Abstract
The tools available to carry out in vivo analysis of Ca2+ dynamics in plants are powerful and mature technologies that still require the proper controls.
Introduction
Calcium (Ca2+) is a well-known second messenger in both unicellular and multicellular organisms (Berridge et al., 2000; Carafoli and Krebs, 2016). In plants, apart from its role as a structural component (White and Broadley, 2003), calcium plays a role in signaling events in response to a multitude of developmental and environmental stimuli (Kudla et al., 2010; Edel et al., 2017; Kudla et al., 2018). Biotic and abiotic challenges affect the cellular Ca2+ homeostasis by triggering transient changes of Ca2+ concentrations in the cytosol as well as in subcellular compartments (McAinsh and Pittman, 2009; Stael et al., 2012; Costa et al., 2018; Pirayesh et al., 2021; Resentini et al., 2021b).
The basis of calcium’s role as a signaling component lies in its peculiar chemistry and the existence of a large electrochemical gradient across the cell’s membranes, maintained by the activity of the proton- and calcium-ATPases (H+-ATPases and Ca2+-ATPases; Palmgren, 2001; Demidchik et al., 2018; Klejchova et al., 2021). The main evolutionary reason for evolving mechanisms that generate and maintain this large gradient is based on the need to keep the cytosolic Ca2+ concentrations ([Ca2+]cyt) low, to prevent the precipitation of organic and inorganic molecules (e.g. phosphates including adenosine triphosphate (ATP)) (Clapham, 2007). Under resting conditions, the [Ca2+]cyt is in the range of hundreds of nanomolar (100–200 nM) whereas in the external spaces and subcellular compartments it can reach up to millimolar ranges (Stael et al., 2012). This steep concentration gradient implies that the opening of a limited number of calcium permeable channels, located on the different cellular membranes, is sufficient to rapidly increase the [Ca2+]cyt with a 10-fold increase compared with the resting concentration (Demidchik et al., 2018).
Within the plant cell, the change in [Ca2+]cyt is sensed by Ca2+ binding proteins acting as primary responders (e.g. calcium dependent kinases) or sensor relays. Among the latter Calmodulin (CaM), CaM-like proteins (CMLs) and calcineurin B-like proteins (CBLs) all bind cytosolic calcium, which triggers a conformational change of the proteins enabling them to interact with different targets modulating their activities (Kudla et al., 2018; Tian et al., 2020). When the Ca2+ sensors are stimulated they become primed to regulate downstream processes, which include ion fluxes, enzymatic activities, transcription, etc. (DeFalco et al., 2010; Kudla et al., 2018; Tian et al., 2020). Importantly, after the perception of a stimulus and the occurrence of the Ca2+ transient, resting [Ca2+]cyt needs to be quickly reestablished to prevent cell death (Clapham, 2007). The molecular mechanisms that are responsible for the recovery of the resting [Ca2+]cyt are Ca2+ buffers and Ca2+ active transporters, such as Ca2+-ATPases and Ca2+/cation exchangers (CAX) that are localized in the plasma membrane and membranes of intracellular compartments (Corso et al., 2018; Costa et al., 2018; Demidchik et al., 2018; Hilleary et al., 2020; Ishka et al., 2021; Resentini et al., 2021b).
The intertwined and coordinated activities of influx and efflux Ca2+ transport systems in the plasma membrane and internal stores (Stael et al., 2012; Costa et al., 2018; Pirayesh et al., 2021; Resentini et al., 2021b) jointly shape the characteristic cellular Ca2+ dynamics, also called Ca2+ signatures (Sanders et al., 2002).
The possibility of visualizing and studying Ca2+ signatures is based on the exploitation of “calcium imaging techniques” that, thanks to continuous improvement, have allowed us to study Ca2+ dynamics with increasing resolution and sensitivity with minimal invasiveness in different organisms, including plants. An important boost for the progression of the calcium imaging techniques is the possibility to extend the use of valuable and innovative tools across the different kingdoms of life. In such a scenario, one driver of innovation is neuroscience studies. In neurons, action potentials (APs) or the activation of ionotropic glutamate receptors trigger large and rapid changes in [Ca2+]cyt (Tian et al., 2009). This has led neuroscientists to develop innovative technologies to study Ca2+ dynamics, which are historically based on the simultaneous and continuous improvement of Ca2+ indicators, and the development and the implementation of the appropriate microscopy instrumentation.
In this update, we will briefly retrace the history of Ca2+ sensitive indicators (Fig. 1) focusing on their use in plants, providing important clues about how to choose among them, and also taking into consideration the most appropriate imaging techniques. We will explain when it is recommended to use ratiometric biosensors and when instead it is reasonable and more convenient to use intensiometric ones. The intention of this update is to guide the readers into the recent developments in the area of Ca2+ biosensing in plants and make them enthusiastic about this amazing and imaginative field of research.
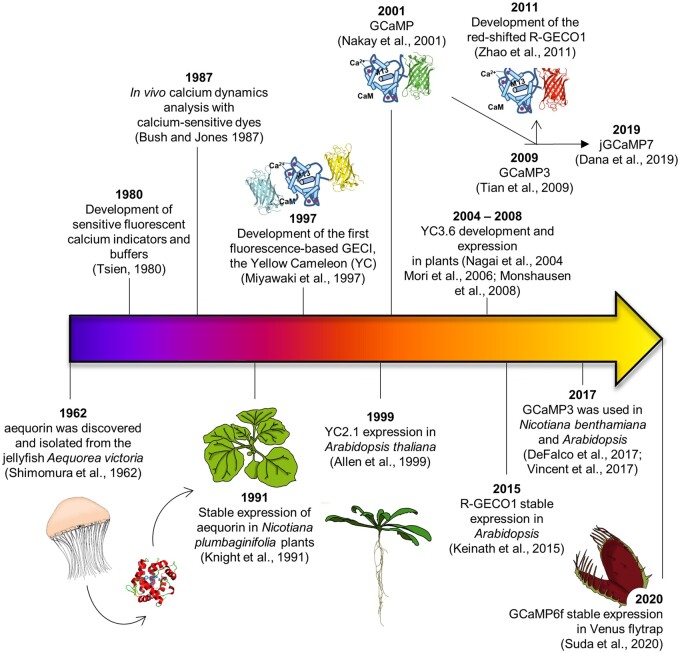
Figure 1.
History of major achievements for the in vivo study of Ca2+ biosensing in plants.
Synthetic dyes for measurements of cytosolic calcium dynamics
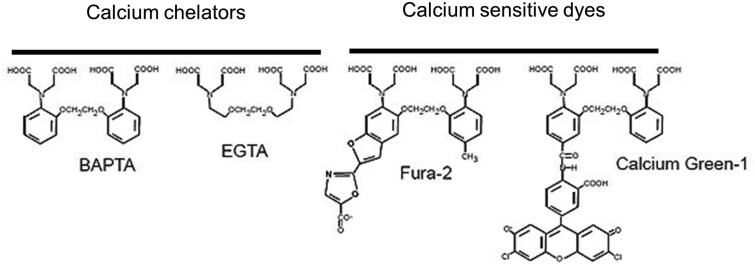
A breakthrough in the field of Ca2+ imaging was the development of sensitive fluorescent Ca2+ indicators (or dyes) and buffers by Tsien (1980). These indicators were the result of the “fusion” between Ca2+-selective chelators like ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) or 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA) with a fluorescent chromophore (Fig. 2).
Figure 2.
Examples of two commonly used Ca2+ chelators and calcium-sensitive dyes derived from them.
In plants, the first attempts to study Ca2+ dynamics in vivo were based on the use of Ca2+ sensitive dyes (e.g. Fura-2, Fura-2 dextran, calcium green dextran, and Indo-1) which were instrumental in measuring [Ca2+] in aleurone protoplasts, guard cells in response to ABA, in growing pollen tubes or in root hairs in response to nodulation factors (Bush and Jones, 1987; McAinsh et al., 1990; Gilroy et al., 1991; McAinsh et al., 1995; Ehrhardt et al., 1996; Holdaway-Clarke et al., 1997). The use of those dyes allowed the measurement, for the first time, of Ca2+ variation within isolated mitochondria from land plants, giving the basic knowledge of the molecular mechanism for Ca2+ uptake in these organelles (Zottini and Zannoni, 1993). Whereas the use of Ca2+-sensitive dyes allowed the making of fundamental discoveries, they have some limitations which primarily include (i) their requirement to be loaded or manually injected into the cells, possibly leading to an unequal amount of dye within different cells; (ii) when accumulated into the cells they can be compartmentalized or sequestered into vacuoles (Bush and Jones, 1990); and (iii) an excessive loading can affect cytosolic Ca2+ availability due to their Ca2+ chelator-backbone (Bush and Jones, 1990; Table 1). Nevertheless, a major advantage of Ca2+-sensitive dyes is the ability to perform Ca2+ imaging analyses without the need to generate transgenic plants (see the next section), as transformation protocols are not available for every species (Table 1). If a reliable Ca2+-sensitive dye for plant cells were to be available, it could be used for example to perform the first screen of mutants, instead of performing a tedious transgenic plants selection (Fichman and Mittler, 2021). Nevertheless, since the use of dyes has still more disadvantages than advantages, in the last 20 years, plant scientists have moved to the use of genetically encoded Ca2+ indicators (GECIs) (Perez Koldenkova and Nagai, 2013) which opened another dimension for the quantitative in vivo imaging of Ca2+ dynamics.
Table 1.
Summary of advantages and disadvantages/limitations of the different available Ca2+ indicators
| Type of indicator | Advantages | Disadvantages/limitations |
|---|---|---|
| Synthetic dyes | (i) Ca2+ imaging analyses without the need to generate transgenic plants. | (i) To be loaded or manually injected into the cells; (ii) can be compartmentalized or sequestered into vacuoles; (iii) an excessive loading can bring to cytosolic Ca2+ buffering. |
| Aequorin | (i) Does not need to be excited with fluorescent light; (ii) long-term experiments; and (iii) it allows a good estimation of Ca2+ concentrations. | (i) Low quantum yield efficiency; (ii) requirement for the external provision of coelenterazine; and (iii) non-linearity of its rate of light emission. |
| Ratiometric GECIs (e.g. Cameleon, YC-Nano) | (i) Reliability; (ii) reduced artifacts; (iii) strong independence from the expression levels; and (iv) good for genetic backgrounds comparison of resting Ca2+. | (i) Large size of the indicators; (ii) tailored strategies for their targeting; (iii) possible silencing issues; and (iv) more sophisticated and expensive imaging equipment is required. |
| Intensiometric GECIs (e.g. GECOs/GCaMPs) | (i) Need simple microscope setups; (ii) sensitivity to detect subtle changes of Ca2+; (iii) they allow the easy measurement of Ca2+ in adult plants; and (iv) an easier combination of different spectral variants for simultaneous imaging of Ca2+ in different compartments. | (i) Possible artifacts related to their intensiometric nature and (ii) Ca2+ buffering with some high affinity and high Hill coefficient variants. |
Genetically encoded calcium indicators
Aequorin
Aequorin is a bioluminescent protein that was discovered and isolated from the jellyfish Aequorea victoria by Shimomura et al. (1962). Shimamura was awarded the Nobel Prize in 2008 together with Martin Chalfie and Roger Tsien for the discovery and exploitation of the green fluorescent protein (GFP). Without the work done by these three visionary scientists this Update, this Focus Issue, and a large field of bio-imaging would not exist.
Aequorin is a 22-kDa holoprotein that contains bound oxygen, a prosthetic group, the coelenterazine, and three Ca2+-binding sites. When aequorin binds Ca2+ ions (Shimomura, 1995), it undergoes a conformational change, converting itself into a luciferase, which then catalyzes the luminescence reaction of coelenterazine. Coelenterazine is oxidized to coelenteramide with the emission of CO2 and blue light (at 465 nm) which is caused by the decay of the coelenteramide from an excited state (Ohmiya and Hirano, 1996). This property offered a useful tool for detecting the concentration of Ca2+ ions in real time. However, the aequorin quantum yield is very low, requiring the simultaneous reactions of hundreds or thousands of proteins to collect enough photons to a level detectable by photon emission measurements (Mithöfer and Mazars, 2002). Nevertheless, identification of the aequorin gene and of its coding sequence (Prasher et al., 1985) allowed the protein to be expressed recombinantly in cells and tissues from different organisms. In the 1980s, the injection of recombinant aequorin in Chara allowed the detection of a transient increase in light emission when the cell generated an AP (Williamson and Ashley, 1982). A real revolution then occurred in 1991 when Marc Knight and colleagues generated the very first transgenic multicellular organism, Nicotiana plumbaginifolia plants, stably expressing aequorin (Knight et al., 1991; Figure 1). The authors reported that in response to touch, cold-shock, and elicitors, the plants showed clear photon emission determined by cytosolic Ca2+ increase. This demonstration opened de facto the modern era of Ca2+ imaging in plants. Indeed, the low quantum yield of aequorin hinders high-resolution imaging and, as a matter of fact, its detection was usually carried out with the use of a luminometer, collecting the emitted photons without creating an image (Mithöfer and Mazars, 2002). However, modern ultrasensitive cameras now allow the detecting of the photons emitted by a single whole plant, providing low-resolution images and averaged responses from different tissues or cells (Kiegle et al., 2000; Zhu et al., 2013; Kiep et al., 2015). Therefore, the recent technological advances in camera sensitivity have revitalized the use of aequorin imaging, posing the basis for the design of powerful and successful genetic screenings (Yuan et al., 2014; Jiang et al., 2019; Chen et al., 2020; Wu et al., 2020).
Even if aequorin is the oldest GECI, it still represents a reliable tool to study Ca2+ dynamics in plants, in particular, to determine quantitatively the magnitude of the responses. In fact, for every single experiment, the measurement of the light expressed as relative luminescence units (RLUs) can be converted into absolute [Ca2+] thanks to a calibration curve that considers the total amount of aequorin molecules present in the sample. This can be done by discharging the reconstituted aequorin with a solution containing Ca2+ and ethanol (100 mM CaCl2, 10% ethanol (v/v)) (Allen et al., 1977; Mithöfer and Mazars, 2002). Specifically, the formula used to perform the conversion is the one reported in Mithöfer and Mazars (2002):
where L0 is the aequorin luminescence intensity per second and Lmax is the total amount of luminescence present in the sample over the experiment. KR and KTR are the dissociation constants for the first and second Ca2+ ions bound by aequorin, respectively (Mithöfer and Mazars, 2002).
As with every technique, aequorin-based Ca2+ analysis presents both advantages and disadvantages (Table 1). An advantage of aequorin is that it does not need to be excited with fluorescent light, which helps when the measurement of Ca2+ levels for long time intervals is required (Sai and Johnson, 2002; Love et al., 2004; Martí et al., 2013; Martí Ruiz et al., 2020) . Among the disadvantages, besides the already cited low quantum yield efficiency, is its requirement for the external provision of coelenterazine (Knight et al., 1991; Mithöfer and Mazars, 2002) which foresees the incubation of the plant material with the prosthetic group for several hours before starting the experiment. Moreover, after its oxidation, coelenterazine is irreversibly consumed, with the consequent decrease of the active aequorin pool. Another disadvantage of aequorin is the non-linearity of light emission rate as a function of the Ca2+ concentration (Robert et al., 2000; Table 1). Within the physiological range of [Ca2+]cyt (10−7–10−5 M), the photon emission rate increases by more than 100-fold for a 10-fold change in [Ca2+]. In practical terms, this means that for a change in [Ca2+] from 0.1 µM (at resting) to 3–4 µM (in response to a stimulus), the rate of aequorin photon emission increases over 1000-fold. As a consequence, the signal coming from 1 activated cell will be the same as that of 1000 cells at resting [Ca2+]. If Ca2+ is not homogeneous in the population of cells, which is indeed the case when using entire seedlings or leaf disks, the overall aequorin light emission is dominated by the most responding cells subpopulation, leading to an averaging of the response. One way that can be followed to study more specifically the contribution of different cell types is the expression of aequorin under the control of tissue-specific promoters or using enhancer trap lines. By following this strategy, it was possible to discover the oscillatory dynamics of Ca2+ signaling in root cells (Kiegle et al., 2000), circadian gating of cold-induced Ca2+ oscillations in guard cells (Dodd et al., 2006), and cell-type and stimulus-specific Ca2+ oscillations (Martí et al., 2013).
Aequorin being a GECI could also be targeted to different subcellular compartments, such as the tonoplast (Knight et al., 1996), the nucleus (van Der Luit et al., 1999), the Golgi apparatus (Ordenes et al., 2012), mitochondria (Logan and Knight, 2003), plastids/chloroplasts (Johnson et al., 1995; Mehlmer et al., 2012; Sello et al., 2016), chloroplast subcompartments like the outer and inner envelope membranes (Mehlmer et al., 2012), and the thylakoid lumen and membrane (Sello et al., 2018; Table 2). Aequorin was also targeted to the apoplastic space (Gao et al., 2004).
Table 2.
Summary of available fluorescent-based GECIs used in plants
| Name | Version | Type | Peaks of excitation/emission (nm) | In vitro Kd for Ca2+a | Subcellular localization | References |
|---|---|---|---|---|---|---|
| Cameleon | YC2.1 | Ratio EYFP/ECFP | Ex 440/Em 480/530 | 0.8 μM/2 μM | Cytosol and nucleus | Allen et al. (1999); Miyawaki et al. (1999) |
| YC3.6 | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 250 nM | Cytosol and nucleus | Nagai et al. (2004); Mori et al. (2006) | |
| NES-YC3.6 | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 250 nM | Cytosol | Krebs et al. (2012) | |
| NLS-YC3.6 | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 250 nM | Nucleus | Krebs et al. (2012) | |
| NUP-YC3.6 | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 250 nM | Nucleus | Costa et al. (2017) | |
| 4mt-YC3.6 | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 250 nM | Mitochondria | Loro et al. (2012) | |
| PM-YC3.6-LTI6b | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 250 nM | Plasma membrane | Krebs et al. (2012) | |
| 2Bam4-YC3.6 | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 250 nM | Chloroplasts and plastids | Loro et al. (2016) | |
| YC-Nano 65 | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 65 nM | Cytosol and nucleus | Horikawa et al. (2010); Choi et al. (2014) | |
| Ratio cpVenus/ECFP | ||||||
| SP-YC4.6-ER | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 58 nM/14.4 μM | Endoplasmic reticulum | Nagai et al. (2004); Iwano et al. (2009) | |
| 2Bam4-YC4.6 | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 58 nM/14.4 μM | Chloroplasts and plastids | Loro et al. (2016) | |
| 4mt-D3cpv | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 600 nM | Mitochondria | Loro et al. (2013) | |
| D3cpv-KVK-SKL | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 600 nM | Peroxisomes | Palmer et al. (2006); Costa et al. (2010) | |
| TP-D3cpv | Ratio cpVenus/ECFP | Ex 440/Em 480/530 | 600 nM | Tonoplast | Krebs et al. (2012) | |
| CRT-D4ER | Ratio citrine/ECFP | Ex 440/Em 480/530 | 195 μM | Endoplasmic reticulum | Palmer et al. (2006); Bonza et al. (2013) | |
| Twitch | Twitch 3 | Ratio cpCit174/ECFP | Ex 440/Em 480/530 | 250 nM | Cytosol and nucleus | Thestrup et al. (2014); Waadt et al. (2017) |
| Twitch 2B | Ratio cpVenus/mCerulean3 | Ex 440/Em 480/530 | 200 nM | Cytosol and nucleus | Thestrup et al. (2014); Waadt et al. (2017) | |
| CerTN-L15 | CerTN-L15 | Ratio citrine/cerulean | Ex 440/Em 480/530 | 1.2 μM | Cytosol and nucleus | Heim et al. (2007); Denninger et al. (2014) |
| GECOs | R-GECO1 | Intensiometric mApple | Ex 561/Em 600 | 482 nM | Cytosol and nucleus | Zhao et al. (2011); Ngo et al. (2014); Keinath et al. (2015) |
| NR-GECO1 | Intensiometric mApple | Ex 561/Em 600 | 482 nM | Nuclear | Zhao et al. (2011); Kelner et al. (2018) | |
| NR-GECO1.2 | Intensiometric mApple | Ex 561/Em 600 | 1.2 μM | Nuclear | Wu et al. (2013); Kelner et al. (2018); Leitão et al. (2019) | |
| CG-GECO1 | Intensiometric cpGFP | Ex 488/Em 515 | 749 nM | Cytosol | Zhao et al. (2011); Kelner et al. (2018) | |
| CG-GECO1.2 | Intensiometric cpGFP | Ex 405/Em 515 | 1.15 μM | Cytosol | Zhao et al. (2011); Kelner et al. (2018); Leitão et al. (2019) | |
| GEM-GECO1 | Intensiometric cpGFP | Ex 405/Em 515 | 340 nM | Cytosol and nucleus | Zhao et al. (2011); Waadt et al. (2017) | |
| B-GECO1-mCherry | Ratio cpGFP/mCherry | Ex 405/561/Em 480/600 | NA | Cytosol and nucleus | Waadt et al. (2017) | |
| G-GECO1.1-mCherry | Ratio cpGFP/mCherry | Ex 488/561/Em 515/600 | NA | Cytosol and nucleus | Waadt et al. (2017) | |
| R-GECO1-mTurquoise | Ratio mApple/mTurqouise | Ex 405/561/Em 480/600 | NA | Cytosol and nucleus | Waadt et al. (2017) | |
| GCaMPs | GCaMP3 | Intensiometric cpGFP | Ex 488/Em 515 | 542 nM | Cytosol and nucleus | Tian et al. (2009); Nguyen et al. (2018) |
| GCaMP5 | Intensiometric cpGFP | Ex 488/Em 515 | NA | Cytosol and nucleus | Akerboom et al. (2012); Diao et al. (2018) | |
| GCaMP6f | Intensiometric cpGFP | Ex 488/Em 515 | 375 nM | Cytosol and nucleus | Chen et al. (2013); Waadt et al. (2017) | |
| GCaMP6s | Intensiometric cpGFP | Ex 488/Em 515 | 144 nM | Cytosol and nucleus | Liu et al. (2017); Shao et al. (2020) | |
| NES-GCaMP6m | Intensiometric cpGFP | Ex 488/Em 515 | 167 nM | Cytosol | Luo et al. (2020) | |
| NLS-GCaMP6m | Intensiometric cpGFP | Ex 488/Em 515 | 167 nM | Nucleus | Luo et al. (2020) | |
| ER-GCaMP6-210 | Intensiometric cpGFP | Ex 488/Em 515 | 210 μM | Endoplasmic reticulum | de Juan-Sanz et al. (2017); Resentini et al. (2021a) | |
| GCaMP6f-mcherry | Ratio cpGFP/mCherry | Ex 488/561/Em 480/600 | NA | Cytosol and nucleus | Waadt et al. (2017) | |
| MatryoshCaMP6s | Ratio cpGFP/LSSmOrange | Ex 440/Em 515/600 | 197 nM | Cytosol and nucleus | Ast et al. (2017) | |
| Case | Case12 | Intensiometric cpGFP | Ex 488/Em 515 | 1 μM | Cytosol and nucleus | Souslova et al. (2007); Zhu et al. (2013) |
| CEPIA | CRT1a-R-CEPIAer | Intensiometric cpGFP | Ex 561/Em 600 | 565 μM | Enoplasmic reticulum | Suzuki et al. (2014); Luo et al. (2020) |
| Aequorin | Aequorin | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Cytosol and nucleus | Knight et al. (1991); Brini et al. (1995) |
| Aequorin | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Nucleus | van Der Luit et al. (1999) | |
| Aequorin | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Chloroplast stroma | Johnson et al. (1995) | |
| Aequorin | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Mitochondria | Logan and Knight (2003) | |
| Aequorin | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Golgi | Ordenes et al. (2012) | |
| Aequorin | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Vacuole/tonoplast | Knight et al. (1996) | |
| YFP-aequorin | CYA | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Cytosol | Mehlmer et al. (2012) |
| NYA | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Nucleus | Mehlmer et al. (2012) | |
| YA | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Plasma membrane | Mehlmer et al. (2012) | |
| CHYA | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Chloroplast/plastid stroma | Mehlmer et al. (2012); Sello et al. (2016) | |
| MYA | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Mitochondria | Mehlmer et al. (2012) | |
| OEYA | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Chloroplast outer envelope | Mehlmer et al. (2012); Sello et al. (2016) | |
| IEYA | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Chloroplast inner envelope | Mehlmer et al. (2012); Sello et al. (2016) | |
| TL-YA | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Chloroplast thylakoid lumen | Sello et al. (2018) | |
| TM-YA | Bioluminescence | No Ex-/Em 465 | 7.2–13 μM | Chloroplast thylakoid membrane | Sello et al. (2018) | |
| GFP5-aequorin | pchitGFP5:AQ | Bioluminescence | No Ex-/Em 465 | NA | Apoplast | Gao et al. (2004) |
| GFP-aequorin | G5A | Bioluminescence resonance energy transfer | No Ex-/Em 515 | NA | Cytosol and nucleus | Baubet et al. (2000); Xiong et al. (2014) |
The in vitro Kd for Ca2+ of the different sensors are those reported in the original works.
In A. victoria, the photons emitted by aequorin excite the GFP which causes the jellyfish to emit fluorescence (Prasher et al., 1992). In laboratory experiments, this property has been exploited to generate a bioluminescence resonance energy transfer (BRET)-based GFP-aequorin reporter (i.e. G5A; Baubet et al., 2000; Rogers et al., 2005). This BRET-based sensor has overcome one of the major limitations of aequorin, that is its low amount of emitted light. This allows easier detection of [Ca2+] variations using a camera by permitting the possibility to reduce the exposure time compared with aequorin imaging. However, this sensor still requires the exogenous administration of coelenterazine. Nevertheless, the G5A sensor expression in Arabidopsis (Arabidopsis thaliana) plants by Xiong et al. (2014) allowed visualization for the first time of long-distance Ca2+ waves propagating from the roots to shoot upon salt treatment.
FRET-based fluorescent sensors
To overcome aequorin limitations, in the late 1990s, scientists started to exploit fluorescent proteins (FPs) (i.e. GFP) and its spectral variants to develop another generation of GECIs. The first ever fluorescence-based GECI, named Yellow Cameleon (YC), was developed by Roger Tsien in 1997 (Miyawaki et al., 1997) quickly followed by the development of the YC2.1 version (Miyawaki et al., 1999) which was shortly after expressed in plants (Allen et al., 1999; Figure 1). This sensor is based on the Ca2+-induced protein–protein interaction between CaM and the CaM-binding peptide M13, a fragment from the myosin light chain kinase (Miyawaki et al., 1997). Both components are fused by a flexible linker and are sandwiched between a FP pair with partial overlap between the emission spectrum of the donor and the absorption spectrum of the acceptor. Upon Ca2+ binding, CaM interacts with M13, bringing the donor and acceptor FPs into sufficiently close proximity to allow for Förster resonance energy transfer (FRET) between them through a nonradiative dipole–dipole coupling. This phenomenon can be measured via fluorescence microscopy. At the level of the acceptor molecule, it results in the emission of fluorescence upon excitation of the donor, allowing the estimation of Ca2+-induced FRET by the ratio of donor and acceptor emission upon excitation of the donor using relatively simple microscopy setups (Miyawaki et al., 1997; Rudolf et al., 2003).
A wide variety of FRET-based GECI has been developed based on FRET pairs with different spectral properties and in combination with a variety of Ca2+ regulated protein–protein interactions (Greenwald et al., 2018; https://biosensordb.ucsd.edu/biosensorDB/bsSearch.php). The spectral range of the FRET pairs is usually cyan/yellow, green/red occupying a wide spectrum, reducing the options for dual imaging with other sensors or reporters.
In plants, the most popular FRET-based GECIs are based on cyan and yellow FRET pairs linked together by the Ca2+-binding protein CaM and the CaM-binding peptide M13, as in the original YC configuration by Roger Tsien. FRET, and thus [Ca2+] increases, can be conveniently measured by the increase in the ratio between the emission intensity of Enhanced Yellow FP (EYFP) and Enhanced Cyan FP (ECFP) upon ECFP excitation. The Cameleon indicators can go back and forth from the bound and unbound Ca2+ state. However, this is limited by the bleaching of the sensor by the excitation light especially when high magnifications are used, which may cause photo-oxidative cellular stress (Laissue et al., 2017). One of the most important properties of Cameleon is its pure ratiometric nature: a single wavelength excitation and a dual emission (Miyawaki et al., 1997). The advantages of having a ratiometric sensor will be further discussed in the next section. The Cameleon’s reliability brought, in the first years of the 2000s, an “explosion” of different variants, with different Ca2+ affinities, different linkers, and different FP pairs (Table 2;Palmer and Tsien, 2006). In 2004, the Miyawaki group developed the Cameleon YC3.60 (often called YC3.6) where the EYFP was replaced with a circularly permuted variant of the Venus FP (cpVenus) (Nagai et al., 2002). The use of the cpVenus as a FRET acceptor greatly increased the energy transfer efficiency from the donor (ECFP), which in practical terms allowed the very reliable performance of in vivo measurements. In fact, in living cells, the simultaneous decrease in ECFP and increase in cpVenus fluorescence emissions, due to FRET, were almost identical to the in vitro analyses (Nagai et al., 2004). This property is of great relevance because it permits ascertaining with high confidence that a change of cpVenus/ECFP ratio, even if small, corresponds to actual FRET levels, thus, a real [Ca2+] change. This latter aspect gains importance when a single cell or single organelle imaging is performed. Moreover, whereas both ECFP and cpVenus might show a pH sensitivity, the FRET ratio is almost unaffected, at least in a narrow change of physiological cytosolic pHs (around pH 7–7.5; Nagai et al., 2004; Behera et al., 2018).
The YC3.6 was expressed in Arabidopsis under the control of the guard cell and pollen-specific promoters, pGC1 and pLat52, respectively, which allowed detecting spontaneous Ca2+ oscillations in these two cell types with an improved FRET efficiency (Mori et al., 2006; Yang et al., 2008; Iwano et al., 2009) in comparison to the first YC versions used in plants, the YC2.1 (Allen et al., 1999) and the YC3.1 (Michard et al., 2008; Iwano et al., 2009). The YC3.6 was then expressed under the control of the CaMV35S promoter, making it possible in Arabidopsis to study, at high spatial and temporal resolution, root hair tip Ca2+ oscillations (Monshausen et al., 2008). The next step in the exploitation of YC3.6 was obtained by Schumacher’s group which placed the sensor with a cytosolic or nuclear localization signal (NES-YC3.6 and NLS-YC3.6) under the control of the pUBQ10 promoter (Grefen et al., 2010; Krebs et al., 2012) that offered a homogeneous expression of the indicator in both Arabidopsis wild-type and mutant backgrounds (Wagner et al., 2015; Teardo et al., 2017; Behera et al., 2018, Corso et al., 2018; Hazak et al., 2019; Wang et al., 2020) as well as in rice (Oryza sativa; Behera et al., 2015). YC3.6 was also successfully expressed in the moss Physcomitrium patens (formerly Physcomitrella patens), allowing the visualization of systemic Ca2+ wave propagations in the absence of vascular tissues (Storti et al., 2018). Another important breakthrough was the expression of YC3.6 under the control of the synergid-specific promoter, pMYB98, which allowed the monitoring of Ca2+ dynamics in this type of cells during the fertilization process (Hamamura et al., 2014; Ngo et al., 2014). The versatility and reliability of YC3.6 were also demonstrated by its use for the analysis of Ca2+ dynamics in different subcellular compartments including mitochondria, chloroplast stroma, and subplasmalemmal space (Table 2;Krebs et al., 2012; Loro et al., 2012, 2016; Storti et al., 2018).
Based on the original work from Nagai et al. (2004) the in vitro Kd of the YC3.6 is 250 nM (Table 2) with a Hill coefficient (Table 3) of 1.7. The knowledge of these two parameters allows the rough conversion of the cpVenus/ECFP ratio into an [Ca2+] if the ratio minimum and ratio maximum are experimentally measured (Palmer and Tsien, 2006; Monshausen et al., 2008; Wagner et al., 2015). One formula that can be used to perform the conversion is the one reported in Monshausen et al. (2008):
where R represents the cpVenus/ECFP ratio measured at any given time during the experiment, n represents the Hill coefficient, and the Kd the in vitro affinity for Ca2+ (Table 3). However, since both the Kd and the Hill coefficients are usually measured in vitro and not in vivo, the ratio conversion into concentration values must be taken with caution. As a matter of fact, a recent work reported for the YC3.6 an in vitro Kd of 719 nM and a Hill coefficient of 2.12 (Li et al., 2021), values that are quite distant from those reported in the original work by Nagai et al. (2004) (250 nM and 1.7, respectively); therefore, pointing out the need for prudence when the conversion is applied (Palmer and Tsien, 2006). Nonetheless, the YC3.6 is indeed a sensor suitable for the analysis of Ca2+ dynamics when a given stimulus can induce an increase that is around and above its Kd value. It is also true that in response to stimuli that induce small Ca2+ increases, YC3.6 shows some limitations in comparison to the more recent generation of ultrasensitive GECIs (Keinath et al., 2015; Waadt et al., 2017), being in fact not efficient at detecting very subtle changes of [Ca2+] (Keinath et al., 2015). However, the beauty of any genetically encoded sensor is that by following rational and random mutagenesis approaches they can be modified to address specific needs. In 2010, the group of Nagai generated a series of Cameleon variants with higher affinity for Ca2+ that were dubbed YC-Nano (Table 2;Horikawa et al., 2010). In particular, the YC-Nano 65 (with an in vitro Kd for Ca2+ of 65 nM; Table 2) was efficiently expressed in Arabidopsis and this was instrumental to demonstrate the existence of a long-distance subtle Ca2+ wave in seedlings locally challenged with salt stress (Choi et al., 2014). The same sensor was used to compare the cytosolic [Ca2+] at resting and in response to wounding between the wild type and fatty acid oxygenation upregulated 2 (fou2) mutant (Lenglet et al., 2017). Very recently, YC-Nano 65 was used to study the response to flg22 in cotyledon and leaf cells in the Arabidopsis wild type, aca4/aca11 and aca1/2/7 autoinhibited Ca2+-ATPase mutants (Hilleary et al., 2020; Ishka et al., 2021).
Table 3.
Brief summary of the principal definitions used to describe GECIs properties
| Biochemical parameter | Description |
|---|---|
| K d (µM) | Apparent dissociation constant for Ca2+ of the sensor, at this concentration, half of the indicators are bound with Ca2+. Given that Hill coefficients are usually higher than 1, the Kd delineates the optimal concentration range at which a GECI should be used. |
| k on (s−1) | Indicates the rate of Ca2+ association shown by the sensor, and thus the speed by which a sensor responds to an increase in Ca2+ levels. |
| k off (s−1) | Indicates the rate of Ca2+ dissociation shown by the sensor, and thus the speed by which a sensor responds to a decrease in Ca2+ levels. |
| Hill coefficient | Indicates the cooperativity of the sensor in the Ca2+ binding process. A value greater than 1 indicates that binding of one Ca2+ ion facilitates the binding of another. The CaM-based GECI bind four Ca2+ ions. The closer to 1 the more linear the output of the reporter. |
| Dynamic range | For non-ratiometric indicators indicate the maximal fluorescence intensity (typically in a Ca2+-bound state) divided by minimal fluorescence intensity (determined in the presence of EGTA). For ratiometric indicators indicate the maximum fluorescence emissions ratio (typically in a Ca2+-bound state) divided by minimal fluorescence emissions ratio (determined in the presence of EGTA). |
A side-by-side in vivo comparison with plants expressing the YC3.6 and the YC-Nano 65 has not been published yet, but we can predict that the higher affinity for Ca2+ of the YC-Nano could saturate in response to different stimuli. Moreover, the different biochemical properties of YC-Nano 65 and YC3.6, like the rate of association (kon) and dissociation (koff) for Ca2+ (Table 3), can determine different sensor dynamics which can be attributed in vivo, as different Ca2+ dynamics. We do, therefore, suggest not comparing the published data obtained with these two sensors, but rather, to perform preliminary independent experiments to see which one offers the best readout in relation to the applied stimulus or developmental program under investigation.
An important aspect that needs to be considered is that plants expressing the Cameleon YC3.6 in the cytosol do not show any obvious gross phenotypes, pointing out that the sensor per se does not alter the Ca2+ homeostasis and the plant physiology in stable mature plants (Waadt et al., 2017). However, possible effects of Ca2+ buffering should be considered in every biological process of interest.
Besides Cameleon and its variants (Table 2), there are other FRET-based sensors that have been successfully used in plants. We can cite here the CerTN-L15 (Heim et al., 2007; Denninger et al., 2014) and the Twitch 2B and 3 (Thestrup et al., 2014; Waadt et al., 2017) that are alternative FRET-based Ca2+ sensors which instead of having the CaM domain, use troponin C, a protein exclusively found in myocytes, to sense and bind Ca2+. The substitution of the CaM domain with troponin C prevents any interference due to the endogenous CaM when present at high concentrations such as in the subplasmalemmal region (Miyawaki et al., 1999; Palmer et al., 2006). Plants expressing these two sensors have been shown to properly report [Ca2+] dynamics in synergids and root cells, but their use has so far been limited (Denninger et al., 2014; Waadt et al., 2017). An alternative to the FRET-based sensors might be represented by the use of dimerization-dependent FPs (ddFPs), which is a technology involving the reversible binding of two dark FP monomers to form a fluorescent heterodimeric complex (Alford et al., 2012).
Single FP GECIs
Single fluorophore-based Ca2+ indicators GECIs are intensiometric Ca2+ sensors, based on a circularly permuted FP (e.g. GFP, YFP, or mApple; Baird et al., 1999; Nakai et al., 2001) fused at its C- and N-termini with the components of a Ca2+ sensing module (i.e. CaM domain and the M13 peptide). In the presence of Ca2+, this causes a tightening of the interaction between C- and N-termini of the fluorophore, protecting the chromophore from the environment and leading to increased brightness. In simple words, this interaction induces a dramatic alteration of the spectral properties of the FP with a strong increase in the fluorescence emitted, thus, making these GECIs suitable to indicate Ca2+ levels in real time (Nakai et al., 2001). Currently, this principle has been also exploited for the development of different types of biosensors, including kinase activity reporters (e.g. ExRAI; Greenwald et al., 2018).
Similarly to Cameleon, for single FP sensors there was a strong development that yielded a family of GECIs with different colors, different Ca2+ affinities, different Hill coefficients, less pH sensitivity, and with improved signal-to-noise ratio (Table 2; Zhao et al., 2011). Just to cite an example, from 2001 to 2019, there has been an evolution that has led from the GCaMP to the jGCaMP7 (Figure 1;Nakai et al., 2001; Tian et al., 2009; Akerboom et al., 2012; Chen et al., 2013; Greenwald et al., 2018; Dana et al., 2019). The aim to improve GCaMPs was driven by the necessity to increase the sensitivity and kinetics of the sensors, trying to make them closer, in terms of properties, to the synthetic Ca2+ dyes that for the needs of neuroscience are still among the most sensitive and rapid Ca2+ indicators. It is worth considering that in plant cells, the kinetics of the sensor response does not represent a big limitation, since in most of the published works the imaging sampling was set at every 2–5 s (e.g. guard cells, pollen tubes and root hair growth, and root tip cells; Allen et al., 1999; Monshausen et al., 2008; Yang et al., 2008; Michard et al., 2011; Candeo et al., 2017; Li et al., 2021; Resentini et al., 2021a). Instead, the ease of use and the sensitivity of single FP GECIs are good properties that have pushed the plant community to move toward their use. The fact that single FP GECIs rely on a single excitation and a single emission makes them particularly suitable to be combined with other fluorescent markers or sensors, and for their use with simple and accessible microscope equipment (Table 1). The first single FP GECI expressed in plant cells were the GFP-based Ca2+ indicator Case12 (Zhu et al., 2013) and the red-shifted R-GECO1 (Ngo et al., 2014; Keinath et al., 2015). The latter is a red fluorescent GECI derived from the GCaMP3 (Tian et al., 2009), where the circularly permuted GFP (cpGFP) was substituted with the cpmApple (Zhao et al., 2011). Importantly, R-GECO1 is excited with green light (e.g. 561 nm; Table 2) which besides offering a greater tissue penetration neither stimulates photosynthesis nor photoreceptors (Taiz et al., 2014), making it particularly suited for use in green tissues. In 2015, R-GECO1 was first used in a series of “classical experiments” in Arabidopsis root meristems, treated with external ATP and pathogen elicitors such as flg22 and chitin, and demonstrating the very high sensitivity of this sensor (Keinath et al., 2015). It must be said that the same sensor was also previously expressed in pollen tubes used to fertilize an Arabidopsis line expressing the Cameleon YC3.6 in synergids, thus enabling Ca2+ imaging in two different tissues (Ngo et al., 2014). The in vitro Kd of R-GECO1 of 482 nM (Table 2) with a Hill coefficient of 2.06 (Zhao et al., 2011) makes this sensor suitable to efficiently detect cytosolic Ca2+ variations, and its constitutive expression under the pUBQ10 promoter in the stable Arabidopsis line does not produce gross and visible phenotypes (Waadt et al., 2017; Resentini et al., 2021a). Instead, when other color variants of the GCaMP3 (e.g. GEM-GECO1, B-GECO1, and G-GECO1.1; Table 2) were stably expressed in Arabidopsis, the plants showed some growth defects (Waadt et al., 2017). This negative effect on plant physiology indicates that these reporters with a higher Hill coefficient for GEM- (2.94) and B-GECO1 (2.64) or a lower Ca2+ affinity for the G-GECO1.1 (Kd 618 nM; Zhao et al., 2011) might potentially act as Ca2+ buffers, but this needs to be demonstrated. Overall, as a good practice, it is advised to compare the phenotype of a chosen sensor line with wild-type plants in the context of a given process of interest, and never to forget that the sensor could have a potential effect on plant physiology.
The GCaMP3 is a relatively old GCaMP version (Tian et al., 2009), and neuroscientists have now moved to the use of the most recent versions (GCaMP6 and jGCaMP7). Nevertheless, GCaMP3 was only recently expressed in Nicotiana benthamiana, Arabidopsis, and P. patens (Figure 1;DeFalco et al., 2017; Kleist et al., 2017; Vincent et al., 2017; Nguyen et al., 2018; Toyota et al., 2018; Krogman et al., 2020) and allowed the detection of bright long-distance leaf-to-leaf Ca2+ waves (Toyota et al., 2018; Nguyen et al., 2018). GCaMP3 has been also expressed under the control of tissue-specific promoters (i.e. phloem and different root tissues; Vincent et al., 2017; Toyota et al., 2018; Krogman et al., 2020).
Similarly to EYFP/ECFP FRET-based sensors, GCaMPs come with some disadvantages, as the blue excitation light can stimulate photosynthesis (Taiz et al., 2014) and can trigger in itself [Ca2+]cyt increase via phototropins (Zhao et al., 2013; Ishka et al., 2021). This could be avoided using R-GECO1, which being excited with green light is possibly more compatible with long-term imaging in the shoot. Other GCaMP versions, such as GCaMP5, GCaMP6f, and GCaMP6s (Table 2;Zhu et al., 2013; Liu et al., 2017; Vincent et al., 2017; Diao et al., 2018; Shao et al., 2020; Suda et al., 2020; Ishka et al., 2021) as well as other green and red variants of GECO1 (CG-GECO1.2 and R-GECO1.2; Kelner et al., 2018; Table 2) have also been used in plant cells.
Such a wealth of available tools might, however, generate some confusion in choosing one version over another. Indeed, the different GCaMP variants have different colors, different Kd for Ca2+ which make them suitable for use in different compartments or, similarly to the use of the YC-Nano 65, to report subtle changes of [Ca2+]. As an example, when subtle changes of [Ca2+] are expected, the high affinity for Ca2+ of GCaMP6s (Kd of 144 nM) is more suitable than GCaMP3 (Kd of 542 nM) or GCaMP6f (Kd of 375 nM; Table 2). However, a complete side by side comparison of all different sensors used in plant cells is not available, with only some tested by Waadt et al. (2017). Possibly an interested researcher should preliminarily test them to identify the best one for each experimental condition. We are currently working with both R-GECO1 and GCaMP3 expressing plants, and overall, they provide similar results. Nevertheless, as anticipated above, the most recent generation of single FP GECIs is continuously improving to increase the sensitivity and the signal-to-noise ratio to perform imaging in neuronal tissues, which might also be useful for plant scientists. As a matter of fact, all the single FP GECIs exploit an off/on response with a negligible fluorescent signal in the Ca2+ unbound state and strong fluorescence with even relatively small changes in [Ca2+]. Such a property represents a big advantage when imaging an entire plant. Indeed, both GCaMP3 and R-GECO1 have demonstrated that they offer enough sensitivity to detect and measure Ca2+ wave propagation between leaves in adult Arabidopsis plants in response to wounding or insect chewing (Nguyen et al., 2018; Toyota et al., 2018; Resentini et al., 2021a; Table 1). This result represents a major achievement in the field that was brought to a new dimension when the GCaMP6f was expressed in the carnivorous plant Dionaea muscipula, also known as Venus flytrap (Figure 1; Suda et al., 2020). The Venus flytrap rapidly closes the valves following mechanical stimulation of “sensitive hairs” and a role of Ca2+ as a second messenger in the closing mechanism was previously hypothesized (Scherzer et al., 2015; Hedrich and Neher, 2018). By using the GCaMP6f, Suda et al. (2020) demonstrated that the stimulation of the sensory hairs had the effect of inducing a Ca2+ wave that propagated throughout the entire leaf blade at a speed greater than 20 mm/s (Suda et al., 2020). Such a propagation rate exceeds at least 20 times the Ca2+ response to leaf injury in Arabidopsis (Toyota et al., 2018; Shao et al., 2020), and thus requires GECI with fast kon-kinetics such as the GCaMP6f.
In conclusion, the use of different single FP GECIs provides reliable data and offers the chance to capture dynamic processes in multicellular adult organisms, opening opportunities to explore new hypotheses. Also, Cameleon sensors allow the performance of large imaging experiments (Beneloujaephajri et al., 2013; Benikhlef et al., 2013; Costa et al., 2017; Behera et al., 2018; Doccula et al., 2018; Hilleary et al., 2020; Ishka et al., 2021), but for their use, a piece of more sophisticated and expensive equipment is required, whereas single FP GECIs need a simple single excitation, single emission fluorescence microscope equipped with a good camera (Table 1; Vincent et al., 2017; Nguyen et al., 2018; Toyota et al., 2018; Resentini et al., 2021a).
A notable disadvantage of the single FP GECI is that the obtained intensities are not only determined by Ca2+ levels but are also dependent on local expression levels and can be quenched at low pH.
Organellar calcium dynamics: state of the art
The contribution of organellar Ca2+ handling in the regulation of signaling processes has been recently reviewed, so we will redirect interested readers to those papers (Stael et al., 2012; Costa et al., 2018; Pirayesh et al., 2021; Resentini et al., 2021b). Nevertheless, here, we briefly summarize what GECIs are available for the study of Ca2+ dynamics in subcellular compartments and which tools, among those recently developed, we foresee for the advancement of the field. Table 2 is an updated version of the table published in Costa et al. (2018) and reports the most available Arabidopsis lines expressing subcellular targeted GECIs.
Aequorin was targeted to the cytosol, nucleus, mitochondria, Golgi apparatus, tonoplast, apoplast, chloroplast, and chloroplasts’ subcompartments (Knight et al., 1991, 1996; Johnson et al., 1995; van Der Luit et al., 1999; Logan and Knight, 2003; Gao et al., 2004; Mehlmer et al., 2012; Ordenes et al., 2012; Sello et al., 2016, 2018). One needs to bear in mind that aequorin can offer only an averaged response of organelles or subcellular compartments from different cells and often from multiple plants. Therefore, in the case of the requirement of single organelles, the use of fluorescent GECIs is required.
Both FRET-based cameleon and single FP sensors were targeted to different subcellular locales, allowing the detection of Ca2+ dynamics within different compartments with, in some cases, a single cell or even single organelle resolution (Iwano et al., 2009; Costa et al., 2010, 2013; Krebs et al., 2012; Loro et al., 2012, 2016; Bonza et al., 2013; Kelner et al., 2018; Leitão et al., 2019; Resentini et al., 2021a). A special mention is needed for the simultaneous expression, in Medicago truncatula and Arabidopsis, of the G-GECO1.1 (and 1.2) and R-GECO1.1 (and 1.2) sensors targeted, respectively, to the cytosol and nucleus (Kelner et al., 2018; Leitão et al., 2019). High-resolution imaging of these sensors in Medicago root hairs revealed that in response to Nod-Factors, the nuclear Ca2+ increase preceded the cytosolic one (Kelner et al., 2018). Similarly, a nuclear spontaneous Ca2+ increase anticipated the cytosolic one in Arabidopsis root tip meristematic cells during growth (Leitão et al., 2019). Moreover, the use of Cameleon YC3.6 targeted to mitochondria and nucleus enabled the simultaneous imaging of Ca2+ in these two compartments in Arabidopsis guard cells challenged with osmotic stress (Loro et al., 2012) or root tip cells in response to ATP (Loro et al., 2013).
At the present time, most of the Ca2+ analyses in subcellular compartments have been carried out with Cameleon variants, and only limited use of recent generation GECIs has been exploited (reviewed in Costa et al., 2018; Luo et al., 2020). The trend is different in animal cells, where more recent GECIs have been used in different subcellular compartments. Modified GCaMP6, CEPIA, and RCaMP sensors (Greenwald et al., 2018) with different spectral features or lower affinities for Ca2+ have shown to be suitable for analyses of Ca2+ dynamics in the endoplasmic reticulum (ER) and mitochondria of single neurons in response to APs (Suzuki et al., 2014; Dana et al., 2016; de Juan-Sanz et al., 2017; Ashrafi et al., 2020). Hence, this is the right time to explore the use of these sensors in plants, as they can offer some advantages compared with Cameleon sensors, although there are other limitations. The use of Cameleon sensors for the analysis of Ca2+ dynamics in subcellular compartments is primarily justified by its ratiometric nature that helps to reduce artifacts when measuring Ca2+ in moving organelles at high magnification or when comparing the steady-state concentration (Table 1;Wagner et al., 2015; Corso et al., 2018). As an example, based on our own experience, the large size of Cameleon (∼73 KDa) may pose some limitations for its proper targeting. In the case of mitochondria, ER, and chloroplasts, the Cameleon targeting required tailored strategies. To prevent a cytosolic mislocalization of the Cameleon (i) the mitochondrial targeting sequence had to be repeated four times (Palmer et al., 2006; Loro et al., 2012), (ii) the targeting to the ER required both a KDEL sequence at the C-terminus end plus a plant-specific calreticulin targeting sequence at the N-terminus end of the sensor (Palmer et al., 2004; Iwano et al., 2009; Bonza et al., 2013), and (iii) the targeting to the chloroplast stroma needed to double the BETA-AMYLASE 4 (Bam4) targeting sequence (Loro et al., 2016). The proper targeting of the Cameleon was indeed obtained, but this was at the expense of different issues, such as a more difficult design of the construct, and silencing. As an example, a strong silencing was relevant when the Cameleon was targeted to chloroplasts (2Bam4-YC3.6). In fact, the generation of stable fluorescent plants could be obtained only in the Arabidopsis RNA-dependent RNA polymerase-6-11 (rdr6-11) mutant background which is compromised in silencing (Peragine et al., 2004; Loro et al., 2016). Nevertheless, with the ER-targeted version of the Cameleon (CRT-D4ER), its expression was also limited to juvenile tissues, with some difficulties found in expressing it in some T-DNA lines (Corso et al., 2018; Shkolnik et al., 2018). We hope that the use of single FP GECIs might reduce silencing issues. A recent publication has reported the use of the ER-localized CRT1a-R-CEPIAer sensor and no issues related to silencing were highlighted (Luo et al., 2020). We have also successfully expressed the ER-GCaMP6-210 sensor localized in the ER (Table 2; de Juan-Sanz et al., 2017; Resentini et al., 2021a) in Arabidopsis wild-type and R-GECO1 backgrounds, offering the possibility to perform the simultaneous Ca2+ imaging in the ER and cytoplasm of adult plants (Resentini et al., 2021a). This result was not reached with the CRT-D4ER, not even in the ER alone, where the fluorescence was lost with aging.
We foresee that in the coming years, the use of single FP GECIs for the analyses of organellar Ca2+ dynamics should accompany the use of Cameleon, in particular for the simultaneous analysis of Ca2+ dynamics in different compartments, by combining sensors with different excitation and emission spectra (Resentini et al., 2021a). Moreover, organellar-targeted single FP GECIs should also be employed to analyze Ca2+ dynamics in adult plants, by exploiting their high sensitivity and dynamic range (Table 3).
Ratiometric versus intensiometric GECIs: a practical guide for their choice
In this Update article, we have referred several times to the advantages offered by using ratiometric FRET-based sensors compared with intensiometric ones. In this section, we want to further discuss this statement. Traditionally, ratiometry-based Ca2+ recordings rely completely on ratio changes, and these measurements are not influenced by the actual amount of the indicator or by changes in the focusing position of the imaging system (Rudolf et al., 2003). This is particularly true with FRET-based sensors since the fluorescence emission from the acceptor depends on the light absorbed by the donor, and a single excitation is used, requiring a single excitation light path. In practical terms, one of the advantages of using a FRET-based ratiometric sensor, like Cameleon, is that there is the possibility to efficiently compare the steady-state ratios among different genotypes or in the same genetic background in response to long-term treatments. This is true for different subcellular compartments such as cytosol, mitochondria, and ER (Laanemets et al., 2013; Wagner et al., 2015; Lenglet et al., 2017; Yang et al., 2017; Behera et al., 2018; Corso et al., 2018; Doccula et al., 2018; Shkolnik et al., 2018; Fasani et al., 2019; Hilleary et al., 2020; Ishka et al., 2021). Another advantage of ratiometric FRET-based sensors is that the results can be presented as raw acceptor/donor fluorescence emissions ratios (e.g. cpVenus/ECFP) without the need for calibration. Our direct experience with intensiometric sensors has revealed that the fluorescence is already variable at resting conditions, even within the same genetic background grown in the same conditions (Figure 3, B). To obviate the fluorescence variability among samples, when R-GECO1 or GCaMPs are used, the data are often presented as normalized fluorescence and not as absolute values (Vincent et al., 2017; Dindas et al., 2018; Nguyen et al., 2018; Toyota et al., 2018; Resentini et al., 2021a). Usually, this does not represent a problem; however, the need to rely on normalization may represent an issue for a correct interpretation of the results. To support this statement, we here present a set of experimental data that help to better clarify this point.
Figure 3.
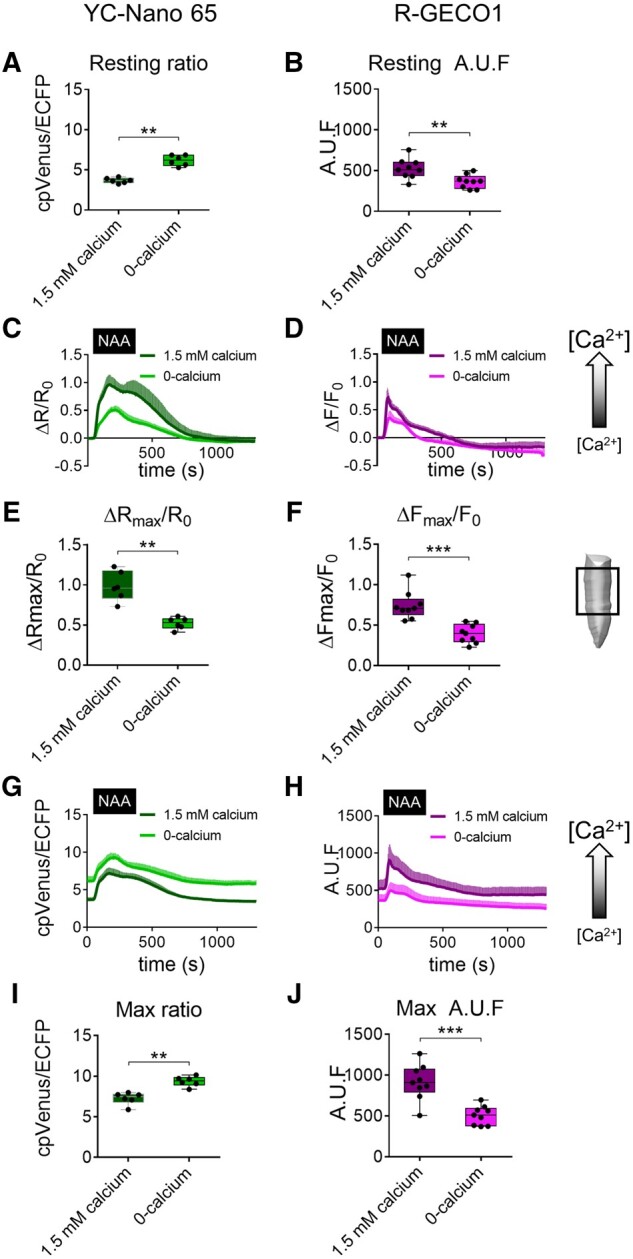
Comparison of the Ca2+ response in root tip cells expressing YC-Nano 65 or R-GECO1. Seedlings were exposed to a pulse of 10 µM NAA and medium containing either 1.5 mM calcium (CaCl2) or 0-calcium. The schematic drawing on the right shows the root tip region examined. A, cpVenus/ECFP ratio at rest in root tip cells of YC-Nano 65 seedling in 1.5 mM calcium and 0-calcium. B, R-GECO1 fluorescence at rest in root tip cells of R-GECO1 seedlings in 1.5 mM calcium and 0-calcium. C, Normalized averaged YC-Nano 65 cpVenus/ECFP ratios (R) (ΔR/R0 = (R−R0)/R0) over time in root tip cells in response to NAA as indicated by the black box on the x-axis. D, Normalized averaged R-GECO1 fluorescence (F) (ΔF/F0 = (F−F0)/F0) over time in root tip cells in response to NAA as indicated by the black box on the x-axis. R0 and F0 are the prestimulus values of R and F, respectively. The shaded right-side arrow indicates the direction of the [Ca2+]cyt increase. E, Maximal relative amplitude as ΔRmax/R0 of cpVenus/ECFP ratio triggered by NAA in 1.5 mM calcium and 0-mM calcium conditions. F, Maximal relative amplitude as ΔFmax/F0 of R-GECO1 fluorescence triggered by NAA in 1.5 mM calcium and 0-calcium conditions. G and H, Same experiments shown in (C and D) without normalization to the pre-stimulus situation. The shaded right-side arrow indicates the direction of the [Ca2+]cyt increase. I, Maximal amplitude of cpVenus/ECFP ratio triggered by NAA application in 1.5 mM calcium and 0-calcium. J, Maximal amplitude of R-GECO1 fluorescence triggered by NAA application in 1.5 mM calcium and 0-calcium. Arbitrary units of fluorescence (AUF). n ≥ 6. Data were plotted as box-and-whisker plots using GraphPad, in which all the experimental points are plotted, and their distribution represented as a box that extends from the 25th to 75th percentiles. The line in the middle of the box is plotted at the median. P values were calculated with an unpaired Student’s t test. Error bars = sd. **P ≤ 0.005, ***P ≤ 0.0005 (t test).
Arabidopsis seedlings expressing the Cameleon YC-Nano 65 and the R-GECO1 were germinated and grown in a standard growth half-strength MS medium and at 6-d-old stage were independently incubated for 10 min in solutions with different concentrations of Ca2+ (0-calcium and 1.5 mM calcium) and imaged with the wide field microscope setup described in Behera et al. (2018) (Supplemental Materials and Methods). After 10-min incubation, the root tip cells of the seedlings were imaged while keeping them in continuous perfusion before a 3-min pulse treatment with 10 μM of the synthetic auxin 1-naphthylacetic acid (NAA). For each experiment, the cpVenus/ECFP ratio for YC-Nano 65 and single fluorescence changes for R-GECO1 were measured over 20 min. The data were plotted over time as both normalized (ΔR/R0 and ΔF/F0) or raw cpVenus/ECFP ratios and R-GECO1 fluorescence (Figure 3, C, D, G, and H). At the end of the experiment, four sets of data could be compared: (i) YC-Nano 65 seedlings treated with 10 μM NAA in 0-calcium; (ii) YC-Nano 65 seedlings treated with 10 μM NAA in 1.5 mM calcium; (iii) R-GECO1 seedlings treated with 10 μM NAA in 0-calcium, and (iv) R-GECO1 seedlings treated with 10 μM NAA in 1.5 mM calcium. The results showed that NAA induced an increase in the cpVenus/ECFP ratio and R-GECO1 fluorescence that corresponds to a [Ca2+]cyt increase (Figure 3, C and D), as previously demonstrated (Behera et al., 2018). Interestingly, by comparing the maximum change of normalized cpVenus/ECFP ratio and R-GECO1 fluorescence, it was clear that the [Ca2+]cyt increase was higher in the medium with 1.5 mM calcium than the one observed with 0-calcium (Figure 3, E and F). Thus, both sensors reported the same result, suggesting that the amplitude of the NAA-induced cytosolic Ca2+ increase depends on the availability of Ca2+ in the medium. However, comparison of the raw cpVenus/ECFP ratio and non-normalized R-GECO1 fluorescence changes provided a different result (Figure 3, G and H). In fact, the incubation of seedlings in 0-calcium had the opposite effects in the two sensor lines. On the one hand, the YC-Nano 65 showed a higher ratio at resting (Figure 3, A and G) with a relatively smaller differential response to NAA treatment as evidenced by the normalized data (Figure 3, E), despite the raw maximum signal being higher than in 1.5 mM calcium (Figure 3, I). On the other hand, the resting R-GECO1 fluorescence (Figure 3, B) was lower in 0-calcium compared with 1.5 mM calcium with corresponding reductions in NAA response (Figure 3, F and J). In conclusion, both sensors reported a decreased NAA response, when Ca2+ was removed from the media, but with different starting points, de facto with opposite results.
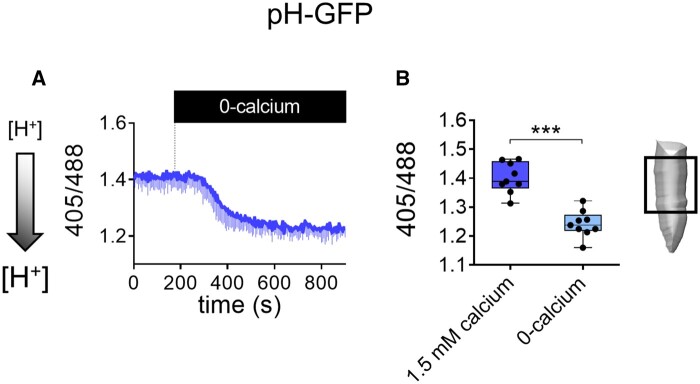
Since it has been reported that R-GECO1 may suffer from a pH sensitivity (Zhao et al., 2011; Keinath et al., 2015), we measured the cytosolic pH in root tip cells of Arabidopsis seedlings using the ratiometric pH sensor pH-GFP (Moseyko and Feldman, 2001; Behera et al., 2018) in response to 0-calcium treatment (Figure 4). The experiment clearly showed that shifting from 1.5 mM calcium to 0-calcium led to a cytosolic acidification (Figure 4, A and B). Thus, the incubation with 0-calcium can have multiple effects, including changing the cytosolic pH. Such acidification may quench the R-GECO1 fluorescence, reducing its dynamic range. On the other hand, even considering that the pH can also affect the cpVenus and ECFP fluorescence of the YC-Nano 65 Cameleon (Supplemental Figure S1) the ratio calculation seems to better correct for this potential issue as previously shown for the YC3.6 (Nagai et al., 2004; Behera et al., 2018). The understanding of how 0-calcium treatment affects both cytosolic Ca2+ and pH is currently under investigation. Nevertheless, our example illustrates that different treatments can potentially alter the properties of the sensors and warranting a careful comparison among them.
Figure 4.
Cytosolic pH dynamics in root tip cells in response to 0-calcium treatment. A, pH dynamics in pH-GFP root tip cells of the region indicated in the schematic drawing, treated with medium with 0-calcium (from 1.5 mM calcium to 0-calcium) as indicated by the black box on the x-axis. The ratio corresponds to the emission of the pH-GFP sensor when excited with light at 405 nm divided by the emission of the sensor when excited at 488 nm. B, The amplitude of 405/488 ratio in 1.5 mM calcium and 0-calcium. n = ≥5. The shaded left-side arrow indicates the direction of the pH decrease. Data were plotted as box-and-whisker plots using GraphPad, in which all the experimental points are plotted, and their distribution represented as a box that extends from the 25th to 75th percentiles. The line in the middle of the box is plotted at the median. P values were calculated with an unpaired Student’s t test. Error bars = sd. ***P ≤ 0.0005 (t test).
This series of experiments demonstrate that intensiometric sensors can be prone to artifacts and that whereas they show superior sensitivity compared with FRET sensors (Krebs et al., 2012; Keinath et al., 2015; Waadt et al., 2017; Resentini et al., 2021a), they might lead to a wrong interpretation of the results. This fact does not signify that intensiometric sensors are not reliable, but that depending on the type of experiment being done, some additional controls should be carried out. In light of this, the use of the recently developed transcriptionally linked dual sensors, such as the CapHensor which allows the simultaneous analysis of Ca2+ and pH dynamics (Li et al., 2021) or the R-GECO1-GSL-E2GFP to simultaneously monitor Ca2+, H+, and Cl– (Waadt et al., 2020) could be an option. Of particular note, the development of different dual sensor reporters to simultaneously monitor the dynamics of Ca2+ with other ions, analytes, or redox potentials of different redox couples will represent an important tool to reach an integrated picture of the physiological response to different stimuli (Wagner et al., 2019; Waadt et al., 2020).
Despite the reservations made on the pH sensitivity of single FP GECIs, to correct for variation in expression levels, in frame, or nested fusions of single FP GECI with other FPs have been developed (Ast et al., 2017; Waadt et al., 2017, 2020). Specifically, Waadt et al. (2017) generated a construct where the mTurquoise FP (Goedhart et al., 2010) was introduced in a vector harboring the R-GECO1 (Table 2). Since mTurquoise fluorescence emission is independent of a change in [Ca2+], this strategy allowed the authors to use it as a reference (Waadt et al., 2017). The ratiometric R-GECO1-mTurquoise allowed the authors to reveal detailed maps of [Ca2+] changes in response to auxin and ATP in root tip cells (Waadt et al., 2017). A different approach aimed at generating a ratiometric GCaMP sensor was followed by the Frommer’s group, which adopted a strategy based on the employment of a single FP-cassette that nests a stable reference FP (large Stokes shift LSSmOrange) within the GCaMP6s reporter that was brought to the generation of the MatryoshCaMP6s sensor (Table 2;Ast et al., 2017). The main difference between the two approaches is that the “MatryoshCaMP6s sensor” requires a single excitation with a dual emission, instead of the two excitations and two emissions required for the R-GECO1 and mTurquoise, respectively. Indeed, similarly to Cameleon, the use of a second FP may limit the use of the Ca2+ sensor with other markers or biosensors. The availability of the R-GECO1-mTurquoise ratiometric sensor or a similar design for a GCaMPs-mCherry (Waadt et al., 2017) can represent a good compromise for the correction of artifacts, potentially also reducing the pH sensitivity. However, the need for dual excitation and dual emission may still limit their use: whereas the MatryoshCaMP6s has again an increased size that could introduce other limitations. Alternatively, dual excitation single FP GECIs could be used, such as GEX-GECO and REX-GECO (Zhao et al., 2011; Wu et al., 2014). Unfortunately, the latter are relatively dim and need further optimization. Interestingly, a kinase activity sensor based on GCaMP3 displayed two excitation peaks with a similar sensitivity to pH changes between 5.6 and 10 (Mehta et al., 2018), suggesting that the ratiometric excitation imaging of this kinase sensor is relatively insensitive to pH changes. Therefore, it seems possible to engineer an excitation ratiometric GECI for normalization to expression levels and pH changes, based on the bright single FP GCaMP3. A reasonably up-to-date database of currently available genetically encoded sensors can be found at BiosensorDB.ucsd.edu.
Microscopy techniques for plant calcium imaging
Traditionally, Ca2+ imaging has been carried out by using standard wide field and confocal fluorescence microscopy, while imaging of aequorin has been usually performed with back-illuminated charge-coupled device (CCD) cameras equipped with a light-tight box. The first Ca2+ analyses performed in single cells, like stomatal guard cells, root hairs, and pollen tubes, were carried out with microscopes using high magnification objectives. However, when Ca2+-sensitive dyes were the only available sensors it was difficult to image more than one cell per experiment (e.g. McAinsh et al., 1995; Ehrhardt et al., 1996; Holdaway-Clarke et al., 1997; Garcia-Mata et al., 2003). Instead, using transgenic plants, stably expressing GECIs, it was possible to measure Ca2+ dynamics both at single cell level (Allen et al., 1999; Monshausen et al., 2008; Loro et al., 2012; Hilleary et al., 2020) and in the entire organs or tissues, by decreasing the magnification (Kiegle et al., 2000; Krebs et al., 2012; Zhu et al., 2013; Kiep et al., 2015; Yuan et al., 2014; Jiang et al., 2019; Chen et al., 2020; Hilleary et al., 2020; Wu et al., 2020; Ishka et al., 2021).
To perform Cameleon-based Ca2+ imaging, a wide field microscope with one filter for the excitation and one for the emission are not sufficient, whereas this configuration is suitable for a single FP GECI. The use of Cameleon requires the detection of the emissions from two FPs obtained by performing a quick change of filters with a filter wheel that therefore needs repetition of the excitation step. An alternative configuration exploits a beam splitter coupled to one or two cameras to simultaneously acquire the two fluorescence signals (e.g. cpVenus and ECFP). The use of a beam splitter configuration for Cameleon detection was recently reported with a stereomicroscope, allowing the performance of a FRET-based analysis in entire Arabidopsis cotyledons and young leaves expressing the YC-Nano 65 (Hilleary et al., 2020; Ishka et al., 2021). With the wide field microscope, single cell resolution can be obtained when imaging guard cells from an epidermal strip preparation or in vitro germinated pollen tubes, but the lack of optical sectioning hinders the analysis of Ca2+ dynamics at single cell level in an entire organ (Table 4). Nevertheless, the use of tissue-specific promoters to guide the expression of the sensor in a given cell type allows the performing of single cell analysis in intact leaves by means of wide field microscopy (Yang et al., 2008). However, in cases where the sensor is ubiquitously expressed, to obtain single cell resolution an optical sectioning microscope is needed. The most widely used optical sectioning technique is the confocal microscope, which makes use of a pinhole to reject the light coming from sample positions that are not in focus. In this configuration, a single laser (445 or 458 nm to excite the acceptor) and two photomultipliers are sufficient for FRET imaging, allowing many plant-biology laboratories to perform Ca2+ imaging experiments at high spatial resolution (Tanaka et al., 2010; Loro et al., 2012; Costa et al., 2013; Krebs and Schumacher, 2013; Choi et al., 2014). When required to perform Ca2+ imaging analysis at a high temporal resolution, the laser scanning confocal microscope shows some limitations in terms of acquisition speed (Table 4). The sampling is in the order of 1 s, with a traditional galvo scanner. As an example, with a pixel number of 1024 × 1024, a typical acquisition frequency is 0.5 Hz, whereas with a half resolution of 512 × 512 it is 1 Hz. For a higher sampling rate, a spinning disk confocal is a possible choice, as it essentially parallelizes the confocal pinhole detection (Table 4). However, in this case, the need for simultaneous acquisition of both fluorescent ECFP and cpVenus emissions as well as the need for a dedicated laser (i.e. 445 nm) increases the complexity and the price of the microscope setup (Table 4). To gain access to a high resolution, high speed FRET acquisition, we developed a tailored fluorescent light sheet fluorescence microscope that allowed us to perform single cell imaging with a fast rate of acquisition as well as long-term developmental analyses (Costa et al., 2013; Candeo et al., 2017; Romano Armada et al., 2019; Alfieri et al., 2020; Table 4). To do experiments with plants expressing single FP GECIs, the same microscopes described above can be used with standard configurations for a single excitation and a single emission, de facto expanding the audience of possible users. The more recent generation of single FP GECIs has largely facilitated Ca2+ imaging analyses since relatively low-cost cameras with good sensitivity exist, which can provide reliable measurements. This is shown in recent papers demonstrating that stereomicroscopes equipped with standard CCD or complementary metal–oxide–semiconductor (CMOS) cameras and common light sources like fluorescent lamps or LED illumination systems are suitable to perform analyses of Ca2+ dynamics in entire adult plants (Vincent et al., 2017; Nguyen et al., 2018; Toyota et al., 2018; Shao et al., 2020; Suda et al., 2020; Resentini et al., 2021a) whereas, traditionally, the Ca2+ analyses were mainly confined to root cells of Arabidopsis, rice seedlings or a limited number of leaf epidermal cells (Krebs et al., 2012; Loro et al., 2012; Behera et al., 2015; Keinath et al., 2015; Waadt et al., 2017; Table 4).
Table 4.
Summary of advantages and limitations of different microscopy systems suitable for Ca2+ imaging
| Type of microscopy | Advantages | Limitations |
|---|---|---|
| Wide field microscopy | (i) Suitable for its use with FRET-based and single FP GECI; (ii) single cell resolution allowed with specific sample preparations (e.g. Guard cells in epidermal strips). | (i) Lack of optical sectioning. |
| Laser scanning confocal microscopy | (i) Optical sectioning allowed; (ii) spectral separation with multiple detectors; (iii) single cell analyses allowed in entire organs; (iv) most of the commercial configurations allow FRET analyses. | (i) Acquisition speed. |
| Spinning disk confocal microscopy | (i) Fast rate of acquisition; (ii) optical sectioning allowed; (iii) single cell analyses allowed in entire organs; (iv) lower phototoxicity. | (i) High price. |
| Light sheet fluorescence microscopy | (i) Fast rate of acquisition; (ii) optical sectioning allowed; (iii) single cell analyses allowed in entire organs; (iv) long-term analyses; (v) tailored design. | (i) Low versatility. |
Future developments
Here, we take the opportunity to foresee possible future developments. Whereas single FP GECIs may be prone to artifacts we still foresee an extensive use of them by harnessing their unique properties. As an example, having GECIs emitting at different wavelengths allows one to study Ca2+ dynamics in different subcellular compartments, within the same cell. This has been largely carried out in animal cells, but the applications in plants are still limited (Greenwald et al., 2018; Kelner et al., 2018; Resentini, 2021a). We anticipate that we could express GCaMP sensors, in mitochondria or in the ER together with the R-GECO1 localized to the cytosol or other combinations. By producing plasmids that harbor both sensors in the same backbone (Waadt et al., 2020; Li et al., 2021) we can transform different (single or multiple) mutants to identify and study the Ca2+ transport mechanisms in the different membranes (e.g. Wagner et al., 2015; Corso et al., 2018; Kelner et al., 2018). R-GECO1 or other red shifted variants (e.g. K-GECO1 or R-CaMPs; Shen et al., 2018) can be expressed together with Cameleon or other FRET-based GECIs. To perform simultaneous Ca2+ imaging analyses in different tissues, different GECIs can be expressed in the same plant under the control of different tissue-specific promoters (Ngo et al., 2014).
At the same time, optical imaging techniques could play a relevant role in the study of Ca2+ dynamics at different spatial scales. Super-resolution microscopy, the field of optical imaging that goes beyond the diffraction limit, is having an increased rate of application in plant biology (Komis et al., 2015; Shaw et al., 2019) even if its use in Ca2+ studies is still limited. The use of high-speed spinning disk confocal and light sheet fluorescence microscopy allows one to image the plant at single cell or even at a single organelle level. We expect future adoption of these techniques aimed at further increasing the spatio-temporal resolution of the analyses of Ca2+ dynamics. A high-end microscopy approach that is also used to monitor FRET-based sensors involves the measurement of fluorescent lifetimes of the donor molecule. This requires a pulsed laser, coupled to a very fast, high resolution photon counting device to determine the time of fluorescence emission of every molecule after excitation. A great advantage of this approach is that the readout is independent of expression levels and that the combination with dark acceptors allows for multiplex analysis. However, given that many photons need to be counted, there are important limitations to the speed by which the imaging can proceed. Current technologies are starting to achieve the required speed (fast-FRET-FLIM) for imaging fast processes such as Ca2+ signaling and will be instrumental for multiplexed imaging of a variety of sensors.
At a larger spatial scale, plants expressing two or more sensors simultaneously, with non-overlapping spectra, can be analyzed with low magnification (Resentini et al., 2021a), for example with a stereomicroscope equipped with an automatized filters wheel and multiple LED light sources. A low magnification approach is particularly well suited for imaging plants in close-to-physiological conditions. However, one must bear in mind that when dealing with large plant organs, one major limitation of optical imaging is the diffusion of light within them. Photoacoustic imaging which combines ultrasonic resolution with high contrast and specificity of light (Xu and Wang, 2006) could solve this problem. In photoacoustic imaging, short laser pulses are used to generate megahertz ultrasound waves in-depth into the tissue, usually referred to as photoacoustic, optoacoustic, or thermoacoustic signals. Dyes, GECIs, and metallochromic Ca2+ indicators (Dean-Ben et al., 2017; Liu et al., 2019; Dana et al., 2016; Roberts et al., 2018) have been proposed for photoacoustic imaging and imaging of the neural dynamics has been demonstrated in the mammalian brain, at depths superior to fluorescence microscopy (Gottschalk et al., 2019). Photoacoustic technology has not yet been used, to our knowledge, for volumetric Ca2+ imaging in plant biology. However, considering that photoacoustic imaging offers possibilities such as pigment identification (Tserevelakis et al., 2016) and chemoselective imaging (Zeng et al., 2019), it could potentially be a key technology in the years to come. The development of additional imaging technologies should allow plants to be kept in their pots without being touched or perturbed until the day of the experiment.
As a further development, additional genetic screens could be designed. As an example, the mutagenesis of plant lines expressing different GECIs in different cellular locales could be developed. The generated mutants could be screened with a fluorescent plate reader (Wagner et al., 2019) to identify those mutants impaired in the Ca2+ accumulation in a given compartment.
Conclusions
We are facing a revolution in terms of imaging that is based on the availability of tools that will permit us to really use this technology to increase our knowledge about how plants cope with and adapt to a changing environment. We are now getting closer to perform imaging experiments in real physiological conditions and not being limited to the use of a specimen mounted on a microscope slide. Imaging technologies are non-destructive and can be used to extract information from the plants at a whole-plant resolution, from one organ or organelle to another within the same plant (Lew et al., 2020).
Advances
GECIs have been efficiently employed in plants.
In vivo Ca2+ imaging at high spatial and temporal resolution is possible at tissue, cell, and organelle levels.
Plants expressing GECIs with different affinities for Ca2+ are available for cytosolic and organellar analyses.
Ratiometric GECIs allow a reliable comparison of Ca2+ levels in different genetic backgrounds and in long-term treatments.
Intensiometric GECIs require simple and cost-effective imaging setups.
Outstanding questions
Can GECIs be used for in vivo analyses of Ca2+ dynamics in real physiological conditions?
Can Ca2+ imaging technology in model plants translate to crops?
Can GECIs be used as environmental biosensors?
Can we design genetic screening based on recent generation GECIs to identify additional components of Ca2+ signaling?
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Single wavelength emissions as arbitrary units of fluorescence of cpVenus (yellow trace) and ECFP (light blue trace) of the YC-Nano 65 used for the ratio calculations of Figure 3, C and G.
Supplementary Material
Acknowledgments
We thank Dr. Melanie Krebs (University of Heidelberg) for providing us the pUBQ10-R-GECO1 and pUBQ10-pH-GFP Arabidopsis lines and Prof. Joerg Kudla (University of Muenster) for providing us the pCaMV35S-YC Nano 65 Cameleon Arabidopsis line.
Funding
This work was supported by Piano di Sviluppo di Ateneo 2019 (University of Milan) (to A.C.), by a PhD fellowship from the University of Milan (to M.G.), by Ministero dell’Istruzione, dell’Università e della Ricerca Fondo per Progetti di ricerca di Rilevante Interesse Nazionale 2017 (PRIN 2017ZBBYNC) for a postdoctoral fellowship (to F.R.); by Ministero dell’Istruzione, dell’Università e della Ricerca Fondo per Progetti di ricerca di Rilevante Interesse Nazionale 2017 (PRIN 2017FBS8YN) (to M.Z.), and by a FWO Research Grant (G002220N) (to S.V.).
Conflict of interest statement. The authors have declared that no conflict of interests exist.
M.G. carried out the experiments reported in Figures 3 and 4 and prepared Figures 1, 3, and 4. F.R. prepared Tables 1 and 4. A.C. conceived the project and wrote the article with feedback from all authors. A.C. serves as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is Alex Costa (alex.costa@unimi.it).
References
- Alfieri A, Doccula FG, Pederzoli R, Grenzi M, Bonza MC, Luoni L, Candeo A, Romano Armada N, Barbiroli A, Valentini G, et al. (2020) The structural bases for agonist diversity in an Arabidopsis thaliana glutamate receptor-like channel. Proc Natl Acad Sci USA 117: 752–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford SC, Ding Y, Simmen T, Campbell RE (2012) Dimerization-dependent green and yellow fluorescent proteins. ACS Synth Biol 21: 569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen DG, Blinks JR, Prendergast FG (1977) Aequorin luminescence: Relation of light emission to calcium concentration—a calcium-independent component. Science 195: 996–998 [DOI] [PubMed] [Google Scholar]
- Allen GJ, Kwak JM, Chu SP, Llopis J, Tsien RY, Harper JF, Schroeder JI (1999) Cameleon calcium indicator reports cytoplasmic calcium dynamics in Arabidopsis guard cells. Plant J 19: 735–747 [DOI] [PubMed] [Google Scholar]
- Ashrafi G, de Juan-Sanz J, Farrell RJ, Ryan TA (2020) Molecular tuning of the axonal mitochondrial Ca2+ uniporter ensures metabolic flexibility of neurotransmission. Neuron 105: 678–687.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast C, Foret J, Oltrogge LM, De Michele R, Kleist TJ, Ho CH, Frommer WB (2017) Ratiometric Matryoshka biosensors from a nested cassette of green- and orange-emitting fluorescent proteins. Nat Commun 8: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY (1999) Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci USA 96: 11241–11246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubet V, Le Mouellic H, Campbell AK, Lucas-Meunier E, Fossier P, Brulet P (2000) Chimeric green fluorescent protein-aequorin as bioluminescent Ca2+ reporters at the single-cell level. Proc Natl Acad Sci USA 97: 7260–7265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera S, Wang N, Zhang C, Schmitz-Thom I, Strohkamp S, Schültke S, Hashimoto K, Xiong L, Kudla J (2015) Analyses of Ca2+ dynamics using a ubiquitin-10 promoter-driven Yellow Cameleon 3.6 indicator reveal reliable transgene expression and differences in cytoplasmic Ca2+ responses in Arabidopsis and rice (Oryza sativa) roots. New Phytol 206: 751–760 [DOI] [PubMed] [Google Scholar]
- Behera S, Zhaolong X, Luoni L, Bonza MC, Doccula FG, De Michelis MI, Morris RJ, Schwarzländer M, Costa A (2018) Cellular Ca2+ signals generate defined pH signatures in plants. Plant Cell 30: 2704–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneloujaephajri E, Costa A, L’Haridon F, Métraux JP, Binda M (2013) Production of reactive oxygen species and wound-induced resistance in Arabidopsis thaliana against Botrytis cinerea are preceded and depend on a burst of calcium. BMC Plant Biol 13: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benikhlef L, L’Haridon F, Abou-Mansour E, Serrano M, Binda M, Costa A, Lehmann S, Métraux JP (2013) Perception of soft mechanical stress in Arabidopsis leaves activates disease resistance. BMC Plant Biol 13: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, Lipp P, Bootman MD (2000) The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol 1: 11–21 [DOI] [PubMed] [Google Scholar]
- Bonza MC, Loro G, Behera S, Wong A, Kudla J, Costa A (2013) Analyses of Ca2+ accumulation and dynamics in the endoplasmic reticulum of Arabidopsis root cells using a genetically encoded Cameleon sensor. Plant Physiol 163: 1230–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brini M, Marsault R, Bastianutto C, Alvarez J, Pozzan T, Rizzuto R (1995) Transfected aequorin in the measurement of cytosolic Ca2+ concentration ([Ca2+]c). A critical evaluation. Journal of Biological Chemistry 270: 9896–9903 [DOI] [PubMed] [Google Scholar]
- Bush DS, Jones RL (1987) Measurement of cytoplasmic calcium in aleurone protoplasts using indo-1 and fura-2. Cell Calcium 8: 455–472 [DOI] [PubMed] [Google Scholar]
- Bush DS, Jones RL (1990) Measuring intracellular Ca levels in plant cells using the fluorescent probes, indo-1 and fura-2: Progress and prospects. Plant Physiol 93: 841–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candeo A, Doccula FG, Valentini G, Bassi A, Costa A (2017) Light sheet fluorescence microscopy quantifies calcium oscillations in root hairs of Arabidopsis thaliana. Plant Cell Physiol 58: 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafoli E, Krebs J (2016) Why calcium? How calcium became the best communicator. J Biol Chem 291: 20849–20857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Gao J, Sun S, Zhang Z, Yu B, Li J, Xie C, Li G, Wang P, Song C-P, et al. (2020) BONZAI proteins control global osmotic stress responses in plants. Current Biol 30: 4815–4825 [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013) Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WG, Toyota M, Kim SH, Hilleary R, Gilroy S (2014) Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc Natl Acad Sci USA 111: 6497–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham DE (2007) Calcium signaling. Cell 14: 1047–1058 [DOI] [PubMed] [Google Scholar]
- Corso M, Doccula FG, de Melo JRF, Costa A, Verbruggen N (2018) Endoplasmic reticulum-localized CCX2 is required for osmotolerance by regulating ER and cytosolic Ca2+ dynamics in Arabidopsis. Proc Natl Acad Sci USA 115: 3966–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Candeo A, Fieramonti L, Valentini G, Bassi A (2013) Calcium dynamics in root cells of Arabidopsis thaliana visualized with selective plane illumination microscopy. PLoS ONE 8: e75646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Drago I, Behera S, Zottini M, Pizzo P, Schroeder JI, Pozzan T, Lo Schiavo F (2010) H2O2 in plant peroxisomes: an in vivo analysis uncovers a Ca2+-dependent scavenging system. Plant J 62: 760–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Luoni L, Marrano CA, Hashimoto K, Köster P, Giacometti S, De Michelis MI, Kudla J, Bonza MC (2017) Ca2+-dependent phosphoregulation of the plasma membrane Ca2+-ATPase ACA8 modulates stimulus-induced calcium signatures. J Exp Bot 68: 3215–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Navazio L, Szabo I (2018) The contribution of organelles to plant intracellular calcium signalling. J Exp Bot 69: 4175–4193 [DOI] [PubMed] [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, Tsegaye G, Holt GT, Hu A, Walpita D, et al. (2016) Sensitive red protein calcium indicators for imaging neural activity. eLife 5: e12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, et al. (2019) High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat Methods 16: 649–657 [DOI] [PubMed] [Google Scholar]
- Dana N, Fowler RA, Allen A, Zoldan J, Suggs L, Emelianov S. ( 2016) In vitro photoacoustic sensing of calcium dynamics with arsenazo III. Laser Phys Lett 13: 075603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan-Sanz J, Holt GT, Schreiter ER, de Juan F, Kim DS, Ryan TA (2017) Axonal endoplasmic reticulum Ca2+ content controls release probability in CNS nerve terminals. Neuron 93: 867–881.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean-Ben XL, Gottschalk S, Sela G, Shoham S, Razansky D (2017) Functional optoacoustic neuro-tomography of calcium fluxes in adult zebrafish brain in vivo. Opt Lett 42: 959–962 [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Bender KW, Snedden WA (2010) Breaking the code: Ca2+ sensors in plant signalling. Biochem J 425: 27–40 [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Toyota M, Phan V, Karia P, Moeder W, Gilroy S, Yoshioka K (2017) Using GCaMP3 to study Ca2+ signaling in Nicotiana species. Plant Cell Physiol 58: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Demidchik V, Shabala S, Isayenkov S, Cuin TA, Pottosin I (2018) Calcium transport across plant membranes: Mechanisms and functions. New Phytol 220: 49–69 [DOI] [PubMed] [Google Scholar]
- Denninger P, Bleckmann A, Lausser A, Vogler F, Ott T, Ehrhardt DW, Frommer WB, Sprunck S, Dresselhaus T, Grossmann G (2014) Male–female communication triggers calcium signatures during fertilization in Arabidopsis. Nat Commun 5: 4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao M, Qu X, Huang S (2018) Calcium imaging in Arabidopsis pollen cells using G-CaMP5. J Integr Plant Biol 60: 897–906 [DOI] [PubMed] [Google Scholar]
- Dindas J, Scherzer S, Roelfsema MRG, von Meyer K, Müller HM, Al-Rasheid KAS, Palme K, Dietrich P, Becker D, Bennett MJ, et al. (2018) AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat Commun 9: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doccula FG, Luoni L, Behera S, Bonza MC, Costa A (2018) In vivo analysis of calcium levels and glutathione redox status in Arabidopsis epidermal leaf cells infected with the hypersensitive response-inducing bacteria Pseudomonas syringae pv. tomato AvrB (PstAvrB). Methods Mol Biol 1743: 125–141 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Jakobsen MK, Baker AJ, Telzerow A, Hou SW, Laplaze L, Barrot L, Poethig RS, Haseloff J, Webb AA (2006) Time of day modulates low-temperature Ca signals in Arabidopsis. Plant J 48: 962–973 [DOI] [PubMed] [Google Scholar]
- Edel KH, Marchadier E, Brownlee C, Kudla J, Hetherington AM (2017) The evolution of calcium-based signalling in plants. Curr Biol 27: R667–R679 [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Fasani E, DalCorso G, Costa A, Zenoni S, Furini A. (2019) The Arabidopsis thaliana transcription factor MYB59 regulates calcium signalling during plant growth and stress response. Plant Mol Biol 99: 517–534 [DOI] [PubMed] [Google Scholar]
- Fichman Y, Mittler R (2021) Integration of electric, calcium, reactive oxygen species and hydraulic signals during rapid systemic signaling in plants. Plant J (doi:10.1111/tpj.15360) [DOI] [PubMed] [Google Scholar]
- Gao D, Knight MR, Trewavas AJ, Sattelmacher B, Plieth C (2004) Self-reporting Arabidopsis expressing pH and [Ca2+] indicators unveil ion dynamics in the cytoplasm and in the apoplast under abiotic stress. Plant Physiol 134: 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata C, Gay R, Sokolovski S, Hills A, Lamattina L, Blatt MR (2003) Nitric oxide regulates K+ and Cl− channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc Natl Acad Sci USA 100: 11116–111121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Fricker MD, Read ND, Trewavas AJ (1991) Role of calcium in signal transduction of Commelina guard cells. Plant Cell 3: 333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J, van Weeren L, Hink MA, Vischer NO, Jalink K, Gadella TW Jr (2010) Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat Methods 7: 137–139 [DOI] [PubMed] [Google Scholar]
- Gottschalk S, Degtyaruk O, McLarney B, Rebling J, Hutter MA, Deán-Ben XL, Shoham S, Razansky D (2019) Rapid volumetric optoacoustic imaging of neural dynamics across the mouse brain. Nat Biomed Eng 3: 392–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald EC, Mehta S, Zhang J (2018) Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem Rev 118: 11707–11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Hamamura Y, Nishimaki M, Takeuchi H, Geitmann A, Kurihara D, Higashiyama T (2014) Live imaging of calcium spikes during double fertilization in Arabidopsis. Nat Commun 5: 4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazak O, Mamon E, Lavy M, Sternberg H, Behera S, Schmitz-Thom I, Bloch D, Dementiev O, Gutman I, Danziger T, et al. (2019) A novel Ca2+-binding protein that can rapidly transduce auxin responses during root growth. PLoS Biol 17: e3000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich R, Neher E (2018) Venus flytrap: how an excitable, carnivorous plant works. Trends Plant Sci 23: 220–234 PMID: 29336976 DOI: 10.1016/j.tplants.2017.12.004 [DOI] [PubMed] [Google Scholar]
- Heim N, Garaschuk O, Friedrich MW, Mank M, Milos RI, Kovalchuk Y, Konnerth A, Griesbeck O (2007) Improved calcium imaging in transgenic mice expressing a troponin C-based biosensor. Nat Methods 4: 127–129 [DOI] [PubMed] [Google Scholar]
- Hilleary R, Paez-Valencia J, Vens C, Toyota M, Palmgren M, Gilroy S (2020) Tonoplast-localized Ca2+ pumps regulate Ca2+ signals during pattern-triggered immunity in Arabidopsis thaliana. Proc Natl Acad Sci USA 117: 18849–18857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdaway-Clarke TL, Feijo JA, Hackett GR, Kunkel JG, Hepler PK (1997) Pollen tube growth and the intracellular cytosolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell 9: 1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa K, Yamada Y, Matsuda T, Kobayashi K, Hashimoto M, Matsu-ura T, Miyawaki A, Michikawa T, Mikoshiba K, Nagai T (2010) Spontaneous network activity visualized by ultrasensitive Ca2+ indicators, yellow Cameleon-Nano. Nat Methods 7: 729–732 [DOI] [PubMed] [Google Scholar]
- Ishka MR, Brown E, Rosenberg A, Romanowsky S, Davis J, Choi W-G, Harper JF (2021) Arabidopsis Ca2+-ATPases 1, 2, and 7 in the endoplasmic reticulum contribute to growth and pollen fitness. Plant Phys 185: 1966–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano M, Entani T, Shiba H, Kakita M, Nagai T, Mizuno H, Miyawaki A, Shoji T, Kubo K, Isogai A, et al. (2009) Fine-tuning of the cytoplasmic Ca2+ concentration is essential for pollen tube growth. Plant Physiol 150: 1322–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Zhou X, Tao M, Yuan F, Liu L, Wu F, Wu X, Xiang Y, Niu Y, Liu F, et al. (2019) Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572: 341–346 [DOI] [PubMed] [Google Scholar]
- Johnson CH, Knight MR, Kondo T, Masson P, Sedbrook J, Haley A, Trewavas A (1995) Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science 269: 1863–1865 [DOI] [PubMed] [Google Scholar]
- Keinath NF, Waadt R, Brugman R, Schroeder JI, Grossmann G, Schumacher K, Krebs M (2015) Live cell imaging with R-GECO1 sheds light on flg22- and chitin-induced transient [Ca2+]cyt patterns in Arabidopsis. Mol Plant 8: 1188–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelner A, Leitao N, Chabaud M, Charpentier M, de Carvalho-Niebel F (2018) Dual color sensors for simultaneous analysis of calcium signal dynamics in the nuclear and cytoplasmic compartments of plant cells. Front Plant Sci 9: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiegle E, Moore CA, Haseloff J, Tester MA, Knight MR (2000) Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J 23: 267–278 [DOI] [PubMed] [Google Scholar]
- Kiep V, Vadassery J, Lattke J, Maass JP, Boland W, Peiter E, Mithofer A (2015) Systemic cytosolic Ca2+ elevation is activated upon wounding and herbivory in Arabidopsis. New Phytol 207: 996–1004 [DOI] [PubMed] [Google Scholar]
- Kleist TJ, Cartwright HN, Perera AM, Christianson ML, Lemaux PG, Luan S (2017) Genetically encoded calcium indicators for fluorescence imaging in the moss Physcomitrella: GCaMP3 provides a bright new look. Plant Biotechnol J 15: 1235–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klejchova M, Silva-Alvim FAL, Blatt MR, Alvim JC (2021) Membrane voltage as a dynamic platform for spatio-temporal signalling, physiological and developmental regulation. Plant Physiol 185: 1523–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ (1991) Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Komis G, Šamajová O, Ovečka M, Šamaj J (2015) Super-resolution microscopy in plant cell imaging. Trends Plant Sci 20: 834–843. [DOI] [PubMed] [Google Scholar]
- Krebs M, Held K, Binder A, Hashimoto K, Den Herder G, Parniske M, Kudla J, Schumacher K (2012) FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J 69: 181–192 [DOI] [PubMed] [Google Scholar]
- Krebs M, Schumacher K (2013) Live cell imaging of cytoplasmic and nuclear Ca2+ dynamics in Arabidopsis roots. Cold Spring Harb Protoc 2013: 776–780 [DOI] [PubMed] [Google Scholar]
- Krogman W, Sparks JA, Blancaflor EB (2020) Cell type-specific imaging of calcium signaling in Arabidopsis thaliana seedling roots using GCaMP3. Int J Mol Sci 21: 6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K (2010) Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Becker D, Grill E, Hedrich R, Hippler M, Kummer U, Parniske M, Romeis T, Schumacher K (2018) Advances and current challenges in calcium signaling. New Phytol 218: 414–431 [DOI] [PubMed] [Google Scholar]
- Laanemets K, Wang YF, Lindgren O, Wu J, Nishimura N, Lee S, Caddell D, Merilo E, Brosche M, Kilk K, et al. (2013) Mutations in the SLAC1 anion channel slow stomatal opening and severely reduce K+ uptake channel activity via enhanced cytosolic [Ca2+] and increased Ca2+ sensitivity of K+ uptake channels. New Phytol 197: 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laissue PP, Alghamdi RA, Tomancak P, Reynaud EG, Shroff H (2017) Assessing phototoxicity in live fluorescence imaging. Nat Methods 14: 657–661 [DOI] [PubMed] [Google Scholar]
- Leitão N, Dangeville P, Carter R, Charpentier M (2019) Nuclear calcium signatures are associated with root development. Nat Commun 10: 4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenglet A, Jaslan D, Toyota M, Mueller M, Muller T, Schonknecht G, Marten I, Gilroy S, Hedrich R, Farmer EE (2017) Control of basal jasmonate signalling and defence through modulation of intracellular cation flux capacity. New Phytol 216: 1161–1169 [DOI] [PubMed] [Google Scholar]
- Lew TTS, Sarojam R, Jang IC, Park BS, Naqvi NI, Wong MH, Singh GP, Ram RJ, Shoseyov O, Saito K, et al. (2020) Species-independent analytical tools for next-generation agriculture. Nat Plants 6: 1408–1417 [DOI] [PubMed] [Google Scholar]
- Li K, Prada J, Damineli DSC, Liese A, Romeis T, Dandekar T, Feijó JA, Hedrich R, Konrad KR (2021) An optimized genetically encoded dual reporter for simultaneous ratio imaging of Ca2+ and H+ reveals new insights into ion signaling in plants. New Phytol (doi:10.1111/nph.17202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Niu Y, Konishi M, Wu Y, Du H, Sun Chung H, Li L, Boudsocq M, McCormack M, Maekawa S, et al. (2017) Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545: 311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WW, Chen SH, Li PC (2019) Functional calcium imaging using optical-resolution photoacoustic microscopy in a 3D tumor cell culture. Proc SPIE ( 10.1117/12.2510936) [DOI] [Google Scholar]
- Logan DC, Knight MR (2003) Mitochondrial and cytosolic calcium dynamics are differentially regulated in plants. Plant Physiol 133: 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loro G, Costa A (2013) Imaging of mitochondrial and nuclear Ca2+ dynamics in Arabidopsis roots. Cold Spring Harb Protoc 8: 781–785 [DOI] [PubMed] [Google Scholar]
- Loro G, Drago I, Pozzan T, Schiavo FL, Zottini M, Costa A (2012) Targeting of Cameleons to various subcellular compartments reveals a strict cytoplasmic/mitochondrial Ca2+ handling relationship in plant cells. Plant J 71: 1–13 [DOI] [PubMed] [Google Scholar]
- Loro G, Wagner S, Doccula FG, Behera S, Weinl S, Kudla J, Schwarzlander M, Costa A, Zottini M (2016) Chloroplast-specific in vivo Ca2+ imaging using Yellow Cameleon fluorescent protein sensors reveals organelle-autonomous Ca2+ signatures in the stroma. Plant Physiol 171: 2317–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love J, Dodd AN, Webb AA (2004) Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell 16: 956–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Chen L, Huang F, Gao P, Zhao H, Wang Y, Han S (2020) Intraorganellar calcium imaging in Arabidopsis seedling roots using the GCaMP variants GCaMP6m and R-CEPIA1er. J Plant Physiol 246–247: 153127. [DOI] [PubMed] [Google Scholar]
- Martí MC, Stancombe MA, Webb AA (2013) Cell- and stimulus type-specific intracellular free Ca2+ signals in Arabidopsis. Plant Physiol 163: 625–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí Ruiz MC, Jung HJ, Webb AAR (2020) Circadian gating of dark-induced increases in chloroplast- and cytosolic-free calcium in Arabidopsis. New Phytol 225: 1993–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh MR, Brownlee C, Hetherington AM (1990) Abscisic acid-induced elevation of guard cell cytosolic Ca2+ precedes stomatal closure. Nature 343: 186–188 [Google Scholar]
- McAinsh MR, Pittman JK (2009) Shaping the calcium signature. New Phytol 181: 275–294 [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Webb A, Taylor JE, Hetherington AM (1995) Stimulus-induced oscillations in guard cell cytosolic free calcium. Plant Cell 7: 1207–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlmer N, Parvin N, Hurst CH, Knight MR, Teige M, Vothknecht UC (2012) A toolset of aequorin expression vectors for in planta studies of subcellular calcium concentrations in Arabidopsis thaliana. J Exp Bot 63: 1751–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Zhang Y, Roth RH, Zhang JF, Mo A, Tenner B, Huganir RL, Zhang J (2018) Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat Cell Biol 20: 1215–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michard E, Dias P, Feijó JA (2008) Tobacco pollen tubes as cellular models for ion dynamics: improved spatial and temporal resolution of extracellular flux and free cytosolic concentration of calcium and protons using pHluorin and YC3.1 CaMeleon. Sex Plant Reprod 21: 169–181 [Google Scholar]
- Michard E, Lima PT, Borges F, Silva AC, Portes MT, Carvalho JE, Gilliham M, Liu L-H, Obermeyer G, Feijó JA (2011) Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332: 434–437 [DOI] [PubMed] [Google Scholar]
- Mithöfer A, Mazars C (2002) Aequorin-based measurements of intracellular Ca2+-signatures in plant cells. Biol Proc Online 4: 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, Griesbeck O, Heim R, Tsien RY (1999) Dynamic and quantitative Ca2+ measurements using improved Cameleons. Proc Natl Acad Sci USA 96: 2135–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY (1997). Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388: 882–887 [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Messerli MA, Gilroy S (2008) Imaging of the Yellow Cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol 147: 1690–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al. (2006) CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseyko N, Feldman LJ (2001) Expression of pH-sensitive green fluorescent protein in Arabidopsis thaliana. Plant Cell Environ 24: 557–563 [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87–90 [DOI] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A (2004) Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA 101: 10554–10559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, Imoto K (2001) A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat Biotechnol 19: 137–141 [DOI] [PubMed] [Google Scholar]
- Ngo QA, Vogler H, Lituiev DS, Nestorova A, Grossniklaus U (2014) A calcium dialog mediated by the FERONIA signal transduction pathway controls plant sperm delivery. Dev Cell 29: 491–500 [DOI] [PubMed] [Google Scholar]
- Nguyen CT, Kurenda A, Stolz S, Chételat A, Farmer EE (2018) Identification of cell populations necessary for leaf-to-leaf electrical signaling in a wounded plant. Proc Natl Acad Sci USA 115: 10178–10183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya Y, Hirano T (1996) Shining the light: The mechanism of the bioluminescence reaction of calcium-binding photoproteins. Chem Biol 3: 337–347 [DOI] [PubMed] [Google Scholar]
- Ordenes VR, Moreno I, Maturana D, Norambuena L, Trewavas AJ, Orellana A (2012) In vivo analysis of the calcium signature in the plant Golgi apparatus reveals unique dynamics. Cell Calcium 52: 397–404 [DOI] [PubMed] [Google Scholar]
- Palmer AE, Giacomello M, Kortemme T, Hires SA, Lev-Ram V, Baker D, Tsien RY (2006) Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem Biol 13: 521–530 [DOI] [PubMed] [Google Scholar]
- Palmer AE, Jin C, Reed JC, Tsien RY (2004) Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc Natl Acad Sci USA 101: 17404–17409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AE, Tsien RY (2006) Measuring calcium signaling using genetically targetable fluorescent indicators. Nat Protoc 1: 1057–1065 [DOI] [PubMed] [Google Scholar]
- Palmgren MG (2001) PLANT PLASMA MEMBRANE H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52: 817–845 [DOI] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Koldenkova V, Nagai T (2013) Genetically encoded Ca2+ indicators: properties and evaluation. Biochim Biophys Acta 1833: 1787–1797 [DOI] [PubMed] [Google Scholar]
- Pirayesh N, Giridhar M, Ben Khedher A, Vothknecht UC, Chigri F (2021) Organellar calcium signaling in plants: An update. Biochim Biophys Acta Mol Cell Res 1868: 118948. [DOI] [PubMed] [Google Scholar]
- Prasher D, McCann RO, Cormier MJ (1985) Cloning and expression of the cDNA coding for aequorin, a bioluminescent calcium-binding protein. Biochem Biophys Res Commun 126: 1259–1268 [DOI] [PubMed] [Google Scholar]
- Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ (1992) Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111: 229–233 [DOI] [PubMed] [Google Scholar]
- Resentini F, Grenzi M, Ancora D, Cademartori M, Luoni L, Franco M, Bassi A, Bonza MC, Costa A (2021a) Simultaneous imaging of ER and cytosolic Ca2+ dynamics reveals long-distance ER Ca2+ waves in plants. Plant Physiol 187: 603–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resentini F, Ruberti C, Grenzi M, Bonza MC, Costa A (2021b) The signatures of organellar calcium. Plant Physiol (doi.org/10.1093/plphys/kiab189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V, Pinton P, Tosello V, Rizzuto R, Pozzan T (2000) Recombinant aequorin as tool for monitoring calcium concentration in subcellular compartments. Methods Enzymol 327: 440–456 [DOI] [PubMed] [Google Scholar]
- Roberts S, Seeger M, Jiang Y, Mishra A, Sigmund F, Stelzl A, Lauri A, Symvoulidis P, Rolbieski H, Preller M, et al. (2018) Calcium sensor for photoacoustic imaging. J Am Chem Soc 140: 2718–2721 [DOI] [PubMed] [Google Scholar]
- Rogers KL, Stinnakre J, Agulhon C, Jublot D, Shorte SL, Kremer EJ, Brulet P (2005) Visualization of local Ca2+ dynamics with genetically encoded bioluminescent reporters. Eur J Neurosci 21: 597–610 [DOI] [PubMed] [Google Scholar]
- Romano Armada N, Doccula FG, Candeo A, Valentini G, Costa A, Bassi A (2019) In vivo light sheet fluorescence microscopy of calcium oscillations in Arabidopsis thaliana. Methods Mol Biol 1925: 87–101 [DOI] [PubMed] [Google Scholar]
- Rudolf R, Mongillo M, Rizzuto R, Pozzan T (2003) Looking forward to seeing calcium. Nat Rev Mol Cell Biol 4: 579–586 [DOI] [PubMed] [Google Scholar]
- Sai J, Johnson CH (2002) Dark-stimulated calcium ion fluxes in the chloroplast stroma and cytosol. Plant Cell 14: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell 14: S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer S, Böhm J, Krol E, Shabala L, Kreuzer I, Larisch C, Bemm F, Al-Rasheid KA, Shabala S, Rennenberg H, et al. (2015) Calcium sensor kinase activates potassium uptake systems in gland cells of Venus flytraps. Proc Natl Acad Sci USA 112: 7309–7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sello S, Moscatiello R, Mehlmer N, Leonardelli M, Carraretto L, Cortese E, Zanella FG, Baldan B, Szabo I, Vothknecht UC, et al. (2018) Chloroplast Ca2+ fluxes into and across thylakoids revealed by thylakoid-targeted aequorin probes. Plant Physiol 177: 38–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sello S, Perotto J, Carraretto L, Szabo I, Vothknecht UC, Navazio L (2016) Dissecting stimulus-specific Ca2+ signals in amyloplasts and chloroplasts of Arabidopsis thaliana cell suspension cultures. J Exp Bot 67: 3965–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Q, Gao Q, Lhamo D, Zhang H, Luan S (2020) Two glutamate- and pH-regulated Ca2+ channels are required for systemic wound signaling in Arabidopsis. Sci Signal 13: eaba1453. [DOI] [PubMed] [Google Scholar]
- Shaw SL, Thoms D, Powers J (2019) Structured illumination approaches for super-resolution in plant cells. Microscopy 68: 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Dana H, Abdelfattah AS, Patel R, Shea J, Molina RS, Rawal B, Rancic V, Chang YF, Wu L, et al. (2018) A genetically encoded Ca2+ indicator based on circularly permutated sea anemone red fluorescent protein eqFP578. BMC Biol 16: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura O (1995) A short story of aequorin. Biol Bull 189: 1–5 [DOI] [PubMed] [Google Scholar]
- Shimomura O, Johnson FH, Saiga Y (1962) Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol 59: 223–239 [DOI] [PubMed] [Google Scholar]
- Shkolnik D, Nuriel R, Bonza MC, Costa A, Fromm H (2018) MIZ1 regulates ECA1 to generate a slow, long-distance phloem-transmitted Ca2+ signal essential for root water tracking in Arabidopsis. Proc Natl Acad Sci USA 115: 8031–8036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souslova EA, Belousov VV, Lock JG, Strömblad S, Kasparov S, Bolshakov AP, Pinelis VG, Labas YA, Lukyanov S, Mayr LM, et al. (2007) Single fluorescent protein-based Ca2+ sensors with increased dynamic range. BMC Biotechnol. 29;7:37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stael S, Wurzinger B, Mair A, Mehlmer N, Vothknecht UC, Teige M (. 2012) Plant organellar calcium signalling: an emerging field. J Exp Bot 63: 1525–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storti M, Costa A, Golin S, Zottini M, Morosinotto T, Alboresi A (2018) Systemic calcium wave propagation in Physcomitrella patens. Plant Cell Physiol 59: 1377–1384 [DOI] [PubMed] [Google Scholar]
- Suda H, Mano H, Toyota M, Fukushima K, Mimura T, Tsutsui I, Hedrich R, Tamada Y, Hasebe M (2020) Calcium dynamics during trap closure visualized in transgenic Venus flytrap. Nat Plants 6: 1219–1224 [DOI] [PubMed] [Google Scholar]
- Suzuki J, Kanemaru K, Ishii K, Ohkura M, Okubo Y, Iino M (2014) Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nat Commun 5: 4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L, Zeiger E, Møller IM, Murphy A (2014) Plant Physiology and Development. Ed 6, Sinauer Associates Inc., Sunderland, Massachusetts, USA. ISBN: 9781605352558
- Tanaka K, Swanson SJ, Gilroy S, Stacey G (2010) Extracellular nucleotides elicit cytosolic free calcium oscillations in Arabidopsis. Plant Physiol 154: 705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teardo E, Carraretto L, Wagner S, Formentin E, Behera S, De Bortoli S, Larosa V, Fuchs P, Lo Schiavo F, Raffaello A, et al. (2017) Physiological characterization of a plant mitochondrial calcium uniporter in vitro and in vivo. Plant Physiol 173: 1355–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thestrup T, Litzlbauer J, Bartholomäus I, Mues M, Russo L, Dana H, Kovalchuk Y, Liang Y, Kalamakis G, Laukat Y, et al. (2014) Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat Methods 11: 175–182 [DOI] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, et al. (2009) Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods 6: 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Wang C, Gao Q, Li L, Luan S. (2020) Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat Plants 6: 750–759 [DOI] [PubMed] [Google Scholar]
- Toyota M, Spencer D, Sawai-Toyota S, Jiaqi W, Zhang T, Koo AJ, Howe GA, Gilroy S (2018) Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Tserevelakis GJ, Tsagkaraki M, Zacharakis G (2016) Hybrid photoacoustic and optical imaging of pigments in vegetative tissues. J Microsc 263: 300–306 [DOI] [PubMed] [Google Scholar]
- Tsien RY (1980) New calcium indicators and buffers with high selectivity against magnesium and protons: design, synthesis, and properties of prototype structures. Biochemistry 27: 2396–2404 [DOI] [PubMed] [Google Scholar]
- van Der Luit AH, Olivari C, Haley A, Knight MR, Trewavas AJ (1999) Distinct calcium signaling pathways regulate calmodulin gene expression in tobacco. Plant Physiol 121: 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent TR, Avramova M, Canham J, Higgins P, Bilkey N, Mugford ST, Pitino M, Toyota M, Gilroy S, Miller AJ, et al. (2017) Interplay of plasma membrane and vacuolar ion channels, together with BAK1, elicits rapid cytosolic calcium elevations in Arabidopsis during aphid feeding. Plant Cell 29: 1460–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Köster P, Andrés Z, Waadt C, Bradamante G, Lampou K, Kudla J, Schumacher K (2020) Dual-reporting transcriptionally linked genetically encoded fluorescent indicators resolve the spatiotemporal coordination of cytosolic abscisic acid and second messenger dynamics in Arabidopsis. Plant Cell 32: 2582–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Krebs M, Kudla J, Schumacher K (2017) Multiparameter imaging of calcium and abscisic acid and high-resolution quantitative calcium measurements using R-GECO1-mTurquoise in Arabidopsis. New Phytol 216: 303–320 [DOI] [PubMed] [Google Scholar]
- Wagner S, Behera S, De Bortoli S, Logan DC, Fuchs P, Carraretto L, Teardo E, Cendron L, Nietzel T, Fussl M, et al. (2015) The EF-hand Ca2+ binding protein MICU choreographs mitochondrial Ca2+ dynamics in Arabidopsis. Plant Cell 27: 3190–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Steinbeck J, Fuchs P, Lichtenauer S, Elsässer M, Schippers JHM, Nietzel T, Ruberti C, Van Aken O, Meyer AJ, et al. (2019) Multiparametric real-time sensing of cytosolic physiology links hypoxia responses to mitochondrial electron transport. New Phytol 224: 1668–1684 [DOI] [PubMed] [Google Scholar]
- Wang R, Himschoot E, Grenzi M, Chen J, Krebs M, Schumacher K, Nowack MK, Van Damme D, De Smet I, Beeckman T, et al. (2020) Pharmacological and genetic manipulations of Ca2+ signaling have contrasting effects on auxin-regulated trafficking. bioRxiv 2020.09.04.283101 (doi: 10.1101/2020.09.04.283101) [DOI] [Google Scholar]
- White PJ, Broadley MR (2003) Calcium in plants. Ann Bot 92: 487–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson RE, Ashley CC (1982) Free Ca2+ and cytoplasmic streaming in the alga Chara. Nature 296: 647–650 [DOI] [PubMed] [Google Scholar]
- Wu F, Chi Y, Jiang Z, Xu Y, Xie L, Huang F, Wan D, Ni J, Yuan F, Wu X, et al. (2020) Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 578: 577–581 [DOI] [PubMed] [Google Scholar]
- Wu J, Abdelfattah AS, Miraucourt LS, Kutsarova E, Ruangkittisakul A, Zhou H, Ballanyi K, Wicks G, Drobizhev M, Rebane A, et al. (2014) A long Stokes shift red fluorescent Ca2+ indicator protein for two-photon and ratiometric imaging. Nat Commun 5: 5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Liu L, Matsuda T, Zhao Y, Rebane A, Drobizhev M, Chang YF, Araki S, Arai Y, March K, et al. (2013) Improved orange and red Ca+ indicators and photophysical considerations for optogenetic applications. ACS Chem Neurosci 4: 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong TC, Ronzier E, Sanchez F, Corratge-Faillie C, Mazars C, Thibaud JB (2014) Imaging long distance propagating calcium signals in intact plant leaves with the BRET-based GFP-aequorin reporter. Front Plant Sci 5: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Wang LV (2006) Photoacoustic imaging in biomedicine. Rev Sci Instrum 77: 041101 [Google Scholar]
- Yang DL, Shi Z, Bao Y, Yan J, Yang Z, Yu H, Li Y, Gou M, Wang S, Zou B, et al. (2017) Calcium pumps and interacting BON1 protein modulate calcium signature, stomatal closure, and plant immunity. Plant Physiol 175: 424–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI (2008) Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, Yang H, Xue Y, Kong D, Ye R, Li C, Zhang J, Theprungsirikul L, Shrift T, Krichilsky B, et al. (2014) OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514: 367–371 [DOI] [PubMed] [Google Scholar]
- Zeng L, Ma G, Xu H, Mu J, Li F, Gao X, Deng Z, Qu J, Huang P, Lin J (2019) In vivo chemoselective photoacoustic imaging of copper(II) in plant and animal subjects. Small 15: e1803866 [Is the journal title adequate?] [DOI] [PubMed] [Google Scholar]
- Zhao X, Wang YL, Qiao XR, Wang J, Wang LD, Xu CS, Zhang X (2013) Phototropins function in high-intensity blue light-induced hypocotyl phototropism in Arabidopsis by altering cytosolic calcium. Plant Physiol 162: 1539–15351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, et al. (2011) An expanded palette of genetically encoded Ca2+ indicators. Science 333: 1888–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Feng Y, Liang G, Liu N, Zhu JK (2013) Aequorin-based luminescence imaging reveals stimulus- and tissue-specific Ca2+ dynamics in Arabidopsis plants. Mol Plant 6: 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zottini M, Zannoni D (1993) The use of fura-2 fluorescence to monitor the movement of free calcium ions into the matrix of plant mitochondria (Pisum sativum and Helianthus tuberosus). Plant Physiol 102: 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.