Abstract
Zika virus is a positive‐sense single‐stranded RNA virus, which can be transmitted across the placenta and has adverse effects on fetal development during pregnancy. The severity of these complications highlights the importance of prevention and treatment. However, no vaccines or drugs are currently available. In this study, we characterize the IFNβ‐mediated anti‐viral response in trophoblast cells in order to identify critical components that are necessary for the successful control of viral replication and determine whether components of the IFN‐induced response can be used as a replacement therapy for ZIKA virus infection during pregnancy. We identify and characterize interferon‐stimulated gene 20 (ISG20) as playing a central role in controlling Zika virus infection in trophoblast cells and successfully establish a recombinant ISG20‐Fc protein that effectively decreases viral titers in vitro and in vivo by maintaining its exonuclease activity and displaying potential immune modulatory functions. Recombinant ISG20‐Fc has thus the potential to be further developed as an anti‐viral treatment against ZIKA viral infection in high‐risk populations, particularly in pregnant women.
Keywords: interferon‐beta, interferon‐stimulated gene, ISG20, placenta trophoblast, ZIKA virus
Subject Categories: Immunology; Microbiology, Virology & Host Pathogen Interaction; Signal Transduction
ISG20 is the key exonuclease to inhibit ZIKA viral replication in trophoblast cells via the degradation of viral RNA. A recombinant ISG20‐Fc protein reduces ZIKA infection in vitro and in vivo.

Introduction
During normal pregnancy, the placenta is capable of mounting a robust immune response and acting as a barrier to control different types of infections. Therefore, the placenta is the most important immune organ that protects the fetus from infectious pathogens, which pose major threats for fetal development (Mor et al, 2017). This protection is mediated by pattern recognition receptors (PRRs) that are expressed at the maternal–fetal interface, such as Toll‐like receptors (TLRs) and the expression of anti‐viral factors, such as type I interferon attributes a central role in viral infection clearance (Koga & Mor, 2010; Cardenas et al, 2011). However, in some cases, like during ZIKA virus (ZIKV) infection, this protective barrier is breached and leads to serious pathologic consequences for the fetus development.
ZIKV is a positive‐sense single‐stranded RNA virus, which belongs to the Flavivirus genus in the Flaviviridae family (Dick et al, 1952). Infection with ZIKV during pregnancy leads to adverse pregnancy outcomes, including preterm birth, stillbirth, and congenital Zika syndrome (CZS) (Sadovsky et al, 2016; Shan et al, 2016; Schwartz, 2017b). Infants with CZS may show microcephaly, abnormal brain development, limb contractures, eye abnormalities, and other neurological manifestations (Johansson et al, 2016; Karimi et al, 2016; Rasmussen et al, 2016). Moreover, a recent study shows that infants exposed to ZIKV infection in utero showed neurodevelopmental delays as toddlers, despite having “normal” brain imaging and head circumference at birth (Honein et al, 2020). This highlights the long‐term effects of ZIKV infection on offspring development and further implicates the need to protect mother and fetus against ZIKV infection during the early stage of pregnancy.
As of July 2019, ZIKV has been documented in 87 countries and territories, (Franca et al, 2016; Organization, 2018, 2019; Brady et al, 2019). In the Americas, the ZIKV outbreak peaked in 2016, but the incidence subsequently declined during 2017 and 2018. However, in 2018, there were still a total of 31,587 suspected, probable, and confirmed cases of ZIKV disease reported (Organization, 2019; Honein et al, 2020; Pattnaik et al, 2020). All areas with prior ZIKV infection have the potential for re‐emergence and re‐introduction. From the facts above, it is clear that ZIKV infection poses a high risk for human reproduction health and results in a massive economic burden and workload on the healthcare systems, which calls for the rapid development of safe and efficacious vaccines and therapeutics.
Although major efforts have been invested on the development of vaccines and anti‐ZIKA therapies (Pattnaik et al, 2020), no one has been materialized and the number of ZIKV‐infected patients continues to grow; therefore, there is a need for a better understanding on the cellular mechanism that provide effective protection during infection that could lead to the development of more effective therapeutic approaches that can prevent ZIKV‐induced developmental abnormalities in fetus.
Type I interferon (IFN) signaling plays a crucial role in controlling ZIKV replication and pathogenesis (Aliota et al, 2016; Lazear et al, 2016; Yockey et al, 2016); particularly during the first trimester of pregnancy (Racicot et al, 2016). As demonstrated in mouse models lacking type I IFN signaling, these IFNAR1−/− mice were more susceptible to ZIKV infection and developed neurological disease (Lazear et al, 2016; Miner et al, 2016; Racicot et al, 2017). Furthermore, in our previous study, we demonstrated the specific fetal/placental‐derived type I IFN signaling plays an important role in the prevention of fetal viral infection and is the key for ensuring maternal survival (Aldo et al, 2016b; Racicot et al, 2017; Kwon et al, 2018). This protective effect depends on the integrity of the IFNβ/IFNAR1 pathway and their downstream interferon‐stimulated genes (ISGs).
Type I IFNs, including IFN‐α and IFN‐β, are the primary IFNs that are generated in most cell types during infections. The canonical IFN response will be initiated upon recognition of viral genomes/transcripts by RNA‐sensing helicases, such as the retinoic acid‐inducible gene I (RIG‐I) and melanoma‐associated differentiation antigen 5 (MDA5) (Kell & Gale, 2015; Said et al, 2018). After binding viral RNA, RIG‐1/MDA5 recruits the adaptor mitochondrial anti‐viral‐signaling protein (MAVS) to trigger the phosphorylation of the anti‐viral transcription factors IFN regulatory factor 3 (IRF3) and NF‐κB, which leads to the induction of type I IFN production (Pichlmair & Reis e Sousa, 2007; Zevini et al, 2017). Secreted type I IFN binds to the heterodimeric transmembrane receptor IFNAR, consisting of IFNAR1 and IFANR2 chains, and signals through the JAK/STAT pathway to induce the expression of hundreds of ISGs (Platanias, 2005; Levy et al, 2011). These ISGs exert a potent anti‐viral response on specific steps of the viral life cycle and inhibit virus replication and shedding (Schneider et al, 2014). An early step in the cellular anti‐viral response is to target the viral RNA before it is able to replicate and produce new viral particles to infect neighboring cells. This process is accomplished through the expression of specific exonucleases (Schneider et al, 2014).
Therefore, the objectives of this study were twofold: (i) to characterize the IFNβ‐mediated anti‐viral response in trophoblast cells in order to identify critical components that are necessary for the successful control of viral replication and (ii) to determine whether we could use the components of the IFN‐induced response as a replacement therapy for ZIKV infection during pregnancy in cases where its function has being affected. We found that ISG20, an interferon‐inducible 3′–5′ exonucleases with potent anti‐viral activity against different viruses (Espert et al, 2005; Zhang et al, 2007; Jiang et al, 2008; Qu et al, 2016; Liu et al, 2017), is one of the earlier ISGs induced by a protective IFNβ‐mediated response to ZIKA infection. Deletion of ISG20 in trophoblast cells decreases their capacity to control ZIKV replication and render these cells more susceptible to the infection.
In this study, we report the characterization of ISG20 expression and function during ZIKV infection in trophoblast cells and its central immune modulatory function. Use of a recombinant ISG20 protein, which conserves its RNase activity, restores protection against ZIKV by inhibiting ZIKV replication in vitro and in vivo. Our findings highlight the role of ISG20 as one of the key ISGs responsible for inhibiting ZIKV infection and demonstrate its potential application for the treatment/prevention of ZIKV infections during pregnancy.
Results
ZIKV infection induces IFNβ and ISG mRNA expression in first‐trimester trophoblast cells
Our first objective was to characterize the cellular components involved during an effective anti‐viral response elicited by first‐trimester trophoblast cells exposed to ZIKV. We used a human first‐trimester trophoblast cell line (Sw.71), which has been well characterized in previous studies (Aldo et al, 2007; Straszewski‐Chavez et al, 2009; Aldo et al, 2010), and primary cultures of human first‐trimester trophoblast cells.
Both cell types were infected with Cambodia ZIKV (MOI = 2) for 1 h, refreshed with regular media, and monitored to determine changes in cell growth and survival. During the infection, Sw.71 cells and primary cultures of first‐trimester trophoblast infected with ZIKV did not show any apoptosis‐related morphological changes or disturbance on cell growth when compared to the control (Appendix Fig S3). We then evaluated viral titers at different time points post‐infection by qRT–PCR and detected ZIKV titers in Sw.71 at 24 h post‐infection (h.p.i.), with higher levels at 48 h.p.i and decrease on viral titers at 72 h.p.i. (Fig 1A). In human primary trophoblast cell cultures (HPC), we observed ZIKV titers increased at 24 h.p.i and stayed at high level of ZIKV titer until 72 h.p.i. and decreased at 96 h.p.i. (Fig 1A). Due to the technical limitations of maintaining primary cultures for longer time, we were not able to further evaluate the change on viral titers at later times (more than 96 h.p.i.), but as indicated above we did not observe any sign of cell death in the primary trophoblast cell cultures.
Figure 1. ZIKV infection induces type I IFNβ expression in human first‐trimester trophoblast cell line Sw.71 and human primary cultures.

- ZIKV titers in Sw.71 and HPC trophoblasts. *P < 0.05 by one‐way ANOVA.
- IFNα is not highly induced in response to ZIKV infection in trophoblast cells. IFNα mRNA expression was inhibited in the first 24 h.p.i. and maintained at low level during ZIKV infection. Difference not significant (n.s.) by one‐way ANOVA.
- ZIKV infection induced a time‐dependent increase in IFNβ mRNA expression in Sw.71 and HPC. *P < 0.05 and **P < 0.01 by one‐way ANOVA.
- Expression levels of secreted IFNβ detected in trophoblast supernatants by ELLA. Supernatants were collected from ZIKV‐infected and control trophoblast cultures at different times, and IFNβ protein secretion was quantified by ELLA. Note the increase of secreted IFNβ in the ZIKV‐infected supernatant in a time‐dependent manner. ****P < 0.0001 by Student’s t‐test against individual time point NT control.
To understand the anti‐viral response generated by trophoblast cells to ZIKV infection, the type I interferon response was first characterized in Sw.71 and HPC infected with ZIKV (MOI = 2) over different time points post‐ZIKV infection. Interestingly, we observed no changes on IFNα mRNA expression by ZIKV infection on trophoblast cells (Fig 1B), but a time‐dependent increase of IFNβ mRNA (Fig 1C), which was consistent with the time‐dependent pattern of ZIKV titers. IFNβ mRNA expression in Sw.71 increased in the first 24 h.p.i., peaked at 48 h.p.i., and further decreased 72 h.p.i. (Fig 1C). In HPC, the IFNβ response followed similar pattern as the viral infection; IFNβ mRNA increased at 48 h.p.i. and remained high at 72 h.p.i. and 96 h.p.i. (Fig 1C). Although the response times were different between the cell line and the primary cultures, both cell types showed significant increase of IFNβ expression in response to ZIKV infection (Fig 1C).
To validate the observed increase of IFNβ mRNA expression and its potential function following ZIKV infection, we tested the presence of secreted IFNβ protein in the supernatant of Sw.71 and HPC trophoblast cells at different time points post‐ZIKV infection. We did not detect secreted IFNβ in the supernatant of control non‐infected Sw.71 cells; however, a substantial concentration of secreted IFNβ protein was detected in the supernatant of ZIKV‐infected Sw.71 cells at 48 and 72 h.p.i. (Fig 1D). While in HPC, IFNβ protein was undetectable in the supernatant from both non‐infected and ZIKV‐infected cells at 24 and 48 h.p.i., a significant increase of secreted IFNβ protein was observed at 72 and 96 h.p.i (Fig 1D). Therefore, these findings confirm that trophoblast cells can recognize ZIKV infection and initiate a type I IFNβ response to ZIKV infection.
IFNβ promotes ISG expression in trophoblast cells in response to ZIKV infection
Following its secretion from cells, IFNβ mediates its anti‐viral activity by inducing the expression of interferon‐stimulated genes (ISGs) (Schneider et al, 2014). Thus, we evaluated the mRNA expression of the anti‐viral ISGs representing an IFNβ‐related gene signature (ISG20, MX1, OAS1, ISG15, CH25H, TRIM22, Tetherin, and Viperin) (Kumaran Satyanarayanan et al, 2019) in Sw. 71 and HPC trophoblast cells following ZIKV infection. Similar to IFNβ expression pattern, we observed a significant increase of the mRNA expression for all these ISGs at 48 h.p.i. followed by decreased expression levels at 72 h.p.i. in Sw.71 cells infected with ZIKV (Fig EV1). HPC showed similar response although at different times, with a peak of ISG mRNA expression at 96 h.p.i., which correlated with the time of IFNβ peak expression (Fig EV2).
Figure EV1. ZIKV infection results in a time‐dependent interferon‐stimulated gene (ISG) response in trophoblast cell line Sw.71.

Human first‐trimester trophoblast cell line Sw.71 was infected with ZIKV (MOI = 2) for 1 h and refreshed with regular media over time. RNA was collected for measuring viral titers and gene expression by qRT–PCR. Data represent as mean ± SEM; n = 3 biological replicates; **P < 0.01 by one‐way ANOVA. Note that ISG (ISG20, MX1, OAS1, ISG15, CH25H, TRIM22, Tetherin, Viperin) mRNA expressions were all induced in trophoblast cell line Sw.71. NT, no treatment group.
Figure EV2. ZIKV infection results in a time‐dependent interferon‐stimulated gene (ISG) response in human primary culture trophoblast cells.

Human primary culture trophoblast cells were infected with ZIKV (MOI = 2) for 1 h and refreshed with regular media over time. RNA was collected for measuring viral titers and gene expression by qRT–PCR. Data represent as mean ± SEM; n = 3 biological replicates; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by one‐way ANOVA. Note that ISG expressions were firstly inhibited in the first 24 h.p.i. and then increased afterward. NT, no treatment group and HPC, human primary culture.
To confirm that these ZIKA‐induced ISGs are downstream of IFNβ, Sw.71 cells were treated with increasing doses of IFNβ (3, 30, 300 ng/ml) for 8 h, and ISG mRNA expression was determined by qRT–PCR. Our data show that IFNβ was able to induce the expression of these ISGs in a dose‐dependent manner (Fig EV3). These results suggest that IFNβ‐induced ISG expression in trophoblast cells is the early responders to ZIKV infection. Interestingly, we did not observe a similar IFNβ/ISG signature in trophoblast cells infected with a DNA virus, herpes simplex virus‐2 (HSV‐2) (Appendix Fig S4), suggestive of the differential responses of trophoblast cells to different virus infection.
Figure EV3. ZIKV‐induced ISGs are the downstream of IFNβ signaling in trophoblast cells.

Sw.71 were treated with different doses of IFNβ (3, 30, 300 ng/ml) for 8 h, and RNA was collected for determining anti‐ZIKA ISG expressions by RT–PCR. Data represent as mean ± SEM; n = 3 biological replicates; *P < 0.05, **P < 0.01, and ***P < 0.001 by one‐way ANOVA. NT, no treatment group. Note that even at the lowest dose (3 ng/ml), IFNβ was able to induce a robust increase of ISG mRNA expression.
Induction of ISG20 expression by IFNβ in response to ZIKV infection
An early step in the cellular anti‐viral response is to target the viral RNA before it is able to replicate and produce new viral particles. This process is accomplished through the expression of exonucleases that specifically degrade viral RNA (Schneider et al, 2014). In our screening of the anti‐viral ISGs, we observed the induction of ISG20 in human first‐trimester trophoblast cells infected with ZIKV (Figs EV1 and EV2). ISG20 functions as an interferon‐inducible 3′–5′ exonuclease and has been shown to exert a potent anti‐viral activity against different viruses (Espert et al, 2005; Zhang et al, 2007; Jiang et al, 2008; Qu et al, 2016; Liu et al, 2017). Therefore, we pursued the characterization of ISG20 because of its early potential interruptive function with the viral cycle.
First, we confirmed that ISG20 expression is directly regulated by IFNβ. Thus, Sw.71 or HPC were treated with increasing concentrations of IFNβ (3, 30, 300 ng/ml) for 8 h and ISG20 mRNA expression was quantified by qRT–PCR. Our data showed that IFNβ was a very strong inducer of ISG20 mRNA expression in trophoblast cells (Sw.71 and HPC) and its effect was dose‐dependent (Fig 2A). Furthermore, the induction of ISG20 by IFNβ was early (Fig 2B), where we observed a significant increase of ISG20 mRNA expression as early as 2 h following exposure to IFNβ (300 ng/ml) and was further enhanced until 16 h and slightly decreased at 24 h (Fig 2B).
Figure 2. IFNβ promotes ISG20 mRNA and protein expression in a dose‐ and time‐dependent manner in trophoblast cells.

Sw.71 (left panel) and HPC cells (right panel) were treated with IFNβ to detect ISG20 mRNA and protein expression.
- Sw.71 and HPC cells were treated with different doses of IFNβ (3, 30, 300 ng/ml) for 8 h, and RNA was collected for determining ISG20 mRNA expressions by qRT–PCR. Note the increase of ISG20 mRNA expression in a dose‐dependent manner. *P < 0.05 and **P < 0.01 by one‐way ANOVA.
- Sw.71 and HPC cells were treated with 300ng/ml IFNβ over time, and RNA was collected for determining ISG20 mRNA expressions by qRT–PCR. Note the increase of ISG20 mRNA expression in a time‐dependent manner. *P < 0.05 by one‐way ANOVA.
- Sw.71 and HPC cells were treated with different doses of IFNβ (3, 30, 300 ng/ml) for 24 h, and protein was collected for determining ISG20 protein expressions by Western blot. Note the increase of ISG20 protein expression in a dose‐dependent manner. β‐actin served as a loading control.
- Sw.71 and HPC cells were treated with 300 ng/ml IFNβ over time, and protein was collected for determining ISG20 protein expressions by Western blot. Note that there was no ISG20 protein expression in the no treatment group in both Sw.71 and HPC, and only after IFNβ treatment, ISG20 protein expression exhibited a time‐dependent manner. β‐actin served as a loading control.
Source data are available online for this figure.
Next, we determined whether the changes in ISG20 mRNA were translated into protein expression. Trophoblast cells were treated with increasing concentrations of IFNβ for 24h, and protein expression was determined by Western blot analysis. Interestingly, we found no protein expression in non‐stimulated trophoblast cells (Fig 2C and D), even though they have high basal mRNA levels as shown by the Cq value ranging from 23 to 26. However, IFNβ was able to induce ISG20 protein expression in a dose‐ and time‐dependent manner in both Sw.71 and HPC (Fig 2C and D). Intriguingly, this protein expression is tightly regulated. We saw ISG20 protein expression as early as 4 h post‐IFNβ treatment and increased up to 16 h in HPC. Afterward, ISG20 protein expression decreased to almost basal levels (Fig 2D). These data demonstrate that ISG20 transcription and translation are both inducible by IFNβ in first‐trimester trophoblast cells.
Poly(I:C) induces IFNβ and ISG20 expression in trophoblast cells
Our next objective was to investigate ISG20 expression in response to a general viral RNA. Therefore, we first used polyinosinic: polycytidylic acid (Poly(I:C)), which is a synthetic double‐stranded RNA that can mimic viral infection when applied in vitro and in vivo (Fortier et al, 2004) and triggers IFNβ expression (Kumar et al, 2006; Li et al, 2012; Dauletbaev et al, 2015). We tested the hypothesis that ISG20 expression could be induced by Poly(I:C) treatment in trophoblast cells. We treated Sw.71 and primary trophoblast cells with different doses of Poly(I:C) (0.25, 2.5, 25 μg/ml). IFNβ and ISG20 mRNA and protein expression were determined by qRT–PCR and Western blot, respectively. Poly(I:C) treatment induced IFNβ, as expected, and ISG20 mRNA expression in a dose‐dependent manner in Sw.71 and HPC trophoblast cells (Fig EV4A). At the protein level, Poly(I:C) also induced ISG20 protein expression in a dose‐dependent manner in both cell types (Fig EV4B). HPC trophoblasts seemed to be more sensitive to Poly(I:C) treatment since we were able to detect increased levels of ISG20 protein expression when cells were treated with Poly(I:C) at its lowest concentration (0.25 μg/ml) (Fig EV4B). Based on these findings, we then determined the earlier time when Poly(I:C) can promote IFNβ and ISG20 expression in trophoblast cells. Accordingly, Sw.71 and primary trophoblast cells were treated with Poly(I:C) (25 μg/ml) and cell pellets were collected at 2, 4, 8, 16, and 24 h post‐treatment for mRNA evaluation and 4, 8, 16, 24, and 48 h post‐treatment for protein evaluation. As we can see in Fig EV4C, Poly(I:C) treatment in Sw.71 cells induced IFNβ mRNA expression as early as 2 h post‐treatment reaching higher levels at 16 h and decreasing at 24 h. ISG20 mRNA expression followed similar pattern of expression as IFNβ, although we detected the earliest increase at 8 h post‐treatment and a major increase at 16 and 24 h (Fig EV4C). In HPC, we observed a similar early response to Poly(I:C) for IFNβ and ISG20. However, contrary to Sw.71 cells, the IFNβ mRNA levels remained higher even at 24 h post‐treatment (Fig EV4C). We saw a comparable response in protein expression when HPC were treated with Poly(I:C). As indicated above, trophoblast cells do not express ISG20 protein in basal conditions. However, following treatment with Poly(I:C) (25 μg/ml), we were able to detect ISG20 protein expression as early as 16 h post‐treatment (Fig EV4D). Altogether, these data suggest that ZIKA viral RNA and synthetic viral RNA such as Poly(I:C) can be sensed by trophoblasts cells, which will lead to the induction of a type I IFNβ response and expression of ISG20.
Figure EV4. Poly(I:C) treatment leads to a dose‐ and time‐dependent increase of IFNβ and ISG20 expression in trophoblast cells.

- Sw.71 and HPC cells were treated with different doses of Poly(I:C) (0.25, 2.5, 25 μg/ml) for 8 h, and RNA was collected for determining ISG20 mRNA expressions by qRT–PCR. Note the increase of IFNβ and ISG20 mRNA expression by Poly(I:C) in a dose‐dependent manner. Data represent as mean ± SEM; n = 3 biological replicates; *P < 0.05 and **P < 0.01 by one‐way ANOVA.
- Sw.71 and HPC cells were treated with different doses of Poly(I:C) (0.25, 2.5, 25 μg/ml) for 24 h, and protein was collected for determining ISG20 protein expressions by Western blot. Note the increase of ISG20 protein expression in a dose‐dependent manner. β‐actin served as a loading control.
- Sw.71 and HPC cells were treated with 25 μg/ml Poly(I:C) over time, and RNA was collected for determining IFNβ and ISG20 mRNA expressions by qRT–PCR. Note the increase of IFNβ and ISG20 mRNA expression in a time‐dependent manner. Data represent as mean ± SEM; n = 3 biological replicates; *P < 0.05, **P < 0.01, and ***P < 0.001 by one‐way ANOVA.
- Sw.71 and HPC cells were treated with 25 μg/ml Poly(I:C) over time, and protein was collected for determining ISG20 protein expressions by Western blot. Note again that there was no ISG20 protein expression in the no treatment group in both Sw.71 and HPC, and only after Poly(I:C) treatment, ISG20 protein expressed in a time‐dependent manner. β‐actin served as a loading control.
Role of ISG20 in controlling ZIKV infection in trophoblast cells
Next, we sought to evaluate whether trophoblasts would elicit a similar ISG20 response to ZIKV infection as the one observed with Poly(I:C). Firstly, we infected Sw.71 with ZIKA virus (MOI = 2) for 1 h and collected cell pellets at 24, 48, 56, and 72 h.p.i. and determined ISG20 protein expression by Western blot. Our results confirmed that ZIKV infection induced ISG20 protein expression (Fig 3A) in a time‐dependent manner, which was correlated to the increase in ISG20 mRNA (Fig EV1). Moreover, ISG20 protein expression was detected at 24, 48, and 56 h.p.i. and decreased at 72 h.p.i. (Fig 3A).
Figure 3. Lack of ISG20 in trophoblast cells leads to enhanced ZIKV replication and viral shedding.

- ISG20 protein expression was increased in ZIKV infection. Sw.71 cells were infected with ZIKV (MOI = 2) over time, and proteins were collected for Western blot analysis. β‐actin served as a loading control.
- No ISG20 protein expression in ISG20−/− Sw.71 in ZIKV infection. Sw.71 and ISG20−/− Sw.71 cells were infected with ZIKV (MOI = 2) for 1 h and refreshed with regular media for 48 h, and proteins were collected for Western blot analysis. β‐actin served as a loading control.
- Higher ZIKA titer was shown in ISG20−/− Sw.71 cells. Sw.71 and ISG20−/− Sw.71 cells were infected with ZIKV (MOI = 2) for 1 h and refreshed with regular media for 48 h, and RNA was collected for determining the viral titer and gene expression by qRT–PCR. Data represent as mean ± SEM; n = 4 biological replicates; *P < 0.05 by Student’s t‐test.
- More viral shedding in ZIKV‐infected ISG20−/− Sw.71 cell culture supernatant. Supernatant from ZIKV‐infected Sw.71 and ISG20−/− Sw.71 cells were collected, and plaque assay was performed using Vero cells. A representative plaque assay picture is presented. Note that more plaques were formed in Vero cells by incubating with ZIKV‐infected ISG20−/− Sw.71 cell culture supernatant. Scale bar = 130 μm.
- Plaques were counted, and plaque‐forming unit (pfu) ratio was calculated for the comparison of the viral titers. Data represent as mean ± SEM; n = 3 biological replicates. *P < 0.05 by Student’s t‐test.
Source data are available online for this figure.
To further elucidate the specific role of ISG20 during ZIKV infection, we established a trophoblast cell line lacking ISG20 (ISG20−/− Sw.71) using the CRISPR‐Cas9 system. The validation of ISG20−/− Sw.71 was evidenced by the lack of ISG20 protein expression following Poly(I:C) treatment (Appendix Fig S2). We then infected wild‐type (wt) Sw.71 and ISG20−/− Sw.71 with ZIKA virus (MOI = 2) for 1 h and incubated with refreshed growth media for 48 h. Afterward, RNA and protein were collected to determine viral titers by qRT–PCR and protein expression by Western blot. In contrast to wt Sw.71 cells, which showed ISG20 protein expression following ZIKV infection, no detectable ISG20 protein expression was observed in ISG20−/− Sw.71 (Fig 3B). More importantly, ISG20−/− Sw.71 exhibited significant higher viral titer levels when compared to wt Sw.71 (Fig 3C), suggesting that the lack of ISG20 rendered trophoblast cells more susceptible to ZIKV infection.
Subsequently, we evaluated whether the lack of ISG20 could have an impact in the process of ZIKV shedding. Vero cells were cultured in the presence of supernatants collected from 48 h ZIKV‐infected wt Sw.71 and ISG20−/− Sw.71 cells, and ZIKV shedding was determined by plaque assay. As shown in Fig 3D and E, more plaques were formed in Vero cells exposed to supernatants from ZIKV‐infected ISG20−/− Sw.71 group compared to supernatants from the ZIKV‐infected wt Sw.71 group.
To further understand the mechanism responsible for the increased viral titer in ISG20−/− Sw.71, we assessed whether the lack of ISG20 would affect the expression of other ISGs necessary for the anti‐ZIKV response. First, we assessed the levels of IFNβ expression after ZIKV infection and observed that, although the levels of induction were different, both wt Sw.71 cells and ISG20−/− Sw.71 cells could significantly induce IFNβ mRNA expression after ZIKV infection (Fig 4A). Next, we evaluated the mRNA expression of several other anti‐viral ISGs (MX1, OAS1, ISG15, CH25H, TRIM22, Tetherin, and Viperin) in ZIKV‐infected wt Sw.71 and ISG20−/− Sw.71 cells. As shown in Fig 4B, both cell lines showed an increased expression of anti‐viral ISGs, while the only major difference between the two cell lines was the lack of ISG20 in the ISG20−/− Sw.71 trophoblast cells (Fig 4C). Interestingly, although we observed increased mRNA levels for the tested ISGs in ISG20−/− Sw.71 trophoblast cells, the fold changes were not as robust as those observed in wt Sw.71 (Fig 4B), which suggests that ISG20 may play a role in regulating the expression of other ISGs during ZIKV infection. This is consistent with previous study which indicated that ISG20 upregulates other interferon response proteins to inhibit translation of the alphavirus genome, such as IFIT1 genes (Weiss et al, 2018). However, the immunological modulatory role of ISG20 on other ISGs is still under investigation.
Figure 4. Lack of ISG20 in trophoblast cells weakens the protection provided by IFNβ against ZIKV infection.

- Induction of IFNβ mRNA in response to ZIKV infection in Sw.71 and ISG20−/− Sw.71 trophoblast cells. Sw.71 and ISG20−/− Sw.71 cells were infected with ZIKV (MOI = 2) for 1 h and refreshed with regular media for 48 h, and RNA was collected for qRT–PCR. Note that ZIKV infection induces IFNβ mRNA expression in Sw.71 as well as in ISG20−/− Sw.71 cells. Data represent as mean ± SEM; n = 3˜4 biological replicates; *P < 0.05 and **P < 0.01 by Student’s t‐test.
- ISG mRNA expression stimulated by IFNβ in Sw.71 and ISG20−/− Sw.71 trophoblast cells. Sw.71 and ISG20−/− Sw.71 cells were treated with 30 ng/ml IFNβ for 24 h, and RNA was collected for determining the ISG gene expression by qRT–PCR. Data represent as mean ± SEM; n = 3 biological replicates; *P < 0.05 and **P < 0.01 by Student’s t‐test against control.
- No ISG20 protein expression in ISG20−/− Sw.71 cells after IFNβ treatment. Sw.71 and ISG20−/− Sw.71 cells were treated with 30 ng/ml IFNβ for 24 h, and proteins were collected for Western blot analysis. β‐actin served as a loading control for Western blot.
- IFNβ pre‐treatment significantly prevented trophoblast cells from ZIKV infection; however, this protection was evidently attenuated due to lack of ISG20. Sw.71 and ISG20−/− Sw.71 cells were pre‐treated with or without 30 ng/ml IFNβ for 24 h, followed by ZIKV infection (MOI = 2) for 1 h, and refreshed with regular media for 24 h, and RNA was collected to determine the viral titers by qRT–PCR. Data represent as mean ± SEM; n = 4 biological replicates; *P < 0.05 by Student’s t‐test.
Source data are available online for this figure.
We then evaluated the response to ZIKV infection in IFNβ pre‐treated trophoblast cells and showed that treatment of wt Sw.71 with IFNβ (30 ng/ml) was able to increase ISG20 protein expression and significantly decreased ZIKV titers (Fig 4C and D). However, similar pre‐treatment with IFNβ in ISG20−/− Sw.71 failed to induce ISG20 and had minimal effect on controlling ZIKV titers (Fig 4C and D). In summary, these data demonstrate that ISG20 is an early and critical component of the anti‐ZIKV response, which functions by inhibiting ZIKV replication and dissemination in trophoblast cells.
Characterization of the anti‐viral effect of recombinant ISG20
We hypothesized that ISG20 could be used as an anti‐viral treatment by blocking early stages of viral replication. To test this hypothesis, we developed a recombinant form of ISG20 by cloning the full length of human ISG20, a linker (Gly‐Ser‐Gly‐Ser‐Gly), and the human Fc domain into pcDNA4 vector. We also added a cell secretion signal and a His tag for purification. The protein structure is shown in Fig 5A and includes the following regions: (i) ISG20 sequence (pink); (ii) linker (orange); (iii) Fc domain (white); (iv) Tev sequence, cleavage between Q and G (cyan); (v) secretory signal sequence (ENPP7) (red); and 6) His Tag (green). The production steps are summarized in Fig 5B and encompass transfection of the ISG20 expression plasmid into Chinese hamster ovary (CHO) cells, followed by selection for plasmid integration by growing the cells in the presence of Zeocin. Positive clones were selected, and the expression of ISG20 was evaluated in the cytosol (endogenous) and supernatant (secreted) by Western blot analysis (Fig 5C). Positive clones were selected based on the intracellular expression of ISG20, but more importantly, the detection of ISG20 expression in the supernatant, confirming the secretion of the protein (Fig 5C). No detectable ISG20 protein was found in the supernatant or cell lysate of the negative clone or non‐transfected CHO cells (Fig 5C).
Figure 5. Design and characterization of the anti‐viral effect of recombinant ISG20 protein.

- Structure of the recombinant ISG20‐Fc protein. The components are (i) ISG20 sequence (pink); (ii) linker (orange); (iii) Fc domain (white); (iv) Tev sequence, cleavage between Q and G (cyan); (v) signal sequence (ENPP7) (red); and (vi) His Tag (green).
- Workflow of clone production and selection.
- Selection and verification of positive clones. In the positive clone, ISG20 protein expressed in both cytosol (endogenous) and supernatant (secreted) compared to the non‐transfected CHO cells and transfected negative clones. CHO, Chinese hamster ovary cell without transfection; β‐actin served as a loading control.
- ISG20 secretion in the positive clone supernatant significantly decreased ZIKV infection in trophoblast cells. Supernatants from negative and positive clones were collected and added to ISG20−/− Sw.71 trophoblast cells together with ZIKA virus (MOI = 2) for 1 h, followed by refreshing with new growth media for 48 h. RNA was then collected to determine viral titers by qRT–PCR. Data represent as mean ± SEM; n = 3 biological replicates; **P < 0.01 by Student’s t‐test.
- Recombinant ISG20‐Fc degrades ZIKA viral RNA and HSV‐2 viral DNA. 50ng purified viral RNA (ZIKV) or DNA (HSV‐2) was incubated with increasing concentrations of rISG20‐Fc (5, 50, 500 ng) in the presence of RNase inhibitor for 90min at 37°C followed by quantification of viral titers by qRT–PCR, and agarose gel was used to evaluate the RNA degradation by electrophoresis. The representative picture of agarose gel is presented. rISG20‐Fc was able to degrade both ZIKV RNA and HSV‐2 DNA in a dose‐dependent manner; however, rISG20‐Fc was more efficient in degrading viral RNA than DNA. Data represent as mean ± SEM; n = 3 biological replicates; **P < 0.01. ***P < 0.001, and ****P < 0.0001 by one‐way ANOVA.
Source data are available online for this figure.
Next, we collected the conditioned media from the positive and negative clones and added that to the ISG20−/− Sw.71 trophoblast cells together with ZIKA virus (MOI = 2) for 1 h, followed by refreshing with new growth media for 48 h. RNA was collected to determine viral titers by qRT–PCR. The conditioned media from the positive clone significantly reduced ZIKV viral titers compared to the negative clone (Fig 5D). These data suggest that the secreted form of ISG20 preserves its RNase activity and shows anti‐ZIKA effect. Consequently, we proceeded to purify the recombinant ISG20‐Fc protein for further characterization (see Materials and Methods).
Anti‐viral activity of recombinant ISG20‐Fc protein
To test whether the exonuclease activity of our recombinant ISG20‐Fc (rISG20‐Fc) was preserved, we first examined its ability to degrade viral RNA or DNA. We used two different viruses as the substrates for this experiment: RNA virus (ZIKA) and DNA virus (HSV‐2). 50 ng purified ZIKV RNA or HSV‐2 DNA was incubated with increasing concentrations of rISG20‐Fc (5, 50, 500 ng) for 90 min at 37°C in the presence of RNase inhibitor to exclude the effect of exogenous RNase, followed by quantification of viral titers by qRT–PCR. rISG20‐Fc was able to degrade both ZIKV RNA and HSV‐2 DNA in a dose‐dependent manner; however, rISG20‐Fc was more efficient in degrading viral RNA than DNA (Fig 5E).
In vivo efficacy of rISG20‐Fc inhibiting ZIKA viral replication in IFNAR1−/− pregnant mice
Having shown that rISG20‐Fc maintained its exonuclease activity, we then tested whether rISG20‐Fc could have an anti‐viral effect by using the IFNAR1−/− mice, which are highly sensitive to ZIKV infection (Lazear et al, 2016). Adult (8–12 weeks of age) IFNAR1−/− pregnant mice were intraperitoneally (i.p.) infected with 1 × 105 pfu ZIKV or 1% FBS DMEM/F12 media (vehicle) at day E8.5 (Fig 6A). Afterward, animals received three doses of rISG20‐Fc (1 mg/kg) i.p. 1 h post‐ZIKV infection and on days E9.5 and E10.5 of their pregnancy. Control animals were injected with PBS at the same time points (Fig 6A). On E14.5, mice were sacrificed and organs were collected to assess viral titer. In control mice, macroscopic evaluation of the uterus showed the presence of fetal death and resorptions in ZIKV‐infected mice (Fig 6B). However, rISG20‐Fc treatment rescued this phenotype by significantly decreasing pregnancy loss (Fig 6C). No differences in the number of implantation sites were found between the two groups, which confirms the protective effect of rISG20‐Fc (Fig EV5).
Figure 6. In vivo efficacy of rISG20‐Fc improving pregnancy outcome in IFNAR1−/− pregnant mice during ZIKV infection.
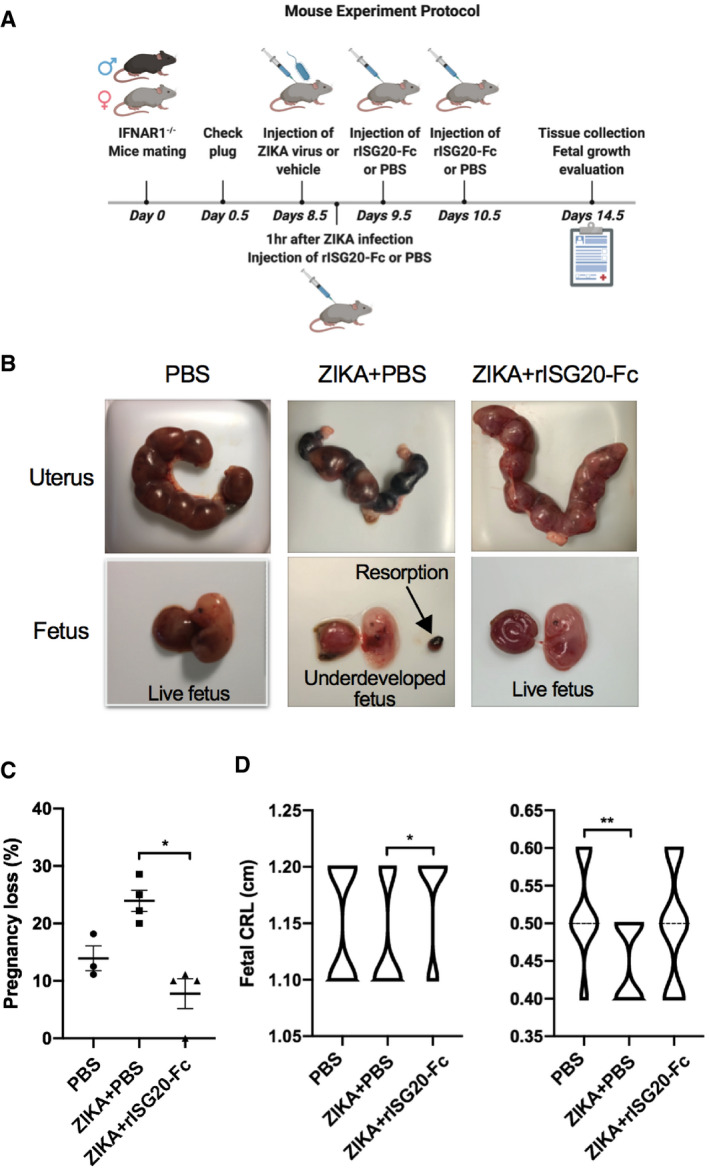
- Mouse experiment protocol. Adult (8–12 weeks of age) IFNAR1−/− pregnant mice were infected i.p. with 1 × 105 pfu ZIKV or 1% FBS DMEM/F12 media (vehicle) on embryo day 8.5 (E8.5). 1 h after ZIKV/vehicle injection, treatment with rISG20‐Fc (1 mg/kg) or PBS (control) was administrated i.p. to the pregnant mice. On E9.5 and E10.5, same protein/PBS injection was performed on the pregnant mice. On E14.5, the mice were sacrificed and organs were collected for viral titer quantification. N = 3 for control PBS group and n = 4 for treatment groups.
- Macroscopic evaluation of the pregnant uterus and fetus. Compared to the fetal death and reabsorptions in ZIKV‐infected mice, rISG20‐Fc treatment rescued this phenotype by decreasing pregnancy loss (resorption).
- Effect of rISG20‐Fc treatment on pregnancy loss. rISG20‐Fc treatment significantly decreased the pregnancy loss caused by ZIKA infection. The pregnancy loss rate (number of dead fetus + resorptions/ total number of implantations ×100) was calculated. Data represent as mean ± SEM; n = 3–4 mice per group; *P < 0.05 and **P < 0.01 by one‐way ANOVA.
- rISG20‐Fc treatment improves fetal development. Measurement of crown‐rump length (CRL) and occipitofrontal diameter (OFD) was determined on E14.5. n = 3–4 mice per group and 6–8 fetuses from each pregnant mouse were evaluated; *P < 0.05 and **P < 0.01 by one‐way ANOVA.
Figure EV5. rISG20‐Fc treatment has no effect on implantation number in ZIKV‐infected IFNAR1−/− pregnant mice.

Effect of rISG20‐Fc treatment on implantation number. No differences were found in terms of the implantation number between groups. N = 3 mice for control PBS group and n = 4 mice for treatment groups. Data represent as mean ± SEM; difference not significant by one‐way ANOVA.
Next, we analyzed the impact of ZIKV infection and treatment with rISG20‐Fc on fetal development. As previously reported (Uraki et al, 2017), ZIKV infection has major negative impacts on fetal development as demonstrated by a significant decrease in crown‐rump length (CRL) and occipitofrontal diameter (OFD). Treatment of ZIKV‐infected pregnant mice with rISG20‐Fc protected fetal development as demonstrated by improved fetal CRL compared to the infected group (Fig 6D).
We then evaluated the anti‐viral effect of rISG20‐Fc treatment by quantifying ZIKV titers on the maternal and fetal side. On the maternal side, we observed a significant decrease in ZIKV titers in the maternal serum of mice treated with rISG20‐Fc (Fig 7A). We also observed decreased titers in the spleen of these animals, although it was not statistically significant (P = 0.075) (Fig 7A). On the fetal side, we saw a significant decrease in ZIKV titers in the fetal brain of rISG20‐Fc‐treated mice (Fig 7B). Interestingly, there was no observed difference in ZIKV titer in the placenta between the two groups. This suggests that rISG20‐Fc may block viral transmission from the placenta to fetus, thus protecting the fetus from the detrimental effects of ZIKV infection.
Figure 7. In vivo efficacy of rISG20‐Fc inhibiting ZIKA viral replication and promoting placental integrity in IFNAR1−/− pregnant mice.

- rISG20‐Fc treatment alleviates maternal viral burden. Note that rISG20‐Fc treatment significantly decreased the maternal serum viral titers. Data represent as mean ± SEM; n = 3 mice for control PBS group and n = 4 mice for treatment groups; *P < 0.05 by Student’s t‐test.
- rISG20‐Fc treatment reduces ZIKV titers in the fetal brain. Although there was no difference of ZIKV titer in the placenta, at the fetal side, there was a significant decrease of viral titer in the fetal brain, suggesting rISG20‐Fc can block viral transmission from placenta to fetus. Data represent as mean ± SEM; n = 4 for each group and 3–4 placentas/fetal brains from every mouse were analyzed; *P < 0.05 by Student’s t‐test.
- rISG20‐Fc treatment decreases eotaxin significantly in maternal circulation. Data represent as mean ± SEM; n = 2 for PBS control group and n = 3 for every treatment group; *P < 0.05 by one‐way ANOVA; difference not significant for IL‐9 and IFNγ by one‐way ANOVA.
- Representative hematoxylin and eosin staining of the mouse placenta on E14.5. Labyrinth layers were marked with a solid line. Note the major alteration on placenta structure characterized by multifocal loss of tissue architecture (necrosis) and the defective blood vessel formation of labyrinth in ZIKV+PBS group, and rISG20‐Fc treatment improves vascularity and decreases decidual edema and cellular fragmentation at the labyrinth. Scale bar = 580 μm (upper images); scale bar = 230 μm (bottom images).
Impact of rISG20‐Fc treatment on placental integrity
Finally, we explored whether treatment with rISG20‐Fc could modulate the immunological response elicited by ZIKV infection on the maternal and fetal side. Thus, we evaluated maternal serum cytokine expression of ZIKV‐infected dams treated with rISG20‐Fc or vehicle control. Interestingly, we observed that rISG20‐Fc treatment had a modulatory effect on the anti‐viral response by significantly reducing potential damaging inflammatory cytokines such as eotaxin and promoting increased expression levels of anti‐viral mediators like IL‐9 and IFNγ (Fig 7C).
Next, histologic analysis of placenta samples from each group was performed to determine the impact of the ZIKV‐induced inflammatory process on the placenta and whether this could be countered with rISG20‐Fc treatment. Thus, uteroplacental units were collected at E14.5 (day 6 post‐infection), and H&E staining was performed. All histologic samples were analyzed in a blinded manner by an independent animal pathologist. As shown in Fig 7D, placentas obtained from animals infected with ZIKV showed major alteration on placenta structure characterized by multifocal loss of tissue architecture (necrosis) in the labyrinth, apparent damage of the vascularity evidenced by decreased blood vessel density, edema observed only in the decidua of infected mice, and necrosis and inflammation foci observed in the labyrinth of infected mice. Additional pathologic changes present within the labyrinth of infected mice included an overall tissue hypereosinophilia, nuclear pyknosis, and cellular fragmentation (Fig 7D). Treatment with rISG20‐Fc reversed some of these changes, including increased vascularity, decreased decidual edema, and cellular fragmentation at the labyrinth (Fig 7D). These data suggest that rISG20‐Fc treatment can contribute to placenta integrity during ZIKV infection and facilitate fetal development.
Discussion
We report for the first time the characterization of an effective IFNβ‐mediated anti‐ZIKV response in trophoblast cells and the identification of ISG20, an endogenous anti‐viral protein, which is naturally produced by the placenta/trophoblast, as an earlier anti‐viral exonuclease that is critical for stopping the viral cycle by promoting viral RNA degradation. Furthermore, we successfully established a recombinant ISG20‐Fc protein that maintains its exonuclease activity and effectively decreases viral titers in vitro and in vivo.
ZIKV infection during pregnancy remains a significant concern due to the major impacts on fetal development, such as microcephaly and other catastrophic fetal malformations (Alvarado & Schwartz, 2017; Schwartz, 2017a). Due to the high sensitivity of the infection during pregnancy, ZIKV is now included in the list of viral infections dangerous for pregnancy known as “TORCH” pathogens (Coyne & Lazear, 2016; Schwartz, 2017b), along with Toxoplasma gondii, rubella virus, cytomegalovirus (CMV), and herpes simplex virus (HSV), which are considered as the major causes of fetal morbidity and mortality. Unfortunately, there is no effective vaccine or treatment for ZIKV infection. Therefore, development of new therapies is needed.
In this study, we characterized the natural anti‐viral response to ZIKV infection and identified early responders that are important for stopping viral replication. The host innate immune response to viral infection is critical for controlling viral replication through the induction of type I IFNs and ISG expression (Espert et al, 2003; Ivashkiv & Donlin, 2014; Schneider et al, 2014). ISGs are considered as the anti‐viral effectors of the type I IFN response (Gifford et al, 2007). These effectors may function by targeting different stages of viral life cycle, such as inhibition of virus entry, viral RNA replication, viral particle assembly, and release (Jiang et al, 2008; Munakata et al, 2008; Sadler & Williams, 2008; Schneider et al, 2014). Hence, with the absence of type I IFN‐ISGs signals, virus will be likely to replicate more and destroy the tissues. For example, in ZIKV infection mouse model, defects in the type I IFN signaling axis such as the lack of IFNAR1 (IFNAR1−/−) or IRF3/5/7 have been shown to increase the sensitivity of ZIKV burden in multiple organs, such as the brain, spinal cord, and testes. In these same models, ZIKV‐infected IFNAR1−/− mice developed neurological diseases and succumbed to ZIKV infection (Lazear et al, 2016). Moreover, ZIKV infection in IFNAR1−/− mice during early pregnancy resulted in fetal demise, which was associated with the infection of placenta and fetal brain (Miner et al, 2016).
During early pregnancy, first‐trimester trophoblast cells are able to mount a strong type I IFNβ response, which is characterized by the expression of multiple ISGs (Kwon et al, 2018). This response is characteristic of first‐trimester trophoblast cells, which differs from third trimester placentas where type III interferons are highly expressed (Bayer et al, 2016). This differential expression may have to do with the different stages of placenta/trophoblast differentiation and function. Although early studies had suggested that type III interferons were functionally redundant with type I interferons (Bierne et al, 2012), several reports indicate that IFN‐λs have specific functions which differ from type I interferons (Sommereyns et al, 2008). Another difference between the type I IFNs and type III IFN pathways is the use of distinct receptors. IFN‐λs specifically interact with a heterodimeric receptor composed of two chains, a specific ligand‐binding chain IFN‐λR1 (or IL‐28Rα) and the IL‐10R2 (or IL‐10Rβ) chain (Marcello et al, 2006). In contrast, type I interferons are ligands of the IFNAR receptor (Nan et al, 2018). The evolutionary adaptation that confers type I IFNβ signaling in the developing placenta may reflect the need of immune regulation and tolerance required for the establishment of the pregnancy (Mor et al, 2017). The type I IFN response is more potent, rapid, and transient, (necessary during early placentation), whereas the type III IFN response is less potent, slower, and sustained (second and third trimester). In addition, type III IFN response is less inflammatory and concentrated at epithelial and barrier surfaces (Lazear et al, 2019). Although type III IFNs are an important component of the anti‐viral response and have being evaluated in several reports, in this study, we focus on the downstream signals of IFNβ in the first‐trimester trophoblast cells, in an area that is poorly understood.
We found here a significant increase in IFNβ transcription and translation during ZIKV infection in the Sw.71 first‐trimester trophoblast cell line and primary trophoblast cell cultures. This was then followed by the expression of a number of critical anti‐viral ISGs involved in this response, including Isg20, Mx1, Oas1, Isg15, CH25H, Trim22, Tetherin, and Viperin. Among these ISGs, ISG20, which is a 3′–5′ exonuclease, stands out for its early disruptive role in viral RNA replication. Prior studies have noted the importance of the anti‐viral effect of ISG20 in several viruses, including hepatitis B virus (HBV), hepatitis C virus (HCV), West Nile virus, dengue virus, and human immunodeficiency virus (HIV) (Espert et al, 2005; Jiang et al, 2008; Liu et al, 2017), and confirmed that ISG20‐mediated anti‐viral activity mainly depends on its exonuclease activity (Zheng et al, 2017). Moreover, apart from the capacity of degrading viral RNA as an exonuclease (Leong et al, 2016; Imam et al, 2020), other anti‐viral mechanisms of ISG20 have also been studied. For example, ISG20 inhibits both replication and transcription levels of the nucleoprotein and matrix protein genes required by influenza A virus (Qu et al, 2016). Interestingly, it is reported that ISG20 upregulates other interferon response proteins to inhibit translation of the alphavirus genome, such as IFIT1 genes (Weiss et al, 2018). In fact, it has been proposed that ISG20 could also target the expression of host microRNA or long noncoding RNA, which regulates many other genes (Liu et al, 2017; Chai et al, 2018). However, the regulatory role of ISG20 on other genes is still unclear and needs more investigation. Therefore, these additional mechanisms of ISG20 emphasize its important role in controlling virus infection through either interrupting viral cycle or/and modulating host immune response, and future studies are needed for elucidating the different underlying mechanisms.
In this study, we focused on characterizing ISG20 expression and function in first‐trimester trophoblast cells and observed that, as previously reported (Espert et al, 2003; Espert et al, 2004; Weiss et al, 2018), IFNβ and Poly(I:C) are both strong inducers of ISG20 expression. Poly(I:C) is a synthetic double‐stranded RNA (dsRNA), which mimics a viral infection and activates the TLR3 and cytoplasmic dsRNA sensors signaling pathways leading to increase of IFNβ expression (Kato et al, 2006; Watanabe et al, 2011). We demonstrate here that exposure of trophoblast cells to Poly(I:C) induces IFNβ and ISG20 expression. These findings further support the interpretation that first‐trimester trophoblast cells are capable of mounting an anti‐viral response, which is characterized by the expression of IFNβ and ISG20. Noteworthy, we did not observe ISG20 protein expression in the cells during regular culture conditions although we did detect high levels of mRNA expression. Only upon viral infection or IFNβ treatment, we can transiently detect the protein expression of ISG20. The regulatory pathways associated with this quick and transient expression are under further investigation.
To determine the role of ISG20 in the anti‐ZIKV response, we established a trophoblast cell line where ISG20 was knocked out. We found that lack of ISG20 weakens the protective effect provided by IFNβ against ZIKV infection, which further confirms the central role of ISG20 in the IFN‐β‐mediated response against ZIKV infection.
In order to repurpose this anti‐viral gene as a therapeutic for ZIKV infection, we designed a recombinant ISG20 protein. To design the ISG20 protein, we added an Fc domain of human neonatal IgG1 that functions to increase protein stability and facilitate the uptake by cells (Czajkowsky et al, 2012; Bell et al, 2013). Since multiple cell types express the Fc receptor, including immune cells, mucosal epithelial cells, and trophoblast cells (Lozano et al, 2018), it is plausible that our recombinant ISG20‐Fc may bind to Fc receptors on the surface of cells to facilitate endocytosis. More importantly, it is well established that IgG transfer from the mother to fetus across the placenta is mediated by neonatal Fc receptors expressed by the placenta (Lozano et al, 2018). Consequently, the presence of the Fc domain in rISG20‐Fc may allow the binding of the recombinant protein to trophoblast cells and facilitate its transport from the maternal circulation into the fetal tissues. Furthermore, we thoroughly examined the exonuclease activity of rISG20‐Fc by incubating the recombinant protein with purified ZIKV RNA and HSV‐2 DNA. rISG20‐Fc is highly effective at degrading ZIKV RNA as well as HSV‐2 DNA. However, as previously reported (Moser et al, 1997; Nguyen et al, 2001), rISG20‐Fc displays a higher efficacy toward the degradation of viral RNA rather than DNA.
The placenta is an extremely important immune organ, which acts as a physical and immunological barrier to block the transmission of infectious agents to fetus during pregnancy (Mor & Cardenas, 2010; Mor et al, 2017). Due to the invasive nature of the placenta in both humans and, more limited, in mice, trophoblasts make direct contact with maternal circulation. Previous studies have shown that ZIKV infects human trophoblasts (Bayer et al, 2016; Lazear et al, 2016; Quicke et al, 2016), and the primitive trophoblasts formed in the early stage of pregnancy are more sensitive to ZIKV infection compared to the later stage after the formation of syncytium (Sheridan et al, 2017). Therefore, we postulated that use of rISG20‐Fc could be an important tool to provide a protective effect in the developing placenta during early pregnancy. Indeed, when tested in vivo using the IFNAR1−/− mouse model, rISG20‐Fc was found to be effective in protecting the fetus from ZIKV infection. While IFNAR1−/− pregnant mice infected with ZIKV showed a high number of resorptions and underdeveloped fetuses, treatment with rISG20‐Fc was able to rescue the fetus and improve their growth by preventing ZIKV‐induced placental damage.
Furthermore, it is likely that rISG20‐Fc has systemic immune modulatory function as it can also decrease the viral titer in maternal circulation by facilitating the secretion of anti‐viral cytokines, such as IL‐9 (Yu et al, 2016) and IFNλ (Kang et al, 2018), in addition to balancing the overexpression of pro‐inflammatory factors, such as eotaxin (Rankin et al, 2000). Unexpectedly, we observed similar levels of ZIKV titers in placental samples from treated and control groups. No observed decrease of viral titers in the placenta suggests that rISG20‐Fc may be transferred from maternal circulation to the fetus through the Fc receptor in placenta and then degrades viral RNA on the fetal side, as demonstrated in Fig 8. Further studies are in process to characterize the transfer of rISG20‐Fc from the maternal to the fetal side.
Figure 8. Mechanism by which rISG20‐Fc inhibits ZIKA viral replication and decreases viral infection in the fetus.

When ZIKA virus infects the pregnant woman, the virus can reach the placenta and transmit to the fetus. In the ZIKV replication cycle, it starts with the virus binding to host cell surface receptors, leading to endocytosis of the virus. Internalized viral particles release the viral RNA into the cytoplasm of the host cell and start replication. During its replication, rISG20‐Fc degrades ZIKV RNA and inhibits the subsequent transcription and translation. Therefore, less new virus particles are released from the placenta to fetus.
Altogether, we identified ISG20 as a central effector in the IFNβ‐induced response to ZIKV infection in trophoblast cells. We have successfully generated a recombinant form of ISG20 that maintains its exonuclease activity and has protective effect on an animal model that is sensitive to ZIKV infection. We show that rISG20‐Fc possess a strong anti‐viral exonuclease activity in addition to potential immune modulatory functions. Thus, it is plausible that our recombinant protein can be applied toward other types of viral infections, especially for RNA virus infection. The findings from this study provide a new insight on the pathways regulating ZIKV infection in first‐trimester trophoblast cells and offer a valuable tool to combat ZIKV infection during pregnancy.
Materials and Methods
Cell culture and infection
Mycoplasma‐free immortalized human trophoblast Sw.71 cells and human endometrial stroma cells (HESC) were cultured in DMEM/ F12 or DMEM supplemented with 10% FBS, 10 mm HEPES, 0.1 mm MEM non‐essential amino acids, 1 mm sodium pyruvate, and 100 U/ml penicillin/streptomycin (Life Technologies; Waltham, MA, USA) under 5% CO2 at 37°C. For viral infections, cells were seeded in 6‐well plates at 1.75 × 105 cells per well; the next day, ZIKV was added to the cells for 1‐h incubation (at indicated MOI) with gentle agitation every 20 min, and after 1 h, the inoculum was removed and the cells were washed twice with phosphate‐buffered saline (PBS) and then cells were maintained in 10% FBS DMEM/F12 media for the duration of the experiment. At indicated time points after infection, cell pellets and conditioned media were collected for downstream analysis.
Human primary trophoblast isolation and culture
Human primary trophoblast cells were isolated from first‐trimester elective terminations as previously described (Straszewski‐Chavez et al, 2009). A signed written consent form was obtained from the patients. The use of placental tissues, specimens, and consent forms was approved by the Yale University Human Investigation Committee (#2000021607). The tissue specimen was collected in cold, sterile phosphate‐buffered saline (PBS) and immediately transported to the laboratory for cell culture preparation. Briefly, first‐trimester placental villous tissues were cut and digested in PBS supplemented with 0.25% Trypsin (Gibco, Grand Island, NY, USA) for 10 min at 37°C with gentle agitation. An equal volume of 10% FBS (Gibco, Grand Island, NY, USA) and Dulbecco's modified Eagle medium (DMEM) (Gibco, Grand Island, NY, USA) was added to inactivate the trypsin. The supernatant was collected and centrifuged at 800 g at room temperature for 10 min. The pellet was resuspended in 5 ml DMEM media supplemented with 10% FBS. This suspension was laid over lymphocyte separation media (ICN Biomedicals, Inc., Aurora, OH, USA) and centrifuged at 1,000 g for 20 min. The interface containing the trophoblast cells was collected and centrifuged at 800 g for 10 min. Cells were resuspended in DMEM with 10% FBS and then plated on a 6‐well plate to grow.
Knockout of ISG20 using CRISPR/Cas9
ISG20 was knocked out in Sw.71 first‐trimester trophoblast cell line using CRISPR/Cas9. The guide RNA for ISG20 was designed using the CRISPR design tool from the Zhang Laboratory at MIT (CRISPR.mit.edu) (Sanjana et al, 2014; Shalem et al, 2014). Two DNA oligos were synthesized as follows: sense: 5′‐CACCGCAGCACCGTGGACGTTCACG‐3′ and antisense: 5′‐AAACCGTGAACGTCCACGGTGCTGC‐3′. Oligos were phosphorylated using T4 polynucleotide kinase and annealed by heating equimolar amounts to 95°C and cooling slowly to room temperature. This resulting guide was introduced to lentiCRISPRv2GFP plasmid using BsmBI restriction sites, and lentiCRISPRv2GFP was a gift from David Feldser (Addgene plasmid # 82416) (Walter et al, 2017). 10 μg of the resulting plasmid was co‐transfected with 8 μg of packaging plasmid pCMV‐VSV‐G and 4 μg of envelope plasmid psPAX2 in the presence of 60 μg of polyethylenimine (PEI) into HEK293T cells in a 100‐mm dish. pCMV‐VSV‐G was a gift from Bob Weinberg (Addgene plasmid # 8454) (Stewart et al, 2003), and psPAX2 was a gift from Didier Trono (Addgene plasmid # 12260). Then, packaged viral particles were collected by ultracentrifugation and were transduced into the Sw.71 cells. The cells were sorted based on the GFP signal by fluorescence activated cell sorting (FACS) following transduction. The deletion of ISG20 was confirmed using Sanger sequencing technique performed by GENEWIZ, and the overall efficiency was 91.1% analyzed by TIDE (Tracking of Indels by Decomposition). Moreover, we verified the protein expression by Western blot (Appendix Fig S2).
Virus
ZIKV strain FSS 13025, which was originally isolated in Cambodia in 2010, was obtained from the World Reference Center for Emerging Viruses and Arboviruses at University of Texas Medical Branch, Galveston as previously described (Aldo et al, 2016b). ZIKA virus was propagated in African green monkey kidney (Vero) cells by infecting the monolayer with viral stock. When the cytopathic effect was observed in the whole monolayer, the infected supernatant was collected and centrifuged. The virus stocks were aliquoted and stored at −80°C. The viral titer of viral stock was determined by plaque assay.
Plaque assay
The infectivity of the virus was determined by plaque assay in Vero cells. Briefly, Vero cells were plated into 12‐well plates at 2.5 × 105 cells/well and inoculated with 200 µl of 10‐fold serial dilutions of viral stocks and incubated at 37°C for 1 h with gentle agitation. After inoculation, Vero cells were overlaid with media containing DMEM (Gibco, #11965‐084), 2% FBS, and 0.6% Avicel (FMC, # CL‐611). Cells were maintained at 37°C in 5% CO2 for 5 days. After 5‐day incubation, overlays were aspirated and the cells were fixed in 4% formaldehyde solution in PBS before staining with 1% crystal violet in 20% methanol. Viral plaques were photographed, and each plaque was counted as a plaque‐forming unit (PFU). Viral titer was calculated as PFU/[volume virus (ml) × (dilution factor)].
Production and purification of recombinant ISG20‐Fc protein
For the production of recombinant ISG20 protein from mammalian cell cultures, we designed the plasmid and transfected it into Chinese Hamster ovary cells (CHO). Briefly, an artificial gene sequence was obtained from Life Technologies, which coded for the following: 5′ BamHI restriction site, cell export signal sequence from hENPP7, 10xHis tag for nickel purification, Tev sequence (to enable cleavage of the protein), full‐length human ISG20, a linker (Gly‐Ser‐Gly‐Ser‐Gly), the Fc domain of human IgG1, and a 3′ EcoRI restriction site. In order to increase the binding affinity of our fusion protein with the Fc receptor, we changed three amino acids within the Fc region by using the QuikChange II site‐directed mutagenesis kits (Agilent, #200523), introducing M257Y, S259T, and T261E mutations simultaneously in the Fc region.
Mutagenesis primers are as follows:
MST Fwd: 5′‐ccccaaagcccaaagacactctgtatatcaccagggagcctgaagttacatgcgtcgttgt‐3′
MST Rev: 5′‐acaacgacgcatgtaacttcaggctccctggtgatatacagagtgtctttgggctttgggg‐3′
All of this sequence had been codon optimized for efficient expression in CHO cells. Using BamHI/EcoRI, this cassette was subcloned into pcDNA4 (Invitrogen, #V102020) vector, because the carrier vector in which the cassette was supplied lacked a promoter. Once completed, the plasmid was transfected into CHO cells using polyethyleneimine, and cells were then selected for plasmid integration with Zeocin treatment.
After transfection and Zeocin selection, the cells were dissociated and serial diluted into a selection media containing 150 μg/ml Zeocin for 2 weeks to establish stable single cell clones. After the expanding of single clones, positive clones were selected by evaluating the protein expression of ISG20‐Fc in the cytosol (endogenous) and supernatant (secreted) by Western blot analysis. Additionally, conditioned media from stable cell clones was used to assess anti‐ZIKA activity in vitro, and the clones with the highest efficacy were expanded and adapted for suspension growth in Pepro AF‐CHO serum‐free media (Peprotech). Six liters of media at 5 × 106 cells/ml was centrifuged at 1,000× g for 30 min, and the secreted protein was purified to homogeneity as previously described (Albright et al, 2015) using an ÄKTA Pure 25 M equipped for multi‐step automated purification with modifications. First, many non‐specific CHO cell secreted proteins were eliminated from the active fraction by ammonium sulfate precipitating, final concentration of 23% (NH4)2SO4. The remaining soluble protein was further purified by sequential nickel column and MabSelect PrismA affinity column purification followed by an S75 sizing column. Purified proteins were stored as frozen stocks in PBS at −80°C.
RNA degradation assay
Purified ZIKV RNA and HSV‐2 DNA were extracted using QIAamp MinElute Virus Spin Kit (Qiagen, catalog no. 57704), and 50 ng purified viral RNA/DNA was incubated with the recombinant protein at different concentrations for 90 min at 37°C in the presence of SUPERase In™ RNase inhibitor (Invitrogen, catalog no. AM2694), and the resulting RNA/DNA was tested for viral copies by qRT–PCR. Subsequently, the PCR product was used for agarose gel electrophoresis to evaluate the degradation of RNA/DNA as described (Aranda et al, 2012).
Mouse experiments
The IFNAR1−/− (B6.129S2‐ Ifnar1tm1Agt/Mmjax) mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and bred in a specific‐pathogen‐free facility at Wayne State University. Adult (8–12 weeks of age) IFNAR1−/− mice were set up for timed‐mating and the plug day was considered as embryonic days E0.5. On embryonic days E8.5, the plug‐positive mice were randomly grouped and inoculated intraperitoneally (i.p.) with either 1 × 105 plaque‐forming units (pfu) of ZIKA virus (in 100 μl volume) or 1% FBS DMEM/F12 media (vehicle). 1 h after ZIKV/vehicle injection, treatment with rISG20‐Fc (1 mg/kg) or PBS (control) was administrated i.p. to the pregnant mice. On E9.5 and E10.5, same protein/PBS injection was performed on the pregnant mice. On E14.5, the mice were sacrificed for tissue collection. At least three animals per group were established to ensure adequate power for statistics. Furthermore, the researchers taking the measures and collecting samples are unaware of which treatment each mouse has received until after the experiment is over.
Pregnancy outcome parameters including the number of implantations and resorptions were recorded, and fetus development was evaluated by comparing the weight, fetal crown‐rump length (CRL), and occipitofrontal diameter (OFD) between groups. Maternal spleen, placentas, and fetal brain were collected and stored in RNAlater™stabilization solution (Invitrogen, catalog no. AM7021) in −80°C for viral titer quantification. Furthermore, maternal serum was used for cytokine expression analysis by Luminex (Bio‐Rad) and some placentas from each group were stored in 4% paraformaldehyde for hematoxylin and eosin staining.
RNA extraction and real‐time PCR analysis
Cells were collected and total RNA was extracted using the RNeasy Mini kit (Qiagen, catalog no. 74106) according to the manufacturer’s instructions. Viral RNA from mice serum was extracted using QIAamp MinElute Virus Spin Kit (Qiagen, catalog no. 57704) according to the manufacturer’s protocol. RNA from mice tissue was extracted as described. Briefly, tissue was homogenized in TRIzol (Ambion; Waltham, MA, USA) in 1.0‐mm zirconium beads (Benchmark; Sayreville, NJ, USA) for two cycles at 400 g for 2 min. Supernatant was then transferred to a new tube and incubated for 5 min at room temperature (RT). 200 μl chloroform/ml of TRIzol was added, shaken for 15 s, and then incubated for 3 min at RT. Samples were centrifuged at 12,000 g for 15 min at 4°C, and aqueous phase was transferred to new tubes. Then, 500 μl of 100% isopropanol was added, incubated at RT for 10 min, and centrifuged at 12,000 g for 10 min at 4°C. Supernatant was then discarded, and cell pellet was washed twice with 500 μl of 100% ethanol and centrifuged at 12,000 g for 5 min. The pellet was then washed twice with 1 ml of 75% ethanol, vortexed briefly, then centrifuged at 7,500 g for 5 min at 4°C, and then air‐dried for 10 min. The pellet was resuspended in 50 μl of RNase‐free water.
RNA concentration and purity were assessed using spectrophotometric analyses of 260/280 ratios, and only samples with values of 1.8 or higher were used for PCR analysis. One microgram of RNA was reverse‐transcribed for each sample using Bio‐Rad (Hercules, CA, USA) iScript cDNA synthesis kit. iTaq Universal SYBR Green Supermix (Bio‐Rad) and gene‐specific primers (Appendix Table S1) were added to the RT reactions that were diluted 1:5 with nuclease‐free water and run on the CFX96, C1000 system qPCR machine (Bio‐Rad). Values were normalized to GAPDH and calculated with 2−ΔΔCt method as described (Livak & Schmittgen, 2001).
Virus titer quantification by qRT–PCR
ZIKA viral titer was quantified by one‐step quantitative reverse transcriptase PCR (qRT–PCR). ZIKA viral RNA was isolated from Zika virus stock using QIAamp MinElute Virus Spin Kit (Qiagen, catalog no. 57704) to create a standard curve using serial 10‐fold dilutions of ZIKV RNA. One microgram of RNA from samples was run on CFX96, C1000 system qPCR machine (Bio‐Rad) using a one‐step PCR mix (Promega, catalog no. A6120). Zika virus was detected using primer pair: forward 5′‐CCGCTGCCCAACACAAG‐3′ and reverse 5′‐CCACTAACGTTCTTTTGCAGACAT‐3′. The probe sequence is 5′‐AGCCTACCTTGACAAGCAGTCAGACACTCAA‐3′ 6‐FAM/ZEN/IBFQ (Lanciotti et al, 2008; Foy et al, 2011). The cycling conditions involved activation at 45°C for 15 min and 95°C for 2 min, followed by 40 amplification cycles of 95°C for 15 s, and 60°C for 1 min.
Viral RNA was quantified by comparing each sample’s threshold cycle (CT ) value with a ZIKV RNA standard curve.
Western blotting
For protein extraction, cells were lysed on ice in cell lysis buffer (1% Triton X‐100, 0.05% SDS, 100 mM Na2 PO4, and 150 mM NaCl) supplemented with protease inhibitor mixture (Roche) and PMSF for 15 min followed by centrifugation at 16,000 g for 15 min at 4°C to remove cell debris. The protein concentration was determined by bicinchoninic acid (BCA) assay (Pierce, catalog no. 23223, Rockford, IL). 30 μg of each protein lysate was electrophoresed on a 12% SDS–polyacrylamide gel. The proteins were then transferred onto polyvinylidene difluoride membranes (EMD Millipore). The membrane was blocked with PBS‐0.05% Tween 20 (PBS‐Tween) containing 5% nonfat milk (Fisher Scientific, Pittsburgh, PA), and the membranes were washed three times and incubated with primary antibody in 1% milk PBS‐Tween at 4°C overnight. The membranes were then washed with PBS‐Tween three times followed by a secondary antibody in 1% milk PBS‐Tween for 2 h at RT. Immunoreactivity was detected using enhanced chemiluminescence (NEN Life Sciences, Waltham, MA) and imaged by Kodak 20000MM Image Station. Antibodies were diluted as follows: 1:1,000 anti‐ISG20 (Proteintech, catalog no. 22097‐1‐AP); 1:10,000 anti‐β‐actin (Sigma, catalog no. A2066); and 1:10,000 peroxidase‐conjugated anti‐rabbit IgG (Cell Signaling Technology, catalog no. 7074).
IFNβ secretion analysis by ELLA
The IFNβ secretion of supernatants from the trophoblast cells with or without ZIKV infection was determined using the Simple Plex immunoassay system (ELLA, Protein Simple, San Jose, CA) as previously described (Aldo et al, 2016a). Briefly, 50 μl of sample was added to sample inlet ports on a cartridge, and then, the sample was split into multiple parallel channels from the sample inlet port. Each channel was specific for one particular analyte and subjected to a typical sandwich immunoassay protocol. The entire immunoassay procedure was automated, and the analyzed results were obtained using the manufacture‐encoded calibration curves.
Statistical analysis
Statistical analyses were performed using Prism software, version 8 (GraphPad, San Diego, CA). All data are presented as means ± SEM. Differences between two groups were analyzed using unpaired Student’s t‐test, and differences among multiple groups were analyzed by one‐way ANOVA. Depending on the distribution of the continuous variables, nonparametric test was used if the data were not normally distributed. A P < 0.05 was considered statistically significant.
Study approval
This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Wayne State University School of Medicine (Assurance Number A3310‐01). For the human primary trophoblast cell culture, a signed written consent form was obtained from the patients. The use of placental tissues, specimens, and consent forms was approved by the Yale University Human Investigation Committee (#2000021607).
Author contributions
Conceptualization, GM, PA, and JD; Methodology, CMR, JD, PS, and YY; Investigation, JD, HL, XQ, and JJ, Resources, BL, DB, AL and PS; Writing‐ Original Draft, JD; Writing‐Review & Editing, CMR, PS, AM and GM; Supervision, GM, DB, and AL; Funding Acquisition, GM.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Source Data for Figure 2
Source Data Figure 3
Source Data for Figure 4
Source Data for Figure 5
Acknowledgement
This work was supported in part by NIH grant NIAID 1R01AI145829‐01.
EMBO reports (2021) 22: e52450.
Data availability
No primary datasets have been generated and deposited.
References
- Albright RA, Stabach P, Cao W, Kavanagh D, Mullen I, Braddock AA, Covo MS, Tehan M, Yang G, Cheng Zet al (2015) ENPP1‐Fc prevents mortality and vascular calcifications in rodent model of generalized arterial calcification of infancy. Nat Commun 6: 10006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldo PB, Krikun G, Visintin I, Lockwood C, Romero R, Mor G (2007) A novel three‐dimensional in vitro system to study trophoblast‐endothelium cell interactions. Am J Reprod Immunol 58: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldo PB, Mulla MJ, Romero R, Mor G, Abrahams VM (2010) Viral ssRNA induces first trimester trophoblast apoptosis through an inflammatory mechanism. Am J Reprod Immunol 64: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldo P, Marusov G, Svancara D, David J, Mor G (2016a) Simple Plex: a novel multi‐analyte, automated microfluidic immunoassay platform for the detection of human and mouse cytokines and chemokines. Am J Reprod Immunol 75: 678–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldo P, You Y, Szigeti K, Horvath TL, Lindenbach B, Mor G (2016b) HSV‐2 enhances ZIKV infection of the placenta and induces apoptosis in first‐trimester trophoblast cells. Am J Reprod Immunol 76: 348–357 [DOI] [PubMed] [Google Scholar]
- Aliota MT, Caine EA, Walker EC, Larkin KE, Camacho E, Osorio JE (2016) Characterization of lethal Zika virus infection in AG129 mice. PLoS Negl Trop Dis 10: e0004682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado MG, Schwartz DA (2017) Zika virus infection in pregnancy, microcephaly, and maternal and fetal health: what we think, what we know, and what we think we know. Arch Pathol Lab Med 141: 26–32 [DOI] [PubMed] [Google Scholar]
- Aranda PS, LaJoie DM, Jorcyk CL (2012) Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis 33: 366–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, Marques ET Jr, Cherry S, Sadovsky Y, Coyne CB (2016) Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe 19: 705–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell MR, Engleka MJ, Malik A, Strickler JE (2013) To fuse or not to fuse: what is your purpose? Protein Sci 22: 1466–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Travier L, Mahlakõiv T, Tailleux L, Subtil A, Lebreton A, Paliwal A, Gicquel B, Staeheli P, Lecuit Met al (2012) Activation of type III interferon genes by pathogenic bacteria in infected epithelial cells and mouse placenta. PLoS One 7: e39080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady OJ, Osgood‐Zimmerman A, Kassebaum NJ, Ray SE, de Araújo VEM, da Nóbrega AA, Frutuoso LCV, Lecca RCR, Stevens A, Zoca de Oliveira Bet al (2019) The association between Zika virus infection and microcephaly in Brazil 2015–2017: an observational analysis of over 4 million births. PLoS Medicine 16: e1002755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas I, Mor G, Aldo P, Lang SM, Stabach P, Sharp A, Romero R, Mazaki‐Tovi S, Gervasi M, Means RE (2011) Placental viral infection sensitizes to endotoxin‐induced pre‐term labor: a double hit hypothesis. AJRI 65: 110–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Li J, Shangguan Q, Liu Q, Li X, Qi D, Tong X, Liu W, Ye X (2018) Lnc‐ISG20 inhibits influenza A virus replication by enhancing ISG20 expression. J Virol 92: e00539‐1–e00539‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne CB, Lazear HM (2016) Zika virus ‐ reigniting the TORCH. Nat Rev Microbiol 14: 707–715 [DOI] [PubMed] [Google Scholar]
- Czajkowsky DM, Hu J, Shao Z, Pleass RJ (2012) Fc‐fusion proteins: new developments and future perspectives. EMBO Mol Med 4: 1015–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauletbaev N, Cammisano M, Herscovitch K, Lands LC (2015) Stimulation of the RIG‐I/MAVS pathway by polyinosinic: polycytidylic acid upregulates IFN‐β in airway epithelial cells with minimal costimulation of IL‐8. J Immunol 195: 2829–2841 [DOI] [PubMed] [Google Scholar]
- Dick GW, Kitchen SF, Haddow AJ (1952) Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 46: 509–520 [DOI] [PubMed] [Google Scholar]
- Espert L, Degols G, Gongora C, Blondel D, Williams BR, Silverman RH, Mechti N (2003) ISG20, a new interferon‐induced RNase specific for single‐stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J Biol Chem 278: 16151–16158 [DOI] [PubMed] [Google Scholar]
- Espert L, Degols G, Lin YL, Vincent T, Benkirane M, Mechti N (2005) Interferon‐induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J Gen Virol 86: 2221–2229 [DOI] [PubMed] [Google Scholar]
- Espert L, Rey C, Gonzalez L, Degols G, Chelbi‐Alix MK, Mechti N, Gongora C (2004) The exonuclease ISG20 is directly induced by synthetic dsRNA via NF‐kappaB and IRF1 activation. Oncogene 23: 4636–4640 [DOI] [PubMed] [Google Scholar]
- Fortier ME, Kent S, Ashdown H, Poole S, Boksa P, Luheshi GN (2004) The viral mimic, polyinosinic:polycytidylic acid, induces fever in rats via an interleukin‐1‐dependent mechanism. Am J Physiol Regul Integr Comp Physiol 287: R759–766 [DOI] [PubMed] [Google Scholar]
- Foy BD, Kobylinski KC, Chilson Foy JL, Blitvich BJ, Travassos da Rosa A, Haddow AD, Lanciotti RS, Tesh RB (2011) Probable non‐vector‐borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis 17: 880–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- França GVA, Schuler‐Faccini L, Oliveira WK, Henriques CMP, Carmo EH, Pedi VD, Nunes ML, Castro MC, Serruya S, Silveira MFet al (2016) Congenital Zika virus syndrome in Brazil: a case series of the first 1501 livebirths with complete investigation. Lancet 388: 891–897 [DOI] [PubMed] [Google Scholar]
- Gifford CA, Racicot K, Clark DS, Austin KJ, Hansen TR, Lucy MC, Davies CJ, Ott TL (2007) Regulation of interferon‐stimulated genes in peripheral blood leukocytes in pregnant and bred, nonpregnant dairy cows. J Dairy Sci 90: 274–280 [DOI] [PubMed] [Google Scholar]
- Honein MA, Woodworth KR, Gregory CJ (2020) Neurodevelopmental abnormalities associated with in utero Zika virus infection in infants and children‐the unfolding story. JAMA Pediatr 174: 237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam H, Kim GW, Mir SA, Khan M, Siddiqui A (2020) Interferon‐stimulated gene 20 (ISG20) selectively degrades N6‐methyladenosine modified Hepatitis B Virus transcripts. PLoS Pathog 16: e1008338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT (2014) Regulation of type I interferon responses. Nat Rev Immunol 14: 36–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block TM, Guo JT (2008) Identification of three interferon‐inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol 82: 1665–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MA, Mier YT‐RL, Reefhuis J, Gilboa SM, Hills SL (2016) Zika and the risk of microcephaly. N Engl J Med 71: 635–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Brown HM, Hwang S (2018) Direct antiviral mechanisms of interferon‐gamma. Immune Netw 18: e33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi O, Goorhuis A, Schinkel J, Codrington J, Vreden SGS, Vermaat JS, Stijnis C, Grobusch MP (2016) Thrombocytopenia and subcutaneous bleedings in a patient with Zika virus infection. Lancet 387: 939–940 [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJet al (2006) Differential roles of MDA5 and RIG‐I helicases in the recognition of RNA viruses. Nature 441: 101–105 [DOI] [PubMed] [Google Scholar]
- Kell AM, Gale M Jr (2015) RIG‐I in RNA virus recognition. Virology 479–480: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga K, Mor G (2010) Toll‐like receptors at the maternal‐fetal interface in normal pregnancy and pregnancy disorders. Am J Reprod Immunol 63: 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Zhang J, Yu FS (2006) Toll‐like receptor 3 agonist poly(I:C)‐induced antiviral response in human corneal epithelial cells. Immunology 117: 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran Satyanarayanan S, El Kebir D, Soboh S, Butenko S, Sekheri M, Saadi J, Peled N, Assi S, Othman A, Schif‐Zuck Set al (2019) IFN‐beta is a macrophage‐derived effector cytokine facilitating the resolution of bacterial inflammation. Nat Commun 10: 3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J‐Y, Aldo P, You Y, Ding J, Racicot K, Dong X, Murphy J, Glukshtad G, Silasi M, Peng Jet al (2018) Relevance of placental type I interferon beta regulation for pregnancy success. Cell Mol Immunol 15: 1010–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR (2008) Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14: 1232–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS (2016) A mouse model of Zika virus pathogenesis. Cell Host Microbe 19: 720–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Schoggins JW, Diamond MS (2019) Shared and distinct functions of type I and type III interferons. Immunity 50: 907–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong CR, Funami K, Oshiumi H, Mengao D, Takaki H, Matsumoto M, Aly HH, Watashi K, Chayama K, Seya T (2016) Interferon‐stimulated gene of 20 kDa protein (ISG20) degrades RNA of hepatitis B virus to impede the replication of HBV in vitro and in vivo . Oncotarget 7: 68179–68193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE, Marie IJ, Durbin JE (2011) Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol 1: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y‐G, Siripanyaphinyo U, Tumkosit U, Noranate N, A‐nuegoonpipat A, Pan Y, Kameoka M, Kurosu T, Ikuta K, Takeda Net al (2012) Poly (I:C), an agonist of toll‐like receptor‐3, inhibits replication of the Chikungunya virus in BEAS‐2B cells. Virol J 9: 114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Nie H, Mao R, Mitra B, Cai D, Yan R, Guo JT, Block TM, Mechti N, Guo H (2017) Interferon‐inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem‐loop structure of viral RNA. PLoS Pathog 13: e1006296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lozano NA, Lozano A, Marini V, Saranz RJ, Blumberg RS, Baker K, Agresta MF, Ponzio MF (2018) Expression of FcRn receptor in placental tissue and its relationship with IgG levels in term and preterm newborns. Am J Reprod Immunol 80: e12972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello T, Grakoui A, Barba‐Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM (2006) Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology 131: 1887–1898 [DOI] [PubMed] [Google Scholar]
- Miner J, Cao B, Govero J, Smith A, Fernandez E, Cabrera O, Garber C, Noll M, Klein R, Noguchi Ket al (2016) Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 165: 1081–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor G, Aldo P, Alvero AB (2017) The unique immunological and microbial aspects of pregnancy. Nat Rev Immunol 17: 469–482 [DOI] [PubMed] [Google Scholar]
- Mor G, Cardenas I (2010) The immune system in pregnancy: a unique complexity. AJRI 63: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser MJ, Holley WR, Chatterjee A, Mian IS (1997) The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res 25: 5110–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munakata K, Yamamoto M, Anjiki N, Nishiyama M, Imamura S, Iizuka S, Takashima K, Ishige A, Hioki K, Ohnishi Yet al (2008) Importance of the interferon‐alpha system in murine large intestine indicated by microarray analysis of commensal bacteria‐induced immunological changes. BMC Genom 9: 192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan J, Wang Y, Yang J, Stark GR (2018) IRF9 and unphosphorylated STAT2 cooperate with NF‐κB to drive IL6 expression. Proc Natl Acad Sci USA 115: 3906–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LH, Espert L, Mechti N, Wilson DM (2001) The human interferon‐ and estrogen‐regulated ISG20/HEM45 gene product degrades single‐stranded RNA and DNA in vitro. Biochemistry 40: 7174–7179 [DOI] [PubMed] [Google Scholar]
- Organization WH (2018) Zika virus
- Organization WH (2019) Zika epidemiology updateZika epidemiology update
- Pattnaik A, Sahoo BR, Pattnaik AK (2020) Current status of Zika virus vaccines: successes and challenges. Vaccines 8: 266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Reis e Sousa C (2007) Innate recognition of viruses. Immunity 27: 370–383 [DOI] [PubMed] [Google Scholar]
- Platanias LC (2005) Mechanisms of type‐I‐ and type‐II‐interferon‐mediated signalling. Nat Rev Immunol 5: 375–386 [DOI] [PubMed] [Google Scholar]
- Qu H, Li J, Yang L, Sun L, Liu W, He H (2016) Influenza A Virus‐induced expression of ISG20 inhibits viral replication by interacting with nucleoprotein. Virus Genes 52: 759–767 [DOI] [PubMed] [Google Scholar]
- Quicke K, Bowen J, Johnson E, McDonald C, Ma H, O’Neal J, Rajakumar A, Wrammert J, Rimawi B, Pulendran Bet al (2016) Zika virus infects human placental macrophages. Cell Host Microbe 20: 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racicot K, Aldo P, El‐Guindy A, Kwon JY, Romero R, Mor G (2017) Cutting edge: fetal/placental type I IFN can affect maternal survival and fetal viral load during viral infection. J Immunol 198: 3029–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racicot K, Kwon JY, Aldo P, Abrahams V, El‐Guindy A, Romero R, Mor G (2016) Type I interferon regulates the placental inflammatory response to bacteria and is targeted by virus: mechanism of polymicrobial infection‐induced preterm birth. Am J Reprod Immunol 75: 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SM, Conroy DM, Williams TJ (2000) Eotaxin and eosinophil recruitment: implications for human disease. Mol Med Today 6: 20–27 [DOI] [PubMed] [Google Scholar]
- Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR (2016) Zika virus and birth defects‐reviewing the evidence for causality. N Engl J Med 374: 1981–1987 [DOI] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR (2008) Interferon‐inducible antiviral effectors. Nat Rev Immunol 8: 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovsky Y, Clifton VL, Knofler M (2016) Editorial: ZIKA virus and placenta. Placenta 40: A1 10.1016/j.placenta.2016.03.008 [DOI] [PubMed] [Google Scholar]
- Said EA, Tremblay N, Al‐Balushi MS, Al‐Jabri AA, Lamarre D (2018) Viruses seen by our cells: the role of viral RNA sensors. J Immunol Res 2018: 9480497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F (2014) Improved vectors and genome‐wide libraries for CRISPR screening. Nat Methods 11: 783–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM (2014) Interferon‐stimulated genes: a complex web of host defenses. Annu Rev Immunol 32: 513–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DA (2017a) Autopsy and postmortem studies are concordant: pathology of Zika virus infection is neurotropic in fetuses and infants with microcephaly following transplacental transmission. Arch Pathol Lab Med 141: 68–72 [DOI] [PubMed] [Google Scholar]
- Schwartz DA (2017b) The origins and emergence of Zika virus, the newest TORCH infection: what's old is new again. Arch Pathol Lab Med 141: 18–25 [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi Xi, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JGet al (2014) Genome‐scale CRISPR‐Cas9 knockout screening in human cells. Science 343: 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C, Xie X, Muruato AE, Rossi SL, Roundy CM, Azar SR, Yang Y, Tesh RB, Bourne N, Barrett ADet al (2016) An infectious cDNA clone of Zika virus to study viral virulence, mosquito transmission, and antiviral inhibitors. Cell Host Microbe 19: 891–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan MA, Yunusov D, Balaraman V, Alexenko AP, Yabe S, Verjovski‐Almeida S, Schust DJ, Franz AW, Sadovsky Y, Ezashi Tet al (2017) Vulnerability of primitive human placental trophoblast to Zika virus. Proc Natl Acad Sci USA 114: E1587–E1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommereyns C, Paul S, Staeheli P, Michiels T (2008) IFN‐Lambda (IFN‐λ) is expressed in a tissue‐dependent fashion and primarily acts on epithelial cells in vivo . PLoS Pathog 4: e1000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PAet al (2003) Lentivirus‐delivered stable gene silencing by RNAi in primary cells. RNA 9: 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straszewski‐Chavez SL, Abrahams VM, Alvero AB, Aldo PB, Ma Y, Guller S, Romero R, Mor G (2009) The isolation and characterization of a novel telomerase immortalized first trimester trophoblast cell line, Swan 71. Placenta 30: 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraki R, Jurado KA, Hwang J, Szigeti‐Buck K, Horvath TL, Iwasaki A, Fikrig E (2017) Fetal growth restriction caused by sexual transmission of Zika virus in mice. J Infect Dis 215: 1720–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter DM, Venancio OS, Buza EL, Tobias JW, Deshpande C, Gudiel AA, Kim‐Kiselak C, Cicchini M, Yates TJ, Feldser DM (2017) Systematic in vivo inactivation of chromatin‐regulating enzymes identifies Setd2 as a potent tumor suppressor in lung adenocarcinoma. Cancer Res 77: 1719–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Tatematsu M, Saeki K, Shibata S, Shime H, Yoshimura A, Obuse C, Seya T, Matsumoto M (2011) Raftlin is involved in the nucleocapture complex to induce poly(I:C)‐mediated TLR3 activation. J Biol Chem 286: 10702–10711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss CM, Trobaugh DW, Sun C, Lucas TM, Diamond MS, Ryman KD, Klimstra WB (2018) The interferon‐induced exonuclease ISG20 exerts antiviral activity through upregulation of type I interferon response proteins. mSphere 3: e00209‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yockey LJ, Varela L, Rakib T, Khoury‐Hanold W, Fink SL, Stutz B, Szigeti‐Buck K, Van den Pol A, Lindenbach BD, Horvath TLet al (2016) Vaginal exposure to Zika virus during pregnancy leads to fetal brain infection. Cell 166: 1247–1256.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Long Q, Li HH, Liang W, Liao YH, Yuan J, Cheng X (2016) IL‐9 inhibits viral replication in coxsackievirus B3‐induced myocarditis. Front Immunol 7: 409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zevini A, Olagnier D, Hiscott J (2017) Crosstalk between cytoplasmic RIG‐I and STING sensing pathways. Trends Immunol 38: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Burke CW, Ryman KD, Klimstra WB (2007) Identification and characterization of interferon‐induced proteins that inhibit alphavirus replication. J Virol 81: 11246–11255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Wang L, Pan J (2017) Interferon‐stimulated gene 20‐kDa protein (ISG20) in infection and disease: review and outlook. Intractable Rare Dis Res 6: 35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]


