ABSTRACT
Claudins (CLDNs) represent major transmembrane proteins of tight junctions and contribute to the barrier function. They also serve as anchors for several signaling proteins, but the underlying molecular basis has yet to be established. The present review covers the recent progress in our understanding of the CLDN signaling pathway in health and disease. We discuss the functional relevance of phosphotyrosine motifs in the C-terminal cytoplasmic domain of CLDNs and define mutual regulation between CLDNs and Src-family kinases (SFKs). In addition, we focus on the crosstalk between CLDN and transcription factor signaling. We also describe how aberrant CLDN–transcription factor signaling promotes or inhibits cancer progression. We propose that a link between various cell adhesion molecules and transcription factors coordinates a range of physiological and pathological events via activation or suppression of target genes.
KEYWORDS: Tight junction, CLDN, cell adhesion signal, signal transduction, PI3K, AKT, nuclear receptor, Src-family kinase, transcription factors
Introduction
The claudin (CLDN) family is capable of forming tight-junction strands and thereby represents the backbone of tight junctions in vertebrate epithelial and endothelial cells, as well as in other types of cells.1,2 It is composed of 27 members in mammals, and a specific combination of CLDNs is expressed in a given cell/tissue type. CLDNs are tetraspan transmembrane proteins that include two extracellular loops (EC1 and EC2) and N- and C-terminal cytoplasmic domains. The CLDN-EC1 creates paracellular barriers or pores for selective ions and solutes,3–7 whereas both CLDN-EC1 and CLDN-EC2 contribute to cis- and trans-interactions between CLDNs.7–11 On the other hand, the C-terminal cytoplasmic domain of many CLDN subtypes contains specific sequence motifs such as PDZ domain-binding motifs and phosphorylation consensus sites5, 12–16 and receives or propagates a magnitude of intracellular signals as platforms; however, it remains poorly defined how CLDN signaling reaches the nucleus and regulates gene expression.17
CLDNs are absolutely required for human health, and their dysregulations are involved in the pathogenesis of diverse diseases.4-6,18-16 For instance, it is well known that mutations in several CLDN genes cause various human hereditary diseases. In addition, CLDNs frequently show aberrant expression and/or localization in a wide variety of cancers, resulting in either promotion or repression of tumor progression, most probably by dysregulated CLDN signaling.19–25,22,26–32 Moreover, recent studies have established the region-selective CLDN5 breakdown in brain disorders, such as schizophrenia, depression, Alzheimer’s disease, and multiple sclerosis.33–39 In this respect, we and others have previously reported how endothelial CLDN5 expression is regionally disrupted in these psychiatric disorders.40,41
In the current review, we focus on the link between CLDN and transcription factor signaling because it is theoretically attributed to the organization of a broad range of cellular processes, including cell growth, survival, differentiation, polarity, migration, and metabolism, via regulation of the expression of corresponding target genes. We also discuss aberrant CLDN–transcription factor signaling in cancer. We do not describe recent progress in our understanding of numerous aspects of other tight-junction players, such as junctional adhesion molecules (JAMs), tight junction-associated MARVEL domain-containing proteins (TAMPs: occludin, tricellulin, and MarvelD3), and the ZO family of scaffolding proteins.42–55
The CLDN/SFK/PI3K/AKT/transcription factor signaling cascade
We previously reported that retinoid X receptor α (RXRα)/retinoic acid receptor γ (RARγ) and another member of the nuclear receptor superfamily, hepatocyte nuclear factor 4α (HNF4α), trigger the formation of mature cell–cell junctions and microvilli, expression of tight-junction markers (CLDN6, CLDN7, occludin, and ZO-1α+ variant) and a microvilli marker (ezrin/radixin/moesin-binding phosphoprotein 50 [EBP50]), as well as morphological differentiation into epithelial cells from stem cells.56–59 These effects are incredibly similar to the CLDN6-triggered ones,60 implying a possible crosstalk between them. Along this line, we have recently identified the CLDN–transcription factor signaling pathway.61
Reciprocal regulation between CLDNs and SFKs
First, we showed, by using the corresponding deletion mutants, that the CLDN6–adhesion signal is transduced through the EC2 domain and the first half C-terminal cytoplasmic domains but not through the EC1 domain (Figure 1). Second, we paid attention to the four tyrosine residues in the C-terminal domain of CLDN6, which are completely conserved among vertebrates, and revealed that Y196/200, but not Y213/218, are definitely required for CLDN6-signaling ability. Third, we disclosed that CLDN6 recruits and activates Src-family kinases (SFKs) in EC2-dependent and Y196/200-dependent manners, and SFKs in turn phosphorylate CLDN6 at Y196/200 in an EC2-dependent fashion. The functional relevance of the EC2 in the CLDN6–adhesion signal was also supported by using the C-terminal half of Clostridium perfringens enterotoxin (C-CPE).60,61 Moreover, we have identified SFK members associated with CLDN6 and obtained evidence showing that recombinant proteins corresponding to the Src homology 2 (SH2) domains of certain SFKs directly bind to the C-terminal cytoplasmic domain of CLDN6 and the Y196/200-containing peptide (our unpublished results).
Figure 1.
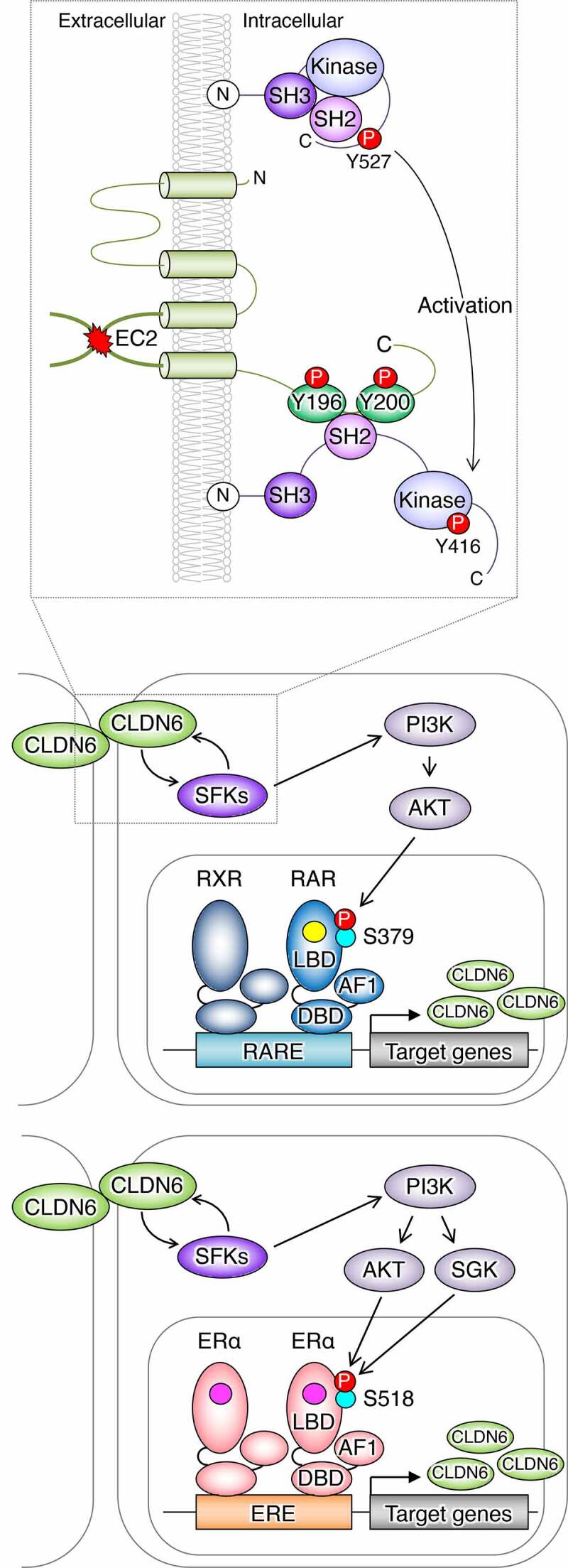
Schematic model for regulation of the nuclear receptor activity by the CLDN–adhesion signaling. The schema is modified from that reported previously (Sugimoto et al., 2019). SH2/3: Src homology 2/3 domain; Kinase: kinase domain; AF1: activation function-1; DBD: DNA-binding domain; LBD: ligand-binding domain; RARE: retinoic acid response element; ERE: estrogen response element; RA: yellow circle; estrogen: pink circle
Through a careful search for amino acid sequence of human and mouse CLDN1–20, both Y196 and Y200 in the C-terminal cytoplasmic domain of CLDN6 are conserved in CLDN2/4/12 (Figures 2 and 3). It is also noteworthy that CLDN6Y196 and CLDN6Y200 are conserved in CLDN3/7/8/10/14/16 and CLDN1/5/9/17/18, respectively.
Figure 2.

Amino acid sequences of a part of the C-terminal cytoplasmic domain in human CLDN1-20. Tyrosine residues corresponding to CLDN6Y196/200 are highlighted and the conserved ones are indicated in red
Figure 3.

Amino acid sequences of a part of the C-terminal cytoplasmic domain in mouse CLDN1-20. Tyrosine residues corresponding to CLDN6Y196/200 are highlighted and the conserved ones are indicated in red
We have found mutual regulation between certain CLDN subtypes (tyrosine residues corresponding to CLDN6Y200) and SFKs (our unpublished results). In addition, Li et al.62 have recently reported that CLDN11 is phosphorylated at two adjacent tyrosine residues, which are positioned at different sites from CLDN6Y196/200, as described above, and activates SRC. The C-terminal domain of CLDN1 is also associated with SRC although the involvement of the pY motifs is not determined.63 It should also be noted that amino acids around pY, in particular 3–5 residues at the C-terminal side, influence the binding specificity of the SH2 domain.64–67 For instance, the C-terminal amino acids of the CLDN6Y200-corresponding tyrosine residue in human and mouse CLDN18 are very different from others; therefore, it does not seem to function as the pY motif. CLDN18 is known to decrease PI3K and AKT activities,68,69 suggesting this notion. Further studies are required to determine whether other CLDN subtypes, similar to CLDN6, couple with SFKs via the pY motifs in the C-terminal cytoplasmic domains.
SFKs are known to be activated by several cell–cell and cell–matrix adhesion proteins lacking intrinsic kinase activity, such as E-cadherin, integrins, and cellular prion protein.62,70–76, In addition, engagement of JAMs, the JAML (junctional adhesion molecule-like) and CAR (coxsackie and adenovirus receptor), stimulates PI3K,77which is the major downstream signal of SFKs. Besides, it is well known that certain signaling proteins, which contain the SH2, phosphotyrosine-binding (PTB), Hakai-tyrosine binding (HYB), C2, and pyruvate kinase M2 domains, bind to the pY motifs.66,67,78–81 Taken together, these findings strongly suggest that the pY motifs in the C-terminal cytoplasmic domains of diverse cell–cell and cell–matrix adhesion molecules generally serve as the signaling landscapes for SFKs and other pY motif-binding proteins.
A link between CLDN/SFK and transcription factor signalings
Importantly, the CLDN6/SFK/PI3K/AKT axis targets the AKT-phosphorylation sites in the RARγ and the estrogen receptor α (ERα) and stimulates their activities, thereby regulating the expression of respective target genes. This conclusion was drawn from the following results: (1) the CLDN6-induced cellular events were hindered in three distinct F9:Rxra–/–:Rarg–/–:Cldn6 cell lines, despite SFKs being activated; (2) AKT formed a complex with either RXRα/RARγ or ERα; and (3) characterization of F9:Rxra–/–:Rarg–/–:Cldn6:iRxra-Rarg2S379A (hereafter, “i” means doxycycline-inducible expression of a give gene) and F9:Rxra–/–:Rarg–/–:Cldn6:iRxra-Rarg2S379E cells, as well as MCF-7:ESR1S518E cells, revealed that CLDN6 signaling directs S379 and S518 in mouse RARγ and human ERα, respectively. In addition, CLDN6-provoked RARγS379 phosphorylation in mice resulted in releasing the nuclear receptor corepressor (NCoR) from several retinoic acid response elements (RAREs) of three distinct RA target genes, including Cldn6. Since RXRα/RARγ heterodimer appears to induce Cldn6 gene expression,56,61 the positive loop of the CLDN6–RARγ cascade could contribute not only to triggering but also to the maintenance of CLDN6-initiated cellular events. Intriguingly, the AKT-consensus phosphorylation motifs are conserved in 14 of 48 members of human nuclear receptors, implying the biological relevance of this phosphorylation site.
Aberrant CLDN–transcription factor signaling in cancer
We have recently reported that aberrant CLDN6 expression in endometrial cancer tissues is significantly associated with several clinicopathological variables, such as surgical stage III/IV, histological type, histological grade 3, lymphovascular space involvement, lymph node metastasis, and distant metastasis.82 Additionally, we showed that the high CLDN6 expression in endometrial cancer represents an independent prognosis marker, and the 5-y survival rate was approximately 30%, which was one-third of that in the low-expression group.
In an additional study, we found that aberrant CLDN6 expression promotes the malignant phenotypes of endometrial cancer in vitro and in vivo via hijacking the CLDN6–ERα axis.83 For instance, we demonstrated that abnormal CLDN6–ERα signaling stimulates not only cell proliferation but also collective cell migration in the leading front of endometrial cancer cells. It is noteworthy that activated SFKs appear to be concentrated at the cell borders together with CLDN6 in Ishikawa:CLDN6 cells but not in parental Ishikawa cells (Figure 4). The EC2 domain and Y196/200 of CLDN6 were required to recruit and activate SFKs and to stimulate malignant phenotypes of endometrial cancer cells. In addition, the CLDN6/SFK/PI3K pathway propagates both AKT and serum- and glucocorticoid-regulated kinase (SGK), which share a high degree of homology and the same consensus phosphorylation motif,84 resulting in targeting S518 in human ERα and activating target genes in a ligand-independent manner. Furthermore, RNAseq and RT-qPCR analyses indicated the presence of not only ERα-dependent but also ERα-independent CLDN6 signaling (Figure 5). The identification of this machinery highlights the regulation of transcription factor activity by cell adhesion to advance tumor progression.
Figure 4.
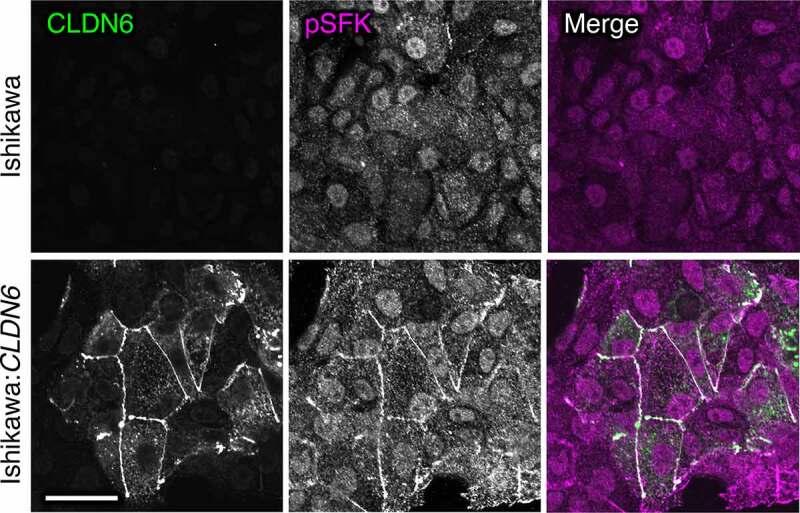
Aberrant SFK activation in human endometrial cancer cells by CLDN6. Scale bar, 20 μm
Figure 5.
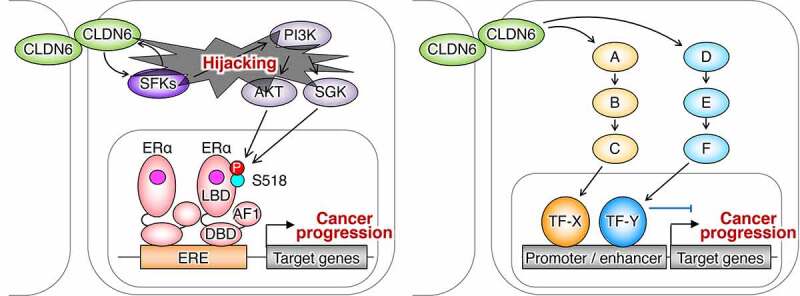
The ERα-dependent and independent CLDN6 signaling in endometrial cancer. A–F indicate signaling proteins. TF, transcription factor
Another issue that should be mentioned is other abnormal CLDN–transcription factor signaling in health and disease. The CLDN18/Yes-associated protein (YAP) pathway regulates the homeostasis of normal lung stem and progenitor cells, and its deficiency promotes tumorigenesis and progression of lung and gastric adenocarcinoma.69,85–87 By contrast, CLDN2 activates YAP, leading to self-renewal of human colorectal cancer stem-like cells.88 Since CLDN2 has two conserved pY in the C-terminal cytoplasmic domain as described above, it should also be verified whether CLDN2/SFK signaling is involved in the self-renewal of various cancer stem-like cells.
Conclusions and future directions
The work discussed in the present review highlights the CLDN–transcription factor signaling pathway, notably the CLDN/SFK/PI3K/AKT/nuclear receptor cascade. Cell–cell and cell–matrix adhesion molecules are indispensable not only for proper tissue integrity but also for signaling properties that coordinate a wide range of cell behaviors. In other words, appropriate tissue formation connected by various cell adhesion proteins should be pre-requested for normal cell-adhesion signal. Since cell–cell and cell–matrix adhesion proteins are broadly expressed in distinct cell types, we propose that various combinations of cell adhesion molecules and transcription factors coordinate diverse physiological and pathological processes, including cancer. The cell-adhesion signals most probably lead to posttranslational modification of transcription factors, thereby regulating their activities. In future, it would be interesting to generalize the cell adhesion–transcription factor signaling pathway in health and disease.
Funding Statement
This work was supported by Japan Agency for Medical Research and Development [JP20lm0203010]; Japan Society for the Promotion of Science [19K16615]; Japan Society for the Promotion of Science [17K08699]; Japan Society for the Promotion of Science [17K17978]; the Uehara Memorial Foundation; the Takeda Science Foundation; and the Ichiro Kanehara Foundation for the Promotion of Medical Sciences and Medical Care.
Author Contributions
H.C. wrote the paper, and H.C. and K.S. prepared figures and agreed to the final version of the manuscript.
Acknowledgments
We thank the former and present members of our laboratory for a helpful discussion. We also thank the English Editing Service of the Medical Research Promotion Office, Fukushima Medical University, for their assistance with the manuscript.
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S.. Claudin-1 and −2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141(7):1–11. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuse M, Sasaki H, Fujimoto K, Tsukita S. A single gene product, claudin-1 or −2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol. 1998;143(2):391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furuse M, Tsukita S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006;16(4):181–188. doi: 10.1016/j.tcb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68(1):403–429. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- 5.Chiba H, Osanai M, Murata M, Kojima T, Sawada N. Transmembrane proteins of tight junctions. Biochim Biophys Acta. 2008;1778(3):588–600. doi: 10.1016/j.bbamem.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Zihni C, Mills C, Matter K, Balda MS. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol. 2016;17(9):564–580. doi: 10.1038/nrm.2016.80. [DOI] [PubMed] [Google Scholar]
- 7.Tsukita S, Tanaka H, Tamura TA. The claudins: from tight junctions to biological systems. Trends Biochem Sci. 2019;44(2):141–152. doi: 10.1016/j.tibs.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochim Biophys Acta. 2008;1778(3):631–645. doi: 10.1016/j.bbamem.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Piontek J, Winkler L, Wolburg H, Müller SL, Zuleger N, Piehl C, Wiesner B, Krause G, Blasig IE. Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J. 2008;22(1):146–158. doi: 10.1096/fj.07-8319com. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, Fujiyoshi Y. Crystal structure of a claudin provides insight into the architecture of tight junctions. Science. 2014;344(6181):304–307. doi: 10.1126/science.1248571. [DOI] [PubMed] [Google Scholar]
- 11.Haseloff RF, Piontek J, Blasig IE. Chapter 5 – the investigation of cis- and trans-interactions between claudins. In: Yu AS L editor. Current topics in membranes. Academic Press, USA; 2010. p. 97–112. [Google Scholar]
- 12.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol. 1999;147(6):1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2(4):285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 14.González-Mariscal L, Garay E, Quirós M. Chapter 6 – regulation of claudins by posttranslational modifications and cell-signaling cascades. In: Yu AS L editor. Current topics in membranes. Academic Press, USA; 2010. p. 113–150. [Google Scholar]
- 15.Van Itallie CM, Anderson JM. Phosphorylation of tight junction transmembrane proteins: many sites, much to do. Tissue Barriers. 2018;6(1):e1382671. doi: 10.1080/21688370.2017.1382671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabariès S, McNulty A, Ouellet V, Annis MG, Dessureault M, Vinette M, Hachem Y, Lavoie B, Omeroglu A, Simon H-G, et al. Afadin cooperates with claudin-2 to promote breast cancer metastasis. Genes Dev. 2019;33(3–4):180–193. doi: 10.1101/gad.319194.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavallaro U, Dejana E. Adhesion molecule signalling: not always a sticky business. Nat Rev Mol Cell Biol. 2011;12(3):189–197. doi: 10.1038/nrm3068. [DOI] [PubMed] [Google Scholar]
- 18.Günzel D, Yu ASL. Claudins and the modulation of tight junction permeability. Physiol Rev. 2013;93(2):525–569. doi: 10.1152/physrev.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawada N. Tight junction-related human diseases. Pathol Int. 2013;63(1):1–12. doi: 10.1111/pin.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barmeyer C, Schulzke JD, Fromm M. Claudin-related intestinal diseases. Semin Cell Dev Biol. 2015;42:30–38. doi: 10.1016/j.semcdb.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Tawar RG, Colpitts CC, Lupberger J, El-Saghire H, Zeisel MB, Baumert TF. Claudins and pathogenesis of viral infection. Semin Cell Dev Biol. 2015;42:39–46. doi: 10.1016/j.semcdb.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Zeisel MB, Dhawan P, Baumert TF. Tight junction proteins in gastrointestinal and liver disease. Gut. 2019;68(3):547–561. doi: 10.1136/gutjnl-2018-316906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Culemann S, Grüneboom A, Nicolás-Ávila JÁ, Weidner D, Lämmle KF, Rothe T, Quintana JA, Kirchner P, Krljanac B, Eberhardt M, et al. Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature. 2019;572(7771):670–675. doi: 10.1038/s41586-019-1471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Z, Yu J, Lin W, Tang H, Zhang W, Lu B. Molecular hydrogen accelerates the reversal of acute obstructive cholangitis‑induced liver dysfunction by restoring gap and tight junctions. Mol Med Rep. 2019;19(6):5177–5184. doi: 10.3892/mmr.2019.10179. [DOI] [PubMed] [Google Scholar]
- 25.Zuo L, Kuo W-T, Turner JR. Tight junctions as targets and effectors of mucosal immune homeostasis. Cell Mol Gastroenterol Hepatol. 2020;10(2):327–340. doi: 10.1016/j.jcmgh.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira SS, Morgado-Díaz JA. Claudins: multifunctional players in epithelial tight junctions and their role in cancer. Cell Mol Life Sci. 2007;64(1):17–28. doi: 10.1007/s00018-006-6314-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valle BL, Morin PJ. Chapter 13 – claudins in cancer biology. In: Yu AS L editor. Current topics in membranes. Academic Press, United States; 2010. p. 293–333. [Google Scholar]
- 28.Turksen K, Troy T-C. Junctions gone bad: claudins and loss of the barrier in cancer. Biochim Biophys Acta. 2011;1816(1):73–79. doi: 10.1016/j.bbcan.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Osanai M, Takasawa A, Murata M, Sawada N. Claudins in cancer: bench to bedside. Pflugers Arch. 2017;469(1):55–67. doi: 10.1007/s00424-016-1877-7. [DOI] [PubMed] [Google Scholar]
- 30.Tabariès S, Siegel PM. The role of claudins in cancer metastasis. Oncogene. 2017;36(9):1176–1190. doi: 10.1038/onc.2016.289. [DOI] [PubMed] [Google Scholar]
- 31.Gowrikumar S, Singh AB, Dhawan P. Role of claudin proteins in regulating cancer stem cells and chemoresistance-potential implication in disease prognosis and therapy. Int J Mol Sci. 2019;21(1):53. doi: 10.3390/ijms21010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhat AA, Syed N, Therachiyil L, Nisar S, Hashem S, Macha MA, Yadav SK, Krishnankutty R, Muralitharan S, Al-Naemi H, et al. Claudin-1, a double-edged sword in cancer. Int J Mol Sci. 2020;21(2):569. Available from: 10.3390/ijms21020569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menard C, Pfau ML, Hodes GE, Kana V, Wang VX, Bouchard S, Takahashi A, Flanigan ME, Aleyasin H, LeClair KB, et al. Social stress induces neurovascular pathology promoting depression. Nat Neurosci. 2017;20(12):1752–1760. doi: 10.1038/s41593-017-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishiura K, Ichikawa-Tomikawa N, Sugimoto K, Kunii Y, Kashiwagi K, Tanaka M, Yokoyama Y, Hino M, Sugino T, Yabe H, et al. PKA activation and endothelial claudin-5 breakdown in the schizophrenic prefrontal cortex. Oncotarget. 2017;8(55):93382–93391. doi: 10.18632/oncotarget.21850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greene C, Campbell M. Tight junction modulation of the blood brain barrier: CNS delivery of small molecules. Tissue Barriers. 2016;4(1):e1138017. doi: 10.1080/21688370.2015.1138017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamazaki Y, Shinohara M, Shinohara M, Yamazaki A, Murray ME, Liesinger AM, Heckman MG, Lesser ER, Parisi JE, Petersen RC, et al. Selective loss of cortical endothelial tight junction proteins during Alzheimer’s disease progression. Brain. 2019;142(4):1077–1092. doi: 10.1093/brain/awz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niu J, Tsai H-H, Hoi KK, Huang N, Yu G, Kim K, Baranzini SE, Xiao L, Chan JR, Fancy SPJ. Aberrant oligodendroglial-vascular interactions disrupt the blood-brain barrier, triggering CNS inflammation. Nat Neurosci. 2019;22(5):709–718. doi: 10.1038/s41593-019-0369-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dudek KA, Dion-Albert L, Lebel M, LeClair K, Labrecque S, Tuck E, Ferrer Perez C, Golden SA, Tamminga C, Turecki G, et al. Molecular adaptations of the blood-brain barrier promote stress resilience vs. Depression Proc Natl Acad Sci U S A. 2020;117(6):3326–3336. doi: 10.1073/pnas.1914655117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greene C, Hanley N, Campbell M. Blood-brain barrier associated tight junction disruption is a hallmark feature of major psychiatric disorders. Transl Psychiatry. 2020;10(1):373. doi: 10.1038/s41398-020-01054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greene C, Hanley N, Campbell M. Claudin-5: gatekeeper of neurological function. Fluids Barriers CNS. 2019;16(1):3. doi: 10.1186/s12987-019-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiba H, Ichikawa-Tomikawa N, Imura T, Sugimoto K. The region-selective regulation of endothelial claudin-5 expression and signaling in brain health and disorders. J Cell Physiol. 2021. Available from 10.1002/jcp.30357 [DOI] [PubMed] [Google Scholar]
- 42.Spadaro D, Le S, Laroche T, Mean I, Jond L, Yan J, Citi S. Tension-dependent stretching activates ZO-1 to control the junctional localization of its interactors. Curr Biol. 2017;27(3783–3795.e8):3783–3795.e8. doi: 10.1016/j.cub.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 43.Zenker J, White MD, Gasnier M, Alvarez YD, Lim HYG, Bissiere S, Biro M, Plachta N. Expanding actin rings zipper the mouse embryo for blastocyst formation. Cell. 2018;173(3):776–791.e17. doi: 10.1016/j.cell.2018.02.035. [DOI] [PubMed] [Google Scholar]
- 44.Bendriem RM, Singh S, Aleem AA, Antonetti DA, Ross ME. Tight junction protein occludin regulates progenitor self-renewal and survival in developing cortex. Elife. 2019;8:664. doi: 10.7554/eLife.49376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beutel O, Maraspini R, Pombo-García K, Martin-Lemaitre C, Honigmann A. Phase separation of zonula occludens proteins drives formation of tight junctions. Cell. 2019;179(4):923–936.e11. doi: 10.1016/j.cell.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 46.Kuo W-T, Shen L, Zuo L, Shashikanth N, Ong MLDM, Wu L, Zha J, Edelblum KL, Wang Y, Wang Y, et al. Inflammation-induced occludin downregulation limits epithelial apoptosis by suppressing caspase-3 expression. Gastroenterology. 2019;157(5):1323–1337. doi: 10.1053/j.gastro.2019.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otani T, Nguyen TP, Tokuda S, Sugihara K, Sugawara T, Furuse K, Miura T, Ebnet K, Furuse FM. Claudins and JAM-A coordinately regulate tight junction formation and epithelial polarity. J Cell Biol. 2019;218(10):3372–3396. doi: 10.1083/jcb.201812157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwayer C, Shamipour S, Pranjic-Ferscha K, Schauer A, Balda M, Tada M, Matter K, Heisenberg C-P. Mechanosensation of tight junctions depends on ZO-1 phase separation and flow. Cell. 2019;179(4):937–952.e18. doi: 10.1016/j.cell.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 49.Barnat M, Capizzi M, Aparicio E, Boluda S, Wennagel D, Kacher R, Kassem R, Lenoir S, Agasse F, Braz BY, et al. Huntington’s disease alters human neurodevelopment. Science. 2020;369(6505):787–793. doi: 10.1126/science.aax3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belardi B, Hamkins-Indik T, Harris AR, Kim J, Xu K, Fletcher DA, Weak A. Link with actin organizes tight junctions to control epithelial permeability. Dev Cell. 2020;54(792–804.e7):792–804.e7. doi: 10.1016/j.devcel.2020.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Citi S. Cell Biology:Tight junctions as biomolecular condensates. Curr Biol. 2020;30(2):R83–6. doi: 10.1016/j.cub.2019.11.060. [DOI] [PubMed] [Google Scholar]
- 52.Hartmann C, Schwietzer YA, Otani T, Furuse M, Ebnet K. Physiological functions of junctional adhesion molecules (JAMs) in tight junctions. Biochim Biophys Acta. 2020;1862(9):183299. doi: 10.1016/j.bbamem.2020.183299. [DOI] [PubMed] [Google Scholar]
- 53.Lauko A, Mu Z, Gutmann DH, Naik UP, Lathia JD. Junctional adhesion molecules in cancer: a paradigm for the diverse functions of cell-cell interactions in tumor progression. Cancer Res. 2020;80(22):4878–4885. doi: 10.1158/0008-5472.CAN-20-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otani T, Furuse M. Tight junction structure and function revisited. Trends Cell Biol. 2020;30(10):805–817. doi: 10.1016/j.tcb.2020.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Zhou T, Lu Y, Xu C, Wang R, Zhang L, Lu P. Occludin protects secretory cells from ER stress by facilitating SNARE-dependent apical protein exocytosis. Proc Natl Acad Sci U S A. 2020;117(9):4758–4769. doi: 10.1073/pnas.1909731117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kubota H, Chiba H, Takakuwa Y, Osanai M, Tobioka H, Kohama G, Mori M, Retinoid SN. X receptor alpha and retinoic acid receptor gamma mediate expression of genes encoding tight-junction proteins and barrier function in F9 cells during visceral endodermal differentiation. Exp Cell Res. 2001;263(1):163–172. doi: 10.1006/excr.2000.5113. [DOI] [PubMed] [Google Scholar]
- 57.Chiba H, Gotoh T, Kojima T, Satohisa S, Kikuchi K, Osanai M, Sawada N. Hepatocyte nuclear factor (HNF)-4alpha triggers formation of functional tight junctions and establishment of polarized epithelial morphology in F9 embryonal carcinoma cells. Exp Cell Res. 2003;286(2):288–297. doi: 10.1016/S0014-4827(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 58.Satohisa S, Chiba H, Osanai M, Ohno S, Kojima T, Saito T, Sawada N. Behavior of tight-junction, adherens-junction and cell polarity proteins during HNF-4alpha-induced epithelial polarization. Exp Cell Res. 2005;310(1):66–78. doi: 10.1016/j.yexcr.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 59.Chiba H, Sakai N, Murata M, Osanai M, Ninomiya T, Kojima T, Sawada N. The nuclear receptor hepatocyte nuclear factor 4alpha acts as a morphogen to induce the formation of microvilli. J Cell Biol. 2006;175(6):971–980. doi: 10.1083/jcb.200608012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugimoto K, Ichikawa-Tomikawa N, Satohisa S, Akashi Y, Kanai R, Saito T, Sawada N, Chiba H. The tight-junction protein claudin-6 induces epithelial differentiation from mouse F9 and embryonic stem cells. PLoS One. 2013;8(10):e75106. doi: 10.1371/journal.pone.0075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugimoto K, Ichikawa-Tomikawa N, Kashiwagi K, Endo C, Tanaka S, Sawada N, Watabe T, Higashi T, Chiba H. Cell adhesion signals regulate the nuclear receptor activity. Proc Natl Acad Sci U S A. 2019;116(49):24600–24609. doi: 10.1073/pnas.1913346116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li C-F, Chen J-Y, Ho Y-H, Hsu W-H, Wu L-C, Lan H-Y, Hsu D-S-S, Tai S-K, Chang Y-C, Yang M-H. Snail-induced claudin-11 prompts collective migration for tumour progression. Nat Cell Biol. 2019;21(2):251–262. doi: 10.1038/s41556-018-0268-z. [DOI] [PubMed] [Google Scholar]
- 63.Singh AB, Sharma A, Dhawan P. Claudin-1 expression confers resistance to anoikis in colon cancer cells in a Src-dependent manner. Carcinogenesis. 2012;33(12):2538–2547. doi: 10.1093/carcin/bgs275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang H, Li L, Wu C, Schibli D, Colwill K, Ma S, Li C, Roy P, Ho K, Songyang Z, et al. Defining the specificity space of the human SRC homology 2 domain. Mol Cell Proteomics. 2008;7(4):768–784. doi: 10.1074/mcp.M700312-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Filippakopoulos P, Müller S, Knapp S. SH2 domains: modulators of nonreceptor tyrosine kinase activity. Curr Opin Struct Biol. 2009;19(6):643–649. doi: 10.1016/j.sbi.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu BA, Engelmann BW, Jablonowski K, Higginbotham K, Stergachis AB, Nash PD. SRC homology 2 domain binding sites in insulin, IGF-1 and FGF receptor mediated signaling networks reveal an extensive potential interactome. Cell Commun Signal. 2012;10(1):27. doi: 10.1186/1478-811X-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mayer BJ. The discovery of modular binding domains: building blocks of cell signalling. Nat Rev Mol Cell Biol. 2015;16(11):691–698. doi: 10.1038/nrm4068. [DOI] [PubMed] [Google Scholar]
- 68.Shimobaba S, Taga S, Akizuki R, Hichino A, Endo S, Matsunaga T, Watanabe R, Yamaguchi M, Yamazaki Y, Sugatani J, et al. Claudin-18 inhibits cell proliferation and motility mediated by inhibition of phosphorylation of PDK1 and Akt in human lung adenocarcinoma A549 cells. Biochim Biophys Acta. 2016;1863(6):1170–1178. doi: 10.1016/j.bbamcr.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 69.Luo J, Chimge N-O, Zhou B, Flodby P, Castaldi A, Firth AL, Liu Y, Wang H, Yang C, Marconett CN, et al. CLDN18.1 attenuates malignancy and related signaling pathways of lung adenocarcinoma in vivo and in vitro. Int J Cancer. 2018;143(12):3169–3180. doi: 10.1002/ijc.31734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brunton VG, Irj M, Frame MC. Cell adhesion receptors, tyrosine kinases and actin modulators: a complex three-way circuitry. Biochim Biophys Acta. 2004;1692(2–3):121–144. doi: 10.1016/j.bbamcr.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 71.McLachlan RW, Kraemer A, Helwani FM, Kovacs EM, Yap AS, Mostov K. E-cadherin adhesion activates c-Src signaling at cell-cell contacts. Mol Biol Cell. 2007;18(8):3214–3223. doi: 10.1091/mbc.e06-12-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cabodi S, Di Stefano P, Leal M, Del Pc TA, Bisaro B, Morello V, Damiano L, Aramu S, Repetto D, Tornillo G, et al. Integrins and signal transduction. Adv Exp Med Biol. 2010;674:43–54. [DOI] [PubMed] [Google Scholar]
- 73.Canel M, Serrels A, Frame MC, Brunton VG. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J Cell Sci. 2013;126(2):393–401. doi: 10.1242/jcs.100115. [DOI] [PubMed] [Google Scholar]
- 74.Xiao R, Xi X-D, Chen Z, Chen S-J S-J, Meng G. Structural framework of c-Src activation by integrin β3. Blood. 2013;121(4):700–706. doi: 10.1182/blood-2012-07-440644. [DOI] [PubMed] [Google Scholar]
- 75.Ochs K, Málaga-Trillo E. Common themes in PrP signaling: the Src remains the same. Front Cell Dev Biol. 2014;2:63. doi: 10.3389/fcell.2014.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamidi H, Ivaska J. Every step of the way: integrins in cancer progression and metastasis. Nat Rev Cancer. 2018;18(9):533–548. doi: 10.1038/s41568-018-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Witherden DA, Verdino P, Rieder SE, Garijo O, Mills RE, Teyton L, Fischer WH, Wilson IA, Havran WL. The junctional adhesion molecule JAML is a costimulatory receptor for epithelial gammadelta T cell activation. Science. 2010;329(5996):1205–1210. doi: 10.1126/science.1192698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yaffe MB. MAGUK SH3 domains–swapped and stranded by their kinases? Structure. 2002;10(1):3–5. doi: 10.1016/S0969-2126(01)00705-5. [DOI] [PubMed] [Google Scholar]
- 79.Lim WA, Pawson T. Phosphotyrosine signaling: evolving a new cellular communication system. Cell. 2010;142(5):661–667. doi: 10.1016/j.cell.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu BA, Nash PD. Evolution of SH2 domains and phosphotyrosine signalling networks. Philos Trans R Soc Lond B Biol Sci. 2012;367(1602):2556–2573. doi: 10.1098/rstb.2012.0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wagner MJ, Stacey MM, Liu BA, Pawson T. Molecular mechanisms of SH2- and PTB-domain-containing proteins in receptor tyrosine kinase signaling. Cold Spring Harb Perspect Biol. 2013;5(12):a008987. doi: 10.1101/cshperspect.a008987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kojima M, Sugimoto K, Tanaka M, Endo Y, Kato H, Honda T, Furukawa S, Nishiyama H, Watanabe T, Soeda S, et al. Prognostic significance of aberrant claudin-6 expression in endometrial cancer. Cancers. 2020;12(10):2748. Available from: 10.3390/cancers12102748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kojima M, Sugimoto K, Kobayashi M, Ichikawa-Tomikawa N, Kashiwagi K, Watanabe T, Soeda S, Fujimori K, Chiba H. Aberrant claudin-6-adhesion signaling promotes endometrial cancer progression via estrogen receptor α. Mol Cancer Res [in press; Available from]. 2021. https://mcr.aacrjournals.org/content/early/2021/03/16/1541-7786.MCR-20-0835 [DOI] [PubMed] [Google Scholar]
- 84.Lang F, Böhmer C, Palmada M, Seebohm G, Strutz-Seebohm N, Vallon V. (Patho)physiological significance of the serum- and glucocorticoid-inducible kinase isoforms. Physiol Rev. 2006;86(4):1151–1178. doi: 10.1152/physrev.00050.2005. [DOI] [PubMed] [Google Scholar]
- 85.Hagen SJ, Ang L-H, Zheng Y, Karahan SN, Wu J, Wang YE, Caron TJ, Gad AP, Muthupalani S, Fox JG. Loss of tight junction protein claudin 18 promotes progressive neoplasia development in mouse stomach. Gastroenterology. 2018;155(6):1852–1867. doi: 10.1053/j.gastro.2018.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou B, Flodby P, Luo J, Castillo DR, Liu Y, Yu F-X, McConnell A, Varghese B, Li G, Chimge N-O, et al. Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. J Clin Invest. 2018;128(3):970–984. doi: 10.1172/JCI90429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Suzuki K, Sentani K, Tanaka H, Yano T, Suzuki K, Oshima M, Yasui W, Tamura A, Tsukita S. Deficiency of stomach-type claudin-18 in mice induces gastric tumor formation independent of H pylori infection. Cell Mol Gastroenterol Hepatol. 2019;8(1):119–142. doi: 10.1016/j.jcmgh.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paquet-Fifield S, Koh SL, Cheng L, Beyit LM, Shembrey C, Mølck C, Behrenbruch C, Papin M, Gironella M, Guelfi S, et al. Tight junction protein claudin-2 promotes self-renewal of human colorectal cancer stem-like cells. Cancer Res. 2018;78(11):2925–2938. doi: 10.1158/0008-5472.CAN-17-1869. [DOI] [PubMed] [Google Scholar]


