Abstract
Purpose: The aim of the present study was to screen differential metabolites of gastric cancer (GC) and identify the key metabolic pathways of GC.
Methods: GC (n=28) and matched paracancerous (PC) tissues were collected, and LC-MS/MS analysis were performed to detect metabolites of GC and PC tissues. Metabolite pathways based on differential metabolites were enriched by MetaboAnalyst, and genes related to metabolite pathways were identified using the KEGGREST function of the R software package. Transcriptomics data from The Cancer Genome Atlas (TCGA) was analyzed to obtain differentially expressed genes (DEGs) of GC. Overlapping genes were acquired from metabonimics and transcriptomics data. Pathway enrichment analysis was performed using String. The protein expression of genes was validated by the Human Protein Atlas (HPA) database.
Results: A total of 325 key metabolites were identified, 111 of which were differentially expressed between the GC and PC groups. Seven metabolite pathways enriched by MetaboAnalyst were chosen, and 361 genes were identified by KEGGREST. A total of 2831 DEGs were identified from the TCGA cohort. Of these, 1317 were down-regulated, and 1636 were up-regulated. Twenty-two overlapping genes were identified between genes related to metabolism and DEGs. Glycerophospholipid (GPL) metabolism is likely associated with GC, of which AGPAT9 and ETNPPL showed lower expressed in GC tissues.
Conclusions: We investigated the tissue-based metabolomics profile of GC, and several differential metabolites were identified. GPL metabolism may affect on progression of GC.
Keywords: Gastric cancer, Glycerophospholipid metabolism, Metabolomics, Transcriptomics
Introduction
Gastric cancer (GC) is one of the most common malignant tumors and ranks fifth as the cause of death among 36 cancers in the world [1]. Approximately 50% of cancer patients in China have gastrointestinal tumors, mainly in GC, and the 5-year survival rate is less than 35% [2]. GC has a multistep progression [3]. The 5-year survival rate of patients with advanced GC is less than 20%, but it may reach more than 90% if it only invades the mucosal or submucosal layer [4]. However, GC has no specific clinical symptoms in the early stage, and most patients are in the middle and advanced stage when diagnosed, which leads to a poor prognosis. Therefore, it is imperative to explore the mechanism of GC and identify biomarkers for early diagnosis.
Metabolomics is a novel technique that explores the biological states of metabolites in tissue extracts and body fluids such as plasma, serum and urine [5,6] and has been used to characterize metabolic disorders and identify potential biomarkers of various cancers [7–9]. Early studies on GC using metabolomics were mainly performed on plasma [10–12], serum [13,14], and urine [15] samples, and there were few studies involved in GC tissues. Metabolic profiling of patients with GC using tissue samples with and without lymph node metastasis (LNM) was performed, and some differential metabolites were identified as potential factors for the diagnosis and prognosis of GC patients with or without LNM [16]. The present study aimed to screen differential metabolites and reveal metabolic pathways related with GC with the help of transcriptomics data from TCGA.
Materials and methods
Patient selection
Patients recruited in the study were diagnosed with GC according to results of a gastroscopy examination and a biopsy. Inclusion criteria included (i) aged between 20 and 80 years old, both male and female; (ii) primary tumor; and (iii) no previous treatment for cancer such as radiation, operation, and chemoradiotherapy. Exclusion criteria included (i) metabolic disease including hyperlipoidemia, diabetes mellitus and gout; (ii) congenital diseases; (iii) severe gastric cancer (survival < 2 months); and (iv) distant metastases.
The present study was approved by the Ethics Committee of Henan Provincial People’s Hospital, and all subjects provided informed consent. The registration number is ChiCTR2100041912.
Sample collection
GC and matched paracancerous (PC) tissues were collected from dissected specimens of patients undergoing radical gastrectomy of the same anatomical region and they were obtained from mucosa to muscle layers but serosa was removed. PC tissues were 5 cm from the cancer tissues. Samples were washed immediately with phosphate buffered saline and frozen in liquid nitrogen and stored at −80°C. The pathological stages were determined based on the tumor node metastasis (TNM) staging system from the American Joint Cancer Committee/Union for International Cancer Control (AJCC/UICC).
Preparation and LC-MS/MS analysis of tissue samples
Prior to analysis, 50 mg of the tissue sample was weighed and 1000 μl of extract solution (acetonitrile: methanol: water = 2:2:1, with an isotope-labeled internal standard mixture) was added. After vortexing for 30 s, samples were grind for 4 min and homogenized by sonicating for 5 min in an ice-water bath. These steps were repeated three more times. Samples were incubated for 1 h at −40°C and centrifuged at 12000 rpm for 15 min at 4°C. The supernatant was transferred to a fresh glass vial for analysis.
LC-MS/MS analysis were performed using an ultra-high-performance liquid chromatography (UHPLC) system (Vanquish, Thermo Fisher Scientific) with a UPLC BEH amide column (2.1 mm * 100 mm, 1.7 μm) coupled to the Q Exactive HFX mass spectrometer (Orbitrap MS, Thermo). The mobile phase was composed of 25 mmol/L ammonium acetate and 25 ammonia hydroxide in water (pH 9.75) and acetonitrile [17]. The injection volume was 3 ml and the auto-sampler temperature was 4°C. The Q Exactive HFX mass spectrometer was used to acquire MS/MS spectra in information-dependent acquisition (IDA) mode in the control panel of the acquisition software (Xcalibur, Thermo). In this mode, the acquisition software continuously evaluated the full-scan MS spectrum. The ESI source conditions were set as follows: sheath gas flow rate: 25 Arb, Aux gas flow rate: 20 Arb, capillary temperature: 350°C, full MS resolution: 60000, MS/MS resolution: 7500, collision energy: 10/30/60 in NCE mode, and spray voltage: 3.6 kV (positive) or -3.2 kV (negative).
Data preprocessing and annotation
The original data were converted to mzXML format by ProteoWizard and R package XCMS (version 3.2) [18]. An in-house program was used for peak detection, extraction, alignment, and integration. Then an in-house MS2 database (BiotreeDB, V2.1) was used for in metabolite annotation [18]. The cutoff for annotation was set at 0.3.
Statistical analysis
The data were processed using the SIMCA 16.0.2 software package (Sartorius Stedim Data Analytics AB, Umea, Sweden) [19,20] for principal component analysis (PCA) and orthogonal projections to latent structures-discriminant analysis (OPLS-DA). As an unsupervised pattern recognition method, PCA shows the distribution of origin data and general separation. OPLS-DA was used to obtain maximal covariance between variables and the sample category in both positive and negative models. Seven-fold cross-validation and 200 permutation tests were used to estimate the robustness and the predictive ability of our model [21].
Variables with variable importance in the projection (VIP) > 0.5 were defined as key metabolites. MetaboAnalyst (https://www.metaboanalyst.ca/) was used to perform a search for metabolite pathways based on key metabolites, and metabolite pathways that satisfied the condition of P<0.05 were chosen for further analysis.
Integration of metabolomics and transcriptomics data
Genes related to metabolite pathways were identified using the KEGGREST function of the R software package. GC transcriptomic data were obtained from The Cancer Genome Atlas (TCGA) and comprised 375 GC and 32 non-tumor tissues. The DESeq2 R package was used to conduct normalization and differential gene expression analysis, and we specified |log2FC| (fold change) > 2 and the P value < 0.05 as cutoffs to identify differentially expressed genes (DEGs). A Venn diagram was generated by VENNY 2.1 to obtain overlapping genes.
Identification of key signaling pathways, genes, and metabolites
String (https://string-db.org/) was used to find Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways indicating the interaction between proteins. KEGG pathways were obtained using String to identify signaling pathways enriched by overlapping genes. The diagram of the metabolic pathway with the minimum value, glycerophospholipid (GPL) metabolism, for the false discovery rate (FDR) was drafted. The protein expression of genes related to GPL metabolism between GC and normal tissues was determined using immunohistochemistry (IHC) from the Human Protein Atlas database (HPA) (https://www.proteinatlas.org/).
Results
Baseline clinical characteristics of patients
A total of 28 GC tissues and 28 matched PC tissues were collected. Characteristics of subjects were showed in Table 1, including age, gender, weight, height, body mass index (BMI), hobbies, types of tumor localization, and pathologic tumor stages.
Table 1. The characteristics summary of subjects.
| Characteristics | Patients (n=28) |
|---|---|
| Age (years, means ± SD) | 60 ± 9 |
| Gender (female/male) | 9/19 |
| Weight (kg, means ± SD) | 58.6 |
| Height (m, means ± SD) | 1.65 |
| BMI (kg/m2, means ± SD) | 21.62 |
| Hobbies | |
| Smoking | 15 |
| Drinking | 10 |
| Tumor localizations, no | |
| Cardia | 5 |
| Fundus of stomach | 1 |
| Body of stomach | 5 |
| Lesser curvature | 8 |
| Antrum | 9 |
| Pathologic tumor stages, no | |
| I (IA, IB) | 1 |
| II (IIA, IIB) | 7 |
| III (IIIA, IIIB) | 10 |
| IV (IVA, IVB) | 10 |
Note: BMI, body mass index.
Metabolic profiles of gastric cancer
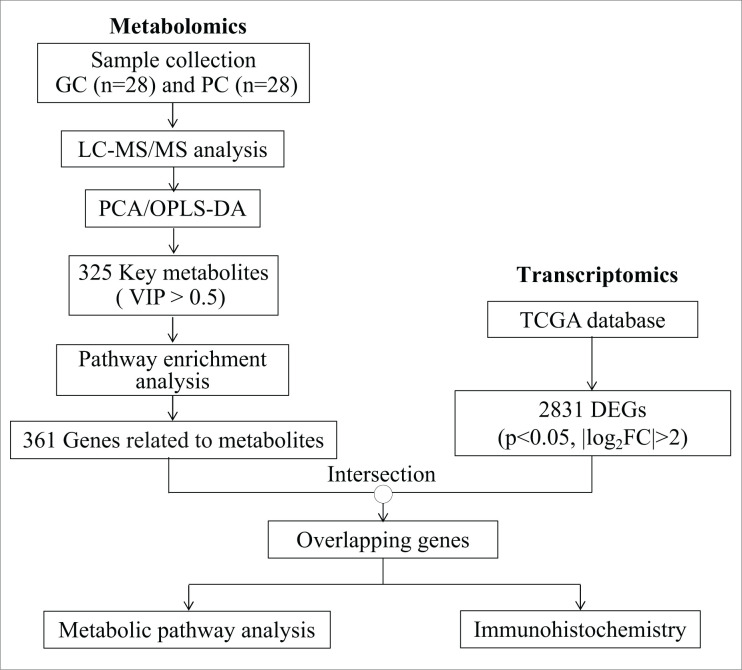
An overview of the study profile is shown in Figure 1. A total of 4893 peaks (negative ion mode: 2163, positive ion mode: 2730) were obtained, and 485 metabolites (negative ion mode: 145, positive ion mode: 340) were identified. There were different mass spectrum peaks identified between the GC and PC groups, as shown in Supplementary Figure S1. Examination of the PCA score plots (Supplementary Figure S2a–d) showed that most of the samples were within a 95% confidence interval (CI) but failed to provide satisfactory separation of data. Permutation tests of the OPLS-DA models for the two groups were carried out to prevent overfit of models (Supplementary Figure S2e and S2f), and the results showed that the models have good predictability and do not overfit.
Figure 1. An overview of study profile for identifying metabolites and metabolic pathways of GC.
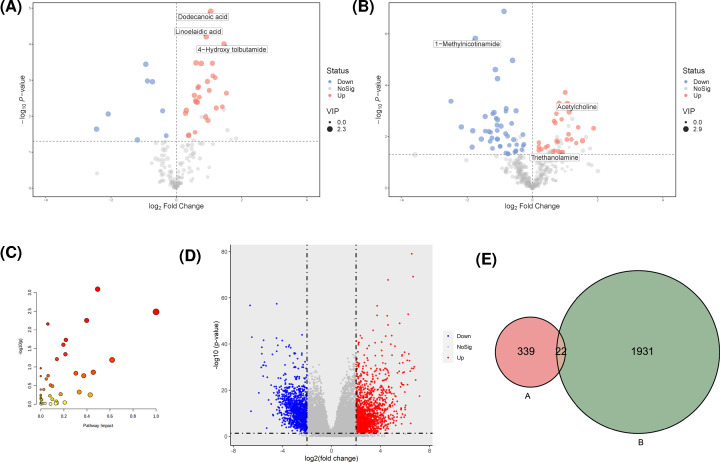
A total of 325 key metabolites (Supplementary Table S1) were identified under the condition of VIP > 0.5. Of these metabolites, there were 35 significantly different peaks (27 up-regulated and 8 down-regulated) in the negative ion model, and 76 significantly different peaks (29 up-regulated and 47 down-regulated) in the positive ion model (P<0.05 and VIP > 1, Figure 2A,B).
Figure 2. Integration analysis of metabolomics and transcriptomics data.
(A) Volcano plots derived from negative ion mode of GC patients. (B) Volcano plot derived from positive ion mode of GC patients. The red and blue points represented up-regulated and down-regulated genes, respectively. (C) Bubble analysis of metabolic pathways between GC and PC groups. Ordinate showed the significance and abscissa represented the impact of pathway. (D) Volcano plot of DEGs from TCGA. The red and blue points represented up-regulated and down-regulated genes, respectively. (E) Venn diagram of metabolic pathways related genes and DEGs. (A) represented genes related with metabolites and (B) indicated DEGs obtained from TCGA.
Pathway enrichment analysis
Pathway enrichment analysis was performed to identify signaling pathways related to key metabolites. A total of 48 metabolic pathways were found (Figure 2C and Table 2); of these, seven were chosen (P<0.05) for further analysis, including alanine, aspartate and glutamate metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, glycerophospholipid metabolism, pantothenate and CoA biosynthesis, purine metabolism, arginine and proline metabolism and sphingolipid metabolism. The KEGGREST function of the R software package was used to identify genes involved in the seven metabolite pathways, and 361 genes were obtained after removal of duplicates (Supplementary Table S2).
Table 2. Details of pathway analyses based on metabolomics data.
| Pathway description | Total | Count | P.value | FDR |
|---|---|---|---|---|
| Alanine, aspartate, and glutamate metabolism | 28 | 9 | 0.0008 | 0.0681 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | 4 | 3 | 0.0033 | 0.1389 |
| Glycerophospholipid metabolism | 36 | 9 | 0.0056 | 0.1459 |
| Pantothenate and CoA biosynthesis | 19 | 6 | 0.0069 | 0.1459 |
| Purine metabolism | 65 | 12 | 0.0187 | 0.3138 |
| Arginine and proline metabolism | 38 | 8 | 0.0251 | 0.3521 |
| Sphingolipid metabolism | 21 | 5 | 0.0451 | 0.5414 |
| Histidine metabolism | 16 | 4 | 0.0610 | 0.5987 |
| Phenylalanine metabolism | 10 | 3 | 0.0641 | 0.5987 |
| D-glutamine and D-glutamate metabolism | 6 | 2 | 0.1077 | 0.9043 |
| Beta-alanine metabolism | 21 | 4 | 0.1380 | 0.9817 |
| Arginine biosynthesis | 14 | 3 | 0.1466 | 0.9817 |
| Butanoate metabolism | 15 | 3 | 0.1710 | 0.9817 |
| Nicotinate and nicotinamide metabolism | 15 | 3 | 0.1710 | 0.9817 |
| Aminoacyl-tRNA biosynthesis | 48 | 7 | 0.1753 | 0.9817 |
| Cysteine and methionine metabolism | 33 | 5 | 0.2088 | 1.0000 |
| Ether lipid metabolism | 20 | 3 | 0.3044 | 1.0000 |
| Pyrimidine metabolism | 39 | 5 | 0.3238 | 1.0000 |
| Glycine, serine, and threonine metabolism | 33 | 4 | 0.3984 | 1.0000 |
| Linoleic acid metabolism | 5 | 1 | 0.3993 | 1.0000 |
| Arachidonic acid metabolism | 36 | 4 | 0.4662 | 1.0000 |
| Pentose and glucuronate interconversions | 18 | 2 | 0.5324 | 1.0000 |
| Ascorbate and aldarate metabolism | 8 | 1 | 0.5579 | 1.0000 |
| Valine, leucine and isoleucine biosynthesis | 8 | 1 | 0.5579 | 1.0000 |
| Taurine and hypotaurine metabolism | 8 | 1 | 0.5579 | 1.0000 |
| Citrate cycle (TCA cycle) | 20 | 2 | 0.5912 | 1.0000 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 9 | 1 | 0.6009 | 1.0000 |
| Propanoate metabolism | 23 | 2 | 0.6688 | 1.0000 |
| Biosynthesis of unsaturated fatty acids | 36 | 3 | 0.6934 | 1.0000 |
| Glycolysis/Gluconeogenesis | 26 | 2 | 0.7341 | 1.0000 |
| Alpha-linolenic acid metabolism | 13 | 1 | 0.7352 | 1.0000 |
| Galactose metabolism | 27 | 2 | 0.7533 | 1.0000 |
| Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 14 | 1 | 0.7610 | 1.0000 |
| Starch and sucrose metabolism | 18 | 1 | 0.8416 | 1.0000 |
| Pentose phosphate pathway | 22 | 1 | 0.8952 | 1.0000 |
| Pyruvate metabolism | 22 | 1 | 0.8952 | 1.0000 |
| Fatty acid degradation | 39 | 2 | 0.9051 | 1.0000 |
| Tyrosine metabolism | 42 | 2 | 0.9262 | 1.0000 |
| Folate biosynthesis | 27 | 1 | 0.9375 | 1.0000 |
| Phosphatidylinositol signaling system | 28 | 1 | 0.9437 | 1.0000 |
| Fatty acid biosynthesis | 47 | 2 | 0.9519 | 1.0000 |
| Inositol phosphate metabolism | 30 | 1 | 0.9542 | 1.0000 |
| Glyoxylate and dicarboxylate metabolism | 32 | 1 | 0.9628 | 1.0000 |
| Amino sugar and nucleotide sugar metabolism | 37 | 1 | 0.9779 | 1.0000 |
| Fatty acid elongation | 39 | 1 | 0.9821 | 1.0000 |
| Valine, leucine, and isoleucine degradation | 40 | 1 | 0.9839 | 1.0000 |
| Primary bile acid biosynthesis | 46 | 1 | 0.9914 | 1.0000 |
| Steroid hormone biosynthesis | 85 | 1 | 0.9999 | 1.0000 |
Note: FDR, false discovery rate.
Differentially expressed genes in GC
To explore genes related to the metabolism of GC and establish relationships between genes and metabolites, DEGs between GC and non-tumor tissues from the TCGA were screened. A total of 2831 DEGs were identified as shown in Figure 2D, of which 1195 were down-regulated and 1636 were up-regulated. 878 DEGs were failed to be identified as gene, then excluded. So, 1953 DEGs were included in further analysis.
Potential biomarker analysis
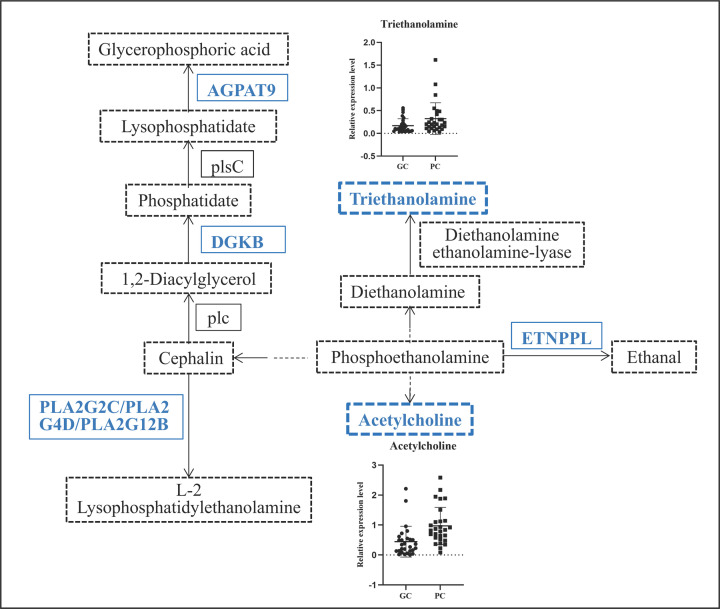
To explore the relationship between metabolites and genes, we looked for overlap between genes related to metabolites and DEGs obtained from the TCGA cohort and found 22 gene were overlapping genes (Figure 2E and Table 3). String was used to explore the interaction networks of 22 overlapping genes, and 23 pathways were enriched in total (Table 4). GPL metabolism with the minimum FDR value contains six genes (PLA2G2C, PLA2G4D, PLA2G12B, ETNPPL, DGKB, and AGPAT9) and two metabolites (acetylcholine and triethanolamine) and is considered a pathway that affects the progression of GC. The six genes and two metabolites in GPL metabolism were down-regulated (Figure 3). Moreover, the IHC staining obtained from the HPA database showed lower expression of ETNPPL and AGPAT9 in GC tissue than in normal tissues (Figure 4).
Table 3. Overlapped genes among metabolic pathways related genes and DEGs.
| Genes | logFC | P.value | Genes | logFC | P.value |
|---|---|---|---|---|---|
| CERS3 | −3.5985 | 6.90E-08 | ENPP3 | −3.1016 | 1.55E-40 |
| ASAH2 | −4.2801 | 4.32E-62 | GPAT3 | −2.8780 | 8.69E-22 |
| ACER1 | −6.3393 | 3.82E-20 | DGKB | −3.2171 | 1.49E-15 |
| ENPP7 | −3.1016 | 9.60E-59 | PLA2G12B | −2.0154 | 4.38E-06 |
| PSAPL1 | 2.0386 | 4.23E-11 | PLA2G2C | −2.0147 | 1.31E-15 |
| CKM | −3.5381 | 7.80E-27 | PLA2G4D | −2.0143 | 5.44E-10 |
| CKMT2 | −3.5353 | 2.49E-25 | ETNPPL | −3.0742 | 1.54E-10 |
| CKB | −3.5423 | 1.22E-17 | PAH | −2.0414 | 5.25E-11 |
| NOS1 | −2.0788 | 6.66E-13 | GPT | −2.8628 | 1.87E-12 |
| NT5C1A | −2.0720 | 8.39E-15 | RIMKLB | 2.0639 | 2.57E-19 |
| AMPD1 | −4.8170 | 3.96E-17 | ASPA | −4.2283 | 2.50E-16 |
Note: FC, fold change.
Table 4. Signaling pathways focused by overlapped genes.
| ID | Pathway description | Observed gene count | FDR | Matching proteins |
|---|---|---|---|---|
| hsa01100 | Metabolic pathways | 21 | 1.08E-22 | CKM, NT5C1A, PLA2G2C, ASPA, CERS3, PLA2G4D, ETNPPL, CKB, ACER1, ENPP7, RIMKLB, PLA2G12B, GPT, ASAH2, DGKB, CKMT2, ENPP3, AMPD1, PAH, NOS1, AGPAT9 |
| hsa00564 | Glycerophospholipid metabolism | 6 | 3.23E-08 | PLA2G2C, PLA2G4D, ETNPPL, PLA2G12B, DGKB, AGPAT9 |
| hsa00330 | Arginine and proline metabolism | 4 | 4.40E-06 | CKM, CKB, CKMT2, NOS1 |
| hsa00600 | Sphingolipid metabolism | 4 | 4.40E-06 | CERS3, ACER1, ENPP7, ASAH2 |
| hsa00592 | Alpha-linolenic acid metabolism | 3 | 4.27E-05 | PLA2G2C, PLA2G4D, PLA2G12B |
| hsa00591 | Linoleic acid metabolism | 3 | 5.37E-05 | PLA2G2C, PLA2G4D, PLA2G12B |
| hsa00250 | Alanine, aspartate, and glutamate metabolism | 3 | 7.80E-05 | ASPA, RIMKLB, GPT |
| hsa00565 | Ether lipid metabolism | 3 | 0.00015 | PLA2G2C, PLA2G4D, PLA2G12B |
| hsa00590 | Arachidonic acid metabolism | 3 | 0.00029 | PLA2G2C, PLA2G4D, PLA2G12B |
| hsa00220 | Arginine biosynthesis | 2 | 0.0015 | GPT, NOS1 |
| hsa04071 | Sphingolipid signaling pathway | 3 | 0.0015 | CERS3, ACER1, ASAH2 |
| hsa04270 | Vascular smooth muscle contraction | 3 | 0.0015 | PLA2G2C, PLA2G4D, PLA2G12B |
| hsa00760 | Nicotinate and nicotinamide metabolism | 2 | 0.0024 | NT5C1A, ENPP3 |
| hsa00230 | Purine metabolism | 3 | 0.0037 | NT5C1A, ENPP3, AMPD1 |
| hsa04975 | Fat digestion and absorption | 2 | 0.0037 | PLA2G2C, PLA2G12B |
| hsa00561 | Glycerolipid metabolism | 2 | 0.0071 | DGKB, AGPAT9 |
| hsa04014 | Ras signaling pathway | 3 | 0.0071 | PLA2G2C, PLA2G4D, PLA2G12B |
| hsa04730 | Long-term depression | 2 | 0.0071 | PLA2G4D, NOS1 |
| hsa01230 | Biosynthesis of amino acids | 2 | 0.0088 | GPT, PAH |
| hsa04972 | Pancreatic secretion | 2 | 0.0142 | PLA2G2C, PLA2G12B |
| hsa00240 | Pyrimidine metabolism | 2 | 0.0143 | NT5C1A, ENPP3 |
| hsa05231 | Choline metabolism in cancer | 2 | 0.0143 | PLA2G4D, DGKB |
| hsa04072 | Phospholipase D signaling pathway | 2 | 0.0275 | PLA2G4D, DGKB |
Note: FDR, false discovery rate.
Figure 3. Regulatory pathway diagram of glycerophospholipid metabolism.
The blue words represent down-regulation of genes or metabolites in GC.
Figure 4. Immunohistochemistry of genes related to glycerophospholipid metabolism from the HPA database.
(A) ETNPPL in GC (right) and normal tissues (left); (B) AGPAT9 in GC (right) and normal tissues (left).
Discussion
In the present study, we performed metabolic profiling of GC using GC and PC tissues and found that certain metabolites were significantly different. Twenty-two genes related to metabolites were identified based on metabolomics analysis with the help of TCGA transcriptomics data. GPL metabolism, which involves in six genes (PLA2G2C, PLA2G4D, PLA2G12B, ETNPPL, DGKB, and AGPAT9) and two metabolites (acetylcholine and triethanolamine), is likely associated with GC. The protein expression of AGPAT9 and ETNPPL were decreased in GC tissues.
The occurrence and development of cancers are dependent on molecular alterations and multiple of omics technologies, including metabolomics, transcriptomics, genomics and proteomics, have been performed to elucidate the mechanisms of cancer [22]. Transcriptomics analysis of cancers provides an orthogonal perspective for the expression of metabolic genes to investigate metabolism [23]. Some studies combined results of single-level omics with bioinformatics data available from websites, such as the TCGA and GEO, to screen biomarkers or explore the pathogenesis of cancer. The use of a combination of metabolomics and transcriptomics data from the TCGA or GEO is relatively frequent. Purine metabolism and related genes AMPD1 and RRM2 were enriched by combining metabolomics and transcriptomics data from the TCGA and GEO in breast cancer [24]. Differential metabolites in non-small cell lung cancer (NSCLC) were identified that were further verified by transcriptomics analysis of TCGA data, and a fully connected network of metabolites and genes in NSCLC was generated [25]. Sphingolipids were identified in breast cancer patients, and genes related to metabolism, such as CERK, SPHK1, and SGMS1, were verified by a TCGA cohort [26]. We performed metabolic profiling of GC and several metabolic pathways, and related genes and metabolites were identified with the help of TCGA transcriptomics data in our study.
Metabolomics research is committed to identifying and developing metabolically active targets in cancer therapeutics and pharmacology [27]. Many studies have shown the tremendous potential of metabolomics in GC. The urinary metabolomic profile of GC was explored and 77 metabolites were identified. A parsimonious biomarker profile of GC was investigated, and model performance was assessed [15]. Serum metabolites in patients with GC and their relationship with the prognosis of GC were investigated. Researchers found that three metabolites, 2,4-hexadienoic acid, 4-methylphenyl dodecanoate, and glycerol tributanoate, were related to the prognosis of GC [28]. Differential metabolites and metabolic pathways were identified in GC and its adjacent tissues in our study. GPL metabolism was enriched, and two metabolites involved in GPL metabolism, acetylcholine and triethanolamine, were differently expressed between the GC and PC groups. Because GPL is the most abundant lipid, its metabolism is closely related to oncogenesis and progression since the purpose of abnormal lipid metabolism is to synthesize more cell membranes lipids to meet the needs of the rapid proliferation of cancer cells and their increased demand for energy [29,30]. GPL metabolism has been reported to be dysregulated in many cancers, including NSCLC [31], melanoma [32], glioma [33], prostate cancer [34], colorectal cancer [35], and oral squamous cell carcinoma [36]. Therefore, GPL metabolism is likely associated with GC.
As one of the hallmarks of cancer [37], altered metabolism is regulated by genetic alterations to meet increased energy demands [38]. Changes in key genes had an impact on metabolic pathways to some extent [39]. A previous study showed that fatty acid metabolism was enriched by PHTF2 in GC and lipid metabolism regulated by PHTF2 significantly affected the tumorigenesis of GC cells [40]. Our study found that six genes (PLA2G2C, PLA2G4D, PLA2G12B, ETNPPL, DGKB, and AGPAT9) were altered in the GPL metabolic pathway. Researchers suggested that AGPAT9 may be correlated with cancer risk [41,42]. AGPAT9 is considered a hub gene that may exhibit significant prognostic potential for clear cell renal cell carcinoma because of its relation to immune infiltration [43]. A study found that AGPAT9 inhibited proliferation, migration, and invasion of breast cancer cell indicated that increasing AGPAT9 expression may be a new approach for breast cancer treatment [44]. ETNPPL is underexpressed in primary glioblastoma (GBM) and shows potential diagnostic implication for GBM [45]. Overexpression of ETNPPL reduced the growth of glioma stem cells and ETNPPL expression was inversely correlated to glioma grade [46]. Summarily, AGPAT9 and ETNPPL are anti-oncogenes of cancer. Our study showed that AGPAT9 and ETNPPL were down-regulated in patients with GC and participated in the regulation of GPL metabolism, and the protein expression of AGPAT9 and ETNPPL was lower in GC than in normal tissues, which is consistent with other studies.
There are some limitations of our study. First, the sample size was relatively small, although the GC and PC tissues were matched. Second, all patients were recruited from a single center, which might limit their generalization. Third, GC is very heterogenous due to anatomic location, molecular classification, and etiologies, but we failed to group those factors when identifying metabolites. Stratified analysis based on anatomic location, molecular classification, and etiologies is worth expecting to identify metabolites and metabolic pathways of GC. Our study may be regarded as a preliminary study exploring metabolic characteristics of GC and PC tissues, and further research with a larger sample, multiple research centers, and verification using other omics technologies is necessary to confirm our results. In conclusion, we investigated the tissue-based metabolomics profile of GC, and several differential metabolites were identified. GPL metabolism may affect the progression of GC with the help of TCGA transcriptomics data.
Supplementary Material
Abbreviations
- BMI
body mass index
- CI
confidence interval
- DEG
differentially expressed gene
- FC
fold change
- FDR
false discovery rate
- GBM
glioblastoma
- GC
gastric cancer
- GPI
glycosylphosphatidylinositol
- GPL
glycerophospholipid
- NSCLC
non-small cell lung cancer
- PC
paracancerous
- PCA
principal component analysis
- TCGA
The Cancer Genome Atlas
- TNM
tumor node metastasis
Contributor Information
Gang Wu, Email: mri_1976@163.com.
Wei Zhang, Email: kuilm0082_we@126.com.
Peizhi Ma, Email: deyue088231@126.com.
Data Availability
Metabonomics data is available in the open-access repository MetaboLights, study ID: MTBLS3303.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by National Natural Science Foundation of China [grant number 81803357].
CRediT Author Contribution
Yaqin Wang: Data curation, Methodology, Writing—original draft. Wenchao Chen: Data curation, Methodology. Kun Li: Data curation, Funding acquisition, Writing—review & editing. Gang Wu: Data curation, Funding acquisition, Writing—review & editing. Wei Zhang: Resources, Writing—review & editing. Peizhi Ma: Resources, Writing—review & editing. Siqi Feng: Resources, Writing—review & editing.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A. and Jemal A. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Feng R.M., Zong Y.N., Cao S.M. and Xu R.H. (2019) Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun. (Lond) 39, 22 10.1186/s40880-019-0368-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H., Zhang H., Deng P., Liu C., Li D., Jie H.et al. (2016) Tissue metabolic profiling of human gastric cancer assessed by (1)H NMR. BMC Cancer 16, 371 10.1186/s12885-016-2356-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung J., Jung Y., Bang E.J., Cho S.I., Jang Y.J., Kwak J.M.et al. (2014) Noninvasive diagnosis and evaluation of curative surgery for gastric cancer by using NMR-based metabolomic profiling. Ann. Surg. Oncol. 4, S736–S742 10.1245/s10434-014-3886-0 [DOI] [PubMed] [Google Scholar]
- 5.Holmes E., Wilson I.D. and Nicholson J.K. (2008) Metabolic phenotyping in health and disease. Cell 134, 714–717 10.1016/j.cell.2008.08.026 [DOI] [PubMed] [Google Scholar]
- 6.Nicholson J.K. and Lindon J.C. (2008) Systems biology: metabonomics. Nature 455, 1054–1056 10.1038/4551054a [DOI] [PubMed] [Google Scholar]
- 7.Cala M.P., Aldana J., Medina J., Sanchez J., Guio J., Wist J.et al. (2018) Multiplatform plasma metabolic and lipid fingerprinting of breast cancer: a pilot control-case study in Colombian Hispanic women. PLoS ONE 13, e0190958 10.1371/journal.pone.0190958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen P.H., Cai L., Huffman K., Yang C., Kim J., Faubert B.et al. (2019) Metabolic diversity in human non-small cell lung cancer cells. Mol. Cell. 76, 838–851 10.1016/j.molcel.2019.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denkert C., Budczies J., Weichert W., Wohlgemuth G., Scholz M., Kind T.et al. (2008) Metabolite profiling of human colon carcinoma–deregulation of TCA cycle and amino acid turnover. Mol. Cancer 7, 72 10.1186/1476-4598-7-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi J.M., Park W.S., Song K.Y., Lee H.J. and Jung B.H. (2016) Development of simultaneous analysis of tryptophan metabolites in serum and gastric juice - an investigation towards establishing a biomarker test for gastric cancer diagnosis. Biomed. Chromatogr. 30, 1963–1974 10.1002/bmc.3773 [DOI] [PubMed] [Google Scholar]
- 11.Kuligowski J., Sanjuan-Herraez D., Vazquez-Sanchez M.A., Brunet-Vega A., Pericay C., Ramirez-Lazaro M.J.et al. (2016) Metabolomic analysis of gastric cancer progression within the correa's cascade using ultraperformance liquid chromatography-mass spectrometry. J. Proteome Res. 15, 2729–2738 10.1021/acs.jproteome.6b00281 [DOI] [PubMed] [Google Scholar]
- 12.Lario S., Ramirez-Lazaro M.J., Sanjuan-Herraez D., Brunet-Vega A., Pericay C., Gombau L.et al. (2017) Plasma sample based analysis of gastric cancer progression using targeted metabolomics. Sci. Rep. 7, 17774 10.1038/s41598-017-17921-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corona G., Cannizzaro R., Miolo G., Caggiari L., De Zorzi M., Repetto O.et al. (2018) Use of metabolomics as a complementary omic approach to implement risk criteria for first-degree relatives of gastric cancer patients. Int. J. Mol. Sci. 19, 750 10.3390/ijms19030750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H.N., Wu H., Tseng Y.J., Chen Y.J., Zhang D.Y., Zhu L.et al. (2018) Serum microRNA signatures and metabolomics have high diagnostic value in gastric cancer. BMC Cancer 18, 415 10.1186/s12885-018-4343-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan A.W., Mercier P., Schiller D., Bailey R., Robbins S., Eurich D.T.et al. (2016) (1)H-NMR urinary metabolomic profiling for diagnosis of gastric cancer. Br. J. Cancer 114, 59–62 10.1038/bjc.2015.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H., Cui L., Liu W., Wang Z., Ye Y., Li X.et al. (2018) (1)H NMR metabolic profiling of gastric cancer patients with lymph node metastasis. Metabolomics 14, 47 10.1007/s11306-018-1344-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi R., Zhang J., Fang B., Tian X., Feng Y., Cheng Z.et al. (2020) Runners’ metabolomic changes following marathon. Nutr. Metab. (Lond.) 17, 19 10.1186/s12986-020-00436-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhl C., Tautenhahn R., Bottcher C., Larson T.R. and Neumann S. (2012) CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 84, 283–289 10.1021/ac202450g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L., Chang R., Pan S., Xu J., Cao Q., Su G.et al. (2020) Plasma metabolomics study of Vogt-Koyanagi-Harada disease identifies potential diagnostic biomarkers. Exp. Eye Res. 196, 108070 10.1016/j.exer.2020.108070 [DOI] [PubMed] [Google Scholar]
- 20.Triba M.N., Le Moyec L., Amathieu R., Goossens C., Bouchemal N., Nahon P.et al. (2015) PLS/OPLS models in metabolomics: the impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 11, 13–19 10.1039/C4MB00414K [DOI] [PubMed] [Google Scholar]
- 21.Deng J., Xia Z., Qiu D., Jiao X., Xiao B., Zhou W.et al. (2019) Nontarget metabolomics profiling of neuromyelitis optica spectrum disorder. Biomed. Chromatogr. 33, e4533 10.1002/bmc.4533 [DOI] [PubMed] [Google Scholar]
- 22.Alyass A., Turcotte M. and Meyre D. (2015) From big data analysis to personalized medicine for all: challenges and opportunities. BMC Med. Genom. 8, 33 10.1186/s12920-015-0108-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakimi A.A., Reznik E., Lee C.H., Creighton C.J., Brannon A.R., Luna A.et al. (2016) An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 29, 104–116 10.1016/j.ccell.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo X., Yu H., Song Y. and Sun T. (2019) Integration of metabolomic and transcriptomic data reveals metabolic pathway alteration in breast cancer and impact of related signature on survial. J. Cell. Physiol. 234, 13021–13031 10.1002/jcp.27973 [DOI] [PubMed] [Google Scholar]
- 25.Ruiying C., Zeyun L., Yongliang Y., Zijia Z., Ji Z., Xin T.et al. (2020) A comprehensive analysis of metabolomics and transcriptomics in non-small cell lung cancer. PLoS ONE 15, e0232272 10.1371/journal.pone.0232272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhadwal P., Dahiya D., Shinde D., Vaiphei K., Math R.G.H., Randhawa V.et al. (2020) LC-HRMS based approach to identify novel sphingolipid biomarkers in breast cancer patients. Sci. Rep. 10, 4668 10.1038/s41598-020-61283-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar A. and Misra B.B. (2019) Challenges and opportunities in cancer metabolomics. Proteomics 19, e1900042 10.1002/pmic.201900042 [DOI] [PubMed] [Google Scholar]
- 28.Wang D., Li W., Zou Q., Yin L., Du Y., Gu J.et al. (2017) Serum metabolomic profiling of human gastric cancer and its relationship with the prognosis. Oncotarget 8, 110000–110015 10.18632/oncotarget.21314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menendez J.A. and Lupu R. (2007) Fatty acid synthase and the lipogenicphenotype in cancer pathogenesis. Nat. Rev. Cancer 7, 763–777 10.1038/nrc2222 [DOI] [PubMed] [Google Scholar]
- 30.Currie E., Schulze A., Zechner R., Walther T.C. and Farese R.V. Jr (2013) Cellular fatty acid metabolism and cancer. Cell Metab. 18, 153–161 10.1016/j.cmet.2013.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y., Ma Z., Zhong J., Li L., Min L., Xu L.et al. (2018) Simultaneous quantifcation of serum monounsaturated and polyunsaturated phosphatidylcholines as potential biomarkers for diagnosing nonsmall cell lung cancer. Sci. Rep. 8, 7137 10.1038/s41598-018-25552-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henderson F., Johnston H.R., Badrock A.P., Jones E.A., Forster D., Nagaraju R.T.et al. (2019) Enhanced fatty acid scavenging and glycerophospholipid metabolism accompany melanocyte neoplasia progression in zebrafifish. Cancer Res. 79, 2136–2151 10.1158/0008-5472.CAN-18-2409 [DOI] [PubMed] [Google Scholar]
- 33.Wagner P.M., Sosa Alderete L.G., Gorné L.D., Gaveglio V., Salvador G., Pasquaré S.et al. (2019) Proliferative glioblastoma cancer cells exhibit persisting temporal control of metabolism and display differential temporal drug susceptibility in chemotherapy. Mol. Neurobiol. 56, 1276–1292 10.1007/s12035-018-1152-3 [DOI] [PubMed] [Google Scholar]
- 34.Chen X., Zhu Y., Jijiwa M., Nasu M., Ai J., Dai S.et al. (2020) Identification of plasma lipid species as promising diagnostic markers for prostate cancer. BMC Med. Inform. Decis. Mak. 20, 223 10.1186/s12911-020-01242-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shan S., Wu C., Shi J., Zhang X., Niu J., Li H.et al. (2020) Inhibitory effects of peroxidase from foxtail millet bran on colitis associated colorectal carcinogenesis by the blockage of glycerophospholipid metabolism. J. Agric. Food Chem. 68, 8295–8307 10.1021/acs.jafc.0c03257 [DOI] [PubMed] [Google Scholar]
- 36.Dickinson A., Saraswat M., Joenväärä S., Agarwal R., Jyllikoski D., Wilkman T.et al. (2020) Mass spectrometry-based lipidomics of oral squamous cell carcinoma tissue reveals aberrant cholesterol and glycerophospholipid metabolism-a pilot study. Transl. Oncol. 13, 100807 10.1016/j.tranon.2020.100807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanahan D. and Weinberg R.A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 38.Zhang G., He P., Tan H., Budhu A., Gaedcke J., Ghadimi B.M.et al. (2013) Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin. Cancer Res. 19, 4983–4993 10.1158/1078-0432.CCR-13-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W., Bai X., Liu Y., Wang W., Han J., Wang Q.et al. (2015) Topologically inferring pathway activity toward precise cancer classification via integrating genomic and metabolomic data: prostate cancer as a case. Sci. Rep. 5, 13192 10.1038/srep13192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chi Y., Wang H., Wang F. and Ding M. (2020) PHTF2 regulates lipids metabolism in gastric cancer. Aging (Albany NY) 12, 6600–6610 10.18632/aging.102995 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Balaban S., Lee L.S., Schreuder M. and Hoy A.J. (2015) Obesity and cancer progression: is there a role of fatty acid metabolism? Biomed. Res. Int. 2015, 274585 10.1155/2015/274585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwal A.K. (2012) Lysophospholipid acyltransferases: 1-acylglycerol-3-phosphate o-acyltransferases. from discovery to disease. Curr. Opin. Lipidol. 23, 290–302 10.1097/MOL.0b013e328354fcf4 [DOI] [PubMed] [Google Scholar]
- 43.Xu W.H., Xu Y., Wang J., Wan F.N., Wang H.K., Cao D.L.et al. (2019) Prognostic value and immune infiltration of novel signatures in clear cell renal cell carcinoma microenvironment. Aging (Albany NY) 11, 6999–7020 10.18632/aging.102233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan S.H., Wang Y.Y., Wu Z.Y., Zhang Z.F., Lu J., Li M.Q.et al. (2015) AGPAT9 suppresses cell growth, invasion and metastasis by counteracting acidic tumor microenvironment through KLF4/LASS2/V-ATPase signaling pathway in breast cancer. Oncotarget 6, 18406–18417 10.18632/oncotarget.4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.González-García N., Nieto-Librero A.B., Vital A.L., Tao H.J., González-Tablas M., Otero Á.et al. (2020) Multivariate analysis reveals differentially expressed genes among distinct subtypes of diffuse astrocytic gliomas: diagnostic implications. Sci. Rep. 10, 11270 10.1038/s41598-020-67743-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leventoux N., Augustus M., Azar S., Riquier S., Villemin J.P., Guelfi S.et al. (2020) Transformation foci in IDH1-mutated gliomas show STAT3 phosphorylation and downregulate the metabolic enzyme ETNPPL, a negative regulator of glioma growth. Sci. Rep. 10, 5504 10.1038/s41598-020-62145-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metabonomics data is available in the open-access repository MetaboLights, study ID: MTBLS3303.