease complexity and heterogeneity in AML [2]; lastly, the tissue and cell-type-specific characteristic of lncRNA [30,31].
Previous studies suggest the expression of lncRNA may be regulated by transcription factors [27,32]. C/EBPβ was upregulated during myeloid differentiation and plays a critical role in myeloid differentiation [23], and mediates the actions of lots of genes during differentiation, not restricted to mRNA mediations (such as PU.1 [33]) but it also mediates lncRNAs (such as NEAT1 [32]). In this study, we used an online database to predict the potential binding site between C/EBPβ and HOTAIR promoter. Furthermore, we confirmed the relationship between C/EBPβ and HOTAIR promoter with ChIP and ChIP-PCR assays, and indicated that ATRA upregulated HOTAIR expression via C/EBPβ. Above all, we enriched the regulation mechanism of HOTAIR expression in AML differentiation.
HOTAIR was first described by Rinn et al, it silences target genes mainly by recruiting and binding EZH2, SUZ12, and LSD1 histone-modifiers to alter histone modifications at H3K27 [34]. Subsequent studies have observed the elevated expression of HOTAIR in lots of cancers, with some suggesting that HOTAIR has an important role in cancer development and progression [14,17,22]. HOTAIR regulating cell cycle progression is reported in numerous cancers [8,16,35]. Besides, AML cell fails to exit the cell cycle, which leads to the failure of terminal myeloid cell differentiation, but ATRA is known to promote myeloid differentiation by inducing cell cycle arrest at the G1 phase [5,24]. However, whether HOTAIR is involved in cell cycle regulation in leukaemia differentiation remains undetermined. In this study, the G0/G1 phase was significantly decreased by HOTAIR silencing while HOTAIR overexpression increased it. Our data identified that HOTAIR may be a cell cycle regulator and indicated HOTAIR is involved in cell cycle progression in leukaemia differentiation.
Briefly, p21 is a cyclin-dependent kinases inhibitor that binds to cyclin/CDK complex to repress its activity; furthermore, p21 is the key regulator during ATRA-induced differentiation, ATRA promotes differentiation through upregulated p21 [5,24]. Recent studies have shown that HOTAIR repressed not only p21 in cigarette smoke extract to induce cell cycle disorder [8] but also suppresses p21 in colorectal [36] and gastric cancers [37]. Meanwhile, HOTAIR could enhance the expression of p21 in nasopharyngeal carcinoma [38]. In this study, we demonstrated that HOTAIR could negatively regulate the level of p21, to modulate the p21/cyclin D1/CDK pathway to arrest cell cycle during myeloid differentiation.
Recent studies showed HOTAIR represses its target gene via miRNA, acting as a ceRNA; for example, HOTAIR promotes osteosarcoma development by sponging miR-217 [15]; HOTAIR modulates c-KIT expression through sponging miR-193a in AML [20]. In this study, the online database showed that miR-17-5p shares complementary sequences with HOTAIR and p21, indicating miR-17-5p may participate in regulating HOTAIR and p21; furthermore, luciferase reporter assays confirmed the association between HOTAIR, miR-17-5p, and p21. Indeed, the correlation between HOTAIR and miR-17-5p was reported in osteogenic differentiation [39], thyroid cancer [40], and cervical cancer [41]; HOTAIR regulated biological behaviour in these diseases by sponging miR-17-5p. In addition, previous studies showed that miR-17-5p negatively regulated p21 in synovial sarcoma [42] and nasopharyngeal carcinoma [43]. Also, miR-17-5p was downregulated in myeloid differentiation [44], which was consistent with our result, as well as miR-17-5p directly targeting p21 in myeloid leukaemia cell differentiation [45]. Collectively, these results indicated that HOTAIR regulated cell differentiation through p21/cyclin D1/CDK4 pathways via sponging miR-17-5p.
Previous data has shown that HOTAIR was upregulated in AML patients and correlated with poor prognosis [20,46–49], Zhang et al. [21] found no significant difference in HOTAIR expression between AML cases and controls, but HOTAIR was increased in AML-M5 patients. In addition, Sayad et al [50]. indicated that HOTAIR is not a prognostic biomarker for AML patients. The expression of HOTAIR in AML patients remains controversial and could be FAB-subtype specific. Here, our data showed lower HOTAIR expression in AML patients than IDA patients, and HOTAIR expression was also different in each subtype; the differences in the control groups used could explain this. Bone marrow samples of healthy donors were used in the previous study, while in this study, we used the bone marrow samples of IDA patients as the controls. The explanation for the varying HOTAIR expression across the different subtypes in our study could be the difference in sample composition ratios among the various studies. This could influence the expression level in the patients, resulting in differences in expressions. Additionally, we did not observe a correlation between HOTAIR and all the AML patients’ parameters analysed; however, HOTAIR correlated with platelet level in AML-M2 patients. These could be attributed to the high heterogeneity of AML [2] and the FAB-subtype specific characteristic of HOTAIR [21]. Nonetheless, a larger cohort study is needed to further confirm the expression levels of HOTAIR in AML patients.
In conclusion, the current study reveals the effects of HOTAIR on myeloid differentiation; and clarifies the C/EBP β/HOTAIR/miR-17-5p/p21 pathway in myeloid differentiation and also found the correlation between HOTAIR and platelet counts in AML-M2 patients. The findings of the present study provide a novel insight into the mechanism of lncRNA-mediated myeloid differentiation and indicated that HOTAIR may be a promising therapeutic target for leukaemia, especially for AML patients with the M2 subtype.
Materials and methods
Patients and samples
Bone marrow samples of 84 newly diagnosed AML patients and 20 iron-deficiency anaemia (IDA) patients were collected between March 2013 and August 2018, and they were diagnosed according to the FAB criteria. The study was performed in accordance with the principles of the Declaration of Helsinki. All the patients’ data were collected according to the approval of the institutional review board, and informed consent was obtained from all patients or guardians.
Cell line and cell culture
AML cell lines including HL-60, NB4, U937, and THP-1 cell lines were ordered from the Shanghai cell bank of the Chinese Academy of Sciences, and cultured in RPMI 1640 (Gibco) supplemented with 10% foetal bovine serum. Cells were maintained at 37°C in a 5% CO2 incubator. ATRA was purchased from Sigma-Aldrich (USA) and a final concentration of 1 μM was used to test for cell differentiation.
Cell transfection
SiRNA target HOTAIR was designed and synthesized by GenePharma (Shanghai, China) and the sequence was: 5ʹ-GCCUUUGCUUCGUGCUGAUTT-3ʹ (forward), R: 5ʹ-AUCAGCACGAAGCAAAGGCTT-3ʹ (reverse). The lncRNA HOTAIR plasmid (LZRS-HOTAIR #26110) was purchased from Addgene (http://www.addgene.org). MiRNA mimics and inhibitor were purchased from Ribobio Inc. (Guangzhou, Guangdong, China). The cells were transfected with the siRNA or plasmid using Lipofectamine 2000 (Life Technologies, Grand Island, NY, USA) according to the manufacturer’s protocol, and the culture was replaced with fresh medium 6–8 h after the transfection.
Flow cytometry analysis
PE-conjugated anti-CD11b and COULTER DNA PREP Reagents Kit were purchased from Beckman Coulter–Immunotech and were used to evaluate the cell differentiation and the cell cycle. Data acquisition and analysis were performed using a flow cytometer (FC500 MPL, Beckman Coulter) and EXPO 32 MultiComp software (Beckman Coulter, Miami, FL, USA). Cell cycle phase distributions were generated by fitting DNA content histograms to applicable models using ModFit LT software for Windows (Version 5.0).
Cell proliferation assay
Cell proliferation assay was measured at 48 hours after ATRA treatment using a Cell Counting Kit-8 (CCK8) assay (Beyotime Institute of Biotechnology, China), according to the manufacturer’s protocol.
Real-time quantitative PCR analysis
Total RNA of cultured cells and bone marrow samples were isolated with TRIZOL reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. cDNA was synthesized with primerScript RT reagent Kit (TaKaRa, Dalian, China) according to the manufacturer’s manual. Quantitative real-time PCR (qRT-PCR) was performed using SYBR Green PCR Mix (TaKaRa, Dalian, China) according to the manufacturer’s manual. The primer sequences are: (1) HOTAIR: GGTAGAAAAAGCAACCACGAAGC (forward), ACATAAACCTCTGTCTGTGAGTGCC (reverse), (2) GAPDH: AGCAAGAGCACAAGAGGAAG (forward), GGTTGAGCACAGGGTACTTT (reverse). The relative expression fold change of lncRNA was calculated by the 2-∆∆Ct method. In addition, miRNA reverse transcription PCR and qRT-PCR primers were ordered from Ribobio Inc. (Guangzhou, Guangdong, China), and PCR tests were performed according to the manufacturer’s manual.
Bioinformatics analysis
We used the online database PROMO (http://alggen.lsi.upc.es/) to predict the transcription factors binding sites that the HOTAIR promoter region possesses and the online database JASPAR (http://jaspar.genereg.net/) was used to confirm the binding sites.
The online database (starbase.sysu.edu.cn/) was used to predict the potential binding sites between miR-17-5p and HOTAIR, as well as the binding sites between miR-17-5p and p21.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assay (ChIP) was performed using the Chromatin IP Kit (Beyotime Institute of Biotechnology, China) according to the manufacturer’s protocol. The following product was used in this experiment: C/EBPβ Polyclonal Antibody (Thermo Fisher Scientific, catalogue # PA5-27244, RRID AB_2544720). We analysed the immunoprecipitated DNA by qRT-PCR. All primers for ChIP-qPCR are listed in Supplementary Material: Table S1.
Luciferase reporter
The cDNA fragment of HOTAIR and synthesized p21 3′untranslated region (3′UTR) fragments were cloned into the psiCHECK2 vector (Promega, Madison, WI) to generate HOTAIR wild-type (WT) and p21-3′UTR WT plasmids. The complementary sequence containing mutation sites of seed sequences was designed based on HOTAIR-WT and p21-3′UTR WT plasmids and inserted into the psiCHECK2 vector to generate HOTAIR mutation-type (MT) and p21-3′UTR MT plasmids.
HEK293T cells were transfected with psiCHECK2-p21-3ʹUTR WT, psiCHECK2-p21-3ʹUTR MT, psiCHECK2-HOTAIR WT, psiCHECK2-HOTAIR MT, and miR-17-5p mimic or negative control (miR-nc) using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol. Luminescence was assayed 48 hours later using the Dual-Luciferase Reporter Assay Kit (Beyotime Institute of Biotechnology, China) according to the manufacturer’s instructions. Results were normalized to the Renilla luminescence from the same vector and are shown as the ratio between the various treatments and cells transfected with the mutant vector.
Western blot
Total proteins isolated from cells were immunoblotted with different antibodies following a published protocol [51]. The primary antibodies used were p21 (1: 3000 dilution), Cyclin D1 (1: 2000 dilution), CDK4 (1: 5000 dilution) (Cell Signalling, Danvers, MA, USA), C/EBPβ (1:1000 dilution) (Thermo Fisher Scientific) and β-actin (1: 3000 dilution) (Santa Cruz Biotechnology, Dallas, TX, USA).
Statistical analysis
The data were expressed as Mean ± SD of three independent experiments, and the statistical analyses were performed by PRISM 5.0 software (GraphPad Software Inc., San Diego, CA, USA). The clinical statistical analyses were carried out using SPSS software version 19.0 for Windows (SPSS Inc., IL, USA). Spearman correlation was used to measure the relationship between HOTAIR and clinical parameters of AML patients. P values less than 0.05 were considered statistically significant. All results were reproduced in at least three independent experiments.
Supplementary Material
Funding Statement
This work was partly supported by the Key Research and Development Plan of Anhui Province, China [201904a07020058], Higher School of Anhui Provincial Natural Science Research Project [KJ2018A0198], Foundation of Anhui Medical University [2019xkj134], National Science Foundation of China [81272259], Scientific Research Foundation of the Institute for Translational Medicine [SRFITMAP, 2017zhyx13KJ]. Major Science and Technology Project of Anhui Province, China [No. 201903a07020030], Basic and Clinical Cooperative Research Promotion Plan of Anhui Medical Uiversity[2020xkjT021].
Disclosure statement
The authors declare that there is no conflict of interests regarding the publication of this paper
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Canaani J, Beohou E, Labopin M, et al. Impact of FAB classification on predicting outcome in acute myeloid leukemia, not otherwise specified, patients undergoing allogeneic stem cell transplantation in CR1: an analysis of 1690 patients from the acute leukemia working party of EBMT. Am J Hematol. 2017;92(4):344–350. . [DOI] [PubMed] [Google Scholar]
- [2].Garzon R, Volinia S, Papaioannou D, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2014;111(52):18679–18684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Khaled S, Al Malki M, Marcucci G.. Acute myeloid leukemia: biologic, prognostic, and therapeutic insights. Oncology (Williston Park). 2016;30(4):318–329. [PubMed] [Google Scholar]
- [4].Wang Y, Jin W, Jia X, et al. Transcriptional repression of CDKN2D by PML/RARalpha contributes to the altered proliferation and differentiation block of acute promyelocytic leukemia cells. Cell Death Dis. 2014;5:e1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang JG, Barsky LW, Davicioni E, et al. Retinoic acid induces leukemia cell G1 arrest and transition into differentiation by inhibiting cyclin-dependent kinase-activating kinase binding and phosphorylation of PML/RARalpha. Faseb J. 2006;20(12):2142–2144. [DOI] [PubMed] [Google Scholar]
- [6].Lo-Coco F, Avvisati G, Vignetti M, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med. 2013;369(2):111–121. [DOI] [PubMed] [Google Scholar]
- [7].Tallman MS. New agents for the treatment of acute myeloid leukemia. Best Pract Res Clin Haematol. 2006;19(2):311–320. [DOI] [PubMed] [Google Scholar]
- [8].Liu Y, Wang B, Liu X, et al. Epigenetic silencing of p21 by long non-coding RNA HOTAIR is involved in the cell cycle disorder induced by cigarette smoke extract. Toxicol Lett. 2016;240(1):60–67. [DOI] [PubMed] [Google Scholar]
- [9].Zeng C, Xu Y, Xu L, et al. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zeng C, Yu X, Lai J, et al. Overexpression of the long non-coding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemia. J Hematol Oncol. 2015;8:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014;11(6):777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen L, Wang W, Cao L, et al. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. 2016;39(4):330–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang X, Li L, Zhao K, et al. A novel LncRNA HITT forms a regulatory loop with HIF-1alpha to modulate angiogenesis and tumor growth. Cell Death Differ. 2020;27(4):1431–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mao T, He C, Wu H, et al. Silencing lncRNA HOTAIR declines synovial inflammation and synoviocyte proliferation and promotes synoviocyte apoptosis in osteoarthritis rats by inhibiting Wnt/beta-catenin signaling pathway. Cell Cycle. 2019;18(22):3189–3205. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [15].Wang B, Qu XL, Liu J, et al. HOTAIR promotes osteosarcoma development by sponging miR-217 and targeting ZEB1. J Cell Physiol. 2019;234(5):6173–6181. [DOI] [PubMed] [Google Scholar]
- [16].Sun X, Du P, Yuan W, et al. Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. 2015;6:e1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Song Y, Wang R, Li LW, et al. Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. Int J Oncol. 2019;54(1):77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Qu X, Alsager S, Zhuo Y, et al. HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett. 2019;454:90–97. [DOI] [PubMed] [Google Scholar]
- [20].Xing CY, Hu XQ, Xie FY, et al. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015;589(15):1981–1987. [DOI] [PubMed] [Google Scholar]
- [21].Zhang YY, Huang SH, Zhou HR, et al. Role of HOTAIR in the diagnosis and prognosis of acute leukemia. Oncol Rep. 2016;36(6):3113–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang S, Chen S, Yang G, et al. Long noncoding RNA HOTAIR as an independent prognostic marker in cancer: a meta-analysis. PLoS One. 2014;9(8):e105538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yan Y, Hanse EA, Stedman K, et al. Transcription factor C/EBP-beta induces tumor-suppressor phosphatase PHLPP2 through repression of the miR-17-92 cluster in differentiating AML cells. Cell Death Differ. 2016;23(7):1232–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hu XT, Zuckerman KS. Role of cell cycle regulatory molecules in retinoic acid- and vitamin D3-induced differentiation of acute myeloid leukaemia cells. Cell Prolif. 2014;47(3):200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kuley-Bagheri Y, Kreuzer KA, Monsef I, et al. Effects of all-trans retinoic acid (ATRA) in addition to chemotherapy for adults with acute myeloid leukaemia (AML) (non-acute promyelocytic leukaemia (non-APL)). Cochrane Database Syst Rev. 2018;8:CD011960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].He X, Wang J, Chen J, et al. lncRNA UCA1 predicts a poor prognosis and regulates cell proliferation and migration by repressing p21 and SPRY1 expression in GC. Mol Ther Nucleic Acids. 2019;18:605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wei S, Zhao M, Wang X, et al. PU.1 controls the expression of long noncoding RNA HOTAIRM1 during granulocytic differentiation. J Hematol Oncol. 2016;9(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen ZH, Wang WT, Huang W, et al. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017;24(2):212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Agarwal A, Bolosky WJ, Wilson DB, et al. Differentiation of leukemic blasts is not completely blocked in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2019;116(49):24593–24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hu L, Li M, Pu L, et al. The different prognostic value of long non-coding RNA BANCR in human cancers. Minerva Med. 2017;108(1):97–100. [DOI] [PubMed] [Google Scholar]
- [31].Heubach J, Monsior J, Deenen R, et al. The long noncoding RNA HOTAIR has tissue and cell type-dependent effects on HOX gene expression and phenotype of urothelial cancer cells. Mol Cancer. 2015;14:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang Y, Fu L, Sun A, et al. C/EBPbeta contributes to transcriptional activation of long non-coding RNA NEAT1 during APL cell differentiation. Biochem Biophys Res Commun. 2018;499(2):99–104. [DOI] [PubMed] [Google Scholar]
- [33].Mueller BU, Pabst T, Fos J, et al. ATRA resolves the differentiation block in t(15;17) acute myeloid leukemia by restoring PU.1 expression. Blood. 2006;107(8):3330–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang K, Sun X, Zhou X, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6(1):537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lin K, Jiang H, Zhang LL, et al. Down-Regulated LncRNA-HOTAIR Suppressed Colorectal Cancer Cell Proliferation, Invasion, and Migration by Mediating p21. Dig Dis Sci. 2018;63(9):2320–2331. [DOI] [PubMed] [Google Scholar]
- [37].Xu Z, Chen H, Yang B, et al. The association of HOTAIR with the diagnosis and prognosis of gastric cancer and its effect on the proliferation of gastric cancer cells. Can J Gastroenterol Hepatol. 2019;2019:3076345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ma DD, Yuan LL, Lin LQ. LncRNA HOTAIR contributes to the tumorigenesis of nasopharyngeal carcinoma via up-regulating FASN. Eur Rev Med Pharmacol Sci. 2017;21(22):5143–5152. [DOI] [PubMed] [Google Scholar]
- [39].Wei B, Wei W, Zhao B, et al. Long non-coding RNA HOTAIR inhibits miR-17-5p to regulate osteogenic differentiation and proliferation in non-traumatic osteonecrosis of femoral head. PLoS One. 2017;12(2):e0169097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Liu X, Liu G, Lu Y, et al. Long non-coding RNA HOTAIR promotes cell viability, migration and invasion in thyroid cancer cells by sponging miR-17-5p. Neoplasma. 2020;67(2):229–237. [DOI] [PubMed] [Google Scholar]
- [41].Ji F, Wuerkenbieke D, He Y, et al. Long Noncoding RNA HOTAIR: an Oncogene in Human Cervical Cancer Interacting With MicroRNA-17-5p. Oncol Res. 2018;26(3):353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Minami Y, Kohsaka S, Tsuda M, et al. SS18-SSX-regulated miR-17 promotes tumor growth of synovial sarcoma by inhibiting p21WAF1/CIP1. Cancer Sci. 2014;105(9):1152–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen C, Lu Z, Yang J, et al. MiR-17-5p promotes cancer cell proliferation and tumorigenesis in nasopharyngeal carcinoma by targeting p21. Cancer Med. 2016;5(12):3489–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dakir EH, Mollinedo F. Genome-wide miRNA profiling and pivotal roles of miRs 125a-5p and 17-92 cluster in human neutrophil maturation and differentiation of acute myeloid leukemia cells. Oncotarget. 2019;10(51):5313–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].He M, Wang QY, Yin QQ, et al. HIF-1alpha downregulates miR-17/20a directly targeting p21 and STAT3: a role in myeloid leukemic cell differentiation. Cell Death Differ. 2013;20(3):408–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hao S, Shao Z. HOTAIR is upregulated in acute myeloid leukemia and that indicates a poor prognosis. Int J Clin Exp Pathol. 2015;8(6):7223–7228. [PMC free article] [PubMed] [Google Scholar]
- [47].Wu S, Zheng C, Chen S, et al. Overexpression of long non-coding RNA HOTAIR predicts a poor prognosis in patients with acute myeloid leukemia. Oncol Lett. 2015;10(4):2410–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].El-Khazragy N, Ghozy S, Matbouly S, et al. Interaction between 12p chromosomal abnormalities and Lnc-HOTAIR mediated pathway in acute myeloid leukemia. J Cell Biochem. 2019;120(9):15288–15296. [DOI] [PubMed] [Google Scholar]
- [49].Wang SL, Huang Y, Su R, et al. Silencing long non-coding RNA HOTAIR exerts anti-oncogenic effect on human acute myeloid leukemia via demethylation of HOXA5 by inhibiting Dnmt3b. Cancer Cell Int. 2019;19:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sayad A, Hajifathali A, Hamidieh AA, et al. RNA is not a Biomarker for Acute Myeloid Leukemia (AML) in Iranian Patients. Asian Pac J Cancer Prev. 2017;18(6):1581–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Xiong SD, Yu K, Liu XH, et al. Ribosome-inactivating proteins isolated from dietary bitter melon induce apoptosis and inhibit histone deacetylase-1 selectively in premalignant and malignant prostate cancer cells. Int J Cancer. 2009;125(4):774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request


