Abstract
Recruitment of transcriptional coactivators following ligand activation is a critical step in nuclear receptor-mediated target gene expression. Upon binding an agonist, the receptor undergoes a conformational change which facilitates the formation of a specific coactivator binding pocket within the carboxyl terminus of the receptor. This permits the α-helical LXXLL motif within some coactivators to interact with the nuclear receptors. Until recently, the LXXLL motif was thought to function solely as a docking module; however, it now appears that sequences flanking the core motif may play a role in determining receptor selectivity. To address this issue, we used a combinatorial phage display approach to evaluate the role of flanking sequences in influencing these interactions. We sampled more than 108 variations of the core LXXLL motif with estradiol-activated estrogen receptor alpha (ERα) as a target and found three different classes of peptides. All of these peptides interacted with ERα in an agonist-dependent manner and disrupted ERα-mediated transcriptional activity when introduced into target cells. Using a series of ERα-mutants, we found that these three classes of peptides showed different interaction patterns from each other, suggesting that not all LXXLL motifs are the same and that receptor binding selectivity can be achieved by altering sequences flanking the LXXLL core motif. Most notable in this regard was the discovery of a peptide which, when overexpressed in cells, selectively disrupted ERβ- but not ERα-mediated reporter gene expression. This novel ERβ-specific antagonist may be useful in identifying and characterizing the ERβ-regulated process in estradiol-responsive cells. In conclusion, using a combinatorial approach to define cofactor-receptor interactions, we have clearly been able to demonstrate that not all LXXLL motifs are functionally equivalent, a finding which suggests that it may be possible to target receptor-LXXLL interactions to develop receptor-specific antagonists.
The nuclear receptor superfamily consists of many sequence-related transcription factors that initiate and coordinate the responses to a wide range of physiological signals (13, 24). A simplified model of transcriptional activation by these receptors involves activation of the receptors by their cognate ligands, recruitment of the receptor homo- or heterodimers to target DNA sequences, and subsequent modulation of gene transcription upon interaction with the general transcription machinery. It now appears, however, that nuclear receptor action is more complicated. For instance, most of these receptors are associated with corepressor proteins that silence their activity in the absence of ligands, and activation therefore involves displacement of the associated corepressors by coactivators, an event that permits the functional interaction of the receptor with the cellular transcription machinery (8, 17). Thus, the nature and abundance of these receptor-associated proteins may be a primary determinant of nuclear receptor pharmacology.
A number of coactivators such as SRC-1/NCoA-1 (5, 30), GRIP-1/TIF-2/NCoA2 (16, 48), p/CIP/AIB-1/ACTR (1, 7, 23, 46), and CBP/p300 (9, 12) have been identified and shown to be important for nuclear receptor transactivation. All of these proteins contain a signature LXXLL motif (NR box) which is necessary and sufficient to permit the interaction between receptors and coactivators (15). Results from cocrystallization studies of LXXLL-containing peptides with the ligand-activated hormone binding domains (HBD) of ER and PPARγ demonstrated that these motifs fit into a groove formed by helices 3, 4, 5, and 12 on the receptor (26, 41). Although these structures provided valuable insight into how coactivators dock with steroid hormone receptors, they did not indicate how selectivity of one receptor for a specific LXXLL motif is achieved. It is clear from previous work that each coactivator has specific receptor preferences (11, 15, 19, 25, 49) and that understanding the basis for this selectivity may permit the design of strategies that could be used to target specific receptor-cofactor interactions with novel pharmaceuticals. Preliminary studies, which focused on this problem, have revealed that the two internal residues flanked by leucines within the NR core do not have direct contact with the receptor and do not appear to be important for receptor binding (15, 26, 41). Classical site-directed and alanine-scanning mutagenesis has been used to evaluate how the LXXLL motif interacts with the nuclear receptors and to identify the sequences within the short motif that govern affinity and specificity (11, 15, 19, 25, 49). These studies revealed that sequences N- and C-terminal to the LXXLL motif appear to have the greatest impact on their receptor selectivity and binding affinity (25). However, because of the limited sampling permitted by traditional mutagenesis approaches, it has not been possible to adequately address the issue of LXXLL specificity and selectivity. For this reason, we have used phage display technology to screen a large combinatorial peptide library in which more than 108 combinations of the LXXLL motif was created. This library was then used to probe the nature of the ER-coactivator interaction with a view to identifying the sequences surrounding the LXXLL core motif that are responsible for receptor selectivity and affinity.
Phage display technology has been used successfully in the past to search for peptide sequences that mimic endogenous protein-protein interactions (20, 35, 44). In a previous study, we used this technology to screen for ER-interacting motifs with random peptide libraries and found that LXXLL-containing peptides formed a major sequence cluster when estradiol-activated ER was used as a target (32). Taken together, these data suggested that (i) the information within a short peptide is sufficient to confer specific protein-protein interactions and (ii) the LXXLL motifs appear to be a dominant feature utilized by coactivators to enable them to interact with ligand-activated nuclear receptors. In this study, we further dissected the mechanisms governing the LXXLL motif-ER interactions. Using a phage library enriched for LXXLL-containing peptides to screen for ER interaction sequences, we identified three different subclasses of peptides. All of these peptides interacted with ER in an agonist-dependent manner and mimicked the interaction of coactivators with ER. They differed, however, in their ability to interact with different ER mutants and with other steroid receptors.
MATERIALS AND METHODS
Abbreviations.
ERα, estrogen receptor alpha; ERβ, estrogen receptor beta; GR, glucocorticoid receptor; PR-A and PR-B, progesterone receptor isoforms A and B; AR, androgen receptor; TRβ, thyroid hormone receptor beta; RARα, retinoic acid receptor alpha; RXRα, retinoid X receptor alpha; VDR, 1,25-(OH)2-vitamin D3 receptor; PPARγ, peroxisome proliferator-activated receptor γ; GRIP-1, glucocorticoid receptor interacting protein 1; SRC-1, steroid receptor coactivator 1; RIP140, receptor-interacting protein 140; TRAP220, the 220-kDa TR-associated protein; DAX-1, dose-sensitive sex reversal-AHC critical region on the X chromosome gene 1; SHP, short heterodimer partner; PGC-1, PPARγ coactivator 1; HBD, hormone binding domain; PBS, phosphate-buffered saline; BSA, bovine serum albumin; SDS, sodium dodecyl sulfate; PAGE, polyacrylamide gel electrophoresis; Gal4DBD, Gal4 DNA binding domain; SERM, selective estrogen receptor modulator.
Chemicals.
17β-estradiol, 4-hydroxytamoxifen, 9-cis-retinoic acid, dexamethasone, diethylstilbesterol, 5α-dihydrotestosterone, T3 (3,3′,5-triiodo-l-thyronine), and progesterone were obtained from Sigma Chemical Co. (St. Louis, Mo.); Δ8,9-dehydroestrone, equilin, and estrone were kindly provided by M. Dey (Wyeth-Ayerst Pharmaceuticals, Radnor, Pa.); ICI 182,780 was a gift from A. Wakeling (Zeneca Pharmaceuticals, Macclesfield, United Kingdom); GW7604 was provided by T. Willson (Glaxo Wellcome Research and Development, Research Triangle Park, N.C.); and 1,25-dihydroxyvitamin D3 was purchased from Duphar Pharmaceuticals (Daweesp, The Netherlands).
Cell culture and transient transfection.
Human cervical cancer (HeLa) and hepatoma (HepG2) cells were cultured in minimum essential medium (Life Technologies, Inc.) supplemented with 10% fetal bovine serum (HyClone), 0.1 mM nonessential amino acids, and 1 mM sodium pyruvate (Life Technologies, Inc.) and maintained in a humidified 37°C incubator with 5% CO2. For transient transfections, cells were split into 24-well plates 24 h before transfection. Lipofectin (Life Technologies, Inc.)-mediated transfection has been described in detail previously (27). A DNA-Lipofectin mixture containing a total of 3,000 ng of plasmid in each of triplicate samples was incubated with cells for 3 to 5 h, and transfection was stopped by replacing the transfection mix with fresh medium (minimal essential medium without phenol red) containing 10% charcoal-stripped serum. Receptor ligands were added to the cells 14 to 16 h before the assay. Luciferase and β-galactosidase activities were measured as described (27). In mammalian two-hybrid assays, for a typical triplicate of transfection, 2,000 ng of 5×Gal4Luc3 reporter plasmid, 400 ng of receptor-VP16 fusion, 400 ng of pM (Gal4DBD)-peptide fusion constructs, and 200 ng of normalization plasmid pCMVβgal were used. For ER transcription disruption assays, 1,600 ng of 3×ERE-TATA-Luc reporter, 200 ng of pCMVβgal, 400 ng of either pRST7ERα, pRST7ERβ, or other receptor mutant constructs, and 0 to 800 ng of pM-peptide fusion plasmids were used as indicated in the figure legend. The parent pM vector (Gal4DBD without peptide fusion) was used in these experiments to balance the amount of input DNA in transfections. All transfections were performed at least three times; data shown are results of representative experiments.
Construction of the phage library.
A focused peptide library in the format of (X)7LXXLL(X)7, where X is any amino acid and L is leucine, was constructed essentially as described previously with the M13 phage-based cloning vector mBAX (43). The top-strand oligonucleotide 5′-AGTGTGTGCCTCGAGA(NNK)7CTG(NNK)2CTGCTG(NNK)7TCTAGACTGTGCAGT-3′ (N = A, C, G, or T; K = C or T) was purchased from Life Technologies, gel purified, and annealed to its complementary-strand oligonucleotide, 5′-ACTGCACAGTCTAGA-3′. The resulting DNA complex was extended with Klenow polymerase in the presence of deoxynucleoside triphosphates to generate double-stranded DNA and was subsequently digested with XhoI and XbaI and ligated into the mBAX vector, previously digested with the same restriction enzymes. The ligated products were electroporated into Escherichia coli JS-5 cells and amplified on 2YT (Life Technologies, Inc.) plates for 6 h to create the (X)7LXXLL(X)7 peptide library. The amplified phage were then eluted from the plates with PBS, concentrated, and finally resuspended in 20% glycerol–PBS and stored at −70°C in 500-μl aliquots. The library has a complexity of 1.5 × 108 different peptide sequences.
Affinity selection of ERα-binding sequences.
Baculovirus-expressed full-length ERα was provided by PanVera Corp. (Madison, Wis.). Approximately 0.25 μg (4 pmol) of ERα was diluted in 100 μl of NaHCO3 (pH 8.5) plus 10−6 M 17β-estradiol, applied to a single well in a 96-well Immulon 4 plate (Dynex Technologies, Inc.), and incubated at room temperature for 3 h. An equal amount of BSA was added to the adjacent well as a control target. The wells were blocked with 150 μl of 0.1% BSA in NaHCO3 for an additional 1 h at room temperature and washed five times with PBST (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4 [pH 7.3], 0.1% Tween 20) to remove excess protein. Then 25 μl of the phage peptide library (with >1010 phage particles) diluted in 125 μl of PBST with 10−6 M 17β-estradiol and 0.1% BSA was added to the wells, and the plate was sealed and incubated for 8 h at room temperature. Nonbinding phage were removed by washing the wells five times with PBST. The bound phage were eluted with 100 μl of prewarmed (50°C) 50 mM glycine-HCl (pH 2.0) followed by 100 μl of 100 mM ethanolamine (pH 11.0). The first eluent was neutralized by adding 200 μl of 200 mM Na2HPO4 (pH 8.5) and combined with the second eluent. Phage eluted from the targets were amplified in E. coli DH5αF′ cells for 8 h, and the supernatant containing amplified phage was collected for use in subsequent rounds of panning. A total of three rounds of panning were performed. Enrichment of ER binding phage was confirmed by enzyme-linked immunosorbent assay as described below. Individual phage were plaque purified after the third panning, and the peptide sequences were deduced by DNA sequencing.
Enzyme-linked immunosorbent assay.
Full-length ERα (0.4 pmol per well) was activated by different ER ligands and coated on 96-well Immulon 4 plates as described above. Then 50 μl of phage stock was applied to the wells and incubated with the targets for 1 h at room temperature. Unbound phage were removed by five washes with PBST. A 1:5,000 dilution of horseradish peroxidase-conjugated anti-M13 antibody (Amersham)–PBST was added to the wells, and the mixture was incubated for 1 h at room temperature and then washed five times with PBST. Bound antibody-enzyme conjugate was detected by ABTS (2′,2′-azino-bis-3-ethylbenzthiazoline-6-sulfonic acid) in the presence of 0.05% H2O2, and the color change was measured at 405 nm on a plate reader (Multiskan MS; Labsystems).
Plasmids.
All the Gal4DBD-peptide fusions were constructed as follows. DNA sequences coding for the peptides were excised from mBAX vector with XhoI and XbaI and subcloned into the pMsx vector (derived from the pM vector [Clontech] with a linker sequence to generate in-frame SalI and XbaI sites for cloning). The fusion constructs expressing two copies of the LXXLL motifs, 2×F6 and 2×293, were derived from their corresponding single-copy peptide-DBD fusion plasmids by adding a linker sequence (adapted from the sequences found between the GRIP-1 NR box 2 and box 3). Subsequently, a second copy of the LXXLL peptide was added, resulting in the two copies of LXXLL motifs being separated by 50 amino acids, the same spacing found between the GRIP-1 NR box 2 and box 3. The pVP16ERα construct was generated by PCR of the full-length human ERα cDNA with primers containing EcoRI sites flanking both 5′ and 3′ ends, and the resulting PCR product was subcloned into the EcoRI site in the pVP16 vector (Clontech). pVP16ERβ, pVP16RARα, and pVP16RXRα were generated in a similar fashion. pVP16VDR was a gift of J. W. Pike (University of Cincinnati, Cincinnati, Ohio); VP16TRβ expression plasmid (pCMX-VP-F-hTRβ) was provided by D. D. Moore (Baylor College of Medicine, Houston, Tex.); and VP16GR, VP16PR-A, VP16PR-B, and VP16AR expression plasmids were gifts from J. Miner (VP16GR), D. X. Wen (VP16PR-A and VP16PR-B), and K. Marschke (VP16AR) (Ligand Pharmaceuticals, San Diego, Calif.). Plasmids expressing VP16-ERα mutants were constructed by excision of mutant ER cDNAs from their corresponding expression plasmids (ER-TAF1 [ERα-3×], ERα-LL, and ERα-535 stop plasmids [28, 47]) and subcloned into the pVP16 vector. The VP16-ERα point mutants (ER-D538N, ER-E542Q, and ER-D545N) were generated by using the QuikChange site-directed mutagenesis kit (Stratagene) with wild type pVP16-ERα as a template. Mammalian expression plasmids for ERα, ERβ, and ER179C, as well as the 3×ERE-TATA-Luc reporter construct, are described elsewhere (47). The 5×Gal4Luc3 plasmid was modified from 5×Gal4-TATA-Luc (a gift from X. F. Wang, Duke University, Durham, N.C.) by replacing the luciferase gene with a modified version of luciferase cDNA from the pGL3 basic vector (Promega). GRIP-1 (NR-box) and SRC-1 (NR-box) constructs were generated by subcloning PCR products corresponding to GRIP-1 amino acids 629 to 760 and SRC-1 amino acids 621 to 765 into the pM vector (13a). All PCR products were sequenced to ensure the fidelity of the resultant constructs. An expression plasmid for TRAP220 (pCIN4-TRAP220) was provided by R. Roeder (Rockefeller University, New York, N.Y.). Full-length GRIP-1 and RIP140 expression plasmids were made in the pcDNA3 vector (Invitrogen) by ligating full-length GRIP-1 and RIP140 cDNAs excised from pGRIP1/fl (provided by M. Stallcup, University of Southern California, Los Angeles, Calif.) and pEF-RIP140 (provided by M. Parker, Imperial Cancer Research Fund, London, United Kingdom), respectively.
Western blot analysis.
Western blotting was performed with nuclear extracts isolated from HeLa cells transfected with each of the Gal4DBD-peptide fusion plasmids together with a green fluorescent protein expression vector (pEGFP-C3) for normalization purposes. Nuclear extracts were prepared as described previously (38). A 20-μg portion of protein from each extract was separated on an SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories). The blots were first probed with an anti-Gal4DBD monoclonal antibody (Santa Cruz Biotechnology, Inc.) to detect peptide fusions and subsequently probed with an anti-green fluorescent protein polyclonal antibody (Clontech) to detect the coexpressed EGFP. The immunocomplexes were visualized by enhanced chemiluminescence (Amersham Corp.) as specified by manufacturer.
Receptor-cofactor in vitro pulldown assays.
A 4-pmol quantity of baculovirus-expressed full-length ERα or ERβ (each obtained from Panvera) was immobilized on Immulon 4 plates and blocked as described above. An equal amount of BSA was added to the adjacent wells as a “no-receptor” control target. 35S-labeled RIP140, GRIP-1, and TRAP220 were translated in vitro with the TNT-coupled reticulocyte lysate system (Promega Corp.) from their mammalian expression plasmids described above. Then 8-μl volumes of the translated proteins were added to 96-well plates containing immobilized ERα, ERβ, or BSA and incubated at 4°C overnight. The wells were washed five times with PBST to remove unbound protein, and the bound protein was eluted by adding prewarmed (80°C) SDS-PAGE sample buffer and incubated at 80°C for 5 min. The supernatant was collected and boiled for 5 min before being separated on an SDS-polyacrylamide gel. The gel was dried, and the signals were detected by autoradiography.
RESULTS
Affinity selection of ligand-dependent ER binding peptides.
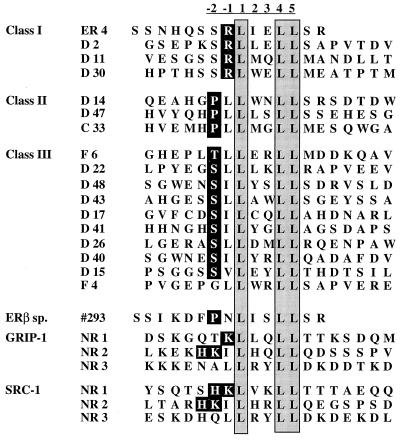
The transcriptional activity of ER within target cells is influenced by its ability to interact with specific factors that decrease (corepressors) or increase (coactivators) its transcriptional activity (42). Over the past few years, the application of various molecular biology approaches has led to the discovery of co-activators that interact with the nuclear receptor HBD through a conserved LXXLL motif in a ligand-dependent manner. In this study, we used a combinatorial phage display approach to determine how flanking sequences influence the LXXLL motif-receptor interactions. The advantages of using this approach are twofold: a vast number of sequences can easily be assessed, and, more importantly, sequences obtained from this type of screening often reflect sequences that can be found in nature (35, 44). Specifically, a 19-mer phage “focused” library in which the LXXLL motif was flanked on each side by seven random amino acid residues was constructed. The resulting phage library was used to select for peptides that bound with high affinity to estradiol-activated ERα. Phage particles that bound specifically to ERα in a ligand-dependent manner were selected and amplified, and the amino acid sequences were deduced following DNA sequencing. Figure 1 shows representative peptide sequences derived from the isolated phage. Based on sequences flanking the core LXXLL motif, three different sequence clusters have emerged. Class I peptides contain a conserved serine at the −2 position and a positively charged residue (R) at the −1 position. Class II peptides have a proline occupying the −2 position and a hydrophobic leucine (L) residue directly preceding the LXXLL motif. Two of the three peptides in class II also contain a charged histidine (H) at the −3 position, and this histidine appears to have an influence on their binding characteristics (see Discussion). Class III peptides share a conserved serine (S) or threonine (T) at the −2 position followed by a hydrophobic leucine (L) or isoleucine (I) at the −1 position. In these initial characterizations, we used the intact bacteriophage to evaluate the ERα binding properties of these peptide sequences. To show that the peptide alone is both necessary and sufficient for ER binding, we subcloned representative members of each class of peptides as fusion proteins to bacterial alkaline phosphatase (50) and demonstrated that the purified recombinant peptide-enzyme fusions interacted specifically with ERα (data not shown).
FIG. 1.
Affinity selection of ERα binding motifs by using phage display technology. Baculovirus-expressed full-length ERα was treated with 10−6 M 17β-estradiol and immobilized on 96-well Immulon 4 plates as a screening target. The LXXLL motif-containing phage peptide library was constructed as described in Materials and Methods. Phage that interacted specifically with estradiol-activated ER were selected, and the peptide sequences were deduced by DNA sequencing. These peptides were classified into three different classes based on sequences flanking the conserved LXXLL motif. Peptide #293 was obtained in a similar manner from random peptide libraries; it bound specifically to estradiol-activated ERβ when analyzed in vitro. Sequences from the center three copies of LXXLL motifs in the SRC-1 and GRIP-1 coactivators are also included for comparison. For reference, we have defined the first conserved leucine as position 1.
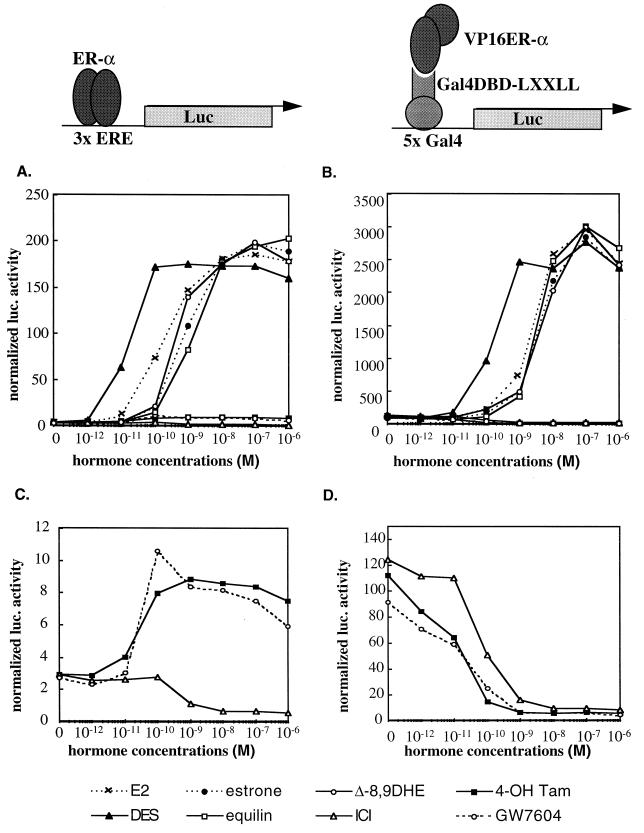
We next developed a series of mammalian two-hybrid assays to confirm that the LXXLL-containing peptides identified could interact with ERα in the context of the intact cell. For this purpose, full-length ERα was expressed as a fusion protein with the VP16 acidic activation domain and the peptide sequences were produced as fusions with the yeast Gal4DBD. Interaction between ERα-VP16 and the LXXLL-Gal4DBD fusions was assessed by using the 5×Gal4Luc3 luciferase reporter gene, which contains five copies of the Gal4 responsive element upstream of a simple TATA box. Shown in Fig. 2 are comparisons of the abilities of different ligands to activate ERα transcription through a classical ER responsive element (Fig. 2A) and their ability to facilitate the interaction of the LXXLL peptide (class I-ER4) with ER (Fig. 2B). All steroidal and nonsteroidal ER agonists strongly activated transcription from the 3×-ERE-TATA-Luc reporter (Fig. 2A), while the SERMs 4-hydroxytamoxifen and GW7604 displayed minimal agonist activity within this promoter context (Fig. 2C). The pure antagonist ICI 182,780, as expected, functioned as an inverse agonist that suppressed the transcription below the basal, no-hormone treatment level (Fig. 2C). When analyzing the interaction between the LXXLL motif and ERα, we observed a low but significant basal level of interaction in the absence of any ligand treatment, indicating that some of the expressed ERα is already in an active conformation, allowing the LXXLL peptide to interact. At present, we do not know whether this basal activity is caused by residual estrogens present in the charcoal-stripped serum or is due to alternative pathways that activate ER-mediated transcription. However, we observed that above the basal level, the interaction of the LXXLL peptide with ERα was entirely ER agonist dependent. The ability of both steroidal and nonsteroidal ER agonists to promote the ERα-LXXLL peptide interaction parallels the ability of these compounds to activate ERα-mediated transcription through a classical ER–ERE-mediated pathway. This indicates that all of these compounds are mechanistically similar, inducing similar conformational changes within ERα, and that within target cells these ligand-receptor complexes are likely to recruit the same coactivators. Interestingly, none of the ER antagonists or SERMs tested were able to facilitate ERα-LXXLL interactions. The pure antagonist ICI 182,780 totally abolished both basal peptide-ERα interactions and ERα-mediated transcription (Fig. 2C and D). In addition, although SERMs such as 4-hydroxytamoxifen and GW7604 can manifest partial agonist activity in certain cell types and promoter contexts (Fig. 2C and data not shown), in this experiment they actually drove the receptor into a conformation which prohibited LXXLL peptide-ERα interactions from occurring. As a result, the basal level of interaction between ERα and peptides containing the LXXLL motif was abolished in the presence of these compounds (Fig. 2D). The crystal structures of raloxifene-, tamoxifen-, and estradiol-activated ERα HBD have recently been solved and indicate that the coactivator binding groove within the receptor is occupied by a mispositioned helix 12 upon antagonist binding (4, 41). Helix 12 of the receptor thus prevents the coactivator LXXLL motif from interacting. Although some of our peptides seem to bind strongly to ERα in the presence of estradiol, none of them were able to interact with ERα in the presence of any of the SERMs tested, including 4-hydroxytamoxifen, nafoxidine, raloxifene, GW7604, and clomiphene (data not shown). Therefore, the partial agonist activity manifested by these compounds in some cells is likely to require cofactors distinct from those required by estradiol-activated ER (29). These data support the notion that the ability to facilitate the interaction of ER with LXXLL-containing coactivators is a fundamental step common to both ligand-dependent and basal transcriptional activity mediated by ERα. The observation that ER-peptide interactions do not occur in the presence of ER antagonists or mixed agonists may explain why compounds like tamoxifen and ICI 182,780 can inhibit both basal and ligand-dependent activation of ER. We also conducted the same analysis with other LXXLL-containing peptides and observed similar results (data not shown).
FIG. 2.
The interaction between LXXLL-containing peptides and ER occurs only in the presence of receptor agonists. The LXXLL-containing ER4 peptide sequence was fused to Gal4DBD, while the full-length ERα was expressed as a VP16 transactivation domain fusion protein. The interaction between ER4 peptide and ERα was assessed by using the 5×Gal4Luc3 reporter gene (B and D). The ability of different ER ligands to facilitate LXXLL peptide-ERα interactions was compared to the ability of these ligands to induce ER-mediated transactivation, as assayed by using the 3×ERE-TATA-Luc reporter (A and C). HepG2 cells were transiently transfected with the ERα expression vector (pRST7ERα) and its reporter 3×ERE-TATA-Luc construct (A and C) or Gal4DBD-ER4, pVP16-ERα, and 5×Gal4Luc3 (B and D) and treated with different ER ligands as indicated in the key. Luciferase (Luc) activity was normalized to the activity of the cotransfected pCMVβgal plasmid. E2, 17β-estradiol; 4-OH Tam, 4-hydroxytamoxifen; ICI, ICI 182,780; DES, diethylstilbesterol; Δ-8,9DHE, delta-8,9-dehydroestrone.
Not all LXXLL motifs are functionally equivalent.
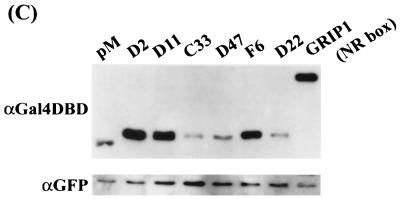
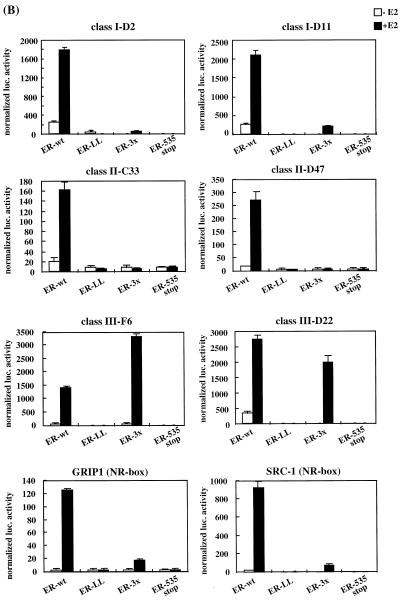
We next examined whether all of the LXXLL-containing peptides selected by using phage display were functionally equivalent. The previously defined ternary structures of the LXXLL motifs cocrystallized with either the ERα or PPARγ HBD indicated that these motifs bind to a hydrophobic groove created by helices 3, 4, 5, and require an intact helix 12 (26, 41). Therefore, the ability of the LXXLL motifs identified to interact with the coactivator binding groove was assessed by using a modified mammalian two-hybrid assay. Several ERα mutants with alterations in helix 12 as well as the wild-type ERα were produced as VP16 fusion proteins to test their ability to recruit LXXLL motifs (Fig. 3A). We found that all of the peptides tested interacted with wild-type ERα in a ligand-dependent fashion. As expected, the middle three copies of the LXXLL motif (NR box) found in the coactivators SRC-1 and GRIP-1 also interacted in a similar fashion (Fig. 3B, ER-wt). Western analysis showed that different classes of peptide-Gal4DBD fusion proteins have different expression levels in the cells; therefore, the data presented in this assay can be used to compare only their binding patterns, not their relative binding affinities (Fig. 3C). For instance, the class II peptides interacted with ERα with relatively higher affinity than did the class I and III peptides in the in vitro binding assays (data not shown). The expression levels of these peptides, however, are much lower than those of the other classes of peptides, which may explain the observed lower readout in the mammalian two-hybrid assays. Regardless, the mammalian two-hybrid assay remains a useful tool to characterize the in vivo interactions between ERα and the peptides.
FIG. 3.
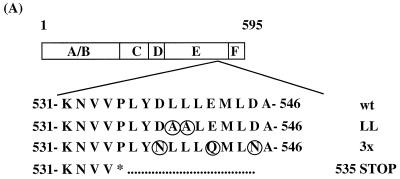
Not all LXXLL peptide-ER interactions require a functional AF-2. The three groups of LXXLL-containing peptides interacted differentially with ER helix 12 mutants. (A) A schematic drawing of the wild-type (wt) ER is shown along with a region of the HBD corresponding to ER activation function 2 (AF-2). Residues that were mutated are indicated by circles. (B) Mammalian two-hybrid assays were used to test whether all the LXXLL motifs interacted with the same region of ER. Peptide sequences representing three LXXLL classes were expressed as fusion proteins to the Gal4DBD. Wild-type (wt) and mutant ERα were expressed as VP16 fusion proteins. The binding capacity of different peptides to wild-type and mutant ER was measured by using a 5×Gal4Luc3 reporter construct. GRIP-1 (NR-box) and SRC-1 (NR-box) constructs contain the center three copies of an LXXLL motif (amino acids 629 to 760 for GRIP-1 and 621 to 765 for SRC-1) fused to Gal4DBD. (C) Western analysis of the expression levels of selected Gal4DBD-peptide fusions. Nuclear extracts were prepared from transfected HeLa cells and analyzed using SDS-PAGE. The peptide-Gal4DBD fusion proteins were detected with a monoclonal antibody raised against Gal4DBD (αGal4DBD). The expression levels of the Gal4DBD fusions were normalized by assaying the levels of EGFP expressed from a cotransfected plasmid (pEGFP-C3). Specifically, the identical blot was reprobed with a polyclonal anti-GFP antibody (αGFP).
Truncation of ER helix 12 (ER535 stop) does not affect ligand binding or dimerization; however, the ability of the receptor to interact with any LXXLL peptides was totally abolished. This was consistent with the observation that helix 12 is required to form the coactivator binding groove, and, more importantly, it implied that all the affinity-selected LXXLL-containing peptides bind to the same coactivator binding groove. Furthermore, mutation of a pair of the hydrophobic residues in helix 12 (L539L540→A539A540) significantly decreased the ERα transcriptional activity and also abolished the interaction of ERα with all of the LXXLL peptides tested (ER-LL in Fig. 3B).
Previously, we and others have demonstrated that alteration of the three charged residues in ERα helix 12 (D538E542D545→N538Q542N545; ER-3×) abolishes ERα transcriptional activity in most cell types (10, 28, 47) and prevents the interaction of GRIP-1- and SRC-1-type coactivators with ERα. Predictably, in our experiments, the interaction of the ERα-3× receptor mutant with the GRIP-1 and SRC-1 NR boxes was significantly lower than that of the wild-type receptor (Fig. 3B, ER-3×). The ability of class I and II peptides to interact with ERα was also prevented by these specific ERα helix 12 mutations, indicating that they may bind to ERα in a manner which is similar to that of the GRIP-1 and SRC-1 LXXLL motifs. In contrast, the interactions between class III peptides and ERα was not affected by these mutations. Importantly, the ERα-3× mutant is fully functional in certain cell types, which is interesting in light of the observed weak interaction of this receptor with coactivators like SRC-1 and GRIP-1. Our observations suggest, however, that the activity exhibited by this mutant receptor might be the result of its interaction with cofactors containing class III type LXXLL motifs. Regardless, however, it appears that the LXXLL motif is not merely a receptor-cofactor docking sequence but also contains information that governs the specificity of these interactions.
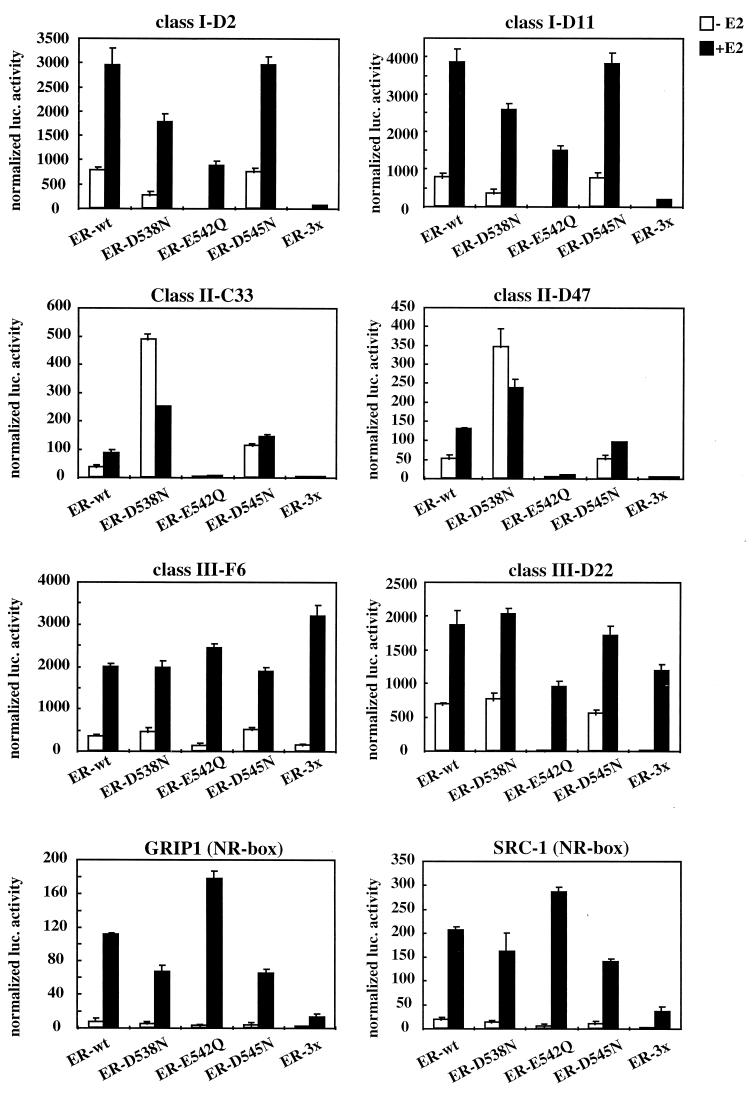
To further characterize the interactions between ERα and these three classes of peptides, we made individual mutations within the ER-3× to evaluate the relative contributions of each of the three charged residues (D538, E542, and D545) in ER-LXXLL motif interactions. This analysis revealed that the diminished interaction of class I peptides with ER-3× seems to be the sum of changing Asp-538 and Glu-542 to their corresponding amides; however, the change of Glu-542 to Gln-542 had the greatest impact on this interaction (Fig. 4). Glu-542 also appears to be the most important residue in determining the interaction between ERα and class II peptides, since mutation of this residue led to a total loss of interaction. Interestingly, changing Asp-538 to Asn-538 increased the binding of ERα with the class II peptides; however, this was observed to occur in a ligand-independent manner. Predictably, none of the mutations appear to have affected the ability of ERα to recruit class III peptides, consistent with the notion that ERα might interact with this class of peptides in a specific manner. The interaction patterns of ERα with GRIP-1 and SRC-1 NR boxes are similar to each other, in that none of the individual residue changes had a significant impact on the strength of the interaction. Replacing all three residues, however, greatly reduced the ability of ERα to bind to these NR boxes. The precise mechanism of interaction of ERα with these peptides can be resolved only by studying the cocrystal structure of these complexes. The results of these assays, nevertheless, once again highlight the fact that not all LXXLL motifs interact with ERα in the same manner.
FIG. 4.
The interaction of ERα with each of the three classes of LXXLL peptides identified is affected differentially by helix 12 mutations. The contributions of each of the three charged residues (D538, E542, D545) within helix 12 to LXXLL motif-ERα interactions were evaluated. Specifically, we created single point mutations of each residue to their corresponding amides and evaluated the impact of these mutations on ERα-LXXLL peptide interactions in a mammalian two-hybrid assay. The mutants indicated were generated by site-directed mutagenesis within the wild-type (wt) VP16-ERα backbone. Selected peptide sequences representing each of the three LXXLL classes were expressed as Gal4DBD fusions. The binding capacity of the different peptides to wild-type and mutant ER was measured by using a 5×Gal4Luc3 reporter construct. GRIP-1 (NR-box) and SRC-1 (NR-box) constructs contain the center three copies of an LXXLL motif (amino acids 629 to 760 for GRIP-1 and 621 to 765 for SRC-1) fused to Gal4DBD.
LXXLL-containing peptides can disrupt ERα transcriptional activity in the target cells.
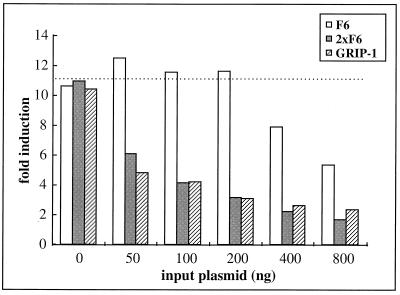
If peptides obtained from phage display are in fact mimicking the interactions between ERα and endogenous cofactors, they should function in a dominant negative manner when coexpressed in cells, disrupting these interactions and blocking the ER transcriptional activity. Coexpression in HeLa cells of the peptide F6-Gal4DBD fusion did indeed decrease the estradiol-induced ER-dependent reporter gene expression to approximately 50% of that without the peptide (Fig. 5, F6). We have also tested other peptides from all three classes and found that all the LXXLL peptides we obtained were able to disrupt ER transcriptional activity in a similar manner (data not shown). It was suggested previously (26) that multiple copies of the NR boxes in GRIP-1 and SRC-1 can bind to ERα in a synergistic manner. Thus, as expected, expression of the center three copies of the NR boxes from GRIP-1 permitted a more effective inhibition of ER-mediated transcription than did expression of a single-copy peptide (Fig. 5, compare F6 and GRIP-1). Based on this result, we examined the inhibitory activity of a construct expressing two copies of the LXXLL motif on ERα transcriptional activity. The linker between the two copies was adapted from sequences found between the GRIP-1 NR box 2 and NR box 3 (see Materials and Methods). When analyzed in target cells, the fusion proteins containing two copies of the F6 peptide were more effective inhibitors of ERα transcriptional activity than were those expressing a single copy. 2×F6 was functionally comparable to the construct expressing the GRIP-1 NR boxes, which contains three copies of the LXXLL motif (Fig. 5, 2×F6). The increased efficacy of 2×F6 as an inhibitor of ER function required each of the two LXXLL motifs, since addition of the GRIP-1 linker sequence to a single copy of F6 did not increase its antagonist efficacy (data not shown).
FIG. 5.
LXXLL-containing peptides disrupt ERα transcriptional activity when overexpressed in target cells. HeLa cells were transfected with the ERα expression plasmid (pRST7ERα), 3×ERE-TATA-Luc reporter, along with increasing amounts of a construct expressing the peptide-Gal4DBD fusions as indicated. F6 contains a single copy of the F6 peptide, 2×F6 contains two copies of the F6 peptide with 50 amino acids separating the two LXXLL motifs, and GRIP-1 contains the center three NR boxes from the coactivator GRIP-1. All these peptides were expressed as fusion proteins to Gal4DBD. In addition, a pCMVβgal plasmid was cotransfected to normalize for transfection efficiency. After transfection, cells were induced with 10−7 M 17β-estradiol for 16 h before assaying. Fold induction represents the ratio of estradiol-induced activity versus no-hormone control for each transfection.
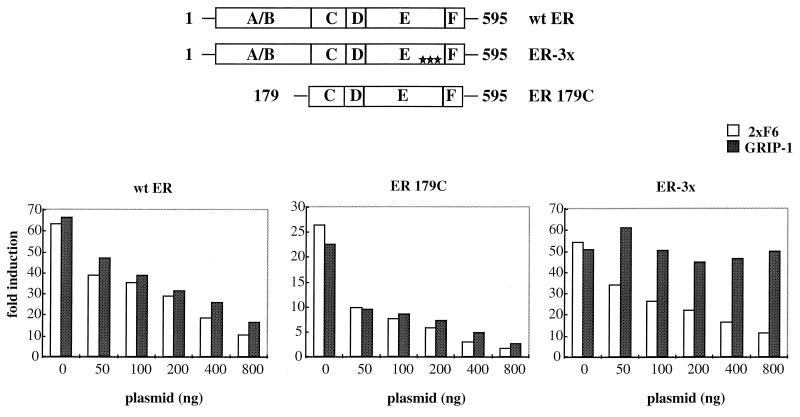
It has been demonstrated by us and others that ER contains two distinct activation function domains, AF-1 and AF-2, whose activities are manifested in a cell-selective manner (3, 34, 45, 47). Both AF-1 and AF-2 functions are required for maximal ER transcriptional activity in HeLa cells, while AF-1 is the dominant activation function in HepG2 cells. Our peptide disruption results closely correlated with these observations. In HeLa cells, overexpression of LXXLL-containing peptides abolished almost 100% of the ER transcriptional activity (Fig. 5), highlighting the obligate role of AF-2 in ER-mediated function and showing that AF-1 is not able to function independently of AF-2 in this background. However, we have observed that the roles of AF-1 and AF-2 in HepG2 cells are different. It was demonstrated in a previous study that mutations in ER-AF2 that block the binding of the coactivators SRC-1 and GRIP-1 with ER have no effect on ER transcriptional activity in HepG2 cells (19, 28, 47). We interpreted these data to mean that either (i) in this context AF-1 is dominant and AF-2 is not required or (ii) in this cell line a cofactor exists whose interaction with ER does not require an intact AF-2. To discriminate between these possibilities, we used the LXXLL-containing peptides to study the role of AF-1 and AF-2 in ER signaling in this background. The results of this analysis are shown in Fig. 6. When either the 2×F6 or GRIP-1 peptides were overexpressed in HepG2 cells, they inhibited wild-type ER transcriptional activity; however, it was not inhibited down to the basal levels (Fig. 6, wt ER). The transcriptional activity was still about 10-fold over the basal levels at the highest dose of input peptide fusion plasmid, indicating that some independent AF-1 activity is possible in this cell context. This hypothesis is supported by the observation that the activity of an ER-mutant lacking AF-1 was inhibited more readily (twofold over the basal level at the highest input plasmid dose) by overexpression of either of the peptide fusions (Fig. 6, ER 179C). The most interesting result, however, was that the class III peptide (2×F6) was an efficient inhibitor of ER-3× transcriptional activity whereas the GRIP-1 NR-box peptide was inefficient (Fig. 6, ER-3×). Taking these results together, we observed that the class III peptide F6 interacted with ER-3× (Fig. 3B) and that overexpression of this peptide inhibited the transcriptional activity of this mutant receptor, suggesting that a cofactor which contains an F6-like LXXLL motif may exist in HepG2 cells and may be important for ER function.
FIG. 6.
The differential ability of LXXLL-containing peptides to disrupt ERα-mediated transactivation function reveals the presence of multiple ER-interacting coactivators. HepG2 cells were transfected with pRST7-ERα (wt), ERα179C, or ERα-3× mutant expression plasmids along with the 3×ERE-TATA-Luc reporter gene and increasing amounts of the Gal4DBD-peptide fusion constructs (as indicated). Fold induction represents the ratio of estradiol-induced (10−7 M) activity versus no-hormone control for each transfection.
Sequences flanking the LXXLL core motif influence receptor selectivity.
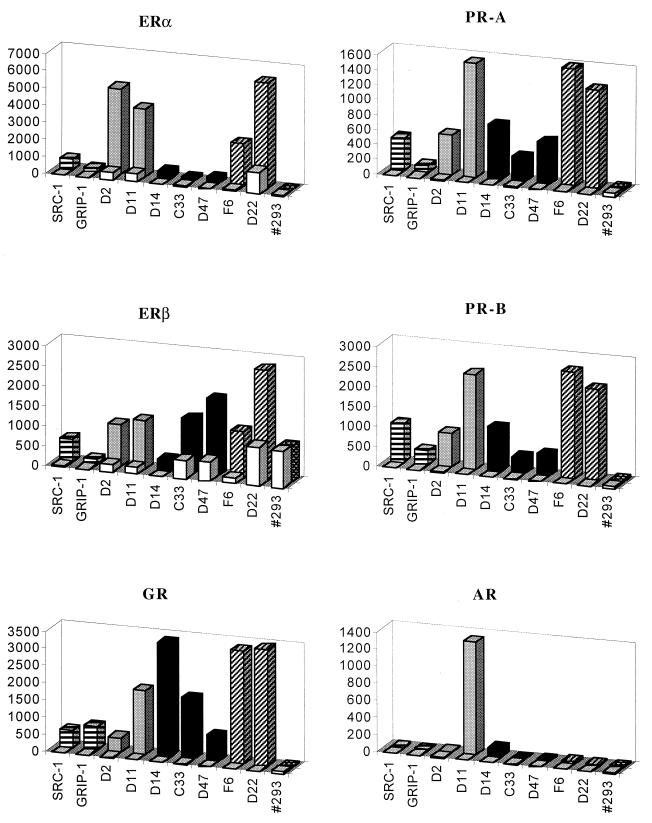
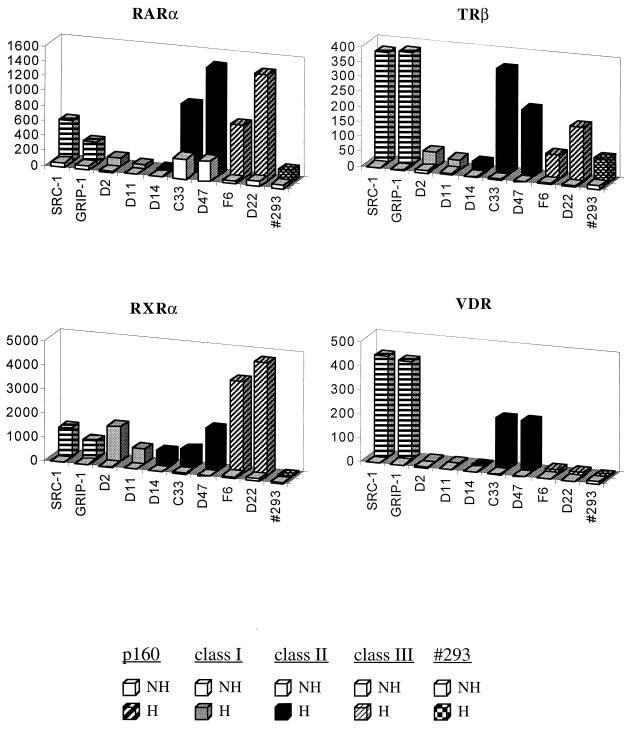
The GRIP-1 and SRC-1 coactivators containing multiple LXXLL motifs interacted with most nuclear receptors. Alterations of residues surrounding these motifs have been shown to affect receptor selectivity; therefore, we next wished to define the sequences within the NR box which enable it to discriminate between receptors by using the LXXLL-containing peptides identified. For this study, we used representative members of each class of LXXLL identified from our focused library along with an LXXLL motif, #293, which was identified previously in screens of random peptide libraries for peptides which interacted with estradiol-activated ERβ (reference 32 and data not shown). This specificity analysis was accomplished by performing mammalian two-hybrid assays, in which the LXXLL-containing peptides were fused to Gal4DBD and the full-length receptors were expressed as VP16 fusion proteins. As shown in Fig. 7, most steroid receptors interacted with all three classes of peptides efficiently. The lower luciferase activity observed with class II peptides is probably related to the lower (∼10-fold) expression level of this class of peptides (Fig. 3C). Regardless, the RXR heterodimerization partners, such as RARα, TRβ, and VDR, demonstrated a strong preference for class II over the other classes of peptides. Interestingly, ERβ also showed the same tendency, preferring to interact with class II motifs, suggesting that the coactivator binding groove in ERα and ERβ may be functionally different. Interestingly, with the exception of D11, the AR interacted weakly with all the LXXLL peptides tested, supporting the hypothesis that alternative coactivator recruitment methods are used by AR and that the N terminus is more important than AF-2 in recruiting coactivators to the receptor (2, 33, 51).
FIG. 7.
Nuclear receptors have distinct preferences for different LXXLL motifs. The interactions between different LXXLL motifs and nuclear receptors were assayed by using a mammalian two-hybrid system. Full-length receptors and selected peptides were expressed as VP16 and Gal4DBD fusion proteins, respectively. The magnitude of these interactions was measured by using a 5×Gal4Luc3 reporter gene. Open bars, no hormone; hatched or filled bars, hormone treatments. The following hormones were used in this experiment: 10−7 M 17β-estradiol for ERα and ERβ, 10−7 M progesterone for PR-A and PR-B, 10−7 M dexamethasone for GR, 10−7 M 9-cis-retinoic acid for RARα and RXRα, 10−7 M T3 for TRβ, 10−7 M 1,25-dihydroxyvitamin D3 for VDR, and 10−6 M 5α-dihydrotestosterone for AR. The luciferase activity was normalized to the activity of the cotransfected pCMVβgal.
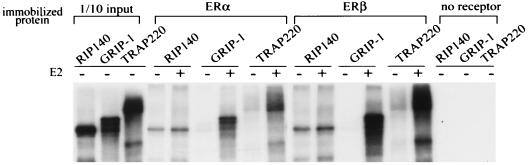
We next compared the sequences of these three classes of LXXLL-motifs with sequences of NR boxes in known coactivators and found that the class I peptides share similar features with two of the LXXLL motifs found in GRIP-1- and SRC-1-type (p160s) cofactors, in which a positively charged residue precedes the LXXLL motif (Table 1). The class II peptides were represented by the two LXXLL motifs found in TRAP220 (52), in which a proline occupies the −2 position. The class III peptides are most abundant in cofactor RIP140 (6), but similar motifs can also be found in PGC-1 (36), and the orphan receptors SHP and DAX-1 (39, 53). Based on our findings, we predicted that each of these cofactors should interact with both isoforms of ER. These factors have already been shown to interact with ERα, whereas minimal information on their ERβ binding properties has yet to be reported. In a pulldown assay with purified full-length ERα and ERβ immobilized on 96-well plates, we were able to confirm that each of these proteins, representing all three LXXLL classes, was able to interact with both ER isoforms in a ligand-dependent manner (Fig. 8).
TABLE 1.
Each of the three classes of ER-interacting LXXLL motifs is found within known coactivators
| Class | Coactivator | Sequencea |
|---|---|---|
| Class I | SRLXXLL | |
| GRIP1 | TKLLQLL | |
| SRC-1 | HKLVKLL | |
| AIB-1 | KKLLQLL | |
| Class II | PΦLXXLL | |
| TRAP220 | PILTSLL | |
| PMLMNLL | ||
| RIP140 | PILYYML | |
| Class III | (S/T)ΦLXXLL | |
| RIP140 | TYLEGLL | |
| TLLASLL | ||
| SLLLHLL | ||
| TLLQLLL | ||
| TVLQLLL | ||
| PGC-1 | SLLKKLL | |
| DAX-1 | SILYNLL | |
| SILYSML | ||
| SILYSLL | ||
| SHP | TILYALL | |
| SILKKIL |
X, any amino acid; Φ, hydrophobic amino acid. Conserved amino acids in each class are in boldface type.
FIG. 8.
LXXLL motif-containing cofactors interact with both ERα and ERβ in vitro in a ligand-dependent manner. Equal amounts of full-length ERα, ERβ, or control BSA were immobilized on 96-well plates in the presence or absence of 1 μM estradiol. Full-length RIP140, GRIP-1, and TRAP220 were translated in vitro and labeled with [35S]methionine. Labeled cofactors were added to the wells containing immobilized protein and incubated at 4°C overnight. Unbound protein was removed by washing, and the bound protein was eluted, separated by SDS-PAGE, and visualized by autoradiography.
Peptide #293 is an ERβ-selective antagonist.
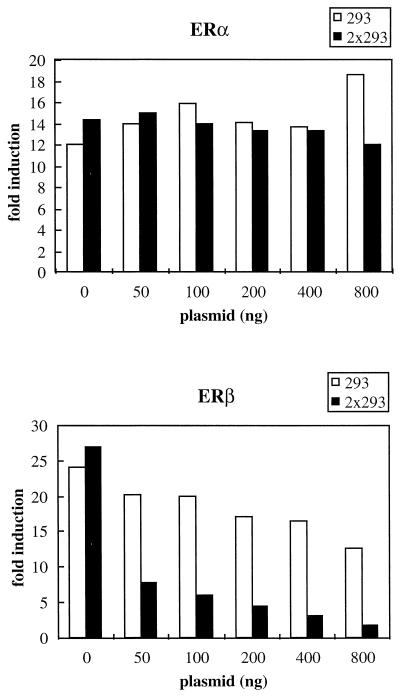
When peptide #293 was screened against a panel of nuclear receptors, it showed a strong preference for ERβ and interacted weakly with TRβ and RARα but did not interact significantly with the other receptors tested (Fig. 7). Thus, receptor specificity can be achieved by altering sequences flanking the core LXXLL motif, and it is possible that ERβ-specific coactivators will be found to contain this or a structurally similar motif. To test whether peptide #293 could specifically target ERβ transcriptional activity, we overexpressed it as a Gal4DBD fusion protein and assayed its ability to disrupt ERβ-dependent reporter gene expression. As shown in Fig. 9, expression of #293 had no effect on ERα-mediated gene expression but the ERβ transcriptional activity was significantly reduced. Similar to the results with ERα, two copies of the #293 motif (2×293) disrupted ERβ function more efficiently than did a single-copy peptide. Nevertheless, ERα transcriptional activity remained unaffected by the expression of 2×293. Clearly, not all LXXLL motifs have the same receptor binding selectivity. Thus, we believe that receptor-specific LXXLL motifs can be found and used to target specific cofactor-receptor interactions.
FIG. 9.
Peptide #293 selectively disrupts ERβ-dependent reporter gene expression without affecting ERα-mediated transcription when expressed in target cells. Peptide #293 containing an LXXLL motif was affinity selected by phage display with estradiol-activated ERβ as a target. Expression of either one copy or two copies of this peptide did not interfere with the transcriptional activity of ERα but disrupted ERβ-mediated transcriptional activity. HeLa cells were transfected with either ERα or ERβ expression plasmids, along with 3×ERE-TATA-Luc reporter, pCMVβgal, and increasing amounts of Gal4DBD-peptide fusion constructs as indicated. Fold induction represents the ratio of activity estradiol-induced activity versus no-hormone control for each transfection.
DISCUSSION
The identification of ER-associated coactivators and corepressors has helped us understand how different ligands acting through the same receptor can manifest different biological activities. The importance of these proteins in mediating ER pharmacology was highlighted by our previous studies, which described the identification of different classes of peptides whose ability to interact with ER is influenced by the nature of the bound ligand (29, 32). All of these interactions represent potential ER-cofactor interactions and suggest that ER pharmacology is more complex than was originally believed. In this study, we have focused on one receptor binding motif, LXXLL, and have demonstrated that even within this specific core there are multiple classes of functionally different LXXLL motifs. Using estradiol-activated ERα, we screened 108 variations of the LXXLL motif and identified three classes of peptides that interact with the coactivator binding pocket within the ERα HBD. The classifications were further substantiated by studies which revealed that each class of peptide displayed specific receptor preferences and that their binding to ERα was differentially affected by ER helix 12 mutations. In spite of their differences, the LXXLL-containing peptides all appear to bind in an agonist-dependent manner to the same coactivator binding groove within ERα HBD. None of the peptides identified interact with ER-535stop (helix 12 deletion) or the LL mutant (L539L540→A539A540). This is not surprising, since the cocrystal structure of ER with NR box 2 of GRIP-1 shows that several residues in helix 12, including L-539, are required to make van der Waals contacts between the coactivator groove and the LXXLL peptide. It is likely that truncation of helix 12 or mutations of the paired hydrophobic residues destabilize such interactions. Furthermore, replacing the three charged residues in helix 12 with their corresponding amides (ER-3×) disrupts the ability of class I and class II peptides to interact with ER. The ternary structure predicted from the cocrystal structure suggests that the conserved glutamic acid (E542) in ER helix 12 plus the lysine residue (K362) in helix 3 cap the LXXLL peptide in the coactivator binding groove through hydrogen bonding to the backbone amides or carbonyls of the residues on the N- or C-terminal turns of the peptide helix. Although the charged side chain is not directly involved in the hydrogen bonding, the positively charged residue preceding the LXXLL motifs is thought to be important for orienting and positioning these motifs within the coactivator binding groove, which is capped on one end by the negatively charged E-542 (26, 41). Consistent with this idea, our results showed that changing the Glu-542 into Gln-542, which neutralizes the charge but still preserves the hydrogen bonding, greatly reduced the ability of this mutant receptor to interact with class I and class II peptides. One of the most surprising findings of our study, however, is that the class III peptides, which do not contain any positively charged residues immediately preceding the LXXLL motif, interact strongly with both wild-type ER and the ER-3× mutant, supporting the hypothesis that this class of peptides binds in a unique manner to the ER AF-2 and that the “charged-clamp” model may not hold for all LXXLL interactions.
Because of the unique properties of the class III LXXLL, we searched the sequences of known nuclear receptor-interacting motifs for analogous sequences. Interestingly, class III-like LXXLL motifs were found to be present in multiple copies in RIP140, where the LXXLL motifs are preceded by a serine or threonine and an isoleucine or leucine. Importantly, RIP140 was shown to interact with ER-3× (6), whereas GRIP-1 and SRC-1 did not, suggesting that the class III peptides represent a biologically relevant LXXLL motif. Similar types of motifs were also found in the orphan receptors DAX-1 and SHP (39, 53), two receptors that are able to interact with estradiol-activated ER and disrupt its ability to activate transcription. Although the domains within DAX-1/SHP responsible for these interactions have not been precisely determined, based on their interaction patterns (induced by estradiol, inhibited by tamoxifen, and insensitive to ER-3× mutations), we anticipate that these interactions are mediated, at least in part, through LXXLL-like motifs. Since both RIP140 and SHP can disrupt wild-type- as well as ER-3× mutant-mediated transactivation (references 18 and 40 and data not shown), it is tantalizing to speculate that class III type motifs might be used by ER inhibitors instead of ER coactivators. We were able to show, however, that the F6 peptide (class III) can compete with endogenous cofactors and suppress estradiol-induced ER activation in target cells. This leaves open the possibility that another class of receptor coactivators that use the class III-like LXXLL motif remains to be found. Clearly, not all LXXLL motifs are the same. However, until each of these motifs is found within a bona fide ER regulator, the functional significance of these different peptides cannot be determined. Regardless, our study highlights a heretofore unanticipated complexity in ER action.
All of the AF-2-interacting coactivators that have been found contain an LXXLL motif. Thus, given the homology in the AF-2 domain among receptors and the simplicity of the LXXLL motif, it was difficult to understand how receptor specificity could occur. Interestingly, with the collection of peptides we obtained, we were able to demonstrate that ERα and ERβ, two highly homologous receptors with similar ligand binding characteristics, showed distinct preferences for different classes of peptides. Previously, we found that the ERβ homodimer is a weaker transcriptional activator than the ERα homodimer and the ERαβ heterodimer (14). It would be interesting to see if the differences in their transcriptional activity are due to their differential association with different cofactors. Although ERα and ERβ have overlapping affinities for their ligands and DNA responsive elements, they are not functionally redundant (22, 31). Their ability to interact differentially with different LXXLL motifs within coactivators might explain how ERα and ERβ manifest different transcriptional activities in target cells.
The PPARγ-binding protein (54) and its human homolog TRAP220 (also called DRIP205) (37, 52) contain LXXLL motifs that have a proline at the −2 position, similar to the class II peptides. These cofactors were identified originally by their ability to interact with PPARγ, TR, and VDR in vivo and were shown to interact with RAR and RXR at high affinity in vitro. A remarkably similar pattern was observed in our study when we demonstrated by mammalian two-hybrid analysis that TR, VDR, RAR, and ERβ appeared to have a stronger preference for the class II peptides, suggesting that the occurrence of a proline at the −2 position might favor these interactions. Based upon alanine scanning studies, McInerney et al. suggested that receptor recognition is most probably contributed by residues C-terminal to the LXXLL motifs (25). In our study, however, we did not find a good consensus in the C terminus in over 50 peptides selected from both random and focused library screening, using either ERα or ERβ as the target (Fig. 1) (reference 32 and data not shown). In contrast, residues at the −2 and −1 positions are dominated by either S(R or K) or S(I or L), which suggests that residues in these positions are important for cofactor-ER interaction through the LXXLL motif and that these sequences are generally accepted by steroid hormone receptors. Moreover, certain receptors such as TR, VDR, RXR, and ERβ appear to favor motifs with a proline at the −2 position, again highlighting the importance of this residue for receptor-cofactor recognition. However, we cannot rule out the possibility that the differences observed reflect a selection bias, since we have used only ER as a target for affinity selection. We would also like to emphasize that although residues occupying the −1 and −2 positions seem to be a critical determinant of LXXLL specificity, sequences outside these regions are also important, since a different receptor binding specificity has also been observed within the same class of peptides. For example, the ERβ-specific #293 peptide may be considered a class II member, because it also contains a proline at the −2 position. Clearly, however, sequences in addition to the proline at −2 are important, since #293 has a unique receptor selectivity.
The identification of novel classes of LXXLL motifs and the finding that they interact with ER in different ways have highlighted the complexity of ER action. As yet, given the limited number of coactivators and corepressors available for analysis, it is not possible to evaluate the full significance of our findings. However, we believe that these studies provide a glimpse of what is to come. In addition to the mechanistic insight offered by these studies, they have provided some novel technology which may be used in drug discovery. Some investigators have used the coactivator receptor ligand assay (CARLA) as a way of screening for compounds which function as receptor agonists and allow the formation of an AF-2/coactivator groove (21). For known receptors, where the cofactor interactions have been well established, this is likely to be useful. However, when studying an orphan receptor for which no ligand has been identified, its success relies on whether the receptor can interact with the coactivator chosen. For this purpose, a “universal” coactivator is desirable. Our studies have illustrated that several different LXXLL motifs interact differentially with different receptors. Therefore, the use of a single peptide in a screening paradigm can be risky, but the chance of success will be increased by incorporating several different classes of peptides in the screen.
Another application of these peptides, validated in our study, is their use as peptide antagonists of receptor function. For instance, peptide #293, when introduced into cells, specifically inhibits ERβ-mediated responses to estrogen. Since a specific small-molecule inhibitor of ERβ has not been identified, we believe that the #293 peptide may allow us to unravel some of the biology of this receptor. We believe that the technology used in our studies will also be useful for the study of orphan receptors. Specifically, we suggest that the identification of peptides which bind specifically to an orphan receptor and which inhibit its transcriptional activity can be used as “peptide antagonists” to study the biology of the receptor when its ligands are not known.
The results presented in this study confirm that the coactivator LXXLL motif is necessary and sufficient for receptor interaction. In addition, they revealed the importance of sequences surrounding the LXXLL core in determining receptor selectivity and in defining the manner in which coactivators interact with the nuclear receptors. The complexity highlighted by these studies suggests that the currently available coactivators and corepressors represent only a fraction of those which will ultimately be found and shown to interact with the nuclear receptors.
ACKNOWLEDGMENTS
We thank J. W. Pike, D. D. Moore, J. Miner, D. X. Wen, K. Marschke, X.-F. Wang, R. G. Roeder, M. G. Parker, M. Stallcup, and P. Giangrande for providing plasmids. We also thank V. Clack for her help in the preparation of the manuscript.
This work was supported in part by an NIH grant (DK48807) to D.P.M., a discovery grant from Duke University Comprehensive Cancer Center to D.J.K., and a research grant from the Chemical Industry Institute of Toxicology to D.P.M.
REFERENCES
- 1.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 2.Berrevoets C A, Doesburg P, Steketee K, Trapman J, Brinkmann A O. Functional interactions of the AF-2 activation domain core region of the human androgen receptor with the amino-terminal domain and with the transcriptional coactivator TIF2 (transcriptional intermediary factor2) Mol Endocrinol. 1998;12:1172–1183. doi: 10.1210/mend.12.8.0153. [DOI] [PubMed] [Google Scholar]
- 3.Berry M, Metzgar D, Chambon P. Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J. 1990;9:2811–2818. doi: 10.1002/j.1460-2075.1990.tb07469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brzozowski A M, Pike A C, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 5.Cavaillès V, Dauvois S, Danielian P S, Parker M G. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavaillès V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 8.Chen J D, Li H. Coactivation and corepression in transcriptional regulation by steroid/nuclear hormone receptors. Crit Rev Eukaryotic Gene Expression. 1998;8:169–190. doi: 10.1615/critreveukargeneexpr.v8.i2.40. [DOI] [PubMed] [Google Scholar]
- 9.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 10.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 12.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 13.Evans R M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Giangrande, P. H. Unpublished data.
- 14.Hall, J. M., and D. P. McDonnell. The ERβ-isoform of the human estrogen receptor modulates ERα transcriptional activity and is a key regulator of cellular sensitivity to estrogens. Endocrinology, in press. [DOI] [PubMed]
- 15.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 16.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 18.Johansson L, Thomsen J S, Damdimopoulos A E, Spyrou G, Gustafsson J-A, Treuter E. The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERα and ERβ. J Biol Chem. 1999;274:345–353. doi: 10.1074/jbc.274.1.345. [DOI] [PubMed] [Google Scholar]
- 19.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kay B K, Kurakin A V, Hyde-DeRuyscher R. From peptides to drugs via phage display. Drug Discov Today. 1998;3:370–378. [Google Scholar]
- 21.Krey G, Braissant O, L’Horset F, Kalkhoven E, Perroud M, Parker M G, Wahli W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol Endocrinol. 1997;11:779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 22.Kuiper G G J M, Carlson B, Grandien K, Enmark E, Haggblad J, Nilsson S, Gustafsson J-A. Comparison of the ligand binding specificity and transcript distribution of estrogen receptors α and β. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 23.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 27.Norris J, Fan D, Aleman C, Marks J R, Futreal P A, Wiseman R W, Iglehart J D, Deininger P L, McDonnell D P. Identification of a new subclass of Alu DNA repeats which can function as estrogen receptor-dependent transcriptional enhancers. J Biol Chem. 1995;270:22777–22782. doi: 10.1074/jbc.270.39.22777. [DOI] [PubMed] [Google Scholar]
- 28.Norris J D, Fan D, Stallcup M R, McDonnell D P. Enhancement of estrogen receptor transcriptional activity by the coactivator GRIP-1 highlights the role of activation function 2 in determining estrogen receptor pharmacology. J Biol Chem. 1998;273:6679–6688. doi: 10.1074/jbc.273.12.6679. [DOI] [PubMed] [Google Scholar]
- 29.Norris J D, Paige L A, Christensen D J, Chang C-Y, Huacani M R, Fan D, Hamilton P T, Fowlkes D M, McDonnell D P. Peptide antagonists of the human estrogen receptor. Science. 1999;285:744–746. doi: 10.1126/science.285.5428.744. [DOI] [PubMed] [Google Scholar]
- 30.Onate S A, Tsai S, Tsai M-J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 31.Paech K, Webb P, Kuiper G G J M, Nilsson S, Gustafsson J-A, Kushner P J, Scanlan T S. Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- 32.Paige L A, Christensen D J, Gron H, Norris J D, Gottlin E B, Padilla K M, Chang C, Ballas L M, Hamilton P T, McDonnell D P, Fowlkes D M. Estrogen receptor modulators each induce distinct conformational changes in ERα and ERβ. Proc Natl Acad Sci USA. 1999;96:3999–4004. doi: 10.1073/pnas.96.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker M G, Bevan C, Cowley S, Heery D, Kalkhoven E, Mak H Y, Needham M, Valentine J, White R. Recruitment of p160/RIP160 coactivators to steroid hormone receptors. The Steroid Receptor Superfamily, AACR Special Conference in Cancer Research. 1999. [Google Scholar]
- 34.Pham T A, Hwung Y P, Santiso-Mere D, McDonnell D P, O’Malley B W. Ligand-dependent and -independent function of the transactivation regions of the human estrogen receptor in yeast. Mol Endocrinol. 1992;6:1043–1050. doi: 10.1210/mend.6.7.1508220. [DOI] [PubMed] [Google Scholar]
- 35.Pirozzi G, McConnell S J, Uveges A J, Carter J M, Sparks A B, Kay B K, Fowlkes D M. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem. 1997;272:14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- 36.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 37.Rachez C, Suldan Z, Ward J, Chang C B, Burakov D, Erdjument-Bromage H, Tempst P, Freeman L P. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seol W, Choi H S, Moore D D. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272:1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- 40.Seol W, Hanstein B, Brown M, Moore D D. Inhibition of estrogen receptor action by the orphan receptor SHP (short heterodimer partner) Mol Endocrinol. 1998;12:1551–1557. doi: 10.1210/mend.12.10.0184. [DOI] [PubMed] [Google Scholar]
- 41.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 42.Smith C L, Nawaz Z, O’Malley B W. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 43.Sparks A B, Adey N B, Cwirla S, Kay B K. Phage display of peptides and proteins: a laboratory manual. San Diego, Calif: Academic Press, Inc.; 1996. [Google Scholar]
- 44.Sparks A B, Hoffman M G, McConnell S J, Fowlkes D M, Kay B K. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- 45.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 46.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 47.Tzukerman M T, Esty A, Santiso-Mere D, Danielian P, Parker M G, Stein R B, Pike J W, McDonnell D P. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- 48.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 49.Voegel J J, Heine M J S, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamabhai M, Kay B K. Examining the specificity of src homolog 3 domain-ligand interactions with alkaline phosphatase fusion proteins. Anal Biochem. 1997;247:143–151. doi: 10.1006/abio.1997.2040. [DOI] [PubMed] [Google Scholar]
- 51.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan C X, Ito M, Fondell J D, Fu Z Y, Roeder R G. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zanaria E, Muscatelli F, Bardoni B, Strom T M, Guioli S, Guo W, Lalli E, Moser C, Walker A P, McCabe E R, et al. An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature. 1994;372:635–641. doi: 10.1038/372635a0. [DOI] [PubMed] [Google Scholar]
- 54.Zhu Y, Qi C, Jain S, Rao M S, Reddy J K. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]