Abstract

Science is full of overlooked and undervalued research waiting to be rediscovered. Proteomics is no exception. In this perspective, we follow the ripples from a 1960 study of Zuckerkandl, Jones, and Pauling comparing tryptic peptides across animal species. This pioneering work directly led to the molecular clock hypothesis and the ensuing explosion in molecular phylogenetics. In the decades following, proteins continued to provide essential clues on evolutionary history. While technology has continued to improve, contemporary proteomics has strayed from this larger biological context, rarely comparing species or asking how protein structure, function, and interactions have evolved. Here we recombine proteomics with molecular phylogenetics, highlighting the value of framing proteomic results in a larger biological context and how almost forgotten research, though technologically surpassed, can still generate new ideas and illuminate our work from a different perspective. Though it is infeasible to read all research published on a large topic, looking up older papers can be surprisingly rewarding when rediscovering a “gem” at the end of a long citation chain, aided by digital collections and perpetually helpful librarians. Proper literature study reduces unnecessary repetition and allows research to be more insightful and impactful by truly standing on the shoulders of giants. All data was uploaded to MassIVE (https://massive.ucsd.edu/) as dataset MSV000087993.
Keywords: historical perspective, molecular phylogenetics, comparative proteomics, mass spectrometry, spectral libraries
Introduction
Some 60 years ago, Zuckerkandl, Jones, and Pauling compared hemoglobins across the animal kingdom by analyzing their tryptic peptide patterns in two-dimensional paper chromatography.1 This work initiated the field of molecular evolution, which has revealed many of the details of the history of life on Earth. Rapid and sensitive analysis of tryptic peptide patterns is a cornerstone of mass spectrometry-based proteomics and remains useful in molecular phylogenetics and species identification. Sadly, however, the evolutionary perspective is almost completely absent in proteomics. In this Perspective, we revisit the pioneering work of Zuckerkandl, Jones, Pauling, and contemporaries in the context of current proteomics methods, highlighting how recent advancements combine molecular phylogenetics and mass spectrometry-based proteomics. In doing so, we place modern proteomics practices in a broader historical context and illustrate the value in reading papers published in the past century.
Humble Beginnings
In 1960, the understanding of genes and protein synthesis was limited, with the first pieces of the genetic code only deciphered the following year. Sequencing of proteins such as hemoglobin was extremely laborious, but peptide mapping or fingerprinting by electrophoresis and chromatography on paper provided a far quicker way to compare related proteins2,3 and detect amino acid variants, such as E7V in hemoglobin subunit beta in sickle cell anemia.4 In their 1960 paper,1 Zuckerkandl, Jones, and Pauling used this method to compare hemoglobins across primates, artiodactyls, and marine vertebrates. Fingerprints of hemoglobin orthologs revealed differences in amino acid composition increasing with evolutionary distance, leading to the molecular clock hypothesis, later described (though not named) in a 1962 book chapter.5 Focusing on hemoglobin allowed Zuckerkandl and Pauling to develop the molecular clock framework, though other proteins, such as cytochrome c,6 fibrinogen,7 and ferredoxin,8 were also used in early studies of molecular phylogenetics. In 1963, Zuckerkandl discussed the bias introduced by only studying a single protein and wistfully acknowledged that “we cannot hope to analyze the sequence of all proteins to be found in a given organism” (ref (9), p. 259). Now, 60 years later and with over 90% of the human proteome blueprint recently completed,10 this hope is no longer aspirational, though achieving this coverage in a single analysis is difficult for any species, and how to best use such dense data for molecular phylogenetics is far from straightforward.
Amino Acid Sequencing
Before DNA sequencing came to dominate in molecular phylogenetics, comparative studies relying on protein sequences were driving the field. Already in 1949, Sanger had compared protein orthologs from cow and sheep with respect to amino acid composition,11 showing that the pig insulin A chain includes a threonine absent in cow and sheep. Building off this work with more species and more complete sequencing12,13 highlighted that amino acid sequence differences between species may say something about evolutionary trends. This concept was formalized by Tuppy in 1959, stating that if assuming that differences in protein structure between species are caused by gene mutations, “proteins should differ from each other in such a way that the more places in which the polypeptide chains differ, through an exchange of amino acid residues, the further the organisms producing them are separated from each other in evolution”.14 As more proteins were studied, their sequences and orthologs across the tree of life were collected, beginning in 1965 in the Atlas of Protein Sequence and Structure,15 which allowed systematic analysis of these sequences.16 Punch cards17 and eventually computers18 were used to piece together protein sequences from short peptide fragments, first addressing the protein inference problem still challenging bottom-up proteomics. This work ultimately resulted in the MASSPEC software for reconstructing complete protein sequences from mass spectra of a single hydrolysate19—a problem similar to deciphering the complex mass spectra acquired in top-down proteomics. In 1966, Dayhoff and Eck aligned ferredoxin sequences to reconstruct their phylogenetic relationships,8 and the first actual phylogenetic trees derived from amino acid sequences—those of cytochrome c—were published by Fitch and Margoliash in 1967.20
Proteomics
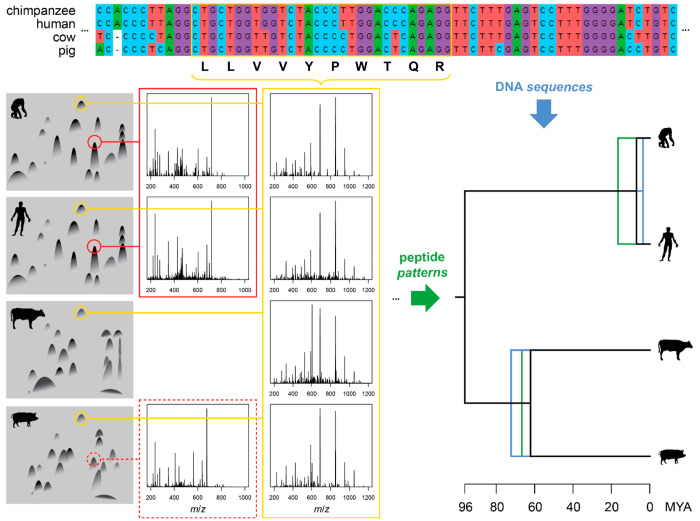
Mass spectrometry had already been used to sequence short peptides the year before Zuckerkandl’s, Jones’s, and Pauling’s 1960 chromatography-based paper,21 showing promise due to its inherent speed and sensitivity. Large-scale peptide analysis by mass spectrometry proved extraordinarily challenging, and it took three decades of hard work and the advent of new ionization techniques22 to realize this promise and pave the way for proteomics. For most of this intervening period, the Edman method23,24 was used to determine amino acid sequences and contributed most of the amino acid sequence data for molecular phylogenetics as well as protein biochemistry. Unlike Edman sequencing, but similar to the peptide fingerprinting of the 1950s and 1960s, mass spectrometry-based bottom-up or “shotgun” proteomics primarily analyzes tryptic peptides. The technology is obviously different, typically using a combination of liquid chromatography with tandem mass spectrometry, where tryptic peptides behave predictably and fragmentation patterns in the latter are used to identify peptides through comparison with predicted25 or previously acquired spectra,26 or constructed de novo, e.g., using graph theory.27 The analyzed tryptic peptides however remain the same (Figure 1).
Figure 1.
Comparison of the technology used in 1960 by Zuckerkandl, Jones, and Pauling1 with contemporary proteomics and genomics. Left: tryptic peptide fingerprints by electrophoresis and chromatography on paper of hemoglobin from chimpanzee, human, cow, and pig, redrawn from the original1 and matched with tandem mass spectra from two hemoglobin beta chain (HBB) peptides: in red VHLTPEEK from chimpanzee and human and VHLSAEEK from pig (dotted), and in yellow LLVVYPWTQR common to all four species. Top: MUSCLE alignment of the HBB orthologs with the region coding for the shared peptide. Right: phylogenetic trees constructed from tandem mass spectra (green) and the HBB gene sequences (blue). Note that the non-identical DNA sequences coding for the identical peptide contain more phylogenetic information than the peptide sequence alone. Details on how the figure was generated are available as Supporting Information, and data used to generate the figure are available in MassIVE (dataset MSV000087993) and with more details on https://osf.io/hm8dq.
Building in part from these early studies of hemoglobin, the clinical relevance and availability have made blood and its components one of the most analyzed biological substances in proteomics. In 1977, Anderson and Anderson (father and son) used a two-dimensional gel electrophoresis method developed by O’Farrell28 to separate and identify 30 intact proteins in human plasma,29 explicitly suggesting that genetic variants altering protein charge or size “should be routinely detectable in at least 20 proteins at once, facilitating studies of human mutation rates”. This emphasis on detecting genetic variants in proteins made sense at the time, since nucleic acid sequencing was still slow and cumbersome, with the first tiny RNA phage genome just completed the year before (bacteriophage MS2)30 and the first DNA genome (bacteriophage ϕX174) published just nine months earlier.31 Since then, the plasma proteome has been extensively studied for single amino acid variants,32 disease biomarkers,33 heritability of expression,34−36 and post-translational modifications (PTMs),37 though searching proteomics data for a large number of rare variants and PTMs remains a statistical challenge. While the new PEFF format (PSI Extended Fasta Format)38 captures some of this existing knowledge, pairwise matching of tandem mass spectra, e.g., from blood sera, can find shared variants without prior assumptions.39 Such direct comparisons of mass spectra are independent of gene predictions or knowledge on genetic variation, thus equally applicable to species with complete reference proteomes as to unsequenced non-human non-model systems. This type of comparison of mass spectra is the contemporary analog of Zuckerkandl’s multi-species peptide fingerprints.
Molecular Phylogenetics
Phylogenetics benefited greatly from the discovery and analysis of DNA, RNA, and proteins. The study by Fitch and Margoliash frequently credited with the advent of molecular phylogenetics compared amino acid sequences of cytochrome c across 20 species,20 work which was largely inspired by the Zuckerkandl, Jones, and Pauling hemoglobin peptide fingerprinting.1 Large-scale molecular phylogenetics in its modern form is probably best exemplified by DNA “barcoding” (e.g., The Barcode of Life40,41), which for animals utilizes a 650 base pair sequence from the 5′ region of mitochondrial cytochrome c oxidase 1. Though useful, critical discoveries have relied on more than a single gene barcode. For instance, the amino acid sequences of αA-crystallin (CRYAA), aquaporin-2 (AQP2), and interphotoreceptor retinol-binding protein (IRBP) led to the delineation of Afrotheria clade of mammals in 2001.42 The original molecular clock hypothesis is elegantly and visually integrated in a broader phylochronology by tools such as TimeTree,43 which combine molecular information with fossil evidence, climate change, and impact events in a holistic evolutionary timeline. Contemporary molecular phylogenetics includes constructing trees using whole genomes,44 though this overabundance of data and overchoice of clustering methods has also increased the complexity and computational cost of the analysis, without always providing clearer answers. Especially in non-humans, analysis of tryptic peptide patterns generated by two-dimensional electrophoresis and chromatography on paper or mass spectrometry is arguably less laborious than current genomic or transcriptomic analyses.
A Return to Peptide-Informed Phylogeny via Mass Spectrometry
Though molecular phylogenetics is dominated by DNA-based identification and barcoding of species, alternative peptide and protein-based methods never completely exited the stage following Zuckerkandl’s, Jones’s, and Pauling’s pioneering use of tryptic peptides. Yates et al.45 compared cytochrome c peptide mass maps from 12 species across the animal kingdom, demonstrating the power of mass spectrometry to distinguish between peptides and that closely related species share more peptide masses, e.g., chimpanzee sharing most tryptic peptides with human, mouse with rat, ostrich with emu, and dog with the other carnivoran (southern elephant seal). Proteomics is currently seeing increasing application in non-model organisms.46 The near universal applicability of mass spectrometry, the relative stability of proteins, and the ability to compare related species allow analysis of virtually any sample, including hair,47,48 artifacts,49,50 or even fossils. A famous—or possibly infamous—phylogenetic example is the analysis of proteins from fossils from the Cretaceous period.51,52 Proteomic data from extinct species are typically matched against sequences from living relatives53 or sequenced de novo and aligned with known sequences. Genome-independent methods such as compareMS239 and DISMS254 compare tandem mass spectra directly against each other, thereby deriving a similarity (or distance) between two datasets based on the peptide sequences, abundances, and post-translational modifications. The same metric can be applied between one dataset from an “unknown” and several previously acquired datasets placed in spectral libraries,55,56 thereby predicting the unknown phylogeny. Molecular phylogenetics can be used to infer the origin of a sample by finding the closest protein sequence or mass spectrum in a reference library.57,58 Targeted and untargeted proteomic methods have been developed for food59,60 and feed analysis61 and shown to be able to detect and quantify mixtures and contamination at or below 1%. Another genome-independent application is the rapid bacterial identification, routinely performed in thousands of clinical laboratories by MALDI-TOF mass spectrometry and comparison of spectra dominated by ribosomal proteins.62 In these examples, the information is biased toward highly abundant proteins, which tend to be more conserved than less abundant proteins (and even more so than non-coding regions of the genome). Proteomics methods now routinely achieve a depth of coverage of several thousand proteins for most biological samples, capturing information from many low-abundance proteins and allowing de novo sequencing of thousands of peptides. More recent data-independent acquisition methods avoid some of the stochastic nature and undersampling of data-dependent acquisition of tandem mass spectra and may prove more robust in generating data for phylogenetics or species identification applications, though possibly at the cost of increased dependency on chromatographic reproducibility. Overall, contemporary mass spectrometry-based methods are effective tools for molecular phylogenetics, and continued advancements will improve existing capabilities and create new applications.
From the Past via the Present to the Future
Based on the history of molecular phylogenetics it is obvious that information from the proteome over time became less valued than that from DNA or RNA. Sequencing proteins in the 1950s and 1960s was extremely laborious, which is why peptide mapping or fingerprinting by electrophoresis and chromatography on paper was used to compare related proteins. Not even state-of-the-art mass spectrometry can compete with the data generated by next-generation DNA sequencing. Even if massively parallel protein sequencing is realized,63 amino acid sequences still miss information from non-coding regions. However, current technological advancements present opportunities for proteomics to be used in ways that nucleic acid sequencing cannot, including analysis of samples with little or no DNA, degraded RNA, differentiating tissues or cell types, and function or environmental inferences in microbial communities, all driving new applications and discovery. The development of miniature mass spectrometers64 capable of using paper spray ionization of blots on paper, fast tryptic digestion,65 and separations enable rapid peptide fingerprinting of biological specimens in the field on a time-scale of seconds to minutes. Potential use cases are innumerable but would include the rapid identification of species and composition of unknown food and feed samples, detection of contaminants and adulterants,56,59,61,66 and more mundane quality control applications. In many of these, the data analysis would involve comparisons between peptide fingerprints similar to how they were used in molecular phylogenetics over 60 years ago. Species not previously analyzed can still be identified by their similarity to analyzed ones, and phylogenetic trees can be constructed directly from the data. Furthermore, nothing rules out combining these very broad analyses with targeted selected-reaction monitoring (SRM) methods for known species (e.g., porcine60) or known contaminants, even in the same analysis. Though mass spectrometry technology continues to evolve, fundamental ideas published many decades ago can remain relevant and still suggest new and potentially rewarding avenues of research.
Conclusions
We hope that our experience connecting peptides, phylogenetics, and mass spectrometry by taking inspiration from a study published 60 years ago can stimulate others to follow literature trails back in time, reading the papers to rediscover ideas and abandoned branches of investigation and seeing these in a new light. Accessing these papers has never been easier. But more specifically, we want to impel students to learn about the history of our field—or any field they study—as well as the evolutionary history of biological molecules and organisms. The first appearance of a term rarely equals the first description of the connoted concept. For less researched topics, there are often gaps in the literature, even if gaps in our field are not as extreme as the 160-year delay between Gauss’s original derivation of the fast Fourier transform in 1805 and its rediscovery by Cooley and Tukey in 1965. Even more importantly, proteomics should not be limited to tallying up protein “parts list”—an exercise akin to stamp collecting. Proteomics should be about the proteins in a biological context. And this context always includes the evolutionary history of the proteins, protein complexes, biochemical mechanisms, and biological systems studied. How conserved are the protein sequences, post-translational modifications, structures, and interactions? What does this all mean for the suitability of model systems and the reusability of biomolecular sequences, structures, spectral libraries, annotations, and pathways across the tree of life? These are important questions, far too rarely asked of proteomics data. As Dobzhansky famously titled his 1973 essay: “Nothing in Biology Makes Sense Except in the Light of Evolution”.67
Acknowledgments
The authors thank the reviewers for their critical feedback and filling some of the gaps in our historical narrative. Tandem mass spectrometry data used for clustering of chimpanzee, human, cow, and pig serum were graciously provided with permission from a collaboration with Dr. Michael G. Janech (College of Charleston) as part of the CoMPARe Program (Comparative Mammalian Proteome Aggregator Resource). Specifically, the cow samples were provided by Dr. Paola Boggiatto and Dr. Randy Sacco at the National Animal Disease Center, ARS, USDA, Ames, IA. The chimpanzee samples were provided by the Chattanooga Zoo, and the pig samples were from the Medical University of South Carolina (Charleston, SC). In addition to institutional permits and approval, data collection was performed under NIST ACUC MML-AR20-0001. The identification of certain commercial equipment, instruments, software, or materials does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the products identified are necessarily the best available for the purpose.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.1c00528.
Details on how Figure 1 was generated (PDF)
The authors declare no competing financial interest.
Notes
All data was uploaded to MassIVE (https://massive.ucsd.edu/) as dataset MSV000087993.
Supplementary Material
References
- Zuckerkandl E.; Jones R. T.; Pauling L. A comparison of animal hemoglobins by tryptic peptide pattern analysis. Proc. Natl. Acad. Sci. U. S. A. 1960, 46 (10), 1349–1360. 10.1073/pnas.46.10.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugaard G.; Kroner T. D. Partition chromatography of amino acids with applied voltage. J. Am. Chem. Soc. 1948, 70 (6), 2135–2137. 10.1021/ja01186a041. [DOI] [PubMed] [Google Scholar]
- Ingram V. M. A specific chemical difference between the globins of normal human and sickle-cell anaemia haemoglobin. Nature 1956, 178 (4537), 792–4. 10.1038/178792a0. [DOI] [PubMed] [Google Scholar]
- Ingram V. M. Abnormal human haemoglobins: I. The comparison of normal human and sickle-cell haemoglobins by “fingerprinting. Biochim. Biophys. Acta 1958, 28, 539–545. 10.1016/0006-3002(58)90516-X. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E.; Pauling L. B.. Molecular disease, evolution, and genetic heterogeneity. In Horizons in Biochemistry; Kasha M., Pullman B., Eds.; Academic Press: New York, 1962; pp 189–225. [Google Scholar]
- Margoliash E. Primary Structure and Evolution of Cytochrome C. Proc. Natl. Acad. Sci. U. S. A. 1963, 50, 672–9. 10.1073/pnas.50.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F.; Blombäck B. Amino-Acid Sequence Investigations of Fibrinopeptides from Various Mammals: Evolutionary Implications. Nature 1964, 202, 147–52. 10.1038/202147a0. [DOI] [PubMed] [Google Scholar]
- Eck R. V.; Dayhoff M. O. Evolution of the structure of ferredoxin based on living relics of primitive amino Acid sequences. Science (Washington, DC, U. S.) 1966, 152 (3720), 363–6. 10.1126/science.152.3720.363. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E.Perspectives in Molecular Anthropology. In Classification and Human Evolution; Washburn S. L., Ed.; Routledge, Taylor & Francis Group: London and New York, 1963; pp 243–272. [Google Scholar]
- Adhikari S.; Nice E. C.; Deutsch E. W.; Lane L.; Omenn G. S.; Pennington S. R.; Paik Y. K.; Overall C. M.; Corrales F. J.; Cristea I. M.; Van Eyk J. E.; Uhlén M.; Lindskog C.; Chan D. W.; Bairoch A.; Waddington J. C.; Justice J. L.; LaBaer J.; Rodriguez H.; He F.; Kostrzewa M.; Ping P.; Gundry R. L.; Stewart P.; Srivastava S.; Srivastava S.; Nogueira F. C. S.; Domont G. B.; Vandenbrouck Y.; Lam M. P. Y.; Wennersten S.; Vizcaino J. A.; Wilkins M.; Schwenk J. M.; Lundberg E.; Bandeira N.; Marko-Varga G.; Weintraub S. T.; Pineau C.; Kusebauch U.; Moritz R. L.; Ahn S. B.; Palmblad M.; Snyder M. P.; Aebersold R.; Baker M. S. A high-stringency blueprint of the human proteome. Nat. Commun. 2020, 11 (1), 5301. 10.1038/s41467-020-19045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F. Species differences in insulins. Nature 1949, 164 (4169), 529. 10.1038/164529a0. [DOI] [PubMed] [Google Scholar]
- Brown H.; Sanger F.; Kitai R. The structure of pig and sheep insulins. Biochem. J. 1955, 60 (4), 556–65. 10.1042/bj0600556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. I.; Naughton M. A.; Sanger F. Species differences in insulin. Arch. Biochem. Biophys. 1956, 65 (1), 427–38. 10.1016/0003-9861(56)90203-X. [DOI] [PubMed] [Google Scholar]
- Tuppy H. Aminosäure-Sequenzen in Proteinen. Naturwissenschaften 1959, 46, 35–43. 10.1007/BF00599080. [DOI] [Google Scholar]
- Dayhoff M. O.; Eck R. V.; Chang M. A.; Sochard M. R.. Atlas of Protein Sequence and Structure; National Biomedical Research Foundation: Silver Spring, MD, 1965; Vol. 1. [Google Scholar]
- Strasser B. J. Collecting, comparing, and computing sequences: the making of Margaret O. Dayhoff’s Atlas of Protein Sequence and Structure, 1954–1965. J. Hist Biol. 2010, 43 (4), 623–60. 10.1007/s10739-009-9221-0. [DOI] [PubMed] [Google Scholar]
- Eck R. V. A simplified strategy for sequence analysis of large proteins. Nature 1962, 193, 241–3. 10.1038/193241a0. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O. Computer aids to protein sequence determination. J. Theor. Biol. 1965, 8 (1), 97–112. 10.1016/0022-5193(65)90096-2. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O.; Eck R. V. MASSPEC: a computer program for complete sequence analysis of large proteins from mass spectrometry data of a single sample. Comput. Biol. Med. 1970, 1 (1), 5–28. 10.1016/0010-4825(70)90013-2. [DOI] [PubMed] [Google Scholar]
- Fitch W. M.; Margoliash E. Construction of phylogenetic trees. Science (Washington, DC, U.S.) 1967, 155 (3760), 279–84. 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Biemann K.; Gapp G.; Seibl J. Application of mass spectrometry to structure problems. I. Amino acid sequence in peptides. J. Am. Chem. Soc. 1959, 81 (9), 2274–2275. 10.1021/ja01518a069. [DOI] [Google Scholar]
- Fenn J. B. Electrospray ionization mass spectrometry: How it all began. J. Biomol. Technol. 2002, 13 (3), 101–18. [PMC free article] [PubMed] [Google Scholar]
- Niall H. D. Automated Edman degradation: the protein sequenator. Methods Enzymol. 1973, 27, 942–1010. 10.1016/S0076-6879(73)27039-8. [DOI] [PubMed] [Google Scholar]
- Edman P. A method for the determination of amino acid sequence in peptides. Arch. Biochem. 1949, 22 (3), 475–6. [PubMed] [Google Scholar]
- Eng J. K.; McCormack A. L.; Yates J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5 (11), 976–89. 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Lam H.; Deutsch E. W.; Eddes J. S.; Eng J. K.; King N.; Stein S. E.; Aebersold R. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 2007, 7 (5), 655–67. 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- Bartels C. Fast algorithm for peptide sequencing by mass spectroscopy. Biol. Mass Spectrom. 1990, 19 (6), 363–8. 10.1002/bms.1200190607. [DOI] [PubMed] [Google Scholar]
- O’Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975, 250 (10), 4007–21. 10.1016/S0021-9258(19)41496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L.; Anderson N. G. High resolution two-dimensional electrophoresis of human plasma proteins. Proc. Natl. Acad. Sci. U. S. A. 1977, 74 (12), 5421–5. 10.1073/pnas.74.12.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers W.; Contreras R.; Duerinck F.; Haegeman G.; Iserentant D.; Merregaert J.; Min Jou W.; Molemans F.; Raeymaekers A.; Van den Berghe A.; Volckaert G.; Ysebaert M. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature 1976, 260 (5551), 500–7. 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- Sanger F.; Air G. M.; Barrell B. G.; Brown N. L.; Coulson A. R.; Fiddes C. A.; Hutchison C. A.; Slocombe P. M.; Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature 1977, 265 (5596), 687–95. 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Madison J.; Galliano M.; Watkins S.; Minchiotti L.; Porta F.; Rossi A.; Putnam F. W. Genetic variants of human serum albumin in Italy: point mutants and a carboxyl-terminal variant. Proc. Natl. Acad. Sci. U. S. A. 1994, 91 (14), 6476–6480. 10.1073/pnas.91.14.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P. E.; Holdt L. M.; Teupser D.; Mann M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 2017, 13 (9), 942. 10.15252/msb.20156297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson Å.; Enroth S.; Palmblad M.; Deelder A. M.; Bergquist J.; Gyllensten U. Identification of genetic variants influencing the human plasma proteome. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (12), 4673–8. 10.1073/pnas.1217238110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Buil A.; Collins B. C.; Gillet L. C.; Blum L. C.; Cheng L. Y.; Vitek O.; Mouritsen J.; Lachance G.; Spector T. D.; Dermitzakis E. T.; Aebersold R. Quantitative variability of 342 plasma proteins in a human twin population. Mol. Syst. Biol. 2015, 11 (1), 786. 10.15252/msb.20145728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon T.; Smith E. N.; Matsui H.; Braekkan S. K.; Wilsgaard T.; Njølstad I.; Mathiesen E. B.; Hansen J. B.; Frazer K. A. Associations Between Common and Rare Exonic Genetic Variants and Serum Levels of 20 Cardiovascular-Related Proteins: The Tromsø Study. Circ.: Cardiovasc. Genet. 2016, 9 (4), 375–83. 10.1161/CIRCGENETICS.115.001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytseva O. O.; Freidin M. B.; Keser T.; Štambuk J.; Ugrina I.; Šimurina M.; Vilaj M.; Štambuk T.; Trbojević-Akmačić I.; Pučić-Baković M.; Lauc G.; Williams F. M. K.; Novokmet M. Heritability of Human Plasma N-Glycome. J. Proteome Res. 2020, 19 (1), 85–91. 10.1021/acs.jproteome.9b00348. [DOI] [PubMed] [Google Scholar]
- Binz P. A.; Shofstahl J.; Vizcaíno J. A.; Barsnes H.; Chalkley R. J.; Menschaert G.; Alpi E.; Clauser K.; Eng J. K.; Lane L.; Seymour S. L.; Sánchez L. F. H.; Mayer G.; Eisenacher M.; Perez-Riverol Y.; Kapp E. A.; Mendoza L.; Baker P. R.; Collins A.; Van Den Bossche T.; Deutsch E. W. Proteomics Standards Initiative Extended FASTA Format. J. Proteome Res. 2019, 18 (6), 2686–2692. 10.1021/acs.jproteome.9b00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmblad M.; Deelder A. M. Molecular phylogenetics by direct comparison of tandem mass spectra. Rapid Commun. Mass Spectrom. 2012, 26 (7), 728–32. 10.1002/rcm.6162. [DOI] [PubMed] [Google Scholar]
- Ratnasingham S.; Hebert P. D. bold: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7 (3), 355–364. 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. Modernizing the tree of life. Science (Washington, DC, U. S.) 2003, 300 (5626), 1692–7. 10.1126/science.300.5626.1692. [DOI] [PubMed] [Google Scholar]
- van Dijk M. A.; Madsen O.; Catzeflis F.; Stanhope M. J.; de Jong W. W.; Pagel M. Protein sequence signatures support the African clade of mammals. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (1), 188–93. 10.1073/pnas.98.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Stecher G.; Suleski M.; Hedges S. B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34 (7), 1812–1819. 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- A comparative genomics multitool for scientific discovery and conservation. Nature 2020, 587 (7833), 240–245. 10.1038/s41586-020-2876-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. R. III; Speicher S.; Griffin P. R.; Hunkapiller T. Peptide mass maps: a highly informative approach to protein identification. Anal. Biochem. 1993, 214 (2), 397–408. 10.1006/abio.1993.1514. [DOI] [PubMed] [Google Scholar]
- Heck M.; Neely B. A. Proteomics in Non-model Organisms: A New Analytical Frontier. J. Proteome Res. 2020, 19 (9), 3595–3606. 10.1021/acs.jproteome.0c00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adav S. S.; Subbaiaih R. S.; Kerk S. K.; Lee A. Y.; Lai H. Y.; Ng K. W.; Sze S. K.; Schmidtchen A. Studies on the Proteome of Human Hair - Identification of Histones and Deamidated Keratins. Sci. Rep. 2018, 8 (1), 1599. 10.1038/s41598-018-20041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim N.; Plott T. J.; Durbin-Johnson B. P.; Rocke D. M.; Salemi M.; Phinney B. S.; Goecker Z. C.; Pieterse M. J. M.; Parker G. J.; Rice R. H. Elucidation of familial relationships using hair shaft proteomics. Forensic Sci. Int.: Genet. 2021, 54, 102564. 10.1016/j.fsigen.2021.102564. [DOI] [PubMed] [Google Scholar]
- Mackie M.; Ruther P.; Samodova D.; Di Gianvincenzo F.; Granzotto C.; Lyon D.; Peggie D. A.; Howard H.; Harrison L.; Jensen L. J.; Olsen J. V.; Cappellini E. Palaeoproteomic Profiling of Conservation Layers on a 14th Century Italian Wall Painting. Angew. Chem., Int. Ed. 2018, 57 (25), 7369–7374. 10.1002/anie.201713020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland T. P.; Newsome G. A.; Hollinger R. E. Proteomic and direct analysis in real time mass spectrometry analysis of a Native American ceremonial hat. Analyst 2019, 144 (24), 7437–7446. 10.1039/C9AN01557D. [DOI] [PubMed] [Google Scholar]
- Asara J. M.; Schweitzer M. H.; Freimark L. M.; Phillips M.; Cantley L. C. Protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry. Science (Washington, DC, U. S.) 2007, 316 (5822), 280–5. 10.1126/science.1137614. [DOI] [PubMed] [Google Scholar]
- Bern M.; Phinney B. S.; Goldberg D. Reanalysis of Tyrannosaurus rex Mass Spectra. J. Proteome Res. 2009, 8 (9), 4328–32. 10.1021/pr900349r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellini E.; Jensen L. J.; Szklarczyk D.; Ginolhac A.; da Fonseca R. A.; Stafford T. W.; Holen S. R.; Collins M. J.; Orlando L.; Willerslev E.; Gilbert M. T.; Olsen J. V. Proteomic analysis of a pleistocene mammoth femur reveals more than one hundred ancient bone proteins. J. Proteome Res. 2012, 11 (2), 917–26. 10.1021/pr200721u. [DOI] [PubMed] [Google Scholar]
- Rieder V.; Blank-Landeshammer B.; Stuhr M.; Schell T.; Biß K.; Kollipara L.; Meyer A.; Pfenninger M.; Westphal H.; Sickmann A.; Rahnenführer J. DISMS2: A flexible algorithm for direct proteome-wide distance calculation of LC-MS/MS runs. BMC Bioinf. 2017, 18 (1), 148. 10.1186/s12859-017-1514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Önder Ö.; Shao W.; Lam H.; Brisson D. Tracking the sources of blood meals of parasitic arthropods using shotgun proteomics and unidentified tandem mass spectral libraries. Nat. Protoc. 2014, 9 (4), 842–50. 10.1038/nprot.2014.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff T.; Nielsen M. E.; Deelder A. M.; Jessen F.; Palmblad M. Authentication of fish products by large-scale comparison of tandem mass spectra. J. Proteome Res. 2013, 12 (11), 5253–9. 10.1021/pr4006525. [DOI] [PubMed] [Google Scholar]
- Downard K. M. Protein phylogenetics with mass spectrometry. A comparison of methods. Anal. Methods 2021, 13 (12), 1442–1454. 10.1039/D1AY00153A. [DOI] [PubMed] [Google Scholar]
- Ng W. Annotation of ribosomal protein mass peaks in MALDI-TOF mass spectra of bacterial species and their phylogenetic significance. bioRxiv Prepr. 2020, 10.1101/2020.07.15.203653. [DOI] [Google Scholar]
- Ohana D.; Dalebout H.; Marissen R. J.; Wulff T.; Bergquist J.; Deelder A. M.; Palmblad M. Identification of meat products by shotgun spectral matching. Food Chem. 2016, 203, 28–34. 10.1016/j.foodchem.2016.01.138. [DOI] [PubMed] [Google Scholar]
- von Bargen C.; Dojahn J.; Waidelich D.; Humpf H. U.; Brockmeyer J. New sensitive high-performance liquid chromatography-tandem mass spectrometry method for the detection of horse and pork in halal beef. J. Agric. Food Chem. 2013, 61 (49), 11986–94. 10.1021/jf404121b. [DOI] [PubMed] [Google Scholar]
- Rasinger J. D.; Marbaix H.; Dieu M.; Fumière O.; Mauro S.; Palmblad M.; Raes M.; Berntssen M. H. G. Species and tissues specific differentiation of processed animal proteins in aquafeeds using proteomics tools. J. Proteomics 2016, 147, 125–131. 10.1016/j.jprot.2016.05.036. [DOI] [PubMed] [Google Scholar]
- Clark A. E.; Kaleta E. J.; Arora A.; Wolk D. M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 2013, 26 (3), 547–603. 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timp W.; Timp G. Beyond mass spectrometry, the next step in proteomics. Sci. Adv. 2020, 6 (2), eaax8978 10.1126/sciadv.aax8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Liu J.; Cooks R. G.; Ouyang Z. Paper spray for direct analysis of complex mixtures using mass spectrometry. Angew. Chem., Int. Ed. 2010, 49 (5), 877–80. 10.1002/anie.200906314. [DOI] [PubMed] [Google Scholar]
- Zhong X.; Chen H.; Zare R. N. Ultrafast enzymatic digestion of proteins by microdroplet mass spectrometry. Nat. Commun. 2020, 11 (1), 1049. 10.1038/s41467-020-14877-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belghit I.; Varunjikar M.; Lecrenier M. C.; Steinhilber A. E.; Niedzwiecka A.; Wang Y. V.; Dieu M.; Azzollini D.; Lie K.; Lock E. J.; Berntssen M. H. G.; Renard P.; Zagon J.; Fumière O.; van Loon J. J. A.; Larsen T.; Poetz O.; Braeuning A.; Palmblad M.; Rasinger J. D. Future feed control – Tracing banned bovine material in insect meal. Food Control 2021, 128, 108183. 10.1016/j.foodcont.2021.108183. [DOI] [Google Scholar]
- Dobzhansky T. Nothing in Biology Makes Sense Except in the Light of Evolution. American Biology Teacher 1973, 35 (3), 125–129. 10.2307/4444260. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.