Abstract
Light is the ultimate source of energy for photosynthetic organisms, but respiration is fundamental for supporting metabolism during the night or in heterotrophic tissues. In this work, we isolated Physcomitrella (Physcomitrium patens) plants with altered respiration by inactivating Complex I (CI) of the mitochondrial electron transport chain by independently targeting on two essential subunits. Inactivation of CI caused a strong growth impairment even in fully autotrophic conditions in tissues where all cells are photosynthetically active, demonstrating that respiration is essential for photosynthesis. CI mutants showed alterations in the stoichiometry of respiratory complexes while the composition of photosynthetic apparatus was substantially unaffected. CI mutants showed altered photosynthesis with high activity of both Photosystems I and II, likely the result of high chloroplast ATPase activity that led to smaller ΔpH formation across thylakoid membranes, decreasing photosynthetic control on cytochrome b6f in CI mutants. These results demonstrate that alteration of respiratory activity directly impacts photosynthesis in P. patens and that metabolic interaction between organelles is essential in their ability to use light energy for growth.
Mosses lacking mitochondrial Complex I show induced activation of chloroplast ATPase, demonstrating the impact of respiration on the metabolism of plant photosynthetic cells.
Introduction
In photosynthetic organisms, sunlight powers the linear electron flow from water to NADP+ catalyzed by two photosystems (PS), PSII and PSI, cytochrome b6f and ATPase, ultimately generating NADPH and ATP to sustain cellular metabolism. In photosynthetic organisms, mitochondrial respiration is also active with its specific electron transport pathway, or oxidative phosphorylation (OXPHOS). OXPHOS transfers electrons from the substrates NADH and succinate to molecular oxygen (O2) through the activity of enzymatic complexes localized in the inner mitochondrial membrane, Complex I (CI), CII, CIII, and CIV. This electron transfer is coupled to the generation of electrochemical transmembrane gradient that drives the synthesis of ATP through ATP-synthase, also called complex V. The NADH dehydrogenase complex (CI) is the main site for electron insertion into the mitochondrial electron transport chain (ETC) and it can provide up to 40% of the protons for mitochondrial ATP formation (Watt et al., 2010; Braun et al., 2014). In plants, ETC electrons can also follow alternative routes bypassing CI via alternative NADH dehydrogenases (Lecler et al., 2012) and CIII/IV via the alternative terminal oxidase (AOX; Dinant et al., 2001). These alternative routes partly uncouple the electron transport and the electrochemical transmembrane gradient, thus reducing the energy yield of respiration with beneficial effects especially in stress conditions (Zalutskaya et al., 2015), reducing reactive oxygen species (ROS) production from mETC (Møller, 2001; Vanlerberghe et al., 2020).
Respiration in photosynthetic organisms is essential to support energy demand during the night in nonphotosynthetic tissues such as roots or in developmental stages where photosynthesis is not active (e.g. seed germination). An increasing set of evidence, however, underlines the importance of respiration also for sustaining optimal photosynthetic activity (Joliot and Joliot, 2008; Noguchi and Yoshida, 2008; Bailleul et al., 2015) with a strong functional link between chloroplasts and mitochondria bioenergetic metabolism (Cardol et al., 2003; Dutilleul et al., 2003; Schönfeld et al., 2004). In the diatom, Phaeodactylum tricornutum metabolite exchange between chloroplast and mitochondria is essential for carbon fixation (Bailleul et al., 2015). In another example of such a functional link, excess reducing power produced via photosynthesis can be routed to mitochondrial respiration, preventing over-reduction and eventual ROS production in the plastid (Noguchi and Yoshida, 2008; Zhang et al., 2012). AOX activity has also been shown to influence photosynthetic metabolism response to stresses when it consumes excess reductant while decreasing mitochondrial ATP synthesis (Cheung et al., 2015) and maintaining redox balance of plastoquinone pool (Yoshida and Noguchi, 2011; Vanlerberghe et al., 2020). Consistently, AOX protein level was shown to be linked to differences in chloroplast energetic balance, being induced under strong irradiance, suggesting the presence of regulation signals originated from photosynthetic electron transport (Dahal et al., 2016).
Imbalances between the relative rates of production and consumption of ATP/NADPH utilization can lead to the build-up of reactive intermediates of electron transfer processes, driving the formation of harmful ROS (Eberhard et al., 2008; Li et al., 2009). Photosynthetic organisms evolved multiple regulatory mechanisms to balance light-dependent processes and metabolic exploitation of photosynthesis products. Examples of such mechanisms are the dissipation of excess excitation energy as heat (nonphotochemical quenching [NPQ]) or the photosynthetic control to reduce electron transport capacity at the level of cytochrome b6f and prevent over-reduction. Both mechanisms are activated by a decrease of lumenal pH that represents a major signal for regulation of photosynthesis (Eberhard et al., 2008; Li et al., 2009).
Consistent with the essential role of mitochondrial respiration in plant metabolism, knockout (KO) mutants completely depleted in CII (Leon et al., 2007), CIII (Colas Des Francs-Small and Small, 2014), and CIV (Radin et al., 2015) are not viable and so far only knockdown (KD) plants have been isolated and studied. Mutants completely lacking mitochondrial CI activity have instead been described in Arabidopsis (Arabidopsis thaliana; Kühn et al., 2015; Fromm et al., 2016a), as well as in tobacco (Nicotiana tabacum; Vidal et al., 2007) where they showed a severe growth phenotype and alterations in germination, fertilization, and pollen development (Fromm et al., 2016a). European mistletoe Viscum album can live without CI, but it is an obligate semi-parasite living on branches of trees, and thus its energy metabolism is likely remodeled (Maclean et al., 2018; Senkler et al., 2018).
Differently from plants, a large number of respiratory mutants have instead been isolated in the green alga Chlamydomonas reinhardtii where they generally show strong phenotypes under heterotrophic conditions but grow similarly to wild-type (WT) under photoautotrophic conditions (Salinas et al., 2014; Larosa et al., 2018).
These differences suggest that the role of respiration on cell metabolism adapted during plant evolution, motivating the investigation of species that diverged at different times during evolution. Nonvascular plants like the moss Physcomitrium patens diverged from vascular plant ancestors early after land colonization and thus their study allows highlighting the first adaptation to the new environmental conditions. To assess how the biological role of respiration adapted during the evolution of photosynthetic organisms in this work, we generated P. patens plants depleted of active respiratory CI and investigated effects on photosynthetic activity. Results show that the absence of CI stimulates photosynthetic transport caused by more active chloroplast ATPase with alteration of pmf and ΔpH across the thylakoid membranes and impairment of photosynthetic control.
Results
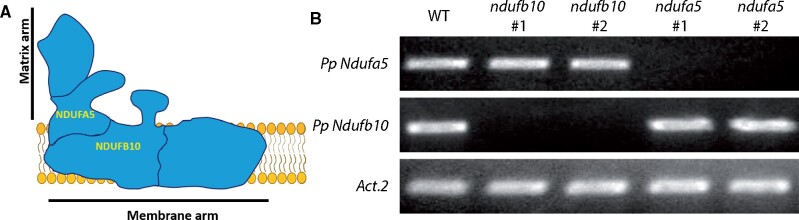
CI in eukaryotes is composed of over 40 subunits and, among them, 14 are highly conserved across kingdoms (Ligas et al., 2019). Plant CI includes nine additional subunits that form a carbonic anhydrase domain (Braun et al., 2014; Fromm et al., 2016a; Subrahmanian et al., 2016). For all CI subunits identified in Arabidopsis, it is possible to identify a putative homolog in P. patens genome, suggesting a general conservation of CI subunits between the two organisms (Supplemental Table S1), consistent with similarities observed between plants and green algae (Klusch et al., 2020). Based on this analysis, NDUFA5 and NDUFB10 (NADH: Ubiquinone Oxidoreductase Subunit A5 and B10) genes were selected to generate P. patens mutants depleted in CI based on two criteria: (1) they encoded for conserved proteins known to be essential for CI activity in C. reinhardtii, A. thaliana, or Homo sapiens (Barbieri et al., 2011; Rak and Rustin, 2014); (2) they were present in a single copy in P. patens nuclear genome, facilitating the generation of mutants and ensuring that any eventual phenotype would be readily assessable. NDUFA5 is localized in the hydrophilic region of the complex binding the substrate NADH (Figure 1A), and it is required for assembly and stability of the matrix arm of CI in human mitochondria (Rak and Rustin, 2014). NDUFB10 deletion in C. reinhardtii impairs the assembly of the distal part of CI membrane module (Figure 1A), responsible for the proton pumping activity coupled with the electron transfer (Barbieri et al., 2011).
Figure 1.
Generation of P. patens plants depleted in CI. A, Schematic representation of CI structure with NDUFA5 and NDUFB10 localization. B, RT-PCR to assess the accumulation of the mRNA encoding for NDUFA5 and NDUFB10 in two independent KO lines.
KO lines for both genes were generated by homologous recombination-mediated gene targeting (Supplemental Figure S1, A and B). Multiple independent KO lines for each gene were isolated, and the insertion of DNA in the expected position of the genome was verified by PCR (Supplemental Figure S1C). The loss of expression of the target gene was also confirmed by reverse transcriptase PCR (RT-PCR; two lines shown in Figure 1B). In the following sections, results from one line per gene are reported, but at least four independent confirmed lines per gene were isolated per each genotype.
All confirmed KO plants showed strongly impaired growth (Figure 2) that was visible upon cultivation on a glucose-enriched medium but also on a mineral media in fully autotrophic conditions. Remarkably, the growth defect was also observed if plants were grown autotrophically under 24 h of continuous illumination, thus avoiding any dark time when respiration is expected to be essential (Figure 2). Glucose presence in the media and continuous illumination stimulated a faster growth in WT plants while the mutants remained unaffected (Supplemental Figure S2).
Figure 2.
Impact of CI inactivation on P. patens growth. WT, ndufa5, and ndufb10 KO lines growth after 28 d under 16 d/8-h light/dark photoperiod either with (top) or without (middle) the addition of 0.5% (w/v) glucose in the media or under 24-h continuous illumination without glucose (bottom).
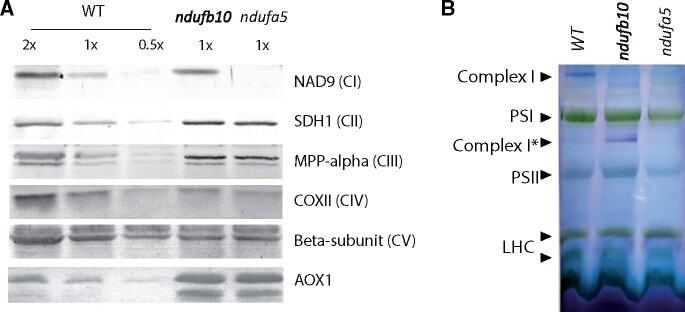
Impact of mutations on respiratory apparatus composition was assessed using specific antibodies (Figure 3A). NAD9, a CI core subunit localized in the Q module of the matrix arm, was missing in ndufa5 mutant, consistently with recently reported evidence that NDUFA5 directly interacts with NAD9 during CI biogenesis (Ligas et al., 2019). NAD9 was instead present in ndufb10 mosses, suggesting that at least part of CI was still present, as observed for other analogous mutants (Ligas et al., 2019). Both ndufa5 and ndufb10 KO plants showed changes in other complexes of the respiratory apparatus and in particular CII and CIII subunits were more abundant in the mutants as compared with WT plants by, respectively, ∼3 and 1.5 times. The most striking difference, however, was that AOX accumulated to ∼10 times higher levels in ndufa5 and ndufb10 than in WT. All differences in protein accumulation were consistent between the two independent mutant lines.
Figure 3.
Impact of CI mutation on the respiratory apparatus composition and CI assembly. A, Immunoblot analysis using antibodies against various subunits of respiratory complexes. Total protein extracts were loaded, and 1× samples correspond to 2 μg of total Chl. B, BN-PAGE separation of crude organelles extracts. Following separation (see Supplemental Figure S3), CI activity was detected with in-gel NADH/NBT staining. Membrane proteins from WT, ndufb10, and ndufa5 were solubilized with 1% (w/v) of β-DM. Main complexes of thylakoid and mitochondrial CI are indicated on the profiles. CI is expected to be ∼1,000 kDa as in Arabidopsis (Klodmann et al., 2010; Oldenkott et al., 2020). SDH, succinate de-hydrogenase; MPP, mitochondrial-processing peptidase; COX, cytochrome c oxidase; CI* indicates an assembly intermediate of CI of ∼850 kDa lacking the distal part of the membrane arm (Schimmeyer et al., 2016); LHC, light-harvesting Chl complexes.
The impact of mutations on CI was also assessed by blue native-polyacrylamide gel electrophoresis (BN-PAGE) on crude membrane extracts, containing both mitochondria and chloroplasts (Supplemental Figure S3). The detection of CI enzymatic activity after electrophoresis showed that in both mutant lines the correct assembly of CI was compromised (Figure 3B). No CI activity was detectable in the case of ndufa5. In the ndufb10 mutant, instead, the complete holoenzyme was absent but a partially assembled CI retaining the NADH dehydrogenase activity was detectable (CI*, Figure 3B) consistent with earlier results in other species (Barbieri et al., 2011).
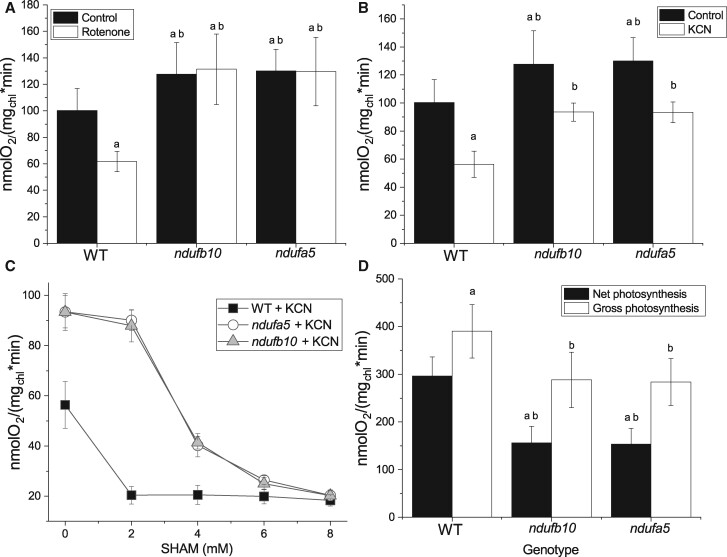
The impact of mutations on respiratory activity was assessed measuring O2 consumption rate. In both ndufa5 and ndufb10 plants, O2 consumption in the dark was higher than that in WT (Figure 4). While this seems counterintuitive for mutants affected in a respiratory complex, this observation can be explained by the compensatory over-accumulation of CII, CIII, and AOX. A similar increase in O2 consumption was indeed also observed in Arabidopsis CI mutants (Kühn et al., 2015). This hypothesis is confirmed by the observation that O2 consumption activity in ndufa5 and ndufb10 was insensitive to the addition of rotenone, a specific CI inhibitor that in WT plants instead reduced O2 consumption by ∼ 40% (Figure 4A).
Figure 4.
O2 consumption and evolution capacity in ndufb10 and ndufa5. A, O2 consumption was monitored with a Clark-type O2 electrode on WT and ndufb10 and ndufa5 plants grown for 10 d in minimal medium. Mosses were maintained in the dark in the absence (black) or presence of 50-µM rotenone (white columns). B, Effect of cyanide (KCN) on O2 consumption. Controls without the inhibitor are in black, samples with addition 1 mM KCN in white. In both A and B, “a” indicates a statistically significant difference compared to untreated WT, “b” indicates significant difference compared to WT treated with the same inhibitor (one-way ANOVA, n > 5, P < 0.01). C, Effect of different concentration of SHAM on O2 consumption in the presence of 1 mM KCN. WT is showed as black square, ndufb10 as gray triangles, ndufa5 as white circles. Average ± sd (n ≥ 5) are reported. D, O2 production of protonema illuminated with a light at 850 µmol photons m−2 s−1 measured with a Clark-type O2 electrode. Net photosynthesis (black) is calculated directly from the O2 evolution rate during illumination. Gross photosynthesis (white) is calculated by adding net photosynthesis with O2 consumption in the dark. Statistically significant differences are indicated by “a” for comparison with WT net O2 evolution, and “b” for comparisons with WT gross O2 evolution (one-way ANOVA, n > 5, P < 0.01). In all panels, average ± sd are reported.
O2 consumption activity was similarly reduced by the addition of KCN, a CIV inhibitor, in both WT and mutants (Figure 4B). The significant residual activity still present was mostly attributable to the presence of alternative oxidases like AOX, as confirmed by the further decrease in O2 evolution induced when its specific inhibitor SHAM was added together with KCN (Figure 4C). While 2 mM SHAM was a saturating dose for WT, a four-fold higher dose was necessary to obtain a full inhibition in the mutants, consistent with their increased AOX content. Calculations as in (Vanlerberghe et al., 2002) show that the mutations did not affect the capacity of the cytochrome pathway, but increased that of the AOX pathway (Supplemental Table S2), supporting its role in the stronger rates of O2 uptake.
Maximal photosynthetic O2 evolution activity was measured in the same samples upon exposure to saturating light. ndufa5 and ndufb10 KO mutants showed a reduction in net photosynthesis of ∼50% compared with the WT (Figure 4D). A large portion of this difference can be explained by the higher O2 consumption rate in ndufa5 and ndufb10 plants, where the difference from WT could be even larger in illuminated plants (Gauthier et al., 2018).
Impact of CI depletion on photosynthesis
Plants depleted of CI showed reduced photoautotrophic growth and alterations in O2 evolution activity, motivating a deeper investigation of the impact of the mutation on photosynthetic activity. Chlorophyll (Chl) content, Chl a/b, and Chl/carotenoid (Car) ratio were unchanged in mutant and WT plants (Table 1). Consistently, the accumulation of all main components of the photosynthetic apparatus (PSI, PSII, cyt b6f, ATPase) showed no major alterations in the mutants except for a slight increase in chloroplast ATPase content (Supplemental Figure S4). Subunits involved in regulatory mechanisms of photosynthesis such as PsbS and LHCSR (Alboresi et al., 2010) were also similarly accumulated in the mutants as in WT plants (Supplemental Figure S4).
Table 1.
Pigment composition and PSII quantum efficiency of P. patens WT and ndufb10 and ndufa5 mutant lines
| Genotype | Chl a/Chl b | Chl/Car | Chl content (µg mg−1 DW) | F v/Fm |
|---|---|---|---|---|
| WT | 2.62 ± 0.14 | 3.73 ± 0.37 | 18.5 ± 2.7 | 0.79 ± 0.01 |
| ndufb10 | 2.59 ± 0.20 | 3.65 ± 0.40 | 18.9 ± 3.0 | 0.78 ± 0.02 |
| ndufa5 | 2.64 ± 0.21 | 3.60 ± 0.42 | 18.7 ± 2.7 | 0.79 ± 0.01 |
Chl a/b ratio, Chl/car ratio, total Chl content and PSII quantum yield (estimated by Fv/Fm) were evaluated in plants cultivated for 10 d in a minimal medium at 50 µmol photons m−2 s−1. For each measurement, average ± sd (n ≥ 5) is reported.
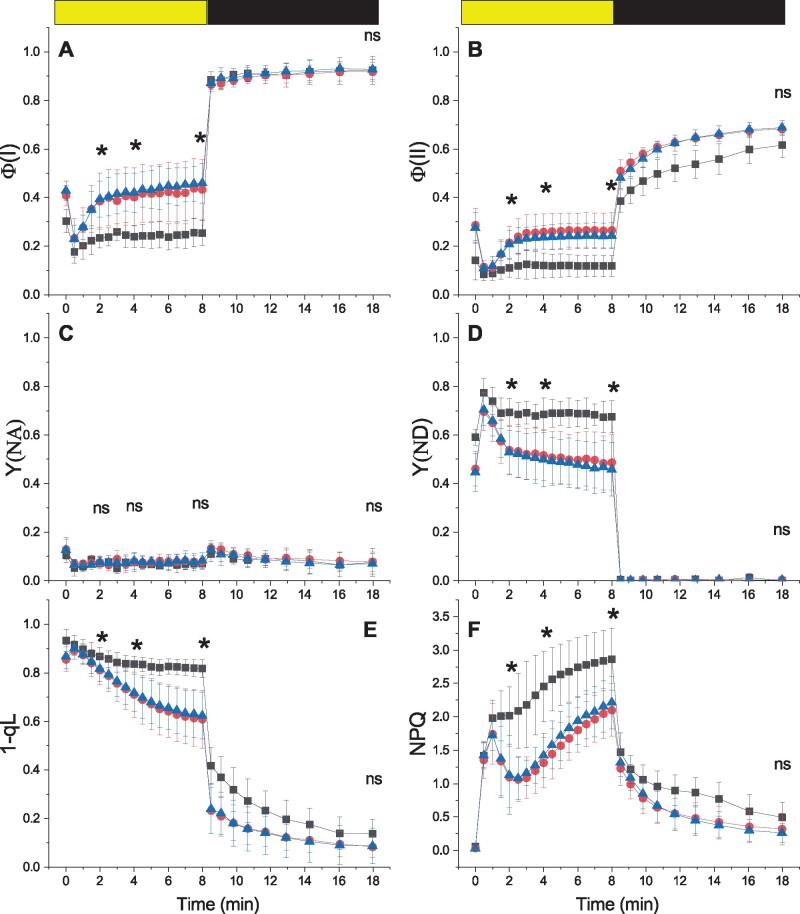
Photosynthetic functionality was analyzed in more detail by fluorescence and Near InfraRed (NIR) spectroscopy analyses. PSII maximum quantum yield (Fv/Fm) in dark-adapted plants was not altered by the mutations (Table 1). However, when plants were exposed to sub-saturating light conditions (330 µmol photons m−2 s−1) both ndufa5 and ndufb10 showed a higher yield for both PSI (Figure 5A) and PSII (Figure 5B), indicative of a higher electron transport rate (ETR). At the level of PSI, the higher yield was attributable to a lower donor side limitation (Y(ND)) while acceptor side limitation (Y(NA)) was negligible in WT and mutant plants (Figure 5, C and D). The lower PSI donor side limitation is consistent with higher rates of electron transport to PSI and with a less reduced quinone A (QA) in the mutants than in the WT, as estimated by 1-qL (Figure 5E). After actinic light was switched off, the estimated maximal quantum yield of PSI recovered to the same level in WT and mutants, whereas the recovery of PSII maximal yield and oxidation of QA was faster in the mutants. NPQ in ndufa5 and ndufb10 was activated as in WT plants but after a few minutes of illumination, it showed a partial relaxation in mutants (Figure 5F).
Figure 5.
Alterations in PS functionality in CI mutants. The yield of PSI (Φ(I), A), PSII (Φ(II), B). PSI acceptor side limitation (Y(NA), C), PSI donor side limitation (Y(ND), D). PQ redox state (1-qL, E) and NPQ (F) were measured with Dual PAM 100 in plants exposed to 330 μmol photons m−2 s−1 of actinic light intensity. Yellow/black bar indicates when actinic light was on/off, respectively. WT, ndufb10, and ndufa5 KO are shown, respectively, with black squares, red circles, and blue triangles. Data are shown as average ± sd (n > 4). Asterisks indicate statistically significant differences of both mutants from WT plants (one-way ANOVA, n > 5, P < 0.01) after 2, 4, and 8 min of illumination and after 10 min in the dark; ns indicates nonstatistically significant differences.
The same analyses showed similar results upon exposure to dim light (50 µmol photons m−2 s−1), corresponding to the illumination in the growth chamber (Supplemental Figure S5). If plants were exposed to 2,000 µmol photons m−2 s−1, a light intensity completely saturating photosynthetic capacity of the plants, WT, and mutants showed instead no significant differences (Supplemental Figure S6).
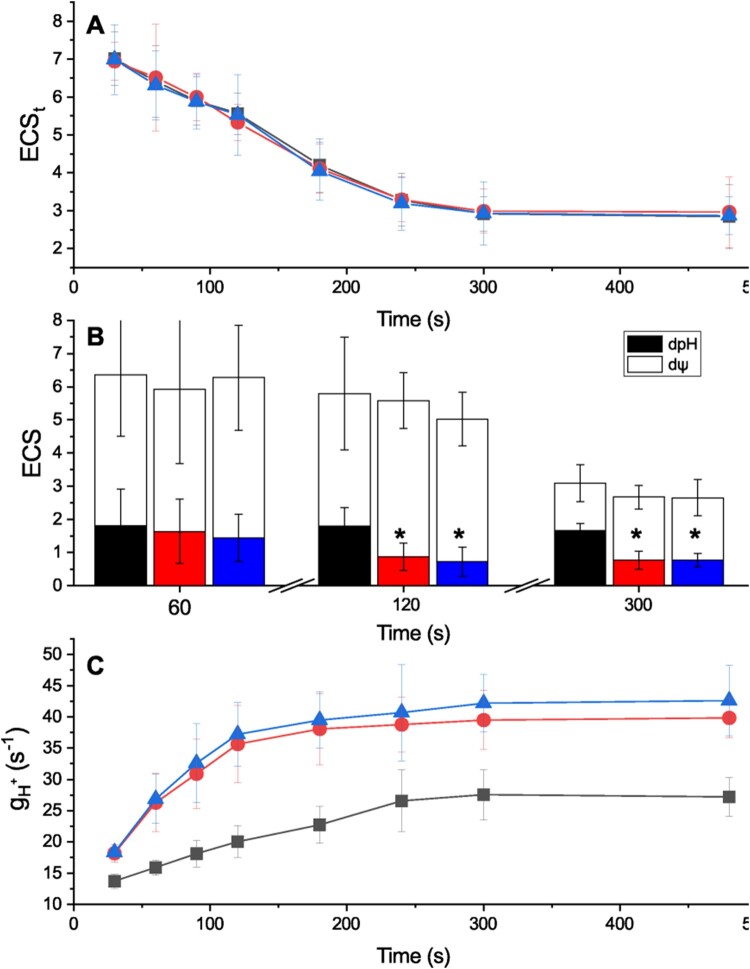
The impact of CI mutations on photosynthetic electron transport was further investigated by measuring electrochromic shift (ECS) caused by the generation of transmembrane potential at the level of the thylakoid membranes (Witt, 1979; Bailleul et al., 2010). The total ECS signal (ECSt), a proxy of the total proton motive force (pmf) generated, was found to be similar in WT and ndufa5 and ndufb10 mutants over different times of illumination with the same actinic light intensity (330 μmol photons m−2 s−1; Figure 6A).
Figure 6.
Impact of CI inactivation on pmf composition and proton conductivity (). A, ECSt generated by illumination with sub-saturating light (300 µmol photons m−2 s−1) at different time points. ECSt was quantified as (ECS520 − ECS546)/PSI Charge Separation and indicative of the pmf generated. WT, ndufb10, and ndufa5 KO are shown, respectively, in black, red, and blue. Average ± sd (n ≥ 5) are reported. B, quantification of total pmf as well as its partition in different components. ΔpH is shown in black, red and blue for WT, ndufb10, and ndufa5 KO, respectively. The electric component (ΔΨ) in shown in white. Averages ± sd (n ≥ 5) are reported and asterisks indicate statistically significant differences (one-way ANOVA, n > 5, P < 0.01). C, measures the proton conductivity of protons across the thylakoid membrane and it reflects the activity of ATPase. This was measured after exposing plants to light with different duration. WT, ndufb10, and ndufa5 KO are shown, respectively, as black squares, red circles, and blue triangles. Averages ± sd (n ≥ 5) are reported.
ECS relaxation kinetics measured after the light was switched off after 300 s of illumination, however, indicated an altered partition between the electric and pH component of the transmembrane potential (Figure 6B; Cruz et al., 2001). These differences were dependent on illumination and became detectable only if plants were exposed to light for >120 s (Figure 6B; Supplemental Figure S7). This suggests that the alterations in pmf partitioning emerged as a result of modifications in the photosynthetic activity (Avenson et al., 2005; Wang and Shikanai, 2019). This difference between genotypes was again light intensity-dependent, and when using a stronger, fully saturating, illumination (1,000 µmol photons m−2 s−1) mutants were indistinguishable from WT (Supplemental Figure S8).
The differences in ΔpH could not be attributed to differential activation of cyclic electron transport, the activity of which appeared to be low in both mutants and WT P. patens (Supplemental Figure S9, see also Kukuczka et al., 2014). To assess the impact of mutations on proton translocation, conductivity () through the ATPase complex was estimated from the decay lifetime of ECS signal after the light was switched off. In both ndufa5 and ndufb10, the ECS decay kinetics was faster than WT, indicating that proton conductivity due to ATPase was higher (Supplemental Figure S10). Monitoring of gH+ using actinic illumination of different duration (between 30 and 480 s) showed that gH+ in WT plants increased with the duration of the illumination reaching a steady state after ∼5 min, consistently with a progressive activation of ATPase (Figure 6C), following a modulation of chloroplast ATPase activity by the sensing of stromal metabolic status and redox state during steady-state photosynthesis (Kramer et al., 1990; Takizawa et al., 2008; Kohzuma et al., 2013; Sekiguchi et al., 2020). Interestingly, in ndufa5 and ndufb10 mutants, the gH+ was higher but also more rapidly activated and it reached the maximal activity after about 120 s of illumination instead of the 300 s required by WT plants (Figure 6C).
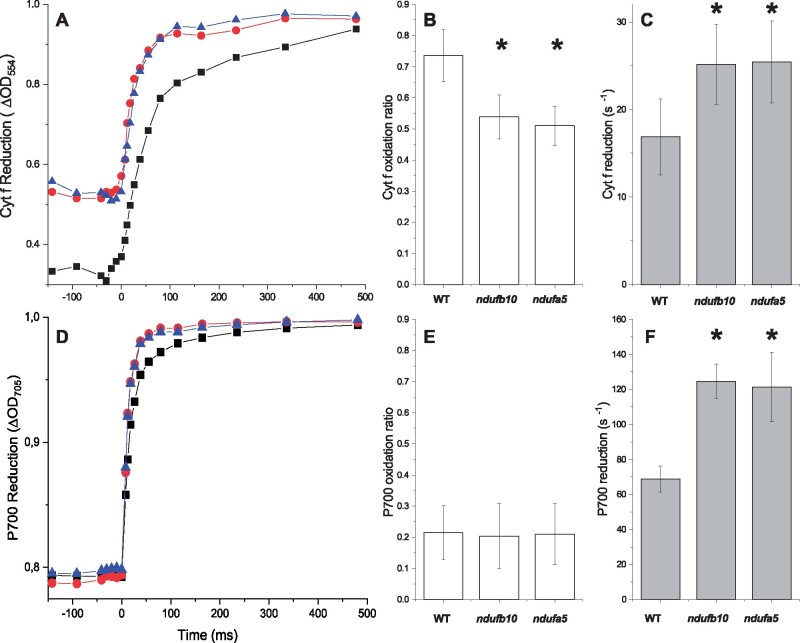
Lumenal pH is a major factor for regulation of photosynthesis and its decrease under strong illumination is known to activate protective mechanisms such as heat dissipation of excess energy (NPQ) and the xanthophyll cycle (Li et al., 2009). Low lumenal pH is also known to inhibit cytochrome b6f activity to avoid over-reduction of PSI, a mechanism known as photosynthetic control (Nishio and Whitmarsh, 1993). When photosynthetic control is active, the ETR from plastoquinol (PQH2) is slower, to reduce possibilities of PSI over-reduction. The impact of CI mutations on Cyt b6f activity was assessed by monitoring Cytochrome f (Cyt f) oxidation state from an absorption signal at 554 nm (Figure 7A; Supplemental Figure S11, see “Materials and methods” for details). The oxidation state in illuminated samples can be estimated by comparing the signal with plants treated with PSII and Cyt b6f inhibitors 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) and 2,5-Dibromo-6-isopropyl-3-methyl-1,4-benzoquinone (DBIMB) where electron transport from PQ is blocked and thus Cyt f is fully oxidized. In ndufb10 and ndufa5 illuminated with sub-saturating light (300 µmol photons m−2 s−1) Cyt f is less oxidized than in WT plants (Figure 7B). When the light was switched off, Cyt f reduction in mutant plants was also faster than in WT (Figure 7C), thus suggesting that in these plants electron transport from PQH2 is faster (Stiehl and Witt, 1969).
Figure 7.
Cytochrome f and PSI oxidation. A, Cytochrome f reduction monitored from ΔOD554 in dark-adapted plants subjected to constant illumination (300 μmol photons m−2 s−1) for 480 s before the light was switched off. WT, ndufb10, and ndufa5 KO are shown, respectively, as black squares, red circles, and blue triangles. B, Oxidation state is expressed as the ratio between Cyt f signals at the end of illumination and the maximal oxidation levels obtained by addition of DCMU/DBMIB to obtain complete oxidation. Reported values are average ± sd (n ≥ 4). C, Cyt f reduction rate in the different genotypes after 480 s actinic light exposure, quantified from half-time reduction. For B and C asterisks, indicate statistically significant differences (one-way ANOVA, n > 4, P < 0.01). D, PSI reduction monitored from ΔOD705 in dark-adapted plants exposed to constant illumination (300 μmol photons m−2 s−1) for 480 s. WT, ndufb10, and ndufa5 KO are shown, respectively, as black squares, red circles, and blue triangles. E, P700 oxidized fraction is expressed as the ratio between P700+ signals at the end of illumination and the maximal oxidation levels obtained by addition of DCMU and DBMIB to block the PSI re-reduction. Reported values are average ± sd (n ≥ 4). F, P700+ reduction rate estimated from half-time reduction after 480 s actinic light exposure in the different genotypes. Average values ± sd are shown. For F, asterisks indicate statistically significant differences (one-way ANOVA, n > 5, P < 0.01).
In the same measuring conditions, the impact on PSI was monitored from P700 oxidation state using a similar spectrophotometric approach, monitoring a differential absorption signal at 705 nm (ΔOD705, Figure 7D). CI mutants showed similar P700 oxidation level compared to WT plants at steady-state illumination (Figure 7E). However, when the light was switched off, the rate of P700 reduction was faster in the mutants, suggesting that electrons flux toward P700 was more rapid (Figure 7F) consistent with a higher PQH2 oxidation rate.
Discussion
Inactivation of CI by targeting two different subunits produced similar photosynthetic phenotypes
In this work, independent KO lines targeting two subunits of CI were generated and verified to have the insertion of the resistance cassette in the expected genome position and to lose the mRNA expression (Figure 1; Supplemental Figure S1). ndufa5 KO showed complete depletion of CI activity in BN-PAGE (Figure 3B) and it did not accumulate NAD9 (Figure 3A), a core subunit of the Q module that is directly involved in the transfer of electrons to the ubiquinone which is required for the complex functionality (Massoz et al., 2013). This is consistent with the previous results showing that NDUFA5 interacts with NAD7 and NAD9 to form an 80 kDa sub-complex that successively is integrated into the CI (Ligas et al., 2019). The second targeted subunit, NDUFB10, is instead integrated later during CI assembly and, in its absence, there is the formation of an incomplete CI of 800 kDa including both the N and Q modules but missing the proton pumping module and thus the biological activity (Barbieri et al., 2011). Despite this partial retention of some subunits (Figure 3), both mutants showed inactive CI as shown by the insensitivity of O2 consumption to the specific inhibitor rotenone (Figure 4A).
In both CI mutant lines, O2 consumption in the dark was enhanced compared to WT plants. This observation can be explained by the increased CII and CIII content but is more likely related to strong over-accumulation of AOX, as evident from the larger fraction of O2 consumption through AOX in the mutants (Figures 3 and 4; Supplemental Table S3). Increased flux through AOX may compensate for the absence of CI activity, which normally accounts for ∼40% of O2 consumption rate in WT (Figure 4A). However, flux through AOX is not expected to translate in a full recovery of the capacity of ATP biosynthesis in the mutants since CII and alternative pathways like AOX do not contribute to the generation of transmembrane potential, whereas flux through CI does (Vanlerberghe et al., 2020).
Similar alterations in respiratory complexes were also observed in CI mutants from Arabidopsis (Fromm et al., 2016b) and Nicotiana (Vidal et al., 2007) that also showed increased AOX accumulation and higher O2 consumption in the dark. In Arabidopsis, it was demonstrated that such increased O2 consumption in the dark is a distinctive feature of complete CI KOs, while this is not observed in CI partial mutants or KD lines (Kühn et al., 2015). Based on this observation, the increase in O2 consumption observed in P. patens ndufa5 and ndufb10 KO is a further confirmation that both lines have fully inactivated CI.
It is remarkable to observe that ndufa5 and ndufb10 KO regardless of the specific mutation showed the same decreases in photosynthetic control, increases in PSI and PSII activities and enhanced proton conductivity (Figures 2–7), even if the targeted genes were different. This similarity strongly suggests that plant responses observed are attributable to the lack of CI activity, while the impact of the absence of specific subunits is minor, at least on the phenotypes analyzed.
CI deficiency alters chloroplast ATP synthase activity and photosynthetic control of electron transport
All P. patens cells in various developmental stages contain chloroplasts and are photosynthetically active even if at different levels (Sakakibara et al., 2003). Here, plants were analyzed 10 d after inoculum on a mineral medium containing no reduced carbon and were mainly composed by chloronema, a developmental stage containing phototrophic cells particularly rich in chloroplasts (Furt et al., 2012). The combined choice of the model organism and developmental stage, thus, enable to assess the impact of respiration on photosynthesis disentangling it from the influence of heterotrophic tissues like roots that are expected to be largely impacted by the lack of respiration, something that is not possible working with vascular plants.
Despite the lack of heterotrophic cells, P. patens plants depleted in CI present a strong phenotype with a drastically impaired growth (Figure 2), demonstrating that respiration plays a major role in photosynthetically active cells in plants. Plants exposed to continuous light regime did not show any growth recovery, suggesting that the impact of respiration is also not limited to sustaining metabolism during the night (Figure 2; Supplemental Figure S2). Even more strikingly, while WT plants exposed to continuous illumination showed faster growth than the ones under 16-h/8-h d/night cycles (Supplemental Figure S2), this is not the case for CI mutants that are not able to exploit the extra light energy available. These observations demonstrate that inactivation of respiration directly impacts photosynthetic metabolism in P. patens plants and their ability to use light energy for growth.
This phenotype is not attributable to drastic changes in photosynthetic apparatus composition since plants depleted in CI did not show any major alteration in pigments (Table 1) nor in the content of protein complexes, PSI, PSII, Cyt b6f with only a slight increase in Chloroplast ATPase (Supplemental Figure S4). Photosynthetic functionality is nevertheless altered, and plants exposed to sub-saturating illumination (300 µmol photons m−2 s−1 and below) show several differences. Both PSI and PSII show a higher yield upon illumination in mutant plants, suggesting a lower saturation level in illuminated plants and thus higher transport rate (Supplemental Figure S9). CI mutants also show increased proton conductivity across the thylakoids membrane (Figure 6C) that, combined with an equivalent pmf, is again consistent with a higher ETR (Supplemental Figure S9).
The higher gH+ observed in the mutants cannot be completely attributed to the observed slight increase accumulation in chloroplast ATPase in the mutants but is more likely caused by a higher activation state or substrate availability, as suggested by the larger changes in gH+ during the dark to light transitions (Figure 6C; Supplemental Figure S10). The turnover rate of the ATPase is known to be limited by the depletion of inorganic phosphate (Pi) or ADP pool through metabolic feedback (Takizawa et al., 2008) and an eventual smaller impact of this metabolic control in CI mutants could also contribute to the higher activity. Consistent with this hypothesis, blocking respiration with various inhibitors strongly affected ATPase activity (Supplemental Figure S12). On the other hand, the presence of inhibitors did not abolish completely the difference between WT and mutants (Supplemental Figure S12), suggesting other mechanisms influencing proton conductivity beyond metabolic feedback must be altered by CI inactivation. A likely candidate is the modulation of the ATPase γ-subunit thiol redox state by thioredoxin (Trx; Kramer et al., 1990; Sekiguchi et al., 2020). The hypothesis of an alteration of Trx control system due to CI inactivation would in fact explain the faster activation observed in ndufa5 and ndufb10 KO (Figure 6C).
The higher ATPase activity in the CI mutants causes a faster translocation of protons in the stroma and an increase in the lumenal pH (Figure 6B), in turn, affecting photosynthesis regulatory mechanisms modulated by lumen acidification. NPQ is induced by a decrease of lumenal pH and the protonation of specific activators PsbS or LHCSR (Li et al., 2002; Alboresi et al., 2010; Liguori et al., 2016). WT and CI mutants accumulated similarly PsbS and LHCSR (Supplemental Figure S4) and, consistently, the maximal NPQ capacity is equivalent in all genotypes (Supplemental Figure S6F). However, mutants exposed to sub-saturating illumination show a larger relaxation of NPQ than WT after a few minutes (Figure 5F) that can be explained by a reduction of ΔpH across thylakoid membrane (Joliot and Finazzi, 2010; Figure 6B). This difference in NPQ does not occur immediately after light is switched on but it only emerges after 2 min of exposition, a time needed for the difference in proton conductivity between WT and mutants to generate a significant separation in ΔpH and consequently NPQ (Figures 5F and 6B).
An altered lumen acidification is also expected to affect photosynthetic control, the modulation of PQH2 oxidation at the Cyt b6f. Mutants exposed to sub-saturating illumination indeed showed a faster rate of Cyt f reduction (Figure 7C), suggesting a higher electron transport activity and a lower photosynthetic control. This hypothesis is consistent with several other differences observed such as the higher efficiency in both PSs (Figure 5, A and B; Supplemental Figure S5, A and B), a less pronounced PSII acceptor limitation (1-qL; Figure 5E; Supplemental Figure S5E) and PSI donor limitation (Y(ND); Figure 5D and Supplemental Figure S5D) compared to WT. PSI also shows faster reduction kinetics (Figure 7F). All the differences observed between the WT and mutants, summarized in Supplemental Figure S13, can be explained with a decreased photosynthetic control and thus a higher Cyt b6f activity, thus increasing the rate of transport of electrons from PQ to PSI as the result of higher ATPase activity and higher lumenal pH (Takizawa et al., 2007, 2008).
It is interesting to observe that when light is in strong excess (1,000–2,000 µmol m−2 s−1; Supplemental Figures S6 and S8), thus 25–50 times higher than the illumination used for plants growth, the mutants become indistinguishable from WT. Under such high light intensities, metabolic processes become equally saturated in WT and mutants, leading to similar ATPase activity and extents of photosynthetic control. The lack of differences at very strong illumination, on the other hand, supports the conclusion that photosynthetic apparatus is not altered in CI mutants. The effects on photosynthesis observed at lower light can thus be attributed to downstream processes, due to respiration alterations.
All these results globally suggest that CI inactivation not only alters respiratory activity in mitochondria but also impacts ATP biosynthesis in the chloroplast. This demonstrates the key role of the metabolic connection between photosynthesis and respiration through the transport of reduced molecules (e.g. malate, citrate, triose-Pi, and glycolate; Shameer et al., 2019) for plants metabolism.
Impact of respiration on photosynthesis adapted during evolution
All CI subunits identified in Arabidopsis are well conserved in P. patens genome (Supplemental Table S1), consistent with the recent observation that CI structure and composition show several similarities between the plant Arabidopsis and the green alga Polytomella sp (Klusch et al., 2020). CI inactivation also have similar effects in both Arabidopsis and P. patens with a strong impact on growth and an increased O2 consumption activity, as the result of changes in composition of respiratory apparatus and an higher AOX content, which ultimately lead to an increase of the electron flow through alternative respiratory pathways (Figures 3A and 4A; Kühn et al., 2015; Fromm et al., 2016a).
Because the P. patens tissues studied here contain only photosynthetically active cells, the observed effects can be attributed to intracellular interactions between chloroplasts and extraplastid within autotrophic cells, excluding any indirect effect from heterotrophic tissues that are instead always a possibility in vascular plants. On the other hand, the similarity of the alterations of respiratory apparatus composition and activity with the ones seen in Arabidopsis (Kühn et al., 2015) and tobacco (Vidal et al., 2007) suggests CI should have a similar influence in metabolism of these vascular plants leaves as well, though there may also be longer-range interactions, including changes in metabolic sinks at in heterotrophic cells like roots.
In contrast with the major impact in P. patens and vascular plants, the role of CI in the green alga C. reinhardtii appears to be more restricted to heterotrophy, since CI KO mutants show similar growth rates under photoautotrophic conditions, but markedly slower growth under heterotrophic conditions (Salinas et al., 2014; Larosa et al., 2018). CI mutants in Chlamydomonas also do not show any reorganization of respiratory apparatus, and showed reduced O2 consumption activity compared to WT (Barbieri et al., 2011; Massoz et al., 2013; Larosa et al., 2018).
While it is not possible to make general conclusions based on comparisons of a few selected species among the highly diverse phototrophic Eukaryotes, these examples suggest the interaction between photosynthesis and respiration changed during evolution. Plants rely on photo-autotrophy and normally cannot complete their developmental cycle in the dark even if reduced carbon is supplied (Neff et al., 2000). The fact that obligate photo-autotrophic organisms rely more on respiration seems counterintuitive, but it can be explained considering that in these organisms mitochondrial respiration activity gradually adapted to work in synergy with photosynthesis, optimizing the metabolite fluxes between organelles (Gardeström and Igamberdiev, 2016; Shameer et al., 2019). Respiration thus could gradually assume a specialized role in essential processes for photosynthesis such as redox balance and photorespiration, becoming itself indispensable for the cells.
On the other hand, Chlamydomonas have a highly flexible metabolism and in their natural environment can growth in autotrophic, heterotrophic, or mixotrophic conditions depending on the presence of light and carbon sources (Harris, 2001; Yang et al., 2015). Chlamydomonas cells can even be exposed to anaerobic conditions when it can activate fermentative metabolism and hydrogen production (Ghirardi et al., 2007; Grossman et al., 2011; Grechanik et al., 2020). In those anaerobic conditions, O2 is absent and respiration is inactivated but still cells can perform photosynthesis (Godaux et al., 2015) that must thus be fully independent of respiratory activity. In organisms with such large metabolic flexibility, and particularly if they are exposed to anaerobic conditions, respiration cannot assume an essential role in the photosynthesis, explaining why they are less impacted by an inactivation. The recent observation that the European mistletoe Viscum album, an obligate semi-parasite living on branches of trees, can live without CI (Maclean et al., 2018; Senkler et al., 2018) is consistent with this idea, suggesting that when the species lost their energy dependence from photosynthesis also its role of respiration for cells could change and become less essential.
Materials and methods
Plant material and growth P. patens
Physcomitrium patens (Gransden) WT, ndufb10 and ndufa5 KO lines were cultivated in the protonemal phase by vegetative propagation on PpNH4 medium (Ashton et al., 1979) and grown under standard conditions: 24°C, long photoperiod (16:8 light: dark) with 50 µmol photons m−2 s−1. Physiological and biochemical characterizations were performed on 10-d-old tissue cultivated in PpNO3 medium (Ashton et al., 1979). The growth rate in all the media and light conditions was evaluated starting from protonema colonies of 2 mm in diameter followed for 21 d. Colony size was measured as in Storti et al. (2019). In brief, high-quality images (600 ppi) were acquired using a Konica Minolta Bizhub C280 scanner. Images were analyzed with FIJI (https://fiji.sc/) using the “threshold color” plugin to subtract the plate background. Colony size was calculated from the integrated density (area × mean density). This strategy was chosen to take into account the development from 2D (chloronema and caulonema) to 3D structures (gametophore and rhizoids) of moss colonies, which are lost when considering only the area.
Moss transformation and mutant selection
The ndufb10 and ndufa5 KO constructs were employed to mutate the Ndufb10 gene and Ndufa5 gene, respectively (Supplemental Figure S1). The transformation was performed through protoplast DNA uptake as described in Alboresi et al. (2010). Six-day-old protonema tissues grown in PPNH4 medium were treated with fungal driselase (Sigma-Aldrich) to break the cell wall. Resulting protoplasts were filtered with 100-μm micro cloth. Protoplasts were washed and resuspended in PEG-4000-containing solution, mixed with digested linear DNA from KO constructs (20 μg) and exposed to heat shock (5 min at 45°C) to open cell membrane. After 1 d of recovery, protoplasts were first immersed in a top layer solution and plated on agarified medium added with mannitol to prevent their lysis. After 7 d, recovered cells were moved to a new plate with antibiotics for transformants selection. The selection was repeated twice. Resulting transformants were homogenized using 3-mm zirconium glass beads (Sigma-Aldrich), and genomic DNA (gDNA) was isolated according to a rapid extraction protocol (Edwards et al., 1991) with minor modification. PCR amplifications of recombination cassette were performed on extracted gDNA (Supplemental Table S3; Figure 1 and Supplemental Figure S1). To confirm that ndufb10 and ndufa5 KO lines were lacking target gene expression RT-PCR was performed on cDNA (RevertAid Reverse Transcriptase; Thermo Scientific) synthesized after RNA extraction.
Western blot analysis
Total protein extracts were obtained by pestled protonema tissue in sample buffer (50 mM TRIS pH 6.8, 100 mM DTT, 2% (w/v) SDS, and 10% (w/v) glycerol). Samples of total Chl were quantified, and every well was loaded accordingly to the quantification. After SDS–PAGE, proteins were transferred to a nitrocellulose membrane (Pall Corporation). Membranes were hybridized with specific primary antibodies: anti-PsaA, Agrisera, catalogue number AS06172; anti-Cyt f, Agrisera, catalogue number AS06119; anti-γ ATPase, Agrisera, catalogue number AS08312, anti-AOX1/2, Agrisera, catalogue number AS04054; custom made anti-NAD9 (Lamattina et al., 1993); custom made anti-SDH1-1 (CII), anti-α-MPP (CIII), anti-COX2 (CIV), anti-β subunit (CV; Peters et al., 2012), and custom made anti-D2, anti-PsbS, and anti-LHCSR (Storti et al., 2019). After hybridization, signals were detected with alkaline phosphatase-conjugated antibody (Sigma Aldrich).
Crude membrane extracts preparation
Crude membrane extracts were prepared as in Pineau et al., (2008) with minor modifications. Approximately 300 mg of fresh or frozen (−80°C) protonema grown in PpNO3 for 10 d were homogenized in 2 mL of 75 mM MOPS-KOH, pH 7.6, 0.6 M sucrose, 4 mM EDTA, 0.2% (w/v) polyvinylpyrrolidone 40, 8 mM cysteine, 0.2% (w/v) bovine serum albumin using a plant Potter glass tissue grinder pound at 0°C. The homogenate was filtrated across one layer of micro cloth with 20-μm pores and centrifuged at 4°C at 1,300 g for 4 min. The supernatant was collected and centrifuged again at 22,000g for 20 min. The resultant pellet, which contained most of the thylakoid and mitochondria membranes, was resuspended in 200 µL of 10 mM MOPS-KOH, pH 7.2, 0.3 M sucrose. The protein concentration on crude membrane extracts was quantified using the BCA protein assay.
Blue native polyacrylamide gel electrophoresis
Gels were cast in 8 × 10 cm plates using the buffer described by Kügler et al., (1997) with an acrylamide gradient of 4%–12% (w/v) in the running gel and 4% (w/v) acrylamide in the stacking gel. A volume of crude membrane extracts corresponding to 100 mg of proteins was washed with three volumes of H2O Milli-Q® and centrifuged at 4°C, 21,470g for 20 min. The pellet was resuspended in 20 µL of ACA buffer 1× (50 mM Bis–Tris, pH 7.0; 750 mM aminocaproic acid; 1 mM EDTA) Järvi et al., (2011). For protein solubilization 20 µL of β-dodecyl maltoside (β-DM) 2% (w/v) prepared in ACA buffer were added to the tube, to reach a final volume of 40 µL, reaching β-DM 1% in ACA buffer. Each tube was vortexed for 30 s, kept on ice for 5 min, and centrifuged at 4°C, 22,000g for 8 min. The supernatant was supplemented with 4 µL of Coomassie Blue 5% solution (20 mM Bis–Tris, 500 mM aminocaproic acid, Coomassie Blue G-250 5% (w/v)). Anode and cathode buffers were the same used by Järvi et al. (2011) for BN gel; the cathode buffer was supplemented with Coomassie Blue G-250 0.02% (w/v). The gel was run at 75 V for 30 min. Then, the cathode buffer was replaced with fresh cathode buffer without Coomassie Blue and the gel was run at 100 V for 30 min, at 125 V for 30 min, at 150 V for 60 min, at 175 V for 30 min, and at 200 V for 60 min. The total running time was about 4 h.
Determination of NAH dehydrogenase in-gel activity
After BN-PAGE, the NADH dehydrogenase activity of CI was revealed by incubation of the gel in the presence of 1 mM nitro blue tetrazolium (NBT) and 0.2 mM NADH in 50-mM potassium Pi buffer (pH 7.0; Barbieri et al., 2011).
Oxygen consumption and oxygen evolution
O2 consumption and evolution were evaluated as in Storti et al. (2020) with a Clark-type O2 electrode (Hansatech, King’s Lynn, UK). In brief, two protonema disks of about 1 cm of diameter coming from 10-d-old plates were introduced in the measurement chamber filled with 2 mL of a solution containing NaCO3 0.1 mM maintained at 23°C. After 10 min in the dark (respiratory rate), the light was turned on and the O2 variation was recorded for other 10 min (photosynthetic rate). O2 consumption and evolution rates were normalized to the total Chl content of each sample. Chl content was evaluated after extraction with 80% (v/v) acetone (Porra et al., 1989). Inhibitors employed for respiratory analysis were 50-µM rotenone (CI), 1-mM KCN (CIV), and from 2 to 8 mM SHAM (AOX). For inhibitor treatments, protonema tissue was incubated 30 min in the dark in medium supplemented with the inhibitors, for the double inhibition with KCN and SHAM the inhibitors were added simultaneously. Inhibitors were also added at the measuring chamber during O2 consumption evaluation. The capacity of cytochrome pathway was measured as O2 consumption that was sensitive to 1 mM KCN in the presence of 8 mM SHAM. The capacity of AOX pathway was measured as O2 consumption that was sensitive to 8 mM of SHAM in the presence of 1 mM KCN (Vanlerberghe et al., 2002).
Pigment analysis
Ten-day-old protonemal tissues were broken with a plastic grinder and pigments were extracted with 80% acetone. Whole-plant extracts in acetone were fitted with those of individual purified pigments to calculate Chla/b and Chl/Car ratios (Croce et al., 2002).
Spectroscopic analyses
In vivo, Chl fluorescence and NIR absorption analyses were performed at room temperature with a Dual-PAM 100 system (Walz) on protonema grown for 10 d in PpNO3 in WT and mutant lines. Before the analysis, the plants were adapted in the dark for 40 min, and the Fv/Fm parameter was calculated as (Fm – Fo)/Fm. Induction curves were obtained setting actinic red light at (approx.) 50, 330, or 2,000 µmol photons m−2s−1, and photosynthetic parameters were recorded every 30 s. At each step, the photosynthetic parameters were calculated as follows: Φ(II) as (Fm′ – Fo)/Fm′, qL as (Fm′ – F)/(Fm′ – Fo′) × Fo′/F, and NPQ as (Fm – Fm′)/Fm′, Φ(I) as 1-Y(ND)-Y(NA); Y(NA) as (Pm – Pm′)/Pm; Y(ND) as (P – Po/Pm; Klughammer and Schreiber, 1994). ECS spectra were recorded with a JTS-10 system (Biologic) in plants that were dark acclimated for 40 min and imbibed with 20 mM HEPES, pH 7.5 and 10 mM KCl. For each measure, the background signal at 546 nm was subtracted from the 520 nm signal, in this way eliminating the contribution of scattering and cytochromes. Functional total PSs quantification was performed by a single flash turnover using a xenon lamp. The light produced by xenon gas can induce PSI double charge separation, and thus PSI content could be overestimated by ∼40% but it does not affect the comparison of different samples (Gerotto et al., 2016). Moreover, samples were incubated with 20 µM DCMU and 4 mM HA (hydroxylamine) to calculate the contribution of PSI alone (Joliot et al., 2004). Total ETR was measured with dark-induced relaxation kinetic; Sacksteder and Kramer, 2000) analysis as in Gerotto et al. (2016). In samples exposed to continuous illumination at 350 μmol photons m−2 s−1, (630 nm LED) light was switched off after 4–5′ to follow relaxation kinetics and evaluate the pmf generated during light treatment as in Storti et al. (2020). The pmf was determined as the difference of the maximum signal at the light steady state and minimum level of ECS in the dark and normalized to total charge separation PSI. The pmf portioning in ΔΨ and ΔpH was performed as in Baker et al. (2007). The ECS relaxation was followed for 30 s after light to dark transition and the ECS signal in this relaxed state was recorded. Difference between the relaxed ECS signal and the maximal signal at light steady state is defined as ECSss, which is proportional to ΔΨ. ΔpH was calculated from the difference between the relaxed ECS signal and a minimum level of ECS in the dark. Proton conductance () was estimated by fitting the first 300 ms of the ECS decay curve with a first-order exponential decay kinetic and indicated as the inverse of the decay time constant as described earlier (Avenson et al., 2005). was calculated after exposure to 30 s, 60 s, 90 s, 120 s, 180 s, 240 s, 300 s, and 480 s of illumination (350 µmol photons m−2 s−1). P700+ absorption kinetics were calculated from the absorption at 705 nm (ΔOD705) and following the re-reduction after plants were illuminated for 5 min at 350 μmol photons m−2 s−1. Oxidized P700 was calculated by comparing the maximum signal from P700+ obtained before and after incubating plants with20 µM DCMU and 150 μM DBIMB. P700+ reduction after the actinic light was switched off was fit to first-order exponential decay to obtain t1/2 values. Cyt f absorption was calculated as for P700+, except for the interference filter at 554 nm. In this case, 546 nm and 573 nm were used as background and removed to 554 nm signal (Kirchhoff et al., 2011). Oxidation status and Cyt f kinetic rate measurements were performed as for P700 measurements.
Accession numbers
Accession numbers of genes analyses in this work are reported in Supplemental Table S1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Generation of P. patens mutant lines lacking respiratory CI.
Supplemental Figure S2 . Growth kinetics of P. patens plants under different conditions.
Supplemental Figure S3 . BN-PAGE separation of crude extracts.
Supplemental Figure S4 . Impact of CI mutations on the photosynthetic apparatus composition.
Supplemental Figure S5 . Photosynthetic efficiency under dim illumination.
Supplemental Figure S6 . Photosynthetic efficiency under saturating illumination.
Supplemental Figure S7 . Proton motive force composition before the steady state.
Supplemental Figure S8 . Photosynthetic proton motive force at saturating illumination.
Supplemental Figure S9 . Photosynthetic electron transfer in P. patens plants.
Supplemental Figure S10 . ATPase activity assessed from ECS relaxation.
Supplemental Figure S11 . Cyt b6f reduction state estimation.
Supplemental Figure S12 . Influence of respiration inhibition on proton conductivity in WT and CI mutants.
Supplemental Figure S13 . Summary of photosynthetic electron transport alterations in CI depleted plants.
Supplemental Table S1 . Identification of conserved CI subunits in P. patens.
Supplemental Table S2 . Cytochrome and AOX Capacity in WT and ndufb10 and ndufa5 plants.
Supplemental Table S3 . Primers employed in this work.
Supplementary Material
Acknowledgments
We thank Dr Etienne Meyer (Institute of Plant Physiology; the Martin-Luther-University, Halle-Wittemberg) for providing the NAD9 antibody and Prof. Hans-Peter Braun (Institute of Plant Genetics; Leibniz University, Hannover) for the SDH1-1, α-MPP, COX2, and ATPase β-subunit antibodies.
Funding
M.M. was funded by the University of Padova. D.M.K. was supported by Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy (DE-FG02-91ER20021).
Conflict of interest statement. The authors declare that the research was conducted in the absence of any commercial orfinancial relationships that could be construed as a potential conflict of interest.
T.M. and A.A. designed the research; M.M., M.S., and A.M.V.V. performed research; M.M., M.S., D.M.K., A.A., and T.M. analyzed data; T.M. and M.M. wrote the article. All authors critically revised the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Tomas Morosinotto (tomas.morosinotto@unipd.it).
References
- Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T (2010) Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc Natl Acad Sci U S A 107: 11128–11133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton NW, Grimsley NH, Cove DJ (1979) Analysis of gametophytic development in the moss, Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta 144: 427–435 [DOI] [PubMed] [Google Scholar]
- Avenson TJ, Cruz JA, Kanazawa A, Kramer DM (2005) Regulating the proton budget of higher plant photosynthesis. Proc Natl Acad Sci U S A 102: 9709–9713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul B, Berne N, Murik O, Petroutsos D, Prihoda J, Tanaka A, Villanova V, Bligny R, Flori S, Falconet D, et al. (2015) Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524: 366–369 [DOI] [PubMed] [Google Scholar]
- Bailleul B, Cardol P, Breyton C, Finazzi G (2010) Electrochromism: A useful probe to study algal photosynthesis. Photosynth Res 106: 179–189 [DOI] [PubMed] [Google Scholar]
- Baker NR, Harbinson J, Kramer DM (2007) Determining the limitations and regulation of photosynthetic energy transduction in leaves. Plant Cell Environ 30: 1107–1125 [DOI] [PubMed] [Google Scholar]
- Barbieri MR, Larosa V, Nouet C, Subrahmanian N, Remacle C, Hamel PP (2011) A forward genetic screen identifies mutants deficient for mitochondrial complex I assembly in Chlamydomonas reinhardtii. Genetics 188: 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun H-P, Binder S, Brennicke A, Eubel H, Fernie AR, Finkemeier I, Klodmann J, König A-C, Kühn K, Meyer E, et al. (2014) The life of plant mitochondrial complex I. Mitochondrion 19 (Pt B): 295–313 [DOI] [PubMed] [Google Scholar]
- Cardol P, Gloire G, Havaux M, Remacle C, Matagne R, Franck F (2003) Photosynthesis and state transitions in mitochondrial mutants of Chlamydomonas reinhardtii affected in respiration. Plant Physiol 133: 2010–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung MCY, Ratcliffe GR, Sweetlove LJ (2015) A method of accounting for enzyme costs in flux balance analysis reveals alternative pathways and metabolite stores in an illuminated Arabidopsis leaf. Plant Physiol 169: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas Des Francs-Small C, Small I (2014) Surrogate mutants for studying mitochondrially encoded functions. Biochimie 100: 234–242 [DOI] [PubMed] [Google Scholar]
- Croce R, Canino G, Ros F, Bassi R (2002) Chromophore organization in the higher-plant photosystem II antenna protein CP26. Biochemistry 41: 7334–7343 [DOI] [PubMed] [Google Scholar]
- Cruz JA, Sacksteder CA, Kanazawa A, Kramer DM (2001) Contribution of electric field (Δψ) to steady-state transthylakoid proton motive force (pmf) in vitro and in vivo. Control of pmf parsing into Δψ and ΔpH by ionic strength. Biochemistry 40: 1226–1237 [DOI] [PubMed] [Google Scholar]
- Dahal K, Martyn GD, Alber NA, Vanlerberghe GC (2016) Coordinated regulation of photosynthetic and respiratory components is necessary to maintain chloroplast energy balance in varied growth conditions. J Exp Bot 68: erw469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant M, Baurain D, Coosemans N, Joris B, Matagne RF (2001) Characterization of two genes encoding the mitochondrial alternative oxidase in Chlamydomonas reinhardtii. Curr Genet 39: 101–108 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Driscoll S, Cornic G, De Paepe R, Foyer CH, Noctor G (2003) Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol 131: 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S, Finazzi G, Wollman F-A (2008) The dynamics of photosynthesis. Annu Rev Genet 42: 463–515 [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm S, Braun H-P, Peterhansel C (2016a) Mitochondrial gamma carbonic anhydrases are required for complex I assembly and plant reproductive development. New Phytol 211: 194–207 [DOI] [PubMed] [Google Scholar]
- Fromm S, Senkler J, Eubel H, Peterhänsel C, Braun H-P (2016b) Life without complex I: proteome analyses of an Arabidopsis mutant lacking the mitochondrial NADH dehydrogenase complex. J Exp Bot 67: 3079–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furt F, Lemoi K, Tüzel E, Vidali L (2012) Quantitative analysis of organelle distribution and dynamics in Physcomitrella patens protonemal cells. BMC Plant Biol 12: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardeström P, Igamberdiev AU (2016) The origin of cytosolic ATP in photosynthetic cells. Physiol Plant 157: 367–379 [DOI] [PubMed] [Google Scholar]
- Gauthier PPG, Battle MO, Griffin KL, Bender ML (2018) Measurement of gross photosynthesis, respiration in the light, and mesophyll conductance using H218O labeling. Plant Physiol 177: 62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerotto C, Alboresi A, Meneghesso A, Jokel M, Suorsa M, Aro E-MEM, Morosinotto T (2016) Flavodiiron proteins act as safety valve for electrons in Physcomitrella patens. Proc Natl Acad Sci U S A 113: 12322–12327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi ML, Posewitz MC, Maness P-C, Dubini A, Yu J, Seibert M (2007) Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Annu Rev Plant Biol 58: 71–91 [DOI] [PubMed] [Google Scholar]
- Godaux D, Bailleul B, Berne N, Cardol P (2015) Induction of photosynthetic carbon fixation in anoxia relies on hydrogenase activity and proton-gradient regulation-like1-mediated cyclic electron flow in Chlamydomonas reinhardtii. Plant Physiol 168: 648–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grechanik V, Romanova A, Naydov I, Tsygankov A (2020) Photoautotrophic cultures of Chlamydomonas reinhardtii: Sulfur deficiency, anoxia, and hydrogen production. Photosynth Res 143: 275–286 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Catalanotti C, Yang W, Dubini A, Magneschi L, Subramanian V, Posewitz MC, Seibert M (2011) Multiple facets of anoxic metabolism and hydrogen production in the unicellular green alga Chlamydomonas reinhardtii. New Phytol 190: 279–288 [DOI] [PubMed] [Google Scholar]
- Harris EH (2001) As a odel rganism. Mol Biol 52: 363–406 [DOI] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Paakkarinen V, Aro E-M (2011) Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem J 439: 207–214 [DOI] [PubMed] [Google Scholar]
- Joliot P, Béal D, Joliot A (2004) Cyclic electron flow under saturating excitation of dark-adapted Arabidopsis leaves. Biochim Biophys Acta 1656: 166–176 [DOI] [PubMed] [Google Scholar]
- Joliot P, Joliot A (2008) Quantification of the electrochemical proton gradient and activation of ATP synthase in leaves. Biochim Biophys Acta-Bioenerg 1777: 676–683 [DOI] [PubMed] [Google Scholar]
- Joliot PA, Finazzi G (2010) Proton equilibration in the chloroplast modulates multiphasic kinetics of nonphotochemical quenching of fluorescence in plants. Proc Natl Acad Sci U S A 107: 12728–12733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H, Hall C, Wood M, Herbstová M, Tsabari O, Nevo R, Charuvi D, Shimoni E, Reich Z (2011) Dynamic control of protein diffusion within the granal thylakoid lumen. Proc Natl Acad Sci U S A 108: 20248–20253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J, Sunderhaus S, Nimtz M, Jänsch L, Braun H-P (2010) Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klughammer C, Schreiber U (1994) An improved method, using saturating light pulses, for the determination of photosystem I quantum yield via P700+-absorbance changes at 830 nm. Planta 192: 261–268 [Google Scholar]
- Klusch N, Senkler J, Yildiz Ö, Kühlbrandt W, Braun H-P (2020) A ferredoxin bridge connects the two arms of plant mitochondrial complex I. bioRxiv 2020.11.23.393975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohzuma K, Bosco CD, Meurer J, Kramer DM (2013) Light- and metabolism-related regulation of the chloroplast ATP synthase has distinct mechanisms and functions. J Biol Chem 288: 13156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Wise RR, Frederick JR, Alm DM, Hesketh JD, Ort DR, Crofts AR (1990) Regulation of coupling factor in field-grown sunflower: A Redox model relating coupling factor activity to the activities of other thioredoxin-dependent chloroplast enzymes. Photosynth Res 26: 213–222 [DOI] [PubMed] [Google Scholar]
- Kügler M, Jänsch L, Kruft V, Schmitz UK, Braun HP (1997) Analysis of the chloroplast protein complexes by blue-native polyacrylamide gel electrophoresis (BN-PAGE). Photosynth Res 53: 35–44 [Google Scholar]
- Kühn K, Obata T, Feher K, Bock R, Fernie AR, Meyer EH (2015) Complete mitochondrial complex I deficiency induces an up-regulation of respiratory fluxes that is abolished by traces of functional complex I. Plant Physiol 168: 1537–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukuczka B, Magneschi L, Petroutsos D, Steinbeck J, Bald T, Powikrowska M, Fufezan C, Finazzi G, Hippler M (2014) Proton gradient regulation5-like1-mediated cyclic electron flow is crucial for acclimation to anoxia and complementary to nonphotochemical quenching in stress adaptation. Plant Physiol 165: 1604–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamattina L, Gonzalez D, Gualberto J, Grienenberger J (1993) Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur J Biochem 217: 831–838 [DOI] [PubMed] [Google Scholar]
- Larosa V, Meneghesso A, La Rocca N, Steinbeck J, Hippler M, Szabò I, Morosinotto T (2018) Mitochondria affect photosynthetic electron transport and photosensitivity in a green alga. Plant Physiol 176: 2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecler R, Vigeolas H, Emonds-Alt B, Cardol P, Remacle C (2012) Characterization of an internal type-II NADH dehydrogenase from Chlamydomonas reinhardtii mitochondria. Curr Genet 58: 205–216 [DOI] [PubMed] [Google Scholar]
- Leon G, Holuigue L, Jordana X (2007) Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant Physiol 143: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-P, Muller-Moule P, Gilmore AM, Niyogi KK (2002) PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc Natl Acad Sci U S A 99: 15222–15227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wakao S, Fischer BB, Niyogi KK (2009) Sensing and responding to excess light. Annu Rev Plant Biol 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Ligas J, Pineau E, Bock R, Huynen MA, Meyer EH (2019) The assembly pathway of complex I in Arabidopsis thaliana. Plant J 97: 447–459 [DOI] [PubMed] [Google Scholar]
- Liguori N, Novoderezhkin V, Roy LM, Van Grondelle R, Croce R (2016) Excitation dynamics and structural implication of the stress-related complex LHCSR3 from the green alga Chlamydomonas reinhardtii. Biochim Biophys Acta-Bioenerg 1857: 1514–1523 [DOI] [PubMed] [Google Scholar]
- Maclean AE, Hertle AP, Ligas J, Bock R, Balk J, Meyer EH (2018) Absence of Complex I is associated with diminished respiratory chain function in European Mistletoe. Curr Biol 28: 1614–1619. e3 [DOI] [PubMed] [Google Scholar]
- Massoz S, Larosa V, Plancke C, Lapaille M, Bailleul B, Pirotte D, Radoux M, Leprince P, Coosemans N, Matagne RF, et al. (2013) Inactivation of genes coding for mitochondrial Nd7 and Nd9 complex I subunits in Chlamydomonas reinhardtii. Impact of complex I loss on respiration and energetic metabolism. Mitochondrion 19: 365–374 [DOI] [PubMed] [Google Scholar]
- Møller IM (2001) P lant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14: 257–271 [PubMed] [Google Scholar]
- Nishio JN, Whitmarsh J (1993) Dissipation of the proton electrochemical potential in intact chloroplasts II. The pH gradient monitored by cytochrome f reduction kinetics. Plant Physiol 101: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi K, Yoshida K (2008) Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion 8: 87–99 [DOI] [PubMed] [Google Scholar]
- Oldenkott B, Burger M, Hein AC, Jörg A, Senkler J, Braun HP, Knoop V, Takenaka M, Schallenberg-Rüdinger M (2020) One C-to-U RNA editing site and two independently evolved editing factors: Testing reciprocal complementation with DYW-Type PPR proteins from the moss physcomitrium (physcomitrella) patens and the flowering plants macadamia integrifolia and Arabidopsis. Plant Cell 32: 2997–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K, Nießen M, Peterhänsel C, Späth B, Hölzle A, Binder S, Marchfelder A, Braun H-P (2012) Complex I–complex II ratio strongly differs in various organs of Arabidopsis thaliana. Plant Mol Biol 79: 273–284 [DOI] [PubMed] [Google Scholar]
- Pineau B, Layoune O, Danon A, De Paepe R (2008) L-galactono-1, 4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. J Biol Chem 283: 32500–32505 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta-Bioenerg 975: 384–394 [Google Scholar]
- Radin I, Mansilla N, Rödel G, Steinebrunner I (2015) The Arabidopsis COX11 homolog is essential for cytochrome c oxidase activity. Front Plant Sci 6: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Rustin P (2014) Supernumerary subunits NDUFA3, NDUFA5 and NDUFA12 are required for the formation of the extramembrane arm of human mitochondrial complex I. FEBS Lett 588: 1832–1838 [DOI] [PubMed] [Google Scholar]
- Sacksteder CA, Kramer DM (2000) Dark-interval relaxation kinetics (DIRK) of absorbance changes as a quantitative probe of steady-state electron transfer. Photosynth Res 66: 145–158 [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Nishiyama T, Sumikawa N, Kofuji R, Murata T, Hasebe M (2003) Involvement of auxin and a homeodomain-leucine zipper I gene in rhizoid development of the moss Physcomitrella patens. Development 130: 4835–4846 [DOI] [PubMed] [Google Scholar]
- Salinas T, Larosa V, Cardol P, Maréchal-Drouard L, Remacle C (2014) Respiratory-deficient mutants of the unicellular green alga Chlamydomonas: A review. Biochimie 100: 207–218 [DOI] [PubMed] [Google Scholar]
- Schimmeyer J, Bock R, Meyer EH (2016) L-Galactono-1,4-lactone dehydrogenase is an assembly factor of the membrane arm of mitochondrial complex I in Arabidopsis. Plant Mol Biol 90: 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönfeld C, Wobbe L, Borgstädt R, Kienast A, Nixon PJ, Kruse O (2004) The nucleus-encoded protein MOC1 is essential for mitochondrial light acclimation in Chlamydomonas reinhardtii. J Biol Chem 279: 50366–50374 [DOI] [PubMed] [Google Scholar]
- Sekiguchi T, Yoshida K, Okegawa Y, Motohashi K, Wakabayashi K, Hisabori T (2020) Chloroplast ATP synthase is reduced by both f-type and m-type thioredoxins. Biochim Biophys Acta-Bioenerg 1861: 148261. [DOI] [PubMed] [Google Scholar]
- Senkler J, Rugen N, Eubel H, Hegermann J, Braun H-P (2018) Absence of Complex I implicates rearrangement of the respiratory chain in European Mistletoe. Curr Biol 28: 1606–1613.e4 [DOI] [PubMed] [Google Scholar]
- Shameer S, Ratcliffe RG, Sweetlove LJ (2019) Leaf energy balance requires mitochondrial respiration and export of chloroplast NADPH in the light. Plant Physiol 180: 1947–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiehl HH, Witt HT (1969) Quantitative treatment of the function of plastoquinone in photosynthesis. Zeitschrift für Naturforsch B 24: 1588–1598 [DOI] [PubMed] [Google Scholar]
- Storti M, Alboresi A, Gerotto C, Aro E-M, Finazzi G, Morosinotto T (2019) Role of cyclic and pseudo-cyclic electron transport in response to dynamic light changes in Physcomitrella patens. Plant Cell Environ 42: 1590–1602 [DOI] [PubMed] [Google Scholar]
- Storti M, Segalla A, Mellon M, Alboresi A, Morosinotto T (2020) Regulation of electron transport is essential for photosystem I stability and plant growth. New Phytol 228: 1316–1326 [DOI] [PubMed] [Google Scholar]
- Subrahmanian N, Remacle C, Hamel PP (2016) Plant mitochondrial Complex I composition and assembly: A review. Biochim Biophys Acta-Bioenerg 1857: 1001–1014 [DOI] [PubMed] [Google Scholar]
- Takizawa K, Cruz JA, Kanazawa A, Kramer DM (2007) The thylakoid proton motive force in vivo. Quantitative, non-invasive probes, energetics, and regulatory consequences of light-induced pmf. Biochim Biophys Acta 1767: 1233–1244 [DOI] [PubMed] [Google Scholar]
- Takizawa K, Kanazawa A, Kramer DM (2008) Depletion of stromal Pi induces high “energy-dependent” antenna exciton quenching (qE) by decreasing proton conductivity at CF O-CF1 ATP synthase. Plant, Cell Environ 31: 235–243 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, Dahal K, Alber NA, Chadee A (2020) Photosynthesis, respiration and growth: A carbon and energy balancing act for alternative oxidase. Mitochondrion 52: 197–211 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, Robson CA, Yip JYH (2002) Induction of mitochondrial alternative oxidase in response to a cell signal pathway down-regulating the cytochrome pathway prevents programmed cell death. Plant Physiol 129: 1829–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal G, Ribas-Carbo M, Garmier M, Dubertret G, Rasmusson AG, Mathieu C, Foyer CH, De Paepe R (2007) Lack of respiratory chain complex I impairs alternative oxidase engagement and modulates redox signaling during elicitor-induced cell death in tobacco. Plant Cell 19: 640–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Shikanai T (2019) Modification of activity of the thylakoid H(+)/K(+) antiporter KEA3 disturbs ΔpH-dependent regulation of photosynthesis. Plant Physiol 181: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt IN, Montgomery MG, Runswick MJ, Leslie AGW, Walker JE (2010) Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci USA 107: 16823–16827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt HT (1979) Energy conversion in the functional membrane of photosynthesis. Analysis by light pulse and electric pulse methods. The central role of the electric field. BBA Rev Bioenerg 505: 355–427 [DOI] [PubMed] [Google Scholar]
- Yang W, Catalanotti C, Wittkopp TM, Posewitz MC, Grossman AR (2015) Algae after dark: Mechanisms to cope with anoxic/hypoxic conditions. Plant J 82: 481–503 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Noguchi K (2011) Interaction between chloroplasts and mitochondria: Activity, function, and regulation of the mitochondrial respiratory system during photosynthesis. In Kempken F, ed, Plant Mitochondria. Springer, New York, pp 383–409 [Google Scholar]
- Zalutskaya Z, Lapina T, Ermilova E (2015) The Chlamydomonas reinhardtii alternative oxidase 1 is regulated by heat stress. Plant Physiol Biochem 97: 229–234 [DOI] [PubMed] [Google Scholar]
- Zhang L-T, Zhang Z-S, Gao H-Y, Meng X-L, Yang C, Liu J-G, Meng Q-W (2012) The mitochondrial alternative oxidase pathway protects the photosynthetic apparatus against photodamage in Rumex K-1 leaves. BMC Plant Biol 12: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.