Abstract
The gene content of the X and Y chromosomes has dramatically diverged during evolution. The ensuing dosage imbalance within the genome of males and females has led to unique chromosome-wide regulatory mechanisms with significant and sex-specific impacts on X-linked gene expression. X inactivation or silencing of most genes on one X chromosome chosen at random in females profoundly affects the manifestation of X-linked diseases, as males inherit a single maternal allele, while females express maternal and paternal alleles in a mosaic manner. An additional complication is the existence of genes that escape X inactivation and thus are ubiquitously expressed from both alleles in females. The mosaic nature of X-linked gene expression and the potential for escape can vary between individuals, tissues, cell types and stages of life. Our understanding of the specialized nature of X-linked genes and of the multilayer epigenetic regulation that influence their expression throughout the organism has been helped by molecular studies conducted by tissue-specific and single-cell-specific approaches. In turn, the definition of molecular events that control X silencing has helped develop new approaches for the treatment of some X-linked disorders. This review focuses on the peculiarities of the X chromosome genetic content and epigenetic regulation in shaping the manifestation of congenital and acquired X-linked disorders in a sex-specific manner.
Introduction
Sex affects the manifestation, epidemiology and pathophysiology of many common diseases such as cardiovascular diseases, cancer, intellectual disabilities, neurodegenerative disorders, metabolic disorders, immune responses and autoimmune diseases (1). One example, relevant to the current Covid-19 pandemic, is that women mount a more robust immune response to infection than males who are often more severely affected by viral infections (2). Conversely, women are more susceptible to late-onset sporadic Alzheimer’s disease than men, although progression may be more rapid in men (3). Sex-stratified differences have been demonstrated in a systematic analysis of all diseases and disease co-occurrences in the Danish population using the ICD-10 and Global Burden of Disease terminologies (4). By compiling sex-specific incidence, risk, temporal aspects of diagnoses and co-occurrence of diagnoses, the authors identified sex-related differences across nearly all major disease types.
Hormones and genetic sex, namely the sex chromosomes, are major biological determinants of sex differences in human disease. In this review, we will show how the sex chromosomes, in particular the X chromosome and its peculiar gene content and modes of regulation, influence the manifestation of diseases in males and females. One important aspect of diseases caused by X-linked mutations relates to the types of genes located on the X chromosome, which have been shaped by evolution. Because of its presence as a single chromosome in males, the X chromosome is a choice location for genes favorable to males. Thus, families of testis-specific genes implicated in male fertility have accumulated on the X chromosome (5–7). Genes expressed in the brain especially in the cortex and the hypothalamus are also enriched on the X chromosome, a phenomenon probably driven by sexual selection (8–11). This is supported by a 3.5-fold higher incidence of X-linked versus autosomal mutations that cause intellectual disability (12). Other types of genes enriched on the X chromosome are genes involved in muscle function and in immune response. Hence, diseases caused by X-linked mutations often affect these tissues/cell types. Following a discussion of the role of X inactivation and escape from X inactivation on the manifestation and potential alleviation of X-linked mutations, we will address the role of X-linked genes in specific disorders including X aneuploidy, immune disorders and cancer, followed by a discussion of potential therapeutic approaches.
Influence of X Chromosome Inactivation on the Manifestation of Congenital Diseases
The human X chromosome carries more than 1100 genes and pathogenic variants cause at least 546 known X-linked diseases (13). Males are usually affected by X-linked pathogenic variants as they lack a second compensatory allele, while females are often asymptomatic carriers of the defect even though only one allele, maternal or paternal, remains expressed in their cells after X chromosome inactivation (XCI) (Fig. 1A) (14). Prior to XCI in pre-implantation human female embryos, single-cell studies have detected expression from both X chromosomes, with evidence of possible dampening of gene expression from each X allele to compensate for the high X versus autosome expression, but this possibility remains controversial (15–17). Following implantation, XCI is initiated by the upregulation of the long non-coding RNA (lncRNA) XIST on the future inactive X chromosome chosen at random. This triggers the recruitment of a number of proteins to silence X-linked genes by implementing layers of repressive epigenetic modifications such as histone modifications, chromatin condensation and methylation at CpG islands (16).
Figure 1 .
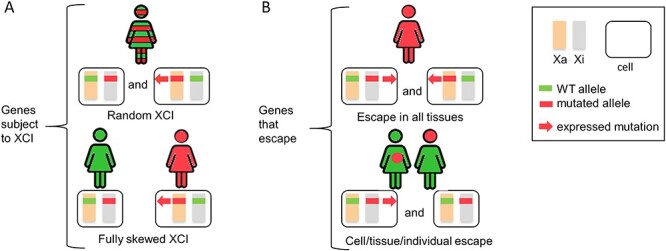
Influence of XCI on phenotypes in X-linked disorders. (A) Phenotypic effects of mutated genes subject to XCI. Following random XCI (top), a mixture of cells expressing either the wild-type allele (green) or the mutated allele (red) is present in female patients (red and green stripes). In case of fully skewed XCI (bottom), female patients will solely express either the wild-type allele (green) and show no disease (green), or the mutant allele (red) and show a disease phenotype (red). (B) Phenotypic effects of mutated genes that escape XCI. Patients with a mutation in a constitutive escape gene (top) will display a disease phenotype in all tissues regardless of random or skewed XCI. Patients with a mutation in a variable escape gene (bottom) will have a disease phenotype in tissues in which the gene escapes (red), even if skewed XCI favors the expression of the normal allele (green) in other tissues (green). This pattern may also differ between individuals. Orange and grey rectangles represent active and inactive X chromosomes. Green and red represent normal and pathogenic genotypes (bars) and phenotypes (symbolic female). Red arrow indicates the expression of the mutant allele.
The manifestation of X-linked mutations depends on the developmental stage considered. Prior to XCI, the presence of two expressed alleles in females may provide a fitness advantage in cases of a heterozygous recessive X-linked mutation (but not a dominant one). However, once XCI has taken place, only a single expressed allele persists in each somatic cell of both sexes. While X-linked-gene hemizygosity in males (except for genes in the pseudoautosomal regions PAR1 and PAR2) results in full expression of deleterious variants, heterozygous females are natural mosaic for cells with either parental X chromosome being active, which confers protection from pathogenic X-linked variants in many conditions (Fig. 1A) (13,18). One of the consequences of random XCI is that cell-to-cell variability for each variant haplotype is doubled in females compared with males, which may enhance the breath of responses to stimuli or injuries, providing further advantage to females in both health and disease (19). Female tissues are a patchwork of cells that express either paternal or maternal X alleles, with patches variable in size and among cell types and tissues due to the timing of silencing during embryogenesis, which occurs when a few hundred cells are present. A surprisingly wide variation in the proportion of cells with maternal or paternal allele expression has been found among tissues of female mice by imaging each allele of a reporter gene labeled with a red or green fluorescent marker (20). A study of healthy human monozygotic female twins has also revealed remarkable differences in X inactivation skewing within pairs, especially in fat and skin tissues (21). However, analyses of gene expression in bulk tissues obscure the variable distribution of cells expressing either paternal or maternal alleles. Single-cell studies in multiple human tissue types, cell types and developmental stages will help determine the extent of XCI variability in women, which could explain variable phenotypes not only in disease but also in healthy individuals.
Although random XCI results in two distinct cell populations, not all women have an overall 50:50 ratio of cells with one or the other X chromosome active. About 10% women display strong XCI skewing, such that greater than 95% of their cells express the same parental allele (Fig. 1A) (22). The proportion of each cell population has profound effects on the manifestation of X-linked diseases in heterozygous females. Monozygotic twin studies reveal that XCI skewing can change with age, a phenomenon often observed in blood and attributed to clonal hematopoiesis (21). Methods to read out skewing based on testing DNA methylation at a single locus, e.g. at the AR gene, have been supplemented by whole X chromosome allele-specific gene expression read-out by exome sequencing (23). Replication analyses can also be used to identify the inactive X chromosome based on its late replication pattern, especially in cases of structural X anomalies (24). Nonrandom XCI can be a chance event during embryogenesis or can be due to a rare mutation in the promoter of the XIST gene, which is essential for the onset of XCI (25). More often, skewing favors the normal allele due to cell selection, in which case a female carrier would usually not display clinical abnormalities (Fig. 1A; Table 1). The rate of cell selection in female tissues varies, which would affect the expression of the disease variant. While such selection mostly favors normal alleles, a rare example of the opposite effect is seen in adrenoleukodystrophy where cells with an ABCD1 mutation have a growth advantage (Table 1) (26). In some cases, complete XCI skewing can result from an undetectable lethal mutation on the so-called ‘normal’ X chromosome, which becomes preferentially silenced, potentially allowing for full expression of a mutation on the other X chromosome. Complete skewing can also be due to confined placental mosaicism in uniparental disomy (27).
Table 1.
Examples of inherited X-linked diseases affected by XCI patterns
| X-linked disease | Gene | XCI status in carriers | Mechanism | Phenotype | References |
|---|---|---|---|---|---|
| Duchenne muscular dystrophy | DMD | Random XCI | Sufficient number of cells expressing cell autonomous protein | N | 28 |
| Duchenne muscular dystrophy | DMD | Skewed XCI toward mutated allele | X:autosome translocation causes skewing | A | 29 |
| Hunter syndrome | IDS | Random XCI | Sufficient amount of secreted protein | N | 30,31 |
| Fabry disease | GLA | Random XCI | Normal protein product not taken by mutant cells | A | 32, 33 |
| Lesh-Nyhan | HPRT | Random XCI in fibroblasts/skewed XCI toward normal in blood | Gap junctions between fibroblasts/cell selection in blood | N | 34 |
| Adrenoleukodystrophy | ABCD1 | Skewed XCI toward mutated allele | Growth advantage of cells expressing mutation | A | 26 |
| Craniofrontonasal syndrome | EFNB1 | Random XCI | No substitution for EFNB1 deficiency | A | 35 |
| Rett syndrome | MECP2 | Variable XCI skewing | Critical protein; mutation lethal in males | A | 36 |
| ICF syndrome | DNMT3B | Aberrant XCI | Hypomethylation of various sequences | A | 39 |
| XLID due to escape genes | e.g. KDM5C, KDM6A | Escape XCI; partial XCI skewing | Haploinsufficiency | A | 61–65 |
| X aneuploidy | Escape genes; e.g. KDM6A | Random XCI | Dosage imbalance; genome-wide expression and DNA methylation effects | A | 74 |
| SLE | TLR7, TLR8, IRAK1, CXORF21 | Eroded XCI | Higher gene expression in B- and T-cells | A | 84–86 |
In some conditions, interactions between mosaic cell populations in female tissues can foster a metabolic cooperation and normal cells may provide essential gene products to correct the defect in mutant cells. In cases of mutations in cell autonomous products, females might not display symptoms if random XCI results in a sufficient number of cells that express the protein, for example in Duchenne muscular dystrophy carriers (Table 1) (28). However, rare cases of carrier women with full expression of the disease occur in individuals with an X; autosome translocation or other structural rearrangement that breaks the DMD gene and causes complete skewing of inactivation of the normal allele (Table 1) (29). In conditions due to mutations in secreted proteins, there may be sufficient product even without skewing of XCI. For example, females with Hunter syndrome, an X-linked lysosomal disease caused by deficiency of iduronic sulfatase (IDS), rarely have any clinical symptoms (Table 1) (30,31). In contrast, females with Fabry disease, another lysosomal disease caused by a deficiency in a-galactosidase (GLA), may have some clinical symptoms also seen in affected males, which can be explained by inability of the lysosome to take up the normal product (Table 1) (32,33). Tissue-specific differences in XCI skewing have also been reported. For example, in asymptomatic women carriers of an HPRT mutation XCI skewing does not occur in fibroblasts where gap junctions allow protein transit between cells but occurs in blood cells that lack junctions (Table 1) (34). Paradoxically, women carriers of an X-linked variant can occasionally manifest more severe symptoms than men. For example, men with a mutation in ephrin-B1 (EFNB1), which causes craniofrontonasal syndrome, are spared severe phenotypes because of redundancy in the essential functions of EFNB1, while women are severely affected because their mosaic state does not permit substitution for EFNB1 deficiency (Table 1) (35). In conditions where the disease is lethal in males, only females are born, for example, in Rett syndrome caused by a loss of function mutation in MECP2 (Table 1) (36). We refer the reader to a recently published comprehensive review of the effects of X-linked mutations on women health for additional examples (13).
Women may also manifest diseases caused by mutations in genes encoding essential components of the XCI machinery itself, a complex process with multiple layers of control (37,38). As mentioned above, a rare mutation in the promoter of the XIST gene results in skewing of inactivation (25). Mutations that affect XCI components often alter many other regulatory mechanisms that employ the same epigenetic processes throughout the genome. An example is afforded by mutations in the DNA methylase DNMT3B, which cause ICF (Immunodeficiency, centromeric region instability, facial anomalies) syndrome (Table 1) (39). This syndrome is characterized by T-cell but not B-cell deficiency and hypomethylation of centromeric repeats, which leads to chromosomal rearrangements throughout the genome. Hypomethylation of some X-linked genes occurs in a subset of ICF female patients, but there is little gene reactivation owing to the multiple layers controlling XCI (39). Few disorders caused by mutations in components of the machinery that specifically control XCI onset and stability have been discovered. One can speculate that such disorders might cause female lethality or complete XCI skewing since silencing of one X chromosome is essential for female survival. Such sex-specific lethality has been reported in mice with a mutation in Smchd1, a gene essential for the onset of XCI and for compaction of the human inactive X chromosome (40,41). In fact, it has been reasoned that the distorted sex ratio at birth (more males than females are born) could be contributed at least in part by female-specific susceptibility to failure of properly silencing one X chromosome (42). Interestingly, stem cell differentiation during embryonic development depends on successful completion of XCI for release of pluripotent factors, as shown by single-cell analyses in which X-linked genes implicated in the MAPK signaling pathway were identified as critical for this process (43–45). Mutations in X factors important for maintenance of XCI have been reported in autoimmune disorders and cancer (see below).
Escape from XCI and Role in Diseases
In human, ~20–30% of X-linked genes are expressed to some extent from the silent X chromosome (46–49). Except for PAR1 genes, which are mostly male-biased, escape genes often have higher expression in women, potentially leading to sexually dimorphic traits in susceptibility to disease (50). However, expression from the allele on the inactive X chromosome is usually lower than that from the active allele, probably due to their embedding within silenced chromatin (49). Genes that escape XCI are often clustered in domains that lack repressive histone marks on both alleles, and they are usually hypomethylated at their CpG island, which has helped their identification (51,52). In addition, the body of some escape genes is marked by non C-G methylation in neuronal cells (53). Escape genes have diverse functions, with those that retain a Y-linked paralog often highly dosage-sensitive and exhibiting critical functions related to the regulation of a number of other genes (54,55). There is a great deal of variability in the extent of escape from XCI among tissues/cell types and individuals, which has led to the definition of constitutive (always escape) and facultative (variable escape) escapees (56,57). Despite potential difficulties with low coverage, single-cell studies of gene expression by RNA-seq have helped identify escape genes based on SNPs. So far, such studies largely validated the previously reported annotations of human escape genes in fibroblast and lymphoblast transcriptomes, but this approach promises to extend our knowledge to many other cell types within human tissues for a better understanding of their role in disease (58). The profiles of escape from XCI vary among mammalian species, which makes it difficult to study them in animal models, except for genes that consistently escape XCI in both human and the model species (44). As an example, recent studies in mouse models have implicated Kdm6a, a gene that escapes XCI in human and mouse, in both Alzheimer disease and immune responses, which makes KDM6A an attractive candidate for a role in the corresponding human conditions (59,60).
Mutations in escape genes located in the pseudoautosomal regions of the sex chromosomes generally behave like mutations in autosomal genes. However, mutations in escape genes located outside the pseudoautosomal regions usually cause abnormal phenotypes in both sexes (Fig. 1B). In carrier females, the severity of phenotypes depends on whether the mutation is dominant, on the expression level from the inactive X and on the level of XCI skewing. Because many escapees are highly dosage sensitive, variants often cause a deficiency, only partially ameliorated by expression from the inactive X allele in women. Skewing of XCI in favor of the normal allele can alleviate the effects of a pathogenic variant and partially protect women from abnormal phenotypes. These effects are exemplified by mutations in the escape genes KDM6A and KDM5C, which cause multiple abnormalities including X-linked intellectual disability (XLID) in both sexes, but with more severity in males than females (Table 1) (61–64). Mutations in escape genes are an especially common cause of XLID (12,65). As discussed below, autoimmune diseases, which are much more common in women, are likely caused by abnormal expression of escape genes. Furthermore, abnormal escape gene dosage due to X aneuploidy contributes to a milieu of deleterious phenotypes including infertility, intellectual disability, immune diseases and cancer (66).
X Chromosome Aneuploidy
Sex chromosome aneuploidies, characterized by the loss or gain of one or more sex chromosomes, disturb the balance of gene products and result in recognizable syndromes such as Turner syndrome (45,X) and Klinefelter syndrome (47,XXY) (Fig. 2A). Turner syndrome is a near-lethal condition during embryogenesis, with individuals who survive presenting an array of abnormal phenotypes including infertility, short stature and heart anomalies (67,68). Klinefelter syndrome is a common cause of male infertility but also presents with autoimmune diseases and cognitive disturbances (69–71). Other X aneuploidy syndromes include triple X syndrome (47,XXX) associated with mild intellectual disability, while a greater number of sex chromosomes, e.g. 49,XXXXX, significantly increases morbidity (72,73). Yet, in all these conditions, only a single X chromosome remains active and all others are silenced regardless of sex. Thus, genes expressed prior to XCI in early development and genes that escape XCI are candidates to explain abnormal phenotypes. Sex-chromosome dosage (SCD) effects on genome-wide expression within a large cohort of individuals with diverse karyotypes (X, XX, XXX, XXXX, XY, XXY, XYY, XXYY and XXXXY) revealed a clear effect of chromosome ploidy on expression of escape genes (Fig. 2B) (74). Interestingly, some X-linked genes were up-regulated on the single X chromosome in 45,X compared with 46,XX or 47,XXX individuals, suggesting a partial compensatory mechanism in Turner syndrome. In addition, an increasing number of sex chromosomes did not increase gene expression linearly, indicating extensive trans-acting inverse effects of SCD on autosomal expression. Other factors may stem from the presence of multiple heterochromatic structures in the nucleus of aneuploid cells, which could disrupt epigenetic marks elsewhere in the genome and affect gene expression and homeostasis. In fact, the 3D structure of the active X chromosome in cells with sex chromosome aneuploidy is disrupted (75). Although dosage imbalance arising from escape genes is believed to contribute significantly to aneuploidy phenotypes, so far few specific genes have been implicated. One potential candidate is KDM6A, which regulates reproduction-related pathways in females and thus may be involved in ovarian dysfunction in Turner syndrome (76,77).
Figure 2 .
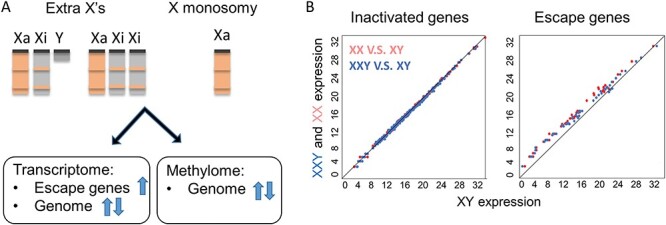
Sex chromosome aneuploidies disturb both the transcriptome and methylome in cells. (A) Extra X chromosome copies in Klinefelter syndrome (XXY) or Triple X syndrome (XXX), and the unique X chromosome in Turner syndrome (45,X) all disturb escape gene transcription, genome-wide gene transcription and DNA methylation. (B) Scatter plots of X-linked gene expression in female (XX, red) or Klinefelter (XXY, blue) versus male (XY). The expression of escape genes (right) is higher in Klinefelter males and normal females compared with normal males, while the expression of genes subject to XCI (left) remains similar between samples. Data reanalyzed from (130).
X chromosome aneuploidy leads to widespread epigenetic changes, including differential DNA methylation in target genes implicated in pathways associated with clinical features. Interestingly, DNA methylation levels on the X chromosomes of 47,XXY patients are distinct from both those of 46,XX and 46,XY controls (Fig. 2A) (78). Furthermore, differences are also evident between 47,XXY (hypermethylated X) and 45,X patients (hypomethylated X) (79). As with expression analyses, DNA methylation changes are not limited to the X chromosomes (80,81). Interestingly, loss of an X chromosome has a distinctive effect on autosomal genes compared to gain of an additional X chromosome. Approximately 80% of the CpG island methylation changes in 45,X individuals represent hypomethylation of autosomal loci, whereas nearly equal numbers of hypo- and hypermethylated CpG sites are observed in autosomes from 47,XXY individuals (82). Importantly, pathway analysis with differentially expressed genes and differentially methylated genes are found to be complementary, rather than overlapping. This finding suggests that DNA methylation as an epigenetic hallmark is not necessary illustrated by the transcriptome. Conversely, reversal of DNA methylation may not lead to a normal transcriptome, which has implications for future therapeutic target screenings (76).
XCI and Immune Diseases
As discussed above, XCI skewing and escape from XCI often vary among individuals, tissues and cell types, which results in phenotypic diversity (21,50,83). Intriguingly, B- and T-cells from women show a less completely inactivated X chromosome, with more escape genes, some immune-related (Fig. 3) (84–86). This may account for a more robust response mounted by women in response to infections, e.g. to COVID-19 (2). The flip side of this sex-specific advantage is that some autoimmune conditions are prevalent in women in whom aberrant escape from XCI has been observed. For instance, multiple sclerosis (MS), a disease in which the immune system attacks the insulating myelin around nerve fibers, is about three times more common in women than men (87). In a recent study, Itoh and colleagues employed an experimental autoimmune encephalomyelitis mouse model of MS to identify the escape gene Kdm6a as the main candidate in MS susceptibility (60).
Figure 3 .
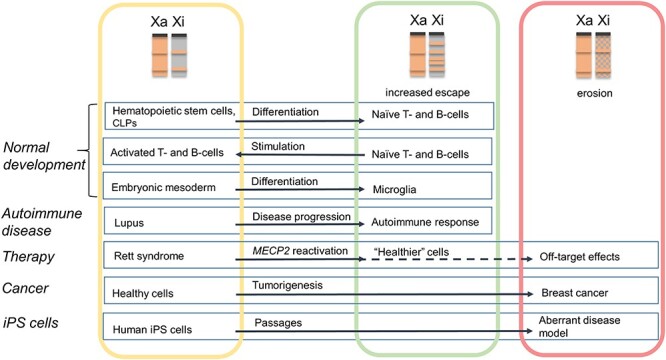
XCI changes in normal development, disease, therapeutic treatment and cell engineering. Cells in the yellow rectangle have normal XCI, cells in the green rectangle have partial X reactivation with an increase in the number of escape genes, and cells in the red rectangle have eroded XCI. CLPs are common lymphoid precursors. The dotted line indicates the potential for erosion due to off-target effects following therapy. Note that X reactivation in cancer cells could be beneficial due to enhanced neoantigens and better response to immunotherapy. Normal reactivation and potential dampening that occur in precursors of germ cells are not depicted.
TLR7, a member of the TLR family important for pathogen recognition and activation of innate immunity, escapes XCI and thus has higher expression in interferon-α producing cell types including B-lymphocytes, monocytes and plasmacytoid dendritic cells of XX and XXY versus XY individuals (88,89). The escape status of TLR7, and of other genes important in immunity including RPS6KA3, CYBB, BTK and IL13RA1, was confirmed in human plasmacytoid dendritic cells, stressing the contribution of multiple X-factors to sex differences in immune responses (90). TLR7 is also more abundant in female than male microglia, which could influence sex differences in brain function (91). A related gene, TLR8, escapes XCI in macrophages, a cell type activated in the inflammatory response to viral infection through the production of granulocyte-macrophage colony-stimulating factor (CSF2) (92). A recent study of human CD11c + atypical memory B-cells, which can expand during aging and cause aberrant responses to infectious diseases as well as female-biased autoimmunity, identified XIST RNA-independent and XIST RNA-dependent X-linked genes critical for maintenance of XCI stability (93). One of the genes whose silencing depends on continuous XIST expression is TLR7, implicating XIST RNA as a critical factor for maintenance of silencing of specific genes. Both TLR7 and TLR8 are highly expressed in systemic lupus erythematosus (SLE), an autoimmune disorder prevalent in women in whose T-cells XIST RNA localization and the XIST RNA interactome are perturbed (Fig. 3). Other escape genes implicated in SLE include IRAK1, and CXORF21, both potential targets for therapy (94,95). Interestingly, XXY males with Klinefelter syndrome are also susceptible to SLE (96).
X Chromosome and Cancer
Abnormal X chromosome copy number and aberrant patterns of XCI can occur in cancer (97,98). Imbalanced dosage of X-linked genes caused by copy number alterations of the whole X chromosome or of regions wherein and by erosion of XCI, has been implicated in oncogenesis. In the early 1950s, Barr and Moore discovered that nuclei of certain breast tumors lacked a Barr body (99). It was later shown that XCI is unstable in some breast tumors, a phenomenon associated with dispersed XIST RNA in the cancer cells (Fig. 3) (100). Translocations involving regions of the X chromosome could also alter gene expression profiles and lead to cancer. For example, relocation of regions of the inactive X chromosome to an autosome could result in reactivation of previously silent X-linked oncogenes. Conversely, loss of expression of an autosomal tumor suppressor can result from translocation to the inactive X chromosome (97).
XIST RNA is a key regulator of dosage compensation and is critical in maintaining XCI. Direct causal relationship between Xist RNA and cancer was confirmed in a mouse model in which Xist deletion in hematopoietic cells induced myeloproliferative neoplasm and myelodysplastic syndrome (101). In human, loss of XIST expression in iPS cells is significantly associated with upregulation of X-linked oncogenes (102). Recently, XIST has been reported to be dysregulated in gastric cancer (103), hepatocellular carcinoma (104), nasopharyngeal carcinoma (105), breast cancer (106) and ovarian cancer (107). XIST appears downregulated in recurrent ovarian tumors, but this is often due to the loss of the inactive X chromosome because of genomic instability in the tumor cells (108). Indeed, loss of either the inactive X chromosome in females or the Y chromosome in males is common in older individuals and may play a role in cancer susceptibility (109–112). Paradoxically, high XIST expression may also be deleterious in patients with a solid tumor, possibly due to sponging of miRNAs leading to upregulation of oncogenes by XIST RNA (113). Another X-linked lncRNA, FIRRE, implicated in XCI is also highly expressed in cancer cells, but its specific role remains to be discovered (114,115).
Acquired somatic mutations in X-linked genes are a frequent cause of cancer. Comparison of 402 whole genomes from a diverse set of childhood and adult tumors revealed hypermutation of the X chromosome (116). Somatic mutations in genes located in repressed chromatin domains including the inactive X chromosome are more frequent than in active domains, which is linked to the 3D chromatin structure (117). The main types of genetic alterations that lead to cancer—tumor-suppressor inactivation and oncogene activation—produce different results when they target X-linked versus autosomal genes. In females, a mutation propagated in a clonal manner would remain silent if it occurs on the silent allele of an X-linked gene. However, males would express the mutation. Accordingly, many cancers with a clear sex difference affect males more than females (118). To note, because XCI is usually stable once established, clonal expansion of a somatic cell in cancer results in a cell population with completely skewed XCI in women. Nonrandom XCI skewing can result in a selective growth advantage of cells with expression of the normal or the pathogenic allele. When selection severely disfavors cells that express the variant, heterozygous females rapidly lose mutant cells and render no symptoms. In contrast, if variant cells have a selective growth advantage, females manifest symptoms of the disease. For example, Foxp3 heterozygous female mice develop breast cancer at an enhanced rate as they age, due to the growth advantage of the mutant cells (119).
For oncogenes that escape XCI, acquired mutations may result in a similar disease susceptibility in men and women. However, for tumor suppressor genes that escape XCI, women would be protected by their second copy, resulting in a lower prevalence compared with men. Indeed, in a large study of about 4100 specimens across 21 tumor types, six escape genes called EXITS (Escape XCI Tumor Suppressor), including ATRX, CNKSR2, DDX3X, KDM5C, KDM6A and MAGEC3, were identified as genes which loss-of-function mutations affected men more frequently than women (120). In some cases, increased expression of an escape gene activated by a carcinogen can increase the risk of cancer in women. For example, GRPR, an escape gene expressed in lung epithelial cells, is associated with an increased risk of lung cancer due to tobacco exposure in women (121).
Therapeutic Induction of X Chromosome Reactivation
As stated above, there is a 3.5-fold increase of XLID disorders, compared with those due to mutations in autosomal genes. Here, we refer the reader to recent reviews on the subject (11,12), and we focus on new research that aims to cure deleterious neurological effects of X-linked disorders by reactivation of the normal allele in female carriers. Reactivation of X-linked genes occurs naturally in germ cells (122,123). However, silencing is highly stable in somatic cells; hence, reactivation is difficult. Rett syndrome caused by a mutation in MECP2 has been the focus of intense searches for compounds that would reactivate the normal allele using inhibitors of XIST expression in cell-based systems (Fig. 3) (124,125). Reactivation has also been achieved in vivo in a mouse model of Rett syndrome, in which small molecule inhibitors of ‘XCI factors’ reactivated Xist and corrected the neurological phenotype. Fortunately, while Mecp2 was reactivated, there did not appear to be widespread off-target effects on overall X-linked gene expression, suggesting compensatory mechanisms to maintain a normal balance of expression between X and autosomes (126). A similar strategy could potentially be applied to other X-linked disorders that affect female carriers, including neurological disorders caused by mutations in CDKL5, HDAC8 and KIAA2022. A new strategy was recently used to reactivate CDKL5 by editing DNA methylation using the demethylase TET1 and dCas9 with guides targeted to the CpG island of the gene (127). Such precise strategy has the advantage of limiting off-target effects on the genome. Female carriers affected by mutations in escape genes, e.g. DDX3X, KDM5C, KDM6A, USP9X or SMC1A, may also benefit from X reactivation to increase expression from the normal allele when it is on the inactive X chromosome. Finally, reactivation of the hypermutated inactive X chromosome in female cancer cells by inhibition of repressive chromatin marks has been proposed as a way to enhance the response to immunotherapy by increasing neoantigens (117).
Conclusions
The content and peculiar regulation of X-linked genes, which were shaped by evolution, profoundly influence the expression of diseases caused by mutations or copy number changes (Table 1). The advent of new methods to examine the onset and stability of X chromosome silencing in females has greatly improved our understanding of X-linked disorders. Future studies conducted in single individual cells promise to add much more information on the modalities of X chromosome regulation, which will help find better approaches to design an effective therapy of congenital and acquired disorders. Such studies can be supplemented by analyses of human induced pluripotent stem (iPS) cell lines to examine sex-specific effects of X-linked mutations in cell types relevant to a disorder. To note, XCI can be eroded in female iPS cells and it is usually skewed toward one allele due to cell cloning, necessitating the analysis of multiple clones together with the verification of the integrity of the inactive X chromosome (Fig. 3) (128,129). Nonetheless, analyses of organoids derived from stem cells obtained from patients with X-linked mutations will help understand their impact during development and in specific tissue/cell types.
Acknowledgements
We thank Di Kim Nguyen (University of Washington) for reanalysis of gene expression data in XXY.
Conflict of Interest statement. The authors have no conflicts of interest to declare.
Contributor Information
He Fang, Department of Laboratory Medicine and Pathology, University of Washington, Seattle, WA 98195, USA.
Xinxian Deng, Department of Laboratory Medicine and Pathology, University of Washington, Seattle, WA 98195, USA.
Christine M Disteche, Department of Laboratory Medicine and Pathology, University of Washington, Seattle, WA 98195, USA; Department of Medicine, University of Washington, Seattle, WA 98195, USA.
Funding
National Institutes of Health (GM131745 to C.M.D., HG011586 to C.M.D., GM1273727 to X.D.).
References
- 1.Mauvais-Jarvis, F., Bairey Merz, N., Barnes, P.J., Brinton, R.D., Carrero, J.J., DeMeo, D.L., De Vries, G.J., Epperson, C.N., Govindan, R., Klein, S.L. et al. (2020) Sex and gender: modifiers of health, disease, and medicine. Lancet, 396, 565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scully, E.P., Haverfield, J., Ursin, R.L., Tannenbaum, C. and Klein, S.L. (2020) Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol., 20, 442–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chouraki, V. and Seshadri, S. (2014) Genetics of Alzheimer's disease. Adv. Genet., 87, 245–294. [DOI] [PubMed] [Google Scholar]
- 4.Westergaard, D., Moseley, P., Sørup, F.K.H., Baldi, P. and Brunak, S. (2019) Population-wide analysis of differences in disease progression patterns in men and women. Nat. Commun., 10, 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller, J.L., Mahadevaiah, S.K., Park, P.J., Warburton, P.E., Page, D.C. and Turner, J.M. (2008) The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet., 40, 794–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinho, A., Barros, A. and Fernandes, S. (2020) Clinical and molecular characterization of Y microdeletions and X-linked CNV67 implications in male fertility: a 20-year experience. Andrology, 8, 307–314. [DOI] [PubMed] [Google Scholar]
- 7.Bellott, D.W., Skaletsky, H., Pyntikova, T., Mardis, E.R., Graves, T., Kremitzki, C., Brown, L.G., Rozen, S., Warren, W.C., Wilson, R.K. and Page, D.C. (2010) Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature, 466, 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zechner, U., Wilda, M., Kehrer-Sawatzki, H., Vogel, W., Fundele, R. and Hameister, H. (2001) A high density of X-linked genes for general cognitive ability: a run-away process shaping human evolution? Trends Genet., 17, 697–701. [DOI] [PubMed] [Google Scholar]
- 9.Ryan, M.J. (2021) Darwin, sexual selection, and the brain. Proc. Natl. Acad. Sci. U. S. A., 118, e2008194118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen, D.K. and Disteche, C.M. (2006) High expression of the mammalian X chromosome in brain. Brain Res., 1126, 46–49. [DOI] [PubMed] [Google Scholar]
- 11.Raznahan, A. and Disteche, C.M. (2021) X-chromosome regulation and sex differences in brain anatomy. Neurosci. Biobehav. Rev., 120, 28–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neri, G., Schwartz, C.E., Lubs, H.A. and Stevenson, R.E. (2018) X-linked intellectual disability update 2017. Am. J. Med. Genet. A, 176, 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Migeon, B.R. (2020) X-linked diseases: susceptible females. Genet. Med., 22, 1156–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LYON, M.F. (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature, 190, 372–373. [DOI] [PubMed] [Google Scholar]
- 15.Petropoulos, S., Edsgärd, D., Reinius, B., Deng, Q., Panula, S.P., Codeluppi, S., Reyes, A.P., Linnarsson, S., Sandberg, R. and Lanner, F. (2016) Single-cell RNA-Seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell, 165, 1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patrat, C., Ouimette, J.F. and Rougeulle, C. (2020) X chromosome inactivation in human development. Development, 147, dev183095. [DOI] [PubMed] [Google Scholar]
- 17.Moreira de Mello, J.C., Fernandes, G.R., Vibranovski, M.D. and Pereira, L.V. (2017) Early X chromosome inactivation during human preimplantation development revealed by single-cell RNA-sequencing. Sci. Rep., 7, 10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Migeon, B. (2014) Females Are Mosaics: X Inactivation and Sex Differences in Disease. OUP, USA. [DOI] [PubMed] [Google Scholar]
- 19.Schurz, H., Salie, M., Tromp, G., Hoal, E.G., Kinnear, C.J. and Möller, M. (2019) The X chromosome and sex-specific effects in infectious disease susceptibility. Hum. Genomics, 13, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu, H., Luo, J., Yu, H., Rattner, A., Mo, A., Wang, Y., Smallwood, P.M., Erlanger, B., Wheelan, S.J. and Nathans, J. (2014) Cellular resolution maps of X chromosome inactivation: implications for neural development, function, and disease. Neuron, 81, 103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zito, A., Davies, M.N., Tsai, P.C., Roberts, S., Andres-Ejarque, R., Nardone, S., Bell, J.T., Wong, C.C.Y. and Small, K.S. (2019) Heritability of skewed X-inactivation in female twins is tissue-specific and associated with age. Nat. Commun., 10, 5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatakeyama, C., Anderson, C.L., Beever, C.L., Peñaherrera, M.S., Brown, C.J. and Robinson, W.P. (2004) The dynamics of X-inactivation skewing as women age. Clin. Genet., 66, 327–332. [DOI] [PubMed] [Google Scholar]
- 23.Vacca, M., Della Ragione, F., Scalabrì, F. and D'Esposito, M. (2016) X inactivation and reactivation in X-linked diseases. Semin. Cell Dev. Biol., 56, 78–87. [DOI] [PubMed] [Google Scholar]
- 24.Leppig, K.A. and Disteche, C.M. (2001) Ring X and other structural X chromosome abnormalities: X inactivation and phenotype. Semin. Reprod. Med., 19, 147–157. [DOI] [PubMed] [Google Scholar]
- 25.Plenge, R.M., Hendrich, B.D., Schwartz, C., Arena, J.F., Naumova, A., Sapienza, C., Winter, R.M. and Willard, H.F. (1997) A promoter mutation in the XIST gene in two unrelated families with skewed X-chromosome inactivation. Nat. Genet., 17, 353–356. [DOI] [PubMed] [Google Scholar]
- 26.Engelen, M., Barbier, M., Dijkstra, I.M., Schür, R., de Bie, R.M., Verhamme, C., Dijkgraaf, M.G., Aubourg, P.A., Wanders, R.J., van Geel, B.M. et al. (2014) X-linked adrenoleukodystrophy in women: a cross-sectional cohort study. Brain, 137, 693–706. [DOI] [PubMed] [Google Scholar]
- 27.Lau, A.W., Brown, C.J., Peñaherrera, M., Langlois, S., Kalousek, D.K. and Robinson, W.P. (1997) Skewed X-chromosome inactivation is common in fetuses or newborns associated with confined placental mosaicism. Am. J. Hum. Genet., 61, 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffman, E.P. and Kunkel, L.M. (1989) Dystrophin abnormalities in Duchenne/Becker muscular dystrophy. Neuron, 2, 1019–1029. [DOI] [PubMed] [Google Scholar]
- 29.Boyd, Y., Buckle, V., Holt, S., Munro, E., Hunter, D. and Craig, I. (1986) Muscular dystrophy in girls with X; autosome translocations. J. Med. Genet., 23, 484–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinto, L.L., Vieira, T.A., Giugliani, R. and Schwartz, I.V. (2010) Expression of the disease on female carriers of X-linked lysosomal disorders: a brief review. Orphanet J. Rare Dis., 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guillén-Navarro, E., Domingo-Jiménez, M.R., Alcalde-Martín, C., Cancho-Candela, R., Couce, M.L., Galán-Gómez, E. and Alonso-Luengo, O. (2013) Clinical manifestations in female carriers of mucopolysaccharidosis type II: a Spanish cross-sectional study. Orphanet J. Rare Dis., 8, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuller, M., Mellett, N., Hein, L.K., Brooks, D.A. and Meikle, P.J. (2015) Absence of α-galactosidase cross-correction in Fabry heterozygote cultured skin fibroblasts. Mol. Genet. Metab., 114, 268–273. [DOI] [PubMed] [Google Scholar]
- 33.Beck, M. and Cox, T.M. (2019) Comment: why are females with Fabry disease affected? Mol Genet Metab Rep, 21, 100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox, R.P., Krauss, M.R., Balis, M.E. and Dancis, J. (1970) Evidence for transfer of enzyme product as the basis of metabolic cooperation between tissue culture fibroblasts of Lesch-Nyhan disease and normal cells. Proc. Natl. Acad. Sci. U. S. A., 67, 1573–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Twigg, S.R., Babbs, C., van den Elzen, M.E., Goriely, A., Taylor, S., McGowan, S.J., Giannoulatou, E., Lonie, L., Ragoussis, J., Sadighi Akha, E. et al. (2013) Cellular interference in craniofrontonasal syndrome: males mosaic for mutations in the X-linked EFNB1 gene are more severely affected than true hemizygotes. Hum. Mol. Genet., 22, 1654–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reichow, B., George-Puskar, A., Lutz, T., Smith, I.C. and Volkmar, F.R. (2015) Brief report: systematic review of Rett syndrome in males. J. Autism Dev. Disord., 45, 3377–3383. [DOI] [PubMed] [Google Scholar]
- 37.Pandya-Jones, A., Markaki, Y., Serizay, J., Chitiashvili, T., Mancia Leon, W.R., Damianov, A., Chronis, C., Papp, B., Chen, C.K., McKee, R. et al. (2020) A protein assembly mediates Xist localization and gene silencing. Nature, 587, 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loda, A. and Heard, E. (2019) Xist RNA in action: past, present, and future. PLoS Genet., 15, e1008333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen, R.S., Wijmenga, C., Luo, P., Stanek, A.M., Canfield, T.K., Weemaes, C.M. and Gartler, S.M. (1999) The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. U. S. A., 96, 14412–14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jansz, N., Keniry, A., Trussart, M., Bildsoe, H., Beck, T., Tonks, I.D., Mould, A.W., Hickey, P., Breslin, K., Iminitoff, M. et al. (2018) Smchd1 regulates long-range chromatin interactions on the inactive X chromosome and at Hox clusters. Nat. Struct. Mol. Biol., 25, 766–777. [DOI] [PubMed] [Google Scholar]
- 41.Nozawa, R.S., Nagao, K., Igami, K.T., Shibata, S., Shirai, N., Nozaki, N., Sado, T., Kimura, H. and Obuse, C. (2013) Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat. Struct. Mol. Biol., 20, 566–573. [DOI] [PubMed] [Google Scholar]
- 42.Migeon, B. (2021) The human female paradox: biological disadvantage preimplantation, biological advantage, thereafter. Curr. Opin. Obstet. Gynecol., 4, 397–404. [Google Scholar]
- 43.Schulz, E.G., Meisig, J., Nakamura, T., Okamoto, I., Sieber, A., Picard, C., Borensztein, M., Saitou, M., Blüthgen, N. and Heard, E. (2014) The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell, 14, 203–216. [DOI] [PubMed] [Google Scholar]
- 44.Genolet, O., Monaco, A.A., Dunkel, I., Boettcher, M. and Schulz, E.G. (2021) Identification of X-chromosomal genes that drive sex differences in embryonic stem cells through a hierarchical CRISPR screening approach. Genome Biol., 22, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonora, G., Ramani, V., Singh, R., Fang, H., Jackson, D., Srivatsan, S., Qiu, R., Lee, C., Trapnell, C., Shendure, J. et al. (2020) Single-cell landscape of nuclear configuration and gene expression during stem cell differentiation and X inactivation. bioRxiv., 11.20.390765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang, H., Disteche, C.M. and Berletch, J.B. (2019) X inactivation and escape: epigenetic and structural features. Front. Cell Dev. Biol., 7, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berletch, J.B., Yang, F., Xu, J., Carrel, L. and Disteche, C.M. (2011) Genes that escape from X inactivation. Hum. Genet., 130, 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balaton, B.P., Cotton, A.M. and Brown, C.J. (2015) Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol. Sex Differ., 6, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carrel, L. and Willard, H.F. (2005) X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature, 434, 400–404. [DOI] [PubMed] [Google Scholar]
- 50.Tukiainen, T., Villani, A.C., Yen, A., Rivas, M.A., Marshall, J.L., Satija, R., Aguirre, M., Gauthier, L., Fleharty, M., Kirby, A. et al. (2017) Landscape of X chromosome inactivation across human tissues. Nature, 550, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balaton, B.P. and Brown, C.J. (2016) Escape artists of the X chromosome. Trends Genet., 32, 348–359. [DOI] [PubMed] [Google Scholar]
- 52.Yang, F., Babak, T., Shendure, J. and Disteche, C.M. (2010) Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res., 20, 614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keown, C.L., Berletch, J.B., Castanon, R., Nery, J.R., Disteche, C.M., Ecker, J.R. and Mukamel, E.A. (2017) Allele-specific non-CG DNA methylation marks domains of active chromatin in female mouse brain. Proc. Natl. Acad. Sci. U. S. A., 114, E2882–E2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bellott, D.W., Hughes, J.F., Skaletsky, H., Brown, L.G., Pyntikova, T., Cho, T.J., Koutseva, N., Zaghlul, S., Graves, T., Rock, S. et al. (2014) Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature, 508, 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cortez, D., Marin, R., Toledo-Flores, D., Froidevaux, L., Liechti, A., Waters, P.D., Grützner, F. and Kaessmann, H. (2014) Origins and functional evolution of Y chromosomes across mammals. Nature, 508, 488–493. [DOI] [PubMed] [Google Scholar]
- 56.Anderson, C.L. and Brown, C.J. (1999) Polymorphic X-chromosome inactivation of the human TIMP1 gene. Am. J. Hum. Genet., 65, 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berletch, J.B., Ma, W., Yang, F., Shendure, J., Noble, W.S., Disteche, C.M. and Deng, X. (2015) Escape from X inactivation varies in mouse tissues. PLoS Genet., 11, e1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wainer Katsir, K. and Linial, M. (2019) Human genes escaping X-inactivation revealed by single cell expression data. BMC Genomics, 20, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis, E.J., Broestl, L., Abdulai-Saiku, S., Worden, K., Bonham, L.W., Miñones-Moyano, E., Moreno, A.J., Wang, D., Chang, K., Williams, G. et al. (2020) A second X chromosome contributes to resilience in a mouse model of Alzheimer’s disease. Sci. Transl. Med., 12, eaaz5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Itoh, Y., Golden, L.C., Itoh, N., Matsukawa, M.A., Ren, E., Tse, V., Arnold, A.P. and Voskuhl, R.R. (2019) The X-linked histone demethylase Kdm6a in CD4+ T lymphocytes modulates autoimmunity. J. Clin. Invest., 129, 3852–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poeta, L., Padula, A., Attianese, B., Valentino, M., Verrillo, L., Filosa, S., Shoubridge, C., Barra, A., Schwartz, C.E., Christensen, J. et al. (2019) Histone demethylase KDM5C is a SAHA-sensitive central hub at the crossroads of transcriptional axes involved in multiple neurodevelopmental disorders. Hum. Mol. Genet., 28, 4089–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adam, M.P., Banka, S., Bjornsson, H.T., Bodamer, O., Chudley, A.E., Harris, J., Kawame, H., Lanpher, B.C., Lindsley, A.W., Merla, G. et al. (2019) Kabuki syndrome: international consensus diagnostic criteria. J. Med. Genet., 56, 89–95. [DOI] [PubMed] [Google Scholar]
- 63.Lederer, D., Shears, D., Benoit, V., Verellen-Dumoulin, C. and Maystadt, I. (2014) A three generation X-linked family with kabuki syndrome phenotype and a frameshift mutation in KDM6A. Am. J. Med. Genet. A, 164A, 1289–1292. [DOI] [PubMed] [Google Scholar]
- 64.Yang, P., Tan, H., Xia, Y., Yu, Q., Wei, X., Guo, R., Peng, Y., Chen, C., Li, H., Mei, L. et al. (2016) De novo exonic deletion of KDM6A in a Chinese girl with kabuki syndrome: a case report and brief literature review. Am. J. Med. Genet. A, 170, 1613–1621. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, Y., Castillo-Morales, A., Jiang, M., Zhu, Y., Hu, L., Urrutia, A.O., Kong, X. and Hurst, L.D. (2016) Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving. Mol. Biol. Evol., 30, 2588–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balaton, B.P., Dixon-McDougall, T., Peeters, S.B. and Brown, C.J. (2018) The eXceptional nature of the X chromosome. Hum. Mol. Genet., 27, R242–R249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Urbach, A. and Benvenisty, N. (2009) Studying early lethality of 45,XO (Turner's syndrome) embryos using human embryonic stem cells. PLoS One, 4, e4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cui, X., Cui, Y., Shi, L., Luan, J., Zhou, X. and Han, J. (2018) A basic understanding of Turner syndrome: incidence, complications, diagnosis, and treatment. Intractable Rare Dis. Res., 7, 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Groth, K.A., Skakkebæk, A., Høst, C., Gravholt, C.H. and Bojesen, A. (2013) Clinical review: Klinefelter syndrome--a clinical update. J. Clin. Endocrinol. Metab., 98, 20–30. [DOI] [PubMed] [Google Scholar]
- 70.Seminog, O.O., Seminog, A.B., Yeates, D. and Goldacre, M.J. (2015) Associations between Klinefelter's syndrome and autoimmune diseases: English national record linkage studies. Autoimmunity, 48, 125–128. [DOI] [PubMed] [Google Scholar]
- 71.Boada, R., Janusz, J., Hutaff-Lee, C. and Tartaglia, N. (2009) The cognitive phenotype in Klinefelter syndrome: a review of the literature including genetic and hormonal factors. Dev. Disabil. Res. Rev., 15, 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leggett, V., Jacobs, P., Nation, K., Scerif, G. and Bishop, D.V. (2010) Neurocognitive outcomes of individuals with a sex chromosome trisomy: XXX, XYY, or XXY: a systematic review. Dev. Med. Child Neurol., 52, 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pirollo, L.M., Salehi, L.B., Sarta, S., Cassone, M., Capogna, M.V., Piccione, E., Novelli, G. and Pietropolli, A. (2015) A new case of prenatally diagnosed pentasomy x: review of the literature. Case Rep. Obstet. Gynecol., 2015, 935202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raznahan, A., Parikshak, N.N., Chandran, V., Blumenthal, J.D., Clasen, L.S., Alexander-Bloch, A.F., Zinn, A.R., Wangsa, D., Wise, J., Murphy, D.G.M. et al. (2018) Sex-chromosome dosage effects on gene expression in humans. Proc. Natl. Acad. Sci. U. S. A., 115, 7398–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jowhar, Z., Shachar, S., Gudla, P.R., Wangsa, D., Torres, E., Russ, J.L., Pegoraro, G., Ried, T., Raznahan, A. and Misteli, T. (2018) Effects of human sex chromosome dosage on spatial chromosome organization. Mol. Biol. Cell, 29, 2458–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trolle, C., Nielsen, M.M., Skakkebæk, A., Lamy, P., Vang, S., Hedegaard, J., Nordentoft, I., Ørntoft, T.F., Pedersen, J.S. and Gravholt, C.H. (2016) Widespread DNA hypomethylation and differential gene expression in Turner syndrome. Sci. Rep., 6, 34220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berletch, J.B., Deng, X., Nguyen, D.K. and Disteche, C.M. (2013) Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A. PLoS Genet., 9, e1003489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Skakkebæk, A., Nielsen, M.M., Trolle, C., Vang, S., Hornshøj, H., Hedegaard, J., Wallentin, M., Bojesen, A., Hertz, J.M., Fedder, J. et al. (2018) DNA hypermethylation and differential gene expression associated with Klinefelter syndrome. Sci. Rep., 8, 13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, X., Hong, D., Ma, S., Ward, T., Ho, M., Pattni, R., Duren, Z., Stankov, A., Bade Shrestha, S., Hallmayer, J. et al. (2020) Integrated functional genomic analyses of Klinefelter and Turner syndromes reveal global network effects of altered X chromosome dosage. Proc. Natl. Acad. Sci. U. S. A., 117, 4864–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharma, A., Jamil, M.A., Nuesgen, N., Schreiner, F., Priebe, L., Hoffmann, P., Herns, S., Nöthen, M.M., Fröhlich, H., Oldenburg, J. et al. (2015) DNA methylation signature in peripheral blood reveals distinct characteristics of human X chromosome numerical aberrations. Clin. Epigenetics, 7, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skakkebaek, A., Viuff, M., Nielsen, M.M. and Gravholt, C.H. (2020) Epigenetics and genomics in Klinefelter syndrome. Am. J. Med. Genet. C Semin. Med. Genet., 184, 216–225. [DOI] [PubMed] [Google Scholar]
- 82.Viuff, M., Skakkebaek, A., Nielsen, M.M., Chang, S. and Gravholt, C.H. (2019) Epigenetics and genomics in Turner syndrome. Am. J. Med. Genet. C Semin. Med. Genet., 181, 68–75. [DOI] [PubMed] [Google Scholar]
- 83.Oliva, M., Muñoz-Aguirre, M., Kim-Hellmuth, S., Wucher, V., Gewirtz, A.D.H., Cotter, D.J., Parsana, P., Kasela, S., Balliu, B., Viñuela, A. et al. (2020) The impact of sex on gene expression across human tissues. Science, 369, eaba3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang, J., Syrett, C.M., Kramer, M.C., Basu, A., Atchison, M.L. and Anguera, M.C. (2016) Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc. Natl. Acad. Sci. U. S. A., 113, E2029–E2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Syrett, C.M., Sindhava, V., Hodawadekar, S., Myles, A., Liang, G., Zhang, Y., Nandi, S., Cancro, M., Atchison, M. and Anguera, M.C. (2017) Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet., 13, e1007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qu, K., Zaba, L.C., Giresi, P.G., Li, R., Longmire, M., Kim, Y.H., Greenleaf, W.J. and Chang, H.Y. (2015) Individuality and variation of personal regulomes in primary human T cells. Cell Syst, 1, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu, G.F. (2019) X-tra X: an escape to autoimmunity. J. Clin. Invest., 129, 3536–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Souyris, M., Cenac, C., Azar, P., Daviaud, D., Canivet, A., Grunenwald, S., Pienkowski, C., Chaumeil, J., Mejía, J.E. and Guéry, J.-C. (2018) TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol., 3, eaap8855. [DOI] [PubMed] [Google Scholar]
- 89.Souyris, M., Mejía, J.E., Chaumeil, J. and Guéry, J.C. (2019) Female predisposition to TLR7-driven autoimmunity: gene dosage and the escape from X chromosome inactivation. Semin. Immunopathol., 41, 153–164. [DOI] [PubMed] [Google Scholar]
- 90.Hagen, S.H., Henseling, F., Hennesen, J., Savel, H., Delahaye, S., Richert, L., Ziegler, S.M. and Altfeld, M. (2020) Heterogeneous escape from X chromosome inactivation results in sex differences in type I IFN responses at the single human pDC level. Cell Rep., 33, 108485–108485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guneykaya, D., Ivanov, A., Hernandez, D.P., Haage, V., Wojtas, B., Meyer, N., Maricos, M., Jordan, P., Buonfiglioli, A., Gielniewski, B. et al. (2018) Transcriptional and translational differences of microglia from male and female brains. Cell Rep., 24, 2773–2783.e2776. [DOI] [PubMed] [Google Scholar]
- 92.McDonald, G., Cabal, N., Vannier, A., Umiker, B., Yin, R.H., Orjalo, A.V., Johansson, H.E., Han, J.H. and Imanishi-Kari, T. (2015) Female bias in systemic lupus erythematosus is associated with the differential expression of X-linked toll-like receptor 8. Front. Immunol., 6, 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu, B., Qi, Y., Li, R., Shi, Q., Satpathy, A.T. and Chang, H.Y. (2021) B cell-specific XIST complex enforces X-inactivation and restrains atypical B cells. Cell, 184, 1790–1803.e1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Odhams, C.A., Roberts, A.L., Vester, S.K., Duarte, C.S.T., Beales, C.T., Clarke, A.J., Lindinger, S., Daffern, S.J., Zito, A., Chen, L. et al. (2019) Interferon inducible X-linked gene CXorf21 may contribute to sexual dimorphism in systemic lupus erythematosus. Nat. Commun., 10, 2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ji, L., Fan, X., Hou, X., Fu, D., Bao, J., Zhuang, A., Chen, S., Fan, Y. and Li, R. (2020) Jieduquyuziyin prescription suppresses inflammatory activity of MRL/lpr mice and their bone marrow-derived macrophages. Front. Pharmacol., 11, 1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scofield, R.H., Bruner, G.R., Namjou, B., Kimberly, R.P., Ramsey-Goldman, R., Petri, M., Reveille, J.D., Alarcón, G.S., Vilá, L.M., Reid, J. et al. (2008) Klinefelter's syndrome (47,XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum., 58, 2511–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spatz, A., Borg, C. and Feunteun, J. (2004) X-chromosome genetics and human cancer. Nat. Rev. Cancer, 4, 617–629. [DOI] [PubMed] [Google Scholar]
- 98.Agrelo, R. and Wutz, A. (2010) ConteXt of change–X inactivation and disease. EMBO Mol. Med., 2, 6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barr, M.L. and Moore, K.L. (1957) Chromosomes, sex chromatin, and cancer. Proc. Can. Cancer Conf., 2, 3–16. [PubMed] [Google Scholar]
- 100.Chaligné, R., Popova, T., Mendoza-Parra, M.A., Saleem, M.A., Gentien, D., Ban, K., Piolot, T., Leroy, O., Mariani, O., Gronemeyer, H. et al. (2015) The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res., 25, 488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yildirim, E., Kirby, J.E., Brown, D.E., Mercier, F.E., Sadreyev, R.I., Scadden, D.T. and Lee, J.T. (2013) Xist RNA is a potent suppressor of hematologic cancer in mice. Cell, 152, 727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Anguera, M.C., Sadreyev, R., Zhang, Z., Szanto, A., Payer, B., Sheridan, S.D., Kwok, S., Haggarty, S.J., Sur, M., Alvarez, J. et al. (2012) Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell, 11, 75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen, D.L., Ju, H.Q., Lu, Y.X., Chen, L.Z., Zeng, Z.L., Zhang, D.S., Luo, H.Y., Wang, F., Qiu, M.Z., Wang, D.S. et al. (2016) Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J. Exp. Clin. Cancer Res., 35, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mo, Y., Lu, Y., Wang, P., Huang, S., He, L., Li, D., Li, F., Huang, J., Lin, X., Li, X. et al. (2017) Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour Biol., 39, 1010428317690999. [DOI] [PubMed] [Google Scholar]
- 105.Song, P., Ye, L.F., Zhang, C., Peng, T. and Zhou, X.H. (2016) Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene, 592, 8–14. [DOI] [PubMed] [Google Scholar]
- 106.Salvador, M.A., Wicinski, J., Cabaud, O., Toiron, Y., Finetti, P., Josselin, E., Lelièvre, H., Kraus-Berthier, L., Depil, S., Bertucci, F. et al. (2013) The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin. Cancer Res., 19, 6520–6531. [DOI] [PubMed] [Google Scholar]
- 107.Huang, K.C., Rao, P.H., Lau, C.C., Heard, E., Ng, S.K., Brown, C., Mok, S.C., Berkowitz, R.S. and Ng, S.W. (2002) Relationship of XIST expression and responses of ovarian cancer to chemotherapy. Mol. Cancer Ther., 1, 769–776. [PubMed] [Google Scholar]
- 108.Kang, J., Lee, H.J., Kim, J., Lee, J.J. and Maeng, L.S. (2015) Dysregulation of X chromosome inactivation in high grade ovarian serous adenocarcinoma. PLoS One, 10, e0118927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Forsberg, L.A., Gisselsson, D. and Dumanski, J.P. (2017) Mosaicism in health and disease - clones picking up speed. Nat. Rev. Genet., 18, 128–142. [DOI] [PubMed] [Google Scholar]
- 110.Pageau, G.J., Hall, L.L., Ganesan, S., Livingston, D.M. and Lawrence, J.B. (2007) The disappearing Barr body in breast and ovarian cancers. Nat. Rev. Cancer, 7, 628–633. [DOI] [PubMed] [Google Scholar]
- 111.Asim, A., Agarwal, S., Avasthi, K.K., Sureka, S., Rastogi, N., Dean, D.D. and Mohindra, S. (2020) Investigation of LOY in prostate, pancreatic, and colorectal cancers in males: a case-control study. Expert. Rev. Mol. Diagn., 20, 1259–1263. [DOI] [PubMed] [Google Scholar]
- 112.Machiela, M.J., Zhou, W., Karlins, E., Sampson, J.N., Freedman, N.D., Yang, Q., Hicks, B., Dagnall, C., Hautman, C., Jacobs, K.B. et al. (2016) Female chromosome X mosaicism is age-related and preferentially affects the inactivated X chromosome. Nat. Commun., 7, 11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou, Q., Hu, W., Zhu, W., Zhang, F., Lin-Lin, L., Liu, C., Songyang, Y.Y., Sun, C.C. and Li, D. (2018) Long non coding RNA XIST as a prognostic cancer marker - a meta-analysis. Clin. Chim. Acta, 482, 1–7. [DOI] [PubMed] [Google Scholar]
- 114.Fang, H., Bonora, G., Lewandowski, J.P., Thakur, J., Filippova, G.N., Henikoff, S., Shendure, J., Duan, Z., Rinn, J.L., Deng, X. et al. (2020) Trans- and cis-acting effects of Firre on epigenetic features of the inactive X chromosome. Nat. Commun., 11, 6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beltrán-Anaya, F.O., Cedro-Tanda, A., Hidalgo-Miranda, A. and Romero-Cordoba, S.L. (2016) Insights into the regulatory role of non-coding RNAs in cancer metabolism. Front. Physiol., 7, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jäger, N., Schlesner, M., Jones, D.T., Raffel, S., Mallm, J.P., Junge, K.M., Weichenhan, D., Bauer, T., Ishaque, N., Kool, M. et al. (2013) Hypermutation of the inactive X chromosome is a frequent event in cancer. Cell, 155, 567–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Akdemir, K.C., Le, V.T., Chandran, S., Li, Y., Verhaak, R.G., Beroukhim, R., Campbell, P.J., Chin, L., Dixon, J.R., Futreal, P.A. et al. (2020) Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. Nat. Genet., 52, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cook, M.B., McGlynn, K.A., Devesa, S.S., Freedman, N.D. and Anderson, W.F. (2011) Sex disparities in cancer mortality and survival. Cancer Epidemiol. Biomark. Prev., 20, 1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zuo, T., Wang, L., Morrison, C., Chang, X., Zhang, H., Li, W., Liu, Y., Wang, Y., Liu, X., Chan, M.W. et al. (2007) FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell, 129, 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dunford, A., Weinstock, D.M., Savova, V., Schumacher, S.E., Cleary, J.P., Yoda, A., Sullivan, T.J., Hess, J.M., Gimelbrant, A.A., Beroukhim, R. et al. (2017) Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet., 49, 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shriver, S.P., Bourdeau, H.A., Gubish, C.T., Tirpak, D.L., Davis, A.L., Luketich, J.D. and Siegfried, J.M. (2000) Sex-specific expression of gastrin-releasing peptide receptor: relationship to smoking history and risk of lung cancer. J. Natl. Cancer Inst., 92, 24–33. [DOI] [PubMed] [Google Scholar]
- 122.Talon, I., Janiszewski, A., Chappell, J., Vanheer, L. and Pasque, V. (2019) Recent advances in understanding the reversal of gene silencing during X chromosome reactivation. Front. Cell Dev. Biol., 7, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chitiashvili, T., Dror, I., Kim, R., Hsu, F.M., Chaudhari, R., Pandolfi, E., Chen, D., Liebscher, S., Schenke-Layland, K., Plath, K. and Clark, A. (2020) Female human primordial germ cells display X-chromosome dosage compensation despite the absence of X-inactivation. Nat. Cell Biol., 22, 1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sripathy, S., Leko, V., Adrianse, R.L., Loe, T., Foss, E.J., Dalrymple, E., Lao, U., Gatbonton-Schwager, T., Carter, K.T., Payer, B. et al. (2017) Screen for reactivation of MeCP2 on the inactive X chromosome identifies the BMP/TGF-β superfamily as a regulator of XIST expression. Proc. Natl. Acad. Sci. U. S. A., 114, 1619–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vacca, M., Tripathi, K.P., Speranza, L., Aiese Cigliano, R., Scalabrì, F., Marracino, F., Madonna, M., Sanseverino, W., Perrone-Capano, C., Guarracino, M.R. and D'Esposito, M. (2016) Effects of Mecp2 loss of function in embryonic cortical neurons: a bioinformatics strategy to sort out non-neuronal cells variability from transcriptome profiling. BMC Bioinformatics, 17(Suppl 2), 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Przanowski, P., Wasko, U., Zheng, Z., Yu, J., Sherman, R., Zhu, L.J., McConnell, M.J., Tushir-Singh, J., Green, M.R. and Bhatnagar, S. (2018) Pharmacological reactivation of inactive X-linked. Proc. Natl. Acad. Sci. U. S. A., 115, 7991–7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Halmai, J.A.N.M., Deng, P., Gonzalez, C.E., Coggins, N.B., Cameron, D., Carter, J.L., Buchanan, F.K.B., Waldo, J.J., Lock, S.R., Anderson, J.D. et al. (2020) Artificial escape from XCI by DNA methylation editing of the CDKL5 gene. Nucleic Acids Res., 48, 2372–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lessing, D., Anguera, M.C. and Lee, J.T. (2013) X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu. Rev. Genomics Hum. Genet., 14, 85–110. [DOI] [PubMed] [Google Scholar]
- 129.Papp, B. and Plath, K. (2013) Epigenetics of reprogramming to induced pluripotency. Cell, 152, 1324–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zitzmann, M., Bongers, R., Werler, S., Bogdanova, N., Wistuba, J., Kliesch, S., Gromoll, J. and Tüttelmann, F. (2015) Gene expression patterns in relation to the clinical phenotype in Klinefelter syndrome. J. Clin. Endocrinol. Metab., 100, E518–E523. [DOI] [PubMed] [Google Scholar]


