Abstract
Enhancers have been conventionally perceived as cis-acting elements that provide binding sites for trans-acting factors. However, recent studies have shown that enhancers are transcribed and that these transcripts, called enhancer RNAs (eRNAs), have a regulatory function. Here, we identified putative eRNAs by profiling and determining the overlap between noncoding RNA expression loci and eRNA-associated histone marks such as H3K27ac and H3K4me1 in hepatocellular carcinoma (HCC) cell lines. Of the 132 HCC-derived noncoding RNAs, 74 overlapped with the eRNA loci defined by the FANTOM consortium, and 65 were located in the proximal regions of genes differentially expressed between normal and tumor tissues in TCGA dataset. Interestingly, knockdown of two selected putative eRNAs, THUMPD3-AS1 and LINC01572, led to downregulation of their target mRNAs and to a reduction in the proliferation and migration of HCC cells. Additionally, the expression of these two noncoding RNAs and target mRNAs was elevated in tumor samples in the TCGA dataset, and high expression was associated with poor survival of patients. Collectively, our study suggests that noncoding RNAs such as THUMPD3-AS1 and LINC01572 (i.e., putative eRNAs) can promote the transcription of genes involved in cell proliferation and differentiation and that the dysregulation of these noncoding RNAs can cause cancers such as HCC.
Keywords: enhancer RNA, hepatocellular carcinoma, long noncoding RNA, RNA polymerase II, transcribed enhancer, transcriptional regulation
INTRODUCTION
Cis-acting regulatory elements, including promoters, enhancers, and insulators, in the genome influence RNA polymerase II (RNA pol II) complex activity during the transcription process by providing binding sites for transcription factors (TFs) and cofactors (Bulger and Groudine, 2010; Maston et al., 2006; Riethoven, 2010). Enhancers, in particular, play central roles in the spatiotemporal regulation of transcription (Banerji et al., 1981; Moreau et al., 1981; Ong and Corces, 2011). It has been shown that enhancers not only provide binding sites for trans-acting factors but also are accessed and transcribed themselves by RNA pol II, leading to the generation of noncoding RNAs (ncRNAs) called enhancer RNAs (eRNAs) (De Santa et al., 2010; Kim et al., 2010). Interestingly, many long ncRNAs (lncRNAs) are known to be transcribed from these enhancer regions (Vučićević et al., 2015), suggesting the role of lncRNAs as eRNAs. The eRNAs transcribed from enhancer regions in the genome are generally short in length (0.2-2 kb), lack a poly A tail, are transcribed bidirectionally, and are pervasively transcribed throughout the genome (although recent studies have found that some eRNAs are long, have a poly A tail, and are transcribed in one direction as either a sense or an antisense transcript) and their expression is positively correlated with that of nearby genes (Andersson et al., 2014; Chen et al., 2018; De Santa et al., 2010; Kim et al., 2010; Melgar et al., 2011; Zhang et al., 2019).
Tens of thousands of eRNAs have been further systematically identified and characterized under the basic definition of ncRNAs transcribed from enhancer regions marked by two histone modification signals, H3K4me1 and H3K27ac, in various human tissues and cell types through the Functional Annotation of the Mammalian Genome (FANTOM) consortium (https://fantom.gsc.riken.jp/) (Andersson et al., 2014; Creyghton et al., 2010). FANTOM has thus far compiled more than 65,000 enhancers that have been identified to produce eRNAs (Andersson et al., 2014). It is becoming clear that active enhancers in the genome are associated with eRNA production, i.e., transcription of eRNAs could be a hallmark for the active state of enhancers (Andersson et al., 2014; De Santa et al., 2010; Hah et al., 2013; Kim et al., 2010; Lam et al., 2013; Li et al., 2013; Melgar et al., 2011; Wu et al., 2014).
Although few findings contradicting the functional importance of eRNAs as trans-acting regulators exist (Alvarez-Dominguez et al., 2017; Tsai et al., 2018), numerous studies have shown that eRNAs function as upstream regulators of adjacent gene expression. For instance, it was shown that depletion of eRNAs was linked to decreased expression levels of neighboring target genes (Arner et al., 2015; Creyghton et al., 2010; Lam et al., 2013; Li et al., 2013; Mousavi et al., 2013; Ørom et al., 2010; Schaukowitch et al., 2014). According to Arner et al. (2015), the transcription of eRNAs is the most upstream event preceding all other downstream transcriptional events during cell differentiation or activation. More than one mechanism of action has also been proposed as to how eRNAs promote target gene expression (Arner et al., 2015). Li et al. (2013) showed that eRNAs could activate target gene expression by forming a chromosomal loop that allows stabilization of enhancer-promoter contact. In contrast, Mousavi et al. (2013) suggested that eRNAs can activate Myod gene expression without forming a chromosomal loop. Furthermore, Schaukowitch et al. (2014) provided another mechanism by which eRNA promote target gene expression, i.e., by releasing the negative elongation factor (NELF) complex from the target promoter to transition paused RNA pol II to active RNA pol II (Schaukowitch et al., 2014).
Recently, the functional importance of eRNAs has been studied in the context of the regulation of inflammatory responses. For instance, the production of eRNAs in the enhancer region of IL-1β induced by LPS contributes to driving proinflammatory responses via the NF-kB signaling pathway in monocytes (Ha et al., 2019). In macrophages, Kdm6a promotes IFN-β transcription via transcription of eRNAs from the IFN-β enhancer region upon innate stimuli (Li et al., 2017). Furthermore, Helicobacter pylori infection can cause resistance to apoptosis by triggering the recruitment of Brd4, which induces the synthesis of BIRC3 eRNA (Chen et al., 2020). In addition, human diseases such as cancers are another context in which the functional importance of eRNAs has been investigated. Zhang et al. (2019) characterized the oncogenic potential of an eRNA (NET1e) located downstream of the oncogene NET1 by finding it in the list of differentially expressed genes (DEGs) in breast cancers. Similarly, Hsieh et al. (2014) have shown that KLK3 eRNA (KLK3e) forms an androgen receptor (AR)-dependent looping complex, leading to transcriptional activation of AR-dependent genes in prostate cancer. Melo et al. (2013) showed that many genomic regions targeted by P53 colocalize with enhancer chromatin marks, leading to enhanced long-distance p53-dependent gene expression regulation in multiple cancer cell lines. Alterations in eRNA expression have also been found to be associated with several other diseases, such as autism spectrum disorders and Huntington’s disease (Bhattarai et al., 2021).
To date, no eRNAs involved in hepatocellular carcinoma (HCC) have been reported. In the present work, we attempted to identify putative eRNAs that are commonly expressed in different human HCC cell lines by integrating two types of chromatin signals, H3K4me1 and H3K27ac, and information about ncRNAs derived from RNA-seq data. Subsequently, we identified putative target mRNAs of each ncRNA identified as a putative eRNA by investigating correlations with the expression levels of the mRNAs. Furthermore, we attempted to prove the function by conducting knockdown (KD) experiments for two selected putative eRNAs: THUMPD3-AS1 and LINC01572.
MATERIALS AND METHODS
Preparation of ChIP-seq and RNA-seq data from seven HCC cell lines
Chromatin immunoprecipitation sequencing (ChIP-seq) and RNA sequencing (RNA-seq) data were obtained from seven hepatocellular cell lines: Huh-7, Huh7.5, FT3-7, PLC-PRF-5, SNU182, SNU387, and SNU449 (Alexander et al., 1976; Ku and Park, 2005; Yi, 2010). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (for Huh-7, Huh7.5, and FT3-7 cells; Gibco, USA) or Roswell Park Memorial Institute-1640 medium (for PLC-PRF-5, SNU182, SNU387, and SNU449 cells; HyClone, USA) containing 10% fetal bovine serum (FBS; Gibco) and 1% antibiotic-antimycotic (Gibco). All of these cell lines were tested for mycoplasma contamination in the Core facility of National Cancer Center, Korea, and verified to be negative.
Total RNA was isolated using Hybrid-R (GeneAll, Korea) for mRNA sequencing, library preparation was performed with DNase I-treated total RNA using a TruSeq kit (Illumina, USA), and paired-end sequencing was performed using the HiSeq platform (Illumina). The total RNA-seq and ChIP-seq reads for each cell line are summarized in Supplementary Tables S1 and S2, respectively. The peak calling results of the two active histone marks, H3K4me1 and H3K27ac, positioned in the genomic regions where the two putative eRNAs located are listed in Supplementary Table S3.
The ChIP-seq and RNA-seq data set analyzed in this study have been deposited in the Gene Expression Omnibus with the project number PRJNA737566 (Reviewer access: https://dataview.ncbi.nlm.nih.gov/object/PRJNA737566?reviewer=r8n75e8rhnls10h1m1nt440f07). ChIP-seq data visualized in UCSC Browser (http://genome.ucsc.edu/s/yeeun/eRNA_study_ChIP%2Dseq).
Extraction of publicly available RNA-seq data for five different cancer types
RNA-seq data for five different cancer types were downloaded from TCGA (https://portal.gdc.cancer.gov/), including liver hepatocellular carcinoma (LIHC), breast cancer (BRCA), colorectal adenocarcinoma (COAD), stomach adenocarcinoma (STAD), and lung adenocarcinoma (LUAD) data. RNA-seq data from 40 LIHC, 102 BRCA, 39 COAD, 16 STAD, and 52 LUAD matched normal-tumor samples of the same patient were used for our analysis.
Bioinformatics pipelines for estimating the expression level of each gene from RNA-seq data
After the quality of raw sequencing reads was checked using ‘FastQC’ (v0.11.8) (Andrews, 2010) for the RNA-seq data generated from the seven hepatocellular cell lines, the contaminating adaptor and unpaired reads were excluded using ‘Trimmomatic’ (v0.38; parameters: PE LEADING, 3; TRAILING, 3; SLIDINGWINDOW, 4:15; MINLEN, 36) (Bolger et al., 2014). The cleaned reads were then aligned to the reference genome (GRCh38/hg38; http://asia.ensembl.org/) using ‘Tophat’ (v2.1.1; parameters: --library-type fr-firststrand) (Trapnell et al., 2009), and the fragments per kilobase of transcript per million mapped reads (FPKM) values were obtained by running ‘Cuffquant’ (v2.2.1; parameters: --library-type fr-firststrand --multi-read-correct --frag-bias-correct) (Trapnell et al., 2010). After normalization of the 7 cell line library sizes using ‘Cuffnorm’ (v2.2.1; parameters: --library-type fr-firststrand --library-norm-method classic-fpkm) (Trapnell et al., 2010), ncRNAs with FPKM ≥ 1 were mapped to the genomic regions where the active enhancer marks H3K27ac and H3K4me1 were located.
Bioinformatics pipeline for layering ChIP-seq data onto the reference genome
The ChIP-seq data generated from the seven hepatocellular cell lines for H3K27ac and H3K4me1 were mapped to the reference genomic regions as follows. The quality check and trimming procedures for raw ChIP-seq reads were the same as those used for the RNA-seq data described above. The cleaned ChIP-seq data were then aligned to the reference genome (GRCh38/hg38) using ‘Bowtie’ (v1.3.0; parameters: -S --fr) (Langmead et al., 2009). Subsequently, the reads mapped to black-list regions from the ENCODE portal (identifier: ENCFF356LFX; https://www.encodeproject.org/) (Sloan et al., 2016) were filtered out using bedtools (2.27.0; parameters: intersect v) (Quinlan and Hall, 2010) and nonuniquely aligned reads and potential PCR duplicates were excluded using ‘MarkDuplicates’ of ‘Picard’ (v2.25.0; REMOVE_DUPLICATES = true, ASSUME_SORTED = true, VALIDATION_STRINGENCY = LENIENT) (Picard Toolkit, 2019), and peak calling and peak annotation were performed using ‘MACS2’ (v2.2.4; parameters: callpeak -f BAMPE -p 0.0001 [--narrow | --broad] -g hs --SPMR [--call-summits | --no-model] -B --keep-dup 1 --mfold 10 100) (Zhang et al., 2008) and ‘annotatePeaks.pl’ of ‘HOMER’ (v4.11.1; parameter: hg38) (Heinz et al., 2010), respectively.
Cell culture and transfection
The human HCC cell line FT3-7 was cultured in DMEM (HyClone) with 10% FBS (HyClone) and penicillin-streptomycin (Gibco) and maintained at 37°C in an incubator containing 5% CO2. To knock down two selected eRNAs, THUMPD3-AS1 (NR_027007.3) and LINC01572 (NR_159370.1), antisense oligonucleotides (ASOs) were designed and purchased from Qiagen (Germany). The sequences of the ASOs were as follows: negative control ASO: 5’-AACACGTCTATACGC-3’, THUMPD3-AS1 ASO: 5’-GACACCTTAGAAAATT-3’, and LINC01572 ASO: 5’-ACATAGAGACAGACTG-3’. The cells were seeded in 6-well plates at a density of 5 × 105 cells/well and transfected with 20 nM ASOs with Lipofectamine RNAiMax (Invitrogen, USA) according to the manufacturer’s instructions. The cells were transfected with ASOs twice for 72 h to increase the KD efficiency.
Cell counting kit-8 assays
Cell proliferation was assessed by cell counting kit-8 (CCK-8; Dojindo, Japan) assays following the manufacturer’s protocol. In brief, cells were seeded in 96-well plates, and 10 µl of CCK-8 reagent was added at the indicated time points (0, 24, 48, and 72 h). Cell viability was measured with a microplate reader (Molecular Devices, USA) at 450 nm.
IncuCyte® cell proliferation assays
The cells were plated in 96-well plates (3595; Corning, USA). The proliferation of cells was monitored for 4 days by an IncuCyte Live-Cell analysis system (Sartorius, Germany).
Colony formation assays
A total of 2,000 cells were plated in 6-well plates and maintained for 2 weeks. To visualize and count colonies grown for 2 weeks, cells were fixed with 4% paraformaldehyde and stained with 1% crystal violet.
Transwell assays for invasion and migration
To evaluate the invasion or migration of cancer cells, transwell plates (8 µm pore size, 3422; Corning) were coated with Matrigel (354234; BD, USA) or gelatin (G1393; Sigma, USA). Cells (3 × 105/ml cells in 100 µl for invasion assays and 1.5 × 105/ml cells in 100 µl for migration assays) in serum-free medium were added to the upper chamber, and 600 µl of 10% FBS medium was added to the lower chamber. After 24-48 h, the invaded and migrated cells were fixed with 4% paraformaldehyde and stained with 1% crystal violet. Six random fields were photographed at ×200 magnification with a microscope (Carl Zeiss, Germany).
Identification of DEGs in KD experiments
Triplicate RNA-seq data were generated from each of the two KD experiments, i.e., the THUMPD3-AS1 KD and LINC01572 KD experiments. Quality checking and cleaning of the raw sequencing reads was performed using ‘FastQC’ and ‘Trimmomatic’ (the same bioinformatics pipelines that were applied to the RNA-seq datasets and ChIP-seq datasets derived from the seven HCC cell lines described above). After the cleaned reads were aligned to the reference genome (GRCh38/hg38) using ‘STAR’ (v2.7.6a; parameters: --runMode alignReads --outFilterMultimapNmax 10 --alignIntronMin 61 --alignIntronMax 265006 --outSAMtype BAM SortedByCoordinate) (Dobin et al., 2013), the raw read counts were estimated using ‘HTSeq’ (v0.11.2; parameters: -s reverse -m intersection-nonempty -r pos -f bam -t exon) (Anders et al., 2015). Subsequently, DEGs were estimated using ‘DESeq2’ (Love et al., 2014) by comparing the normalized read counts between the control (i.e., wild-type cell lines) and the case (i.e., KD cell lines) groups after removing batch effects using the ‘ComBat’ package (v3.36.0; Leek et al., 2012) of R (4.0.3).
Survival analysis of TCGA-LIHC data
Kaplan–Meier survival plots and hazard ratios (HRs) for the two selected ncRNAs, THUMPD3-AS1 and LINC01572, were estimated using Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html) (Tang et al., 2017). The two eRNAs and their target gene names were put into the search engine by setting parameters as follows: ‘Methods’, disease-free survival; ‘Group Cutoff’, 75% and 25% for both ‘Cutoff-High’ and ‘Cutoff-Low’; ‘Hazard Ratio’, Yes; ‘95% Confidence Interval’, No; ‘Axis Units’, Months; and ‘Dataset Selection’, LIHC.
Data analysis
All statistical tests and plots were conducted using the R (4.0.3) and Python (3.6.8) languages. Kendall’s correlation analysis and linear regression analysis were performed with the ‘scipy.stats.stats’ and ‘sklearn.linear_model’ packages, respectively, of Python (3.6.8). Gene ontology (GO) analysis was performed using the DAVID tool (https://david.ncifcrf.gov/home.jsp) (Jiao et al., 2012). Other batch works were performed with home-built Python scripts.
RESULTS
Workflow
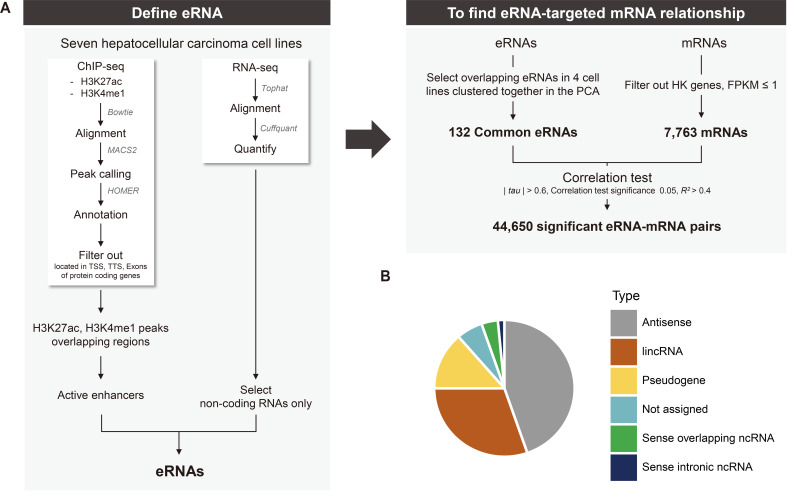
Active enhancers with two histone modification signals, H3K4me1 (i.e., an enhancer chromatin mark) and H3K27ac (i.e., an active chromatin mark), are known to produce eRNAs (Creyghton et al., 2010; Heintzman et al., 2007; 2009). Figure 1 shows our workflow used for identifying and detecting eRNAs: we used two types of ChIP-seq signals marking active enhancers and total RNA-seq data. Our rationale for identifying eRNAs was to select only ncRNAs transcribed from the genomic regions that are supposed to be active enhancer regions.
Fig. 1. Workflow of our study and types of defined putative eRNAs.
(A) Overall workflow. To identify putative eRNAs transcribed from genomic locations with two active enhancer chromatin marks, H3K4me1 and H3K27ac, the overlap of genomic positions onto which ChIP-seq data mapped and positions where ncRNAs are transcribed was determined. Only those ncRNAs commonly transcribed in the active enhancer regions in four different HCC cell lines (FT3-7, Huh7.5, PLC-PRF-5, and SNU182) were defined to be putative eRNAs. Subsequently, putative mRNA targets were searched for by investigating correlations between each putative eRNA and mRNA transcripts expressed in seven HCC cell lines. Please refer to the main text for details. (B) Pie chart of the types of defined putative 132 eRNAs. The types of eRNAs were determined by the annotation information in the GTF file (GRCh38/hg38).
Briefly, only those ncRNAs transcribed from genomic positions at which the two types of ChIP-seq signals were commonly located were selected after excluding other RNAs transcribed from the genomic regions mapped to transcription start sites (TSSs), transcription termination sites (TTSs), and exons of protein-coding genes (Fig. 1A). A list of the ncRNAs that were considered to be transcribed from active enhancer regions was generated for each of seven different HCC cell lines: FT3-7, Huh-7, Huh7.5, PLC-PRF-5, SNU182, SNU387, and SNU449. Subsequently, those ncRNAs commonly identified from four different cell lines were defined as putative eRNAs with high confidence (Fig. 1A).
We decided to overlap the lists of eRNAs from four cell lines, Huh7.5, SNU182, FT3-7, and PLC-PRF-5, rather than from all seven cell lines because principal component analysis (PCA) showed that the gene expression profiles of three cell lines, SNU387, SNU449, and Huh-7, were substantially different from those of the remaining four cell lines (Supplementary Fig. S1). As expected, the list of overlapping eRNAs from all seven cell lines that included the three cell lines with very different gene expression profiles had a substantially reduced number of confident eRNAs such that further exploration would have been difficult. The numbers of ChIP-seq signals, transcribed ncRNAs, and ncRNAs detected in the active enhancer regions are summarized in Supplementary Table S4.
Consequently, a total of 132 ncRNAs that were commonly detected in the four cell lines were defined to be putative eRNAs (Supplementary Table S5), i.e., ncRNAs transcribed from active enhancer regions. Interestingly, all 132 ncRNAs were lncRNAs by definition with lengths > 200 nt. Small ncRNAs, such as miRNAs, piRNAs, and siRNAs, were also present in the individual cell lines we used to produce the ChIP-seq and RNA-seq data, but all disappeared when the ncRNAs from the four different cell types overlapped. Subsequently, by referencing the annotation information in GTF files (GRCh38/hg38), the lncRNAs were subcategorized into antisense ncRNAs, long intergenic noncoding RNA (lincRNAs), pseudogenes, sense overlapping ncRNAs and sense intronic ncRNAs, of which antisense ncRNAs and lincRNAs were the most frequent types (Fig. 1B). No chromosomal bias seemed to be present in the ncRNAs identified as putative eRNAs (Supplementary Table S5). Approximately 56% (74/132) of the genomic positions that were found to produce ncRNAs identified as putative eRNAs also overlapped with the ‘transcribed eRNAs’ identified with FANTOM, producing a significant enrichment of FANTOM eRNAs in the list of eRNAs detected in the present work (chi-squared test, P < 0.01). Only 13% of the ncRNAs overlapped with the FANTOM eRNAs. Note that the entire regions where ncRNAs co-localizing with the two active enhancer marks are produced were defined as ‘eRNA-transcribing regions’. According to previous studies, ‘eRNA-transcribing regions’ are wider than ChIP-seq peaks; Zhang et al. (2019) defined ± 3 kb (6 kb around regions) while Hauberg et al. (2019) defined ± 1.5 kb (3 kb around regions) from the middle site of the enhancers as ‘eRNA-transcribing regions’.
Expression analysis of the putative eRNAs in TCGA liver cancer data
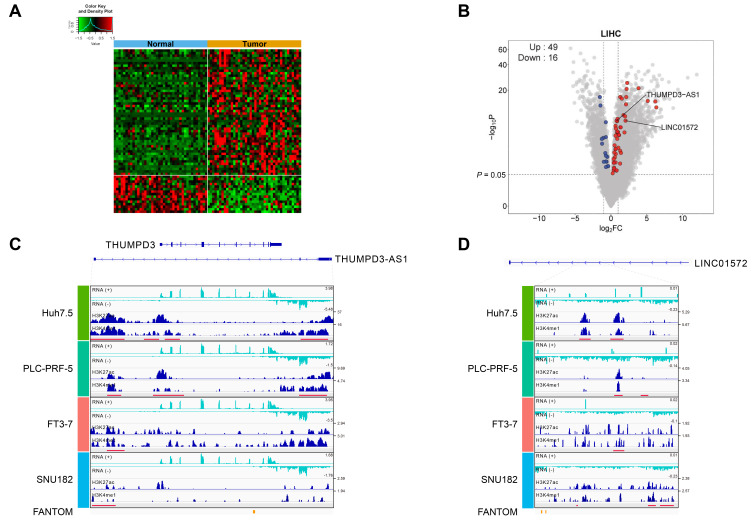
Next, to determine whether the expression of 132 ncRNAs identified as putative eRNAs is altered during tumor formation, we investigated the expression profiles of these ncRNAs in TCGA LIHC data. Approximately 49.2% (65/132) of the 132 ncRNAs were found to be significantly differently expressed (i.e., DEGs) between normal and tumor tissues in the TCGA-LIHC dataset (Fig. 2A). The volcano plot in Fig. 2B shows that the magnitude of the changes in the expression levels of these ncRNA genes was relatively weak but still significant. Notably, the number of upregulated ncRNAs was greater than that of downregulated ncRNAs (49 upregulated and 16 downregulated). The ChIP-seq peak locations and shapes for two putative eRNAs, THUMPD3-AS1 (an antisense ncRNA) and LINC01572 (a lincRNA), are shown in Fig. 2C (THUMPD3-AS1) and Fig. 2D (LINC01572). In addition, in the other four TCGA cancer types, we found that changes in the expression levels of these 132 putative eRNAs substantially varied in terms of the number of overlaps with DEGs and the ratio between upregulated and downregulated genes (Supplementary Fig. S2).
Fig. 2. Characteristics of the 132 putative eRNAs.
(A) Heat map constructed after comparing the 132 putative eRNAs to the list of DEGs from the TCGA-LIHC dataset. A total of 65 out of the 132 eRNAs overlapped with the DEGs estimated by comparing gene expression between normal and tumor samples from the TCGA-LIHC dataset (P < 0.05), and 49 and 16 eRNAs were found to be upregulated and downregulated, respectively, in tumor tissues. (B) A volcano plot of the DEGs estimated from the TCGA-LIHC dataset in (A). All the DEGs (gray), 49 upregulated eRNAs (red), and 16 downregulated eRNAs (blue) are depicted accordingly. (C and D) Integrative Genomics Viewer (IGV) views showing the genomic locations of ChIP-seq signals and RNA-seq data estimated in the four different hepatocellular cell lines for the two selected eRNAs, THUMPD3-AS1 (C) and LINC01572 (D). The red bars in the figure boxes indicate putative enhancer regions where the two active chromatin peaks are located.
Identification of putative eRNA-target mRNA pairs by correlation analysis
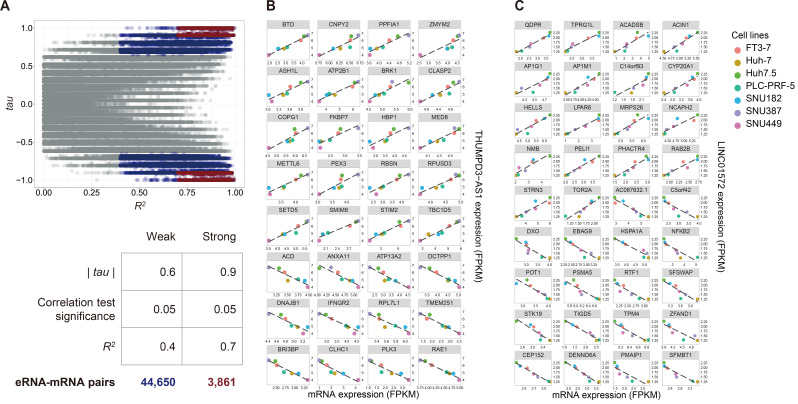
We next tried to identify putative eRNA-target mRNA pairs by correlation analysis using information on the expression levels of these genes from the seven different HCC cell lines. The rationale for this approach was that the expression levels of an eRNA should be correlated (either positively or negatively, although a positive correlation seems to be more likely assuming that enhancers act as trans-acting regulatory factors) with the expression levels of its target mRNA if the putative eRNA does indeed target the promoter of the mRNA.
As a result, we found a total of putative 44,650 eRNA-target mRNA pairs (highlighted in dark blue in Fig. 3A) using weak thresholds (R2 > 0.4 [i.e., criterion to evaluate the goodness of fit from the linear regression analysis], |tau| > 0.6 [i.e., correlation coefficient estimated from Kendall’s correlation analysis], and P < 0.05), and a total of 3,861 putative eRNA-target mRNA pairs using more stringent thresholds (R2 > 0.7, |tau| > 0.9, and P < 0.05) (highlighted in dark red in Fig. 3A). For instance, a total of 343 and 400 mRNAs were found to be significantly correlated with THUMPD3-AS1 and LINC01572, respectively, as listed in Supplementary Tables S6 and S7, and the putative eRNA-target mRNA pairs obtained by the stringent thresholds are shown in Figs. 3B and 3C, respectively. The number of positively correlated pairs were slightly more than that of negatively correlated pairs for both THUMPD3-AS1 (i.e., 53.6%, 184 positively correlated pairs vs 159 negatively correlated pairs) and LINC01572 (i.e., 56.8%, 227 positively correlated pairs vs 173 negatively correlated pairs). The percentage of positively correlated pairs was increased for THUMPD3-AS1 when more stringent thresholds were applied (62.5%, 20 positively correlated pairs vs 12 negatively correlated pairs), whereas no significant changes were found for LINC01572 when the stringent thresholds were used. Interestingly, GO analysis showed that the mRNAs correlated with THUMPD3-AS1 showed enrichment of genes involved in DNA repair, whereas the mRNAs correlated with LINC01572 showed enrichment of genes involved in the RNA-pol II transcription process.
Fig. 3. Example correlations of putative eRNAs and their target mRNAs.
(A) Scatter plot of R 2 and Kendall’s tau values. The relationship between each of the 132 eRNAs and each of 7,763 mRNAs was investigated using Kendall’s tau correlation analysis and linear regression analysis. A scatter plot was then constructed for all the R 2 and Kendall’s tau values estimated from the two analyses. The x-axis represents the R 2 values estimated by the linear regression analysis, and the y-axis represents tau values calculated by Kendall’s tau correlation test. Two thresholds were set to determine putative mRNA targets, as shown in the table below (A), i.e., the weak targets (colored blue on plot A) were identified by R 2 > 0.4, |tau| > 0.6, and P < 0.05, and the strong targets (colored red on plot A) were identified by R 2 > 0.7, |tau| > 0.9, and P < 0.05. (B) Example scatter plot of THUMPD3-AS1 and its 32 putative strong target mRNAs. (C) Example scatter plot of LINC01572 and its 36 putative strong targets.
Subsequently, we investigated whether these correlated genes were located in the same topology associating domains (TADs) where each of the ncRNAs identified as putative eRNAs was positioned. Surprisingly, there were few genes in the same TADs as THUMPD3-AS1 and LINC01572, only 13 and 3 genes, respectively. The two ncRNAs and the correlated genes present in the same TADs as the two ncRNAs will be discussed in more detail in the Discussion section.
KD experiments for the two putative eRNAs THUMPD3-AS1 and LINC01572
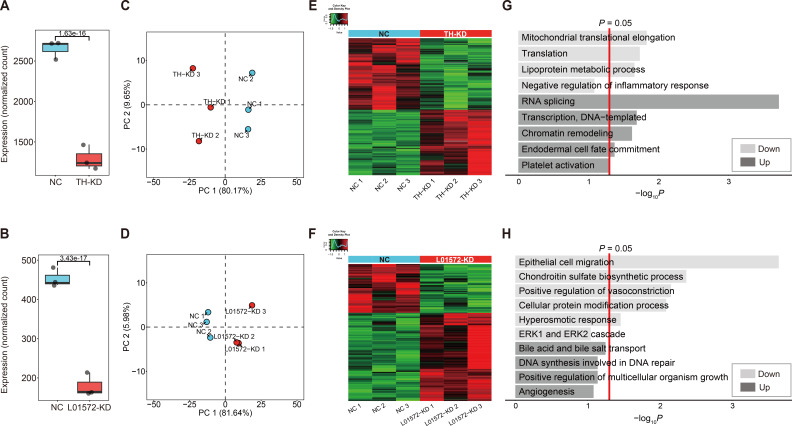
We next investigated which genes had their expression altered by knocking down these two ncRNAs under the assumption that the expression of target genes should be altered by KD of their upstream regulator eRNAs. For this purpose, ASOs were employed to KD expression in the FT3-7 HCC cell line. One ASO was designed to target exon 3 of THUMPD3-AS1, and another was designed to target exon 6 of LINC01572 (Supplementary Fig. S3). These two ncRNAs were selected for the KD experiment because they are relatively novel lncRNAs of two different categories of eRNA, and both were upregulated in the tumor tissues compared to normal tissues of TCGA LIHC. We first determined whether the KD of both THUMPD3-AS1 and LINC01572 was successful by confirming that the expression levels of these ncRNAs were significantly reduced (Figs. 4A and 4B). PCA showed that the gene expression profiles of the cells before and after ncRNA KD were significantly changed, and the cells could be clustered into two different states (Figs. 4C and 4D).
Fig. 4. KD analysis of the two selected putative eRNAs (THUMPD3-AS1 and LINC01572).
(A and B) box plots of the expression levels of the eRNAs THUMPD3-AS1 (A) and LINC01572 (B) before and after knock down. (C and D) PCA plots of the DEGs identified before and after knock down of the two eRNAs: (C) THUMPD3-AS1 KD and (D) LINC01572 KD. NC, negative control; TH-KD, THUMPD3-AS1 KD; L01572-KD, LINC01572 KD. (E and F) Heat map of the DEGs identified before and after eRNA knock down: (E) THUMPD3-AS1 KD and (F) LINC01572 KD. (G and H) GO analysis of the DEGs identified in each eRNA knock down experiment: (G) GO analysis of the DEGs from THUMPD3-AS1 knock down and (H) GO analysis of the DEGs from LINC01572 knock down.
DEGs before and after KD were then identified using the threshold P < 0.01, and a total of 379 (181 upregulated and 198 downregulated) and 188 (121 upregulated and 67 downregulated) DEGs were identified for the THUMPD3-AS1 and LINC01572 KD experiments, respectively (Figs. 4E and 4F). The numbers of DEGs were significantly reduced to 79 and 53 for THUMPD3-AS1 and LINC01572, respectively, by adding another threshold, |fold change| ≥ 2, indicating that the fold changes in target gene expression caused by KD of these ncRNAs were relatively small, i.e., less than 2% of genes seemed to have their expression levels affected by the KD. Notably, the proportions of upregulated or downregulated DEGs were significantly different in the two KD experiments: 52.2% downregulated vs 47.8% upregulated DEGs for THUMPD3-AS1 and 35.6% downregulated vs 64.4% upregulated DEGs for LINC01572. Consistently, GO analysis showed that some genes altered by KD of these two putative eRNAs were enriched in tumorigenesis and angiogenesis functions, but these genes were mostly assigned to different specific functional categories. Notably, genes involved in translational regulation and lipoprotein metabolic processes and inflammatory genes were downregulated with THUMPD3-AS1 KD, whereas genes involved in cell migration, vasoconstriction, and the ERK1/2 cascade were downregulated with LINC01572 KD (Figs. 4G and 4H).
Validation of putative eRNA-target mRNA pairs in the TCGA-LIHC dataset
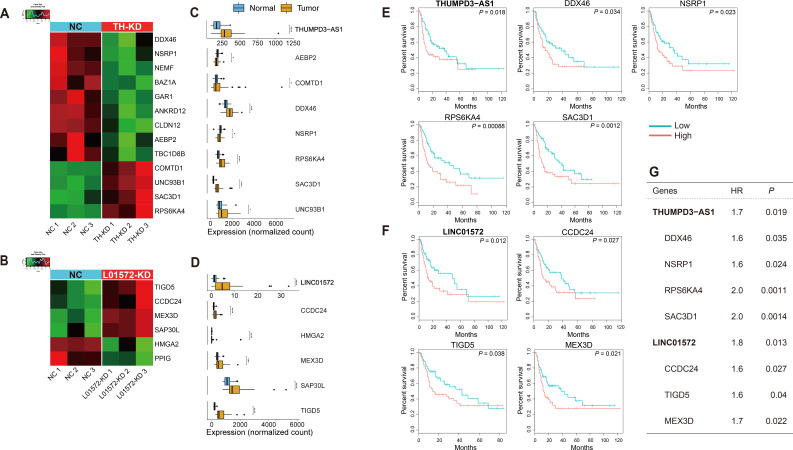
We next analyzed which genes in the lists from the THUMPD3-AS1 and LINC01572 KD experiments overlapped with the putative targets defined by the correlation analyses shown in Fig. 3. A total of thirteen (out of the 379 DEGs) and six (out of the 188 DEGs) genes overlapped with THUMPD3-AS1 and LINC01572 KD-driven DEGs, respectively, showing that approximately 3%-4% of the KD-driven DEGs were in the lists of genes highly correlated with the two putative eRNAs (Figs. 5A and 5B). Interestingly, not only the two ncRNAs but also most of the putative target genes shown in Figs. 5A and 5B were found to be significantly upregulated in tumors compared to normal tissues in the TCGA-LIHC dataset (Figs. 5C and 5D).
Fig. 5. Two putative eRNAs and their validated mRNA targets involved in patient survival.
(A and B) Heat map of the putative target mRNAs overlapping with the lists of the two eRNA KD experiments: (A) putative target mRNAs overlapping with the list of DEGs generated in the THUMPD3-AS1 KD experiment and (B) putative target mRNAs overlapping with the list of DEGs generated in the LINC01572 KD experiment. NC, negative control; TH-KD, THUMPD3-AS1 KD; L01572-KD, LINC01572 KD. (C and D) Boxplots of the expression values of eRNAs and their target mRNAs estimated with the TCGA-LIHC dataset: (C) expression values of THUMPD3-AS1 and its target mRNAs and (D) expression values of LINC01572 and its target mRNAs. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. (E and F) Kaplan–Meier plots of the two eRNAs and their validated target mRNAs. Note that the validated target mRNAs were defined as those eRNA-target mRNA pairs that were confirmed in the analysis in (A-D); (A) and (C) show the THUMPD3-AS1 results, and (B) and (D) show the LINC01572 results. Survival analysis was performed using the clinical information of the TCGA-LIHC dataset (see Materials and Methods section). (G) Table of the hazard ratios (HR) and P values of each gene identified by Cox regression analysis.
Seven genes, AEBP2, COMTD1, DDX46, NSRP1, RPS6KA4, SAC3D1, and UNC93B1, shown in Fig. 5C are particularly interesting because their expression levels were significantly correlated with the expression of THUMPD3-AS1; the expression of these genes was also altered by THUMPD3-AS1 KD and during tumorigenesis in liver cancers. Interestingly, DDX46 (which encodes DEAD-box helicase) and NSRP1 (which encodes nuclear speckle splicing regulator) are involved in transcription regulation, and AEBP2 encodes adipocyte enhancer binding protein 2, which seems to be consistent with the idea that THUMPD3-AS1 is actually an eRNA that regulates transcription. Furthermore, the three remaining genes, RPS6KA4, SAC3D1, and UNC93B1, are known to be involved in cell proliferation, which explains why they were found to be significantly upregulated in the TCGA-LIHC dataset.
Similarly, five genes, CCDC24, HMGA2, MEX3D, SAP30L, and TIGD5, shown in Fig. 5D were detected in the LINC01572 correlation analysis and KD experiment and in the list of TCGA-LIHC DEGs; the HMGA2 and SAP30L proteins are involved in histone modification, and MEX-3 is an RNA binding protein, and the functions of the proteins they encode support the idea that these genes are possible targets of LINC01572.
Furthermore, the expression levels of some of these genes were found to be associated with the overall survival of LIHC patients (Figs. 5E and 5F): upregulation of the genes conferred an approximately two times more likely chance of poor survivals (odds ratio: approximately 1.6-2.0) (Fig. 5G).
THUMPD3-AS1 and LINC01572 are required for the proliferation and migration of HCC cells
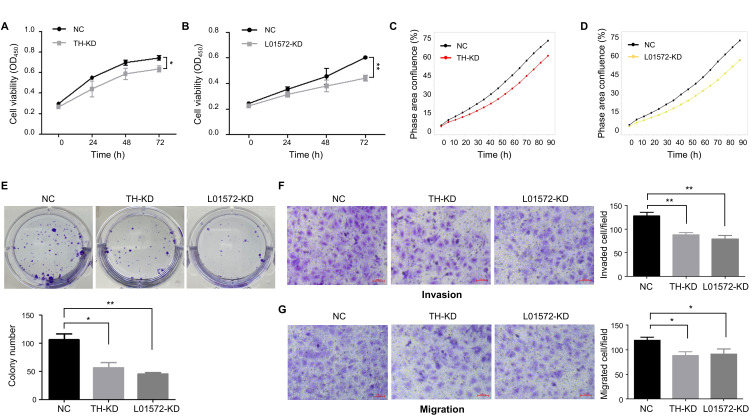
As mentioned in the Introduction, several eRNAs are known to have oncogenic potential. Consistently, we showed (Fig. 5) that the two putative eRNAs, THUMPD3-AS1 and LINC01572, were upregulated in the tumor tissues of the TCGA-LIHC dataset, and the genes affected by KD experiments were found to be involved in cell proliferation and migration. Therefore, we attempted to investigate whether these two ncRNAs could promote the progression of HCC cells using CCK-8 assays and IncuCyte proliferation assays. As shown in Figs. 6A-6D, we found that KD of THUMPD3-AS1 or LINC01572 inhibited the proliferation of cells in proliferation assays. Consistently, the colony formation assays also showed a more than 40% reduction in colony numbers in KD cells compared to control cells (Fig. 6E).
Fig. 6. Roles of the two putative eRNAs in cancer progression.
(A and B) CCK-8 assays were conducted to assess cell viability at the indicated time points: (A) THUMPD3-AS1 KD and (B) LINC01572 KD. OD, optical density; NC, negative control; TH-KD, THUMPD3-AS1 KD; L01572-KD, LINC01572 KD. (C and D) IncuCyte proliferation assays were carried out to determine cell viability at the indicated times: (C) THUMPD3-AS1 KD and (D) LINC01572 KD. The graph shows the mean values of technical triplicates. (E) Representative images of colony formation assays. (F and G) Representative images of transwell assays: (F) invasion assays and (G) migration assays. Scale bars = 100 µm. All data were obtained from at least three independent experiments, and P values were determined by two-tailed unpaired t-test analysis. *P ≤ 0.05; **P ≤ 0.01.
Subsequently, we examined whether these putative eRNAs could affect the invasion and migration of HCC cells using transwell assays. Invaded and migrated cells were detected after allowing the cells to migrate for 24-48 h. As shown in Figs. 6F and 6G, the invasion and migration abilities of THUMPD3-AS1 KD and LINC01572 KD cells were reduced by 20%-30% compared to those in the control group. Taken together, these results suggest that both THUMPD3-AS1 and LINC01572 are required for the proliferation of HCC cells, supporting oncogenic roles of these two putative eRNAs in HCC cells.
DISCUSSION
In the present work, a total of 132 putative eRNAs were identified by intersecting the genomic positions identified from three types of information: (i) enhancer chromatin marks (H3K4me1), (ii) active chromatin marks (H3K27ac), and (iii) transcribed ncRNAs. Subsequently, we identified putative target mRNAs for each of the 132 ncRNAs and tested the hypothesis that the ncRNAs identified as putative eRNAs could regulate the expression of downstream target genes by knocking down two selected eRNAs, THUMPD3-AS1 and LINC01572.
The fact that enhancer regions in the genome are transcribed by RNA pol II to produce ncRNAs has been confirmed by numerous studies (Andersson et al., 2014; Arner et al., 2015; De Santa et al., 2010; Kim et al., 2010; Melgar et al., 2011) since the first reports in 2010 (Kim et al., 2010), which have integrated gene expression information with data regarding active chromatin and open chromatin marks such as H3K27ac, DNase hypersensitivity sites, and enhancer chromatin marks such as H3K4me1 and RNA pol II marks. Here, we integrated two active enhancer chromatin marks, H3K4me1 and H3K27ac, along with transcribed ncRNA data. Only those ncRNAs transcribed from supposedly active enhancer regions commonly detected in four different HCC cell lines were defined to be putative eRNAs with high confidence. Putative target mRNAs of each ncRNA defined to be an eRNA were identified by selecting eRNA-mRNA pairs with significantly high correlation in seven HCC cell lines (Supplementary Tables S6 and S7) because we assumed that such a correlation indicates eRNA regulation of the target mRNA. Several other studies have already successfully estimated the relationship between eRNAs and their target mRNAs (Carullo et al., 2020; Chen et al., 2018; Hsieh et al., 2014; Mousavi et al., 2013; Schaukowitch et al., 2014; Zhang et al., 2019).
We also investigated how often the putative target mRNAs defined by the correlation analysis were located within the TADs of THUMPD3-AS1 or LINC01572 using TAD information derived from human tissues in the human liver STL011 dataset downloaded from (http://3dgenome.fsm.northwestern.edu/) (Wang et al., 2018). Thirteen genes out of the 343 putative targets of THUMPD3-AS1 and only three of the 400 putative targets of LINC01572 were found to be located in the same TADs (Supplementary Tables S6 and S7). It seems that this low overlap between the correlated genes and the TAD-localized genes was primarily because the TAD information derived from the human liver STL011 dataset may not be suitable for estimating chromosomal contact points in HCC, given the high tissue specificity of known TAD genes. According to Andersson et al. (2014), different cell types have different enhancer/gene ratios and cell type-specific eRNA transcription; for example, immune cells and hepatocytes have relatively higher enhancer/gene ratios and higher cell type-specific eRNA transcription, whereas fibroblasts and epithelial cells have lower enhancer/gene ratios and lower cell type-specific eRNA transcription (Andersson et al., 2014).
Interestingly, the thirteen genes located within the THUMPD3-AS1 TAD were found to have significantly higher R 2 values than the R 2 values estimated for the other non-TAD genes in a permutation analysis estimating the R 2 values for 343 genes randomly chosen from the total genes during 10,000 iterations (Supplementary Fig. S4), indicating that the expression of these genes is actually likely to be regulated by THUMPD3-AS1. In contrast, the analysis of LINC01572 showed no statistical differences in R 2 values between genes within the TAD and the other non-TAD genes. This result indicates that the two putative eRNAs THUMPD3-AS1 and LINC01572 might exert different mechanisms of action in regulating target genes and that some putative LINC01572 target mRNAs might not be directly regulated by a transcription regulator function of LINC01572.
While extensive transcription in enhancer genomic regions may no longer be a novel aspect of genomic metabolism, the mechanisms of action by which eRNAs are involved remain to be further studied. As seen in Fig. 1B, we showed that all the putative eRNAs identified in the present work were lncRNAs, and the action mechanism of eRNAs can be inferred based on the known mechanisms of action of lncRNAs. As summarized by Ali and Grote (2020), lncRNAs can regulate gene expression not only through transcriptional products but also through other mechanisms of action. Namely, lncRNAs are known (i) to function as trans-acting factors that bind to target DNAs to regulate target gene expression, (ii) to provide regulatory elements (located within the transcriptional units of lncRNAs), and (iii) to control gene expression through the transcription process itself. Studies have concluded that roles (ii) and (iii) exist, as the levels of target mRNAs are altered by deleting lncRNA genomic regions or by blocking the transcriptional processes that occur at the promoters or TSSs upstream of the lncRNA genes but not by lowering the levels of the lncRNAs (Ali and Grote, 2020). Previous studies have concluded that the role (i) works, when the expression of target mRNAs is affected by blocking eRNAs (i.e., the transcriptional products of enhancer regions). Given that the result shown in Fig. 4 shows that the expression of downstream genes was affected by knock down of either of the two selected putative eRNAs, we can exclude the (ii) and (iii) mechanisms of action, at least for these two ncRNAs.
Taken together, our results confirm that eRNAs are widely transcribed in active enhancer regions, consistent with many other previous studies. Our results also suggest that the conclusion that eRNAs are trans-acting transcriptional regulators is more reasonable than the conclusion that eRNAs are just markers representing active enhancers in the genome. In addition, we provide some experimental evidence that upregulation of the two selected putative eRNAs, THUMPD3-AS1 and LINC01572, may have an oncogenic function.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This research was supported by National Research Foundation of Korea (NRF) (NRF-2019R1A5A6099645, NRF-2017M3A9G7073033) and by the Ministry of Education, Science, and Technology (MEST) (2019R1A2C 1002350).
Footnotes
AUTHOR CONTRIBUTIONS
L.K.K., T.K., and S.S.C. conceived and designed the experiments. Y.E.L., J.L., Y.S.L., J.J.J., H.W., and H.I.C. performed experiments and data analyses. Y.S.L., Y.G.C., and T.K.K. contributed reagents and materials. T.K., L.K.K., T.K.K., and S.S.C. wrote the paper. All authors read and approved the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Alexander J.J., Bey E.M., Geddes E.W., Lecatsas G. Establishment of a continuously growing cell line from primary carcinoma of the liver. S. Afr. Med. J. 1976;50:2124–2128. [PubMed] [Google Scholar]
- Ali T., Grote P. Beyond the RNA-dependent function of LncRNA genes. Elife. 2020;9:e60583. doi: 10.7554/eLife.60583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Dominguez J.R., Knoll M., Gromatzky A.A., Lodish H.F. The super-enhancer-derived alncRNA-EC7/Bloodlinc potentiates red blood cell development in trans. Cell Rep. 2017;19:2503–2514. doi: 10.1016/j.celrep.2017.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T., et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. [Retrieved January 20, 2020];FastQC: a quality control tool for high throughput sequence data. 2010 from https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Arner E., Daub C.O., Vitting-Seerup K., Andersson R., Lilje B., Drablos F., Lennartsson A., Ronnerblad M., Hrydziuszko O., Vitezic M., et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Bhattarai S., Akella A., Gandhi A., Dharap A. Modulation of brain pathology by enhancer RNAs in cerebral ischemia. Mol. Neurobiol. 2021;58:1482–1490. doi: 10.1007/s12035-020-02194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M., Groudine M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev. Biol. 2010;339:250–257. doi: 10.1016/j.ydbio.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carullo N.V., Phillips Iii R.A., Simon R.C., Soto S.A.R., Hinds J.E., Salisbury A.J., Revanna J.S., Bunner K.D., Ianov L., Sultan F.A., et al. Enhancer RNAs predict enhancer-gene regulatory links and are critical for enhancer function in neuronal systems. Nucleic Acids Res. 2020;48:9550–9570. doi: 10.1093/nar/gkaa671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Li C., Peng X., Zhou Z., Weinstein J.N., Cancer Genome Atlas Research Network , Liang H. A pan-cancer analysis of enhancer expression in nearly 9000 patient samples. Cell. 2018;173:386–399.e12. doi: 10.1016/j.cell.2018.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Sheppard D., Dong X., Hu X., Chen M., Chen R., Chakrabarti J., Zavros Y., Peek R.M., Chen L. H. pylori infection confers resistance to apoptosis via Brd4-dependent BIRC3 eRNA synthesis. Cell Death Dis. 2020;11:667. doi: 10.1038/s41419-020-02894-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A., et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F., Barozzi I., Mietton F., Ghisletti S., Polletti S., Tusi B.K., Muller H., Ragoussis J., Wei C., Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S., Cho W., DeKoter R.P., Kim S.O. The transcription factor PU. 1 mediates enhancer-promoter looping that is required for IL-1β eRNA and mRNA transcription in mouse melanoma and macrophage cell lines. J. Biol. Chem. 2019;294:17487–17500. doi: 10.1074/jbc.RA119.010149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N., Murakami S., Nagari A., Danko C.G., Kraus W.L. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauberg M.E., Fullard J.F., Zhu L., Cohain A.T., Giambartolomei C., Misir R., Reach S., Johnson J.S., Wang M., Mattheisen M., et al. Differential activity of transcribed enhancers in the prefrontal cortex of 537 cases with schizophrenia and controls. Mol. Psychiatry. 2019;24:1685–1695. doi: 10.1038/s41380-018-0059-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W., et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., Fei T., Chen Y., Li T., Gao Y., Wang X., Sun T., Sweeney C.J., Lee G.S., Chen S., et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X., Sherman B.T., Huang D.W., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S., et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku J.L., Park J.G. Biology of SNU cell lines. Cancer Res. Treat. 2005;37:1–19. doi: 10.4143/crt.2005.37.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M.T., Cho H., Lesch H.P., Gosselin D., Heinz S., Tanaka-Oishi Y., Benner C., Kaikkonen M.U., Kim A.S., Kosaka M., et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A.Y., Merkurjev D., Zhang J., Ohgi K., Song X., et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang Q., Shi Q., Liu Y., Zhao K., Shen Q., Shi Y., Liu X., Wang C., Li N., et al. Demethylase Kdm6a epigenetically promotes IL-6 and IFN-β production in macrophages. J. Autoimmun. 2017;80:85–94. doi: 10.1016/j.jaut.2017.02.007. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston G.A., Evans S.K., Green M.R. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- Melgar M.F., Collins F.S., Sethupathy P. Discovery of active enhancers through bidirectional expression of short transcripts. Genome Biol. 2011;12:R113. doi: 10.1186/gb-2011-12-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo C.A., Drost J., Wijchers P.J., van de Werken H., de Wit E., Oude Vrielink J.A., Elkon R., Melo S.A., Léveillé N., Kalluri R., et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981;9:6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K., Zare H., Dell’Orso S., Grontved L., Gutierrez-Cruz G., Derfoul A., Hager G.L., Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C., Corces V.G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q., et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard Toolkit. [Retrieved January 20, 2020];Broad Institute, GitHub repository. 2019 from http://broadinstitute.github.io/picard/
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethoven J.M. Regulatory regions in DNA: promoters, enhancers, silencers, and insulators. Methods Mol. Biol. 2010;674:33–42. doi: 10.1007/978-1-60761-854-6_3. [DOI] [PubMed] [Google Scholar]
- Schaukowitch K., Joo J., Liu X., Watts J.K., Martinez C., Kim T. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M., Lee B.T., et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44(D1):D726–D732. doi: 10.1093/nar/gkv1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., Van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P., Dell’Orso S., Rodriguez J., Vivanco K.O., Ko K., Jiang K., Juan A.H., Sarshad A.A., Vian L., Tran M., et al. A muscle-specific enhancer RNA mediates cohesin recruitment and regulates transcription in trans. Mol. Cell. 2018;71:129–141.e8. doi: 10.1016/j.molcel.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vučićević D., Corradin O., Ntini E., Scacheri P.C., Ørom U.A. Long ncRNA expression associates with tissue-specific enhancers. Cell Cycle. 2015;14:253–260. doi: 10.4161/15384101.2014.977641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Li D., Choudhary M.N., Li Y., Hu M., et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19:151. doi: 10.1186/s13059-018-1519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Nord A.S., Akiyama J.A., Shoukry M., Afzal V., Rubin E.M., Pennacchio L.A., Visel A. Tissue-specific RNA expression marks distant-acting developmental enhancers. PLoS Genet. 2014;10:e1004610. doi: 10.1371/journal.pgen.1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M. Hepatitis C virus: propagation, quantification, and storage. Curr. Protoc. Microbiol. 2010;19:15D.1.1–15D.1.11. doi: 10.1002/9780471729259.mc15d01s19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lee J., Ruan H., Ye Y., Krakowiak J., Hu Q., Xiang Y., Gong J., Zhou B., Wang L., et al. Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat. Commun. 2019;10:4562. doi: 10.1038/s41467-019-12543-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.