Abstract
Vesicle-associated membrane proteins 721 and 722 (VAMP721/722) are secretory vesicle-localized arginine-conserved soluble N-ethylmaleimide-sensitive factor attachment protein receptors (R-SNAREs) to drive exocytosis in plants. They are involved in diverse physiological processes in plants by interacting with distinct plasma membrane (PM) syntaxins. Here, we show that synaptotagmin 5 (SYT5) is involved in plant defense against Pseudomonas syringae pv tomato (Pst) DC3000 by regulating SYP132-VAMP721/722 interactions. Calcium-dependent stimulation of in vitro SYP132-VAMP722 interaction by SYT5 and reduced in vivo SYP132-VAMP721/722 interaction in syt5 plants suggest that SYT5 regulates the interaction between SYP132 and VAMP721/722. We interestingly found that disease resistance to Pst DC3000 bacterium but not to Erysiphe pisi fungus is compromised in syt5 plants. Since SYP132 plays an immune function to bacteria, elevated growth of surface-inoculated Pst DC3000 in VAMP721/722-deficient plants suggests that SYT5 contributes to plant immunity to Pst DC3000 by promoting the SYP132-VAMP721/722 immune secretory pathway.
Keywords: plant immunity, Pst DC3000, SYP132, SYT5, VAMP721/722
INTRODUCTION
Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) are now regarded as the minimal core factors to drive the vesicle fusion in eukaryotes including plants (Jahn and Scheller, 2006; Lipka et al., 2007; Yun and Kwon, 2017). To overcome energetically unfavorable lipid fusion, they form a SNARE complex in which four α-helices are bundled. Based on the conserved central amino acid, SNAREs are classified into the glutamine-conserved Q-SNARE group that is localized to a target membrane, and the arginine-conserved R-SNARE group that is located on a vesicle membrane (Jahn and Scheller, 2006; Lipka et al., 2007; Yun and Kwon, 2017). Based on their positions in a SNARE complex, Q-SNAREs are further subgrouped into Qa-SNARE, Qb-SNARE, Qc-SNARE, and Qbc-SNARE (which contains two SNARE domains) (Jahn and Scheller, 2006; Lipka et al., 2007; Yun and Kwon, 2017). In general, three distinct SNAREs (Qa + Qbc + R) form a ternary SNARE complex for exocytosis, whereas four SNAREs do a quaternary SNARE complex for intracellular vesicle fusion (Jahn and Scheller, 2006; Kwon et al., 2020a; Lipka et al., 2007; Yun and Kwon, 2017).
The first identified SNARE components to form a biologically relevant SNARE complex in plants are the plasma membrane (PM)-localized SYP121 (syntaxin of plant 121, also called PEN1) Qa-SNARE, the PM-attached SNAP33 Qbc-SNARE and the vesicle-residing VAMP721/722 (vesicle-associated membrane proteins 721 and 722) R-SNAREs (Collins et al., 2003; Kwon et al., 2008). The SYP121-SNAP33-VAMP721/722 ternary SNARE complex drives an immune exocytosis. Interestingly, while SYP121 function is limited to defense against fungal pathogens, VAMP721/722 are additionally required for resistance to oomycete pathogens (Kwon et al., 2008). VAMP721/722 are also required for growth, cell division and abiotic stress responses (El Kasmi et al., 2013; Ichikawa et al., 2014; Kim et al., 2019; Kwon et al., 2008; Yi et al., 2013; Yun et al., 2013). Promiscuous SNARE complex formation of VAMP721/722 with distinct Qa-SNAREs such as SYP121, SYP111 (KNOLLE), SYP123 and SYP132 (El Kasmi et al., 2013; Ichikawa et al., 2014; Kwon et al., 2008; Yun et al., 2013) strongly suggests that at least one regulatory protein should control the SNARE complex formation of VAMP721/722 with a specific Qa-SNARE in a particular biological process in plants. This can be additionally supported by promiscuous SNARE complex formation of SYP121 with members in the VAMP72 R-SNARE group (Kwon et al., 2008).
In plants, three groups of regulatory proteins such as Sec1/Munc18 (SM), small GTPase and synaptotagmin (SYT) have been studied for modulating the trafficking functionality of SNAREs. The KEULE (also called SEC11) SM protein controls the SNARE complex formation of VAMP721/722 with KNOLLE for cytokinesis (Karnahl et al., 2018; Park et al., 2012), but with SYP121 for potassium uptake and growth (Karnik et al., 2013; 2015; Waghmare et al., 2019; Zhang et al., 2019) by binding to the N-terminus of respective Qa-SNARE. Recently, a KEULE paralog, SEC1B, was found to be predominantly engaged in regulating the SYP132-dependent secretion during pollen growth (Karnahl et al., 2018). The plant-specific ARA6 GTPase is regarded to switch the interaction of SYP121 between VAMP721/722 and VAMP727 (Ebine et al., 2011). SYP121 is known to continuously cycle between the PM and endosomes (Reichardt et al., 2011). Focal accumulation of endocytosed SYP121 or its barley ortholog, ROR2, to fungal entry sites was found to require the GNOM ADP-ribosylation factor-guanine nucleotide exchange factor (ARF-GEF) in Arabidopsis or ARFA1b/c GTPases in barely, respectively (Bohlenius et al., 2010; Nielsen et al., 2012). SYT1 preferentially residing in the endoplasmic reticulum (ER) (Levy et al., 2015; Perez-Sancho et al., 2015) was originally found to repair damaged PM by abiotic stresses and control endocytosis often resulting in promoting cell-to-cell movement of viral movement proteins (Lewis and Lazarowitz, 2010; Schapire et al., 2008; Yamazaki et al., 2008). SYT1 together with SYT5 was recently found to regulate the intactness and rearrangement of ER-PM contact sites for stress responses to rare earth elements and viral pathogens in plants (Ishikawa et al., 2020; Lee et al., 2020).
We previously found that SYT1 additionally down-regulates SYP121 abundance possibly via endocytosis to control plant disease resistance to fungal pathogens (Kim et al., 2016). We therefore examined whether SYT5 also functions in plant immunity. Based on specific impairment of resistance to Pseudomonas syringae pv tomato (Pst) DC3000 bacterium but not to Erysiphe pisi pea powdery mildew fungus in syt5 plants, we found that SYT5 interacts with the PM-localized SYP132 which is required for plant defense against bacteria but not fungi (Kalde et al., 2007). Elevated growth of surface-inoculated Pst DC3000 in VAMP721/722-depleted plants indicates that VAMP721/722 are required for previously unrevealed plant immunity to epiphytic bacteria. In vitro stimulation of SYP132-VAMP722 interaction by SYT5, and reduced SYP132-VAMP721/722 interaction in syt5 plants suggest that SYT5 positively regulates the SYP132-VAMP721/722 immune exocytosis to bacteria.
MATERIALS AND METHODS
Plant materials
Plants used for experiments were grown at 22°C with 10-h light/14-h dark photoperiod. To isolate T-DNA-inserted syt5-1 and syt5-2, SALK_036961 and GABI_679H10 were obtained from ABRC and GABI-Kat. Homozygous mutant plants were selected by genomic DNA polymerase chain reaction (PCR) with T-DNA-specific (5’-GCGTGGACCGCTTGCTGCAACT and 5’-ATATTGACCATCATACTCATTGC) and SYT5-specific primers (5’-GTGATATTTCAGCAATTCCTGGAC and 5’-TAGTCTTTTCACGCAAAGGGC), and further analyzed by immunoblot with anti-SYT5 antibody.
Purification of recombinant proteins
To express recombinant proteins, cDNAs corresponding to SYP132, VAMP722, SYT5∆TM, SYP111, SYP121, and SYP123 were amplified by PCR and introduced into the pGEX-6p-1 vector (GE Healthcare Life Sciences, USA). To express HA-SYP132, HA-corresponding DNA sequence was added to a primer to amplify SYP132 cDNA. All GST-fused recombinant proteins were expressed in Escherichia coli BL21-CodonPlus (Agilent Technologies, USA), and purified by affinity chromatography using glutathione-Sepharose 4B (GE Healthcare Life Sciences). To obtain GST-free proteins, GST moiety was removed by PreScission Protease (GE Healthcare Life Sciences). GST-removed SYT5∆TM was used to generate anti-SYT5 antibody in a rabbit, and GST-free SYP132 to generate anti-SYP132 antibody in a chicken. Purified GST-removed SYP111, SYP121, SYP123 and SYP132 were used to test the specificity of the generated anti-SYP132 antibody.
In vitro protein interaction assay
To test the interaction between SYP132 and SYT5, equimolar recombinant HA-SYP132 and GST-SYT5∆TM were incubated in the absence or presence of 1 mM CaCl2. To assess the effect of Ca2+ on SYT5-SYP132 interaction, 1 mM EDTA was added during incubation. To test the interaction between SYP132 and VAMP722, equimolar recombinant HA-SYP132 and GST-VAMP722 were mixed together with GST-free SYT5∆TM in the absence or presence of 1 mM CaCl2. To analyze whether the Ca2+-promoted SYT5∆TM-SYP132 interaction affects SYP132-VAMP722 interaction, 1 mM EDTA was added during incubation. Interacted proteins were then precipitated with glutathione-Sepharose 4B and the precipitates were analyzed by immunoblot with anti-HA antibody to detect HA-SYP132 in the precipitates.
In vivo protein interaction assay
Proteins were extracted from the indicated genotype plants by suspending ground plant materials in 1× phosphate-buffered saline (PBS) containing 1% Triton X-100. Protein amounts were measured by the Bio-Rad protein assay (Bio-Rad, USA). Protein extracts were first pre-cleared with Protein A/G-agarose beads (Santa Cruz Biotechnology, USA) and incubated with anti-SYT5 or anti-VAMP721/722 antibody. Anti-SYT5 antibody-bound or anti-VAMP721/722 antibody-bound proteins were then retrieved by precipitation with Protein A/G-agarose beads. The immunoprecipitates were finally analyzed by immunoblot with anti-SYP132 antibody. A part (3%) of protein extracts used for immunoprecipitation were subject to immunoblot with anti-SYT5, anti-SYP121, anti-SYP132, anti-VAMP721/722 antibody, or anti-SNAP33 antiserum for showing steady state levels of SYT5, SYP121, SYP132, VAMP721/722, or SNAP33, respectively.
Pathogenicity test
Plants were grown in soil for 4-5 weeks to be inoculated with E. pisi or dip-inoculated with Pst DC3000 (1 × 107 cfu). To be liquid-inoculated with Pst DC3000 (1 × 105 cfu), plants were grown for 10 days in liquid Murashige and Skoog (MS) medium containing 1% sucrose. For measuring E. pisi entry rate, inoculated leaves were stained with 0.6% Coomassie blue in 100% ethanol and subject to light microscopy. Conidiospores with secondary epiphytic hyphae were counted as successful fungal entry. Bacterial numbers (in liquid-inoculated seedlings at 2 days post-inoculation [dpi] or in dip-inoculated leaves at 3 hours post-inoculation [hpi] and 3 dpi) were counted by serial dilution plating.
Confocal microscopy
For immunostaining, 7-day-old transgenic Arabidopsis seedlings expressing either GFP-SYP132 or mRFP-VAMP722 were fixed in MTSB buffer (50 mM PIPES, 5 mM EGTA and 5 mM MgSO4, pH 7.0 adjusted with KOH) containing 4% paraformaldehyde for 1 h at room temperature under vacuum infiltration (Sauer et al., 2006). Fixed seedlings were placed onto poly-L-Lys-coated glass slides, and washed with MTSB buffer containing 0.1% Triton X-100 and with deionized water. Fixed seedlings were then incubated with 2% driselase in MTSB buffer for 40 min to digest their cell wall, and incubated with PBS containing 20% DMSO and 3% NP40 for 1 h. Following washing with MTSB buffer containing 0.1% Triton and with deionized water, fixed seedlings were pre-incubated with a blocking buffer (PBS containing 5% BSA) at 37°C for 1 h and incubated with anti-SYT5 antibody in the blocking buffer at 4°C overnight. After washing with PBS containing 0.1% Triton X-100, fixed seedlings were incubated with either Alexa 488- or Alexa 546-conjugated rabbit igG antibody (Invitrogen, USA) in the blocking buffer at room temperature for 3 h. Following washing with PBS containing 0.1% Triton X-100, these seedlings were finally transferred into a mounting medium (100 mM Tris [pH 8.5] containing 25% glycerol) containing Mowiol 4-88 (Calbiochem, USA). All fluorescent images were taken by LSM780 confocal microscope (Zeiss, Germany) equipped with a 40× objective (C-Apochromat 40×/1.1 W) and processed by Zen 2011 software (Zeiss) and Adobe Photoshop CS5 (Adobe, USA). GFP and Alexa 488 were excited with an argon laser at 488 nm, and mRFP and Alexa 546 were at 561 nm. For two-color imaging, multitracking was configured to avoid cross-talk between fluorescence channels.
RESULTS
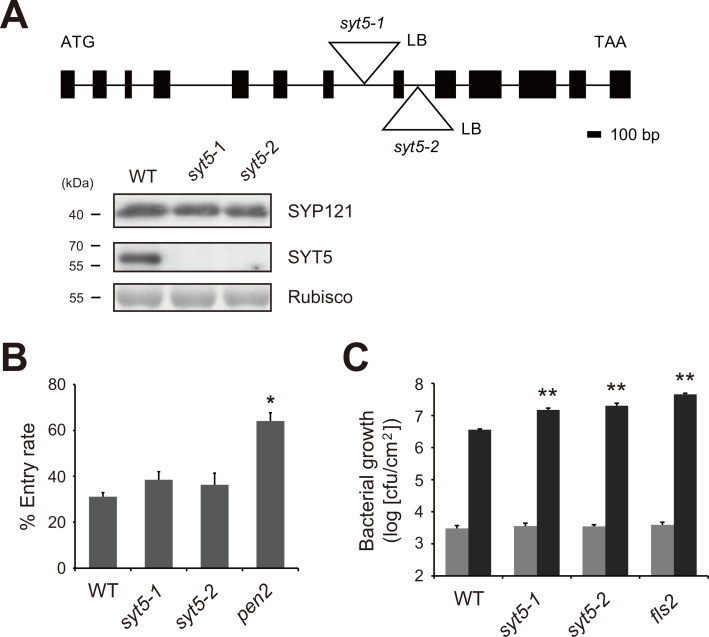
It was reported that transiently expressed SYT5-GFP is localized to endomembrane compartments in Arabidopsis protoplasts (Yamazaki et al., 2010). SYT5-GFP was also reported to be localized to the ER/PM contact sites by transient expression in Nicotiana benthamiana (Ishikawa et al., 2020). SYT5 together with SYT1 is suggested to regulate the ER-PM interactions and plasmodesmata (PD) conductance in Arabidopsis (Ishikawa et al., 2020; Lee et al., 2020). We previously reported that SYT1, which is localized to the PM and ER/PM contact sites, negatively controls plant disease resistance to powdery mildew fungi by modulating SYP121 levels (Kim et al., 2016). Therefore, we tested whether SYT5 is also required for immune responses to phytopathogens. For this, we isolated two independent T-DNA insertion syt5 mutant plants, syt5-1 and syt5-2. Although T-DNAs are inserted in introns in both syt5-1 and syt5-2 (Supplementary Fig. S1), we found that SYT5 proteins were barely detectable in those mutant plants by immunoblot with anti-SYT5 antibody (Fig. 1A). We then inoculated syt5-1 and syt5-2 plants with the Arabidopsis-nonadapted E. pisi pea powdery mildew fungus. However, we found no significant difference in fungal entry rates between wild-type (WT) and syt5 plants (Fig. 1B), indicating no requirement of SYT5 for plant immunity to E. pisi. We next dip-inoculated the above-mentioned mutant plants with the Arabidopsis-pathogenic Pst DC3000 bacterium. At 3 dpi, we interestingly found more bacterial growth in both syt5-1 and syt5-2 plants than in WT (Fig. 1C). This suggests that SYT5 is involved in plant immunity to Pst DC3000 bacterium but not to E. pisi fungus.
Fig. 1. SYT5 is required for defense against Pst DC3000 bacterium but not for E. pisi fungus.
(A) A schematic view of SYT5 gene. Two independent T-DNA insertion sites for syt5-1 (SALK_036961) and syt5-2 (GABI_679H10) were marked by triangles. LB, left boarder of inserted T-DNA. Box, exon; line, intron. Homozygosity of syt5-1 and syt5-2 plants were tested by immunoblot. Proteins extracted from the indicated genotype plants were subject to immunoblot with anti-SYT5 and anti-SYP121 antibodies. Equal loading was visualized by staining Rubisco with Coomassie blue. (B) Dispensability of SYT5 for resistance to E. pisi. The indicated genotype plants were inoculated with E. pisi conidiospores. At 2 dpi, inoculated leaves were stained with Commassie blue and analyzed for fungal entry rates. Conidiospores with secondary epiphytic hyphae were regarded as successfully entered fungi. Values are presented as mean ± SE from three biological replications. (C) Elevated Pst DC3000 growth in syt5 plants. The indicated genotype plants were dip-inoculated with Pst DC3000 (1 × 107 cfu). The in-leaf bacterial growth was measured at 3 hpi (gray) or at 3 dpi (black). Values are presented as mean ± SE from four biological replications. *P < 0.05; **P < 0.01 in comparison to WT (Student t-test).
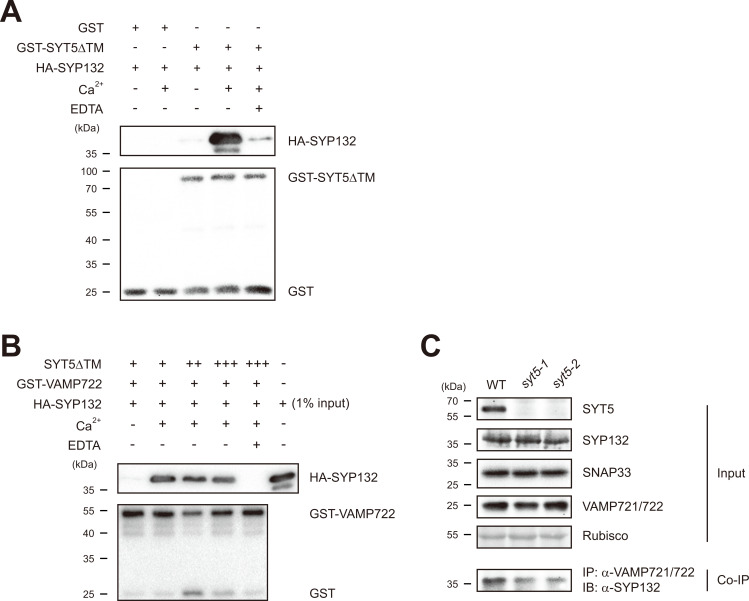
We previously reported that SYT1 interacts with the PM-residing SYP121 syntaxin to modulate plant immune responses by controlling SYP121 level in plants (Kim et al., 2016). We therefore tested whether SYT5 is also able to interact with a PM syntaxin. It is known that SYP121 is required for defense against powdery mildew fungi, whereas SYP132 is for resistance to bacteria (Kalde et al., 2007; Kwon et al., 2008). We found that SYP121 levels in both syt5-1 and syt5-2 are indistinguishable from that in WT by immunoblot with anti-SYP121 antibody (Fig. 1A). We therefore investigated an interaction between SYT5 and SYP132, because SYT5 is required for immunity to Pst DC3000 bacterium (Fig. 1C). We first tested their direct interaction in vitro, using hemagglutinin-tagged SYP132 (HA-SYP132) and transmembrane motif-lacking SYT5 fused with glutathione-S-transferase (GST-SYT5ΔTM) that were purified from E. coli. After mixing HA-SYP132 and GST- SYT5ΔTM, we precipitated GST- SYT5ΔTM with glutathione-agarose and analyzed the precipitates by immunoblot with anti-HA antibody. Detection of HA-SYP132 in the GST-SYT5ΔTM precipitates (Fig. 2A) indicates that SYP132 and SYT5 directly interact with each other. Interestingly, their interaction was dramatically enhanced by Ca2+ addition (Fig. 2A). Abolishment of such Ca2+-induced elevation of their interaction by adding the cation chelator EDTA (Fig. 2A) indicates that Ca2+ stimulates SYP132-SYT5 interaction most likely via binding to the Ca2+-binding C2 domains of SYT5. No detection of HA-SYP132 in the GST precipitates regardless of Ca2+ (Fig. 2A) indicates that SYP132 does not nonspecifically interact with GST even in the presenece of Ca2+.
Fig. 2. SYT5 stimulates SYP132-VAMP721/722 interaction.
(A) Ca2+-stimulated direct interaction between SYT5 and SYP132 in vitro. Equimolar recombinant HA-tagged SYP132 (HA-SYP132) and the TM-lacking GST-fused SYT5 (GST-SYT5ΔTM) were incubated in the presence or absence of 1 mM CaCl2. Their interaction was analyzed by immunoblot with anti-HA antibody to detect HA-SYP132 in the precipitates with GST-SYT5ΔTM. To test a Ca2+ effect on their interaction, 1 mM EDTA was added during incubation. Equal loading was visualized by immunoblot with anti-GST antibody to detect GST-SYT5ΔTM. GST was used as a negative control. (B) SYT5-induced interaction between SYP132 and VAMP722 in the presence of Ca2+ in vitro. Equimolar recombinant HA-SYP132 and GST-VAMP722 were incubated with increasing amounts of SYT5ΔTM (+, equimolar; ++, 5-fold more; +++, 10-fold more amounts) in the presence or absence of 1 mM CaCl2. The SYP132-VAMP722 interaction was analyzed by immunoblot with anti-HA antibody to detect HA-SYP132 in the precipitates with GST-VAMP722. To show the size of HA-SYP132, 1% of used HA-SYP132 was subject to immunoblot with anti-HA antibody. To test a Ca2+ effect, 1 mM EDTA was added during incubation. Equal loading was visualized by immunoblot with anti-GST antibody to detect GST-VAMP722. (C) Reduced SYP132-VAMP721/722 interaction in syt5 plants. Protein extracts from the indicated genotype plants leaves were precipitated (IP) with anti-VAMP721/722 antibody and the precipitates (Co-IP) were analyzed by immunoblot (IB) with anti-SYP132 antibody. To show expression levels of SYT5, SYP132, SNAP33, and VAMP721/722 in the indicated genotype plants, 3% of protein extracts used for Co-IP were subject to immunoblot with anti-SYT5, anti-SYP132, anti-VAMP721/722 antibody or anti-SNAP33 antiserum. Equal loading was visualized by staining Rubisco with Ponceau S.
We next tested whether SYT5 and SYP132 also interact in plants. For this, we generated anti-SYT5 and anti-SYP132 antibodies in a rabbit and a chicken, respectively. The anti-SYT5 antibody detects endogenous SYT5 in WT but not in syt5-1 and syt5-2 plants in immunoblot (Fig. 1A), indicative of its specificity to SYT5. Unlike syt5 plants, syp132 plants are lethal (Kalde et al., 2007), leading to difficulty in testing an in vivo specificity of anti-SYP132 antibody. Therefore, to test the specificity of anti-SYP132 antibody, four different PM syntaxins belonging to the SYP1 group (SYP111, SYP121, SYP123, and SYP132) were purified from E. coli (Supplementary Fig. S1) and subject to immunoblot. Immunoblot results with anti-SYP132 antibody also indicate that the generated anti-SYP132 antibody can discriminate SYP132 from the other tested PM syntaxins (Supplementary Fig. S1). In spite of this specificity of anti-SYP132 antibody, the chicken IgY nonspecifically reacts with SYT5 and VAMP721/722 (data not shown). Therefore, we performed a co-immunoprecipitation (Co-IP) assay with WT protein extracts using anti-SYT5 antibody for immunoprecipitation and anti-SYP132 antibody for immunoblot. Detection of SYP132 in the SYT5 precipitates but not in the rabbit IgG ones (Supplementary Fig. S2A) indicates that endogenous SYT5 and SYP132 interact in plants.
SYP132 specifically interacts with VAMP721/722 that reside in the trans-Golgi network (TGN) and secretory vesicles (Ichikawa et al., 2014; Yun et al., 2013). Since SYT5 interacts with SYP132 (Fig. 2A, Supplementary Fig. S2A), we then tested whether SYT5 is able to regulate SYP132-VAMP721/722 interaction. We first examined the effect of SYT5 on SYP132-VAMP722 interaction in vitro using E. coli-purified recombinant proteins, SYT5ΔTM, HA-SYP132 and GST-fused VAMP722 (GST-VAMP722). We failed to detect HA-SYP132 in the GST-VAMP722 precipitates even in the presence of SYT5ΔTM (Fig. 2B). Similarly, we previously reported that SYP121 alone scarcely interacts with VAMP722 in vitro (Kwon et al., 2008). When Ca2+ was added during their incubation, we unexpectedly detected significant amount of SYP132 in the GST-VAMP722 precipitates (Fig. 2B). Interestingly, this Ca2+-aided SYP132-VAMP722 interaction by SYT5 was completely abolished by EDTA addition (Fig. 2B). Since Ca2+ greatly increased SYT5-SYP132 interaction (Fig. 2A), these results suggest that Ca2+-bound SYT5 induces SYP132-VAMP722 complex. This additionally suggests that SYT5 may stimulate a secretory pathway involving SYP132 and VAMP721/722 in plants. Similar levels of HA-SYP132 in the GST-VAMP722 precipitates with more amounts of SYT5ΔTM (Fig. 2B) suggest that SYT5 affects SYP132-VAMP722 interaction with equimolar ratio.
Since all reported in planta interactions between SYP132 and VAMP721/722 were tested in transgenic plants or protoplasts expressing heterogeneous proteins with tags (El Kasmi et al., 2013; Ichikawa et al., 2014; Yun et al., 2013), we investigated endogenous SYP132-VAMP721/722 interaction with our generated antibodies. In a co-IP assay with WT protein extracts using anti-SYT5 antibody for immunoprecipitation and anti-SYP132 antibody for immunoblot, we detected SYP132 in the VAMP721/722 precipitates (Supplementary Fig. S2B), indicating their in planta interaction. We then compared SYP132 amounts in the VAMP721/722 precipitates between WT and syt5 plants to examine an in vivo SYT5 function to regulate SYP132-VAMP721/722 interaction. By immunoblot, we found that SYP132, SNAP33 and VAMP721/722 levels are indistinguishable between WT and both syt5 lines (Fig. 2C), indicating that SYT5 does not affect the steady-state abundance of SYP132, SNAP33 and VAMP721/722. However, we detected reduced SYP132 levels in the VAMP721/722 precipitates in both syt5-1 and syt5-2 plants compared to WT (Fig. 2C). Together with the in vitro results (Fig. 2B), this suggests that SYT5 can positively control SYP132-VAMP721/722 interactions in plants. Since SYP132 is required for defense against bacteria (Kalde et al., 2007), elevated bacterial growth in syt5 plants (Fig. 1C) can be explained by reduced SYP132-VAMP721/722 interactions in the absence of SYT5 (Fig. 2C). Similar levels of SYP132, SNAP33 and VAMP721/722 between WT and syt5 plants (Fig. 2C) suggest that the reduced SYP132-VAMP721/722 interactions in syt5 plants does not result from their reduced levels.
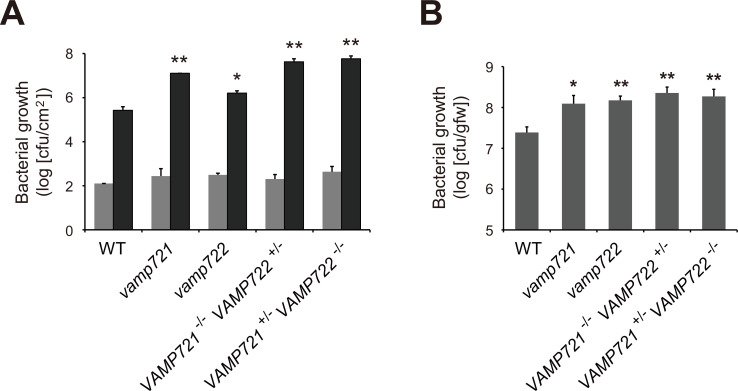
We previously reported that VAMP721/722 are indispensable for plant resistance to Pst DC3000 inoculated by vacuum-infiltration (Kwon et al., 2008). However, this type of inoculation allows bacteria to directly enter the leaf tissue to bypass most plant immune responses to epiphytic bacteria. Therefore, to understand an immune function of VAMP721/722 to epiphytic bacteria, we dip-inoculated plants grown in soil with Pst DC3000 and counted the number of multiplied bacteria in leaves. Since the vamp721 vamp722 double null mutant is lethal, we inoculated plants with differentially lowered VAMP721/722 gene dosage (vamp721, vamp722, VAMP721 -/- VAMP722 +/-, and VAMP721 +/- VAMP722 -/-) (Kwon et al., 2008). At 3 dpi, we found more bacterial growth in all four mutant genotypes than in WT plants (Fig. 3A). Since VAMP721 and VAMP722 are regarded to be immunologically redundant, the elevated bacterial growth in vamp721 and vamp722 plants (Fig. 3A) was unexpected. Hence, we additionally examined bacterial growth in VAMP721/722-lowered genotypes by a different inoculation approach. For this, we inoculated liquid-grown plants by adding bacterial suspension to the medium and counted the number of bacteria within plants (Schreiber et al., 2008). At 2 dpi, we again found increased bacterial growth in vamp721 and vamp722 plants as well as in VAMP721 -/- VAMP722 +/- and VAMP721 +/- VAMP722 -/- plants (Fig. 3B). Recent secretomic analyses revealed that at least a part of cargos transported by VAMP721 and VAMP722 are distinct (Uemura et al., 2019). Taken together, it is therefore likely that at least a part of antibacterial molecules transported by VAMP721 and VAMP722 vesicles might be distinct, which are secreted by SYP132-VAMP721/722 interactions.
Fig. 3. VAMP721/722 are required for plant resistance to surface-inoculated Pst DC3000.
(A) Elevated bacterial growth in all four VAMP721/722-related mutant plants (vamp721, vamp722, VAMP721 -/- VAMP722 +/-, and VAMP721 +/- VAMP722 -/-). The indicated genotype plants grown in soil were dip-inoculated with Pst DC3000 (1 × 107 cfu). The in-leaf bacterial growth was measured at 3 hpi (gray) or at 3 dpi (black). Values are presented as mean ± SE from three biological replications. (B) Increased bacterial growth in all four VAMP721/722-related mutant seedlings. The indicated genotype plants grown in liquid MS medium were liquid-inoculated with Pst DC3000 (1 × 105 cfu). The in-seedling bacterial growth was measured at 2 dpi. Values are presented as mean ± SE from three biological replications. *P < 0.05; **P < 0.01 in comparison to WT (Student t-test).
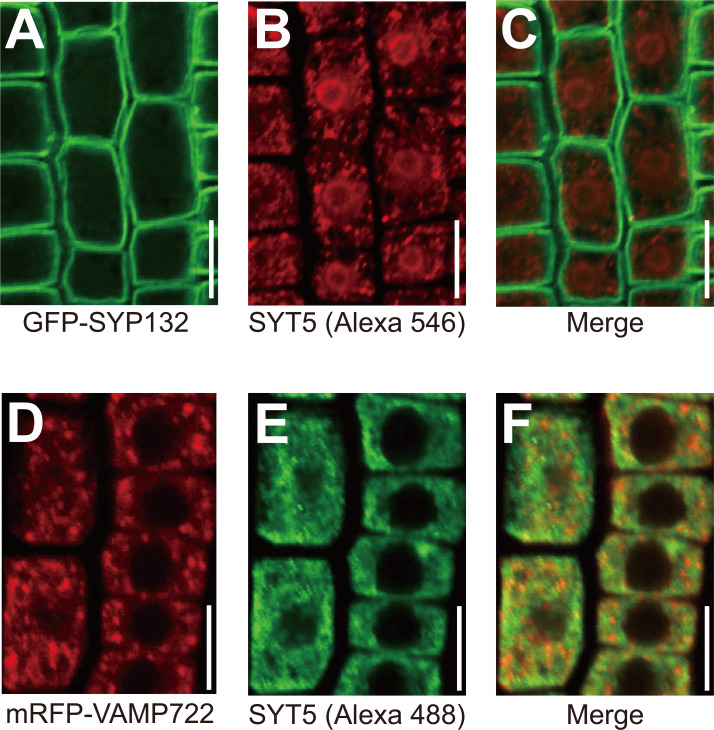
We next examined subcellular localization of endogenous SYT5 in plants. We immuno-fluorescently stained endogenous SYT5 with two distinct secondary antibodies (Alexa 546 with red fluorescence and Alexa 488 with green one) in roots of transgenic plants expressing either the PM-localized GFP-SYP132 or the TGN/vesicle-located mRFP-VAMP722 (Ichikawa et al., 2014). Due to the difficulty in immunostaining root elongating cells leading to smear images (data not shown), we microscopically observed meristematic root cells (Supplementary Fig. S3, Fig. 4). As previously reported (Lee et al., 2020), we detected immuno-fluorescently marked SYT5 in perinuclear area (Figs. 4B and 4E). Distinct localization patterns of SYT5 from GFP-SYP132 (Figs. 4A-4C) and mRFP-VAMP722 (Figs. 4D-4F) indicate that SYT5 is largely localized to the ER. In a dividing root meristematic cell, we interestingly found that SYT5 is not detected in the developing cell plate (Supplementary Fig. S3D-S3F), where SYT1 was reported to be localized (Yamazaki et al., 2010). Since SYP121 level is elevated in syt1 plants (Kim et al., 2016) but not in syt5 plants (Fig. 1A), this implies that SYT5 may play an additional cellular activity to maintaining the ER-PM contact sites together with SYT1.
Fig. 4. Endogenous SYT5 is localized to the perinuclear region in Arabidopsis roots.
(A-C) Cellular localization of endogenous SYT5 in roots of transgenic plants expressing the PM-localized GFP-SYP132. Endogenous SYT5 was detected by anti-SYT5 antibody and Alexa 546 (red fluorescence)-conjugated anti-rabbit IgG antibody. Fluoresent images were analyzed by confocal microscopy. Scale bars = 10 μm. (D-F) Cellular localization of endogenous SYT5 in roots of transgenic plants expressing the TGN/secretory vesicle-localized mRFP-VAMP722. Endogenous SYT5 was detected by anti-SYT5 antibody and Alexa 488 (green fluorescence)-conjugated anti-rabbit IgG antibody. Fluoresent images were analyzed by confocal microscopy. Scale bars = 10 μm.
DISCUSSION
Together with SYT1, SYT5 was recently reported to be engaged in stress responses to rare earth elements and viral pathogens by regulating the rearrangement and intactness of ER-PM contact sites (Ishikawa et al., 2020; Lee et al., 2020). We previously found that SYT1 has an additional immune activity to its ER-PM contact-controlling function (Kim et al., 2016). Elevated SYP121 abundance in syt1 plants and reduced resistance to powdery mildew fungi by SYP121 deletion in syt1 plants suggest that SYT1 negatively functions in immune responses to fungal pathogens by regulating SYP121 levels. We in this study show that SYT5 is also involved in plant resistance to Pst DC3000 bacterium. Compromised resistance to Pst DC3000 bacterium but WT-like defense against E. pisi fungus in syt5 plants (Figs. 1B and 1C) suggest that SYT5 plays an important function in immune responses to bacterial pathogens but not to fungal ones. Comparable SYP121 levels between WT and syt5 plants (Fig. 1A) and no detection of SYT5 in the SYT1-detected cell plate (Supplementary Figs. S3D-S3F) support that SYT5 has a distinct function from SYT1.
VAMP721/722 are the major exocytosis-associated R-SNAREs in Arabidopsis. They participate in a number of physiological processes such as cell division, growth, and biotic/abiotic stress responses even including symbiosis in plants (El Kasmi et al., 2013; Ichikawa et al., 2014; Ivanov et al., 2012; Kim et al., 2019; Kwon et al., 2008; Sogawa et al., 2019; Yi et al., 2013; Yun et al., 2013). In contrast to VAMP721/722, a plant PM syntaxin is involved rather in a specific biological process. Although how VAMP721/722 can be engaged in such diverse processes is largely unknown, their interactions with distinct PM syntaxins such as SYP111, SYP121, SYP122, SYP123, and SYP132 (El Kasmi et al., 2013; Ichikawa et al., 2014; Kwon et al., 2008; Pajonk et al., 2008; Yun et al., 2013) implicate that VAMP721/722 may work for a cellular activity by interacting with a respective PM syntaxin. It is regarded that SYP121 immune function is restricted to fungal pathogens, whereas SYP132 defense activity is limited to resistance to bacterial pathogens (Kalde et al., 2007; Kwon et al., 2008). Based on elevated bacterial growth in syt5 plants (Fig. 1C), we found that SYT5 interacts with SYP132 both in vitro and in vivo (Fig. 2A, Supplementary Fig. S2A). In addition, we found that SYP132-VAMP721/722 interactions are diminished in syt5 plants (Fig. 2C). Greatly increased in vitro interaction between SYP132 and VAMP722 by SYT5, which is otherwise rarely detectable in the absence of SYT5 (Fig. 2B), suggests that SYT5 stimulates the interaction between SYP132 and VAMP721/722 in plants. Since VAMP721/722 drive the immune exocytosis, reduced SYP132-VAMP721/722 interactions in syt5 plants (Fig. 2C) may explain why immune responses in syt5 plants to Pst DC3000 are impaired, likely due to disrupted secretion of extracellular immune molecules.
Unlike typical SYTs, all known plant SYTs contain an additional SYT-like mitochondrial lipid-binding (SMP) domain (Saheki and De Camilli, 2017). In animals, these SYTs called extended-SYTs (E-SYTs) are responsible for tethering the ER to the PM to generate the ER-PM contact sites at which lipids are transported through the SMP domain between two distinct membranes (Saheki and De Camilli, 2017). Likewise, plant SYTs, especially SYT1, SYT5, and SYT7 (also called Ca2+- and lipid-binding protein 1 [CLB1]) were found to play a critical role in maintaining the ER-PM contact sites for responses to various biotic/abiotic stresses (Ishikawa et al., 2020; Lee et al., 2020; Levy et al., 2015; Lewis and Lazarowitz, 2010; Perez-Sancho et al., 2015; Schapire et al., 2008; Yamazaki et al., 2008). Interestingly, it was recently reported that a part of endocytosis-associated autophagy pathway is initiated at the ER-PM contact sites (Wang et al., 2019), suggesting that the ER-PM contact sites may control vesicle trafficking in plants. Indeed, SYT2 is suggested to play a role in secretion in Arabidopsis (Wang et al., 2015; Zhang et al., 2011). In addition, E-SYTs were recently reported to regulate membrane trafficking even in animals (El Kasmi et al., 2018; Kikuma et al., 2017). In response to pathogen attack, plant cells reorganize all subcellular compartments near to the pathogen-attempted area (Bestwick et al., 1997; Koh et al., 2005; Takemoto et al., 2003), likely for rapid and efficient production and secretion of immune molecules including secondary metabolites (Khare et al., 2020). In this scenario, SYT5 may contribute to plant immunity to bacteria by promoting the SYP132/VAMP721/722-associated exocytosis at the ER-PM contact sites. The ‘kiss-and-run’ exocytosis may explain the observed distinct localization of SYT5, SYP132, and VAMP722 in plant cells (Fig. 4, Supplementary Fig. S3), in spite of their interactions (Fig. 2, Supplementary Fig. S2).
Although we previously reported that VAMP721/722 are dispensable for plant immune responses to Pst DC3000 bacterium (Kwon et al., 2008), we here present that they are required for defense against Pst DC3000 (Fig. 3). Such difference is due to different ways of bacterial inoculation; we infiltrated bacteria in the previous study, whereas surface-inoculated in the present study. Interestingly, it was reported that epiphytic (on-leaf) and apoplastic (in-leaf) bacteria express distinct sets of genes (Yu et al., 2013), likely for dissimilar virulent strategies in considerably different leaf-exterior and -interior environments. Therefore, it is likely that VAMP721/722 are important for plant immune responses to epiphytic bacteria but not to apoplastic ones. Interestingly, we observed elevated bacterial growth even in vamp721 and vamp722 single mutant plants in two distinctly surface-inoculated experiments (Fig. 3). Although VAMP721 and VAMP722 are functionally redundant in general, recent studies revealed their differential activities to deliver cell wall-related proteins and an immune protein, RPW8.2 (Kim et al., 2014; Uemura et al., 2019). Indeed, depending on a pathogen type, plant resistance requires differential dosages of VAMP721 and VAMP722 genes (Kwon et al., 2008). This supports that VAMP721 and VAMP722 vesicles transport at least partly distinct as well as common cargos. No more elevation of bacterial growth in VAMP721 -/- VAMP722 +/- and VAMP721 +/- VAMP722 -/- plants than in vamp721 and vamp722 single mutant plants (Fig. 3) suggests that immune molecules differentially secreted by VAMP721 and VAMP722 vesicles might be interchangeable but not additive for defense against epiphytic bacteria.
Although VAMP721/722 are required for immune responses in plants, any other direct immune molecules but RPW8.2 and phospholipase Dδ (PLDδ) were not identified so far to be transported by VAMP721/722 vesicles (Kim et al., 2014; Xing et al., 2019). Proteomic approaches using VAMP721/722-depleted plants found that mostly cell wall-associated proteins are likely to be delivered by VAMP721/722 vesicles in plants (Kwon et al., 2020b; Uemura et al., 2019). Enhanced resistance to fungal pathogens but not to Pst DC3000 in RPW8.2-expressing plants (Li et al., 2018), and impaired immune responses to E. pisi fungus but not to Pst DC3000 in PLDδ-deficient plants (Johansson et al., 2014) suggest that plants may modify the cell wall to defend against bacterial pathogens. Indeed, it was reported that the MYB15-governed lignin biosynthetic pathway is important for plant resistance to surface-inoculated Pst DC3000 (Chezem et al., 2017). In addition, extracellular lignin accumulation was recently reported to limit bacterial motility within plant leaves, ultimately resulting in pathogenesis termination (Lee et al., 2019). Interestingly, bacteria on the leaf surface preferentially express the movement-related genes, compared to leaf-interior bacteria (Yu et al., 2013). This suggests that the growth of epiphytic bacteria depends on their motility. Therefore, it is implied that plants may secrete cell wall-modifying proteins via VAMP721/722 vesicles to restrict bacterial growth on leaf surface likely by disturbing bacterial movement.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by a 2019 Research Grant (520190026 to H.K.) from Kangwon National University, Korea, a grant (PJ01477001 to C.K.) from Rural Development Administration, Korea, a grant (P0016045 to C.K.) from Korea Institute for Advancement of Technology, Korea, and grants (2016R1D1A1B02007322 to C.K.; 2021R1F1A1063111 to H.S.Y.) from National Research Foundation, Korea.
We thank Hae Ri Kwon and Yunjin Choi for technical assistance.
Footnotes
AUTHOR CONTRIBUTIONS
All authors (S.K., H.K., K.P., D.J.C., M.K.K., C.K., and H.S.Y.)conceived and performed experiments. H.K., C.K., and H.S.Y. wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Bestwick C.S., Brown I.R., Bennett M.H., Mansfield J.W. Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseolicola . Plant Cell. 1997;9:209–221. doi: 10.1105/tpc.9.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlenius H., Morch S.M., Godfrey D., Nielsen M.E., Thordal-Christensen H. The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell. 2010;22:3831–3844. doi: 10.1105/tpc.110.078063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chezem W.R., Memon A., Li F.S., Weng J.K., Clay N.K. SG2-Type R2R3-MYB transcription factor MYB15 controls defense-induced lignification and basal immunity in Arabidopsis. Plant Cell. 2017;29:1907–1926. doi: 10.1105/tpc.16.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N.C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J.L., Hückelhoven R., Stein M., Freialdenhoven A., Somerville S.C., et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- Ebine K., Fujimoto M., Okatani Y., Nishiyama T., Goh T., Ito E., Dainobu T., Nishitani A., Uemura T., Sato M.H., et al. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 2011;13:853–859. doi: 10.1038/ncb2270. [DOI] [PubMed] [Google Scholar]
- El Kasmi F., Krause C., Hiller U., Stierhof Y.D., Mayer U., Conner L., Kong L., Reichardt I., Sanderfoot A.A., Jürgens G. SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol. Biol. Cell. 2013;24:1593–1601. doi: 10.1091/mbc.E13-02-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi I., Khadivjam B., Lackman M., Duron J., Bonneil E., Thibault P., Lippé R. Extended synaptotagmin 1 interacts with herpes simplex virus 1 glycoprotein M and negatively modulates virus-induced membrane fusion. J. Virol. 2018;92:e01281–17. doi: 10.1128/JVI.01281-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M., Hirano T., Enami K., Fuselier T., Kato N., Kwon C., Voigt B., Schulze-Lefert P., Baluška F., Sato M.H. Syntaxin of plant proteins SYP123 and SYP132 mediate root hair tip growth in Arabidopsis thaliana . Plant Cell Physiol. 2014;55:790–800. doi: 10.1093/pcp/pcu048. [DOI] [PubMed] [Google Scholar]
- Ishikawa K., Tamura K., Fukao Y., Shimada T. Structural and functional relationships between plasmodesmata and plant endoplasmic reticulum-plasma membrane contact sites consisting of three synaptotagmins. New Phytol. 2020;226:798–808. doi: 10.1111/nph.16391. [DOI] [PubMed] [Google Scholar]
- Ivanov S., Fedorova E.E., Limpens E., De Mita S., Genre A., Bonfante P., Bisseling T. Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8316–8321. doi: 10.1073/pnas.1200407109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., Scheller R.H. SNAREs--engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Johansson O.N., Fahlberg P., Karimi E., Nilsson A.K., Ellerstrom M., Andersson M.X. Redundancy among phospholipase D isoforms in resistance triggered by recognition of the Pseudomonas syringae effector AvrRpm1 in Arabidopsis thaliana . Front. Plant Sci. 2014;5:639. doi: 10.3389/fpls.2014.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalde M., Nuhse T.S., Findlay K., Peck S.C. The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11850–11855. doi: 10.1073/pnas.0701083104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnahl M., Park M., Krause C., Hiller U., Mayer U., Stierhof Y.D., Jürgens G. Functional diversification of Arabidopsis SEC1-related SM proteins in cytokinetic and secretory membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 2018;115:6309–6314. doi: 10.1073/pnas.1722611115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik R., Grefen C., Bayne R., Honsbein A., Kohler T., Kioumourtzoglou D., Williams M., Bryant N.J., Blatt M.R. Arabidopsis Sec1/Munc18 protein SEC11 is a competitive and dynamic modulator of SNARE binding and SYP121-dependent vesicle traffic. Plant Cell. 2013;25:1368–1382. doi: 10.1105/tpc.112.108506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik R., Zhang B., Waghmare S., Aderhold C., Grefen C., Blatt M.R. Binding of SEC11 indicates its role in SNARE recycling after vesicle fusion and identifies two pathways for vesicular traffic to the plasma membrane. Plant Cell. 2015;27:675–694. doi: 10.1105/tpc.114.134429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S., Singh N.B., Singh A., Hussain I., Niharika K., Yadav V., Bano C., Yadav R.K., Amist N. Plant secondary metabolites synthesis and their regulations under biotic and abiotic constraints. J. Plant Biol. 2020;63:203–216. [Google Scholar]
- Kikuma K., Li X., Kim D., Sutter D., Dickman D.K. Extended synaptotagmin localizes to presynaptic ER and promotes neurotransmission and synaptic growth in Drosophila . Genetics. 2017;207:993–1006. doi: 10.1534/genetics.117.300261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Kwon H., Kim S., Kim M.K., Botella M.A., Yun H.S., Kwon C. Synaptotagmin 1 negatively controls the two distinct immune secretory pathways to powdery mildew fungi in Arabidopsis. Plant Cell Physiol. 2016;57:1133–1141. doi: 10.1093/pcp/pcw061. [DOI] [PubMed] [Google Scholar]
- Kim H., O'Connell R., Maekawa-Yoshikawa M., Uemura T., Neumann U., Schulze-Lefert P. The powdery mildew resistance protein RPW8.2 is carried on VAMP721/722 vesicles to the extrahaustorial membrane of haustorial complexes. Plant J. 2014;79:835–847. doi: 10.1111/tpj.12591. [DOI] [PubMed] [Google Scholar]
- Kim S., Choi Y., Kwon C., Yun H.S. Endoplasmic reticulum stress-induced accumulation of VAMP721/722 requires CALRETICULIN 1 and CALRETICULIN 2 in Arabidopsis. J. Integr. Plant Biol. 2019;61:974–980. doi: 10.1111/jipb.12728. [DOI] [PubMed] [Google Scholar]
- Koh S., Andre A., Edwards H., Ehrhardt D., Somerville S. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J. 2005;44:516–529. doi: 10.1111/j.1365-313X.2005.02545.x. [DOI] [PubMed] [Google Scholar]
- Kwon C., Lee J.H., Yun H.S. SNAREs in plant biotic and abiotic stress responses. Mol. Cells. 2020a;43:501–508. doi: 10.14348/molcells.2020.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C., Neu C., Pajonk S., Yun H.S., Lipka U., Humphry M., Bau S., Straus M., Kwaaitaal M., Rampelt H., et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–840. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- Kwon H., Cho D.J., Lee H., Nam M.H., Kwon C., Yun H.S. CCOAOMT1, a candidate cargo secreted via VAMP721/722 secretory vesicles in Arabidopsis. Biochem. Biophys. Res. Commun. 2020b;524:977–982. doi: 10.1016/j.bbrc.2020.02.029. [DOI] [PubMed] [Google Scholar]
- Lee E., Santana B.V.N., Samuels E., Benitez-Fuente F., Corsi E., Botella M.A., Perez-Sancho J., Vanneste S., Friml J., Macho A., et al. Rare earth elements induce cytoskeleton-dependent and PI4P-associated rearrangement of SYT1/SYT5 endoplasmic reticulum-plasma membrane contact site complexes in Arabidopsis. J. Exp. Bot. 2020;71:3986–3998. doi: 10.1093/jxb/eraa138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.H., Jeon H.S., Kim S.H., Chung J.H., Roppolo D., Lee H.J., Cho H.J., Tobimatsu Y., Ralph J., Park O.K. Lignin-based barrier restricts pathogens to the infection site and confers resistance in plants. EMBO J. 2019;38:e101948. doi: 10.15252/embj.2019101948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A., Zheng J.Y., Lazarowitz S.G. Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr. Biol. 2015;25:2018–2025. doi: 10.1016/j.cub.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.D., Lazarowitz S.G. Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2491–2496. doi: 10.1073/pnas.0909080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Y., Wang Q.X., Wang T.T., Cao X.L., Zhao Z.X., Zhao S.L., Xu Y.J., Xiao Z.Y., Li J.L., et al. RESISTANCE TO POWDERY MILDEW8.1 boosts pattern-triggered immunity against multiple pathogens in Arabidopsis and rice. Plant Biotechnol. J. 2018;16:428–441. doi: 10.1111/pbi.12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V., Kwon C., Panstruga R. SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu. Rev. Cell Dev. Biol. 2007;23:147–174. doi: 10.1146/annurev.cellbio.23.090506.123529. [DOI] [PubMed] [Google Scholar]
- Nielsen M.E., Feechan A., Bohlenius H., Ueda T., Thordal-Christensen H. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc. Natl. Acad. Sci. U. S. A. 2012;109:11443–11448. doi: 10.1073/pnas.1117596109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajonk S., Kwon C., Clemens N., Panstruga R., Schulze-Lefert P. Activity determinants and functional specialization of Arabidopsis PEN1 syntaxin in innate immunity. J. Biol. Chem. 2008;283:26974–26984. doi: 10.1074/jbc.M805236200. [DOI] [PubMed] [Google Scholar]
- Park M., Touihri S., Muller I., Mayer U., Jurgens G. Sec1/Munc18 protein stabilizes fusion-competent syntaxin for membrane fusion in Arabidopsis cytokinesis. Dev. Cell. 2012;22:989–1000. doi: 10.1016/j.devcel.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Perez-Sancho J., Vanneste S., Lee E., McFarlane H.E., Esteban Del Valle A., Valpuesta V., Friml J., Botella M.A., Rosado A. The Arabidopsis synaptotagmin1 is enriched in endoplasmic reticulum-plasma membrane contact sites and confers cellular resistance to mechanical stresses. Plant Physiol. 2015;168:132–143. doi: 10.1104/pp.15.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt I., Slane D., El Kasmi F., Knoll C., Fuchs R., Mayer U., Lipka V., Jürgens G. Mechanisms of functional specificity among plasma-membrane syntaxins in Arabidopsis . Traffic. 2011;12:1269–1280. doi: 10.1111/j.1600-0854.2011.01222.x. [DOI] [PubMed] [Google Scholar]
- Saheki Y., De Camilli P. The extended-synaptotagmins. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:1490–1493. doi: 10.1016/j.bbamcr.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M., Paciorek T., Benkova E., Friml J. Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat. Protoc. 2006;1:98–103. doi: 10.1038/nprot.2006.15. [DOI] [PubMed] [Google Scholar]
- Schapire A.L., Voigt B., Jasik J., Rosado A., Lopez-Cobollo R., Menzel D., Salinas J., Mancuso S., Valpuesta V., Baluska F., et al. Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell. 2008;20:3374–3388. doi: 10.1105/tpc.108.063859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber K., Ckurshumova W., Peek J., Desveaux D. A high-throughput chemical screen for resistance to Pseudomonas syringae in Arabidopsis. Plant J. 2008;54:522–531. doi: 10.1111/j.1365-313X.2008.03425.x. [DOI] [PubMed] [Google Scholar]
- Sogawa A., Yamazaki A., Yamasaki H., Komi M., Manabe T., Tajima S., Hayashi M., Nomura M. SNARE proteins LjVAMP72a and LjVAMP72b are required for root symbiosis and root hair formation in Lotus japonicus. Front. Plant Sci. 2019;9:1992. doi: 10.3389/fpls.2018.01992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D., Jones D.A., Hardham A.R. GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. Plant J. 2003;33:775–792. doi: 10.1046/j.1365-313x.2003.01673.x. [DOI] [PubMed] [Google Scholar]
- Uemura T., Nakano R.T., Takagi J., Wang Y., Kramer K., Finkemeier I., Nakagami H., Tsuda K., Ueda T., Schulze-Lefert P., et al. A Golgi-released subpopulation of the trans-Golgi network mediates protein secretion in Arabidopsis. Plant Physiol. 2019;179:519–532. doi: 10.1104/pp.18.01228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghmare S., Lefoulon C., Zhang B., Liliekyte E., Donald N., Blatt M.R. K+ Channel-SEC11 binding exchange regulates SNARE assembly for secretory traffic. Plant Physiol. 2019;181:1096–1113. doi: 10.1104/pp.19.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Han S., Siao W., Song C., Xiang Y., Wu X., Cheng P., Li H., Jásik J., Mičieta K., et al. Arabidopsis synaptotagmin 2 participates in pollen germination and tube growth and is delivered to plasma membrane via conventional secretion. Mol. Plant. 2015;8:1737–1750. doi: 10.1016/j.molp.2015.09.003. [DOI] [PubMed] [Google Scholar]
- Wang P., Pleskot R., Zang J., Winkler J., Wang J., Yperman K., Zhang T., Wang K., Gong J., Guan Y., et al. Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat. Commun. 2019;10:5132. doi: 10.1038/s41467-019-12782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J., Li X., Wang X., Lv X., Wang L., Zhang L., Zhu Y., Shen Q., Baluška F., Šamaj J., et al. Secretion of phospholipase D functions as a regulatory mechanism in plant innate immunity. Plant Cell. 2019;31:3015–3032. doi: 10.1105/tpc.19.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., Kawamura Y., Minami A., Uemura M. Calcium-dependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1. Plant Cell. 2008;20:3389–3404. doi: 10.1105/tpc.108.062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., Takata N., Uemura M., Kawamura Y. Arabidopsis synaptotagmin SYT1, a type I signal-anchor protein, requires tandem C2 domains for delivery to the plasma membrane. J. Biol. Chem. 2010;285:23165–23176. doi: 10.1074/jbc.M109.084046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C., Park S., Yun H.S., Kwon C. Vesicle-associated membrane proteins 721 and 722 are required for unimpeded growth of Arabidopsis under ABA application. J. Plant Physiol. 2013;170:529–533. doi: 10.1016/j.jplph.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Yu X., Lund S.P., Scott R.A., Greenwald J.W., Records A.H., Nettleton D., Lindow S.E., Gross D.C., Beattie G.A. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E425–E434. doi: 10.1073/pnas.1221892110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H.S., Kwaaitaal M., Kato N., Yi C., Park S., Sato M.H., Schulze-Lefert P., Kwon C. Requirement of vesicle-associated membrane protein 721 and 722 for sustained growth during immune responses in Arabidopsis. Mol. Cells. 2013;35:481–488. doi: 10.1007/s10059-013-2130-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H.S., Kwon C. Vesicle trafficking in plant immunity. Curr. Opin. Plant Biol. 2017;40:34–42. doi: 10.1016/j.pbi.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Zhang B., Karnik R., Alvim J., Donald N., Blatt M.R. Dual sites for SEC11 on the SNARE SYP121 implicate a binding exchange during secretory traffic. Plant Physiol. 2019;180:228–239. doi: 10.1104/pp.18.01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhang L., Gao B., Fan H., Jin J., Botella M.A., Jiang L., Lin J. Golgi apparatus-localized synaptotagmin 2 is required for unconventional secretion in Arabidopsis . PLoS One. 2011;6:e26477. doi: 10.1371/journal.pone.0026477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.