Abstract
Mdj1p, a homolog of the bacterial DnaJ chaperone protein, plays an essential role in the biogenesis of functional mitochondria in the yeast Saccharomyces cerevisiae. We analyzed the role of Mdj1p in the inheritance of mitochondrial DNA (mtDNA). Mitochondrial genomes were rapidly lost in a temperature-sensitive mdj1 mutant under nonpermissive conditions. The activity of mtDNA polymerase was severely reduced in the absence of functional Mdj1p at a nonpermissive temperature, demonstrating the dependence of the enzyme on Mdj1p. At a permissive temperature, the activity of mtDNA polymerase was not affected by the absence of Mdj1p. However, under these conditions, intact [rho+] genomes were rapidly converted to nonfunctional [rho−] genomes which were stably propagated in an mdj1 deletion strain. We propose that mtDNA polymerase depends on Mdj1p as a chaperone in order to acquire and/or maintain an active conformation at an elevated temperature. In addition, Mdj1p is required for the inheritance of intact mitochondrial genomes at a temperature supporting optimal growth; this second function appears to be unrelated to the function of Mdj1p in maintaining mtDNA polymerase activity.
The inheritance of functional mitochondria depends on faithful replication and transmission of mitochondrial DNA (mtDNA). The mitochondrial genome of the yeast Saccharomyces cerevisiae encodes eight major proteins. Seven mitochondrially encoded proteins are components of respiratory chain complexes, and one is a subunit of mitochondrial ribosomes. All other mitochondrial proteins are encoded in the nucleus, synthesized on cytosolic ribosomes, and imported into mitochondria. An intact mitochondrial genome is necessary for respiratory competence but not for the viability of yeast cells (1). Several proteins required for inheritance of mtDNA are known in yeast. These include mtDNA polymerase, Mip1p (17); a single-stranded DNA binding protein, Rim1p (9); a histone-like protein, Abf2p (43); mitochondrial RNA polymerase, Rpo41p (19); a DNA-binding protein of the mitochondrial inner membrane, Yhm2p (5); and several other proteins of unclear function. The molecular mechanism of mtDNA replication is only poorly understood. It was suggested that mitochondrial RNA polymerase synthesizes an RNA transcript which is then used as a primer for mtDNA polymerase to synthesize a new DNA strand (42). Alternatively, yeast mtDNA might be primarily replicated by a rolling circle-like mechanism (36). In addition, recombination and the conversion of recombination intermediates into replication forks have been proposed to be important during the replication of yeast mtDNA (13).
Several cases are known where molecular chaperones play a key role in DNA replication processes. The bacterial dnaK and dnaJ genes, for example, were originally identified and named based on their essential role in the replication of bacteriophage λ DNA, and the purification of the DnaK and DnaJ proteins was facilitated by their activity in DNA synthesis (for a review, see references 49 and 50). Both chaperones perform essential and direct functions in the replication of bacteriophage λ DNA by the activation of a prepriming complex (50). A direct function of molecular chaperones in DNA replication, however, is known only for bacteriophages (λ, P1, and Mu) or plasmids (F and RK2) (4, 29). Propagation of the bacterial chromosome is affected by the inactivation either of DnaK or its partner proteins DnaJ or GrpE. In this case, the chaperones appear to play an indirect role by protecting the DnaA initiator protein against aggregation and inactivation (3, 25). Similarly, eukaryotic homologs of DnaK and DnaJ have been shown to protect calf thymus DNA polymerase ɛ against thermal inactivation in vitro (48). So far, there is no evidence for a direct role of molecular chaperones in either bacterial or eukaryotic chromosomal DNA replication. It is still an open question whether the direct involvement of molecular chaperones in the synthesis of plasmids and phage DNA is a specific adaptation, or whether it is a more common feature that characterizes also processes of genomic DNA propagation.
Molecular chaperones of the Hsp70 class play an important role in mitochondrial biogenesis (reviewed in references 7 and 32). Mitochondrial Hsp70 (mtHsp70, also termed Ssc1p), a homolog of bacterial DnaK, is an essential constituent of the mitochondrial protein import machinery. Moreover, mtHsp70 is involved in folding and assembly of newly imported and mitochondrially synthesized proteins, in the degradation of misfolded proteins in the matrix, and in the protection of proteins against heat-induced aggregation (22, 27, 45). Apparently, all functions of mtHsp70 depend on the activity of its cofactor Mge1p, a homolog of bacterial GrpE (24, 33, 46). Since both mtHsp70 and Mge1p are indispensable for protein import, the encoding genes are essential in yeast. In contrast, Mdj1p, a homolog of bacterial DnaJ, is not essential for mitochondrial protein import (40). Mdj1p has been shown to cooperate with mtHsp70 during protein folding and degradation and in the protection of proteins against heat stress (39, 45, 47). Deletion of the MDJ1 gene leads to a temperature-sensitive growth phenotype. It was reported earlier that cells lacking Mdj1p become [rho0], i.e., they completely lose mtDNA (40). This observation implies that Mdj1p directly or indirectly is involved in the maintenance of mtDNA.
The key proteins involved in mtDNA replication are encoded in the nucleus and synthesized on cytosolic ribosomes (6). After translocation across the mitochondrial membranes, they have to acquire their native conformation in the mitochondrial matrix, a process that might require the assistance of chaperones (21, 26). Furthermore, mitochondrial chaperones might play a role in the inheritance of mtDNA beyond the step of folding of the required proteins.
In the present study, we investigated the role of Mdj1p in the replication of mtDNA. We show that, under restrictive conditions, the activity of mtDNA polymerase is substantially reduced in a conditional mdj1 mutant, resulting in the rapid loss of mtDNA. Furthermore, we find that the activity of mtDNA polymerase does not depend on Mdj1p at low temperature. Under these conditions, the absence of Mdj1p leads to a rapid conversion of functional [rho+] genomes to nonfunctional [rho−] genomes which are then stably maintained. Taken together, our results suggest a dual role of Mdj1p in the maintenance of mtDNA.
MATERIALS AND METHODS
Strains, growth conditions, and isolation of mitochondria.
Standard genetic techniques were used for the growth and manipulation of yeast strains (20). The conditional mdj1-5 mutant strain (47), the isogenic wild-type and [rho0] strains, and the Δmdj1 deletion strain (40) were described previously. For construction of the pYesMIP1-myc plasmid, the entire MIP1 coding region was cloned into pBluescript II KS vector by using the procedure described previously (12) and plasmid YEpT7-3 (14) (kindly provided by F. Foury, Universite Catolique de Louvain, Louvain, Belgium). Then, a sequence encoding the 9E10 c-myc epitope was introduced immediately upstream of the stop codon by PCR by using the Quik Change site-directed mutagenesis kit (Stratagene). The presence of the myc epitope was confirmed by sequencing the MIP1-myc coding sequence. This gene was subcloned into the pYES 2.0 vector (Invitrogen), yielding plasmid pYesMIP1-myc. The hyper-suppressive [rho−] genome [HS3324] was transferred to [rho0] recipient strains by cytoduction (31) via the [rho0] kar1 mutant strain 10507-15-4b (MATa) as described previously (35).
Mitochondria were isolated as described elsewhere (23). During incubation with zymolyase 20T (Seikagaku Corp.), cycloheximide (Sigma) was added at a concentration of 150 μg/ml.
Analysis of mtDNA.
4′,6′-Diamidino-2-phenylindole (DAPI) staining was used to visualize mtDNA in cells essentially as described previously (2). Briefly, 1 ml of cells (optical density at 600 nm [OD600] of 1.0) were resuspended in 1 ml buffer P (40 mM KH2PO4, pH 6.5; 0.5 mM MgCl2) containing 4% formaldehyde and fixed for 1 h at 30°C. After three washes in buffer P and one in buffer PS (buffer P containing 1 M sorbitol), the cells were spheroplasted with 2 mg of zymolyase 20T per ml in PS at 30°C for 20 min. Spheroplasts were washed three times in PS and incubated for 5 min in 1 μg of DAPI per ml in phosphate-buffered saline (PBS; 0.8% NaCl; 0.02% KCl; 0.114% Na2HPO4; 0.02% KH2PO4, pH 7.4). Cells were washed in PBS and placed on poly-l-lysine-coated multiwell slides, air dried, and mounted. Microscopy was performed with a Zeiss Axioplan microscope equipped for epifluorescence by filter sets G365/FT395/LP420.
For the purification of mtDNA, total cellular DNA was isolated. mtDNA was separated from nuclear DNA essentially as described (16). Briefly, 1-liter cultures were grown to an OD600 of 2.0 to 3.0, and the cells were harvested by centrifugation. After treatment with zymolyase, the resulting spheroplasts were washed with 100 ml of 1.2 M sorbitol, resuspended in 100 ml of 20 mM Tris-HCl (pH 8)–50 mM EDTA–1% (wt/vol) sodium dodecyl sulfate, and incubated for 30 min at 65°C. Then, 50 ml of 5 M potassium acetate was added and incubated on ice for 45 min. After centrifugation at 9,000 × g for 10 min, 150 ml of isopropanol was added to the supernatant. Precipitated DNA was resuspended in 10 ml of 10 mM Tris-HCl (pH 8.0)–1 mM EDTA. Then, 1.16 g of CsCl and 24 μl of (10 mg/ml) bisbenzimide (Hoechst 33258; Sigma) were added for 1 ml of DNA solution. The mixture was centrifuged in an NVTi 65 rotor (Beckman Instruments) for 16 h at 60,000 rpm. DNA was visualized with long-wavelength UV light, and an image was recorded with a UVP Gel Documentation System. Standard methods were used for the analysis of isolated mtDNA with restriction enzymes (41).
Assay of DNA synthesis in isolated mitochondria.
For the analysis of mtDNA synthesis in organello, isolated mitochondria (0.6 mg/ml [total protein]) were incubated in 50 μl of medium containing 10 mM morpholinepropanesulfonic acid (MOPS)-KOH, pH 7.4; 220 mM sucrose; 60 μg of bovine serum albumin (BSA) per ml; 100 mM KCl; 10 mM MgCl2; 10 mM creatinine phosphate; 100 μg of creatinine kinase per ml; 2 mM ATP; 10 μM concentrations each of GTP, CTP, and UTP; 50 μM concentrations each of dCTP, dGTP, dATP, dTTP, and [methyl-3H]dTTP (30 to 50 cpm/pmol of deoxynucleoside triphosphate [dNTP]); and 50 μg of aphidicolin (Sigma) per ml. The rate of incorporation of radioactive dTTP into mtDNA was measured after incubation for 30 min at 30°C (or as indicated) by precipitating the labeled mtDNA with 750 μl of 10% trichloroacetic acid–100 mM sodium pyrophosphate. The precipitates were collected on glass fiber filters (GF/C-Whatman) and then washed three times with 1 ml of 1 M hydrochloric acid–100 mM sodium pyrophosphate and once with 2 ml of ethanol. The filters were dried, transferred to scintillation fluid, and counted.
Assay of mtDNA polymerase activity in soluble mitochondrial extracts.
For the preparation of mitochondrial extracts containing mtDNA polymerase, isolated mitochondria were pelleted by centrifugation in a microcentrifuge at 10,000 rpm for 5 min at 2°C. Then, an extraction medium containing 20 mM Tris-HCl (pH 8.0)–5 mM dithiothreitol (DTT)–1 mM phenylmethylsulfonyl fluoride was added at a volume equal to the volume of the mitochondrial pellet. Mitochondria were dissolved by strong vortexing for 1 min. NaCl was added to a final concentration of 0.5 M, and samples were vortexed for additional 2 min. The mitochondrial extract was centrifuged at 100,000 × g for 1 h in TLA45 rotor (Beckman). The supernatant containing mtDNA polymerase was collected.
Mitochondrial DNA polymerase activity was measured in 25 μl of reaction mixture containing 50 mM Tris-HCl (pH 8.0); 100 μg of BSA per ml; 2.5 mM DTT; 50 mM NaCl; 10 mM MgCl2; 50 μg of aphidicolin per ml; and 25 μM concentrations each of dATP, dCTP, dTTP, dGTP, and [methyl-3H]dTTP (30 to 50 cpm/pmol dNTP). Double-stranded salmon sperm DNA activated by DNase I treatment was prepared as described previously (14) and used as a substrate at 0.12 mg/ml. After the addition of substrate DNA to the reaction solution with mitochondrial extract (6 to 8 μg of protein or as indicated), the samples were incubated for 30 min at 30°C. The rate of DNA synthesis was analyzed by counting the radioactivity present in acid-insoluble material as described above. For all experiments, both the amount of extract and the substrate were titrated in order to ensure that the substrate was not limiting the reaction rate.
RESULTS
Loss of mitochondrial DNA in the conditional mdj1-5 mutant.
We reported earlier that deletion of the MDJ1 gene leads to a temperature-sensitive growth phenotype and to the complete loss of mtDNA, i.e., the cells become [rho0] (40). In order to study the mechanism by which Mdj1p is involved in the maintenance of mtDNA, we employed a conditional mutant of Mdj1p harboring a deletion of amino acids 269 to 285, Mdj1-5p. The mdj1-5 strain exhibits a temperature-sensitive growth phenotype on nonfermentable carbon sources; however, it is still able to grow at 37°C as long as a fermentable carbon source is provided (47). First, we tested the kinetics of loss of mtDNA in this temperature-sensitive mutant in vivo. A preculture grown at a permissive temperature (25°C) was divided into one half which was kept continuously at 25°C, and a second half which was shifted to the nonpermissive temperature, 37°C. Cells were grown in liquid culture on a fermentable carbon source (glucose) to allow loss of mtDNA. The cultures were always kept at the logarithmic growth phase. At the time points indicated in Fig. 1, aliquots were collected from both cultures, and cells were plated at an appropriate dilution onto glucose-containing medium. After an incubation for 24 h at permissive temperature, colonies were replica plated onto medium containing a nonfermentable carbon source (glycerol), and the percentage of colonies able to grow was determined. The numbers directly reflect the percentage of cells harboring functional mitochondria in liquid culture (Fig. 1A). As a control, cells were replica plated onto medium selective for the auxotrophic marker of the plasmid containing the mdj1-5 gene in order to exclude possible effects due to the loss of the plasmid (not shown). When the mdj1-5 strain was grown at a permissive temperature, the number of cells maintaining their respiratory functions was close to 100% throughout the time course of the experiment. In contrast, when the culture was shifted to a nonpermissive temperature the number of cells containing functional mitochondria dropped immediately. After cultivation for 24 h at 37°C (i.e., eight generations), less than 10% of the cells were able to grow on a nonfermentable carbon source (Fig. 1A), whereas wild-type cells remained respiratory competent under both growth conditions (not shown).
FIG. 1.
mtDNA is lost in the temperature-sensitive mdj1-5 mutant strain at an elevated temperature. (A) Loss of respiratory competence of mdj1-5 cells grown at elevated temperature. A preculture of mdj1-5 cells was grown for 24 h at 25°C in glucose-containing synthetic complete medium which was selective for the auxotrophic marker of the plasmid carrying the mdj1-5 allele. This culture was diluted with fresh medium to an OD600 of 0.4 and divided into two halves. One half was incubated at the permissive temperature, 25°C (●), whereas the second half was shifted to the nonpermissive temperature, 37°C (○). After 3 h, an aliquot of the 37°C culture was returned to 25°C (▵; the shift is indicated by an arrow). At the indicated time points, aliquots of the cultures were plated on glucose-containing medium, and the remainder of the culture was diluted to an OD600 of 0.4 in order to keep the culture in the logarithmic growth phase. The next day, colonies were replica plated to glycerol-containing medium, and the percentage of respiratory-competent colonies was determined. (B) Loss of mtDNA in mdj1-5 cells grown at an elevated temperature. Cells from the aliquots obtained as described above were stained with DAPI and analyzed by fluorescence microscopy. The percentage of cells containing at least four chondrolites was plotted for each time point. (C) Detection of mtDNA in CsCl gradients. mdj1-5 cells were grown for 24 h at the indicated temperatures in glucose-containing medium. As a control, wild-type [rho+] and [rho0] cells were grown at 25°C. Total cellular DNA was isolated and stained with bisbenzimide, mtDNA was separated from nuclear DNA by centrifugation in CsCl gradients, and DNA was visualized under UV light.
The complete loss of functional mitochondria was observed only when cells were cultivated at elevated temperature throughout the experiment. When cultures maintained at 37°C for 3 h were returned to 25°C for the remaining time of the experiment, an initial drop of the number of respiratory-competent cells was observed for the samples taken at elevated temperature. Upon return to permissive temperature, the percentage of cells containing functional mitochondria was stabilized, but at a reduced level (Fig. 1A), indicating that the loss of respiratory competence was irreversible.
To test whether the lack of respiratory functions in mdj1-5 cells cultivated at a nonpermissive temperature correlates with the loss of mtDNA, aliquots of cultures were stained with DAPI, and the presence of chondrolites (small bodies in the cell) was taken as evidence for the presence of mtDNA as determined by using fluorescence microscopy (2). For each time point, at least 100 individual cells were inspected. The number of cells containing mtDNA remained constant when the culture was grown under permissive conditions (ca. 95% of cells contained visible chondrolites). This correlated well with the number of cells which were respiratory-competent (Fig. 1B). In contrast, the number of chondrolites dropped significantly in cells which were grown at a nonpermissive temperature. After 24 h of growth at 37°C, ca. 80% of the cells did not contain any visible mtDNA (Fig. 1B). These cells were indistinguishable from a control strain lacking mtDNA (not shown). Notably, the loss of mtDNA occurred within a few hours after the loss of respiratory competence.
The loss of mtDNA in a population of mdj1-5 cells upon growth at nonpermissive conditions was also monitored in another, independent experiment. Total cellular DNA was isolated from a wild-type [rho+] strain, a [rho0] control strain, and mdj1-5 cells grown under permissive or nonpermissive conditions in glucose-containing medium. mtDNA was separated from nuclear DNA by centrifugation in a cesium chloride density gradient containing the dye bisbenzimide (16). mdj1-5 cells grown at a permissive temperature (25°C) for 24 h contained roughly the same amount of mtDNA as the wild type, whereas mdj1-5 cells grown at the nonpermissive temperature (37°C) were indistinguishable from the [rho0] control strain (Fig. 1C).
These data show that mtDNA is rapidly lost when a yeast strain lacking functional Mdj1p is grown at elevated temperature. One possible explanation for the loss of mtDNA could be an improper segregation of the mitochondrial genomes during cell division (34). In the case of mdj1-5, we consider this unlikely since fluorescence microscopy of DAPI-stained mitotic cells indicated that mtDNA was always evenly partitioned between dividing cells (data not shown). Alternatively, a block of mtDNA synthesis might be responsible for the loss of the mitochondrial genome. Assuming that an average yeast cell contains about 30 copies of mtDNA (1) and that mtDNA synthesis is blocked in mdj1-5 cells upon shift to 37°C, eight cell divisions are more than enough to result in loss of mtDNA by simple dilution.
Mdj1p is required for mitochondrial DNA synthesis at elevated temperature.
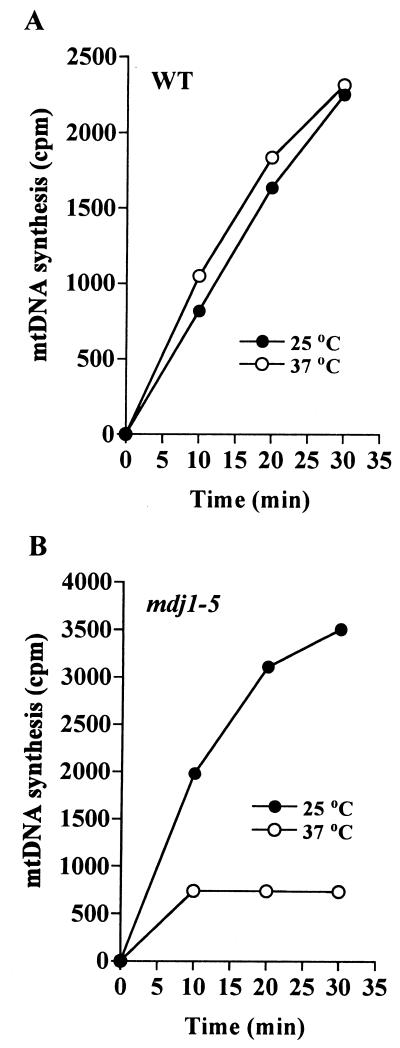
In order to test whether functional Mdj1p is required for efficient synthesis of mtDNA at elevated temperature, we measured the rate of DNA synthesis in isolated organelles. Mitochondria were isolated from wild-type and mdj1-5 strains grown at 25°C or shifted to 37°C and grown for 3 h at this temperature. Under the latter conditions, the phenotype of the mdj1-5 strain was induced, i.e., the number of respiratory-competent cells was significantly decreased (Fig. 1A); however, almost 100% of the cells still contained mtDNA (Fig. 1B). Mitochondria were incubated for the indicated time periods at 30°C in the presence of [3H]dTTP, DNA was precipitated, and the amount of incorporated radioactivity was determined. No difference in the rate of mtDNA synthesis was observed for wild-type mitochondria isolated from cells grown at either 25 or 37°C. In contrast, in mdj1-5 mitochondria mtDNA synthesis was strongly reduced when the cells had been exposed to the nonpermissive temperature (Fig. 2). These results indicate that even a relatively short exposure of mdj1-5 cells to nonpermissive conditions results in a substantial inhibition of mtDNA synthesis. Since cells were exposed to the elevated temperature for no more than one generation time, we conclude that a defect in segregation of mtDNA cannot be responsible for this effect. We propose that Mdj1p is required for the synthesis of mtDNA at an elevated temperature.
FIG. 2.
Mdj1p is required for efficient DNA synthesis in mitochondria isolated from cells grown at elevated temperature. (A) Wild-type yeast cells were grown in glucose-containing medium in a 10-liter fermenter for 12 h at 25°C. At an OD600 of 2.0, 5 liters of the culture was harvested, and mitochondria were isolated (●). The remaining culture was diluted twofold with fresh medium, the temperature was raised to 37°C, cells were grown for another 3 h to an OD600 of 2.0, and the mitochondria were isolated (○). Mitochondria were incubated for the indicated time periods at 30°C in the presence of radioactive nucleotides. Synthesis of mtDNA was measured by the incorporation of radioactivity into acid-insoluble material (for details, see Materials and Methods). (B) Synthesis of mtDNA was measured in mitochondria isolated from mdj1-5 cells, as described for panel A.
Mdj1p is required for mitochondrial DNA polymerase activity at elevated temperature.
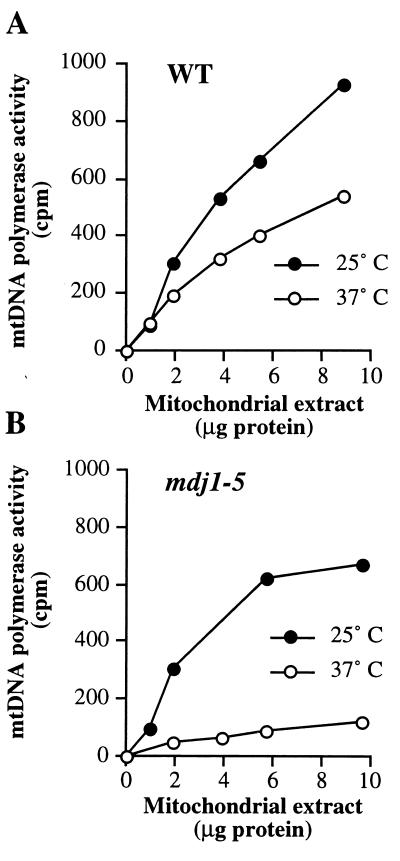
Mdj1p plays an important role in folding of newly imported mitochondrial proteins and in their protection against heat inactivation (39, 40, 47). Thus, proteins involved in DNA synthesis in mitochondria might require Mdj1p for proper folding and/or protection against heat stress at an elevated temperature. We asked whether inactivation of Mdj1-5p at the nonpermissive temperature directly affects the activity of mitochondrial DNA polymerase, Mip1p, the sole enzyme catalyzing synthesis of mtDNA (14, 42). Wild-type and mdj1-5 cells were grown at 25°C or shifted to 37°C and then grown at this temperature for 3 h. Mitochondrial extracts were prepared, and Mip1p activity was measured according to a published procedure (14). Briefly, mitochondria were opened by vigorous vortexing and extracted with 0.5 M sodium chloride. To this extract, nicked salmon sperm DNA was added as a template, and the incorporation of [3H]dTTP was measured in the presence of aphidicolin, an inhibitor of nuclear DNA polymerases (for details, see Materials and Methods). This assay measures the activity of mtDNA polymerase independently of other factors, since it was shown that purified yeast mtDNA polymerase fills gaps in double-stranded DNA without the action of any other replication proteins (12). Heat treatment of cells resulted in a decrease of mtDNA polymerase activity to ca. 60% in wild-type extracts. In contrast, mtDNA polymerase activity was much more strongly reduced in extracts prepared from mdj1-5 cells grown at 37°C (Fig. 3). We conclude that functional Mdj1p is required for the activity of mtDNA polymerase at elevated temperature.
FIG. 3.
Mdj1p is required for mtDNA polymerase activity in mitochondrial extracts from cells grown at elevated temperature. (A) Mitochondria were isolated from wild-type cells as described for Fig. 2 and extracted with 0.5 M sodium chloride. The indicated amounts of extracts were incubated with activated double-stranded salmon sperm DNA in the presence of radioactive nucleotides for 30 min at 30°C. The activity of mtDNA polymerase was measured by the incorporation of radioactivity into acid-insoluble material. (B) The activity of mtDNA polymerase was measured in mitochondrial extracts isolated from mdj1-5 cells.
In order to exclude the possibility that the observed decrease of mtDNA polymerase activity in the mdj1-5 strain was due to a defect in protein import, we tested whether the amount of mtDNA polymerase present in mitochondria was affected at a nonpermissive temperature. An epitope-tagged version of mtDNA polymerase with a C-terminal myc tag was expressed from a plasmid (pYesMIP1-myc) under control of the inducible GAL promoter in mdj1-5 cells (mdj1-5 [MIP-myc]). A single major band was detected in a Western blot of total cell lysates isolated from transformed mdj1-5 cells (Fig. 4A). This band was not present in nontransformed cells, indicating that monoclonal antibodies against the myc epitope did not cross-react with other yeast proteins (Fig. 4A). Next, mitochondria were isolated from the mdj1-5 [MIP-myc] strain grown at 25°C or shifted to 37°C and then grown at an elevated temperature for 3 h. Similar amounts of mtDNA polymerase were detected in both preparations, indicating that inactivation of Mdj1p does not affect the import of mtDNA polymerase into mitochondria (Fig. 4B). mtDNA polymerase activity measured in extracts prepared from mdj1-5 [MIP-myc] cells grown at 25°C was 10-fold higher than in extracts prepared from nontransformed mdj1-5 cells. However, the activity measured in mitochondrial extracts obtained from mdj1-5 [MIP-myc] cells grown for 3 h at 37°C was reduced by 83% (data not shown), confirming that the myc-tagged mtDNA polymerase was as heat sensitive as wild-type enzyme as shown in Fig. 3B.
FIG. 4.
The amount of mtDNA polymerase is not reduced in mitochondria of mdj1-5 cells grown at a nonpermissive temperature. (A) Two independent clones of mdj1-5 harboring plasmid pYesMIP1-myc (lanes 1 and 2) and mdj1-5 without this plasmid (lane 3) were grown for 24 h at 25°C in galactose-containing synthetic complete medium selective for the auxotrophic markers of the plasmids carrying the mdj1-5 and MIP1-myc alleles. Cells corresponding to 1 OD600 unit were collected, resuspended in 40 μl of Laemmli lysis buffer, and lysed by vortexing in the presence of glass beads and subsequent boiling for 10 min. Then, 10 μl of each lysate was analyzed by Western blotting by employing anti-c-myc monoclonal antibodies (Boehringer Mannheim). (B) The mdj1-5 strain harboring plasmid pYesMIP-myc was grown at 25°C as described in panel A. At an OD600 of 1.3 half of the culture was harvested, and mitochondria were isolated. The temperature of the remaining culture was raised to 37°C, and the cells were grown for another 3 h before the mitochondria were isolated. Epitope-tagged mtDNA polymerase was detected in isolated mitochondria as described in panel A. A polyclonal antiserum against subunit β of the F1 ATPase was used to standardize the amount of mitochondrial proteins loaded on each lane.
Loss of mitochondrial DNA synthesis activity and decreased mitochondrial DNA polymerase activity in mdj1-5 are reversible.
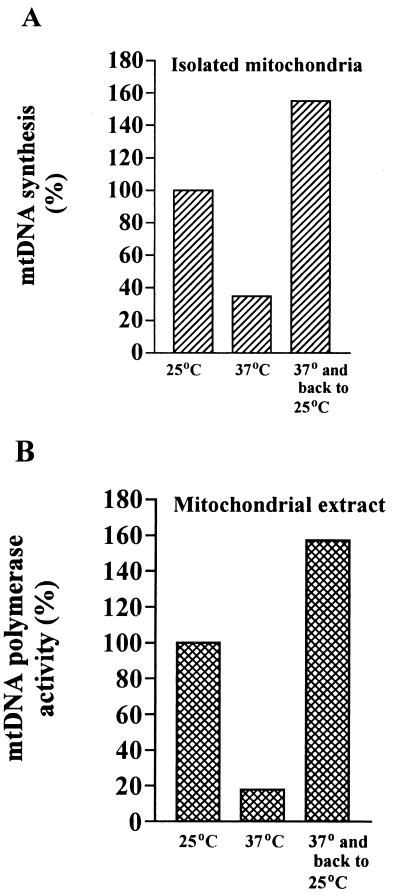
The experiments in vivo indicated that cultivation of mdj1-5 cells at 37°C leads to the complete loss of mtDNA, an irreversible event. If decreased mtDNA polymerase activity was the primary cause for the observed loss of mtDNA, it can be predicted that this effect is reversible as long as a DNA template is present and mtDNA polymerase activity can be restored at permissive temperature. A culture of mdj1-5 cells grown at 25°C was subsequently shifted to 37°C for 3 h, i.e., one generation time, and returned to 25°C for another 3 h. Mitochondria were isolated, and DNA synthesis in the organelles and mtDNA polymerase activity in extracts were measured. mtDNA synthesis and polymerase activity were strongly reduced by cultivation at 37°C; however, mtDNA synthesis resumed an even higher than the original level after return to the permissive temperature (Fig. 5). These findings are compatible with the view that cultivation of yeast cells lacking functional Mdj1p leads to a rapid loss of mtDNA polymerase activity at elevated temperature. This would be followed by the loss of mtDNA by dilution, a process that would take longer than one generation time, as used in this experiment. Upon transfer of cells back to 25°C, newly synthesized Mip1p can be imported from the cytosol into mitochondria. At the permissive temperature, the enzyme can fold properly, and mtDNA synthesis activity is restored.
FIG. 5.
Thermal inactivation of DNA synthesis is reversible. (A) DNA synthesis in isolated mitochondria of mdj1-5. The mdj1-5 strain was grown in glucose-containing medium in a 10-liter fermenter for 12 h at 25°C. At an OD600 of 2.0, 5 liters of the culture was harvested, and the mitochondria were isolated. The remaining culture was diluted twofold with fresh medium, the temperature was raised to 37°C, the cells were grown for another 3 h to an OD600 of 2.0, 5 liters of culture was harvested, and mitochondria were isolated. The remaining culture was diluted with fresh medium, and the temperature was shifted back to 25°C. After another 3 h, the third batch of mitochondria was prepared. DNA synthesis in mitochondria was measured as described for Fig. 2. (B) mtDNA polymerase activity in extracts of mdj1-5. Cells were grown as described above, and mtDNA polymerase activity was measured as described for Fig. 3.
Mdj1p is required for the maintenance of a hypersuppressive [HS3324] mitochondrial genome at elevated temperature.
Maintenance and replication of the mitochondrial genome is a complex process dependent on many cellular activities. It has been shown that transcription and translation of mitochondrial genes are required for the stable maintenance of an intact wild-type [rho+] genome (37). However, a class of mtDNA deletion [rho−] mutants, the hypersuppressive mitochondrial genomes (10), is stably maintained in cells lacking the gene encoding mitochondrial RNA polymerase (13, 35). Maintenance of hypersuppressive genomes is also independent of factors such as the histone-like protein Ab2fp or the mitochondrial transcription factor Mtf1p (44). One such [rho−] mutant, termed [HS3324], consists of 960 bp containing an active origin of mtDNA replication (ori5) (15). The [HS3324] genome, which does not encode any mitochondrial proteins, does not show any mitochondrial translation activity. Thus, its inheritance is not dependent on active mitochondrial translation. Its replication, however, must be dependent on the function of mtDNA polymerase. Thus, the [HS3324] genome allowed us to investigate the role of Mdj1p in mtDNA synthesis independent of mitochondrial transcription and translation activities.
We introduced mitochondria containing [HS3324] mtDNA into a mdj1-5 [rho0] strain by cytoduction (31). Next, mtDNA was isolated from mdj1-5 cells grown at permissive and nonpermissive temperatures and separated from nuclear DNA in a cesium chloride gradient (Fig. 6A). As with [rho+] DNA, the [HS3324] mtDNA was stably maintained at 25°C but was lost after 24 h growth at 37°C (compare to Fig. 1C). Similarly, upon exposure to elevated temperature (37°C), both mtDNA synthesis activity in isolated mitochondria and the activity of mtDNA polymerase in mitochondrial extracts were observed to be decreased to the same extent as in [rho+] mitochondria (compare Fig. 6B and C with Fig. 2B and 3B). These results support our hypothesis that the lack of mtDNA polymerase activity is the primary reason for the loss of mtDNA in the absence of functional Mdj1p at elevated temperature.
FIG. 6.
Mdj1p is required for replication of the hypersuppressive [HS3324] genome at elevated temperature. (A) Loss of mtDNA in mdj1-5 [HS3324] at elevated temperature. The mdj1-5 strain harboring [HS3324] mtDNA was cultivated for 24 h in 1 liter of glucose-containing medium at the indicated temperatures. Total cellular DNA was isolated and visualized as described for Fig. 1C. (B) DNA synthesis in mitochondria isolated from mdj1-5 [HS3324]. The mdj1-5 [HS3324] strain was grown, and mtDNA synthesis measured as in Fig. 2. (C) mtDNA polymerase activity in mitochondrial extracts isolated from mdj1-5 [HS3324]. The mdj1-5 [HS3324] strain was grown and mtDNA polymerase activity was measured as in Fig. 3.
Mdj1p is not required for mitochondrial DNA polymerase activity at low temperature.
We asked whether in the absence of Mdj1p, mtDNA polymerase activity would be also decreased at a low temperature. It was shown previously that [rho0] mitochondria isolated from either yeast or mammalian cells contain active DNA polymerase, indicating that the presence of mtDNA is not a prerequisite for the biogenesis of the functional enzyme (8, 17). We isolated mitochondria from Δmdj1 [rho0], wild-type [rho0], and wild-type [rho+] strains grown on glucose-containing medium at 25°C. mtDNA polymerase activity was measured in mitochondrial extracts. Surprisingly, similar levels of polymerase activities were detected, whether or not Mdj1p was present (Fig. 7). We conclude that functional mtDNA polymerase can be made at a low temperature without the assistance of Mdj1p and that Mdj1p is required for mtDNA polymerase activity only at elevated temperature.
FIG. 7.
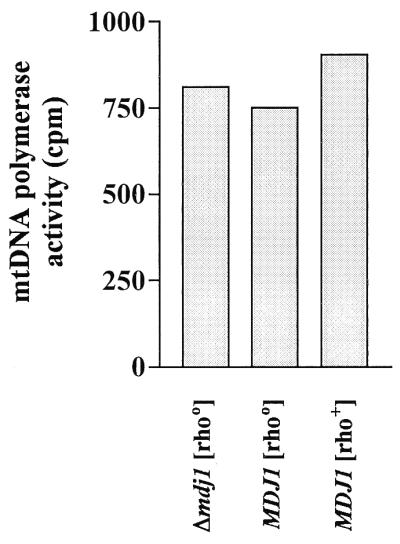
mtDNA polymerase activity does not depend on Mdj1p at a low temperature. A Δmdj1 [rho0] strain, a wild-type [rho0] strain, and a wild-type [rho+] strain were cultivated in glucose-containing medium at 25°C. mtDNA polymerase activity was measured at 30°C as described for Fig. 3.
The hypersuppressive [HS3324] mitochondrial genome can be stably maintained in the absence of Mdj1p at low temperature.
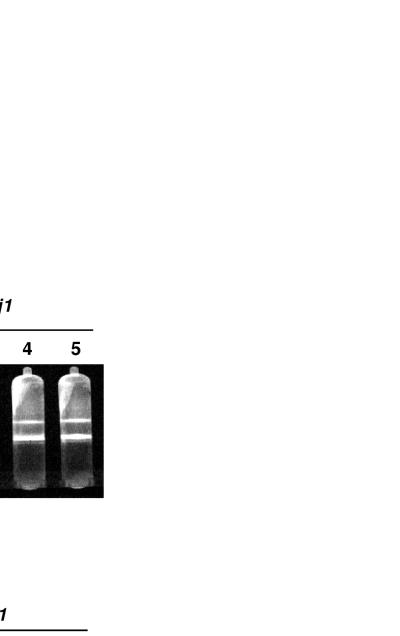
We asked whether Mdj1p would be required for the maintenance of the hypersuppressive [HS3324] genome under conditions where active mtDNA polymerase is made. We selected clones of a mdj1-5 [HS3324] strain which had lost the plasmid harboring the mdj1-5 gene, i.e., clones that carried only a chromosomal disrupted allele of mdj1. From these, we randomly picked four Δmdj1 clones for further investigation. The absence of Mdj1p in mitochondria isolated from all four clones was confirmed by immunoblot analysis (data not shown). The strains were grown in liquid culture at 25°C and kept at logarithmic growth for one week, i.e., for approximately 50 generations. Then, total cellular DNA was isolated and mtDNA was separated by centrifugation in a cesium chloride gradient. Interestingly, all four clones contained mtDNA (Fig. 8A). We isolated mtDNA from the cesium chloride gradients and confirmed its identity with the [HS3324] genome by restriction analysis and agarose gel electrophoresis (Fig. 8B). These results show that at 25°C, the [HS3324] mtDNA can be stably maintained in the absence of Mdj1p, implying that Mdj1p is not required for the synthesis of mtDNA at a low temperature.
FIG. 8.
At a low temperature the hypersuppressive [HS3324] genome can be stably maintained in the absence of Mdj1p. (A) Detection of mtDNA in CsCl gradients. A MDJ1 strain harboring the [HS3324] genome, a MDJ1 [rho0] strain, and four clones of the Δmdj1 strain harboring [HS3324] were grown for 24 h at 25°C in glucose-containing medium. Total cellular DNA was isolated and visualized as described for Fig. 1C. (B) Analysis of mitochondrial genomes by restriction digestion. Mitochondrial [rho+] DNA obtained from a wild-type strain, [HS3324] DNA obtained from a wild-type strain, and mitochondrial DNAs harvested from the gradients described above were digested with EcoRV, separated by agarose gel electrophoresis, stained with ethidium bromide, and visualized under UV light. The asterisk indicates the EcoRV fragment of the [HS3324] genome. MW, molecular weight standard.
Deletion of MDJ1 leads to transition of mitochondrial [rho+] genomes to [rho−] genomes at low temperature.
Since the [HS3324] mtDNA could be stably maintained in Δmdj1 cells at a low temperature, we investigated the inheritance of mitochondrial [rho+] genomes under the same conditions. Cells of a strain that carries [rho+] mtDNA, an inactivated chromosomal mdj1 allele, and an extrachromosomal copy of MDJ1 on a plasmid (YBW16 [47]) were selected for loss of the plasmid harboring the MDJ1 gene. Five of these Δmdj1 clones were further analyzed, after the absence of Mdj1p was confirmed by immunoblotting (data not shown). All five clones were unable to grow on a medium containing a nonfermentable carbon source (data not shown). Cells were grown for 24 h in glucose-containing medium at 25°C, and mtDNA was isolated by cesium chloride gradient centrifugation of total cellular DNA. Surprisingly, all five clones contained clearly visible mtDNA (Fig. 9A). Next, mtDNA was isolated from the cesium chloride gradients and subjected to restriction analysis. The restriction patterns of mtDNA obtained from all five Δmdj1 clones were clearly different from the [rho+] control (Fig. 9B). To confirm that these genomes were nonfunctional, the Δmdj1 strains were crossed with a [rho0] tester strain harboring a wild-type copy of MDJ1. The resulting diploids were all unable to grow on nonfermentable carbon sources (data not shown). Similar results were obtained after cells were cultivated at 25°C for up to 1 week (data not shown). These results indicate that at a low temperature the absence of Mdj1p leads to a transition of mitochondrial [rho+] genomes to nonfunctional [rho−] genomes. The [rho−] genomes can be stably maintained and propagated. Thus, we conclude that Mdj1p is not only required for mtDNA polymerase activity at an elevated temperature but also for a thus-far-unknown process(es) during maintenance of a functional genome at low temperature.
FIG. 9.
Mitochondrial [rho+] genomes are converted at a low temperature to [rho−] genomes in the absence of Mdj1p. (A) Detection of mtDNA in CsCl gradients. A MDJ1 strain harboring a [rho+] genome, a MDJ1 [rho0] strain, and five freshly made clones of the Δmdj1 strain were grown for 24 h at 25°C in glucose-containing medium. Total cellular DNA was isolated and visualized as described for Fig. 1C. (B) Analysis of mtDNA by restriction digestion was as in Fig. 8.
DISCUSSION
It was shown previously that the mitochondrial chaperone Mdj1p plays an essential role in the biogenesis of functional mitochondria (40). In the present study we investigated its function in the inheritance of mtDNA. Several lines of evidence indicate that mtDNA polymerase is a direct target of Mdj1p at elevated temperature. Most importantly, in the absence of functional Mdj1p, the growth of cells at elevated temperature led to a substantial loss of mtDNA polymerase activity. The assay used in these experiments employs artificial DNA template and measures the activity of mtDNA polymerase independently of other replication proteins. This implies that mtDNA polymerase activity depends on functional Mdj1p at an elevated temperature. Furthermore, when DNA synthesis was measured in intact mitochondria, a strong decrease was seen under nonpermissive conditions, indicating that a mtDNA synthesis defect could be also observed in the presence of the native mitochondrial replication complex. Finally, mitochondrial genomes, including hypersuppressive [HS3324] mtDNA, were rapidly lost in vivo in the mdj1-5 mutant grown under nonpermissive conditions, probably by dilution of preexisting mtDNA molecules. Such a phenotype is to be expected when mtDNA synthesis is inhibited due to an inactivated mtDNA polymerase.
How might Mdj1p affect mtDNA polymerase activity at an elevated temperature? After synthesis on cytosolic ribosomes, mtDNA polymerase must be translocated across the mitochondrial membranes in an unfolded state (38). During this process, hydrophobic surfaces of the translocating polypeptide chain become exposed to the cellular milieu, rendering the protein prone to aggregation as long as the translocation of folding-competent domains is not completed. Similarly, upon cultivation of the cells at elevated temperature, thermal unfolding of the native enzyme may lead to the loss of catalytic activity. Molecular chaperones are known to shelter unfolded proteins against misfolding and aggregation by transiently shielding hydrophobic patches (21, 26). By using artificial substrate proteins, we reported earlier that Mdj1p acts as a chaperone in both processes and that its action is especially important at elevated temperature (39, 40, 47). It is conceivable that mtDNA polymerase requires Mdj1p as a chaperone in order to acquire and/or maintain its functional conformation at an elevated temperature. We suggest that mtDNA polymerase is one of the few as-yet-known examples of a natural target protein whose folding is dependent on molecular chaperones, in our case Mdj1p.
At a low temperature, mtDNA polymerase activity does not depend on Mdj1p. However, under these conditions, deletion of MDJ1 leads to the rapid conversion of [rho+] to [rho−] genomes which then are stably propagated in the Δmdj1 strain. Conversion of [rho+] to [rho−] genomes is observed even in wild-type yeast strains at a frequency of 1% (11). The molecular mechanism of this phenomenon, however, is not well understood. We consider four possibilities to explain a role of Mdj1p in this process. First, Mdj1p might assist the folding of a yet-unknown factor(s) which is required for the faithful propagation of [rho+] genomes. In this case, Mdj1p would act as a chaperone for a newly imported protein even at a low temperature. This would be in accordance with the observation that the artificial substrate protein firefly luciferase requires Mdj1p for activity even at 25°C after import into isolated mitochondria (40). Second, Mdj1p might affect properties of mtDNA polymerase independently of the DNA synthesis activity measured in our assay. Such properties could include for example the processivity of the enzyme (30), i.e., the time the polymerase is bound to the DNA template once it has started the synthesis of a new DNA strand. A decreased processivity might result in the formation of free 3′ ends of DNA. This in turn might increase the rate of recombination which might be responsible for transition from [rho+] to [rho−] genomes. Interestingly, we observed that the stability of purified mtDNA polymerase at high temperature depends on its binding to DNA (17a). It is possible that, vice versa, Mdj1p affects the stability of the enzyme, and this might influence its binding to DNA. Third, Mdj1p might be involved in mtDNA inheritance by a mechanism that is linked to the biogenesis of mitochondrial translation products. It was shown that mitochondrial protein synthesis is required for the maintenance of a [rho+] genome (37). On the other hand, Mdj1p is involved in the biogenesis of mitochondrially encoded proteins; it interacts with nascent chains in mitochondria and protects the mitochondrially synthesized var1 protein against aggregation (47). It is possible that in the absence of Mdj1p, abnormal mitochondrial translation products accumulate, and this then might lead to defects in mtDNA replication by a yet-unknown mechanism. Fourth, Mdj1p might play a direct role in the activation of mtDNA replication complexes. Such a role of Mdj1p might be analogous to the function of Escherichia coli DnaJ which is required for the activation of prepriming complexes in bacteriophage λ DNA replication (49, 50). In this system the phage appears to employ the host’s chaperone system in order to adapt its own replication factors to the bacterial DNA replication machinery (28). Interestingly, mtDNA polymerase is similar to eukaryotic but not to prokaryotic DNA polymerases (42). Thus, it can be speculated that Mdj1p might be required for the functional interaction of mtDNA polymerase with other replication factors stemming from the prokaryotic endosymbiotic ancestors of mitochondria (18).
In conclusion, Mdj1p is an important factor for the faithful replication of mtDNA in vivo and in vitro. We have identified mtDNA polymerase as a target of Mdj1p, at least at an elevated temperature. Further possible interacting proteins and the exact function of Mdj1p in the maintenance of [rho+] genomes at optimal temperature remain to be identified.
ACKNOWLEDGMENTS
We are grateful to W. L. Fangman for the gift of strains harboring [HS3324] mtDNA and kar1 mutation. We thank F. Foury for plasmid YEpT7 and the laboratory protocol for preparation of mitochondrial extracts. We thank E. A. Craig, J. M. Herrmann, I. Konieczny, K. Liberek, and M. Zylicz for discussions and critical reading of the manuscript.
This work was supported by the Polish State Committee for Scientific Research Project 6P04A01712 given to M. Zylicz, and the work of M.D. was partially supported by Project 6P04A03915. At the beginning of this work, a stay of J.M. in the laboratory of W.N. was supported by an EMBO short-term fellowship (EE 3-1995). This work was supported by the Deutsche Forschungsgemeinschaft through the Schwerpunktprogramm Molekulare Zellbiologie der Hitzestressantwort, SCHW375/2-1, and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 2.Azpiroz R, Butow R A. Patterns of mitochondrial sorting in yeast zygotes. Mol Biol Cell. 1993;4:21–36. doi: 10.1091/mbc.4.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banecki B, Kaguni J M, Marszalek J. Role of adenine nucleotides, molecular chaperones and chaperonins in stabilization of active monomer form of DnaA initiator protein of Escherichia coli. Biochim Biophys Acta. 1998;1442:39–48. doi: 10.1016/s0167-4781(98)00118-3. [DOI] [PubMed] [Google Scholar]
- 4.Chattoraj D K. Role of molecular chaperones in the initiation of plasmid DNA replication. Genet Eng. 1995;17:81–98. [PubMed] [Google Scholar]
- 5.Cho J H, Ha S J, Kao L R, Megraw T L, Chae C B. A novel DNA-binding protein bound to the mitochondrial inner membrane restores the null mutation of mitochondrial histone Abf2p in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5712–5723. doi: 10.1128/mcb.18.10.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clayton D A. Nuclear gadgets in mitochondrial DNA replication and transcription. Trends Biochem Sci. 1991;16:107–111. doi: 10.1016/0968-0004(91)90043-u. [DOI] [PubMed] [Google Scholar]
- 7.Craig E A, Gambill B D, Nelson R J. Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol Rev. 1993;57:402–414. doi: 10.1128/mr.57.2.402-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis A F, Ropp P A, Clayton D A, Copeland W C. Mitochondrial DNA polymerase gamma is expressed and translated in the absence of mitochondrial DNA maintenance and replication. Nucleic Acids Res. 1996;24:2753–2759. doi: 10.1093/nar/24.14.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diffley J F, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Zamaroczy M, Marotta R, Faugeron-Fonty G, Goursot R, Mangin M, Baldacci G, Bernardi G. The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature. 1981;292:75–78. doi: 10.1038/292075a0. [DOI] [PubMed] [Google Scholar]
- 11.Dujon B. Mitochondrial genes and function. In: Broach J, Jones E, Strathern J, editors. The molecular biology of yeast Saccharomyces cerevisiae. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 505–635. [Google Scholar]
- 12.Eriksson S, Xu B, Clayton D A. Efficient incorporation of anti-HIV deoxynucleotides by recombinant yeast mitochondrial DNA polymerase. J Biol Chem. 1995;270:18929–18934. doi: 10.1074/jbc.270.32.18929. [DOI] [PubMed] [Google Scholar]
- 13.Fangman W L, Henly J W, Brewer B J. RPO41-independent maintenance of [rho−] mitochondrial DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:10–15. doi: 10.1128/mcb.10.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foury F. Cloning and sequencing of the nuclear gene MIP1 encoding the catalytic subunit of the yeast mitochondrial DNA polymerase. J Biol Chem. 1989;264:20552–20560. [PubMed] [Google Scholar]
- 15.Foury F, Roganti T, Lecrenier N, Purnelle B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998;440:325–331. doi: 10.1016/s0014-5793(98)01467-7. [DOI] [PubMed] [Google Scholar]
- 16.Fox T D, Folley L S, Mulero J J, McMullin T W, Thorsness P E, Hedin L O, Costanzo M C. Analysis and manipulations of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- 17.Genga A, Bianchi L, Foury F. A nuclear mutant of Saccharomyces cerevisiae deficient in mitochondrial DNA replication and polymerase activity. J Biol Chem. 1986;261:9328–9332. [PubMed] [Google Scholar]
- 17a.Germaniuk, A., and J. Marszalek. Unpublished data.
- 18.Gray M W, Burger G, Lang B F. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 19.Greenleaf A L, Kelly J L, Lehman I R. Yeast RPO41 gene product is required for transcription and maintenance of the mitochondrial genome. Proc Natl Acad Sci USA. 1986;83:3391–3394. doi: 10.1073/pnas.83.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. 194. Methods in enzymology. San Diego, Calif: Academic, Press, Inc.; 1991. [PubMed] [Google Scholar]
- 21.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 22.Herrmann J M, Stuart R A, Craig E A, Neupert W. Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J Cell Biol. 1994;127:893–902. doi: 10.1083/jcb.127.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrmann J M, Fölsch H, Neupert W, Stuart R A. Isolation of yeast mitochondria and study of mitochondrial protein translation. In: Celis J E, editor. Cell biology: a laboratory handbook. San Diego, Calif: Academic Press, Inc.; 1994. pp. 538–544. [Google Scholar]
- 24.Horst M, Oppliger W, Rospert S, Schonfeld H-J, Schatz G, Azem A. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 1997;16:1842–1849. doi: 10.1093/emboj/16.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang D S, Crooke E, Kornberg A. Aggregated DnaA protein is dissociated and activated for DNA replication by phospholipase or dnaK protein. J Biol Chem. 1990;26:19244–19248. [PubMed] [Google Scholar]
- 26.Johnson J L, Craig E A. Protein folding in vivo: unraveling complex pathways. Cell. 1997;90:201–204. doi: 10.1016/s0092-8674(00)80327-x. [DOI] [PubMed] [Google Scholar]
- 27.Kang P J, Ostermann J, Shilling J, Neupert W, Craig E A, Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- 28.Konieczny I, Marszalek J. The requirement for molecular chaperones in λDNA replication is reduced by the mutation π in λP gene which weakens the interaction between λP protein and DnaB helicase. J Biol Chem. 1995;270:9792–9799. doi: 10.1074/jbc.270.17.9792. [DOI] [PubMed] [Google Scholar]
- 29.Konieczny, I., and M. Zylicz. Role of bacterial chaperones in DNA replication. Genet. Eng., in press. [DOI] [PubMed]
- 30.Kornberg A, Baker A. DNA replication. W. H. New York, N.Y: Freeman; 1992. [Google Scholar]
- 31.Lancashire W E, Mattoon J R. Cytoduction a tool for mitochondrial genetic studies in yeast. Mol Gen Genet. 1979;170:333–344. doi: 10.1007/BF00267067. [DOI] [PubMed] [Google Scholar]
- 32.Langer T, Neupert W. Chaperoning mitochondrial biogenesis. In: Morimoto R I, Tissieres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 53–83. [Google Scholar]
- 33.Laloraya S, Gambill B D, Craig E A. A role for a eukaryotic GrpE-related protein, Mge1p, in protein translocation. Proc Natl Acad Sci USA. 1994;91:6481–6485. doi: 10.1073/pnas.91.14.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lockshon D, Zweifel S G, Freeman-Cook L L, Lorimer H E, Brewer B J, Fangman W L. A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell. 1995;16:947–955. doi: 10.1016/0092-8674(95)90014-4. [DOI] [PubMed] [Google Scholar]
- 35.Lorimer H E, Brewer B J, Fangman W L. A test of the transcription model for biased inheritance of yeast mitochondrial DNA. Mol Cell Biol. 1995;15:4803–4809. doi: 10.1128/mcb.15.9.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maleszka R, Skelly P J, Clark-Walker G D. Rolling circle replication of DNA in yeast mitochondria. EMBO J. 1991;10:3923–3929. doi: 10.1002/j.1460-2075.1991.tb04962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers A M, Pape L K, Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 39.Prip-Buus C, Westermann B, Schmitt M, Langer T, Neupert W, Schwarz E. Role of the mitochondrial DnaJ homologue, Mdj1p, in the prevention of heat-induced protein aggregation. FEBS Lett. 1996;380:142–146. doi: 10.1016/0014-5793(96)00049-x. [DOI] [PubMed] [Google Scholar]
- 40.Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, Neupert W. Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell. 1994;77:249–259. doi: 10.1016/0092-8674(94)90317-4. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Schmitt M E, Clayton D A. Conserved features of yeast and mammalian mitochondrial DNA replication. Curr Opin Genet Dev. 1993;3:769–774. doi: 10.1016/s0959-437x(05)80097-8. [DOI] [PubMed] [Google Scholar]
- 43.Van Dyck E, Foury F, Stillman B, Brill S J. A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J. 1992;11:3421–3430. doi: 10.1002/j.1460-2075.1992.tb05421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Dyck E, Clayton D A. Transcription-dependent DNA transactions in the mitochondrial genome of a yeast hypersuppressive petite mutant. Mol Cell Biol. 1998;18:2976–2985. doi: 10.1128/mcb.18.5.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner I, Arlt H, van Dyck L, Langer T, Neupert W. Molecular chaperones cooperate with PIM1 protease in the degradation of misfolded proteins in mitochondria. EMBO J. 1994;13:5135–5145. doi: 10.1002/j.1460-2075.1994.tb06843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westermann B, Prip-Buus C, Neupert W, Schwarz E. The role of the GrpE homologue, Mge1p, in mediating protein import and protein folding in mitochondria. EMBO J. 1995;14:3452–3460. doi: 10.1002/j.1460-2075.1995.tb07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westermann B, Gaume B, Herrmann J M, Neupert W, Schwarz E. Role of the mitochondrial DnaJ homolog Mdj1p as a chaperone for mitochondrially synthesized and imported proteins. Mol Cell Biol. 1996;16:7063–7071. doi: 10.1128/mcb.16.12.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziemienowcz A, Zylicz M, Floth C, Hubscher U J. Calf thymus Hsc70 protein protects and reactivates prokaryotic and eukaryotic enzymes. J Biol Chem. 1995;270:15479–15484. doi: 10.1074/jbc.270.26.15479. [DOI] [PubMed] [Google Scholar]
- 49.Zylicz M. The Escherichia coli chaperones involved in DNA replication. Phil Trans R Soc Lond B. 1992;339:271–278. doi: 10.1098/rstb.1993.0025. [DOI] [PubMed] [Google Scholar]
- 50.Zylicz M, Wawrzynow A, Marszalek J, Liberek K, Baneck B, Konieczny I, Blaszczak A, Barski P, Jakobkiewicz J, Gonciarz-Swiatek M, Duchniewicz M, Puzewicz J, Krzewska J. Role of chaperones in replication of bacteriophage lambda DNA. In: Bukau B, editor. Molecular chaperones and folding catalysts: Regulation, cellular function and mechanisms. New York, N.Y: Harwood Academic Publishers; 1999. pp. 295–311. [Google Scholar]