Abstract
Modern genome-scale methods that identify new genes, such as proteogenomics and ribosome profiling, have revealed, to the surprise of many, that overlap in genes, open reading frames and even coding sequences is widespread and functionally integrated into prokaryotic, eukaryotic and viral genomes. In parallel, the constraints that overlapping regions place on genome sequences and their evolution can be harnessed in bioengineering to build more robust synthetic strains and constructs. With a focus on overlapping protein-coding and RNA-coding genes, this Review examines their discovery, topology and biogenesis in the context of their genome biology. We highlight exciting new uses for sequence overlap to control translation, compress synthetic genetic constructs, and protect against mutation.
Subject terms: Genomics, Synthetic biology, Genome
The authors review overlapping sequences as fundamental features of prokaryotic, eukaryotic and viral genomes, discussing the diverse topologies and functions of overlapping genes, open reading frames and coding sequences. Moreover, they highlight the potential of harnessing sequence overlaps for synthetic biology approaches.
Introduction
When the first DNA genome was sequenced by Frederick Sanger in 1977, the results solved a perplexing mystery that had bothered scientists for some time. Previous analysis of the proteins produced by bacteriophage φX174 during infection seemed to require coding sequences (CDSs) longer than the measured length of the phage genome1. The mystery was solved when analysis of the genome sequence revealed extensive overlap between coding regions, with the internal scaffolding gene overlapping the genome replication gene and the lysis gene embedded entirely within the external scaffolding gene1,2. The compressed nature of these viral genes led to the conclusion that hidden within the genome could be other undiscovered sites of polypeptide synthesis2. Further refinement of the φX174 gene model showed an alternative start site within the genome replication gene A that produced a truncated protein with an identical CDS to the C-terminus of the A protein but holding a distinct function3,4. Thus, overlapping genes have been observed from the very beginning of sequencing and genomics. Since then, overlapping genes, and more specifically open reading frames (ORFs) and CDSs, have become a common genetic feature described during viral genome annotation5, including within the SARS-CoV-2 genome6. However, until recently, their true abundance and importance was overlooked outside of the realm of viral genomics7 and their discovery and annotation within cellular genomes have generally been treated as unique and idiosyncratic.
Today, we are seeing a renaissance of the field owing to the rapid advancement of genome-scale protein and RNA measurement tools and increasingly advanced prediction algorithms (Box 1), which have collectively revealed an abundance of overlapping genes and ORFs within cellular genomes. Recent work on the human genome has placed estimates of overlapping features much higher than previously thought8,9, encompassing 26% of all protein-coding genes10. This estimate will likely increase in the future as small ORFs (sORFs) encoding microproteins are increasingly being found in the human genome within previously annotated genes11–13.
In this Review, we define a gene overlap in eukaryotes when at least one nucleotide is shared between the outermost boundaries of the primary transcripts of two or more genes, such that a DNA base mutation at the point of overlap would affect transcripts of all genes involved in the overlap (Fig. 1a, top). Thus, overlapping genes as defined here include 5′ and 3′ untranslated regions (UTRs) as well as introns. Overlapping ORFs and CDSs, which are components of genes, are distinctly defined here as when the overlap occurs in a sequence region of two or more genes that encode protein in the mature transcript such that a DNA base mutation at the point of overlap would alter a codon and potentially the protein sequence of one or more members of the overlap. We define a gene overlap in prokaryotes and viruses as when the CDSs of two genes share a nucleotide either on the same or opposite strands (Fig. 1a, bottom). These definitions are compatible with a recently updated, community-driven effort to create consensus classifications of non-canonical ORFs, of which overlaps are one example14.
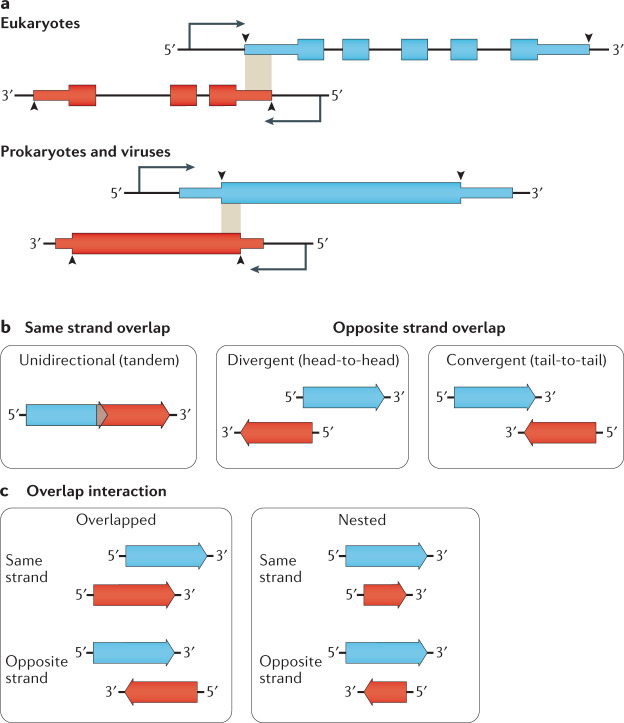
Fig. 1. Overlapping gene definition and topologies.
a | Gene overlap definitions differ between prokaryotes and eukaryotes. (Top) Eukaryote overlaps are most frequently defined as overlaps between the boundaries of the primary transcript, shown here in the shaded region. Often, the overlap is only between the 5′ untranslated region (UTR) or 3′ UTR of both transcripts (5′ UTR overlap shown)10,170. (Bottom) In contrast, prokaryote and virus genes are only considered to overlap if their coding sequences overlap5,27. Thin boxes denote 5′ and 3′ UTRs while thick boxes are coding sequences. Arrowheads indicate the extent of the consensus definition of gene boundaries within studies referenced in this review. b | Genes and open reading frames (ORFs) can be overlapped in one of three topologies. Unidirectional (also called tandem) overlaps occur between genes and ORFs on the same strand. Divergent (also called head-to-head) overlaps occur between genes and ORFs on opposite strands that overlap at their 5′-ends. Convergent (also called tail-to-tail) overlaps occur between genes and ORFs on opposite strands that overlap at the 3′-ends27. c | Gene and ORF interactions can be either overlapped, where only limited portions of each gene or ORF are overlapping, or nested, where the entire sequence of one partner falls within the boundaries of the other.
Here, we review overlapping genes as fundamental features of both cellular and viral genomes. We first discuss the diverse topologies and functions of overlapping genes in natural genomes across prokaryotes, eukaryotes and viruses. We then highlight their importance for synthetic biology approaches, as bioengineers are both faced with disentangling CDSs to refactor gene clusters and whole genomes and inspired to implement these features in synthetic genetic constructs to control protein expression and slow evolution. We limit our discussion to protein-coding and RNA-coding regions within genomes that partially or completely overlap at least one other gene. For information on ORFs localized entirely within 5′ or 3′ UTRs, which itself is a rapidly evolving field, we direct readers to other works15,16.
Box 1 Identifying overlapping genes and ORFs.
Genome annotation is the bedrock against which genome-scale measurements are compared, with most bioinformatics pipelines today annotating genomes through a combination of sequence alignments and hidden Markov modelling. However, many of these standardized methods may be inappropriate for the discovery of overlapping genes because they are reliant on already curated genes, where overlapping genes are poorly represented and contain atypical sequence composition40,41,176. For example, the RAST177 pipeline uses both ab initio (GLIMMER) and sequence homology steps (SEED genome database) to annotate genomes178 but markedly penalizes overlaps between predicted open reading frames (ORFs), which potentially misses vital features177. Furthermore, genome annotation standards are biased against feature overlaps, especially genes “completely contained in another gene”179. The solution may be custom algorithms tailored for overlap mapping that have been created specifically for viral genome annotations (for example, OLGenie180) and annotation pipelines based on hidden Markov models trained on databases of experimentally confirmed overlapping genes181. Some tools, such as Glimmer3 and BG7, are more tolerant of overlapping ORFs by retaining candidate ORFs even if they overlap other predicted ORFs182,183. New annotation databases, such as OpenProt184, are being created in response to the growing realization that eukaryotic gene models need to include polycistronic transcripts with non-AUG initiation sites185.
Proteogenomic methods, including bottom-up proteomics and ribosome profiling, in combination with DNA sequencing and perturbation, have been critical for the identification of overlapping genes. Mass spectrometry-based proteomic techniques are used mainly to confirm the expression of gene products based on genomic sequence annotation and are notionally limited by the quality of annotations. Most commonly, proteomics is performed using shotgun tandem mass spectrometry, whereby proteolytic peptide digests are ionized and sequenced based on peptide fragment ion mass-to-charge ratios, thus providing primary evidence of translated gene products. However, for large-scale studies, MS data must be computationally matched to in silico digests of the theoretical proteome. Unbiased six-frame genome translations can be used to maximize the proteome ‘search space’ but are rarely implemented due to expanded computational analysis time and high false-discovery rates186. In addition, recent studies have shown unexpectedly strong non-AUG translation initiation187,188, which are not accounted for in standard six-frame AUG translations. N-terminal peptide enrichment strategies can be used to identify sites of translation initiation, regardless of start codon used189,190, but the database needs to already include these candidates. Despite these considerations, proteomic measurements can be powerful, with one study identifying 1,259 alternative proteins produced from previously annotated human transcripts191.
Complementary to mass spectrometry proteomics, ribosome profiling (Ribo-Seq) is a method that involves capturing ribosomes as they decode mRNA and sequencing the section of the transcript bound by the ribosome192. In particular, the translation initiation site Ribo-Seq variant, which uses inhibitors to pause ribosomes on the start codon, has revealed an abundance of new translation initiation sites within transcripts in prokaryotic29, eukaryotic11 and viral genomes193–195.
RNA sequencing alone can also identify genomic regions with overlapping transcripts. For example, 180,000 alternate ORFs within previously annotated coding regions were found in humans66, and a transcription start site profiling study in Helicobacter pylori identified pervasive transcription on the opposite strand of canonical genes (that is, antisense transcription)196.
Overlapping ORFs discovered using the above methods have been verified using a variety of reverse genetics approaches, including CRISPR–Cas9 and catalytically dead Cas9 (dCas9) disruption11,12,65, as well as an attempt at proof-by-synthesis to establish the absence of any undiscovered overlapping genes197.
Overlapping gene topology and function
Studying overlapping genes across cellular and viral genomes reveals different patterns of overlap topologies that vary in frequency between prokaryotes and eukaryotes8,17. The reasons for these observed patterns are either more frequent biogenesis of certain types, evolutionary selection for retention of certain topologies or a combination of the two. At the moment, no consensus exists for the relative importance of these two factors, that is, creation versus retention. Overlap is thought to arise from at least six mechanisms that result in one gene becoming entangled with another, either through sequence extension9,18,19, re-arrangement of existing genes20,21, or de novo gene and ORF creation within an existing gene22.
Three directional overlap topologies are possible (Fig. 1b). Unidirectional overlaps (→→) occur between genes encoded on the same strand and may be further categorized according to the reading frame for overlapping ORFs. The remaining two topologies occur between genes on opposite strands and are called convergent (→←) and divergent (←→) (Fig. 1b). Unidirectional overlaps are more frequent in genomes of viruses and bacteria5,17, whereas the divergent and convergent overlaps are more frequent in eukaryote genomes10,23. The way the two genes interact can be described as either overlapped, with only part of each gene sequence occupying the same genomic region, or nested (Fig. 1c), whereby the entire extent of one gene is enclosed within the borders of a larger gene. The relationship between overlapping and nested genes has been described in other ways, including ‘internal–external’20 or ‘mother–daughter’ genes24.
The different ways that genes are defined in prokaryotes and eukaryotes in the literature has possibly biased estimates of the prevalent types of overlaps between these groups. For example, in prokaryotic and virus literature, gene overlaps are only considered when the CDSs of the genes overlap5,17, whereas in eukaryotic literature overlaps are more often considered between the primary transcript boundaries10,25 (Fig. 1a). The effect of these different definitions is that certain types of overlap seem to be more prevalent in eukaryotes versus prokaryotes but, if the same definitions were used for both, these apparent differences could in fact disappear. For instance, overlapping CDSs have certain constraints on relative reading frame and sequence composition26,27 that overlaps between 5′ and 3′ UTR do not. Within the limitations posed by the way overlapping genes are described in the literature, we compare and discuss prokaryotic and eukaryotic gene overlap from both their idiosyncratic aspects as well as their similarities, where present.
Prokaryotes
Overlapping CDSs within prokaryotic genomes have been reported in both bacteria28–30 and archaea31 and, on average, 27% of CDSs in these groups are involved in at least one instance of overlap19. Across prokaryotes, the frequency of CDS overlap within a genome seems to be constant regardless of genome size17,32, although certain groups can deviate sharply from this pattern. For example, intracellular microbial parasites show a weak correlation between genome size and the number of overlapping CDSs33.
In prokaryotic genomes, 84% of CDS overlaps are unidirectional17 (→→) and produced through start codon or stop codon loss, resulting in one member of a pair of adjacent non-overlapped CDSs expanding their coding sequence into their adjacent partner (Fig. 2a,b). Sequence analysis shows that stop codon loss of the upstream partner is the most frequent mechanism for unidirectional overlap creation32,34. Start codon loss of the downstream partner and de novo start codon creation within an existing CDS (Fig. 2c) also generate unidirectional overlaps18,32. Over 98% of currently identified unidirectional overlaps are less than 60-bp long, with the vast majority of these short overlaps either 1 bp or 4 bp overlapping start and stop codons (TA[A]TG, TG[A]TG, or [ATGA])17,35. This overlap motif may be intimately tied to prokaryotic operons, where clusters of related genes are under the regulatory control of a single promoter, and overlapping start and stop codons of their respective CDSs may facilitate enhanced regulatory control through translational coupling between adjacent partners36.
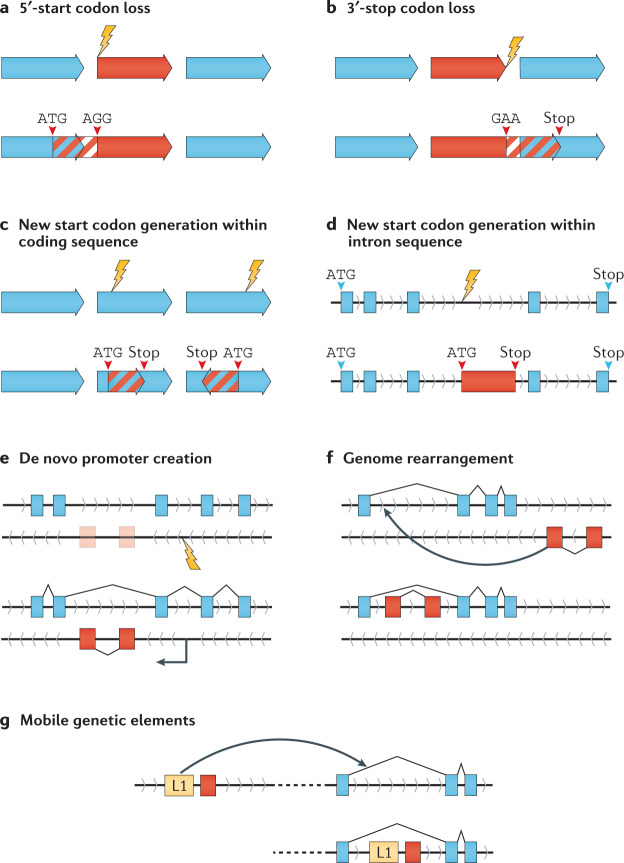
Fig. 2. Mechanisms of gene and ORF overlap creation.
New overlaps can be created through a range of mechanisms and likely require numerous complementary developments to produce the appropriate sequence context for retention of gene or open reading frame (ORF) functionality. a | Mutations removing the start codon of a downstream ORF may result in the next available upstream start codon being utilized, which could be within an upstream ORF18. b | Mutational loss of a stop codon may result in the extension of an ORF. Similar to start codon loss, the next available stop codon may be utilized, which could be within a downstream ORF19. c | De novo generation of an ORF may begin with the creation of a start codon within an existing coding region through mutation and, in conjunction with a downstream stop codon, produces an overlapping ORF18. d | Non-coding intron sequences may acquire a start codon through mutation and, in conjunction with a downstream stop codon, produce a nested ORF20. e | Mutations that result in the de novo development of a sequence capable of recruiting transcriptional machinery (such as a promoter or enhancer) may result in a new overlapping gene171. f | Genome rearrangements, such as inversions and translocations, may result in distant non-overlapping genes becoming overlapped. This mechanism has been seen within human cancers. g | Mobile genetic elements carrying genes (such as transposons or proviral genes) may localize to within a gene, generating a new gene overlap172,173.
Convergent (→←) and divergent (←→) overlaps (Fig. 1b) are observed at lower frequencies in prokaryotes compared with eukaryotes, and similar to unidirectional overlaps, are biased towards short overlap lengths35. Short convergent overlaps are strongly biased towards 4-bp stop codon overlaps owing to the incompatibility of forward-strand stop codons (TAA, TAG, TGA) with reverse-strand stop codons (TTA, CTA, TCA) in any other configuration37. Divergent overlaps (Fig. 1b) do not have strong phase biases but are substantially rarer than convergent overlaps38, which is likely due to the presence of critical sequence structures in the 5′-end of CDSs that impose additional evolutionary constraints on the successful retention of these overlap topologies.
It is currently unclear whether the commonness of short tandem start–stop overlaps compared to long nested overlaps (Fig. 1b) is a result of biology or merely reflects our ease to detect them. Despite increasing numbers of fully nested CDSs within prokaryotes being discovered due to a convergence of proteomic and ribosome profiling methods (Box 1), the idea that many more long nested overlaps within prokaryotes remain to be discovered is contentious19,35 and genome annotation pipelines are biased against their existance39. The unusual sequence characteristics of long overlapping CDSs may have also contributed to the difficulty of their discovery, resulting in undercounting40,41. One reason put forward to explain why long nested overlaps should be rare includes the evolutionary burden of maintaining larger overlaps, although evidence to the contrary showing positive selection at overlaps27,42,43 shows that this explanation may be too simplistic. Selection for long convergent overlaps has been shown to have a strong reading frame bias and it has been suggested that retention involves positive selection at the birth of the overlap, followed by purifying selection afterwards27. Recently, an overlapping protein-encoding CDS with extensive 603 bp overlap has been discovered embedded in the highly conserved ompA gene in enterohaemorrhagic Escherichia coli44, showing that, with improved measurement tools, more of these long nested overlaps may be discovered42.
While the precise selective forces governing the retention of long unidirectional CDS overlaps in prokaryotes are unknown, the selective forces governing the retention of some short stop–start overlaps likely act through their enhancing effect on gene expression36 (Fig. 3a,b). Furthermore, overlapping CDS frequency is higher in fast-growing thermophilic organisms, which suggests that genome streamlining is an adaptive strategy for fast growth at high temperatures45,46. Mechanistically, overlaps between start and stop codons of adjacent unidirectional CDSs provide additional benefits for translational coupling47–50 and ribosome re-initiation48,50 (Fig. 3a,b) in addition to benefits already provided by operons51,52. The menaquinone biosynthesis pathway in E. coli is an example of multiple gene members connected via overlapped stop–start sites within a single operon across all three reading frames (Fig. 4a).
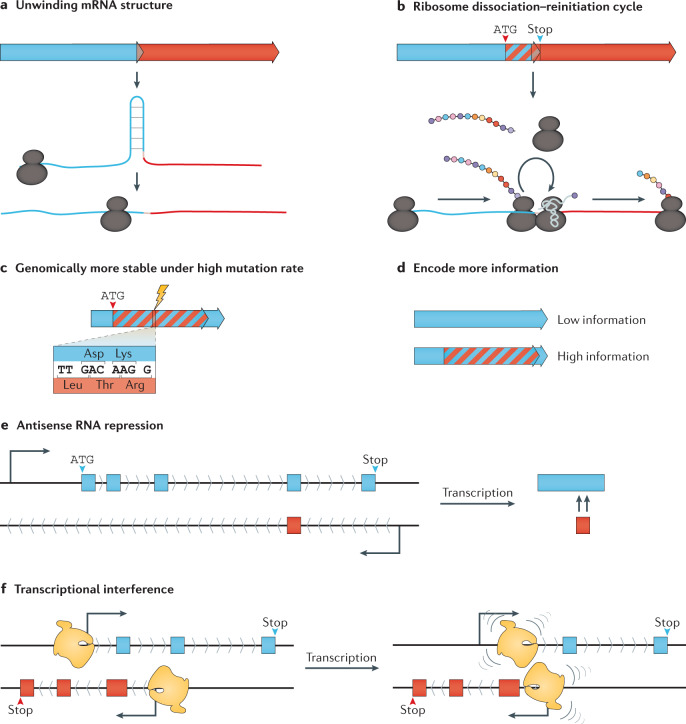
Fig. 3. Selective pressures involved in retaining gene and ORF overlaps.
a,b | Overlapping start and stop codons cause translation coupling between unidirectional overlapping open reading frames (ORFs) through unwinding of mRNA secondary structure around the ribosome binding site and start codon and by enhancing ribosome re-initiation48. c | Overlapping sequence regions cause mutations to affect more than one ORF, increasing fitness cost and preserving overlapped sequences under mutational pressure71,75. d | Encoding more ORFs in the same sequence region allows genetic novelty with reduced genome changes, which is particularly advantageous for viruses that have spatial constraints on genome size76,77. e | Sense–antisense gene and ORF overlap is frequently involved with gene expression regulation, including non-coding RNA and long non-coding RNA96. f | Transcriptional tuning from convergent overlapping genes and ORFs as a result of interactions between RNA polymerase collisions (transcriptional interference174,175).
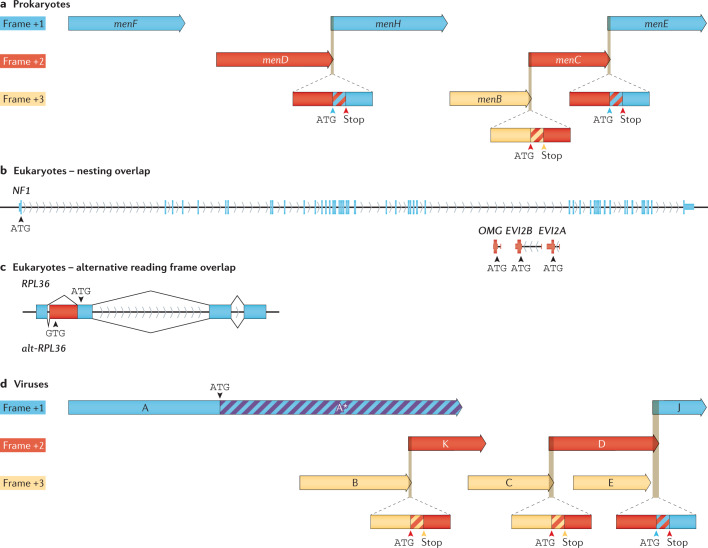
Fig. 4. Gene and ORF overlap across prokaryotes, eukaryotes and their viruses.
a | Escherichia coli menaquinone biosynthesis operon contains three short stop–start coding sequence (CDS) overlaps. b | The large human gene NF1 and internal nested protein-coding ORFs OMG, EVI2B and EVI2A are located within NF1 introns. c | Recently described alt-RPL36 (bottom) overlaps the human ribosomal protein gene RPL36 (ref.65) through an out-of-frame GTG start codon within a 5′-extended RPL36 exon present on RPL36 transcript variant 2. The alt-RPL36 CDS generates a longer protein with an entirely different sequence from RPL36 (ref.65). d | The virus φX174 contains overlaps in all three reading frames: three short unidirectional stop–start CDS overlaps, two nested CDSs, and one in-frame start generating an N-terminally trunkated protein.
Functional entanglement of overlapped CDSs can act on their retention over evolutionary selection beyond gene expression levels. For example, the overlapping drrA/drrB genes encode an efflux pump for the anticancer agent doxorubicin in the production strain Streptomyces peucetius. When the overlap was disrupted, the expression levels of DrrA and DrrB proteins remained unchanged and membrane trafficking was unaffected but functional assembly of the protein complex was lost47. Correct protein complex assemply has been revealed to be spatially regulated at the translation level for genes linked in operons, which may explain the DrrA/DrrB finding47,53. Overlap functions such as this are likely to be prevalent for overlapped CDSs given the functional assortment of genes involved in overlap54.
Eukaryotes
In eukaryotic genomes, the prevalence of overlapping genes is difficult to assess because of the inconsistent nomenclature that is used to describe the relationship between the genes, their 5′ or 3′ UTRs, and CDSs. Unlike prokaryotes, classifications and studies of overlapping genes in eukaryotes are as varied as their genome size and complexity. The predominant type of overlap is convergent8,10,23 (Fig. 1b), although generalization within eukaryotes is less useful given their genome diversity, which ranges from unicellular eukaryotes with compact, intron-poor genomes to complex, multicellular eukaryotes with expanded genomes and high intron densities55,56.
Most overlapping genes in eukaryotes are classified as such because their 5′ or 3′ UTRs overlap57. Of those with overlaps between the start and stop codon boundaries of either member (Fig. 1a), introns provide an additional non-coding location for gene transcripts to overlap. When an entire ORF is contained within an overlapping gene’s intron it is referred to as intron nesting20,58. True exon–exon overlaps make up the minority of transcript overlap in eukaryotes8,23 but new technologies (Box 1) suggest that they may be more common than currently appreciated11,12.
Nested gene overlaps in eukaryotes occur most frequently within an intron of the larger partner as is the case for three antisense nested genes, EVI2A, EVI2B and OMG, within intron 27b of the human NF1 gene (Fig. 4b). Nested overlaps are thought to be created through four processes: (1) mobilization of a distal gene into the intron of another gene (for example, through retrotransposition), (2) de novo creation of an ORF within an intron of an existing gene, (3) one ORF is internalized after an adjacent gene acquires additional exons and (4) two external genes flanking another gene fuse, thus internalizing the other gene20 (Fig. 2). The introns that harbour nested genes are considerably longer than other introns, suggesting acquisition of an existing gene through retrotransposition, among other mechanisms, is a dominant process rather than de novo evolution21,59. However, evidence from metazoans shows that several de novo genes have emerged from introns in that lineage20,60. The extent of the nesting can vary from an internal gene with a single exon residing within the intron of an external gene (for example, H2BFS within HSF2BP in humans21) to multiple layered ‘Russian doll-like’ nestings in Drosophila melanogaster20.
Eukaryotic overlapping protein-coding genes are implicated in lineage-specific groups. For example, the majority of vertebrate genes with overlapping transcripts are not conserved across species9,57 likely because overlapping genes tend to be young and frequently lost during evolutionary time57. A broad study of five well-described metazoan genomes (Caenorhabditis elegans, Danio rerio, Drosophila melanogaster, Mus musculus and human) found that, for protein-coding genes, transcript overlap is selected against and mainly species specific and the majority of new overlaps are in terminal non-coding exons25. Overlap between opposite strand exons containing coding sequence is also lineage specific, with the mammalian genes THRA (which encodes thyroid hormone receptor alpha) and NR1D1 (which encodes nuclear receptor subfamily 1 group D member 1) displaying convergent overlap in the coding sequence portion of their 3′ exons, whereas marsupials seem to have lost this feature since their divergent evolution over 90 million years ago. This change results in an absence, during marsupial development, of the TRα2 protein, a variant of the receptor unable to bind the hormone61.
Although rare, eukaryotes contain genes with CDS overlaps8,9,62,63 as well as overlaps that span exon–intron boundaries57,64. A community-driven roadmap on translated ORFs has proposed that these overlapping CDSs be annotated as novel genes despite the shared locus14. The recently described alt-RPL36 ORF65 (Fig. 4c) is one such example of a gene possessing two distinct and functional CDSs overlapping the same genomic sequence. These alternative ORFs66 are often functionally related and implicated in a range of human diseases12,67. For example, the cyclin-dependent kinase inhibitor 2A (p16INK4a) and tumour suppressor ARF, which regulate the tumour suppressors retinoblastoma protein (RB) and p53 transcription factor68, are produced as alternatively spliced transcripts from what is now considered the same gene (CDKN2A), even though the proteins do not share sequence or structural similarity, and the E1b exon that produces the ARF protein is ~20 kb upstream of the other CDKN2A exons68. Similarly, a recently discovered nested overlapping ORF within the FUS ORF (alt-FUS) is associated with neurodegeneration69 and alt-Ataxin is mutated in spinocerebellar ataxia type 1 (ref.64).
Viruses
The topology of overlapping genes in viruses is determined both by the host cell type as well as by constraints unique to viruses. Despite viruses having diverse genomes (RNA or DNA in single-stranded or double-stranded form) and lifestyles, overlapping CDSs are found across all known virus groups5,70. The proportion of viruses with overlapping CDSs within their genomes varies from double-stranded RNA viruses having fewer than a quarter to almost three-quarters of retroviridae (single-stranded RNA using reverse transcriptase) and single-stranded DNA genomes containing overlapping CDSs5. Segmented viruses, those with the genome split into separate pieces and packaged either all in the same capsid or in separate capsids, are more likely to contain an overlap than non-segmented viruses5. The retention of overlapping CDSs in viruses has been attributed to enabling evolutionary rate reduction and increasing mutational robustness71,72 as well as being a result of capsid size limitations73.
The role of overlapping genes in reducing the rate of viral evolution has been most intensively examined in RNA viruses, which have higher mutation rates, smaller genomes and less CDS overlap than DNA viruses of comparable length5,73,74. Studies have supported the notion that CDS overlap increases hypersensitivity to mutation (as a mutation on average would affect more than one CDS)26 but that genome (or population) mutational robustness is increased overall71 (Fig. 3c). This has been eloquently demonstrated with the overlapping rev and tat genes of the RNA virus HIV1 (ref.75). Functional segregation is observed between the overlapped regions, facilitating the purging of possible deleterious mutations; that is, important nucleotide or amino acid regions of one gene overlap regions subject to fewer constraints in the other75.
Thus, given that gene overlap regions are likely protective and increase fitness, why then do RNA viruses have fewer overlapping genes than DNA viruses with lower mutation rates and less restrictive genome sizes?5,73 The answer may lie in the balancing of different selection pressures. For instance, the lower mutation rate of DNA viruses facilitates greater genomic novelty and evolutionary exploration within a structurally constrained genome and may therefore be the primary driver of gene overlaps76,77 (Fig. 3d). By contrast, in RNA viruses, overlaps may primarily be a means for maintaining mutational robustness in the face of higher mutational rates (Fig. 3c)71,75 as exemplified with the population fitness advantage conferred by the rev and tat overlap of HIV1 (ref.75).
Virus capsid size restrictions driving the evolution of gene overlaps has been a focal point of investigation due to early observations of dramatic viability loss in viruses with genomes engineered to be longer than wild type78. For instance, increasing the single-stranded DNA genome length of ΦX174 by >1% results in almost complete loss of infectivity79. This is thought to be the result of the strict physical constraints imposed by the finite capsid volume and, as such, any evolutionary innovation must be facilitated in the existing sequence space (Fig. 3d) rather than by increasing genome length. This idea is supported by work with adeno-associated viruses as gene delivery vectors, where viral packaging is constrained by genetic cargo size limits80, necessitating the use of multiple vectors to deliver large human genes such as CFTR81. Studies have shown a strong prevalence of overlapping CDS births in the +2 frame over the +3 frame40,77, which is likely due to two factors: mutational bias, whereby start codons are more prevalent in the +2 reading frame relative to known CDSs40,74, and recent evidence suggesting that the sequence of known CDSs in the +2/–2 reading frames preserves key physicochemical properties of the original sequence82.
The seemingly simple relationship between genome and capsid has also been questioned. Combined structural and genomic data have shown that most viruses do not fully utilize the available internal space of the capsid76. Furthermore, viruses are highly biased towards short overlaps, with the vast majority less than 50 nt (ref.5) in length, overall negatively correlated with genome length70, with absolute nucleotide overlap summed across the genome rarely exceeding 1,500 nt (ref.76). This distribution of overlap length within viruses points towards overlaps being favoured for several different reasons, with short CDS overlaps enabling translational coupling, whereas long overlaps being retained mainly when they generate genetic novelty that increases fitness. For example, a 4-nt (ATGA) stop–start overlap within a Totivirus directs coupled translation of the CDSs83, whereas a 276-nt overlap in phage ΦX174 between its recently evolved lysis gene E and scaffolding gene D (Fig. 4d) enables the phage to lyse its host and release virions more efficiently1,2.
Overlap of ncRNA with protein-coding genes
Another important and highly abundant type of overlap within genomes is between non-coding RNA (ncRNA) genes and those of protein-coding genes. Shared sequence overlap may be between the mature ncRNA transcript region and the CDS region of mature protein-coding mRNA or it may only occur between 5′ and/or 3′ UTR regions of the transcripts.
In prokaryotic genomes, ncRNAs are an increasingly identified feature84, with cis-encoded antisense RNA regulation being a major player in physiological responses84,85. Examples of these pairings have demonstrated tight-knit regulation of expression of the protein-coding gene such as in type I toxin/antitoxin systems86 and in Mg2+ tolerance and virulence87. Interestingly, examples of unusually long antisense RNA have also been found, which likely hold greater regulatory control functions (such as regulation of entire operons) and have acquired their own designation as ‘excludons’84,88. Overlapping regulatory RNAs embedded within the coding sequence of bacterial genes can act in diverse regulatory roles89–91. Evidence is also emerging that ncRNAs in prokaryotes can contain protein-coding ORFs92,93. For more information on prokaryotic overlapping ncRNAs, we refer readers to another review84.
In eukaryotes, the sense–antisense overlapping transcripts are called cis-natural antisense transcripts (cis-NATs) and this type of overlap topology is frequently found in eukaryotic genomes in convergent or divergent relationships (Fig. 1b). Cis-NATs have regulatory functions at the RNA level25,94 and the most frequent combination is one protein-coding transcript paired with an antisense non-protein-coding transcript95 enabling enhanced transcriptional and post-transcriptional gene regulation96 (Fig. 3e,f). The regulatory roles of cis-NATs span major biological functions97 but can be generalized into protein expression regulation98, splice site masking99,100, double-stranded RNA-dependent mechanisms101,102 and chromatin remodelling103,104. Furthermore, due to the cis-acting mechanism and shared genetic loci, the evolutionary trajectories of both genes are closely entwined105,106. As such, interesting questions surround their evolution and acquisition, such as whether one member of the pair arose de novo through the acquisition of a promoter or by other mechanisms (Fig. 2). Recently, some overlapping ncRNA antisense transcripts have been found to also encode proteins11,107, further increasing the complexity and constraints of these overlapping interactions.
Many cis-NATs have been associated with human disease, including cancer progression108–111. For example, the convergently overlapping WDR83 and DHPS genes both encode proteins; together, RNA duplexing of their 3′ UTRs results in the concordant increase in their transcript stability and protein expression, ultimately resulting in increased cell proliferation in gastric cancer cells102. In a subpopulation of patients with α-thalassaemia, the disorder is caused by a chromosomal deletion that creates a new gene overlap between HBA2 and LUC7L, resulting in antisense transcripts from LUC7L silencing the otherwise intact copy of HBA2 through CpG island methylation112.
An emerging feature of many ncRNAs is the presence of internal translationally active sequences termed sORFs. These sORFs are commonly defined as an ORF that spans no more than 300 nt that, owing to these small lengths, have lain hidden within previously described ncRNA transcripts113. In humans, 30% of sORF-derived proteins (also called microproteins) identified by mass spectrometry were mapped internally to annotated genes114. Subsequent studies have expanded this number using a variety of methods115,116, including recent work that systematically uncovered hundreds of sORFs. The sORFs were found overlapping both internal sequences as well as the start codons of annotated ORFs11. Investigations into the functionality of the overlapping sORFs have implicated many in human disease pathology107,117. Furthermore, it is likely that many of the ncRNAs found to possess sORFs are in fact misannotated and should be re-defined as mRNA; however, there are examples of RNA that possess dual functionality (non-coding and coding), thereby complicating classifications118,119. More information on this developing area can be found in recent reviews120,121, including the in-motion and recent community-driven initiative to comprehensively define and catalogue these classes of non-canonical ORFs in major databases14.
Overlapping genes in bioengineering
As we have outlined, gene overlaps in natural genomes are complex and their true number is only beginning to emerge. However, in synthetic biology, the re-engineering of natural genomes is well under way. Synthetic biology uses raw genetic material from diverse sources within heterologous systems to create new metabolic pathways122, enzymatic activities123, orthogonal transcription124,125 and translation initiation systems126,127, and complex genetic devices128,129. As such, the functional characteristics of overlapped genetic elements are becoming increasingly important to understand. Furthermore, the field of synthetic genomics is rapidly rebuilding entire genomes from the ground up (for example, E. coli130 or the yeast Saccharomyces cerevisiae131), with important choices to be made during the design stage for how to deal with overlapping sequences132.
Refactoring overlapping genes
Genome refactoring is a process of reorganizing gene architecture by reformatting the underlying sequences while maintaining functionality133. With the aim of increasing modularity, refactoring is often used to remove overlaps between genes so each is encoded on a separate piece of DNA. The effects of removing overlaps by encoding CDSs into their own distinct sequence regions may disrupt regulatory elements, such as promoters, or important RNA secondary structure elements as well as translational coupling from stop–start overlaps. Genome refactoring was pioneered with the bacteriophage T7 (ref.133) but is now commonly applied to biosynthetic gene clusters, where the aim is to exert transcriptional and translational control over the cluster in a heterologous host122.
Over the past 15 years, a number of genome engineering projects that modified overlapping CDSs and gene 5′ or 3′ UTRs have resulted in losses in viability and efficiency in the final bioengineered product133–137. For example, removing CDS overlaps in the bacteriophage T7 resulted in infectious virus yet significantly reduced fitness133. Subsequent work using serial passaging and selection for high growth rate over 100 generations was able to show substantial fitness increases similar to pre-adapted wild-type levels138. Similarly, a project to ‘decompress’ bacteriophage φX174 had the explicit aim to test the essentiality of CDS overlaps134. While coding potential was retained (Fig. 5a), this refactoring led to numerous phenotypic defects, including a substantial reduction in burst size and lower attachment efficiency, along with large changes in levels of several essential assembly and replication proteins produced during the infection cycle139.
Fig. 5. Disruptions to overlapping genes within a refactored phage genome and a complex biosynthetic gene cluster.
a | Creation of φX174.1f, also known as decompressed φX174, disrupted four unidirectional stop–start coding sequence (CDS) overlaps and two fully nested overlapping CDSs134. b | Refactoring the nitrogen fixation cluster from Klebsiella oxytoca disrupted four stop–start CDS overlaps and CDS overlaps varying from 1–14 bp (ref.140).
The first complete refactoring of a complex biosynthetic cluster involving overlapping CDSs involved moving the nitrogen fixation cluster of Klebsiella oxytoca into E. coli140. This process involved rebuilding the entire gene cluster from the bottom up, with the removal of non-essential CDSs, codon optimization and disruption of six CDS overlaps (Fig. 5b). In a subsequent, larger project, the group refactored the Salmonella pathogenicity island 1 to isolate and control production of the type III secretion system141. The refactoring disrupted eight CDS overlaps potentially involved in translational coupling and totalling 90 bp in length. Interestingly, the team discovered that the spaO gene contained an in-frame alternative start site at a GTG codon, essentially an in-frame overlapped CDS141. In both the nitrogen fixation cluster and the type III secretion system, potential functional deficiencies caused by the removal of CDS overlaps and translational coupling were compensated through careful empirical tuning of the individual ribosome binding sites (RBSs) and transcriptional regulation140,141.
Other smaller-scale refactoring projects have targeted overlapping CDSs specifically to remove engineering limitations. For example, the gene overlaps in the dbz operon in Rhodococcus erythropolis, which is used to remove sulfur and upgrade petroleum, were removed to relieve a bottleneck in the efficiency of the process. Through rational design targeting the rate-limiting enzyme of this operon (DszB), removal of the overlap of the start and stop codons of dszA and dszB CDSs resulted in a 12-fold increase in desulfurization activity over the wild-type operon142. Similarly, M13 phage CDSs VII and IX are naturally overlapped, limiting our ability to use the P9 protein for phage display. Removal of the CDS overlap solved this problem, although it resulted in a 1.4-fold decrease in phage infectivity143.
Beyond refactoring gene clusters, entire cellular genomes have now been refactored. During the design of the synthetic yeast chromosomes in the Yeast2.0 project, 15 instances of ORF overlap were identified where the desired TAG>TAA stop codon swap would have altered the codons of a verified ORF; however, details of how each instance was specifically addressed was not reported144. An E. coli genome engineering project to replace all 321 instances of TAG stop codons with TAA encountered several instances of CDS overlap where replacement might affect one of the partners. The first instance was the convergent overlapping yegV and yegW CDSs (both contained TAG stop codons in the overlapped region). Fortuitously, conversion of both overlapping TAGs to TAA conserved amino acid identity of the opposite CDS145.
A more extensive refactoring project to create an E. coli with a 61-codon genome via the removal of two sense (TCG and TCA) and one stop codon (TAG) encountered 91 instances of where these codons occurred in a region overlapping two CDSs132. If the overlapping CDSs were convergent, either silent mutations were incorporated or, if otherwise unavailable, the CDSs were separated by duplicating the overlap region followed by their independent recoding. In instances of unidirectional overlap, the CDSs were separated by duplicating the overlap region plus 20 bp upstream for a synthetic insert. At the start of this insert, an in-frame stop codon (TAA) was added to terminate translation from the original RBS. The result of this sophisticated refactoring process produced a viable E. coli albeit with a doubling time 1.6x longer than the parent strain under standard conditions132. Due to the vast number of changes across the genome, it is not possible at this time to attribute the slowed growth rate to CDS overlap disruption, although translational coupling between unidirectional overlaps would likely be disrupted by the RBS duplication protocol.
Conversely, some studies have taken a more cautious approach towards overlapping genes. For example, in the construction of the widely used E. coli K-12 single knockout library (Keio collection), deletions of dual coding regions were avoided by conserving overlap regions146. Similarly, in the minimal Mycoplasma mycoides genome, instances in which a retained CDS (essential or quasi-essential) was partially overlapped with a CDS to be deleted (non-essential) resulted in the overlapping region being retained136.
Applications of engineered overlapping genes
With increasing recognition that gene overlaps are functionally important and play vital roles within natural organisms, the construction of new overlapping genes has begun to be exploited in bioengineering. Theoretical work has previously shown that the genetic code is flexible enough to accommodate artificial overlap of protein domains147,148, and even artificial proteins149, often with the stated aim to protect the overlapping CDS from genetic drift150 in similar ways to that found in viruses71,75,151.
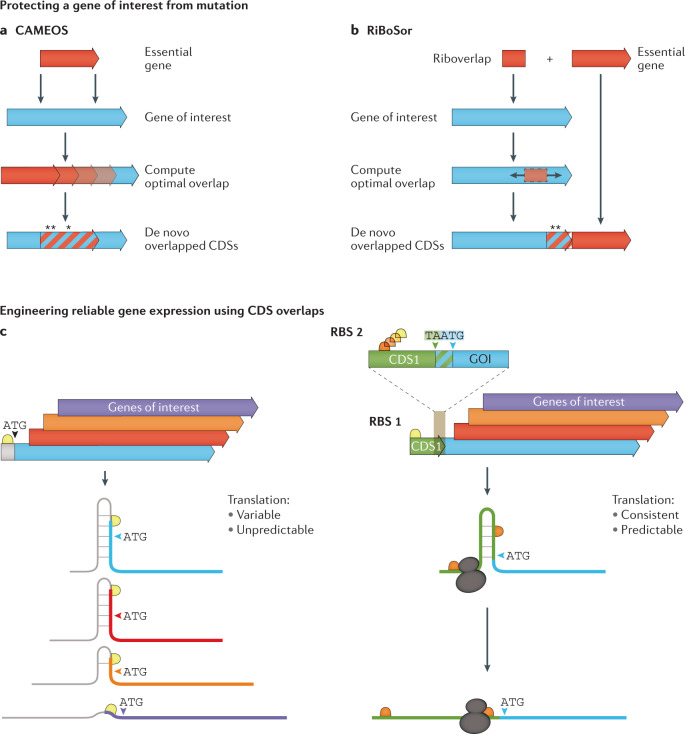
Recently, two methods for generating artificial CDS overlaps between a gene of interest and an essential gene have been described and empirically tested152,153. The Constraining Adaptive Mutations using Engineered Overlapping Sequences (CAMEOS) method152 searches for available overlaps between the CDSs of an essential gene and a gene of interest to be shielded from mutation152 (Fig. 6a). The algorithm uses a two-step process that relies on pre-existing or newly computed statistical models of the protein families that are being assessed for overlap. Furthermore, the CAMEOS dynamic programming algorithm searches for optimal solutions that consider both short-range (local codon usage) and long-range (epistatic) interactions while minimizing amino acid changes of the encoded proteins. CAMEOS was capable of creating a synthetic amino acid biosynthetic gene containing two additional out-of-frame nested essential CDSs. Protein functionality was maintained in the encoded enzymes despite up to 50% non-conservative amino acid changes and runs of up to six consecutive amino acid changes. Assessments of mutational robustness in the first 30 codons of the new CDS overlaps showed that the recodings were able to prevent any sequence changes to the non-essential CDS over 150 generations of growth, whereas the control CDS without overlap mutated by generation 50. The method was also shown to have some promise in the biocontainment of engineered constructs by overlapping a toxin gene with a gene of interest. If the engineered CDS is transferred to another organism, the toxin CDS will either kill the host or there will be a mutation in the toxin CDS that also inactivates the engineered CDS, thereby ensuring that the enhanced bioengineered phenotype is not transferred into the environment152.
Fig. 6. Exploiting CDS overlap for applications in bioengineering.
a | The Constraining Adaptive Mutations using Engineered Overlapping Sequences (CAMEOS) method searches for available overlaps between an essential gene and a gene of interest to be shielded from mutation while minimizing sequence changes152. Asterisks identify amino acid modifications to accommodate coding sequence (CDS) overlap. b | The RiBoSor algorithm searches for places within a gene of interest to silently create a ribosome binding site followed by a start codon in a different reading frame than the existing CDS. This generates a CDS that extends to the 3′ end of the existing CDS. An essential gene is then fused in-frame to the newly created CDS just 3′ of the stop codon of the original CDS153. Asterisks identify amino acid modifications to accommodate overlap. c | Reliable and tunable expression of a gene of interest can be facilitated by the bicistronic device155. (Left) A single ribosome binding site upstream of a variety of different CDSs can result in different interactions with the RBS and the coding sequence, causing variable translation initation rates that are difficult to predict. (Right) In the bicistronic device, the binding of a ribosome to the upstream ribosome binding site 1 of CDS 1 and its translation towards the gene of interest will disrupt inhibitory sequence structures. The ribosome will recognize the ribosome binding site (RBS2) of the downstream gene of interest and re-initiate translation providing a platform for reliable expression of the gene of interest.
The RiBoSor method153 takes a distinct approach to create a synthetic CDS overlap to protect a CDS of interest from mutation. The algorithm searches for locations within a CDS to silently create an out-of-frame RBS and start codon (Fig. 6b). The objective is to create a CDS in a different reading frame, called a Riboverlap, that runs uninterrupted to the 3′ end of the CDS of interest. If stop codons occur that would interrupt the new synthetic overlapped CDS, the algorithm tries to silently change them. An essential CDS is then fused in-frame to the newly created CDS just 3′ of the stop codon of the CDS of interest. Theoretically, this method should be both computationally simpler and more flexible than CAMEOS but also potentially less effective at constraining mutational pressure on the CDS of interest.
Another engineered genetic architecture taking inspiration from natural genomes features a CDS of interest directly downstream and overlapping a short translated CDS. Importantly, within this short coding sequence is the RBS site of the gene of interest that leads to a stop–start overlapping codon junction, facilitating coupled translation. Originally implemented by placing the trpE/trpD translationally coupled stop–start sequence upstream of the human γ-interferon gene154, a standardized bicistronic device architecture was recently created155. The bicistronic architecture results in robust and tunable protein expression regardless of the gene of interest (Fig. 6c). The success of this approach has been demonstrated in several studies155–157. Translational coupling in eukaryotes is currently less amenable to exploitation due to the mainly monocistronic mRNAs. However, there are increasing numbers of polycistronic transcripts being documented that suggest that this architecture may be useful in eukaryotes if correctly implemented158.
Creating organisms with new genetic codes will have a profound effect on the exploitation of overlapping genes in both positive and negative ways. For example, removing synonymous coding capacity within a genome to free up codons for encoding unnatural amino acids132,159,160 will make it difficult or impossible to retain existing CDS overlaps that rely on the degeneracy of the second and third codon positions. Conversely, engineering ribosomes to decode 4-nt codons161,162 will also expand the potential for synonymous codons and overlapping CDSs. New six-letter and eight-letter genetic codes163,164 could provide many additional synonymous codons for extensive overlapping CDS possibilities. However, substantial effort would be needed to create the multitude of tRNA–aminoacyl synthetase pairs126 to make this a reality. A 256-codon genetic code would allow up to 12 synonymous codons per amino acid, greatly expanding CDS overlap opportunities.
Conclusions and future perspectives
In this Review, we sought to highlight gene overlaps from a wide variety of genomes across the diversity of biology. There has been a vigorous renewal of interest in overlapping genes that can be directly attributed to recent advances in bioinformatics, sequencing and allied proteogenomic technologies. Overlapping genes, transcripts and ORFs have been a part of genome biology from the first sequenced RNA and DNA-based genomes2,165; however, their abundance and ubiquity have only just come into focus for eukaryotic genomes with the advent of recent genome-scale measurement technologies. From past and present literature, it seems clear that the definitions and assessments of overlap topology between eukaryotic, prokaryotic and viral genomes have been disconnected. It is unclear how this discordance arose; however, differing genome architecture, biology and researcher fields of interest are likely notable contributors. As new technologies, such as ribosome profiling, are showcasing, eukaryotes (and in particular humans) seem to encode an abundance of small and alternative overlapping ORFs6,11,12,14,166 spurring excitement in this genome biology. Future work will show whether the majority are true functional overlaps between protein-coding ORFs, non-coding translational regulatory regions or a result of measurement biases.
Going forwards, it would be highly desirable to harmonize the definition of gene overlap between eukaryotes, prokaryotes and viruses. This would enable true comparisons of overlap topology prevalence, more robust evolutionary studies, and highlight any domain-specific mechanisms contributing to overlap birth and fixation. One likely reason for the differences in gene overlap definition is the transcript-centric gene definition currently dominant in eukaryotes, which has not yet been adopted in prokaryote biology. This is undoubtedly due to the technical difficulty in defining prokaryotic transcripts compared to eukaryotic transcripts as well as to its lower emphasis in the field (until recently167,168).
In addition to different definitions of gene overlap, the way overlapping CDSs in the same loci are treated in prokaryote and eukaryote genome annotation is distinctly different. For example, p16INK4a and tumour suppressor ARF in humans are considered splice variants of the CDKN2A gene despite not sharing any sequence identity whereas, if these were found in a prokaryotic genome, they would be annotated as different genes. For eukaryotes, this is possibly changing with new proposals to annotate these overlapping ORFs as different genes are currently proposed14.
Lastly, the conventional idea of the monocistronic eukaryotic transcript is slowly being eroded64,169 with the advent of new research demonstrating transcripts harbouring multiple CDSs66,67. Moving away from this out-of-date convention will encourage researchers to pursue new lines of enquiry, such as the biological significance of polycistronic arrangements (well-known as operons in prokaryotes), or expanded insights into translation initiation.
We also discussed instances where engineered systems can take inspiration from natural overlapped gene systems for a variety of applications. Synthetic biology and genetic refactoring methods are frequently testing the limits of modifying and reformatting gene architectures in heterologous and endogenous hosts. We also identified future bioengineering research in expanded genetic codes that could make engineered gene overlaps more accessible and exploitable.
The rapidly advancing area of synthetic genomics, where entire genomes are being constructed anew, often with radically different topologies and overlapping genes disrupted or removed entirely, will require a much deeper understanding of genotype–phenotype relationships than we currently enjoy. Alternative and expanded genetic codes and codon-decoding capacity will open up new exciting possibilities for the design of extensively overlapped genetic systems to resist evolutionary drift and add additional functionality to new biotechnological applications of engineered overlapping genes not yet envisioned.
Acknowledgements
The authors thank R. Walker, H. Kroukamp, H. Goold, T. Williams, R. Willows, M. Maselko (Macquarie University), E. Holmes (University of Sydney), Y. Chen (UCLA), J. T. Beatty (UBC), V. Mutalik (Lawrence Berkeley National Lab), J. Chen (UT Southwestern Medical Centre), K. Y. Wei (Amgen), A. S. Khalil (Boston University), Y. Shen (BGI-Shenzhen), and J. Calles (Stanford University) for helpful discussions. P. R. J. thanks D. Endy for introducing him to gene overlaps of terrestrial origin and possibly beyond. The authors apologize to those whose work was not cited due to space limitations. The authors acknowledge funding support from Macquarie Research Excellence PhD Scholarship (BWW). P. R. J. is supported by NHMRC Ideas Grant APP1185399. M. P. M. acknowledges funding support from Bowel Cancer Research Foundation Australia.
Glossary
- Coding sequences
(CDSs). A continuous stretch of nucleotides that are bounded by a start and stop codon and undergo translation.
- Overlapping genes
In eukaryotes, a gene overlap is when at least one nucleotide on either the same or opposite strand is shared between the primary transcripts of two or more genes. In prokaryotes and viruses, it is when at least two different coding sequences share a nucleotide either on the same or opposite strands.
- Open reading frames
(ORFs). A continuous stretch of nucleotides, on genome or transcript, that are bounded by a start and stop codon.
- Small ORFs
(sORFs). ORFs that are equal to or less than 300 nt in length.
- Primary transcripts
A transcribed RNA molecule, containing both exons and introns, prior to undergoing post-transcriptional processing to yield a final, mature transcript.
- Overlapping ORFs
When at least one nucleotide on either the same or opposite strand is shared between two sequences that consist of a length divisible by three and begin with a translation start codon and end at a stop codon.
- Operons
Regions in the genome of prokaryotes that encode multiple adjacent CDSs under the control of a single promoter.
- Translational coupling
The interdependence of translation efficiency of overlapping CDSs, in particular for those with overlapping start–stop codons.
- Positive selection
The process whereby the frequency of an allele in a population increases as a result of an increase of fitness of their carrier.
- Purifying selection
The process whereby harmful alleles are eliminated from a population. Also known as negative selection.
- Retrotransposition
The movement of genetic information from one genomic location to another through an RNA intermediary.
- Alternative ORFs
Also called non-canonical ORFs, are ORFs that occupy a shared sequence region with a canonical CDS, often in a different reading frame.
- Non-coding RNA
(ncRNA). A strand of RNA that has been transcribed from DNA but does not undergo translation. This RNA will typically have a regulatory function.
- Cis-natural antisense transcripts
(cis-NATs). Transcribed products from the DNA strand complementary to a region harbouring a sense transcript of either protein-coding or non-coding genes.
- Refactoring
The reorganization of biological systems or pathways with the goal of improving the ease and predictability of future engineering efforts. The process often involves removing CDS overlaps and changing regulatory sequences.
- Biosynthetic gene clusters
A physically clustered group of two or more genes that encode a biosynthetic pathway for the production of a specialized metabolite.
- Bicistronic
A transcript (mRNA) that encodes two CDSs.
- Monocistronic
An mRNA transcript that contains a single CDS.
- Polycistronic
An mRNA transcript that contains two or more CDSs.
Author contributions
B. W. W. and P. R. J. researched the literature and wrote the article. All authors provided substantial contributions to discussions of the content, and reviewed and/or edited the manuscript before submission.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Genetics thanks K. Neuhaus and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barrell BG, Air GM, Hutchison CA., 3rd Overlapping genes in bacteriophage phiX174. Nature. 1976;264:34–41. doi: 10.1038/264034a0. [DOI] [PubMed] [Google Scholar]
- 2.Sanger F, et al. Nucleotide sequence of bacteriophage φX174 DNA. Nature. 1977;265:687. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 3.Linney E, Hayashi M. Intragenic regulation of the synthesis of ΦX174 gene A proteins. Nature. 1974;249:345. doi: 10.1038/249345a0. [DOI] [PubMed] [Google Scholar]
- 4.Roznowski AP, Doore SM, Kemp SZ, Fane BA. Finally, a role befitting Astar: the strongly conserved, unessential microvirus A* proteins ensure the product fidelity of packaging reactions. J. Virol. 2020;94:e01593-19. doi: 10.1128/JVI.01593-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlub TE, Holmes EC. Properties and abundance of overlapping genes in viruses. Virus Evol. 2020;6:veaa009. doi: 10.1093/ve/veaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson CW, et al. Dynamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic. eLife. 2020;9:e59633. doi: 10.7554/eLife.59633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Normark S, et al. Overlapping genes. Annu. Rev. Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- 8.Sanna CR, Li W-H, Zhang L. Overlapping genes in the human and mouse genomes. BMC Genomics. 2008;9:169. doi: 10.1186/1471-2164-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veeramachaneni V, Makalowski W, Galdzicki M, Sood R, Makalowska I. Mammalian overlapping genes: the comparative perspective. Genome Res. 2004;14:280–286. doi: 10.1101/gr.1590904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C-H, Pan C-Y, Lin W-C. Overlapping protein-coding genes in human genome and their coincidental expression in tissues. Sci. Rep. 2019;9:13377. doi: 10.1038/s41598-019-49802-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, et al. Pervasive functional translation of noncanonical human open reading frames. Science. 2020;367:1140–1146. doi: 10.1126/science.aay0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prensner JR, et al. Noncanonical open reading frames encode functional proteins essential for cancer cell survival. Nat. Biotechnol. 2021;39:697–704. doi: 10.1038/s41587-020-00806-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B, et al. Identification and analysis of small proteins and short open reading frame encoded peptides in Hep3B cell. J. Proteom. 2021;230:103965. doi: 10.1016/j.jprot.2020.103965. [DOI] [PubMed] [Google Scholar]
- 14.Mudge JM, et al. A community-driven roadmap to advance research on translated open reading frames detected by Ribo-seq. bioRxiv. 2021 doi: 10.1101/2021.06.10.447896. [DOI] [Google Scholar]
- 15.Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Q, et al. Translation of small downstream ORFs enhances translation of canonical main open reading frames. EMBO J. 2020;39:e104763. doi: 10.15252/embj.2020104763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson ZI, Chisholm SW. Properties of overlapping genes are conserved across microbial genomes. Genome Res. 2004;14:2268–2272. doi: 10.1101/gr.2433104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cock PJA, Whitworth DE. Evolution of relative reading frame bias in unidirectional prokaryotic gene overlaps. Mol. Biol. Evol. 2009;27:753–756. doi: 10.1093/molbev/msp302. [DOI] [PubMed] [Google Scholar]
- 19.Lillo F, Krakauer DC. A statistical analysis of the three-fold evolution of genomic compression through frame overlaps in prokaryotes. Biol. Direct. 2007;2:22–22. doi: 10.1186/1745-6150-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Assis R, Kondrashov AS, Koonin EV, Kondrashov FA. Nested genes and increasing organizational complexity of metazoan genomes. Trends Genet. 2008;24:475–478. doi: 10.1016/j.tig.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu P, Ma D, Xu M. Nested genes in the human genome. Genomics. 2005;86:414–422. doi: 10.1016/j.ygeno.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Van Oss SB, Carvunis AR. De novo gene birth. PLoS Genet. 2019;15:e1008160. doi: 10.1371/journal.pgen.1008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makalowska I, Lin C-F, Makalowski W. Overlapping genes in vertebrate genomes. Comput. Biol. Chem. 2005;29:1–12. doi: 10.1016/j.compbiolchem.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Wichmann S, Ardern Z. Optimality in the standard genetic code is robust with respect to comparison code sets. Biosystems. 2019;185:104023. doi: 10.1016/j.biosystems.2019.104023. [DOI] [PubMed] [Google Scholar]
- 25.Soldà G, et al. Non-random retention of protein-coding overlapping genes in Metazoa. BMC Genomics. 2008;9:174. doi: 10.1186/1471-2164-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krakauer DC. Stability and evolution of overlapping genes. Evol. Int. J. Org. Evol. 2000;54:731–739. doi: 10.1111/j.0014-3820.2000.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 27.Rogozin IB, et al. Purifying and directional selection in overlapping prokaryotic genes. Trends Genet. 2002;18:228–232. doi: 10.1016/s0168-9525(02)02649-5. [DOI] [PubMed] [Google Scholar]
- 28.Hamoen LW, Eshuis H, Jongbloed J, Venema G, van Sinderen D. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol. Microbiol. 1995;15:55–63. doi: 10.1111/j.1365-2958.1995.tb02220.x. [DOI] [PubMed] [Google Scholar]
- 29.Meydan S, et al. Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Mol. Cell. 2019;74:481–493.e6. doi: 10.1016/j.molcel.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feltens R, Gossringer M, Willkomm DK, Urlaub H, Hartmann RK. An unusual mechanism of bacterial gene expression revealed for the RNase P protein of Thermus strains. Proc. Natl Acad. Sci. USA. 2003;100:5724–5729. doi: 10.1073/pnas.0931462100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones CE, Fleming TM, Cowan DA, Littlechild JA, Piper PW. The phosphoglycerate kinase and glyceraldehyde-3-phosphate dehydrogenase genes from the thermophilic archaeon Sulfolobus solfataricus overlap by 8-bp. Isolation, sequencing of the genes and expression in Escherichia coli. Eur. J. Biochem. 1995;233:800–808. doi: 10.1111/j.1432-1033.1995.800_3.x. [DOI] [PubMed] [Google Scholar]
- 32.Fukuda Y, Nakayama Y, Tomita M. On dynamics of overlapping genes in bacterial genomes. Gene. 2003;323:181–187. doi: 10.1016/j.gene.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 33.Sakharkar KR, Sakharkar MK, Verma C, Chow VTK. Comparative study of overlapping genes in bacteria, with special reference to Rickettsia prowazekii and Rickettsia conorii. Int. J. Syst. Evol. Microbiol. 2005;55:1205–1209. doi: 10.1099/ijs.0.63446-0. [DOI] [PubMed] [Google Scholar]
- 34.Fonseca MM, Harris DJ, Posada D. Origin and length distribution of unidirectional prokaryotic overlapping genes. G3. 2013;4:19–27. doi: 10.1534/g3.113.005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palleja A, Harrington ED, Bork P. Large gene overlaps in prokaryotic genomes: result of functional constraints or mispredictions? BMC Genomics. 2008;9:335. doi: 10.1186/1471-2164-9-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price MN, Arkin AP, Alm EJ. The life-cycle of operons. PLoS Genet. 2006;2:e96. doi: 10.1371/journal.pgen.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delcher AL, Kingsford C, Salzberg SL. A unified model explaining the offsets of overlapping and near-overlapping prokaryotic genes. Mol. Biol. Evol. 2007;24:2091–2098. doi: 10.1093/molbev/msm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huvet M, Stumpf MPH. Overlapping genes: a window on gene evolvability. BMC Genomics. 2014;15:721. doi: 10.1186/1471-2164-15-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatusova T, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willis S, Masel J. Gene birth contributes to structural disorder encoded by overlapping genes. Genetics. 2018;210:303–313. doi: 10.1534/genetics.118.301249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pavesi A, et al. Overlapping genes and the proteins they encode differ significantly in their sequence composition from non-overlapping genes. PLoS One. 2018;13:e0202513. doi: 10.1371/journal.pone.0202513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreitmeier M, et al. Shadow ORFs illuminated: long overlapping genes in Pseudomonas aeruginosa are translated and under purifying selection. bioRxiv. 2021 doi: 10.1101/2021.02.09.430400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zehentner B, Ardern Z, Kreitmeier M, Scherer S, Neuhaus K. Evidence for numerous embedded antisense overlapping genes in diverse E. coli strains. bioRxiv. 2020 doi: 10.1101/2020.11.18.388249. [DOI] [Google Scholar]
- 44.Zehentner B, Ardern Z, Kreitmeier M, Scherer S, Neuhaus K. A novel pH-regulated, unusual 603 bp overlapping protein coding gene pop is encoded antisense to ompA in Escherichia coli O157:H7 (EHEC) Front. Microbiol. 2020;11:377. doi: 10.3389/fmicb.2020.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabath N, Ferrada E, Barve A, Wagner A. Growth temperature and genome size in bacteria are negatively correlated, suggesting genomic streamlining during thermal adaptation. Genome Biol. Evol. 2013;5:966–977. doi: 10.1093/gbe/evt050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saha D, Panda A, Podder S, Ghosh TC. Overlapping genes: a new strategy of thermophilic stress tolerance in prokaryotes. Extremophiles. 2015;19:345–353. doi: 10.1007/s00792-014-0720-3. [DOI] [PubMed] [Google Scholar]
- 47.Pradhan P, Li W, Kaur P. Translational coupling controls expression and function of the DrrAB drug efflux pump. J. Mol. Biol. 2009;385:831–842. doi: 10.1016/j.jmb.2008.11.027. [DOI] [PubMed] [Google Scholar]
- 48.Huber M, et al. Translational coupling via termination-reinitiation in archaea and bacteria. Nat. Commun. 2019;10:4006. doi: 10.1038/s41467-019-11999-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das A, Yanofsky C. Restoration of a translational stop-start overlap reinstates translational coupling in a mutant trpB’-trpA gene pair of the Escherichia coli tryptophan operon. Nucleic acids Res. 1989;17:9333–9340. doi: 10.1093/nar/17.22.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das A, Yanofsky C. A ribosome binding site sequence is necessary for efficient expression of the distal gene of a translationally-coupled gene pair. Nucleic acids Res. 1984;12:4757–4768. doi: 10.1093/nar/12.11.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price MN, Huang KH, Arkin AP, Alm EJ. Operon formation is driven by co-regulation and not by horizontal gene transfer. Genome Res. 2005;15:809–819. doi: 10.1101/gr.3368805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osbourn AE, Field B. Operons. Cell. Mol. Life Sci. 2009;66:3755–3775. doi: 10.1007/s00018-009-0114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shieh YW, et al. Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science. 2015;350:678–680. doi: 10.1126/science.aac8171. [DOI] [PubMed] [Google Scholar]
- 54.Meydan S, Vázquez-Laslop N, Mankin AS. Genes within genes in bacterial genomes. Microbiol Spectr. 2018 doi: 10.1128/microbiolspec.RWR-0020-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeffares DC, Mourier T, Penny D. The biology of intron gain and loss. Trends Genet. 2006;22:16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 56.Williams BA, Slamovits CH, Patron NJ, Fast NM, Keeling PJ. A high frequency of overlapping gene expression in compacted eukaryotic genomes. Proc. Natl Acad. Sci. USA. 2005;102:10936–10941. doi: 10.1073/pnas.0501321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Makałowska I, Lin C-F, Hernandez K. Birth and death of gene overlaps in vertebrates. BMC Evolut. Biol. 2007;7:193. doi: 10.1186/1471-2148-7-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar A. An overview of nested genes in eukaryotic genomes. Eukaryot. Cell. 2009;8:1321. doi: 10.1128/EC.00143-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee YCG, Chang H-H. The evolution and functional significance of nested gene structures in Drosophila melanogaster. Genome Biol. Evol. 2013;5:1978–1985. doi: 10.1093/gbe/evt149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heames B, Schmitz J, Bornberg-Bauer E. A continuum of evolving de novo genes drives protein-coding novelty in Drosophila. J. Mol. Evol. 2020;88:382–398. doi: 10.1007/s00239-020-09939-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rindfleisch BC, Brown MS, VandeBerg JL, Munroe SH. Structure and expression of two nuclear receptor genes in marsupials: insights into the evolution of the antisense overlap between the α-thyroid hormone receptor and Rev-erbα. BMC Mol. Biol. 2010;11:97. doi: 10.1186/1471-2199-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loughran G, et al. Unusually efficient CUG initiation of an overlapping reading frame in POLG mRNA yields novel protein POLGARF. Proc. Natl Acad. Sci. USA. 2020;117:24936–24946. doi: 10.1073/pnas.2001433117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khan YA, et al. Evidence for a novel overlapping coding sequence in POLG initiated at a CUG start codon. BMC Genet. 2020;21:25. doi: 10.1186/s12863-020-0828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brunet MA, Levesque SA, Hunting DJ, Cohen AA, Roucou X. Recognition of the polycistronic nature of human genes is critical to understanding the genotype-phenotype relationship. Genome Res. 2018;28:609–624. doi: 10.1101/gr.230938.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao X, et al. Alt-RPL36 downregulates the PI3K-AKT-mTOR signaling pathway by interacting with TMEM24. Nat. Commun. 2021;12:508. doi: 10.1038/s41467-020-20841-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Samandi S, et al. Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins. eLife. 2017;6:e27860. doi: 10.7554/eLife.27860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karginov TA, Pastor DPH, Semler BL, Gomez CM. Mammalian polycistronic mRNAs and disease. Trends Genet. 2017;33:129–142. doi: 10.1016/j.tig.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 69.Brunet MA, et al. The FUS gene is dual-coding with both proteins contributing to FUS-mediated toxicity. EMBO Rep. 2021;22:e50640. doi: 10.15252/embr.202050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muñoz-Baena L, Poon AFY. Using networks to analyze and visualize the distribution of overlapping reading frames in virus genomes. bioRxiv. 2021 doi: 10.1101/2021.06.10.447953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simon-Loriere E, Holmes EC, Pagán I. The effect of gene overlapping on the rate of RNA virus evolution. Mol. Biol. Evol. 2013;30:1916–1928. doi: 10.1093/molbev/mst094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krakauer DC, Plotkin JB. Redundancy, antiredundancy, and the robustness of genomes. Proc. Natl Acad. Sci. USA. 2002;99:1405–1409. doi: 10.1073/pnas.032668599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chirico N, Vianelli A, Belshaw R. Why genes overlap in viruses. Proc. Biol. Sci. 2010;277:3809–3817. doi: 10.1098/rspb.2010.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Belshaw R, Pybus OG, Rambaut A. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 2007;17:1496–1504. doi: 10.1101/gr.6305707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fernandes JD, et al. Functional segregation of overlapping genes in HIV. Cell. 2016;167:1762–1773.e12. doi: 10.1016/j.cell.2016.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brandes N, Linial M. Gene overlapping and size constraints in the viral world. Biol. Direct. 2016;11:26. doi: 10.1186/s13062-016-0128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pavesi A. New insights into the evolutionary features of viral overlapping genes by discriminant analysis. Virology. 2020;546:51–66. doi: 10.1016/j.virol.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feiss M, Fisher RA, Crayton MA, Egner C. Packaging of the bacteriophage λ chromosome: Effect of chromosome length. Virology. 1977;77:281–293. doi: 10.1016/0042-6822(77)90425-1. [DOI] [PubMed] [Google Scholar]
- 79.Aoyama A, Hayashi M. Effects of genome size on bacteriophage phi X174 DNA packaging in vitro. J. Biol. Chem. 1985;260:11033–11038. [PubMed] [Google Scholar]
- 80.Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vaidyanathan S, et al. Targeted replacement of full-length CFTR in human airway stem cells by CRISPR-Cas9 for pan-mutation correction in the endogenous locus. Mol. Ther. 2021 doi: 10.1016/j.ymthe.2021.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bartonek L, Braun D, Zagrovic B. Frameshifting preserves key physicochemical properties of proteins. Proc. Natl Acad. Sci. USA. 2020;117:5907. doi: 10.1073/pnas.1911203117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li H, Havens WM, Nibert ML, Ghabrial SA. RNA sequence determinants of a coupled termination-reinitiation strategy for downstream open reading frame translation in Helminthosporium victoriae virus 190S and other victoriviruses (family Totiviridae) J. Virol. 2011;85:7343–7352. doi: 10.1128/JVI.00364-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toledo-Arana A, Lasa I. Advances in bacterial transcriptome understanding: From overlapping transcription to the excludon concept. Mol. Microbiol. 2020;113:593–602. doi: 10.1111/mmi.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gelsinger DR, DiRuggiero J. Transcriptional landscape and regulatory roles of small noncoding RNAs in the oxidative stress response of the Haloarchaeon haloferax volcanii. J. Bacteriol. 2018;200:e00779-17. doi: 10.1128/JB.00779-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Choi JS, Park H, Kim W, Lee Y. Coordinate regulation of the expression of SdsR toxin and its downstream pphA gene by RyeA antitoxin in Escherichia coli. Sci. Rep. 2019;9:9627. doi: 10.1038/s41598-019-45998-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee E-J, Groisman EA. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol. Microbiol. 2010;76:1020–1033. doi: 10.1111/j.1365-2958.2010.07161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sesto N, Wurtzel O, Archambaud C, Sorek R, Cossart P. The excludon: a new concept in bacterial antisense RNA-mediated gene regulation. Nat. Rev. Microbiol. 2013;11:75–82. doi: 10.1038/nrmicro2934. [DOI] [PubMed] [Google Scholar]
- 89.Dar D, Sorek R. Bacterial noncoding RNAs excised from within protein-coding transcripts. mBio. 2018;9:e01730-18. doi: 10.1128/mBio.01730-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Adams PP, Storz G. Prevalence of small base-pairing RNAs derived from diverse genomic loci. Biochim. Biophys. Acta Gene Regul. Mech. 2020;1863:194524. doi: 10.1016/j.bbagrm.2020.194524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Adams PP, et al. Regulatory roles of Escherichia coli 5′ UTR and ORF-internal RNAs detected by 3′ end mapping. eLife. 2021;10:e62438. doi: 10.7554/eLife.62438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Neuhaus K, et al. Differentiation of ncRNAs from small mRNAs in Escherichia coli O157:H7 EDL933 (EHEC) by combined RNAseq and RIBOseq – ryhB encodes the regulatory RNA RyhB and a peptide, RyhP. BMC Genomics. 2017;18:216. doi: 10.1186/s12864-017-3586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vanderpool CK, Balasubramanian D, Lloyd CR. Dual-function RNA regulators in bacteria. Biochimie. 2011;93:1943–1949. doi: 10.1016/j.biochi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jin H, Vacic V, Girke T, Lonardi S, Zhu J-K. Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol. Biol. 2008;9:6. doi: 10.1186/1471-2199-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 97.Werner A. Biological functions of natural antisense transcripts. BMC Biol. 2013;11:31. doi: 10.1186/1741-7007-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Matsuda E, Garfinkel DJ. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc. Natl Acad. Sci. USA. 2009;106:15657–15662. doi: 10.1073/pnas.0908305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chu J, Dolnick BJ. Natural antisense (rTSα) RNA induces site-specific cleavage of thymidylate synthase mRNA. Biochim. Biophys. Acta. 2002;1587:183–193. doi: 10.1016/s0925-4439(02)00081-9. [DOI] [PubMed] [Google Scholar]
- 100.Morrissy AS, Griffith M, Marra MA. Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res. 2011;21:1203–1212. doi: 10.1101/gr.113431.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Su W-Y, et al. Bidirectional regulation between WDR83 and its natural antisense transcript DHPS in gastric cancer. Cell Res. 2012;22:1374–1389. doi: 10.1038/cr.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jeon Y, Sarma K, Lee JT. New and Xisting regulatory mechanisms of X chromosome inactivation. Curr. Opin. Genet. Dev. 2012;22:62–71. doi: 10.1016/j.gde.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 2005;21:326–329. doi: 10.1016/j.tig.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 106.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 107.Wu P, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer. 2020;19:22. doi: 10.1186/s12943-020-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reis EM, et al. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004;23:6684–6692. doi: 10.1038/sj.onc.1207880. [DOI] [PubMed] [Google Scholar]
- 109.Yin J, et al. UXT-AS1-induced alternative splicing of UXT is associated with tumor progression in colorectal cancer. Am. J. Cancer Res. 2017;7:462–472. [PMC free article] [PubMed] [Google Scholar]
- 110.Tu Q, et al. CDKN2B deletion is essential for pancreatic cancer development instead of unmeaningful co-deletion due to juxtaposition to CDKN2A. Oncogene. 2018;37:128–138. doi: 10.1038/onc.2017.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tufarelli C, et al. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 113.Jackson R, et al. The translation of non-canonical open reading frames controls mucosal immunity. Nature. 2018;564:434–438. doi: 10.1038/s41586-018-0794-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Slavoff SA, et al. Peptidomic discovery of short open reading frame–encoded peptides in human cells. Nat. Chem. Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ma J, et al. Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue. J. Proteome Res. 2014;13:1757–1765. doi: 10.1021/pr401280w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bazzini AA, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guo B, et al. Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO J. 2020;39:e102190. doi: 10.15252/embj.2019102190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee CQE, et al. Coding and non-coding roles of MOCCI (C15ORF48) coordinate to regulate host inflammation and immunity. Nat. Commun. 2021;12:2130. doi: 10.1038/s41467-021-22397-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nam J-W, Choi S-W, You B-H. Incredible RNA: dual functions of coding and noncoding. Mol. Cell. 2016;39:367–374. doi: 10.14348/molcells.2016.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schlesinger D, Elsässer SJ. Revisiting sORFs: overcoming challenges to identify and characterize functional microproteins. FEBS J. 2021 doi: 10.1111/febs.15769. [DOI] [PubMed] [Google Scholar]
- 121.Ruiz-Orera J, Villanueva-Cañas JL, Albà MM. Evolution of new proteins from translated sORFs in long non-coding RNAs. Exp. Cell Res. 2020;391:111940. doi: 10.1016/j.yexcr.2020.111940. [DOI] [PubMed] [Google Scholar]
- 122.Smanski MJ, et al. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 2016;14:135–149. doi: 10.1038/nrmicro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bayer TS, et al. Synthesis of methyl halides from biomass using engineered microbes. J. Am. Chem. Soc. 2009;131:6508–6515. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- 124.Segall-Shapiro TH, Meyer AJ, Ellington AD, Sontag ED, Voigt CA. A ‘resource allocator’ for transcription based on a highly fragmented T7 RNA polymerase. Mol. Syst. Biol. 2014;10:742. doi: 10.15252/msb.20145299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rhodius VA, et al. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol. Syst. Biol. 2013;9:702. doi: 10.1038/msb.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cervettini D, et al. Rapid discovery and evolution of orthogonal aminoacyl-tRNA synthetase–tRNA pairs. Nat. Biotechnol. 2020;38:989–999. doi: 10.1038/s41587-020-0479-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aleksashin NA, et al. A fully orthogonal system for protein synthesis in bacterial cells. Nat. Commun. 2020;11:1858. doi: 10.1038/s41467-020-15756-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tang T-C, et al. Materials design by synthetic biology. Nat. Rev. Mater. 2021;6:332–350. [Google Scholar]
- 129.Chen Y, et al. Genetic circuit design automation for yeast. Nat. Microbiol. 2020;5:1349–1360. doi: 10.1038/s41564-020-0757-2. [DOI] [PubMed] [Google Scholar]
- 130.Robertson WE, et al. Creating custom synthetic genomes in Escherichia coli with REXER and GENESIS. Nat. Protoc. 2021;16:2345–2380. doi: 10.1038/s41596-020-00464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pretorius I, Boeke J. Yeast 2.0 — connecting the dots in the construction of the world’s first functional synthetic eukaryotic genome. FEMS Yeast Res. 2018;18:foy032. doi: 10.1093/femsyr/foy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fredens J, et al. Total synthesis of Escherichia coli with a recoded genome. Nature. 2019;569:514–518. doi: 10.1038/s41586-019-1192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Mol. Syst. Biol. 2005;1:2005.0018. doi: 10.1038/msb4100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jaschke PR, Lieberman EK, Rodriguez J, Sierra A, Endy D. A fully decompressed synthetic bacteriophage ΦX174 genome assembled and archived in yeast. Virology. 2012;434:278–284. doi: 10.1016/j.virol.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 135.Gimpel JA, Nour-Eldin HH, Scranton MA, Li D, Mayfield SP. Refactoring the six-gene photosystem II core in the chloroplast of the green algae Chlamydomonas reinhardtii. ACS Synth. Biol. 2016;5:589–596. doi: 10.1021/acssynbio.5b00076. [DOI] [PubMed] [Google Scholar]
- 136.Hutchison CA, III, et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 137.Venetz JE, et al. Chemical synthesis rewriting of a bacterial genome to achieve design flexibility and biological functionality. Proc. Natl Acad. Sci. USA. 2019;116:8070. doi: 10.1073/pnas.1818259116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Springman R, Molineux IJ, Duong C, Bull RJ, Bull JJ. Evolutionary stability of a refactored phage genome. ACS Synth. Biol. 2012;1:425–430. doi: 10.1021/sb300040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wright BW, Ruan J, Molloy MP, Jaschke PR. Genome modularization reveals overlapped gene topology is necessary for efficient viral reproduction. ACS Synth. Biol. 2020;9:3079–3090. doi: 10.1021/acssynbio.0c00323. [DOI] [PubMed] [Google Scholar]
- 140.Temme K, Zhao D, Voigt CA. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc. Natl Acad. Sci. USA. 2012;109:7085–7090. doi: 10.1073/pnas.1120788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Song M, et al. Control of type III protein secretion using a minimal genetic system. Nat. Commun. 2017;8:14737. doi: 10.1038/ncomms14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li G-Q, et al. Improvement of dibenzothiophene desulfurization activity by removing the gene overlap in the dsz operon. Biosci. Biotechnol. Biochem. 2007;71:849–854. doi: 10.1271/bbb.60189. [DOI] [PubMed] [Google Scholar]
- 143.Ghosh D, Kohli AG, Moser F, Endy D, Belcher AM. Refactored M13 bacteriophage as a platform for tumor cell imaging and drug delivery. ACS Synth. Biol. 2012;1:576–582. doi: 10.1021/sb300052u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Richardson SM, et al. Design of a synthetic yeast genome. Science. 2017;355:1040. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- 145.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wichmann S, Scherer S, Ardern Z. Computational design of genes encoding completely overlapping protein domains: Influence of genetic code and taxonomic rank. bioRxiv. 2020 doi: 10.1101/2020.09.25.312959. [DOI] [Google Scholar]
- 148.Opuu V, Silvert M, Simonson T. Computational design of fully overlapping coding schemes for protein pairs and triplets. Sci. Rep. 2017;7:15873. doi: 10.1038/s41598-017-16221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Inouye M, Ishida Y, Inouye K. Designing of a single gene encoding four functional proteins. J. Theor. Biol. 2017;419:266–268. doi: 10.1016/j.jtbi.2017.01.042. [DOI] [PubMed] [Google Scholar]
- 150.Frénoy A, Taddei F, Misevic D. Genetic architecture promotes the evolution and maintenance of cooperation. PLoS Comput. Biol. 2013;9:e1003339. doi: 10.1371/journal.pcbi.1003339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bull JJ, Barrick JE. Arresting evolution. Trends Genet. 2017;33:910–920. doi: 10.1016/j.tig.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Blazejewski T, Ho H-I, Wang HH. Synthetic sequence entanglement augments stability and containment of genetic information in cells. Science. 2019;365:595. doi: 10.1126/science.aav5477. [DOI] [PubMed] [Google Scholar]
- 153.Decrulle AL, et al. Engineering gene overlaps to sustain genetic constructs in vivo. bioRxiv. 2019 doi: 10.1101/659243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Makoff AJ, Smallwood AE. The use of two-cistron constructions in improving the expression of a heterologous gene in E.coli. Nucleic Acids Res. 1990;18:1711–1718. doi: 10.1093/nar/18.7.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mutalik VK, et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013;10:354–360. doi: 10.1038/nmeth.2404. [DOI] [PubMed] [Google Scholar]
- 156.Roy V, et al. A bicistronic vector with destabilized mRNA secondary structure yields scalable higher titer expression of human neurturin in E. coli. Biotechnol. Bioeng. 2017;114:1753–1761. doi: 10.1002/bit.26299. [DOI] [PubMed] [Google Scholar]
- 157.Claassens NJ, et al. Bicistronic design-based continuous and high-level membrane protein production in Escherichia coli. ACS Synth. Biol. 2019;8:1685–1690. doi: 10.1021/acssynbio.9b00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stallmeyer B, Drugeon G, Reiss J, Haenni AL, Mendel RR. Human molybdopterin synthase gene: identification of a bicistronic transcript with overlapping reading frames. Am. J. Hum. Genet. 1999;64:698–705. doi: 10.1086/302295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ostrov N, et al. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353:819. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- 160.Calles J, Justice I, Brinkley D, Garcia A, Endy D. Fail-safe genetic codes designed to intrinsically contain engineered organisms. Nucleic Acids Res. 2019;47:10439–10451. doi: 10.1093/nar/gkz745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Anderson JC, et al. An expanded genetic code with a functional quadruplet codon. Proc. Natl Acad. Sci. USA. 2004;101:7566. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang K, Schmied WH, Chin JW. Reprogramming the genetic code: from triplet to quadruplet codes. Angew. Chem. Int. Ed. 2012;51:2288–2297. doi: 10.1002/anie.201105016. [DOI] [PubMed] [Google Scholar]
- 163.Malyshev DA, et al. Efficient and sequence-independent replication of DNA containing a third base pair establishes a functional six-letter genetic alphabet. Proc. Natl Acad. Sci. USA. 2012;109:12005–12010. doi: 10.1073/pnas.1205176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hoshika S, et al. Hachimoji DNA and RNA: a genetic system with eight building blocks. Science. 2019;363:884–887. doi: 10.1126/science.aat0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fiers W, et al. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976;260:500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- 166.van Heesch S, et al. The translational landscape of the human heart. Cell. 2019;178:242–260.e29. doi: 10.1016/j.cell.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 167.James K, Cockell SJ, Zenkin N. Deep sequencing approaches for the analysis of prokaryotic transcriptional boundaries and dynamics. Methods. 2017;120:76–84. doi: 10.1016/j.ymeth.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 168.Güell M, Yus E, Lluch-Senar M, Serrano L. Bacterial transcriptomics: what is beyond the RNA horiz-ome? Nat. Rev. Microbiol. 2011;9:658–669. doi: 10.1038/nrmicro2620. [DOI] [PubMed] [Google Scholar]
- 169.Mouilleron H, Delcourt V, Roucou X. Death of a dogma: eukaryotic mRNAs can code for more than one protein. Nucleic Acids Res. 2016;44:14–23. doi: 10.1093/nar/gkv1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ho M-R, Tsai K-W, Lin W-C. A unified framework of overlapping genes: towards the origination and endogenic regulation. Genomics. 2012;100:231–239. doi: 10.1016/j.ygeno.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 171.Majic P, Payne JL. Enhancers facilitate the birth of de novo genes and gene integration into regulatory networks. Mol. Biol. Evol. 2020;37:1165–1178. doi: 10.1093/molbev/msz300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat. Rev. Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Brophy JAN, Voigt CA. Antisense transcription as a tool to tune gene expression. Mol. Syst. Biol. 2016;12:854. doi: 10.15252/msb.20156540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shearwin KE, Callen BP, Egan JB. Transcriptional interference–a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pavesi A, Magiorkinis G, Karlin DG. Viral proteins originated de novo by overprinting can be identified by codon usage: application to the “gene nursery” of Deltaretroviruses. PLoS Comput. Biol. 2013;9:e1003162. doi: 10.1371/journal.pcbi.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.National Center for Biotechnology Information. NCBI Prokaryotic Genome Annotation Pipelinehttps://www.ncbi.nlm.nih.gov/genome/annotation_prok/standards/ (2021).
- 180.Nelson CW, Ardern Z, Wei X. OLGenie: estimating natural selection to predict functional overlapping genes. Mol. Biol. Evol. 2020;37:2440–2449. doi: 10.1093/molbev/msaa087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.McCauley S, Hein J. Using hidden Markov models and observed evolution to annotate viral genomes. Bioinformatics. 2006;22:1308–1316. doi: 10.1093/bioinformatics/btl092. [DOI] [PubMed] [Google Scholar]
- 182.Pareja-Tobes P, Manrique M, Pareja-Tobes E, Pareja E, Tobes R. BG7: a new approach for bacterial genome annotation designed for next generation sequencing data. PLoS One. 2012;7:e49239. doi: 10.1371/journal.pone.0049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Brunet MA, et al. OpenProt: a more comprehensive guide to explore eukaryotic coding potential and proteomes. Nucleic Acids Res. 2019;47:D403–D410. doi: 10.1093/nar/gky936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Brunet MA, et al. OpenProt 2021: deeper functional annotation of the coding potential of eukaryotic genomes. Nucleic Acids Res. 2020;49:D380–D388. doi: 10.1093/nar/gkaa1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteom. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Firnberg E, Labonte JW, Gray JJ, Ostermeier M. A comprehensive, high-resolution map of a gene’s fitness landscape. Mol. Biol. Evol. 2014;31:1581–1592. doi: 10.1093/molbev/msu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hecht A, et al. Measurements of translation initiation from all 64 codons in E. coli. Nucleic Acids Res. 2017;45:3615–3626. doi: 10.1093/nar/gkx070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Berry IJ, Steele JR, Padula MP, Djordjevic SP. The application of terminomics for the identification of protein start sites and proteoforms in bacteria. Proteomics. 2016;16:257–272. doi: 10.1002/pmic.201500319. [DOI] [PubMed] [Google Scholar]
- 190.Willems P, et al. N-terminal proteomics assisted profiling of the unexplored translation initiation landscape in Arabidopsis thaliana. MCP. 2017;16:1064–1080. doi: 10.1074/mcp.M116.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Vanderperre B, et al. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS One. 2013;8:e70698. doi: 10.1371/journal.pone.0070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Finkel Y, et al. Comprehensive annotations of human herpesvirus 6A and 6B genomes reveal novel and conserved genomic features. eLife. 2020;9:e50960. doi: 10.7554/eLife.50960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Machkovech HM, Bloom JD, Subramaniam AR. Comprehensive profiling of translation initiation in influenza virus infected cells. PLoS Pathog. 2019;15:e1007518. doi: 10.1371/journal.ppat.1007518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Liu X, Jiang H, Gu Z, Roberts JW. High-resolution view of bacteriophage lambda gene expression by ribosome profiling. Proc. Natl Acad. Sci. USA. 2013;110:11928–11933. doi: 10.1073/pnas.1309739110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 197.Jaschke PR, Dotson GA, Hung KS, Liu D, Endy D. Definitive demonstration by synthesis of genome annotation completeness. Proc. Natl Acad. Sci. USA. 2019;116:24206–24213. doi: 10.1073/pnas.1905990116. [DOI] [PMC free article] [PubMed] [Google Scholar]