ABSTRACT
Overexpression of DTYMK is related with tumorigenesis and progression in several human tumors. However, the role of upregulated DTYMK in hepatocellular carcinoma (HCC) patients still remains unclear. In this study, the DTYMK expression in HCC tumors was evaluated in three GEO series (GSE14520, GSE54236, GSE63898), TCGA-LIHC, and ICGC-IRLR-JP cohorts. Survival analysis of DTYMK based on TCGA-LIHC and ICGC-LIRI-JP cohorts was conducted. We found that DTYMK was dramatically upregulated in tumor tissue compared with that in adjacent liver tissue. Kaplan-Meier analysis revealed that high expression of DTYMK in HCC patients’ tumor tissue was significantly corresponded to worse overall survival (OS) (P < 0.05). Further analysis showed that overexpressing DTYMK led to poor relapse free survival (RFS) and disease-specific survival (DSS) (all P < 0.05). In conclusion, DTYMK is upregulated in tumors and correlated with poor prognosis in HCC patients. In our report, DTYMK is higher expression in HCC cancer tissue and cell line than tumor adjacent tissue and normal liver cell line. Knocking down DTYMK can inhabit tumor cell proliferation by interfering cell cycle, whereas overexpression of DTYMK can promote tumor cell proliferation. These findings indicate that upregulation of DTYMK enhances tumor growth and proliferation by promoting cell cycle.
KEYWORDS: DTYMK, hepatocarcinoma, cell proliferation, cell cycle
Introduction
Primary liver cancer is one of the most common malignant tumors in the world, and approximately 90% of liver cancer is hepatocellular carcinoma (HCC). Every year more than 800,000 new cases and deaths are reported. HCC is highly proliferative, invasive and metastatic potential with a low survival rate [1,2]. A large number of studies have explored the mechanism of occurrence, recurrence and metastasis, tumorigenesis, and treatment of HCC. However, many problems remain to be clarified [3,4]. One group found that genetic aberrations have a high correlation with the occurrence, development, and progression of HCC[5]. It’= is urgent to find the original drive force and identify promising prognostic biomarkers for clinical diagnosis as well as targets for drug design [6].
DTYMK is known to catalyze the phosphorylation of dTMP to dTDP [7]. This enzyme plays a vital role in DNA synthesis in vivo and is a main intermediate enzyme in many pyrimidines analog drugs pathways [8,9]. DTYMK is highly expressed in the bone marrow, lymph node, colon, and appendix which cell proliferation and division are vigorous. Therefore, it may also be essential for the proliferation and division of tumor cells. Upregulation of DTYMK has been reported in nonsmall cell lung carcinoma [10], correlated with cell proliferation and LKB1-mutant related lung cancer metabolic disorder [11]. Other studies indicate that DTYMK is highly expressed in oral cancer, 5-FU relative drug-resistance colon cancer has functions in tumor occurrence and development. The role DTYMK plays in HCC, however, is controversially discussed. One research revealed that DTYMK is a poor predictor for well-differential HCC and promotes cancer stem cell character of HCC cells [12]. Another study shows that DTYMK participates in immune cell infiltration and the high expression of DTYMK is a wicked factor of overall survival (OS) and disease-free survival (DFS) in HCC patients [13]. However, the specific mechanism of DTYMK in HCC patients is still required to further identification.
For the purpose of interpreting the relationship between DTYMK expression level and HCC outcomes, we identified the DTYMK mRNA expression in Gene Expression Omnibus (GEO), Liver Cancer – RIKEN, JP Project from International Cancer Genome Consortium (ICGC-LIRI-JP), and The Cancer Genome Atlas (TCGA) databases and conducted a survival analysis based on TCGA, ICGC-LIRI-JP cohorts, and Kaplan-Meier Plotter database. We draw a conclusion that DTYMK is highly expressed in HCC patients and is a certain poor indicator of OS and PFS. Subsequent analysis reveals that DTYMK is correlated with HCC cell division and cell proliferation of P53-related patients. We examined the expression of DTYMK by immunohistochemistry in tumor tissues and paired precancerous liver tissue in 60 HCC patients. Meanwhile we conducted CCK8 proliferation assay in different cellar cycles to verify the subsequent outcomes. Our findings prove that DTYMK is a HCC proliferation malignant biomarker and a potent target for HCC diagnosis and therapy.
Materials and methods
Data acquisition and process
The expression matrix and clinical characters of cohorts included TCGA-LIHC, ICGC-LIRI-JP, GSE14520, GSE54236, and GSE63898 were downloaded from HCCDB database (http://lifeome.net/database/hccdb/home.html) [14]. For GEO cohorts, the probe signal intensity values were transformed by log2. For TCGA-LIHC and ICGC LIRI-JP cohorts, the normalized read counts were transformed by log2. The comparisons of mRNA expression level between cancer and noncancer tissues in 5 cohorts were conducted with unpaired t test with Welch’s correction by GraphPad Prism 7.04 software. P < 0.05 was considered to be statistically significant.
Survival analysis
The mRNA expression of DTYMK and survival characters of patients in TCGA-LIHC and ICGC-LIRI-JP cohorts were extracted for Kaplan-Meier overall survival (OS) analysis. The liver cancer patients were divided into DTYMK-high and DTYMK-low group by the median mRNA expression value of DTYMK in cohorts independently. In addition, Kaplan-Meier Plotter [liver cancer] (http://kmplot.com/analysis/index.php?p=service&cancer=liver_rnaseq) was used to identify the correlation of DTYMK and OS, Relapse free survival (RFS), and disease specific survival (DSS) in liver cancer patients [15]. The log-rank test was applied for survival analysis and P < 0.05 was considered to be statistically significant.
The correlation of clinical parameters and DTYMK in liver cancer
The mRNA expression of DTYMK and clinical parameters of patients in TCGA-LIHC, ICGC-LIRI-JP cohorts were extracted. The patients were divided into two groups according to the single clinical parameter. The mRNA level of DTYMK was compared between two groups. In addition, tumor double time was investigated between DTYMK-high and DTYMK-low liver cancer patients in GSE54236 cohort. Unpaired t test with Welch’s correction was applied for static analysis and P < 0.05 was considered to be statistically significant.
Functional enrichment analysis
Coexpressed genes with DTYMK were computed and displayed according to the guideline in the HCCDB database website. Meta scape (http://metascape.org) was used for the annotation and functional and disease-related enriched analysis of coexpressed genes, such as gene ontology and Kyoto encyclopedia of genes [16]. The Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/) was used for confirming the correlation of DTYMK with PCNA and MKi67[17].
Human samples
The experiment with human liver cancer tissues was authorized by the Human Ethics Committee. All subjects were provided written informed consent.
Cell lines and culture conditions
Human HCC cell lines HepG2, HepG3B, LM3, and Huh7 and human normal liver cell line LO-2 were acquired from ATCC and cultivated in DMEM medium (Gibco) with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin (Gibco) at 37°C with 5% CO2. All cell lines were certificated by STR profiling and there was no mycoplasma contamination.
Western blotting
Cell proteins were extracted by denatured buffer and then quantified by pierce BCA protein assay (Thermo Scientific). The HCC cell line protein lysate were separated on SDS-PAGE, transferred to NC membranes (Millipore), blocked, and then detected by primary antibody DTYMK (Abcam, ab154867, 1:2000) and HRP-conjugated secondary antibody (Sigma), followed by being exposed with enhanced chemiluminescence (ECL). Housekeep gene β-Actin (Abcam, ab8226,1:5000) was used as a loading control.
Plasmids and lentivirus production
DTYMK cDNA were cloned into the viral skeleton plasmid PCDH-3*flag-tagged vector for stable expression in HCC cell lines. Annealing and connection of the shRNA were performed and then constructed into the modified plasmid pLKO.1. The well-constructed vectors were transinfected into HEK293T cells by lentivirus packaging plasmids psPAX and pMD2.0 G. The shRNA sequences and DTYMK primer sequence and DTYMK plasmid PCR primer in this research are shown below: DYMKT-sh1-F:CCGGTGGGAACAAGTGCCGTTAATTCTCGAGAATTAACGGCACTTGTTCCCATTTTTTG; DYMKT-sh1-R:
AATTCAAAAAATGGGAACAAGTGCCGTTAATTCTCGAGAATTAACGGCACTTGTTCCCA;
DYMKT-sh2-F: CCGGGTCGTGGACAGATACGCATTTCTCGAGCAAATGCGTATCTGTCCACGACTTTTTTG;
DYMKT-sh2-R: AATTCAAAAAAGTCGTGGACAGATACGCATTTCTCGAGAAATGCGTATCTGTCCACGAC;
DTYMK-F:
TCGAGCTCAAGCTTCGAATTCATGGCGGCCCGGCGCGGG;
DTYMK-R:
GTCATCCTTGTAGTCGGATCCCTTCCATAGCTCCCCCAGCG.
Cell growth assay
Lentivirus-infected stable cells were seeded into 96-well plates and cultured in 10% FBS DMEM (2000 cells per well, five parallel wells). Then, the cells were collected at different points in time, and cell number in each well was counted by CCK-8 reagent. Absorbance at 450 nm was employed to determine of the number of viable cells.
Immunohistochemistry and tissue array
The human HCC tissue samples were harvested by pathology department after obtaining consent. Tumor and normal adjacent tissues 2 cm away from the tumor was prepared. The detailed clinicopathological data (show in supplementary table 2) were scored according to the tumor classification of the American Joint Committee on Cancer (AJCC)/International Union Against Cancer (UICC) tumor staging system. For the tissue microarray (TMA) construction, all cancers and adjacent tissue samples were reevaluated and the representative areas were chosen. IHC was operated by standard IHC protocol as described before. Anti-DTYMK rabbit polyclonal antibody (1:200, Abcam, ab154867) and anti-PCNA rabbit polyclonal antibody (1:2000, ProteinTech, 15,910-1-AP) were used in this study. DTYMK expression level in tissues was evaluated by two pathologists independently. The methods of scores for staining frequency and intensity were added to obtain an overall staining score (OSS) was described previously [18]. A staining index (values, 0–12) was determined by multiplying the score for staining intensity with the score for both cytoplasmic and nuclear positive. Staining intensity grade is classified as: 0, negative; 1, weak; 2, moderate; 3, strong. The frequency of positive cells is defined as: less than 5%, 0 point; 5%–25%, 1 point; 26%–50%, two points; 51%–75%, 3 points; More than 75%, 4 points 1[9].
Cell cycle assay
Cell Cycle and Apoptosis Analysis Kit was used to determine cell cycle based on the specifications. HepG2 and Huh-7 cells were cultured in 24 wells plate for 24 h. Next, cells were digested with trypsin and suspended into single cell, and fixed with 70% ethanol at 4°C at least 2 hours. Then cells were resuspended and washed by PBS. 0.2 mg/mL RNase A and 50 μg/mL PI in PBS was applied for staining cells for 30 min. The percentages of cells of G0/G1, S, and G2/M were counted by Beckman Cytoflex.
Colony formation assay
Cells were seeded into 6-well plate at a density of 100 cells per well with three parallel wells. After 3 weeks of cultivation DMEM with 10%FBS, formed clones were fixed by 10% formalin then stained with 0.1% crystal violet. Numbers of colonies were counted attentively.
Statistical analysis
All data were statistically analyzed with GraphPad Prism 8.0. Two-tailed t test was applied to analyze the difference between the two groups. Data were presented as mean ± SD or SEM. Differences at P < 0.05 were considered statistically significant.
Results
Expression pattern and prognostic implication ofDTYMK in HCC
In our study, TCGA-LIHC, ICGC-LIRI-JP, and three GEO (GSE14520, GSE54236, and GSE63898) cohorts were utilized to analyzed the difference of DTYMK mRNA expression between HCC and normal hepatic tissues. It showed that DTYMK mRNA expression level was higher in HCC samples than corresponding adjacent normal samples with P < 0.05 (Figure 1(a-e)). Moreover, we found that high DTYMK expression was significantly associated with later stages (stages III + IV) (Figure 1(f,g)), vascular invasion (Figure 1(h,i)), more severe fibrosis (F3-4) (Figure 1(j)), and shorter tumor doubling time (Figure 1(k)). Besides, TCGA-LIHC and ICGC-LIRI-JP cohorts and Kaplan-Meier Plotter [liver cancer] online tool were utilized to explore the correlation between DTYMK and survival in HCC patients. Our results indicated that HCC patients with high DTYMK expression had poor OS (Figure 1(l,n)), RFS (Figure 1(o)), and DSS (Figure 1(p)) compared to those with low DTYMK expression.
Figure 1.
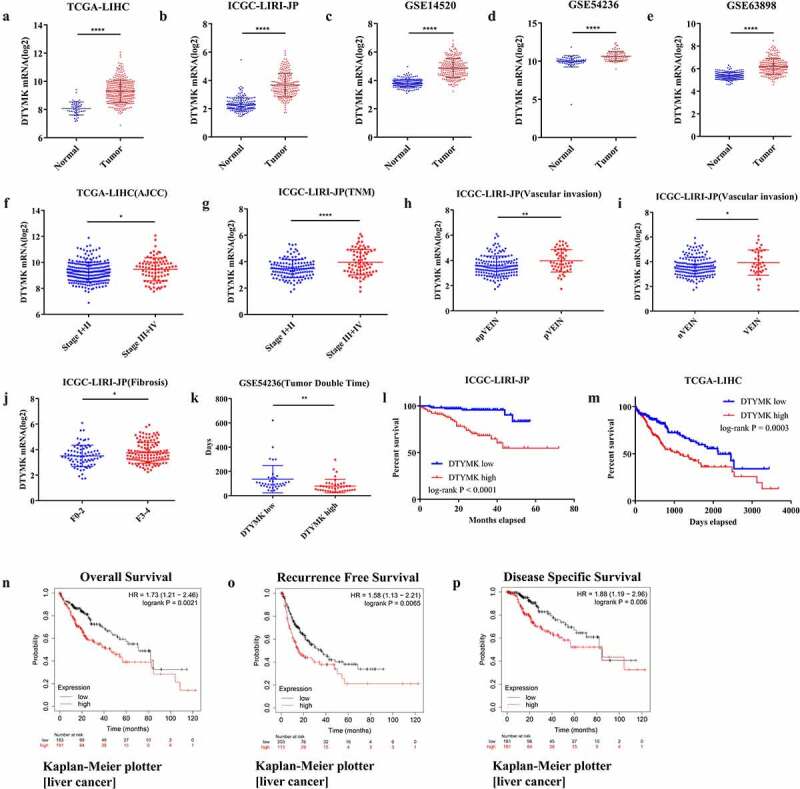
Expression pattern and prognostic implication of DTYMK in HCC
(a) DTYMK mRNA expression between HCC and normal hepatic tissues in TCGA-LIHC cohort; (b) DTYMK mRNA expression between HCC and normal hepatic tissues in ICGC-LIRI-JP cohort; (c) DTYMK mRNA expression between HCC and normal hepatic tissues in GSE14520 cohort; (d) DTYMK mRNA expression between HCC and normal hepatic tissues in GSE54236 cohort; (e) DTYMK mRNA expression between HCC and normal hepatic tissues in GSE63898 cohort; (f) DTYMK mRNA expression between stage I+ II HCC and stage III+IV HCC samples in TCGA-LIHC cohort; (g) DTYMK mRNA expression between stage I+ II HCC and stage III+IV HCC samples in ICGC-LIRI-JP cohort; (h) DTYMK mRNA expression between HCC without portal vein invasion and HCC with portal vein invasion samples in ICGC-LIRI-JP cohort; (i) DTYMK mRNA expression between HCC without vascular invasion and HCC with vascular invasion samples in ICGC-LIRI-JP cohort; (j) DTYMK mRNA expression between fibrosis grade F0-2 HCC and fibrosis grade F3-4 HCC samples in ICGC-LIRI-JP cohort; (k) tumor doubling time between high DTYMK mRNA expression samples and low DTYMK mRNA expression samples in GSE54326 cohort; (l) Kaplan-Meier curve of overall survival for DTYMK expression in ICGC-LIRI-JP cohort; (M) Kaplan-Meier curve of overall survival for DTYMK expression in TCGA-LIHC cohort; (N) Kaplan-Meier curve of overall survival for DTYMK expression in Kaplan-Meier Plotter; (o) Kaplan-Meier curve of recurrence free survival for DTYMK expression in Kaplan-Meier Plotter; (p) Kaplan-Meier curve of disease specific survival for DTYMK expression in Kaplan-Meier Plotter.
DTYMK is correlated with proliferation and cell cycle of HCC
Co-expressed genes with DTYMK were annotated and functional enriched analyzed. Our results revealed that DTYMK and its coexpressed genes play a critical role in cell cycle, mitotic nuclear division and signal transduction driven by p53 class mediator etc. (Figure 2(a,b)). In addition, we utilized GEPIA online tool to explore the correlation of DTYMK with MKI67 and PCNA based on the TCGA-LIHC cohort. The results showed that DTYMK mRNA expression was positively correlated with MKI67 expression and PCNA expression in HCC samples (Figure 2(c,d)). Besides, we found that DTYMK expression was upregulated in TP53 mutated HCC samples compared to that without TP53 mutation (Figure 2(e)).
Figure 2.
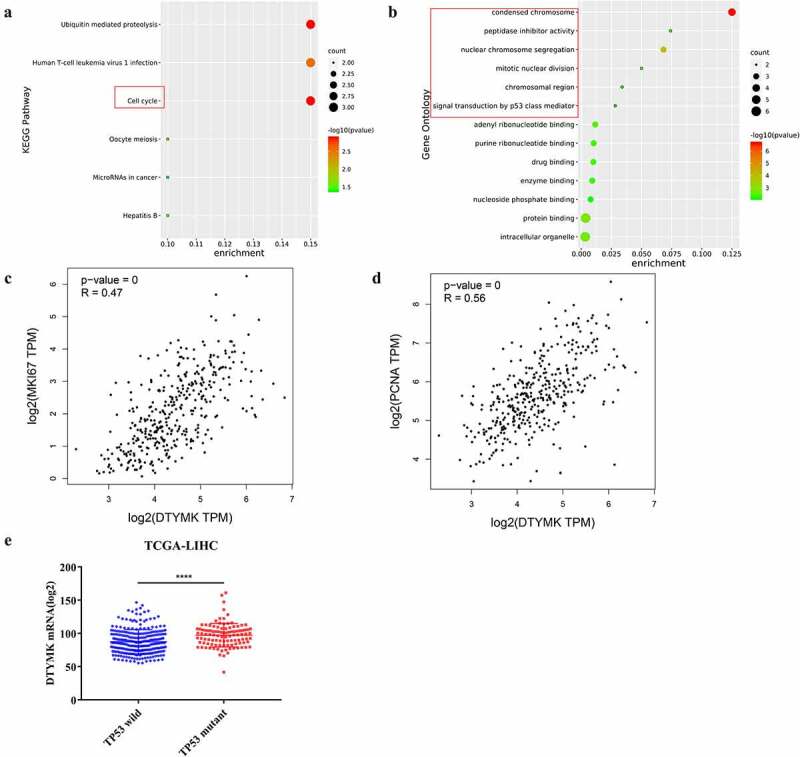
Functional analysis of DTYMK and its coexpressed genes
(a) Functional enriched analysis of DTYMK and its coexpressed genes (KEGG); (b) functional enriched analysis of DTYMK and its coexpressed genes (GO); (c) correlation of DTYMK with MKI67; (d) correlation of DTYMK with PCNA; (e) DTYMK mRNA expression between TP53 mutant and TP53 wild HCC patients. KEGG, Kyoto encyclopedia of genes and genomes; GO; gene ontology.
Validation the DTYMK protein expression in HCC cell lines and HCC patients
We tested protein expression level of DTYMK by Western blot in HCC cell lines compared to normal liver cell line L-02. IHC was performed in tissue sections with HCC (TMA) containing 60 pairs of primary liver samples. The results showed that DTYMK protein expression was mainly confined to the cell plasma and dramatically upregulated in HCC samples when compared to the adjacent normal liver cells (Figure 3(a,c)). We next investigated DTYMK expression levels in patients with different TNM stages. We found that there was an increase in DTYMK expression gradually with higher HCC grades. The average DTYMK expression level increased in the term of TNM stage I to IV and pathology grade I to III (Figure 3(d,e)). This finding implied a possible role of DTYMK upregulation in promoting HCC development. To verify the potential clinical meaning of DTYMK upregulation according to our TCGA analysis, we investigated the correlation of DTYMK expression levels and the overall survival time of HCC patients. Importantly, upregulation of DTYMK was observed remarkably associated with a worse OS rate of the HCC patients (Figure 3(f)).
Figure 3.
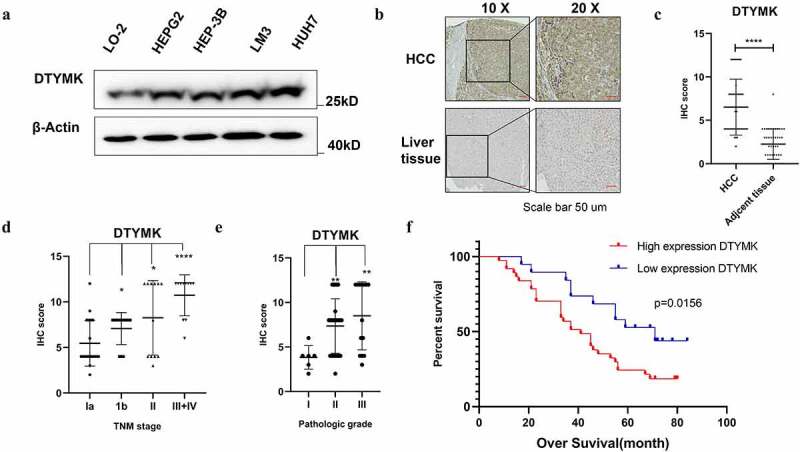
Validation the DTYMK expression in HCC cell lines and HCC patients
(a) Western blot analysis of DTYMK in HCC cells including LO-2, HEPG2, HEP3B, LM-3, and Huh-7. β-Actin was used as a loading control; (b) expression of DTYMK protein by IHC. DTYMK protein was predominantly detected at the cytoplasm. Arepresentative case shows the upregulation of DTYMK protein in HCC compared to that in its adjacent normal liver tissue; (c) semi-quantitative analyses of IHC staining of DTYMK in 60 HCC patients TMAs with tumor sample and adjacent tissue; (D) semi-quantitative analyses of IHC staining of DTYMK in 60 HCC patients TMAs with different TNM-stage patient; (e) semi-quantitative analyses of IHC staining of DTYMK in 60 HCC patients TMAs with different pathology grade patients (f) Kaplan–Meier of survival of 60 patients with HCC (two groups stratified by DTYMK expression level. Differences between the groups were shown by a log-rank test.)
DTYMK promotes HCC cell proliferation
To investigate functional roles of DTYMK in HCC, we constructed DTYMK overexpression cell line HepG2 and the DTYMK stable knockdown Huh7 cell lines with two independent shRNA sequences (shSETDB#1 and #2). Successful overexpression and knockdown of DTYMK was confirmed with western blotting (Figure 4(a,b)). It was shown that the overexpression of DTYMK promoted the proliferation of HepG2 cells while the knockdown of DTYMK significantly led to the suppression of Huh7 cell proliferation, which was demonstrated by CCK8 growth curve analysis (Figure 4(c,d)). Consistent results were obtained that the overexpression of DTYMK promoted the ability of colony formation of HCC cells while the knockdown of DTYMK suppressed that. (Figure 4(e,g)).
Figure 4.
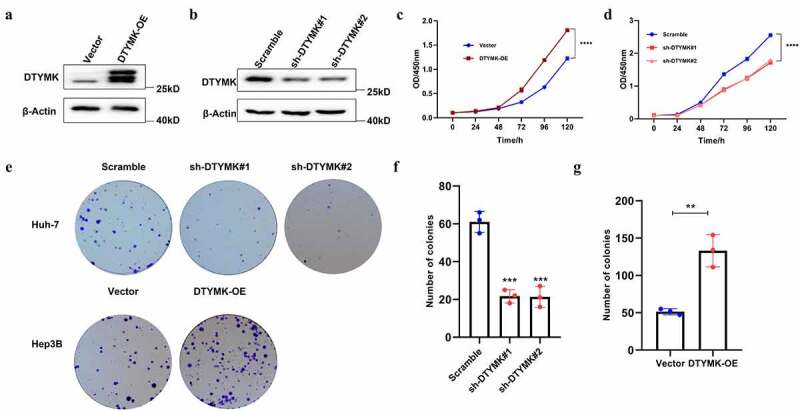
DTYMK promotes HCC cell proliferation
(a) DTYMK-overexpressed HepG2 cells was confirmed with Western blot; (b) DTYMK-knockdowned Huh7 cells was confirmed with western blot; (c) CCK-8 assay was performed in DTYMK-overexpressed HepG2 cells; (d) CCK-8 assay was performed in DTYMK-knockdowned Huh7 cells; (e) colony formation assay with DTYMK overexpression or DTYMK knock down of two indicated cell lines; (f) histograms show the mean numbers of formed colon in DTYMK-overexpressed HepG2 cells; (g) histograms show the mean numbers of formed colon DTYMK-knockdowned Huh7 cells.
DTYMK regulate HCC cell cycle progression
The TCGA database analysis results showed that DTYMK participates in cell cycles in HCC cells. To further validate this idea, we knocked down DTYMK by shRNA in stable Huh-7 cell line which highly expressed DTYMK. We applied a standard cell cycle assay by flow cytometry using propidium iodide staining. An approximately 20% increase of cells in the G0/G1 phase was observed in Huh-7 cells. Meanwhile, the knockdown of DTYMK led to a 14% decrease in the number of cells in the G2/M phase, compared to cells transfected with the scramble shRNA (Figures 5(a,b)). Besides, overexpression of DTYMK decreased HepG2 cells in the G0/G1 phase while increased the number of cells in the G2/M phase (Figure 5(c,d)).
Figure 5.
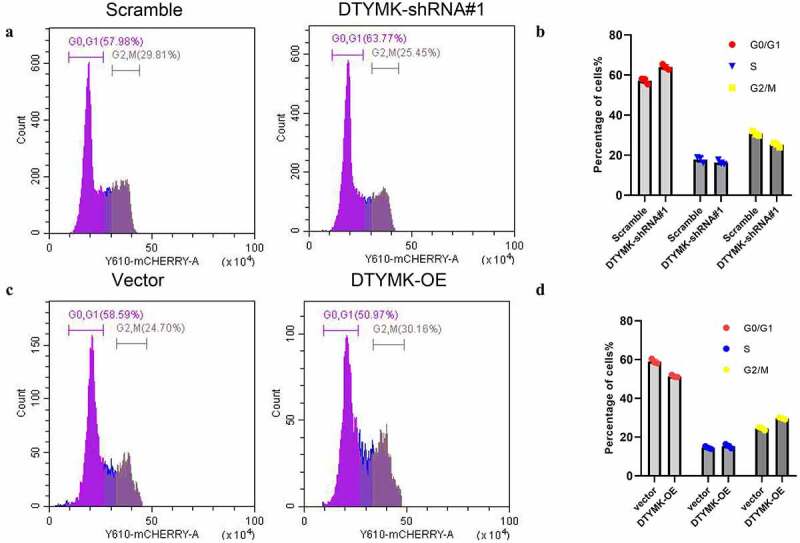
DTYMK regulate HCC cell cycle progression
(a) Flow cytometry assays were conducted to analyze cell cycle in Huh-7-NC and Huh-7 DTYMK-shRNA cells; (b) quantification of G0/1, S and G2/M phase in Huh-7-NC and Huh-7 DTYMK-shRNA cells from three independent experiments; (c) flow cytometry assays were conducted to analyze cell cycle in HepG2-NC and HepG2 DTYMK-overexpression cells; (d) quantification of G0/1, S, and G2/M phase in HepG2-NC and HepG2 DTYMK-overexpression cells from three independent experiments.
Discussion
In this research, we first systematically analyzed the expression pattern and prognostic prediction value of DTYMK in HCC patients by integrating cohorts from three GEO series, TCGA, and ICGC. The results show that DTYMK is upregulated in tumor tissue compared to adjacent normal hepatic tissues in all cohorts, and a higher level of DTYMK indicates a poorer prognosis in HCC patients. In addition, we found that DTYMK may play a critical role in tumorigenesis and progression by regulating cell cycle and cell proliferation.
DTYMK functions in de novo pyrimidine pathway, which is related to the dTTP biosynthesis, part of Pyrimidine metabolism [7,8]. Overexpression of DTYMK in vivo leads to increasing the production of nucleotides which is necessary for DNA synthesis and repair [20]. It can be inferred that DTYMK could be related with cell cycle and drug resistant in physiological or pathological process by influencing thymidylate synthesis [21–24]. In recent years, DTYMK is reported to be upregulated accompanied by drug resistance in several cancers such as lung cancer, colon cancer, hepatic carcinoma and esophagus cancer [25], but there is no further investigation for the detailed mechanism.
In another experiment in HCC, DTYMK is dramatically highly expressed in HCC than in adjacent liver tissue. We demonstrated that higher expression of DTYMK in tumor is significantly associated with worse over survival in HCC patients. Besides, the expression level of DTYMK is also in correlated with TNM stage and pathology grade in HCC patients. The expression of DTYMK was validated in HCC cell lines HepG2, Hep3B, LM-3, and Huh-7 and human normal liver cell line LO-2. Knocking down DTYMK by shRNA silencing in Huh-7 cells decreased cell proliferation and colony formation ability. Conversely, the overexpression of DTYMK in HepG2 increased cell proliferation and colony formation ability. The ontogenetic functional study in HCC cell line was consistent with the result in clinical analysis that the expression of DTYMK was positively correlated with proliferation marker PCNA. PCNA is known as a biomarker of cell proliferation, which can effectively reflect the proliferation level of tumor cells, especially in liver cancer, nose pharyngeal cancer, and other poorly differentiated malignant tumors [26,27]. Progression of the cell cycle is tightly related with cell proliferation and growth [28,29]. The dysfunction of DTYMK induces cell cycle arrest in Huh-7 cell line while overexpressing DTYMK increase M phase cell percent, which demonstrated that DTYMK induced cancer cell proliferation by promoting cell division.
Finally, we proposed that DTYMK presented oncogenic properties in HCC cells. The overexpression of DTYMK promotes HCC cell proliferation. Specifically, knocking down DTYMK led to cell cycle arrest while overexpressing promoting cell cycle. As a result, DTYMK is a promising target for HCC diagnosis and treatment [30–32]. Whereas some limitations in this study need consideration as well. First, we did not explore the explicit mechanism that DTYMK drives HCC cell growth and changes cell cycle division, there were evidences that DTYMK promoting the cell cycle progression by increasing the pool of dTTP which was the raw material of DNA synthesis in colon cancer cells [33]. During the cell cycle progression, the production of dTTP is highly regulated to coordinate with DNA replication [34]. Besides, it was reported that Raltitrexed (RTX), an inhibitor of thymidylate synthase (TS), arrested HepG2 cells at G0/G1 via downregulation of cyclin A and CDK2 [35]. Second, the reason why DTYMK is upregulated in HCC cells is still unknown. Future research is required to explore the detailed mechanism between distinct DTYMK and carcinogenesis of HCC and to reveal the mechanism of DTYMK in other carcinomas, which may be critical to the development of inhibitors in liver cancer patients.
Supplementary Material
Acknowledgments
The results of this study are based on the data from TCGA (https://www.cancer.gov/tcga), Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo) and ICGC-LIRI-JP (https://icgc.org/). We thank the authors who provided the data for this study.
Funding Statement
This research is supported by Doctoral Research Fund, Affiliated Hospital of Jining Medical University, BS-007; Jining Medical University [BS-007].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Forner A, Reig M, Bruix J.. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. [DOI] [PubMed] [Google Scholar]
- [2].Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–1462. [DOI] [PubMed] [Google Scholar]
- [3].Diaz-Gonzalez A, Reig M, Bruix J. Treatment of hepatocellular carcinoma. Dig Dis. 2016;34(5):597–602. [DOI] [PubMed] [Google Scholar]
- [4].Clark T, Maximin S, Meier J, et al. Review of epidemiology, screening, imaging diagnosis, response assessment, and treatment. Curr Probl Diagn Radiol. 2015;44(6):479–486. [DOI] [PubMed] [Google Scholar]
- [5].Kim SK, Takeda H, Takai A, et al. Comprehensive analysis of genetic aberrations linked to tumorigenesis in regenerative nodules of liver cirrhosis. J Gastroenterol. 2019;54(7):628–640. [DOI] [PubMed] [Google Scholar]
- [6].Villanueva A, Hoshida Y, Toffanin S, et al. New strategies in hepatocellular carcinoma: genomic prognostic markers. Clin Cancer Res. 2010;16(19):4688–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee LS, Cheng Y. Human thymidylate kinase. Purification, characterization, and kinetic behavior of the thymidylate kinase derived from chronic myelocytic leukemia. J Biol Chem. 1977;252(16):5686–5691. [PubMed] [Google Scholar]
- [8].Huang SH, Tang A, Drisco B, et al. Human dTMP kinase: gene expression and enzymatic activity coinciding with cell cycle progression and cell growth. DNA Cell Biol. 1994;13(5):461–471. [DOI] [PubMed] [Google Scholar]
- [9].Caspi R, Billington R, Fulcher CA, et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46(D1):D633–D639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang H, Wang X, Xu L, et al. High expression levels of pyrimidine metabolic rate-limiting enzymes are adverse prognostic factors in lung adenocarcinoma: a study based on the cancer genome atlas and gene expression omnibus datasets. Purinergic Signal. 2020;16(3):347–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu Y, Marks K, Cowley GS, et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-Mutant lung cancer. Cancer Discov. 2013;3(8):870–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yeh HW, Lee SS, Chang CY, et al. Pyrimidine metabolic rate limiting enzymes in poorly-differentiated hepatocellular carcinoma are signature genes of cancer stemness and associated with poor prognosis. Oncotarget. 2017;8(44):77734–77751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang D, Liu J, Liu S, et al. Identification of crucial genes associated with immune cell infiltration in hepatocellular carcinoma by weighted gene Co-expression network analysis. Front Genet. 2020;11:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lian Q, Wang S, Zhang G, et al. HCCDB: a database of hepatocellular carcinoma expression atlas. Genomics Proteomics Bioinformatics. 2018;16(4):269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Menyhart O, Nagy A, Gyorffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5(12):181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kaemmerer D, Peter L, Lupp A, et al. Comparing of IRS and Her2 as immunohistochemical scoring schemes in gastroenteropancreatic neuroendocrine tumors. Int J Clin Exp Pathol. 2012;5:187–194. [PMC free article] [PubMed] [Google Scholar]
- [19].Min KW, CHAE SW, Kim D-H, et al. Fascin expression predicts an aggressive clinical course in patients with advanced breast cancer. Oncol Lett. 2015;10(1):121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].De Angelis PM, Fjell B, Kravik KL, et al. Molecular characterizations of derivatives of HCT116 colorectal cancer cells that are resistant to the chemotherapeutic agent 5-fluorouracil. Int J Oncol. 2004;24:1279–1288. [PubMed] [Google Scholar]
- [21].Saxena P, Severi L, Santucci M, et al. Conformational propensity and biological studies of proline mutated LR peptides inhibiting human thymidylate synthase and ovarian cancer cell growth. J Med Chem. 2018;61(16):7374–7380. [DOI] [PubMed] [Google Scholar]
- [22].Peters GJ, van der Wilt CL, van Triest B, et al. Thymidylate synthase and drug resistance. Eur J Cancer. 1995;31A(7–8):1299–1305. [DOI] [PubMed] [Google Scholar]
- [23].Kim S, Park DH, Shim J. Thymidylate synthase and dihydropyrimidine dehydrogenase levels are associated with response to 5-fluorouracil in caenorhabditis elegans. Mol Cells. 2008;26:344–349. [PubMed] [Google Scholar]
- [24].Guo FY, Yang J, Xiong SM, et al. [Clinical significance of epidermal growth factor receptor and thymidylate synthase expression in primary liver cancer]. Zhonghua Gan Zang Bing Za Zhi. 2018;26:666–669. [DOI] [PubMed] [Google Scholar]
- [25].Peters GJ, van Triest B, Backus HHJ, et al. Molecular downstream events and induction of thymidylate synthase in mutant and wild-type p53 colon cancer cell lines after treatment with 5-fluorouracil and the thymidylate synthase inhibitor raltitrexed. Eur J Cancer. 2000;36(7):916–924. [DOI] [PubMed] [Google Scholar]
- [26].Mailand N, Gibbs-Seymour I, Bekker-Jensen S. Regulation of PCNA-protein interactions for genome stability. Nat Rev Mol Cell Biol. 2013;14(5):269–282. [DOI] [PubMed] [Google Scholar]
- [27].Evison BJ, Actis ML, Wu SZ, et al. A site-selective, irreversible inhibitor of the DNA replication auxiliary factor proliferating cell nuclear antigen (PCNA). Bioorg Med Chem. 2014;22(22):6333–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Moon J, Yu SJ, Kim HS, et al. Induction of G(1) cell cycle arrest and p27(KIP1) increase by panaxydol isolated from Panax ginseng. Biochem Pharmacol. 2000;59(9):1109–1116. [DOI] [PubMed] [Google Scholar]
- [29].Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–348. [DOI] [PubMed] [Google Scholar]
- [30].Wilson PM, Danenberg PV, Johnston PG, et al. Standing the test of time: targeting thymidylate biosynthesis in cancer therapy. Nat Rev Clin Oncol. 2014;11(5):282–298. [DOI] [PubMed] [Google Scholar]
- [31].Shiina T, Saito G, Sakaizawa T, et al. Higher tissue levels of thymidylate synthase determined by ELISA are associated with poor prognosis of patients with lung cancer. Tohoku J Exp Med. 2017;242(4):303–316. [DOI] [PubMed] [Google Scholar]
- [32].Shimizu A, Kaira K, Okubo Y, et al. Expression of thymidylate synthase (TS) and its prognostic significance in patients with cutaneous angiosarcoma. Neoplasma. 2017;64(6):916–921. [DOI] [PubMed] [Google Scholar]
- [33].Hu CM, Chang ZF. Synthetic lethality by lentiviral short hairpin RNA silencing of thymidylate kinase and doxorubicin in colon cancer cells regardless of the p53 status. Cancer Res. 2008;68(8):2831–2840. [DOI] [PubMed] [Google Scholar]
- [34].Hu CM, Chang ZF. Mitotic control of dTTP pool: a necessity or coincidence? J Biomed Sci. 2007;14(4):491–497. [DOI] [PubMed] [Google Scholar]
- [35].Zhao H, Zhang Y, Sun J, et al. Raltitrexed inhibits HepG2 cell proliferation via G0/G1 cell cycle arrest. Oncol Res. 2016;23(5):237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


