Abstract
According to the World Health Organization (WHO), novel coronavirus (COVID-19) is an infectious disease and has a significant social and economic impact. The main challenge in fighting against this disease is its scale. Due to the outbreak, medical facilities are under pressure due to case numbers. A quick diagnosis system is required to address these challenges. To this end, a stochastic deep learning model is proposed. The main idea is to constrain the deep-representations over a Gaussian prior to reinforce the discriminability in feature space. The model can work on chest X-ray or CT-scan images. It provides a fast diagnosis of COVID-19 and can scale seamlessly. The work presents a comprehensive evaluation of previously proposed approaches for X-ray based disease diagnosis. The approach works by learning a latent space over X-ray image distribution from the ensemble of state-of-the-art convolutional-nets, and then linearly regressing the predictions from an ensemble of classifiers which take the latent vector as input. We experimented with publicly available datasets having three classes: COVID-19, normal and pneumonia yielding an overall accuracy and AUC of 0.91 and 0.97, respectively. Moreover, for robust evaluation, experiments were performed on a large chest X-ray dataset to classify among Atelectasis, Effusion, Infiltration, Nodule, and Pneumonia classes. The results demonstrate that the proposed model has better understanding of the X-ray images which make the network more generic to be later used with other domains of medical image analysis.
Keywords: COVID-19, X-ray, Feature extraction, Image processing, Classification, Machine learning, Deep learning
Introduction
In December 2019, the first case of a novel coronavirus (COVID-19) was found in Wuhan, China. It was thought that this virus originated from zoonotic-like species, however, the cause of this virus has still not been determined [1–3]. Coronavirus, officially known as Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) causes an infectious disease that has spread to over 215 countries [4]. This deadly disease is contagious which is making it a significant threat to human beings. As per the current statistics [5], number of confirmed COVID-19 cases have crossed 103, 338, 512 and more than 2,233,559 deaths across the world [6]. People with weak immune systems and already suffering from other diseases like sugar, blood pressure, etc. are more prone to the infection [7]. It is spreading through close contact or the droplets when the infected coughs, sneezes or talks [8]. For a vaccine to develop for COVID-19, diagnosis through real-time reverse transcription-polymerase chain reaction (RT-PCR) is confirmed [9]. However, RT-PCR still lacks time management, and there are still lying a long chain of trials from animals to humans, and the virus is regularly mutating. Due to this, it may arise that the infected person may not be recognized and may not get the suitable treatment and may spread the virus to a healthy population and cannot be acceptable in this pandemic situation. It has been observed that X-rays and CT images are sensitive to the screening of the COVID-19 among the patients. Initially, lung X-ray is performed for suspected or confirmed patients through specific circuits. It proves to be a discriminating element wherein the patient’s further diagnosis depends on the clinical situation and the chest X-ray film results. Many recent works depending on the elaborated image features with clinical and diagnostic results may help in early detection of COVID-19 [10–12]. Technological advancement using the machine and deep learning applications for automatic prediction of COVID-19 traces in people has taken over people’s minds [13].
A work on X-ray image classification is introduced by Tulin et al. [14]. They used the DarkNet model for classifying the detected YOLO-based object in the input images. Another work on X-ray images, called Decompose, Transfer, and Compose (DeTraC), is introduced by Asmaa et al. in [15]. DeTraC used class decomposition mechanism to deal with class boundaries leading to an accuracy of 0.931 and sensitivity of 1. A work by Taban et al. on chest X-ray images used the 12-off-the-shelf CNN architectures in transfer learning [16]. GANs have also been used in the process of COVID-19 disease analysis by Mohamed et al. [17]. It used deep transfer learning based on GANs for 3-class classification among COVID-19, Normal and Pneumonia X-ray images. They utilized the dataset created by Joseph et al. [18] which yielded an accuracy, sensitivity, and F1-score of 0.8148, 0.8148, and 0.8146, respectively. Another work on the classification task of COVID-19 among the three classes of COVID-19, Normal and Pneumonia X-ray images was performed by Enzo et al. [19]. They used the datasets in [18, 20, 21] with the Pneumonia images 1 for achieving considerable performance. Wang et al. has introduced a COVID-19 predictive classification technique by using 453 chest CT images and achieved an accuracy of [22]. Khaled et al. proposed another work for COVID-19 detection using the transfer learning-based hybrid 2D/3D CNN architecture, VGG16, for screening chest X-rays [23]. Yifan et al. has introduced the use of CNN in the classification process of COVID-19 [24]. Their model achieved an acceptable recall and precision values of 0.75 and 0.64, respectively. Depth-wise separable convolution layer and a spatial pyramid pooling module (SPP) modules are combined with VGG16 for obtaining better results. Similar work by N.Narayan et al. [2] with the use of Inception (Xception) model on chest X-ray images was also performed.
In a work by Rahmatizadeh et al., they introduced a three-step AI-based model to improve the critical care of patients [25]. An intensive care unit (ICU) is performed where the input evidence includes surgical, paraclinical, customized medicine (OMICS) and epidemiology data. The performance included assessment, therapy; the stratification of threats, prognosis, and direction, and finally, it concludes that the health care system’s efforts to overcome the problem of COVID-19 detection in patients with the help of today’s AI-based technology can easily and effectively tackle the patients, particularly for the patients in ICU suffering from COVID-19.
Similarly, many such works on COVID-19 diagnosis have sprung up which helped the experts in effective classification of the disease. In Tables 1, 2 and 3, a comprehensive review for the use of deep learning for COVID-19 classification is tabulated. Inspired by the dire need to develop means to fight COVID-19, and intrigued by the works of open access support of the research community, this work leverages the variational aspects of encoding the radiological image features into classification scores for predicting the COVID-19 related lung opacities. Here, a deep learning-based stacked pipeline incorporating deep CNNs is encouraged. DenseNet and GoogLeNet are used for feature extraction with the involvement of a variational environment to further extract the latent space of the extracted features. This model was trained on the publically available COVID-19 datasets for performing diagnostic tests. The main contributions of this paper are as follows:
A framework for COVID-19 detection based on X-ray images is proposed, which outperforms state-of-the-art CNN models.
We propose a two-step ensemble approach under a variational setting for learning the joint representation of representations generated by fine-tuned CNNs.
We perform extensive experiments on COVID-19 dataset along with a challenging multi-class benchmark containing X-ray images.
Table 1.
Overview of prediction models for diagnosis of COVID-19
| Author Year | Dataset | Classification type | Method | Performance metrics | |||||
|---|---|---|---|---|---|---|---|---|---|
| Acc | Sens | Spec | Precision | F1-score | AUC | ||||
| Abbas et al. 2021 [15] | X-ray | Multi | Transfer Learning on AlexNet, VGG19, ResNet, GoogleNet, SqueezeNet | ✗ | ✗ | ✗ | |||
| Mohamed et al. 2020 [17] | X-ray | Multi | Transfer Learning on GAN using Alexnet, Googlenet, and Restnet18 | ✗ | ✗ | ||||
| Enzo et al. 2020 [19] | X-ray | Binary | Transfer Learning using ResNet-18 and ResNet-50 | ✗ | ✗ | ||||
| Tulin et al. 2020 [14] | X-ray | Binary Multi | DarkNet model with YOLO for multi-class classification | ✗ | |||||
| Taban et al. 2020 [16] | X-ray | Multi | 12-off-the-shelf CNN architectures (transfer learning) with clinical advice | ✗ | ✗ | ||||
| Wang et al. 2020 [22] | CT | Binary | Transfer-learning on Inception Net | ✗ | ✗ | ✗ | |||
| Khaled et al. 2020 [23] | X-ray | Binary | VGG16 with SPP module (transfer learning) | ✗ | ✗ | ✗ | |||
| N.Narayan et al. 2020 [2] | X-ray | Multi | Transfer learning using Inception (Xception) model | ✗ | |||||
| Rahmatizadeh et al. 2020 [25] | X-ray | Multi | 3-step decision-making system to improve the critical care of COVID-19 patients | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ |
Table 2.
DATASET-1 ): Class-wise bifurcation of Pneumonia dataset
| Dataset bifurcation | Classes | ||
|---|---|---|---|
| Normal | Pneumonia | Total | |
| Train set | 1341 | 3875 | 4216 |
| Test set | 234 | 390 | 624 |
| Total | 1575 | 4265 | 5840 |
Table 3.
DATASET-2 ): Class-wise bifurcation of COVID-19 dataset
| Dataset bifurcation | Classes | |||
|---|---|---|---|---|
| COVID-19 | Normal | Pneumonia | Total | |
| Train set | 196 | 210 | 210 | 616 |
| Test set | 112 | 113 | 113 | 338 |
| Total | 308 | 323 | 323 | 954 |
Method
Dataset specification
The 2019 novel coronavirus has shown numerous unique symptoms to detect the viral disease COVID-19 from the patients. It has been deduced that the COVID-19 patterns can be perceived from either CT or chest X-ray images. Considering the seriousness of the situation worldwide, the publically available datasets have been collected and worked upon.
Initially, we have taken Chest X-ray images (Pneumonia) [26], which is represented as , which contains a total of 5840 images. The training set contains 4216 images which are further bifurcated into classes, Pneumonia and Normal with 1341 and 3875 images, respectively. Similarly, the test set contains 624 images subdivided into Pneumonia and Normal with 390 and 234 image samples, respectively.
Another dataset for COVID-19 images is collected from three COVID-19 datasets which include COVID-19 Radiography Database [27] containing approximately 219 images with another dataset [28] containing a total of 69 images with 60 samples in training and 9 samples in testing. The third dataset has been taken from an open-source repository [29] which contained a total of 35 COVID-19 images. All the datasets are combined to form a collection of 308 images that are further used for experimentation on the proposed framework. All in all, 308 images have been segregated as 196 for training and the rest 112 for testing, approximately in the ratio of 2:1.
We have comprehensively performed experiments on the NIH Chest X-ray images (dataset ) 2 containing 14 categories, out of which we have selected 5 categories namely, Atelectasis, Effusion, Infiltration, Nodule, and Pneumonia which have higher correlation among them (tabulated in Table 4). Considering the dataset specifications, we found that the Pneumonia subset of images was limited in number and mixed with other classes (upon initial experimental trials). Therefore, to remove this apprehension, we amalgamated the Pneumonia images from dataset in the dataset. The snag about the dataset is that the images have been extracted based on the query utilizing the natural language processing (NLP), which has confused the reader with multiple classes for a single image. Therefore, to avoid confusion, we have taken the diseases with minimal overlapping and maximum images in the concerned image category. Some of the sample images from the dataset and are shown in Figs. 1 and 2, respectively. From a clinical point of view, we have referenced many literature studies which have used the similar dataset that is used in our proposed model. This dataset only contains the images of the lungs of different patients irrespective of their demographic and individual information. Therefore, we have only added lung chest X-ray images for the experiments.
Table 4.
DATASET-3 ): Class-wise bifurcation of NIH Chest X-ray dataset
| Dataset bifurcation | Classes | |||||
|---|---|---|---|---|---|---|
| Atelectasis | Effusion | Infiltration | Nodule | Pneumonia | Total | |
| Train set | 3414 | 2788 | 7327 | 2248 | 3875 | 19,625 |
| Test set | 801 | 1167 | 2220 | 457 | 390 | 5035 |
| Total | 4215 | 3955 | 9547 | 2705 | 4265 | 24,660 |
Fig. 1.
Sample images from and dataset used for the experimentation purpose in the course of this study; a COVID-19 images, b Normal images, and c Pneumonia images
Fig. 2.
Sample images from dataset used for the experimentation purpose in the course of this study; a Atelectasis b Effusion, c Infiltration, d Nodule and e Pneumonia
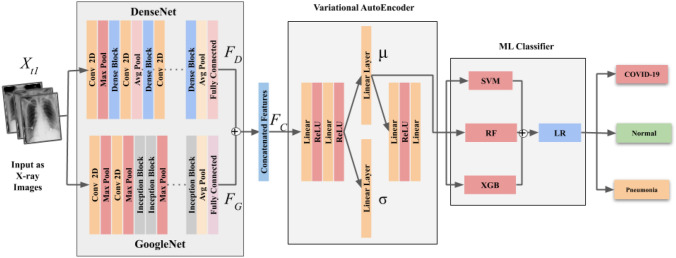
Network architecture
In this section, we propose the detailed involvement of deep learning modules for estimating the feasibility of using the chest X-rays to diagnose COVID-19. The concept of the stacked architecture of DenseNet and GoogleNet is explained here. Later, variational autoencoder is operated upon the output of stacked architecture. The deep modules are trained on dataset, which includes the set of images and their labels for Normal and Pneumonia patients. It contains a total of 4265 Pneumonia with 1575 Normal image samples.
DenseNet [30], in its basic architecture, is a deep CNN that helps eradicate the issue of vanishing-gradient efficiently using cross-connection by allowing the use of previously extracted features by the network [31]. This advantage is captured to achieve better performance with lesser computation. For the binary class classification, two fully connected (FC) layers with 1024 hidden features are added, which resulted in a final output of two classes. To fully utilize the network gradients, the ReLU activation function is used between the two FC layers, which does not activate all neurons at once and enhances the network for elevating the performance. At the approaching end of the model, logsoftmax function instead of the softmax function is encouraged to provide better numerical computation and gradient optimization. For the DenseNet neural network, the convolution layer’s key task is to extract the image features, preventing the error of manually extracting features. It consists of a series of feature maps. A convolution kernel holds these feature maps, and consists of weights as parameters, and is often called convolution filter.
The above convolution kernel consists of learnable parameters and is transformed using cross-connections with function maps of the previous layer. To further obtain the extracted function map, the resulting elements are processed, followed by an offset, and finally moved through a nonlinear ReLU activation.
An assortment of different convolution kernels allows the extraction of more complex features. The formula for calculation shall be as shown in Eq. 1.
| 1 |
where denotes the activation function with non-linearity. denotes the feature maps input to the convolution kernel selected by the layer, and signifies the convolution operation performed using kernel with represents the offset added to extracted kernel features. L denotes the number of layers in our convolution neural network, and means the convolution kernel used over input of the feature map between the L th layer and the feature map of the layer. After training, the output layer, last FC layer, and ReLU activation function are removed from the DenseNet model and the remaining part is used as a feature extractor. The model gives 1024 features as an output.
GoogleNet [32], is a state-of-the-art deep learning architecture that allows better and more computationally efficient results. This network consists of inception modules where every penultimate layer and the next layer is joined with four connections. First, a convolution is performed on the output of the previous layer. This is done because the output of the previous layer consists of some useful extracted features which further need to be used, then a and a convolutions are performed on output from the previous layer.
For dimensionality reduction, a filter is used before performing a and convolution. Also, a max-pooling layer is followed by a filter convolution, which helps achieve an optimal sparse structure. The model with pre-trained weights is trained on ImageNet [33] dataset, followed by replacing the last FC layer of the network with the addition of two FC layers having 1024 hidden features and final output of class probabilities. The activation function is amalgamated between the added layers. The network’s output layer is of the logsoftmax function with the ‘Adam’ optimizer to finally train the network. The individually trained DenseNet and GoogLeNet are now fused to form an ensemble acting as a feature extractor. The X-ray images of COVID-19, Normal, and Pneumonia constitute dataset , which consists of images and their corresponding labels as , respectively. The images are further divided into train and test sets where every image being resized to a dimension of . These images are then given as input to the trained model of GoogLeNet and DenseNet extracting 1024 features, represented as and , respectively, at the output, giving us and which contains the features and , respectively, as depicted in Eq. 2.
| 2 |
where are the total extracted output features with and and denoting the concatenation operation. and are then concatenated to form features with a dimension of 2048, which is used for training the variational autoencoder followed by ML-based classification features. Prior sending the feature vector from the aforementioned ensemble into the variational architecture, normalization is performed on to make it compatible with the autoencoder. For this, mean and standard deviation is calculated which is used to normalize to provide features .
The concatenated features have a very high dimension which brings out a requirement to use dimensionality reduction. For learning better distributions and sparse features from , we integrated a variational autoencoder. The variational aspect of the pipeline consists of an encoder-decoder assembly which rather than giving the latent space features, provides us with the distribution of the latent features in the form of its mean and standard deviation as:
| 3 |
The reparametrization of and is then performed by associating a constant with and adding the resultant to to give features .
| 4 |
The latent space encoder is then used to give the output feature space of 100 feature. A specialized loss function, , consists of two factors, one which penalizes the reconstruction error and another which allows the learned distribution to be similar to our predefined distribution which is assumed to be a Gaussian distribution. The loss function is a sum of binary cross-entropy loss i.e. and KL divergence loss i.e. as given:
| 5 |
where and denote the mean and standard deviation. However, in the proposed model (as shown in Fig. 3), and are used in Eq. 5 for and thereby giving us . In a nutshell, a VAE with and have regularised encoding distribution during training which gives mean and the standard deviation to understand the distribution of the latent space. Therefore, for increasing overall model robustness, are passed through to extract and which adequately represent the latent space distribution.
Fig. 3.
Schematic representation of the proposed framework depicting different modules for the classification of COVID-19 v/s Normal v/s Pneumonia classes
Machine learning based classification At the last stage, we employ the ML predictive classifiers wherein is used to train the stacked arrangement of three classifiers, namely Support Vector Machine (SVM) [34], Random Forest (RF) [35], and XGBoost [36]. The output of this ensembling is later predicted by logistic regression (LR). The framework gives the output, , which is the final label to the X-ray image which either belongs to COVID-19, Normal or Pneumonia patient. The above-proposed network architecture is described in Algorithm 1.
Experiments and results
Experimental setup
Implementation details The efficacy of the network performance is rigorously investigated using Python 3.8 with a processor of Intel Xeon(R) Gold 5120 CPU @ 2.20GHz with 93.1 GiB RAM on Ubuntu 18.04.2 LTS with NVIDIA Quadro P5000 with 16GB graphics memory.
Evaluation criteria
For evaluating the network robustness, confusion matrix with area under curve (AUC) property [37, 38] for ROC curves are estimated. They provide a detailed understanding of how well the model fits for final classification. AUC helps in checking how well a classifier is able to distinguish among various classes. Model’s performance is measured using the traditional metrics of Accuracy (Ac), Sensitivity (Sen), and Specificity (Spe) as in Eqs. 6, 7 and 8, respectively. as given in Eq. 9 is a measure that reports the balance between precision and recall.
ROC-AUC ROC curve is a graph that uses the parameters of true positive rate (TPR) and false positive rate (FPR). It shows the performance of a classification model at all classification thresholds. Area under the curve for ROC [37, 38] is an effective measure to check the efficacy of ML classifiers.
| 6 |
| 7 |
| 8 |
| 9 |
TP, TN, FP, and FN are true positive, true negative, false positive, and false negative, respectively.
Result analysis
To ethically make a comparative analysis and selection of the best CNN model for feature extraction, we initially displayed the baseline results on the in-depth modules, DenseNet, ResNet, and GoogleNet on the dataset. The baseline result is produced by training the models on the Normal and Pneumonia dataset, as shown in Fig. 4. The three chosen models are compared using the Area Under the Curve (AUC) property in Receiver Operating Characteristics (ROC) curves, as shown in Fig. 5. From the comparison, the two networks DenseNet and GoogLeNet, are selected and are worked upon for further analysis. We observed that the three deep learning modules’ baseline approach did not perform satisfactorily at the training stage (whose evaluated metrics are depicted in Table 5) and it is deduced that the performance of GoogleNet and DenseNet is better than ResNet. Therefore, to tackle the issue of appropriate and efficient feature extraction, only DenseNet and GoogLeNet are taken for further assessment.
Fig. 4.
Confusion matrix of the baseline results on DenseNet, ResNet and GoogLeNet
Fig. 5.
ROC curves of the baseline results on DenseNet, ResNet and GoogLeNet
Table 5.
Baseline training-based performance on the GoogLeNet, ResNet and DenseNet
| Module | Performance metrics | ||||
|---|---|---|---|---|---|
| Ac | Sen | Spe | F1-score | AUC | |
| GoogleNet | 0.876 | 0.837 | 0.837 | 0.857 | 0.957 |
| ResNet | 0.862 | 0.823 | 0.823 | 0.841 | 0.952 |
| DenseNet | 0.924 | 0.908 | 0.908 | 0.917 | 0.969 |
Keeping the above conditions in mind, we extend our experiments on dataset using the stacked architecture of DenseNet and GoogLeNet. The extracted features are then tested using SVM, whose results in the form of the confusion matrix and AUC in the ROC curves are depicted in Figs. 6 and 7, respectively. The test results have shown that some of the COVID-19 and Pneumonia samples are closely correlated. Therefore, the predicted results in the confusion matrix have mislabeled categories of both the classes. From the above results, we observed that the models with such high dimensions are highly computational. Also, features correlate with each other spatially with some semantic discrepancies among them. Therefore, for dimensionality reduction and to enrich the process with robust features, a better feature recognition segment, variational autoencoder, is used that will have better generalization ability.
Fig. 6.

Confusion Matrix of the DenseNet and GoogleNet feature Extraction modules with SVM-based classification
Fig. 7.

ROC curves of the DenseNet and GoogleNet feature Extraction modules with SVM-based classifiers
Variational autoencoder (VAE) To further upgrade the accuracy with fewer dimensions, we need to extract more sparsity among the features which will help us to generate more meaningful latent space. Variational autoencoder (VAE) is used for this purpose. The concatenated features are passed through VAE incorporating a non-linear dimensionality reduction technique, t-SNE [39], which is applied to extract the latent space of 100 features. t-SNE representation calculates a similarity match between the data instances in both high and low dimensions and later optimize them. The latent space visualization of 100 features is as shown in Fig. 9. From the figure, it is inferred that the Normal cases have easily been segregated while the COVID-19 and the Pneumonia cases are being overlapped which can be understood in a way that the pneumonia is observed in early stages of COVID-19.
Fig. 9.
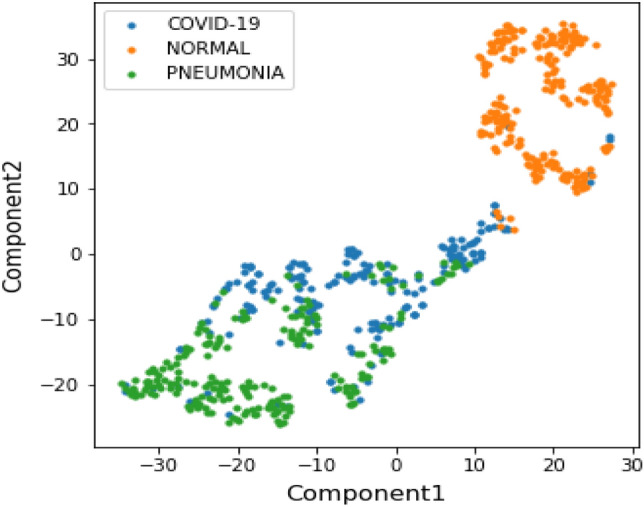
Visualization of latent space of VAE with 2 components using t-SNE
Quantitative analysis
We are now in a state of understanding the latent space distribution of the extracted features, which are further classified using the ML-based predictive classifiers. Considering the tactfulness of ML classifiers, we chose three basic classifiers, namely, SVM, RF, and XGBoost. These classifiers have successfully and efficiently classified the latent features into their basic classes. Finally, the probabilities predicted from these classifiers are used as meta-features for Logistic Regression to give the final label to our input images.
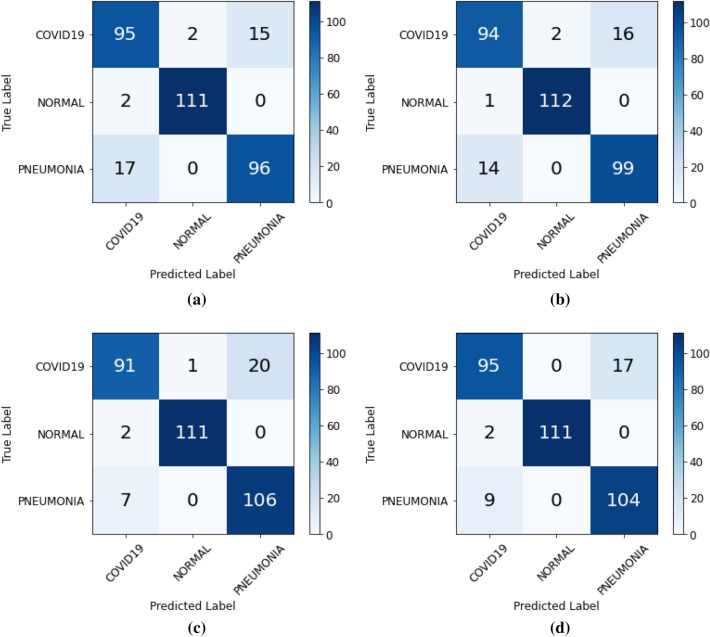
The classification results over dataset for XGB, RF and SVM have been visualized in the confusion matrix as shown in Fig. 8a–c, respectively. Figure 8d depicts the classification results for the ensemble of XGB, RF, and SVM classifiers with a final estimator of Logistic Regression. From these confusion matrices, various evaluation metrics were calculated for each classifier, as shown in Table 6. The achieved Ac using SVM, RF, and XGB is 0.911, 0.902, and 0.893, respectively. The accuracy of SVM is highest in comparison to the other two classifiers. The maximum AUC for the three classifiers is recognized RF with a value of 0.974. The combined ROC curves for the three ML-based classifiers and the combined final estimator of LR is also shown in Fig. 11. These curves helps us to find the most optimal model out of all four machine learning classifiers. The ensemble of ML classifiers with the LR as a final estimator is the most optimal and robust model with the maximum area under the ROC curve being 0.976. The ensemble classifier gave the best results compared to other machine learning classifiers with an Ac, Sen, Spe, F1-score, and AUC of 0.917, 0.916, 0.958, 0.917 and 0.976, respectively. The ensemble classifier outperforms other ML classifiers.
Fig. 8.
Confusion matrix for a XGB, b RF, c SVM, and d Ensemble of XGB, RF and SVM classifiers displaying the final classification among the three classes of COVID-19, Normal and Pneumonia
Table 6.
Performance evaluation of ML-based classifiers on X-ray images for classification between COVID-19, Normal, and Pneumonia patients
| Module | Performance metrics | ||||
|---|---|---|---|---|---|
| Ac | Sen | Spe | F1-score | AUC | |
| SVM | 0.911 | 0.910 | 0.955 | 0.910 | 0.971 |
| RF | 0.902 | 0.902 | 0.951 | 0.901 | 0.974 |
| XGB | 0.893 | 0.893 | 0.946 | 0.893 | 0.972 |
| Ensemble | 0. 917 | 0.916 | 0.958 | 0.917 | 0.976 |
The results of our proposed approach are shown in bold
Fig. 11.

ROC for Machine Learning based classification among the three classes of COVID-19, Normal and Pneumonia
Classification of COVID-19 using CT images
In this study, we have also experimented with the Computer Tomographic (CT) images of COVID-19.
Dataset description The Dataset comprises of two categories, namely, COVID-19 and Normal. The training and test sets consisted of 498 and 200 images, respectively, which in turn contains equal proportions of COVID-19 and Normal images [40]. The results of the experimentation are shown in Table 7. Due to the sparse availability of the CT pneumonia images’, deep networks are trained on CT image samples of COVID-19.
Table 7.
Performance evaluation of ML-based classifiers for classification between COVID-19, Normal, Pneumonia using CT images
| Module | Performance metrics | ||||
|---|---|---|---|---|---|
| Ac | Sen | Spe | F1-score | AUC | |
| SVM | 0.805 | 0.805 | 0.805 | 0.805 | 0.805 |
| RF | 0.825 | 0.825 | 0.825 | 0.825 | 0.825 |
| XGB | 0.810 | 0.810 | 0.810 | 0.810 | 0.810 |
| Ensemble | 0.820 | 0.820 | 0.820 | 0.820 | 0.820 |
The results of our proposed approach are shown in bold
Model performance Table 7 demonstrates the quantitative results on the CT images for COVID-19 detection among COVID-19, Normal, and Pneumonia patient images. It is inferred that the proposed model has performed well on the corresponding COVID-19 CT images. The final classification accuracy of the assemblage of ML classifiers is , which is satisfactory but less compared to the results obtained on the X-ray COVID-19 images as mentioned in Table 6. It is a known fact that CT images are detailed when compared to the X-ray images, which helps in providing aid to radiologists for diagnostic purposes. Nevertheless, during this pandemic crisis, the easy availability of the CT images was quite troublesome. Therefore, the readily available X-ray images are taken, showing better results for the proposed framework.
Multi-class classification results on a challenging dataset
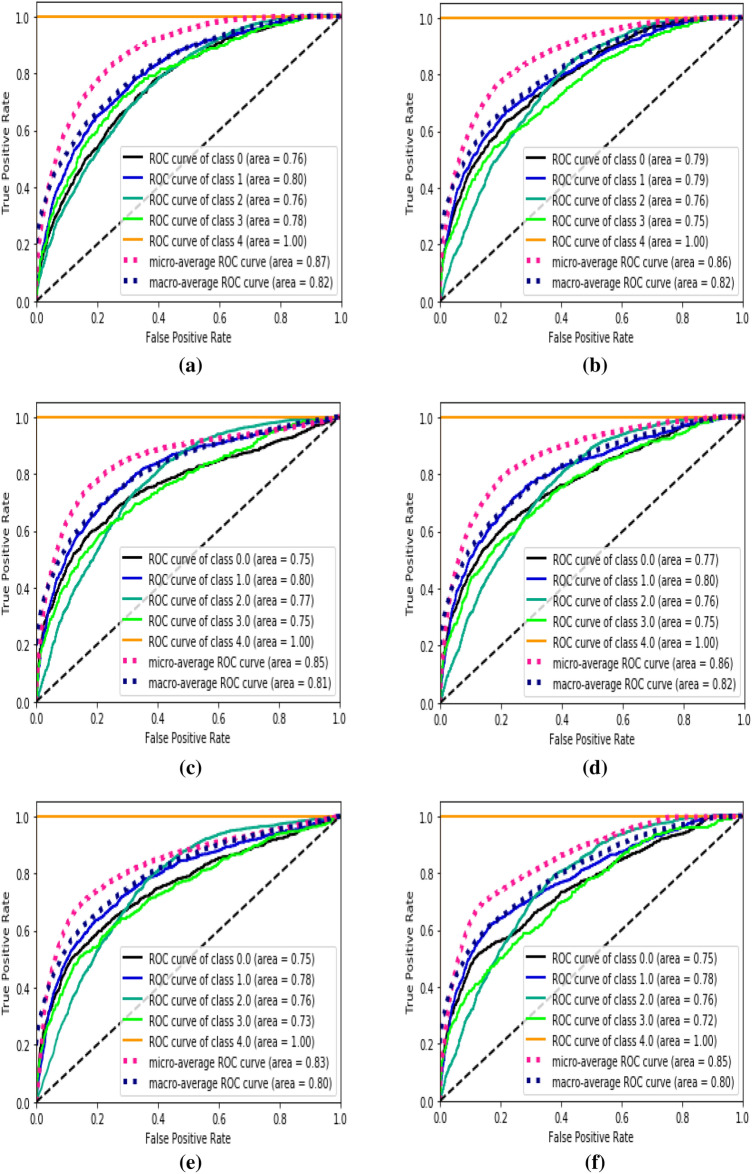
To further analyze the viability of our proposed framework, we implement the framework on dataset. For this, we have reported the results for multi-class classification results for Atelectasis, Effusion, Infiltration, Nodule, and Pneumonia image samples. Figure 10 shows the corresponding ROC curves for the different stage results on the aforementioned dataset. Figure 10a and b show the ROC curves for the initial training of the DenseNet and GoogLeNet for the above mentioned five classes with Fig. 10c–e showing the corresponding final 5-label classification by SVM, RF, and XGB classifiers, respectively.
Fig. 10.
ROC curves for a DenseNet, b GoogLeNet, c SVM, d RF, e XGB and f Ensemble of XGB, RF and XGB, displaying the multi-class classification results on the NIH chest X-ray images
The extracted latent space from the VAE is converted into components using t-SNE, which is visualized in Fig. 12. It can be seen from the figure that many data points are distorted from the cluster of their labeled classes which in turn affects the performance of the chosen classifiers which is clearly visible in Table 8. From the table, it can be observed that the quantitative performance varies greatly, in which the proposed pipeline has performed superior to the works done by Xiaosong et al. [20]. The comparative study for the AUC for multi-class classification for four classes (leaving out Pneumonia) is as depicted in Table 9 where the proposed method has outperformed the state-of-the-art results.
Fig. 12.

Visualization of latent space of VAE with 2 components using t-SNE for Dataset
Table 8.
Performance evaluation of ML-based classifiers for multi-class classification on dataset
| Module | Performance metrics | ||||
|---|---|---|---|---|---|
| Ac | Sen | Spe | F1-score | AUC | |
| GoogleNet | 0.603 | 0.586 | 0.882 | 0.591 | 0.818 |
| DenseNet | 0.614 | 0.597 | 0.886 | 0.605 | 0.813 |
| SVM | 0.614 | 0.597 | 0.886 | 0.605 | 0.813 |
| RF | 0.611 | 0.589 | 0.884 | 0.598 | 0.815 |
| XGB | 0.609 | 0.594 | 0.884 | 0.600 | 0.803 |
| Ensemble | 0.616 | 0.597 | 0.886 | 0.605 | 0.802 |
The results of our proposed approach are shown in bold
Table 9.
Comparative analysis of the AUC for multi-class classification for the proposed model setting
| Study | Classes | |||
|---|---|---|---|---|
| Atelectasis | Effusion | Infiltration | Nodule | |
| Wang et al. [20] | 0.70 | 0.73 | 0.61 | 0.71 |
| Our approach | 0.75 | 0.78 | 0.76 | 0.72 |
The results of our proposed approach are shown in bold
Discussion
The recent developments in the DL-based techniques using feature extraction and image processing for COVID-19 classification have marked new opportunities in the field of medical imaging. Automatic classification of COVID-19 among COVID-19, Normal and Pneumonia image samples proves to be a significant step for clinical interpretation and treatment planning by the CADx systems. The proposed model is divided into image feature extraction using DL models, enhancing features using VAE with final classification using ML-based predictive classifiers. The input images are passed through the CNN models: DenseNet and GoogleNet, to generate features individual to each architecture, which are later concatenated to form a feature vector. This feature vector is then fed to a VAE that learns meaningful latent space for easy and effective patterns of the input features. The output of the VAE are then used as input for ML-based predictive classifiers for classification. This multi-stage classification process has enabled the network to effectively identify the image patterns and helps improve the accuracy and robustness of the model.
For comparison, we have investigated the existing works which are as tabulated in Table 10. Mohamed et al. [17] demonstrated the use of GAN with deep transfer learning among COVID-19, Normal and Pneumonia X-ray image samples using the dataset created by Joseph et al. [18] that achieved an Ac, Sen, and F1-score of 0.8148, 0.8148, and 0.8146, respectively. Another work on the classification task of COVID-19 was performed by Enzo et al. in [19] utilizing the datasets [18, 20, 21] with the Pneumonia images 3 have achieved considerable performance. Yifan et al. [24] has also introduced the use of CNN in the classification of COVID-19 with the fine-tuning performance with recall and precision of 0.75 and 0.64, respectively. However, the results produced by the proposed model outperformed the state-of-the-art (SOTA) techniques [17] with an enhancement of , and on Ac, Sen, and F1-score, respectively. Based on the proposed architecture, the method has outshown the overall classification process.
Table 10.
Performance comparison of the proposed classification scheme among COVID-19, Normal and Pneumonia with the state-of-the-art methodologies
| Study | Dataset type | Performance metrics | ||||
|---|---|---|---|---|---|---|
| Ac | Sen | Spe | F1-score | AUC | ||
| Mohamed et al. [17] | X-ray | 0.81 | 0.81 | – | 0.84 | – |
| Enzo et al. [19] | X-ray | 0.65 | 0.71 | 0.52 | 0.73 | – |
| Yifan et al. [24] | X-ray | – | 0.75 | – | 0.64 | 0.94 |
| Our approach | X-ray | 0.91 | 0.91 | 0.95 | 0.91 | 0.97 |
The results of our proposed approach are shown in bold
Conclusion
This study proposes a novel framework to classify diseased images of people using their chest X-rays among COVID-19, Normal, and Pneumonia. It is the need of the hour to have such a technique for COVID-19 classification that is cost-effective and practically accurate . This study uses state-of-the-art DL architectures for feature extraction and image processing. The input images are passed through the CNN models, DenseNet and GoogleNet, to generate individual features which are later concatenated. These concatenated feature vectors are sent to a variational autoencoder that learns a meaningful latent space from the features and further inputs the extracted features to ML-based classifiers. ML-based predictive classifiers perform final predictions that helps improve the accuracy and robustness of the model. Thus, the proposed study helps to achieve an overall accuracy and AUC of 0.91 and 0.97, respectively. Further, we have tested the proposed framework’s scalability and efficacy on a challenging dataset to classify between Atelectasis, Effusion, Infiltration, Nodule, and Pneumonia and to check for viability and the diverse nature of the framework in applicant fields of medical image classification. The results of this dataset showed significant improvement over the state-of-the-art methodologies. Future aspects may extend this study to the datasets having more number of images with some other biological and physical parameters, which will help improve the results and the model’s viability in the field of AI-based detection of COVID-19 and other lungs related diseases.
Acknowledgements
We acknowledge Consulate General of India, Osaka-Kobe for constant support and encouragement for this Bilateral India-Japan Artificial Intelligence (INJA-AI) module between the researchers of Indian Institute of Technology Roorkee (IITR), India and Kyoto University, Japan to better manage COVID-19 by non-invasive prediction.
Funding
There is no funding provided in the course of this study.
Data availability
This article does not contain any studies with human participants or animals performed by any of the authors. All the database is acquired from the public logging system (Internet source) whose appropriate references are added in the sections above.
Declarations
Conflict of interest
All authors declare that they have no conflict of interest, financial or otherwise.
Code availability
Any public available code is cited in the text at appropriate places and the novel code for the work can taken from the authors upon request.
Ethical approval
Not applicable.
Informed Consent
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ridhi Arora, Vipul Bansal, Himanshu Buckchash, Rahul Kumar and Narayanan Narayanan contributed equally to this work.
Contributor Information
Ridhi Arora, Email: rarora@cs.iitr.ac.in.
Vipul Bansal, Email: vbansal@me.iitr.ac.in.
Himanshu Buckchash, Email: hbuckchash@cs.iitr.ac.in.
Rahul Kumar, Email: rkumar9@cs.iitr.ac.in.
Vinodh J. Sahayasheela, Email: vinodh@chemb.kuchem.kyoto-u.ac.jp
Narayanan Narayanan, Email: nn@annalakshmi.net.
Ganesh N. Pandian, Email: ganesh@kuchem.kyoto-u.ac.jp
Balasubramanian Raman, Email: bala@cs.iitr.ac.in.
References
- 1.Boopathi S, Poma AB, Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J Biomol Struct Dyn. 2021;39(9):3409–3418. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das NN, Kumar N, Kaur M, Kumar V, Singh D (2020) Automated deep transfer learning-based approach for detection of COVID-19 infection in chest x-rays. Irbm [DOI] [PMC free article] [PubMed]
- 3.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I, et al. Broad-spectrum antiviral gs-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9(396):eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization; Naming the coronavirus disease (COVID-19) and the virus that causes it. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it. Accessed 6 June 2020
- 5.World Health Organization; WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/. Accessed 17 Jan 2020
- 6.Worldometers: COVID-19 CORONAVIRUS PANDEMIC. https://www.worldometers.info/coronavirus/. Accessed 6 June 2020
- 7.World Health Organization; WHO releases guidelines to help countries maintain essential health services during the COVID-19 pandemic. https://www.who.int/news-room/detail/30-03-2020-who-releases-guidelines-to-help-countries-maintain-essential-health-services-during-the-covid-19-pandemic. Accessed 6 June 2020
- 8.Centers for Disease Control and Prevention; Symptoms of Coronavirus. cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html. Accessed 6 June 2020
- 9.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. Correlation of chest ct and rt-pcr testing in coronavirus disease, (2019) (covid-19) in china: a report of 1014 cases. Radiology. 2020;296:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong W, Agarwal PP. Chest imaging appearance of covid-19 infection. Radiol Cardiothorac Imaging. 2020;2(1):e200028. doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, Fan Y, Zheng C. Radiological findings from 81 patients with covid-19 pneumonia in wuhan, china: a descriptive study. Lancet Infect Dis. 2020;20:425. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santosh KC. Ai-driven tools for coronavirus outbreak: need of active learning and cross-population train/test models on multitudinal/multimodal data. J Med Syst. 2020;44(5):1–5. doi: 10.1007/s10916-020-01562-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbas A, Abdelsamea MM, Gaber MM. Classification of covid-19 in chest x-ray images using detrac deep convolutional neural network. Appl Intell. 2021;51:854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majeed T, Rashid R, Ali D, Asaad A. Issues associated with deploying cnn transfer learning to detect covid-19 from chest x-rays. Phys Eng Sci Med. 2020;43(4):1289–1303. doi: 10.1007/s13246-020-00934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loey M, Smarandache F, Khalifa NEM. Within the lack of chest covid-19 x-ray dataset: a novel detection model based on gan and deep transfer learning. Symmetry. 2020;12(4):651. doi: 10.3390/sym12040651. [DOI] [Google Scholar]
- 18.Cohen JP, Morrison P, Dao L (2020) Covid-19 image data collection. arXiv:2003.11597
- 19.Tartaglione E, Barbano CA, Berzovini C, Calandri M, Grangetto M (2020) Unveiling covid-19 from chest x-ray with deep learning: a hurdles race with small data. arXiv preprint http://arxiv.org/abs/2004.05405 [DOI] [PMC free article] [PubMed]
- 20.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE conference on computer vision and pattern recognition. pp 2097–2106
- 21.Kermany D, Zhang K, Goldbaum M. Labeled optical coherence tomography (oct) and chest x-ray images for classification. Mendeley Data. 2018 doi: 10.17632/rscbjbr9sj.2. [DOI] [Google Scholar]
- 22.Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X, et al. A deep learning algorithm using ct images to screen for corona virus disease (covid-19) Eur Radiol. 2021;24(1–9):2020. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayoudh K, Hamdaoui F, Mtibaa A. Hybrid-covid: a novel hybrid 2d/3d cnn based on cross-domain adaptation approach for covid-19 screening from chest x-ray images. Phys Eng Sci Med. 2020;43(4):1415–1431. doi: 10.1007/s13246-020-00957-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Niu S, Qiu Z, Wei Y, Zhao P, Yao J, Huang J, Wu Q, Tan M (2020) Covid-da: deep domain adaptation from typical pneumonia to covid-19. arXiv preprint arXiv:2005.01577,
- 25.Rahmatizadeh S, Valizadeh-Haghi Sh, Dabbagh A. The role of artificial intelligence in management of critical covid-19 patients. J Cell Mol Anesth. 2020;5(1):16–22. [Google Scholar]
- 26.Chest X-Ray Images (Pneumonia). https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia. Accessed 6 June 2020
- 27.COVID-19 Radiography Database. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database. Accessed 6 June 2020
- 28.COVID Dataset. https://drive.google.com/a/cs.iitr.ac.in/uc?id=1coM7x3378f-Ou2l6Pg2wldaOI7Dntu1a. Accessed 6 June 2020
- 29.Figure1-COVID-chestxray-dataset. https://github.com/agchung/Figure1-COVID-chestxray-dataset. Accessed 6 June 2020
- 30.Huang G, Liu Z, Van DML, Weinberger KQ (2017) Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
- 31.Liu Y, Pan J, Su Z, Tang K. Robust dense correspondence using deep convolutional features. Vis Comput. 2019;36:1–15. [Google Scholar]
- 32.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 1–9
- 33.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In Adv Neural Inf Process Syst. 2012;25:1097–1105. [Google Scholar]
- 34.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273–297. [Google Scholar]
- 35.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 36.Chen T, Guestrin C (2016) Xgboost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, pp 785–794
- 37.Zweig Mark H, Gregory C. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39(4):561–577. doi: 10.1093/clinchem/39.4.561. [DOI] [PubMed] [Google Scholar]
- 38.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 39.van der Maaten L, Hinton G. Visualizing data using t-sne. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 40.Zhao J, Zhang Y, He X, Xie P (2020) Covid-ct-dataset: a ct scan dataset about covid-19. arXiv preprint arXiv:2003.13865
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article does not contain any studies with human participants or animals performed by any of the authors. All the database is acquired from the public logging system (Internet source) whose appropriate references are added in the sections above.