Abstract
The paired-homeodomain transcription factor PAX4 is expressed in the developing pancreas and along with PAX6 is required for normal development of the endocrine cells. In the absence of PAX4, the numbers of insulin-producing β cells and somatostatin-producing δ cells are drastically reduced, while the numbers of glucagon-producing α cells are increased. To gain insight into PAX4 function, we cloned a full-length Pax4 cDNA from a β-cell cDNA library and identified a bipartite consensus DNA binding sequence consisting of a homeodomain binding site separated from a paired domain binding site by 15 nucleotides. The paired half of this consensus sequence has similarities to the PAX6 paired domain consensus binding site, and the two proteins bind to common sequences in several islet genes, although with different relative affinities. When expressed in an α-cell line, PAX4 represses transcription through the glucagon or insulin promoters or through an isolated PAX4 binding site. This repression is not simply due to competition with the PAX6 transcriptional activator for the same binding site, since PAX4 fused to the unrelated yeast GAL4 DNA binding domain also represses transcription through the GAL4 binding site in the α-cell line and to a lesser degree in β-cell lines and NIH 3T3 cells. Repressor activity maps to more than one domain within the molecule, although the homeodomain and carboxyl terminus give the strongest repression. PAX4 transcriptional regulation apparently plays a role only early in islet development, since Pax4 mRNA as determined by reverse transcriptase PCR peaks at embryonic day 13.5 in the fetal mouse pancreas and is undetectable in adult islets. In summary, PAX4 can function as a transcriptional repressor and is expressed early in pancreatic development, which may allow it to suppress α-cell differentiation and permit β-cell differentiation.
During development, the mammalian pancreas arises from the epithelial cells of the embryonic gut at the foregut-midgut junction and differentiates into two distinct compartments: the exocrine tissue, which produces digestive enzymes, and the endocrine islets of Langerhans, which produce specific hormones. The islets are arranged into a core of insulin-producing β cells surrounded by a mantle of glucagon-producing α cells, and smaller numbers of somatostatin- and pancreatic polypeptide-producing cells (δ and PP cells, respectively) (34).
The coordinated regulation of gene expression required for normal pancreatic development is not completely understood but clearly requires the orderly activation of nuclear transcription factors by both intracellular and extracellular signals. Several transcription factors (PDX1, ISL1, PAX6, PAX4, BETA2/NeuroD, and NKX2.2) are required for normal pancreatic endocrine development, and many of these factors also regulate gene expression in mature islet cells (1, 2, 19, 24, 26, 32, 35–37). However, one of these factors, PAX4, has been identified only as a regulator of endocrine development, and its target genes are unknown (35).
PAX4 belongs to the PAX family of transcription factors and contains both a paired domain and a homeodomain (18, 42) which are potential DNA binding domains (DBDs). In the normal murine embryo, its mRNA is detected at embryonic day 9.5 (e9.5) in ventral spinal cord and pancreas (35). Indirect evidence from mice containing the β-galactosidase coding sequence inserted into the Pax4 gene suggests that at birth PAX4 expression is restricted to the β cells within the pancreas (35). Its critical role in pancreatic endocrine development is demonstrated by the fact that mice homozygous for a null mutation in the Pax4 gene have a marked decrease in β and δ cells and an increase in α cells, although the mechanism for these changes is undefined (35). Importantly, insulin-expressing cells are detected in the null mutants at e10.5, suggesting that insulin transcription can occur in the absence of PAX4. Ultimately however, the null mutants die within a few days of birth, apparently as a consequence of insulin deficiency. Heterozygotes containing a single mutated Pax4 allele are normal. It is interesting that PAX6, which is highly related to PAX4, is also required for normal endocrine pancreatic development (36), although its absence reduces all four endocrine lineages (32). Furthermore, double null mutants for both Pax4 and Pax6 fail to produce any mature pancreatic endocrine cells (36), suggesting that these two factors together are required for endocrine cell differentiation.
To gain insight into the mechanisms of PAX4 function in the endocrine pancreas, we determined where it binds and how it regulates transcription. We identified a consensus DNA binding site for PAX4 and showed that PAX4 can bind to various sequences in the rat insulin I, somatostatin, and glucagon promoters, all of which have previously been shown to bind PAX6 (32). We found that PAX4 can act as a transcriptional repressor and showed that the homeodomain and carboxyl portion of the molecule confers the greatest repressive activity. Finally, by reverse transcriptase PCR (RT-PCR), we demonstrate that PAX4 expression peaks early in pancreatic development and that PAX4 is not expressed in mature islets.
MATERIALS AND METHODS
Cloning of murine Pax4.
The Pax4 cDNA was cloned from a βTC3 cell line plasmid library (a generous gift from D. Hanahan, University of California, San Francisco) by PCR, using a high-fidelity system (Expand PCR; Boehringer Mannheim, Indianapolis, Ind.), producing two overlapping clones. The 5′ clone was produced by PCR using the SP6 forward primer (located in the vector) and a reverse primer beginning at the 3′ end of the paired box (5′-CGGAATTCCTGAAGTGCCCGAAGTACTCG-3′). This clone extends 18 bp 5′ of the initiator ATG codon. The 3′ clone was produced by using a forward primer from the 5′ end of the paired box (5′-CGCGGATCCCTCAGCAGTGTGAATCAGCT-3′) and the T7 reverse primer (located in the vector). The two clones span 1.1 kb of the Pax4 cDNA and include the entire predicted coding sequence.
In vitro protein expression.
To produce a PAX4 expression vector, the two overlapping clones were digested and then religated at an FseI site located between 43 to 50 bp into the paired box. The full-length coding sequence was inserted into the pSP72 vector (Promega, Madison, Wis.), enabling protein expression with the bacterial SP6 promoter. A truncated coding sequence, terminating at codon 274, was also created in the same vector because the full-length PAX4 (349 amino acids [aa]) was not expressed with adequate efficiency. The PAX6 expression vector (pmPAX6) was a gift from M. Busslinger, Research Institute of Molecular Pathology, Vienna, Austria (4). PAX4 and PAX6 proteins were produced by in vitro transcribed/translated (IVTT) reaction, using the TNT coupled reticulocyte lysate system (Promega) according to the manufacturer’s instructions.
PAX4 binding site selection.
To select for PAX4 binding sequences by using a published technique (3), a 76-mer sequence with 30 central random bases was used as a probe in an electrophoretic mobility shift assay (EMSA). The sequence of the top strand of the 76-mer was 5′-GTGACCAGATCTAATCGTGGTCCTN30ACGGTCGACGAGTACGCGTTAC-3′. Radiolabeling of the second strand with [α-32P]dCTP was performed with Klenow polymerase, deoxynucleoside triphosphates, and the primer 5′-GTAACGCGTACTCGTCGACCGT-3′ (REV). The labeled, double-stranded random sequences were incubated with in vitro-produced truncated PAX4 and electrophoresed under nondenaturing conditions. The specific retarded complex was cut out of the gel, and the selected 76-mers were eluted. The eluted 76-mers were then amplified and relabeled by using 16 cycles of PCR with [α-32P]dCTP, the REV primer, and the forward primer 5′-GTGACCAGATCTAATCGTGGTCCT-3′ (FWD). This PCR product was used as probe for the next round of EMSA selection. A total of 13 selection cycles were performed, and the selected 76-mers were subcloned into pCR2.1 (Invitrogen) and sequenced after the 10th and 13th cycles.
EMSA.
Single-stranded oligonucleotide probes were 5′ end labeled with [γ-32P]ATP by using T4 polynucleotide kinase. Labeled oligonucleotides were column purified and annealed to an excess of the complementary strand. EMSA buffers and electrophoresis conditions were as previously described (13). Where in vitro-synthesized PAX4 or PAX6 was used, 0.05 to 1 μl of the 50-μl reaction volume was added.
The oligonucleotide probes used in EMSA, and their locations in relation to the transcriptional start site for the gene, are as shown (top strand): rat insulin I C2 element (rInsIC2), bp −328 to −304, 5′-CTGGGAAATGAGGTGGAAAATGCTC-3′; rat somatostatin upstream enhancer (SMS.UE), bp −102 to −76, 5′-GCGAGGCTAATGGTGCGTAAAAGCACT-3′; rat glucagon G3 (GluG3), bp −265 to −231, 5′-TTTTTCACGCCTGACTGAGATTGAAGGGTGTATTT-3′; and rat glucagon G1 (GluG1), bp −100 to −52, 5′-CAAAACCCCATTATTTACAGATGAGAAATTTATATTGTCAGCGTAATAT-3′.
Cell culture and transient transfections.
A PAX4 eukaryotic expression vector was produced by subcloning the full-length Pax4 cDNA into a vector (pBAT12) containing the human cytomegalovirus (CMV) immediate-early gene promoter. pBAT12 is identical to pBAT7 (14) except for a modification in the polylinker and replacement of the early simian virus 40 polyadenylation signal with the late simian virus 40 polyadenylation signal. The reporter constructs were created by using either the −410 bp rat insulin I promoter, the −482 bp rat glucagon promoter, the Rous sarcoma virus (RSV) long terminal repeat (LTR), the CMV promoter, or the isolated rInsIC2 fragment linked to the herpes simplex virus thymidine kinase (HSV-TK) promoter to drive the chloramphenicol acetyltransferase (CAT) gene in the pFOXCAT2 vector (25).
One-hybrid expression vectors encoding PAX4- or PAX6-GAL4 DBD fusion proteins were made by amplifying the appropriate coding fragments of PAX4 or PAX6 by PCR and then ligating the fragment in frame into the EcoRI and BamHI sites of the GAL4 DBD vector (pM; Clontech). The GAL4-TK reporter plasmids were created in the pFOX2 plasmid backbone (25) by ligating a single copy of the GAL4 upstream activating sequence (UAS) to the HSV-TK promoter driving the CAT gene (plasmid pFOXCAT2.TK.1GAL) or five copies upstream of the TK promoter driving the fire fly luciferase gene (plasmid pFOXLuc2.TK.5GAL). The other GAL4 reporter plasmid, with a much lower basal promoter activity, consists of five GAL4 binding sites linked to the minimal adenovirus E1b promoter driving CAT expression (pG5CAT; Clontech). The CMV green fluorescent protein plasmid (pFOXEGFP2.CMV) was constructed with the CMV promoter upstream of the enhanced green fluorescent protein (EGFP) cDNA (Clontech).
NIH 3T3, αTC1.6, βTC3, and HIT-T15 M2.2.2 cells were cultured, transfected with calcium phosphate, and harvested as previously described (13). CAT assays and luciferase assays were performed as previously described, using 10 μg of cell protein extract (13, 14).
Mouse pancreatic buds were isolated from fetal mice at e12.5, and the mesenchyme and epithelium were separated as previously described (15). The epithelium from each bud was individually transfected with 3 μg of the plasmid DNAs indicated and 9 μg of the cationic lipid transfection reagent Transfast (Promega). After transfection, the epithelium was washed and recombined with the mesenchyme on a surface of Matrigel (Collaborative Research). After culturing for 24 h, the buds were harvested and RT-PCR was performed.
RT-PCR.
Total RNA was purified from the indicated tissue source by using an RNAqueous kit (Ambion, Austin, Tex.) and quantified by spectrophotometry. Using a modification of the method described by Gittes and Rutter (16), 250 ng of total RNA was then used as template to synthesize first-strand cDNA by the action of avian myeloblastosis virus reverse transcriptase primed by a poly(dT) primer. The cDNA produced was used as a template for the indicated number of cycles of PCR using primers that amplify fragments of the indicated cDNA (PAX4 primers 1 [GTGTTGGCTCCAGTTCTTCC] and 2 [AACCAAACCCTCACCGTGTC]; β-actin primers 1 [TGAGAGGGAAATCGTGCGTG] and 2 [TGCTTGCTGATCCACATCTGC]; mouse insulin II primers 1 [GCCCTAAGTGATCCGCTACAATC] and 2 [GCAGCACTGATCTACAATGCCAC]; luciferase and EGFP primer 1 [TGGGCAGGCTGCTGGTCTGAG]; luciferase primer 2 [TGCTCTCCAGCGGTTCCATC]; EGFP primer 2 [TCAGGGTCAGCTTGCCGTAGG]) Each set of primers was designed to include at least one intron in order to allow the discrimination of contaminating genomic or plasmid DNA from cDNA. The cycling program used for all samples was 95°C for 2 min and then 25 to 40 cycles of 95°C for 20 s, 55°C for 30 s, and 72°C for 30 s. Samples were analyzed on a 6% polyacrylamide gel and stained with ethidium bromide. Identity of the amplified products was confirmed by subcloning and DNA sequencing. PCR performed for luciferase and EGFP in the transfected pancreatic buds included 100,000 cpm of [α-32P] dCTP. PCR products were quantified with a phosphorimager and expressed as the ratio of luciferase cDNA to EGFP cDNA.
RESULTS
Cloning of Pax4.
To study the function of PAX4, we cloned the Pax4 cDNA from a β-cell tumor line (βTC3) plasmid cDNA library. The predicted amino acid sequence of PAX4 is identical to a recently published sequence derived from the MIN6 insulinoma cell line (18), except for a proline at position 277 in our sequence, which has been replaced with a serine. PAX4 is therefore a 349-aa protein with its 128-aa paired domain beginning at the fifth residue after the initiator methionine. A 36-aa intervening sequence separates the paired domain from the 60-aa homeodomain, and a further 121 aa extend from the homeodomain to the carboxyl terminus.
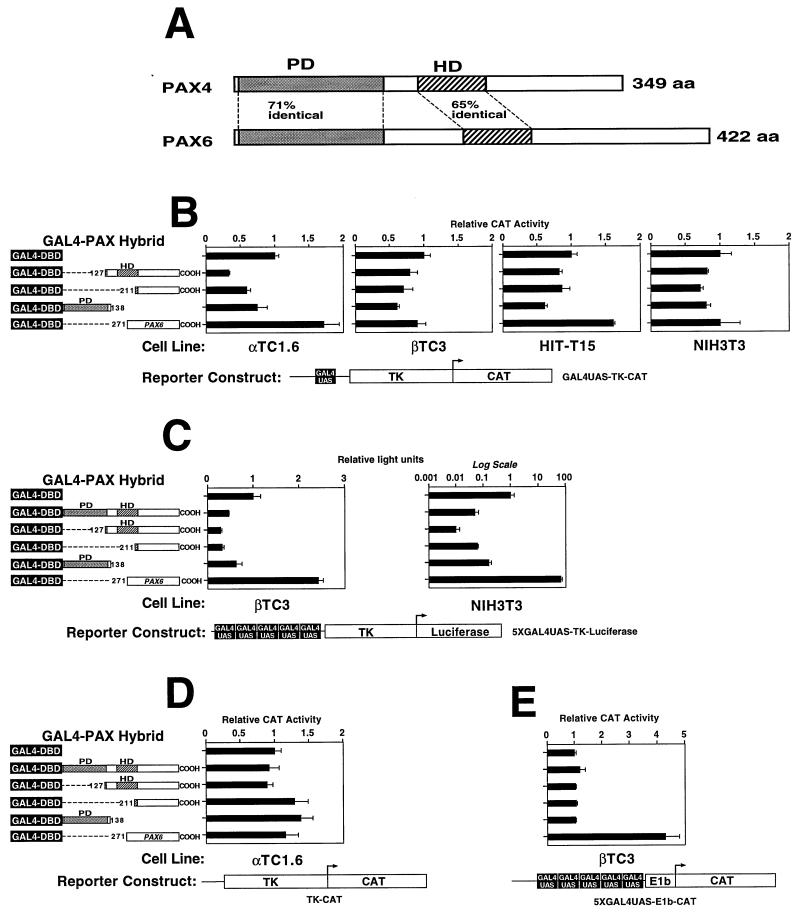
Compared to PAX6, the paired domains of both proteins begin 4 to 5 aa from the start codon. PAX6 is a substantially longer protein than PAX4 (422 versus 349 aa), due to a longer intervening sequence between the paired and homeodomains (77 versus 36 aa) and a longer C-terminal tail beyond the homeodomain (154 versus 121 aa). The two proteins are 70% identical in their paired domains and 65% identical in their homeodomains (see Fig. 6A). Outside the highly conserved paired and homeodomains, PAX4 and PAX6 share no obvious sequence similarity.
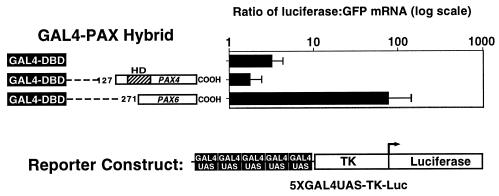
FIG. 6.
PAX4 transcriptional repression in fetal pancreatic epithelium. Transient transfections were performed in mouse e11.5 pancreatic epithelium with expression vectors containing the GAL4 DBD coding sequence fused with the PAX4 and PAX6 cDNA fragments shown, the pFOXLuc2.TK.5GAL reporter plasmid, and the pFOXEGFP.CMV internal standard. Transfections were performed in triplicate on individual fetal pancreatic buds. Luciferase mRNA levels were measured by RT-PCR and standardized to EGFP mRNA levels. The graph shows mean ± standard error of the mean.
PAX4 binding site selection.
To gain insight into PAX4 function, we attempted to determine PAX4 recognition sequences by using a previously described strategy (3) and ultimately to identify its downstream target genes. To select oligonucleotides with high PAX4 binding affinity, we incubated a pool of radiolabeled oligonucleotide probes containing a central core of 30 random bases with in vitro-produced PAX4 and performed an EMSA. We used a C-terminally truncated in vitro-produced PAX4 because the synthesis of full-length PAX4 was less efficient (data not shown). The truncated protein is 274 aa instead of 349 aa but contains both the paired and homeodomains.
After the first EMSA, the specific PAX4-DNA retarded complex was cut from the gel and the bound oligonucleotides were eluted. PCR was performed to amplify and to radiolabel the selected oligonucleotides, which were then used for the next round of EMSA selection. Oligonucleotide products were subcloned for sequencing after the 10th and 13th rounds of selection. All of the sequences obtained from the 13th round of selection were subjected to an ungapped alignment using the multiple sequence alignment function in the MacVector 6.5 software package (Oxford Molecular Ltd.). This alignment reveals a bipartite consensus (Fig. 1A). The 3′ sequence generally conforms to the consensus DNA binding sequences for other paired domains (20) and shares a six-nucleotide overlap with the PAX6 consensus binding sequence (Fig. 1E) obtained by using the paired domain alone (7). The 5′ sequence contains the overlapping TAAT sequence found in many binding sites for homeodomain transcription factors (39).
FIG. 1.
PAX4 binding site selection. In vitro-produced truncated PAX4 protein including the paired domain and homeodomain was used in EMSA to select sequences with high binding affinity from a random pool of oligonucleotides containing 30 degenerate base pairs flanked by PCR primer sites. (A) Alignment of sequences of oligonucleotide products subcloned and sequenced after 13 rounds of selection. Sequences are aligned by the MacVector multiple-sequence alignment function to show the best fit. (B) Alignment of homeodomain type sequences after 13 rounds of selection. (C) Alignment of paired domain-type sequences after 13 rounds of selection. (D) Alignment of oligonucleotide products subcloned and sequenced after 10 rounds of selection. TAAT sequences are underlined. (E) The consensus PAX4 recognition sequence established from the aligned sequences compared to the PAX6 consensus recognition sequence, based on paired domain binding alone (7). Matching bases are in boldface; rat hormone gene regulatory elements with possible PAX4 binding sites are underlined.
Optimally alignment of the sequences for the paired domain site (Fig. 1C) reveals a 6-bp consensus, ANNN(C/T)CACCC, that varies by only one base from the core of the PAX6 consensus binding sequence obtained by using the paired domain alone (Fig. 1E) (7). Similarly, when the sequences are optimally aligned for the homeodomain site (Fig. 1B), an 8-bp consensus, AA(T/A)AATTA, that conforms well with common homeodomain binding motifs (20, 39, 43) is apparent.
Given the wide space between the sites, we cannot rule out the possibility that optimal homeodomain or paired domain sites might include additional bases on the 5′ or 3′ ends, respectively, that we cannot detect because our degenerate binding site was 30 bp long. The paired domain actually includes two DBDs the PAI and RED domains (4, 8, 20, 44). Our paired domain consensus sequence lacks some of the bases on the 3′ end that are usually associated with RED binding. We cannot be certain if this is due to differences in the RED domain in PAX4, the presence of the homeodomain, or the limitations of our oligonucleotide design. It should be noted that not all paired-homeodomain proteins contact DNA through the RED domain; the intact Drosophila Paired protein (Prd) binds only with its homeodomain and PAI domain (20).
Comparison of the round 13 consensus with the sequences obtained in round 10 demonstrates that most of the round 10 sequences also contain both the paired binding site and the homeodomain binding site (Fig. 1D). As in round 13, the TAAT motifs are 5′ relative to the paired site, but the exact position varies. It should be kept in mind that the distance between the homeodomain and paired domain binding sites is constrained by the size of the degenerate oligonucleotides, leaving open the possibility of a longer intervening sequence as well.
PAX4 binds to hormone gene promoters.
Comparison of the consensus PAX4 binding sequence with the insulin promoter sequence shows that a sequence matching five of the seven bases in the consensus is found in rInsIC2 (Fig. 1E). As this element has been shown to bind strongly to PAX6 (32), and as PAX4 and PAX6 recognition sequences share some overlap, we compared the PAX4 consensus sequence to sequences of other known PAX6 binding sites on islet hormone gene promoters, namely, rat SMS.UE and rat GluG3 (32). In both SMS.UE and GluG3, there are sites matching the PAX4 consensus (Fig. 1E). We also tested for PAX4 and PAX6 binding to rat GluG1, as it shares sequence similarity to GluG3 (21, 29) and binds similar protein complexes (29). The GluG1 sequence contains two potential PAX4 paired binding sites, although with only four matching bases at each site, and two TAAT sequences (Fig. 1E).
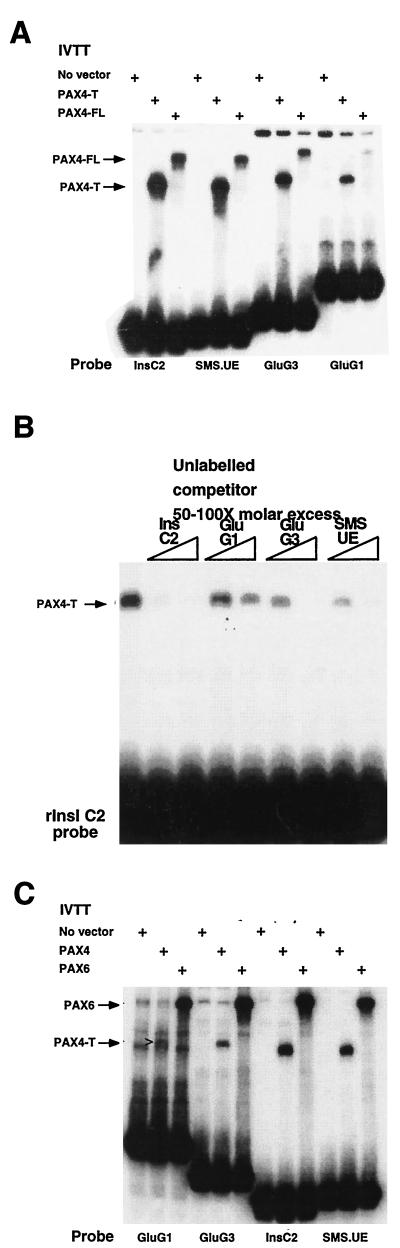
Oligonucleotides including these potential binding sites were end labeled and used as probes in EMSA, initially to compare the DNA binding characteristics of the truncated and full-length PAX4. PAX4 binds with highest affinity to rInsIC2, followed by SMS.UE and GluG3 (Fig. 2A). Binding to GluG1 is substantially weaker than binding to the other oligonucleotides (Fig. 2A). Importantly, we show that the truncated and full-length proteins have the same relative affinities for the different oligonucleotide probes.
FIG. 2.
PAX4 binding to islet gene promoters. (A) EMSA using truncated (PAX4-T) and full-length (PAX4-FL) PAX4 to bind to hormone gene regulatory elements. 32P-end-labeled oligonucleotides (10,000 cpm) were incubated with 1 μl of a 50-μl IVTT reaction in each lane and then electrophoresed. (B) EMSA using PAX4-T to bind to 32P-end-labeled rInsIC2 probe (10,000 cpm). A 50- to 100-fold molar excess of unlabeled competitor oligonucleotides was added to the lanes indicated; 0.05 μl of a 50-μl IVTT reaction was used in each lane. (C) EMSA comparing the binding of PAX4-T and PAX6 to 32P-end-labeled hormone gene regulatory elements; 0.1 μl of a 50-μl IVTT reaction was used in each lane. The open arrowhead (>) points to a weak PAX4-T–GluG1 complex.
To confirm the relative binding affinities of PAX4 to these elements, EMSA was performed with rInsIC2 as the probe, as it has the highest affinity for PAX4, and the other elements were used as unlabeled competitors. The most effective competitor is rInsIC2, followed by SMS.UE, GluG3, and finally GluG1 (Fig. 2B). These results confirm that PAX4 has the following relative binding affinities: rInsIC2 > SMS.UE > GluG3 > GluG1.
We then compared binding of PAX4 and PAX6 to the same oligonucleotide probes. An equal amount of each protein, assessed by [35S]Met incorporation (not shown), was used in each EMSA binding reaction. EMSA shows that PAX6 has a substantially higher affinity for all of the oligonucleotides than does the truncated PAX4 and has the following relative affinities: GluG3 > rInsIC2 > GluG1 = SMS.UE (Fig. 2C and reference 32). We also tested but could not demonstrate PAX4 binding to several other β-cell promoter elements, including the human insulin (hIns) C2 element, the rIns and hIns C1 elements, rInsI A3/4 element, the hIns Z element, and the rat β-cell-specific glucokinase promoter Pal1 and Pal2 sites (11, 12, 31, 33) (data not shown).
PAX4 represses transcription.
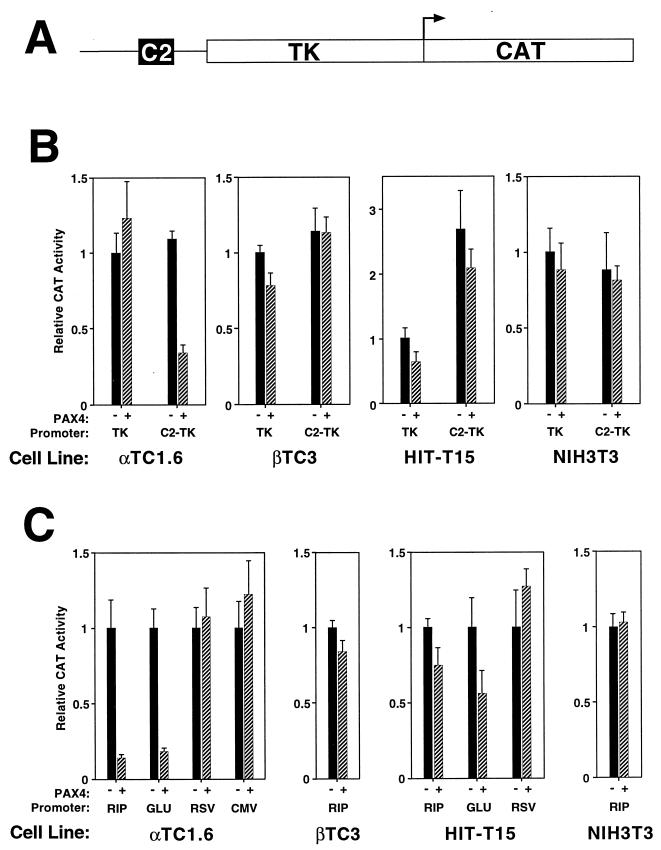
We used the high affinity PAX4 C2 binding site to assess the transcriptional effect of PAX4 in cultured cells. A single copy of the isolated C2 element linked to the HSV-TK promoter was used to drive CAT gene expression. PAX4 expression was driven by the CMV immediate-early promoter in the eukaryotic expression vector pBAT12-PAX4. Expression and reporter constructs were cotransfected into α (αTC1.6), β (βTC3 and HIT-T15 M2.2.2), and nonislet (NIH 3T3) cell lines, and CAT activity was assessed.
In the β-cell lines and NIH 3T3 cells, PAX4 has little effect on the transcriptional activity of the isolated C2 element (Fig. 3B). In contrast, in αTC1.6 cells, PAX4 represses transcription from the isolated C2 element approximately threefold but has no effect on the TK promoter in the absence of the C2 element.
FIG. 3.
PAX4 transcriptional repression. Transient transfections of the indicated cell lines were performed with the PAX4 expression vector and reporter constructs consisting of the HSV-TK promoter alone (TK) or the isolated rInsIC2 element linked to TK-HSV (C2-TK) (A) and the −410-bp rat insulin I promoter (RIP), −482-bp rat glucagon promoter (GLU), RSV LTR (RSV), or CMV promoter (CMV) driving CAT expression. Transfections were performed in duplicate on at least three separate occasions. Relative CAT activity of the TK-CAT reporter in the absence of PAX4 expression is set arbitrarily at +1. In panel B, relative CAT activity of each construct in the absence of PAX4 expression is set arbitrarily at +1. Graphs show mean ± standard error of the mean.
Confirming the results with the isolated C2 binding site, PAX4 also represses transcription from the intact insulin and glucagon promoters (Fig. 3C). Repression is again seen predominantly in the αTC1.6 cells but does not occur with the unrelated RSV and CMV promoters (Fig. 3C). These results demonstrate that PAX4 represses transcription through its binding site and that this effect may depend on the cellular context.
PAX4 contains transcriptional repression domains.
Repression by PAX4 could result from competition for binding at the rInsIC2 site with a strong activator (such as PAX6) or could reflect an intrinsic repressor function of the PAX4 molecule. To distinguish between these two possibilities, we made chimeric proteins by fusing portions of PAX4 to the yeast GAL4 DBD. We also used the C-terminal region of PAX6 (codon 271 to COOH) as a known positive activating domain (17, 32). To allow direct comparison to the experiments using the C2 site, an analogous reporter construct was constructed by simply replacing the C2 element in the C2-TK-CAT plasmid with a single GAL4 DNA-binding site (the GAL4 UAS). The fusion protein constructs and reporter constructs were cotransfected into cultured cells. This approach allows for DNA binding through the GAL4 DBD, independent of the PAX4 paired and homeodomains.
In αTC1.6 cells, the region of PAX4 C terminal to the paired domain (codon 127 to COOH) represses CAT activity to approximately 40% of baseline (Fig. 4B), a degree of repression similar to that produced by intact PAX4 acting through an isolated C2 site. The PAX4 carboxyl terminus beyond the homeodomain (codon 211 to COOH) can also repress CAT activity. While there is some weak repression in the other cell lines, the α-cell line gives the strongest repression, consistent with the α-cell repression demonstrated by the intact PAX4 protein on the C2 element (Fig. 3B). When a reporter construct that contains multiple GAL4 UAS binding sites is used, the repression by GAL4-PAX4 fusion proteins is more pronounced and can be detected in NIH 3T3 and βTC3 cells as well (Fig. 4C).
FIG. 4.
PAX4 repressor domains. (A) Comparison of PAX4 and PAX6 structure. PD, paired domain; HD, homeodomain. (B) Transient transfections of the indicated cell lines were performed with the chimeric PAX4-GAL4 DBD fusion protein constructs shown and a reporter construct consisting of the GAL4 binding site linked to HSV-TK driving CAT expression (GAL4UAS-TK-CAT). (C) Transient transfections of βTC3 and NIH3T3 cells were performed with a reporter construct consisting of five copies of the GAL4 GAL4 UAS. (D) Transient transfections of αTC1.6 cells were performed with a reporter construct consisting of HSV-TK driving CAT expression without a GAL4 binding site (TK-CAT). (E) Transient transfections of βTC3 cells were performed with a reporter consisting of five GAL4 binding sites linked to the minimal adenovirus E1b promoter driving CAT expression (5XGAL4UAS-E1b-CAT). Note that the lowermost expression construct consists of the PAX6 C-terminal domain fused to the GAL4 DBD. Transfections were performed in duplicate on at least three separate occasions. Relative CAT activity of the GAL4-DBD expression construct alone is set arbitrarily at +1. Graphs show mean ± standard error of the mean.
To demonstrate that PAX4 repression in this assay requires specific DNA binding between the GAL4 DBD and its cognate recognition sequence (GAL4 UAS), we used a reporter construct consisting of the HSV-TK promoter driving CAT but without a GAL4 UAS. None of the PAX4-GAL4 fusion constructs produced a repressive effect when cotransfected with this reporter in αTC1.6 cells (Fig. 4D). Thus, the repressive effects seen in this system can occur only when the expressed chimeric PAX4-GAL4 DBD proteins bind specifically to a promoter containing the GAL4 UAS.
To ensure that the transcriptional activation potential of PAX4 was not hidden by the high basal activity of the HSV-TK promoter, we also tested the same fusion constructs with a construct containing five GAL4 UASs linked to the adenovirus E1b promoter driving CAT expression (pG5CAT; Clontech). Even with this minimal promoter, the PAX4 constructs do not activate in βTC3 cells, whereas stimulatory activity of the PAX6 carboxyl terminus is obvious (Fig. 4E).
PAX4 is not expressed in mature β cells.
Transgenic experiments have previously suggested that PAX4 is expressed specifically in pancreatic β cells, making its role as a transcriptional repressor, especially of the insulin promoter, difficult to understand. Because of this inconsistency and our inability to detect PAX4 protein in mature β cells by EMSA or Western blot analysis (data not shown), we tested for the expression of Pax4 mRNA by RT-PCR.
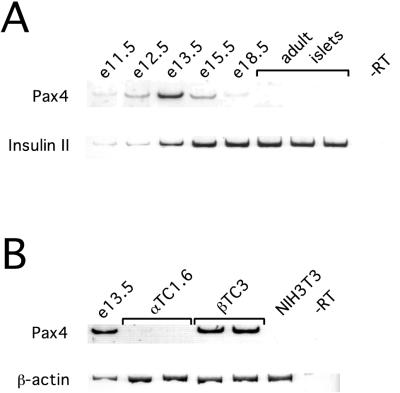
Pax4 mRNA can be detected in the fetal mouse pancreas, peaking at e13.5, but no Pax4 mRNA can be detected in mature mouse islets (Fig. 5A), even when PCR is extended to 40 cycles (data not shown). Pax4 mRNA can also be detected in βTC3 cells, but not in the NIH 3T3 fibroblast cells (Fig. 5B). Pax4 mRNA is expressed at much lower level in αTC1.6 cells, and a faint band can be detected in Fig. 5B, although this is more obvious with additional cycles of PCR (data not shown).
FIG. 5.
Expression of Pax4 mRNA. RT-PCR was performed with 250 ng of total RNA from the tissues shown. PCR products were removed and separated by polyacrylamide gel electrophoresis after 30 cycles for PAX4 and β-actin, and 25 cycles for insulin II. Lanes labeled-RT show the product of identical samples performed without reverse transcriptase using RNA from e13.5 fetal mouse pancreas (A) and βTC3 cells (B).
Given its expression in the fetal pancreas, we tested the transcriptional function of PAX4 in the fetal pancreas. Pancreatic epithelium from E12.5 mouse fetuses was cotransfected with the GAL4-PAX4 expression vector containing the region of PAX4 C-terminal to the paired domain (codon 127 to COOH) and a reporter plasmid containing five copies of the GAL4 UAS upstream of the TK promoter driving the firefly luciferase gene. A plasmid with the CMV promoter driving an EGFP cDNA was included as an internal standard. After 24 h, transcriptional activity was gauged by measuring luciferase mRNA levels by RT-PCR and normalizing with EGFP mRNA levels. The results demonstrate that as in the cell lines, the PAX4 carboxy terminus represses transcription, unlike PAX6, which activates transcription (Fig. 6).
DISCUSSION
In these studies, we selected DNA oligonucleotides that bind to a PAX4 protein that contains both the paired domain and the homeodomain. Sequences of the selected oligonucleotides reveal the presence of two linked sites: a consensus homeodomain binding site and a consensus paired domain binding site. The relative position of these two sites appears to be important, since in the more highly selected set of oligonucleotides the spacing becomes fixed. Both PAX3 (28) and Drosophila Prd (20) have also been found to bind with high affinity to sequences that include both paired and homeodomain binding sites. Interestingly, both on a natural site in the even-skipped gene (9) and in sites selected by a method similar to the one used here (20), the two binding sites for Prd are much closer than we found for PAX4, a difference of approximately one helical turn. For both PAX4 and Prd, the paired and homeodomain binding sites lie on opposite sides of the DNA.
The paired-like half of the bipartite DNA binding sequence for Pax4 is similar but not identical to the consensus binding site for the PAX6 paired domain (7). The differences from the PAX6 binding sequence could be due to the fact that we used a PAX4 protein containing both the paired and homeodomains for binding site selection, while only the paired domain was used for determining the PAX6 consensus sequences. The presence of both domains may alter the specificity of each, as is the case for PAX3 (40). However, differences in DNA binding specificity were confirmed by using a set of paired domain binding sites from islet genes that demonstrate different relative affinities for the full-length PAX4 and PAX6 proteins. The presence and exact position of a linked homeodomain binding site could potentially increase these differences. Furthermore, in the intranuclear environment, within the context of chromatin structure and other interacting proteins, the differences in binding affinities may be further accentuated. Nonetheless, these studies suggest that PAX4 and PAX6 could potentially compete for some binding sites.
Our transient transfection experiments show that PAX4 can repress transcription through a single isolated binding site linked to a promoter or through a binding site in the context of an intact promoter. Furthermore, the experiments with GAL4 DBD fusion constructs mapped this repressor function predominantly to the region of PAX4 C terminal to the paired domain. DNA binding is necessary for repression but may be mediated by a heterologous DBD, such as the yeast GAL4 DBD, in place of the paired and homeodomains. Transcriptional repression is not unique within the PAX family, having been shown with PAX5 (5, 41) and the Drosophila paired domain family member, Goosecoid (23). More recently, PAX6 has also been shown to act as a transcriptional repressor, requiring both its paired domain and homeodomain for this effect on β-crystallin genes in the lens (6). This finding suggests that PAX6 repression may be due to competition with transcriptional activators for DNA binding sites. However, chimeric proteins consisting of the GAL4 DBD fused to various regions of PAX6 show that some regions of PAX6 may also have modest intrinsic repressor activity (38).
It is becoming increasingly recognized that some transcription factors can act as either repressors or activators, depending on the cell type, promoter context, and concentration of the factor (reviewed in references 22, 27, and 30). Indeed, within the PAX family, both PAX5 (BSAP) and PAX6 have recently been shown to have such a dual function (6, 41). However, it is important to note that in many studies, transcriptional effects are assessed with DNA regulatory elements removed from their native chromosomal locations, and using highly efficient vectors to express the transcription factors, potentially resulting in supraphysiological intranuclear concentrations of the factors. In addition, differential splicing of the Pax4 mRNA within the region coding the carboxy end of the protein has been detected and could affect the balance of repressor/activator function. Thus, our studies do not exclude the possibility that PAX4 can function as a transcriptional activator in some contexts. In fact, Fujitani et al. (10) have now shown that in certain cellular contexts, portions of PAX4 may function as transcriptional activators.
Even if PAX4 can function as a transcriptional activator, it seems unlikely that it is an important regulator of islet hormone gene expression in vivo. Despite the high affinity of PAX4 for rInsIC2 and the evidence that PAX4 may be expressed in early β cells (35), insulin transcription persists in animals lacking PAX4 (35), and we have failed to detect activation of the insulin promoter by PAX4 in any context. PAX4 could potentially suppress insulin transcription in α cells, but PAX4 expression has not been detected in α cells in vivo (35). Finally, its absence from adult islets rules out a role for PAX4 in maintaining the differentiated phenotype of the mature endocrine cells.
Could opposing effects on transcription by PAX4 and PAX6 modulate the expression of target genes? While PAX4 and PAX6 could potentially compete for some binding sites, their differences in binding specificity suggest that the two proteins may have different target genes. There also may be little or no overlap in the expression of the two proteins since PAX4 is expressed in developing islet cells, and PAX6 is expressed in the fully differentiated islet cells (32).
On the other hand, the repressor function of PAX4 may play an essential role during pancreatic development, by suppressing the α-cell gene expression program and committing developing endocrine cells to the β- and δ-cell lineages. The loss of PAX4 repression would explain the marked increase in α cells and decrease in β and δ cells seen in animals homozygous for a targeted disruption of the Pax4 gene (35). Identifying the target genes for PAX4 could help explain how the β cell phenotype is established and maintained.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank J. Wang, J. Leong, Y. Zhang, and J. Lau for technical assistance, members of the Michael German laboratory for helpful comments and criticisms, and Gabriele Bergers for advice on PCR strategies for cloning Pax4. H.C.E. and S.B.S. are recipients of Juvenile Diabetes Foundation International postdoctoral fellowships. This work was supported by National Institutes of Health grant DK41822.
REFERENCES
- 1.Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 2.Ahlgren U, Pfaff S L, Jessell T M, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- 3.Blackwell T K, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 4.Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- 5.Dorfler P, Busslinger M. C-terminal activating and inhibitory domains determine the transactivation potential of BSAP (Pax-5), Pax-2 and Pax-8. EMBO J. 1996;15:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan M K, Haynes II J I, Cvekl A, Piatigorsky J. Dual roles for Pax-6: a transcriptional repressor of lens fiber cell-specific β-crystallin genes. Mol Cell Biol. 1998;18:5579–5586. doi: 10.1128/mcb.18.9.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 8.Epstein J A, Glaser T, Cai J, Jepeal L, Walton D S, Maas R L. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 9.Fujioka M, Miskiewicz P, Raj L, Gulledge A A, Weir M, Goto T. Drosophila Paired regulates late even-skipped expression through a composite binding site for the paired domain and the homeodomain. Development. 1996;122:2697–2707. doi: 10.1242/dev.122.9.2697. [DOI] [PubMed] [Google Scholar]
- 10.Fujitani Y, Kajimoto Y, Yasada T, Matsuoka T, Kaneto H, Umayahara Y, Fujita N, Watada H, Miyazaki J, Yamasaki Y, Hori M. Identification of a portable repression domain and an E1A-responsive activation domain in Pax4: a possible role of Pax4 as a transcriptional repressor in the pancreas. Mol Cell Biol. 1999;19:8281–8291. doi: 10.1128/mcb.19.12.8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.German M, Ashcroft S, Docherty K, Edlund H, Edlund T, Goodison S, Imura H, Kennedy G, Madsen O, Melloul D, Moss L, Olson K, Permutt M A, Philippe J, Robertson R P, Rutter W J, Serup P, Stein R, Steiner D, Tsai M-J, Walker M D. The insulin gene promoter. Diabetes. 1995;44:1002–1004. doi: 10.2337/diab.44.8.1002. [DOI] [PubMed] [Google Scholar]
- 12.German M S. Insulin gene structure and regulation. In: Draznin B, LeRoith D, editors. Molecular biology of diabetes, part 1. Totawa, N.J: Humana Press Inc.; 1994. pp. 91–117. [Google Scholar]
- 13.German M S, Moss L G, Wang J, Rutter W J. The insulin and islet amyloid polypeptide genes contain similar cell-specific promoter elements that bind identical β-cell nuclear complexes. Mol Cell Biol. 1992;12:1777–1788. doi: 10.1128/mcb.12.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.German M S, Wang J, Chadwick R B, Rutter W J. Synergistic activation of the insulin gene by a LIM-homeo domain protein and a basic helix-loop-helix protein: building a functional insulin minienhancer complex. Genes Dev. 1992;6:2165–2176. doi: 10.1101/gad.6.11.2165. [DOI] [PubMed] [Google Scholar]
- 15.Gittes G K, Galante P E, Hanahan D, Rutter W J, Debas H T. Lineage-specific morphogenesis in the developing pancreas: role of mesenchymal factors. Development. 1996;122:439–447. doi: 10.1242/dev.122.2.439. [DOI] [PubMed] [Google Scholar]
- 16.Gittes G K, Rutter W J. Onset of cell-specific gene expression in the developing mouse pancreas. Proc Natl Acad Sci USA. 1992;89:1128–1132. doi: 10.1073/pnas.89.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glaser T, Jepeal L, Edwards J G, Young S R, Favor J, Maas R L. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 18.Inoue H, Nomiyama J, Nakai K, Matsutani A, Tanizawa Y, Oka Y. Isolation of full-length cDNA of mouse PAX4 gene and identification of its human homologue. Biochem Biophys Res Commun. 1998;243:628–633. doi: 10.1006/bbrc.1998.8144. [DOI] [PubMed] [Google Scholar]
- 19.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 20.Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- 21.Knepel W, Vallejo M, Chafitz J A, Habener J F. The pancreatic islet-specific glucagon G3 transcription factors recognize control elements in the rat somatostatin and insulin-I genes. Mol Endocrinol. 1991;5:1457–1466. doi: 10.1210/mend-5-10-1457. [DOI] [PubMed] [Google Scholar]
- 22.Lefstin J A, Yamamoto K R. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 23.Mailhos C, Andre S, Mollereau B, Goriely A, Hemmati-Brivanlou A, Desplan C. Drosophila Goosecoid requires a conserved heptapeptide for repression of paired-class homeoprotein activators. Development. 1998;125:937–947. doi: 10.1242/dev.125.5.937. [DOI] [PubMed] [Google Scholar]
- 24.Naya F J, Huang H P, Qiu Y, Mutoh H, DeMayo F J, Leiter A B, Tsai M J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odagiri H, Wang J, German M S. Function of the human insulin promoter in primary cultured islet cells. J Biol Chem. 1996;271:1909–1915. doi: 10.1074/jbc.271.4.1909. [DOI] [PubMed] [Google Scholar]
- 26.Offield M F, Jetton T L, Labosky P A, Ray M, Stein R W, Magnuson M A, Hogan B L M, Wright C V E. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 27.Ogbourne S, Antalis T M. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem J. 1998;331:1–14. doi: 10.1042/bj3310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phelan S A, Loeken M R. Identification of a new binding motif for the paired domain of Pax-3 and unusual characteristics of spacing of bipartite recognition elements on binding and transcription activation. J Biol Chem. 1998;273:19153–19159. doi: 10.1074/jbc.273.30.19153. [DOI] [PubMed] [Google Scholar]
- 29.Philippe J, Morel C, Cordier-Bussat M. Islet-specific proteins interact with the insulin-response element of the glucagon gene. J Biol Chem. 1995;270:3039–3045. doi: 10.1074/jbc.270.7.3039. [DOI] [PubMed] [Google Scholar]
- 30.Roy S, Garges S, Adhya S. Activation and repression of transcription by differential contact: two sides of a coin. J Biol Chem. 1998;273:14059–14062. doi: 10.1074/jbc.273.23.14059. [DOI] [PubMed] [Google Scholar]
- 31.Sander M, Griffen S C, Huang J, German M S. A novel glucose-responsive element in the human insulin gene functions uniquely in primary cultured islets. Proc Natl Acad Sci USA. 1998;95:11572–11577. doi: 10.1073/pnas.95.20.11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sander M, Neubuser A, Kalamaras J, Ee H C, Martin G R, German M S. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 33.Shelton K D, Franklin A J, Khoor A, Beechem J, Magnuson M A. Multiple elements in the upstream glucokinase promoter contribute to transcription in insulinoma cells. Mol Cell Biol. 1992;12:4578–4589. doi: 10.1128/mcb.12.10.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slack J M W. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 35.Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The Pax4 gene is essential for differentiation of insulin-producing β cells in the mammalian pancreas. Nature. 1997;386:399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- 36.St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Pax6 is required for differentiation of glucagon-producing α-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 37.Sussel L, Kalamaras J, Hartigan O C D J, Meneses J J, Pedersen R A, Rubenstein J L, German M S. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 38.Tang H K, Singh S, Saunders G F. Dissection of the transactivation function of the transcription factor encoded by the eye developmental gene PAX6. J Biol Chem. 1998;273:7210–7221. doi: 10.1074/jbc.273.13.7210. [DOI] [PubMed] [Google Scholar]
- 39.Treisman J, Harris E, Wilson D, Desplan C. The homeodomain: a new face for the helix-turn-helix? Bioessays. 1992;14:145–150. doi: 10.1002/bies.950140302. [DOI] [PubMed] [Google Scholar]
- 40.Underhill D A, Gros P. The paired-domain regulates DNA binding by the homeodomain within the intact Pax-3 protein. J Biol Chem. 1997;272:14175–14182. doi: 10.1074/jbc.272.22.14175. [DOI] [PubMed] [Google Scholar]
- 41.Wallin J J, Gackstetter E R, Koshland M E. Dependence of BSAP repressor and activator functions on BSAP concentration. Science. 1998;279:1961–1964. doi: 10.1126/science.279.5358.1961. [DOI] [PubMed] [Google Scholar]
- 42.Walther C, Guenet J-L, Simon D, Deutsch U, Jostes B, Goulding M D, Plachov D, Balling R, Gruss P. Pax: a multigene family of paired box-containing genes. Genomics. 1991;11:424–434. doi: 10.1016/0888-7543(91)90151-4. [DOI] [PubMed] [Google Scholar]
- 43.Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Cooperative dimerization of paired class homeo domains on DNA. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- 44.Xu W, Rould M A, Jun S, Desplan C, Pabo C O. Crystal structure of a paired domain-DNA complex at 2.5 Å resolution reveals structural basis for Pax developmental mutations. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]