ABSTRACT
Intervertebral disc degeneration (IVDD) is a complicated pathological condition accompanying with low back pain. This study was designed to figure out the mechanism of lncRNA XIST in IVDD. Abnormally expressed lncRNAs in IVDD patients were measured. The correlations among XIST, miR-19 and PTEN were identified. Overexpression and silencing of XIST, miR-19 and PTEN were introduced and their roles in NPC autophagy in vitro were detected. The potential signaling pathway involved in these events was identified. Consequently, high expression of XIST was found in IVDD patients. It induced NPC autophagy and reduced NPC viability. XIST could serve as a competing endogenous RNA (ceRNA) for miR-19 and upregulate PTEN expression. The overexpression of XIST reduced miR-19 expression, which was followed by enhanced PTEN expression. Upregulation of miR-19 increased NPC viability and proliferation, while decreased NPC autophagy that regulated by XIST, while overexpressed PTEN reversed the above changes. Moreover, overexpression of XIST inactivated the PI3k/Akt signaling pathway.
KEYWORDS: Intervertebral disc degeneration, long non-coding RNA XIST, microRNA-19, PTEN, competing endogenous RNA, nucleus pulposus cells, autophagy
Introduction
Low back pain (LBP) is one of the commonest complicated disorders and a main cause of disability worldwide [1], while intervertebral disc degeneration (IVDD) is a main cause of LBP [2,3]. IVDD is a complex disease of the spine, resulting in musculoskeletal impairment and reduced life quality for patients [4]. Recently, the prevalence of IVDD has risen every year with the changes of human lifestyle and working habits, affecting human body health and bringing huge economic burdens to the modern society [5]. However, there are still boundaries in the clinical treatment of IVDD since the current treatments can only partially alleviate the symptoms but cannot thoroughly reverse the trend of IVDD [6,7]. Hence, an intensive understanding of the pathology of IVDD and developing novel therapeutic options for IVDD is urgently needed.
Non-coding RNAs, including long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), are diverse transcriptional outcomes of mammalian genomes which are unaccompanied by protein-coding function and are participating in diverse processes coordinating gene expression [8]. lncRNAs have been well recognized as key regulators in diverse biological functions such as genome imprinting, gene expression, chromatin modification,and epigenetic regulation [9]. miRNAs are a class of short noncoding RNAs composed of 20 ~ 25 nucleotides that work as negative regulators of post-transcription gene expression by binding to target mRNAs and thus contributing to transcript degradation [10]. Recently, emerging studies have suggested that lncRNAs [6,9] and miRNAs [11,12] are implicated in the pathogenesis and growth of IVDD. Interestingly, it has been demonstrated that lncRNAs can serve as competing endogenous RNAs (ceRNAs) for miRNAs and reverse the repressive effects of the miRNAs on other transcripts [13,14]. A small number of ceNRA networks have been raised in IVDD development [15]. For instance, lncRNA HOTAIR was demonstrated to reduce nucleus pulposus cell (NPC) apoptosis and to alleviate IVDD via sponging miR-34a-5p [16]. LncRNA SNHG1 was suggested to promote NPC proliferation through the SNHG1/miR-326/CCND1 network [17]. However, unlike the large number of investigations other diseases, the ceRNAs concerning progression of IVDD has just begun with largely uncovered. Taken together, this study was designed to figure out a potential ceRNA network implicated in IVDD progression. Differentially expressed lncRNAs were identified using microarray analysis, and then the possible target molecules and pathways were predicted and validated through a couple of cell experiments.
Material and methods
Ethics statement
The study got the approval of the Clinical Ethical Committee of the First People’s Hospital of Yongkang (Approval No. 201706–016). Signed informed consent was acquired from each eligible participant.
Clinical samples collection
A total of 36 intervertebral disc (IVD) tissue samples from IVDD patients who were diagnosed and admitted in the First People’s Hospital of Yongkang and another 12 samples from non-IVDD patients who underwent surgeries for other causes from September 2017 to March 2018 were collected. Involved subjects were 24 male cases and 24 female cases and their detail baseline clinical characteristics were provided in Table 1.
Table 1.
Baseline clinical information of the included patients
| Patients No. | Diagnosis | Disc level | Modified Pfirrmann grade | Gender | Age |
|---|---|---|---|---|---|
| 1 | Lumbar disc herniation | L5-S1 | 1 | M | 51 |
| 2 | Lumbar disc herniation | L3-L4 | 5 | M | 39 |
| 3 | Spinal stenosis | L3-L4 | 1 | F | 62 |
| 4 | Lumbar disc herniation | L5-S1 | 2 | M | 49 |
| 5 | Spondylolisthesis | L4-L5 | 2 | M | 58 |
| 6 | Lumbar disc herniation | L4-L5 | 1 | F | 52 |
| 7 | Spinal stenosis | L3-L4 | 2 | F | 59 |
| 8 | Spinal stenosis | L3-L4 | 3 | M | 53 |
| 9 | Spondylolisthesis | L5-S1 | 4 | M | 56 |
| 10 | Lumbar disc herniation | L4-L5 | 5 | M | 46 |
| 11 | Spondylolisthesis | L4-L5 | 4 | M | 51 |
| 12 | Lumbar disc herniation | L4-L5 | 3 | M | 55 |
| 13 | Spinal stenosis | L5-S1 | 2 | F | 45 |
| 14 | Spinal stenosis | L3-4 | 4 | M | 54 |
| 15 | Lumbar disc herniation | L3-L4 | 1 | M | 47 |
| 16 | Lumbar disc herniation | L4-L5 | 3 | M | 52 |
| 17 | Spondylolisthesis | L4-L5 | 2 | F | 53 |
| 18 | Lumbar disc herniation | L3-L4 | 5 | F | 59 |
| 19 | Spondylolisthesis | L4-L5 | 3 | F | 61 |
| 20 | Lumbar disc herniation | L4-L5 | 2 | M | 52 |
| 21 | Spondylolisthesis | L4-L5 | 1 | F | 45 |
| 22 | Lumbar disc herniation | L5-S1 | 5 | F | 44 |
| 23 | Lumbar disc herniation | L3-L4 | 2 | F | 39 |
| 24 | Lumbar disc herniation | L4-L5 | 4 | M | 53 |
| 25 | Spondylolisthesis | L4-L5 | 2 | F | 51 |
| 26 | Lumbar disc herniation | L3-L4 | 5 | F | 59 |
| 27 | Lumbar disc herniation | L4-L5 | 3 | F | 60 |
| 28 | Spondylolisthesis | L3-L4 | 4 | F | 50 |
| 29 | Lumbar disc herniation | L5-S1 | 3 | M | 48 |
| 30 | Spinal stenosis | L3-L4 | 5 | F | 46 |
| 31 | Spinal stenosis | L3-L4 | 3 | F | 52 |
| 32 | Spondylolisthesis | L4-L5 | 4 | M | 43 |
| 33 | Lumbar disc herniation | L5-S1 | 5 | F | 52 |
| 34 | Lumbar disc herniation | L4-L5 | 3 | M | 51 |
| 35 | Spondylolisthesis | L4-L5 | 4 | M | 55 |
| 36 | Lumbar disc herniation | L4-L5 | 5 | M | 42 |
| 37 | Vertebral fracture | L3 | 1 | M | 38 |
| 38 | Vertebral fracture | L4 | 1 | M | 34 |
| 39 | Vertebral fracture | L4, L5 | 1 | F | 41 |
| 40 | Vertebral fracture | L2 | 1 | M | 27 |
| 41 | Vertebral fracture | L1 | 1 | F | 22 |
| 42 | Vertebral fracture | L4 | 1 | M | 43 |
| 43 | Vertebral fracture | L5 | 1 | M | 35 |
| 44 | Vertebral fracture | L2-L3 | 1 | M | 37 |
| 45 | Vertebral fracture | L3 | 1 | M | 33 |
| 46 | Vertebral fracture | L3 | 1 | F | 26 |
| 47 | Vertebral fracture | L3 | 1 | M | 39 |
| 48 | Vertebral fracture | L4 | 1 | F | 30 |
Microarray analysis
A microarray analysis was performed as per a previous report [18] to measure the differentially expressed lncRNAs in IVDD and normal tissues. Total RNA of lumbar disc tissues from 3 healthy controls and 3 IVDD patients was extracted. Next, 0.5 μg RNA was used for cDNA synthesis using a GeneChip 3'Invitro Transcription Express Kit (902789, Thermo Fisher Scientific Inc., Waltham, MA, USA). Then the cDNA was fragmented and hybridized with a human lncRNA expression array (AS-LNC-H-V4.0, Arraystar Inc., Rockville, MD, USA). Thereafter, the microarray was washed and scanned using a GeneChipTM Scanner 3000 7 G system (000213, Thermo).
Construction of XIST and PTEN overexpressing or silencing vectors
After amplification, the cDNA of XIST and PTEN was cloned to construct pCDNA3.1-XIST or pCDNA3.1-PTEN vectors (Invitrogen Inc., Carlsbad, CA, USA) using specific primers (Sangon Biotech Co., Ltd, Shanghai, China). The XIST-specific small interfering RNA (siRNA) was synthesized by TianGen Biotech Co., Ltd. (Beijing, China), and the corresponding pcDNA3.1 empty vector and scramble siRNA were constructed as negative controls (NC).
RT-qPCR
Total RNA of cells and tissues was collected using TRIzol LS Reagent (Takara Bio, Otsu, Shiga, Japan). Formaldehyde denaturing gel electrophoresis was applied to identify qualified RNA for subsequent experiments. Next, RNA reverse transcription PCR (RT-PCR) was conducted as per the instructions of a Prime ScriptTM RT assay kit (Takara Bio, Otsu, Shiga, Japan). Quantitation of mRNA was performed using a SYBR Premix Ex Taq (Takara Bio, Otsu, Shiga, Japan)) based RT-qPCR with glyceraldehyde-3-phosphate dehydrogenase set as an internal reference. The primer sequences were shown in Table 2.
Table 2.
Primer sequences for RT-qPCR
| Primer | Sequence (5’- 3’) |
|---|---|
| XIST | F: GAATATGAGTTGTAAGTGGTAG AGTTT |
| R: TACAAACTTAACAAAA AAAAATCATACT | |
| MIR4435-2HG | F: AGGTTGAGAAAGGCGAGACA |
| R: AATTGTGAGGACACCCAAGC | |
| lncRNA:iab8 | F: GCTCCTGGTTCCTCTTGGTC |
| R: GGTTGAGTCAAGTTGCGCTG | |
| ATXN8OS | F: GCGCCGAATTCATCCTTCACCTGTT |
| R: CAAAAGCTTCTCAGCAGCCAGCCA | |
| ceRNA2 | F: CTCAATCCGGGAGCCATGTT |
| R: CTGGGTTTAGGCCAGACAGG | |
| NKILA | F: AACCAAACCTACCCACAACG |
| R: ACCACTAAGTCAATCCCAGGTG | |
| Linc00511 | F: CGCAAGGACCCTCTGTTAGG |
| R: GAAGGCGGATCGTCTCTCAG | |
| miR-19 | F: GCGTGTGCAAATCTATGCAA |
| R: GTGCAGGGTCCGAGGT | |
| U6 | F: GCTTCGGCAGCACATATACTAA |
| R: AACGCTTCACGAATTTGCGT | |
| β-actin | F: ACAGTCAGCCGCATCTTCTT |
| R: GACAAGCTTCCCGTTCTCAG |
Note: RT-qPCR, reverse transcription-quantitative polymerase chain reaction; XIST, X Inactive-Specific Transcript; lncRNA, long non-coding RNA; miR, microRNA; ATXN8OS, ataxin 8 opposite strand; NKILA, NF-ĸB interacting long noncoding RNA; F, forward; R, reverse.
NPC culture and transfection
Rat NPCs purchased from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academic of Science (Shanghai, China) were cultured in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum (FBS) (all purchased from Gibco Company, Grand Island, NY, USA) for 48 h and then passaged. Thereafter, well-growing NPCs of passage 3 were transfected with pCDNA3.1-XIST, pCDNA-3.1 empty vector, scramble-siRNA, XIST-siRNA-1, XIST-siRNA-2, and XIST-siRNA-3, respectively. Forty-eight hours after transfection, the RNA of cells was extracted and the transfection efficacies were evaluated based on the XIST expression measured by RT-qPCR, and then the cells were harvested for the subsequent experiments. Next, the NPCs which transfected with pCDNA3.1-XIST, or XIST-siRNA vectors were further transfected with miR-NC, miR-19 mimics, pCDNA3.1 empty vector or pCDNA3.1-PTEN (each 100 ng/mL), respectively, after which the transfections were identified as aforementioned, and the cells were collected.
Senescence-associated β-galactosidase (SA-β-gal) staining
The activity of β-galactosidase in NPCs was evaluated using a SA‐β‐gal staining kit (Beyotime Biotechnology Co. Ltd., Chengdu, Sichuan, China) following the kit’s instructions. The staining was observed under an inverted microscope (Nikon) with 5 fields randomly selected, under which the senescent cells were stained in blue-green.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
The viability of NPCs was measured as per the instructions of an MTT assay kit (SSigma–AldrichChemical Company, Shanghai, China), and the optical density (OD) value at 490 nm was evaluated using a microplate reader (Multiscan, Thermo).
Immunofluorescence assay
NPCs cultured on cover glasses were rinsed 3 times with phosphate buffer saline (PBS), fixed in 4% polyformaldehyde at 4°C for 15 min, and treated with 0.5% Triton-100 X for 20 min. Next, the cells were incubated with the primary antibody against LC3 (1:100, ab48394) at 4°C overnight. Then the cell slides were washed with PBS and incubated with fluorescein isothiocyanate-labeled secondary antibody against goat-anti rabbit (1:5000, ab150088) at 37°C for 1 h. The cells were then counter-stained using 4', 6-diamidino-2-phenylindole and observed under a microscope (DM3000, Leica, Solms, Germany). All the antibodies were purchased from Abcam.
Immunocytochemistry
NPC slides were incubated with the primary antibodies against Col II (1:1000, ab34712, Abcam) and aggrecan (1:5000 ab186414, Abcam) at 4°C overnight. On the second day, the cell slides were washed with PBS and incubated with the secondary antibody against IgG for 30 min and visualized using DAB. The staining was observed under the inverted microscope (Nikon) at 200 × magnification with 5 fields randomly selected.
Western blot analysis
Total proteins in NPCs and IVD tissues were extracted using radio-immunoprecipitation assay cell lysis buffer containing protease inhibitor (Roche Ltd, Basel, Switzerland), and the protein concentration was detected using a bicinchoninic acid kit. Next, the extracted proteins were separated with 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Sangon Biotech Co., Ltd, Shanghai, China) using the semi-dry method. Afterward, the membranes were treated with blocking buffer to block nonspecific bindings and incubated with the primary antibodies against the antigens in Table 3 at 15°C for 1 h. After 3 PBST (PBS + 0 · 1% Tween-20) washes, the membranes were incubated with secondary antibody horseradish peroxidase-labeled goat-anti rabbit for 2 h, and the bands were visualized using enhanced chemiluminescence and imaged using BioSpectrum system (Bio-Rad Inc., Hercules, CA, USA). The signal intensity of target bands was analyzed using ImageJ v1.48 u software (National Institutes of Health, Bethesda, Maryland, USA).
Table 3.
Antibodies used in western blot analysis
| Antibodies | Art. No. | Dilution ratio | Reference |
|---|---|---|---|
| LC3 | ab48394 | 1:2000 | PMID: 30384327 |
| Beclin | ab207612 | 1:2000 | PMID: 30384327 |
| Atg4B | ab154843 | 1:10000 | PMID: 30384327 |
| p62 | ab91526 | 1:100 | PMID: 30384327 |
| β-actin | ab179467 | 1:5000 | PMID: 30384327 |
| PI3K | ab32089 | 1:1000 | PMID: 30384327 |
| p-PI3K | ab182651 | 1:1000 | PMID: 30384327 |
| Akt | ab8805 | 1:500 | PMID: 30384327 |
| p-Akt | ab8933 | 1:500 | PMID: 30384327 |
| PTEN | ab32199 | 1:10000 | PMID: 30313146 |
Note: All antibodies were purchased from Abcam Inc. (Cambridge, MA, USA); LC3, microtubule-associated protein 1 light chain3; PI3K, phosphatidylinositol 3-kinase; p-PI3K, phosphorylated PI3K; Akt, protein kinase B; p-Akt, phosphorylated Akt; PTEN, phosphatase and tensin homologue deleted on chromosome 10.
Transmission electron microscope (TEM) observation
NPCs were successively fixed in a dry environment for 20 min, washed with PBS, incubated in glutaraldehyde for 5 min, washed with distilled water for 8 times (2 min each time), incubated in uranyl oxalate (pH = 7) for 5 min, and then incubated with hydroxypropyl methylcellulose on ice for 10 min. Next, the remaining liquid was discarded and the NPCs were allowed to stand for 5 ~ 10 min, after which the autophagosome formation in NPCs was observed under a TEM (Olympus).
Dual luciferase reporter gene assay
The computer-based miR target detection programs miRcode (http://www.mircode.org/) and TargetScan (http://www.targetscan.org/vert_72/) were used to predict the binding sites between miR-19 and XIST, and between miR-19 and the 3'untranslated region (3'UTR) of PTEN. Then the pCMV-REPORTTM-based luciferase reporter vectors (Thermo) containing wild-type (WT) XIST (pCMV-XIST-WT) or mutated type (MT) XIST at the mutative miR-19 binding sites (pCMV-XIST-MT) and the corresponding pCMV-PTEN-WT and pCMV-PTEN-WT vectors were constructed. The constructed vectors along with either miR-19 mimics or miR NC were delivered into HEK-29 cells (Shanghai Gaining Biotechnology Co., Ltd., Shanghai, China) using LipofectamineTM 2000 (Invitrogen). Cells were lysed 24 h after transfection and then the relative luciferase activity was determined using the dual-luciferase reporter assay system according to the kit’s instructions (Promega Corporation, WI, USA).
RNA pull-down assay
The specific binding relation between RNA-RNA was identified using a Pierce™ Magnetic RNA-Protein Pull-Down Kit (20,164, Thermo) as per the manufacturer’s protocol. Briefly, the cells were lysed and the lysis was filled with RNase-free DNase I (SSigma–Aldrich and then incubated with biotin-labeled miR-19 sequence (1 μg) and streptomycin-coated magnetic beads (SSigma–Aldrich at 4°C for 3 h. The RNA-RNA compound was then detected using RT-qPCR, with cell lysis as Input and miR-19-MT as NC.
Statistical analysis
SPSS 21.0 (IBM Corp. Armonk, NY, USA) was applied for data analysis. Kolmogorov–Smirnov checked data in normal distribution. Measurement data were described as mean ± standard deviation. Differences between every two groups were compared using the unpaired t-test while the differences among multiple groups were analyzed using one-way analysis of variance (ANOVA). Tukey’s multiple comparisons test was used for the pairwise comparison after ANOVA analysis. The correlation analysis between the two groups was performed with Pearson’s correlation coefficient test. Log rank test was used for ppoststatisticalanalysis. P was obtained from a two-tailed test, and p < 0.05 was regarded statistically significant.
Results
LncRNA-XIST is up-regulated in IVDD patients
Aberrant expression of several lncRNAs has been documented to participate in the IVDD progression [2,19]. In line with these studies, we first measured differentially expressed lncRNAs between 10 IVD tissue samples from 5 IVDD patients and 5 non-IVDD patients using microarray analysis with 40,173 lncRNA probes. Consequently, a total of 211 lncRNAs were differentially expressed, using |logFC > 2| and p < 0.05 as screening conditions, among which 116 lncRNAs were upregulated while 95 were downregulated in the IVDD patients. The heatmap is shown in Figure 1a. Among them, lncRNA XIST presented a most significant elevated expression (by 67.6 folds). To validate the accuracy of the analysis outcomes, RT-qPCR was further performed to detect the expression of six randomly selected lncRNAs MIR4435-2HG, lncRNA: iab8, ATXN8OS, CERNA2, NKILA and Linc00511 in the IVD of 36 IVDD patients and 12 controls. Their expressions showed the same trends as microarray analysis (p < 0.05) (Figure 1b). Thereafter, we further assessed lncRNA XIST expression in 36 IVDD patients with different Pfirrmann grades (ranging 1–5) and 12 normal IVD controls using RT-qPCR, which suggested that XIST presented Pfirrmann grade-dependent high expression in IVDD patients (p < 0.05) (Figure 1c).
Figure 1.
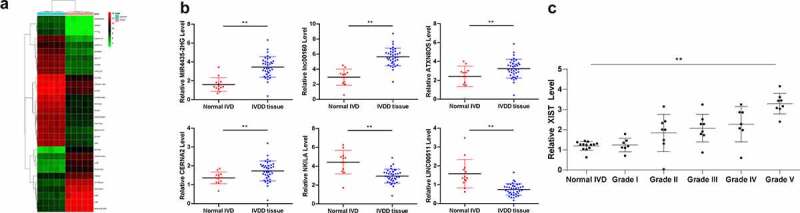
LncRNA XIST is upregulated in IVDD patients in a Pfirrmann grade-dependent manner. (a), heatmap for differentially expressed lncRNAs between 10 IVD tissue samples from 5 IVDD patients and 5 non-IVDD patients using microarray analysis; (b), randomly selected 6 differentially expressed lncRNAs (MIR4435-2HG, lncRNA:iab8, ATXN8OS, CERNA2, NKILA and LINC00511) identified using RT-qPCR; (c), lncRNA XIST expression in 36 IVDD patients with different Pfirrmann grades (1–5) and 12 normal IVD controls (0) measured using RT-qPCR. Data were analyzed by the t test or one-way ANOVA, followed by Tukey’s multiple comparison test; * p < 0.05, compared to the IVD normal controls
Silencing XIST improves NPC viability and inhibits autophagy in vitro
In vitro experiments were performed to investigate the role of XIST in NPCs, where the NPCs were transfected with pCDNA3.1-XIST or XIST-siRNA vectors (si-XIST-1, si-XIST-2, si-XIST-3), respectively. Forty-eight hours later, XIST expression in NPCs was detected using RT-qPCR. As shown in Figure 2a, the transfections were successfully conducted, and among siRNA vectors, siRNA-2 presented the best interference efficiency (p < 0.05).
Figure 2.
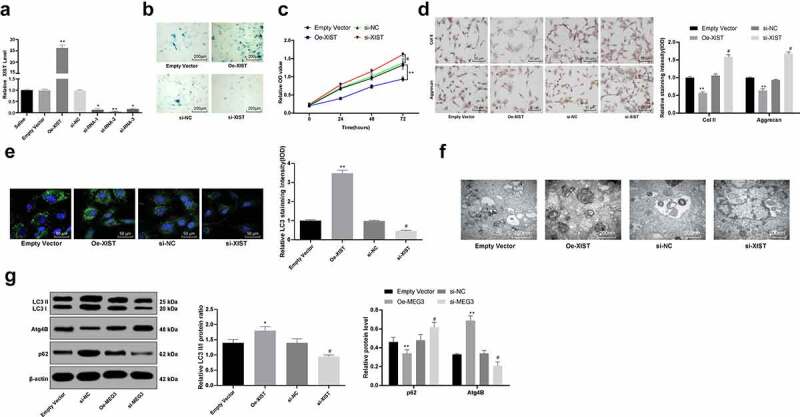
Silencing XIST enhances NPC viability and inhibits autophagy in vitro. (a), XIST expression in NPCs measured using RT-qPCR after transfection; (b), senescence of each group of NPCs detected by SA-β-gal staining; (c), viability of NPCs assessed using MTT assay; (d), levels of Col II and aggrecan in NPCs assessed using immunocytochemistry; (e), LC3 expression in NPCs confirmed by immunofluorescence staining; (f), formation of autophagosomes in NPCs observed under a TEM; (g), protein levels of LC3II, LC3I, p62 and Atg4B evaluated using western blot analysis. Each experiment was performed three times independently. Data are expressed as the mean ± standard deviation (n = 3); Data were analyzed by one-way, followed by Tukey’s multiple comparison test; *, compared to the empty vector group, p < 0.05; # compared to the si-NC group, p < 0.05
The NPC senescence was measured using SA‐β‐gal staining, and the result suggested that overexpression of XIST promoted NPC senescence (p < 0.05) (Figure 2b). Meanwhile, MTT assay was conducted to identify the role of XIST in NPC viability, which suggested that overexpression of XIST inhibited NPC viability, while silencing XIST presented opposite trends (p < 0.05) (Figure 2c-d).
Reduction in Col II and aggrecan secretion by NPCs may exacerbate IVDD [1]. Thus, immunocytochemistry was applied to measure the impact of XIST on NPC functions. As shown in Figure 2e, overexpression of XIST significantly reduced the levels of Col II and aggrecan in NPCs. Correspondingly, silencing XIST led to opposite trends (all p < 0.05). Regarding the autophagy of NPCs in vitro, immunofluorescence staining suggested that overexpression of XIST obviously enhanced LC3 level in NPCs (figure 2f). While the TEM observation showed that the formation of autophagosomes in cells was enhanced by overexpression of XIST (Figure 2g). Likewise, the overexpression of XIST increased LC3II/LC3I and the protein level of Atg4B, but decreased the p62 level according to western blot analysis (Figure 2h), while silencing XIST presented opposite trends (all p < 0.05) (Figure 2e-g).
XIST functions as a ceRNA for miR-19 to regulate PTEN expression
We further explored the potential mechanisms involved in those events. We first looked up the literature and found that lncRNA can often play a role as a ceRNA to competitive binding to miR. The bioinformatics analysis website Starbase (http://starbase.sysu.edu.cn/index.php) predicted that XIST can bind to multiple miRs, such as miR-146, miR-19, miR-34a and miR-27a, many of which are related to the NPCs. miR-34a is obviously expressed in the degenerative NP tissues and cells of IVDD patients, and it can be regulated by HOTAIR to participate in the apoptosis of NPCs [20]. Regulating the expression of miR-146 can affect the repair and regeneration of NPCs [21]. miR-27a may play a promoting role in the development of IDD, and inhibiting miR-27a inhibited the release of proinflammatory factors from intervertebral disc cells through the regulation of p38/MAPK signaling pathway [22]. However, miR-19 has not been reported in the intervertebral disc, so we chose miR-19 for the study. According to this binding sequence between XIST sequence and miR-19 (Figure 3a), the corresponding pCMV-XIST-WT and pCMV-XIST-MT vectors were co-transfected along with either miR-19 mimics or miR-NC into NPCs, and it was found that XIST directly bound to miR-19 (p < 0.05) (Figure 3b). Moreover, this binding relation was further confirmed using RNA pull-down assay (Figure 3c). Besides, the expression of miR-19 presented a negative correlation with the expression of XIST in the IVD tissues of the 36 included IVDD patients according to RT-qPCR (p < 0.05) (Figure 3d). Moreover, the fact that the overexpression of XIST inhibited miR-19 expression (or conversely low-expression of XIST enhanced miR-19 expression) also identified that XIST could sponge miR-19 (p < 0.05) (Figure 3e).
Figure 3.
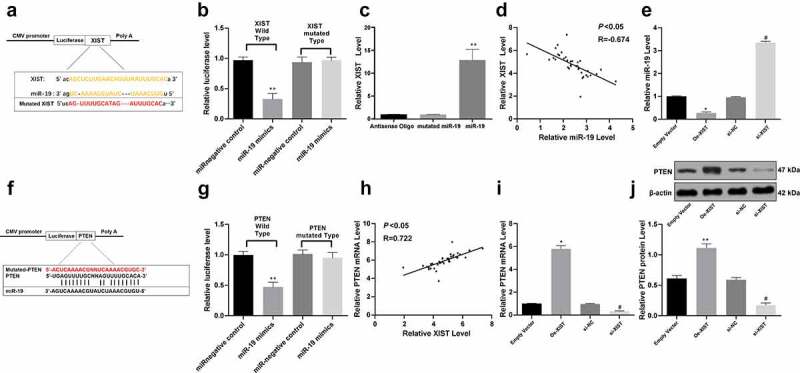
XIST serves as a ceRNA for miR-19 to regulate PTEN expression. (a), binding relation between miR-19 and XIST predicted on a bio-information website (http://www.mircode.org/); (b,c), binding relation between miR-19 and XIST identified by dual luciferase reporter gene assay (b) and RNA-pull down assay (c); (d), plot analysis of the correlation between XIST and miR-19 in 36 IVDD patients; (e), relative expression of miR-19 determined by RT-qPCR; (f), the binding relation between miR-19 and PTEN predicted via TargetScan (http://www.targetscan.org/vert_72/); (g), binding relation between miR-19 and PTEN identified using dual luciferase reporter gene assay; (h), plot analysis of the correlation between miR-19 and PTEN in 36 IVDD patients; (i,j), relative mRNA expression (i) and protein level of PTEN (j) measured using RT-qPCR and western blot analysis, respectively; Each experiment was performed three times independently. Data were analyzed by one-way, followed by Tukey’s multiple comparison test. In panels (d) and (h), Pearson’s correlation coefficient test was utilized for statistical analysis, n = 45, *, compared to the empty vector group, p < 0.05, #, compared to the si-NC group, p < 0.05
Next, the target genes of miR-19 were predicted via the TargetScan online program. Among the abundant potential genes, PTEN aroused our concerns since PTEN plays an important role in the occurrence and development of various diseases and it has been revealed to be correlated with the IVDD development [23]. Meanwhile, miR-19 could bind to the 3'UTR of PTEN according to the TargetScan (figure 3f). Likewise, the pCMV-PTEN-WT and pCMV-PTEN-MT vectors were constructed and co-transfected with either miR-19 mimics or miR-NC into NPCs, which suggested the specific binding between miR-19 and PTEN (p < 0.05) (Figure 3g). Next, the PTEN mRNA expression in 36 IVDD patients was found to be positively correlated with XIST expression (p < 0.05) (Figure 3h). Moreover, artificial overexpression of XIST (or downregulation) enhanced (or correspondingly decreased) the PTEN expression in NPCs according to RT-qPCR and western blot analysis (all p < 0.05) (Figure 3i-j).
Overexpression of miR-19 increases NPC viability reduced by XIST
Next, the NPCs with up- or down-regulated XIST were further transfected with miR-19 mimics, miR-NC, pCDNA3.1-PTEN or pCDNA3.1 empty vector, respectively, and the transfection was successfully performed according to the changes of miR-19 and PTEN expression determined using RT-qPCR (all p < 0.05) (Figure 4a).
Figure 4.
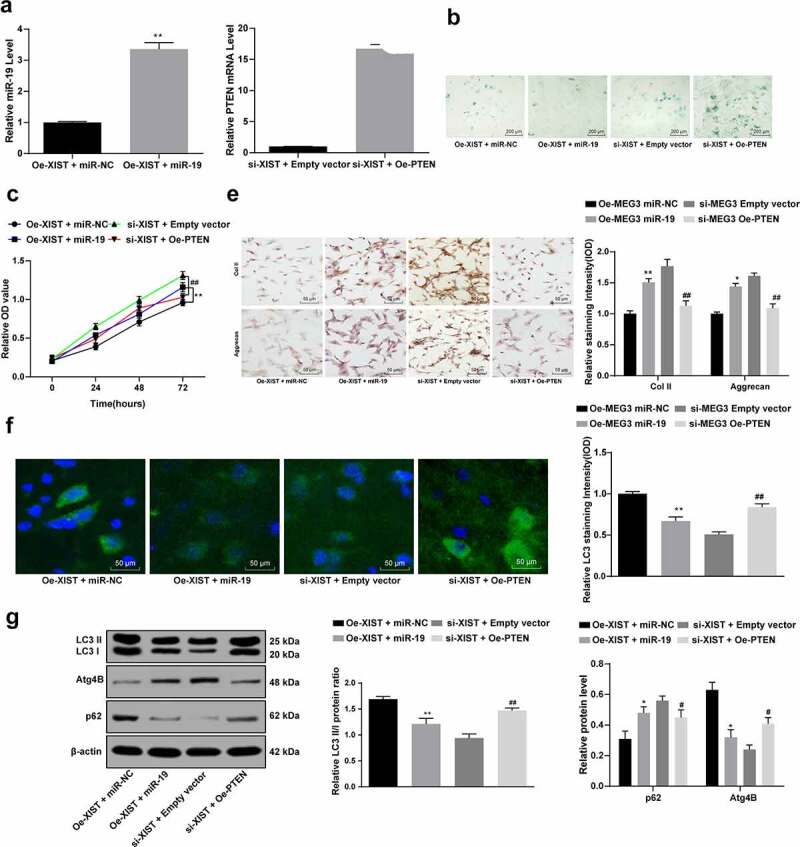
Overexpression of miR-19 increases NPC viability and decreases cell autophagy regulated by XIST. (a), the expression of miR-19 and PTEN mRNA measured using RT-qPCR after transfection; (b), senescence of NPCs detected with SA‐β‐gal staining; (c), viability of NPCs assessed using MTT assay; (d), levels of Col II and aggrecan in NPCs assessed using immunocytochemistry; (e), LC3 expression in NPCs confirmed with immunofluorescence staining; (f), protein levels of LC3I, LC3II, p62 and Atg4B evaluated using western blot analysis. Data are expressed as the mean ± standard deviation. Each experiment was performed three times independently. Data were analyzed by the t test or one-way ANOVA, followed by Tukey’s multiple comparison test. *, compared to the empty vector group, p < 0.05, # compared to the si-NC group, p < 0.05; Repetition = 3
Next, the senescence, viability and autophagy of NPCs were evaluated. SA‐β‐gal staining suggested that overexpression of miR-19 blocked NPC senescence (Figure 4b). MTT assay suggested that miR-19 promoted NPC proliferation (Figure 4c). Moreover, immunocytochemistry results told that overexpression of miR-19 increased the levels of Col II and aggrecan in NPCs (Figure 4d). Immunofluorescence staining suggested that overexpression of miR-19 decreased LC3 level in NPCs (Figure 4e). According to western blot analysis, overexpression of miR-19 decreased LC3II/LC3I and the protein level of Atg4B, but increased the p62 level in NPCs (figure 3f), while the overexpression of PTEN presented opposite trends (all p < 0.05) (Figure 4b-f).
Overexpression of XIST inactivates the PI3K/Akt signaling pathway
Activation of PI3K/Akt axis has been suggested to decrease NPC damage and to alleviate IVDD progression [24], while PTEN was suggested to inactivate the PI3K/Akt pathway [23]. Given the above findings that XIST could promote PTEN expression, we hypothesized that the PI3K/Akt signaling might be involved in the above events. Then the protein levels of PI3K/Akt were measured using western blot analysis, which suggested that overexpressed XIST inactivated the PI3K/Akt signaling pathway in NPCs, evidenced by significantly reduced phosphorylation of p-Akt and p-PI3k (all p < 0.05) (Figure 5a-b).
Figure 5.
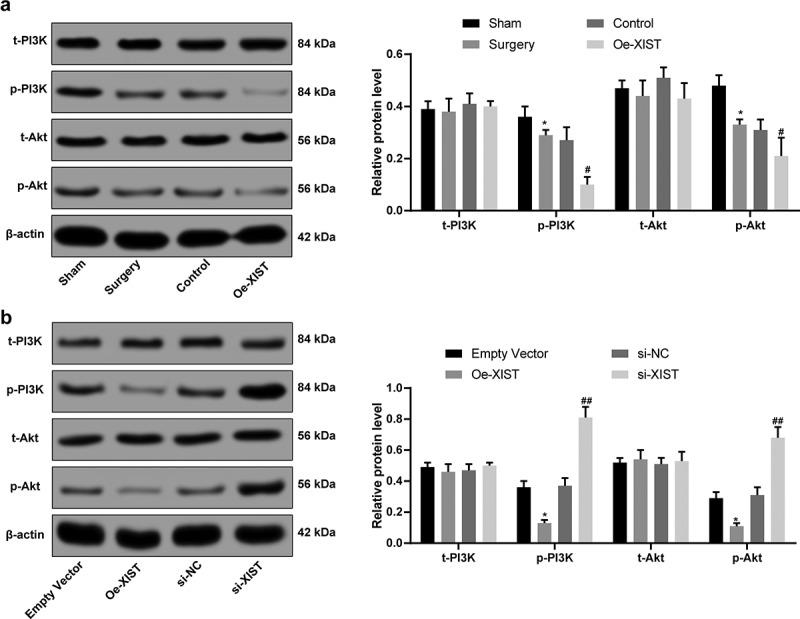
XIST inactivates the PI3K/Akt signaling pathway. (a), PI3K and Akt levels in IVD tissues measured using western blot analysis; (b), protein levels of PI3K and Akt in NPCs measured using western blot analysis; Each experiment was performed three times independently. The data are expressed as the mean ± standard deviation. Data were analyzed by one-way ANOVA, followed by Tukey’s multiple comparison test. *, compared to the sham or empty vector group, p < 0.05, # compared to the control group, p < 0.05, ## compared to the si-NC group, p < 0.01
Discussion
IVDD is a main predisposing factor for LBP which imposes considerable physiological and economic burdens to modern society [25]. In recent years, the lncRNA/miRNA/mRNA ceRNA networks by which lncRNAs communicate with mRNAs by rivaling with shared miRNAs have aroused wide concerns [13,26]. This study was carried out to probe the roles of XIST in the development of IVDD and found that XIST could increase PTEN expression via sponging miR-19, thus promoting IVDD progression with the inactivation of the PI3K/Akt signaling pathway.
Emerging studies suggest that lncRNAs may play critical roles in IVDD [27,28]. The initial finding of the study was that XIST was aberrantly highly expressed in the IVD tissues of IVDD patients. Though the role of XIST in IVDD has never been investigated before, it has been documented to promote apoptosis and inflammatory lesion of microglia cells following spinal cord injury [29]. Moreover, our study found that
silencing of XIST obviously increased levels of Col II and aggrecan, and decreased NPC autophagy with decreased LC3II/LC3I while increased p62 expression in NPCs.During IVDD progress, NPC apoptosis increases, while extracellular matrix (ECM) degrades and aberrant cell proliferation results in cell clusters, and then the level ofvarious inflammatory cytokines elevates [25,30]. Col II and aggrecan are key proteins synthesized by NPCs [31]. The degeneration of the disc results in a shift from Col II to Col I by NPCs and a decline in aggrecan synthesis, leading to dehydrated matrix cells and ultimately loss ofswelling pressure that requiring mechanical support [32]. Autophagy is a type of programmed cell death that has been recognized to be implicated in diverse pathological and physiological processes including the degenerative diseases [33]. A basal level of autophagy is elemental for normal disc cell survival, while during the process of IVDD, the autophagy was aberrantly increased in NP cells and AF cells, with a significant upregulation of autophagy related-proteins including LC3 and Atg12 [34]. Aberrant activation of autophagy has been documented to induce apoptosis of NPCs and to promote IVDD [35,36]. Autophagy is activated in IVDD NP cells [37]. Jiang et al. suggested that senescence and autophagy were upregulated in diabetic IVDD patients, and autophagy may serve as a response mechanism toward the changes of NP cells [38]. Inactivating overdue autophagy has been suggested to reduce the severity of IVDD [39].
The promoting effect of XIST on degenerative NPCs triggered us to find out the underlying mechanisms in it. Our study identified that XIST directly bound to miR-19, while miR-19 negatively targeted the 3'UTR of PTEN. Meanwhile, the overexpression of miR-19 reversed the inhibitory role of XIST in NPC viability, while the upregulation of XIST also enhanced PTEN expression in NPCs. miR-19 has been demonstrated to be downregulated in multiple human replicative and organismal aging models [40]. Meanwhile, a recent study documented the promoting effect of miR-19 on skeletal muscle cell differentiation [41]. PTEN is a well-established major brake of the PI3K/Akt signaling pathway [42]. Likewise, PTEN has been suggested to cause aberrant NPC proliferation by inactivating the Akt signaling pathway [43]. XIST could enhance PTEN expression, while PTEN has been revealed to inactivate the PI3K/Akt axis thus leading to IVDD [23,43]. We measured the PI3K/Akt activation following XIST overexpression, and validated that XIST inactivated the PI3K/Akt signaling pathway. PI3K/Akt activation has been suggested to prevent against IDD via increasing ECM concentration, blocking cell apoptosis, promoting cell pproliferation,and maintaining the balance of cell autophagy [44]. Moreover, inhibition of PTEN or activation of PI3K/Akt has also demonstrated to promote the angiogenesis in the degenerative disc [45]. Herein, it could be inferred that XIST elevates PTEN expression via miR-19, thus leading to PI3k/Akt inactivation and the deterioration of IVDD.
To sum up, this study provided evidence that XIST could work as a ceRNA for miR-19, and the XIST/miR-19/PTEN crosstalk could exacerbate IVDD development via reducing NPC viability, promoting NPC ddegeneration,and inducing NPC autophagy in IVD tissues through the inactivation of PI3K/Akt signaling pathway. Hopefully, this study could provide new insights into the application of ceRNA in IVDD and provide novel therapeutic option for IVDD treatment. However, more studies related in this field are required to identify our findings and to develop more intensive understanding of IVDD prevention and treatment. Due to our current experimental conditions and funding constraints, we have not been able to use different si-XIST#1 and si-XIST #2 both in animal and cell experiments. In the future, we will strive to create sufficient experimental conditions to carry out more in-depth experiments.
Disclosure statement
The authors declared that they have no competing interests.
Availability of data and materials
All the data generated or analyzed during this study are included in this published article.
References
- [1].Zhang Z, Xu T, Chen J, et al. Parkin-mediated mitophagy as a potential therapeutic target for intervertebral disc degeneration. Cell Death Dis. 2018;9:980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Li X, Lou Z, Liu J, et al. Upregulation of the long noncoding RNA lncPolE contributes to intervertebral disc degeneration by negatively regulating DNA polymerase epsilon. Am J Transl Res. 2019;11:2843–2854. [PMC free article] [PubMed] [Google Scholar]
- [3].Sampara P, Banala RR, Vemuri SK, et al. Understanding the molecular biology of intervertebral disc degeneration and potential gene therapy strategies for regeneration: a review. Gene Ther. 2018;25:67–82. [DOI] [PubMed] [Google Scholar]
- [4].Yang Q, Guo XP, Cheng YL, et al. MicroRNA-143-5p targeting eEF2 gene mediates intervertebral disc degeneration through the AMPK signaling pathway. Arthritis Res Ther. 2019;21:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu P, Chang F, Zhang T, et al. Downregulation of microRNA-125a is involved in intervertebral disc degeneration by targeting pro-apoptotic Bcl-2 antagonist killer 1. Iran J Basic Med Sci. 2017;20:1260–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen WK, Yu XH, Yang W, et al. lncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang K, Chen T, Ying X, et al. Ligustilide alleviated IL-1beta induced apoptosis and extracellular matrix degradation of nucleus pulposus cells and attenuates intervertebral disc degeneration in vivo. Int Immunopharmacol. 2019;69:398–407. [DOI] [PubMed] [Google Scholar]
- [8].Wan ZY, Song F, Sun Z, et al. Aberrantly expressed long noncoding RNAs in human intervertebral disc degeneration: a microarray related study. Arthritis Res Ther. 2014;16:465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mi D, Cai C, Zhou B, et al. Long noncoding RNA FAF1 promotes intervertebral disc degeneration by targeting the Erk signaling pathway. Mol Med Rep. 2018;17:3158–3163. [DOI] [PubMed] [Google Scholar]
- [10].Sun L, Zhao M, Wang Y, et al. Neuroprotective effects of miR-27a against traumatic brain injury via suppressing FoxO3a-mediated neuronal autophagy. Biochem Biophys Res Commun. 2017;482:1141–1147. [DOI] [PubMed] [Google Scholar]
- [11].Li Z, Yu X, Shen J, et al. MicroRNA in intervertebral disc degeneration. Cell Prolif. 2015;48:278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhao K, Zhang Y, Kang L, et al. Epigenetic silencing of miRNA-143 regulates apoptosis by targeting BCL2 in human intervertebral disc degeneration. Gene. 2017;628:259–266. [DOI] [PubMed] [Google Scholar]
- [13].Smillie CL, Sirey T, Ponting CP.. Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit Rev Biochem Mol Biol. 2018;53:231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhao Y, Wang H, Wu C, et al. Construction and investigation of lncRNA-associated ceRNA regulatory network in papillary thyroid cancer. Oncol Rep. 2018;39:1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhu J, Zhang X, Gao W, et al. lncRNA/circRNAmiRNAmRNA ceRNA network in lumbar intervertebral disc degeneration. Mol Med Rep. 2019;20:3160–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shao T, Hu Y, Tang W, et al. The long noncoding RNA HOTAIR serves as a microRNA-34a-5p sponge to reduce nucleus pulposus cell apoptosis via a NOTCH1-mediated mechanism. Gene. 2019;715:144029. [DOI] [PubMed] [Google Scholar]
- [17].Tan H, Zhao L, Song R, et al. The long noncoding RNA SNHG1 promotes nucleus pulposus cell proliferation through regulating miR-326 and CCND1. Am J Physiol Cell Physiol. 2018;315:C21–C7. [DOI] [PubMed] [Google Scholar]
- [18].Wang K, Song Y, Liu W, et al. The noncoding RNA linc-ADAMTS5 cooperates with RREB1 to protect from intervertebral disc degeneration through inhibiting ADAMTS5 expression. Clin Sci (Lond). 2017;131:965–979. [DOI] [PubMed] [Google Scholar]
- [19].Wang XB, Wang H, Long HQ, et al. LINC00641 regulates autophagy and intervertebral disc degeneration by acting as a competitive endogenous RNA of miR-153-3p under nutrition deprivation stress. J Cell Physiol. 2019;234:7115–7127. [DOI] [PubMed] [Google Scholar]
- [20].Yu Y, Zhang X, Li Z, et al. LncRNA HOTAIR suppresses TNF-alpha induced apoptosis of nucleus pulposus cells by regulating miR-34a/Bcl-2 axis. Biomed Pharmacother. 2018;107:729–737. [DOI] [PubMed] [Google Scholar]
- [21].Yang RS, Wang YH, Ding C, et al. MiR-146 regulates the repair and regeneration of intervertebral nucleus pulposus cells via Notch1 pathway. Eur Rev Med Pharmacol Sci. 2019;23:4591–4598. [DOI] [PubMed] [Google Scholar]
- [22].Cao Z, Chen L. Inhibition of miR-27a suppresses the inflammatory response via the p38/MAPK pathway in intervertebral disc cells. Exp Ther Med. 2017;14:4572–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang B, Wang D, Yan T, et al. MiR-138-5p promotes TNF-alpha-induced apoptosis in human intervertebral disc degeneration by targeting SIRT1 through PTEN/PI3K/Akt signaling. Exp Cell Res. 2016;345:199–205. [DOI] [PubMed] [Google Scholar]
- [24].Tan Y, Yao X, Dai Z, et al. Bone morphogenetic protein 2 alleviated intervertebral disc degeneration through mediating the degradation of ECM and apoptosis of nucleus pulposus cells via the PI3K/Akt pathway. Int J Mol Med. 2019;43:583–592. [DOI] [PubMed] [Google Scholar]
- [25].Wu X, Liu Y, Guo X, et al. Prolactin inhibits the progression of intervertebral disc degeneration through inactivation of the NF-kappaB pathway in rats. Cell Death Dis. 2018;9:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang H, Niu L, Jiang S, et al. Comprehensive analysis of aberrantly expressed profiles of lncRNAs and miRNAs with associated ceRNA network in muscle-invasive bladder cancer. Oncotarget. 2016;7:86174–86185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ruan Z, Ma H, Li J, et al. The long non-coding RNA NEAT1 contributes to extracellular matrix degradation in degenerative human nucleus pulposus cells. Exp Biol Med (Maywood). 2018;243:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xi Y, Jiang T, Wang W, et al. Long non-coding HCG18 promotes intervertebral disc degeneration by sponging miR-146a-5p and regulating TRAF6 expression. Sci Rep. 2017;7:13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Qin Z, Fei L, Qinglun S, et al. Knockdown of long noncoding RNA XIST mitigates the apoptosis and inflammatory injury of microglia cells after spinal cord injury through miR-27a/Smurf1 axis. Neurosci Lett. 2019;134649. [DOI] [PubMed] [Google Scholar]
- [30].Wang X, Lv G, Li J et al. LncRNA-RP11-296A18.3/miR-138/HIF1A Pathway Regulates the Proliferation ECM Synthesis of Human Nucleus Pulposus Cells (HNPCs). J Cell Biochem 2017; 118:4862–4871 [DOI] [PubMed] [Google Scholar]
- [31].Preradovic A, Kleinpeter G, Feichtinger H et al. Quantitation of collagen I, collagen II and aggrecan mRNA and expression of the corresponding proteins in human nucleus pulposus cells in monolayer cultures. Cell Tissue Res 2005;321:459–464 [DOI] [PubMed] [Google Scholar]
- [32].Urits I, Capuco A, Sharma M et al. Stem Cell Therapies for Treatment of Discogenic Low Back Pain: a Comprehensive Review. Curr Pain Headache Rep 2019; 23:65. [DOI] [PubMed] [Google Scholar]
- [33].Ding F, Shao ZW, Xiong LM. Cell death in intervertebral disc degeneration. Apoptosis. 2013;18:777–785. [DOI] [PubMed] [Google Scholar]
- [34].Zhang SJ, Yang W, Wang C, et al. Autophagy: a double-edged sword in intervertebral disk degeneration. Clin Chim Acta. 2016;457:27–35. [DOI] [PubMed] [Google Scholar]
- [35].Lin Y, Jiao Y, Yuan Y, et al. Propionibacterium acnes induces intervertebral disc degeneration by promoting nucleus pulposus cell apoptosis via the TLR2/JNK/mitochondrial-mediated pathway. Emerg Microbes Infect. 2018;7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Luo R, Liao Z, Song Y, et al. Berberine ameliorates oxidative stress-induced apoptosis by modulating ER stress and autophagy in human nucleus pulposus cells. Life Sci. 2019;228:85–97. [DOI] [PubMed] [Google Scholar]
- [37].Ma KG, Shao ZW, Yang SH, et al. Autophagy is activated in compression-induced cell degeneration and is mediated by reactive oxygen species in nucleus pulposus cells exposed to compression. Osteoarthritis Cartilage. 2013;21:2030–2038. [DOI] [PubMed] [Google Scholar]
- [38].Jiang L, Zhang X, Zheng X, et al. Apoptosis, senescence, and autophagy in rat nucleus pulposus cells: implications for diabetic intervertebral disc degeneration. J Orthop Res. 2013;31:692–702. [DOI] [PubMed] [Google Scholar]
- [39].Jin LY, Lv ZD, Wang K, et al. Estradiol alleviates intervertebral disc degeneration through modulating the antioxidant enzymes and inhibiting autophagy in the model of menopause rats. Oxid Med Cell Longev. 2018;2018:7890291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Grillari J, Hackl M, Grillari-Voglauer R. miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology. 2010;11:501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kong D, He M, Yang L, et al. MiR-17 and miR-19 cooperatively promote skeletal muscle cell differentiation. Cell Mol Life Sci. 2019;76:5041–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Georgescu MM. PTEN tumor suppressor network in PI3K-Akt pathway control. Genes Cancer. 2010;1:1170–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu H, Huang X, Liu X, et al. miR-21 promotes human nucleus pulposus cell proliferation through PTEN/AKT signaling. Int J Mol Sci. 2014;15:4007–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ouyang ZH, Wang WJ, Yan YG, et al. The PI3K/Akt pathway: a critical player in intervertebral disc degeneration. Oncotarget. 2017;8:57870–57881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang H, Wang P, Zhang X, et al. SDF1/CXCR7 signaling axis participates in angiogenesis in degenerated discs via the PI3K/AKT pathway. DNA Cell Biol. 2019;38:457–467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data generated or analyzed during this study are included in this published article.


