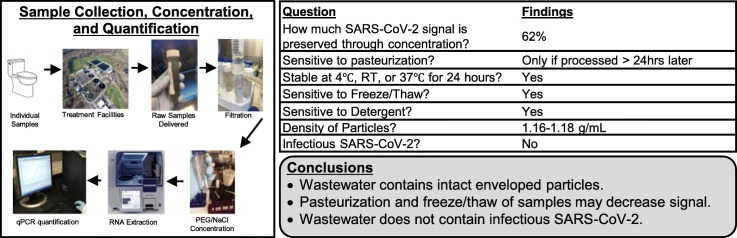
Abstract
SARS-CoV-2 genetic material has been detected in raw wastewater around the world throughout the COVID-19 pandemic and has served as a useful tool for monitoring community levels of SARS-CoV-2 infections. SARS-CoV-2 genetic material is highly detectable in a patient's feces and the household wastewater for several days before and after a positive COVID-19 qPCR test from throat or sputum samples. Here, we characterize genetic material collected from raw wastewater samples and determine recovery efficiency during a concentration process. We find that pasteurization of raw wastewater samples did not reduce SARS-CoV-2 signal if RNA is extracted immediately after pasteurization. On the contrary, we find that signal decreased by approximately half when RNA was extracted 24–36 h post-pasteurization and ~90% when freeze-thawed prior to concentration. As a matrix control, we use an engineered enveloped RNA virus. Surprisingly, after concentration, the recovery of SARS-CoV-2 signal is consistently higher than the recovery of the control virus leading us to question the nature of the SARS-CoV-2 genetic material detected in wastewater. We see no significant difference in signal after different 24-hour temperature changes; however, treatment with detergent decreases signal ~100-fold. Furthermore, the density of the samples is comparable to enveloped retrovirus particles, yet, interestingly, when raw wastewater samples were used to inoculate cells, no cytopathic effects were seen indicating that wastewater samples do not contain infectious SARS-CoV-2. Together, this suggests that wastewater contains fully intact enveloped particles.
Keywords: Coronavirus, COVID-19, Wastewater, Sewage, Surveillance, Pasteurization
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), was first identified in Wuhan, China in December 2019 and was declared a global pandemic by the World Health Organization (WHO) in March 2020. To date, SARS-CoV-2 has produced >201 million cases and >4.2 million COVID-19 related deaths worldwide (WHO, August 5th 2021). SARS-CoV-2 has been shown to be spread primarily by respiratory droplets and occasionally by aerosols (Liu et al., 2020; Port et al., 2021; Crawford et al., 2021; Wang and Du, 2020).
SARS-CoV-2 has 75–80% nucleotide similarity to severe acute respiratory syndrome coronavirus (SARS-CoV) that was responsible for outbreaks of severe acute respiratory syndrome in 2002 and 2003 in Guangdong Province, China (Zhu et al., 2020; Zhong et al., 2003; Ksiazek et al., 2003; Drosten et al., 2003). Both SARS-CoV and SARS-CoV-2 use the cellular receptor Angiotensin-converting enzyme 2 (ACE2) which is highly expressed in the lung and oral mucosa and expressed at lower levels in the digestive tract (Yan et al., 2020; Hamming et al., 2004; Xu et al., 2020). Due partially to population sizes, material shortages, inaccessibility to laboratory equipment, a vast array of disease severity, and healthcare coverage concerns, it has not been possible to test every individual regularly for a SARS-CoV-2 infection. Even as testing becomes more and more available, asymptomatic individuals may not get tested and can unknowingly continue spread. These limits cause difficulty in monitoring community spread.
It has been reported that SARS-CoV, SARS-CoV-2, and other coronavirus RNA is detectable in feces of infected patients up to 6 to 10 days before symptom onset (Corman et al., 2016; Leung et al., 2003; Gu et al., 2020; Holshue et al., 2020; Song et al., 2020; Chen et al., 2020a). Additionally, screening of sewage, both community and hospital, for the detection and prevalence of viruses including SARS-CoV-2, Poliovirus, noroviruses, adenoviruses, rotaviruses, polyomaviruses, Hepatitis A virus, and gastroenteritis viruses have been documented and can correlate closely with the occurrence of cases in the community (Katayama et al., 2008; Fumian et al., 2010a; Wang et al., 2005a; Kroiss et al., 2018; Peccia et al., 2020; Asghar et al., 2014; Bofill-Mas et al., 2000; Fumian et al., 2010b; Pintó et al., 2007; Rodríguez-Díaz et al., 2009; Victoria et al., 2010; Villena et al., 2003). This supports wastewater surveillance as a useful method for monitoring community levels of SARS-CoV-2 infections (Peccia et al., 2020; Baldovin et al., 2021; Agrawal et al., 2021; Cao and Francis, 2021; Bivins et al., 2020; Venugopal et al., 2020).
Unlike from sputum, samples collected from feces of infected patients do not generally appear to contain infectious viral particles despite high levels of detectable viral RNA (Wolfel et al., 2020a; Chen et al., 2020b; Ling et al., 2020; Wolfel et al., 2020b); however, infectious particles cultured from feces has been reported before (Xiao et al., 2020). To date and to the best of our knowledge, there have been no confirmed cases of COVID-19 linked directly to wastewater treatment plants. Several groups have examined the survival rate of various other coronaviruses in raw, unpasteurized wastewater, and generally conclude that after a maximum of 3 days there is a 99.9% decrease in infectivity (Gundy et al., 2009; Wang et al., 2005b; Ye et al., 2016; La Rosa et al., 2020; Carducci et al., 2020).
During periods of lower community infection rates, it was necessary to concentrate raw wastewater samples to reliably detect SARS-CoV-2 genetic material by quantitative reverse transcription polymerase chain reaction PCR (qPCR). Viral concentration from wastewater has been done using several different methods, and comparison of methods has been the focus of several manuscripts since early 2020 (Ahmed et al., 2020; Corpuz et al., 2020; Philo et al., 2021). Our lab used a well-known Polyethylene glycol (PEG) and NaCl method for viral concentration that has been used for over 50 years and was first used with SARS-CoV-2 in 2020 (Kanarek and Tribe, 1967; Vajda, 1978; Kohno et al., 2002; Wu et al., 2020). As a control for viral recovery throughout concentration, we used a unique enveloped RNA virus containing an artificial gene sequence not found in nature. Interestingly, we noticed a disparity in recovery rates between our control virus and SARS-CoV-2 signal, leading us to question the difference in make-up of the genetic material detectable in wastewater in comparison to our control virus. In this innovative manuscript, we examine recovery, temperature resistance, density, detergent resistance, and infectivity and conclude, for the first time, that the genomic material detected in wastewater is enveloped and non-infectious.
2. Methods
2.1. Plasmids and Puro Virus production
The NL4-3 derived HIV containing the CMV driven Puromycin resistance gene and lacking the accessory genes Vif, Vpr, Nef, and Env was engineered using InFusion Cloning (TaKara). To make this construct, we used a previously described NL4-3 derived HIV-CMV-GFP provided by Vineet Kewal Rammani (National Cancer Institute (NCI) – Frederick) (Zufferey et al., 1997; Lucas et al., 2010). This proviral vector lacks the accessory genes vif, vpr, nef, and env and contains a CMV promoter driven GFP in the place of nef. The NL4-3 derived HIV-CMV-GFP was digested using Stu1 and Xma1 (New England Biolabs (NEB)) to remove the GFP gene, and a gBlock fragment (Integrated DNA Technologies (IDT)) of a uniquely codon optimized Puromycin resistance gene was put in its place. The unique puromycin resistance gene sequence is as follows:
ATGACAGAGTATAAGCCAACCGTCCGGCTCGCAACGAGAGACGATGTCCCGAGGGCAGTGCGCACGCTCGCCGCGGCCTTTGCGGACTACCCTGCAACAAGACACACTGTGGATCCCGATCGCCACATAGAGCGCGTGACTGAGCTGCAAGAACTGTTCCTTACCAGGGTGGGTCTCGATATCGGTAAGGTTTGGGTCGCCGACGACGGAGCGGCAGTGGCAGTCTGGACCACTCCTGAGAGCGTAGAAGCAGGCGCAGTGTTTGCAGAAATTGGCCCTAGAATGGCCGAATTGTCCGGTAGCCGGCTCGCTGCTCAGCAGCAGATGGAAGGCCTGCTCGCACCTCACAGACCCAAAGAACCCGCGTGGTTCCTGGCGACAGTGGGAGTCAGTCCAGACCATCAGGGCAAAGGTCTCGGCTCAGCAGTTGTACTGCCTGGGGTAGAGGCCGCAGAAAGGGCAGGGGTGCCGGCCTTCCTGGAAACATCTGCACCCAGAAACTTGCCTTTCTACGAGAGGCTGGGATTCACCGTTACCGCCGACGTGGAGGTGCCCGAAGGACCGCGCACTTGGTGCATGACGAGAAAGCCCGGGGCTTGA
To create the control sequence for qPCR standard curve, the plasmid described above was digested with EcoR1. The primer pair (COVID19-N 5p: 5′ ATGTCTGATAATGGACCCCAAAATCAGCG 3; COVID19-N 3p: 5′ TTAGGCCTGAGTTGAGTCAGCACTGC 3′) was used to amplify the N ORF fragment from IDT's 2019-nCoV_N_Positive Control plasmid and cloned in a pSC-A of StrataClone PCR cloning kit of Agilent for sequence check. Later, the N ORF fragments were infused using an InFusion kit (Takara) as described above.
The control, Puro Virus, was made via stable cell line. To make the cell line, The above plasmid and Vesicular Stomatitis Virus protein G (VSV-G) expression plasmid were transfected into HEK293FT cells (Invitrogen, Carlsbad, CA, USA). After 48 h, viral supernatant was collected and used to transduce fresh HEK293FT cells which were selected with puromycin 48 h post-transduction. HEK293FT cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 7.5% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, and 10 mM nonessential amino acids. Virus was quantified using qPCR (described below).
2.2. Quantitative RT-qPCR assay
The TaqMan probe (VIC-5′ CGGTAAGGTTTGGGTCGCCGAC 3′-QSY) and the primer pair (puro Forward: 5′ CCCGATCGCCACATAGAGC 3′; puro Reverse: 5′ CCATTCTAGGGCCAATTTCTGC 3′) were designed and used to target the puro RNA described above. Primers and probe specificity was tested by BLAST analysis (NCBI) to prevent known nonspecific binding targets that could be obtained in a human specimen. The choice of VIC fluorescent dye for the puro TaqMan probe is for the application in the multiplex reactions with the SARS-CoV-2 N gene TaqMan probe utilizing the FAM reporter. The TaqMan probe (FAM-5′ ACCCCGCATTACGTTTGGTGGACC 3′ BHQ1) and the primer pair (2019-nCoV_N1-F: 5′ GACCCCAAAATCAGCGAAAT 3′; 2019-nCoV_N1-R: 5′ TCTGGTTACTGCCAGTTGAATCTG 3′) for N1 detection, and The TaqMan probe (FAM 5′ ACAATTTGCCCCCAGCGCTTCAG 3′ BHQ1) and the primer pair (2019-nCoV_N2-F: 5′ TTACAAACATTGGCCGCAAA 3′; 2019-nCoV_N2-R: 5′ GCGCGACATTCCGAAGAA 3′) for N2 detection were purchased from Integrated DNA Technologies (IDT), based on the CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel (Acceptable Alternative Primer and Probe Sets) https://www.cdc.gov/coronavirus/2019-ncov/downloads/List-of-Acceptable-Commercial-Primers-Probes.pdf.
A plasmid (described above) carrying a unique puro resistance gene fragment along with a N gene fragment was constructed, purified from Escherichia coli, and used as standards for the RT-qPCR assay to ensure an equal molar ratio of puro and N gene detection. A standard curve was constructed at concentrations of 200,000 through 2 gene copies μL−1 and utilized to determine the copy number of the target puro gene in the wastewater samples that had the puro control virus added prior to concentration as an internal control of RNA extraction rate.
Final RT-qPCR one step mixtures consisted of 5 μL TaqPath 1-step RT-qPCR Master Mix (Thermo Fisher cat# A15299), 500 nM each of puro or SARS-CoV-2 primers, 125 nM of TaqMan probe, 5 μL of wastewater RNA extract and RNase/DNase-free water to reach a final volume of 20 μL. All RT-qPCR assays were performed using a 7500 Fast real-time qPCR machine (Applied Biosystems). The reactions were initiated with 1 cycle of Uracil N-glycosylase (UNG) incubation at 25 °C for 2 min to eliminate carryover and then 1 cycle of reverse transcription at 50 °C for 15 min, followed by 1 cycle of activation of DNA polymerase at 95 °C for 2 min and then 45 cycles of 95 °C for 3 s for DNA denaturation and 55 °C for 30 s for anneal and extension. The data is collected at the 55 °C extension step.
2.3. Sample acquisition
Wastewater treatment plants from around Missouri collected at least 1 L of 24-hour composite sample of raw sewage from influent wastewater intake. Data such as influent flow rate, pH, water temperature, chemical oxygen demand (COD), and total suspended solids (TSS) was collected. From the 1 L composite sample, 3 × 50 mL aliquots of sample were packaged in coolers containing ice packs and delivered to the University of Missouri within 24 h from collection.
2.4. Concentration and recovery
Nine samples in duplicate containing 50 mL of raw wastewater were stored at 4 °C (18 total 50 mL samples). Raw samples were spun at 2000 ×g for 5 min to remove large particulates, then vacuum filtered through a 0.22 μm filter (Millipore cat# SCGPOO525), mixed with a 50% (wt/v) Polyethylene glycol (PEG)(Research Products International (RPI) cat# P48080) and 1.2 M NaCl solution for a final concentration of 12% PEG and 0.3 M NaCl. NL4-3 derived HIV containing the CMV driven Puromycin resistance gene and lacking the accessory genes Vif, Vpr, Nef, and Env were added to the filtered sample/PEG/NaCl mixture at a concentration of 5.1 × 107 viral particles per sample (50 mL). Samples were mixed thoroughly and kept at 4 °C for 1 h, then spun at 12,000 ×g at 4 °C for 2 h. All but 140 μL of supernatant was removed, and the entire volume was extracted for RNA purification. RNA was extracted from the samples using the Qiagen QIAmp Viral RNA mini kit (cat# 52906) in a QIAcube Connect (Qiagen cat# 9002864). Additionally, 140 μL of wastewater was collected prior to filtering the sample, after filtration (before addition of PEG solution); these samples were multiplied to be comparable to the full 50 mL volume. After concentration of virus. Viral recovery was determined by qPCR as described above. Importantly, for samples containing high copy numbers of SARS-CoV-2 genetic material (>1.5 million copies/L), it was not necessary to concentrate samples, so for some experiments, unconcentrated wastewater was used to remove variables that may be introduced in concentration.
2.5. Stability assessment
Six samples containing 50 mL of raw wastewater were stored at 4 °C. Samples were mixed gently and split into 3 × 16.7 mL aliquots. Aliquots were stored at either 4 °C, Room Temperature (RT) (~22 °C), or 37 °C for 24 h. After 24 h, 140 μL of sample was collected, RNA was extracted, and viral recovery was determined as previously described. As the samples had a concentration of SARS-CoV-2 signal over 1.5 million copies/L, the concentration step was not necessary and removed any further variables that would be introduced from this step. Statistics are a paired, two tailed Student's t-test run on Microsoft Excel. The function used was =t-test, array1, array2, tails, type where array1 and array2 are the values of each sample at each condition, respectively, and options for two tails and paired tests were selected.
2.6. Pasteurization
Six duplicate samples containing 50 mL of raw wastewater were stored at 4 °C (12 total samples). One 50 mL tube of each duplicate sample was kept at 4 °C, and the other 50 mL tube of sample was incubated at 60 °C for 2 h then the entire sample was concentrated as described above. RNA extraction from the entire pellet, RT-qPCR, and statists were performed as described above in 2.4, 2.2, 2.5, respectively.
For pasteurization effect on signal 24 h later, six duplicate samples were pasteurized and RNA extraction was done either immediately after the 2-hour incubation at 60 °C or 24–36 h after pasteurization (12 total samples). RNA extraction, RT-qPCR, and statistics were performed as described above in 2.4, 2.2, 2.5, respectively.
2.7. Freeze-thaw sensitivity
Six duplicate samples each containing 50 mL of raw wastewater collected from a wastewater facility as described above, the same week, were stored at 4 °C. At the start of the study, one 50 mL tube of each duplicate sample was kept at 4 °C, and the other 50 mL tube of sample was stored at −80 °C for 36 h then thawed and concentrated as described above. RNA extraction, RT-qPCR, and statists were performed as described above in 2.4, 2.2, 2.5, respectively.
2.8. Detergent sensitivity
Six duplicate samples containing 50 mL of raw wastewater were collected as described above and stored at 4 °C (12 total samples). One of each duplicate sample was treated with either 1% Triton X 100 or PBS for 2 h at 37 °C. From unconcentrated, raw wastewater samples, RNA extraction, RT-qPCR, and statists were performed as described above in 2.4, 2.2, 2.5, respectively.
2.9. Density
Three samples containing 50 mL of raw wastewater were stored at 4 °C. The Puro Virus (NL4-3 derived HIV containing CMV driven Puromycin resistance and lacking the accessory genes Vif, Vpr, Nef, and Env) was added to raw samples at a concentration of 5.1 × 107 viral particles per sample. Samples were concentrated as described above. Concentrated samples were then added to a density gradient ranging from 0% to 28% iodixanol in a 0.25 M sucrose dilutant according to the Optiprep protocol (Sigma cat# 92339-11-2). Gradients were spun in a Sorvall Discovery 100SE ultracentrifuge at 31,000 RPM for 3 h at 4 °C. After centrifugation, the gradient was fractioned. RNA extraction and RT-qPCR were performed as described above in 2.4, 2.2, respectively. Density of each fraction was confirmed using a densitometer (Abbe, model: C10).
2.10. Infectivity
Ten raw wastewater samples from the week of June 28th 2021 were collected and brought to University of Missouri as described in Section 2.3. Fresh samples were filtered through a 0.22 mm filter (Millipore cat# SCGPOO525). For the cell maintenance, the Vero E6 cells (CRL1586™, ATCC) were maintained in Dulbecco's Modified Eagle Medium (GIBCO, Grand Island, NY) containing 10% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA) at 37 °C with 5% CO2. For the first recovery, 200 ml of each sample was inoculated to Vero E6 cells in 6-well plates at a confluence of ~90%. After 1 h of adsorption, the inoculum was removed, and the cells were washed with PBS and covered with fresh optiMEM (Gibco, Thermo Fisher Scientific) containing 1× Antibiotic-Antimycotic (Gibco, Thermo Fisher Scientific). Three days post inoculation, 1 mL of the supernatant from the last virus recovery was centrifuged and inoculated to fresh Vero E6 cells for the second and third virus recovery and cytopathic effect was observed. A clinical isolate from Missouri, SARS-CoV-2/human/USA/20 × 1003/2020, (Full genome available at GenBank Accession ID: MW521470.1) was used as the positive control at a multiplicity of infection (MOI) of 0.001. This isolate has a D614G mutation.
3. Results
3.1. SARS-CoV-2 signal recovery through concentration
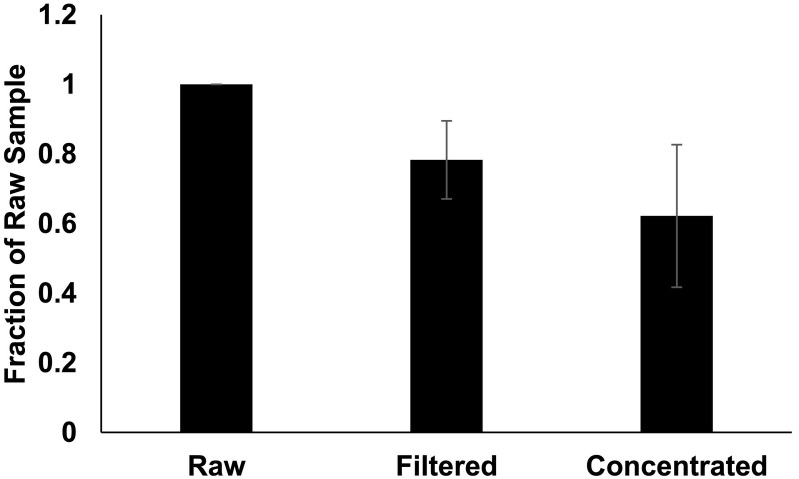
Generally, it has been necessary to concentrate wastewater samples for reliable detection of SARS-CoV-2 by qPCR. Because concentration is sometimes necessary, it is important to know the rate of recovery throughout the process. During a COVID-19 outbreak at a Missouri prison, we were able to reliably detect signal in raw, unconcentrated wastewater as signal was high in this wastewater collection facility at the time. This allowed us to compare signal before concentration and after each subsequent step (Fig. 1 ). We extracted RNA from samples before filtration, after filtration, and after concentration for qPCR quantification. Unconcentrated sample numbers were multiplied based on the volume of original sample to ensure that recovery could be compared throughout the concentration process. Filtering preserved an average of 78% of signal and concentration preserved an average of 62% signal when compared to raw samples.
Fig. 1.
Recovery. N = 9. Raw samples were spun at 2000 ×g for 5 min to remove large particulates, then vacuum filtered through a 0.22 μm filter, and mixed with Polyethylene glycol (PEG) and NaCl solution for a final concentration of 12% PEG and 0.3 M NaCl. Samples were mixed thoroughly and kept at 4 °C for 1 h, then spun at 12,000 ×g at 4 °C for 2 h. RNA was extracted from pellet, and viral recovery was determined by qPCR. Wastewater was collected prior to filtering the sample, after filtration (before addition of PEG solution), and after concentration of virus. Signal from unconcentrated samples was multiplied based on the total volume of sample to be concentrated to allow for equal comparison at each step. Error bars represent standard deviation.
3.2. Pasteurization and freeze-thaw can reduce SARS-CoV-2 signal
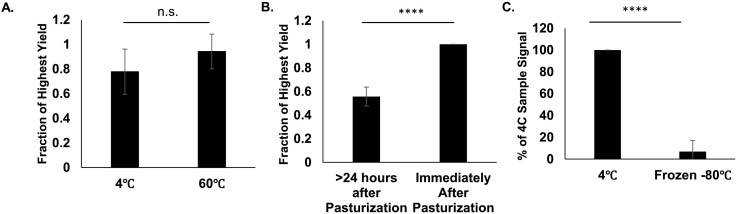
Although wastewater is not thought to contain infectious SARS-CoV-2, raw wastewater contains a variety of other pathogens. Filtration through a 0.22 μm filter should remove many of these pathogens, but some have suggested pasteurizing samples at 60 °C for 1 to 2 h to inactivate potential pathogens (Pecson et al., 2021). To test the effect on signal due to pasteurization, duplicate samples were kept at either 4 °C or 60 °C for 2 h prior to RNA extraction (Fig. 2A). We found no significant difference between pasteurized and non-pasteurized samples (P value = 0.23) when RNA was extracted immediately after pasteurization; however, it is important to note that signal dropped significantly if samples were returned to 4 °C after pasteurization and RNA was collected 24–36 h after pasteurization was completed (Fig. 2B). A 24–36-hour period between pasteurization resulted in a 44% reduction in signal (P value = 2.05 × 10−6). Additionally, in a separate experiment, when samples were frozen at −80 °C for 36 h and thawed prior to concentration, over 90% of the SARS-CoV-2 signal was lost (Fig. 2C).
Fig. 2.
Impact of Pasteurization and Freeze-Thaw. A) N = 6. Duplicate samples were kept at 4 °C or incubated at 60 °C for 2 h. Raw samples were concentrated, and RNA was extracted from the pellet. Viral recovery was determined by qPCR. Fraction of Highest yield was calculated by the ratio signal between treatments. Error bars represent standard deviation. P-Value = 0.23 B) N = 6. Duplicate samples were kept at 4 °C or incubated at 60 °C for 2 h. RNA was extracted from raw samples either immediately after 2-hour incubation at 60 °C or 24–36 h after pasteurization. Viral recovery was determined by qPCR. Fraction of Highest yield was calculated by the ratio signal between treatments. Error bars represent standard deviation. P-Value = 2.05E−6. C) N = 6. Duplicate raw samples were kept at kept at 4 °C or −80 °C for 48 h. Raw Samples were concentrated, and RNA was extracted from the pellet. Viral recovery was determined by qPCR. Error bars represent standard deviation. P-Value = 3.13E−6.
3.3. SARS-CoV-2 signal is higher than enveloped virus control
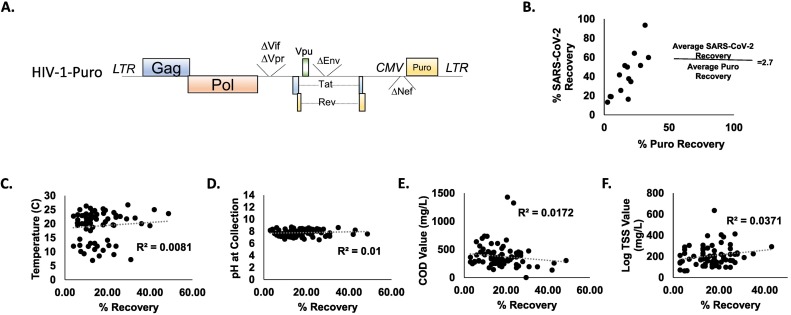
As a control throughout wastewater screening, an NL4-3 derived HIV virus containing a CMV driven Puromycin resistance gene and lacking the accessory genes Vif, Vpr, Nef, and Env (Henceforth called ‘Puro Virus’) was added to raw wastewater samples at a concentration of 5.1 × 107 viral particles per sample (Fig. 3A). Importantly, the Puro Virus contains a uniquely codon optimized puromycin resistance gene. This vector was chosen because the unique sequence present in the Puro Virus ensures that any signal detected throughout our experiments with this probe is from our internal matrix control and not the environment ensuring that nothing environmental will be amplified in our control. Additionally, Both SARS-CoV-2 and the Puro Virus are positive-sense RNA contained in an envelope at a similar size. Interestingly, upon comparison of signal detected in highly potent samples (>1.5 million copies/L of SARS-CoV-2 signal) before and after concentration, SARS-CoV-2 recovery was consistently higher than the Puro Virus recovery (Fig. 3B). On average, SARS-CoV-2 recovery was 2.7-fold higher than Puro Virus recovery (N = 14). The consistent disparity between the SARS-CoV-2 recovery and the Puro Virus recovery led us to hypothesize that the SARS-CoV-2 signal was coming from a different source than enveloped RNA, such as non-enveloped ribonuclear complexes, as we would expect that most enveloped particles would interact similarly with PEG during concentration.
Fig. 3.
A) Schematic of Puro Virus Control. NL4-3 derived HIV containing CMV driven, uniquely codon optimized, Puromycin resistance and lacking the accessory genes Vif, Vpr, Nef, and Env. B) Relative Recovery of Puro Virus Signal and SARS-CoV-2 Signal. Samples were spiked with Puro Virus at a concentration of 5.1 × 107 viral particles per sample. RNA was extracted from samples both before and after concentration, and viral recovery was determined by qPCR. C–F) Parameters at time of Collection in Relation to Control Recovery C) N = 68. Temperature D) N = 97 pH value E) N = 84. COD F) N = 69. log TSS value of wastewater at the end of 24-hour composite sample collection. R2 values for linear regression lines are 0.0081, 0.01, 0.0172, and 0.0371, respectively.
Using the Puro Virus control, we also examined parameters at time of wastewater collection including Temperature, pH value, Chemical Oxygen Demand (COD) value, and Total Suspended Solids (TSS) value and the effect these parameters had on the control puro virus recovery (Fig. 3C–F). This data was analyzed two ways; first, R2 values were calculated for each dataset as shown in Fig. 3C–F. In addition, to determine if chemical or physical parameters in combination could affect the viral recovery, multiple linear regression analysis was performed using SAS 9.4 (SAS Institute Inc., Cary, NC, USA). Two separate full-model analyses were performed as not every sample had all four parameters collected. One analysis included TSS, COD, and pH (n = 55) and as independent variables and one which included temperature, COD, and pH as independent variables (n = 29); puro recovery (%) served as the dependent variable in both tests. Neither the model including TSS nor the model including temperature were significant (P = 0.061, r2 = 0.133 and P = 0.458, r2 = 0.097, respectively) (Data not shown). Following the finding that the Puro Virus had a consistently lower signal than SARS-CoV-2 signal and that chemical parameters at collection do not impact recovery, we aimed to characterize the SARS-CoV-2 genetic material detected in wastewater.
3.4. SARS-CoV-2 signal stability
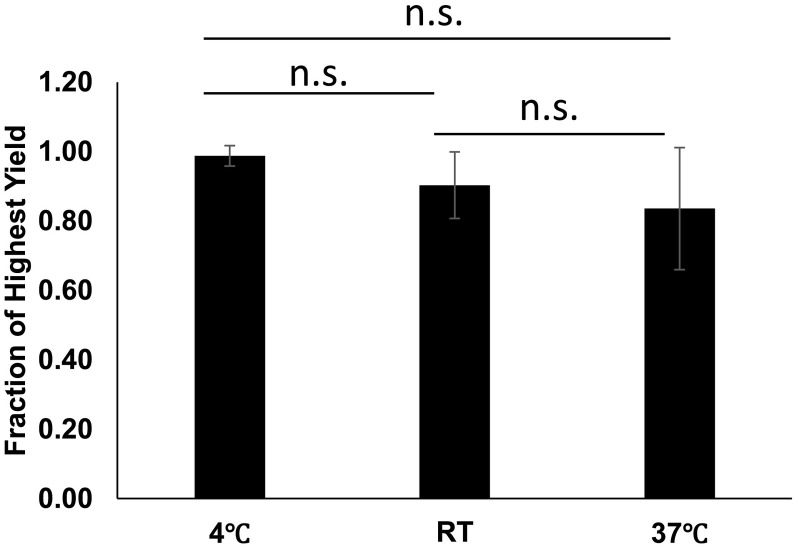
After collection from a wastewater treatment plant as described above, samples are stored at 4 °C until they are processed; however, we wanted to test the stability of samples at a variety of temperatures to determine if temperature control made a large impact on SARS-CoV-2 signal. A portion of each sample was kept at either 4 °C, RT, or 37 °C for 24 h as described in methods. RNA was extracted from each sample, and SARS-CoV-2 signal was quantified using qPCR (Fig. 4 ). Although samples that were maintained at 4 °C had the least variability, there was no significant difference in signal from samples kept at any temperature (P values range 0.097 to 0.363). This finding is interesting as an increase in temperature from 4 °C to either RT or 37 °C could impact the activity of various enzymes that could be present in raw wastewater and impact rates of degradation of genetic material.
Fig. 4.
Temperature Stability. N = 6. Samples were mixed gently and split into 3 aliquots. Aliquots were stored at either 4 °C, RT, or 37 °C for 24 h. After 24 h, RNA was extracted, and viral recovery was determined by qPCR. Error bars represent standard deviation. P-Values: 4 °C to RT = 0.097, 4 °C to 37 °C = 0.108, RT to 37 °C = 0.363.
3.5. Detergent removes SARS-CoV-2 signal
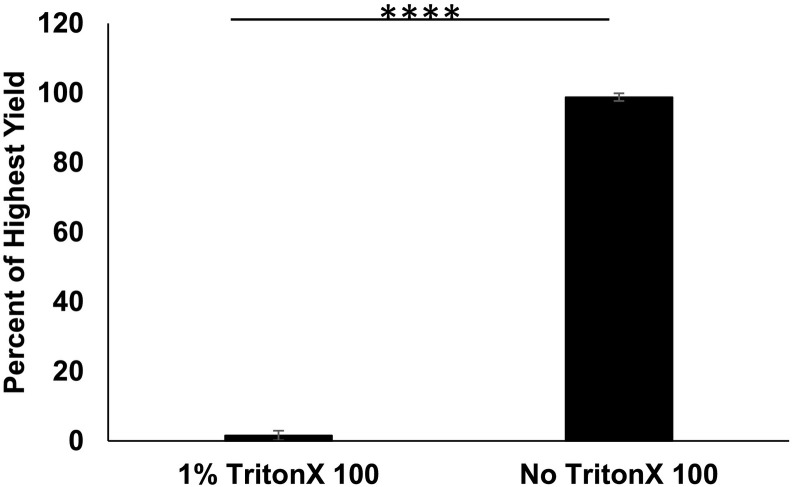
Because SARS-CoV-2 is an enveloped virus, it is very likely to be sensitive to detergent because of a disruption of the lipids composing the envelope. Presence of an envelope may protect genomic material from enzymes that may quickly degrade vulnerable genetic material. We were curious as to whether the SARS-CoV-2 signal detected in wastewater was sensitive to detergent. To answer this question, duplicate samples of raw wastewater were treated with either 1% TritonX-100 or PBS and kept at 37 °C for 2 h (Fig. 5 ). RNA was extracted from 140 μL of unconcentrated wastewater, and samples were quantified using qPCR. Treatment with TritionX-100 reduced signal about 100-fold in comparison to samples treated with PBS alone. Contrary to our original hypothesis, this indicates that SARS-CoV-2 signal detected in wastewater is likely protected by a lipid bilayer, but this finding could also have other causes.
Fig. 5.
Detergent Sensitivity. N = 6. Duplicate samples were treated with either 1% Triton X 100 or PBS for 2 h at 37 °C. RNA was extracted from raw samples, and viral recovery was determined by qPCR. Error bars represent standard deviation. P-Value = 2.10E−20.
3.6. Density of SARS-CoV-2
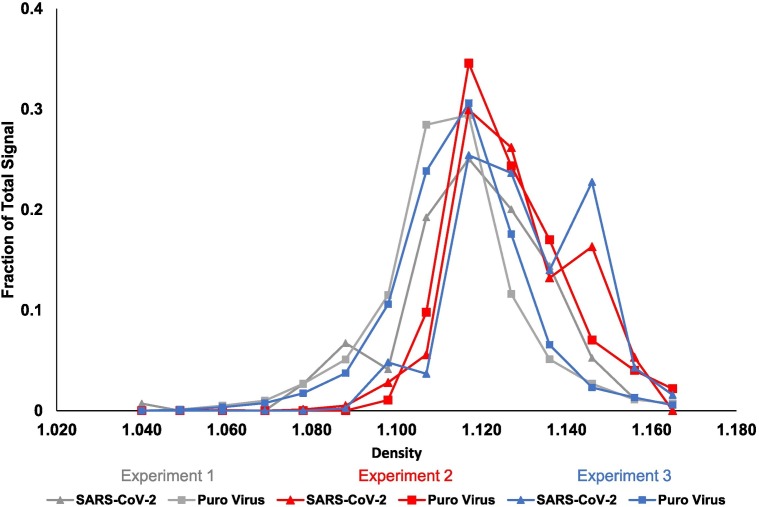
A loss of signal following treatment with detergent could be caused by a variety of things including breaking up of protein complexes. Because of this, we wanted to know if the density of SARS-CoV-2 particles was comparable to densities of other known enveloped viruses. To test density, concentrated wastewater samples also containing the Puro Virus at a concentration of 5.1 × 107 viral particles per sample were run through a density gradient containing 0% to 28% iodixanol in a 0.25 M sucrose dilutant and RNA was extracted from each fraction (Fig. 6 ). Each fraction was then probed by qPCR for SARS-CoV-2 signal and the Puro Virus control. The fraction containing the highest signal for both the Puro Virus and SARS-CoV-2 correlates with a density (ρ) between 1.16 and 1.18 g·mL−1 as calculated according to the Optiprep protocol and confirmed by refractometer. This finding is in agreement with the expected density of retroviruses and further supports that the genetic material in wastewater is similar to an enveloped viral particle (Poiesz et al., 1980).
Fig. 6.
Density. N = 3. Puro Virus was added to raw wastewater samples at a concentration of 5.1 × 107 viral particles per sample. Raw samples were concentrated then added to a density gradient ranging from 0% to 28% iodixanol in a 0.25 M sucrose dilutant and spun in a Sorvall Discovery 100SE ultracentrifuge at 31,000 RPM for 3 h at 4 °C. RNA was extracted from fractions, and viral recovery was determined by qPCR.
3.7. No cytopathic effects from wastewater samples
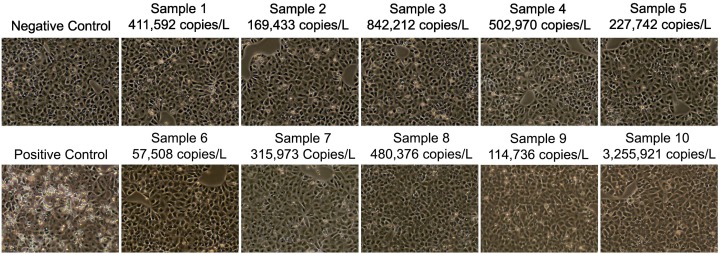
The similarities in density along with detergent sensitivity suggest that the genetic material is enveloped. We were curious to know if wastewater samples contained infectious particles. To examine this question, aliquots of 10 raw wastewater samples with SARS-CoV-2 signals ranging from 169,433 to 3,255,921 Copies/L were collected and used to inoculate Vero E6 cells within a week of collection. Seventy-two hours after inoculation, supernatant was collected and used to inoculate fresh Vero E6 cells. At each passage, cells were examined for cytopathic effects. No cytopathic effects were seen at any point during the experiment (Fig. 7 ). The absence of cytopathic effects suggests that wastewater samples do not contain infectious SARS-CoV-2 Particles. As a positive control, cells were infected with SARS-CoV-2/human/USA/20 × 1003/2020 (GenBank Accession ID: MW521470.1) at a MOI of 0.001. These cells showed significant cytopathic effects whereas the negative control cells had no cytopathic effects.
Fig. 7.
Infectivity. CPE of Vero E6 cells 3 days post inoculation with fresh wastewater samples. Images taken 3 days after the 3rd inoculation. Ten total samples were tested. Numbers above each picture represent the copy number per liter from that sample as measured by qPCR.
4. Discussion
COVID-19 causes a wide variety of symptoms and disease severity. This combined with inaccessibility to testing due partially to supply costs, availability, and varied access to healthcare makes accurately tracking cases of COVID-19 difficult. Like several other viruses, SARS-CoV-2 was shown to be present in the COVID-19 patient feces and therefore wastewater despite being primarily transmitted via respiratory droplets (Corman et al., 2016; Leung et al., 2003; Gu et al., 2020; Holshue et al., 2020; Song et al., 2020). To date, there have been no confirmed cases of SARS-CoV-2 infection from wastewater treatment plants, and infectious virus has not been able to be reliably cultured from wastewater (Gundy et al., 2009; Wang et al., 2005b; Ye et al., 2016). In early stages of the pandemic and during periods of lower community spread of SARS-CoV-2, it has been necessary to concentrate wastewater samples to reliably detect genetic material using qPCR. Using samples from wastewater treatment facilities around Missouri from areas with high community levels of COVID-19, we have shown that filtration and concentration of wastewater samples reliably reserves ~60% of signal from raw wastewater, supporting that this method of filtering and concentration is a consistent method for concentrating and comparing community viral loads (Fig. 1). While the method described here includes filtration through a 0.22 μm filter to remove other debris and bacterial pathogens, other groups have suggested pasteurizing wastewater samples to inactivate any pathogens present in wastewater (Wu et al., 2020; Weidhaas et al., 2021). We have shown here that pasteurizing samples for 2 h does not impact signal if RNA is extracted from samples immediately; however, this does not remain true for samples that have been pasteurized and returned to 4 °C for RNA extraction between 24 and 36 h post-pasteurization (Fig. 2). While the decrease in signal is approximately 2-fold, it remains an important note that pasteurization may make a consistent experimental timeline of higher importance for those who are quantifying viral loads after pasteurization. Some groups such as Wu et al. (2020) and Weidhaas et al. (2021) report no significant difference between pasteurized and unpasteurized samples; however, others, like Palmer et al. (2021) report a 50–55% decrease in SARS-CoV-2 following pasteurization (Wu et al., 2020; Weidhaas et al., 2021; Palmer et al., 2021). The studies done by Wu et al. (2020), Weidhaas et al. (2021), nor Palmer et al. (2021) do not specifically discuss time after pasteurization, so this may be the cause of contradictory data.
Interestingly, we noticed that recovery of the Puro Virus control was consistently about 3-fold lower than the recovery of SARS-CoV-2 through concentration, and that chemical properties of wastewater at time of collection did not impact Puro Virus Recovery (Fig. 3). Initially, this led us to believe that the SARS-CoV-2 signal present in wastewater was coming from a different source than an enveloped particle, but our data suggests the contrary and led us to investigate the nature of the genomic material producing signal.
From the time it is deposited into a sewer system to its arrival at a wastewater treatment plant, a fecal sample may go through a variety of temperature changes. Additionally, as temperatures get closer to 37 °C, enzymes that degrade genetic material may become more active. We show that in a 24-hour period, temperature changes are tolerated as there is no significant difference in signals from samples kept at 4 °C, room temperature, or 37 °C (Fig. 4). This finding is interesting as it suggests some sort of protection of genetic material from degradation and suggests that outdoor temperatures may not impact reliability of signal detection when levels of SARS-CoV-2 genetic material are monitored over time.
Because of genomic material found in feces, it was a high concern early in the pandemic that feces and wastewater may be a source infectious virus; however, efforts to culture infectious virus from fecal or wastewater samples have failed (Chen et al., 2020a; Wolfel et al., 2020a; Ling et al., 2020; Wolfel et al., 2020b). Here we demonstrate that genomic RNA collected from wastewater samples is sensitive to detergent (Fig. 5). This finding suggests that the genomic material is protected by a lipid bilayer as the inactivation of viral particles by detergents has been well documented (Welch et al., 2020; Hellstern and Solheim, 2011; Horowitz et al., 1998; Prince et al., 1986; Roberts, 2008). Additionally, the material concentrated from wastewater samples has a very similar density to non-infectious retroviral particles again supporting that the material concentrated from wastewater shares similarities to an enveloped viral particle (Fig. 6). In agreement with prior findings discussed above, wastewater samples did not contain any infectious SARS-CoV-2 particles (Fig. 7). It is feasible that enzymes present in the digestive tract, such as Trypsin, cleave much of the Spike glycoprotein from virus present in the digestive tract resulting in viral particles that are enveloped yet not infectious. Further studies need to be done to determine whether the genomic material is full length SARS-CoV-2 genomic material and to investigate why the genomic material found in wastewater may be non-infectious while also remaining to be protected by a lipid bilayer.
Funding acknowledgement
Funding for the project was administered by the Missouri Department of Health and Senior Services (DHSS). Research reported in this publication was supported by funding from the Centers for Disease Control and the National Institute on Drug Abuse of the National Institutes of Health under award number U01DA053893-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Centers for Disease Control.
CRediT authorship contribution statement
Carolyn A. Robinson: Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Formal analysis, Writing – original draft. Hsin-Yeh Hsieh: Methodology, Investigation. Shu-Yu Hsu: Investigation. Yang Wang: Investigation. Braxton T. Salcedo: Investigation, Writing – review & editing. Anthony Belenchia: Conceptualization, Data curation, Funding acquisition. Jessica Klutts: Conceptualization, Investigation. Sally Zemmer: Conceptualization, Investigation. Melissa Reynolds: Conceptualization, Data curation, Funding acquisition, Writing – review & editing. Elizabeth Semkiw: Conceptualization, Data curation. Trevor Foley: Conceptualization, Investigation. XiuFeng Wan: Conceptualization, Funding acquisition, Writing – review & editing. Chris G. Wieberg: Conceptualization, Project administration, Funding acquisition, Writing – review & editing. Jeff Wenzel: Conceptualization, Project administration, Funding acquisition, Writing – review & editing. Chung-Ho Lin: Conceptualization, Funding acquisition. Marc C. Johnson: Conceptualization, Project administration, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Editor: Warish Ahmed
References
- Agrawal S., Orschler L., Lackner S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in southern Germany. Sci. Rep. 2021;11(1):5372. doi: 10.1038/s41598-021-84914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar H., et al. Environmental surveillance for polioviruses in the global polio eradication initiative. J. Infect. Dis. 2014;210(Suppl. 1):S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldovin T., et al. SARS-CoV-2 RNA detection and persistence in wastewater samples: an experimental network for COVID-19 environmental surveillance in Padua, Veneto region (NE Italy) Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., et al. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54(13):7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Bofill-Mas S., Pina S., Girones R. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl. Environ. Microbiol. 2000;66(1):238–245. doi: 10.1128/aem.66.1.238-245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Francis R. On forecasting the community-level COVID-19 cases from the concentration of SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carducci A., et al. Making waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92(7):833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Chen Y., et al. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020;525(1):135–140. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., et al. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin. Infect. Dis. 2016;62(4):477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpuz M.V.A., et al. Viruses in wastewater: occurrence, abundance and detection methods. Sci. Total Environ. 2020;745 doi: 10.1016/j.scitotenv.2020.140910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford C., et al. Modeling of aerosol transmission of airborne pathogens in ICU rooms of COVID-19 patients with acute respiratory failure. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-91265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Fumian T.M., et al. Detection of rotavirus a in sewage samples using multiplex qPCR and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J. Virol. Methods. 2010;170(1–2):42–46. doi: 10.1016/j.jviromet.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Fumian T.M., et al. Molecular detection, quantification and characterization of human polyomavirus JC from waste water in Rio De Janeiro,Brazil. 2010;8(3):438–445. doi: 10.2166/wh.2010.090. [DOI] [PubMed] [Google Scholar]
- Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020;158(6):1518–1519. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1(1):10. [Google Scholar]
- Hamming I., et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. a first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstern P., Solheim B.G. The use of Solvent/Detergent treatment in pathogen reduction of plasma. Transfus. Med. Hemother. 2011;38(1):65–70. doi: 10.1159/000323552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz B., et al. Virus inactivation by solvent/detergent treatment and the manufacture of SD-plasma. Vox Sang. 1998;74(Suppl. 1):203–206. doi: 10.1111/j.1423-0410.1998.tb05473.x. [DOI] [PubMed] [Google Scholar]
- Kanarek A.D., Tribe G.W. Concentration of certain myxoviruses with polyethylene glycol. Nature. 1967;214(5091):927–928. doi: 10.1038/214927a0. [DOI] [PubMed] [Google Scholar]
- Katayama H., et al. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res. 2008;42(6–7):1441–1448. doi: 10.1016/j.watres.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Kohno T., et al. A new improved method for the concentration of HIV-1 infective particles. J. Virol. Methods. 2002;106(2):167–173. doi: 10.1016/s0166-0934(02)00162-3. [DOI] [PubMed] [Google Scholar]
- Kroiss S.J., et al. Assessing the sensitivity of the polio environmental surveillance system. PLoS One. 2018;13(12) doi: 10.1371/journal.pone.0208336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- La Rosa G., et al. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W.K., et al. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125(4):1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y., et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020;133(9):1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582(7813):557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Lucas T.M., et al. Pseudotyping incompatibility between HIV-1 and gibbon ape leukemia virus Env is modulated by Vpu. J. Virol. 2010;84(6):2666–2674. doi: 10.1128/JVI.01562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer E.J., et al. Development of a reproducible method for monitoring SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;799 doi: 10.1016/j.scitotenv.2021.149405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., et al. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. Environ. Sci. (Camb) 2021;7:504–520. doi: 10.1039/d0ew00946f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo S.E., et al. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.144215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintó R., et al. Hepatitis a virus in urban sewage from two Mediterranean countries. Epidemiol. Infect. 2007;135(2):270–273. doi: 10.1017/S0950268806006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poiesz B.J., et al. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 1980;77(12):7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port J.R., et al. Nat.Commun. 2021;12(1) doi: 10.1038/s41467-021-25156-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince A.M., Horowitz B., Brotman B. Sterilisation of hepatitis and HTLV-III viruses by exposure to tri(n-butyl)phosphate and sodium cholate. Lancet. 1986;1(8483):706–710. doi: 10.1016/s0140-6736(86)91101-3. [DOI] [PubMed] [Google Scholar]
- Roberts P.L. Virus inactivation by solvent/detergent treatment using Triton X-100 in a high purity factor VIII. Biologicals. 2008;36(5):330–335. doi: 10.1016/j.biologicals.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Díaz J., et al. Detection and characterization of waterborne gastroenteritis viruses in urban sewage and sewage-polluted river waters in Caracas,Venezuela. 2009;75(2):387–394. doi: 10.1128/AEM.02045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., et al. SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 2020;69(6):1143–1144. doi: 10.1136/gutjnl-2020-320891. [DOI] [PubMed] [Google Scholar]
- Vajda B.P. Concentration and purification of viruses and bacteriophages with polyethylene glycol. Folia Microbiol. (Praha) 1978;23(1):88–96. doi: 10.1007/BF02876605. [DOI] [PubMed] [Google Scholar]
- Venugopal A., et al. Novel wastewater surveillance strategy for early detection of coronavirus disease 2019 hotspots. Curr. Opin. Environ. Sci. Health. 2020;17:8–13. doi: 10.1016/j.coesh.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria M., et al. One year monitoring of norovirus in a sewage treatment plant in Rio de Janeiro,Brazil. 2010;8(1):158–165. doi: 10.2166/wh.2009.012. [DOI] [PubMed] [Google Scholar]
- Villena C., et al. Group a rotavirus in sewage samples from Barcelona and Cairo: emergence of unusual genotypes. Appl. Environ. Microbiol. 2003;69(7):3919–3923. doi: 10.1128/AEM.69.7.3919-3923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Du G. COVID-19 may transmit through aerosol. Ir. J. Med. Sci. 2020;189(4):1143–1144. doi: 10.1007/s11845-020-02218-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., et al. Excretion and detection of SARS coronavirus and its nucleic acid from digestive system. World J. Gastroenterol. 2005;11(28):4390–4395. doi: 10.3748/wjg.v11.i28.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., et al. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126(1–2):171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J., et al. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775 doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch S.R., et al. Analysis of inactivation of SARS-CoV-2 by specimen transport media, nucleic acid extraction reagents, detergents, and fixatives. J. Clin. Microbiol. 2020;58(11):e01713–e01720. doi: 10.1128/JCM.01713-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfel R., et al. Author correction: virological assessment of hospitalized patients with COVID-2019. Nature. 2020;588(7839):E35. doi: 10.1038/s41586-020-2984-3. [DOI] [PubMed] [Google Scholar]
- Wolfel R., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., et al. SARS-CoV-2 titers in wastewater are higher than expected fromclinically confirmed cases. 2020;5(4) doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833 e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral. Sci. 2020;12(1):8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., et al. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhong N.S., et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362(9393):1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R., et al. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15(9):871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]