Abstract
The paraflagellar rod (PFR) of the African trypanosome Trypanosoma brucei represents an excellent model to study flagellum assembly. The PFR is an intraflagellar structure present alongside the axoneme and is composed of two major proteins, PFRA and PFRC. By inducible expression of a functional epitope-tagged PFRA protein, we have been able to monitor PFR assembly in vivo. As T. brucei cells progress through their cell cycle, they possess both an old and a new flagellum. The induction of expression of tagged PFRA in trypanosomes growing a new flagellum provided an excellent marker of newly synthesized subunits. This procedure showed two different sites of addition: a major, polar site at the distal tip of the flagellum and a minor, nonpolar site along the length of the partially assembled PFR. Moreover, we have observed turnover of epitope-tagged PFRA in old flagella that takes place throughout the length of the PFR structure. Expression of truncated PFRA mutant proteins identified a sequence necessary for flagellum localization by import or binding. This sequence was not sufficient to confer full flagellum localization to a green fluorescent protein reporter. A second sequence, necessary for the addition of PFRA protein to the distal tip, was also identified. In the absence of this sequence, the mutant PFRA proteins were localized both in the cytosol and in the flagellum where they could still be added along the length of the PFR. This seven-amino-acid sequence is conserved in all PFRA and PFRC proteins and shows homology to a sequence in the flagellar dynein heavy chain of Chlamydomonas reinhardtii.
Flagella and cilia are found in diverse eukaryotic cells, ranging from protists to mammals, and the axoneme is remarkably conserved throughout evolution. This cylindrical structure is composed of nine peripheral microtubule doublets surrounding a central pair of single microtubules. Simple flagella contain only an axoneme, whereas others exhibit additional extraaxonemal structures (1, 15). These structures vary from outer fibers, whose symmetry reflects the axonemal ninefold symmetry, to components such as the paraflagellar rod (PFR) of kinetoplastid protozoans, which is as large as the axoneme and runs along it in a particular plane (2).
Axonemal assembly has been examined in the green alga Chlamydomonas reinhardtii by mutational analysis and by exploiting the flagellum regeneration characteristics of this organism (20, 26). To assemble a flagellum, particular conditions must be fulfilled: a large number of protein precursors is required concurrently (at least 250 different peptides for the axoneme [33]); these peptides must be imported, as there are no ribosomes in the flagellum; and they must be assembled correctly. In Chlamydomonas, deflagellation induces a rapid increase of the transcription of axonemal genes and of the half-life of their mRNAs (20). The proteins are then transported to the distal tip of the flagellum, where assembly occurs (19, 34, 39, 56). This transport step has been extensively characterized over recent years. The bidirectional intraflagellar transport of granules underneath the flagellar membrane was first demonstrated by Kozminski and coworkers (23). These granules can be seen as electron-dense units present between the B tubule of the outer doublet and the flagellar membrane (24, 36). Complexes of 13 to 15 proteins present in such structures have been identified (9, 32, 33), and partial amino acid sequencing of two of them revealed homologies with proteins in nematode and in mouse (9). These complexes are moved towards the tip of the flagellum by the action of at least one motor protein, the kinesin-like protein FLA10 (24, 34), and are brought back to the base of the flagellum by at least one other motor protein, the dynein DHC1b (32, 32a). FLA-10 is found in low concentrations along the length of the flagellum but is localized mostly around the basal body area (9). Homologues of FLA10 are expressed in other organisms, and alteration of their function leads to defects in assembly of cilia in sea urchin (29), assembly of sensory neurons in nematodes (45, 51), and assembly of mouse embryonic cilia (31), suggesting that a conserved mechanism could exist for construction of these structures.
The flagellum of Trypanosoma brucei represents another interesting system that has the particular advantage of forming a new flagellum whilst retaining the old one. The trypanosome cell is actively motile and possesses a single flagellum, with its basal body close to the posterior end. It is attached to the cell body, throughout most of its length, via the flagellum attachment zone (FAZ). This complex network of filaments underlies the plasma membrane and the flagellum membrane that are adpressed at this point (21, 47). The PFR lies parallel to the conventional axoneme and exhibits approximately the same diameter (47, 53). It is present along the whole length of the flagellum with the exception of the flagellar pocket, the area where the flagellum emerges from the cell body. Electron microscopic analysis showed that the lattice-like structure of the PFR is composed of filaments crossing each other at defined angles (11, 13, 41). Three domains are recognized and are defined by their position relative to the axoneme as proximal, intermediate, and distal (2). The proximal domain is connected via some V- or Y-shaped fibers to doublets 4 to 7 of the axoneme and to the FAZ via another set of filaments (13, 16, 17, 47). In Leishmania mexicana (42) and in T. brucei (4), ablation of a major PFR protein leads to the disappearance of the distal and intermediate domains of the PFR with only a rudimentary structure remaining, resembling the proximal domain. As a consequence, the mutant cells exhibited a dramatic reduction in their velocity, showing that the PFR plays a major role in flagellar and cellular motility (4a).
Trypanosomes represent an excellent model in which to study the targeting of flagellar proteins and their assembly into axonemal and nonaxonemal structures such as the PFR. The two major protein constituents, termed PFRA and PFRC, as well as their corresponding genes, have been identified (10, 43). PFRA and PFRC are abundant, and their localization is restricted to the flagellum. During progression through the cell cycle, trypanosomes grow a new flagellum, always originating at the posterior end of the cell, but the old flagellum is not disassembled and remains present in a more anterior position (47). Therefore, this system provides us the opportunity to compare growing and nongrowing flagella in the same cell.
To study assembly of the PFR, we have introduced the sequence of the Ty-1 epitope tag within the PFRA gene (3) and cloned the tagged gene in an inducible expression vector (7, 55). This construct was transformed (12, 25, 52) into trypanosomes expressing the tetracycline repressor such that expression of the tagged PFRA gene was tightly dependent on the presence of tetracycline. Expression of the tagged PFRA protein whilst a trypanosome was growing its new flagellum provided the opportunity to study PFR protein targeting and assembly sites. We show that new PFR subunits are added at two different sites: a major site at the distal tip of the flagellum and a minor site along the length of the structure. Moreover, we demonstrate that, in such experiments, a small incorporation of PFRA protein takes place in the old flagellum, a structure that was assembled before the tagged protein was expressed and available, suggesting the existence of a system for turnover of flagellar components. Truncated epitope-tagged PFRA proteins were expressed, and their localization fell into three groups. A first series of truncations behaved as the full-length protein. A second group localized to the cytosol and the flagellum but were added only along the length of the PFR. The last group of truncated proteins was exclusively cytosolic and was not detected in the flagellum. Analysis of the deleted sequences in PFRA identified regions that are necessary for flagellum localization.
MATERIALS AND METHODS
Trypanosomes and transfection.
Procyclic T. brucei brucei 427 was used throughout this study. Trypanosomes were grown at 27°C in semidefined medium 79 containing 10% fetal calf serum. For transfection, T. brucei was grown to a density of 4 × 106 to 8 × 106 cells per ml, and 3 × 107 ice-cold cells were electroporated with 20 μg of linearized DNA (6) and were put back in 10 ml of warm medium. The next day, appropriate amounts of antibiotics (25 μg of hygromycin and 2.5 μg of phleomycin per ml) were added, and the cells were dispensed in 24-well plates and were incubated at 27°C. After 7 to 21 days, all the transformants were screened by immunofluorescence with anti-PFR or anti-epitope tag antibodies, and the interesting ones were subcloned by limiting dilution. A tetracycline-repressor-expressing cell line (PTH) was obtained after transformation of T. brucei brucei 427 with the pHD360 vector as described (55). For control of expression, plasmids were transformed in PTH trypanosomes and were inserted by homologous recombination into a transcriptionally silent site in the inverted rDNA spacer (55). For time course induction, cells were grown in normal medium containing 1 μg of tetracycline per ml, and, for long incubations, induced and noninduced populations were diluted to and maintained at a density of 1 × 106 to 8 × 106 cells per ml.
Plasmids.
Plasmids pHD360 and pHD430 have been described (55). The full-length, epitope-tagged PFRA gene was extracted from the pPFRATAG-SK (3) after digestion with HindIII and BglII and were ligated in matching sites HindIII and BamHI of pHD430. Truncations were generated by PCR using the common forward primer 5′-AAGCTTATGAGTGGAAAGGAA-3′ (the HindIII site is underlined and the start codon is in bold) and the following specific reverse primers: 5′-GAGATCTCTAGCGATCCATGTTGCC-3′ (full length, bp 1 to 1800 encoding amino acids (aa) 1 to 600), GAGATCTCTATTCGTTCGCCTTC (truncation T1, bp 1 to 651 encoding aa 1 to 217), GAGATCTCTAATGGCGTGACTT (truncation T2, bp 1 to 1275 encoding aa 1 to 425), GAGATCTCTACATCTCCAACTC (truncation T3, bp 1 to 1539 encoding aa 1 to 513), AGATCTCTAGTGTGCACGGTACTCCAC (truncation T10, bp 1 to 1584 encoding aa 1 to 528), GAGATCTCTAGCGATCCATGTTGCC (truncation T5, bp 1 to 1662 encoding aa 1 to 554), AGATCTCTAAGTAGGTCCAAACATCTCC (truncation T11, bp 1 to 1689 encoding aa 1 to 563), GAGATCTCTACACCTCCTCCTGCTTC (truncation T9, bp 1 to 1710 encoding aa 1 to 570), or GAGATCTCTACAGGAGCATCTTAGATCG (truncation T6, bp 1 to 1758 encoding aa 1 to 586) (BglII sites are underlined, and in-frame stop codons are in boldface), using the pPFRATAG-SK as template. The PCR products were cloned in a pBluescript T vector, were cut out with HindIII and BglII, and were inserted into the matching sites of pHD430. For GFP-PFRA fusion constructs, the green fluorescent protein (GFP) gene was amplified by PCR by using the forward primer 5′-GGACTAGTATGAGTAAAGGAGAAG-3′ (the SpeI site is underlined, and the start codon is in boldface) and the reverse primer 5′-GGAAGCTTTTTGTATAGTTCATCC-3′ (the HindIII site is underlined and the GFP 3′ end without its stop codon is italicized) to delete the stop codon and cloned in a pBluescript T vector. To clone the fusion genes, we generated the p430L, a modified version of pHD430 in which the luciferase gene was removed after digestion with HindIII and BamHI to introduce a new linker composed of an XbaI site, a HindIII site, and a BamHI site. First, the GFP gene (without a stop codon) was digested with SpeI and HindIII and was ligated in the compatible sites XbaI and HindIII to generate the plasmid pGFPNS430. Then, the full-length, epitope-tagged PFRA gene or the truncated T1-PFRA gene was removed from its respective vector after digestion with HindIII and BglII and was inserted in the matching sites of pGFPNS430 to create pGFPPFRATAG430 and pGFPT1TAG430, respectively. The linking HindIII site adds the amino acids Lys and Leu. The putative flagellar localization sequence (bp 1540 to 1710, encoding aa 514 to 570) was amplified by PCR by using the 5′-TTCGAAGAAGCTTCCTACTGAGGATGCGCTGAAC-3′ sequence as forward primer (HindIII site underlined) and 5′-GAGATCTCTACACCTCCTCCTGCTTC-3′ as reverse primer (BglII site underlined and in-frame stop codon in boldface) and was cloned into the matching sites of pGFPNS430L, creating the same Lys-Leu linker. For transfections, DNA was purified from cesium chloride gradients or Qiagen resins.
Flagellum extraction.
Trypanosomes were harvested by centrifugation and were washed once in PEM {100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.9, 2 mM EGTA, and 1 mM MgSO4}. Cells were resuspended in PEM containing 1% Nonidet P-40 to extract the cytoskeleton. After 2 min, the mixture was spun down, and the pellet, containing the cytoskeleton, was resuspended in 1 M NaCl. The solution was spread on poly-l-lysine-coated slides and was allowed to settle for 15 to 30 min in a humid atmosphere. Samples were fixed in −20°C methanol for at least 15 min and were processed for immunofluorescence. Regular microscope examination throughout the process confirmed proper extraction.
Antibodies, immunofluorescence, and immunoblotting.
Two different anti-PFR monoclonal antibodies (22), L13D6 (recognizing PFRA and PFRC) and L8C4 (recognizing exclusively PFRA), and the monoclonal antibody BB2 against the Ty epitope tag (3) were used as hybridoma supernatants. For immunofluorescence, trypanosomes were spread on poly-l-lysine-coated slides, were fixed in methanol, and were processed as described (46). Slides were examined with a Zeiss Axioskop or a Leica DMRXA microscope. Images were captured with a cooled, slow-scan charge-coupled device camera and were processed by using Adobe Photoshop. For immunoblotting, 107 trypanosomes were washed twice in phosphate-buffered saline, were resuspended in Laemmli buffer in the presence of protease inhibitors, and were boiled before loading. Proteins (equivalent to 106 cells or 10 μg of protein per lane) were transferred to nitrocellulose membranes and were stained with India ink before processing. The following dilutions of primary antibodies were used: L13D6, 1:20; L8C4, 1:20; and BB2, 1:50. Final detection was carried out by using an ECL kit according to manufacturer’s instructions (Amersham).
RESULTS
Rapid and efficient induction of epitope-tagged PFRA protein.
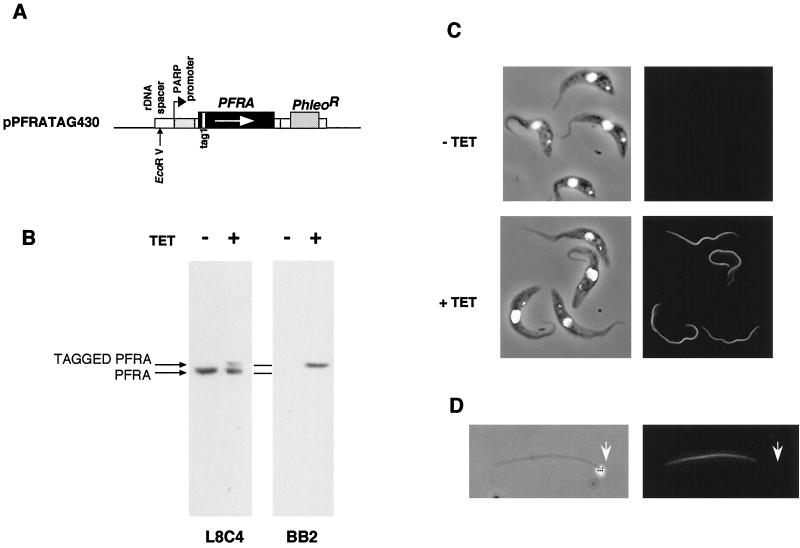
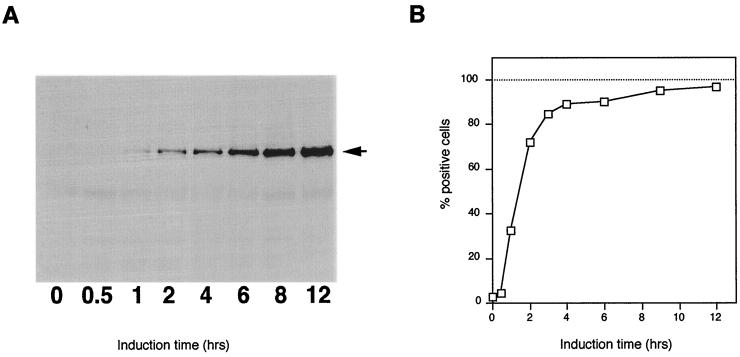
An epitope-tagged full-length copy of the PFRA gene (3) was cloned in the inducible expression vector pHD430 (kindly donated by C. Clayton, Heidelberg, Germany) to generate the plasmid pPFRATAG430 (Fig. 1A) and was transformed into trypanosomes expressing the tetracycline repressor. The resulting transformants were cloned by limiting dilution, and the cell line named PFRAtag was selected for further experiments. When these cells were grown in the absence of tetracycline, the epitope-tagged PFRA protein could not be detected by immunoblotting nor by immunofluorescence with the antitag antibody BB2 (Fig. 1B and C). However, after incubation with tetracycline for a week (more than 20 doubling times), a protein corresponding to the expected mass of the epitope-tagged PFRA was detected by immunoblotting with both the antitag BB2 and the anti-PFRA antibody L8C4 (Fig. 1B). As expected, the tagged protein was slightly larger than the endogenous one. Immunofluorescence analysis showed a bright signal restricted to the PFR within the flagella of all induced cells (Fig. 1C). Double immunofluorescence with antitag and anti-PFR antibodies revealed perfect colocalization of both proteins (data not shown). Flagella from such trypanosomes were isolated by treatment of detergent-extracted cytoskeletons with 1 M NaCl (44). Immunofluorescence revealed the presence of the epitope-tagged PFRA protein (Fig. 1D). The proximal end of the flagellum can be easily identified by the presence of the kinetoplast, the mitochondrial genome of the trypanosome cell, that remains connected to the basal body in these preparations (37). The short, unstained region at the proximal end corresponds to the region normally present inside the flagellar pocket where PFR is absent (2). The doubling times of induced and noninduced PFRAtag trypanosomes were identical to that of wild-type untransformed trypanosomes, indicating no deleterious effects of the tagged PFRA protein. Finally, expression of the epitope-tagged PFRA protein in the snl-1 trypanosome mutant lacking endogenous PFRA was able to complement the paralysis phenotype and to reconstitute the PFR structure, demonstrating that the tagged PFRA protein was functionally equivalent to the wild type (4b). We then analyzed the kinetics of expression of the tagged PFRA protein and its location (Fig. 2). At the population level on a Western blot (Fig. 2A), the tagged PFRA band was detected as early as 1 h after the addition of tetracycline, and its concentration increased rapidly to reach a plateau after 10 to 12 h. During the same experiment, samples were fixed and processed for immunofluorescence by using the antitag antibody. Cells exhibiting a positive signal were scored and plotted versus the period of induction (Fig. 2B). After 1 h, 35% of the trypanosomes were already positive, a value that rose to 70% after 2 h before quickly approaching 100%. These kinetic parameters are perfectly suited for studies of PFR growth, which normally takes about 4.5 h for completion within an 8.5-h cell cycle (47).
FIG. 1.
Expression of full-length, epitope-tagged PFRA in PFRAtag trypanosomes. (A) Construct used for inducible expression of full-length, epitope-tagged PFRA. Large boxes represent coding sequences, small boxes represent promoters, 5′ or 3′ untranslated regions, and targeting sequences. Thin lines represent the pGEM bacterial sequence. The EcoRV restriction site used for linearization and insertion into the trypanosome genome (12, 25, 52) is indicated by a vertical arrow. (B) Immunoblot of PFRAtag trypanosome total protein samples grown with or without tetracycline. Membranes were probed with either the anti-PFRA L8C4 monoclonal antibody (left) or with the antitag BB2 monoclonal antibody (right). (C) Immunofluorescence of PFRAtag trypanosomes grown with or without tetracycline. Left, DAPI images (white), merged to phase contrast images; right, immunofluorescence signal. (D) Detergent and salt-extracted flagellum from PFRAtag trypanosomes grown with tetracycline and stained with the BB2 monoclonal antibody. Left, DAPI image (white), merged to phase contrast image, showing the presence of the kinetoplast (arrowhead) that remains tightly connected to the basal body (37); right, immunofluorescence signal.
FIG. 2.
Induction kinetics of expression of tagged PFRA. PFRAtag trypanosomes were grown for the indicated periods in the presence of tetracycline. (A) Immunoblotting analysis with the antitag BB2 monoclonal antibody, showing the presence of the epitope-tagged PFRA protein at the expected molecular weight as early as 1 h after addition of the inducer. (B) Immunofluorescence analysis with the antitag BB2. Positive cells were scored and plotted versus time of induction (at least 1,000 cells for each time point).
New PFRA subunits are added at two different sites in the new flagellum.
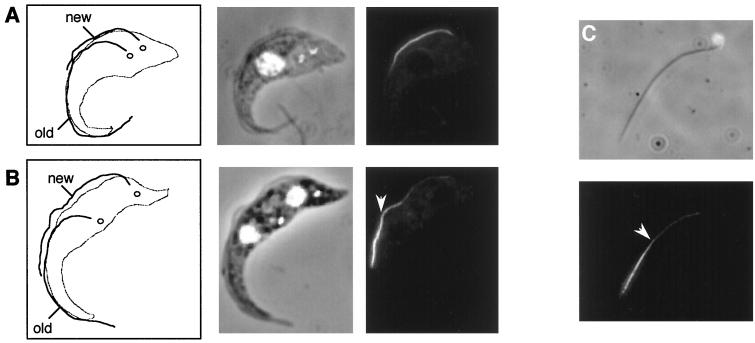
To determine the location of the PFR assembly site, PFRAtag trypanosomes were induced for 2 h with tetracycline and were analyzed by immunofluorescence with the BB2 antitag monoclonal antibody. When tagged PFRA expression was induced in exponentially growing cultures, trypanosomes were found at every stage of the cell cycle. This offers the opportunity to study construction of the PFR within the new flagellum, which always originates at the posterior end of the trypanosome (47). Trypanosomes that are early in the cell cycle and have a short flagellum, which will have formed entirely in the presence of tagged PFRA protein, show homogeneous staining of this new PFR (Fig. 3A). However, in cells where the new flagellum had elongated to more than half of the old flagellum length (Fig. 3B), the distal end was brightly labelled, whereas the proximal part only showed a weak signal. A discrete breakpoint between the two regions could be identified. This bipartite staining pattern was extremely reproducible and was observed in all such cells from at least five separate experiments. The bipartite signal remained after isolation of flagella by treatment of detergent-extracted cytoskeletons with high concentrations of salt (Fig. 3C), demonstrating that the epitope-tagged protein was stably incorporated in the PFR. This indicates that the minor proximal label was not merely PFR proteins in the process of being transported to the distal tip. Our interpretation of these results is that there are two addition sites for PFRA during PFR construction: a major one at the distal tip and a minor one along the whole length of the PFR structure, adding subunits to the partially completed structure.
FIG. 3.
The pattern of PFR assembly. PFRAtag trypanosomes were grown for 2 h in the presence of tetracycline and were analyzed by immunofluorescence with the antitag BB2 monoclonal antibody. The left panels of A and B show drawings of the trypanosome cell indicating the new and old flagella and the separated kinetoplasts (47). Note that the PFR is only present inside the flagellum from the point where the axoneme exits the cell body (2). The central panels show the DAPI images (white) merged to phase contrast images, and the right panels show the immunofluorescence signal. (A) Trypanosome with a short new flagellum whose assembly was initiated during the period of induction of tagged PFRA protein. This exhibits homogeneous PFR staining along the length of the PFR. (B) Trypanosome whose growth of the new flagellum was initiated before the induction show bipartite staining. The distal end is brightly labelled, but the proximal end is not. (C) Detergent and salt-extracted flagellum from a PFRAtag trypanosome induced for 2 h with tetracycline. The proximal end of the flagellum can be identified by the attached mitochondrial DNA (37). The bipartite staining is still present, with more intense labelling at the distal end. The arrowhead indicates the break point between the bright distal and the less intense proximal staining.
PFR growth appears to be linear.
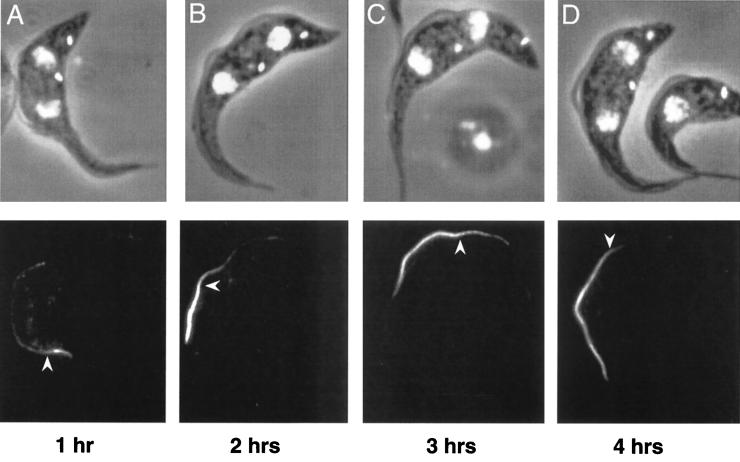
To assess the rate of construction of the PFR, PFRAtag trypanosomes were induced to express the epitope-tagged PFRA protein for 1, 2, 3 or 4 h and were analyzed by immunofluorescence (Fig. 4A to D). In the new flagella of biflagellated cells, the antitag antibody produced bright staining of the distal tip and a weak signal along the length of the PFR, as observed before. However, the length of the brightly labelled segment increased with the induction time (Fig. 4). When cells were induced for 5 h or longer, the PFR in the new flagellum was brightly stained with the antitag antibody throughout its length and did not show the bipartite signal. To estimate the growth rate of the PFR, we measured the length of the bright segment of flagella exhibiting bipartite staining in PFRAtag trypanosomes induced for 1 (3.4 ± 0.8 μm), 2 (7.2 ± 0.9 μm) or 3 h (10.6 ± 1.5 μm) (at least 100 cells for each time point). Determination of the breakpoint was not easy for cells induced for 4 h, and these data were, therefore, not included. These results suggested a linear growth rate for at least the first 3 h, calculated at 3.6 μm per h.
FIG. 4.
Pattern and timing of PFR assembly. PFRAtag trypanosomes were grown for 1 (A), 2 (B), 3 (C) or 4 h (D) in the presence of tetracycline and were analyzed by immunofluorescence with the antitag BB2 monoclonal antibody. The top panels show the DAPI images (white) merged to the phase contrast images, and the bottom panels show the immunofluorescence signal. The arrowhead indicates the break point between the bright distal and the less intense proximal staining.
PFRA protein is incorporated in the old flagellum.
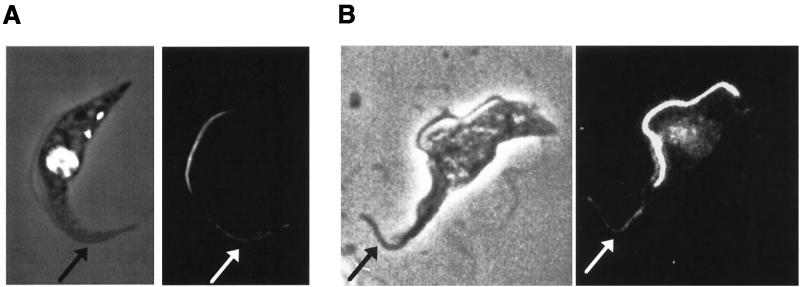
When PFRAtag trypanosomes were induced to express the tagged PFRA for 1 to 4 h, many dividing cells exhibited weak signals with the antitag in the old flagellum in addition to the bright staining of the new flagellum (Fig. 5A). This was observed in about 60% of dividing cells and was resistant to detergent extraction (data not shown). This was somewhat surprising, since the PFR present in this flagellum was assembled in the previous cell cycle, before the tagged PFRA protein was available. To assess whether or not this incorporation of the tagged PFRA protein was due to slight leakiness of the tetracycline repressor system, the plasmid pKMPFRATAG (3), expressing the tagged PFRA protein, was transiently transfected in wild-type trypanosomes. Samples were fixed 2 h after electroporation, and immunofluorescence analysis with the antitag antibody showed that cells growing a new flagellum exhibited the weak signal along the length of their old flagellum, in addition to the bright staining in the new flagellum (Fig. 5B). Thus, tagged PFRA protein could be incorporated into the PFR of the old flagellum, even though this was constructed prior to transient transfection and, hence, in the absence of the tagged PFRA protein.
FIG. 5.
PFRA incorporation into the old flagellum of PFRAtag trypanosomes. (A) PFRAtag trypanosomes were grown for 2 h in the presence of tetracycline and were analyzed by immunofluorescence with the antitag BB2 monoclonal antibody. The left panel shows the DAPI image (white) merged to the phase contrast image, and the right panel shows the immunofluorescence signal. The tagged PFRA protein is present mostly in the new flagellum as expected but also in the old flagellum (indicated by the arrow). (B) Wild-type trypanosomes were transiently transfected with the plasmid pKMPFRATAG (3) to express the tagged PFRA protein and were then fixed 2 h after electroporation. Positive cells show the presence of the tagged PFRA protein in the new flagellum but also in the old flagellum (arrow). Transient transfection often leads to considerable overexpression, explaining the difference in signal intensity between the two experiments.
The carboxy-terminal end of the PFRA protein is necessary for flagellar localization.
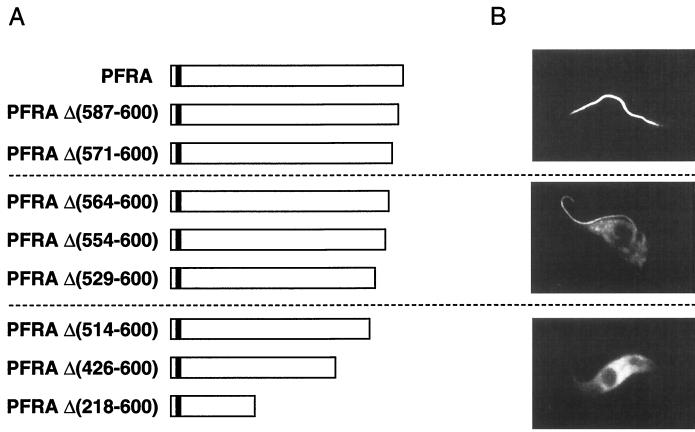
Various truncated, epitope-tagged, PFRA genes were generated and cloned in trypanosome expression vectors for transfection into wild-type trypanosomes. Therefore, the truncated proteins were expressed in presence of the wild-type, endogenous, PFRA. Immunolocalization was carried out by fluorescence microscopy, and the truncated proteins were classified in three groups according to their localization (Fig. 6). In the first series [PFRAΔ(587-600) and -Δ(571-600)], truncated PFRA proteins behaved exactly the same as the full-length protein and exhibited the classic flagellar localization (Fig. 6). In a second group [PFRAΔ(564-600), -Δ(554-600), and -Δ(529-600)], the recombinant proteins showed dual localization in both the flagellum and the cytosol (Fig. 6). Finally, the last group of truncated proteins [PFRAΔ(514-600), -Δ(426-600), and -Δ(218-600)] were localized only in the cytosol and were excluded from the flagellum. When cytosolic accumulation occurred, the truncated proteins were excluded from the nucleus. In all cases, these transformants behaved normally (in terms of motility, cell cycle timings, and growth rate), and no obvious modifications of the PFR structure were detected. These data suggested that the sequence between aa 514 [after PFRAΔ(514-600)] and 570 [before PFRAΔ(571-600)] contains a motif necessary for localization to the flagellum.
FIG. 6.
Immunolocalization of truncated PFRA proteins. A series of epitope-tagged truncated PFRA proteins were expressed in wild-type trypanosomes. (A) Map of the different PFRA truncations. The deleted segment is shown by the numbering of amino acids in the PFRA sequence (43). White boxes represent PFRA sequences, and the small black box indicates the sequence of the epitope tag. (B) Immunofluorescence localization of the epitope-tagged truncated proteins revealed by the antitag BB2 monoclonal antibody. Only one representative image is shown for each group.
PFRA aa 514 to 570 are not sufficient for localization of a reporter protein to the flagellum.
To assess the importance of the domain between aa 514 and 570 for protein localization, we fused its coding DNA sequence to the 3′ end of the GFP gene and cloned the fusion gene into a trypanosome expression vector. As controls, the GFP gene was fused to the full-length, epitope-tagged, PFRA gene or to the truncated PFRA gene encoding PFRAΔ(218-600). The constructs were transfected into wild-type trypanosomes, and the transformants were analyzed by direct fluorescence microscopy on live cells. As expected, GFP::PFRATAG localized to the flagellum and GFP::PFRAΔ(218-600) localized to the cytosol only, excluding the nucleus and the flagellum (data not shown). These localizations are identical to these of tagged full-length PFRA or tagged PFRAΔ(218-600) without GFP (Fig. 6). Both results were confirmed with anti-epitope tag antibodies (data not shown). When GFP was fused to aa 514 to 570 of PFRA, the sequence necessary for flagellum localization, it was found everywhere inside the cell, including the nucleus and the flagellum. Hence, the presence of this sequence alone was not sufficient to confer exclusive flagellum localization.
PFRA aa 563 to 570 are necessary for incorporation at the major assembly site.
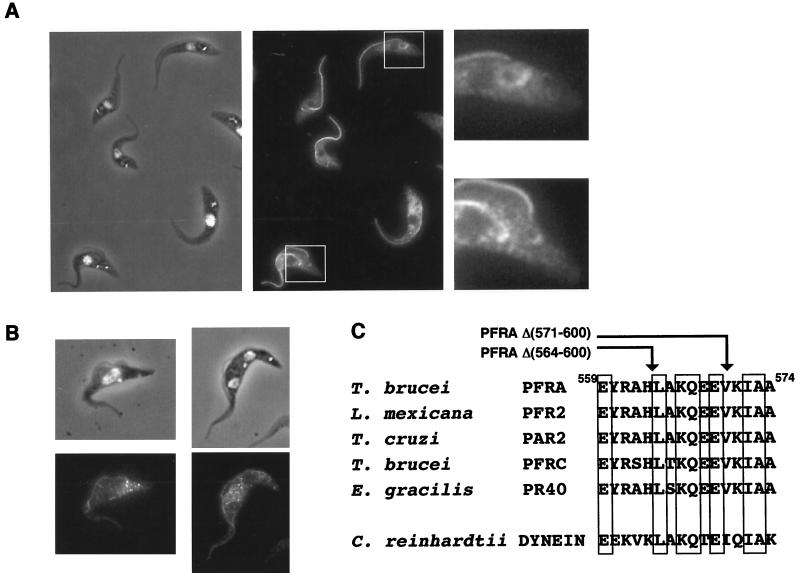
To understand the dual localization (flagellum and cytosol) of some truncated PFRA mutant proteins, we cloned a representative, the truncated PFRA gene encoding PFRAΔ(554-600), into the inducible expression vector pHD430. This was transformed in trypanosomes expressing the tetracycline repressor to confer controllable expression. In cells induced for 24 h, the truncated protein showed dual localization in the flagellum and the cytosol but was more abundant in the basal body and flagellar pocket area of both the new and the old flagella (areas enlarged on Fig. 7A). The assembly of the PFRAΔ(554-600) was followed in time course experiments similar to those described above for the full-length PFRA protein. After 1 to 2 h of incubation with tetracycline, PFRAΔ(554-600) localized along the length of the new flagellum, from its exit from the flagellar pocket to its distal tip. The staining did not show the bipartite pattern that was seen for the full-length protein (Fig. 7B). Therefore, this truncated PFRAΔ(554-600) protein was still incorporated at the minor site of PFR assembly along the length of the flagellum but did not appear to be intensively incorporated at the distal tip. The same result was obtained for truncation PFRAΔ(564-600), whereas truncations PFRAΔ(571-600) and PFRAΔ(587-600) (Fig. 6) were found at the distal tip of the new flagellum (data not shown). The short critical sequence separating PFRAΔ(564-600) from PFRAΔ(571-600) was compared to other proteins using database searches, and an interesting homology to the dynein β heavy chain of the outer arm of the flagellum of Chlamydomonas (27) was detected. This sequence (Fig. 7C) is also conserved in PFRA and PFRC proteins of T. brucei, Trypanosoma cruzi, L. mexicana, and Euglena gracilis.
FIG. 7.
The truncated PFRAΔ(554-600) shows dual localization and is only added along the length of the growing PFR. (A) Trypanosomes were induced to express the truncated and epitope-tagged PFRAΔ(554-600) for 2 days and were processed for immunofluorescence with the anti-tag BB2. The left panel illustrates DAPI staining (white) merged to the phase contrast image, and the central panel shows the immunofluorescence signal. There is a concentration of truncated protein around the basal body and kinetoplast area of both growing and old flagella (areas enlarged on the right panel). (B) Time course induction of PFRAΔ(554-600) expression. Cells were grown for 2 h in the presence of tetracycline. The truncated protein is localized in both the new and the old flagellum but without any polarity. (C) Sequence comparison of the PFRA region required for addition at the distal tip of the flagellum. The arrows indicate the carboxy-terminal end of the truncated PFRA that is not added at the distal tip [PFRAΔ(564-600)] and of the PFRAΔ(571-600) that behaves normally. Indicated numbers correspond to the T. brucei PFRA sequence (43). Conserved residues are boxed. A high degree of conservation is observed in PFRA homologues in T. cruzi (PAR2 [5]), L. mexicana (PFR2 [28]), E. gracilis (PR-40 [30]), and PFRC (10), but homology is also found with the Chlamydomonas heavy chain of axonemal β-dynein (27).
DISCUSSION
When considering the ontogeny and subsequent possible remodelling of the major flagellar components such as the axoneme and the PFR, one can envisage seven critical steps. These would encompass (i) synthesis of flagellar proteins in the cytosol, (ii) transfer to the base of the flagellum, (iii) entry into the flagellum compartment, (iv) anterograde transport to the assembly site(s), (v) assembly in a structure, (vi) retrograde transport for recycling, and (vii) maintenance or turnover. Our study provides evidence for the existence of at least some of these steps in the construction of the T. brucei PFR.
Transfer and import of PFRA proteins into the flagellum.
Given their restricted localization to the flagellum, the PFR proteins represent an excellent model to study flagellum targeting. No ribosomes are present in the flagellum, and all proteins must, therefore, be imported from the cytoplasm (40). Different truncations of the PFRA protein were epitope tagged and expressed in wild-type trypanosomes in the presence of wild-type PFRA proteins. Truncation of the last 30 aa (aa 571 to 600) did not affect the fully flagellar location of the recombinant protein that was also transported to the distal tip. However, when 87 aa (or more) were deleted from the carboxy terminus, the truncated proteins were present exclusively in the cytosol. Therefore, the region from aa 514 to 570 contains a sequence that is necessary for flagellum localization. When this sequence was fused to the carboxy-terminal end of GFP as a reporter, the fusion protein localized to the flagellum but also everywhere else in the cell, including the nucleus. Thus, this sequence is necessary but not sufficient to confer specific flagellum localization. This sequence could act as a flagellum-targeting signal, maybe functioning as part of a more complex system involving different regions of the protein or a particular conformation to endow full flagellar targeting. However, at present, we cannot distinguish other mechanisms from this form of directed targeting of proteins to the flagellum. One such mechanism could involve relatively free access to the flagellum with the assembly process being the driving force for differential localization of only flagellar proteins. If this mechanism was operative, the region from aa 514 to 570 may contain a domain necessary for PFRA to bind to the PFR. One would then suggest that progressive deletions of this domain first reduced the binding capacity of the tip and then reduced the side binding capacity of the truncated PFRA protein to the PFR structure. A consequence would then be a progressive shift of the truncated PFRA proteins to the free cytosolic form. This would appear to be localized solely in the cell body because of the low cytosolic volume of the flagellum.
Truncation of the region from aa 514 to 570 produces mutant PFRA proteins that are localized to both the flagellum and the cytosol, where they show higher concentration around the basal body area. This would suggest that a proportion of these mutant proteins is still transferred to the base of the flagellum. Interestingly, in Chlamydomonas, proteins involved in intraflagellar transport (see below) are also localized around the basal body (9).
Transport of PFRA proteins to their assembly sites.
Time course induction of expression of an epitope-tagged PFRA protein allowed us to visualize recently synthesized subunits during the assembly of the PFR in vivo. Two different sites of addition were identified: a major polar site at the distal end of the PFR and a minor, nonpolar site present along the length of the PFR. A group of PFRA mutant proteins were only added along the length of the flagellum and not specifically at the distal tip. Since these mutant proteins were expressed in the presence of the wild-type PFRA, they provide insight into how these proteins are incorporated into the PFR.
Distal addition of subunits of the trypanosome axoneme has been illustrated by the use of a monoclonal antibody recognizing tyrosinated α-tubulin, a marker of newly assembled microtubules. This revealed a discrete gradient along the growing new flagellum, with the highest concentration at the distal tip (48). Incorporation of newly labelled proteins occurs mostly at the distal tip of the growing flagellum (39, 56), and similar patterns have been directly demonstrated for several individual components of the axoneme in Chlamydomonas, including α-tubulin, the radial spoke protein 3 (19), and the inner dynein arm protein p28 (34). These data imply that newly synthesized flagellum proteins have to be transported to this distal site before incorporation into their respective structures. This process has been directly visualized in Chlamydomonas flagella, where bidirectional intraflagellar transport of granules (also called rafts) can be observed by light microscopy (23, 40).
The axonemal outer dynein arm IC69 protein does not seem to be transported by this mechanism. In dikaryon rescue experiments where wild-type Chlamydomonas cells were mated with mutants lacking IC69, the rescue of the outer dynein arm protein occurred throughout the length of the axoneme (34) and did not require the activity of FLA10. This is the opposite of what was observed for the inner dynein arm component IDA4 mutant, which was rescued via the distal tip, but only in presence of an active FLA10. Piperno and coworkers (34) suggested that different modes of transfer may be used according to protein localization within the axonemal shaft.
The addition sites of the new PFRA subunits in the growing PFR and the modified locations of the truncated PFRA mutant proteins in T. brucei reveal intriguing parallels with the assembly model of the axoneme in Chlamydomonas described above. This raises questions of how the PFR precursors are transported to their respective addition sites and of how the two phenomena are related in space (in the different domains of the PFR) and in time (in the order of addition site used). Structures morphologically resembling the rafts are present in the flagellum of T. brucei (for an example, see Fig. 7 in reference 47), suggesting that its axoneme might be assembled by a similar process. However, it is not clear whether PFR proteins use the same or a separate but related transport system.
Preferential incorporation of some truncated PFRA proteins [PFRAΔ(564-600), -Δ(554-600), and -Δ(529-600)] at the distal tip of the growing flagellum was not observed, but addition along the length of the PFR still occurred. The seven-amino-acids sequence missing from PFRAΔ(564-600), compared with PFRAΔ(571-600) which behaves normally, are highly conserved in both PFRA and PFRC proteins in T. brucei, T. cruzi, L. mexicana, and E. gracilis. The short sequence was compared to other proteins using database searches, and an interesting homology to the dynein β heavy chain of the outer arm of the flagellum of Chlamydomonas (27) was detected. It is not known whether this sequence is required for flagellar localization of dynein. This sequence did not share any identity with the flagellar-membrane-targeting sequence of the glucose transporter identified by Snapp and Landfear (49) in Leishmania parasites. In that case, two different isoforms of the glucose transporter differed only in their localization (flagellum and plasma membrane, respectively) and by their amino-terminal sequence. Truncations of this sequence in the flagellar isoform identified a short region necessary and sufficient for flagellum targeting.
Assembly of PFRA proteins.
Specific proteins are likely to be required to orchestrate proper incorporation of precursors at the major assembly sites of both the PFR and the axoneme. In Chlamydomonas, HSP70 localizes at the distal end of the flagellum (8). In T. brucei, a monoclonal antibody stains the distal end of the axoneme in both old and growing flagella, suggesting material that may be involved in capping of the axoneme (57). There is, however, no such identification of polar structures or proteins in the PFR.
Turnover of PFRA proteins in the old flagellum.
The epitope-tagged PFRA was incorporated into the old flagellum of biflagellated trypanosomes, even though this was assembled at least one cell cycle prior to the tagged protein being available. This suggests that the PFR structure can be remodelled. Electron microscopy of both negatively stained and thinly sectioned old and new flagella show that the PFR is completely constructed by the end of the cell cycle. Hence, we can exclude the possibility that this incorporation is a late stage of construction. Rosenbaum and Child (38) have observed that nongrowing, nonregenerating flagella of different protists were able to incorporate as much as 50% of the total radiolabelled protein of growing flagella. Turnover of tektin has been identified in the cilia of sea urchin during embryonic development (50). The filaments that constitute the PFR are morphologically related to intermediate filaments, although no sequence similarity has ever been detected (43). Interestingly, intermediate filaments such as vimentin are themselves subject to subsequent remodelling (54).
The PFR growth rate appears linear.
We have estimated the growth rate of the PFR, and it appears to be linear at ∼3.6 μm per h. This contrasts with the flagellum regeneration in Chlamydomonas that shows a fast initiation (12 μm per h) before a reduction in speed, presumably because of the longer distance required to transfer precursor proteins to the assembly site (reviewed in reference 26). Our system did not allow us to measure the growth rate for the last hour of the PFR elongation, but measurement of the size of the PFR in postmitotic cells (16.8 ± 0.99 μm, n = 46) compared to the expected size from a constant linear growth rate did not hint at any reduction in growth rate (data not shown). The PFR growth rate is relatively low compared to that of the flagellum of Chlamydomonas (26). This may be because of the requirement to assemble three physically connected structures: the axoneme, the PFR, and the FAZ filament system (22, 47).
The tetracycline-inducible expression is a system of choice to study the assembly of organelles.
Our results showed that the tetracycline-inducible gene expression system, combined with epitope tagging, allowed rapid expression of proteins that can be detected at both the population and individual cell levels. Cell behavior and growth of the induced and noninduced PFRAtag trypanosomes were identical, and the two populations could only be discriminated by the presence of the tagged PFRA protein. This system facilitates the study of localization of mutant proteins and will certainly be a valuable tool for the examination of organelle assembly in trypanosomes and other organisms.
ACKNOWLEDGMENTS
This work was supported by a Programme and Equipment Grant from the Wellcome Trust. T.H.M. was funded by the Natural Sciences and Engineering Research Council of Canada and the Underwood Fund of the Biotechnology and Biological Sciences Research Council, United Kingdom. K.R.M. is a Dunkerly Fellow. We thank Christine Clayton (ZMBH, Heidelberg, Germany) for providing the expression vectors pHD360 and pHD430, and Iain Hagan (University of Manchester) for providing the GFP gene.
We also thank Linda Kohl (University of Manchester) and unknown referees for critical and constructive comments on the work and the manuscript.
REFERENCES
- 1.Baccetti B. Evolutionary trends in sperm structure. Comp Biochem Physiol. 1986;85:29–36. doi: 10.1016/0300-9629(86)90457-3. [DOI] [PubMed] [Google Scholar]
- 2.Bastin P, Matthews K R, Gull K. The paraflagellar rod of Kinetoplastida: solved and unsolved questions. Parasitol Today. 1996;12:302–307. doi: 10.1016/0169-4758(96)10031-4. [DOI] [PubMed] [Google Scholar]
- 3.Bastin P, Bagherzadeh A, Matthews K R, Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- 4.Bastin P, Sherwin T, Gull K. Paraflagellar rod is vital for trypanosome motility. Nature. 1998;391:548. doi: 10.1038/35300. [DOI] [PubMed] [Google Scholar]
- 4a.Bastin P, Gull K. Assembly and function of complex flagellar structures illustrated by the paraflagellar rod of trypanosomes. Protist. 1999;150:113–123. doi: 10.1016/S1434-4610(99)70015-5. [DOI] [PubMed] [Google Scholar]
- 4b.Bastin, P., T. Pullen, T. Sherwin, and K. Gull. Protein transport and flagellum assembly dynamics revealed by analysis of the paralysed trypanosome mutant snl-1. J. Cell Sci., in press. [DOI] [PubMed]
- 5.Beard C A, Saborlo J L, Tewari D, Krieglstein K G, Henschen A H, Mannings J E. Evidence for two distinct major protein components, PAR 1 and PAR 2, in the paraflagellar rod of Trypanosoma cruzi. J Biol Chem. 1992;267:21656–21662. [PubMed] [Google Scholar]
- 6.Beverley S M, Clayton C E. Transfection of Leishmania and Trypanosoma brucei by electroporation. In: Hyde E, editor. Methods in molecular biology of parasites. Totowa, N.J: Humana Press Inc.; 1993. pp. 333–348. [DOI] [PubMed] [Google Scholar]
- 7.Biebinger S, Wirtz L E, Lorentz P, Clayton C E. Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol Biochem Parasitol. 1997;85:99–112. doi: 10.1016/s0166-6851(96)02815-0. [DOI] [PubMed] [Google Scholar]
- 8.Bloch M, Johnson K A. Identification of a molecular chaperone in the eukaryotic flagellum and its localization to the site of microtubule assembly. J Cell Sci. 1995;108:3541–3545. doi: 10.1242/jcs.108.11.3541. [DOI] [PubMed] [Google Scholar]
- 9.Cole D, Diener D R, Himelblau A L, Beech P L, Fuster J C, Rosenbaum J. Chlamydomonas kinesin II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deflorin J, Rudolf M, Seebeck T. The major components of the paraflagellar rod of Trypanosoma brucei are two similar but distinct proteins which are encoded by two different gene loci. J Biol Chem. 1994;269:28745–28751. [PubMed] [Google Scholar]
- 11.De Souza W, Souto-Padron T. The paraxial structure of the flagellum of Trypanosomatidae. J Parasitol. 1980;66:229–235. [PubMed] [Google Scholar]
- 12.Eid J, Sollner-Webb B. Stable integrative transformation of Trypanosoma brucei that occurs exclusively by homologous recombination. Proc Natl Acad Sci USA. 1991;88:2118–2121. doi: 10.1073/pnas.88.6.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farina M, Attias M, Souto-Padron T, De Souza W. Further studies on the organization of the paraxial rod of trypanosomatids. J Protozool. 1986;33:552–557. [Google Scholar]
- 14.Fok A K, Wang H, Katayama A, Aihira M S, Allen R D. 22S axonemal dynein is preassembled and functional prior to being transported and attached on the axonemes. Cell Motil Cytoskelet. 1994;29:215–224. doi: 10.1002/cm.970290304. [DOI] [PubMed] [Google Scholar]
- 15.Fouquet J P, Kann M L. The cytoskeleton of mammalian spermatozoa. Biol Cell. 1994;81:89–93. doi: 10.1016/s0248-4900(94)80001-4. [DOI] [PubMed] [Google Scholar]
- 16.Hemphill A, Lawson D, Seebeck T. The cytoskeletal architecture of Trypanosoma brucei. J Parasitol. 1991;77:603–612. [PubMed] [Google Scholar]
- 17.Hemphill A, Seebeck T, Lawson D. The Trypanosoma brucei cytoskeleton: ultrastructure and localization of microtubule-associated and spectrin-like proteins using quick-freeze deep-etch, immunogold electron microscopy. J Struct Biol. 1991;107:211–220. doi: 10.1016/1047-8477(91)90046-y. [DOI] [PubMed] [Google Scholar]
- 18.Hyams J. The Euglena paraflagellar rod: structure, relationship to other flagellar components and preliminary biochemical characterization. J Cell Sci. 1982;55:199–210. doi: 10.1242/jcs.55.1.199. [DOI] [PubMed] [Google Scholar]
- 19.Johnson K A, Rosenbaum J L. Polarity of flagellar assembly in Chlamydomonas. J Cell Biol. 1992;119:1605–1611. doi: 10.1083/jcb.119.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson K A, Rosenbaum J L. Flagellar regeneration in Chlamydomonas: a model system for studying organelle assembly. Trends Cell Biol. 1993;3:156–161. doi: 10.1016/0962-8924(93)90136-o. [DOI] [PubMed] [Google Scholar]
- 21.Kohl L, Gull K. Molecular architecture of the trypanosome cytoskeleton. Mol Biochem Parasitol. 1998;93:1–9. doi: 10.1016/s0166-6851(98)00014-0. [DOI] [PubMed] [Google Scholar]
- 22.Kohl L, Sherwin T, Gull K. Assembly of the paraflagellar rod and of the flagellum attachment zone complex in Trypanosoma brucei. J Eukaryot Microbiol. 1999;46:105–109. doi: 10.1111/j.1550-7408.1999.tb04592.x. [DOI] [PubMed] [Google Scholar]
- 23.Kozminski K G, Johnson K A, Forscher P, Rosenbaum J L. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozminski K G, Beech P, Rosenbaum J L. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee M G S, Van der Ploeg L H T. Homologous recombination and stable transfection in the parasitic protozoan Trypanosoma brucei. Science. 1990;250:1583–1587. doi: 10.1126/science.2177225. [DOI] [PubMed] [Google Scholar]
- 26.Lefevbre P A, Rosenbaum J L. Regulation of the synthesis and assembly of ciliary and flagellar proteins during regeneration. Ann Rev Cell Biol. 1986;2:517–546. doi: 10.1146/annurev.cb.02.110186.002505. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell D R, Brown K S. Sequence analysis of the Chlamydomonas alpha and beta dynein genes. J Cell Sci. 1994;107:635–644. doi: 10.1242/jcs.107.3.635. [DOI] [PubMed] [Google Scholar]
- 28.Moore L, Santrich C, LeBowitz J H. Stage-specific expression of the Leishmania mexicana paraflagellar rod protein PFR-2. Mol Biochem Parasitol. 1996;80:125–135. doi: 10.1016/0166-6851(96)02688-6. [DOI] [PubMed] [Google Scholar]
- 29.Morris R L, Scholey J M. Heterotrimeric kinesin II is required for the assembly of motile 9+2 ciliary axonemes on sea urchin embryo. J Cell Biol. 1997;138:1009–1022. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ngo H M, Bouck G B. Heterogeneity and a coiled-coil prediction of trypanosomatid-like flagellar rod proteins in Euglena. J Eukaryot Microbiol. 1998;45:323–333. doi: 10.1111/j.1550-7408.1998.tb04543.x. [DOI] [PubMed] [Google Scholar]
- 31.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of a nodal cilia regenerating leftward flow of extraembryonic fluid in mice lacking KIF-3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 32.Pazour G J, Wilkerson C G, Witman G B. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport. J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Pazour G J, Dickert B L, Witman G B. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piperno G, Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagellum. Proc Natl Acad Sci USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piperno G, Mead K, Henderson S. Inner dynein arms but not outer dynein arms require the activity of the kinesin homologue protein KHP1Fla10 to reach the distal part of the flagella in Chlamydomonas. J Cell Biol. 1996;133:371–379. doi: 10.1083/jcb.133.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piperno G, Huang B, Luck D. Two dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1977;74:1600–1604. doi: 10.1073/pnas.74.4.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ringo D L. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J Cell Biol. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson D R, Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 1991;352:731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- 38.Rosenbaum J L, Child F M. Flagellar regeneration in protozoan flagellates. J Cell Biol. 1967;34:345–364. doi: 10.1083/jcb.34.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenbaum J L, Moulder J E, Ringo D L. Flagellar elongation and shortening in Chlamydomonas. J Cell Biol. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenbaum J L, Cole D G, Diener D R. Intraflagellar transport: the eyes have it. J Cell Biol. 1999;144:385–388. doi: 10.1083/jcb.144.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell D G, Gull K. Structural and biochemical characterization of the paraflagellar rod structure of Crithidia fasciculata. Eur J Cell Biol. 1983;30:137–143. [PubMed] [Google Scholar]
- 42.Santrich C, Moore L, Bastin P, Sherwin T, Brokaw C, Gull K, Lebowitz J H. A motility function for the paraflagellar rod in Leishmania parasites revealed by PFR-2 gene knockouts. Mol Biochem Parasitol. 1997;90:95–109. doi: 10.1016/s0166-6851(97)00149-7. [DOI] [PubMed] [Google Scholar]
- 43.Schlaeppi K, Deflorin J, Seebeck T. The major component of the paraflagellar rod of Trypanosoma brucei is a helical protein that is encoded by two identical, tandemly linked genes. J Cell Biol. 1989;109:1695–1709. doi: 10.1083/jcb.109.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider A, Sherwin T, Sasse R, Russell D G, Gull K, Seebeck T. Subpellicular and flagellar microtubules of Trypanosoma brucei contain the same α-tubulin isoforms. J Cell Biol. 1987;104:431–438. doi: 10.1083/jcb.104.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shakir M A, Fukushige T, Yasuda H, Miwa J, Siddiqui S S. C. elegans osm-3 gene mediating osmotic avoidance behaviour encodes a kinesin-like protein. Neuroreport. 1993;4:891–894. doi: 10.1097/00001756-199307000-00013. [DOI] [PubMed] [Google Scholar]
- 46.Sherwin T, Schneider A, Sasse R, Seebeck T, Gull K. Distinct localisation and cell cycle dependence of COOH terminally tyrosinolated α-tubulin in the microtubules of Trypanosoma brucei. J Cell Biol. 1987;104:439–446. doi: 10.1083/jcb.104.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherwin T, Gull K. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeleton modifications. Phil Trans R Soc London Ser B. 1989;323:575–588. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- 48.Sherwin T, Gull K. Visualization of detyrosination along single microtubules reveals novel mechanisms of assembly during cytoskeletal duplication in trypanosomes. Cell. 1989;57:211–221. doi: 10.1016/0092-8674(89)90959-8. [DOI] [PubMed] [Google Scholar]
- 49.Snapp E L, Landfear S M. Cytoskeletal association is important for differential targeting of glucose transporter isoforms in Leishmania. J Cell Biol. 1997;139:1775–1783. doi: 10.1083/jcb.139.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephens R E. Tubulin and tektin in sea urchin embryonic cilia: pathways of protein incorporation during turnover and regeneration. J Cell Sci. 1994;107:683–692. [PubMed] [Google Scholar]
- 51.Tabish M, Siddiqui Z K, Nishikawa K, Siddiqui S S. Exclusive expression of C. elegans osm-3 kinesin gene in chemosensory neurons open to external environment. J Mol Biol. 1995;247:377–389. doi: 10.1006/jmbi.1994.0146. [DOI] [PubMed] [Google Scholar]
- 52.ten Asbroek A L M A, Oueliette M, Borst P. Targeted insertion of the neomycin phosphotransferase gene into the tubulin gene cluster of Trypanosoma brucei. Nature. 1990;348:174–175. doi: 10.1038/348174a0. [DOI] [PubMed] [Google Scholar]
- 53.Vickerman K. The mechanism of cyclical development in trypanosomes of the Trypanosoma brucei subgroup: a hypothesis based on ultrastructural observations. Trans R Soc Trop Med Hyg. 1962;56:487–495. doi: 10.1016/0035-9203(62)90072-x. [DOI] [PubMed] [Google Scholar]
- 54.Vickstrom K L, Lim S S, Goldman R D, Borisy G G. Steady state dynamics of intermediate filament networks. J Cell Biol. 1992;118:121–129. doi: 10.1083/jcb.118.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wirtz L E, Clayton C E. Inducible gene expression in trypanosomes mediated by a procaryotic repressor. Science. 1995;268:1179–1183. doi: 10.1126/science.7761835. [DOI] [PubMed] [Google Scholar]
- 56.Witman G B. The site of in vivo assembly of flagellar microtubules. Ann N Y Acad Sci. 1975;253:178–191. doi: 10.1111/j.1749-6632.1975.tb19199.x. [DOI] [PubMed] [Google Scholar]
- 57.Woodward R, Carden M J, Gull K. Immunological characterization of cytoskeletal proteins associated with the basal body, axoneme and flagellum attachment zone of Trypanosoma brucei. Parasitology. 1995;111:77–85. doi: 10.1017/s0031182000064623. [DOI] [PubMed] [Google Scholar]