Abstract
Viruses are the second leading cause of cancer worldwide, and human papillomavirus (HPV)–associated head and neck cancers are increasing in incidence in the United States. HPV preferentially infects the crypts of the tonsils rather than the surface epithelium. The present study sought to characterize the unique microenvironment within the crypts to better understand the viral tropism of HPV to a lymphoid-rich organ. Laser-capture microdissection of distinct anatomic areas (crypts, surface epithelium, and germinal centers) of the tonsil, coupled with transcriptional analysis and multiparameter immunofluorescence staining demonstrated that the tonsillar crypts are enriched with myeloid populations that co-express multiple canonical and noncanonical immune checkpoints, including PD-L1, CTLA-4, HAVCR2 (TIM-3), ADORA2A, IDO1, BTLA, LGALS3, CDH1, CEACAM1, PVR, and C10orf54 (VISTA). The resident monocytes may foster a permissive microenvironment that facilitates HPV infection and persistence. Furthermore, the myeloid populations within HPV-associated tonsil cancers co-express the same immune checkpoints, providing insight into potential novel immunotherapeutic targets for HPV-associated head and neck cancers.
The Waldeyer tonsillar ring serves as the first line of defense against foreign pathogens entering the respiratory tract and/or oral cavity,1 including high-risk human papillomavirus (HPV) that causes up to 80% of tonsil cancers diagnosed in the United States.2,3 The surface squamous epithelium of the tonsil invaginates into the underlying lymphoid stroma to form tonsillar crypts that are lined by a highly specialized lymphoepithelium that lacks a contiguous basement membrane.1,4 Tonsillar crypts represent an immune-privileged site because of localized expression of immune checkpoint programmed death ligand-1 (PD-L1).5
The present study aimed to further elucidate the natural regulatory mechanisms that support immune privilege and viral tropism within the tonsillar crypts. For the first time, myeloid populations are reported to be enriched within the tonsillar crypts compared with the germinal centers or surface epithelium of the tonsil. Furthermore, the myeloid cells co-express multiple immune checkpoint pathways fostering an immune permissive microenvironment. These findings provide insight into the viral tropism of the virus to target a particular region of a lymphoid-rich organ and have potential implications in identifying novel immunotherapeutic targets in HPV-associated tonsil cancers.
Materials and Methods
Human Specimens
Fresh tonsil tissue was obtained from 12 patients undergoing a tonsillectomy for benign recurrent tonsilitis. Normal tonsil samples were confirmed to be HPV negative by both p16 immunohistochemical (IHC) staining and HPV in situ hybridization. A post-hoc power calculation was performed to determine whether the study was sufficiently powered to observe differences in expression of the immune checkpoint markers of interest. The difference in PD-L1 expression was selected as an estimation for effect size because PD-L1 has previously been shown to be specifically expressed in the crypts compared with other tonsil regions.5 At an ⍺ = 0.05, power calculations estimated needing two samples to have 80% power and four samples to have >99% power of observing similar differences in other checkpoints. All data acquisition was performed blinded (T.M.S., T.J.D.). Formalin-fixed, paraffin-embedded tissue from HPV-16–associated head and neck cancer patients was also obtained. Research approval on human subjects was obtained from the Johns Hopkins University Institutional Review Boards (number NA_00012214) and the Massachusetts General Hospital (Institutional Review Board number 2014P000559). This study was performed in accordance with the Declaration of Helsinki.
Laser-Capture Microdissection and RNA Isolation
Fresh human tonsils were embedded in OCT medium, frozen, and cut into sections (7 μm thick) on Arcturus PEN membrane glass slides (Applied Biosystems, Foster City, CA); then, they were stained with a modified hematoxylin and eosin protocol. Briefly, sectioned slides were air dried for 15 minutes, then transferred to 70% ethanol (VWR, Radnor, PA; catalog number BDH1156-1LP) for 5 minutes, rinsed with deionized water (VWR; catalog number BDH1168-5GL) for 5 minutes, and then stained with hematoxylin (Thermo Fisher Scientific, Waltham, MA; catalog number 72611) for 3 minutes. After rinsing with tap water for 1 minute, slides were stained with Bluing Reagent (RICCA Chemical, Pocomoke City, MD; catalog number 6697-1) for 1.5 minutes and then again rinsed for 1 minute with tap water. Slides were transferred to 95% ethanol for 1 minute then stained with 1:1 eosin/70% ethanol (Thermo Fisher Scientific; catalog number 6766007) for 30 seconds. Finally, slides were dehydrated with 95% ethanol for 1 minute, 100% ethanol for 2 minutes twice, and then xylene (VWR; catalog number 89370-088) for 5 minutes twice.
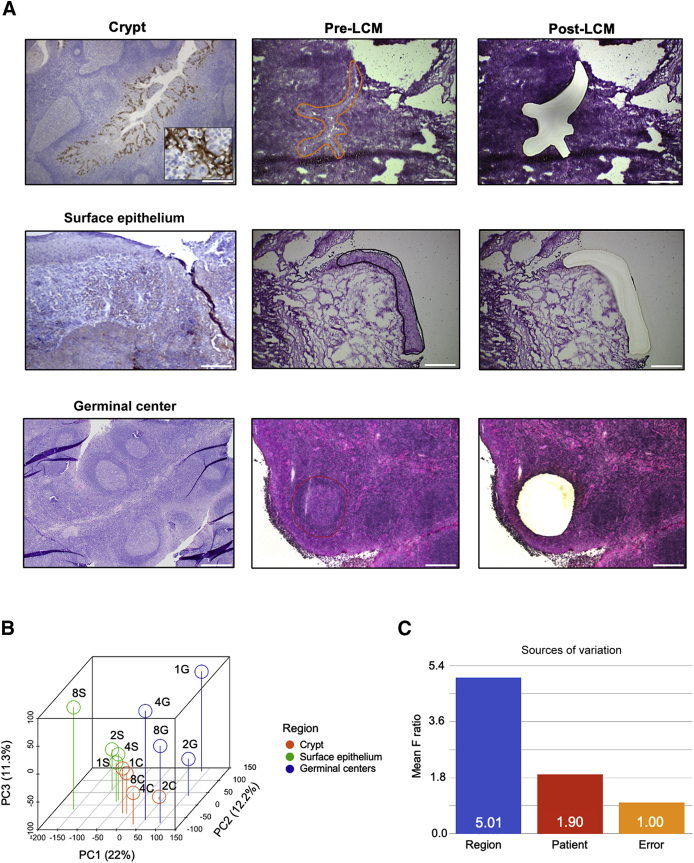
Laser-capture microdissection was performed using a Leica LMD6000 microscope (Wetzlar, Germany). Tonsillar crypts were identified both histologically and by PD-L1 IHC, as previously described.5 Three regions were targeted for laser-capture microdissection: i) surface squamous epithelium, ii) crypt epithelium, and iii) germinal centers (Figure 1).
Figure 1.
Laser-capture microdissection (LCM) of the tonsillar crypts, germinal centers, and surface epithelium was used to transcriptionally profile these three distinct regions. A: Programmed death ligand-1 (PD-L1) immunohistochemical staining and pre-LCM and post-LCM images of the crypts, surface epithelium, and germinal centers. Inset: High-power magnification of PD-L1 staining in the crypt. B and C: Principal component (PC) analysis shows segregation of samples by tonsillar region (B) and source of variation from analysis of variance models (C). Scale bar = 500 μm (A).
RNA Labeling and Illumina Microarray
RNA quality was assessed using an Agilent RNA 6000 Pico Kit (Agilent, Santa Clara, CA; catalog number 5067-1513). Microarray was done using Ovation PicoSL WTA System (Redwood City, CA; part number 3310-48) and Encore BiotinIL Module (Redwood City, CA; part number 4210-48), per the manufacturer's instructions, except the biotin-labeled single-strand DNA was hybridized to beadchips at 48°C. Signal was detected with streptavidin-Cy3 and quantified using an Illumina (San Diego, CA) iScan.
Illumina probe expression values were obtained using Illumina GenomeStudio with standard settings and no background correction (Supplemental Tables S1 and S2). Expression values for all probes for each sample were median scaled and log2 transformed. Probes for the same gene transcript were aggregated by calculation of the mean (n = 31,427 transcripts). Principal component analysis was performed on the complete expression matrix using the R statistics function prcomp.
The data discussed in this publication have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus6 and are accessible through Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo; accession number GSE155139).
Integrative Transcriptomic Analysis
To identify transcripts differentially expressed between the tonsillar crypts compared with germinal centers, and crypts compared with surface epithelium, three linear models were fitted using limma (R package: limma_3.34.9). The design matrix for these linear models included tissue region. Genes with a nominal P value of <0.005 and a Benjamini-Hochberg false discovery rate of 0.1 were considered to be differentially expressed genes (DEGs).
DEGs between regions and corresponding limma statistics (log fold change, P value, false discovery rate, and adjusted P values) were uploaded to Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA). For DEGs between tonsillar crypts and germinal centers (Supplemental Table S3), 1425 analysis-ready molecules were submitted for analysis, whereas 1536 were submitted for the crypts versus the surface epithelium, and 276 were submitted for the crypts versus both regions.
Canonical pathway analysis in IPA calculates the percentage of DEGs up-regulated or down-regulated in each pathway and predicts the activation state of the implicated biological functions reflected by the activation z score. The bases of this inferred activation state are literature-derived relationships between genes and the corresponding biological function. Canonical pathways consist of activating and inhibiting genes. If a gene is up-regulated in a given comparison for a certain pathway, but has an inhibitory function, it will contribute negatively to the activation z score. For some canonical pathways, no literature-derived information can robustly estimate the activation state. For genes with fewer findings or ambiguous directions of regulation (either activating or inhibiting), the IPA prediction algorithm puts less weight on relationship to estimate the likely activation state of biological functions. The full lists of up-regulated and down-regulated genes used for IPA analysis are found in Supplemental Table S4. Heat maps for immune checkpoint DEGs were plotted in R using the Bioconductor package ComplexHeatmap version 3.8. Boxplots were obtained using ggplot2.
Single-sample gene set enrichment analysis was performed on median-scaled and log2-transformed expression data. Enrichment scores were visualized using ComplexHeatmap version 3.8. Boxplots of enrichment scores were generated using the ggboxplot function of R package ggpubr_0.1.8. Differentially enriched gene sets were identified by linear models using the limma package (Supplemental Table S5).
CIBERSORTx
CIBERSORTx, an analytical tool to quantify relative levels of distinct cell types within complex gene expression mixtures,7 was applied to the gene expression data of the different anatomic subsites of benign tonsil tissue. The gene signature matrix of head and neck cell subsets derived from single-cell RNA sequencing, as shared in Newman et al,7 was used as signature matrix file in the Impute Cell Fractions analysis module with the following settings: run mode, relative; batch correction, disabled; and disable quantile normalization, true. Estimates for healthy epithelial cell and fibroblast levels were derived from malignant cells and cancer-associated fibroblasts estimates, respectively.
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue sections and antigen retrieval were performed as previously described.5 IHC antibodies included anti-CD14 (AbCam, Cambridge, UK; rabbit–anti-human; catalog number ab133503; 1:500); anti-CD68 (AbCam; mouse–anti-human; catalog number ab201340; 1:200); anti–carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1; AbCam; rabbit–anti-human; catalog number ab108397; 1:200); anti–PD-L1 (Cell Signaling, Danvers, MA; rabbit–anti-human; catalog number 13864; 1:200); anti–poliovirus receptor (PVR; Cell Signaling; rabbit–anti-human; catalog number 81254; 1:200); anti–V-domain Ig suppressor of T-cell activation (VISTA; Cell Signaling; rabbit–anti-human; catalog number 64953; 1:200); and anti–indoleamine 2,3-dioxygenase 1 (IDO1; R&D Systems, Minneapolis, MN; mouse–anti-human; catalog number MAB6030; 1:200).
Multiplex Immunofluorescence Staining
Formalin-fixed, paraffin-embedded tissue sections were deparaffinized and dehydrated in xylene and graded ethanol solutions. Slides were subjected to heat-induced epitope retrieval in Envision FLEX Target Retrieval Solution, pH 9.0, and endogenous peroxidases were quenched by incubating in 3% H2O2 according to the manufacture's manual (Dako, An Agilent Technologies, Santa Clara, CA). Immunofluorescence (IF) antibodies included anti-CD14 (AbCam; rabbit–anti-human; catalog number ab133503; 1:500); anti-CD1a (Cell Marque, Rocklin, CA; rabbit–anti-human; catalog number CD1a_EP3622); anti-CD68 (AbCam; mouse–anti-human; catalog number ab201340; 1:200); anti–CEACAM1 (AbCam; rabbit–anti-human; catalog number ab108397; 1:200); anti–cytokeratin 5 and 6 (MilliporeSigma, Burlington, MA; mouse–anti-human; catalog number MAB1620; 1:100); anti–PD-L1 (Cell Signaling; rabbit–anti-human; catalog number 13684; 1:200); anti-PVR (Cell Signaling; rabbit–anti-human; catalog number 81254; 1:200); and anti-VISTA (Cell Signaling; rabbit–anti-human; catalog number 64953; 1:200). Mouse IgG1, κ (BD Biosciences, Franklin Lakes, NJ; catalog number 557273; 1:100 and 1:200), and rabbit IgG (BD Biosciences; catalog number 550875; 1:200 and 1:500) were used as isotype controls.
Antigen-antibody binding was visualized using the FLEX+ polymer system (Dako, An Agilent Technologies), and the signal was amplified with AlexaFluor 488, AlexaFluor 555, and AlexaFluor 647 Tyramide Super Boost kits (Invitrogen, Carlsbad, CA), AlexaFluor 750 conjugated anti-mouse IgG (Invitrogen), or AlexaFluor 750 anti-rabbit IgG (Invitrogen). Fluorescent signals were protected, and nuclei were visualized with Prolong Diamond Antifade Reagent with DAPI (Invitrogen).
Digital Image Acquisition and Analysis
IF images were acquired using a Zeiss (Pleasanton, CA) 20× Plan-Neofluor 0.5 numerical aperture air objective lens on the TissueFAXS slide scanning system (TissueGnostics, GmbH, Vienna, Austria) based on a Zeiss Axio Imager Z2 upright epifluorescence microscope. Analysis was performed using TissueQuest (TissueGnostics, GmbH). Analysis parameters were set to identify CD14, CD68, CEACAM1, PD-L1, PVR, and VISTA on a per-cell basis utilizing the nuclei as cellular reference. Cells were quantified using TissueQuest version 6.0.1.136 (TissueGnostics, GmbH) using stitched 2048 × 2048, eight-bit gray-scale raw images. Nuclei were segmented on the basis of DAPI fluorescence intensity. Cells were delineated from each other using a watershed algorithm that identifies borders between adjacent cells (each cell being compared with surrounding neighbor cells), with the nuclei of each cell serving as the cell center. From each nucleus, the markers specified were segmented on a whole-cell basis in a process proceeding radially outwards from the center of each nucleus. Whole-cell masks for each marker were then quantified on the basis of average pixel intensity across the entire cell area. Cells positive for a particular marker were identified on the basis of thresholding for signal intensity above both background and non-specific control fluorescence. Percentage of positive cells was then determined relative to total cell (DAPI-positive) counts. Statistical comparisons were made with a two-tailed t-test.
Flow Cytometry
Frozen human tonsil samples were thawed and rested in complete RPMI 1640 medium (RPMI 1640 medium + 10% fetal bovine serum + 1% penicillin/streptomycin/l-glutamine + 10 mmol/L HEPES) for 1 hour in an incubator. Cells were then seeded in a 96-well U-bottom plate and washed twice with phosphate-buffered saline and stained with viability dye, Zombie NIR (Biolegend, Dedham, MA), for 20 minutes on ice in the dark. Subsequently, cells were washed with fluorescence-activated cell sorting buffer (2 mmol/L EDTA + 0.05% sodium azide + 4% fetal bovine serum in phosphate-buffered saline), and then stained with surface antibody cocktail for 30 minutes on ice in the dark, including BV421-PVR (clone SKII.4; Biolegend), BV480-CD3 (clone UCHT1; BD Bioscience), BV510-CD11b (clone ICRF44; Biolegend), BV570–human leukocyte antigen-DR (cloneL243; Biolegend), BV605-CEACAM1/5/6 (CD66a/c/e; clone ASL-32; Biolegend), BV650-CD11c (clone 3.9; Biolegend), BV711–T-cell immunoglobulin and mucin-domain containing-3 (clone F38-2E2; Biolegend), BV750-CD1a (clone SK9; BD Bioscience), BV785 cytotoxic T-lymphocyte-associated protein 4 (clone BNI3; Biolegend), peridinin-chlorophyll-protein-CD16 (clone 3G8; Biolegend), peridinin-chlorophyll-protein-CD19 (clone HIB19; Biolegend), phycoerythrin–PD-L1 (clone 29E.2A3; Biolegend), phycoerythrin–eFluor 610–CD68 (clone eBioY1/82A; eBioscience, San Diego, CA), PE-Cy5–B- and T-lymphocyte attenuator (BTLA; clone MIH26; Biolegend), PE-Cy7–VISTA (clone B7H5DS8; eBioscience), AlexaFluor 700–CD14 (clone HCD14; Biolegend), and Allophycocyanin Fire 750–E-cadherin (clone 67A4; Biolegend). After the surface antibody incubation, cells were washed twice with fluorescence-activated cell sorting buffer, resuspended with 200 μL of Forkhead box protein P3 fixation/permeabilization buffer (eBioscience), and incubated for 30 minutes on ice in the dark. Cells were then washed with Perm wash buffer (eBioscience) and stained with intranuclear antibody Allophycocyanin-IDO (clone eyedio; eBioscience) for 30 minutes on ice in the dark. After two washes using fluorescence-activated cell sorting buffer, cells were resuspended in 200 μL of fluorescence-activated cell sorting buffer and then acquired using the spectral flow cytometry Cytek Aurora 4-laser system (Fremont, CA). Data were analyzed by using FlowJo version 10 (TreeStar, Ashland, OR).
Results
The Tonsillar Crypts Are Enriched with Myeloid Cells Compared with the Surface Epithelium and Germinal Centers
Laser-capture microdissection was used to transcriptionally profile and compare various regions within the tonsils, specifically the crypts, germinal centers, and surface epithelium (Figure 1A). Principal component analysis of the gene expression matrix of these regions demonstrated that human tonsils segregated by region rather than patient (Figure 1, B and C), suggesting that the tonsil has distinct transcriptional and functional anatomic regions.
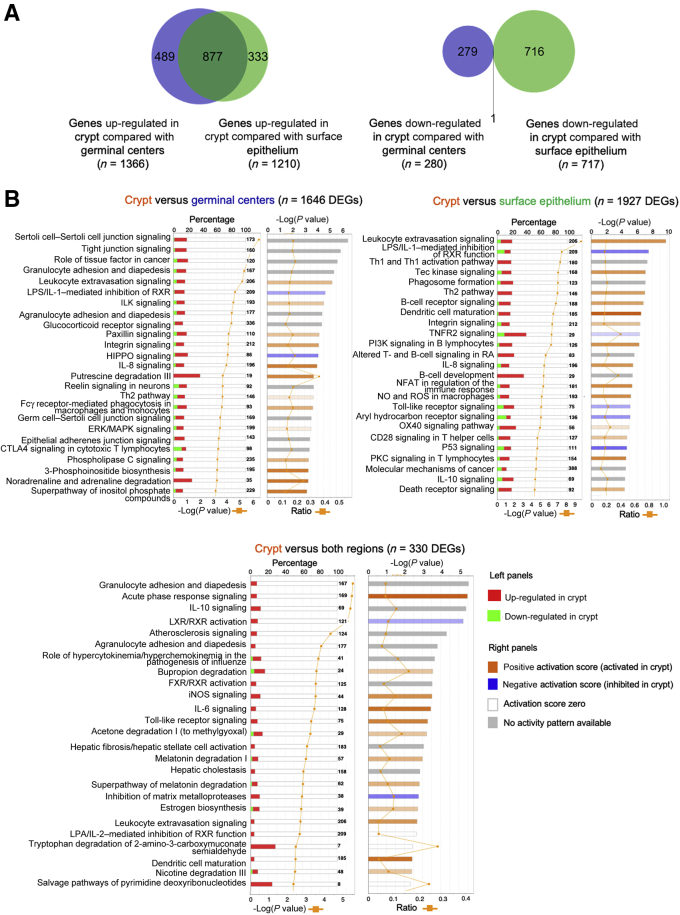
To further investigate this, differential gene expression patterns between the tonsillar crypts and the germinal centers and surface epithelium were determined. The crypts had 1646 DEGs (1366 up-regulated and 280 down-regulated) compared with germinal centers, and 1927 DEGs compared with surface epithelium (1210 up-regulated and 717 down-regulated). Compared with the combined germinal center and surface epithelium, 287 genes were up-regulated and 43 were down-regulated in the crypt. More importantly, a high number of DEGs (n = 877) overexpressed in the crypt compared with germinal centers overlapped with the DEGs of the crypt versus the surface epithelium (Figure 2A). This confirmed that the tonsil has distinct transcriptional compartments that serve unique functions.
Figure 2.
Transcriptomic pathway analysis demonstrates that the tonsillar crypts are a functionally distinct compartment within the tonsil. A: A high number of differentially expressed genes (DEGs) overexpressed in the tonsillar crypts compared with germinal centers overlap with the DEGs of the crypts versus the surface epithelium. B: The left side of the chart displays the percentage of DEGs present in each canonical pathway and reflects their up-regulation (red) or down-regulation (green) in the crypt. The orange line indicates the –log (P value). The right side displays the predicted activation state of the implicated biological functions reflected by the activation z score. The bases of this inferred activation state are literature-derived relationships between genes and the corresponding biological function. Pathways that are activated in the crypts are marked with an orange bar, indicating a positive activation score, whereas pathways inhibited in the crypts are marked with a blue bar, indicating a negative activation score. For canonical pathways with a gray bar, no literature-derived information exists to estimate the activation state. An orange line plots the ratio of the percentage of genes up-regulated/down-regulated in each pathway. CTLA-4, cytotoxic T-lymphocyte-associated protein 4; ERK, extracellular signal-regulated kinase; ILK, integrin-linked kinase; iNOS, inducible NO synthase; LPS, lipopolysaccharide; LXR, liver X receptors; MAPK, mitogen-activated protein kinase; NFAT, nuclear factor of activated T-cells; NO, nitric oxide; PI3K, phosphatidylinositol 3-kinase; PKC, protein kinase C; RA, retinoid acid; RXR, retinoid X receptor; ROS, reactive oxygen species; Th1, type 1 helper T cell; Th2, type 2 helper T cell; TNFR, tumor necrosis factor receptor.
IPA demonstrates that the tonsillar crypt has an active immune microenvironment driven by dendritic cells, macrophages, and monocyte activity. Genes up-regulated in the crypt, compared with germinal centers, clustered into pathways specific to the unique epithelial lining of the crypt and immune-related pathways (Figure 2B). Immune pathways differentially up-regulated in the crypt, compared with germinal centers, included granulocyte and agranulocyte (lymphocyte, monocyte, and macrophage) adhesion and diapedesis, leukocyte extravasation signaling, IL-8 signaling, the type 2 helper T-cell pathway, and FcƔ-receptor–mediated phagocytosis in macrophages and monocytes. Up-regulated pathways in the crypt compared with the surface epithelium consisted of a wider range of immune-related pathways, indicating T-helper 1 and 2 interactions, B-cell receptor signaling paired with suppressive macrophage/monocyte activities, such as production of the immune-suppressive nitric oxide and IL-10, and dysregulation of toll-like receptor signaling. When assessing the differentially up-regulated pathways within the tonsillar crypts compared with both the surface epithelium and the germinal centers, there was an enrichment of genes associated with canonical monocyte/macrophage pathways in addition to leukocyte extravasation signaling and dendritic cell maturation, suggesting a high level of monocyte-macrophage activity within the crypts (Figure 2B).
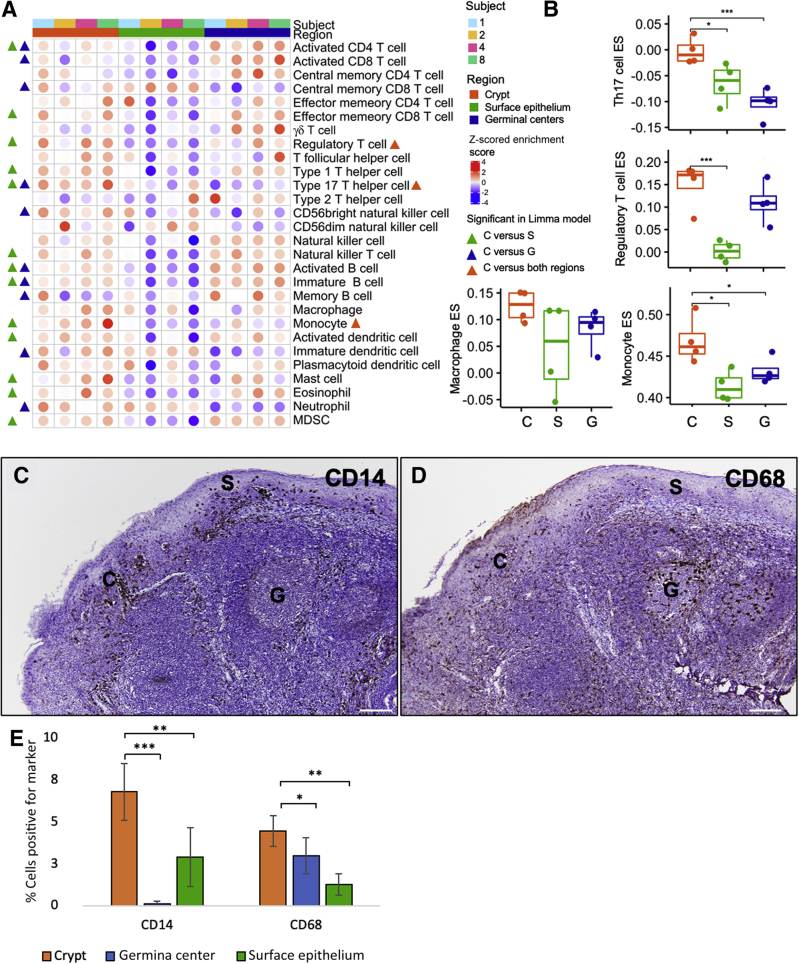
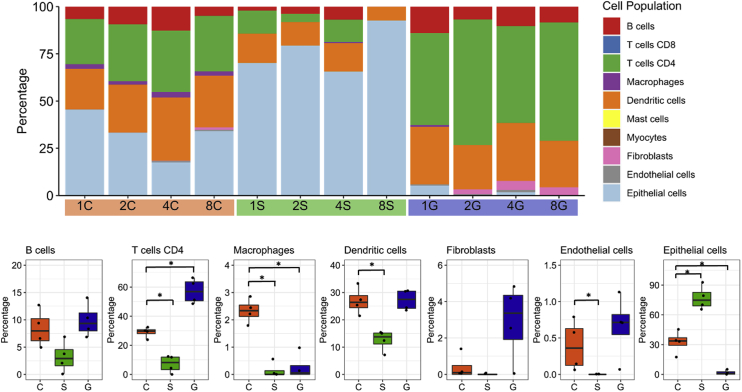
Next, the gene expression data set was also deconvoluted to assess which immune cell subsets were enriched in the tonsillar crypts (Figure 3, A and B). Although enrichment scores for immune cells were high in both the tonsillar crypts and the germinal centers, the tonsillar crypts had a greater enrichment of monocyte (limma, P = 0.014), regulatory T cell (limma, P = 0.024), and type 17 helper T cell (limma, P = 0.0028) related transcripts compared with the other compartments (Figure 3B). Thus, the crypts have a highly immune-regulated microenvironment and are likely the region within the tonsil where foreign antigens that enter through the oral cavity (such as food, bacteria, and viruses) are sampled by the innate immune system. Last, using a separate deconvolution algorithm, CIBERSORTx,7 the proportion of macrophages was significantly increased in tonsillar crypts compared with the germinal centers (P = 0.029) and surface epithelium (P = 0.027) (Supplemental Figure S1). The transcriptional findings of localized immune cell populations within the crypts were confirmed by CD14 and CD68 IHC staining of normal tonsil (Figure 3, C–E).
Figure 3.
The tonsillar crypts (C) have an active immune environment characterized by type 17 helper T cell (Th17), regulatory T cell (Treg), and monocyte and macrophage activity. A and B: Deconvolution of gene expression data shows enrichment for immune cells (namely, Th17, Tregs, monocytes, and macrophages) in the tonsillar crypts compared with the germinal centers (G) and surface epithelium (S). A: Significant immune cell signatures in fitted limma linear models (P < 0.05) are annotated by triangles [crypt versus surface epithelium (green), crypt versus germinal centers (blue), and crypt versus both regions (orange)] in the heat map. C and D: Immunohistochemical staining for CD14 (C) and CD68 (D) confirms presence of these cells in the normal tonsillar crypts. E: Bar graph demonstrating the quantitative detection of CD14 and CD68 cells within the crypts, germinal centers, and surface epithelium of normal tonsil, as assessed by multiplex immunofluorescence. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P ≤ 0.001. Scale bar = 200 μm (C and D). Original magnification, ×100 (C and D). ES, enrichment score; MDSC, myeloid-derived suppressor cell.
Monocytes within the Tonsillar Crypts Co-Express Multiple Inhibitory Immune Checkpoints
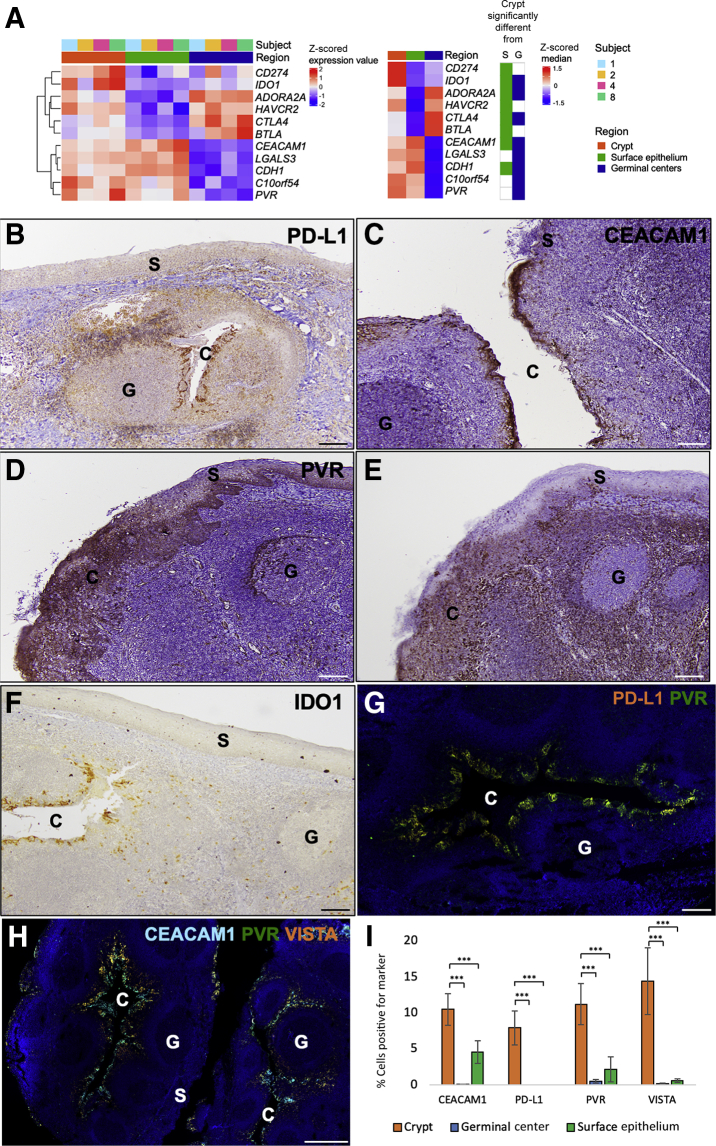
In addition to differential immune cell gene signature expression, several canonical immune checkpoints, including PD-L1, CTLA-4, TIM-3 (HAVCR2), IDO1, adenosine A2A receptor (ADORA2A), and BTLA were found to be differentially up-regulated genes in the crypt compared with the surface epithelium. There were several immune checkpoints that were also down-regulated in the crypts, including E-cadherin (CDH1) and CEACAM1 (Figure 4A). The relative decrease in expression of CDH1 and CEACAM1 in the crypt compared with the surface epithelium is likely due to the role of these genes in epithelial cell-cell adhesion. Although the tonsillar crypts and germinal centers are both regions of high immune cell activity, several immune checkpoints were identified that were differentially up-regulated in the crypts compared with the germinal centers. These included CDH1, CEACAM1, PVR, VISTA (or C10orf54), galectin 3 (LGALS3), and IDO1. Both CTLA-4 and ADORA2A were down-regulated in the crypts compared with the germinal centers, likely as a result of increased T-cell–B-cell interaction inherent to the germinal centers (Figure 4A).
Figure 4.
Multiple immune checkpoint markers are differentially up-regulated and co-expressed on monocytes in the tonsillar crypts (C). A: Heat map demonstrating differentially expressed immune checkpoints in tonsillar crypts compared with surface epithelium (S) or germinal centers (G) for each sample (right). Median expression value in each region (left heat map) and significant difference in crypts compared with surface epithelium or germinal centers is indicated. B–F: Immunohistochemistry staining of programmed death ligand-1 (PD-L1; B), carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1; C), poliovirus receptor (PVR; D), V-domain Ig suppressor of T-cell activation (VISTA; E), and indoleamine 2,3-dioxygenase 1 (IDO1; F) within the tonsillar crypts, germinal centers, and surface epithelium of normal tonsils shows that multiple immune checkpoint markers are preferentially localized to the tonsillar crypts. G: Immunofluorescence images of normal tonsillar crypts (serial sections of similar regions for each panel) show colocalization of immune checkpoints PD-L1 (orange) and PVR (green). H: Immunofluorescence images of normal tonsillar crypts (serial sections of similar regions for each panel) show colocalization of immune checkpoints CEACAM1 (aqua blue), VISTA (orange), and PVR (green) within the crypts compared with both the germinal centers and the surface epithelium. I: Bar graph demonstrating the quantitative detection of the various immune checkpoints within the crypts, germinal centers, and surface epithelium within normal tonsil, as assessed by multiplex immunofluorescence. ∗∗∗P ≤ 0.001. Scale bar = 200 μm (B–H). Original magnification, ×100 (B–F).
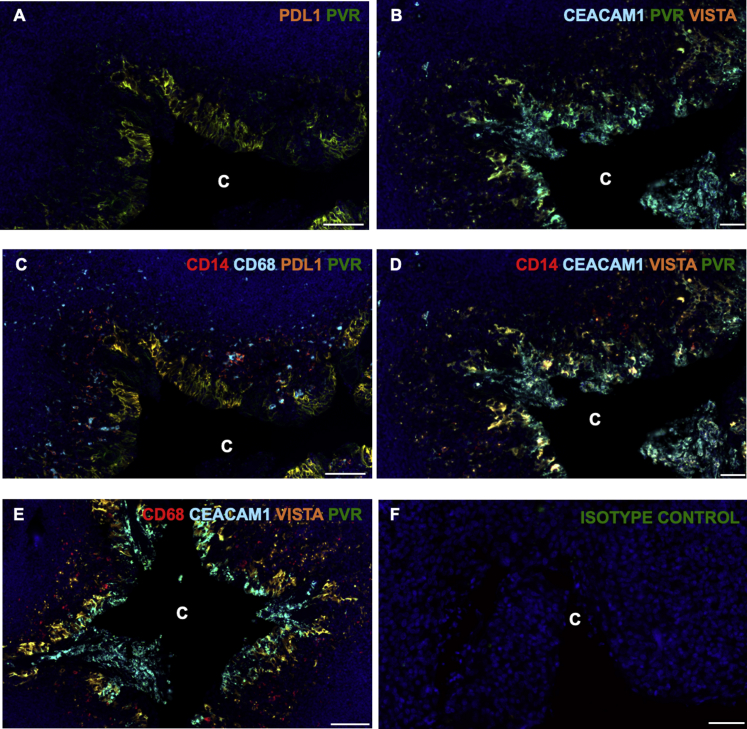
Spatial context of immune checkpoint proteins was then assessed using IHC and multiplex IF staining. IHC demonstrated differential expression of five of the immune checkpoints, PD-L1, CEACAM1, PVR, VISTA, and IDO1, within the tonsillar crypts (Figure 4, B–F). Although PVR was also expressed in both the germinal center and surface epithelium, it was predominately expressed in the crypt. Quantitative multiplex IF confirmed the differential colocalization of PD-L1, PVR, CEACAM1, and VISTA within the crypts compared with both the germinal centers and the surface epithelium (P ≤ 0.001) (Figure 4, G–I, Supplemental Figure S2 A-F, and Supplemental Table S6).
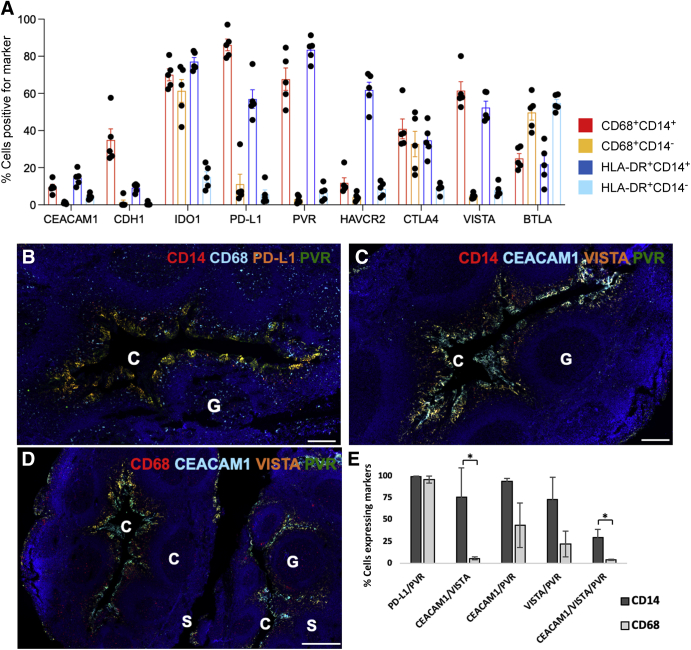
Flow cytometry was performed to attribute immune checkpoint expression on the monocytes and macrophages in tonsil tissue (Figure 5A and Supplemental Figure S3). PD-L1, PVR, and VISTA were expressed on 86.1%, 67.6%, and 61.5% of the tonsillar CD14+CD68+ monocyte/macrophages, respectively, whereas 61.3% of CD68+CD14–monocyte/dendritic cells expressed IDO1. Human leukocyte antigen-DR+CD14+ resident macrophages most commonly expressed PVR (83.4%), IDO1 (77.1%), Hepatitis A virus cellular receptor 2 (61.8%), PD-L1 (57.0%), and VISTA (52.4%), whereas other human leukocyte antigen-DR+CD14– dendritic cells expressed BTLA (54.8%). These results confirmed that a high frequency of monocytes and macrophages differentially express immune checkpoint markers within normal tonsil tissue. Utilizing multiplex IF, CD14 and CD68 monocytes and macrophages were assessed for co-expression of the immune checkpoints (PD-L1, PVR, CEACAM1, and VISTA) within the crypts (Figure 5, B–E, and Supplemental Table S7). As a control, other nonmyeloid cells, such as keratinocytes, were found not to be major contributors to immune checkpoint expression (Supplemental Figure S4). In fact, >96% of the CD14/PD-L1 double-positive cells and CD68/PD-L1 double-positive cells co-expressed PVR (Figure 5B). Similarly CEACAM1, VISTA, and PVR were differentially co-expressed by monocytes and macrophages localized within the crypts (Figure 5, C and D, and Supplemental Figure S5). Similar to the co-expression of PD-L1 and PVR, >73% of CD14 cells within tonsillar crypts co-expressed other immune checkpoints, including CEACAM1/VISTA, CEACAM1/PVR, and VISTA/PVR (Figure 5E, Supplemental Figure S5, and Supplemental Table S7). CD68 cells also co-expressed multiple immune checkpoints, primarily PD-L1/PVR (96%), VISTA/PVR (22%), and CEACAM1/PVR (44%) (Figure 5E, Supplemental Figure S5, and Supplemental Table S7).
Figure 5.
Tonsillar crypts (C) show differential protein expression of immune checkpoints by flow cytometry and multiplex immunofluorescence. A: Expression levels of nine immune checkpoint markers on myeloid cells isolated from five human tonsils using flow cytometry. Each dot is one individual tonsil sample. B–D: Wide-field views of immunofluorescence images scanned from normal tonsillar crypts (serial sections of similar regions for each panel), showing localization of monocytes based on immune checkpoint receptors. B: Differential colocalization of CD14 (red) and CD68 (aqua blue) in monocytes, programmed death ligand-1 (PD-L1; orange), and poliovirus receptor (PVR; green) within crypts. When the fluorescent channels overlap significantly, the cells appear orange-yellow. There are discrete patches of cells located on the inner boundaries, corresponding possibly to stratified reticulated epithelium. C: CD14-positive monocytes (red) showed high levels of colocalization of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1; aqua blue), V-domain Ig suppressor of T-cell activation (VISTA; orange), and PVR (green). D: CD68-positive monocytes showed overlapping CEACAM1 (aqua blue), VISTA (orange), and PVR (green) localization, which also appeared in a subset of germinal centers (G). E: Many CD14- or CD68-positive cells in the normal tonsil crypts expressed two or more immune checkpoint markers. Data indicate means ± SEM (A). ∗P < 0.05. Scale bars: 200 μm (B and C); 500 μm (D). BTLA, B- and T-lymphocyte attenuator; CDH1, E-cadherin; CTLA, cytotoxic T-lymphocyte-associated protein4; HAVCR2, hepatitis A virus cellular receptor 2; HLA, human leukocyte antigen; IDO1, indoleamine 2,3-dioxygenase 1; S, surface epithelium.
Myeloid Populations Co-Expressing PD-L1, CEACAM1, VISTA, and PVR Are Found in HPV-Associated Head and Neck Cancer
Next, the oncologic relevance of these myeloid populations in HPV-associated tonsil cancers was explored (Figure 6 and Supplemental Table S7). Greater than 50% of tumor-infiltrating CD14 and CD68 cells expressed a combination of PD-L1, CEACAM1, PVR, and VISTA in HPV-associated tonsil cancers (Supplemental Figures S6 and S7 and Supplemental Table S7), suggesting that myeloid cells co-expressing these same immune checkpoint pathways may contribute to host immune evasion in head and neck cancers.
Figure 6.
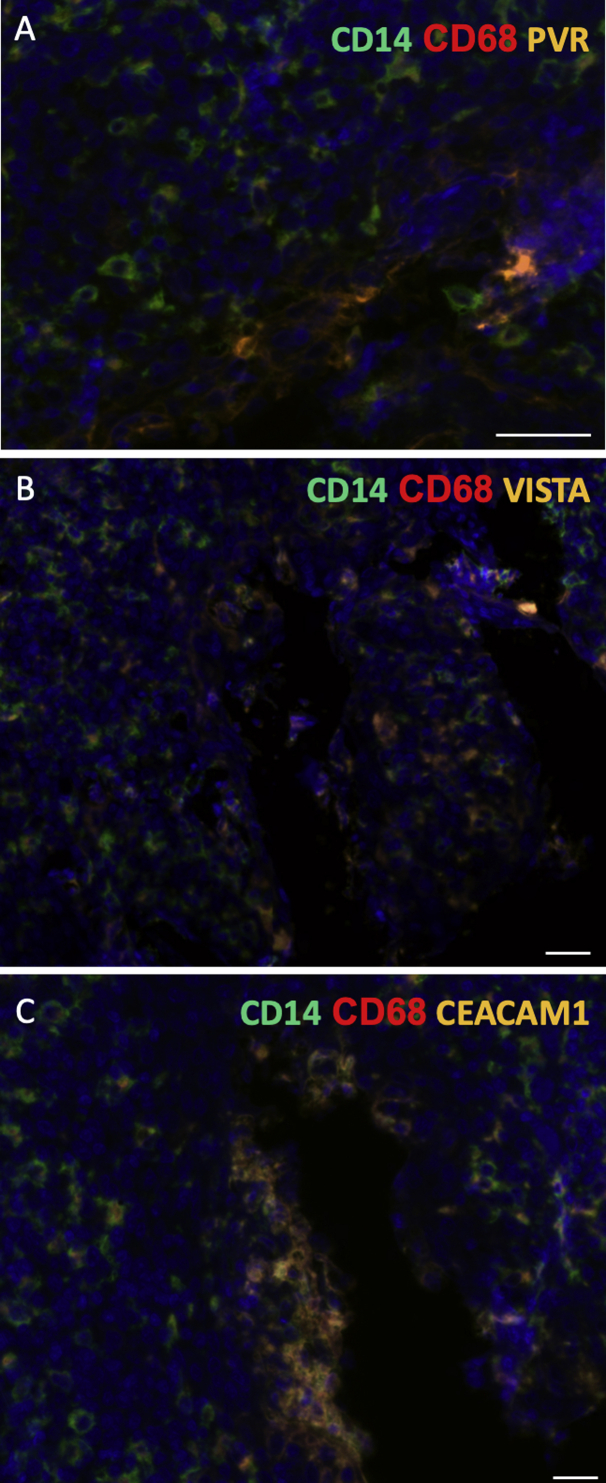
Wide-field views of immunofluorescence images scanned from normal tonsillar crypts (serial sections of similar regions for each panel), showing localization of monocytes based on immune checkpoint receptors. A and B: Immune checkpoint markers carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1; aqua blue), V-domain Ig suppressor of T-cell activation (VISTA; orange), and poliovirus receptor (PVR; green) are expressed in tonsillar cancer. A: Low power. B: High power. C–F: Immune checkpoint markers CEACAM1 (aqua blue), VISTA (orange), and PVR (green) colocalize with CD14 (red; C) and CD68 (red; D) monocytes and macrophages (E and F). When fluorescent channels overlap significantly, the cells appear orange-yellow. Scale bars: 100 μm (A, C, and E); 50 μm (B, D, and F).
Discussion
Viruses are the second leading cause of cancer worldwide.8 Despite the expression of immunogenic foreign viral antigens, viruses are able to evade host immune defense mechanisms to establish chronic infection and subsequent malignant transformation. The present study reports that various immune cells are compartmentalized within the tonsil to form distinct phenotypic and functional regions. The surface epithelium is stratified squamous epithelium and serves as a barrier characterized by tight junctions and cell-cell interactions. The germinal centers are areas of adaptive immune activity with a predominance of B- and T-cell activity. Within the tonsillar crypts, innate immunity predominates with a mononuclear phagocyte system with differential co-expression of multiple immune checkpoints PD-L1, CEACAM1, VISTA, PVR, galectin-3, IDO1, and CDH1. The localized expression of several immune checkpoints by infiltrating monocytes contributes to an immune permissive microenvironment for HPV infection, and these same immunosuppressive myeloid populations persist in HPV-associated tonsil cancers.
Several differentially up-regulated immune checkpoints identified in the tonsillar crypts (ie, PD-L1, TIM3/CEACAM1, VISTA, and PVR) are actively being investigated in cancer therapeutics. VISTA, known as C10orf54, contains an extracellular domain that is most similar to PD-L1.9 In humans, VISTA is predominately expressed on myeloid cells10 and has also been shown to suppress T-cell activation, proliferation, and cytokine production.10,11 Other immune checkpoints (CEACAM1 and PVR) that were overexpressed in the tonsillar crypts are known to directly inhibit natural killer (NK) cell activation. NK cells play a crucial role in innate immunity through their direct lysis of target cells and their secretion of cytokines,12 whereas T cells play a pivotal role in adaptive immunity through their recognition of specific tumor antigens. CEACAM1 expression has been shown to dampen anti-tumor immunity by down-regulating NKG2D ligand expression on tumor cells, which leads to reduced natural killer cell–mediated lysis and increased tumor escape of immune surveillance.9, 10, 11,13 PVR, which is expressed on dendritic cells, has been shown to bind a surface protein, TIGIT, which contains an immunoreceptor tyrosine-based inhibitory motif on NK and T cells.14 PVR binding to activated T cells and NK cells appears to enhance production of IL-10, thereby decreasing the activity of immune cells in the tumor microenvironment.14
Blockade of the PD1/PD-L1 axis alone achieves 15% to 20% clinical response rates in recurrent/metastatic head and neck cancer. Thus, targeting multiple immune checkpoints, beyond single targeting of the PD1/PD-L1 axis, could be required to successfully re-activate both T and NK anti-tumor immune responses. Further functional T and/or NK cell assays are needed to precisely elucidate the phenotypic effect of multiple immune checkpoint inhibition on these populations. Notably, T-cell immunoglobulin and mucin-domain containing-3, CEACAM1, VISTA, and PVR have blocking antibodies in clinical trials, and combinatorial targeting of these immune checkpoints may be required to treat HPV-associated head and neck cancers. Alternatively, selective depletion or reprogramming of the immunosuppressive monocytic population that co-expresses these immune checkpoint ligands may be needed.
LGALS3 is a relatively novel immune checkpoint, and the LGALS3 gene encodes for galectin-3, which plays an important role in cell-cell adhesion, cell-matrix interactions, macrophage activation, and angiogenesis.15 LGALS3 is also likely to regulate adaptive and innate immune responses. Extracellular LGALS3 has been shown to induce the apoptosis of lymphocytes,16,17 and binding of LGALS3 to Mucin 1 (MUC1) has been shown to drive loss of NK-mediated cytolysis.18,19 Altered galectin-3 expression can impact immune cell chemoattraction, immunosuppression, apoptosis, adhesion, invasion, and metastasis. Multiple studies have explored the role of galectin-3 inhibitors in preclinical and clinical settings,20, 21, 22, 23 showing that small-molecule antagonists can induce cancer cell death20 and augment response to PD-L1 blockade.21 Phase 1 clinical studies are currently underway for a galectin-3 inhibitor, GR-MD-02 in melanoma in combination with ipilimumab (https://clinicaltrials.gov, trial number NCT02117362) and GR-MD-02 in combination with pembrolizumab (trial number NCT02575404). These findings, in combination with the identification of differential expression of LGALS3 in generating immune privilege within the normal tonsillar crypts, suggest that LGALS3 may be an important immune checkpoint that merits further investigation in HPV-associated head and neck cancers.
The present study was designed to better understand the viral tropism of HPV to the tonsillar crypts by characterizing different anatomic regions within the tonsils. For a virus to display tropism for a specific host tissue, it must first have the means to break through the host organism's first line of defense, known as the innate immune system. HPV targets its initial pathogenic entry within the upper respiratory tract and eventual residence in an area concentrated with monocytes and macrophages that co-express multiple immune checkpoints. Because chronic HPV infection can lead to malignant transformation, similar mechanisms of immune evasion were identified in HPV-associated tonsil cancers. Further comprehensive characterization of immune checkpoint pathway activation and identification of key immune cell populations contributing to the immunosuppressive microenvironment in head and neck cancers are currently underway utilizing single-cell RNA-sequencing analysis. The results of the study suggest that multiple immune checkpoints may need to be targeted and/or the immunosuppressive monocyte population, which co-expresses these multiple immune checkpoints, reprogrammed to overcome adaptive immune resistance to immunotherapy in head and neck cancer patients.
Acknowledgment
We sincerely thank our patients for their participation in our study.
Footnotes
Supported by NIH/National Institute of Dental and Craniofacial Research1R01 DE025340 (S.I.P.) and NIH/National Cancer InstituteP01 CA240239 (W.C.F., M.J.P., and S.I.P.).
A.K.M. and J.R. contributed equally to this work.
Disclosures: F.M.M. is an employee of Gilead Sciences, Inc., and serves on the advisory board at Calidi Biotherapeutics and Immunovative Therapies Corp. S.I.P. has received consultancy payments from Abbvie, Astrazeneca/MedImmune, Cue Biopharma, Fusion Pharmaceuticals, MSD/Merck, Newlink Genetics, Oncolys Biopharma, Replimmune, Scopus Biopharma, and Sensei Bio. S.I.P. has received grants/research support from Abbvie, Astrazeneca/MedImmune, Cue Biopharma, and Tesaro. M.J.P. has received consultancy payments from Aileron Therapeutics, AstraZeneca, Elstar Therapeutics, KSQ Therapeutics, MPM Capital, Siamab Therapeutics, Third Rock Ventures, and Tidal Therapeutics. All other authors declare no potential conflicts of interest.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2021.06.012.
Author Contributions
A.K.M., J.R., T.S., Y.C., D.R., W.H., A.E.B., W.H.W., D.B., and S.I.P. designed experiments and analyzed the data. G.D.Y., T.D., J.A.B., and W.C.F. collected samples and analyzed the data. F.M.M., L.J.W., D.G.D., D.T.L., and M.J.P. analyzed the data. S.I.P. managed the project and funding. All authors wrote and edited the manuscript.
Supplemental Data
Supplemental Figure S1.
Stacked bar plots and boxplots of percentage of specific cell subsets across crypts (C), surface epithelium (S), and germinal centers (G) of benign tonsil tissue, as determined by the CIBERSORTx deconvolution algorithm. Macrophages were most commonly found in the tonsillar crypts. ∗P < 0.05.
Supplemental Figure S2.
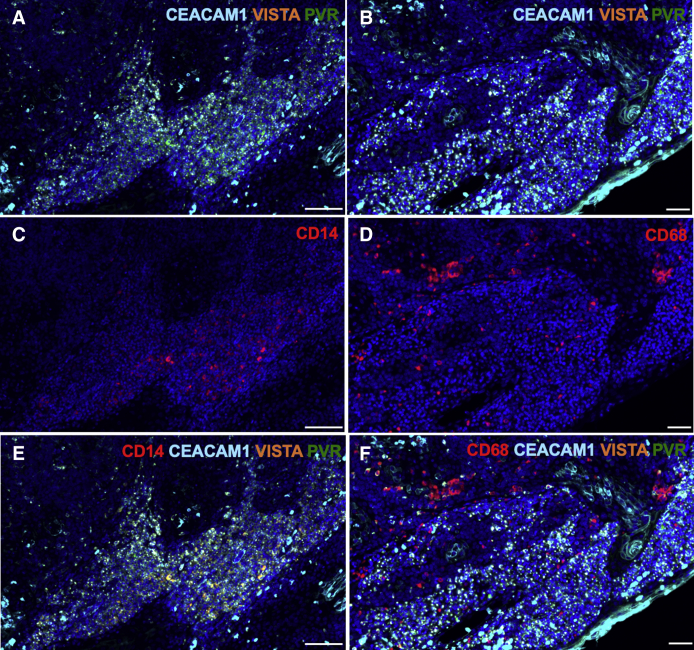
Tonsillar crypts (C) show differential protein expression of immune checkpoints by CD14- and CD68-positive monocytes and macrophages. A and B: Higher-magnification views of scanned tonsillar crypts, showing distribution and degrees of colocalization of programmed death ligand-1 (PD-L1; orange) and poliovirus receptor (PVR; green; A), and carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1; aqua blue), PVR (green), and V-domain Ig suppressor of T-cell activation (VISTA; orange; B). C–E: Higher-magnification views of scanned tonsillar crypts, showing distribution and degrees of colocalization of CD14/CD68/PD-L1/PVR, CD14/CEACAM/VISTA/PVR, and CD68/CEACAM/VISTA/PVR. C: Note the smaller inner clusters of CD14-positive cells that are further away from the crypt boundary and are not co-expressors, whereas at the crypt epithelial boundary there is significantly more overlap. It is also apparent that immune checkpoint inhibitors are more abundant on CD14-positive versus CD68-positive monocytes. D: CD14-positive cells co-express CEACAM1 (aqua blue), VISTA (orange), and PVR (green). E: In contrast to abundant CEACAM1 colocalization in CD14-positive monocytes, fewer CD68 positive monocytes showed significant CEACAM1/VISTA/PVR triple expression. F: Isotype control staining, showing no background autofluorescence. Scale bars: 100 μm (A, C, and E); 50 μm (B, D, and F).
Expression levels of nine immune checkpoint markers on myeloid cells isolated from five human tonsils using flow cytometry. A: Gating strategy for flow cytometry data. Lineage-negative population was defined as CD3–, CD19–, and CD16– cells. The gating strategy of four populations was shown, including human leukocyte antigen (HLA)-DR+CD14+ (blue), HLA-DR+CD14– (cyan), CD68+CD14+ (red), and CD68+CD14– (orange). The numbers indicate the frequency of parent population. B: The expression levels of poliovirus receptor (PVR), programmed death ligand-1 (PD-L1), cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), T-cell immunoglouglin and mucin-domain containg-3 [TIM-3, also knowns as hepatitis A virus cellular receptor 2 (HAVCR2)], B- and T-lymphocyte attenuator (BTLA), V-domain Ig suppressor of T-cell activation (VISTA), E-cadherin (CDH1), carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), and indoleamine 2,3-dioxygenase 1 (IDO1) are shown for four different monocyte and macrophage populations defined in Supplemental Figure S1A. The unstained controls (top row of panel B, black box) are shown. The numbers in the quadrant are the frequencies of each gate. FSC, forward scatter; SSC, side scatter.
Supplemental Figure S4.
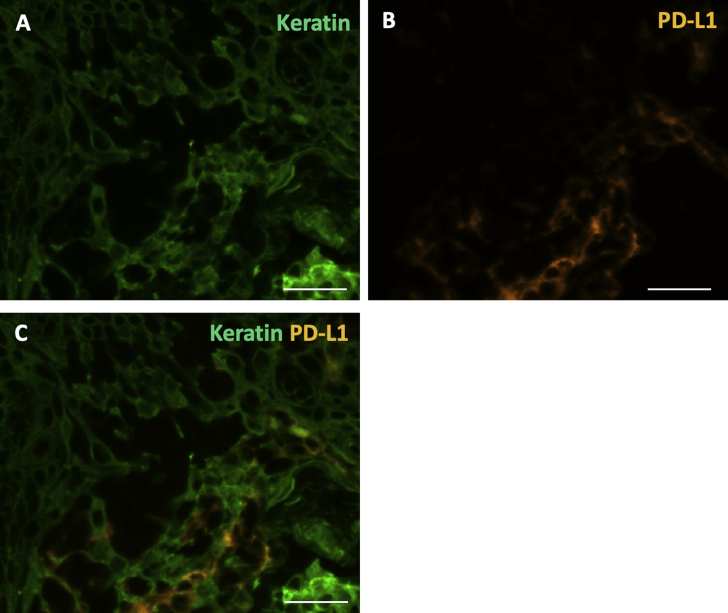
Programmed death ligand-1 (PD-L1) expression is not predominantly derived from epithelial cells. A and B: Higher-magnification immunofluorescence, showing the distribution of keratin (green) expressing cells (A) and PD-L1 (orange) expressing cells (B). C: Few cells co-express both keratin and PD-L1, suggesting that epithelial cells are not the predominant contributors of PD-L1 expression. Scale bar = 20 μm (A–C).
Supplemental Figure S5.
High-resolution views of immunofluorescence images scanned from normal tonsillar crypts (serial sections of similar regions for each panel), showing localization of monocytes and macrophages based on immune checkpoint receptors. Colocalization of CD14 (green) and CD68 (red) with poliovirus receptor (PVR; orange; A), V-domain Ig suppressor of T-cell activation (VISTA; orange; B), or carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1; orange; C). Scale bar = 20 μm (A–C).
Supplemental Figure S6.
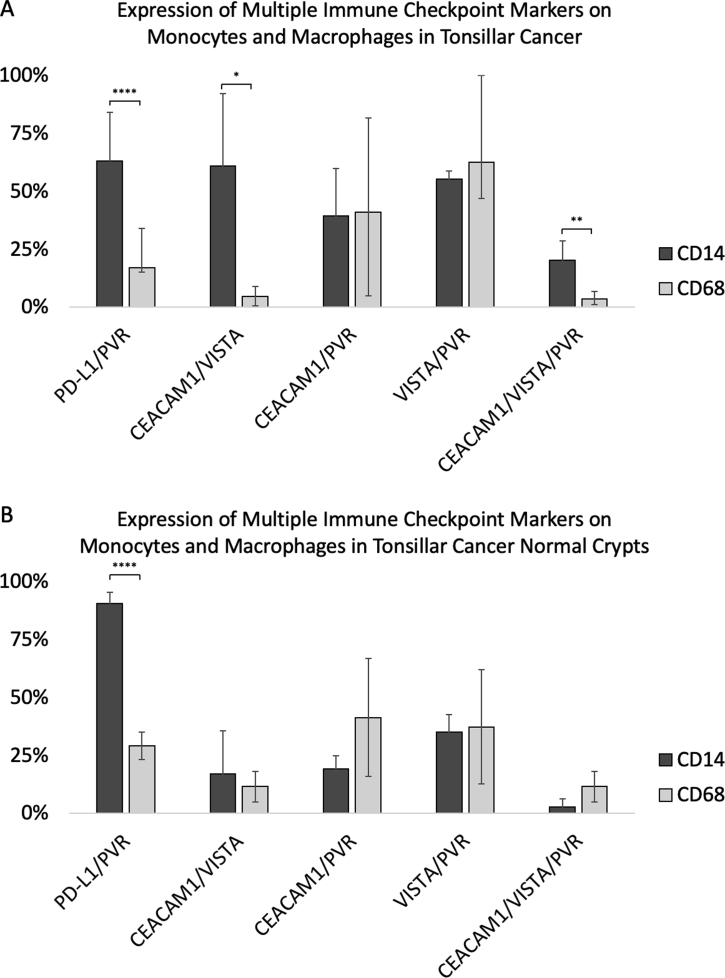
CD14 and CD68 immune cells in tonsillar cancer often express multiple immune checkpoint markers. A: Monocytes and macrophages in tonsil cancer express multiple immune checkpoint markers. B: Similar to tonsillar cancer, noncancerous crypts juxtaposed to tonsil cancer also co-express two or more immune checkpoint markers. For certain combinations of immune checkpoint marker co-expression, CD14-positive cells were more likely to express multiple markers compared with CD68-positive cells. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗∗P < 0.0001. CEACAM1, carcinoembryonic antigen-related cell adhesion molecule 1; PD-L1, programmed death ligand-1; PVR, poliovirus receptor; VISTA, V-domain Ig suppressor of T-cell activation.
Supplemental Figure S7.
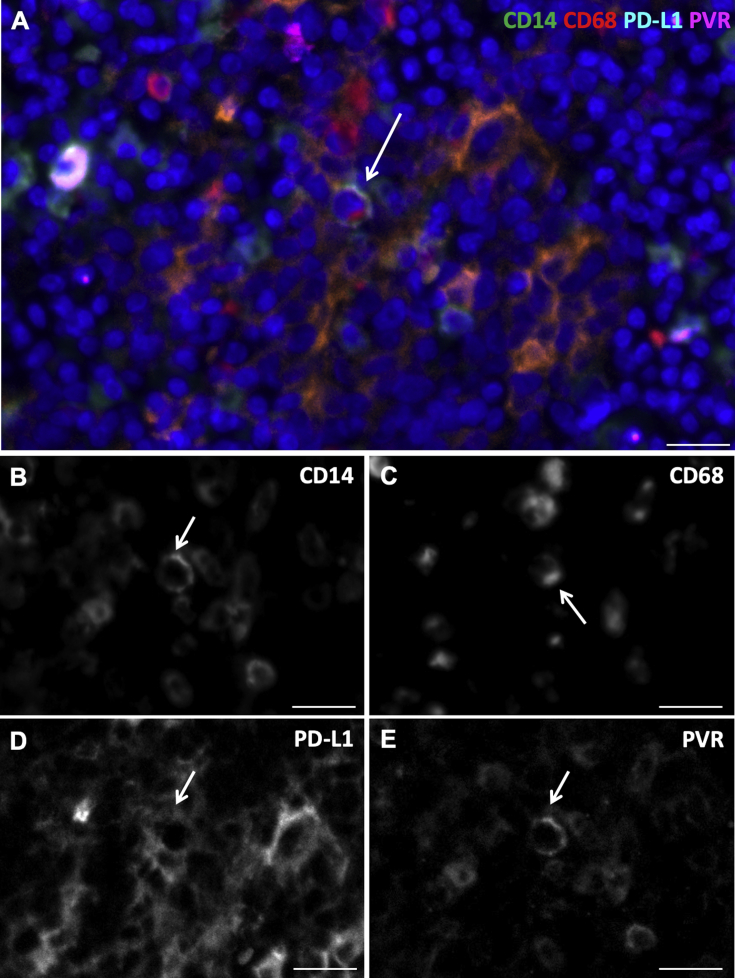
A: High-magnification immunofluorescence image of a cell (white arrow) with more prominent cytoplasmic/membrane immunostaining of CD14 (green), CD68 (red), programmed death ligand-1 (PD-L1; aqua blue), and poliovirus receptor (PVR; violet) within a patch of differentially co-expressing monocytes (arrow). B–E: The black and white images represent each raw channel as captured before look-up table color application, showing unequivocal presence of these markers on the cell with prominent cytoplasmic/membrane immunostaining (white arrows). CD68 (red) was not entirely localized with CD14 (green) in this cell, but instead offset to one side of the cell cytoplasm (more polarized distribution). Scale bar = 20 μm (A–E).
References
- 1.Pai S.I., Westra W.H. Molecular pathology of head and neck cancer: implications for diagnosis, prognosis, and treatment. Annu Rev Pathol. 2009;4:49–70. doi: 10.1146/annurev.pathol.4.110807.092158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehanna H., Beech T., Nicholson T., El-Hariry I., McConkey C., Paleri V., Roberts S. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer--systematic review and meta-analysis of trends by time and region. Head Neck. 2013;35:747–755. doi: 10.1002/hed.22015. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi A.K., Engels E.A., Pfeiffer R.M., Hernandez B.Y., Xiao W., Kim E., Jiang B., Goodman M.T., Sibug-Saber M., Cozen W., Liu L., Lynch C.F., Wentzensen N., Jordan R.C., Altekruse S., Anderson W.F., Rosenberg P.S., Gillison M.L. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howard J.D., Chung C.H. Biology of human papillomavirus-related oropharyngeal cancer. Semin Radiat Oncol. 2012;22:187–193. doi: 10.1016/j.semradonc.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyford-Pike S., Peng S., Young G.D., Taube J.M., Westra W.H., Akpeng B., Bruno T.C., Richmon J.D., Wang H., Bishop J.A., Chen L., Drake C.G., Topalian S.L., Pardoll D.M., Pai S.I. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013;73:1733–1741. doi: 10.1158/0008-5472.CAN-12-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., Diehn M., Alizadeh A.A. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plummer M., de Martel C., Vignat J., Ferlay J., Bray F., Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016;4:e609–e616. doi: 10.1016/S2214-109X(16)30143-7. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z., Chen L., Baker K., Olszak T., Zeissig S., Huang Y.H., Kuo T.T., Mandelboim O., Beauchemin N., Lanier L.L., Blumberg R.S. CEACAM1 dampens antitumor immunity by down-regulating NKG2D ligand expression on tumor cells. J Exp Med. 2011;208:2633–2640. doi: 10.1084/jem.20102575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lines J.L., Sempere L.F., Broughton T., Wang L., Noelle R. VISTA is a novel broad-spectrum negative checkpoint regulator for cancer immunotherapy. Cancer Immunol Res. 2014;2:510–517. doi: 10.1158/2326-6066.CIR-14-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lines J.L., Pantazi E., Mak J., Sempere L.F., Wang L., O'Connell S., Ceeraz S., Suriawinata A.A., Yan S., Ernstoff M.S., Noelle R. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 2014;74:1924–1932. doi: 10.1158/0008-5472.CAN-13-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biron C.A., Nguyen K.B., Pien G.C., Cousens L.P., Salazar-Mather T.P. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 13.Le Mercier I., Chen W., Lines J.L., Day M., Li J., Sergent P., Noelle R.J., Wang L. VISTA regulates the development of protective antitumor immunity. Cancer Res. 2014;74:1933–1944. doi: 10.1158/0008-5472.CAN-13-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu X., Harden K., Gonzalez L.C., Francesco M., Chiang E., Irving B., Tom I., Ivelja S., Refino C.J., Clark H., Eaton D., Grogan J.L. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 15.Kindt N., Journe F., Ghanem G.E., Saussez S. Galectins and carcinogenesis: their role in head and neck carcinomas and thyroid carcinomas. Int J Mol Sci. 2017;18:2745. doi: 10.3390/ijms18122745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukumori T., Takenaka Y., Yoshii T., Kim H.R., Hogan V., Inohara H., Kagawa S., Raz A. CD29 and CD7 mediate galectin-3-induced type II T-cell apoptosis. Cancer Res. 2003;63:8302–8311. [PubMed] [Google Scholar]
- 17.Peng W., Wang H.Y., Miyahara Y., Peng G., Wang R.F. Tumor-associated galectin-3 modulates the function of tumor-reactive T cells. Cancer Res. 2008;68:7228–7236. doi: 10.1158/0008-5472.CAN-08-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki Y., Sutoh M., Hatakeyama S., Mori K., Yamamoto H., Koie T., Saitoh H., Yamaya K., Funyu T., Habuchi T., Arai Y., Fukuda M., Ohyama C., Tsuboi S. MUC1 carrying core 2 O-glycans functions as a molecular shield against NK cell attack, promoting bladder tumor metastasis. Int J Oncol. 2012;40:1831–1838. doi: 10.3892/ijo.2012.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W., Guo H., Geng J., Zheng X., Wei H., Sun R., Tian Z. Tumor-released galectin-3, a soluble inhibitory ligand of human NKp30, plays an important role in tumor escape from NK cell attack. J Biol Chem. 2014;289:33311–33319. doi: 10.1074/jbc.M114.603464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Streetly M.J., Maharaj L., Joel S., Schey S.A., Gribben J.G., Cotter F.E. GCS-100, a novel galectin-3 antagonist, modulates MCL-1, NOXA, and cell cycle to induce myeloma cell death. Blood. 2010;115:3939–3948. doi: 10.1182/blood-2009-10-251660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vuong L., Kouverianou E., Rooney C.M., McHugh B.J., Howie S.E.M., Gregory C.D., Forbes S.J., Henderson N.C., Zetterberg F.R., Nilsson U.J., Leffler H., Ford P., Pedersen A., Gravelle L., Tantawi S., Schambye H., Sethi T., MacKinnon A.C. An orally active galectin-3 antagonist inhibits lung adenocarcinoma growth and augments response to PD-L1 blockade. Cancer Res. 2019;79:1480–1492. doi: 10.1158/0008-5472.CAN-18-2244. [DOI] [PubMed] [Google Scholar]
- 22.Stegmayr J., Zetterberg F., Carlsson M.C., Huang X., Sharma G., Kahl-Knutson B., Schambye H., Nilsson U.J., Oredsson S., Leffler H. Extracellular and intracellular small-molecule galectin-3 inhibitors. Sci Rep. 2019;9:2186. doi: 10.1038/s41598-019-38497-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farhad M., Rolig A.S., Redmond W.L. The role of galectin-3 in modulating tumor growth and immunosuppression within the tumor microenvironment. Oncoimmunology. 2018;7:e1434467. doi: 10.1080/2162402X.2018.1434467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression levels of nine immune checkpoint markers on myeloid cells isolated from five human tonsils using flow cytometry. A: Gating strategy for flow cytometry data. Lineage-negative population was defined as CD3–, CD19–, and CD16– cells. The gating strategy of four populations was shown, including human leukocyte antigen (HLA)-DR+CD14+ (blue), HLA-DR+CD14– (cyan), CD68+CD14+ (red), and CD68+CD14– (orange). The numbers indicate the frequency of parent population. B: The expression levels of poliovirus receptor (PVR), programmed death ligand-1 (PD-L1), cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), T-cell immunoglouglin and mucin-domain containg-3 [TIM-3, also knowns as hepatitis A virus cellular receptor 2 (HAVCR2)], B- and T-lymphocyte attenuator (BTLA), V-domain Ig suppressor of T-cell activation (VISTA), E-cadherin (CDH1), carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), and indoleamine 2,3-dioxygenase 1 (IDO1) are shown for four different monocyte and macrophage populations defined in Supplemental Figure S1A. The unstained controls (top row of panel B, black box) are shown. The numbers in the quadrant are the frequencies of each gate. FSC, forward scatter; SSC, side scatter.