Abstract
To investigate the mechanism of action of volatile anesthetics, we are studying mutants of the yeast Saccharomyces cerevisiae that have altered sensitivity to isoflurane, a widely used clinical anesthetic. Several lines of evidence from these studies implicate a role for ubiquitin metabolism in cellular response to volatile anesthetics: (i) mutations in the ZZZ1 gene render cells resistant to isoflurane, and the ZZZ1 gene is identical to BUL1 (binds ubiquitin ligase), which appears to be involved in the ubiquitination pathway; (ii) ZZZ4, which we previously found is involved in anesthetic response, is identical to the DOA1/UFD3 gene, which was identified based on altered degradation of ubiquitinated proteins; (iii) analysis of zzz1Δ zzz4Δ double mutants suggests that these genes encode products involved in the same pathway for anesthetic response since the double mutant is no more resistant to anesthetic than either of the single mutant parents; (iv) ubiquitin ligase (MDP1/RSP5) mutants are altered in their response to isoflurane; and (v) mutants with decreased proteasome activity are resistant to isoflurane. The ZZZ1 and MDP1/RSP5 gene products appear to play important roles in determining effective anesthetic dose in yeast since increased levels of either gene increases isoflurane sensitivity whereas decreased activity decreases sensitivity. Like zzz4 strains, zzz1 mutants are resistant to all five volatile anesthetics tested, suggesting there are similarities in the mechanisms of action of a variety of volatile anesthetics in yeast and that ubiquitin metabolism affects response to all the agents examined.
Although volatile inhaled anesthetics are most widely known for their ability to induce anesthesia rendering patients unconscious and insensitive to pain, these compounds also induce effects in all cells and tissues that have been carefully examined. For example, in mammals, volatile anesthetics directly depress cardiac contractility (33), dilate vascular smooth muscle (25), and relax skeletal muscle (3, 22) independent of their anesthetic action. In plants, they inhibit protoplasm flow and CO2 decomposition (29), and we find that in the yeast Saccharomyces cerevisiae, volatile anesthetics arrest cell division (23).
In the late 1800s it was demonstrated that there is a correlation between the lipophilicity of volatile anesthetics and their potency for inducing effects in a variety of different organisms (for recent reviews, see references 27 and 28). This correlation also holds for the growth-inhibitory effect of these compounds in yeast (23). The correlation between lipophilicity and potency has become central to all discussions of the activity of volatile anesthetics, and it has led to speculation that these compounds could directly affect the activity of one or a few critical membrane-bound proteins by binding to hydrophobic portions of the protein(s) or by interacting with lipids surrounding the protein(s) (for a recent review, see reference 24). However, the actual mechanism of action for any effects of volatile anesthetics remains unknown.
Molecular genetic analysis in model organisms provides a powerful tool for elucidating the mechanism of action of drugs. Although yeasts are less sensitive to anesthetics than mammals (23), in several important properties the parallels between these compounds as yeast growth inhibitors (23, 44) and mammalian anesthetics (24) are quite striking. These parallels include (i) rapid and reversible effects; (ii) very sharp dose-response curve; (iii) direct correlation between potency and lipophilicity; (iv) additivity of effective concentrations of mixtures of compounds; and (v) lack of effect in yeast of lipophilic compounds that are nonanesthetic in mammals. To further investigate the mechanism(s) of response to volatile anesthetics, we are isolating and characterizing yeast mutants with altered sensitivity to these compounds. Here we report that ubiquitin metabolism affects the cellular response to anesthetics and plays a role in determining the effective anesthetic dose for yeast.
MATERIALS AND METHODS
Strains and media.
Yeast strains used in these studies are listed in Table 1. Double-stranded plasmids were propagated in Escherichia coli MC1066 [leuB trpC pyrF::Tn5 (Kanr) araT lacX74 del strA hsdR hsdM (obtained from M. Casadaban)]. For γδ mutagenesis, E. coli HB101 F′ lac pro and HB101 pyrA::Tn5 (Kanr) (F−) were used. Yeast (26) and bacterial (34) media were prepared as described previously. Standard methods were used for yeast genetic analysis (31).
TABLE 1.
S. cerevisiae strains and zzz1 deletions
| Strain or deletion | Genotype or description | Reference |
|---|---|---|
| Strains | ||
| RLK88-3C | MATa his4-260 leu2-3,112 ura3-52 ade2-1 trp1-HIII lys2ΔBX can1R | 26 |
| K1784 | MATa his4-260 leu2-3,112 ura3-52 ade2-1 trp1-HIII lys2ΔBX can1R rDNA::ADE2 rDNA::URA3 | This study |
| P107 | MATa/MATα his4-260/his4-260 leu2-3,112/leu2-3,112 ura3-52/ura3-52 ade2-1/ade2-1 trp1-HIII/trp1-HIII lys2ΔBX/lys2ΔBX-CAN1-LYS2 can1R/can1R rDNA::ADE2/rDNA | This study |
| T8-1D | MATα SUP11 ade2-1 mod5-1 ura3-1 lys2-1 leu2-3,112 his4-519 | 48 |
| T7-7B | MATα SUP11 ade2-1 mod5-1 ura3-1 lys2-1 leu2-3,112 his4-519 mdp1-1 | 48 |
| TZ26 | MATα SUP11 ade2-1 mod5-1 ura3-1 lys2-1 leu2-3,112 his4-519 mdp1-16 | 48 |
| WCG4a | MATa his3-11,15 leu2-3,112 ura3 | 14 |
| WCG4a-11/22a | MATa his3-11,15 leu2-3,112 ura3 pre1-1 pre2-2 | 14 |
| WCG4a-11/21a | MATa his3-11,15 leu2-3,112 ura3 pre1-1 pre2-1 | 14 |
| Deletions | ||
| zzz1Δ-0 | Lacks translation initiation site and all ZZZ1 coding sequence except carboxy-terminal 10 amino acids | |
| zzz1Δ-122 | Potentially encodes a peptide containing amino-terminal 122 residues of ZZZ1 | |
| zzz1Δ-174 | Potentially encodes a peptide containing amino-terminal 174 residues of ZZZ1 | |
| zzz1Δ dsk2Δ | Lacks all of ZZZ1 as well as the amino terminus of the adjacent gene, DSK2 |
Anesthetic exposure and mutant isolation.
Isoflurane (Anaquest), halothane without thymol as a preservative (kindly provided by Halocarbon, North Augusta, S.C.), sevoflurane (kindly provided by Maruishi Pharmaceutical Co., Osaka, Japan), methoxyflurane (kindly provided by Abbott Laboratories, King of Prussia, Pa.), and enflurane (Anaquest) were used for these studies. Exposure to these agents and determination of the concentration of the compounds were performed as described previously (23, 44). Spontaneous mutants that grew on solid media in the presence of 12% isoflurane, a concentration that inhibits growth of wild-type RLK88-3C (Table 1), were isolated. Sensitivity or resistance to anesthetics and ability to grow at various temperatures were determined by diluting freshly saturated cultures of strains 50-fold and spotting 5 μl of this dilution onto solid medium and incubating the plates for 3 to 4 days in the appropriate conditions. Quantitative growth assays to generate dose-response curves were performed by spotting approximately 4,000 cells onto synthetic complete (SC) medium (26) and incubating the plates for 72 h at 30°C in the absence or presence of various concentrations of isoflurane. Growth from the initial 4,000 cells was removed on agar cores and resuspended in sterile water. Dilutions were spread on SC medium and incubated at 30°C to determine viable cell counts. Percent growth was determined by comparing viable cells at each concentration of isoflurane to viable cells obtained in the absence of isoflurane (defined as 100% growth). Quadruplicate spots were assayed at each concentration of anesthetic.
DNA manipulations and plasmid constructions.
Restriction and modification enzymes were purchased from several sources and used according to the instructions of the manufacturers. Standard procedures for the purification of plasmid (34) and yeast (31) DNA were used. Southern hybridizations were performed as described previously (34).
Plasmid pL1777, a derivative of YCp50 (30), contains a 12.3-kb fragment of yeast genomic DNA from chromosome XIII that includes ZZZ1. To initially localize the sequences encoding ZZZ1, various portions of pL1777 were excised by cutting with appropriate restriction enzymes that cleaved in at least two different locations on the plasmid. Following inactivation of the restriction enzyme, the restricted DNA was diluted and ligated to produce the corresponding deletion derivative. Among the deletion derivatives constructed, the ZZZ1 gene remained functional only when an XbaI fragment of pL1777 was deleted. The plasmid containing this XbaI deletion, pL1896, was used in γδ transposon mutagenesis (12) to further localize the ZZZ1 gene.
To determine the DNA sequence of ZZZ1, the 5.2-kb SnaBI-XbaI fragment containing ZZZ1 (Fig. 1) was isolated from pL1896, treated with the Klenow fragment of DNA polymerase I to make it blunt ended, and ligated into pTZ19U (Pharmacia). Derivatives containing the two orientations of this fragment, pL1992 and pL1995, were isolated and sequenced by the dideoxy-mediated chain termination method (35). The XhoI-ClaI deletion of ZZZ1, in pL2150, was constructed by digesting pL1992 with XhoI and ClaI, ligating with the 2-kb HpaI-SalI fragment containing LEU2, treating this ligation reaction with the Klenow fragment of DNA polymerase I, and then continuing the ligation. As anticipated, deletions obtained from this mixture contained a hybrid SalI/XhoI site at the junction between the ZZZ1 and LEU2 sequences. The EcoRV deletion of ZZZ1, in pL2177, was constructed by digesting pL1992 with EcoRV and ligating in the 1.2-kb HindIII fragment containing URA3 that had been made blunt ended by incubation with the Klenow fragment of DNA polymerase I. The NcoI-NsiI deletion mutation, in pL2178, was constructed by digesting pL1992 with NcoI and NsiI, treating with the Klenow fragment of DNA polymerase I, and ligating in the blunt-ended HindIII fragment containing URA3. These three deletion mutations were targeted for gene disruption (32) by digesting with XbaI and transforming (36) the linearized DNA into P107 (Table 1). The HincII-EcoRV deletion mutation was constructed by digesting pL1992 with HincII and EcoRV and ligating in a NotI linker. The resulting plasmid was digested with NotI, and a hisG-URA3-hisG fragment (1) modified to have NotI linkers at both ends was inserted. This deletion derivative was targeted for gene disruption by digesting with EcoRI and transforming the linearized DNA into P107 (Table 1). Diploid transformants with appropriate deletions on one copy of the chromosome were identified by Southern analysis (34). These strains were sporulated, and the resulting tetrads were dissected to obtain haploid strains containing the deletions. Haploids that potentially contained these deletions were confirmed by Southern hybridization.
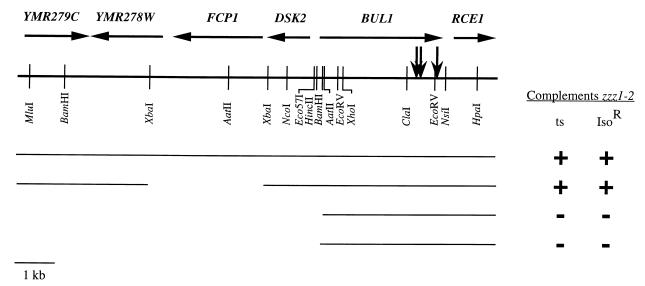
FIG. 1.
ZZZ1 gene and flanking genomic DNA. ZZZ1 was initially cloned on a 12.3-kb fragment of yeast genomic DNA in plasmids pL1777 and pL1779. The horizontal arrows indicate open reading frames present in this fragment. The vertical arrows indicate γδ transposons that inactivate the ZZZ1 gene. The thin horizontal lines below the restriction map of this fragment indicate the sequences present in several of the deletion derivatives that we constructed. Not all HincII and Eco57I sites present in the fragment are shown. +, complements; −, fails to complement.
To construct single-copy and multicopy plasmids that contain only ZZZ1, an XbaI linker was inserted at the NsiI site between ZZZ1 and RCE1 (Fig. 1) to create pL3773. The 3.2-kb Eco57I-XbaI fragment of this plasmid that contains ZZZ1 was inserted into both YCplac33 and YEplac195 (10) that were cleaved with XbaI and SmaI. The vector and ZZZ1 fragments were initially ligated to permit the XbaI sites to ligate, then the Eco57I site was made blunt ended by treatment with the Klenow fragment of DNA polymerase I, and the ligation was continued. The resulting YCpZZZ1 and YEpZZZ1 plasmids are named pL3900 and pL3832, respectively.
Single-copy and multicopy plasmids containing the MDP1/RSP5 ubiquitin ligase gene, YCp33MDP1 (47) and RB1 (48), were kindly provided by A. Hopper. We refer to these plasmids as YCpMDP1 and YEpMDP1, respectively.
The entire protein encoding sequences of PEP4 or PRB1 were deleted and replaced with loxP-kanMX-loxP from pUG6 (11) by using appropriate PCR-generated gene disruption cassettes. Appropriate disruptions were verified by PCR. To construct strains containing deletions of both PEP4 and PRB1, pep4Δ::loxP-kanMX-loxP strains were transformed with pSH47, which contains a galactose-regulated Cre recombinase (11), and derivatives that had excised kanMX and one of the loxP sites were identified based on their sensitivity to G418. Since the wild-type RLK88-3C (Table 1) strain does not induce gene expression in response to galactose, it was necessary to screen several hundred colonies to identify a few that were G418 sensitive. pep4Δ::loxP derivatives were then transformed with the prb1Δ::loxP-kanMX-loxP disruption cassette.
RESULTS
Analysis of Isor zzz1 mutants.
Spontaneous mutants resistant to the growth-inhibitory effects of isoflurane arise at a fairly high frequency. Seven of fifteen spontaneous mutants characterized to date contain mutations in the same nuclear gene, ZZZ1. The M1 and M3 mutants of ZZZ1 (Table 2) were isolated from 105 cells of a single culture of RLK88-3C (Table 1) plated on solid medium and incubated in the presence of 12% isoflurane. Phenotypic differences between these mutants indicate that they arose from independent events (Table 2; see below). Except for M15, the zzz1 mutants listed in Table 2 were obtained by spotting approximately 105 cells from a series of independently established cultures onto solid medium and incubating the plates in the presence of 12% isoflurane. Since only a single mutant colony was picked and characterized from each culture, these mutants are also of independent origin. M15 is an unselected isoflurane-resistant (Isor) mutant from yeast strain K1784 (Table 1) that was identified among colonies being used as isoflurane-sensitive (Isos) controls. The K1784 strain was derived from RLK88-3C by two separate transformations and is thus essentially isogenic with the other mutants.
TABLE 2.
Characterization of zzz1 mutants
| Original mutant designation, current allele name | Isoflurane resistance | MICa (% isoflurane) | Temperature responseb |
|---|---|---|---|
| M1, zzz1-1 | Slightly semidominant | 15.5 | Slightly ts, recessive |
| M3, zzz1-2 | Recessive | 13.5 | ts, recessive |
| M6, zzz1-5 | Slightly semidominant | 15.0 | ts, recessive |
| M7, zzz1-3 | Semidominant | 15.0 | ts, recessive |
| M9, zzz1-6 | Slightly semidominant | 14.0 | Slightly ts, recessive |
| M14, zzz1-7 | Slightly semidominant | 14.0 | ts, semidominant |
| M15, zzz1-4 | Slightly semidominant | 15.0 | tr |
The MIC for the wild type, RLK88-3C (Table 1), is 12%.
ts, temperature sensitive; tr, temperature resistant.
While the MIC (the minimum concentration needed to prevent visible growth on solid medium after 3 days [23]) of isoflurane in the normal strain, RLK88-3C (Table 1), is 12%, the MICs for these seven mutants range from 13.5 to 15.5%, indicating that the different mutations affect the activity of the ZZZ1 gene product to various extents (Table 2). Heterozygous zzz1/+ diploids for six of these seven mutants, M1, M6, M7, M9, M14, and M15, grow to some extent at the isoflurane MIC. The amount of growth is variable but in all cases is less than that obtained with the comparable homozygous diploid mutant, indicating that these mutants are semidominant with respect to altered response to isoflurane. In contrast, the M3 mutant is recessive (Table 2). Examination of at least six four-spore viable tetrads from heterozygous diploids for each of these seven mutants yielded two resistant and two sensitive spores in every case, showing that each mutant contains either a single nuclear mutation or closely linked nuclear mutations that render cells Isor.
In addition to altered response to isoflurane, six of these mutants, M1, M3, M6, M7, M9, and M14, are temperature sensitive since they either fail to grow (M3, M6, M7, and M14) or grow little (M1 and M9) at 37.5°C in the absence of isoflurane (Table 2). Only M15 grows well at elevated temperatures. Temperature sensitivity is recessive for M1, M3, M6, M7, and M9 but is semidominant for M14, based on the amount of growth observed at 37.5°C for the various heterozygous diploids compared to appropriate homozygous mutant diploids. In tetrads derived from heterozygous diploids of the six temperature-sensitive mutants, the temperature sensitivity segregates 2:2 and cosegregates with the Isor phenotype, indicating that the same nuclear mutation plays a critical role in both phenotypes.
The M1 mutant, defined as zzz1-1, was crossed to each of the other six mutants in Table 2. At least 32 spores from each cross were examined, and all spores derived from these six crosses were Isor, indicating that all of these mutants contain mutations in ZZZ1 (or in a gene that is at most 3 map units from ZZZ1).
Molecular analysis of ZZZ1.
To obtain additional insight regarding the molecular basis of anesthetic resistance, we cloned the ZZZ1 gene from a centromeric yeast DNA library based on its ability to complement the recessive temperature sensitivity of M3 (zzz1-2). Two temperature-resistant transformants that were also Isos were recovered. Loss of the plasmids from these transformants resulted in reversion to the temperature-sensitive and Isor phenotypes of zzz1-2. The plasmids from the transformants were recovered into E. coli. Reintroduction of these plasmids, designated pL1777 and pL1779, into a zzz1-2 strain gave rise to temperature-resistant and Isos transformants. These plasmids also partially complemented the Isor phenotype of zzz1-1 (M1) strains as would be expected for the semidominant phenotype of zzz1-1. Taken together, these results indicate that a plasmid-borne gene in pL1777 and pL1779 complements the mutant phenotypes of the zzz1-1 and zzz1-2 strains.
Restriction analysis showed that plasmids pL1777 and pL1779 contain identical 12.3-kb inserts. Deletion analysis and γδ transposon mutagenesis were used to further localize the sequences in pL1777 that complement the zzz1 phenotypes. Removal of the XbaI fragment produced a construct that complemented the zzz1-2 phenotypes (Fig. 1). All other deletions tested, such as AatII and BamHI deletions (Fig. 1), remove extensive portions of insert DNA and abolish complementing activity. Transposon mutagenesis localized the complementing gene more precisely. From more than 100 transposon mutants isolated, three disrupted the complementing gene. All three of these transposons mapped within a 500-bp region near the right-hand end of the insert (Fig. 1). In combination, the deletion and transposon analyses indicate that one end of the complementing gene is located between the AatII and XbaI sites located in the right half of Fig. 1, while the other end of the gene extends past the EcoRV site near the right end of the insert.
To determine if the complementing gene is ZZZ1 or a second-site suppressor of zzz1 mutations, the XhoI-ClaI fragment near the site of insertion of the γδ transposons (Fig. 1) was replaced by LEU2. Gene replacement transformation (32) was used to place this LEU2-tagged insert on one chromosome of the Zzz+ diploid P107 (Table 1). Spore viability among tetrads derived from this diploid was 97%. In tetrads with all four spores viable, two spores were Leu+ Isor and two spores were Leu− Isos, indicating that the gene disrupted by LEU2 is involved in normal anesthetic response of yeast. Southern blot analysis showed that the LEU2 gene was properly inserted on the chromosome of the Leu+ Isor spores. A Leu+ Isor spore was crossed to a zzz1-1 and a zzz1-2 haploid. Among 25 tetrads with four viable spores from each of these crosses, all of the spores were Isor, indicating that the LEU2 gene was inserted in the ZZZ1 gene.
ZZZ1 is identical to BUL1.
Our sequence of ZZZ1, which is identical to the sequence determined for the YMR275C open reading frame on chromosome XIII by the yeast genome sequencing project, indicates that this gene encodes a 976-amino-acid protein. This gene has been identified and independently sequenced in two other mutant hunts and named BUL1 (binds ubiquitin ligase; GenBank accession no. D50083 [46]) and RDS1 (respiration deficiency suppressor; GenBank accession no. X88901). The predicted protein sequences for Bul1p and Rds1p each differ by one amino acid from that predicted for Zzz1p (and YMR275C) from our sequencing of ZZZ1: Zzz1p has a valine at amino acid 35 whereas Bul1p has an alanine; and Zzz1p has an alanine at amino acid 963 whereas Rds1p has an arginine.
The XhoI-ClaI deletion of ZZZ1 described above, called zzz1Δ-174 (Table 1), has the potential to encode a 177-amino-acid peptide composed of 174 amino acids from the amino terminus of ZZZ1 and 3 amino acids encoded by the fragment containing the LEU2 gene. Haploids containing this deletion are viable and Isor. It is possible that this truncated peptide still performs some functions of wild-type Zzz1p but is resistant to isoflurane. To determine whether a null mutation of ZZZ1 is viable and Isor, we tested three additional deletion alleles (Table 1) of this gene: (i) an NcoI-NsiI deletion (Fig. 1) that removes all of ZZZ1 as well as the amino terminus of the adjacent gene, DSK2 (4) (called zzz1Δ dsk2Δ); (ii) a HincII-EcoRV deletion that removes the translation initiation site and all but the carboxy-terminal 10 amino acids of ZZZ1 (called zzz1Δ-0); and (iii) an EcoRV deletion in which the amino-terminal 122 amino acids of Zzz1p remain (called zzz1Δ-122). Each of these deletion constructs was transformed into the Zzz+ diploid P107 as described above. Transformants in which one chromosomal copy of the ZZZ1 gene was replaced by the deletion were identified by Southern analysis. Tetrads from these heterozygous diploids were dissected; in all cases spore viability was greater than 82%, with two of the spore colonies containing the deletion allele and two containing the wild-type gene as confirmed by Southern analysis. Thus, ZZZ1 is not an essential gene since strains in which this gene is completely deleted are viable and in fact show no noticeable difference from wild-type strains with respect to growth at 30°C in the absence of anesthetic.
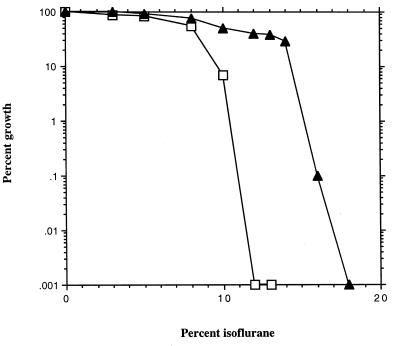
Haploid strains containing each of these zzz1Δ mutations are Isor. The MIC for zzz1Δ-0 strains is 16% isoflurane, which is higher than the MIC for any spontaneous mutant (Table 1), suggesting that each spontaneous mutant retains partial Zzz1p activity. The dose-response curves for both wild-type and zzz1Δ-0 strains are parallel and very sharp (Fig. 2). The sharpness of this response is exemplified by finding that for both wild-type and zzz1Δ strains, growth is largely unaffected by a concentration of isoflurane that is one-half the MIC. While it is not clear why the dose-response curve is so steep, the fundamental phenomenon for response to isoflurane is similar in wild-type and zzz1Δ-0 mutants and is reminiscent of the sharp dose-response observed for the activity of these compounds in mammalian anesthesia (6).
FIG. 2.
Isoflurane dose-response curve for growth of wild-type and zzz1Δ strains. Growth of wild-type (□) and zzz1Δ (▴) strains in the presence of various concentrations of isoflurane was assessed by number of viable cells following 3 days of incubation on solid SC medium. Growth is expressed as [(number of viable cells at a given isoflurane concentration)/(number of viable cells in the absence of isoflurane)] × 100.
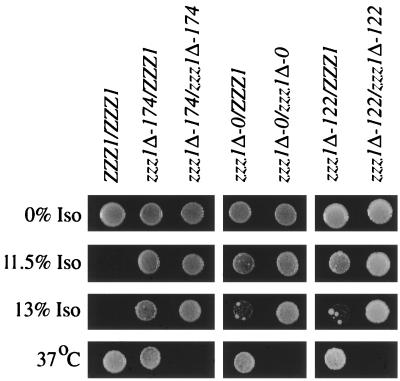
Diploids heterozygous for any one of the four zzz1 deletions are able to grow in the presence of concentrations of isoflurane that are growth inhibitory to homozygous diploid Zzz+ strains (Fig. 3). Thus, these deletion mutations are semidominant as are many of the spontaneous mutations (Table 2). Heterozygotes of the zzz1Δ-174 mutation, which has the potential to encode the longest amino-terminal zzz1 peptide of the deletions tested, grow substantially better in the presence isoflurane than do heterozygotes containing the zzz1Δ-0 (Fig. 3), zzz1Δ-122 (Fig. 3), or zzz1Δ dsk2Δ (not shown) mutation. All of the deletion heterozygotes are temperature resistant, and thus these deletions behave as recessive mutations for the temperature sensitivity phenotype (Fig. 3).
FIG. 3.
zzz1Δ mutations are semidominant. Five microliters of a 1:50 dilution of freshly saturated cultures of appropriate diploid strains was spotted on SC medium and incubated in the indicated conditions for 3 days. Genetic analysis showed that the large papillae observed in spots of zzz1Δ-0/ZZZ1 and zzz1Δ-122/ZZZ1 heterozygous strains at 13% isoflurane (Iso) are zzz1Δ/zzz1Δ homozygotes likely derived from recombination during mitotic growth.
Temperature sensitivity of zzz1 mutants is a synthetic phenotype.
In haploids, each of the zzz1Δ mutants is temperature sensitive. Yashiroda et al. (46) found that the temperature sensitivity of a bul1 disruptant was suppressed by SSD1 on a single-copy vector. Transformants of temperature-sensitive, Isor zzz1-2 or zzz1Δ::0 strains that contain a YCpSSD1 plasmid [pTW1007 (43)] remain Isor but are now temperature resistant. Since temperature sensitivity of these zzz1 strains is not altered by the presence of a control YCp vector, it is likely that SSD1 is responsible for suppression of this phenotype. The SSD1 gene has been shown to be polymorphic in yeast (40), and some strains that are classified as wild type contain an ssd1 allele that does not produce Ssd1p (41). Thus, temperature sensitivity of zzz1 mutants may be a synthetic phenotype due to an ssd1 mutation in our wild-type strain RLK88-3C combined with particular zzz1 mutations. Alternatively, the presence of a functional chromosomal SSD1 gene along with YCpSSD1 may suppress the temperature sensitivity of zzz1 mutations. Regardless, these results indicate that temperature sensitivity and isoflurane resistance are separable.
Overexpression of ZZZ1 increases sensitivity to isoflurane.
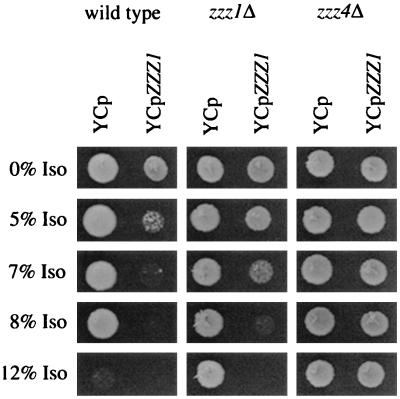
Finding that even complete deletions of ZZZ1 have a semidominant phenotype suggests that decreased cellular levels of Zzz1p affect the response to isoflurane. In contrast, transformation of a stable, single-copy plasmid containing ZZZ1, YCpZZZ1, into wild-type cells, which presumably increases Zzz1p levels, decreases the isoflurane MIC from 12% to 7% (Fig. 4). Even at 5% isoflurane, growth of the wild-type/YCpZZZ1 transformants is dramatically decreased. The ZZZ1 gene is responsible for the decreased MIC since it is the only gene present on this plasmid (see Materials and Methods) and the control YCp vector does not affect the isoflurane MIC of the wild-type strain (Fig. 4). It appears that this YCp-borne ZZZ1 gene is expressed at higher levels than the chromosomal ZZZ1 gene since the isoflurane MIC for a zzz1Δ-0 strain containing YCpZZZ1 is unexpectedly low, 9% instead of 12% (Fig. 4). In the absence of isoflurane, wild-type or zzz1Δ-0 strains containing YCpZZZ1 grow normally (Fig. 5A). However, we find that transformants of these same strains containing ZZZ1 on a multicopy plasmid, YEpZZZ1, grow very slowly even in the absence of anesthetic (Fig. 5A), indicating that substantial overexpression of ZZZ1 is detrimental to yeast growth. Yashiroda et al. (46) found no effect on growth of their wild-type cells when a multicopy plasmid containing BUL1 was introduced. The difference in behavior of multicopy ZZZ1 and BUL1 plasmids may be strain dependent, multicopy vector dependent, or due to the apparent amino acid difference at position 35 of the BUL1 and ZZZ1 genes used in the plasmids.
FIG. 4.
Effect of YCpZZZ1 on MIC. Wild-type, zzz1Δ, and zzz4Δ strains transformed with plasmid YCp or YCpZZZ1 were tested for growth in the presence of various concentrations of isoflurane (Iso).
FIG. 5.
Effect of overexpression of ZZZ1 on growth. Wild-type (Wt), zzz1Δ, and zzz4Δ strains transformed with plasmid YCp, YCpZZZ1, YEp, or YEpZZZ1 were streaked on medium that selects for retention of the plasmid and incubated at 30°C in the absence of anesthetic for 48 h.
Interaction of ZZZ1 and ZZZ4.
Double-mutant studies indicate that the ZZZ1 and ZZZ4 (23) gene products participate in a common pathway. As single mutants, zzz4Δ::LEU2 strains, which contain a null mutation of ZZZ4 (23), grow more slowly in the presence of isoflurane and have a lower MIC than zzz1Δ-0 strains (Fig. 6). Double zzz1Δ-0 zzz4::LEU2 mutants show the same isoflurane response as zzz4Δ::LEU2 single-mutant strains (Fig. 6). This type of interaction in which the altered response of the double mutant is no greater than that of either single mutant parent suggests that these genes identify components involved in a single pathway rather than in multiple pathways (7, 8).
FIG. 6.
Interaction between zzz1Δ and zzz4Δ mutations. Strains isogenic except for the appropriate zzz mutation(s) were isolated from tetrads derived from a zzz1Δ/ZZZ1 zzz4Δ/ZZZ4 diploid and tested for growth in the presence of various concentrations of isoflurane (Iso).
Two lines of evidence from studies of overexpression of ZZZ1 in zzz4Δ::LEU2 strains also support this contention. First, in contrast to wild-type and zzz1Δ-0 strains, the isoflurane resistance of zzz4Δ::LEU2 strains transformed with YCpZZZ1 is not altered (Fig. 4). Second, growth of zzz4Δ::LEU2 strains is not affected by the presence of YEpZZZ1 (Fig. 5C). Both findings indicate that ZZZ4 encodes a product necessary for the effects imparted by plasmid-borne copies of ZZZ1.
Ubiquitin ligase affects response to isoflurane.
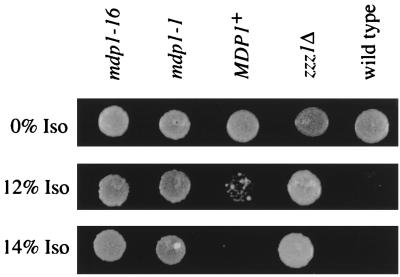
While the function of the BUL1/ZZZ1 gene product is unknown, two-hybrid analysis indicates that it interacts with the yeast ubiquitin ligase encoded by MDP1/RSP5 (20, 48), suggesting that Bul1p/Zzz1p may play a role in ubiquitin metabolism (46). Additional support for this possibility comes from finding that ZZZ4, another gene that affects volatile anesthetic response (23), is identical to DOA1/UFD3, which is involved in ubiquitin-dependent protein degradation (9, 16). Thus, the anesthetic resistance of zzz1 and zzz4 mutants may result from altered ubiquitin metabolism. To further test this possibility, we examined the response of ubiquitin ligase (mdp1/rsp5) mutants to isoflurane. Strains containing either of the two mdp1 mutations tested, mdp1-1 and mdp1-16 (Table 1), are resistant to the growth-inhibitory effects of isoflurane compared to an isogenic wild-type strain, MDP1+ (Fig. 7). The isogenic wild-type strain, MDP1+ (Table 1), for these mutants is slightly more resistant to isoflurane than RLK88-3C, the wild-type strain that we normally use, as shown by the slight growth of the MDP1+ strain at 12% isoflurane (Fig. 7).
FIG. 7.
Ubiquitin ligase mutants are resistant to isoflurane. Isogenic MDP1+ and mdp1 mutants were tested for ability to grow in the presence of isoflurane (Iso). For comparison, growth of RLK88-3C, the standard wild-type strain used in our studies (Table 1), and that of a zzz1Δ mutant isogenic to RLK88-3C are shown in the same conditions.
Transformants of wild-type RLK88-3C containing a single extra copy of MDP1/RSP5 on a centromeric vector, YCpMDP1, have increased sensitivity to isoflurane. Growth of these transformants is dramatically decreased in 10% isoflurane, and the MIC for these transformants is 11%, rather than compared to 12% for wild-type/YCp transformants (Fig. 8). Transformants of RLK88-3C that overexpress ubiquitin ligase from the multicopy plasmid YEpMDP1 show a further decrease in the isoflurane MIC to 10% (Fig. 8). In the absence of isoflurane, growth of wild-type RLK88-3C transformants containing either the YCpMDP1 or YEpMDP1 vector is not noticeably affected in either spots (Fig. 8) or streaks (data not shown). In zzz1Δ or zzz4Δ strains, neither the growth nor the level of anesthetic resistance is affected by plasmid YCpMDP1 or YEpMDP1 (Fig. 8 and data not shown).
FIG. 8.
Overexpression of ubiquitin ligase increases sensitivity to isoflurane. Wild-type (Wt) and zzz1Δ strains were transformed with YCp, YCpMDP1(RSP5), YEp, or YEpMDP1(RSP5) and tested for response to isoflurane (Iso).
Proteasome function affects cellular response to isoflurane.
To ascertain whether degradation of ubiquitinated proteins is involved in cellular response to volatile anesthetics, mutants that affect the proteolytic activity of the proteasome or vacuole were examined for response to isoflurane. pre1 pre2 mutants that have decreased proteasomal activity (14) are more resistant to the growth-inhibitory effects of isoflurane at 30°C (Fig. 9A) than an isogenic wild-type control. The pre1-1 pre2-2 strain, which grows better than the pre1-1 pre2-1 strain in the absence of isoflurane (14), grew better than the pre1-1 pre2-1 strain at elevated concentrations of isoflurane. At 24°C, the MIC of isoflurane for all three strains is 12% (data not shown). To test for effects of vacuolar degradation on the response to volatile anesthetics, we determined the responses of pep4Δ, prb1Δ, and pep4Δ prb1Δ mutants isogenic to RLK88-3C to isoflurane. All three of these vacuolar mutants show a pattern of growth inhibition identical to that of the wild type (Fig. 9B), indicating that vacuolar degradation does not play a role in cellular response to volatile anesthetics. To assess the effect of vacuolar degradation on anesthetic response in cells with impaired proteasome function, we examined pre1-1 pre2-2 pep4Δ triple mutants for growth in the presence of isoflurane. As seen in the RLK88-3C background, the pep4Δ mutation does not affect anesthetic response in the WCG4a background (Fig. 9A). The response of the pre1-1 pre2-2 pep4Δ mutant to isoflurane is identical to that of the pre1-1 pre2-2 mutant (Fig. 9A), indicating that vacuolar function does not affect anesthetic response even when proteasome function is deficient.
FIG. 9.
Proteasome mutants affect volatile anesthetic response. Strains isogenic except for various proteasome and/or vacuole mutations, pre1 pre2, pep4Δ, and pre1 pre2 pep4Δ, were examined for growth in the presence of various concentrations of isoflurane (Iso) (A). The wild-type (Wt) strain RLK88-3C and vacuolar mutants that are isogenic except for the indicated mutations, pep4Δ, prb1Δ, and pep4Δ prb1Δ, were tested for response to isoflurane (B).
zzz1 mutants are cross-resistant to other volatile anesthetics.
A wide variety of compounds can function as volatile anesthetics (for a recent review, see reference 24). We find that all seven of the spontaneous zzz1 mutants isolated based on their resistance to isoflurane as well as the four zzz1Δ mutants that we constructed are resistant to the volatile anesthetics methoxyflurane, halothane, enflurane, and sevoflurane, as well as isoflurane (data not shown). We have previously shown that zzz4 mutations, also initially identified as being Isor, render cells cross-resistant to these additional volatile anesthetics (23). These findings raise the possibility that ubiquitin metabolism is involved in a general way in the cellular response to volatile anesthetics.
DISCUSSION
Mutations in ZZZ1, which is identical to BUL1, render yeast resistant to the growth-inhibitory effects of volatile anesthetics. Bul1p/Zzz1p is proposed to be involved in protein ubiquitination since it interacts with the ubiquitin ligase encoded by MDP1/RSP5 (46). The finding that another gene we have identified, ZZZ4/DOA1/UFD3 (23), is involved in ubiquitin-dependent proteolysis (9, 18) and that ubiquitin ligase (MDP1/RSP5) mutations and proteasome (PRE) mutations also affect anesthetic sensitivity in yeast suggests that ubiquitin metabolism and proteasomal degradation of ubiquitinated proteins could play key roles in cellular response to volatile anesthetics. Our previous unexplained results that zzz1 and zzz4 mutants are sensitive to cadmium (44) are consistent with the finding of Jungmann et al. (21) that some mutants defective in ubiquitin metabolism are cadmium hypersensitive.
Effective anesthetic dose in yeast.
Although there are numerous similarities between the actions of volatile compounds as yeast growth inhibitors and mammalian anesthetics (44), one difference is that MICs of volatile anesthetics in yeast are approximately ninefold higher than the concentration required for anesthesia in mammals (23). The MIC of isoflurane for yeast growth is substantially reduced by the presence of even a single extra copy of ZZZ1 (YCpZZZ1) or MDP1/RSP5 (YCpMDP1) in wild-type strains (Fig. 4 and 8), indicating that ubiquitin metabolism, involving ZZZ1 and MDP1/RSP5 in particular, may play a key role in determining the basal state of anesthetic sensitivity in cells. Completely or partially defective mutations of either of these genes increases anesthetic resistance. A further indication that ZZZ1 levels substantially affect anesthetic response comes from the finding that zzz1Δ/+ heterozygotes, including ones containing the zzz1Δ-0 mutation, which lack only a single copy of ZZZ1 compared to +/+ diploids, have increased resistance to isoflurane (Fig. 3). Thus, it appears that increased activity of Zzz1p or Mdp1(Rsp5p) leads to increased sensitivity while decreased activity results in decreased sensitivity.
Dominant negative zzz1Δ.
While zzz1Δ/+ heterozygotes that contain a complete deletion of ZZZ1 (zzz1Δ-0 or zzz1Δ dsk2Δ [Table 1]) have a low level of isoflurane resistance, heterozygotes that contain the zzz1Δ-174 mutation, which has the potential to encode a peptide containing the amino-terminal 174 residues of Zzz1p, grow much better in the presence of isoflurane. This suggests that zzz1Δ-174 is a dominant negative mutation (15, 42). The zzz1Δ-122 allele, which has the potential to encode a 122-residue amino-terminal peptide, does not behave as a dominant negative mutation. Finding that Bul1p (Zzz1p) interacts with Rsp5p (46) suggests an intriguing potential explanation for this difference. Rsp5p is a homologue of the mammalian Nedd4 protein (13, 19, 20). Both Rsp5 and Nedd4 contain WW domains (for a review, see reference 39) a module of approximately 30 to 40 amino acids containing two strictly conserved tryptophans (2, 5, 19). The WW domains of Nedd4 have been shown to bind to PY domains in subunits of the epithelial Na+ channel (37, 38). The potential amino-terminal peptide of Zzz1p encoded by the zzz1Δ-174 deletion contains a PY consensus sequence (XPPXY) at amino acids 156 to 160 (FPPSY) and thus may be able to bind and form nonfunctional complex. The potential zzz1Δ-122 product does not contain this PY domain and thus may be unable to bind and form complex. Support for this suggestion comes from finding that the PY consensus sequence of Bul1p/Zzz1p is necessary for interaction with Rsp5p in the two-hybrid assay (45).
Role of ubiquitin metabolism.
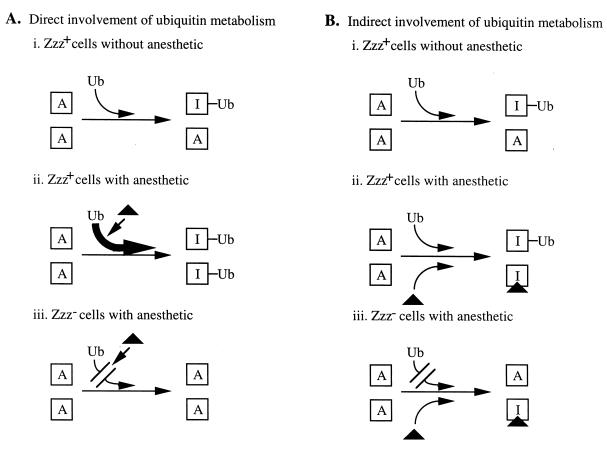
The role of ubiquitin metabolism in anesthetic response may be direct or indirect. Direct involvement would indicate that this is possibly a type of stress response with anesthetic exposure altering ubiquitin metabolism. For example, in Zzz+ (wild-type) cells, anesthetic exposure could lead to increased ubiquitination that increases degradation, alters regulation, and/or affects endocytosis (17) of cellular proteins, some of which are critical for division (compare Fig. 10Ai and Aii). In such a scenario, Zzz− mutants are unable to alter ubiquitin metabolism when exposed to anesthetics (Fig. 10Aiii) and thus would continue to divide when exposed to these compounds.
FIG. 10.
Models for involvement of ubiquitin metabolism in anesthetic response. See Discussion for descriptions of the details of the models. Squares, proteins critical for cell growth; A, active protein; I, inactive protein, Ub, ubiquitin; triangles, anesthetic.
In contrast, ubiquitin metabolism may indirectly affect anesthetic response. For instance, in Zzz+ cells the concentration of the active form of one or more proteins necessary for cell growth may normally be controlled by a ubiquitin-dependent process (Fig. 10Bi). In addition, binding of anesthetic may also inactivate the protein in Zzz+ cells (Fig. 10Bii), causing growth arrest. In Zzz− cells grown in the absence of anesthetic, there may be increased cellular concentrations of active protein due to defective ubiquitin metabolism. Only some of this larger pool of active protein is inactivated when the mutant cells are exposed to the MIC of anesthetic (Fig. 10Biii), thus permitting the cells to continue to grow in the presence of anesthetic.
Finding that proteasome mutants affect response to volatile anesthetics suggests that ubiquitin-dependent degradation of proteins plays a role in the normal activity of volatile anesthetics. However, it is possible that ubiquitination affects anesthetic response in other ways such as altering regulation or endocytosis of critical proteins (17). Continued analysis of additional yeast anesthetic-response mutants and of the cellular effects of volatile anesthetics in yeast should provide further insights regarding the mechanism of action of these critical drugs.
ACKNOWLEDGMENTS
We thank Ross Shiman for his continued enthusiasm and intense interest in this work and Ross Shiman, Anita K. Hopper, and Reeta Prusty for helpful discussions about this work and critical comments regarding the manuscript.
This work was supported by grant GM57822 to R.L.K. from the National Institutes of Health.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andre B, Springael J Y. WWP, a new amino acid motif present in single or multiple copies in various proteins including dystrophin and the SH3-binding Yes-associated protein YAP65. Biochem Biophys Res Commun. 1994;205:1201–1205. doi: 10.1006/bbrc.1994.2793. [DOI] [PubMed] [Google Scholar]
- 3.Auer J, Meltzer S J. /1914. The effect of ether inhalation upon the skeletal motor mechanism. J Pharmacol Exp Ther. 1913;5:521–522. [Google Scholar]
- 4.Biggins S, Ivanovska I, Rose M D. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J Cell Biol. 1996;133:1331–1346. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 6.Franks N P, Lieb W R. Molecular and cellular mechanisms of general anaesthesia. Nature. 1994;367:607–614. doi: 10.1038/367607a0. [DOI] [PubMed] [Google Scholar]
- 7.Game J C, Cox B S. Epistatic interactions between four rad loci in yeast. Mutat Res. 1972;16:353–362. doi: 10.1016/0027-5107(72)90203-5. [DOI] [PubMed] [Google Scholar]
- 8.Game J C, Cox B S. Synergistic interactions between rad mutations in yeast. Mutat Res. 1973;20:35–44. doi: 10.1016/0027-5107(73)90095-x. [DOI] [PubMed] [Google Scholar]
- 9.Ghislain M, Dohmen R J, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 11.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guyer M S. Uses of the transposon γδ in the analysis of cloned genes. Methods Enzymol. 1983;101:362–369. doi: 10.1016/0076-6879(83)01027-7. [DOI] [PubMed] [Google Scholar]
- 13.Hein C, Springael J Y, Volland C, Haguenauer-Tsapis R, Andre B. NPI1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 14.Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf D H. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- 15.Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 16.Hochstrasser, M. Personal communication.
- 17.Hochstrasser M. Protein degradation or regulation: Ub the judge. Cell. 1996;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- 18.Hochstrasser M, Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast α2 repressor. Cell. 1990;61:697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann K, Bucher P. The rsp5-domain is shared by proteins of diverse functions. FEBS Lett. 1995;358:153–157. doi: 10.1016/0014-5793(94)01415-w. [DOI] [PubMed] [Google Scholar]
- 20.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jungmann J, Reins H A, Schobert C, Jentsch S. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature. 1993;361:369–371. doi: 10.1038/361369a0. [DOI] [PubMed] [Google Scholar]
- 22.Karis J H, Gissen A J, Nastuk W L. Mode of action of diethyl ether in blocking neuromuscular transmission. Anesthesiology. 1966;27:42–51. doi: 10.1097/00000542-196601000-00008. [DOI] [PubMed] [Google Scholar]
- 23.Keil R L, Wolfe D, Reiner T, Peterson C J, Riley J L. Molecular genetic analysis of volatile-anesthetic action. Mol Cell Biol. 1996;16:3446–3453. doi: 10.1128/mcb.16.7.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koblin D D. Mechanisms of action. In: Miller R D, editor. Anesthesia. 4th ed. Vol. 1. New York, N.Y: Churchill Livingstone; 1994. pp. 67–99. [Google Scholar]
- 25.Larach D R, Schuler H G, Skeehan T M, Peterson C J. Direct effects of myocardial depressant drugs on coronary vascular tone: anesthetic vasodilation by halothane and isoflurane. J Pharmacol Exp Ther. 1990;254:58–64. [PubMed] [Google Scholar]
- 26.Lin Y-H, Keil R L. Mutations affecting RNA polymerase I-stimulated exchange and rDNA recombination in yeast. Genetics. 1991;127:31–38. doi: 10.1093/genetics/127.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipnick R L. Charles Ernest Overton: narcosis studies and a contribution to general pharmacology. Trends Pharmacol Sci. 1986;7:161–164. [Google Scholar]
- 28.Lipnick R L. Hans Horst Meyer and the lipoid theory of narcosis. Trends Pharmacol Sci. 1989;10:265–269. doi: 10.1016/0165-6147(89)90025-4. [DOI] [PubMed] [Google Scholar]
- 29.Overton C E. Studies of narcosis. Jena, Germany: Gustav Fischer; 1901. . (English translation, 1991; Chapman and Hall, London ed.) [Google Scholar]
- 30.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 31.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 32.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 33.Rusy B F, Komai H. Anesthetic depression of myocardial contractility: a review of possible mechanisms. Anesthesiology. 1987;67:745–766. doi: 10.1097/00000542-198711000-00020. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 37.Schild L, Lu Y, Gautschi I, Schneeberger E, Lifton R P, Rossier B C. Identification of a PY motif in the epithelial Na channel subunits as a target sequence for mutations causing channel activation found in Liddle syndrome. EMBO J. 1996;15:2381–2387. [PMC free article] [PubMed] [Google Scholar]
- 38.Staub O, Dho S, Henry P C, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 39.Sudol M. Structure and function of the WW domain. Prog Biophys Mol Biol. 1996;65:113–132. doi: 10.1016/s0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- 40.Sutton A, Immanuel D, Arndt K T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uesono Y, Fujita A, Toh-e A, Kikuchi Y. The MCS1/SSD1/SRK1/SSL1 gene is involved in stable maintenance of the chromosome in yeast. Gene. 1994;143:135–138. doi: 10.1016/0378-1119(94)90618-1. [DOI] [PubMed] [Google Scholar]
- 42.Wilkie A O M. The molecular basis of genetic dominance. J Med Genet. 1994;31:89–98. doi: 10.1136/jmg.31.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson R B, Brenner A A, White T B, Engler M J, Gaughran J P, Tatchell K. The Saccharomyces cerevisiae SRK1 gene, a suppressor of bcy1 and ins1, may be involved in protein phosphatase function. Mol Cell Biol. 1991;11:3369–3373. doi: 10.1128/mcb.11.6.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolfe D, Hester P, Keil R L. Volatile anesthetic additivity and specificity in Saccharomyces cerevisiae: implications for yeast as a model system to study mechanisms of anesthetic action. Anesthesiology. 1998;89:174–181. doi: 10.1097/00000542-199807000-00024. [DOI] [PubMed] [Google Scholar]
- 45.Yashiroda H, Kaida D, Toh-e A, Kikuchi Y. The PY-motif of Bul1 protein is essential for growth of Saccharomyces cerevisiae under various stress conditions. Gene. 1998;225:39–46. doi: 10.1016/s0378-1119(98)00535-6. [DOI] [PubMed] [Google Scholar]
- 46.Yashiroda H, Oguchi T, Yasuda Y, Toh-e A, Kikuchi Y. Bul1, a new protein that binds to the Rsp5 ubiquitin ligase in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3255–3263. doi: 10.1128/mcb.16.7.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zoladek, T. Personal communication.
- 48.Zoladek T, Tobiasz A, Vaduva G, Boguta M, Martin N C, Hopper A K. MDP1, a Saccharomyces cerevisiae gene involved in mitochondrial/cytoplasmic protein distribution, is identical to the ubiquitin-protein ligase gene RSP5. Genetics. 1997;145:595–603. doi: 10.1093/genetics/145.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]