Abstract
Activation of Rac1, a member of the Rho family of GTPases, is associated with multiple cellular responses, including membrane ruffling and focal complex formation. The mechanisms by which Rac1 is coupled to these functional responses are not well understood. It was recently shown that ARF6, a GTPase implicated in cytoskeletal alterations and a membrane recycling pathway, is required for Rac1-dependent phagocytosis in macrophages (Q. Zhang et al., J. Biol. Chem. 273:19977–19981, 1998). To determine whether ARF6 is required for Rac1-dependent cytoskeletal responses in macrophages, we expressed wild-type (WT) or guanine nucleotide binding-deficient alleles (T27N) of ARF6 in macrophages coexpressing activated alleles of Rac1 (Q61L) or Cdc42 (Q61L) or stimulated with colony-stimulating factor 1 (CSF-1). Expression of ARF6 T27N but not ARF6 WT inhibited ruffles mediated by Rac1 Q61L or CSF-1. In contrast, expression of ARF6 T27N did not inhibit Rac1 Q61L-mediated focal complex formation and did not impair Cdc42 Q61L-mediated filopodial formation. Cryoimmunogold electron microscopy demonstrated the presence of ARF6 in membrane ruffles induced by either CSF-1 or Rac1 Q61L. Addition of CSF-1 to macrophages led to the redistribution of ARF6 from the interior of the cell to the plasma membrane, suggesting that this growth factor triggers ARF6 activation. Direct targeting of Rac1 to the plasma membrane did not bypass the blockade in ruffling induced by ARF6 T27N, indicating that ARF6 regulates a pathway leading to membrane ruffling that occurs after the activation and membrane association of Rac. These data demonstrate that intact ARF6 function is required for coupling activated Rac to one of several effector pathways and suggest that a principal function of ARF6 is to coordinate Rac activation with plasma membrane-based protrusive events.
Members of the Rho family of GTPases are capable of triggering distinct cytoskeletal responses in a variety of cell types. For example, activated alleles of Rac trigger membrane ruffling in fibroblasts (39) and macrophages (2, 15) while activated alleles of Cdc42 trigger filopodium formation in fibroblasts (26). In contrast, activated Rho induces stress fiber formation in fibroblasts (39). The molecular requirements underlying these distinct shape changes are only partly understood. Rho-dependent stress fiber formation occurs through activation of Rho-kinase by Rho-GTP (3, 27). Activated Rho-kinase phosphorylates myosin II and myosin phosphatase, activating the former and inactivating the latter (4, 25). Cdc42-mediated filopodial formation requires the participation of N-WASP, a member of the Wiskott-Aldrich Syndrome Protein (WASP) family, which induces free barbed ends of actin filaments in the presence of Cdc42-GTP (31). For Rac1, induction of membrane ruffles is accompanied by activation of LIM-kinase 1, which phosphorylates cofilin at Ser 3, leading to a reversal of cofilin-induced actin depolymerization (6, 47). Actin reorganization by Rac and Cdc42 may also require participation of WAVE/Scar1, a recently identified WASP family member that interacts with the p21 subunit of the Arp 2/3 complex (29) and induces actin assembly through profilin (32). All three Rho family members are capable of binding phosphoinositide kinases (14, 42) that may promote cytoskeletal assembly by inducing actin filament uncapping (23). However, Rac-induced uncapping may also be phosphoinositide independent (7), and many other components of the Rac effector response have yet to be defined.
ARF6 is a brefeldin A-insensitive member of the ARF family of GTPases (12, 35, 37). In HeLa cells overexpressing ARF6, incubation with aluminum fluoride, an activator of heterotrimeric G proteins, leads to formation of membrane protrusions enriched in both ARF6 and F-actin (37). In CHO cells, expression of an activated allele of ARF6 leads to F-actin-rich plasma membrane protrusions (18). These studies suggest that ARF6 participates in cytoskeletal assembly, although the spectrum of receptors to which it is coupled and its precise mechanism of action are unknown. In the latter study, evidence was presented that ARF6 triggers cytoskeletal changes independently of Rac, and Rac-mediated cytoskeletal alterations appeared to be independent of ARF6 function (18).
Studies by Radhakrishna and colleagues (36) and D’Souza-Schorey and colleagues (17) support a model of ARF6 function in which the GTPase cycles between the plasma membrane and an intracellular recycling compartment, depending on its nucleotide status. In HeLa (36) and CHO (17) cells, expression of ARF6 T27N, a guanine nucleotide binding-deficient allele, leads to its accumulation in a pericentriolar vesicular compartment, whereas expression of ARF6 Q67L, a GTPase-deficient binding allele, leads to accumulation of the GTPase at the plasma membrane or to membrane invaginations contiguous with the plasma membrane. ARF6 participates in a recycling pathway of a subset of transmembrane receptors, since expression of ARF6 T27N leads to inhibition of the reappearance at the cell surface of receptors lacking AP-2 binding sequences, following their downregulation by monoclonal antibodies (MAbs) (37). In addition, ARF6 colocalizes with cellubrevin in an intracellular vesicular compartment, suggesting that this compartment may fuse with syntaxin-containing compartments, such as the plasma membrane (17).
We recently demonstrated that normal guanine nucleotide cycling on ARF6 is required for FcγR-mediated phagocytosis in macrophages (48). Inhibition of either Rac1 or ARF6 function produced the same phenotype in macrophages challenged with immunoglobulin G (IgG)-coated particles: ablation or marked impairment of focal F-actin accumulations beneath nascent pseudopods (15, 48). These results suggest that, in macrophages, ARF6 and Rac1 may lie on the same pathway leading to cytoskeletal alterations and pseudopodial formation. Since inhibition of Rac1 function leads to impaired cytoskeletal responses mediated by multiple structurally unrelated surface receptors in macrophages (15), all of which result in plasma membrane-based protrusions, we wondered whether ARF6 participates in Rac-dependent cytoskeletal responses in general. In this study, we addressed whether inhibition of ARF6 function affects functional responses of Rac1 and Cdc42 in macrophages.
MATERIALS AND METHODS
Cells and reagents.
RAW LacR/FMLPR.2 cells, a clone derived from the RAW 246.7 cell line (American Type Culture Collection, Bethesda, Md.), were maintained in RPMI medium containing 10% fetal bovine serum, 100 U of penicillin G per ml, and 100 μg of streptomycin per ml at 37°C in a CO2 incubator. Rabbit antiserum against ARF6 and pXS vectors containing various ARF6 constructs were from Julie Donaldson (National Heart, Lung, and Blood Institute, Bethesda, Md.). Myc-Rac1 Q61L and Myc-Cdc42 Q61L in pRK5 were from Gary Bokoch (Scripps Research Institute, La Jolla, Calif.). 16:7:Syk in pCDM8 was from Brian Seed (Massachusetts General Hospital, Boston, Mass.). Rhodamine-phalloidin was from Molecular Probes (Eugene, Oreg.). MAb 9E10 against Myc was from Oncogene Science (Cambridge, Mass.). MAb 12CA5 against hemagglutinin (HA) was from Boehringer Mannheim (Indianapolis, Ind.). Rabbit antiserum against HA was from Medical and Biological Laboratories (Nagoya, Japan). F(ab′)2 fragments of fluorescein isothiocyanate-, AMCA-, and rhodamine-conjugated anti-rabbit and anti-mouse IgG were from Jackson ImmunoResearch (West Grove, Pa.). Rabbit anti-mouse IgG was from DAKO (Carpinteria, Calif.). Gold-conjugated goat anti-rabbit and anti-mouse IgG were from Amersham Nederland (’s-Hertogenbosch, The Netherlands). Superfect transfection reagent was from Qiagen (Santa Clarita, Calif.).
A plasma membrane-targeted fusion protein consisting of the extracellular domain of human CD16, the transmembrane domain of CD7, and an intracellular domain consisting of Myc-Rac1 Q61L was constructed by PCR. A MluI site was engineered at the 5′ end of Myc-Rac1 Q61L and ligated in frame with 16:7:Syk in pCDM8 digested with MluI and ApaI to remove the Syk coding sequence. All other constructs were transfected as noted above, except Myc-Cdc42 Q61L was first subcloned into pCMV3R (15). A fusion protein containing maltose binding protein and POR1 was kindly provided by Crislyn D’Souza Schorey (University of Notre Dame, Notre Dame, Ind.).
Fluorescence microscopy.
Adherent RAW LacR/FMLPR.2 cells were transfected with the indicated constructs. Sixteen hours following transfection, cells were serum starved for 1 h and incubated in the presence or absence of 10 ng of colony-stimulating factor 1 (CSF-1) per ml for 5 min at 37°C and fixed in 0.1 M phosphate buffer containing 3.7% formaldehyde. Indirect immunofluorescence was performed as described (48). A ruffling index was quantitated as described (48); the index varied from 0 (no ruffling present) to 200 (diffuse ruffling over 75% or more of the dorsal surfaces of 100 adherent macrophages). Filopodium formation was quantitated in an analogous fashion (i.e., each cell was assigned a score of 0 (three or fewer filopodia), 1 (three to six filopodia), or 2 (more than six filopodia), and the filopodium index was calculated as the sum of the filopodium scores of 100 cells. For each experiment, at least 50 to 100 cells in five or more microscope fields were scored. All cells in a field were independently scored for the presence of ruffling and filopodia.
Cryoimmunogold electron microscopy.
Adherent RAW LacR/FMLPR.2 cells were transfected with the indicated constructs. Sixteen hours following transfection, cells were serum starved for 1 h and incubated in the presence or absence of 10 ng of CSF-1 per ml for 5 min at 37°C. Transfected cells were fixed for 2 h in a mixture of 0.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.2) and pelleted in 10% (wt/vol) gelatin in phosphate-buffered saline. Ultrathin frozen sections were incubated at room temperature with the indicated antibodies and 5- and/or 10-nm-diameter gold conjugates as described previously (11). In cells transfected with the HA-ARF6 wild type (WT) alone, cryosections were incubated with polyclonal anti-HA followed by anti-rabbit IgG–10-nm-diameter gold. In cells transfected with Myc-Rac Q61L alone, cryosections were incubated with mouse anti-Myc and anti-mouse IgG–10-nm-diameter gold. In cells cotransfected with Myc-Rac Q61L and HA-ARF6 WT, cells were labeled sequentially with mouse anti-HA, rabbit anti-mouse IgG, and anti-rabbit IgG–5-nm-diameter gold, incubated with 1% glutaraldehyde for 10 min followed by incubation with mouse anti-Myc and anti-mouse IgG–10-nm-diameter gold. After immunolabeling, cryosections were embedded in a mixture of methylcellulose and uranyl acetate and examined with a CM10 electron microscope (Philips, Eindhoven, The Netherlands).
Quantitation of immunogold labeling.
We quantitated the relative distribution of HA-ARF6 in ruffling and nonruffling areas of plasma membrane in cells transfected with HA-ARF6 WT and incubated in the presence or absence of CSF-1. We also quantitated the relative distribution of Myc-Rac Q61L in ruffling and nonruffling areas of plasma membrane in cells transfected with Myc-Rac Q61L alone or together with ARF6 T27N. In cells transfected with HA-ARF6 WT alone, cross sections of cells that were lightly labeled with rabbit polyclonal anti-HA and anti-rabbit IgG–10-nm-diameter gold were chosen and photographed. The number of gold particles associated with 3 to 7 μm of ruffling plasma membrane and the same lengths of adjacent, nonruffling regions of plasma membrane in each cell were counted. Because we could not obtain a cotransfection efficiency of Myc-Rac Q61L and HA-ARF6 T27 that was sufficiently high for quantitation, we transfected cells with either Myc-Rac Q61L alone or Myc-Rac Q61L together with ARF6 T27N. Parallel immunofluorescence studies demonstrated that the cotransfection efficiency of Myc-Rac Q61L and ARF6 T27N was >90%. In cells transfected with Myc-Rac Q61L alone or together with ARF6 T27N, cross sections of cells that were lightly labeled with 5-nm-diameter gold particles were chosen and photographed.
We also evaluated the relative plasma membrane enrichment of (i) HA-ARF6 on cryosections of cells transfected with HA-ARF6 and incubated in the presence or absence of CSF-1 and (ii) Myc-Rac Q61L in cells expressing both Myc-Rac Q61L and ARF6 T27N. To determine the relative labeling of HA-ARF6 associated with either the plasma membrane or the cytosol and intracellular vesicles, gold particles associated with 7.7 μm of plasma membrane were counted and gold particles associated with 7.25 μm2 of cytosol and associated intracellular vesicles were counted. These numbers were chosen for ease of counting. The resultant ratio was calculated and defined as the plasma membrane labeling index. This parameter thus normalized for cell-to-cell variations in either levels of expression of the individual constructs or cell volume and surface areas.
For all quantitative studies, n refers to the number of cells quantitated. Student t tests were performed on quantitative immunogold data.
RESULTS
Effects of expression of a guanine nucleotide binding-deficient allele of ARF6 on Rac1- and Cdc42-mediated cytoskeletal changes in macrophages.
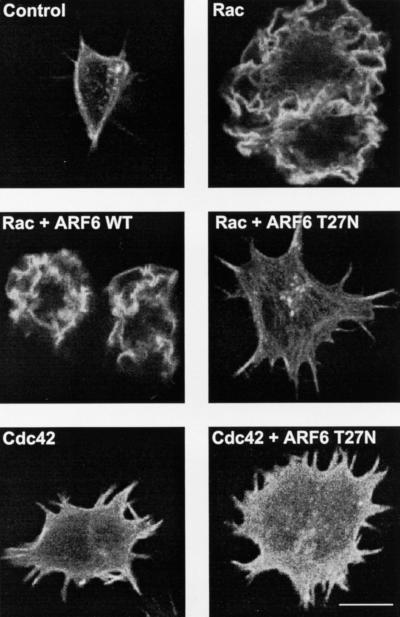
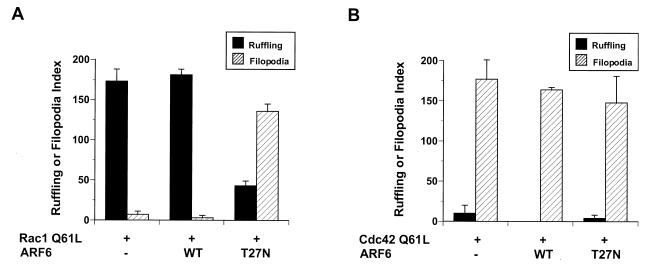
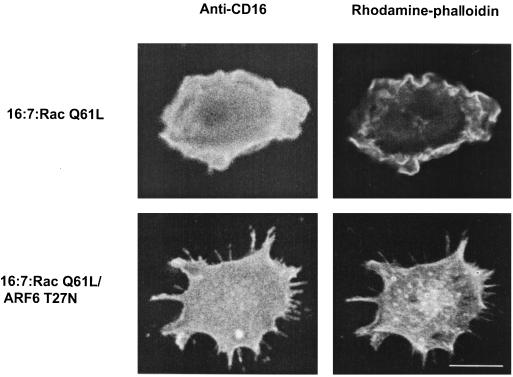
Untransfected cells displayed no membrane ruffles and contained only rudimentary filopodia. Expression of Rac1 Q61L led to the appearance of prominent membrane ruffles over the entire dorsal surfaces of RAW LacR/FMLPR.2 cells. Coexpression of ARF6 did not alter the appearance of the Rac-mediated ruffles. In contrast, coexpression of ARF6 T27N led to cell spreading, the conspicuous absence of membrane ruffles, and an increase in the number and prominence of filopodia present in untransfected cells (Fig. 1). This appearance resembled that of cells transfected with Cdc42 Q61L (Fig. 1) and was due to coexpression of both Rac and ARF6 T27N, because expression of ARF6 T27N alone in these cells did not lead to altered morphology (reference 47 and data not shown). Expression of ARF6 T27N did not impair the induction of filopodia in cells transfected with Cdc42 Q61L (Fig. 1). The inhibition of Rac1-mediated ruffling but not Cdc42-mediated filopodium formation by ARF6 T27N (Fig. 2) indicates that inhibition of ARF6 function specifically affects Rac1- rather than Cdc42-mediated shape changes in macrophages.
FIG. 1.
Effect of ARF6 T27N on Rac1- and Cdc42-mediated cytoskeletal alterations in macrophages. Adherent RAW LacR/FMLPR.2 cells transfected with Myc-Rac1 Q61L (Rac), Myc-Cdc42 Q61L (Cdc42), ARF6 WT, or ARF6 T27N were fixed and stained with a MAb against Myc (to detect either Rac or Cdc42), rabbit antiserum against ARF6, and rhodamine-phalloidin (to detect F-actin). Confocal fluorescence micrographs showing rhodamine-phalloidin fluorescence micrographs of representative cells are shown. Bar = 10 μm.
FIG. 2.
Quantitation of membrane ruffling or filopodia formation of Rac1 Q61L (A)- or Cdc42 61L (B)-expressing macrophages also expressing the indicated ARF6 alleles. Data represent the means ± SEM (n = 4).
Rac1-mediated focal complex formation is not inhibited by expression of a guanine nucleotide binding-deficient allele of ARF6.
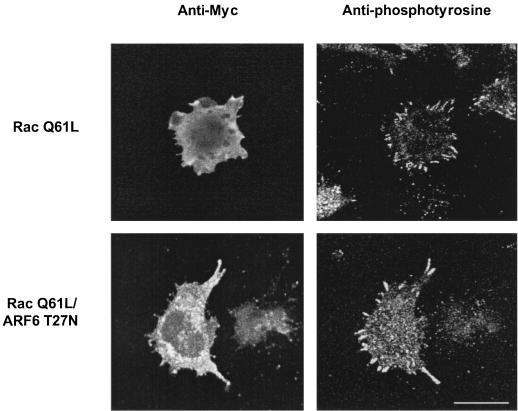
The above data support a role for ARF6 in Rac-mediated cytoskeletal alterations that culminate in membrane ruffling. To determine whether another function of Rac is affected by inhibition of ARF6, we examined the effect of expression of ARF6 T27N on Rac-induced focal complex formation. Expression of an activated allele of Rac1 induced the formation of focal complexes. These were readily apparent in cells stained for phosphotyrosine, which accumulates in these structures. Rac1 induced the appearance of well-formed focal complexes which were otherwise rudimentary or absent in macrophages not expressing the activated Rac allele (Fig. 3). In contrast to Rac-induced focal complexes in Swiss 3T3 fibroblasts (34), the morphology of Rac-induced focal complexes in macrophages was less punctuate and resembled focal adhesions in other cell types, confirming results in another macrophage cell line (2). Coexpression of ARF6 T27N did not inhibit the appearance of these structures and often accentuated them (Fig. 3). Thus, expression of ARF6 T27N inhibits Rac-induced membrane ruffling but not Rac-mediated focal complex formation in macrophages.
FIG. 3.
Effect of ARF6 T27N on focal complex formation induced by Rac1 Q61L in macrophages. Adherent RAW LacR/FMLPR.2 cells transfected with the indicated constructs were fixed and stained with a MAb against Myc (to detect Myc-Rac1 Q61L), rabbit antiserum against ARF6 (to detect cells expressing ARF6 T27N), and MAb PY-99 (to detect phosphotyrosine). Representative confocal micrographs showing Myc (left panels) and PY-99 staining (right panels) are shown. Note prominent phosphotyrosine-rich focal complexes in cells expressing Myc-Rac1 Q61L but not in surrounding nonexpressors. Bar = 10 μm.
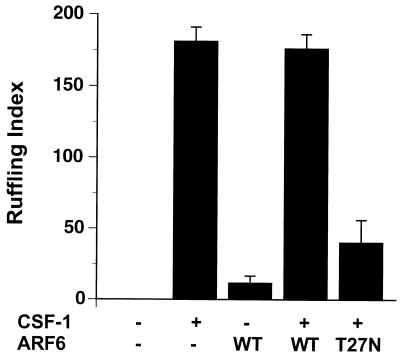
Inhibition of ARF6 function blocks Rac1-dependent ruffling by CSF-1.
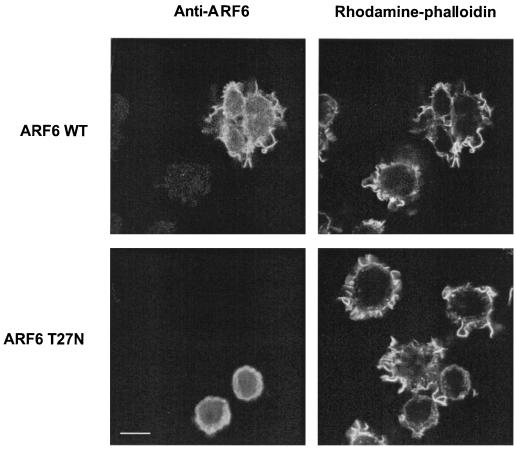
c-Fms, the receptor for CSF-1 on myeloid cells, is structurally homologous to the platelet-derived growth factor receptor. Both are receptor tyrosine kinases that trigger the formation of membrane ruffles when ligated by their respective ligands (9, 29, 32). Membrane ruffling induced by either platelet-derived growth factor or CSF-1 is inhibited by expression of guanine nucleotide binding-deficient alleles of Rac1 (2, 15, 39) or expression of a GAP domain for Rac1 (15). To determine whether intact ARF6 function is required for membrane ruffling mediated by a physiological stimulus in macrophages, we expressed either ARF6 or ARF6 T27N in RAW LacR/FMLPR.2 cells and determined the extent of membrane ruffling induced by CSF-1. Addition of CSF-1 induced the formation of F-actin-rich membrane ruffles in nearly all of the untransfected control macrophages (reference 15 and data not shown). Expression of ARF6 T27N led to inhibition of CSF-1-induced membrane ruffling, whereas expression of ARF6 WT did not affect the extent of CSF-1-induced membrane ruffling (Fig. 4 and 5). Thus, in mouse macrophages, intact ARF6 function is required for Rac1-dependent cytoskeletal alterations mediated by at least two structurally unrelated receptors, FcγR III (15) and c-Fms.
FIG. 4.
Effect of ARF T27N on CSF-1-induced membrane ruffling in macrophages. Adherent RAW LacR/FMLPR.2 cells transfected with the indicated constructs were incubated in the presence or absence of 10 ng of CSF-1 per ml for 5 min at 37°C, fixed, and stained with anti-ARF6 and rhodamine-phalloidin. Representative confocal fluorescence micrographs are shown. Bar = 10 μm.
FIG. 5.
Quantitation of membrane ruffling induced by CSF-1 in macrophages expressing the indicated constructs in nonexpressing controls. Data represent the means ± SEM (n = 5).
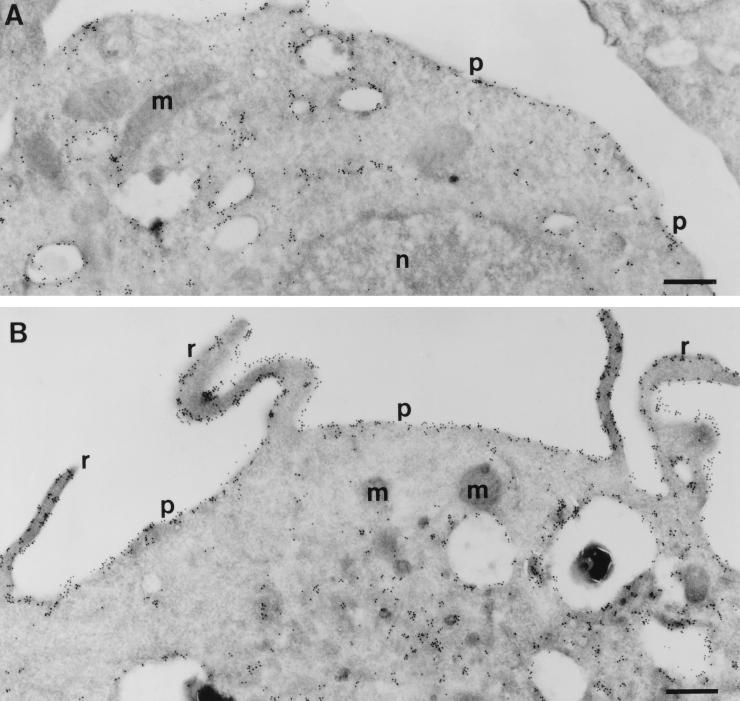
Effect of CSF-1 on the subcellular distribution of ARF6 in macrophages.
Activation of many cellular signaling pathways leads to alterations in the distribution of multiple components that participate in these pathways. Activation of Rho family GTPases correlates with enhanced binding of GTP and translocation to the membrane (1, 8, 19) and, in the case of Cdc42, the cytoskeleton (16). For ARF6, expression of the activated allele (ARF6 Q67L) correlates with localization at the plasma membrane, whereas expression of a GTP binding-deficient allele (ARF6 T27N) leads to accumulation of ARF6 in a juxtanuclear compartment (17, 36). To determine whether CSF-1 induces an alteration in the subcellular localization of ARF6, we expressed epitope-tagged versions of ARF6 in RAW LacR/FMLPR.2 cells, incubated the transfectants in the presence or absence of CSF-1, and processed cells for cryoimmunogold electron microscopy. In unstimulated cells, ARF6 was localized to the plasma membrane and to various intracellular membrane compartments, including those adjacent to the nucleus and others that were more peripheral in location (Fig. 6A). Some of these resembled macropinosomes, which are plentiful in these and other macrophages. Addition of CSF-1 led to the accumulation of membrane ruffles which stained prominently for ARF6 (Fig. 6B). There was no enrichment of ARF6 on the ruffles when compared with adjacent areas of the plasma membrane not involved in ruffling (Fig. 6B and Table 1). However, the relative accumulation of ARF6 on the plasma membrane was increased by 30% in cells incubated with CSF-1 (Table 1). These data indicate that ARF6 undergoes dynamic redistribution to the plasma membrane upon addition of CSF-1, suggesting that CSF-1 triggers ARF6 activation. In addition, intact ARF6 function is required for CSF-1-mediated membrane ruffling.
FIG. 6.
Effect of CSF-1 on the localization of ARF6 in macrophages. Adherent RAW LacR/FMLPR.2 cells transfected with HA-ARF6 were incubated in the absence (A) or presence (B) of 10 ng of CSF-1 for 5 min at 37°C. Ultrathin cryosections were labeled with monoclonal anti-HA followed by rabbit anti-mouse IgG and anti-rabbit IgG–10-nm-diameter gold. ARF6 localized to the plasma membrane (p) and intracellular vesicles and scattered throughout the cytosol. There is also some labeling in the mitochondria (m). Membrane ruffles (r) are apparent in cells incubated with CSF-1 (B). Note that the density of ARF6 labeling is the same for the ruffles and the adjacent areas of the plasma membrane. n, nucleus. Bars = 500 nm.
TABLE 1.
Quantitation of labeling density of HA-ARF6 WT in transfected macrophagesa
| Parameter | Absence of CSF-1 (n) | Presence of CSF-1 (n) |
|---|---|---|
| Gold particles per μm of nonruffling plasma membrane | ND | 39.2 ± 5.4 (5) |
| Gold particles per μm of ruffling plasma membrane | ND | 37.5 ± 6.0 (5)b |
| Plasma membrane labeling indexc | 0.57 ± 0.004 (41) | 0.74 ± 0.006 (41)d |
Adherent macrophages transfected with HA-ARF6 were serum starved for 1 h and incubated in the presence or absence of 10 ng of CSF-1 per ml. Cells were fixed and processed for cryoimmunogold electron microscopy as described in Materials and Methods. Data represent the means ± SEM. ND, not determined.
P = 0.84.
Relative distribution of gold label on the plasma membrane versus intracellular vesicles and cytosol. For details, see Materials and Methods.
P < 0.02 (presence of CSF-1 versus absence of CSF-1).
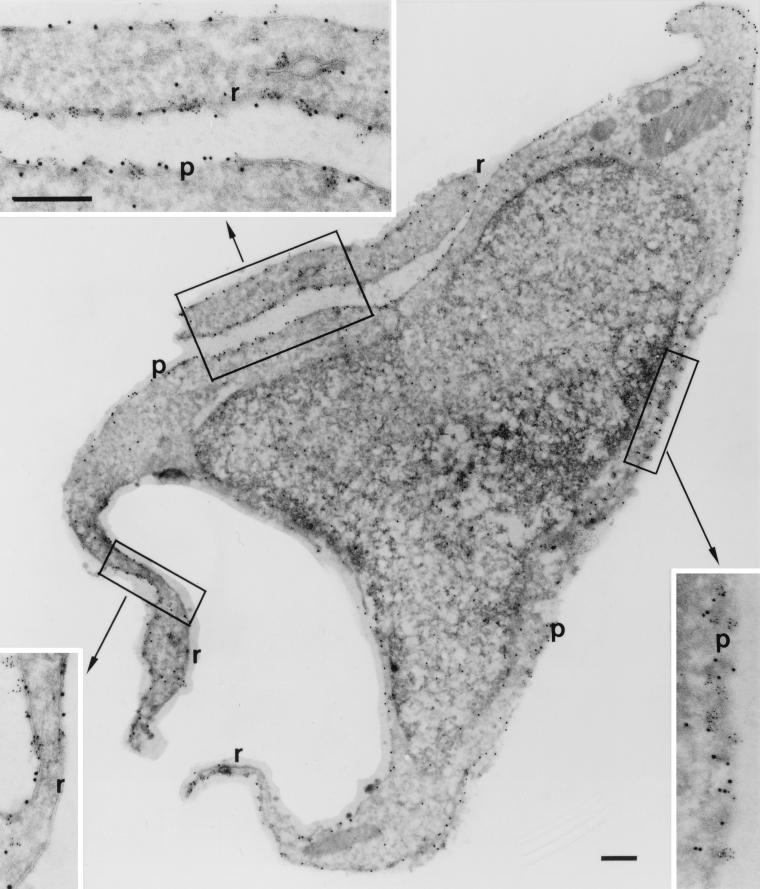
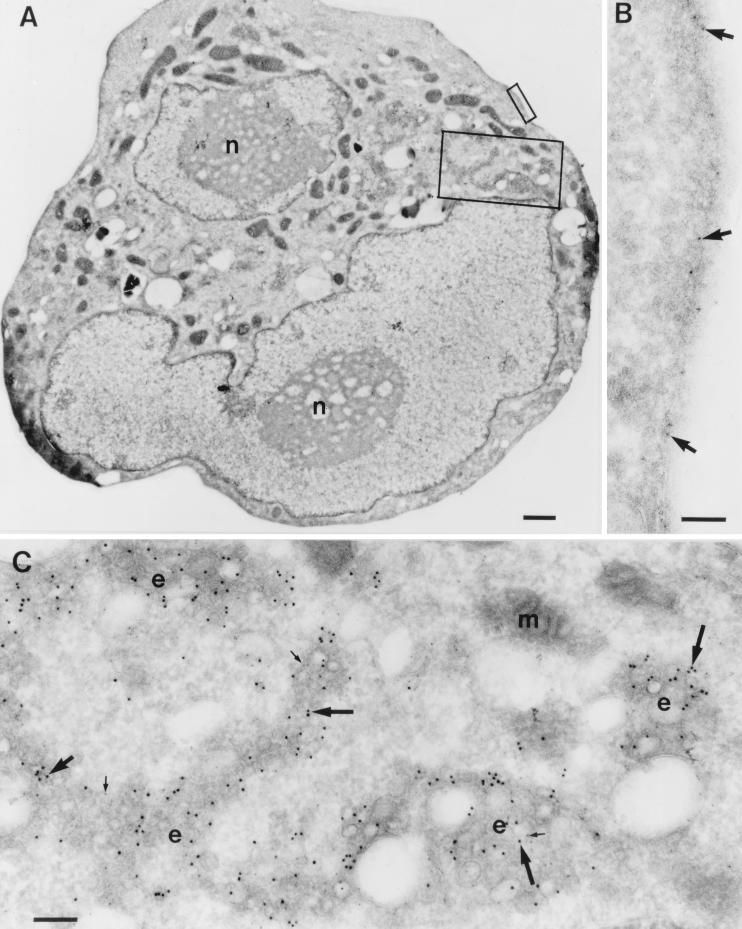
Effect of expression of a GTPase-deficient allele of Rac on the distribution of ARF6.
To determine whether expression of activated Rac1 alters the subcellular distribution of ARF6, we transfected epitope-tagged versions of either ARF6 WT or ARF6 T27N in the presence or absence of Myc-Rac1 Q61L and performed cryoimmunogold electron microscopy. Expression of Myc-Rac1 Q61L led to the appearance of prominent plasma membrane ruffles of variable size. Labeling for both Myc-Rac1 Q61L and HA-ARF6 WT was detected in the ruffles and in adjacent areas of the plasma membrane (Fig. 7). The extent of labeling of Myc-Rac Q61L on areas of plasma membrane which demonstrated ruffling was no different from that of adjacent areas of nonruffling plasma membrane (6.50 ± 0.74 versus 6.75 ± 0.93 particles/μm; means ± standard error of the means (SEM), n = 6). Similarly, the extent of labeling of HA-ARF6 on areas of plasma membrane which demonstrated ruffling was no different from that of adjacent areas of nonruffling plasma membrane (36.2 ± 10.5 versus 38.9 ± 10 particles/μm; means ± SEM, n = 4). Thus, expression of Myc-Rac1 Q61L led to the appearance of ruffles which contained ARF6, but there was no apparent enrichment in the ruffles for either Rac1 Q67L or ARF6. In cells expressing both Myc-Rac1 Q61L and HA-ARF6 T27N, membrane ruffles were conspicuously absent. Labeling for Myc-Rac1 Q61L appeared at the plasma membrane, while labeling for HA-ARF6 T27N appeared mainly in intracellular vesicles (Fig. 8). This distribution of ARF6 T27N is similar to results obtained in 293 (35) and CHO (17) cells. The extent of plasma membrane localization of Myc-Rac1 Q61L was not affected by coexpression of ARF6 T27N; the plasma membrane labeling index for Myc-Rac1 Q61L in the absence or presence of ARF6 T27N was 1.4 ± 0.08 and 1.4 ± 0.10, respectively (means ± SEM, n = 10).
FIG. 7.
Localization of ARF6 and Rac1 in cells coexpressing Myc-Rac1 Q61L and HA-ARF6 WT. Cryosections of adherent RAW LacR/FMLPR.2 cells transfected with the indicated constructs were double labeled with mouse anti-HA followed by rabbit anti-mouse IgG and anti-rabbit IgG–5-nm-diameter gold and with mouse anti-Myc followed by anti-mouse IgG–10-nm-diameter gold. Labeling with Rac1 (10-nm-diameter gold) can be seen on the plasma membrane (p) and on the membrane of the ruffles (r). The insets show a higher magnification of the marked areas showing the labeling with ARF6 (5-nm-diameter gold) and Rac1 (10-nm-diameter gold). The intensity of both labels on the membrane ruffles and on adjacent nonruffling areas of the plasma membrane is equivalent. Bars = 300 nm.
FIG. 8.
Localization of HA-ARF6 T27N in cells coexpressing Myc-Rac1 Q61L. Cryosections of adherent RAW LacR/FMLPR.2 cells transfected with the indicated constructs were double labeled with a mixture of rabbit anti-HA and mouse anti-Myc followed by a mixture of goat anti-rabbit IgG–10-nm-diameter gold and goat anti-mouse IgG–5-nm-diameter gold. (A) Low magnification of a cell expressing both constructs. n, nucleus. (B) Higher magnification of marked area in panel A showing the plasma membrane labeled only with Rac1 (5-nm-diameter gold) (arrows). (C) Higher magnification of the marked area in panel A showing endocytic vesicles (e) labeled for both ARF6 (10-nm-diameter gold) (large arrows) and Rac1 (5-nm-diameter gold) (small arrows). m, mitochondria. Bars = 1,000 nm (A), 100 nm (B), and 200 nm (C).
Targetting of Rac to the plasma membrane does not bypass the blockade of ARF6 T27N of membrane ruffling.
Since inhibition of ARF6 function inhibits Rac-mediated ruffling in macrophages (Fig. 1 and 2) and ARF6 regulates a recycling pathway in other cells (17, 36), we considered the possibility that ARF6 functions to recruit Rac to the plasma membrane. To eliminate the contribution of cytosol-to-membrane recruitment of Rac1 in Rac-mediated ruffling, we constructed a plasma membrane-targeted version of Rac1 Q61L which effectively bypasses changes in subcellular distribution as a contributing factor to its function. We expressed a fusion protein bearing a CD16 ectodomain and a cytosolic domain encoding Rac1 Q61L in macrophages and assessed surface expression of the fusion protein and cellular morphology. In cells expressing the construct 16:7:Myc-Rac1 Q61L, surface expression was confirmed by staining with MAb 3G8 in unpermeabilized cells; these cells displayed diffuse membrane ruffles that resembled those of cells expressing Myc-Rac1 Q61L (compare Fig. 9 with Fig. 1). Membrane ruffling induced by the fusion protein required Rac in the cytosolic domain because substitution with other proteins, such as Lck or Fyn, did not induce membrane ruffling (data not shown). Ruffling mediated by 16:7:Myc-Rac1 Q61L was not due to proteolytic fragments of the fusion protein containing Rac1 because immunoblotting demonstrated only intact 16:7:Myc-Rac1 Q61L (data not shown). Coexpression of ARF6 T27N but not ARF6 WT led to a phenotype characterized by the absence of membrane ruffles and the appearance of well-spread processes that resembled filopodia (compare Fig. 9 with cells expressing either Cdc42 or Rac plus ARF6 T27N in Fig. 1). Therefore, regardless of whether ARF6 contributes to the subcellular distribution of endogenous Rac1, inhibition of ARF6 function interferes with events that culminate in membrane ruffling downstream of Rac activation and membrane targeting. These data suggest that ARF6 regulates one or more events that couple activated Rac to protrusive and cytoskeletal events occurring at the plasma membrane.
FIG. 9.
Effect of ARF6 T27N on membrane ruffles induced by expression of a plasma membrane-targeted fusion protein containing Rac1 Q61L in macrophages. RAW LacR/FMLPR.2 cells were transfected with either 16:7:Myc-Rac1 Q61L or 16:7:Myc-Rac1 Q61L and ARF6 T27N. Expression of 16:7:Myc-Rac1 Q61L at the plasma membrane was confirmed by cell surface staining of CD16 (left panels); F-actin was visualized following subsequent permeabilization and staining with rhodamine-phalloidin (right panels). Bar = 10 μm.
DISCUSSION
Numerous studies have documented an essential role for Rac1 in cytoskeletal responses triggered by many structurally unrelated agonists (reviewed in reference 22). Collectively, these studies assign a central role to Rac1 in many cellular events that require plasma membrane-based cell motility, such as chemotaxis, phagocytosis, and formation of cadherin-dependent cell-cell contact in polarized cells (5, 10, 15, 46). Progress in understanding of Rac function is hampered by a lack of cell-free systems that reconstitute Rac-based motility, similar to those for ActA (42) and Cdc42 (28, 49). However, clues to Rac1 function are suggested by the distinct morphology of Rac-based protrusions. They appear as extended linear accumulations of F-actin-rich cortical cytoplasm underlying plasma membrane protrusions in a pattern often referred to as ruffles. This appearance suggests that Rac-triggered actin assembly is coordinated with alterations in the distribution of plasma membrane constituents. In contrast, Cdc42-directed filopodia are associated with only limited areas of plasma membrane deformation (i.e., at the tips of filopodia). The regulation of plasma membrane protrusions caused by either GTPase is likely to be highly complex, involving elements of the actin-based cytoskeleton and proteins that govern specific membrane trafficking events. The importance of ARF proteins in membrane trafficking events in general and the subcellular distribution of ARF6 identify ARF6 as a candidate that regulates elements in common between the cytoskeleton and plasma membrane. The data presented here are consistent with a role for ARF6 in Rac1-mediated ruffling but not Cdc42-mediated filopodium formation. Our data differ from those of an earlier study (18) in which the authors found that expression of ARF6 T27N did not inhibit membrane ruffling induced by an activated allele of Rac in CHO cells. There are several possible explanations for these differences. First, CHO cells may utilize ARF6-independent pathways for Rac activation, although a recent study (36) found that ARF6 T27N inhibited Rac-mediated ruffling in CHO cells, similar to results of the present study. Alternatively, the levels of expression of ARF6 T27N with Sindbis virus (18) may be less than those we obtained in the present study, leading to less effective inhibition of endogenous ARF6 function. Regardless of the reasons for these discrepancies, there are clear differences between CHO cells and macrophages, not the least of which is the inability of activated alleles of ARF6 to induce in macrophages (48) actin rearrangements similar to those seen in CHO cells (18).
One of the questions that arises from this study is whether ARF6 GTPase activity is triggered or constitutive. To date, there is no evidence that any physiological stimulus is capable of activating GTP exchange activity on any ARF protein. We could not address this question directly for ARF6, since studies of GTP exchange with epitope-tagged ARF6 in 32P orthophosphate-labeled cells have been unsuccessful, possibly due to the intrinsically high constitutive GTPase activity of ARF6 (data not shown). However, we have demonstrated that addition of CSF-1 leads to a redistribution of epitope-tagged ARF6 from an intracellular source to the plasma membrane. Together with evidence presented here and elsewhere that the subcellular localization of ARF6 depends on its nucleotide status (i.e., that GTP-bound ARF6 localizes to the plasma membrane and that GDP-bound or nucleotide-free ARF6 localizes to an intracellular vesicular compartment), these data suggest that CSF-1 directly or indirectly activates ARF6. Similar to other GTPases, activation of ARF6 in vivo is likely to be accomplished by specific guanine nucleotide exchange factors (GEFs). Two candidate GEFs for ARF6 have been identified to date; one is ARNO, a protein containing a Sec7 domain and a PH domain capable of binding PIP3 in vitro (13, 21, 45). Another GEF for ARF6 is EFA6, which has a similar domain structure to ARNO and is even more efficient at catalyzing guanine nucleotide exchange on ARF6 (20). Whether or not ARNO or EFA6 serve to activate ARF6 following addition of CSF-1 in macrophages is unknown, but it is interesting to note that inhibitors of phosphatidylinositol 3-kinase block CSF-1-mediated actin assembly in mouse macrophages (14a). Perhaps lipid products of phosphatidylinositol 3-kinase couple ligated CSF-1 receptors to ARF6 activation through recruitment and/or activation of ARNO or EFA6. One confounding feature of this model, which will require experimental verification, is that ARNO and EFA6 are localized predominantly to the plasma membrane (20, 21), which would be expected to contain ARF6-GTP, rather than ARF6-GDP, the predicted target of ARF6 GEFs.
A potential criticism of the approach used here to identify the ultrastructural distribution of ARF6 and Rac1 (i.e., using epitope-tagged constructs) is that these constructs may not reflect the normal function or distribution of endogenous proteins. We could not address the ultrastructural localization of either endogenous protein because available antisera and MAbs against Rac or ARF6 do not stain cells fixed with aldehydes used in cryoimmunogold electron microscopy (data not shown). However, immunofluorescence studies in NRK cells (41) and mouse macrophages (21a) are consistent with ARF6 localizing to intracellular membrane vesicles and the plasma membranes, similar to what is reported here with an epitope-tagged version of ARF6. Furthermore, the very fact that CSF-1 induced a redistribution of HA-ARF6 to the plasma membrane argues that this fusion protein is responsive to a physiologically relevant stimulus. The simultaneous recruitment of HA-ARF6 to the plasma membrane by CSF-1 but its apparent lack of enrichment in membrane ruffles suggests that mere localization of ARF6 to the plasma membrane is insufficient to promote membrane ruffling. Likewise, the diffuse plasma membrane distribution of Myc-Rac Q61L corresponded to ruffling and nonruffling regions of the plasma membrane. Together, these data indicate that Rac and ARF6 are necessary but not sufficient for plasma membrane ruffling.
One unexplained finding of this study is the induction of filopodia by coexpression of Myc-Rac Q61L and ARF6 T27N. Since ARF6 T27N alone does not induce filopodia in RAW LacR/FMLPR.2 cells (48), this suggests that Rac1 has the capacity to induce filopodia in macrophages under some conditions (e.g., when ARF6 function is inhibited). We have also observed rudimentary filopodia in unstimulated macrophages (Fig. 1) and well-formed filopodia in a small percentage of macrophages expressing grossly elevated levels of activated alleles of Rac in the absence of ARF6 T27N (unpublished results). At this point, we can only speculate that the cytoskeletal rearrangements triggered by Rac1 may require coupling to dynamic changes in trafficking to the plasma membrane and that ARF6 may participate in this trafficking by promoting membrane recycling (37). Of course, filopodium induction by coexpression of Myc-Rac Q61L and ARF6 T27N may well be a very indirect effect of Rac expression.
An important finding of this study is that the forced localization of Rac1 to the plasma membrane does not bypass the blockade in membrane ruffling induced by ARF6 T27N. While this does not directly address whether Rac1 distribution is itself influenced by the state of ARF6 activation or whether the distribution of Rac1 and ARF6 overlap in vivo, it points to a more central role for ARF6 in events occurring downstream of Rac activation. We have not identified the precise molecular targets of ARF6 in the cell but predict that one or more of these are localized to the plasma membrane in an ARF6-dependent fashion and are essential components of a Rac1- but not a Cdc42-directed pathway leading to cytoskeletal assembly. One candidate protein is POR1, a protein identified in a yeast two-hybrid screen that interacts with both Rac1 and ARF6 in vitro (18, 24, 44). It is not known whether these two GTPases compete with each other for binding POR1 nor is it known whether POR1 interacts with either protein in vivo. Using POR1-containing fusion proteins, we have been unable to demonstrate an association between POR1 and activated alleles of either ARF6 or Rac1 derived from transfected cells (data not shown). This may be due to a relatively low affinity of POR1 for both GTPases. Cell lysates may contain multiple proteins that interact more effectively with POR1 than ARF6 or Rac1. We are currently attempting to identify other components of the putative ARF6 compartment that participate in Rac-dependent cytoskeletal functions.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institutes of Health. S. Greenberg is an established investigator of the American Heart Association.
We thank Nico Ong for the help in preparing the micrographs and Julie Donaldson and Crislyn D’Souza-Schorey for helpful discussions.
REFERENCES
- 1.Abo A, Webb M R, Grogan A, Segal A W. Activation of NADPH oxidase involves the dissociation of p21rac from its inhibitory GDP/GTP exchange protein (rhoGDI) followed by its translocation to the plasma membrane. Biochem J. 1994;3:585–591. doi: 10.1042/bj2980585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen W E, Jones G E, Pollard J W, Ridley A J. Rho, Rac and Cdc42 regulate actin organization and cell adhesion in macrophages. J Cell Sci. 1997;110:707–720. doi: 10.1242/jcs.110.6.707. [DOI] [PubMed] [Google Scholar]
- 3.Amano M, Chihara K, Kimura K, Fukata Y, Nakamura N, Matsuura Y, Kaibuchi K. Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science. 1997;275:1308–1311. doi: 10.1126/science.275.5304.1308. [DOI] [PubMed] [Google Scholar]
- 4.Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 5.Anand-Apte B, Zetter B R, Viswanathan A, Qiu R G, Chen J, Ruggieri R, Symons M. Platelet-derived growth factor and fibronectin-stimulated migration are differentially regulated by the Rac and extracellular signal-regulated kinase pathways. J Biol Chem. 1997;272:30688–30692. doi: 10.1074/jbc.272.49.30688. [DOI] [PubMed] [Google Scholar]
- 6.Arber S, Barbayannis F A, Hanser H, Schneider C, Stanyon C A, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 7.Arcaro A. The small GTP-binding protein Rac promotes the dissociation of gelsolin from actin filaments in neutrophils. J Biol Chem. 1998;273:805–813. doi: 10.1074/jbc.273.2.805. [DOI] [PubMed] [Google Scholar]
- 8.Bokoch G M, Bohl B P, Chuang T-H. Guanine nucleotide exchange regulated membrane translocation of Rac/Rho GTP-binding proteins. J Biol Chem. 1994;269:31674–31679. [PubMed] [Google Scholar]
- 9.Boocock C A, Jones G E, Stanley E R, Pollard J W. Colony-stimulating factor-1 induces rapid behavioural responses in the mouse macrophage cell line, BAC1.2F5. J Cell Sci. 1989;93:447–456. doi: 10.1242/jcs.93.3.447. [DOI] [PubMed] [Google Scholar]
- 10.Braga V M M, Machesky L M, Hall A, Hotchin N A. The small GTPases Rho and Rac are required for the establishment of cadherin-dependent cell-cell contacts. J Cell Biol. 1997;137:1421–1431. doi: 10.1083/jcb.137.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calafat J, Janssen H, Stahle-Backdahl M, Zuurbier A E, Knol E F, Egesten A. Human monocytes and neutrophils store transforming growth factor-α in a subpopulation of cytoplasmic granules. Blood. 1997;90:1255–1266. [PubMed] [Google Scholar]
- 12.Cavenagh M M, Whitney J A, Carroll K, Zhang C J, Boman A L, Rosenwald A G, Mellman I, Kahn R A. Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J Biol Chem. 1996;271:21767–21774. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- 13.Chardin P, Paris S, Antonny B, Robineau C L, Beraud-Dufour S, Jackson C L, Chabre M. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature. 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- 14.Chong L D, Traynor-Kaplan A, Bokoch G M, Schwartz M A. The small GTP-binding protein Rho regulates a phosphatidylinositol 4-phosphate 5-kinase in mammalian cells. Cell. 1994;79:507–513. doi: 10.1016/0092-8674(94)90259-3. [DOI] [PubMed] [Google Scholar]
- 14a.Cox, D., and S. Greenberg. Unpublished data.
- 15.Cox D, Chang P, Zhang Q, Reddy P G, Bokoch G M, Greenberg S. Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J Exp Med. 1997;186:1487–1494. doi: 10.1084/jem.186.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dash D, Aepfelbacher M, Siess W. Integrin αIIbβ3-mediated translocation of CDC42Hs to the cytoskeleton in stimulated human platelets. J Biol Chem. 1995;270:17321–17326. doi: 10.1074/jbc.270.29.17321. [DOI] [PubMed] [Google Scholar]
- 17.D’Souza-Schorey C, van Donselaar E, Hsu V W, Yang C, Stahl P D, Peters P J. ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J Cell Biol. 1998;140:603–616. doi: 10.1083/jcb.140.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Souza-Schorey C, Boshans R L, McDonough M, Stahl P D, Van Aelst L. A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO J. 1997;16:5445–5454. doi: 10.1093/emboj/16.17.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleming I N, Elliott C M, Exton J H. Differential translocation of Rho family GTPases by lysophosphatidic acid, endothelin-1, and platelet-derived growth factor. J Biol Chem. 1996;271:33067–33073. doi: 10.1074/jbc.271.51.33067. [DOI] [PubMed] [Google Scholar]
- 20.Franco M, Peters P J, Boretto J, van Donselaar E, Neri A, D’Souza-Schorey C, Chavrier P. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank S, Upender S, Hansen S H, Casanova J E. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J Biol Chem. 1998;273:23–27. doi: 10.1074/jbc.273.1.23. [DOI] [PubMed] [Google Scholar]
- 21a.Greenberg, S. Unpublished results.
- 22.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–513. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 23.Hartwig J H, Bokoch G M, Carpenter C L, Janmey P A, Taylor L A, Toker A, Stossel T P. Thrombin receptor ligation and activated Rac uncap actin filament barbed ends through phosphoinositide synthesis in permeabilized human platelets. Cell. 1995;82:643–653. doi: 10.1016/0092-8674(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 24.Kanoh H, Williger B T, Exton J H. Arfaptin 1, a putative cytosolic target protein of ADP-ribosylation factor, is recruited to Golgi membranes. J Biol Chem. 1997;272:5421–5429. doi: 10.1074/jbc.272.9.5421. [DOI] [PubMed] [Google Scholar]
- 25.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J H, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 26.Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leung T, Chen X Q, Manser E, Lim L. The p160 RhoA-binding kinase ROKα is a member of a kinase family and is involved in the reorganization of the cytoskeleton. Mol Cell Biol. 1996;16:5313–5327. doi: 10.1128/mcb.16.10.5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma L, Cantley L C, Janmey P A, Kirschner M W. Corequirement of specific phosphoinositides and small GTP binding protein Cdc42 in inducing actin assembly in Xenopus egg extracts. J Cell Biol. 1998;140:1125–1136. doi: 10.1083/jcb.140.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Machesky L M, Insall R H. Scar 1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 30.Mellstrom K, Heldin C H, Westermark B. Induction of circular membrane ruffling on human fibroblasts by platelet-derived growth factor. Exp Cell Res. 1988;177:347–359. doi: 10.1016/0014-4827(88)90468-5. [DOI] [PubMed] [Google Scholar]
- 31.Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 32.Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nister M, Hammacher A, Mellstrom K, Siegbahn A, Ronnstrand L, Westermark B, Heldin C H. A glioma-derived PDGF A chain homodimer has different functional activities from a PDGF AB heterodimer purified from human platelets. Cell. 1988;52:791–799. doi: 10.1016/0092-8674(88)90421-7. [DOI] [PubMed] [Google Scholar]
- 34.Nobes C D, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 35.Peters P J, Hsu V W, Ooi C E, Finazzi D, Teal S B, Oorschot V, Donaldson J G, Klausner R D. Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. J Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radhakrishna H, Al-Awar O, Khachikian Z, Donaldson J G. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J Cell Sci. 1999;112:855–866. doi: 10.1242/jcs.112.6.855. [DOI] [PubMed] [Google Scholar]
- 37.Radhakrishna H, Donaldson J G. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radhakrishna H, Klausner R D, Donaldson J G. Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. J Cell Biol. 1996;134:935–947. doi: 10.1083/jcb.134.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridley A J, Hall H. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 40.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 41.Song J, Khachikian Z, Radhakrishna H, Donaldson J G. Localization of endogenous ARF6 to sites of cortical actin rearrangement and involvement of ARF6 in cell spreading. J Cell Sci. 1998;111:2257–2267. doi: 10.1242/jcs.111.15.2257. [DOI] [PubMed] [Google Scholar]
- 42.Theriot J A, Rosenblatt J, Portnoy D A, Goldschmidt-Clermont P J, Mitchison T J. Involvement of profilin in the actin-based motility of L. monocytogenes in cells and in cell-free extracts. Cell. 1994;76:505–517. doi: 10.1016/0092-8674(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 43.Tolias K F, Cantley L C, Carpenter C L. Rho family GTPases bind to phosphoinositide kinases. J Biol Chem. 1995;270:17656–17659. doi: 10.1074/jbc.270.30.17656. [DOI] [PubMed] [Google Scholar]
- 44.Van Aelst L, Joneson T, Bar-Sagi D. Identification of a novel Rac1-interacting protein involved in membrane ruffling. EMBO J. 1996;15:3778–3786. [PMC free article] [PubMed] [Google Scholar]
- 45.Venkateswarlu K, Oatey P B, Tavare J M, Cullen P J. Insulin-dependent translocation of ARNO to the plasma membrane of adipocytes requires phosphatidylinositol 3-kinase. Curr Biol. 1998;8:463–466. doi: 10.1016/s0960-9822(98)70181-2. [DOI] [PubMed] [Google Scholar]
- 46.Weber K S C, Klickstein L B, Weber P C, Weber C. Chemokine-induced monocyte transmigration requires cdc42-mediated cytoskeletal changes. Eur J Immunol. 1998;28:2245–2251. doi: 10.1002/(SICI)1521-4141(199807)28:07<2245::AID-IMMU2245>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 47.Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Q, Cox D, Tseng C-C, Donaldson J G, Greenberg S. A requirement for ARF6 in Fcγ receptor-mediated phagocytosis in macrophages. J Biol Chem. 1998;273:19977–19981. doi: 10.1074/jbc.273.32.19977. [DOI] [PubMed] [Google Scholar]
- 49.Zigmond S H, Joyce M, Yang C S, Brown K, Huang M Z, Pring M. Mechanism of cdc42-induced actin polymerization in neutrophil extracts. J Cell Biol. 1998;142:1001–1012. doi: 10.1083/jcb.142.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]