Abstract
We have studied the non-covalent interaction between PF-07321332 and SARS-CoV-2 main protease at the atomic level using a computational approach based on extensive molecular dynamics simulations with explicit solvent. PF-07321332, whose chemical structure has been recently disclosed, is a promising oral antiviral clinical candidate with well-established anti-SARS-CoV-2 activity in vitro. The drug, currently in phase III clinical trials in combination with ritonavir, relies on the electrophilic attack of a nitrile warhead to the catalytic cysteine of the protease. Nonbonded interaction between the inhibitor and the residues of the binding pocket, as well as with water molecules on the protein surface, have been characterized using two different force fields and the two possible protonation states of the main protease catalytic dyad HIS41-CYS145. When the catalytic dyad is in the neutral state, the non-covalent binding is likely to be stronger. Molecular dynamics simulations seems to lend support for an inhibitory mechanism in two steps: a first non-covalent addition with the dyad in neutral form and then the formation of the thiolate-imidazolium ion pair and the ligand relocation for finalising the electrophilic attack.
Keywords: COVID-19, SARS-CoV-2, Coronavirus, 3CL-PRO, Coronavirus main protease, Molecular dynamics, 3CL-PRO inhibitor, Pfizer, PF-07321332
Graphical abstract
1. Introduction
In the ongoing COVID-19 pandemic, viral infectivity is known to have been recently boosted by at least four different variants in England, Brazil, South Africa and India [1], all involving mutations on the Spike (S) structural protein. It seems reasonable to expect that SARS-CoV-2 mutational activity on S will wane to some extent vaccine efficacy as the pandemics evolve. The need for an effective antiviral drug against COVID-19 is hence more urgent than ever. In this respect, the SARS-CoV-2 main protease (3CLpro) is probably one of the most promising biological targets [2]. While the mutation rate is high on S, the non structural 3CLpro is highly conserved in the coronaviridae family [3], and is responsible for the generation of the entire virus replication machinery, by cleaving the long polyproteins expressed by the virus RNA upon cell entry.
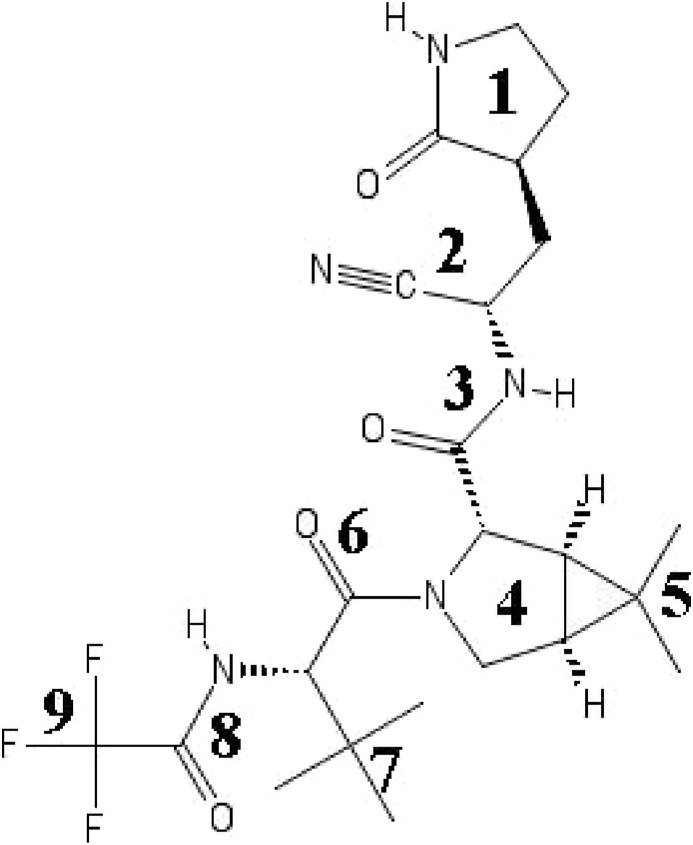
Considerable efforts have been devoted to the identification an effective antiviral agent for SARS-CoV-2 via computational approaches [[4], [5], [6], [7], [8], [9], [10], [11], [12], [13]], [[4], [5], [6], [7], [8], [9], [10], [11], [12], [13]] [[4], [5], [6], [7], [8], [9], [10], [11], [12], [13]] combing different methodologies and analyzing also well-known chemical compounds. On the April 6, 2021, Pfizer disclosed the structure of a 3CLpro inhibitor (Fig. 1 ), named PF-07321332, exhibiting nanomolar affinity and capable of suppressing virus replication in human cells at submicromolar concentrations [14]. The design of PF-07321332 is similar to that of the ML1000 peptidomimetic molecule [15], a covalent reversible Michael 3CLpro inhibitor with submicromolar activity bearing an alpha-keto-amide moiety flanked by two proline-mimetic groups. In PF-07321332, the alpha-keto-amide has been replaced by a nitrile group, acting as a Michael acceptor [16].
Fig. 1.
Structure of PF-07321332 and labelling of groups used for the analysis.
The catalytic mechanism of the cysteine proteases depends on a HIS/CYS catalytic dyad (HIS41-CYS145 in SARS-CoV-2 3CLpro). In Chymotrypsin-like proteases, such as 3CLpro, the nucleophilic attack of the cysteine thiolate is believed to be a concerted process whereby the covalent binding of the substrate is assisted by the histidine, functioning as a proton acceptor from the SH group. In the serine Chymotrypsin protease, the N deprotonated catalytic HIS57 is an H-bond donor on the Nδ1 protonated site for a vicinal ASP102 residue, making HIS57 more basic, hence favouring the proton transfer (PT) from SER195 upon ligand docking [17]. In SARS-CoV-2 3CLpro, such role is apparently played by a water molecule, tightly H-bonded to Nδ1 of HIS41, and revealed in X-ray [18,19] and neutron scattering experiments at room temperature [20]. Papain-like proteases, on the other hand, are believed to act by a thiolate-imidazolium ion pair mechanism, with the dyad already in the zwitterionic form in the native unligated state [16,21]. In this regard, neutron scattering experiments [20] conducted at pH 6.6 and room temperature have shown that HIS41 is protonated and CYS145 is deprotonated on the SARS-CoV-2 3CLpro protein also. This is at variance with the results presented on a previous study by the same authors [18], in agreement with the consensus base catalysis mechanism [22] and with the results obtained in Ref. [23] on the homologous SARS-CoV-1 3CLpro, where the measured pKa of CYS (8.3) and HIS (6.4) are consistent with a general base catalysis chymotrypsin mechanism and cannot be explained by a thiolate-imidazolium ion pair model. Most recently, a thorough MD study on the dimer [24] showed that the ion-pair configuration of the dyad is not compatible with a catalytically competent binding mode for peptide substrates. Substrate docking on the 3CLpro with the dyad in neutral form is hence supposed to favor the CYS to HIS PT [25] preceding the acylation step in the catalysis.
Recently, the catalytic mechanism in 3CLpro was investigated using hybrid QM/MM multi-scale hybrid methods [25,26]. Both these studies agree on the fact that in the apo (unligated) form, the neutral state for the dyad is significantly more stable than the zwitterionic state. Regarding the role of the ligand in the holo form, however, these studies strikingly predict an opposite effect on the ion-pair state, stabilizing in Ref. [25] and destabilizing in Ref. [26] One weak point of these QM/MM approaches for the holo forms is that the latter are prepared [[25], [26], [27]] starting from the X-ray structures where the ligand is already covalently bound to CYS and that were presumably obtained from protein stock pre-incubated with the ligand. The X-ray structure, on the other hand, is not necessarily similar to the actual initial non- covalent bonding pose as the transition state involving the substrate and the binding site must be the result of a concerted process with a C145 to H41 PT transfer induced by the ligand non - covalent docking, followed by a structural modification involving both the ligand and the nearby residues (including the oxyanion hole [28]) to arrive at the covalently bonded form seen in the ligand incubated samples. In other words, what is important for 3CLpro inhibition is the strength of such unknown non-covalent association. This non-covalent affinity, 1 /K m, of the initial Michaelis-Menten complex involving the inhibitor must be stronger than that of a typical 3CLpro substrate corresponding to a substrate-enzyme dissociation constant of the order of −4: 5 kcal/mol [23].
Covalent inhibitors such as PF-07321332 are hence generally designed incorporating the electrophilic group into peptidomimetic substrates that are already known to bind non-covalently with micromolar affinity [29]. The structural stability of the binding site as well as the modulation of the affinity for Michael inhibitors in 3CLpro with different protonation states of the dyad residues was recently assessed by molecular dynamics (MD) simulations [30]. It is hence of interest to study the effect of the protonation states of the dyad in the conformational states of the PF-07321332 compound and to characterise its modality of binding preceding to the nucleophilic attack. To this end we have performed docking calculations and extensive MD simulations of 3CLpro with the dyad in neutral and zwitterionic form. MD calculations were done using two popular force fields (FFs) for protein simulation, namely AMBER [31] and OPLS-AA [32]. We have found that both FFs predict a higher conformational activity of the binding site when the dyad is in the neutral form. Non-covalent binding, as inferred from the ligand-receptor distance distributions, seems to be stronger when 3CLpro catalytic dyad is the neutral form. Water penetration in the catalytic pocket is found to be highly enhanced when HIS41 and CYS145 are in their charged states, irrespective of the FF used. Although the binding modality have some significant differences when using AMBER or OPLS-AA, both FFs agree on the fact that the binding pattern of PF-07321332 is dependent of the protonation state of the dyad, with the nitrile group (Michael acceptor) closer to the dyad when the latter is in the zwitterionic state.
2. Methods
3CLpro is composed of two loosely coupled units that fold independently [33], namely the chymotrypsin-like domains I + II (residues 1–197), hosting the catalytic site, and the cluster of helices domain III (residues 198–304). It is well known that efficient viral polyproteins cleavage occurs when 3CLpro is in the dimeric form [34]. The 3CLpro dimer has two symmetric extended clefts for optimal (linear) peptide chain adhesion [35]. The dimer interface involves the N-terminus of domain I + II and the C-terminus of domain III with no participation of the distal and solvent exposed catalytic site [22,35]. When expressed independently, while domain III has no role in the catalysis [33], the isolated domain I + II is still capable of cleaving a 14-mer peptidic substrate mimicking the N-terminal autocleavage sites of the SARS 3CLpro, although with a much smaller turnover number with respect to the 3CLpro dimer [33]. The inhibition power of small (4-mer) peptidomimetic compounds such as PF-07321332, fitting comfortably in the binding pocket, can hence be reasonably assessed using domain I + II only [35].
Docking calculation were performed using AutoDock Vina [36]. Vina is known to improve the average accuracy of the (non-covalent) binding mode predictions on the well established DUD-E benchmark set [37] of more than 50% compared to AutoDock4, and it was found to be a strong competitor against popular commercial programs, resulting at the top of the pack in many cases. In a recent paper [38], the ability to correctly reproduce the binding modes in SARS-CoV-2 3CLpro of popular docking programs, including Vina, was questioned. In this study [38], 85 co-crystal 3CLpro structures with non-covalently bound ligands were examined. The authors found that in only 15% of these structures, Vina was able to identify the correct crystallographic pose (within a rather conservative 2 Å RMSD tolerance) with the lowest scoring function. Examining their data, we found that in about 60% of the 85 co-crystal structures, the ligand was outside the binding sub-sites S1 and S1’ of the catalytic pocket of the peptidiase, either bound in the S2–S4 sub-sites or at the distal dimer interface or on the surface of domain I + II and III. Most ligands were small and weakly active (especially when far from catalytic site). As all heteroatoms except the ligand were removed prior to docking, the binding modality of small ligands, possibly cohabiting with cavity water molecules in the binding pocket, might can be harder to predict when the latter are not accounted. We hence decided to repeat their analysis using Vina on the 28 3CLpro co-crystal structures where the ligand had at least one moiety in the S1 sub-site, where the 3CLpro substrate P1(GLN) specifically binds. We have found that in about ≅35% of these 28 structures Vina is able to identify the correct (crystallographic) binding pose with a tolerance of less than 3 Å RMSD, and in ≅70% of the cases the crystallographic binding mode was found among the poses within 0.3 kcal/mol of the best scoring pose. As observed in Ref. [38], Vina performances tend to improve for bulky ligands such as PF-07321332. Full results of this analysis can be found in the ESI.
PF-07321332 was docked to the 3CLpro binding site (6LU7 [39]) of domain I + II, considering both states (neutral and zwitterionic) of the catalytic dyad. The calculation runs docking experiments using a so-called gradient optimization algorithm with the box center placed at the midpoint of the vector connecting S γ of C145 to N of H41 with a cubic box size of 1.8 nm side length. The protonation state of all residues except H41 and C145 was assigned using the PropKa program [40]. Using a set of 28 co-crystal structures, we checked that predicted Vina binding poses are essentially unchanged when docking the catalytic site on the single domain I + II and on the full protomer (see Figs. S1 and S2 in ESI).
MD simulations were carried out for the chymotrypsin-like catalytic domain I + II [22,41] of the two isoforms using, in turn, the Amber99sb-ildn [42] and the OPLS-AA/M FFs [32] with the GROMACS code [43] (version 2018.8). In the H–C isoform, H41 was assigned to the tautomer with the protonated N δ [25]. The potential parametrization for the PF-07321332 ligand was generated using two web interfaces: PrimadORAC [44] for AMBER FF and LigParGen [45] for OPLS-AA FF. Starting from the best scoring ligand pose, we have performed extensive molecular dynamics simulations [43,46] with explicit solvent [47,48]. Further methodological aspects are provided in ESI.
3. Results
3.1. Docking calculations
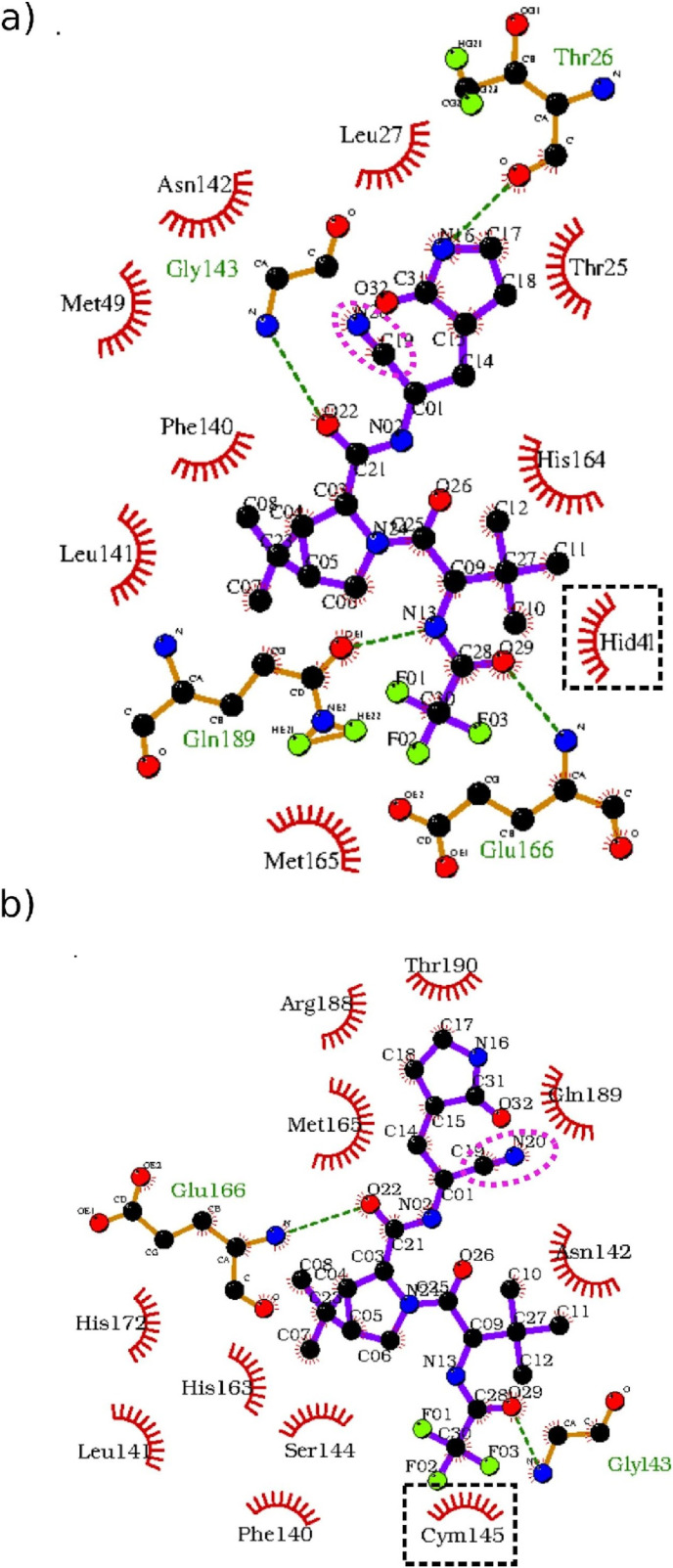
In Fig. 2 we show the best score poses of the ligand obtained in the two protein isoforms. The binding pattern appears to be strongly affected by the protonation state. For the neutral dyad (Fig. 2a), CYS145 (not included in the LigPlot diagram) and HIS41 are approached by the hydrophobic ligand group (4)–(5), and (7), respectively (for groups numbering see Fig. 1). In the zwitterionic state, the dyad (HIS41 not included in LigPlot diagram) is again approached by a weakly hydrophobic moiety (trifluoro group (9)).
Fig. 2.
2D representation of the best docking poses in the binding site obtained with LigPlot [49]. The nitrile group is circled in magenta. The dyad residues are indicated with black dashed squares and the hydrogen bonds with dashed green lines. a) Dyad in neutral state. b) Dyad in zwitterionic state.
In the best Vina pose, the nitrile warhead is far from the dyad in both forms: –CN points to GLY143 with the oxo-pyrrolidine group being close to the neutral dyad H41–C145; when H41–C145 is in the zwitterionic state, –CN is pointing to GLN189 and two hydrogen bonds (involving GLU166 and GLY143) are present. Strikingly, Vina yields for the two isoforms nine poses within 1:1.5 kcal/mol score only, with root mean square deviations (RMSDs) from the best mode ranging from 2 to 5 Å, indicating that binding can occur in quite disparate modalities in both isoforms and/or that the ligand might experience significant conformational activity in the binding site. As shown in Table S4 of the ESI, the minimum distance between the nitrile carbon of PF-07321332 and the Sγ of CYS145 is found in the neutral isoform at 4.3 Å for pose number 7 with a binding free energy 1.3 kcal/mol higher than that of the best Vina pose.
3.2. MD simulations
3.2.1. Protein motions
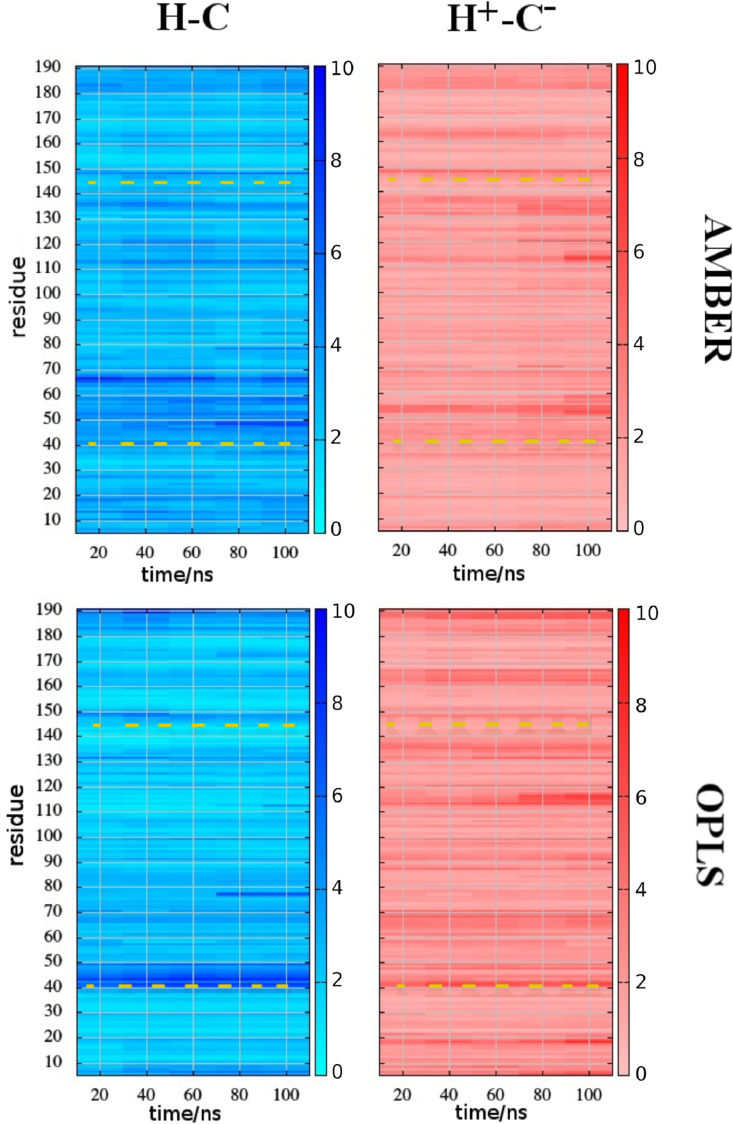
In Fig. 3 , we report the time record of the heat-maps of the RMSD of the protein backbone atoms along the primary sequence of domain I + II. The reference structure is the starting docking structure. The corresponding time records of the overall backbone RMSD are reported in ESI (Fig. S3). The protein fold, after an initial assessment, appears to be stable in all four simulations. In going from the neutral to the zwitterionic form, a remarkable RMSD increase around the catalytic histidine is observed with AMBER, possibly induced by rapid rearrangement of the ligand. At the same time the alpha-helical region 50–70 seems to decrease its mobility in the charged form. The same features, albeit somewhat attenuated, can be observed using OPLS-AA.
Fig. 3.
Heat-maps of RMSD of the protein backbone (domain I + II) for the MD simulations using the two FFs and the two states of the catalytic dyad (H41–C145). The position of the catalytic dyad in the primary sequence is highlighted in yellow. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
A heat-map based on about 100 PDB crystal structures of 3CLpro (free and inhibited with covalent and non-covalent ligands) was reported in Figure 8 of Ref. [22] The structural differences in the map were assumed to depend more on crystal packing than on the presence or absence of ligands or the temperature of the diffraction experiment. Strikingly, the region of maximum experimental variability of the I + II domain is comprised precisely in the 40–70 residue range including the residues H41, M49, Y45 of the S2 region, as observed in our AMBER simulation in the neutral isoform (Fig. 3). Given that more than 50% of the X-ray structures examined in Ref. [22] refer to ligand-free (apo) proteins or proteins with a non-covalent ligand, it is tempting to infer that the neutral isoform is the most likely dyadic state in physiological conditions as resulted with the Amber99sb-ildn FF. The latter FF was found to be more reliable than OPLS-AA on the long time-scale in a recent comparative MD study on ubiquitin an G3 proteins [50].
3.2.2. Ligand motions
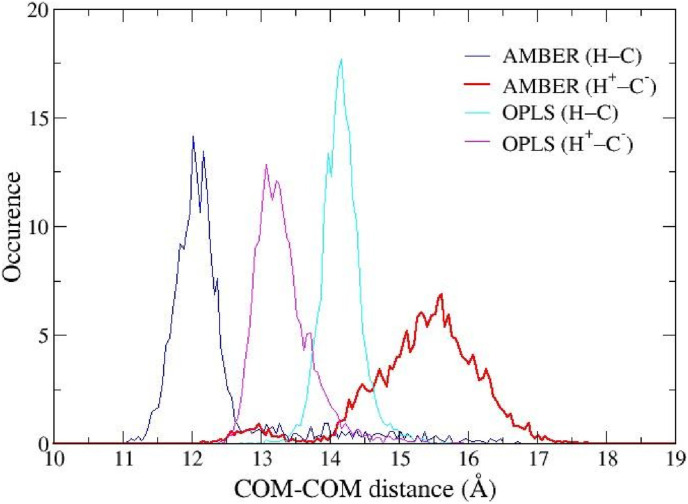
In Fig. 4 we show the distribution of the distances between the centers of mass (COM) of the ligand and the protein. With the AMBER FF, we observe a marked increase of the average COM-COM distance with a significant widening of the distribution in going from the neutral to the zwitterionic isoform. With OPLS-AA, again we observe a widening of the distribution in the zwitterionic isoform with the average COM-COM distance exhibiting a minor shift of about 1 Å in the opposite direction. As the ligand-protein COM-COM distribution is strictly related to the equilibrium constant via the associated potential of mean force [51,52], the sharp and narrow distribution observed in the H–C state seems to indicate that PF-07321332 has a higher non-covalent affinity for this form rather than for the H+−C− isoform where the ligand COM-COM distribution is wider. The increased spread of the distributions seen with both AMBER and OPLS-AA indicates that PF-07321332 is more conformationally active when the catalytic dyad is in the charged state. This fact is further confirmed by the time record of the RMSD of the ligand heavy atoms (Fig. S4 of ESI) and by the histogram of the dihedral angle connecting groups (2) and (3) (Fig. S5 of ESI).
Fig. 4.
COM-COM distribution calculated from MD simulations. In blue AMBER FF and neutral dyad, in red AMBER FF and zwitterionic dyad, in cyan OPLS-AA FF and neutral dyad and in magenta OPLS-AA FF and zwitterionic dyad. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2.3. Solvent behaviour
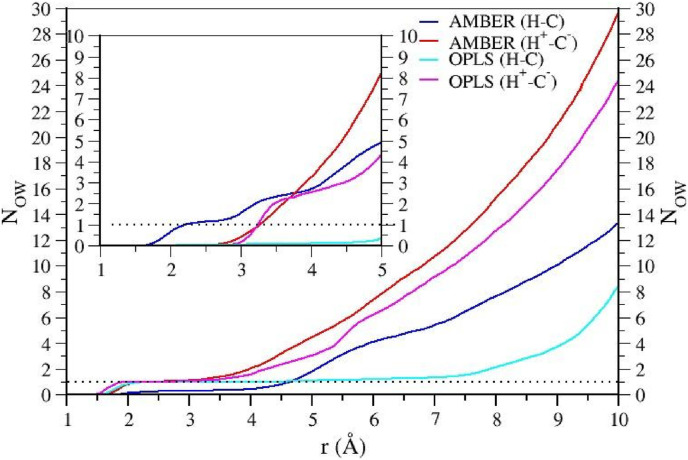
A water molecule (detected in most of the recent crystal structures of free and inhibited 3CLpro22) has been suggested to surrogate the role of ASP102 in the canonical chymotrypsin catalytic reaction by assisting in PT from CYS145 to HIS41 and by stabilizing the positive charge accumulated on HIS41 in the chymotrypsin acid-base mechanism. In 3CLpro, this water is believed to function as an H-bond acceptor from the protonated Nδ1 of HIS41 [25]. In Fig. 5 , we report the integrated radial distribution function, g Hδ1−OW(r), for the contact Hδ1(H41)–OW(H2O) (with OW labelling the oxygen in water molecules) obtained in the simulations. According to both AMBER and OPLS-AA, water molecules appear to penetrate the PF-07321332 engaged binding site region much more effectively when the dyad is in the zwitterionic state. Within a distance of 10 Å from Hδ1(H41), about 30 and 25 solvent molecules are found with OPLS-AA and AMBER, respectively. The g Hδ1−OW(r) function integrates at the same distance of 10 Å to about 1/3 of the molecules when the dyad is in the neutral state. This fact appears to be consistent with a weaker binding in the H+−C− isoform as inferred by the COM-COM distance distributions (see Fig. 4): water tends to occupy more the cavity when the dyad is the zwitterionic state, competing with the ligand. In the OPLS-AA simulations, a water molecule acts as an H-bond acceptor to Nδ1-Hδ1 of H41 for the entire 100 ns in both isoforms. In AMBER, while in H+−C− such H-bonded water is always present, in the H–C state, a water molecule acts as an H-bond donor to the N2 of H41, while the Hδ1(H41) hydrogen is shared by a water oxygen and a carbonyl oxygen of H163. The higher occupancy of solvent molecules in the zwitterionic form has an impact on the solvent exposure of the bound ligand, which, according to both FFs, has an average number of surrounding water molecules N w within a distance of 6 Å of N w≅30 and N w≅25 using the OPLS-AA and AMBER FF, respectively.
Fig. 5.
Integral of the radial distribution function for the Hδ1(H41)–OW(H2O) contact in the four simulations. In the inset, the integral of the N2(H41)–H(H2O) contact.
3.2.4. Ligand-protein interactions
From the RMSD time record of the ligand heavy atoms (Fig. S4 of ESI), we can see that PF-07321332 evolves rapidly to a stable pose about 2.5 Å away from the initial docking pose in the protein with the neutral dyad. This behaviour is similar when using OPLS-AA or AMBER and is reflected in the narrow ligand-protein COM-COM distance distribution observed for the neutral dyad state in both FFs (Fig. 4). For the isoform with H+−C−, both FFs predict an enhanced mobility of the ligand in the binding site. With OPLS-AA, we observe an abrupt rearrangement of the ligand pose during the 100 ns simulation with the RMSD raising to 3.5 Å and back to 2.5 Å (magenta curve of Fig. S4 in ESI), rather than the frequent conformational transitions in the subnanosecond time-scale observed when using the AMBER FF (red curve of Fig. S4 in ESI).
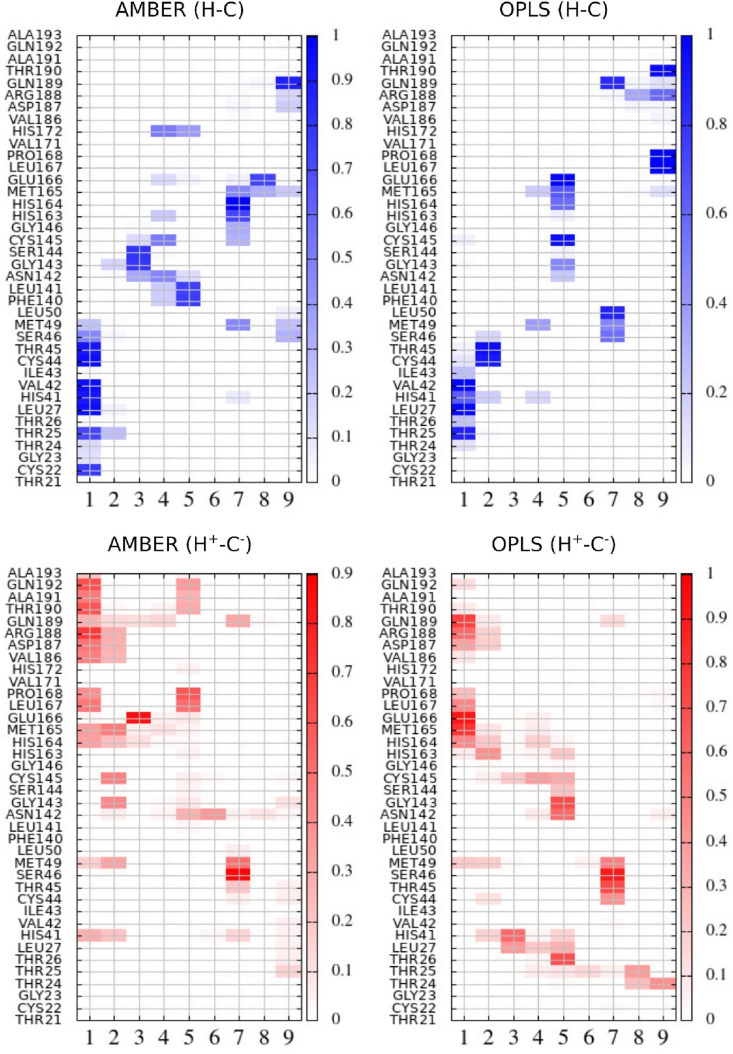
Concerning the detailed ligand-protein interaction, again OPLS-AA and AMBER yield similarities as well as important differences. In Fig. 6 we show the ligand-residue contact map computed in the four simulations. The ligand has been sectioned into 9 groups (see Fig. 1 for groups labeling) with the index (2) referring to the nitrile moiety. A ligand group is assumed to be in contact with a given protein residue if any group-residue atom-atom distance is found below 4.5 Å threshold. The heat-bar on the right of the maps refers to the contact probability.
Fig. 6.
Contact maps between ligand groups (labeling in Fig. 1) and protein residues obtained from MD simulations with AMBER and OPLS-AA FFs and neutral dyad (H–C), AMBER and OPLS-AA FFs and zwitterionic dyad (H+−C−). The heat-bar on the right of the plot represents a contact probability. Values of 1 for the contact probability imply that the ligand group has been found in contact with the given residue in all configurations during the 100 ns simulation.
The contact maps of Fig. 6 inherit some features from Vina-generated corresponding starting structures. Concerning the H–C isoform, the oxo-pyrrolidine group (1), mimicking the glutamine P1 residue of the 3CLpro substrate, is found mostly near the triad THR25, THR26, LEU27 according to both FFs, as in the docked LigPlot structure reported in Fig. 2. The same applies to the terminal trifluoro group (9) found to insist in the region R188, Q189, T190 in the H–C form using both FFs, as in the docked structure of Fig. 2a. For the H+−C− protein, the oxo-pyrrolidine group (1) of the ligand is this time in contact with the terminal end of the domain I + II (187–194) according to both AMBER and OPLS-AA, a feature that is also observed in the starting Vina structure (Fig. 2), while the trifluoro group (9) remains mostly solvent-exposed in AMBER and, to a less extent, in OPLS-AA, drifting away from the Vina position where –CF3 is found near the catalytic negatively charged C145. In the MD simulations of the H+−C− isoform, the nitrile group relocates significantly with respect to the starting Vina pose, approaching, especially for the AMBER case, to the catalytic C145.
As a general trend, the PF-07321332 contact matrix obtained in the simulations appears to be roughly diagonal and anti-diagonal when the dyad is in the zwitterionic and neutral state, respectively. In the anti-diagonal form (blue maps, neutral dyad), groups (7) to (9) of the ligand tend to move in the proximity of the region 163–194 of domain II, while group (1) moves towards the terminal region of the domain I (region 20–50). The ligand orientation in the H+−C− form generates an approximately diagonal pattern with the ligand terminal groups (1) and (7)–(9) on domain II and I, respectively. These non bonded conformations are at variance with the available X-ray structures of similar covalently bound peptidomimetic compounds with the oxo-pyrrolidine moiety (surrogating a GLN) near S1 protein sub-site (defined by the residues [53] S144, H163, E166 and H172).
Neutral dyad (H–C) isoform: According to AMBER (top left panel in Fig. 6), the oxopyrrolidine group (1) insists in the region 22–49 including the catalytic H41, but apparently not in strict contact with C145. The neighboring nitrile group (2) remains mostly solvent exposed with loose contacts with GLY143.
Tighter contacts in the protein region 142–145 (hence including C145) are seen with the central amide group (3). The trifluoro group (9) lies mostly near the polar residues Q189, R188 and D187 while the trimethyl hydrophobic moiety (7) is engaging in stable contacts with H163 and H164. When using OPLS-AA, the contact map shows similarities as well as important differences. For example, the nitrile group is rather far from the neutral dyad according to both FF. However, when using OPLS-AA, this group is stably in contact with T45 and C44 rather than mostly solvent-exposed as with AMBER. OPLS-AA predicts a very tight contact between the trifluoro terminal group and the hydrophobic residues P168 L167, an interaction that is not present when using AMBER, where the –CF3 is strongly interacting with GLN189.
Zwitterionic dyad (H+−C − ) isoform: In this case the FFs show quite important discrepancies especially concerning the nitrile group (2). With AMBER, at variance with the form H–C, the reactive nitrile is found with high probability near the catalytic dyad with the lactame moiety (group (1)) this time bordering the S1 sub-site, while in OPLS-AA, the groups (1,2) are apparently unable to approach S1 and hence C145 or H41. Also, the orientation of the central proline mimetic hydrophobic group (4)–(5) is markedly different in OPLS-AA and AMBER. While in the former case, the groups (4)–(5) lingers mostly within domain I, in the latter case this group is shifted towards domain II. Finally, according to AMBER, the terminal trifluoro-group and flanking amide are solvent-exposed and not engaging in stable contacts with protein residues, while with OPLS-AA we find such group to be in strict contact with T24 and T25 in domain I.
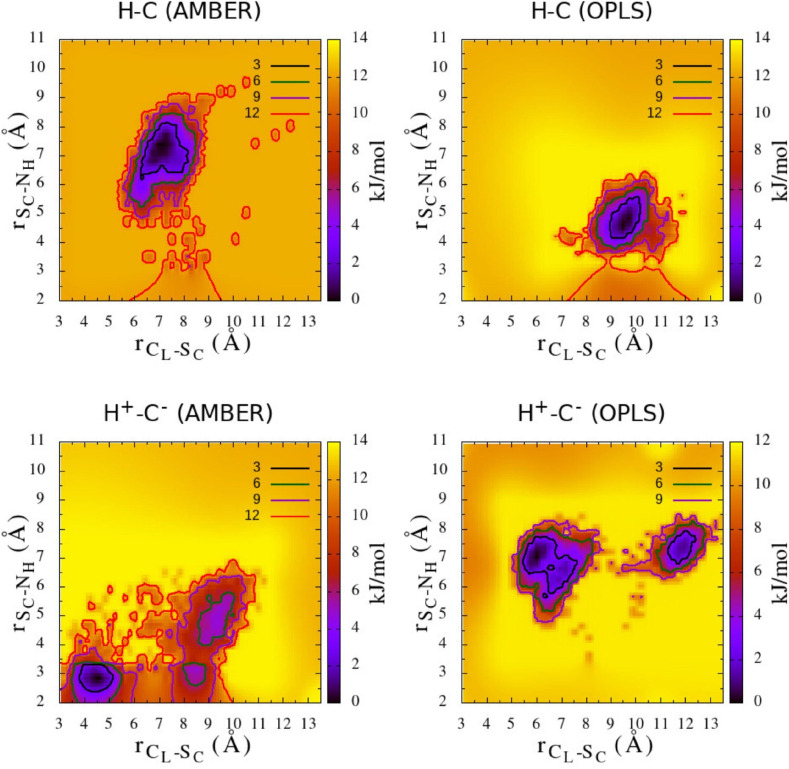
In Fig. 7 we show the 2-dimensional free energy surface (FES) with respect to the i) distance (d x) between the C of ligand nitrile group and the S of C145 and ii) the distance (d y) between the S of C145 and the Nδ1 of H41.
Fig. 7.
2D free energy surface, f(dx,dy) computed as f(dx,dy) = −RT ln(P(dx,dy)/MAX[P(dx,dy)], where P(dx,dy) denotes the joint probability of the two collective variables dx and dy. dx = CL − SC is the distance between the C of ligand nitrile group and the S of C145 and dy = SC − NH is the distance between the S of C145 and the Nδ1 of H41.
Apparently in the H–C isoform the intercalation of PF-07321332 in the binding site tends to push apart the H41–C145 pair, with the nitrile group far from C145 in both AMBER and OPLS-AA cases. According to the AMBER FF, H41 is approached by the oxo-pyrrolidine group that may help render H41 more basic and hence prone to acquire a proton from a solvating water molecule (e.g. Fig. 5). Such event could trigger the formation of the thiolate-imidazolium ion pair with relocation of the ligand dyad, followed by the electrophilic attack of the nitrile Michael acceptor on the thyolate C145 and a PT from the protonated H41 to form the imine derivative. This hypothesis seems to be supported by the FES obtained with AMBER for the H+−C− isoform (lower left panel in Fig. 7) where H41 comes closer to C145 and the electrophilic reactive carbon of the nitrile group is displaced toward C145. Such a mechanism does not seem to be confirmed when using OPLS-AA. In this case, the H41–C145 separation in the H–C state is less pronounced than in AMBER with the FES exhibiting a single deep minimum at d x≅10 Å and d y≅4 Å. In the H+−C− the mutual spatial arrangement of H41/C145 and the –CN group seems to further disfavour the Michael addition. The H-bond occupancy is reported in Tables S5 and S6 of ESI allowing to properly describe the PF-07321332-3CLpro interactions indicated in Fig. 6.
4. Discussion
The X-ray structure of the PF-07321332-3CLpro complex is currently not available on the PDB. The X-ray structure of a strictly related alpha-keto amide 3CLpro-inhibitor (Boceprevir, PDB 7BPR) [54] refers to the covalently bound complex. There, the cyclobutyl moiety (replaced by the group 1, oxo-pyrrolidine, in PF-07321332) in the S1 pocket (S144, H163, E166), while the di-methyl-azabicyclo group (groups 4,5 in PF-07321332) is in S2 (M48, H41 Q189) with the electrophilic carbon of the intercalated dioxo warhead group bound to the cysteinate sulphur atom. Such experimental structure of a PF-07321332 analog is quite different from that of the non-covalent complex emerged from our simulations (defined and detailed in Fig. 6), especially when the protease is in the native neutral isoform H–C. As the unknown structure of the intermediate non-covalent complex can be quite different from the final covalent holo form, the picture provided by the AMBER MD simulations of the two isoforms (see Fig. 6, Fig. 7) seems to lend support to an inhibitory mechanism whereby the initial non-covalent addition of PF-07321332 to the main protease with the dyad in neutral form induces the formation of the thiolate-imidazolium ion pair and the ligand relocation exposing the –CN group in close contact with the C145 thiolate for finalising the electrophilic attack. The formation of the zwitterionic form the non- covalent neutral holo state is possibly mediated by the interaction between the oxo-pyrrolidine moiety of the ligand and H41 with the assistance of H41-bound water molecule. In the MD simulations, PF-07321332 after an initial fast assessment, seems to stabilise in the binding site of both isoforms (see Figs. S4 and S5 of the ESI) in an orientation not too far from that of the starting docking pose. We cannot exclude that in longer simulations major conformational or reorientational rearrangements of the ligand can occur. Such abrupt events, however, on a single complex are rare and generally not observed even in simulations extending in the microseconds time scale [26]. For example, in the classical MD simulations reported in Refs. [[25], [26], [27]] (extending from a minimum of 0.1 to a maximum of 8 μs), the ligand orientation consistently remained similar (RMSD≅ 2.5: 3 Å) to that of the starting pose, prepared using the X-ray with the ligand covalently bound to the cysteine, despite the former was not covalently bound to the protein and the cysteine was protonated.
In systems where the precise structural features of non-covalent binding is unknown, a reasonable approach should be based on the use of reliable docking scoring functions (such as those in Vina, see Methods section and ESI) for non-covalent binding prediction, supplemented with replicates MD simulations [25,35] for pose characterization. The relative stability of multiple docking poses can be examined by evaluating independently their absolute binding free using powerful nonequilibrium techniques [35]. At present, the totally binding free energy has been evaluated using the molecular mechanics Poisson–Boltzmann surface area (MMPBSA) method giving a value of −102.002 ± −21.336 kJ/mol [55].
5. Conclusions
We have studied the non-covalent interaction between PF-07321332 and the domain I + II of the SARS-CoV-2 main protease using MD simulations. The ligand was initially docked to the binding site of the two isoforms of 3CLpro with the catalytic dyad in the neutral and zwitterionic state using the Vina program. The resulting structures were subsequently simulated for 100 ns in the NPT ensemble in standard conditions using the Amber99sb-ildn [42] and the OPLS-AA/M FFs, producing a total simulation time of 0.4 μs. Both FFs indicate that PF-07321332 non-covalent binding is likely to be stronger when the catalytic dyad is in the neutral state. When 3CLpro has the H41–C145 dyad in the ionic form, while with OPLS-AA the reactive nitrile group PF-07321332 undergoes a minor rearrangement in the binding site, the AMBER FF predicts a dramatic change in the binding pattern with a close interaction between the –CN moiety, the thiolate C145 and the protonated H41. The AMBER simulations of the neutral and zwitterionic non-covalent complexes are suggestive of a mechanism whereby the initial non-covalent addition with the dyad in neutral form induces the formation of the thiolate-imidazolium ion pair and the ligand relocation for finalising the electrophilic attack. To further confirm the proposed mechanism, in a follow-up study, we aim at evaluating using an accurate methodology based on enhanced sampling simulations and nonequilibrium alchemy [35], the absolute binding free energies of PF-07321332 non-covalent binding modes in both 3CLpro isoforms.
Author contributions
Marina Macchiagodena: Visualization, Investigation. Marco Pagliai: Visualization, Data Curation. Piero Procacci: Conceptualization, Methodology, Visualization, Investigation. The authors: Writing- Reviewing and Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank MIUR-Italy (“Progetto Dipartimenti di Eccellenza 2018–2022” allocated to Department of Chemistry “Ugo Schiff”) and Dr. Claudia Bello for helpful discussions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmgm.2021.108042.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Mallapaty S. India's massive COVID surge puzzles scientists. Nature. 2021;592:667–668. doi: 10.1038/d41586-021-01059-y. [DOI] [PubMed] [Google Scholar]
- 2.COVID moonshot. 2020. https://postera.ai/covid accessed June 18 2020.
- 3.Macchiagodena M., Pagliai M., Procacci P. Identification of potential binders of the main protease 3CLpro of the COVID-19 via structure-based ligand design and molecular modeling. Chem. Phys. Lett. 2020;750:137489. doi: 10.1016/j.cplett.2020.137489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma J., Kumar Bhardwaj V., Singh R., Rajendran V., Purohit R., Kumar S. An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against non structural protein-15 of SARS-CoV-2. Food Chem. 2021;346:128933. doi: 10.1016/j.foodchem.2020.128933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh R., Bhardwaj V.K., Sharma J., Purohit R., Kumar S. In-silico evaluation of bioactive compounds from tea as potential SARS-CoV-2 nonstructural protein 16 inhibitors. J. Tradit. Complement. Med. 2021 doi: 10.1016/j.jtcme.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhardwaj V.K., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput. Biol. Med. 2021;128:104117. doi: 10.1016/j.compbiomed.2020.104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2021;39:3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R., Bhardwaj V.K., Das P., Purohit R. A computational approach for rational discovery of inhibitors for non-structural protein 1 of SARS-CoV-2. Comput. Biol. Med. 2021;135:104555. doi: 10.1016/j.compbiomed.2021.104555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Bioactive molecules of tea as potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2. Front. Med. 2021;8:645. doi: 10.3389/fmed.2021.684020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhai T., Zhang F., Haider S., Kraut D., Huang Z. An integrated computational and experimental approach to identifying inhibitors for SARS-CoV-2 3CL protease. Front. Mol. Biosci. 2021;8:267. doi: 10.3389/fmolb.2021.661424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries M., Mohamed A.S., Prescott R.A., Valero-Jimenez A.M., Desvignes L., O'Connor R., Steppan C., Devlin J.C., Ivanova E., Herrera A., Schinlever A., Loose P., Ruggles K., Koralov S.B., Anderson A.S., Binder J., Dittmann M., Pfeiffer J.K. A comparative analysis of SARS-CoV-2 antivirals characterizes 3CLpro inhibitor PF-00835231 as a potential new treatment for COVID-19. J. Virol. 2021;95:e01819–e01820. doi: 10.1128/JVI.01819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan A., Ali S.S., Khan M.T., Saleem S., Ali A., Suleman M., Babar Z., Shafiq A., Khan M., Wei D.-Q. Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS-CoV-2 main protease (3CLpro) J. Biomol. Struct. Dyn. 2021;39:4659–4670. doi: 10.1080/07391102.2020.1779128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan A., Heng W., Wang Y., Qiu J., Wei X., Peng S., Saleem S., Khan M., Ali S.S., Wei D.-Q. Silico and in vitro evaluation of kaempferol as a potential inhibitor of the SARS-CoV-2 main protease (3CLpro) Phytother Res. 2021;35:2841–2845. doi: 10.1002/ptr.6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halford B. Pfizer unveils its oral SARS-CoV-2 inhibitor. Chem. Eng. News. 2021:99. [Google Scholar]
- 15.Westberg M., Su Y., Zou X., Ning L., Hurst B., Tarbet B., Lin M.Z. Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases. bioRxiv. 2020 [Google Scholar]
- 16.Dos Santos A.M., Cianni L., De Vita D., Rosini F., Leitão A., Laughton C.A., Lameira J., Montanari C.A. Experimental study and computational modelling of cruzain cysteine protease inhibition by dipeptidyl nitriles. Phys. Chem. Chem. Phys. 2018;20:24317–24328. doi: 10.1039/c8cp03320j. [DOI] [PubMed] [Google Scholar]
- 17.Protein Structure and Function. Oxford University Press; Oxford (UK): 2008. [Google Scholar]
- 18.Kneller D.W., Phillips G., O'Neill H.M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L., Kovalevsky A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020;11:3202. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kneller D.W., Phillips G., O'Neill H.M., Tan K., Joachimiak A., Coates L., Kovalevsky A. Room-temperature X-ray crystallography reveals the oxidation and reactivity of cysteine residues in SARS-CoV-2 3CL Mpro: insights into enzyme mechanism and drug design. IUCrJ. 2020;7:1028–1035. doi: 10.1107/S2052252520012634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kneller D.W., Phillips G., Weiss K.L., Pant S., Zhang Q., O'Neill H.M., Coates L., Kovalevsky A. Unusual zwitterionic catalytic site of SARS–CoV-2 main protease revealed by neutron crystallography. J. Biol. Chem. 2020;295:17365–17373. doi: 10.1074/jbc.AC120.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mechanisms of Protease Action. 1989. pp. 123–125. [Google Scholar]
- 22.Jaskolski M., Dauter Z., Shabalin I.G., Gilski M., Brzezinski D., Kowiel M., Rupp B., Wlodawer A. Crystallographic models of SARS-CoV-2 3CLpro: in-depth assessment of structure quality and validation. IUCrJ. 2021;8:238–256. doi: 10.1107/S2052252521001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang C., Wei P., Fan K., Liu Y., Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43:4568–4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- 24.Diaz N., Suarez D. Influence of charge configuration on substrate binding to SARS-CoV-2 main protease. Chem. Commun. 2021;57:5314–5317. doi: 10.1039/d1cc01449h. [DOI] [PubMed] [Google Scholar]
- 25.Zanetti-Polzi L., Smith M.D., Chipot C., Gumbart J.C., Lynch D.L., Pavlova A., Smith J.C., Daidone I. Tuning proton transfer thermodynamics in SARS-CoV-2 main protease: implications for catalysis and inhibitor design. J. Phys. Chem. Lett. 2021;12:4195–4202. doi: 10.1021/acs.jpclett.1c00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramos-Guzmán C.A., Ruiz-Pernia J.J., Tuñon I. Unraveling the SARS-CoV-2 main protease mechanism using multiscale methods. ACS Catal. 2020;10:12544–12554. doi: 10.1021/acscatal.0c03420. [DOI] [PubMed] [Google Scholar]
- 27.Ramos-Guzman C., Ruiz-Pernia J. Inhibition mechanism of SARS-CoV-2 main protease with ketone-based inhibitors unveiled by multiscale simulations. ChemRxiv. 2021 doi: 10.1002/anie.202110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Q., Li S., Xue F., Zou Y., Chen C., Bartlam M., Rao Z. Structure of the main protease from a global infectious human coronavirus. HCoV-HKU1. J. Virol. 2008;82:8647–8655. doi: 10.1128/JVI.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanzlik R.P., Thompson S.A. Vinylogous amino acid esters: a new class of inactivators for thiol proteases. J. Med. Chem. 1984;27:711–712. doi: 10.1021/jm00372a001. [DOI] [PubMed] [Google Scholar]
- 30.Pavlova A., Lynch D.L., Daidone I., Zanetti-Polzi L., Smith M.D., Chipot C., Kneller D.W., Kovalevsky A., Coates L., Golosov A.A., Dickson C.J., Velez-Vega C., Duca J.S., Vermaas J.V., Pang Y.T., Acharya A., Parks J.M., Smith J.C., Gumbart J.C. Inhibitor binding influences the protonation states of histidines in SARS-CoV-2 main protease. Chem. Sci. 2021;12:1513–1527. doi: 10.1039/d0sc04942e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponder J.W., Case D.A. Vol. 66. Academic Press; 2003. pp. 27–85. (Protein Simulations; Advances in Protein Chemistry). [DOI] [PubMed] [Google Scholar]
- 32.Robertson M.J., Tirado-Rives J., Jorgensen W.L. Improved peptide and protein torsional energetics with the OPLS-AA force field. J. Chem. Theor. Comput. 2015;11:3499–3509. doi: 10.1021/acs.jctc.5b00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi J., Wei Z., Song J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. J. Biol. Chem. 2004;279:24765–24773. doi: 10.1074/jbc.M311744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graziano V., McGrath W.J., Yang L., Mangel W.F. SARS CoV main proteinase: the monomer-dimer equilibrium dissociation constant. Biochemistry. 2006;45:14632–14641. doi: 10.1021/bi061746y. [DOI] [PubMed] [Google Scholar]
- 35.Macchiagodena M., Pagliai M., Karrenbrock M., Guarnieri G., Iannone F., Procacci P. Virtual double-system single-box: a nonequilibrium alchemical technique for absolute binding free energy calculations: application to ligands of the SARS-CoV-2 main protease. J. Chem. Theor. Comput. 2020;16:7160–7172. doi: 10.1021/acs.jctc.0c00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mysinger M.M., Carchia M., Irwin J.J., Shoichet B.K. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J. Med. Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zev S., Raz K., Schwartz R., Tarabeh R., Gupta P.K., Major D.T. Benchmarking the ability of common docking programs to correctly reproduce and score binding modes in SARS-CoV-2 protease mpro. J. Chem. Inf. Model. 2021;61:2957–2966. doi: 10.1021/acs.jcim.1c00263. [DOI] [PubMed] [Google Scholar]
- 39.Liu X., Zhang B., Jin Z., Yang H., Rao Z. RSCB PDB; 2020. The Crystal Structure of 2019-nCoV Main Protease in Complex with an Inhibitor N3. pdbcode: 6LU7. [Google Scholar]
- 40.Li H., Robertson A.D., Jensen J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 41.Procacci P., Macchiagodena M., Pagliai M., Guarnieri G., Iannone F. Interaction of hydroxychloroquine with SARS-CoV2 functional proteins using all-atoms nonequilibrium alchemical simulations. Chem. Commun. 2020;56:8854–8856. doi: 10.1039/d0cc03558k. [DOI] [PubMed] [Google Scholar]
- 42.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 44.Procacci P. PrimaDORAC: a free web interface for the assignment of partial charges, chemical topology, and bonded parameters in organic or drug molecules. J. Chem. Inf. Model. 2017;57:1240–1245. doi: 10.1021/acs.jcim.7b00145. [DOI] [PubMed] [Google Scholar]
- 45.Dodda L.S., Cabeza de Vaca I., Tirado-Rives J., Jorgensen W.L. LigParGen web server: an automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res. 2017;45:W331–W336. doi: 10.1093/nar/gkx312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., Kasson P.M., van der Spoel D., Hess B., Lindahl E. Gromacs 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 48.Pagliai M., Macchiagodena M., Procacci P., Cardini G. Evidence of a low–high density turning point in liquid water at ordinary temperature under pressure: a molecular dynamics study. J. Phys. Chem. Lett. 2019;10:6414–6418. doi: 10.1021/acs.jpclett.9b02724. [DOI] [PubMed] [Google Scholar]
- 49.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 50.Martin-Garcia F., Papaleo E., Gomez-Puertas P., Boomsma W., Lindorff-Larsen K. Comparing molecular dynamics force fields in the essential subspace. PLoS One. 2015;10:1–16. doi: 10.1371/journal.pone.0121114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilson M.K., Given J.A., Bush B.L., McCammon J.A. The StatisticalThermodynamic basis for computation of binding affinities: a critical review. Biophys. J. 1997;72:1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Procacci P., Chelli R. Statistical mechanics of ligand-receptor noncovalent association, revisited: binding site and standard state volumes in modern alchemical theories. J. Chem. Theor. Comput. 2017;13:1924–1933. doi: 10.1021/acs.jctc.6b01192. [DOI] [PubMed] [Google Scholar]
- 53.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.) 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmad B., Batool M., Ain Q.u., Kim M.S., Choi S. Exploring the binding mechanism of PF-07321332 SARS-CoV-2 protease inhibitor through molecular dynamics and binding free energy simulations. Int. J. Mol. Sci. 2021:22. doi: 10.3390/ijms22179124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.