Abstract
Cytokinins are phytohormones that regulate plant development, growth, and responses to stress. In particular, cytokinin has been reported to negatively regulate plant adaptation to high salinity; however, the molecular mechanisms that counteract cytokinin signaling and enable salt tolerance are not fully understood. Here, we provide evidence that salt stress induces the degradation of the cytokinin signaling components Arabidopsis (Arabidopisis thaliana) response regulator 1 (ARR1), ARR10 and ARR12. Furthermore, the stress‐activated mitogen‐activated protein kinase 3 (MPK3) and MPK6 interact with and phosphorylate ARR1/10/12 to promote their degradation in response to salt stress. As expected, salt tolerance is decreased in the mpk3/6 double mutant, but enhanced upon ectopic MPK3/MPK6 activation in an MKK5DD line. Importantly, salt hypersensitivity phenotypes of the mpk3/6 line were significantly alleviated by mutation of ARR1/12. The above results indicate that MPK3/6 enhance salt tolerance in part via their negative regulation of ARR1/10/12 protein stability. Thus, our work reveals a new molecular mechanism underlying salt‐induced stress adaptation and the inhibition of plant growth, via enhanced degradation of cytokinin signaling components.
Keywords: ARR1/10/12, cytokinin, MPK3/6, phosphorylation, salt stress
Subject Categories: Plant Biology, Post-translational Modifications & Proteolysis, Signal Transduction
MKK5‐MPK3/6 signaling positively regulates salt tolerance in Arabidopsis, via phosphorylation and degradation of cytokinin signaling components ARR1/10/12.
Introduction
Excess salinity is harmful to most plants, limiting plant growth, and crop production (Yang & Guo, 2018). Plants have evolved multiple defense mechanisms to sense and respond to salinity stress via altered gene expression, physiology, and metabolism (Zhu, 2002, 2016; Deinlein et al, 2014; Julkowska & Testerink, 2015; Kudla et al, 2018). Stress phytohormones such as abscisic acid (ABA), ethylene (ET), jasmonate (JA), and salicylic acid (SA) play important roles in mediating plant adaption to salinity stress (Verma et al, 2016; Niu et al, 2018). For example, ABA level rapidly increased in response to salt stress (Duan et al, 2013; Geilfus et al, 2015), which maintains water balance by promoting stomata closure and Na+/K+ homeostasis.
Growth‐promoting hormones also appear to play important roles in plant adaptations to salt stress. Recent work suggests that auxin acts downstream of ABA to regulate root growth under salt stress (Xing et al, 2016). In addition, salt stress restricts plant growth by reducing bioactive gibberellin (GA) levels due to accumulation of GA‐deactivating enzyme ELONGATED UPPERMOST INTERNODE (EUI) (Wang et al, 2020). BRI1 KINASE INHIBITOR1 (BKI1) positively regulates salt tolerance by antagonizing 14‐3‐3 and facilitating BRASSINAZOLE RESISTANT 1 (BZR1) and BRI1‐EMSSUPPRESSOR 1 (BES1) translocation into nucleus to trigger brassinosteroids (BRs) responses (Zhang et al, 2017). Thus, multiple growth‐promoting hormones help plants adapt to stress.
The growth‐promoting phytohormone cytokinin plays an important role in various plant stress responses, including drought, heat, and salt (Werner & Schmulling, 2009; Guo & Gan, 2011; Muller & Leyser, 2011; Mackova et al, 2013; Cortleven et al, 2014; Zwack & Rashotte, 2015; Kien et al, 2016; Verma et al, 2016; Zwack et al, 2016; Todaka et al, 2017; Yang et al, 2017; Kieber & Schaller, 2018; Ramireddy et al, 2018; Cortleven et al, 2019). Cytokinin has been reported to negatively regulate salinity stress, and exposure to high salt concentrations decreases the production and transport of cytokinin, enabling stress adaptation (Tran et al, 2007; Nishiyama et al, 2011). For instance, cytokinin oxidases/dehydrogenases (CKXs) regulate cytokinin metabolism and can effectively reduce cytokinin concentration in plants. Overexpressing CKXs increases the tolerance to salt stress and drought stress in Arabidopsis or in Physcomitrella patens (von Schwartzenberg et al, 2007; Nishiyama et al, 2011).
In plants such as Arabidopsis, cytokinin binds Arabidopisis (Arabidopisis thaliana) histidine kinase (AHK) receptors, resulting in autophosphorylation and a subsequent phosphorelay to type‐A and type‐B Arabidopisis response regulators (ARRs), which regulate the expression of downstream target genes (El‐Showk et al, 2013; Kieber & Schaller, 2018). Salt stress reduces the transcript level of AHK1 and AHK2 as well as Arabidopsis histidine phosphotransfer proteins (AHPs) (Tran et al, 2007). However, to what extent salinity stress directly modulates components of the cytokinin signaling pathway to diminish cytokinin signaling and to promote the response to stress is poorly understood. Here, we provide evidence that salt stress represses cytokinin signaling through MPK3/6‐mediated phosphorylation and degradation of type‐B ARR1/10/12. These results uncover a novel regulatory mechanism, whereby salinity stress inhibits plant growth via the regulation cytokinin signaling.
Results
Salt stress induces ARR1/10/12 protein degradation
Cytokinin‐deficient mutant plants display a high tolerance to salt, supporting the idea that cytokinin signaling negatively regulates plant growth under salinity stress (Tran et al, 2007; Mason et al, 2010; Nishiyama et al, 2011). Consistent with the previous results (Mason et al, 2010), we observed that arr1/10, arr10/12, and arr1/12 double mutants showed increased salt tolerance compared to wild‐type Col‐0, as measured by primary root growth, survival rates, and fresh weights of Arabidopsis seedlings (Appendix Fig S1A–D). Consistent with this observation, the salt‐induced expression of the abiotic stress response genes RD29B, MYB15, and ZAT10 was significantly higher in the arr1/12 mutant compared to Col‐0, consistent with its increased salt tolerance (Appendix Fig S2A–C). To further investigate the function of type‐B ARR1/10/12 in salt stress response, over‐expressors of ARR1/10/12 were also employed. As expected, compared with WT, 35S:ARR1:MYC (Huang et al, 2018), 35S:ARR10:YFP and 35S:ARR12:YFP transgenic plants all showed hypersensitivity to cytokinin 6‐Benzylaminopurine (6‐BA; Appendix Figs S3A–C, S4A–C, and S5A–C). Next, 35S:ARR1:MYC (Huang et al, 2018), 35S:ARR10:YFP #8, and 35S:ARR12:YFP #6 transgenic plants which displayed similar sensitivity to 6‐BA were selected for further experiments. We then examined the salt tolerance response of over‐expressors of ARR1/10/12 and found that all these lines displayed enhanced root‐growth inhibition, reduced survival rate, and reduced fresh weight compared to WT (Fig EV1A–D). Collectively, our studies indicate that type‐B ARR1/10/12 negatively regulate salt tolerance responses in plants.
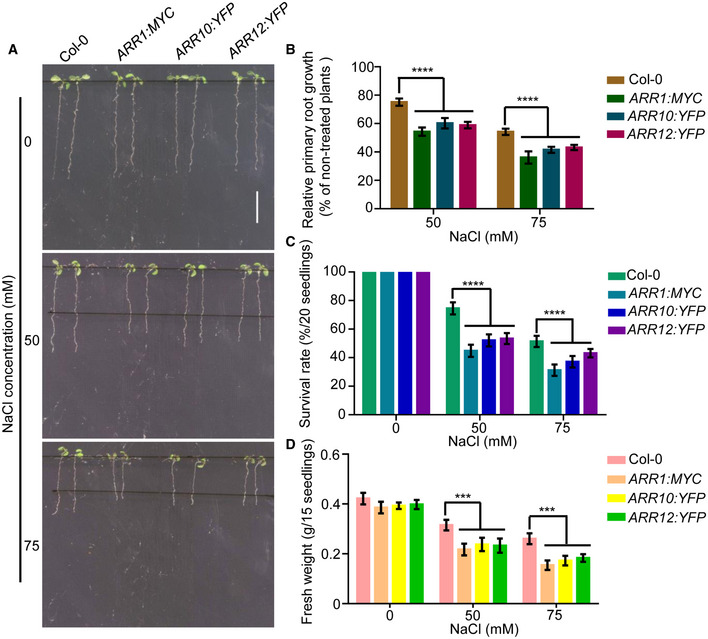
Figure EV1. Transgenic plants overexpressing ARR1/10/12 all show hypersensitivity to salt stress compared with that of wild type.
-
A–DFour‐day‐old seedlings of 35S:ARR1:MYC, 35S:ARR10:YFP, and 35S:ARR12:YFP were transferred to 1/2 MS medium supplemented with 50 mM or 75 mM NaCl. Pictures were taken, and the elongated root length was determined 3 days later (A and B). Changes in survival rates (C) and fresh weights (D) were examined after 10 days of salt treatment. Scale bar, 1 cm.
Data information: In (B–D), data are means of three biological replicates ± SD (n = 60 for (B), and n = 12 for (C) and (D)). ***, and **** indicate significant difference to the corresponding controls with P < 0.001 and P < 0.0001, respectively (Student's t‐test).
To further investigate the role of ARR1/10/12 in salt tolerance, we examined whether their expression changes in response to salt treatment. Results from quantitative RT–PCR analysis indicated that the transcriptional levels of ARR1/10/12 were not significantly affected upon salt stress treatment (Fig EV2A–C). Next, we explored whether salt stress affected protein levels of ARR1/10/12 using 35S:ARR1:MYC, 35S:ARR10:YFP, and 35S:ARR12:YFP seedlings. Western blot results showed that the protein levels of ARR1/10/12 decreased under salt treatment in a time‐dependent manner (Fig 1A–F). Moreover, we also examined whether salt stress affected protein levels of ARR1/10/12 under the control of their native promoters using ARR1p:ARR1g‐GFP, ARR10p:ARR10g‐GFP, and ARR12p:ARR12g‐GFP transgenic lines. The reduced GFP signals of ARR1p:ARR1g‐GFP, ARR10p:ARR10g‐GFP, and ARR12p:ARR12g‐GFP after NaCl treatment also further confirmed the effect of salt stress on the protein levels of ARR1/10/12 (Fig EV2D–I). Next, we did the co‐treatment with the protein synthesis inhibitor cycloheximide (CHX) and found that salt treatment still led to reduced ARR1/10/12 protein levels (Appendix Fig S6A–F), indicating that salt treatment reduces the protein levels of ARR1/10/12 through promoting their degradation. However, the co‐treatment with the 26S proteasome inhibitor MG132 prevented the reduction of ARR1/10/12 protein levels in the presence of salt, revealing that salt stress‐induced ARR1/10/12 degradation via the 26S proteasome pathway (Appendix Fig S7A–C). In summary, this result suggests that salt stress may repress the cytokinin signaling through reducing ARR1/10/12 protein level.
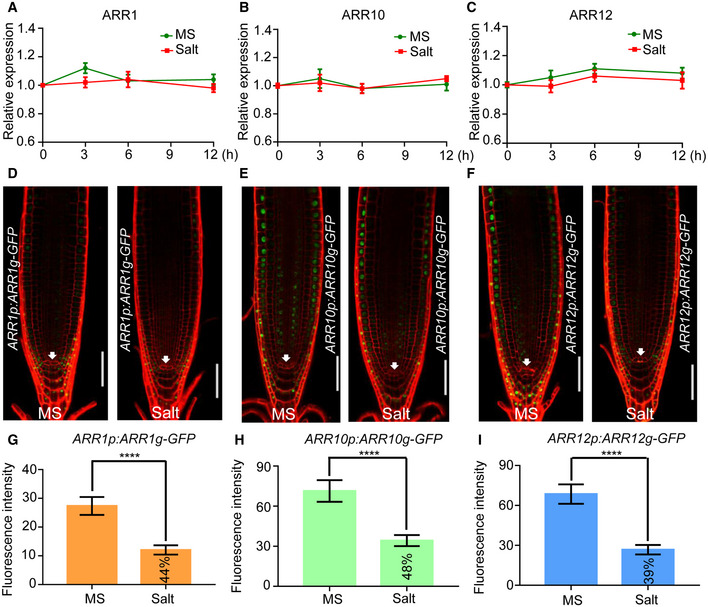
Figure EV2. Salt treatment decreases the protein levels of ARR1/10/12.
-
A–CExpression analysis of ARR1, ARR10, and ARR12 under salt stress. Five‐day‐old wild‐type Col‐0 seedlings were treated with or without 200 mM NaCl for the indicated periods of time. Transcription levels were determined by qRT‐PCR analysis of the indicated genes and normalized to the expression levels of Actin2 and UBQ1.
-
D–FFive‐day‐old seedlings of ARR1p:ARR1g‐GFP, ARR10p:ARR10g‐GFP, and ARR12p:ARR12g‐GFP transgenic plants were transferred to 1/2 MS plates with or without 50 mM NaCl for 12 h. Cell boundaries appear red following propidium iodide staining. The white arrowheads mark the position of the QC (Quiescent center) cells. Scale bar, 100 μm.
-
G–IQuantification of the fluorescence intensity in ARR1p:ARR1g‐GFP, ARR10p:ARR10g‐GFP, and ARR12p:ARR12g‐GFP seedlings in (D–F). The fluorescence intensity of NaCl treated seedlings/fluorescence intensity of NaCl untreated seedlings ratios were labeled in the columns in (G–I).
Data information: In (A–C), data shown are means ± SD of three biological replicates. In (G–I), data are means of three biological replicates ± SD (n = 45). **** indicates significant difference to the corresponding controls with P < 0.0001 (Student's t‐test).
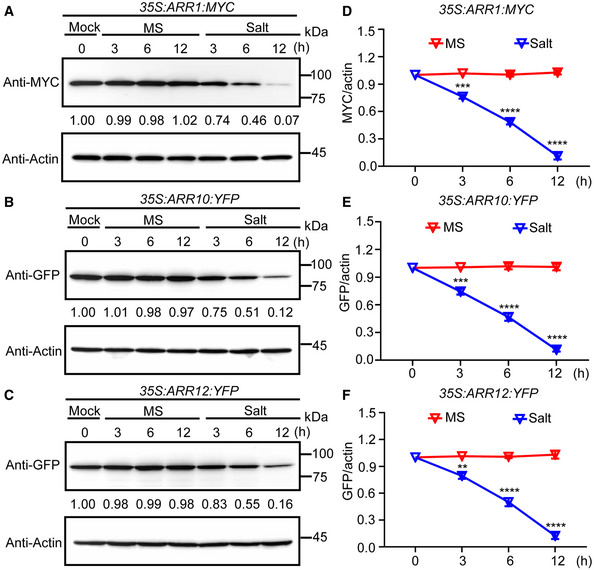
Figure 1. The protein levels of ARR1/10/12 decrease under salt treatment in a time‐dependent manner.
-
A–CARR1/10/12 protein levels were examined by western blot when 5‐day‐old seedlings of 35S:ARR1:MYC, 35S:ARR10:YFP, and 35S:ARR12:YFP were treated with or without 200 mM NaCl for the indicated time points. Actin was used as the internal reference. The relative intensity of band detected by anti‐MYC or anti‐GFP antibody to that by anti‐Actin antibody without treatment was set to 1.0.
-
D–FQuantification analysis of the protein levels of ARR1/10/12 in (A–C).
Data information: In (D–F), the statistical analyses of the related density of western blotting bands were performed using ImageJ software based on three independent biological replicates. Values are means ± SD (n = 3). **, ***, and **** indicate significant difference to the corresponding controls with P < 0.01, P < 0.001, and P < 0.0001, respectively (Student's t‐test).
Source data are available online for this figure.
ARR1/10/12 interact with stress‐activated kinases MPK3 and MPK6
To address how salinity stress reduces ARR1/10/12 protein levels, we performed a yeast two hybrid screen to identify protein interactors using ARR12 as a bait. Using this approach, we found that the stress‐activated mitogen‐activated protein kinases MPK3/6 (Teige et al, 2004; Colcombet & Hirt, 2008; Liu et al, 2010; Rodriguez et al, 2010; Kim et al, 2011; Li et al, 2017) interacted with ARR12 (Table EV1). MPK signaling modules play key regulatory roles in plant development and various stress responses (Danquah et al, 2014; Xu & Zhang, 2015; de Zelicourt et al, 2016; Bigeard & Hirt, 2018). Our data suggest a crosstalk between the MPK signaling modules and the cytokinin signaling during the response to salt stress.
Given that MPK4 belongs to the closest homolog of MPK3/6 and cooperates with MPK3/6 to participate in plant salt or cold stress response (Teige et al, 2004; Ichimura et al, 2010; Yang et al, 2010; Kim et al, 2011; Kim et al, 2012). We performed yeast two hybrid to reexamine the interactions between MPK3/4/6 and ARR1/10/12, and found that, consistent with the yeast two‐hybrid screening, MPK3/6 interacted with ARR12 but not ARR1/10, and MPK4 failed to interact with ARR1/10/12 (Fig 2A). Then, we performed bimolecular fluorescence complementation (BiFC) assays and found that ARR1/10/12‐YFPC all exhibited a strong yellow fluorescent protein (YFP) fluorescence in Arabidopsis mesophyll protoplasts transiently co‐expressing MPK3/6‐YFPN, but not in those cells transformed with empty vector controls or MPK4‐YFPN (Fig 2B and Appendix Fig S8), indicating that ARR1/10/12 specifically interact with MPK3/6 but not MPK4 in planta. In addition, we also found that MPK3/6‐YFP but not MPK4‐YFP can be co‐immunoprecipitated along with ARR1/10/12‐MYC from Arabidopsis protoplasts (Fig 2C). We also identified the functional domains of ARR1/10/12 required for interaction with MPK3/6. According to the UniProt website (http://uniprot.org), ARR1/10/12 contain a response regulatory domain (DDK), a DNA‐binding domain (B‐motif) in the N terminus, and a glutamine‐rich region (Q rich) for the transactivation activity at the C terminus (Appendix Fig S9A). Combined results of yeast two hybrid analysis and a coimmunoprecipitation (CoIP) assay indicated that the glutamine‐rich region (Q rich) responsible for the transactivation activity of ARR1/10/12 was necessary and sufficient to sustain the interaction with MPK3/6 (Appendix Fig S9B–E). Together, these results suggest that ARR1/10/12 interact with MPK3/6.
Figure 2. MPK3/6 interact with ARR1/10/12.
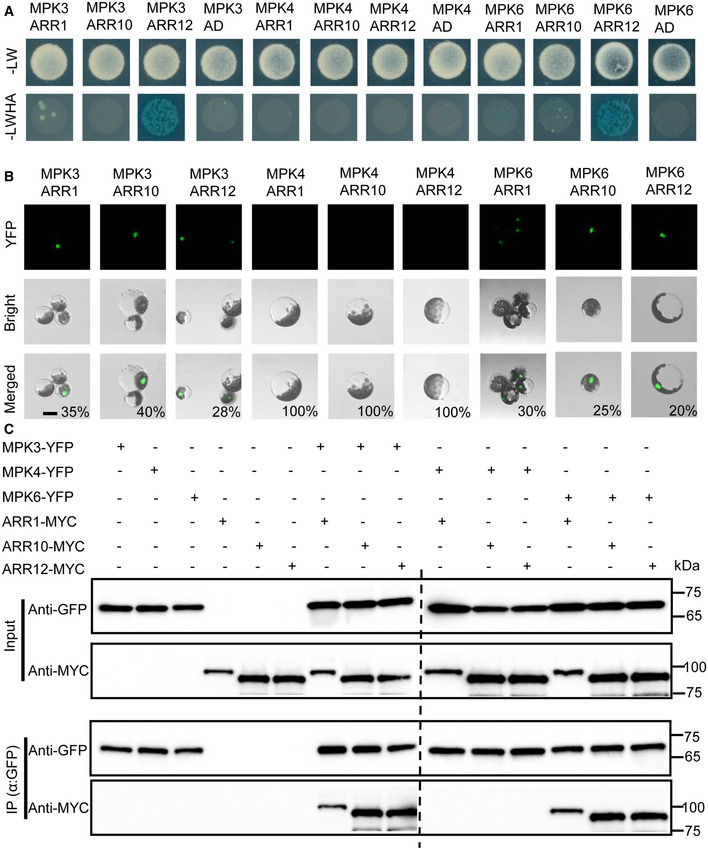
- Yeast two hybrid analysis showing the interaction between MPK3/6 and ARR12. Yeast cells were plated on selection medium: SD‐LW (‐Leu/Trp) or SD‐LWHA (‐Leu/Trp/His/Ade) supplemented with AbA (Aureobasidin A) and X‐α‐gal (5‐bromo‐4‐chloro‐3‐indolyl‐b‐d‐galactopyranoside). MPK3/4/6 coding sequences were fused with the GAL4 DNA‐binding domain (Bait proteins), and ARR1/10/12 coding sequences were fused with the GAL4 activation domain (Prey proteins).
- BiFC analysis of interaction between MPK3/4/6 and ARR1/10/12. Partial yellow fluorescence protein (YFP) constructs were fused to MPK3/4/6 or ARR1/10/12. The fusion constructs were co‐expressed transiently in Arabidopsis protoplasts cells. The interaction between MPK3/4/6 and ARR1/10/12 were visualized by fluorescence microscopy. The percentages labeled in the images mean the probability of present cell in cells observed. Scale bar, 50 μm.
- In vivo coimmunoprecipitation assay of MPK3/4/6 and ARR1/10/12. MPK3/4/6‐YFP and ARR1/10/12‐MYC were co‐expressed in Arabidopsis protoplast cells. Protein extracts (Input) were immunoprecipitated with GFP Trap magnetic agarose beads. Immunoblots were performed with anti‐MYC antibody to detect ARR1/10/12 and with anti‐GFP antibody to detect MPK3/4/6.
Source data are available online for this figure.
Salt stress induces MPK3/6‐dependent phosphorylation of ARR1/10/12
Given that MPK3/6 are known to be activated by salt stress (Ichimura et al, 2000; Droillard et al, 2002; Teige et al, 2004; Kim et al, 2011; Kim et al, 2012) and our observations that these kinases interact with ARR1/10/12, we hypothesized that salt stress triggers MPK3/6‐dependent phosphorylation of ARR1/10/12. To test this hypothesis, we first conducted in vitro phosphorylation assays to test whether ARR1/10/12 could be phosphorylated by MPK3/6. As shown in Fig 3A–C, ARR1/10/12 were specifically and directly phosphorylated by MPK3/6 in the presence of MKK5DD, which is a constitutively active form of MKK5 that activates MPK3/6 (Ren et al, 2002; Liu & Zhang, 2004). To investigate the potential MPK3/6 phosphorylation sites in ARR1/10/12, we performed a liquid chromatography–tandem mass spectrometry (LC‐MS/MS) analysis on ARR1/10/12‐His recombinant proteins that were phosphorylated by MPK3/6 in vitro. The LC‐MS/MS analysis identified Thr553 in ARR1 as a MPK6 phosphorylation site (Appendix Fig S10A). Thr552 or Thr553 in ARR1 was identified as a MPK3 candidate phosphorylation site, in which, a y11 fragment ion with phosphorylation modification (TTPSYDMFTTR) was detected, causing the failure to distinguish the exact phosphorylation site between Thr552 and Thr553 (Appendix Fig S10B). Ser322 or Ser323 in ARR12 was identified as a MPK6 candidate phosphorylation site, in which, a b4 fragment ion with phosphorylation modification (SSPPAGMFLQNQTDIGK) was detected, causing the failure to distinguish the exact phosphorylation site between Ser322 and Ser323 (AppendixFig S10C). However, no phosphorylation site was identified in ARR10 by MPK3 or MPK6 and also ARR12 by MPK3, since the corresponding peptides were not detected by the LC‐MS/MS analysis (Dataset EV1). Furthermore, we also analyzed the amino acid sequence of ARR1/10/12 and found two potential MPK phosphorylation sites (Thr277, Thr553) in ARR1, three potential MPK phosphorylation sites (Ser356, Ser383, and Ser426) in ARR10, and five potential MPK phosphorylation sites (Thr312, Ser323, Thr419, Ser462, and Ser494) in ARR12 based on bioinformatics predictions (http://kinasephos.mbc.nctu.edu.tw) and previous reports (Sharrocks et al, 2000; Enslen & Davis, 2012). We therefore mutated these sites from Ser or Thr to Ala (to mimic the non‐phosphorylated form) and performed in vitro phosphorylation assays with these mutant ARR1/10/12 proteins. The phosphorylation of ARR1/10/12 by MPK3/6 was significantly reduced when we used ARR1T553A, ARR10S383A, and ARR12S323A as substrates (Fig 3A–C). Collectively, these results indicated that T553 is the major residue of ARR1 phosphorylated by MPK3/6, S383 is the major residue of ARR10 phosphorylated by MPK3/6, and S323 is the major residue of ARR12 phosphorylated by MPK3/6.
Figure 3. MPK3/6‐mediated ARR1/10/12 phosphorylation in vitro .
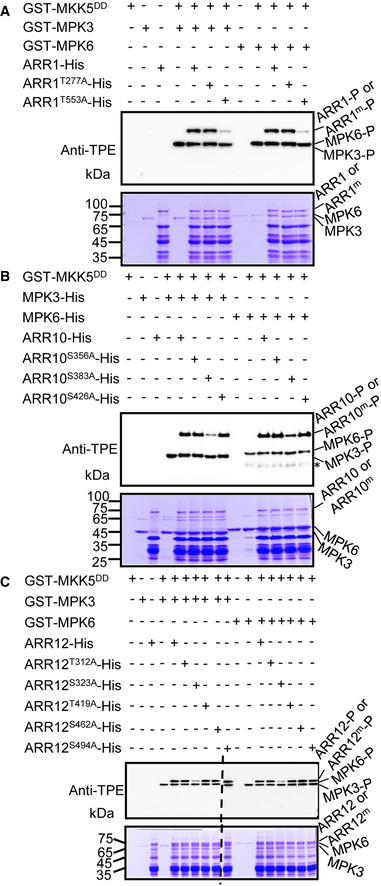
-
A–CThiophosphorylation assays of wild‐type and mutated forms ARR1/10/12 by MPK3/6 in vitro. Potential phosphorylated Ser residues or Thr residues of ARR1, ARR10, and ARR12 were mutated to Ala (ARR1m/ARR10m/ARR12m). Recombinant ARR1/10/12‐His and ARR1m/10m/12m‐His proteins were incubated with MPK3 and MPK6 which were activated by MKK5DD. Phosphorylated ARR1/10/12, ARR1m/10m/12m, and MPK3/6 were visualized by an anti‐thiophosphate ester‐specific antibody (α‐TPE) (upper panel). Asterisk in (B) indicates non‐specific bands. Recombinant MKK5DD, MPK3, MPK6, ARR1/10/12, and ARR1m/10m/12m were detected by coomassie brilliant blue (CBB) staining as loading controls (bottom panel).
Data information: Two independent biological repeats showed similar results.
Source data are available online for this figure.
Then, we performed phosphorylation assays in vivo by separating proteins in a phos‐tag gel followed by immunoblot analysis using 35S:ARR1:MYC, 35S:ARR10:YFP, and 35S:ARR12:YFP transgenic plants (Bethke et al, 2009). As expected, a time‐dependent increase in phosphorylation of these ARR fusion proteins upon salt treatment was observed, which was abolished by calf intestinal alkaline phosphatase (CIAP) treatment (Fig 4A–C). To further confirm that salt‐induced phosphorylation of ARR1/10/12 requires MPK3/6, we generated 35S:ARR1:MYC, 35S:ARR10:YFP, and 35S:ARR12:YFP transgenes in the mpk3/6 double mutant containing a PMPK3:MPK3TG transgene (abbreviated as MPK3SR) (Xu et al, 2014; Su et al, 2017). The small molecule 4‐amino‐1‐tert‐butyl‐3‐(1′‐naphthyl) pyrazolo[3,4‐d]pyrimidine (NA‐PP1) inhibits MPK3TG, therefore NA‐PP1‐treated MPK3SR plants mimic the mpk3/6 double mutant. The result showed that the salt‐induced phosphorylation of ARR1/10/12 was strongly reduced in MPK3SR background (Fig 4A–C). Similar results were observed in another conditional mpk3/6 double mutant, named MPK6SR (mpk3/6 PMPK6:MPK6YG ) (Xu et al, 2014; Su et al, 2017) treated with NA‐PP1 (Appendix Fig S11A–C). Overall, these results suggest that MPK3/6 directly phosphorylate ARR1/10/12 in response to salt stress.
Figure 4. MPK3/6‐mediated ARR1/10/12 phosphorylation in vivo .
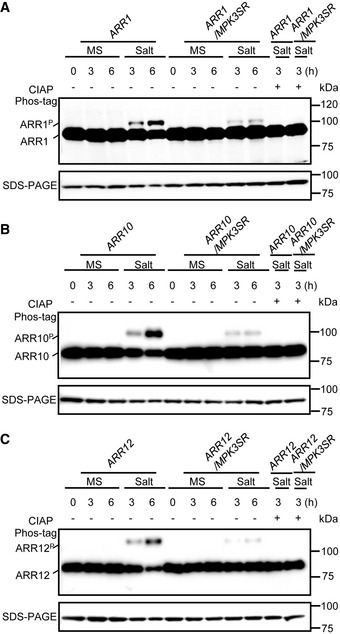
-
A–CImmunoblot analysis with anti‐MYC antibody or anti‐GFP antibody showing the upshift of ARR1, ARR10, and ARR12 in a phos‐tag gel. The phosphorylated ARR1, ARR10, and ARR12 were abolished after treatment with calf intestinal alkaline phosphatase (CIAP). Seven‐day‐old seedlings of 35S:ARR1:MYC (A), 35S:ARR10:YFP (B), and 35S:ARR12:YFP (C) in Col and MPK3SR backgrounds were treated with or without 200 mM NaCl for indicated time points. Total proteins were extracted and subjected to immunoblot analysis. ARR1, ARR10, and ARR12 proteins separated on SDS–PAGE gel without phos‐tag were detected as loading controls.
Data information: Similar results were obtained with three biological repeats.
MPK3/6 mediate salt stress signals to promote ARR1/10/12 degradation
To investigate whether MPK3/6‐dependent phosphorylation of ARR1/10/12 correlate with the salt‐induced reduction in protein levels, we examined how salt treatment affects the total levels of ARR1/10/12 in Col‐0 and NA‐PP1‐treated MPK3SR or MPK6SR. Salt stress led to a gradual, time‐dependent reduction of ARR1/10/12 proteins in Col‐0, but not in the NA‐PP1‐treated MPK3SR or MPK6SR seedlings (Fig 5A–I and Appendix Fig S12A–I). Thus, MPK3/6 activity is required for the salt‐induced decrease in ARR1/10/12 protein levels. To confirm MPK3/6‐dependent regulation of ARR1/10/12, we transiently co‐expressed these proteins in Arabidopsis protoplast cells. Co‐expression of MPK3/6 with MKK5DD, resulted in substantially reduced levels of ARR1/10/12 in protoplasts (Appendix Fig S13A–C).
Figure 5. Salt stress‐induced ARR1/10/12 degradation is dependent on MPK3/6.
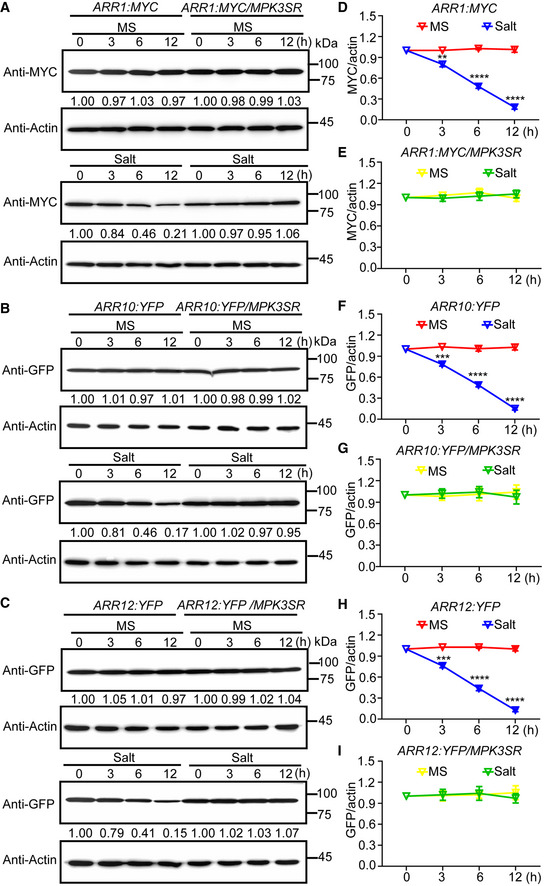
-
A–CImmunoblot analysis the effects of MPK3/6 on salt stress‐induced ARR1/10/12 protein degradation. Seven‐day‐old seedlings of 35S:ARR1:MYC, 35S:ARR10:YFP, and 35S:ARR12:YFP in Col‐0 and MPK3SR backgrounds were treated with or without 200 mM NaCl for indicated time points. Actin was used as the internal reference. The relative intensity of band detected by anti‐MYC or anti‐GFP antibody to that by anti‐Actin antibody without treatment was set to 1.0.
-
D–IQuantification analysis of MPK3/6 effect on salt stress‐induced ARR1/10/12 protein degradation in (A–C).
Data information: In (D–I), the statistical analyses of the related density of Western blotting bands were performed using ImageJ software based on three independent biological replicates. Values are means ± SD (n = 3). **, ***, and **** indicate significant difference to the corresponding controls with P < 0.01, P < 0.001, and P < 0.0001, respectively (Student's t‐test).
To further examine whether MPK3/6 promote ARR1/10/12 degradation dependent on the phosphorylation modification, we generated a subset of transgenic plants overexpressing non‐phosphorylatable forms of ARR1/10/12 (ARR1T553A, ARR10S383A, and ARR12S323A) under the 35S promoter, and selected representative transgenic plants which showed comparable protein expression levels to corresponding wild‐type ARR1/10/12 for protein degradation analysis (Appendix Fig S14A–C). Treatment with the protein synthesis inhibitor cycloheximide (CHX) led to significantly reduced protein levels of ARR1/10/12, which was partially restored by the co‐treatment with the 26S proteasome inhibitor MG132 (Fig EV3A–C). However, ARR1T553A, ARR10S383A, and ARR12S323A were more stable compared with the corresponding normal ARR1/10/12 controls under the same treatment (Fig EV3A–C). These data suggest that MPK3/6‐mediated phosphorylation destabilizes ARR1/10/12.
Figure EV3. Dephosphorylated form of ARR1/10/12 have a higher protein stability.
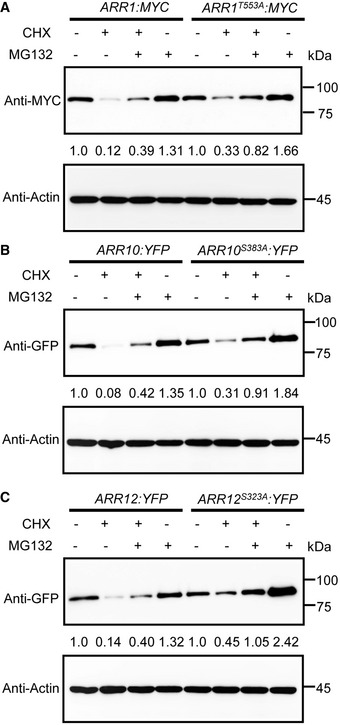
-
A–CImmunoblot analysis the stability of wild‐type and dephosphorylated forms of ARR1, ARR10, and ARR12 proteins in vivo. Five‐day‐old 35S:ARR1:MYC, 35S:ARR10:YFP, 35S:ARR12:YFP, 35S:ARR1T553A:MYC, 35S:ARR10S383A:YFP, and 35S:ARR12S323A:YFP transgenic plants were treated with 200 μM cycloheximide (CHX), 50 μM MG132, or 200 μM CHX, together with 50 μM MG132 for 3 h. ARR1, ARR10, and ARR12 were detected with anti‐MYC or anti‐GFP antibody. Actin was used as a control.
Data information: Similar results were obtained with three biological repeats.
To investigate whether phosphorylation of ARR1/10/12 is required for their degradation in response to salt stress, we examined the protein stability of wild‐type ARR1/10/12 and non‐phosphorylatable forms of ARR1/10/12 proteins under salt stress. As shown in Fig 6A–I, unlike normal ARR1/10/12, ARR1T553A, ARR10S383A, and ARR12S323A were stably maintained after salt treatment, indicating that MPK3/6‐mediated ARR1/10/12 phosphorylation enhances ARR1/10/12 protein degradation under salt stress.
Figure 6. MPK3/6 promote ARR1/10/12 protein degradation under salt stress.
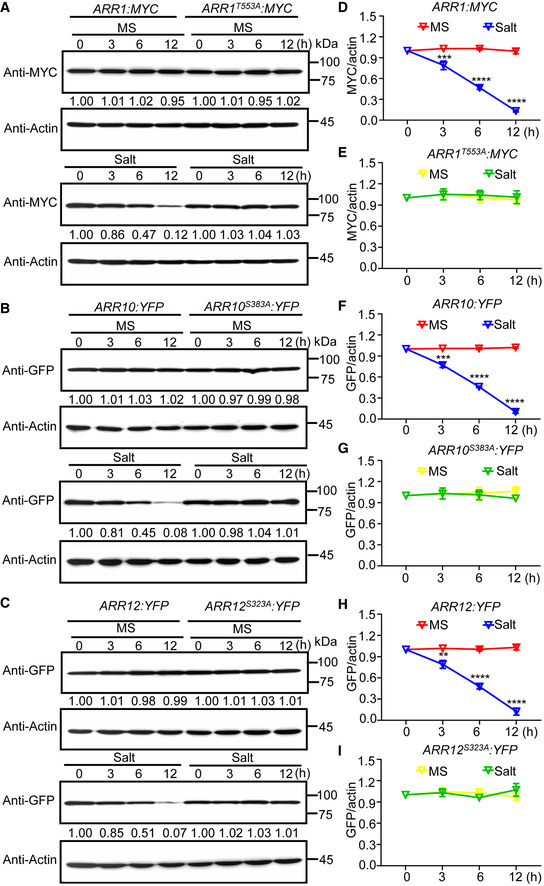
-
A–CImmunoblot analysis stability of ARR1/10/12 and ARR1T553A/10S383A/12S323A proteins under salt stress. Seven‐day‐old seedlings of 35S:ARR1:MYC, 35S:ARR10:YFP, 35S:ARR12:YFP, 35S:ARR1T553A:MYC, 35S:ARR10S383A:YFP, and 35S:ARR12S323A:YFP were treated with or without 200 mM NaCl for indicated time points. Actin was used as the internal reference. The relative intensity of band detected by anti‐MYC or anti‐GFP antibody to that by anti‐Actin antibody without treatment was set to 1.0.
-
D–IQuantification analysis of stability of ARR1/10/12 and ARR1T553A/10S383A/12S323A proteins under salt stress in (A–C).
Data information: In (D–I), the statistical analyses of the related density of Western blotting bands were performed using ImageJ software based on three independent biological replicates. Values are means ± SD (n = 3). **, ***, and **** indicate significant difference to the corresponding controls with P < 0.01, P < 0.001, and P < 0.0001, respectively (Student's t‐test).
Source data are available online for this figure.
To further determine the biological significance of MPK3/6‐mediated ARR1/10/12 phosphorylation in salt stress response, we also generated a subset of transgenic plants overexpressing phospho‐mimic forms of ARR1/10/12 (ARR1T553D, ARR10S383D, and ARR12S323D) under the 35S promoter and selected representative transgenic plants which showed comparable protein expression levels to wild‐type and non‐phosphorylatable forms of ARR1/10/12 for further analysis (Appendix Fig S14A–C). We compared the salt stress response of Col‐0, over‐expressors of wild‐type ARR1/10/12, and over‐expressors of non‐phosphorylatable forms of ARR1/10/12 or phospho‐mimic forms of ARR1/10/12. As shown in Figs 7A–D, 8A–D, and 9A–D, consistent with our previous observations, over‐expressors of wild‐type ARR1/10/12 all showed hypersensitivity to salt stress compared to Col‐0. Moreover, transgenic plants overexpressing non‐phosphorylatable forms of ARR1/10/12 (ARR1T553A, ARR10S383A, and ARR12S323A) displayed increased sensitivity to salt stress compared to corresponding transgenic plants overexpressing wild‐type ARR1/10/12, whereas over‐expressors of phospho‐mimic forms of ARR1/10/12 behaved like wild‐type Col‐0 in response to salt stress. These results collectively suggest that MPK3/6‐mediated ARR1/10/12 phosphorylation decreases the ability of ARR1/10/12 proteins in repressing salt tolerance.
Figure 7. MPK3/6‐mediated ARR1 phosphorylation decreases the ability of ARR1 protein in repressing salt tolerance.
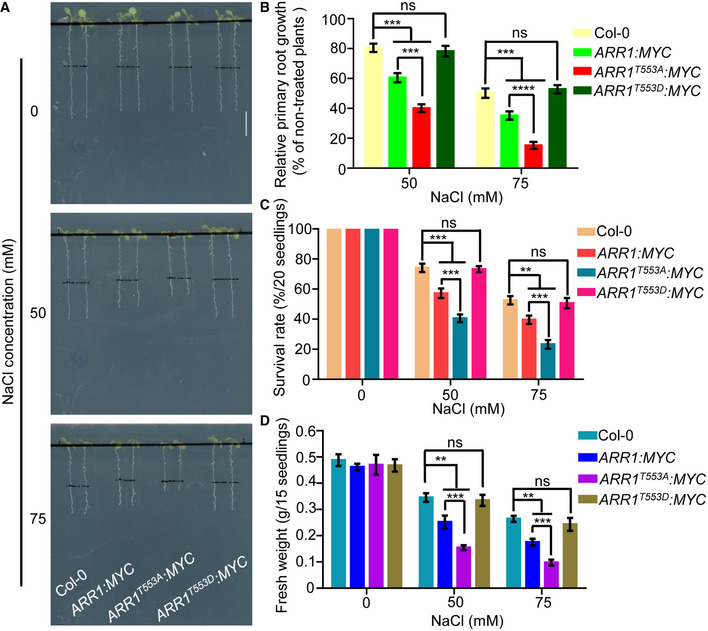
-
A–DFour‐day‐old seedlings of 35S:ARR1:MYC, 35S:ARR1T553A:MYC, and 35S:ARR1T553D:MYC were transferred to 1/2 MS medium supplemented with or without 50 mM or 75 mM NaCl. After 3 days of salt treatment, plant phenotypes (A) and relative primary root lengths (B) were determined. After 10 days of salt treatment, changes in survival rates (C) and fresh weights (D) were examined. Scale bar, 1 cm.
Data information: In (B–D), data are means of three biological replicates ± SD (n = 60 for (B), and n = 12 for (C) and (D)). ns indicates no significant difference to the corresponding controls. **, ***, and **** indicate significant difference to the corresponding controls with P < 0.01, P < 0.001, and P < 0.0001, respectively (Student's t‐test).
Figure 8. MPK3/6‐mediated ARR10 phosphorylation decreases the ability of ARR10 protein in repressing salt tolerance.
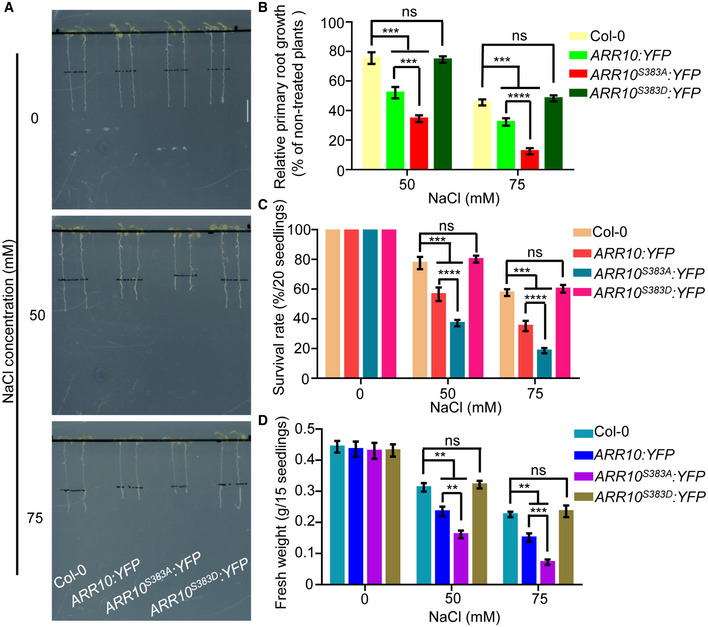
-
A–DFour‐day‐old seedlings of 35S:ARR10:YFP, 35S:ARR10S383A:YFP, and 35S:ARR10S383D:YFP were transferred to 1/2 MS medium supplemented with or without 50 or 75 mM NaCl. After 3 days of salt treatment, plant phenotypes (A) and relative primary root lengths (B) were determined. After 10 days of salt treatment, changes in survival rates (C) and fresh weights (D) were examined. Scale bar, 1 cm.
Data information: In (B–D), data are means of three biological replicates ± SD (n = 60 for (B), and n = 12 for (C) and (D)). ns indicates no significant difference to the corresponding controls. **, ***, and **** indicate significant difference to the corresponding controls with P < 0.01, P < 0.001, and P < 0.0001, respectively (Student's t‐test).
Figure 9. MPK3/6‐mediated ARR12 phosphorylation decreases the ability of ARR12 protein in repressing salt tolerance.
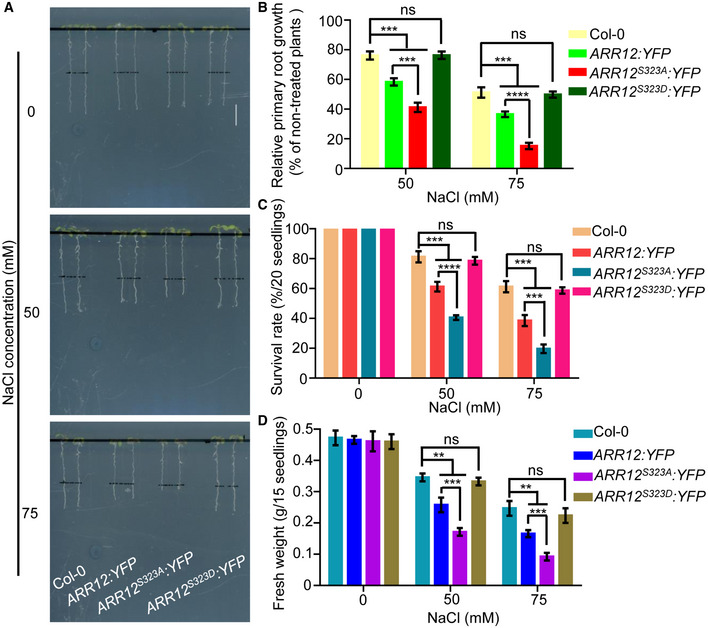
-
A–DFour‐day‐old seedlings of 35S:ARR12:YFP, 35S:ARR12S323A:YFP, and 35S:ARR12S323D:YFP were transferred to 1/2 MS medium supplemented with or without 50 mM or 75 mM NaCl. After 3 days of salt treatment, plant phenotypes (A) and relative primary root lengths (B) were determined. After 10 days of salt treatment, changes in survival rates (C) and fresh weights (D) were examined. Scale bar, 1 cm.
Data information: In (B–D), data are means of three biological replicates ± SD (n = 60 for (B), and n = 12 for (C) and (D)). ns indicates no significant difference to the corresponding controls. **, ***, and **** indicate significant difference to the corresponding controls with P < 0.01, P < 0.001, and P < 0.0001, respectively (Student's t‐test).
Taken together, our data suggest that salinity stress results in MPK3/6‐mediated phosphorylation and degradation/destabilization of ARR1/10/12 proteins.
MPK3/6 act upstream of ARR1/10/12 in the response to salt stress
To investigate whether the interaction between MPK3/6 and ARR1/10/12 affects the responses of plants to salt stress, we first examined the role of MPK3/6 in this stress response. After cultured on 0.5 μM NA‐PP1‐containing 1/2 MS medium for 2 d, Col‐0, mpk3‐1, mpk6‐3, and MPK3SR were then transferred to 0.5 μM NA‐PP1‐containing 1/2 MS medium supplemented with or without 50 or 75 mM NaCl. We found that mpk3‐1, mpk6‐3, and Col‐0 seedlings displayed similar primary root lengths, survival rates, and fresh weights with salt treatment (Appendix Fig S15A–D). However, NA‐PP1‐treated MPK3SR seedlings treated with salt showed significantly enhanced root‐growth inhibition, and their survival rates and fresh weights were strongly reduced (Appendix Fig S15A–D). Moreover, the expression of the salt stress responsive genes RD29B, MYB15, and ZAT10 was significantly reduced in NA‐PP1‐treated MPK3SR seedlings compared to the Col‐0 control seedlings after salt treatment (Appendix Fig S2A–C). Similar results were observed in the MPK6SR seedlings after NA‐PP1 treatment (Appendix Figs S16A–D and S17A–C). These results again confirmed the positive role of MPK3/6 in plant salt tolerance (Yu et al, 2010; Pitzschke et al, 2014).
Additionally, we examined the impact of salt stress on a dexamethasone (DEX)‐inducible MKK5DD transgenic line, which ectopically activates MPK3/6 upon the inducible expression of the gain‐of‐function MKK5DD . Compared to Col‐0, the MKK5DD transgenic seedlings showed significantly less root‐growth inhibition, as well as improved survival rates and fresh weights in salt condition (Appendix Fig S18A–C), consistent with a positive role for MKK5 in plant salt tolerance (Xing et al, 2015). Accordingly, the salt‐induced expression of RD29B, MYB15, and ZAT10 was significantly increased in MKK5DD compared to Col‐0 (Appendix Fig S2A–C). In summary, MKK5‐MPK3/6 signaling module positively regulates plant salt tolerance.
To explore the genetic relationship between MPK3/6 and ARR1/10/12 in the plant response to salinity stress, we generated a MPK3SR arr1/12 quadruple mutant. The hypersensitive salt response of the NA‐PP1‐treated MPK3SR mutant was highly relieved in the arr1/12 background, inferred from primary root length, survival rates, and fresh weights (Fig 10A–D). Consistently, the salt‐induced expression of RD29B, MYB15, and ZAT10 was higher in MPK3SR arr1/12 than that in MPK3SR under salt treatment (Appendix Fig S19A–C). Collectively, the genetic data suggest that MPK3/6 act upstream of ARR1/10/12 to promote the plant adaption to high‐salinity.
Figure 10. MPK3/6 act upstream of ARR1/10/12 in response to salt stress.
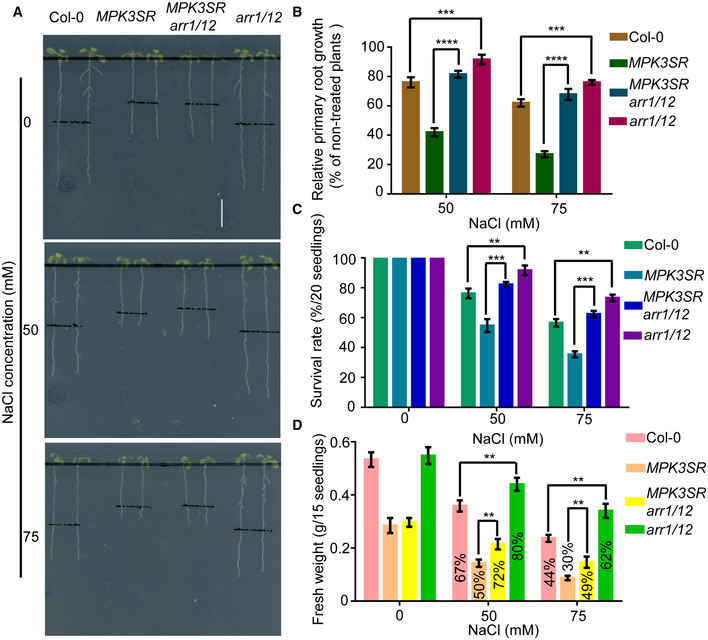
-
A–DThree‐day‐old seedlings of Col‐0, MPK3SR, MPK3SRarr1/12, and arr1/12 were transferred to 0.5 μM NA‐PP1‐containing 1/2 MS medium for 2 days to inhibit MPK3TG in MPK3SR and MPK3SRarr1/12, and then, the plants were transferred to 0.2 μM NA‐PP1‐containing 1/2 MS medium supplemented with or without 50 mM or 75 mM NaCl. Pictures were taken, and the elongated root length was determined 3 days later (A–B). Changes in survival rates (C) and fresh weights (D) were examined 10 days later. The fresh weight of NaCl treated seedlings/fresh weight of NaCl untreated seedlings ratios were labeled in the columns in (D). Scale bar, 1 cm.
Data information: In (B–D), data are means of three biological replicates ± SD (n = 60 for (B), and n = 12 for (C) and (D)). **, ***, and **** indicate significant difference to the corresponding controls with P < 0.01, P < 0.001, and P < 0.0001, respectively (Student's t‐test).
Finally, to investigate whether the stability of ARR1/10/12 or cytokinin signaling were also influenced by MPK3/6 under normal (non‐stressed) conditions, we treated Col‐0, MPK3SR, and arr1/12 with NA‐PP1 and 6‐BA. Consistent with the previous results (Mason et al, 2005), arr1/12 showed significantly reduced sensitivity to 6‐BA, whereas MPK3SR displayed a similar sensitivity to 6‐BA compared to WT (Fig EV4A and B). Consistent with this observation, cytokinin‐responsive genes (ARR5/6) were also similarly induced in MPK3SR and Col‐0 upon 6‐BA treatment (Fig EV4C and D). These results suggest that MPK3/6 are not involved in the regulation of cytokinin signaling under normal growth condition. Taken together, this study supports a model whereby salt stress activates an MKK5‐MPK3/6‐ARR1/10/12 signaling module to promote salt tolerance (Fig 11). Under normal growth conditions, MPK3/6 are less active and ARR1/10/12 maintains optimal cytokinin response to control plant growth and development. Exposing plants to high salinity activates MPK3/6, which phosphorylate and destabilize ARR1/10/12, eventually repressing the cytokinin response and leading to growth inhibition.
Figure EV4. Cytokinin signal transduction is independent on mitogen‐activated protein kinases MPK3/6 under normal growth conditions.
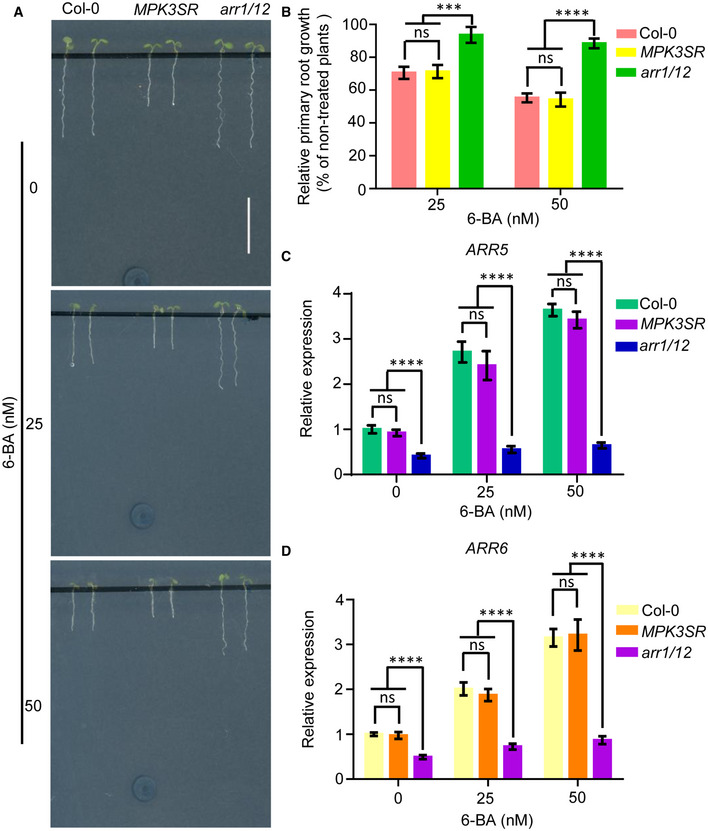
-
A, BPrimary root phenotype (A) and primary root length (B) of Col‐0, MPK3SR, and arr1/12 seedlings grown on 0.25 μM NA‐PP1‐containing 1/2 MS medium supplemented with or without 25 or 50 nM 6‐BA for 5 days. Scale bar, 1 cm.
-
C, DTranscript levels of ARR5 and ARR6 in Col‐0, MPK3SR, and arr1/12 were examined by qRT–PCR. Gene expression levels in wild‐type control plants without 6‐BA treatment were set to 1.0. Actin2 gene and UBQ1 gene were analyzed as internal controls.
Data information: In (B), data are means of three biological replicates ± SD (n = 60). In (C), and (D), values are means ± SD of three biological replicates. ns indicates no significant difference to the corresponding controls. *** and **** indicate significant difference to the corresponding controls with P < 0.001 and P < 0.0001, respectively (Student's t‐test).
Figure 11. Salt stress‐activated MKK5‐MPK3/6‐ARR1/10/12 signaling module to mediate plant growth response to salinity.
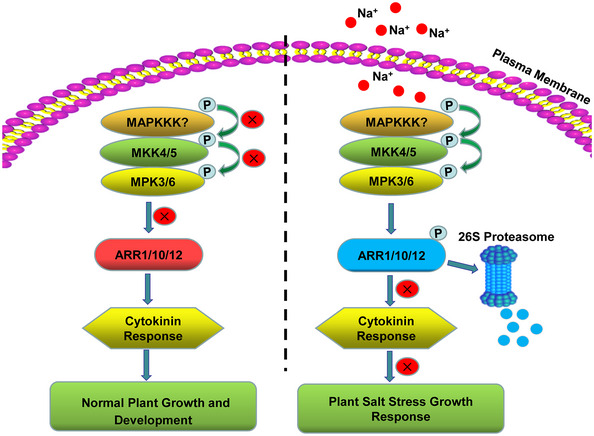
Under normal growth conditions, MPK3/6 are less active and ARR1/10/12 maintains optimal cytokinin response to control plant growth and development. Exposing plants to high salinity activates MPK3/6, which interact with and phosphorylate ARR1/10/12. Phosphorylated ARR1/10/12 are degraded through the 26S proteasome‐dependent pathway, eventually repressing the cytokinin response and leading to growth inhibition. X represents inhibited signaling or process.
Discussion
Cytokinin biosynthetic mutants and signaling mutants have a higher tolerance to salinity stress (Tran et al, 2007; Mason et al, 2010; Nishiyama et al, 2011), indicating that cytokinin negatively regulates plant salt tolerance. Previous studies showed that salt stress alters the transcription of genes that are involved in cytokinin signaling or homeostasis (Tran et al, 2007; Mason et al, 2010; Nishiyama et al, 2011). Here, we provide evidence that salt stress reduces the levels of cytokinin type‐B transcription factors ARR1/10/12, due to a decreased protein stability. As stated in the latest review (Yu et al, 2020), cytokinin promotes and maintains plant growth under normal growth conditions, but its role in salt stress is slightly different. Under mild salt stress, plants have sufficient capacity to cope with this situation and maintain growth, but excessive stress can seriously disrupt or even destroy the plant defense system. Plants delicately slow down their growth to cope with various stress. Cytokinin or auxin induced rapid plant growth will not be helpful to adaptive growth under high‐salinity, and the self‐sacrifice of cytokinin (reduction of synthesis in response to salt stress) has been reported as an important compromised strategy (Nishiyama et al, 2011). This study suggest that, in response to salt stress, besides the synthesis of cytokinin is regulated (Nishiyama et al, 2011), cytokinin signaling is also finely regulated through MPK3/6‐mediated phosphorylation and degradation of ARR1/10/12.
In plants, including all crops, there have been many reports on the relationship between “MPK and salt stress” (Xiong & Yang, 2003; Gu et al, 2010; Wang et al, 2014; Zhu et al, 2020) or “Cytokinin and salt stress” (Cortleven et al, 2019; Wang et al, 2019; Liu et al, 2020b), but there were no reports about the relationship between MPK and cytokinin in plant salt response. The family members of MPK vary in various plants, but their functions are generally similar. The MPK3/6‐ARR1/10/12 regulatory modules revealed in Arabidopsis might be also conserved in other plants including crops, given the high homology between MPKs or ARRs in different plants. Therefore, this study may provide important reference value and important candidate genes for the genetic improvement of crops in salt stress adaptation.
Consistent with previous work, we confirmed a positive role of MPK3/6 in plant salt tolerance. Furthermore, we demonstrated that MKK5‐mediated activation of MPK3/6 can improve salt tolerance by decreasing ARR1/10/12 proteins stability. It will be of interest to investigate whether other MKKs, including MKK2, MKK7, and MKK9, which act upstream of MPK3/6 in plant salt stress responses (Teige et al, 2004; Shen et al, 2019) participate in this pathway. In addition, other MPKs besides MPK3/6 are reported to regulate plant salt stress responses (Xu et al, 2008). Whether other MPKs or even MKKs can directly interact with and phosphorylate ARR1/10/12 remains unknown. It will also be of interest to determine whether MPK3/6 regulate proteins involved in cytokinin biosynthesis and metabolism, and the potential crosstalk between cytokinin signaling and other MPK signaling cascades during the response to salt stress (Rodriguez et al, 2010; Li et al, 2014; Zhu, 2016).
Xie et al (2018) and Zubo et al (2017) made use of chromatin immunoprecipitation sequencing (ChIP‐seq) to provide in vivo DNA‐binding site profiles for ARR1/10/12. Considering the negative role of ARR1/10/12 in salt stress and their nature as transcription activators, we examined the negative regulators of salt tolerance which might be potential binding targets of ARR1/10/12. By mining ChIP‐seq data, three negative regulators, EST1 (Liu et al, 2020a), ANT (Meng et al, 2015), and AtGSTU17 (Chen et al, 2012) were identified and selected for further investigation. Next, we examined whether EST1, ANT, and AtGSTU17 are direct ARR1/10/12 binding targets and how phosphorylation of ARR1/10/12 by MPK3/6 affects the expression of their downstream targets. Compared with WT, the expression of EST1, ANT, and AtGSTU17 genes was significantly upregulated in transgenic plants overexpressing wild‐type, non‐phosphorylatable, and phospho‐mimic ARR1/10/12 without salt treatment (Fig EV5A–C). Furthermore, the expression of EST1, ANT, and AtGSTU17 was dramatically repressed by salt in both WT and transgenic plants overexpressing wild‐type or phospho‐mimic ARR1/10/12, and meanwhile, the repression of these genes in over‐expressors of phospho‐mimic ARR1/10/12 was more pronounced compared with over‐expressors of wild‐type ARR1/10/12 (Fig EV5A–C). However, the expression of EST1, ANT, and AtGSTU17 was weakly repressed upon salt treatment in over‐expressors of non‐phosphorylatable ARR1/10/12 (Fig EV5A–C). Collectively, these results suggest that EST1, ANT, and AtGSTU17 are direct ARR1/10/12 binding targets and phosphorylation of ARR1/10/12 by MPK3/6 negatively regulates the expression of these downstream targets under salt stress.
Figure EV5. Expression analysis of direct downstream targets of ARR1/10/12.
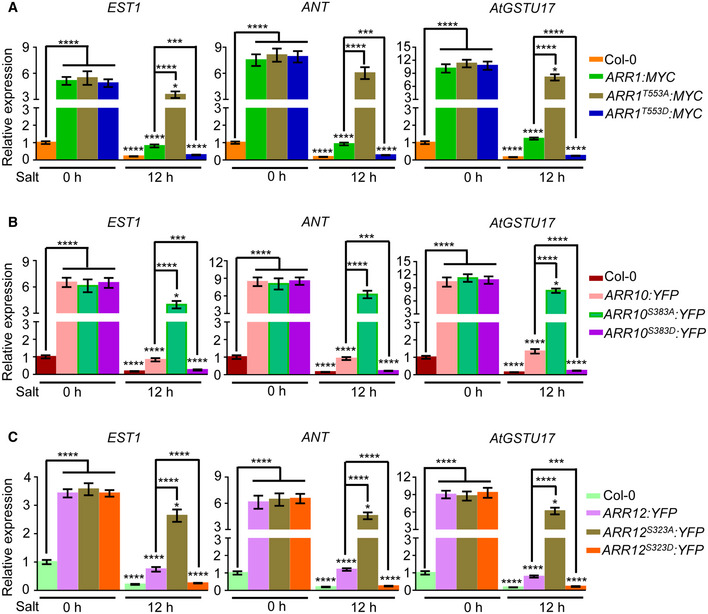
-
A–CThe relative expression of direct downstream targets of ARR1/10/12 such as EST1, ANT, and AtGSTU17 in 7‐day‐old seedlings treated with 200 mM NaCl for given time. Actin2 gene and UBQ1 gene were analyzed as internal controls. Gene expression levels in wild‐type control plants treated with NaCl for 0 h were set to 1.0.
Data information: Data shown are means ± SD of three biological replicates. *P < 0.05, ***P < 0.001, ****P < 0.0001 (Student's t‐test).
Previous studies have uncovered the molecular mechanism of EST1, ANT, and AtGSTU17 in negatively regulating salt tolerance (Chen et al, 2012; Meng et al, 2015; Liu et al, 2020a). The F‐box protein EST1 modulates salt tolerance in Arabidopsis by regulating plasma membrane Na+/H+ antiport activity (Liu et al, 2020a), ANT mediates salt tolerance through regulating the Salt Overly Sensitive (SOS) pathway (Meng et al, 2015), and AtGSTU17 was reported to regulate salt tolerance by affecting the levels of glutathione (GSH) and ABA (Chen et al, 2012). Since EST1, ANT, and AtGSTU17 are direct targets of ARR1/10/12, it will be interesting to study whether ARR1/10/12‐mediated cytokinin signaling regulates plant growth response to salt stress through directly regulating plasma membrane Na+/H+ antiport activity, the SOS signaling cascade or the levels of glutathione (GSH) and ABA.
Materials and Methods
Plant materials and growth conditions
All of the Arabidopsis mutants and/or transgenes used in this study are in the Col‐0 background; the following have been described elsewhere: ARR1p:ARR1g‐GFP (Meng et al, 2017), ARR10p:ARR10g‐GFP (Meng et al, 2017), ARR12p:ARR12g‐GFP (Meng et al, 2017), arr1/10 (Mason et al, 2005; Argyros et al, 2008), arr10/12 (Mason et al, 2005; Argyros et al, 2008), arr1/12 (Mason et al, 2005; Argyros et al, 2008), mpk3‐1 (Li et al, 2017), mpk6‐3 (Xu et al, 2008), MPK3SR (Xu et al, 2014; Su et al, 2017), MPK6SR (Xu et al, 2014; Su et al, 2017), MKK5DD (Ren et al, 2002; Liu & Zhang, 2004), and 35S:ARR1:MYC (Huang et al, 2018). MPK3SR was introduced into arr1/12 mutant background by genetic crosses. Harvested seeds were surface‐sterilized, planted on 1/2 Murashige and Skoog (MS) medium (Sigma‐Aldrich) with 0.8% agar, imbibed at 4°C for 3 days, and then the seeds were germinated vertically at 22°C under 16/8 h (light/dark) illumination.
Hormone treatment
To assay responses to cytokinin, seedlings were grown on 1/2 MS medium supplemented with or without 25 or 50 nM of 6‐BA. Relative primary root length was analyzed using Image J.
Salt treatment
For phenotype analysis of Col‐0, arr mutants and transgenic plants overexpressing wild‐type ARR1/10/12, non‐phosphorylated ARR1T553A/ARR10S383A/ARR12S323A, and phospho‐mimic ARR1T553D/ARR10S383D/ARR12S323D, 4‐day‐old seedlings were transferred to 1/2 MS medium supplemented with or without 50 mM NaCl and 75 mM NaCl. Three days later, primary root length was measured; 10 days later, survival rate and fresh weight were determined. For NA‐PP1 (TRC) experiments, 3‐day‐old seedlings were transferred to plates containing 0.5 μM NA‐PP1 and grown for 2 days, following salt treatment. For dexamethasone (DEX; Sigma‐Aldrich) experiments, 3 days old seedlings were transferred to plates containing 0.2 μM dexamethasone (DEX) and grown for days, following salt treatment. For protein content analysis, 5‐day‐old transgenic plants overexpressing wild‐type ARR1/10/12 or non‐phosphorylated ARR1T553A/ARR10S383A/ARR12S323A mutants were exposed to liquid MS medium with or without 200 mM NaCl for indicated time points; 5‐day‐old seedlings of ARR1p:ARR1g‐GFP, ARR10p:ARR10g‐GFP, and ARR12p:ARR12g‐GFP were transferred to 1/2 MS plates with or without 50 mM NaCl for 12 h.
Construction of transgenic plants
The construct harboring 35S:ARR1T553A:MYC and 35S:ARR1T553D:MYC were produced using the pSuper1300‐MYC vector (Huang et al, 2018). Constructs for expressing wild‐type, non‐phosphorylated, and phospho‐mimic ARR10/12, 35S:ARR10:YFP, 35S:ARR10S383A:YFP, 35S:ARR10S383D:YFP, 35S:ARR12:YFP, 35S:ARR12S323A:YFP, and 35S:ARR12S323D:YFP under the control of 35S promoter were produced using the pCHF3‐YFP vector (Wang et al, 2005; Yan et al, 2017). Transgenic plants were generated by the floral‐dipping method and screened with 50 μg/ml kanamycin on 1/2 MS medium (except for 35S:ARR1T553A:MYC and 35S:ARR1T553D:MYC which were screened with hygromycin). T3 homozygous lines were used for further study. Primers are listed in Table EV2.
Confocal microscopy
Confocal imaging of Arabidopsis seedlings was performed using a Zeiss LSM880 confocal microscope (Germany). Briefly, 5‐day‐old seedlings of ARR1p:ARR1g‐GFP, ARR10p:ARR10g‐GFP, and ARR12p:ARR12g‐GFP were transferred to plates with or without 50 mM NaCl for 12 h, and the root tips of GFP lines were then imaged in water supplemented with propidium iodide (PI, 10 mg/l). The excitation wavelengths of propidium iodide and green fluorescent protein (GFP) were 488 nm and 561 nm, respectively. The emission wavelengths of propidium iodide and green fluorescent protein (GFP) were 575 nm and 500–530 nm, respectively. Photographs of at least 15 seedlings with or without NaCl treatment were taken and analyzed. Similar results were obtained with three biological repeats. Quantitative measurement of GFP signal intensity was performed using ZEN 2 (blue edition).
Yeast two hybrid assay
Yeast two hybrid (Y2H) assay was carried out using Matchmaker Gold Yeast Two Hybrid System according to the manufacturer's protocol (Clontech). To prepare constructs for the yeast two hybrid assay, the coding sequences of MPK3, MPK4, and MPK6 were cloned into pGBKT7 vector, and meanwhile, the coding sequences of ARR1, ARR10, ARR12, and truncated versions of ARR12 were cloned into pGADT7 vector. Interactions allowed growth on SD‐Trp‐Leu‐His‐Ade containing AbA (Aureobasidin A) and X‐α‐gal (5‐bromo‐4‐chloro‐3‐indolyl‐b‐d‐galactopyranoside). Primers used for Y2H are listed in Table EV2.
Bimolecular fluorescence complementation assays
The BiFC assay was performed as previously descried (Yan et al, 2017). Coding sequences of MPK3/4/6 were amplified by PCR and cloned into pSAT6‐nYFP (Tzfira et al, 2005), and coding sequences of ARR1/10/12 were cloned into pSAT6‐cYFP (Tzfira et al, 2005). Fluorescence analysis was performed on an LSM 880 confocal laser‐scanning system (Zeiss, Germany). Primers used for BiFC are listed in Table EV2.
Coimmunoprecipitation assays
The Co‐IP assays were performed as previously described (Lv et al, 2020). Coding sequences of MPK3/4/6, ARR1/10/12, and truncated versions of ARR1/10/12 were amplified by PCR and cloned into pENTRTM /SD/D‐TOPOTM vectors (Thermo Fisher) and then recombined with destination vector cloned into pX‐YFP (35S:C‐YFP) (Tian et al, 2018), and pCAMBIA1390‐7MYC‐6His (Tian et al, 2018). The corresponding constructs were transformed into Arabidopsis protoplasts according to the design of experiment. After incubated overnight, Arabidopsis mesophyll protoplast cells were harvested and lysed in lysis buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP‐40, and 1 mM PMSF) on ice for 30 min with vortexing every 10 min. Cell lysates were then centrifuged at 13,523 g for 10 min, and then, the supernatant was incubated with GFP Trap magnetic agarose beads (ChromoTek, catalog number gtma‐20) for 1 h. The beads were then washed 3 times with dilution buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA), followed by resuspended in 2× SDS sample buffer. The resuspended beads were then boiled for 10 min at 95°C, followed by Western blotting using anti‐MYC (ABclonal, catalog number AE010) antibody or anti‐GFP (TransGen Biotech, catalog number HT801‐02) antibody. Primers used for Co‐IP are listed in Table EV2.
Western blot assay
Total proteins were extracted with lysis buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP‐40, and 1 mM PMSF) from corresponding plant materials. After separation on sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS–PAGE), the proteins were transferred to PVDF membranes (Millipore, catalog number IPVH00010). The membranes were blocked with 1× TBST (0.02% tween in TBS) with 5% non‐fat milk for 1 h at room temperature and then immunoblotted with indicated antibodies at 4°C overnight. After washed with 1× TBST for three times, the membranes were incubated with horseradish peroxidase‐conjugated secondary antibodies at room temperature for 2 h. The chemiluminescent signal was detected by SuperSignal West Pico Luminol/Enhancer Solution (Thermo Fisher Scientific). After exposure, the density of corresponding band was then calculated using the ImageJ software. Acquired data were analyzed using GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA).
Expression analysis by quantitative RT–PCR
Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. One microgram of total RNA were then reversely transcribed using a Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland). qRT–PCR was performed on MyiQTM Real‐time PCR Detection System (Bio‐Rad, Hercules, CA, USA) using FastStart Universal SYBR Green Master Mix (Roche, Basel, Switzerland). ACTIN2 (At3g18780) and UBQ1 (At3g52590) were used as reference genes. Three technical replicates were performed per experiment, and three independent biological experiments were performed. Primers used for qRT–PCR are listed in Table EV2.
Phosphorylation assays
In vitro kinase assay
For in vitro protein expression, the coding sequences of MKK5DD, MPK3, and MPK6 were amplified by PCR and cloned into vector pGEX‐4T‐1 containing a GST tag, and the coding sequences of MPK3, MPK6, and ARRs were amplified by PCR and cloned into vector pET30a containing a His tag. Relative constructs were transformed into E. coli strain BL21 (DE3). For each transformant, a single clone was selected and incubated in 5 ml LB liquid medium. The overnight‐incubated E. coli strains were then transferred into 200 ml LB liquid medium and incubated at 37°C for 2–3 h until OD600 reached 0.4–0.6. The isopropyl β‐d‐1‐thiogalactopyranoside (0.1 mM of final concentration) was added to the cultures, and the strains were incubated at 16°C for a further 12 h. Recombinant GST‐MKK5DD/MPK3/MPK6 proteins were purified using Glutathione Sepharose™ 4 Fast Flow (GE Healthcare, catalog number 17‐5132‐01), and recombinant MPK3/MPK6/ARRs‐His proteins were purified using Ni Sepharose™ High Performance (GE Healthcare, catalog number 17‐5268‐01) according to the manufacturer's protocol. The in vitro kinase assay was performed as previously described (Leissing et al, 2016; Yu et al, 2019). In brief, 0.1 μg of recombinant GST‐MKK5DD, 0.2 μg of recombinant GST‐MPK3/6 or MPK3/6‐His, and 2 μg of recombinant ARRs‐His (substrate protein) were incubated in 30 μl of reaction buffer (50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, and 1 mM DTT) including 1 mM ATPγS (ABclonal, catalog number 138911) at 30°C for 1 h. The reaction was stopped by 20 mM EDTA, and the thiophosphorylated substrates were alkylated with 1.5 mM p‐nitrobenzyl mesylate (ABclonal, catalog number 138910) for 1 h at room temperature. The alkylation reaction was then mixed with 2 × SDS sample buffer before samples were subjected to SDS–PAGE and transferred to PVDF membranes (Millipore, catalog number IPVH00010). Thiophosphorylated kinase substrates were detected with the rabbit monoclonal antibody (Abclonal, catalog number 92570) against the thiophosphate ester. Blots were visualized by SuperSignal West Pico Luminol/Enhancer Solution (Thermo Fisher Scientific). Primers used for in vitro kinase assay are listed in Table EV2.
Sample preparation for mass spectrometry
To prepare samples for mass spectrometry, 0.1 μg of GST‐MKK5DD, 0.2 μg of GST‐MPK3/6 or MPK3/6‐His, and 2 μg of ARR1/10/12‐His were incubated in 30 μl of reaction buffer (50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, and 1 mM DTT) with 50 mM ATP at 30°C for 1 h.
Protein digestion was performed with FASP method described by Wisniewski (Wisniewski et al, 2009). Proteins were reduced with dithiothreitol (DTT) and alkylated with iodoacetamide (IAM). The samples were transferred into 10 kDa ultrafiltration centrifuge tubes (Microcon units). Subsequently, the protein was washed with 50 mM NH4HCO3 buffer three times. Then, the protein suspension was digested with 2 μg trypsin (Promega) overnight at 37°C. Following centrifugation at 16,000 g for 15 min, the filtrate was collected. After adding trifluoroacetic acid (TFA) solution, the digested peptides were desalted with a C18 Stage Tip for further LC‐MS analysis. LC‐MS/MS experiments were performed on a Q Exactive HF‐X mass spectrometer that was coupled to Easy nLC (Thermo Scientific). The raw files were imported into MaxQuant software (version 1.6.1.0, Max Planck Institute of Biochemistry, Martinsried, Germany) for the protein identification against the UniProtKB Arabidopsis thaliana database (http://uniprot.org). The enzyme was set as trypsin, and the maximum missed cleavages were set to 2. The main search peptide tolerance, first search peptide tolerance, and MS/MS tolerance were 4.5, 20, and 20 ppm, respectively. The peptide spectral matching FDR and the protein FDR were both set as 0.01. Razor and unique peptides were used for the protein quantification.
In vivo phosphorylation assay
ARR1/10/12 phosphorylation in vivo was detected by a mobility shift assay using the Phos‐tag reagent as described previously (Li et al, 2017). Briefly, proteins were extracted from 35S:ARR1:MYC, 35S:ARR10:YFP, and 35S:ARR12:YFP in wild‐type Col‐0 and MPK3SR or MPK6SR backgrounds and were separated in a 6% SDS–PAGE gel containing 50 μM Phos‐tag and 200 μM MnCl2 with a constant voltage at 90 V. After electrophoresis, the Mn2+ in the gel was chelated through incubating in the transfer buffer containing 10 mM EDTA, and then, the gel was washed in transfer buffer (50 mM Tris, 40 mM Glycine) for 10 min followed by transferred to a PVDF membrane (Millipore, catalog number IPVH00010). ARR1:MYC was detected with the anti‐MYC antibody (ABclonal, catalog number AE010), and ARR10:YFP and ARR12:YFP were detected with the anti‐GFP antibody (TransGen Biotech, catalog number HT801‐02).
Statistical analysis
Student's t‐test was performed to analyze two‐group datasets. ns indicates no significant difference to the corresponding controls; *, **, ***, and **** indicate significant difference to the corresponding controls with P < 0.05, P < 0.01, P < 0.001, and P < 0.0001, respectively. All values were presented as means ± SD.
Author contributions
ZhY, HT, CL, and ZD designed research; ZhY, JW, FW, CX, BL, XL, and ZiY performed research; ZhY, JW, FW, CX, BL, ZiY, XL, SD, GX, and ZD analyzed data; ZhY, HT, CL, and ZD wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Dataset EV1
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 6
Acknowledgements
We thank Prof. Shuhua Yang, Prof. Shuqun Zhang, and Prof. Xiansheng Zhang for sharing published materials. This work is supported by Youth Interdisciplinary Science and Innovative Research Groups of Shandong University (Grant No. 2020QNQT014), by the National Natural Science Foundation of China (Projects 31470371, 31770305, 31900249, and 32000225), and by Qingdao's Leading Technology Innovator Project. We also thank Jie Dai at Shanghai Bioprofile Technology Company Ltd. for his technical support in mass spectroscopy.
EMBO reports (2021) 22: e52457.
Contributor Information
Huiyu Tian, Email: tianhuiyu@sdu.edu.cn.
Cuiling Li, Email: cuilingli@sdu.edu.cn.
Zhaojun Ding, Email: dingzhaojun@sdu.edu.cn.
Data availability
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/) with the dataset identifier PXD027365 (http://www.ebi.ac.uk/pride/archive/projects/PXD027365).
References
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G, Unthan T, Uhrig JF, Poschl Y, Gust AA, Scheel D, Lee J (2009) Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc Natl Acad Sci USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Hirt H (2018) Nuclear signaling of plant MAPKs. Front Plant Sci 9: 469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JH, Jiang HW, Hsieh EJ, Chen HY, Chien CT, Hsieh HL, Lin TP (2012) Drought and salt stress tolerance of an Arabidopsis glutathione S‐transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol 158: 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Cortleven A, Nitschke S, Klaumunzer M, Abdelgawad H, Asard H, Grimm B, Riefler M, Schmulling T (2014) A novel protective function for cytokinin in the light stress response is mediated by the Arabidopsis histidine kinase2 and Arabidopsis histidine kinase3 receptors. Plant Physiol 164: 1470–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortleven A, Leuendorf JE, Frank M, Pezzetta D, Bolt S, Schmulling T (2019) Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ 42: 998–1018 [DOI] [PubMed] [Google Scholar]
- Danquah A, de Zelicourt A, Colcombet J, Hirt H (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv 32: 40–52 [DOI] [PubMed] [Google Scholar]
- Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI (2014) Plant salt‐tolerance mechanisms. Trends Plant Sci 19: 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droillard MJ, Boudsocq M, Barbier‐Brygoo H, Laurière C (2002) Different protein kinase families are activated by osmotic stresses in Arabidopsis thaliana cell suspensions. Involvement of the MAP kinases AtMPK3 and AtMPK6. FEBS Lett 527: 43–50 [DOI] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, Dinneny JR (2013) Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Showk S, Ruonala R, Helariutta Y (2013) Crossing paths: cytokinin signalling and crosstalk. Development 140: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Enslen H, Davis RJ (2012) Regulation of MAP kinases by docking domains. Biol Cell 93: 5–14 [DOI] [PubMed] [Google Scholar]
- Geilfus CM, Mithofer A, Ludwig‐Muller J, Zorb C, Muehling KH (2015) Chloride‐inducible transient apoplastic alkalinizations induce stomata closure by controlling abscisic acid distribution between leaf apoplast and guard cells in salt‐stressed Vicia faba . New Phytol 208: 803–816 [DOI] [PubMed] [Google Scholar]
- Gu L, Liu Y, Zong X, Liu L, Li DP, Li DQ (2010) Overexpression of maize mitogen‐activated protein kinase gene, ZmSIMK1 in Arabidopsis increases tolerance to salt stress. Mol Biol Rep 37: 4067–4073 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S (2011) AtMYB2 regulates whole plant senescence by inhibiting cytokinin‐mediated branching at late stages of development in Arabidopsis . Plant Physiol 156: 1612–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Hou L, Meng J, You H, Li Z, Gong Z, Yang S, Shi Y (2018) The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis . Mol Plant 11: 970–982 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases AtMPK4 and AtMPK6. Plant J 24: 655–665 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2010) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24: 655–665 [DOI] [PubMed] [Google Scholar]
- Julkowska MM, Testerink C (2015) Tuning plant signaling and growth to survive salt. Trends Plant Sci 20: 586–594 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE (2018) Cytokinin signaling in plant development. Development 145: dev149344 [DOI] [PubMed] [Google Scholar]
- Kien H, Nguyen C, Van HA, Rie N, Yasuko W (2016) Arabidopsistype B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc Natl Acad Sci USA 113: 3090–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Woo DH, Kim JM, Lee SY, Chung WS, Moon YH (2011) Arabidopsis MKK4 mediates osmotic‐stress response via its regulation of MPK3 activity. Biochem Biophys Res Commun 412: 150–154 [DOI] [PubMed] [Google Scholar]
- Kim JM, Woo DH, Kim SH, Lee SY, Park HY, Seok HY, Chung WS, Moon YH (2012) Arabidopsis MKKK20 is involved in osmotic stress response via regulation of MPK6 activity. Plant Cell Rep 31: 217–224 [DOI] [PubMed] [Google Scholar]
- Kudla J, Becker D, Grill E, Hedrich R, Hippler M, Kummer U, Parniske M, Romeis T, Schumacher K (2018) Advances and current challenges in calcium signaling. New Phytol 218: 414–431 [DOI] [PubMed] [Google Scholar]
- Leissing F, Nomoto M, Bocola M, Schwaneberg U, Tada Y, Conrath U, Beckers GJ (2016) Substrate thiophosphorylation by Arabidopsis mitogen‐activated protein kinases. BMC Plant Biol 16: 48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Wang G, Zhao JL, Zhang LQ, Ai LF, Han YF, Sun DY, Zhang SW, Sun Y (2014) The receptor‐like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 26: 2538–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ding Y, Shi Y, Zhang X, Zhang S, Gong Z, Yang S (2017) MPK3‐ and MPK6‐mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis . Dev Cell 43: 630–642.e634 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1‐aminocyclopropane‐1‐carboxylic acid synthase by MPK6, a stress‐responsive mitogen‐activated protein kinase, induces ethylene biosynthesis in Arabidopsis . Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X‐M, Kim KE, Kim K‐C, Nguyen XC, Han HJ, Jung MS, Kim HS, Kim SH, Park HC, Yun D‐Jet al (2010) Cadmium activates Arabidopsis MPK3 and MPK6 via accumulation of reactive oxygen species. Phytochemistry 71: 614–618 [DOI] [PubMed] [Google Scholar]
- Liu J, Lin QF, Qi SL, Feng XJ, Han HL, Xu T, Hua XJ (2020a) The F‐box protein EST1 modulates salt tolerance in Arabidopsis by regulating plasma membrane Na(+)/H(+) antiport activity. J Plant Physiol 251: 153217 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang M, Meng Z, Wang B, Chen M (2020b) Research progress on the roles of cytokinin in plant response to stress. Int J Mol Sci 21: 6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv B, Yu Q, Liu J, Wen X, Yan Z, Hu K, Li H, Kong X, Li C, Tian Het al (2020) Non‐canonical AUX/IAA protein IAA33 competes with canonical AUX/IAA repressor IAA5 to negatively regulate auxin signaling. EMBO J 39: e101515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macková H, Hronková M, Dobrá J, Turečková V, Novák O, Lubovská Z, Motyka V, Haisel D, Hájek T, Prášil ITet al (2013) Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. J Exp Bot 64: 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE (2005) Multiple type‐B response regulators mediate cytokinin signal transduction in Arabidopsis . Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Jha D, Salt DE, Tester M, Hill K, Kieber JJ, Schaller GE (2010) Type‐B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J 64: 753–763 [DOI] [PubMed] [Google Scholar]
- Meng LS, Wang YB, Yao SQ, Liu A (2015) Arabidopsis AINTEGUMENTA mediates salt tolerance by trans‐repressing SCABP8. J Cell Sci 128: 2919–2927 [DOI] [PubMed] [Google Scholar]
- Meng WJ, Cheng ZJ, Sang YL, Zhang MM, Rong XF, Wang ZW, Tang YY, Zhang XS (2017) Type‐B Arabidopsis response regulators specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29: 1357–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D, Leyser O (2011) Auxin, cytokinin and the control of shoot branching. Ann Bot 107: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, Vankova R, Yamaguchi‐Shinozaki K, Shinozaki K, Kakimoto Tet al (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu M, Xie J, Chen C, Cao H, Sun J, Kong Q, Shabala S, Shabala L, Huang Y, Bie Z (2018) An early ABA‐induced stomatal closure, Na+ sequestration in leaf vein and K+ retention in mesophyll confer salt tissue tolerance in Cucurbita species. J Exp Bot 69: 4945–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Datta S, Persak H (2014) Salt stress in Arabidopsis: lipid transfer protein AZI1 and its control by mitogen‐activated protein kinase MPK3. Mol Plant 7: 722–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramireddy E, Hosseini SA, Eggert K, Gillandt S, Gnad H, von Wiren N, Schmulling T (2018) Root engineering in barley: increasing cytokinin degradation produces a larger root system, mineral enrichment in the shoot and improved drought tolerance. Plant Physiol 177: 1078–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S (2002) Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis . J Biol Chem 277: 559–565 [DOI] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J (2010) Mitogen‐activated protein kinase signaling in plants. Annu Rev Plant Biol 61: 621–649 [DOI] [PubMed] [Google Scholar]
- von Schwartzenberg K, Nunez MF, Blaschke H, Dobrev PI, Novak O, Motyka V, Strnad M (2007) Cytokinins in the bryophyte Physcomitrella patens: analyses of activity, distribution, and cytokinin oxidase/dehydrogenase overexpression reveal the role of extracellular cytokinins. Plant Physiol 145: 786–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks AD, Yang SH, Galanis A (2000) Docking domains and substrate‐specificity determination for MAP kinases. Trends Biochem Sci 25: 448–453 [DOI] [PubMed] [Google Scholar]
- Shen L, Zhuang B, Wu Q, Zhang H, Nie J, Jing W, Yang L, Zhang W (2019) Phosphatidic acid promotes the activation and plasma membrane localization of MKK7 and MKK9 in response to salt stress. Plant Sci 287: 110190 [DOI] [PubMed] [Google Scholar]
- Su J, Zhang M, Zhang L, Sun T, Liu Y, Lukowitz W, Xu J, Zhang S (2017) Regulation of stomatal immunity by interdependent functions of a pathogen‐responsive MPK3/MPK6 cascade and abscisic acid. Plant Cell 29: 526–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Doczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis . Mol Cell 15: 141–152 [DOI] [PubMed] [Google Scholar]
- Tian Y, Fan M, Qin Z, Lv H, Wang M, Zhang Z, Zhou W, Zhao NA, Li X, Han Cet al (2018) Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE‐RESISTANT1 transcription factor. Nat Commun 9: 1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D, Zhao YU, Yoshida T, Kudo M, Kidokoro S, Mizoi J, Kodaira K‐S, Takebayashi Y, Kojima M, Sakakibara Het al (2017) Temporal and spatial changes in gene expression, metabolite accumulation and phytohormone content in rice seedlings grown under drought stress conditions. Plant J 90: 61–78 [DOI] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi‐Shinozaki K (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis . Proc Natl Acad Sci USA 104: 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner‐Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Verma V, Ravindran P, Kumar PP (2016) Plant hormone‐mediated regulation of stress responses. BMC Plant Biol 16: 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li X, Meisenhelder J, Hunter T, Yoshida S, Asami T, Chory J (2005) Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell 8: 855–865 [DOI] [PubMed] [Google Scholar]
- Wang F, Jing W, Zhang W (2014) The mitogen‐activated protein kinase cascade MKK1‐MPK4 mediates salt signaling in rice. Plant Sci 227: 181–189 [DOI] [PubMed] [Google Scholar]
- Wang WC, Lin TC, Kieber J, Tsai YC (2019) Response regulators 9 and 10 negatively regulate salinity tolerance in rice. Plant Cell Physiol 60: 2549–2563 [DOI] [PubMed] [Google Scholar]
- Wang J, Qin H, Zhou S, Wei P, Zhang H, Zhou Y, Miao Y, Huang R (2020) The ubiquitin‐binding protein OsDSK2a mediates seedling growth and salt responses by regulating gibberellin metabolism in rice. Plant Cell 32: 414–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Schmulling T (2009) Cytokinin action in plant development. Curr Opin Plant Biol 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Xie M, Chen H, Huang L, O'Neil RC, Shokhirev MN, Ecker JR (2018) A B‐ARR‐mediated cytokinin transcriptional network directs hormone cross‐regulation and shoot development. Nat Commun 9: 1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Chen WH, Jia W, Zhang J (2015) Mitogen‐activated protein kinase kinase 5 (MKK5)‐mediated signalling cascade regulates expression of iron superoxide dismutase gene in Arabidopsis under salinity stress. J Exp Bot 66: 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Zhao Y, Gao J, Xiang C, Zhu JK (2016) The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci Rep 6: 27177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid‐inducible mitogen‐activated protein kinase. Plant Cell 15: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li Y, Wang Y, Liu H, Lei L, Yang H, Liu G, Ren D (2008) Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis . J Biol Chem 283: 26996–27006 [DOI] [PubMed] [Google Scholar]
- Xu J, Xie J, Yan C, Zou X, Ren D, Zhang S (2014) A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase‐mediated oxidative burst are two independent signaling events in plant immunity. Plant J 77: 222–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang S (2015) Mitogen‐activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci 20: 56–64 [DOI] [PubMed] [Google Scholar]
- Yan Z, Liu X, Ljung K, Li S, Zhao W, Yang F, Wang M, Tao Y (2017) Type B response regulators act as central integrators in transcriptional control of the auxin biosynthesis enzyme TAA1. Plant Physiol 175: 1438–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Shad Ali G, Yang L, Du L, Reddy ASN, Poovaiah BW (2010) Calcium/calmodulin‐regulated receptor‐like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal Behav 5: 991–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZB, Liu G, Liu J, Zhang B, Meng W, Muller B, Hayashi KI, Zhang X, Zhao Z, De Smet Iet al (2017) Synergistic action of auxin and cytokinin mediates aluminum‐induced root growth inhibition in Arabidopsis . EMBO Rep 18: 1213–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Guo Y (2018) Unraveling salt stress signaling in plants. J Integr Plant Biol 60: 796–804 [DOI] [PubMed] [Google Scholar]
- Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, Liu J, Xiao Y, Liang Y, Zhang W (2010) Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol 188: 762–773 [DOI] [PubMed] [Google Scholar]
- Yu Z, Zhang DI, Xu Y, Jin S, Zhang L, Zhang S, Yang G, Huang J, Yan K, Wu Cet al (2019) CEPR2 phosphorylates and accelerates the degradation of PYR/PYLs in Arabidopsis . J Exp Bot 70: 5457–5469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Duan X, Luo L, Dai S, Ding Z, Xia G (2020) How plant hormones mediate salt stress responses. Trends Plant Sci 25: 1117–1130 [DOI] [PubMed] [Google Scholar]
- de Zelicourt A, Colcombet J, Hirt H (2016) The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci 21: 677–685 [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhao J, Li L, Gao Y, Zhao L, Patil SB, Fang J, Zhang W, Yang Y, Li Met al (2017) The Arabidopsis U‐box E3 ubiquitin ligase PUB30 negatively regulates salt tolerance by facilitating BRI1 kinase inhibitor 1 (BKI1) degradation. Plant Cell Environ 40: 2831–2843 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhang N, Liu X, Wang S, Li S, Yang J, Wang F, Si H (2020) StMAPK3 controls oxidase activity, photosynthesis and stomatal aperture under salinity and osmosis stress in potato. Plant Physiol Biochem 156: 167–177 [DOI] [PubMed] [Google Scholar]
- Zubo YO, Blakley IC, Yamburenko MV, Worthen JM, Street IH, Franco‐Zorrilla JM, Zhang W, Hill K, Raines T, Solano Ret al (2017) Cytokinin induces genome‐wide binding of the type‐B response regulator ARR10 to regulate growth and development in Arabidopsis . Proc Natl Acad Sci USA 114: E5995–E6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwack PJ, Rashotte AM (2015) Interactions between cytokinin signalling and abiotic stress responses. J Exp Bot 66: 4863–4871 [DOI] [PubMed] [Google Scholar]
- Zwack PJ, De Clercq I, Howton TC, Hallmark HT, Hurny A, Keshishian EA, Parish AM, Benkova E, Mukhtar MS, Van Breusegem Fet al (2016) Cytokinin response factor 6 represses cytokinin‐associated genes during oxidative stress. Plant Physiol 172: 1249–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Dataset EV1
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 6
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/) with the dataset identifier PXD027365 (http://www.ebi.ac.uk/pride/archive/projects/PXD027365).


