Abstract
The type 1 angiotensin II (AngII) receptor (AT1R) transactivates the epidermal growth factor receptor (EGFR), which leads to pathological remodeling of heart, blood vessels and kidney. End-point assays are used as surrogates of EGFR activation, however these downstream readouts are not applicable to live cells, in real-time. Herein, we report the use of a bioluminescence resonance energy transfer (BRET)-based assay to assess recruitment of the EGFR adaptor protein, growth factor receptor-bound protein 2 (Grb2), to the EGFR. In a variety of cell lines, both epidermal growth factor (EGF) and AngII stimulated Grb2 recruitment to EGFR. The BRET assay was used to screen a panel of 9 G protein-coupled receptors (GPCRs) and further developed for other EGFR family members (HER2 and HER3); the AT1R was able to transactivate HER2, but not HER3. Mechanistically, AT1R-mediated ERK1/2 activation was dependent on Gq/11 and EGFR tyrosine kinase activity, whereas the recruitment of Grb2 to the EGFR was independent of Gq/11 and only partially dependent on EGFR tyrosine kinase activity. This Gq/11 independence of EGFR transactivation was confirmed using AT1R mutants and in CRISPR cell lines lacking Gq/11. EGFR transactivation was also apparently independent of β-arrestins. Finally, we used additional BRET-based assays and confocal microscopy to provide evidence that both AngII- and EGF-stimulation promoted AT1R-EGFR heteromerization. In summary, we report an alternative approach to monitoring AT1R-EGFR transactivation in live cells, which provides a more direct and proximal view of this process, including the potential for complexes between the AT1R and EGFR.
Keywords: Angiotensin II, epidermal growth factor receptor, EGFR transactivation, bioluminescence resonance energy transfer, G protein-coupled receptor, receptor heteromerization
1. Introduction
Angiotensin II (AngII) is the major bioactive peptide of the Renin-Angiotensin System (RAS) and influences a broad range of homeostatic and modulatory processes, including cardiovascular and renal physiology. Dysregulation of the RAS is associated with disease via actions on cardiac (1-4), vascular (5,6) and renal (7,8) growth and remodeling, modulation of sympathetic nervous system activity (9-11), endothelial dysfunction (12), angiogenesis (13,14) and inflammation (15-17). AngII acts primarily through the type 1 AngII receptor (AT1R1), a G protein-coupled receptor (GPCR), to mediate an array of intracellular signals, including calcium mobilization and generation of reactive oxygen species (ROS), modulation of receptor and non-receptor tyrosine kinases, mitogen-activated protein kinases (MAPK) (including the extracellular-regulated kinase 1/2 (ERK1/2)) and various ion channels (18,19). We and others have demonstrated that the AT1R can transactivate the epidermal growth factor receptor (EGFR), which in turn modulates cellular growth, tissue remodeling and cellular hypertrophy (20-23).
The EGFR belongs to the ErbB family of receptor tyrosine kinases (RTKs), comprised of four members: EGFR (ErbB1, HER1), HER2 (ErbB2), HER3 (ErbB3) and HER4 (ErbB4) (24). HER receptors are activated by a group of epidermal growth factor (EGF) ligands and undergo homo- or hetero-dimerization during their activation, with HER2 being the preferred dimer partner (25). Activated and tyrosine-phosphorylated HER receptors recruit growth factor receptor-bound protein 2 (Grb2) as an initial step in a complex cascade of growth signaling activation. HER receptors can also be ‘transactivated’ following prior activation of other receptors. Most notably, Ullrich and colleagues first described GPCR-mediated EGFR transactivation, and subsequently proposed a model, whereby activated GPCRs stimulated matrix metalloproteases (MMP), including the ADAMs (A Disintegrin and Metalloprotease), to cleave inactive membrane-bound EGF ligands to activate EGFR (26,27). This model has been broadly accepted, although others have suggested a more complex mechanism (perhaps cell-specific) involving non-receptor tyrosine kinases (e.g., src and Pyk2) (28-32) and the activation of additional second messengers including calcium (33-35), protein kinase C (32,33,36,37), ROS (23,38) and β-arrestin (36,39). Definitive resolution of the precise mechanism of GPCR-EGFR transactivation requires approaches to monitor this process in living cells, in real-time, as well as the capacity to identify key proteins and interactions involved. While some progress has been made on the latter problem – a siRNA screen was recently used to identify novel mediators of AT1R-EGFR transactivation (40) – to date most readouts of EGFR transactivation have been biochemical, end-point assays that do not capture live cell dynamics or kinetics.
Commonly, GPCR-mediated EGFR transactivation has been defined utilizing the activation of ERK1/2 that is inhibited by the small molecule antagonist of EGFR (i.e., AG1478). However, there are several limitations in using ERK1/2 as a surrogate readout of transactivation. EGFR signal transduction is complex with ERK1/2 phosphorylation representing a signal quite distal from the initial step of EGFR activation. Arguably, a more direct readout would be to examine upstream events, including activation and autophosphorylation of the EGFR. Although a direct readout of phospho-EGFR may mitigate the limitations of the ERK1/2 assay, both phospho-EGFR and phospho-ERK1/2 assays are endpoint approaches and only provide a snapshot of cellular events. More recently, proximity-based assays, such as resonance energy transfer (RET) assays and fluorescein arsenical hairpin (FlAsH) biosensors have evolved to enable dynamic analysis of protein-protein and intra-molecular interactions in real-time (41,42). The development and validation of proximity-based assays has enabled their wide use to characterize receptor interactions. They have become extremely useful tools for studying GPCR biology, and specifically the interaction of GPCRs with G proteins and arrestins (43-46).
In this study, we employed a Bioluminescence-RET (BRET)-based assay to monitor the most proximal event in EGFR activation, namely Grb2 interaction with the activated EGFR. We demonstrate, in live cells and in real-time, that both AngII- and EGF-stimulation can promote Grb2 translocation to the activated EGFR, indicating AT1R transactivation of the EGFR. Having established this approach, we extend it to investigate the molecular processes involved and report evidence for the formation of AT1R and EGFR complexes.
2. Materials and Methods
2.1. Materials
[125I]-Labelled AngII was obtained from ProSearch. HEK293, Chinese hamster ovary (CHO-K1), NIH-3T3, cells were obtained from American Type Culture Collection. HEK293FT were obtained by Thermo Fisher Scientific. HEK-adherent-293 cell line was a generous gift from Dr Dominic Ng, University of Queensland, Australia. AngII was obtained from Auspep or Sigma Aldrich and EGF from R&D Systems or Peprotech. Inhibitors used were YM-254890 (Wako Pure Chemical Industries), Candesartan (AstraZeneca), AG1478 (Merck Millipore) with PP2 and BAPTA obtained from Sigma Aldrich.
2.2. Plasmids
This study used a rat AT1a receptor clone (referred to as AT1R). Preparation of the following cDNA constructs has been previously described: Grb2-Venus (47), β-arrestin2-Venus (48), AT1R-Vn, β-arrestin2-Rluc8 (49), pcDNA3.1 β-arrestin1-Flag and β-arrestin2-Flag (50), and AT1R, [TK325]AT1R and [Y215F]AT1R (20). EGFR-Vc was supplied from Ichi N. Maruyama (Okinawa Institute of Science and Technology, Okinawa, Japan) (51). The following cDNA constructs were gifts from collaborators: Venus-K-ras from Prof Nevin Lambert (Georgia Regents University) and HA-V2R from Dr Thierry Durroux (Functional Genomics Institute, Montpellier, France). HA-β1AR was generated by GeneArt (Thermo Fisher Scientific, Regensburg, Germany) and α1bAR, α2aAR, β2AR, HA-β2R and HA-D1R cDNAs were obtained from the Missouri S&T cDNA Resource Center (www.cdna.org). AT1R-Rluc8 was generated by inserting AT1R from AT1R-Venus into pcDNA3-Rluc8, prepared previously from cDNA kindly provided by Andreas Loening and Sanjiv Gambhir (Stanford University, CA). EGFR-Rluc8, HER2-Rluc8 and HER3-Rluc8 were generated by inserting EGFR, HER2, and HER3, obtained from Origene (Rockville, MD, USA) into pcDNA3-Rluc8. EGFR-Venus was subsequently generated from EGFR-Rluc8 by replacing the Rluc8 coding region with Venus cDNA from pcDNA3-Venus prepared previously from pcC2-Venus kindly provided by Atsushi Miyawaki (RIKEN Brain Science Institute, Wako-city, Japan). Internally EE-tagged Gαq and Gα12 and untagged DN Gαq Q209L/D277N mutant, Gα11 and Gα13 were from UMR cDNA resource center (University of Missouri-Rolla (www.cdna.org)). A HEK 293 cell line that lacks both arrestin2 and 3 (β-arrestin 1 and 2) was used as previously described (52).
2.3. Cell Culture & Transfection
NIH-3T3, HEK293, HEK293FT cells and HEK-adherent-293 cell line, which is a derivative of HEK293 with increased adherence, were grown in Dulbecco’s Modified Eagle’s Medium (DMEM, Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Life Technologies) and antibiotic-antimycotic solution and HEK293FT cells contained 0.3mg/ml glutamine. Stably expressing NHA-AT1R -HEK293 and HEK293FT cells were maintained as above in DMEM/10% FBS with the addition of 200μg/ml of media of Geneticin (G-148, Gibco). CHO-K1 cells were maintained in α-MEM (Life Technologies) supplemented with 10% FBS and antibiotic-antimycotic solution. All cells were maintained at 37°C in 5% CO2. All cells were used between passages 5 to 30. Cells were transiently transfected using FuGene (Promega) according to manufacturer’s instructions or with Lipofectamine 2000 (Invitrogen) as previously described (41). All cells were used 48 hours post-transfection.
2.4. Isolation of VSMC
VSMC isolation and procedures were performed with prior approval from Temple University Institutional Animal Care and Use Committee and in accordance with National Institute of Health Guide for the Care and Use of Laboratory animals. Male Sprague-Dawley rats (350g; Charles River Breeding Laboratory Inc., Wilmington, MA) were used for the preparation of primary VSMC cultures. Rodents were euthanized by CO2 inhalation followed by cervical dislocation. VSMC were prepared from thoracic aorta of rats by the explant method as previously described (53,54). VSMCs were subcultured in DMEM containing 10% FBS, penicillin and streptomycin. VSMC were used between passages 3 to 12.
2.5. Protein Extraction & BCA Assays
NHA-AT1R-HEK293 were seeded onto 6-well plates at 600,000 cells/well in DMEM/10% FBS. Cells were serum starved for 24 hours and then stimulated with 100nM AngII, 10nM EGF or vehicle for 5 minutes at 37°C. After treatment, cells were washed in ice-cold PBS (Gibco) (pH 7.4) and lysed with 200μL/well of ice-cold RIPA lysis buffer [50mM Tris-HCl pH 7.5, 100mM NaCl, 2mM EDTA, 50mM sodium fluoride, 0.1% (w/v) SDS, 0.5% (w/v) sodium deoxycholate, 1% (v/v) Triton X-100, 10mM sodium pyrophosphate, 10mM sodium orthovanadate] containing Complete EDTA-free protease inhibitor cocktail (Roche) and PhosSTOP phosphatase inhibitor tablet (Roche). Cells were gently lysed for 1 hour at 4°C and centrifuged at 15,000g for 15 minutes at 4°C. Lysates were scraped from the wells and protein concentration determined using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). For the resolution of all proteins, protein samples were mixed with Laemmli sample buffer (BioRad) containing 8% β-mercaptoethanol and heated at 95°C for 5 minutes.
2.6. SDS-PAGE and Western Blot Analysis
Samples were resolved by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE). Protein samples and a molecular weight standard (Precision Plus Protein™ Dual Color standards, BIO-RAD) were electrophoresed at 100V for approximately 2 hours. Proteins were then transferred electrophoretically onto polyvinylidene fluoride (PVDF) membrane (Immobilon-P, Merck Millipore). Membranes were incubated with blocking buffer (Odyssey) at room temperature for 1 hour and then with primary rabbit polyclonal anti-p44/42 ERK1/2 MAPK and primary mouse monoclonal anti phospho-p44/42 MAPK P-ERK1/2 (Cell Signaling, Cat #4695 and #9106 respectively) (1:1000 dilution in blocking buffer) at 4°C overnight. Membranes were washed with washing buffer (PBS with 0.1% Tween20) 4 times for 5 minutes each and incubated with a goat anti mouse IRDye 800CW (LI-COR Biosciences, Cat# 926-32210) and donkey anti rabbit IRDye 680RD (LI-COR Biosciences, Cat# 926-32222) for 1 hour at room temperature (1:10,000 dilution in blocking buffer). After further washing, membranes were scanned using an Odyssey Licor Scanner (LI-COR Biosciences). Densitometry analysis of membrane was performed using Image Studio Version 5.2 software.
2.7. Ligand-induced BRET Assays & Receptor-Heteromer Investigation Technology
HEK293 and HEK293FT cells (700,000 cells/well), NIH-3T3 (600,000 cells/well), and CHO-K1 cells (400,000 cells/well), were seeded into 6-well plates and maintained at 37°C and 5% CO2 in complete media supplemented with 10% FBS. 24 hours after seeding, cells were transiently transfected using FuGene (Promega) according to manufacturer’s recommendations. 24 hours post-transfection, cells were washed with PBS, and detached using 0.25% trypsin (Gibco), resuspended in FluoroBrite phenol red-free Complete media (Gibco) containing 5% FBS or phenol-red free DMEM containing 25mM HEPES, 0.3mg/ml glutamine, 100IU/ml penicillin and 100μg/ml streptomycin supplemented with 5% FBS and added to poly-D-Lysine-coated (Sigma) 96-well white polystyrene microplate (Nunc or Gibco) at 100,000 cells/well. Early the following day, cells were serum starved with FluoroBrite phenol red-free media + 10% L-Glutamine or 25mM HEPES with 0.3mg/ml glutamine, 100IU/mL penicillin and 100μg/ml streptomycin for at least 6 hours. 48 hours post-transfection, media was replaced with PBS or Hanks’ balanced salt solution (HBSS) containing coelenterazine-h (Promega) at a final concentration of 5μM or furimazine (Promega) at a final concentration of 10μM. BRET1 measurements were taken at 37°C every 30 seconds using: a M1000 PRO Tecan plate reader (Tecan, Life Sciences) with 430-485 nm and 505-590 nm filters; a CLARIOstar Microplate Reader (BMG Labtech) with 420-480 nm and 520-620 nm filters; or a LUMIstar Omega Microplate Reader (BMG Labtech) with 450-480 nm and 520-550 nm filters. Following baseline readings, cells were treated with vehicle, 10μM AngII or 1μM EGF. The BRET signal observed between interacting proteins was normalized by subtracting the background BRET ratio as follows: the long wavelength emission over the short wavelength emission for a cell sample treated with vehicle was subtracted from the same ratio for a second aliquot of the same cell sample treated with 10μM AngII or 1μM EGF (49). For these BRET kinetic assays, the arrow indicates the first BRET reading following treatment of agonist/vehicle (time of ligand/vehicle addition). BRET assessment using parental HEK293 and knockout HEK293 has been described previously (41). Ligand- and vehicle-treated wells were assayed in triplicate wells in each independent experiment.
2.8. Bimolecular Fluorescence Complementation (BiFC)
CHO-K1 cells were plated on collagen-coated 35mm glass bottom dishes (MatTek) at a density of approximately 400,000 cells/dish. 24 hours post-seeding, cells were transiently transfected with 0.15μg DNA of Vn-tagged protein and 0.15μg DNA of Vc-tagged protein, to a total of 0.3μg of protein (compensated with empty vector when expressing DNA constructs individually) using FuGene. 24 hours post transfection, cells were serum starved using FluoroBrite phenol red-free media + 10% L-Glutamine for 24 hours. Cells were assayed 48 hours after transfection. Localization of complemented Venus fluorescence was viewed with an Olympus FV1000 inverted confocal (FV1000, Olympus, Tokyo, Japan) 60x oil immersion objective on a heated (37°C) chamber stage.
2.9. Amaxa Nucleofector Transfection
Isolated VSMC (1.0x106) were transfected by Amaxa Nucleofector program U-25 in basic Smooth Muscle Cells Nucleofector™ solution (Lonza) containing 1.5μg EGFR-Rluc8, 0.5μg Grb2-Venus with 0.15μg NHA-AT1R. 24 hours post-transfection, cells were incubated with serum-free DMEM for 24 hours. Cells were assayed 48 hours post-transfection.
2.10. Generation of CRISPR Gene Deletion Cell lines
Gq/11- and β-arrestin1/2-deleted HEK 293 cell lines were generated using the CRISPR/Cas9 system, as described previously (55,56).
2.11. Data Analysis
All data represent the means ± S.E.M of at least 3 independent experiments. Statistical analysis and curve fitting were carried out using GraphPad Prism version 7.0 (GraphPad Software Inc., CA, USA). Statistical analysis, indicated in figure legends, was determined using ANOVA with appropriate Dunnett’s tests for multiple comparisons or unpaired tests. Differences are considered significant with p < 0.05.
3. Results
3.1. BRET readout of AT1R-EGFR transactivation
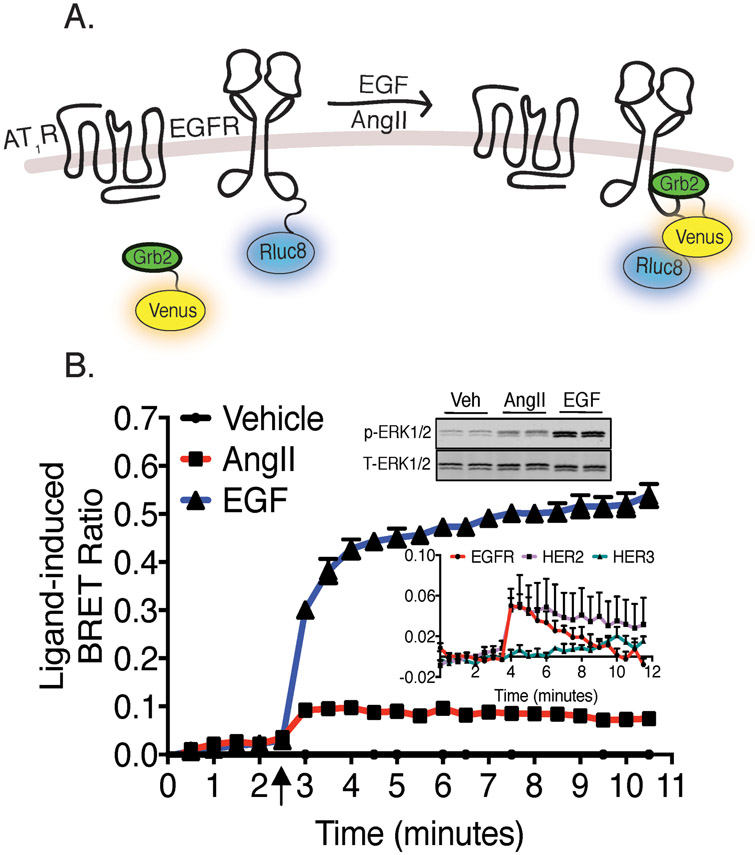
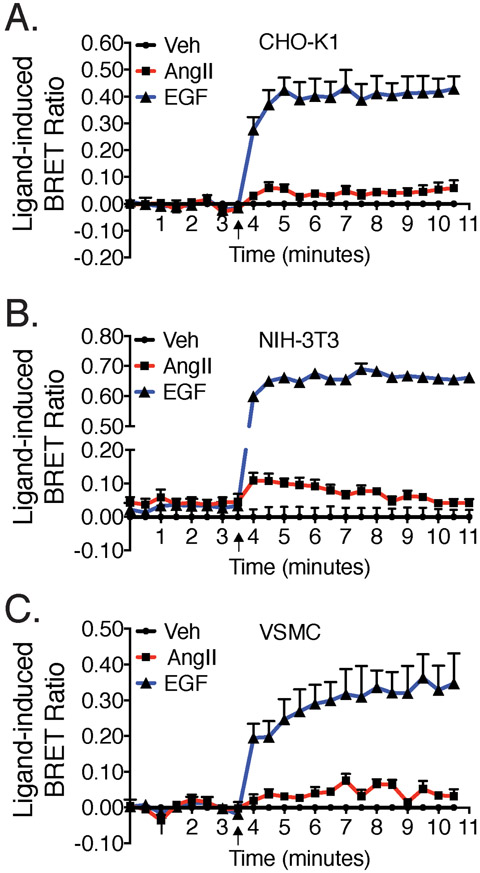
To investigate AT1R-mediated EGFR transactivation, we employed a BRET-based assay to quantitatively monitor, in living HEK293 cells, the recruitment of Grb2 to the EGFR. EGFR-Rluc8 (Renilla luciferase) and Grb2-Venus constructs were co-expressed with the AT1R, as shown in Fig. 1A. EGF stimulation produced a robust and sustained ligand-dependent BRET signal, indicative of Grb2 translocation and interaction with the EGFR (Fig. 1B). Consistent with the idea that GPCR-mediated transactivation engages only a small percentage of the total EGFR signaling capacity, maximal AngII-mediated Grb2 recruitment and the downstream activation of ERK1/2 represent approximately 20% of that stimulated by the EGF ligand (Fig. 1B). We also investigated whether AngII stimulation recruited Grb2 to other members of the EGFR family, specifically HER2 (preferred dimer partner for EGFR, which is not ligand-activated) and HER3 (which is ligand-activated, but is kinase-deficient). HER2, but not HER3, complexes with Grb2 following AngII-stimulation (inset to Fig. 1B). The BRET-based assay for AT1R-mediated transactivation extends to additional immortalized cell lines (CHO-K1 and NIH-3T3 cells, Fig. 2, A and B), as well as isolated primary vascular smooth muscle cells (VSMC) (Fig. 2C).
Fig. 1. Development of a BRET assay to study AT1R-mediated EGFR transactivation.
(A) Schematic of AT1R-EGFR BRET-based transactivation. EGFR fused to a BRET donor (Rluc8), is co-transfected with Grb2 adaptor protein tagged with a BRET acceptor (Venus) and the AT1R. Stimulation of the AT1R promotes activation of the EGFR and recruitment of Grb2. (B) HEK293 cells expressing AT1R, EGFR-Rluc8, and Grb2-Venus were treated with 10μM AngII, 1μM EGF or vehicle. Quantification of ligand-induced BRET ratio (maximum-minimum) between EGFR-Rluc8 and Grb2-Venus following AngII- and EGF-stimulation. Insert is HEK293 cells (stably expressing AT1R) stimulated with 100nM AngII, 10nM EGF or vehicle for 5 minutes before processing for phospho-ERK1/2:total-ERK1/2 (p-ERK:T-ERK) western blots. Blots are representative of 3 independent experiments (B inset) Cells expressing AT1R, Grb2-Venus and either EGFR-Rluc8, HER2-Rluc8 or HER3-Rluc8 and stimulated with 10μM AngII. Agonist stimulation is indicated by arrow. Data represent mean ± SEM of 3 independent experiments.
Fig. 2. BRET-based transactivation occurs in a variety of cell lines.
AT1R, EGFR-Rluc8 and Grb2-Venus were co-transfected into (A) CHO-K1 cells, (B) NIH-3T3 cells, and (C) primary isolated VSMCs and treated with 10μM AngII, 1μM EGF or vehicle. Agonist stimulation is indicated by arrows. Data represent mean ± SEM of 3 independent experiments.
3.2. Screening for EGFR transactivation by multiple GPCRs
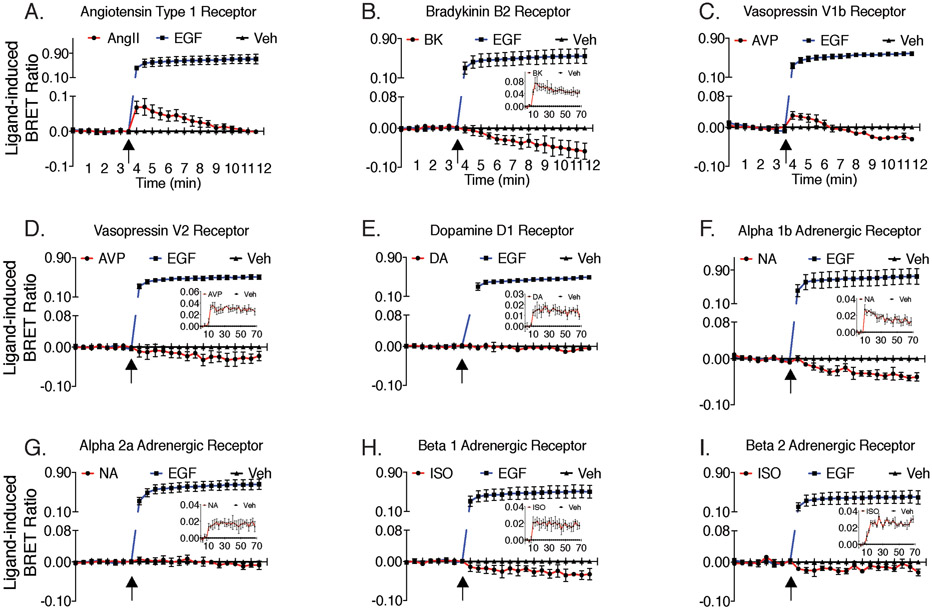
A number of GPCRs have been reported to transactivate the EGFR, including the adrenergic, dopamine, bradykinin, vasopressin and adenosine receptors, although the exact mechanisms remain unresolved (18,57). A focused screen of 9 separate GPCRs and their capacity to recruit Grb2 to the EGFR was performed in HEK293FT, as shown in Fig. 3, A-I. Of the subset of GPCRs tested, we only observed an increase in the EGFR-Grb2 BRET ratio for the AT1R and vasopressin 1b receptor (V1bR). For the other GPCRs, their activation led to no change or a slight decrease in the EGFR-Grb2 BRET ratio. We substantiated that these GPCRs were functional in a BRET-based arrestin trafficking assay (insets to Fig. 3 B, D-I), and all assays included EGF activation of the EGFR as an internal control.
Fig. 3. Screen of multiple GPCRs using the BRET-based transactivation assay.
As per the title of each graph, panels (A-I) show 9 different GPCRs expressed in HEK293FT cells and their ability to induce Grb2-Venus interaction with EGFR-Rluc8 following 1μM EGF or 10μM GPCR agonist stimulation. (Inset) shows each GPCR and their ability to induce β-arrestin2-Rluc8 proximity to plasma membrane marker K-ras-Venus, following 10μM GPCR agonist-stimulation. Agonist stimulation is indicated by arrows. Data represent mean ± SEM of 3-5 independent experiments.
3.3. Pharmacological Characterization of GPCR-EGFR transactivation
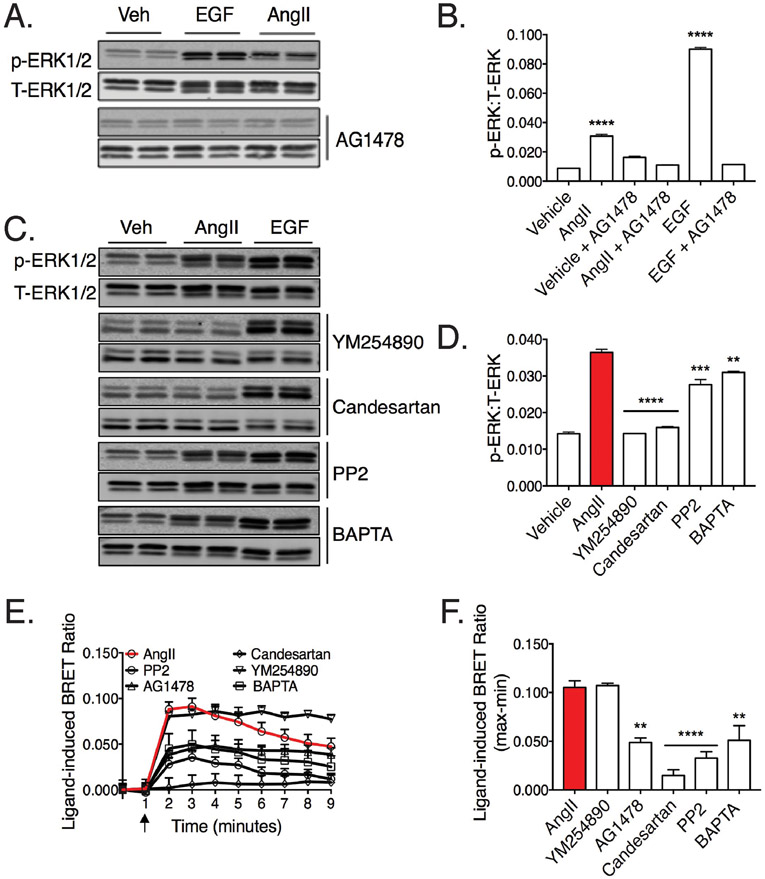
In order to examine the molecular mechanisms underlying AT1R-EGFR transactivation, we first examined ERK1/2 activation as an indirect readout. HEK293 cells expressing the AT1R were stimulated with AngII, leading to the robust and reproducible activation of ERK1/2 (Fig. 4A). To confirm that ERK1/2 activation by AngII was due to EGFR transactivation, cells were pretreated with the EGFR kinase inhibitor AG1478 (Fig. 4, A and B), or the Gq/11 inhibitor YM-254890, the AT1R inverse agonist candesartan, the src inhibitor PP2 or calcium chelator BAPTA (Fig. 4, C and D). Consistent with previous studies (34,40), AngII-induced ERK1/2 activation was attenuated by antagonism of the AT1R, inhibition of the EGFR kinase as well as the inhibition of Gq/11 (Fig. 4, C and D). Furthermore, AT1R-EGFR transactivation was blunted by pharmacological inhibition of calcium signaling and src activity, however to a lesser extent. Taken together, EGFR transactivation (as measured by ERK1/2 activation) by the AT1R appears to be a Gq/11-mediated pathway and dependent on EGFR kinase activity.
Fig. 4. Analysis of AT1R-EGR transactivation when comparing ERK1/2 phosphorylation and Grb2 recruitment to the EGFR.
(A-B) HEK293 cells stably expressing AT1R were treated with 500nM AG1478 (EGFR inhibitor) for 30 minutes prior to stimulation with 100nM AngII, 10nM EGF or vehicle for 5 minutes before processing for phospho-ERK1/2:total-ERK1/2 (p-ERK:T-ERK) western blots. Blots are representative of 3 independent experiments. (B) Quantification of western blot data using densitometry analysis (ImageJ) illustrates p-ERK1/2 changes. **** p<0.001 vs. vehicle.. (C-D) HEK293 stably expressing AT1R were treated with 1μM YM-254890 (Gq/11 inhibitor), 1μM Candesartan (AT1R inverse agonist), 10μM BAPTA (Calcium chelator) and 5μM PP2 (src inhibitor) for 30 minutes prior to stimulation with 100nM AngII, 10nM EGF or vehicle for 5 minutes before processing for ERK/p-ERK western blots. Blots are representative of 3 independent experiments. (D) Quantification of western blot data using densitometry analysis (ImageJ) illustrates phospho-ERK1/2 changes. **** p<0.001, *** p<0.005, ** p<0.01 vs. AngII treated cells. (E-F) HEK293 cells transiently expressing AT1R, EGFR-Rluc8 and Grb2-Venus were treated with 1μM AG1478, 1μM YM-254890, 1μM Candesartan, 10μM BAPTA and 5μM PP2 for 30 minutes prior to stimulation with 10μM AngII, 1μM EGF or vehicle. (F) Quantification of ligand-induced BRET ratio (maximum-minimum) between EGFR-Rluc8 and Grb2-Venus following AngII-treatment and in the presence of inhibitors. Agonist stimulation is indicated by arrows. Data represents mean ± SEM of 3 independent experiments. Statistical analysis by a one-way ANOVA with a Dunnett’s post-test for multiple comparisons; ** p<0.01; *** p<0.005; **** p<0.001 vs. AngII treated cells.
We next examined whether this pharmacological profile was recapitulated when the BRET based Grb2 recruitment assay was used as a readout for EGFR transactivation. Similar to AngII-mediated ERK1/2 activation, candesartan completely inhibited AngII-stimulated Grb2 recruitment to the EGFR, indicating that transactivation of the EGFR is due to ligand activation of the AT1R (Fig. 4E). However, AngII-mediated recruitment of Grb2 to the EGFR was not inhibited by pre-treatment with the Gq/11 inhibitor, YM-254890, which is in stark contrast to the outcome of the ERK1/2-based readout mentioned above. Indeed, YM-254890 treatment led to a sustained BRET signal (Fig. 4E). Additionally, Grb2 recruitment to the EGFR in response to AngII was only partially dependent upon the EGFR kinase activity, as well as src and calcium (Fig. 4, E and F).
3.4. The role of Gq/11 and β-arrestin in AT1R-EGFR transactivation
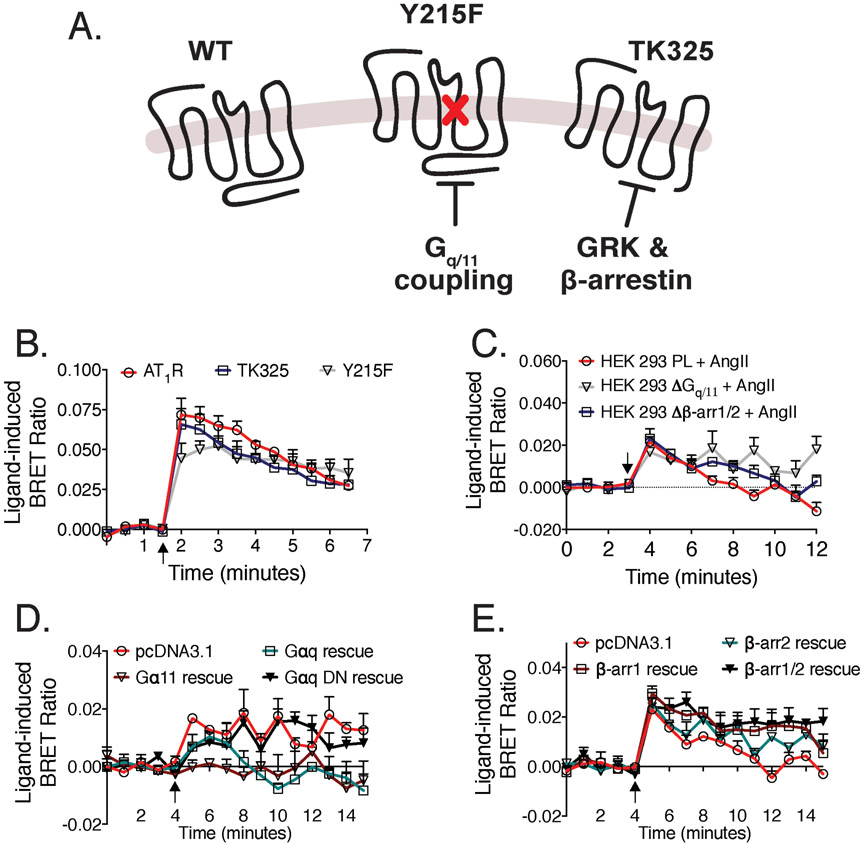
Given, the disparity in the apparent contribution of Gq/11 to EGFR transactivation, we next sought to interrogate a single point mutant of the AT1R, [Y215F]AT1R, previously reported to retain high affinity for AngII, but an inability to couple to Gq/11 mediated signaling (58) (Fig. 5A). Stimulation of the [Y215F]AT1R promoted Grb2 recruitment to the EGFR approximately equivalent to the wild type AT1R (Fig. 5B). Similarly, a truncated mutant of the AT1R, [TK325]AT1R, lacking 34 carboxyl terminal amino acids and deficient in ligand-mediated phosphorylation and recruitment of β-arrestins (59,60) also showed similar levels of Grb2 recruitment as the wild type receptor.
Fig. 5. Dependence on β-arrestin and Gq/11 in AngII-mediated Grb2 recruitment to the EGFR.
(A) Schematic of wild type AT1R and mutant AT1Rs - [TK325]AT1R (truncated AT1R deficient in recruiting β-arrestins) and [Y215F]AT1R (inability to couple to Gq/11 signaling pathways). (B) HEK293 cells expressing EGFR-Rluc8 and Grb2-Venus, alongside wild type AT1R, [Y215F]AT1R or [TK325]AT1R, were treated with 10μM AngII, 1μM EGF or vehicle. (C) EGFR-Rluc8, Grb2-Venus and AT1R were transfected into the HEK293 parental cell line, as well as Gq/11- and β-arrestin1/2-knockout CRISPR cell lines. (D) BRET constructs were expressed in the absence (pcDNA3.1) or presence of either G11, Gq or Gq dominant-negative (DN) and were treated with 10μM AngII. (E) BRET constructs were expressed in the absence (pcDNA3.1) or presence of either β-arrestin1, β-arrestin2 or β-arrestin1/2 and were treated with 10μM AngII. All cell lines were treated with 10μM AngII. Agonist stimulation is indicated by arrows. Data represent mean ± SEM of 3 independent experiments.
To extend our observations that AT1R-EGFR transactivation may be independent of Gq/11 and β-arrestin, we also performed EGFR-Grb2 BRET assays in CRISPR-mediated Gq/11 and β-arrestin knockout HEK293 cell lines (41). The BRET-based sensors were transfected with the wild type AT1R into a HEK293 parental (PL) cell line, a Gq/11 knockout (ΔGq/11) cell line or a β-arrestin1/2 knockout (Δβ-arr1/2) cell line. Comparable AngII-induced EGFR-Grb2 BRET ratios were observed in all three cell lines (Fig. 5C). Rescue experiments (i.e., transfection separately of either Gq, G11 or a dominant negative Gq back into the ΔGq/11 cells); or β-arrestin1/2 (transfected separately or together) into the Δβ-arr1/2 cell line were also performed (Fig. 5, D and E). Restoration of Gq/11 proteins inhibited AngII-mediated transactivation, suggesting, if anything, that Gq/11 coupling disfavors AngII-mediated transactivation. Reintroduction of β-arrestin1 and β-arrestin2 into the Δβ-arr HEK 293 cells did not affect AngII-mediated EGFR transactivation. Together, these data confirm the independence of Gq/11 and β-arrestin for AT1R-EGFR transactivation.
3.5. AT1R and EGFR exist as complexes
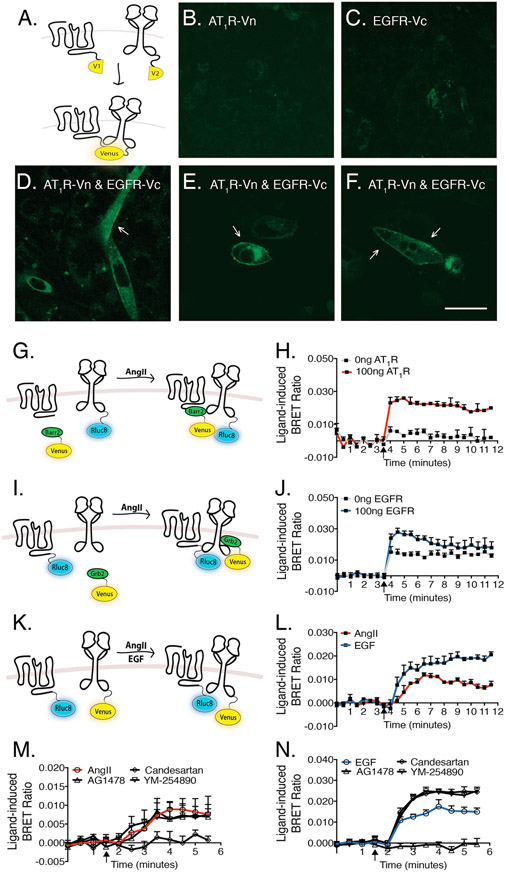
We speculated as to whether the mechanism of GPCR-EGFR transactivation may involve heteromerization of the receptors. In order to study this, we initially used a biomolecular fluorescence complementation (BiFC) assay, whereby complexing between AT1R and EGFR would result in the complementation of YFP fragments attached separately to the AT1R (AT1R-Vn) and the EGFR (EGFR-Vc) (Fig. 6A). AT1R-Vn and EGFR-Vc were not fluorescent when expressed separately in cells (Fig. 6, B and C). Co-expression leads to complemented fluorescence with clear membrane localization (Fig. 6, D-F).
Fig. 6. Detection of AT1R and EGFR receptor complexes using BiFC and Receptor-HIT.
(A) The Venus fluorophore is split into N-terminal (Vn) and C-terminal (Vc) fragments and genetically fused to AT1R and EGFR, respectively. Therefore, if AT1R and EGFR that bear C-terminal Vn and Vc tags form heteromers, fluorescence will be generated via the reconstitution of the Venus fluorophore. (B, C) CHO-K1 cells expressing AT1R-Vn or EGFR-Vc constructs separately displayed very low levels of autofluorescence. (D-F) Co-expression of AT1R-Vn and EGFR-Vc in CHO-K1 cells. Scale bar, 30μm. (G) If the AT1R and EGFR exist as a heteromers, AngII-stimulation will activate β-arrestin2 translocation to the AT1R, enabling BRET to occur between EGFR-Rluc8 and β-arrestin2-Venus. (H) HEK293 cells expressing β-arrestin2-Venus and EGFR-Rluc8 in the presence or absence of AT1R were stimulated with 10μM AngII and the BRET ratio determined. (I) In contrast to EGFR, the AT1R does not interact with Grb2 in a ligand-dependent manner. However, if the AT1R and EGFR exist as a heteromers, AngII stimulation resulting in transactivation of the EGFR and initiation of Grb2 translocation to the activated EGFR should enable BRET to occur between AT1R-Rluc8 and Grb2-Venus. (J) HEK293 cells expressing AT1R-Rluc8 and Grb2-Venus in the presence or absence of transfected EGFR, was stimulated with 10μM AngII. (K) A direct readout of ligand-induced changes in AT1R-Rluc8 proximity to EGFR-Venus. (L) Co-expression of AT1R-Rluc8 and EGFR-Venus in HEK293 cells stimulated with 10μM of AngII or 1μM EGF. (M-N) HEK293 cells expressing AT1R-Rluc8 and EGFR-Venus were treated with 1μM YM-254890, 1μM Candesartan or 1μM AG1478 for 30 minutes prior to stimulation with (M) 10μM AngII only or (N) 1μM EGF only. Agonist stimulation is indicated by arrows. Data represent mean ± SEM of 3-4 independent experiments.
To examine whether these complexes are ligand-dependent, we utilized Receptor-Heteromer Investigation Technology (Receptor-HIT), which is most often used on the BRET platform to offer insights into specific ligand-dependent recruitment of proteins to heteromers (61,62). AngII-induced recruitment of β-arrestin2-Venus to unlabeled AT1R, resulting in BRET with EGFR-Rluc8, is indicative of heteromerization (Fig. 6G). Upon AngII stimulation, an increase in BRET ratio between β-arrestin2-Venus and EGFR-Rluc8 was observed only when the AT1R was co-expressed (Fig. 6H). We also used a complementary variation of the Receptor-HIT assay, whereby AT1R-Rluc8, unlabeled EGFR, and Grb2-Venus were co-expressed in living cells, as shown in Fig. 6I. In contrast to the EGFR, the AT1R does not interact with Grb2 in a ligand-dependent manner. However, if AT1R and EGFR exist as a heteromer, AngII stimulation will transactivate the EGFR and initiate translocation of Grb2 to the activated EGFR, facilitating BRET between AT1R-Rluc8 and Grb2-Venus. Even in the absence of exogenous EGFR transfection, some AngII-mediated BRET was observed, indicating the engagement of endogenous EGFR to recruit Grb2-Venus to AT1R-Rluc8 (Fig. 6J). This effect was enhanced by the ectopic expression of additional unlabeled EGFR (Fig. 6J).
As a final confirmation of ligand-dependent complexing between the AT1R and the EGFR, we co-transfected cells with AT1R-Rluc8 and EGFR-Venus, and stimulated with AngII or EGF. Both AngII and EGF appeared to modulate the proximity of the receptors, as shown in Fig. 6, K and L. Together, these experiments indicate that AT1R and EGFR not only exist in preformed complexes, but that agonist stimulation can modulate their association and/or relative conformations.
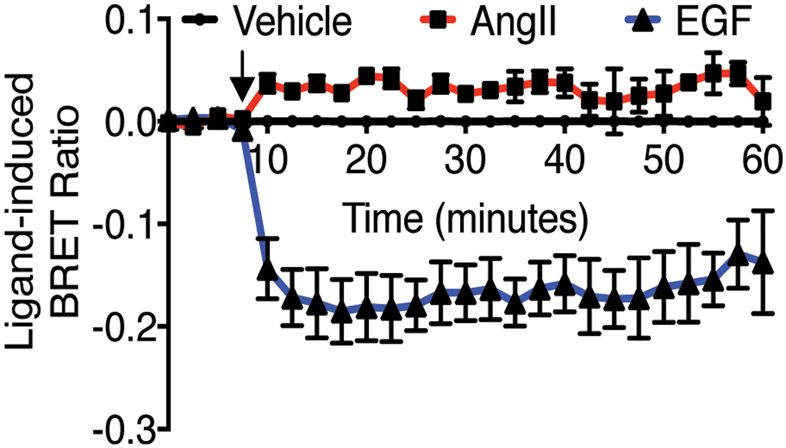
AT1R -EGFR heteromers are unlikely to merely reflect agonist-induced clustering of these proteins in the membrane and/or their concentration and co-trafficking in endosomes, because the association of the EGFR with a membrane marker, K-ras, was decreased following EGF-stimulation, but not AngII-treatment (Fig. 7).
Fig. 7: EGFR localization and trafficking.
HEK293 cells expressing AT1R, EGFR-Rluc8 and K-ras-Venus were treated with 10μM AngII, 1μM EGF and vehicle. Agonist stimulation is indicated by arrows. Data represents mean ± SEM of 5 independent experiments.
3.6. Pharmacological characterization of AT1R-EGFR complexes
Ligand-induced changes in proximity between AT1R-Rluc8 and EGFR-Venus, following AngII- and EGF-stimulation, are shown in Fig. 6, M and N. AngII-mediated increases in BRET between AT1R and EGFR were inhibited by candesartan, but not AG1478 or YM-254890, indicating that ligand-dependent modulation does not require Gq/11 coupling or EGFR kinase activity (Fig. 6M). In contrast, EGF modulation of AT1R-EGFR complexes was abrogated by pretreatment with AG1478 (Fig. 6N). Interestingly, pretreatment with candesartan or YM-254890 further increased the EGF-mediated BRET between AT1R-Rluc8 and EGFR-Venus (Fig. 6N). These data provide evidence for distinct mechanisms controlling AngII- and EGF-induced modulation of the heteromer.
4. Discussion
GPCR-mediated EGFR transactivation is commonly defined in terms of the activation of ERK1/2, which although informative, is an indirect and distal readout. Herein, we describe a BRET assay, based on the recruitment of Grb2 to the EGFR to quantitatively monitor, in living cells and in real-time, the proximal activation of EGFR. Importantly, we identified that the molecular requirements for EGFR transactivation differed depending on the readout used. For the Grb2-EGFR assay, we showed that EGFR transactivation is independent of Gq/11 coupling and does not apparently involve the AT1R carboxy-terminal tail or interaction with arrestin. Using a series of additional BRET-based assays, we provide evidence of preformed complexes between the AT1R and EGFR that show different attributes, depending on which receptor is activated. In summary, the capacity to interrogate proximal EGFR transactivation provides a platform to better define the complex molecular processes involved.
The development of novel approaches to study GPCR-mediated transactivation in live cells is important, and this study describes a unique BRET-based quantification of Grb2 recruitment to the EGFR as a direct readout of GPCR-mediated transactivation. We have previously used the recruitment of Grb2 to RTKs (EGFR and HER3) as a measure of EGF receptor family activation in HEK293 cells following stimulation with EGF and Heregulin (47). Moreover, others have also identified such BRET approaches as bona fide readouts of EGFR activation (63,64). In this study, we initially aimed to determine whether stimulation of the AT1R could promote Grb2 translocation and interaction with EGFR in a ligand-dependent manner in live cells. AT1R-mediated EGFR transactivation was readily demonstrable, but was consistently lower in magnitude compared to EGF-stimulation. The selective activation of EGFR by the AT1R is consistent with many observations that the degree of signaling elicited by GPCRs appears to be only a subset of that obtained following full activation by EGF ligands (27,35,65,66). EGFR transactivation was readily detected in HEK293 cells, which is interesting because a previous study reported that ERK1/2 activation was independent of the EGFR in these cells (65). EGFR-Grb2 recruitment was also recapitulated in other standard cell lines (CHO-K1 and NIH-3T3) and in primary isolated VSMCs, indicating the versatility of this approach to measure EGFR transactivation.
We observed that the AT1R and V1bR showed consistent recruitment of Grb2 to the EGFR, but we were surprised that other GPCRs (previously reported to cause EGFR transactivation (67-70)) did not. It is important to note that a negative response in our assay does not necessarily mean that the GPCR is not transactivating nor should it call into question the veracity of our assay. There are a number of plausible explanations: firstly, a trivial argument might be that these ‘non-transactivating’ GPCRs were non-functional – however, we confirmed their activation by using a readout of ligand-dependent β-arrestin trafficking. Secondly, these GPCRs may not be recruiting Grb2 as part of their activation process or, alternatively, there is an association but it is different in nature to that generated by the AT1R and V1bR. Indeed, for a number of the GPCRs that did not show enhanced EGFR-Grb2 association, we actually observed decreased BRET ratios following ligand stimulation. It could be reasonably argued that this reflects dynamic changes in an equilibrium between association, dissociation and reconfiguration of putative EGFR-Grb2 complexes – where the balance may favor dissociation or reconfiguration that leads to an increased distance between the donor and acceptor moieties. Thirdly, as our assay only reports on the Grb2 recruitment to the EGFR, the possibility exists that receptors other than the EGFR (HER2, HER3 or HER4) and or combinations of dimers between those receptors, are selectively used by these other GPCRs for transactivation. In this regard, it is important to acknowledge that most studies of EGFR transactivation typically use end-point assays, where specificity is inferred via inhibition by the small molecule inhibitor of the EGFR kinase, AG1478. However, high micromolar concentrations of AG1478 (as commonly used) inhibit receptors other than the EGFR (71,72). Importantly, we report the recruitment of Grb2 to HER2, but not HER3, in response to AngII stimulation. Whether a specific GPCR is able to transactivate EGFR, HER2, HER3 or HER4 will require further investigation, using the approaches we have developed. It will be important to consider that HER2 is the preferred dimerization partner for other EGFR family members, and therefore potentially acting in cis to modulate other HER receptors. Indeed, HER2 has been shown to be transactivated and/or modulate transactivation (73,74).
Another major observation of our study was the apparent disconnect between pharmacological inhibitors and their relative effect on the readout of transactivation vis-à-vis ERK1/2 versus BRET-based Grb2 recruitment assays. We clearly showed that candesartan completely inhibited AngII-induced ERK1/2 phosphorylation and recruitment of Grb2 to the EGFR, indicating that EGFR transactivation is due to ligand activation of the AT1R in both systems. Interestingly, cells treated with AG1478 showed a complete inhibition of ERK1/2 activation in response to AngII, however only a partial reduction of AngII-mediated transactivation as measured by BRET-based Grb2 recruitment assay, indicating a possible disconnect between proximal recruitment of Grb2 and the eventual activation of ERK1/2. We would argue that Grb2 recruitment is necessary, but not in itself sufficient, to drive ERK1/2 activation following AT1R activation. Importantly, EGF-mediated recruitment of Grb2 to the EGFR was completely inhibited with pretreatment of AG1478, confirming the efficacy of AG1478.
Even more unambiguous was the relative contribution of Gq/11 signaling in AngII-mediated transactivation as measured by the ERK1/2 and the BRET-based readout. Although Gq/11 was absolutely required for ERK1/2 phosphorylation following AngII-stimulation, in contrast, we observed a sustained EGFR-Grb2 interaction when cells were pretreated with YM-254890. One potential confounding issue was that different concentrations of ligands were used to achieve maximal activation in the ERK1/2 and BRET assays (refer to the legend of Figure 4 for detail) and this may have influenced the pharmacological inhibitory profile. However, we observed similar patterns of inhibition of the ERK1/2 readout when cells were stimulated with higher concentration (i.e., 10 μM AngII and 1 μM EGF, data not shown), indicating the differential contribution of Gq/11 is unlikely to be a mere artefact of ligand concentration.
Previously, it was assumed that transactivation was a consequence of Gq/11 activation (75), but our data using the proximal assay suggests a re-evaluation of this paradigm might be required. We further adapted the BRET experiments to incorporate mutant versions of the AT1R, specifically a G protein-uncoupled AT1R, [Y215F]AT1R, that does not couple efficiently to Gq/11 (20,58). Consistent with previous results, activation of [Y215F]AT1R resulted in wild type-like Grb2 recruitment to the EGFR. Interestingly, others have reported β-arrestin-mediated ERK1/2 signaling, with an involvement in cardiomyocyte survival and hypertrophy (76-78). Here, we used a carboxy-terminal truncated AT1R, [TK325]AT1R, to demonstrate that AT1R C-tail phosphorylation and β-arrestin interaction is not crucial for Grb2 recruitment. Furthermore, using CRISPR knockout HEK293 cell lines we were also able to show that transactivation (indicated by Grb2 recruitment to the EGFR) appears to be Gq/11- as well as β-arrestin1/2-independent. This disparity warrants further study and clearly emphasizes the importance of interrogating EGFR transactivation directly at the most proximal point in the process. It is possible that other G proteins might be involved or that the cell background is critical, and this requires further study.
Based on the above observations, a final consideration for the current study was the possibility that GPCR-mediated EGFR transactivation involved receptor heteromerization (79). We initially used a BiFC assay to provide preliminary evidence for heteromers between the AT1R and EGFR. Although positive, the limitations of this technique in defining ligand-driven heteromer formation meant that we then progressed to Receptor-HIT assays and a BRET-based assay measuring direct association of AT1R-EGFR. Using all three approaches, we provide evidence that AT1R and EGFR can exist as preformed complexes that occur constitutively (i.e. in the absence of ligand) and are predominantly localized to the plasma membrane. In addition to preformed complexes, AT1R-EGFR heteromers appear to be responsive to agonist stimulation and receptor activation. Ligand-induced modulation of the AT1R-EGFR heteromer was validated using two complementary Receptor-HIT approaches, with both demonstrating that AngII leads to a rapid and robust increase in BRET that required both the AT1R and EGFR. Lastly, we demonstrate a more ‘direct’ readout, tagging both receptors and monitoring ligand-induced changes in BRET due to modulation of donor-acceptor proximity. Together, these data strongly indicate that both AngII- and EGF-stimulation can induce the formation of multi-receptor complexes and/or alter the relative conformations of AT1R and EGFR in pre-existing complexes. An alternative explanation (that these data merely reflect a bystander effect or the co-accumulation of the receptors in endosomes) is not supported by our observation (see Fig. 7) that the trafficking of the EGFR from the membrane occurs with EGF stimulation, but not with AngII.
The concept of GPCRs and EGFRs existing as heteromers is consistent with previous studies. Co-immunoprecipitation studies have shown that agonist stimulation (both AngII and EGF) promotes a multi-protein complex containing the AT1R and EGFR, however these endpoint assays were not in live cells or in real-time (79). The β1- and β2-adrenergic receptors associate with the EGFR (66,80), and while isoprenaline enhances β1-adrenergic receptor association with EGFR, this is disrupted by EGF-stimulation. Others have also used BRET to provide evidence for the bile acid receptor, TGR5, forming preassembled heterodimers with the EGFR in lipid rafts, however agonist stimulation showed no further enhancement in receptor association (81). Complementary to our studies, Bhattacharya and colleagues showed G protein-coupled receptor 54 interacts with EGFR at the plasma membrane in the absence of agonist. However, only stimulation with its endogenous ligand, kisspeptin, significantly increased their association, while EGF-treatment did not affect the receptor complex (82). Although it appears that many GPCRs can transactivate EGFRs, heteromerization has only been indicated for a subset. It is still unknown how this interaction affects signaling of either receptor, with the mechanism and/or binding domains that facilitate heteromer formation largely unknown. One possible mechanism that has not been tested directly is the reported contribution of reactive oxygen species (83,84) and their potential oxidation of the EGFR on dimers formation (85).
Whether the complex subserves actual transactivation is difficult to reconcile because specific inhibitors that prevent complex formation are not available. Remarkably, we observed that the modulation of BRET signals associated with AT1R-EGFR heteromerization differed depending on whether the complex was modulated by activation of the AT1R or the EGFR. The AngII-induced complex modulation is apparently independent of both Gq/11 and EGFR kinase activity. G protein-independent transactivation has been reported for β1-adrenergic receptor and EGFR continued association (66). In contrast, in our hands, EGF-mediated modulation of the AT1R-EGFR complex was absolutely dependent on the EGFR kinase domain. Moreover, antagonism of the AT1R or inhibition of Gq/11 did not inhibit complex modulation, but rather enhanced the EGF-mediated increase in BRET signal between AT1R-Rluc8 and EGFR-Venus. Whether this enhancement plays any role in receptor activation is unclear, but it is interesting to note that traditional models of monomeric EGFR dimer formation as the active component have been superseded by models that suggest that up to 40% of EGFRs are present as inactive dimers that form higher order active receptor signaling complexes (86-89). This negative modulation of EGFR may be mirrored by the AT1R, whereby the AT1R and its associated G protein negatively modulate monomeric/dimeric/oligomeric EGFR.
In summary, the combination of different BRET approaches emphasizes the importance of monitoring the most proximal events of EGFR activation, in live cells and in real-time. Using this approach, we propose a Gq/11-independent mechanism of EGFR recruitment, instead providing evidence of ligand-modulated AT1R-EGFR complexes. This work provides a platform to better interrogate the molecular mechanisms underpinning AT1R-EGFR transactivation, and provides powerful new insights into this underappreciated process.
Acknowledgements
We thank Ichiro Maruyama (Okinawa Institute of Science & Technology, Okinawa, Japan) for provision of the EGFR-Vc BiFC plasmid. Funding was provided by NHMRC Project Grant 1085996 awarded to W.G.T. and K.D.G.P. K.D.G.P. is an NHMRC RD Wright Fellow (1085842). This work was also funded by Japan Science and Technology Agency (JST; Grant Number JPMJPR1331), Japan Agency for Medical Research and Development (AMED; JP17gm5910013) and Japan Society for the Promotion of Science (JSPS) KAKENHI (17K08264) (to A.I.).
Competing Interests
K.D.G.P. receives funding from Promega, B.M.G. Labtech and Dimerix as participating organizations of Australian Research Council Linkage Grant LP160100857. These organizations played no role in any aspect of the conception or design of the research, collection, analysis and interpretation of results, or writing and editing of the manuscript. K.D.G.P. is Chief Scientific Advisor of Dimerix, of which he maintains a shareholding. Dimerix has proprietary rights to the Receptor-HIT assay.
Abbreviations
- AngII
Angiotensin II
- AT1R
type 1 angiotensin II receptor
- EGFR/HER1/ErbB1
epidermal growth factor receptor
- BRET
bioluminescence resonance energy transfer
- Grb2
growth factor receptor-bound protein 2
- EGF
epidermal growth factor
- GPCR
G protein-coupled receptor
- HER2/ErbB2
Human epidermal growth factor receptor 2
- HER3/ErbB3
Human epidermal growth factor receptor 3
- HER4/ErbB4
Human epidermal growth factor receptor 4
- RAS
renin angiotensin system
- ROS
reactive oxygen species
- MAPK
mitogen-activated protein kinases
- ERK1/2
extracellular-regulated kinase 1/2
- RTK
receptor tyrosine kinase
- MMP
matrix metalloproteases
- ADAM
a disintegrin and metalloprotease
- RET
resonance energy transfer
- FlAsH
fluorescein arsenical hairpin
- Rluc
Renilla luciferase
- VSMC
vascular smooth muscle cells
- V1bR
vasopressin 1b receptor
- PL
parental
- BiFC
biomolecular fluorescence complementation
- Receptor-HIT
receptor-heteromer investigation technology
- DMEM
Dulbecco’s Modified Eagle’s Medium
- FBS
fetal bovine serum
- HBSS
Hanks’ balanced salt solution
- SDS-PAGE
sodium dodecyl sulphate polyacrylamide gel electrophoresis
- PVDF
polyvinylidene fluoride
- DN
dominant-negative
- AUC
area under the curve
Footnotes
Nomenclature as per International Union of Basic & Clinical Pharmacology (IUPHAR)
REFERENCES
- 1.Lu H, Rateri DL, Cassis LA, and Daugherty A (2008) The role of the renin-angiotensin system in aortic aneurysmal diseases. Current hypertension reports 10, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crowley SD, Gurley SB, Herrera MJ, Ruiz P, Griffiths R, Kumar AP, Kim H-S, Smithies O, Le TH, and Coffman TM (2006) Angiotensin II causes hypertension and cardiac hypertrophy through its receptors in the kidney. Proceedings of the National Academy of Sciences 103, 17985–17990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKinney CA, Fattah C, Loughrey CM, Milligan G, and Nicklin SA (2014) Angiotensin-(1–7) and angiotensin-(1–9): function in cardiac and vascular remodelling. Clinical science 126, 815–827 [DOI] [PubMed] [Google Scholar]
- 4.Weiss D, Sorescu D, and Taylor WR (2001) Angiotensin II and atherosclerosis. The American journal of cardiology 87, 25–32 [DOI] [PubMed] [Google Scholar]
- 5.Van Thiel BS, van der Pluijm I, te Riet L, Essers J, and Danser AJ (2015) The renin–angiotensin system and its involvement in vascular disease. European journal of pharmacology 763, 3–14 [DOI] [PubMed] [Google Scholar]
- 6.Li JF, Yang YH, Zhai ZG, Gu S, Liu Y, Miao R, Zhong PP, Wang Y, Huang XX, and Wang C (2017) Renin-angiotensin system regulates pulmonary arterial smooth muscle cell migration in chronic thromboembolic pulmonary hypertension. American Journal of Physiology-Lung Cellular and Molecular Physiology, ajplung. 00515.02016 [DOI] [PubMed] [Google Scholar]
- 7.Hartono SP, Knudsen BE, Lerman LO, Textor SC, and Grande JP (2014) Combined effect of hyperfiltration and renin angiotensin system activation on development of chronic kidney disease in diabetic db/db mice. BMC nephrology 15, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slomka T, Lennon ES, Akbar H, Gosmanova EO, Bhattacharya SK, Oliphant CS, and Khouzam RN (2016) Effects of renin-angiotensin-aldosterone system blockade in patients with end-stage renal disease. The American journal of the medical sciences 351, 309–316 [DOI] [PubMed] [Google Scholar]
- 9.Guo X, Namekata K, Kimura A, Harada C, and Harada T (2017) The Renin-Angiotensin System Regulates Neurodegeneration in a Mouse Model of Optic Neuritis. The American Journal of Pathology [DOI] [PubMed] [Google Scholar]
- 10.Bessaguet F, Danigo A, Magy L, Sturtz F, Desmoulière A, and Demiot C (2017) Candesartan prevents resiniferatoxin-induced sensory small-fiber neuropathy in mice by promoting angiotensin II-mediated AT2 receptor stimulation. Neuropharmacology 126, 142–150 [DOI] [PubMed] [Google Scholar]
- 11.Miners S, Ashby E, Baig S, Harrison R, Tayler H, Speedy E, Prince JA, Love S, and Kehoe PG (2009) Angiotensin-converting enzyme levels and activity in Alzheimer's disease: differences in brain and CSF ACE and association with ACE1 genotypes. American journal of translational research 1, 163. [PMC free article] [PubMed] [Google Scholar]
- 12.Li G, Zhang H, Zhao L, Zhang Y, Yan D, and Liu Y (2017) Angiotensin-converting enzyme 2 activation ameliorates pulmonary endothelial dysfunction in rats with pulmonary arterial hypertension through mediating phosphorylation of endothelial nitric oxide synthase. Journal of the American Society of Hypertension : JASH [DOI] [PubMed] [Google Scholar]
- 13.Xuan H, Xu B, Wang W, Tanaka H, Fujimura N, Miyata M, Michie SA, and Dalman RL (2017) Inhibition or deletion of angiotensin II type 1 receptor suppresses elastase-induced experimental abdominal aortic aneurysms. Journal of Vascular Surgery [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwai M, and Horiuchi M (2009) Role of renin–angiotensin system in adipose tissue dysfunction. Hypertension Research 32, 425–427 [DOI] [PubMed] [Google Scholar]
- 15.Chen F, Chen D, Zhang Y, Jin L, Zhang H, Wan M, Pan T, Wang X, Su Y, and Xu Y (2017) Interleukin-6 deficiency attenuates angiotensin II-Induced cardiac pathogenesis with increased myocyte hypertrophy. Biochemical and biophysical research communications [DOI] [PubMed] [Google Scholar]
- 16.Husain K, Hernandez W, Ansari RA, and Ferder L (2015) Inflammation, oxidative stress and renin angiotensin system in atherosclerosis. World journal of biological chemistry 6, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinh QN, Drummond GR, Kemp-Harper BK, Diep H, De Silva TM, Kim HA, Vinh A, Robertson AA, Cooper MA, and Mansell A (2017) Pressor response to angiotensin II is enhanced in aged mice and associated with inflammation, vasoconstriction and oxidative stress. Aging (Albany NY) 9, 1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrester SJ, Kawai T, O'Brien S, Thomas W, Harris RC, and Eguchi S (2016) Epidermal Growth Factor Receptor Transactivation: Mechanisms, Pathophysiology, and Potential Therapies in the Cardiovascular System. Annual review of pharmacology and toxicology 56, 627–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai T, Forrester SJ, O’Brien S, Baggett A, Rizzo V, and Eguchi S (2017) AT1 receptor signaling pathways in the cardiovascular system. Pharmacological Research [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith NJ, Chan HW, Qian H, Bourne AM, Hannan KM, Warner FJ, Ritchie RH, Pearson RB, Hannan RD, and Thomas WG (2011) Determination of the exact molecular requirements for type 1 angiotensin receptor epidermal growth factor receptor transactivation and cardiomyocyte hypertrophy. Hypertension 57, 973–980 [DOI] [PubMed] [Google Scholar]
- 21.Thomas WG, Brandenburger Y, Autelitano DJ, Pham T, Qian HW, and Hannan RD (2002) Adenoviral-directed expression of the type 1A angiotensin receptor promotes cardiomyocyte hypertrophy via transactivation of the epidermal growth factor receptor. Circulation Research 90, 135–142 [DOI] [PubMed] [Google Scholar]
- 22.Eguchi S, Dempsey PJ, Frank GD, Motley ED, and Inagami T (2001) Activation of MAPKs by angiotensin II in vascular smooth muscle cells. Metalloprotease-dependent EGF receptor activation is required for activation of ERK and p38 MAPK but not for JNK. The Journal of biological chemistry 276, 7957–7962 [DOI] [PubMed] [Google Scholar]
- 23.Mifune M, Ohtsu H, Suzuki H, Nakashima H, Brailoiu E, Dun NJ, Frank GD, Inagami T, Higashiyama S, Thomas WG, Eckhart AD, Dempsey PJ, and Eguchi S (2005) G protein coupling and second messenger generation are indispensable for metalloprotease-dependent, heparin-binding epidermal growth factor shedding through angiotensin II type-1 receptor. Journal of Biological Chemistry 280, 26592–26599 [DOI] [PubMed] [Google Scholar]
- 24.Grassot J, Mouchiroud G, and Perrière G (2003) RTKdb: database of receptor tyrosine kinase. Nucleic acids research 31, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Kofler M, Jorissen RN, Nice EC, and Burgess AW (2003) The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Molecular cell 11, 495–505 [DOI] [PubMed] [Google Scholar]
- 26.Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, and Ullrich A (1999) EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature 402, 884–888 [DOI] [PubMed] [Google Scholar]
- 27.Daub H, Weiss FU, Wallasch C, and Ullrich A (1996) Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature 379, 557–560 [DOI] [PubMed] [Google Scholar]
- 28.Touyz RM, Wu XH, He G, Salomon S, and Schiffrin EL (2002) Increased Angiotensin II-Mediated Src Signaling via Epidermal Growth Factor Receptor Transactivation Is Associated With Decreased C-Terminal Src Kinase Activity in Vascular Smooth Muscle Cells From Spontaneously Hypertensive Rats. Hypertension 39, 479–485 [DOI] [PubMed] [Google Scholar]
- 29.Luttrell LM, DellaRocca GJ, vanBiesen T, Luttrell DK, and Lefkowitz RJ (1997) G beta gamma subunits Src-dependent phosphorylation of the epidermal growth factor receptor - A scaffold for G protein-coupled receptor-mediated Ras activation. Journal of Biological Chemistry 272, 4637–4644 [DOI] [PubMed] [Google Scholar]
- 30.Eguchi S, Iwasaki H, Inagami T, Numaguchi K, Yamakawa T, Motley ED, Owada KM, Marumo F, and Hirata Y (1999) Involvement of PYK2 in Angiotensin II Signaling of Vascular Smooth Muscle Cells. Hypertension 33, 201–206 [DOI] [PubMed] [Google Scholar]
- 31.Andreev J, Galisteo ML, Kranenburg O, Logan SK, Chiu ES, Okigaki M, Cary LA, Moolenaar WH, and Schlessinger J (2001) Src and Pyk2 mediate G-protein-coupled receptor activation of epidermal growth factor receptor (EGFR) but are not required for coupling to the mitogen-activated protein (MAP) kinase signaling cascade. Journal of Biological Chemistry 276, 20130–20135 [DOI] [PubMed] [Google Scholar]
- 32.Shah BH, and Catt KJ (2002) Calcium-independent activation of extracellularly regulated kinases 1 and 2 by angiotensin II in hepatic C9 cells: Roles of protein kinase c delta, Src/proline-rich tyrosine kinase 2, and epidermal growth factor receptor trans-activation. Molecular pharmacology 61, 343–351 [DOI] [PubMed] [Google Scholar]
- 33.Olson ER, Shamhart PE, Naugle JE, and Meszaros JG (2008) Angiotensin II-induced extracellular signal-regulated kinase 1/2 activation is mediated by protein kinase C delta and intracellular calcium in adult rat cardiac fibroblasts. Hypertension 51, 704–711 [DOI] [PubMed] [Google Scholar]
- 34.Ohtsu H, Higuchi S, Shirai H, Eguchi K, Suzuki H, Hinoki A, Brailoiu E, Eckhart AD, Frank GD, and Eguchi S (2008) Central role of Gq in the hypertrophic signal transduction of angiotensin II in vascular smooth muscle cells. Endocrinology 149, 3569–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eguchi S, Numaguchi K, Iwasaki H, Matsumoto T, Yamakawa T, Utsunomiya H, Motley ED, Kawakatsu H, Owada KM, Hirata Y, Marumo F, and Inagami T (1998) Calcium-dependent Epidermal Growth Factor Receptor Transactivation Mediates the Angiotensin II-induced Mitogen-activated Protein Kinase Activation in Vascular Smooth Muscle Cells. Journal of Biological Chemistry 273, 8890–8896 [DOI] [PubMed] [Google Scholar]
- 36.Wei HJ, Ahn S, Shenoy SK, Karnik SS, Hunyady L, Luttrell LM, and Lefkowitz RJ (2003) Independent beta-arrestin 2 and G protein-mediated pathways for angiotensin II activation of extracellular signal-regulated kinases 1 and 2. Proceedings of the National Academy of Sciences of the United States of America 100, 10782–10787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greco S, Muscella A, Elia MG, Salvatore P, Storelli C, Mazzotta A, Manca C, and Marsigliante S (2003) Angiotensin II activates extracellular signal regulated kinases via protein kinase C and epidermal growth factor receptor in breast cancer cells. Journal of cellular physiology 196, 370–377 [DOI] [PubMed] [Google Scholar]
- 38.Hunyady L. s., and Catt KJ (2006) Pleiotropic AT1 receptor signaling pathways mediating physiological and pathogenic actions of angiotensin II. Molecular endocrinology 20, 953–970 [DOI] [PubMed] [Google Scholar]
- 39.Kim J, Ahn S, Rajagopal K, and Lefkowitz RJ (2009) Independent β-arrestin2 and Gq/protein kinase Cζ pathways for ERK stimulated by angiotensin type 1A receptors in vascular smooth muscle cells converge on transactivation of the epidermal growth factor receptor. Journal of Biological Chemistry 284, 11953–11962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.George AJ, Purdue BW, Gould CM, Thomas DW, Handoko Y, Qian H, Quaife-Ryan GA, Morgan KA, Simpson KJ, Thomas WG, and Hannan RD (2013) A functional siRNA screen identifies genes modulating angiotensin II-mediated EGFR transactivation. J. Cell Sci 126, 5377–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devost D, Sleno R, Pétrin D, Zhang A, Shinjo Y, Okde R, Aoki J, Inoue A, and Hebert TE (2017) Conformational Profiling of the AT1 Angiotensin II Receptor Reflects Biased Agonism, G Protein Coupling, and Cellular Context. Journal of Biological Chemistry 292, 5443–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bourque K, Pétrin D, Sleno R, Devost D, Zhang A, and Hébert TE (2017) Distinct Conformational Dynamics of Three G Protein-Coupled Receptors Measured Using FlAsH-BRET Biosensors. Frontiers in endocrinology 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turu G, Szidonya L, Gaborik Z, Buday L, Spat A, Clark AJ, and Hunyady L (2006) Differential beta-arrestin binding of AT1 and AT2 angiotensin receptors. FEBS Lett 580, 41–45 [DOI] [PubMed] [Google Scholar]
- 44.Shukla AK, Violin JD, Whalen EJ, Gesty-Palmer D, Shenoy SK, and Lefkowitz RJ (2008) Distinct conformational changes in β-arrestin report biased agonism at seven-transmembrane receptors. Proceedings of the National Academy of Sciences 105, 9988–9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang W, Strachan RT, Lefkowitz RJ, and Rockman HA (2014) Allosteric modulation of β-arrestin-biased angiotensin II type 1 receptor signaling by membrane stretch. Journal of Biological Chemistry 289, 28271–28283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adjobo-Hermans MJ, Goedhart J, van Weeren L, Nijmeijer S, Manders EM, Offermanns S, and Gadella TW (2011) Real-time visualization of heterotrimeric G protein Gq activation in living cells. BMC biology 9, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ayoub MA, See HB, Seeber RM, Armstrong SP, and Pfleger KD (2013) Profiling epidermal growth factor receptor and heregulin receptor 3 heteromerization using receptor tyrosine kinase heteromer investigation technology. PloS one 8, e64672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kocan M, See HB, Sampaio N. l. G., Eidne KA, Feldman BJ, and Pfleger KD (2009) Agonist-independent interactions between β-arrestins and mutant vasopressin type II receptors associated with nephrogenic syndrome of inappropriate antidiuresis. Molecular endocrinology 23, 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porrello ER, Pfleger KDG, Seeber RM, Qian H, Oro C, Abogadie F, Delbridge LMD, and Thomas WG (2011) Heteromerization of angiotensin receptors changes trafficking and arrestin recruitment profiles. Cellular signalling 23, 1767–1776 [DOI] [PubMed] [Google Scholar]
- 50.Manglik A, Kim TH, Masureel M, Altenbach C, Yang Z, Hilger D, Lerch MT, Kobilka TS, Thian FS, and Hubbell WL (2015) Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell 161, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tao R-H, and Maruyama IN (2008) All EGF (ErbB) receptors have preformed homo-and heterodimeric structures in living cells. J. Cell Sci 121, 3207–3217 [DOI] [PubMed] [Google Scholar]
- 52.Alvarez-Curto E, Inoue A, Jenkins L, Raihan SZ, Prihandoko R, Tobin AB, and Milligan G (2016) Targeted elimination of G proteins and arrestins defines their specific contributions to both intensity and duration of G protein-coupled receptor signaling. Journal of Biological Chemistry 291, 27147–27159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takaguri A, Shirai H, Kimura K, Hinoki A, Eguchi K, Carlile-Klusacek M, Yang B, Rizzo V, and Eguchi S (2011) Caveolin-1 negatively regulates a metalloprotease-dependent epidermal growth factor receptor transactivation by angiotensin II. Journal of molecular and cellular cardiology 50, 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takayanagi T, Forrester SJ, Kawai T, Obama T, Tsuji T, Elliott KJ, Nuti E, Rossello A, Kwok HF, and Scalia R (2016) Vascular ADAM17 as a Novel Therapeutic Target in Mediating Cardiovascular Hypertrophy and Perivascular Fibrosis Induced by Angiotensin IINovelty and Significance. Hypertension 68, 949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schrage R, Schmitz A-L, Gaffal E, Annala S, Kehraus S, Wenzel D, Büllesbach KM, Bald T, Inoue A, and Shinjo Y (2015) The experimental power of FR900359 to study Gq-regulated biological processes. Nature communications 6, 10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Hayre M, Eichel K, Avino S, Zhao X, Steffen DJ, Feng X, Kawakami K, Aoki J, Messer K, and Sunahara R (2017) Genetic evidence that β-arrestins are dispensable for the initiation of β2-adrenergic receptor signaling to ERK. Sci. Signal 10, eaal3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tilley DG (2011) G protein–dependent and G protein–independent signaling pathways and their impact on cardiac function. Circulation research 109, 217–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hunyady L, Bor M, Balla T, and Catt KJ (1995) Critical role of a conserved intramembrane tyrosine residue in angiotensin II receptor activation. Journal of Biological Chemistry 270, 9702–9705 [DOI] [PubMed] [Google Scholar]
- 59.Thomas WG, Motel TJ, Kule CE, Karoor V, and Baker KM (1998) Phosphorylation of the angiotensin II (AT1A) receptor carboxyl terminus: a role in receptor endocytosis. Molecular Endocrinology 12, 1513–1524 [DOI] [PubMed] [Google Scholar]
- 60.Qian H, Pipolo L, and Thomas WG (2001) Association of β-arrestin 1 with the type 1A angiotensin II receptor involves phosphorylation of the receptor carboxyl terminus and correlates with receptor internalization. Molecular Endocrinology 15, 1706–1719 [DOI] [PubMed] [Google Scholar]
- 61.See HB, Seeber RM, Kocan M, Eidne KA, and Pfleger KD (2011) Application of G protein-coupled receptor-heteromer identification technology to monitor beta-arrestin recruitment to G protein-coupled receptor heteromers. Assay and drug development technologies 9, 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnstone EK, and Pfleger KD (2012) Receptor-Heteromer Investigation Technology and its application using BRET. Frontiers in endocrinology 3, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan PK, Wang J, Littler P-LH, Wong KK, Sweetnam TA, Keefe W, Nash NR, Reding EC, Piu F, and Brann MR (2007) Monitoring interactions between receptor tyrosine kinases and their downstream effector proteins in living cells using bioluminescence resonance energy transfer. Molecular pharmacology 72, 1440–1446 [DOI] [PubMed] [Google Scholar]
- 64.Schiffer HH, Reding EC, Fuhs SR, Lu Q, Piu F, Wong S, Littler P-LH, Weiner DM, Keefe W, and Tan PK (2007) Pharmacology and signaling properties of epidermal growth factor receptor isoforms studied by bioluminescence resonance energy transfer. Molecular pharmacology 71, 508–518 [DOI] [PubMed] [Google Scholar]
- 65.Shah BH, Yesilkaya A, Olivares-Reyes JA, Chen HD, Hunyady L, and Catt KJ (2004) Differential pathways of angiotensin II-induced extracellularly regulated kinase 1/2 phosphorylation in specific cell types: Role of heparin-binding epidermal growth factor. Molecular Endocrinology 18, 2035–2048 [DOI] [PubMed] [Google Scholar]
- 66.Tilley DG, Kim IM, Patel PA, Violin JD, and Rockman HA (2009) beta-Arrestin mediates beta1-adrenergic receptor-epidermal growth factor receptor interaction and downstream signaling. The Journal of biological chemistry 284, 20375–20386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang Y, Benaich N, Tape C, Kwok HF, and Murphy G (2014) Targeting the sheddase activity of ADAM17 by an anti-ADAM17 antibody D1 (A12) inhibits head and neck squamous cell carcinoma cell proliferation and motility via blockage of bradykinin induced HERs transactivation. International journal of biological sciences 10, 702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fuentes LQ, Reyes CE, Sarmiento JM, Villanueva CI, Figueroa CD, Navarro J, and González CB (2008) Vasopressin up-regulates the expression of growth-related immediate-early genes via two distinct EGF receptor transactivation pathways. Cellular signalling 20, 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Volpi S, Liu Y, and Aguilera G (2006) Vasopressin increases GAGA binding activity to the V1b receptor promoter through transactivation of the MAP kinase pathway. Journal of molecular endocrinology 36, 581–590 [DOI] [PubMed] [Google Scholar]
- 70.Inoue A, Ishiguro J, Kitamura H, Arima N, Okutani M, Shuto A, Higashiyama S, Ohwada T, Arai H, and Makide K (2012) TGF [alpha] shedding assay: an accurate and versatile method for detecting GPCR activation. Nature methods 9, 1021–1029 [DOI] [PubMed] [Google Scholar]
- 71.Chan HW, Jenkins A, Pipolo L, Hannan RD, Thomas WG, and Smith NJ (2006) Effect of dominant-negative epidermal growth factor receptors on cardiomyocyte hypertrophy. Journal of receptor and signal transduction research 26, 659–677 [DOI] [PubMed] [Google Scholar]
- 72.Levitzki A, and Gazit A (1995) Tyrosine kinase inhibition: an approach to drug development. SCIENCE-NEW YORK THEN WASHINGTON-, 1782–1782 [DOI] [PubMed] [Google Scholar]
- 73.Negro A, Brar BK, Gu Y, Peterson KL, Vale W, and Lee KF (2006) erbB2 is required for G protein-coupled receptor signaling in the heart. Proceedings of the National Academy of Sciences of the United States of America 103, 15889–15893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin J, and Freeman MR (2003) Transactivation of ErbB1 and ErbB2 receptors by angiotensin II in normal human prostate stromal cells. The Prostate 54, 1–7 [DOI] [PubMed] [Google Scholar]
- 75.Smith NJ, Chan H-W, Qian H, Bourne AM, Hannan KM, Warner FJ, Ritchie RH, Pearson RB, Hannan RD, and Thomas WG (2011) Determination of the Exact Molecular Requirements for Type 1 Angiotensin Receptor Epidermal Growth Factor Receptor Transactivation and Cardiomyocyte Hypertrophy. Hypertension 57, 973–U217 [DOI] [PubMed] [Google Scholar]
- 76.Aplin M, Christensen GL, Schneider M, Heydorn A, Gammeltoft S, Kjølbye AL, Sheikh SP, and Hansen JL (2007) Differential Extracellular Signal - Regulated Kinases 1 and 2 Activation by the Angiotensin Type 1 Receptor Supports Distinct Phenotypes of Cardiac Myocytes. Basic & clinical pharmacology & toxicology 100, 296–301 [DOI] [PubMed] [Google Scholar]
- 77.Esposito G, Perrino C, Cannavo A, Schiattarella GG, Borgia F, Sannino A, Pironti G, Gargiulo G, Di Serafino L, and Franzone A (2011) EGFR trans-activation by urotensin II receptor is mediated by β-arrestin recruitment and confers cardioprotection in pressure overload-induced cardiac hypertrophy. Basic research in cardiology 106, 577–589 [DOI] [PubMed] [Google Scholar]
- 78.Kim J, Ahn S, Rajagopal K, and Lefkowitz RJ (2009) Independent beta-Arrestin2 and G(q)/Protein Kinase C zeta Pathways for ERK Stimulated by Angiotensin Type 1A Receptors in Vascular Smooth Muscle Cells Converge on Transactivation of the Epidermal Growth Factor Receptor. Journal of Biological Chemistry 284, 11953–11962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Olivares-Reyes JA, Shah BH, Hernandez-Aranda J, Garcia-Caballero A, Farshori MP, Garcia-Sainz JA, and Catt KJ (2005) Agonist-induced interactions between angiotensin AT1 and epidermal growth factor receptors. Molecular pharmacology 68, 356–364 [DOI] [PubMed] [Google Scholar]
- 80.Maudsley S, Pierce KL, Zamah AM, Miller WE, Ahn S, Daaka Y, Lefkowitz RJ, and Luttrell LM (2000) The β2-adrenergic receptor mediates extracellular signal-regulated kinase activation via assembly of a multi-receptor complex with the epidermal growth factor receptor. Journal of Biological Chemistry 275, 9572–9580 [DOI] [PubMed] [Google Scholar]
- 81.Jensen DD, Godfrey CB, Niklas C, Canals M, Kocan M, Poole DP, Murphy JE, Alemi F, Cottrell GS, and Korbmacher C (2013) The bile acid receptor TGR5 does not interact with β-arrestins or traffic to endosomes but transmits sustained signals from plasma membrane rafts. Journal of Biological Chemistry 288, 22942–22960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zajac M, Law J, Cvetkovic DD, Pampillo M, McColl L, Pape C, Di Guglielmo GM, Postovit LM, Babwah AV, and Bhattacharya M (2011) GPR54 (KISS1R) transactivates EGFR to promote breast cancer cell invasiveness. PLoS One 6, e21599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cattaneo F, Iaccio A, Guerra G, Montagnani S, and Ammendola R (2011) NADPH-oxidase-dependent reactive oxygen species mediate EGFR transactivation by FPRL1 in WKYMVm-stimulated human lung cancer cells. Free radical biology and medicine 51, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 84.Myers TJ, Brennaman LH, Stevenson M, Higashiyama S, Russell WE, Lee DC, and Sunnarborg SW (2009) Mitochondrial Reactive Oxygen Species Mediate GPCR–induced TACE/ADAM17-dependent Transforming Growth Factor-α Shedding. Molecular biology of the cell 20, 5236–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rafikova O, Rafikov R, Kangath A, Qu N, Aggarwal S, Sharma S, Desai J, Fields T, Ludewig B, and Yuan JX-Y (2016) Redox regulation of epidermal growth factor receptor signaling during the development of pulmonary hypertension. Free Radical Biology and Medicine 95, 96–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu X, Sharma KD, Takahashi T, Iwamoto R, and Mekada E (2002) Ligand-independent dimer formation of epidermal growth factor receptor (EGFR) is a step separable from ligand-induced EGFR signaling. Molecular biology of the cell 13, 2547–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hofman EG, Bader AN, Voortman J, Van den Heuvel DJ, Sigismund S, Verkleij AJ, Gerritsen HC, and en Henegouwen PM v. B. (2010) Ligand-induced EGF receptor oligomerization is kinase-dependent and enhances internalization. Journal of Biological Chemistry 285, 39481–39489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clayton AH, Walker F, Orchard SG, Henderson C, Fuchs D, Rothacker J, Nice EC, and Burgess AW (2005) Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. Journal of Biological Chemistry 280, 30392–30399 [DOI] [PubMed] [Google Scholar]
- 89.Clayton AH, Orchard SG, Nice EC, Posner RG, and Burgess AW (2008) Predominance of activated EGFR higher-order oligomers on the cell surface. Growth Factors 26, 316–324 [DOI] [PubMed] [Google Scholar]