Abstract
Oral squamous cell carcinoma (OSCC) is the most common malignant tumor in the head and neck, and radiotherapy is the main approach for this disease, while irradiation resistance is a huge challenge that influences radiosensitivity. This study aims to determine the role and function of miR-29a-3p and ADAM12 in the radiosensitivity of OSCC cells. The expression pattern of ADAM12 in OSCC cells was searched in TCGA database. The binding of miR-29a-3p and ADAM12 was predicted by Starbase and verified using dual luciferase reporter gene assay. The RNA or protein expressions of miR-29a-3p and ADAM12 were measured by RT-qPCR or western blot. OSCC cell lines were treated by various γ-ray irradiation dosages before the alteration on miR-29a-3p expression and on the cell viability, proliferation, migration and cell apoptosis was detected. ADAM12 was highly expressed in OSCC cells, whose expression in resistant cells was positively correlated with irradiation dosage. Overexpression of ADAM12 in OSCC cells lead to increased cell proliferation and migration ability as well as inhibited cell apoptosis. miRNAs potentially binding ADAM12 in PITA, microT, miRmap and targetscan were screened, among which miR-29a-3p had the maximum differential expression levels in OSCC cells determined by RT-qPCR. Overexpression of miR-29a-3p resulted in suppressed cell viability, proliferation, migration ability and increased cell apoptosis, while this expression pattern can be partially counteracted by ADAM12 overexpression in OSCC cells. miR-29a-3p through targeting and inhibiting AMDM12 enhances the radiosensitivity of OSCC cells.
Key words: miR-29a, ADAM12, oral squamous cell carcinoma, radio-resistance, cell viability
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most common malignancies and accounts for the majority of cases in head and neck squamous cell carcinomas (HNSCCs).1 The OSCC related mortality ranks the top eight of cancer related death with an increasing incidence of more than 300,000 new cases being newly diagnosed each year.2,3 The prognosis for this disease is extremely poor and less than half of OSCC patients can survive the first 5 years,4,5 which can be partially ascribed into metastasis and resistance. 6,7 The treatment options of OSCC include surgical resection for locally OSCC and adjuvant treatment (chemotherapy or radiotherapy) for advanced OSCC.8,9 Especially for patients in stage IIIIV, adjuvant thermotherapy is the most effective choice, but despite the advances in recent years, the prognosis for patients in advanced stage is still far from satisfactory.10 The major obstacle is chemo-resistance.11,12 Therefore, decreasing chemotherapy or radiotherapy resistance is of great importance for improving the overall survival of OSCC patients.
ADAM12 is a protein which was involved in cell adhesion and whose expression is showed to be correlated with poor survival period of patients with pancreatic cancer after chemotherapy.13 Additionally, ADAM12 was proved as a novel molecular marker of laryngeal carcinoma14 and the serum level of ADAM12 is of predictable value for neoadjuvant chemoradiation resistance in esophageal adenocarcinoma patients.15 ADAM12 has also been reported for its upregulated expression in OSCC-derived cell lines and was associated with TNM classification in OSCC patients.16 Nevertheless, no current data supported the implication of ADAM12 in chemoresistance of OSCC. After searching several databases to identify the possible upstream targets of ADAM12, we identified five microRNAs (miRNAs or miRs), including hsamiR- 29a-3p, hsa-miR-29b-3p, hsa-miR-217, hsa-miR-29c-3p and hsa-miR-582-5p, among which miR-29a-3p was finally selected.
miRNAs have been well proved for their implication in cell proliferation, migration, invasion and apoptosis.17 The dysregulated expression of multiple miRNAs has been reported in OSCC,18 indicating miRs have certain role to play in the progression of OSCC. miR-29a-3p enclosed by OSCC-derived exosomes (OSCC-Exo) was showed to promote M2 subtype macrophage polarization through regulating SOCS1/STAT6 signal pathway.19 In addition to that, miR-29a-3p, as a tumor suppressor, was found to be lowly expressed in radio-resistant human nasopharyngeal carcinoma tissues.20 The alternation of miR-29a-3p expression in a pro-fibrosis model indicates a higher risk for radiation therapy, suggesting miR-29a-3p may implicated in biological tumor response (or resistance) to radiation.21 However, to our best knowledge, there is no current study reporting the regulation of miR-29a- 3p in the radiosensitivity of OSCC cells. This study aims to determine the possible role of miR-29a-3p/ADAM12 axis in OSCC cell radiotherapy with the expectation to provide certain theoretic basis for overcoming clinical radio-resistance for OSCC patients.
Method and Materials
Cell culture, transfection and radiation
Normal human oral keratinocytes (NHOK) and OSCC cell lines (HSC2, HSC4 and SCC4) were purchased from ATCC and cultured in Roswell Park Memorial Institute 1640 (RPMI 1640, 12633012, Gibco, New York, NY, USA) which contains 10% fetal bovine serum (FBS, 16140071, Gibco) + 1% double antibiotics (15240062, 100., Gibco) in an incubator at 37°C with 5% CO2.
ADAM12 overexpression plasmid (OE-ADAM12), ADAM12 suppression plasmid (sh-ADAM12), miR-29a-3p mimic, miR- 29a-3p inhibitor and their corresponding negative controls (OENC, sh-NC, mimic-NC, inhibitor-NC) were purchased from GeneChem (Shanghai, China). Cells were seeded in 60 mm culture disk at the concentration of 3.0.105 cells/disk for cell culture of 24 h before 3 μg plasmid, Lipofectamine 2000 (11668019, Invitrogen, Carlsbad, CA, USA), Opti-MEM I (31985062, Gibco) serumreduced medium and 8 ng/mL polybrene (TR-1003, Sigma- Aldrich, St. Louis, MO, USA) were added for cell transfection for 48 h.
The transfected cells were subjected to radiation at the dose of 5 Gy, 10 Gy and 20 Gy using 137Cs γ ray (Best Theratronics Ltd., Kanata, ON, Canada) before cell phenotypes were identified.
Cell counting Kit-8 (CCK-8) assay
CCK-8 kit (Beyotime, Shanghai, China) was used to measure cell viability. The transfected cells were seeded into a 96-well plate (3000 cells/well) with culture medium refreshed every day. After cell culture for 48 h, the proliferation ability of cells was measured based on following procedure. CCK-8 (10 μL) was added into each well and incubated with cells at 37°C for 2 h. The optical density at 450 nm was measured using SpectraMax M5 microplate (Molecular Devices, San Jose, CA, USA). The cell viability of cells in control group was set as 1, based on which the cell viability in experimental groups was calculated accordingly.
Clone formation assay
HSC2 cells at logarithmic phase were digested with 0.25% trypsin and gently flapped into single cells. The collected cells were suspended in culture medium with 10% FBS and the cell suspension was seeded into a culture disk at the concentration of 100 cells per disk in which 10 mL of 37°C-pre-warmed culture medium was supplemented. The culture disk was gently shaken to allow cells to evenly distribute and cells were then cultured for 2 to 3 weeks at 37°C with 5% CO2 and saturated humidity. Cell culture was terminated when visible clones were identified. The supernatant was abandoned and cells were washed with phosphate buffered saline (PBS) for twice before being fixed with 1:3 acetic acid/methanol for 15 min. Then Giemsa solution was used for staining for 10 to 30 min before the Giemsa staining was washed away with running water. Cells were dried and the cell disk was inverted in a transparent film with gridding. The number of cell clones (more than 10 cells) was counted by naked eyes or under a microscope (low power lens) to calculate the clone formation rate. The clone formation rate of cells in control group was set as 100% and the cell viability in experimental groups was calculated accordingly.
Cell scratch assay
Cells (1.105) in control group and experimental group were seeded into a 12-well plate. Once cell confluence reaches 100%, a 10 μL pipette tip was used to make a scratch on the bottom of the plate. Dulbecco phosphate buffered saline (DPBS, 14190250, Gibco) was used to washed the detached cells for three times and the culture medium was replaced with Dulbecco’s Modified Eagle Medium (DMEM) containing 2% FBS. After cell culture for 0 h and 24 h, the cell scratch distance under the Olympus inverted microscope within the same field was observed. The migration rate = (0 h scratch distance -24 h scratch distance)/0 h scratch distance. Three duplicates were set for each group.
Annxin V-FITC
Cells at the confluence of 80% were counted for cell numbers. Cells (1.106) were washed in pre-cold PBS for twice and suspended in 1 . Annexin buffer before incubation with 5 μL Annexin-VFITC (Becton Dickinson) at room temperature for 10 min without light exposure. Then cells were washed in pre-cold PBS and suspended in 300 μL 1 . Annexin. Cell apoptosis rate was calculated using flow cytometry (Guava easyCyte HT from Millipore, Burlington, MA, USA).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
miR-29a-3p Oligod (T) specific RT primers were designed. Cells were dissolved in 1 mL Trizol (Thermo Fisher Scientific, Waltham, MA, USA) to extract the total RNA based on instructions. RNA samples in which the miR-29a-3p expression was detected were treated with 3’ poly-A tail. RNAs were reversed transcript into cDNA using M-MLV reverse transcriptase (D7160L, Beyotime, Shanghai, China) and random primers. The PCR reaction system and reaction conditions were prepared based on the instructions of Premix Ex Taq™II kit (Takara, Dalian, China). ABI7500 quantitative PCR apparatus (Applied Biosystems, Shanghai, China) was used for RT-PCR with U6 as internal control of miRNA and GAPDH of mRNA. Data were analyzed using 2-ΔΔCt method 22: ΔΔCt = [Ct(target gene)-Ct(internal gene)]experimental group -[Ct(target gene)-Ct(internal gene)]control group. The primers for gene are listed in Table 1.
Western blot
Cells were treated by lysis buffer, in which the concentration of total protein was measured using bicinchoninic acid (BCA) kit (23227, Thermo Fisher Scientific). Then proteins were diluted with 5 . sample buffer and separated in 12% separating gel for 90 min before being blocked in PBS blocking buffer with 5% (w / v) skimmed milk powder at room temperature for 1 h. After that, cells were incubated with primary antibody of ADAM12 (1:500, ab223476, Abcam, Cambridge, UK) and GAPDH (1:500, ab9485, Cambridge, UK) at 4°C for overnight. The membranes were washed and incubated with secondary antibody (1: 500, ab114610, Abcam) at room temperature for 1 h. The images were photographed and saved at BioSpectrum Imaging System (UVP, Upland, CA, USA).
Dual luciferase reporter gene assay
The potential binding sites between ADAM12 and miR-29a-3p were predicted by TargetScan and the wide and mutant sequences were accordingly designed. The sequence segments were cloned into pGL3-Basic (Promega, Madison, WI, USA) and accordingly named as MT-ADAM12 group and WT-ADAM12 group. MTADAM12 or WT-ADAM12 with miR-29a mimic or mimic-NC was transfected into HSC2 cells. Forty-eight hours after transfection, the luciferase activity in each group was detected by luciferase detection kit (RG051S, Beyotime).
Table 1.
Primers of genes for RT-qPCR.
| Name of primer | Sequences (5’-3’) |
|---|---|
| ADAM12-F | GCTCGACCAAGTTTCACAGG |
| ADAM12-R | GGGGCCATGCCAGTGATAGA |
| miR-29a-3p-F | ATTGGCTAAAGTCTAC |
| miR-29a-3p-R | TGGTGTCGTGGAGTCG |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
| GAPDH-F | GTGGCTGGCTCAGAAAAAGG |
| GAPDH-R | GGGGAGATTCAGTGTGGTGG |
F, forward primer; R, reverse primer.
Figure 1.
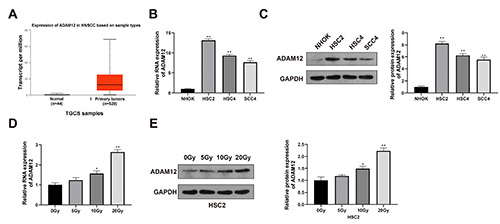
Elevated ADAM12 expression in radio-resistant OSCC cells. Expression profile of ADAM12 in patients with head and neck squamous cell carcinomas was searched in TCGA database (A). RT-qPCR (B) and Western blot (C) were applied to determine the expression level of ADAM12 in OSCC cell lines (HSC2, HSC4 and SCC4) and NHOK cells. The alternation on ADAM12 expression in HSC2 cell after radiation under 5 Gy, 10 Gy and 20 Gy was determined by RT-qPCR (D) and western blot (E). n=3, *p<0.05; **p<0.01. OSCC, oral squamous cell carcinoma.
Statistical analysis
All experiments were performed in triplicates, unless indicated otherwise. SPSS 18.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 8.0 (GraphPad Software Inc.) were used for data analysis. Measurement data were expressed as mean + SD. Comparisons between two groups were analyzed using t-test while comparisons among multiple groups were determined by One-way analysis of variance with Tukey’s for post hoc analysis. A p-value <0.05 was considered to have significant difference.
Results
Elevated ADAM12 expression in radio-resistant OSCC cells
Data in TCGA database showed elevated expression of ADAM12 in patients with HNSCC (Figure 1A). Detection on ADAM12 expression in OSCC cell lines (HSC2, HSC4 and SCC4) and NHOK cells showed that OSCC cell lines had higher expression level of ADAM12 than those in NHOK cells (Figure 1 B,C; p<0.01). HSC2 cells were selected for presentation in following experiments.
Once cell confluence reaches 90%, HSC2 cells were treated by radiation of 0 Gy, 5 Gy, 10 Gy and 20 Gy at the frequency of 137 Cs 0.5 Gy/min. The measurement on ADAM12 expression showed the expression of ADAM12 in survived cells was increased along with the increased radiation dose (Figure 1D; p 0.01), indicating the possible regulatory role of ADAM12 in radio-resistance of OSCC cells. In this regard, radiation at the dose of 20 Gy was used for following experiments.
Figure 2.
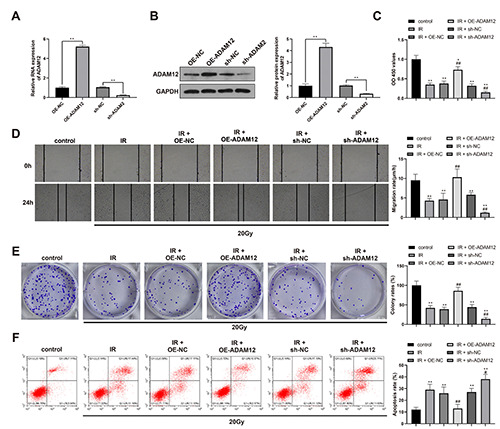
ADAM12 increases the radio-resistance in OSCC cells. ADAM12 knockdown or overexpression was achieved in HSC2 cells. A,B) RT-qPCR (A) and Western blot (B) were used to verify the transfection efficiency of ADAM12. Cell viability, migration, proliferation and apoptosis were determined by CCK-8 (C), scratch assay (D), clone formation assay (E) and flow cytometry (F). n=3; *p<0.05 compared with control group; **p<0.01 compared with control group; #p<0.05 compared with IR group; ##p<0.01 compared with IR group; OSCC, oral squamous cell carcinoma.
Knockdown of ADAM12 enhances the radio-sensitivity of OSCC cells
ADAM12 knockdown was achieved in HSC2 cells and the verification on cell transfection efficiency showed that ADAM12 expression was elevated in OE-ADAM12 group compared with OE-NC group, but decreased in sh-ADAM2 group compared with sh-NC group (Figure 2 A,B; p<0.01).
After radiation of 20Gy, cells were measured for cell viability, migration, proliferation and apoptosis. The results showed that compared with control group, cells in IR group had suppressed cell viability, proliferation and migration ability as well as increased apoptosis rate. Compared with IR group, the cell viability, proliferation and migration ability were elevated and cell apoptosis was suppressed in IR + OE-ADAM12 group, while different expression pattern was found in IR + sh-ADAM12 group (Figure 2 C-F; p<0.01). Those results showed that knockdown of ADAM12 could increase the radio-sensitivity of OSCC cells and overexpression of ADAM12 can decrease the radio-sensitivity of OSCC cells.
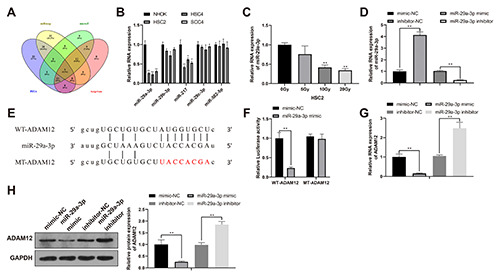
miR-29a-3p targets and negatively regulates ADAM12
To explore the miRNA that can regulate ADAM12 in OSCC, Starbase was used to predict the possible miRNAs binding to ADAM12. The overlapped miRNAs in PITA, microT, miRmap, TargetScan were selected by Venn diagram: hsa-miR-29a-3p, hsamiR- 29b-3p, hsa-miR-217, hsa-miR-29c-3p, hsa-miR-582-5p (Figure 3A). Then the expressions of those five miRNAs were detected in OSCC cell lines which showed that miR-29a-3p had the lowest expression in OSCC cell lines among the above miRNAs (Figure 3B; p<0.01). Therefore miR-29a-3p was selected for following experiments and for cell transfection. Then the expressions of miR-29a-3p in HSC2 cells after different radiation dosage were measured and the results showed that the expression levels of miR-29a-3p was decreased along with the increased radiation dosage (Figure 3C; p<0.01), indicating miR-29a-3p was associated with the radiation resistance of HSC2 cells. The measurement for transfection efficiency showed that compared with mimic-NC group, miR-29a-3p mimic group had elevated miR- 29a-3p expression, while compared with inhibitor-NC group, miR- 29a-3p inhibitor group had suppressed miR-29a-3p expression (Figure 3D).
Dual luciferase reporter gene assay was applied to verify the possible binding of miR-29a-3p with ADAM12 and the binding sites are designed in Figure 3E. The measurement showed that compared with co-transfection with mimic-NC, co-transfection of WT-ADAM12 with miR-29a-3p mimic resulted in decreased luciferase activity (Figure 3F; p<0.01). No significant difference was noticed for cells co-transfected with MT-ADAM12 and miR- 29a-3p mimic. Above results showed miR-29a-3 can target ADAM12.
To determine the regulatory role of miR-29a-3p on ADAM12, miR-29a-3p overexpression or miR-29a-3p inhibitor was transfected into HSC2 cells to detect the expression level of ADAM12. Compared with mimic-NC group, miR-29a-3p mimic group had reduced ADAM12 expression, while compared with inhibitor-NC group, miR-29a-3p inhibitor group had elevated expression level of ADAM12 (Figure 3 G,H; p<0.01). Above results showed that miR-29a-3p can negatively target and regulate ADAM12 expression in HSC2 cells.
Figure 3.
ADAM12 increases the radio-resistance in OSCC cells. ADAM12 knockdown or overexpression was achieved in HSC2 cells. A,B) RT-qPCR (A) and Western blot (B) were used to verify the transfection efficiency of ADAM12. Cell viability, migration, proliferation and apoptosis were determined by CCK-8 (C), scratch assay (D), clone formation assay (E) and flow cytometry (F). n=3; *p<0.05 compared with control group; **p<0.01 compared with control group; #p<0.05 compared with IR group; ##p<0.01 compared with IR group; OSCC, oral squamous cell carcinoma.
miR-29a regulates ADAM12 to enhance the radio-sensitivity of OSCC cells
To determine whether the regulatory effect of miR-29a-3p in ADAM12 affects the radio-sensitivity of OSCC, HSC2 cells were transfected or co-transfected with miR-29a-3p or ADAM12 related plasmids before the cell biological function was detected. The detection showed that compared with IR group, cells in IR + miR- 29a-3p mimic group had suppressed cell viability, proliferation and migration ability in addition to enhanced apoptosis rate, while different expression pattern was found in cells in IR + miR-29a-3p inhibitor group. Compared to IR + miR-29a-3p mimic group, IR + miR-29a-3p mimic + OE-ADAM12 group had enhanced cell viability, proliferation and migration ability, and suppressed cell apoptosis rate, while compared with IR + miR-29a-3p inhibitor group, cells in IR + miR-29a-3p inhibitor + sh-ADAM12 group had decreased cell viability, proliferation and migration ability, and elevated cell apoptosis rate (Figure 4 A-D; p<0.01). Collectively, miR-29a-3p can negatively regulate ADAM12 expression to enhance the radio-sensitivity of OSCC cells.
Figure 4.
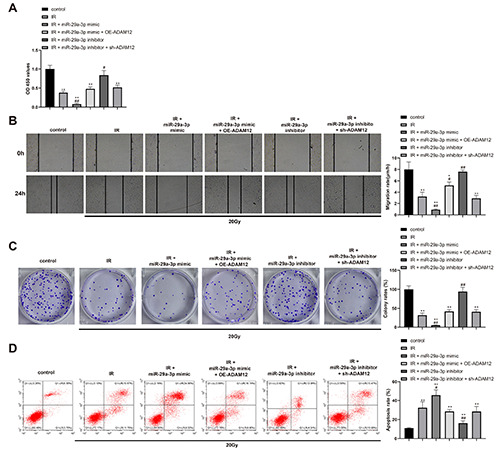
miR-29a-3p by binding ADAM12 regulate the radio-sensitivity of OSCC cells. HSC2 cells were transfected or co-transfected with miR-29a-3p or ADAM12 related plasmids. Cell viability, migration, proliferation and apoptosis were determined by CCK-8 (A), scratch assay (B), clone formation assay (C) and flow cytometry (D). *p<0.05 compared with control group; **p<0.01 compared with control group; #p<0.05 compared with IR group; ##p<0.01 compared with IR group.
Discussion
Chemo-resistance is a common cause for poor prognosis and how to improve the chemo- or radio-sensitivity of tumor cells is of great importance to improve survivals of patients under chemo or radiotherapy. In this study, we identified that ADAM12 is elevated in OSCC cells and associated with radio-resistance of OSCC cells. Additionally, miR-29a-3p was found to target and regulate ADAM12 expression in OSCC cells, through which miR-29a-3p regulates the radio-sensitivity of OSCC cells.
The major finding of this study showed that HSC2, HSC4 and SCC4 cell lines had increased expression of ADAM12 when compared with NHOK cells, suggesting ADAM12 may associate with OSCC progression. Consistent with this finding, elevated ADAM12 expression can be found in several tumors, including breast cancer (BC)23 and esophageal cancer24 by increasing the proliferation, metastasis and endocrine resistance of tumor cells. In a study published in 2012, elevated expression of ADAM12 was showed to have certain relationship with TNM classification and enhanced cell malignant behavior of OSCC cells,16 but no current study highlighted the possible role of ADAM12 in radio-resistance of OSCC cells. Further irradiation treatment by 0Gy~20Gy in HSC2 cells demonstrated that ADAM12 expression was increased along with the increased irradiation dosage. The above finding showed that ADAM12 may have a certain role to play in the radioresistance of OSCC cells. In BC cells, suppression on ADAM12 expression was showed to inhibit p-Akt expression and restore 5- FU sensitivity in 5-FU-resistant BC cells, which contributes to suppressed BC cell survival and invasive abilities.25 In a study exploring the cancer stem cell phenotype in BC cells, knockdown of ADAM12 contributed to suppressed cell migration ability and reduced anoikis resistance.26 Nevertheless, less evidence is available concerned the mechanism of how ADAM12 is regulated in cancer cells.
Online software searched the possible upstream targets of ADAM12 and five miRNAs (hsa-miR-29a-3p, hsa-miR-29b-3p, hsa-miR-217, hsa-miR-29c-3p and hsa-miR-582-5p) were identified. After further detection, miR-29a-3p was eventually selected. The binding of miR-29a-3p with ADAM12 was confirmed by dual luciferase reporter gene assay. Gain- and loss-of-function showed cells transfected with miR-29a-3p overexpression had decreased expression level of ADAM12, while miR-29a-3p inhibition in HSC2 cells led to increased expression level of ADAM12. Those results highlighted the regulatory role of miR-29a-3p in ADAM12 expression in OSCC cells. ADAM12 is well explored in BC cells and a previous study revealed a negative correlation between ADAM12-L and miR-29b/miR-200c.27 In the same study, ADAM12-L 3’UTR was proved to be a direct target gene of miR- 29 and miR-200 family members. miR-29a is also a well identified miR involved in virus replication, metastasis and drug resistance.28 In OSCC, down-regulation of miR-29a was associated with lymph node metastasis and TNM stage, in addition to that overexpression of miR-29a was showed to suppress the activity of Wnt/β-catenin pathway.2. Decreased expression of miR-29a was associated with the progression of nasopharyngeal carcinoma, whose up-regulation in nasopharyngeal carcinoma cell line can increase cell sensitivity to Taxol through inhibiting the transcription (STAT) signal pathway and regulating the oncogenic gene Bcl-2.30 The association between miR-29a and drug resistance can also be found in glioblastoma, which highlighted the regulatory role of miR-29a in ameliorating CD133-mediated chemoresistance in glioblastoma stem cells.31 In this study, HSC2 cells transfected with miR-29a-3p overexpression had suppressed cell proliferation and migration ability as well as increased cell apoptosis, while different expression pattern can be found in cells transfected with miR-29a-3p inhibitor. Moreover, co-transfection with miR-29a-3p overexpression and ADAM12 up-regulation can counteract the effect of miR- 29a-3p on HSC2 cells, suggesting the regulatory role of miR-29a- 3p in mediating cell sensitivity through regulating ADAM12 expression. miR-29a suppression in colorectal cancer was proved to weak drug resistance and increased PTEN expression.32 A similar study showed that overexpression of miR-29a can lead to enhanced sensitivity of HT29/DOX cells to doxorubicin-treatment by targeting membrane transporters P-glycoprotein.33 Although there are studies investigating the mechanism of miR-29a in drug resistance in cancers, mainly in solid tumors, including colon cancer and BC, limited information can be found regarding the possible role and mechanism of miR-29a in mediating cell sensitivity of OSCC.
Taken together, this study highlighted the involvement of miR- 29a-3p in mediating cell sensitivity of OSCC. The collected evidence in this study demonstrated that miR-29a-3p enhances the radiosensitivity of OSCC cells through inhibiting AMDM12 expression. Drug resistance involves a complicated mechanism and in study, we only investigated the possible involvement of miR-29a-3p/ADAM12 axis. To better understand the mechanism of OSCC irradiation resistance, a vast number of further studies are needed to enlighten our understating and to facilitate the proposal of better clinical applications to overcome chemoradiation resistance in OSCC.
References
- 1.Wang J, Wang Y, Kong F, Han R, Song W, Chen D, et al. Identification of a six-gene prognostic signature for oral squamous cell carcinoma. J Cell Physiol 2020;235:3056-68. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [DOI] [PubMed] [Google Scholar]
- 3.Peng QS, Cheng YN, Zhang WB, Fan H, Mao QH, Xu P.circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR-31 to inhibit Hippo signaling pathway. Cell Death Dis 2020;11:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.J SH, Hysi D.Methods and risk of bias in molecular marker prognosis studies in oral squamous cell carcinoma. Oral Dis 2018;24:115-9. [DOI] [PubMed] [Google Scholar]
- 5.Matsuura D, Valim TD, Kulcsar MAV, Pinto FR, Brandao LG, Cernea CR, et al. Risk factors for salvage surgery failure in oral cavity squamous cell carcinoma. Laryngoscope 2018;128:1113-9. [DOI] [PubMed] [Google Scholar]
- 6.Sharma A, Boaz K, Natarajan S.Understanding patterns of invasion: a novel approach to assessment of podoplanin expression in the prediction of lymph node metastasis in oral squamous cell carcinoma. Histopathology 2018;72:672-8. [DOI] [PubMed] [Google Scholar]
- 7.Song JM, Woo BH, Lee JH, Yoon S, Cho Y, Kim YD, et al. Oral administration of Porphyromonas gingivalis, a major pathogen of chronic periodontitis, promotes resistance to paclitaxel in mouse xenografts of oral squamous cell carcinoma. Int J Mol Sci 2019;20;2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fridman E, Na'ara S, Agarwal J, Amit M, Bachar G, Villaret AB, et al. The role of adjuvant treatment in early-stage oral cavity squamous cell carcinoma: An international collaborative study. Cancer 2018;124:2948-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao C, Chang EI, Lai SY. Contemporary approach to locally advanced oral cavity squamous cell carcinoma. Curr Oncol Rep 2019;21:99. [DOI] [PubMed] [Google Scholar]
- 10.Lindemann A, Takahashi H, Patel AA, Osman AA, Myers JN. Targeting the DNA damage response in OSCC with TP53 mutations. J Dent Res 2018;97:635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng X, Luo Q, Zhang H, Wang H, Chen W, Meng G, et al. The role of NLRP3 inflammasome in 5-fluorouracil resistance of oral squamous cell carcinoma. J Exp Clin Cancer Res 2017;36:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokota T, Homma A, Kiyota N, Tahara M, Hanai N, Asakage T, et al. Immunotherapy for squamous cell carcinoma of the head and neck. Jpn J Clin Oncol 2020;50:1089-96. [DOI] [PubMed] [Google Scholar]
- 13.Veenstra VL, Damhofer H, Waasdorp C, van Rijssen LB, van de Vijver MJ, Dijk F, et al. ADAM12 is a circulating marker for stromal activation in pancreatic cancer and predicts response to chemotherapy. Oncogenesis 2018;7:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markowski J, Tyszkiewicz T, Jarzab M, Oczko-Wojciechowska M, Gierek T, Witkowska M, et al. Metal-proteinase ADAM12, kinesin 14 and checkpoint suppressor 1 as new molecular markers of laryngeal carcinoma. Eur Arch Otorhinolaryngol 2009;266:1501-7. [DOI] [PubMed] [Google Scholar]
- 15.Ebbing EA, van der Zalm AP, Steins A, Creemers A, Hermsen S, Rentenaar R, et al. Stromal-derived interleukin 6 drives epithelial-to-mesenchymal transition and therapy resistance in esophageal adenocarcinoma. Proc Natl Acad Sci USA 2019;116:2237-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uehara E, Shiiba M, Shinozuka K, Saito K, Kouzu Y, Koike H, et al. Upregulated expression of ADAM12 is associated with progression of oral squamous cell carcinoma. Int J Oncol 2012;40:1414-22. [DOI] [PubMed] [Google Scholar]
- 17.Pei Y, Yao Q, Li Y, Zhang X, Xie B.microRNA-211 regulates cell proliferation, apoptosis and migration/invasion in human osteosarcoma via targeting EZRIN. Cell Mol Biol Lett 2019;24:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aali M, Mesgarzadeh AH, Najjary S, Abdolahi HM, Kojabad AB, Baradaran B.Evaluating the role of microRNAs alterations in oral squamous cell carcinoma. Gene 2020;757:144936. [DOI] [PubMed] [Google Scholar]
- 19.Cai J, Qiao B, Gao N, Lin N, He W.Oral squamous cell carcinoma- derived exosomes promote M2 subtype macrophage polarization mediated by exosome-enclosed miR-29a-3p. Am J Physiol Cell Physiol 2019;316:C731-40. [DOI] [PubMed] [Google Scholar]
- 20.Guo Y, Zhai J, Zhang J, Ni C, Zhou H.Improved radiotherapy sensitivity of nasopharyngeal carcinoma cells by miR-29-3p targeting COL1A1 3'-UTR. Med Sci Monit 2019;25:3161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinh TK, Fendler W, Chalubinska-Fendler J, Acharya SS, O'Leary C, Deraska PV, et al. Circulating miR-29a and miR- 150 correlate with delivered dose during thoracic radiation therapy for non-small cell lung cancer. Radiat Oncol 2016;11:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burja B, Kuret T, Janko T, Topalovic D, Zivkovic L, Mrak- Poljsak K, et al. Olive leaf extract attenuates inflammatory activation and DNA damage in human arterial endothelial cells. Front Cardiovasc Med 2019;6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy R, Dagher A, Butterfield C, Moses MA. ADAM12 is a novel regulator of tumor angiogenesis via STAT3 signaling. Mol Cancer Res 2017;15:1608-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo ML, Zhou Z, Sun L, Yu L, Sun L, Liu J, et al. An ADAM12 and FAK positive feedback loop amplifies the interaction signal of tumor cells with extracellular matrix to promote esophageal cancer metastasis. Cancer Lett 2018;422:118-28. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Wang Y, Gu J, Zhou D, He Z, Wang X, et al. ADAM12-L confers acquired 5-fluorouracil resistance in breast cancer cells. Sci Rep 2017;7:9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duhachek-Muggy S, Qi Y, Wise R, Alyahya L, Li H, Hodge J, et al. Metalloprotease-disintegrin ADAM12 actively promotes the stem cell-like phenotype in claudin-low breast cancer. Mol Cancer 2017;16:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duhachek-Muggy S, Zolkiewska A.ADAM12-L is a direct target of the miR-29 and miR-200 families in breast cancer. BMC Cancer 2015;15:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang JY, Zhang Q, Wang DD, Yan W, Sha HH, Zhao JH, et al. MiR-29a: a potential therapeutic target and promising biomarker in tumors. Biosci Rep 2018;38:BSR20171265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang C, Wang L, Song H, Wu C.MiR-29a inhibits the progression of oral squamous cell carcinoma by targeting Wnt/beta-catenin signalling pathway. Artif Cells Nanomed Biotechnol 2019;47:3037-42. [DOI] [PubMed] [Google Scholar]
- 30.Gao J, Shao Z, Yan M, Fu T, Zhang L, Yan Y.Targeted regulationof STAT3 by miR-29a in mediating Taxol resistance of nasopharyngeal carcinoma cell line CNE-1. Cancer Biomark 2018;22:641-8. [DOI] [PubMed] [Google Scholar]
- 31.Yang L, Li N, Yan Z, Li C, Zhao Z.MiR-29a-mediated CD133 expression contributes to cisplatin resistance in CD133(+) glioblastoma stem cells. J Mol Neurosci 2018;66:369-77. [DOI] [PubMed] [Google Scholar]
- 32.Yuan LL, Li L, Liu JN, Mei J, Lei CJ. Down-regulation of miR-29a facilitates apoptosis of colorectal carcinoma cell SW480 and suppresses its Paclitaxel resistance. Eur Rev Med Pharmacol Sci 2018;22:5499-507. [DOI] [PubMed] [Google Scholar]
- 33.Shi X, Valizadeh A, Mir SM, Asemi Z, Karimian A, Majidina M, et al. miRNA-29a reverses P-glycoprotein-mediated drug resistance and inhibits proliferation via up-regulation of PTEN in colon cancer cells. Eur J Pharmacol 2020;880:173138. [DOI] [PubMed] [Google Scholar]


