Abstract
It has long been thought that microtubule disassembly, one of the earliest cellular events, contributes to neuronal pruning and neurodegeneration in development and disease. However, how microtubule disassembly drives neuronal pruning remains poorly understood. Here, we conduct a systematic investigation of various microtubule‐destabilizing factors and identify exchange factor for Arf6 (Efa6) and Stathmin (Stai) as new regulators of dendrite pruning in ddaC sensory neurons during Drosophila metamorphosis. We show that Efa6 is both necessary and sufficient to regulate dendrite pruning. Interestingly, Efa6 and Stai facilitate microtubule turnover and disassembly prior to dendrite pruning without compromising the minus‐end‐out microtubule orientation in dendrites. Moreover, our pharmacological and genetic manipulations strongly support a key role of microtubule disassembly in promoting dendrite pruning. Thus, this systematic study highlights the importance of two selective microtubule destabilizers in dendrite pruning and substantiates a causal link between microtubule disassembly and neuronal pruning.
Keywords: dendrite pruning, exchange factor for Arf6, metamorphosis, microtubule disassembly, Stathmin
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Neuroscience
Systematic investigation of microtubule‐destabilizing factors identifies Efa6 and Stathmin as two new regulators of dendrite pruning in Drosophila sensory neurons. Genetic and pharmacological manipulations reveal a causal link between microtubule disassembly and neuronal pruning.

Introduction
During animal development, the formation of mature nervous systems requires both progressive and regressive events. Neurons often elaborate exuberant axons and dendrites at early developmental stages. Following extensive neurite outgrowth, excessive or incorrect processes are often refined or eliminated at late stages. These regressive events occur without causing the death of parental neurons, a process known as neuronal pruning (Luo & O'Leary, 2005; Riccomagno & Kolodkin, 2015; Schuldiner & Yaron, 2015). Neuronal pruning, a precisely regulated developmental process, is essential for the establishment of functional circuits during the maturation of the nervous systems. In mammalian brains, defective neuronal pruning in layer V pyramidal neurons can result in increased dendritic spine density, which is associated with autism spectrum disorders (Tang et al, 2014). On the contrary, excessive neuronal pruning during adolescence highly correlates with early‐onset schizophrenia (Lewis & Levitt, 2002; Sekar et al, 2016). Therefore, understanding the mechanisms of neuronal pruning would provide important insights into pathogenesis of neuronal disorders in humans.
In the holometabolous insect Drosophila, many larval‐born neurons undergo large‐scale remodelling during metamorphosis, a transition stage between larva and adult animals (Yu & Schuldiner, 2014). Mushroom body (MB) γ neurons in the central nervous system (CNS) prune away both axons and dendrites during the first day of metamorphosis (Lee et al, 1999). In the peripheral nervous system (PNS), dorsal dendrite arborization (da) neurons, such as ddaD/E (class I) and ddaC (class IV) neurons, undergo dendrite‐specific pruning without eliminating their axons and subsequently regenerate their dendrites into the adult nervous system (Kuo et al, 2005; Williams & Truman, 2005; Shimono et al, 2009). By contrast, ddaB (class II) and ddaA/F (class III) neurons undergo apoptosis at the early pupal stage (Williams & Truman, 2005). The complex larval dendrites of ddaC neurons (also known as C4da neurons) provide an excellent opportunity to systematically unravel the molecular and cellular mechanisms of dendrite‐specific pruning.
Dendrite pruning of ddaC neurons is manifested with a series of stereotyped events: thinning and severing of proximal dendrites at 5–8 h after puparium formation (APF), fragmentation of detached dendrites at 10‐ to 12‐h APF and debris clearance at 16‐ to 18‐h APF (Fig 1A). The initiation of dendrite pruning is triggered by a late‐larval pulse of the steroid hormone ecdysone and its receptor heterodimer, such as EcR‐B1 and Ultraspiracle (Usp; Kuo et al, 2005; Williams & Truman, 2005). Upon activation, the EcR‐B1‐Usp heterodimer induces the expression of its downstream targets Sox14 (Kirilly et al, 2009; Kirilly et al, 2011) and Headcase (Loncle & Williams, 2012). The transcription factor Sox14 in turn activates the expression of its target effectors, such as the actin‐disassembly regulator Mical (Kirilly et al, 2009; Rode et al, 2018) and a Cullin1‐Slimb E3 ubiqutin ligase complex (Wong et al, 2013). Other ecdysone‐dependent genes and pathways include caspases (Kuo et al, 2006; Williams et al, 2006), calcium signalling (Kanamori et al, 2013; Kanamori et al, 2015) and JNK signalling (Zhu et al, 2019). Apart from these pathways, endocytic pathways (Zhang et al, 2014; Loncle et al, 2015; Zong et al, 2018; Kramer et al, 2019) and secretory pathways (Wang et al, 2017; Wang et al, 2018) also regulate dendrite pruning by modulating endo‐lysosomal degradation of the cell‐adhesion molecule Neuroglian.
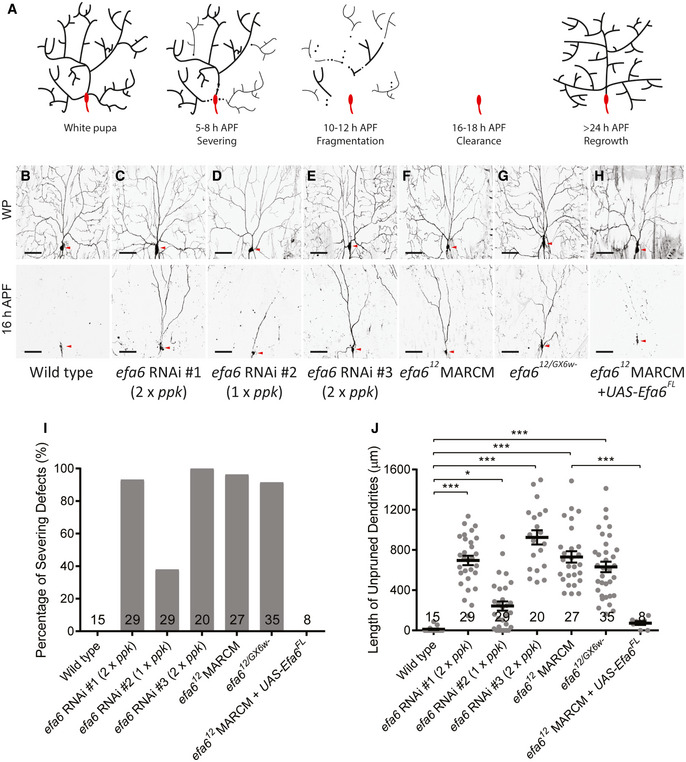
Figure 1. Efa6 is required for dendrite pruning of ddaC sensory neurons.
-
AA schematic representation of dendrite pruning in ddaC neurons.
-
B–HLive confocal images of ddaC neurons expressing mCD8‐GFP by ppk‐Gal4 at WP stage or 16‐h APF. Neurons expressing efa6 RNAi #1‐#3 (C–E), efa612 mutant MARCM clones (F) and efa612/GX6w‐ transheterozygous mutant neurons (G) showed consistent dendrite pruning defects at 16‐h APF, as compared to the wild‐type neurons (B). The dendrite pruning defects were fully rescued when overexpressing full‐length Efa6 in the efa612 mutant clones (H). Red arrowheads point to the ddaC somas.
-
I, JQuantification of dendrite severing defects and unpruned dendrite lengths at 16‐h APF.
Data information: In (I–J), the number of samples (n) in each group is shown above the x‐axis. *P < 0.05, ***P < 0.001, as assessed by one‐way ANOVA with Bonferroni test. All error bars represent SEM. Three independent experiments were conducted. The scale bars in (B–H) represent 50 µm.
Source data are available online for this figure.
During neurite pruning, local microtubule disassembly in degenerating axons/dendrites is one of the earliest cellular alterations in ddaC and MB γ neurons (Watts et al, 2003; Williams & Truman, 2005; Lee et al, 2009) and is also essential for axonal pruning and degeneration in mammalian neurons (Maor‐Nof et al, 2013; Brill et al, 2016). In ddaC neurons, dendritic microtubules become more dynamic at the onset of metamorphosis (Herzmann et al, 2017). Concomitant with this, local microtubule disassembly can be observed in proximal dendrites at 4‐ to 6‐h APF (Williams & Truman, 2005; Lee et al, 2009; Herzmann et al, 2018). Katanin p60‐like 1 (Kat‐60L1), an AAA ATPase analogous to the microtubule‐severing enzyme Katanin‐60 (Kat‐60), was first shown to regulate dendrite pruning and morphology of ddaC neurons (Lee et al, 2009; Stewart et al, 2012), and its potential microtubule‐severing function in these neurons remains unclear. Par‐1 kinase has also been reported to regulate the microtubule breakdown process probably via inhibitory phosphorylation of microtubule stabilizer, Tau (Herzmann et al, 2017). We previously reported unexpected findings that excessive microtubule depolymerization/disassembly, via either pharmacological treatment of microtubule‐destabilizing drug or overexpression of Kat‐60, did not accelerate dendrite pruning, instead, resulted in inhibition of dendrite pruning in ddaC neurons (Tang et al, 2020). Via an unknown mechanism, excessive microtubule disassembly impaired minus‐end‐out microtubule orientation in major dendrites, which contributes to the dendrite pruning defects (Wang et al, 2019; Rui et al, 2020). However, whether and how microtubule disassembly also positively regulates the dendrite pruning process remained poorly understood. Moreover, a link between microtubule disassembly and dendrite pruning has not been established.
In an RNA interference (RNAi) screen for isolating novel microtubule‐destabilizing factors in dendrite pruning, we identified exchange factor for Arf6 (Efa6), a conserved microtubule collapse factor (O'Rourke et al, 2010), as a new regulator of dendrite pruning. Worm and fly Efa6s were found to induce catastrophe or pause from the growing plus ends and eliminate microtubules that grow towards the cortex (O'Rourke et al, 2010; Qu et al, 2019). Moreover, Efa6s play an inhibitory role in axonal outgrowth and regeneration of invertebrate neurons (Chen et al, 2011; Qu et al, 2019). Here, we show that Efa6 plays an important role in dendrite pruning of ddaC neurons. Efa6 does not regulate microtubule orientation but promotes microtubule turnover/disassembly in the dendrites. Moreover, we also identified a second microtubule‐destabilizing factor Stathmin (Stai) as an important pruning regulator. Like Efa6, Stai also promotes microtubule disassembly and thereby dendrite pruning. In addition, we confirm that the microtubule‐severing enzymes, katanin, spastin and fidgetin, are dispensable for dendrite pruning. Overall, our study reveals the selectivity of microtubule‐destabilizing factors during pruning and further supports an important link between microtubule disassembly and neuronal pruning.
Results
Efa6 is cell‐autonomously required for dendrite pruning in ddaC neurons
In order to systematically interrogate potential roles of microtubule‐destabilizing factors in dendrite pruning, we conducted a candidate‐based RNAi screen by knocking down various microtubule regulators, including Efa6, Stai, spastin, katanin, fidgetin and microtubule‐destabilizing kinesins (Appendix Table S1). We used the class IV da‐specific driver, pickpocket (ppk)‐Gal4, to express their transgenic RNAi constructs and examine potential effects. From this screen, we isolated Efa6 and Stai as new regulators of dendrite pruning. However, the microtubule‐severing enzymes (spastin, katanin and fidgetin) are not important for dendrite pruning, similar to the previous studies (Lee et al, 2009; Tao et al, 2016). Three independent RNAi lines (#1 v330083, #2 v42321 and #3 BL57449) targeting the same gene efa6 were isolated. efa6 encodes a conserved microtubule collapse factor that negatively regulates microtubule polymerization and dynamics in nematodes and flies (O'Rourke et al, 2010; Chen et al, 2011; Chen et al, 2015; Qu et al, 2019). Wild‐type ddaC neurons completely eliminated their larval dendrites at 16‐h APF (Fig 1B, I,J). By contrast, expression of these efa6 RNAi lines showed consistent dendrite pruning defects at the same time point (Fig 1C–E, I,J). To confirm these RNAi results, we took advantage of CRISPR‐Cas9 technology (Port et al, 2014) and generated a new allele, efa612 , which deletes almost all the coding region and thus is likely a null allele (Appendix Fig S1A). By generating homozygous mosaic analysis with a repressible cell marker (MARCM) clones (Lee & Luo, 1999), we observed consistent dendrite pruning defects at 16‐h APF in almost all efa612 mutant clones (Fig 1F, I,J) with an average of 730‐µm dendrites unpruned. Moreover, the trans‐heterozygotes between efa612 and the previously reported allele efa6GX6w‐ also exhibited a similar penetrance of dendrite pruning defects in ddaC neurons with 631 µm dendrites attached (Fig 1G, I,J). Importantly, when the full‐length Efa6 protein was overexpressed in efa612 mutant clones, the pruning defects were fully rescued (Fig 1H, I,J). Thus, these results demonstrate that the conserved microtubule collapse factor Efa6 is cell‐autonomously required for dendrite pruning in ddaC neurons. In addition, the Sholl analysis indicates slightly simplified dendrite arbours observed in efa612 MARCM clones at the wandering third instar larval (wL3) stage (Fig EV1A). To ascertain whether Efa6 plays a role in promoting dendrite pruning at the prepupal stages, we temporally induced the expression of efa6 RNAi construct from the 3rd instar larval stage via the gene‐switch system. After RU486 treatment, the majority of efa6 RNAi neurons exhibited prominent pruning defects (Fig EV1B), similar to those from continuous efa6 knockdown (Fig 1C, I,J). Thus, the gene‐switch data suggest a requirement of Efa6 for dendrite pruning at the prepupal stage when dendrite pruning starts to occur.
Figure EV1. Loss of efa6 function slightly affected dendrite arborization, and temporal knockdown of efa6 led to dendrite pruning defects and increased Futsch levels in dendrites.
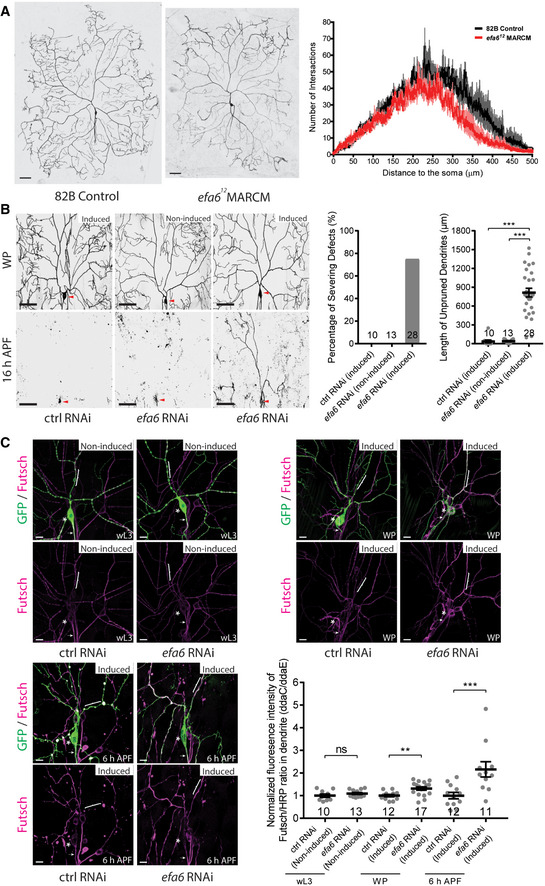
- Live confocal images of wild‐type and efa612 mutant ddaC MARCM clones at wL3 stage (left and middle panels, respectively) and Sholl analysis (right panel).
- Live confocal images of ddaC neurons labelled by ppk‐tdGFP at WP and 16‐h APF. The expression of RNAi is driven by GeneSwitch‐Gal4 induced by RU486 treatment. Induction of efa6 RNAi expression at the late 3rd instar larval stage resulted in significant dendrite pruning defects at 16‐h APF compared to the control RNAi or non‐induced condition. Red arrowheads point to the ddaC somas (left panels). Quantification of dendrite severing defects (middle panel) and unpruned dendrite lengths at 16‐h APF (right panel).
- Confocal images of ddaC neurons expressing ppk‐tdGFP immunostained with anti‐Futsch at wL3, WP and 6‐h APF. Expression of efa6 RNAi construct via RU486‐induced GeneSwitch‐Gal4 slightly increased dendritic Futsch level at WP stage and dramatically increased dendritic Futsch level at 6‐h APF compared to control RNAi. The ddaC somas in are labelled by asterisks, the axons by arrows and the dendrites by brackets. Quantification of Futsch fluorescence intensity in ddaC dendrites (bottom right panel).
Data information: In (B–C), the number of samples (n) in each group is shown on the x‐axis. ns, not significant, **P < 0.01, ***P < 0.001, as assessed by one‐way ANOVA with Bonferroni test or two‐tailed Student’s t‐test. All error bars represent SEM. Three independent experiments were conducted. The scale bars in (A–B) and (C) represent 50 µm and 10 µm, respectively.
Source data are available online for this figure.
To further confirm the RNAi results for the microtubule‐severing enzymes, we generated new indel mutants for katanin and fidgetin (Appendix Fig S1A) via the CRISPR‐Cas9 method (Kondo & Ueda, 2013; Port et al, 2014). Mutant ddaC neurons derived from kat‐60C48 , kat‐80E3 , fignSK1 or fignSK6 showed no pruning defect at 16‐h APF (Appendix Fig S1B). Likewise, MARCM mutant clones from the previously published null allele spas5.75 exhibited normal dendrite pruning (Appendix Fig S1B). This mutant analysis, in line with the previous RNAi data, strongly strengthens the conclusion that katanin, fidgetin and spastin are indeed dispensable for dendrite pruning in ddaC neurons. Formally, we cannot rule out the possibility that these microtubule‐severing enzymes may play a redundant or combinatory role in dendrite pruning. In addition, we also generated FRT82B‐associated mutant allele, kat‐60L1F1 (Appendix Fig S1A), via CRISPR/Cas9‐mediated gene editing. kat‐60L1F1 exhibited dendrite pruning defects in 42% of its mutant MARCM clones (Appendix Fig S1B), comparable to 51% penetrance in the homozygous P‐insertion (kat‐60L1c01236 ) mutant neurons (Lee et al, 2009). However, none of our data show that Kat‐60L1 functions as a microtubule‐severing enzyme in ddaC neurons (see Discussion and Fig EV5).
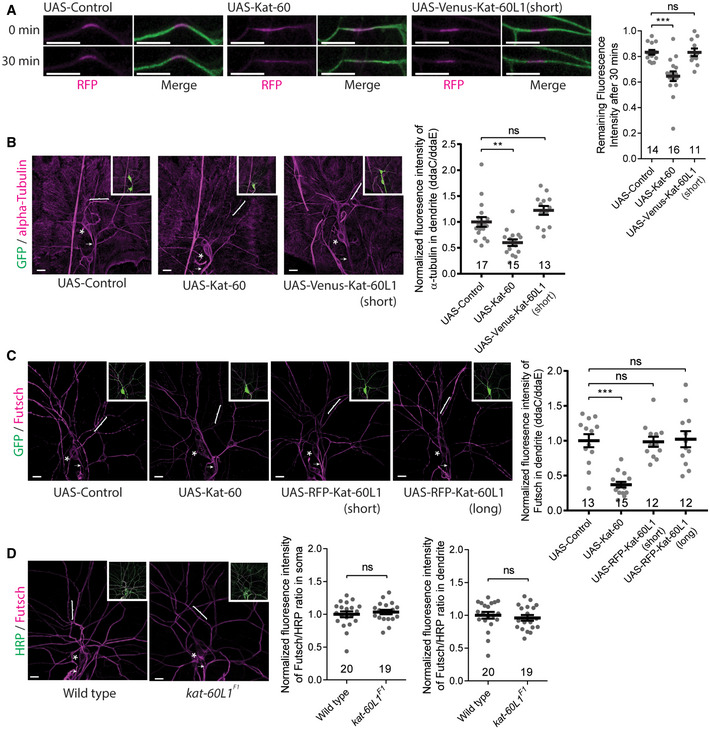
Figure EV5. Kat‐60L1 did not regulate dendritic microtubule turnover and levels in ddaC neurons.
-
ALive confocal images of ddaC dendrites expressing tdEOS::α‐tubulin at wL3 stage. Kat‐60 overexpression increased microtubule turnover rate whereas Kat‐60L1 (short isoform) overexpression did not (left panels). Quantification of microtubule turnover rate as indicated by the remaining fluorescence intensity of converted EOS at 30 mins after photoconversion (right panel).
-
B, CConfocal images of ddaC neurons expressing mCD8‐GFP stained by anti‐α‐tubulin (B) or anti‐Futsch (C). Kat‐60 overexpression reduced α‐tubulin levels in the dendrites while Kat‐60L1 (short isoform) overexpression did not (B). Kat‐60 overexpression reduced Futsch levels in the dendrites while Kat‐60L1 (either short or long isoform) overexpression did not (C). The ddaC somas in are labelled by asterisks, the axons by arrows and the dendrites by brackets (left panels). Quantification of α‐tubulin or Futsch fluorescence intensity in proximal dendrites (right panel).
-
DConfocal images of da sensory neurons immune‐stained by anti‐Futsch and anti‐HRP. The ddaC somas are labelled by asterisks, the axons by arrows and the dendrites by brackets (left panels). Quantification of Futsch/HRP fluorescence intensity ratio in ddaC soma (middle panel) or dendrite (right panel) indicate that microtubule level did not alter in kat‐60L1F1 mutant animals, compared to wild type.
Data information: In (A–D), the number of samples (n) in each group is shown on the x‐axis. ns, not significant, **P < 0.01, ***P < 0.001, as assessed by two‐tailed Student’s t‐test or one‐way ANOVA with Bonferroni test. All error bars represent SEM. Three independent experiments were conducted. The scale bars in (A–D) represent 10 µm.
Source data are available online for this figure.
The microtubule eliminating domain is essential for Efa6’s function in dendrite pruning
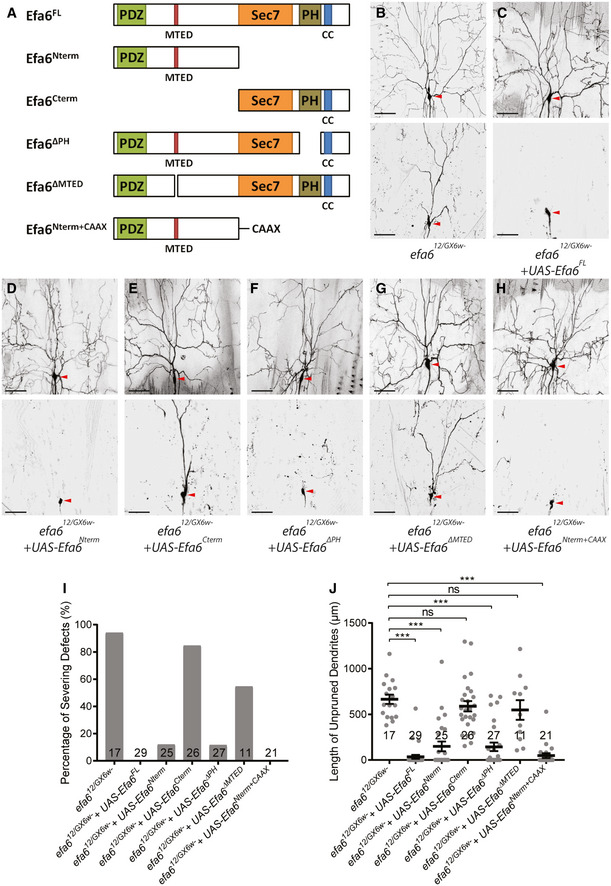
Drosophila Efa6 contains a PDZ domain and an 18‐aa microtubule eliminating domain (MTED) at its amino terminus, a Sec7 GDP‐GTP exchange (GEF) domain, a pleckstrin homology (PH) domain and a coiled‐coil (CC) domain at its carboxyl terminus (Fig 2A) (Qu et al, 2019). To understand the mechanism of Efa6 action in dendrite pruning, we next determined which protein domains of Efa6 are essential for its function. To this end, we generated various transgenes that express different truncated Efa6 proteins (Fig 2A). As controls, overexpression of Efa6 and its variants in ddaC neurons did not impair normal dendrite pruning at 16‐h APF (Appendix Fig S2A). We then expressed these transgenes in the background of efa612/GX6w‐ trans‐heterozygotes, to examine their ability to rescue the dendrite pruning defects. While the majority of efa612/GX6w‐ mutant neurons failed to prune their larval dendrites at 16‐h APF (Fig 2B, I,J), overexpression of full‐length Efa6 (Efa6FL) fully rescued the efa612/GX6w‐ mutant phenotypes (Fig 2C, I,J). Importantly, the expression of a N‐terminal half (Efa6Nterm; Fig 2D, I,J), a PH‐domain‐deleted variant (Efa6ΔPH; Fig 2F, I,J) or a N‐terminal half fused with the membrane‐associated CAAX motif (Efa6Nterm+CAAX; Fig 2H, I,J), almost fully rescued the dendrite pruning defects of efa612/GX6w‐ mutant neurons at 16‐h APF, resembling Efa6FL (Fig 2C, I,J). However, the expression of a C‐terminal half (Efa6Cterm, Fig 2E, I,J) or a MTED‐deleted variant (Efa6ΔMTED, Fig 2G, I,J) failed to rescue majority of pruning defects in efa612/GX6w‐ mutant neurons (Fig 2B, I,J). Thus, the MTED domain in the N‐terminal portion of Efa6 is essential for its function to promote dendrite pruning in ddaC sensory neurons, which is consistent with its role in axon growth of CNS neurons (Qu et al, 2019).
Figure 2. The MTED domain is essential for Efa6 to regulate dendrite pruning.
-
AA schematic illustration of the protein structure of Drosophila Efa6, and various truncated Efa6 proteins.
-
B–HLive confocal images of efa612/GX6w‐ mutant ddaC neurons at WP stage and 16‐h APF. Expression of Efa6FL (C), Efa6Nterm (D), Efa6ΔPH (F) and Efa6Nterm+CAAX (H), but not Efa6Cterm (E) or Efa6ΔMTED (G), rescued the pruning defects in efa612/GX6w‐ mutant ddaC neurons (B). Red arrowheads point to the ddaC somas.
-
I, JQuantification of dendrite severing defects and unpruned dendrite lengths at 16‐h APF.
Data information: In (I–J), the number of samples (n) in each group is shown on the bars. ns, not significant, ***P < 0.001, as assessed by one‐way ANOVA with Bonferroni test. All error bars represent SEM. Three independent experiments were conducted. The scale bars in (B–H) represent 50 µm.
Source data are available online for this figure.
In addition, during Drosophila metamorphosis, ddaD/E neurons also pruned away their dendrites by 19‐h APF (Appendix Fig S2B), whereas ddaF neurons were removed via apoptosis (Appendix Fig S2C) (Williams & Truman, 2005). In efa6 RNAi‐expressing ddaD/E neurons, dendrite pruning was also inhibited (Appendix Fig S2B). In efa6 mutant animals, ddaF neurons, however, were eliminated (100%, n = 37; Appendix Fig S2C). Therefore, Efa6 is required for ddaD/E dendrite pruning but dispensable for ddaF apoptosis.
Overexpression of Efa6 induces precocious dendrite pruning
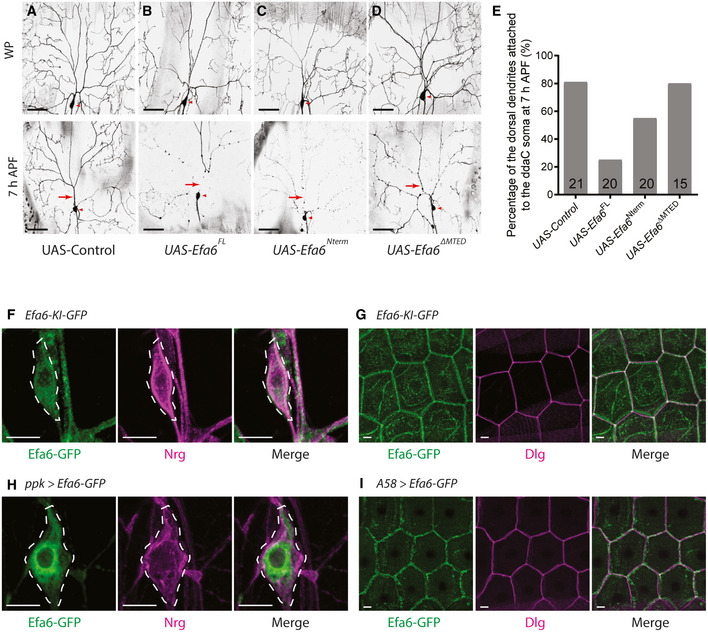
Since loss of efa6 function resulted in the dendrite pruning defects, we next assessed whether elevated expression of Efa6 can accelerate dendrite pruning at an earlier time point. We focused on the dorsal dendrites of ddaC neurons, as most of them in the wild‐type neurons remained attached to the soma at 7‐h APF (81%, Fig 3A and E). In Efa6FL‐overexpressing neurons, however, only 25% of the neurons remained their dorsal dendrites connected to the soma (Fig 3E), whereas the rest showed disconnected dorsal dendrites (Fig 3B). We next tested if the MTED domain is essential to promote precocious pruning. Like Efa6FL, Efa6Nterm overexpression also accelerated the severing of proximal dendrites at 7‐h APF (Fig 3C and E). By contrast, Efa6ΔMTED expression did not promote premature dendrite pruning. 80% of Efa6ΔMTED‐expressing neurons still remained their dorsal dendrites attached (Fig 3D and E), similar to the controls (Fig 3A and E), suggesting that MTED domain is required for Efa6‐mediated dendrite pruning. Thus, Efa6 overexpression promotes premature dendrite pruning via its MTED domain. Taken together, Efa6 is both necessary and sufficient to promote dendrite pruning. Moreover, our data also suggest that MTED‐dependent microtubule depolymerization is important for dendrite pruning during early metamorphosis.
Figure 3. Overexpression of Efa6 induces premature dendrite pruning.
-
A–DLive confocal images of ddaC neurons at WP stage and 7‐h APF. Efa6FL or Efa6Nterm‐overexpressing neurons (B, C) showed precocious pruning phenotype at 7‐h APF, compared to the control and Efa6ΔMTED‐overexpressing neurons (A, D) in which most dendrites remained attached to the soma at the same time point. Red arrowheads point to the ddaC somas. Dorsal dendrites are marked by red arrows.
-
EQuantification of precocious pruning defects as indicated by the percentage of dorsal dendrites attached to the soma at 7‐h APF.
-
F–IConfocal images of da sensory neurons expressing Efa6‐Ki‐GFP (F), or UAS‐Efa6‐GFP driven by ppk‐Gal4 (H) were co‐stained with GFP (green) and Nrg (magenta) at wL3 stage. (G, I) Confocal images of the epidermal cells expressing Efa6‐Ki‐GFP (G) or UAS‐Efa6‐GFP driven by A58‐Gal4 (I) were co‐stained with GFP (green) and Dlg (magenta) at wL3 stage. The ddaC soma is marked by dashed lines.
Data information: In (E), the number of samples (n) in each group is shown on the bars. Three independent experiments were conducted. The scale bars in (A–D) and (F–I) represent 50 µm and 10 µm, respectively.
Source data are available online for this figure.
We next examined the expression of Efa6 in ddaC neurons by using an efa6 knock‐in line (efa6‐Ki‐GFP) during the larval‐to‐pupal transition. efa6‐Ki‐GFP is a genomically engineered knock‐in line, which contains a GFP tag inserted into the last amino acid at the efa6 endogenous locus; moreover, it is homozygous viable and fertile (Huang et al, 2009). By using the efa6‐Ki‐GFP line, we observed that endogenous Efa6 was expressed in ddaC neurons, other sensory neurons (Fig 3F, Appendix Fig S3A) as well as their neighbouring epidermal cells (Fig 3G, Appendix Fig S3A). However, its expression appeared unaltered in ddaC neurons with a trend of increased intensity in epidermal cells from early L3 (eL3) stage to 6‐h APF (Appendix Fig S3A). We have further confirmed that efa6‐Ki‐GFP is functional in ddaC neurons, as its homozygotes pruned away their larval dendrites at 16‐h APF, similar to wild‐type animals (Appendix Fig S3B). Interestingly, endogenous Efa6 showed weak overlapping with the plasma membrane protein Neuroglian (Nrg) in ddaC somas (Fig 3F), although it predominantly co‐localized with the plasma membrane protein Discs large (Dlg) around the cell cortex of epidermal cells (Fig 3G). Like the endogenous protein, overexpressed Efa6‐GFP was also distributed weakly on the plasma membrane of ddaC somas (Fig 3H) but primarily on the plasma membrane of epidermal cells (Fig 3I). Given that Efa6Nterm+CAAX overexpression rescued the efa612/GX6w‐ mutant phenotype (Fig 2H, I,J), we assessed its distribution in ddaC neurons by using an anti‐Efa6 antibody. Similar to overexpressed Efa6‐FL, Efa6Nterm+CAAX showed a non‐plasma membrane distribution in ddaC somas (Appendix Fig S3C).
Efa6 is dispensable for microtubule orientation but important for microtubule growth in the dendrites of ddaC neurons
Microtubule‐associated proteins have been recently reported to regulate dendritic microtubule polarity and dendrite pruning in ddaC neurons (Herzmann et al, 2018; Wang et al, 2019; Rui et al, 2020; Tang et al, 2020). To examine the potential involvement of Efa6 in microtubule orientation, we first examined the distribution of dendritic and axonal microtubule markers, Nod‐β‐Gal and Kin‐β‐Gal, respectively. Nod‐β‐Gal is the chimeric protein that consists of the motor domain from the minus‐end‐directed kinesin Nod, the coiled‐coil domain of kinesin‐1 and the β‐Galactosidase tag and was used as a minus‐end microtubule marker in Drosophila (Clark et al, 1997). Like wild‐type neurons, efa6 RNAi neurons showed normal distribution of Nod‐β‐Gal in the dendrites (Appendix Fig S4A). Kin‐β‐Gal is a plus‐end microtubule marker that consists of the motor domain and coiled‐coil domain of kinesin‐1 with the β‐Galactosidase tag (Clark et al, 1997). In efa6 RNAi neurons, Kin‐β‐Gal remained present in the axons but absent in the dendrites (Appendix Fig S4B), similar to that in wild‐type neurons (Appendix Fig S4B). Thus, efa6 is dispensable for proper distribution of dendritic and axonal microtubule markers in ddaC neurons.
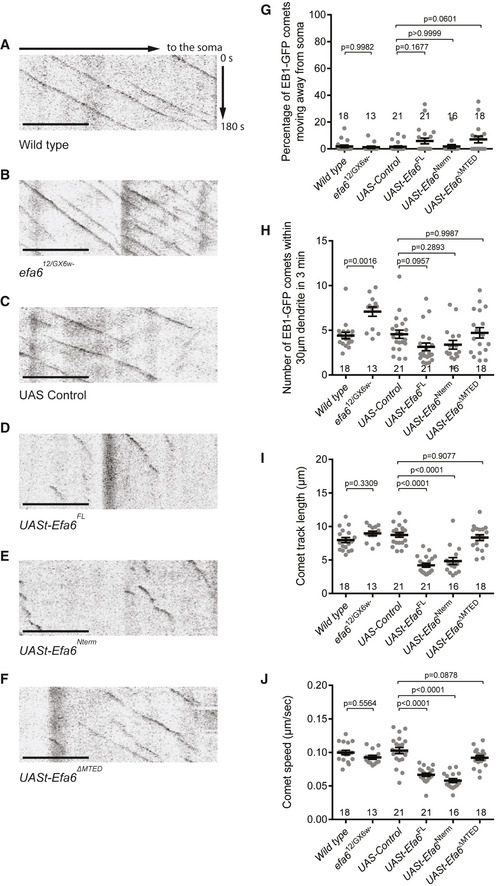
Efa6 has been shown to directly block microtubule polymerization in vitro and function as a microtubule collapse factor in the axons of cultured Drosophila neurons (Qu et al, 2019). We utilized the EB1‐GFP maker to label growing microtubule plus ends and monitor dynamics/orientation of dendritic microtubules in ddaC neurons in vivo. In Drosophila da neurons, the vast majority of dendritic EB1‐GFP comets (> 95%) move towards the soma (retrograde), whereas almost all axonal EB1‐GFP comets migrate away from the soma (anterograde), suggesting a nearly uniform minus‐end‐out MT orientation in major dendrites and a plus‐end‐out MT orientation in axons (Stone et al, 2008; Mattie et al, 2010). In wild‐type ddaC neurons, we found that almost all the dendritic EB1‐GFP comets moved towards the soma in an average speed of 0.10 µm/s (Fig 4A,G,J). The average number of EB1‐GFP comets was 4.40 per 30 µm dendrites (Fig 4A and H), and the average track length was 7.96 µm within 3 min (Fig 4A and I). In Efa6‐depleted neurons, however, the average number of EB1 comets increased significantly to 7.08 per 30 µm dendrites (Fig 4B and H), while the orientation, track length and speed showed no significant alteration (Fig 4B,G,I,J). These data suggest a major role of endogenous Efa6 in microtubule nucleation in ddaC dendrites. In contrast, the directionality, number, track length and speed of EB1‐GFP in the proximal axons of Efa6‐depleted neurons remained unaltered, compared to those in the wild‐type neurons (Appendix Fig S4C), which resembles a previous finding that Efa6 is dispensable for microtubule polymerization in axon shafts of CNS neurons; loss of Efa6 does not affect the polymerisation of microtubules within the axon core but rather at the axon cortex (Qu et al, 2019). Thus, these data suggest that Efa6 might regulate microtubule growth mainly in the dendrites of ddaC neurons. Previous studies show that cytoskeletal alterations can cause a neuronal stress response via JNK signalling (Massaro et al, 2009; Xiong et al, 2010; Feng et al, 2019). This enhanced microtubule growth phenotype, however, is independent of JNK pathway, as the expression levels of the JNK reporter puc‐lacZ showed no significant difference between efa6 RNAi and control RNAi neurons (Appendix Fig S4D). In contrast to those in the control neurons (Fig 4C,I,J), the EB1 track length was significantly decreased to 4.21 µm, and the EB1 speed decreased to 0.07 µm/s in the dendrites of Efa6FL‐overexpressing neurons (Fig 4D,I,J). Similarly, in Efa6Nterm‐expressing neurons, dendritic EB1 track length and speed were decreased to 4.84 µm and 0.06 µm/s (Fig 4E,I,J). Efa6FL‐ or Efa6Nterm‐overexpressing neurons also showed a trend to reduce the comet number in their dendrites (Fig 4D,E,H). These data suggest that Efa6 overexpression can impact mainly on microtubule polymerization and less significantly on nucleation. However, the microtubule orientation was not significantly affected in Efa6FL‐ or Efa6Nterm‐overexpressing neurons (Fig 4D,E, G). The same effects of Efa6FL and Efa6Nterm overexpression on EB1‐GFP behaviour support the notion that Efa6 exerts its function in the cytoplasm. Moreover, in Efa6ΔMTED‐expressing neurons, dendritic microtubule growth and dynamics resembled those in wild‐type neurons (Fig 4F,G–J). Therefore, these results indicate that Efa6 negatively regulates microtubule growth. However, unlike other microtubule regulators, such as Patronin, mini‐spindles and TACC (Feng et al, 2019; Wang et al, 2019; Tang et al, 2020), Efa6 does not modulate minus‐end‐out microtubule orientation in the dendrites in ddaC neurons.
Figure 4. Efa6 inhibits microtubule polymerization in the proximal dendrites of ddaC neurons.
-
A–FRepresentative kymographs of EB1‐GFP comets in the proximal dendrites of ddaC neurons at 96 h AEL. The horizontal arrow indicates the direction towards the soma, and the vertical arrow indicates the time. In contrast to the wild type (A), the number of EB1‐GFP comets was significantly increased in efa612/GX6w‐ mutant neurons (B). The comet length and speed were significantly decreased in Efa6FL‐ and Efa6Nterm‐overexpressing neurons, but not in Efa6ΔMTED‐overexpression neurons (C‐F).
-
G–JQuantification of EB1‐GFP orientation, number, track length and speed.
Data information: In (G–J), the number of samples (n) in each group is shown above the x‐axis. ns, not significant, **P < 0.01, ***P < 0.001, as assessed by two‐tailed Student's t‐test or one‐way ANOVA with Bonferroni test. All error bars represent SEM. Three independent experiments were conducted. The scale bars in (A–F) represent 10 µm.
Source data are available online for this figure.
Efa6 promotes microtubule turnover and disassembly prior to dendrite pruning
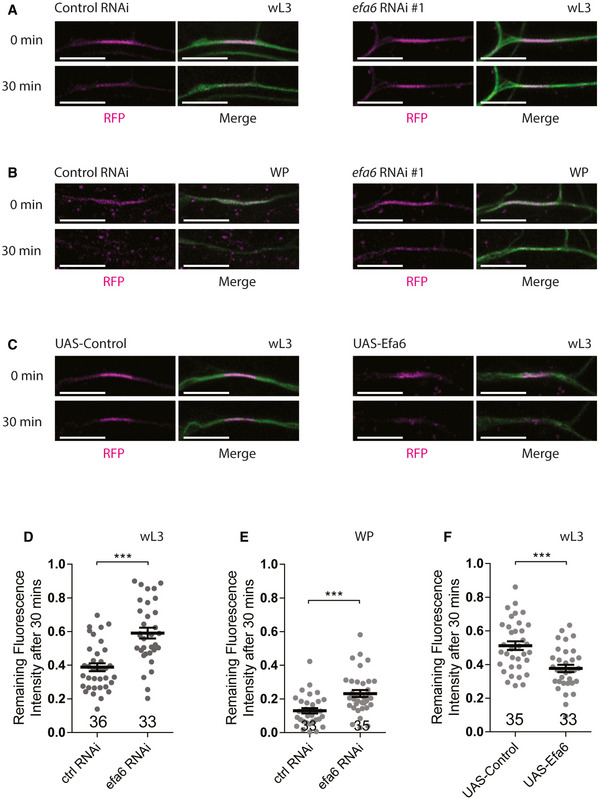
To assess if Efa6 promotes dendrite pruning via microtubule turnover, we next attempted to take advantage of a photoconversion approach by expressing a chimera containing a tandem photoconvertible EOS dimer fused to α‐tubulin (tdEOS::α‐tubulin; Barlan et al, 2013; Lu et al, 2013; Tao et al, 2016). Upon its photoconversion from green to red in a 7‐µm segment of proximal dendrites, we assessed the remaining intensity of the converted EOS after 30‐min recovery. In the dendrites of efa6 RNAi ddaC neurons, we observed higher amounts of the photoconverted tdEOS::α‐tubulin remaining at wL3 (Fig 5A,D) or white prepupal (WP) stages (Fig 5B,E), as compared to those in control neurons (Fig 5A,B, D,E), suggesting reduced microtubule turnover upon Efa6 knockdown. In contrast, the decay of converted tdEOS::α‐tubulin signals accelerated significantly in Efa6‐overexpressing neurons than that in the control neurons (Fig 5C,F). Thus, these data suggest that Efa6 promotes microtubule turnover to enhance microtubule depolymerization, a possible prerequisite for the execution of neuronal pruning.
Figure 5. Efa6 promotes microtubule turnover in the proximal dendrites of ddaC neurons prior to dendrite pruning.
-
A–CLive confocal images of ddaC dendrites expressing tdEOS::α‐tubulin driven by ppk‐Gal4. The microtubule turnover rate was significantly reduced in efa6 RNAi neurons at wL3 (A, right panels) and WP stages (B, right panels) and increased in Efa6‐overexpressing neurons at wL3 stage (C, right panels), compared to the control neurons (A–C, left panels).
-
D–FQuantification of microtubule turnover rate as indicated by the remaining fluorescence intensity of converted EOS after 30 min.
Data information: In (D–F), the number of samples (n) in each group is shown above the x‐axis. ***P < 0.001, as assessed by two‐tailed Student’s t‐test. All error bars represent SEM. Three independent experiments were conducted. The scale bars in (A–C) represent 10 µm.
Source data are available online for this figure.
To investigate whether Efa6 is essential for microtubule disassembly in the dendrites, we examined the levels of α‐tubulin, acetylated α‐tubulin or the microtubule‐associated protein Futsch (22C10) in ddaC neurons. Microtubules in neurons were visualized by staining with the 22C10 antibody against Futsch (Roos et al, 2000). In contrast to wild‐type neurons, efa6 RNAi neurons exhibited a mild but significant increase in polymerized microtubules in the proximal dendrites at WP stage (Appendix Fig S5A), as shown by Futsch staining. Knockdown of Efa6 also resulted in enhanced levels of acetylated α‐tubulin (Appendix Fig S5B), which labels stable microtubules (Janke & Bulinski, 2011). Conversely, Efa6 overexpression led to strong decreases in the levels of Futsch and α‐tubulin in the dendrites, compared to those in the control neurons (Appendix Fig S5C,D). Therefore, Efa6 governs microtubule mass in the dendrites of ddaC neurons, via promoting microtubule turnover and disassembly. To ascertain a role of Efa6 in disassembly of dendritic microtubules at the onset of dendrite pruning, we measured the levels of Futsch‐positive microtubules upon gene‐switch‐induced efa6 RNAi knockdown. Compared to no alterations at the 3rd instar larval stage before RU486 treatment, Futsch levels were slightly upregulated at WP stage and more significantly increased at 6‐h APF (Fig EV1C) in the ddaC dendrites of RU486‐fed animals. Thus, this gene‐switch results suggest that Efa6 regulates disassembly of dendritic microtubules at the prepupal stage and, in turn, dendrite pruning.
Efa6 promotes dendrite pruning via microtubule destabilization
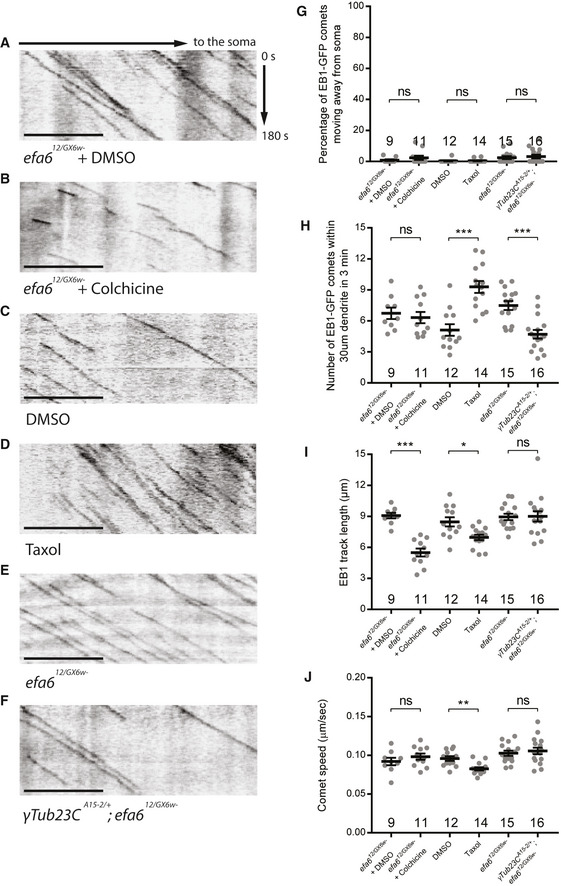
We next ascertained if impaired microtubule destabilization leads to the dendrite pruning defects in efa6 mutant neurons. To this end, we first destabilized or stabilize microtubules by treatment with the microtubule‐depolymerizing drug colchicine or microtubule‐stabilizing drug taxol, respectively. 72‐h AEL larvae were fed with a low concentration of colchicine (1 μg/ml) or taxol (30 μM), which enabled proper head eversion and survival to the adulthood. One‐day colchicine treatment at a low concentration caused microtubule depolymerization without disturbing the minus‐end‐out microtubule orientation in the proximal dendrites of efa612/GX6w‐ ddaC neurons (Fig EV2A,B,G). Average length of dendritic EB1‐GFP comets was significantly shortened (Fig EV2I), consistent with its role in inducing microtubule catastrophe at the plus ends (Mohan et al, 2013). By contrast, one‐day taxol treatment enhanced microtubule polymerization with normal minus‐end‐out orientation in the dendrites (Fig EV2C,D,G). Upon taxol treatment, the number of EB1‐GFP comets and overall Futsch levels were significantly increased in the dendrites (Fig EV2H, Appendix Fig S6A), whereas the track length and speed were reduced (Fig EV2, EV3, EV4, EV5I–J). Importantly, compared to the efa6 RNAi neurons alone (Fig 6A,I), two‐day colchicine treatment significantly attenuated the dendrite pruning defects in efa6 RNAi neurons at 16‐h APF (Fig 6B,I). Conversely, two‐day taxol treatment, which caused partial dendrite pruning defects in wild‐type neurons (Fig 6J), exacerbated the dendrite pruning defects in efa6 RNAi neurons at 16‐h APF (Fig 6C,D,J). Neither colchicine nor taxol treatment altered the average number of primary and secondary dendrites in ddaC neurons at WP stage (Appendix Fig S6B). Thus, these data support the notion that excess microtubule polymerization contributes to the dendrite pruning defects in efa6 mutant neurons.
Figure EV2. Colchicine treatment or reduced γ‐tubulin inhibited dendritic microtubule polymerization in efa6 loss‐of‐function neurons, whereas taxol treatment enhanced microtubule polymerization.
-
A–FRepresentative kymographs of EB1‐GFP comets in the proximal dendrites ddaC neurons. Colchicine treatment significantly reduced comet length in efa612/GX6w‐ mutant background (A–B). Taxol treatment significantly increased comet number in wild‐type ddaC neurons (C–D). Removal of one copy of γTub23C significantly reduced EB1‐GFP comet numbers in the efa612/GX6w‐ mutant ddaC dendrites (E–F).
-
G–JQuantification of EB1 comet orientation, number, length and speed.
Data information: In (G–J), the number of samples (n) in each group is shown on the x‐axis. ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, as assessed by two‐tailed Student’s t‐test. All error bars represent SEM. Three independent experiments were conducted. The scale bars in (A–F) represent 10 µm.
Source data are available online for this figure.
Figure EV3. Stathmin promotes microtubule turnover and negatively regulates overall microtubule mass in the dendrites of ddaC neurons.
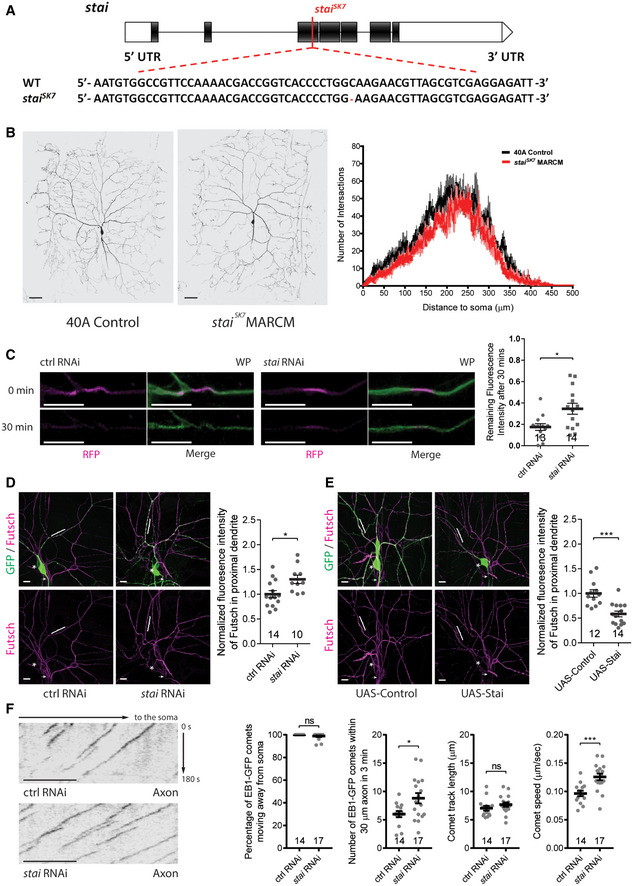
-
AA schematic diagram of the stai gene structure and the lesion site of staiSK7 allele. The red colour highlights the mutated DNA sequence.
-
BLive confocal images of wild‐type and staiSK7 mutant ddaC MARCM clones at wL3 stage (left and middle panels, respectively) and Sholl analysis (right panel).
-
CLive confocal images of control and stai RNAi ddaC dendrites expressing tdEOS::α‐tubulin at WP stage (left and middle panels, respectively). The microtubule turnover rate was significantly reduced in stai RNAi ddaC dendrites. Quantification of microtubule turnover rate as indicated by the remaining fluorescence intensity of converted EOS at 30 min after photoconversion (right panel).
-
D, EConfocal images of ddaC neurons expressing ppk‐Gal4‐driven mCD8‐GFP stained with anti‐Futsch. stai RNAi (D) significantly increased Futsch staining, whereas Stai overexpression (E) significantly reduced Futsch staining in the dendrites. The ddaC somas are labelled by asterisks, the axons by arrows and the dendrites by brackets (left panels). Quantification of Futsch fluorescence intensity in proximal dendrites (right panel).
-
FRepresentative kymographs of EB1‐GFP comets in the proximal axons of ddaC neurons at 96 h AEL (left panels) and quantification of comet orientation (anterograde movement), number, length and speed (right panels).
Data information: In (C–F), the number of samples (n) in each group is shown on the x‐axis. ns, not significant, *P < 0.05, ***P < 0.001, as assessed by two‐tailed Student’s t‐test. All error bars represent SEM. Three independent experiments were conducted. The scale bars in (B) and (C–F) represent 50 µm and 10 µm, respectively.
Source data are available online for this figure.
Figure EV4. Temporal knockdown of stai upregulated dendritic microtubule level and inhibited dendrite pruning.
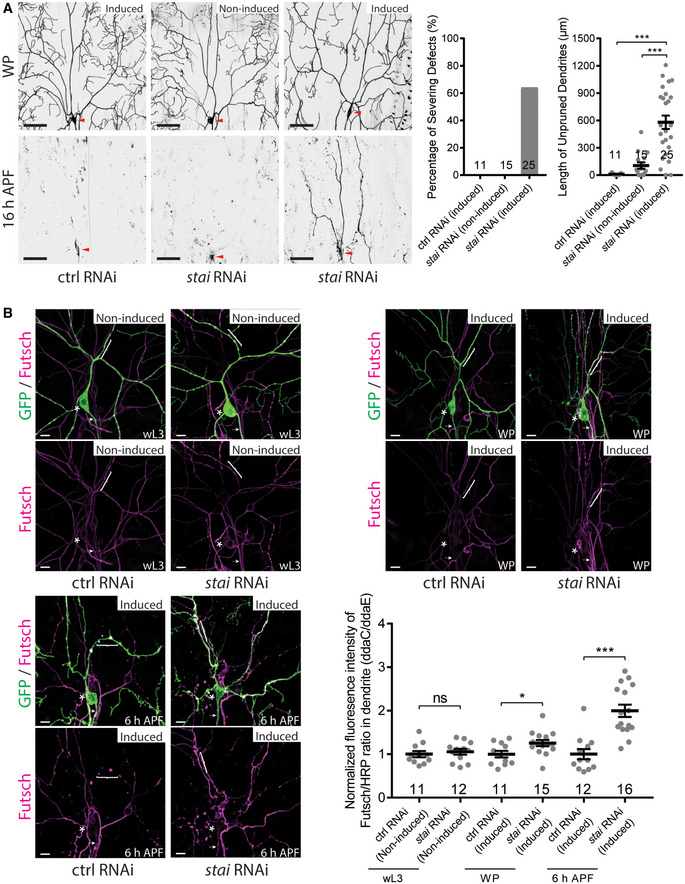
- Live confocal images of ddaC neurons labelled by ppk‐tdGFP at WP and 16‐h APF. Expression of stai RNAi construct via RU486‐induced GeneSwitch‐Gal4 significantly inhibited dendrite pruning at 16‐h APF, compared to the control RNAi or non‐induced condition. Red arrowheads point to the ddaC somas (left panels). Quantification of dendrite severing defects (middle panel) and unpruned dendrite lengths at 16‐h APF (right panel).
- Confocal images of ddaC neurons expressing ppk‐tdGFP immunostained with anti‐Futsch at wL3, WP and 6‐h APF. Induction of stai RNAi expression resulted in elevated levels of dendritic Futsch at WP and 6‐h APF, compared to the control RNAi or non‐induction condition at wL3. The ddaC somas in are labelled by asterisks, the axons by arrows and the dendrites by brackets. Quantification of Futsch fluorescence intensity in ddaC dendrites (bottom right panel).
Data information: In (A–B), the number of samples (n) in each group is shown on the x‐axis. ns, not significant, *P < 0.05, ***P < 0.001, as assessed by one‐way ANOVA with Bonferroni test or two‐tailed Student’s t‐test. All error bars represent SEM. Three independent experiments were conducted. The scale bars in (A) and (B) represent 50 µm and 10 µm, respectively.
Source data are available online for this figure.
Figure 6. Efa6 promotes dendrite pruning via microtubule destabilization.
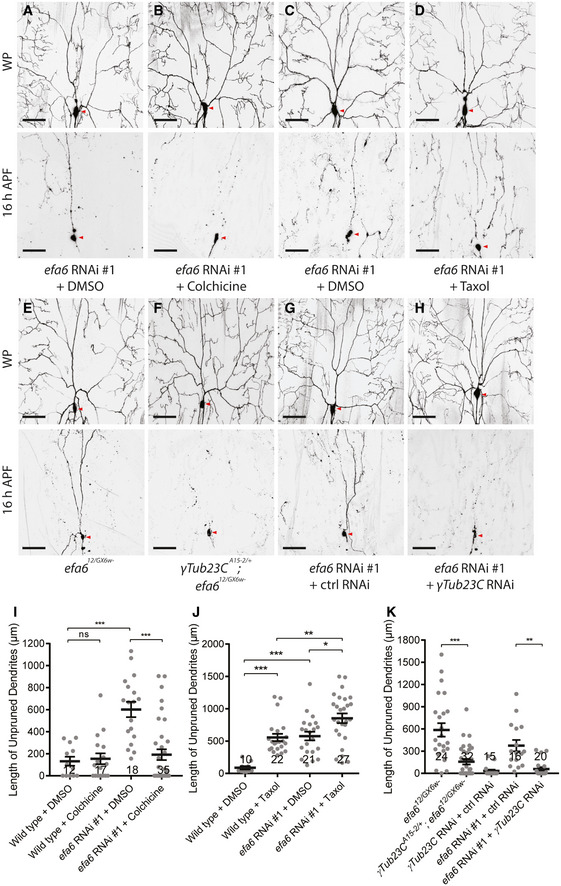
-
A–HLive confocal images of ddaC neurons at WP stage and 16‐h APF. Treatment with colchicine significantly suppressed the pruning defects of efa6 RNAi neurons (A, B), while taxol treatment enhanced the pruning defects (C, D). Removal of one copy of γTub23C (F) or expression of γTub23C RNAi (H) almost fully rescued the dendrite pruning defects in efa6 mutant (E) or RNAi neurons (G), respectively. Red arrowheads point to the ddaC somas.
-
I–KQuantification of unpruned dendrite lengths at 16 h APF.
Data information: In (I–K), the number of samples (n) in each group is shown above the x‐axis. ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, as assessed by two‐tailed Student’s t‐test or one‐way ANOVA with Bonferroni test. All error bars represent SEM. Three independent experiments were conducted. The scale bars in (A–H) represent 50 µm.
Source data are available online for this figure.
Treatment with microtubule‐depolymerizing drugs can cause transcriptional changes in neurons (Nechipurenko & Broihier, 2012). We next genetically knocked down γTub23C, the major Drosophila somatic γ‐tubulin, or removed one copy of γTub23C to attenuate microtubule nucleation ability and thereby compromise microtubule polymerization levels in efa6 RNAi or mutant ddaC neurons. Removing one copy of γTub23C (γTub23CA15‐2/+ ) indeed led to a significant reduction in the number of dendritic EB1‐GFP comets without disturbing their orientation in efa612/GX6w‐ mutant neurons (Fig EV2E–H), suggesting reduced microtubule nucleation and in turn downregulated polymerization. Importantly, removing one copy of γTub23C significantly rescued the dendrite pruning defects of efa612/GX6w‐ mutant neurons (Fig 6E,F,K). Likewise, when γTub23C was knocked down via a functional RNAi line (Ori‐McKenney et al, 2012), the dendrite pruning defects of efa6 RNAi neurons were almost fully rescued (Fig 6G,K). Thus, reduction of γTub23C function, which likely leads to reduced microtubule nucleation and thereby decreased microtubule polymerization in dendrites, attenuates the dendrite pruning defects in efa6 mutant ddaC neurons.
Taken together, these pharmacological and genetic interaction data suggest that microtubule reduction drives dendrite pruning in ddaC neurons.
The microtubule‐destabilizing factor Stathmin promotes dendrite pruning via facilitating microtubule depolymerization and turnover
In our systematic RNAi screen, we also identified the microtubule‐destabilizing factor Stai as another new regulator of dendrite pruning. Stai and its family proteins are known to disassemble microtubules via either sequestrating free tubulins or directly binding microtubules to trigger catastrophe (Belmont & Mitchison, 1996; Gigant et al, 2000). A stai RNAi line (BL36902) exhibited dendrite pruning defects upon ppk‐Gal4‐mediated expression. While wild‐type neurons completely eliminated their dendrites at 16‐h APF (Fig 7A,G,H), 68% of stai RNAi neurons failed to sever their larval dendrites at the same time point (Fig 7B,G,H). To confirm the RNAi phenotype, we generated a new stai allele, staiSK7 , by CRISPR/Cas9 mutagenesis (Fig EV3A; Kondo & Ueda, 2013). staiSK7 ddaC clones also exhibited the dendrite pruning defects with a similar penetrance (Fig 7C,G,H). In addition, slightly simple dendrite arbours were observed in larval staiSK7 ddaC clones (Fig EV3B). Similar to staiSK7 , 72% of mutant clones from the previously published null allele staiKO failed to sever their larval dendrites (Fig 7D,G,H). Thus, Stai is important for dendrite pruning in ddaC neurons. Importantly, the dendrite pruning defects were significantly enhanced in stai and efa6 double RNAi neurons (Fig 7E,G,H), as compared to those in their individual RNAi neurons (Fig 7G,H). Likewise, compared to efa6 mutant or stai RNAi neurons alone, stai RNAi + efa612/GX6w‐ double‐mutant neurons exhibited more severe dendrite pruning defects (Fig 7F,G,H). These additive effects suggest that Efa6‐dependent and Stai‐dependent pathways may converge to promote microtubule disassembly and thereby dendrite pruning.
Figure 7. Stathmin regulates dendrite pruning via inhibiting microtubule polymerization.
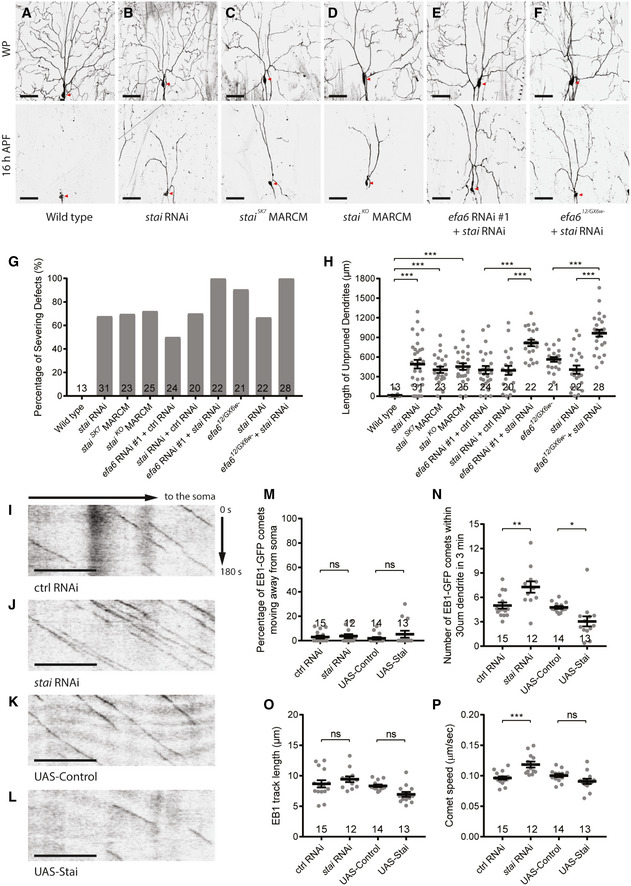
-
A–FLive confocal images of ddaC neurons at WP stage and 16 h APF. stai RNAi neurons (B), staiSK7 (C), staiKO (D) mutant MARCM clones showed similar dendrite pruning defects, compared to the wild‐type ddaC neurons (A). Double RNAi of stai and efa6 significantly enhanced the dendrite pruning defects (E), compared to their individual RNAi knockdown. Expression of stai RNAi in efa612/GX6w‐ mutant background also significantly enhanced the dendrite pruning defects (F), compared to the RNAi or mutant alone. Red arrowheads point to the ddaC somas.
-
G, HQuantification of dendrite severing defects and unpruned dendrite lengths at 16 h APF.
-
I–LRepresentative kymographs of EB1‐GFP comets driven by Gal44‐77 in the proximal dendrites of ddaC neurons at 96 h AEL.
-
M–PQuantification of EB1‐GFP comet orientation, number, track length and speed.
Data information: In (G–H) and (M–P), the number of samples (n) in each group is shown on the x‐axis. ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, as assessed by one‐way ANOVA with Bonferroni test or two‐tailed Student’s t‐test. All error bars represent SEM. Three independent experiments were conducted. The scale bars in (A–F) and (I–L) represent 50 µm and 10 µm, respectively.
Source data are available online for this figure.
We then investigated if Stai regulates microtubule orientation and polymerization in the dendrites. Like those in wild‐type neurons (Fig 7I,K,M), almost all dendritic EB1‐GFP comets migrated retrogradely towards the soma in stai RNAi or Stai‐overexpressing ddaC neurons (Fig 7J,L,M). The average number of EB1‐GFP comets per 30‐µm dendrites significantly increased from 5.0 to 7.3 in stai RNAi ddaC neurons (Fig 7N), however, reduced from 4.8 to 3.0 in Stai‐overexpressing neurons (Fig 7N). stai RNAi knockdown did not affect the EB1‐GFP comet track length (Fig 7O) but caused an increase in the speed in the dendrites (Fig 7P). These data suggest that like Efa6, Stai is important for proper growth of microtubules, but not for their minus‐end‐out orientation in the dendrites. Similar to those in the dendrites (Fig 7M–P), knockdown of stai led to significant increases in EB1‐GFP comet number and speed in ddaC axons (Fig EV3F), suggesting its global effects in both dendrites and axons. Consistently, Stai promotes microtubule turnover in the dendrites, as knockdown of stai led to higher amounts of the photoconverted tdEOS::α‐tubulin remaining at WP stage (Fig EV3C). Finally, Stai is also required to maintain proper microtubule levels in the dendrites. stai RNAi neurons exhibited a significant increase in Futsch staining in the dendrites (Fig EV3D), whereas its overexpression led to a significant reduction in Futsch levels in the dendrites (Fig EV3E). To ascertain whether Stai is required for disassembly of dendritic microtubules at the onset of dendrite pruning, we then examined polymerized microtubule levels in ddaC neurons upon gene‐switch‐induced stai RNAi knockdown. We temporally induced the expression of stai RNAi construct from the 3rd instar larval stage via the gene‐switch system. After RU486 treatment, the majority of stai RNAi neurons failed to prune away their larval dendrite (Fig EV4A), like continuous stai knockdown (Fig 7B,7G,H). We found that Futsch levels were not altered at the 3rd instar larval stage before RU486 treatment. However, after RU486 treatment, Futsch levels were significantly upregulated at WP and 6‐h APF (Fig EV4B), suggesting a role of Stai in promoting disassembly of dendritic microtubules at the prepupal stage and subsequently facilitating dendrite pruning.
Collectively, Stai, another negative regulator of microtubule growth, promotes dendrite pruning via facilitating microtubule turnover and depolymerizing microtubules in dendrites, further strengthening the conclusion that microtubule disassembly drives the ensuing dendrite pruning process in ddaC neurons.
Discussion
Microtubule disassembly, one of the earliest cellular alterations in pruning dendrites or axons, has been thought to be a prerequisite for the execution of neuronal pruning (Watts et al, 2003; Williams & Truman, 2005; Lee et al, 2009). Here, our systematic study reveals that dendrite pruning selectively requires two microtubule destabilizers, Efa6 and Stai. Moreover, our pharmacological and genetic manipulations strongly support a causative role of microtubule disassembly in promoting dendrite pruning of ddaC sensory neurons.
Efa6 promotes dendrite pruning via disassembling dendritic microtubules
The Efa6 protein family is conserved from yeast to mammals and was originally identified in mammals to regulate endosomal membrane recycling and actin cytoskeletal rearrangement via its C‐terminal Sec7 GEF domain (Franco et al, 1999). In the mammalian nervous systems, Efa6, via the GEF domain, regulates dendritic spine formation and axon regeneration in an Arf6‐dependent manner (Choi et al, 2006; Eva et al, 2017). However, in worms and flies, Efa6 orthologs act independently of Arf6 to inhibit microtubule polymerization via their respective N‐terminal MTED domains (O'Rourke et al, 2010; Qu et al, 2019). The MTED domain is conserved in worms and flies, but not in mammals (O'Rourke et al, 2010; Qu et al, 2019). Through their MTEDs, Efa6s function as negative regulators of developmental axon growth/branching and axonal regeneration after injury (Chen et al, 2015; Qu et al, 2019). In this study, we report an important role of Efa6 in promoting dendrite pruning of sensory neurons via its MTED domain. Multiple genetic manipulations with loss or gain of Efa6 function demonstrate that Efa6 is both necessary and sufficient to promote dendrite pruning. Moreover, the structure–function analysis reveals that the MTED domain is essential for Efa6 to promote dendrite pruning, whereas the C‐terminal Sec7 GEF domain is dispensable. Overexpression of the MTED‐deleted Efa6 variant was unable to induce precocious dendrite pruning phenotype. Unlike other microtubule regulators, such as Patronin and Msps/TACC (Feng et al, 2019; Wang et al, 2019; Tang et al, 2020), Efa6 does not modulate the minus‐end‐out microtubule orientation in ddaC dendrites, but rather inhibits microtubule growth and promotes microtubule disassembly at the proximal regions of the dendrites.
How does Efa6 disassemble microtubules in the dendrites during pruning? We found that loss of Efa6 function resulted in increased number of EB1‐GFP‐labelled plus ends of microtubules and reduced microtubule depolymerization/turnover at the proximal dendrites, whereas gain of Efa6 function led to enhanced microtubule depolymerization and disassembly. We therefore prefer the model that Efa6 may directly disassemble microtubules in vivo at the proximal dendrites, leading to initial severing of dendrites. In line with this model, fly Efa6 directly interacts with tubulins and inhibits microtubule polymerization per se (Qu et al, 2019). In the case of axon injury and regeneration in nematodes, Efa6 was reported to indirectly inhibit microtubule dynamics by suppressing its binding partners, TAC‐1 (TACC in Drosophila) and ZYG‐8 (Doublecortin‐like kinase, CG17528 in Drosophila; Chen et al, 2015). However, fly TACC and Efa6 may function independently during dendrite pruning. Our previous study reported that TACC regulates the minus‐end‐out microtubule orientation to promote dendrite pruning in ddaC dendrites (Tang et al, 2020). By contrast, Efa6 does not affect dendritic microtubule orientation in these neurons. Moreover, we generated mutants of the fly zyg8/CG17528 which did not show any dendrite pruning defect, and double mutant of efa6 and zyg8/CG17528 displayed no genetic interaction (Appendix Fig S7A–D). Efa6 has a cortical localization pattern in worm neurons/embryos (O'Rourke et al, 2010; Chen et al, 2015) and mouse fibroblasts (Qu et al, 2019). Interestingly, we found the cytoplasmic distribution of fly Efa6 in ddaC neurons despite its membrane association in epidermal cells. This finding raises the possibility that Efa6 might be released from the plasma membrane to the cytoplasm where it might more potently eliminate microtubules during dendrite pruning. In support of this notion, worm Efa6 can relocalize from the plasma membrane to microtubule minus ends and inhibit microtubule dynamics in axons upon injury (Chen et al, 2015).
A systematic study of microtubule destabilizers in dendrite pruning
In this study, we have systematically interrogated various microtubule‐destabilizing factors for their possible roles in dendrite pruning using both RNAi and clonal approaches. We demonstrate that dendrite pruning requires two microtubule destabilizers, Efa6 and Stai. Stai and its mammalian homologues negatively regulate microtubule dynamics and disassemble microtubules via a mechanism distinct from Efa6. Stai can either sequester α/β‐tubulin dimers and prevent their incorporation into growing microtubules or directly interact with microtubules to promote their disassembly (Cassimeris, 2002). Stai has been involved in diverse models of neurodegeneration, neuronal migration axonal transport, neuronal polarization and regeneration as well as plasticity (Chauvin & Sobel, 2015). Drosophila Stai regulates axonal microtubule integrity, synapse stability and neuronal functions (Graf et al, 2011; Duncan et al, 2013). Here, we identified an additive function of Stai and Efa6 in dendrite pruning of ddaC neurons. Like Efa6, Stai promotes microtubule turnover/disassembly but does not regulate microtubule orientation in the dendrites of ddaC sensory neurons. Loss of Stai function resulted in an increase in polymerized microtubules; conversely, overexpression of Stai caused strong reductions in microtubule growth and mass. Thus, Stai is essential for inhibiting microtubule polymerization and promoting the disassembly during dendrite pruning. However, in contrast to our finding, a previous study has also reported that loss of Stai function results in decreased microtubule levels in the axons of Drosophila CNS neurons (Duncan et al, 2013). It is possible that differential roles of Stai in PNS and CNS neurons might be due to its different phosphorylation (Voelzmann et al, 2016). In Xenopus, the microtubule‐destabilizing activity of Stai is dependent on its phosphorylation state, which is regulated by the serine/threonine type‐2A phosphatase (Tournebize et al, 1997).
The microtubule‐severing enzymes (katanin, fidgetin and spastin), which hydrolyse ATP to sever microtubules into small pieces in vitro (Kuo & Howard, 2021), are apparent candidates that disassemble dendritic microtubules to promote neuronal pruning. These enzymes were also reported to sever microtubules and regulate the neuromuscular junction development or dendrite arborization in Drosophila and mammals (Ahmad et al, 1999; Sherwood et al, 2004; Trotta et al, 2004; Jinushi‐Nakao et al, 2007; Mao et al, 2014; Leo et al, 2015). Moreover, spastin was reported to promote axon pruning at the neuromuscular junctions in postnatal mice by locally destabilizing microtubules (Brill et al, 2016). However, RNAi knockdown of these severing enzymes did not cause any prominent dendrite pruning defects in Drosophila ddaC neurons in the previous studies (Lee et al, 2009; Tao et al, 2016) and our RNAi screen (Appendix Table S1). To exclude the possibility that RNAi knockdown was inefficient for these genes, we generated several mutant alleles for these genes and conducted their clonal analysis in this study. Our results further substantiate that these individual enzymes are indeed dispensable for dendrite pruning. Kat‐60L1, an AAA ATPase analogous to Kat‐60, is involved in ddaC dendrite pruning (Lee et al, 2009) (also this study). However, we found that Kat‐60L1 has no apparent microtubule‐disassembly function in ddaC neurons. Unlike Kat‐60, overexpression of the Kat‐60L1 short isoform neither impaired microtubule turnover (Fig EV5A) nor affected the microtubule levels in the dendrites of ddaC neurons (Fig EV5B). We also analysed the effect of the long isoform of Kat‐60L1 in ddaC neurons, as the long isoform, but not the short isoform, was reported to possess microtubule‐disassembly function in adult mechanosensory neurons (Sun et al, 2021). Unexpectedly, similar to the short isoform, we did not observe reduced Futsch‐positive microtubules in the dendrites when the long isoform of Kat‐60L1 was overexpressed in ddaC neurons (Fig EV5C). Moreover, we did not observe any significant alteration in the levels of polymerized microtubules in dendrites and soma of kat‐60L1F1 mutant ddaC neurons (Fig EV5D). Thus, none of our data suggest that Kat‐60L1 acts as a microtubule‐severing enzyme in ddaC neurons, which contrasts with a recent finding showing that purified Kat‐60L1 protein possesses the microtubule‐severing activity in vitro (Sun et al, 2021). However, our data cannot rule out the possibility that Kat‐60L1 promotes dendrite pruning via severing dendritic microtubules. Further studies will be necessary to determine its potential microtubule‐severing function during ddaC dendrite pruning.
A link between microtubule disassembly and dendrite pruning
Microtubule disassembly precedes membrane scission during neuronal pruning (Williams & Truman, 2005; Lee et al, 2009; Brill et al, 2016). Here, we isolated two microtubule‐destabilizing factors, Efa6 and Stai, whose mutants exhibited excessive microtubule polymerization without affecting microtubule orientation in dendrites. These findings further provided us with the opportunity to investigate a relationship between microtubule disassembly and dendrite pruning. First, we treated efa6 RNAi larvae with colchicine, a microtubule‐destabilizing drug. A low concentration of colchicine not only reduced microtubule polymerization but also fully rescued the dendrite pruning defects in efa6 loss‐of‐function neurons. Second, the treatment with the microtubule‐stabilizing drug taxol caused excessive microtubule polymerization in dendrites and inhibited dendrite pruning in wild‐type neurons. Third, taxol treatment significantly enhanced dendrite pruning defects in the efa6 RNAi background. Finally, reduced γ‐tubulin in efa6 mutant neurons, which likely downregulates microtubule nucleation and in turn polymerization, almost fully rescued the pruning defects. Taken together, multiple lines of genetic and pharmacological evidence support that microtubule disassembly drives dendrite pruning in ddaC sensory neurons.
In summary, this systematic study highlights important roles of two negative regulators of microtubule polymerization, Efa6 and Stai, in facilitating dendrite pruning via microtubule disassembly. Moreover, our study supports a causal relationship between microtubule disassembly and dendrite pruning during early metamorphosis.
Materials and Methods
Fly strains
ppk‐Gal4 on chromosomes II and III (Grueber et al, 2003), SOP‐flp (#42) (Matsubara et al, 2011), UAS‐MicalNterm (Terman et al, 2002), UAS‐EB1‐GFP (Stone et al, 2008), UAS‐Kin‐β‐Gal (Clark et al, 1997), UAS‐Stai (Yang et al, 2012), A58‐Gal4 (Galko & Krasnow, 2004), puc‐lacZ (Martin‐Blanco et al, 1998), spas5.75 (Sherwood et al, 2004), UAS‐Venus‐Kat‐60L1(short) (Stewart et al, 2012), UAS‐RFP‐Kat‐60L1(short), UAS‐RFP‐Kat‐60L1(long) (Sun et al, 2019 ), efa612 , UAS‐Efa6FL , UAS‐Efa6Nterm , UAS‐Efa6Cterm , UAS‐Efa6ΔPH , UAS‐Efa6ΔMTED , UAS‐Efa6Nterm+CAAX , UAS‐Efa6‐GFP, kat‐60C48 , kat‐80E3 , kat‐60L1F1 , zyg841 , zyg867 , staiSK7 , fignSK1 , fignSK6 (this study).
The following stocks were obtained from Bloomington Stock Center (BSC): UAS‐mCD8::GFP, UAS‐mCD8::mCherry, UAS‐Dicer2, FRT40A, FRT82B, tubP‐Gal80, Gal4109(2)80 , Gal44‐77 (BL#8737), nos‐Cas9 (BL#54591), ppk‐CD4‐tdGFP (BL#35843), GSG2295‐Gal4 (BL#40266), UAS‐Nod‐β‐Gal (BL#9912), γ‐tub23CA15‐2 (BL#7042), efa6 RNAi #3 (BL#57449), UASp‐αTub84B‐tdEOS (BL#51313, 51314), γtub23C RNAi (BL#31204), staiKO (BL#58438), stai RNAi (BL#36902), efa6GX6w‐ (BL#60587), efa6‐Ki‐GFP (BL#60588), UAS‐Kat‐60 (BL#64115), Df(2L)Excel8008 (BL#7786).
The following stocks were obtained from Vienna Drosophila RNAi Center (VDRC): efa6 RNAi #1 (v330083), efa6 RNAi #2 (v42321), control RNAi (v25271).
CRISPR/Cas9 mutagenesis
For efa612 , kat‐60C48 , kat‐80E3 , kat‐60L1F1 , zyg841 and zyg867 mutants, guide RNAs were cloned into the pCFD4 plasmid via Gibson assembly (Port et al, 2014). staiSK7 , fignSK1 and fignSK6 mutants were generated as previously described (Kondo & Ueda, 2013). gRNA sequences used are as follows: GACGCTAACGTTCTTGCCAG for stai and GCCGTTAAGAGCAGACCGGA for fign. Transgenic gRNA flies were crossed to nos‐Cas9 flies to generate mutants. All the mutants were confirmed by DNA sequencing.
Generation of efa6 transgenes
The full‐length cDNAs of Drosophila efa6 were amplified from EST IP15395 (intron removed) into pENTR/D‐TOPO vector (Invitrogen), followed by cloning into the pTW or pTWG destination vectors (DGRC) via LR reaction. Various efa6 truncates were generated by either PCR or QuickChange mutagenesis (Agilent Tech) using pENTR‐efa6 as a template. All the embryo microinjection services were provided by BestGene Inc.
Generation of anti‐Efa6 antibody
The cDNA sequence corresponding to the aa81‐330 fragment of Efa6 was amplified from pENTR‐Efa6 into the GST expression vector (pGEX 4T‐1, Pharmacia). The GST‐fused protein was then purified to immunize mice to generate antibodies against Efa6.
MARCM and RNAi analysis of da neurons
MARCM and RNAi analysis of da neurons’ dendrites was carried out as previously described (Kirilly et al, 2009). Embryos were collected and cultivated on normal food at 25°C. To image da neurons, WPs or pupae at 7‐h APF were washed in PBS briefly and mounted with 90% glycerol. For pupae at 16‐h or 19‐h APF, WPs were first collected onto moisturized tissue paper and kept at 25°C overnight. The pupal cases were carefully removed, and the pupae were mounted with 90% glycerol just before imaging. Live confocal images of mCD8::GFP or mCD8::mCherry in a 275 × 275 µm area were acquired by Leica SPE‐II confocal microscope with 40x oil lens. Dorsal is up in all images.
The severing defect is defined by the presence of dendrites that remain attached to the soma at 16‐h APF. Total length of unpruned dendrites was measured using ImageJ plugin Simple Neurite Tracer. Scatter plot graphs with SEM were generated by GraphPad Prism software.
Live imaging of EB1‐GFP comet
Larvae expressing UAS‐EB1‐GFP driven by Gal44‐77 at 96 h AEL were mounted with a tiny drop of halocarbon oil (Santa Cruz, sc‐250077) onto slides for time‐lapse imaging. EB1‐GFP were visualized using Olympus FV3000 confocal with 60× oil lens, 3× zoom. 125 frames of 6‐z‐step images were acquired within 3 min (at 1.45‐s intervals). The ImageJ plugin KymographBuilder was used to generate kymographs from z‐projected time‐lapse images.
Microtubule photoconversion assay
wL3 or WP expressing 2 copies of UASp‐αTub84B‐EOS driven by ppk‐Gal4 were mounted with halocarbon oil onto slides for imaging using Olympus FV3000 confocal (60x oil lens, 3x zoom). A segment (˜7 µm) of dorsal proximal dendrites was stimulated by 405 nm laser to photoconvert the αTub‐EOS from green to red. Photoconverted neurons were immediately imaged and imaged again at 30 min using 488 nm and 561 nm lasers. The remaining fluorescence intensity (FI) was calculated as (FI[converted] ‐ FI[neighbouring])30min/(FI[converted] − FI[neighbouring])0min.
Colchicine, taxol and RU486 treatment
Embryos were collected at 12‐h intervals and reared on normal food. The 2nd instar larvae were transferred to the food containing 5 µg/ml colchicine (Sigma‐Aldrich, C9754) or 30 µM taxol (Bio‐Techne, 1097). wL3 or WPs were collected after 2‐day drug treatment for confocal imaging or microtubule staining. For EB1‐GFP experiments, 1 µg/ml colchicine or 30 µM taxol was used for 1‐day treatment. For gene‐switch experiments, the 3rd instar larvae (about 96 h after egg laying, AEL) were fed with 240 µg/ml RU486/mifepristone. White prepupae were picked up after 24 h treatment, subject to further experimental analyses. Please note that the actual feeding period was as short as 6–8 h, as the 3rd instar larvae started to crawl out from the RU486 food after several hours’ treatment.
Immunofluorescence and antibodies
The following antibodies were used in this study: Mouse anti‐Nrg (1:20, DSHB, BP104), mouse anti‐Dlg (1:50, DSHB, 4F3), mouse anti‐Efa6 (1: 250; Yu lab), mouse anti‐Futsch (1:50, DSHB, 22C10), mouse anti‐α‐Tubulin (1:500, Sigma, T9026), mouse anti‐Acetylated Tubulin (1:500, Sigma, T6793), mouse anti‐Galactosidase (1:1,000, Promega, Z3781), rabbit anti‐GFP (1:1,000, Invitrogen, A‐11122), Cy3‐conjugated goat anti‐mouse antibody, 488‐conjugated goat anti‐rabbit antibody and 649‐conjugated goat anti‐HRP antibody (1:500, Jackson Laboratories, 115‐165‐003, 111‐545‐003, 123‐495‐021).
For normal immunostaining, larvae or pupae for each set of experiments were dissected simultaneously in cold PBS and fixed with 4% formaldehyde for 20 min. For microtubule staining, samples were dissected in Ca2+ free HL3.1 saline, followed by removal of the muscles. The fillets were then fixed in freshly prepared PHEM fixation buffer with 0.25% glutaraldehyde, 4% formaldehyde, and 0.1% Triton X‐100 for 15 min. The samples were then quenched by 50 mM ammonium chloride for 5 min (Witte et al, 2008). Mounting was performed using VectaShield mounting medium. The samples were directly visualized by Leica SPE‐II confocal microscope. Images were taken from projected z‐stacks (at 1.5 µm intervals) to cover the entire volume of ddaC/E neurons.
To quantify the normal immunostaining images, the mean fluorescence intensity in the cell nuclei (puc‐lacZ) or whole soma (Efa6‐Ki‐GFP) of ddaC/ddaE neurons was measured using ImageJ, after subtracting background (Rolling Ball Radius = 30). The ddaC/ddaE ratios were then calculated and normalized to the average of the control group. To quantify the microtubule staining in the dendrites, we measure the α‐tubulin/acetylated α‐tubulin/Futsch/HRP fluorescence intensity in 20 µm of major dorsal dendrites that were 30 µm away from the centre of the soma.
Statistics
Two‐tailed Student’s t‐test was applied to determine statistical significance for pairwise comparison. One‐way ANOVA with Bonferroni test was applied when multiple groups were present. Statistical significance was defined as P < 0.001, P < 0.01, P < 0.05, ns, not significant. Error bars in all graphs represent standard error of the mean (SEM). The number of samples (n) in each group is shown on the bars.
Author contributions
SB and FY conceived and designed the study. SB performed most of the experiments. WLY and BJWL conducted some of Efa6 and katanin experiments. SK provided staiSK7 , fignSK1 and fignSK6 mutant lines. SB and FY analysed the data and wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We thank Y. Jan, A. L. Kolodkin, A. Martinez‐Arias, M.M. Rolls, N.T. Sherwood, T. J.C. Pastor‐Pareja, X. Liang, Uemura, K. Zinn, the Bloomington Stock Center (BSC), DSHB (University of Iowa) and VDRC (Austria) for generously providing antibodies and fly stocks. We thank Dr. Quan Tang and Ms. Alice Tan for generating the pCFD4‐kat60/80 and efa6 constructs, respectively, and the Yu laboratory members for helpful discussion. This work was funded by Temasek Life Sciences Laboratory Singapore (TLL‐2040) (to F.Y.).
EMBO reports (2021) 22: e52679.
Data availability
This study includes no data deposited in external repositories.
References
- Ahmad FJ, Yu W, McNally FJ, Baas PW (1999) An essential role for katanin in severing microtubules in the neuron. J Cell Biol 145: 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlan K, Lu W, Gelfand VI (2013) The microtubule‐binding protein ensconsin is an essential cofactor of kinesin‐1. Curr Biol 23: 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont LD, Mitchison TJ (1996) Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell 84: 623–631 [DOI] [PubMed] [Google Scholar]
- Brill MS, Kleele T, Ruschkies L, Wang M, Marahori NA, Reuter MS, Hausrat TJ, Weigand E, Fisher M, Ahles Aet al (2016) Branch‐specific microtubule destabilization mediates axon branch loss during neuromuscular synapse elimination. Neuron 92: 845–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L (2002) The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol 14: 18–24 [DOI] [PubMed] [Google Scholar]
- Chauvin S, Sobel A (2015) Neuronal stathmins: a family of phosphoproteins cooperating for neuronal development, plasticity and regeneration. Prog Neurogibol 126: 1–18 [DOI] [PubMed] [Google Scholar]
- Chen L, Chuang M, Koorman T, Boxem M, Jin Y, Chisholm AD (2015) Axon injury triggers EFA‐6 mediated destabilization of axonal microtubules via TACC and doublecortin like kinase. Elife 4: e08695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang Z, Ghosh‐Roy A, Hubert T, Yan D, O'Rourke S, Bowerman B, Wu Z, Jin Y, Chisholm AD (2011) Axon regeneration pathways identified by systematic genetic screening in C. elegans . Neuron 71: 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Ko J, Lee JR, Lee HW, Kim K, Chung HS, Kim H, Kim E (2006) ARF6 and EFA6A regulate the development and maintenance of dendritic spines. J Neurosci 26: 4811–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Jan LY, Jan YN (1997) Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development 124: 461–470 [DOI] [PubMed] [Google Scholar]
- Duncan JE, Lytle NK, Zuniga A, Goldstein LS (2013) The microtubule regulatory protein stathmin is required to maintain the integrity of axonal microtubules in Drosophila . PLoS One 8: e68324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eva R, Koseki H, Kanamarlapudi V, Fawcett JW (2017) EFA6 regulates selective polarised transport and axon regeneration from the axon initial segment. J Cell Sci 130: 3663–3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Thyagarajan P, Shorey M, Seebold DY, Weiner AT, Albertson RM, Rao KS, Sagasti A, Goetschius DJ, Rolls MM (2019) Patronin‐mediated minus end growth is required for dendritic microtubule polarity. J Cell Biol 218: 2309–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M, Peters PJ, Boretto J, van Donselaar E, Neri A, D'Souza‐Schorey C, Chavrier P (1999) EFA6, a sec7 domain‐containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J 18: 1480–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galko MJ, Krasnow MA (2004) Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol 2: E239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigant B, Curmi PA, Martin‐Barbey C, Charbaut E, Lachkar S, Lebeau L, Siavoshian S, Sobel A, Knossow M (2000) The 4 A X‐ray structure of a tubulin:stathmin‐like domain complex. Cell 102: 809–816 [DOI] [PubMed] [Google Scholar]
- Graf ER, Heerssen HM, Wright CM, Davis GW, DiAntonio A (2011) Stathmin is required for stability of the Drosophila neuromuscular junction. J Neurosci 31: 15026–15034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Moore AW, Jan LY, Jan YN (2003) Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol 13: 618–626 [DOI] [PubMed] [Google Scholar]
- Herzmann S, Gotzelmann I, Reekers LF, Rumpf S (2018) Spatial regulation of microtubule disruption during dendrite pruning in Drosophila . Development 145: dev156950 [DOI] [PubMed] [Google Scholar]
- Herzmann S, Krumkamp R, Rode S, Kintrup C, Rumpf S (2017) PAR‐1 promotes microtubule breakdown during dendrite pruning in Drosophila . EMBO J 36: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhou W, Dong W, Watson AM, Hong Y (2009) From the Cover: Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci USA 106: 8284–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Bulinski JC (2011) Post‐translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol 12: 773–786 [DOI] [PubMed] [Google Scholar]
- Jinushi‐Nakao S, Arvind R, Amikura R, Kinameri E, Liu AW, Moore AW (2007) Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron 56: 963–978 [DOI] [PubMed] [Google Scholar]
- Kanamori T, Kanai MI, Dairyo Y, Yasunaga K, Morikawa RK, Emoto K (2013) Compartmentalized calcium transients trigger dendrite pruning in Drosophila sensory neurons. Science 340: 1475–1478 [DOI] [PubMed] [Google Scholar]
- Kanamori T, Yoshino J, Yasunaga K, Dairyo Y, Emoto K (2015) Local endocytosis triggers dendritic thinning and pruning in Drosophila sensory neurons. Nat Commun 6: 6515 [DOI] [PubMed] [Google Scholar]
- Kirilly D, Gu Y, Huang Y, Wu Z, Bashirullah A, Low BC, Kolodkin AL, Wang H, Yu F (2009) A genetic pathway composed of Sox14 and Mical governs severing of dendrites during pruning. Nat Neurosci 12: 1497–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilly D, Wong J, Lim E, Wang Y, Zhang H, Wang C, Liao Q, Wang H, Liou Y‐C, Wang Het al (2011) Intrinsic epigenetic factors cooperate with the steroid hormone ecdysone to govern dendrite pruning in Drosophila . Neuron 72: 86–100 [DOI] [PubMed] [Google Scholar]
- Kondo S, Ueda R (2013) Highly improved gene targeting by germline‐specific Cas9 expression in Drosophila . Genetics 195: 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer R, Rode S, Rumpf S (2019) Rab11 is required for neurite pruning and developmental membrane protein degradation in Drosophila sensory neurons. Dev Biol 451: 68–78 [DOI] [PubMed] [Google Scholar]
- Kuo CT, Jan LY, Jan YN (2005) Dendrite‐specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin‐proteasome, and ecdysone signaling. Proc Natl Acad Sci USA 102: 15230–15235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Zhu S, Younger S, Jan LY, Jan YN (2006) Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron 51: 283–290 [DOI] [PubMed] [Google Scholar]
- Kuo YW, Howard J (2021) Cutting, amplifying, and aligning microtubules with severing enzymes. Trends Cell Biol 31: 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Jan LY, Jan YN (2009) Drosophila IKK‐related kinase Ik2 and Katanin p60‐like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc Natl Acad Sci USA 106: 6363–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L (1999) Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 126: 4065–4076 [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461 [DOI] [PubMed] [Google Scholar]
- Leo L, Yu W, D’Rozario M, Waddell E, Marenda D, Baird M, Davidson M, Zhou B, Wu B, Baker Let al (2015) Vertebrate fidgetin restrains axonal growth by severing labile domains of microtubules. Cell Rep 12: 1723–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Levitt P (2002) Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci 25: 409–432 [DOI] [PubMed] [Google Scholar]
- Loncle N, Agromayor M, Martin‐Serrano J, Williams DW (2015) An ESCRT module is required for neuron pruning. Sci Rep 5: 8461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncle N, Williams DW (2012) An interaction screen identifies headcase as a regulator of large‐scale pruning. J Neurosci 32: 17086–17096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Fox P, Lakonishok M, Davidson MW, Gelfand VI (2013) Initial neurite outgrowth in Drosophila neurons is driven by kinesin‐powered microtubule sliding. Curr Biol 23: 1018–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, O'Leary DD (2005) Axon retraction and degeneration in development and disease. Annu Rev Neurosci 28: 127–156 [DOI] [PubMed] [Google Scholar]
- Mao CX, Xiong Y, Xiong Z, Wang Q, Zhang YQ, Jin S (2014) Microtubule‐severing protein Katanin regulates neuromuscular junction development and dendritic elaboration in Drosophila . Development 141: 1064–1074 [DOI] [PubMed] [Google Scholar]
- Maor‐Nof M, Homma N, Raanan C, Nof A, Hirokawa N, Yaron A (2013) Axonal pruning is actively regulated by the microtubule‐destabilizing protein kinesin superfamily protein 2A. Cell Rep 3: 971–977 [DOI] [PubMed] [Google Scholar]
- Martin‐Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez‐Arias A (1998) puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila . Genes Dev 12: 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaro CM, Pielage J, Davis GW (2009) Molecular mechanisms that enhance synapse stability despite persistent disruption of the spectrin/ankyrin/microtubule cytoskeleton. J Cell Biol 187: 101–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara D, Horiuchi SY, Shimono K, Usui T, Uemura T (2011) The seven‐pass transmembrane cadherin Flamingo controls dendritic self‐avoidance via its binding to a LIM domain protein, Espinas, in Drosophila sensory neurons. Genes Dev 25: 1982–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattie FJ, Stackpole MM, Stone MC, Clippard JR, Rudnick DA, Qiu Y, Tao J, Allender DL, Parmar M, Rolls MM (2010) Directed microtubule growth, +TIPs, and kinesin‐2 are required for uniform microtubule polarity in dendrites. Curr Biol 20: 2169–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan R, Katrukha EA, Doodhi H, Smal I, Meijering E, Kapitein LC, Steinmetz MO, Akhmanova A (2013) End‐binding proteins sensitize microtubules to the action of microtubule‐targeting agents. Proc Natl Acad Sci USA 110: 8900–8905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechipurenko IV, Broihier HT (2012) FoxO limits microtubule stability and is itself negatively regulated by microtubule disruption. J Cell Biol 196: 345–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori‐McKenney KM, Jan LY, Jan YN (2012) Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron 76: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Christensen SN, Bowerman B (2010) Caenorhabditis elegans EFA‐6 limits microtubule growth at the cell cortex. Nat Cell Biol 12: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, Bullock SL (2014) Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila . Proc Natl Acad Sci USA 111: E2967–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Hahn I, Lees M, Parkin J, Voelzmann A, Dorey K, Rathbone A, Friel CT, Allan VJ, Okenve‐Ramos Pet al (2019) Efa6 protects axons and regulates their growth and branching by inhibiting microtubule polymerisation at the cortex. Elife 8: e50319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccomagno MM, Kolodkin AL (2015) Sculpting neural circuits by axon and dendrite pruning. Annu Rev Cell Dev Biol 31: 779–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode S, Ohm H, Anhäuser L, Wagner M, Rosing M, Deng X, Sin O, Leidel SA, Storkebaum E, Rentmeister Aet al (2018) Differential requirement for translation initiation factor pathways during ecdysone‐dependent neuronal remodeling in Drosophila . Cell Rep 24: 2287–2299 [DOI] [PubMed] [Google Scholar]
- Roos J, Hummel T, Ng N, Klambt C, Davis GW (2000) Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron 26: 371–382 [DOI] [PubMed] [Google Scholar]
- Rui M, Ng KS, Tang Q, Bu S, Yu F (2020) Protein phosphatase PP2A regulates microtubule orientation and dendrite pruning in Drosophila . EMBO Rep 21: e48843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O, Yaron A (2015) Mechanisms of developmental neurite pruning. Cell Mol Life Sci 72: 101–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren Vet al (2016) Schizophrenia risk from complex variation of complement component 4. Nature 530: 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood NT, Sun Q, Xue M, Zhang B, Zinn K (2004) Drosophila spastin regulates synaptic microtubule networks and is required for normal motor function. PLoS Biol 2: e429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono K, Fujimoto A, Tsuyama T, Yamamoto‐Kochi M, Sato M, Hattori Y, Sugimura K, Usui T, Kimura K, Uemura T (2009) Multidendritic sensory neurons in the adult Drosophila abdomen: origins, dendritic morphology, and segment‐ and age‐dependent programmed cell death. Neural Dev 4: 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A, Tsubouchi A, Rolls MM, Tracey WD, Sherwood NT (2012) Katanin p60‐like1 promotes microtubule growth and terminal dendrite stability in the larval class IV sensory neurons of Drosophila . J Neurosci 32: 11631–11642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MC, Roegiers F, Rolls MM (2008) Microtubules have opposite orientation in axons and dendrites of Drosophila neurons. Mol Biol Cell 19: 4122–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Cui L, Liu Z, Wang Q, Xue Z, Wu M, Sun T, Mao D, Ni J, Pastor‐Pareja JCet al (2021) Katanin p60‐like 1 sculpts the cytoskeleton in mechanosensory cilia. J Cell Biol 220: e202004184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Song Y, Dai J, Mao D, Ma M, Ni JQ, Liang X, Pastor‐Pareja JC (2019) Spectraplakin Shot Maintains Perinuclear Microtubule Organization in Drosophila Polyploid Cells. Dev Cell 49: 731–747 [DOI] [PubMed] [Google Scholar]
- Tang G, Gudsnuk K, Kuo S‐H, Cotrina M, Rosoklija G, Sosunov A, Sonders M, Kanter E, Castagna C, Yamamoto AIet al (2014) Loss of mTOR‐dependent macroautophagy causes autistic‐like synaptic pruning deficits. Neuron 83: 1131–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Rui M, Bu S, Wang Y, Chew LY, Yu F (2020) A microtubule polymerase is required for microtubule orientation and dendrite pruning in Drosophila . EMBO J 39: e103549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Feng C, Rolls MM (2016) The microtubule‐severing protein fidgetin acts after dendrite injury to promote their degeneration. J Cell Sci 129: 3274–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman JR, Mao T, Pasterkamp RJ, Yu HH, Kolodkin AL (2002) MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin‐mediated axonal repulsion. Cell 109: 887–900 [DOI] [PubMed] [Google Scholar]
- Tournebize R, Andersen SS, Verde F, Doree M, Karsenti E, Hyman AA (1997) Distinct roles of PP1 and PP2A‐like phosphatases in control of microtubule dynamics during mitosis. EMBO J 16: 5537–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta N, Orso G, Rossetto MG, Daga A, Broadie K (2004) The hereditary spastic paraplegia gene, spastin, regulates microtubule stability to modulate synaptic structure and function. Curr Biol 14: 1135–1147 [DOI] [PubMed] [Google Scholar]
- Voelzmann A, Hahn I, Pearce SP, Sánchez‐Soriano N, Prokop A (2016) A conceptual view at microtubule plus end dynamics in neuronal axons. Brain Res Bull 126: 226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wang Y, Yu F (2018) Yif1 associates with Yip1 on Golgi and regulates dendrite pruning in sensory neurons during Drosophila metamorphosis. Development 145: dev164475 [DOI] [PubMed] [Google Scholar]
- Wang Y, Rui M, Tang Q, Bu S, Yu F (2019) Patronin governs minus‐end‐out orientation of dendritic microtubules to promote dendrite pruning in Drosophila . Elife 8: e39964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Shi M, Liou YC, Lu L, Yu F (2017) Sec71 functions as a GEF for the small GTPase Arf1 to govern dendrite pruning of Drosophila sensory neurons. Development 144: 1851–1862 [DOI] [PubMed] [Google Scholar]
- Watts RJ, Hoopfer ED, Luo L (2003) Axon pruning during Drosophila metamorphosis. Neuron 38: 871–885 [DOI] [PubMed] [Google Scholar]
- Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW (2006) Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci 9: 1234–1236 [DOI] [PubMed] [Google Scholar]
- Williams DW, Truman JW (2005) Cellular mechanisms of dendrite pruning in Drosophila: insights from in vivo time‐lapse of remodeling dendritic arborizing sensory neurons. Development 132: 3631–3642 [DOI] [PubMed] [Google Scholar]
- Witte H, Neukirchen D, Bradke F (2008) Microtubule stabilization specifies initial neuronal polarization. J Cell Biol 180: 619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJL, Li S, Lim EKH, Wang Y, Wang C, Zhang H, Kirilly D, Wu C, Liou Y‐C, Wang Het al (2013) A Cullin1‐based SCF E3 ubiquitin ligase targets the InR/PI3K/TOR pathway to regulate neuronal pruning. PLoS Biol 11: e1001657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Wang X, Ewanek R, Bhat P, Diantonio A, Collins CA (2010) Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J Cell Biol 191: 211–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Inaki M, Cliffe A, Rorth P (2012) Microtubules and Lis‐1/NudE/dynein regulate invasive cell‐on‐cell migration in Drosophila . PLoS One 7: e40632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Schuldiner O (2014) Axon and dendrite pruning in Drosophila . Curr Opin Neurobiol 27: 192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang Y, Wong JJ, Lim KL, Liou YC, Wang H, Yu F (2014) Endocytic pathways downregulate the L1‐type cell adhesion molecule neuroglian to promote dendrite pruning in Drosophila . Dev Cell 30: 463–478 [DOI] [PubMed] [Google Scholar]
- Zhu S, Chen R, Soba P, Jan YN (2019) JNK signaling coordinates with ecdysone signaling to promote pruning of Drosophila sensory neuron dendrites. Development 146: dev163592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W, Wang Y, Tang Q, Zhang H, Yu F (2018) Prd1 associates with the clathrin adaptor alpha‐Adaptin and the kinesin‐3 Imac/Unc‐104 to govern dendrite pruning in Drosophila . PLoS Biol 16: e2004506 [DOI] [PMC free article] [PubMed] [Google Scholar]


