Abstract

Here, we present an integrated multivariate, univariate, network reconstruction and differential analysis of metabolite–metabolite and metabolite–lipid association networks built from an array of 18 serum metabolites and 110 lipids identified and quantified through nuclear magnetic resonance spectroscopy in a cohort of 248 patients, of which 22 died and 82 developed a poor functional outcome within 3 months from acute ischemic stroke (AIS) treated with intravenous recombinant tissue plasminogen activator. We explored differences in metabolite and lipid connectivity of patients who did not develop a poor outcome and who survived the ischemic stroke from the related opposite conditions. We report statistically significant differences in the connectivity patterns of both low- and high-molecular-weight metabolites, implying underlying variations in the metabolic pathway involving leucine, glycine, glutamine, tyrosine, phenylalanine, citric, lactic, and acetic acids, ketone bodies, and different lipids, thus characterizing patients’ outcomes. Our results evidence the promising and powerful role of the metabolite–metabolite and metabolite–lipid association networks in investigating molecular mechanisms underlying AIS patient’s outcome.
Keywords: lipidome, metabolomics, multivariate exploratory analysis, nuclear magnetic resonance, thrombolysis
Introduction
Acute ischemic stroke (AIS) is caused by thrombosis or embolism that occludes a cerebral vessel cutting the blood flow to an area of the brain; this results in a loss of neurological function. Usually, there is permanent and irreversible damage to part of the affected brain area and an area of penumbra where the function is lost, but the brain is not irreversibly damaged.1 AIS is the leading cause of long-term disability in developing countries and one of the most common causes of mortality worldwide.1 It is estimated that more than 13.7 million people suffer an AIS (i.e., one in six people will have an AIS in their life) and 5.8 million die as a consequence.1
Substantial progress has been made in recent years in both diagnosis and treatment to minimize the impact of AIS on the patients,1 and treatments such as intravenous thrombolysis and endovascular clot retrieval, aiming to remove blood clots and restore blood flow, have been shown to improve outcomes of AIS patients that concern mortality and disability.2−4
Metabolic perturbations are believed to be fundamental events that contribute to the ischemic stroke and to its progression and subsequent unfavorable outcomes,5−12 and comprehensive analytical techniques can provide a great chance to identify key metabolic features involved in the onset and progression of this disease.
Nuclear magnetic resonance (NMR)-based metabolomics allows a high-throughput analysis of various types of samples (e.g., blood, urine, cells, and tissue), providing information of hundreds of different metabolites and lipid features present in biological matrices.13,14 Multivariate and univariate analyses proved to be efficient in characterizing the metabolic signature of diseases15−17 and in the context of molecular epidemiology,18,19 but integrative systems biology techniques offer a comprehensive representation of the structural and functional characteristics of a certain living organism, helping in the understanding of the inter-relationships among metabolic features on the basis of the system behavior.20
Association networks can provide interesting information to describe the status of the biological system under study or to compare the same across different conditions, and correlation among metabolite and lipid levels measured in blood can be used to model and infer, at least partially, the structure of the underlying biological network.21 In this light, network analysis has proven to be an impressive and powerful tool to deepen the knowledge and interpret the complexity of metabolomics data.22−26 In particular, for metabolomics studies, the exploration of association networks was revealed to be more efficient when different conditions are compared in the context of a differential network analysis.22,25,27
In this work, using data from the Italian multicenter observational MArker bioloGici nell’Ictus Cerebrale (MAGIC) study,28,29 we performed a systems biology investigation (combining network analysis with standard univariate, multivariate analysis and classification, see Figure 1) of serum circulating metabolites, lipoproteins, and lipid fractions measured 24 h post-thrombolysis with the aim of providing insights into possible molecular mechanisms associated with the 3-month (3 M) functional outcome (FO) and mortality in a group of 248 patients with AIS treated with intravenous thrombolytic treatment with recombinant tissue plasminogen activator (rt-PA). Because 80% of acute infarctions show arterial occlusions,30 thrombolytic canalization of occluded arteries may reduce the degree of injury to the brain if it is done before the process of infarction has been completed; treatment with rt-PA is the approved treatment for AIS.31−33 The rt-PA is an ∼70 kDa serine protease protein, which is found in several mammalian cells, especially in endothelial cells, the cells lining the blood vessels. It specifically cleavages the arginine–valine bond in plasminogen to form plasmin, another serine protease, which is the major enzyme that dissolves fibrin blood clots.
Figure 1.
Graphical illustration of the analysis followed to explore differences of metabolite and serum profiles of patients who suffered AIS. The AIS is recorded at time t0, while serum samples were collected at t1, that is, 24 h after the thrombolytic intervention (post rt-PA samples). Patient’s survival and patient’s FO were evaluated after 3 months. Subjects/samples were retrospectively divided into four groups of interest samples, according to the outcome: (i) 3M-nD: patients who were alive (nondeceased nD) at three months (3 M) after AIS (n = 226); (ii) 3M-D: patients who were not alive (deceased D) at three months (3 M) after AIS (n = 22); (iii) 3M-GFO: patients with a GFO at three months (3 M) after AIS (n = 166); and (iv) 3M-PFO: patients with a PFO at three months (3 M) after AIS (n = 82). Groups were compared using a combination of univariate analysis, an unsupervised exploratory approach (PCA), classification modeling (Random Forest) and network inference, and analysis of metabolite–lipid association networks for 3M-PFO3M-GFO.
Our analysis shows that dysregulations in the connectivity of triglycerides, high-density lipoprotein (HDL), low-density lipoprotein (LDL), and very-low-density lipoprotein (VLDL) fractions and related subfractions, leucine, glycine, glutamine, tyrosine, phenylalanine, citrate, acetate, lactate, acetone, and 3-hydroxybutyrate are relevant molecular features for the characterization of post-AIS outcomes.
Materials and Methods
Study Population
The subjects and study samples considered are from the MAGIC study,28,29 which originally comprises 327 subjects: only subjects for whom serum specimen was available for metabolomics analysis are considered here (n = 248).
The study group consists of patients who had an AIS and were admitted for thrombolysis treatment with rt-PA in 14 different Italian centers, registered in the Safe Implementation of Thrombolysis in Stroke-International Stroke Thrombolysis Register (SITS-ISTR, www.sitsinternational.org), according to SITS-Monitoring Study criteria.34 Serum samples were collected 24 h after rt-PA (t1), and outcomes were defined at evaluation as follows: (i) mortality at 3 months after AIS (t2 = t0 + 3 months) and (ii) disability (impairment) at 3 months after AIS (t2). See Figure 1 for a graphical overview.
The 248-patient group constitutes a random subset of the original cohort: all demographic and clinical characteristics and risk factors, known to affect the poststroke poor/adverse outcomes, do not statistically differ (P-value > 0.05) between the original cohort and the group considered in this study (see Table 1).
Table 1. Comparison between Demographic and Clinical Characteristics of Patients Enrolled in the Original MAGIC Study (n = 327)28,29 and Those Analyzed in the Present Study where Metabolomics Analysis Was Performed (n = 248).
| demographic and clinical characteristics at baseline (mean ± standard deviation SD) | this study (n = 248) | original study group (n = 327) | P-value |
|---|---|---|---|
| age (years) | 68.8 ± 11.9 | 68.9 ± 12.0 | 0.88 |
| sex (male), n | 137 | 190 | 0.68 |
| onset to treatment time (minutes) | 163.4 ± 83.7 | 163.5 ± 75.7 | 0.86 |
| baseline National Institute of Health stroke scale | 11.9 ± 6.1 | 11.9 ± 6.0 | 0.94 |
| baseline systolic blood pressure (mmHg) | 147.5 ± 21.3 | 148.2 ± 21.7 | 0.69 |
| baseline diastolic blood pressure (mmHg) | 79.7 ± 12.7 | 80.1 ± 12.7 | 0.71 |
| blood glucose (mg/dL) | 130.2 ± 49.5 | 130.2 ± 47.9 | 0.99 |
| risk factors | |||
| hypertension (m) | 143 | 197 | 0.77 |
| diabetes (m) | 36 | 50 | 0.89 |
| hyperlipidemia (m) | 56 | 81 | 0.68 |
| current smoking (m) | 35 | 51 | 0.70 |
| atrial fibrillation (m) | 56 | 73 | 0.83 |
| congestive heart failure (m) | 26 | 35 | 0.98 |
Definition of Patient Groups
Study subjects were divided and analyzed in four groups of interest.
(i) 3M-GFO: patients with a good functional outcome (GFO) at 3 months (3 M) after AIS (n = 166).
(ii) 3M-PFO: patients with a poor functional outcome (PFO) at 3 months (3 M) after AIS (n = 82).
(iii) 3M-nD: patients who were alive (nondeceased, nD) at 3 months (3 M) after AIS (n = 226).
(iv) 3M-D: patients who were not alive (deceased, D) at 3 months (3 M) after AIS (n = 22).
The patient FO was defined according to the modified ranking scale (MRS).35,36 The MRS measures a patient’s independence rather than performance of specific tasks and incorporates mental as well as physical adaptations to the neurological deficits.37 The scale consists of six grades, from 0 to 5, indicating (adapted from Table 1 from refs (35, 36)):
0: No symptoms.
1: No significant disability, despite symptoms; able to perform all usual duties and activities.
2: Slight disability; unable to perform all previous activities but able to look after own affairs without assistance.
3: Moderate disability; requires some help, but able to walk without assistance.
4: Moderately severe disability; unable to walk without assistance and unable to attend to own bodily needs without assistance.
5: Severe disability; bedridden, incontinent, and requires constant nursing care and attention.
Study subjects were dichotomized into good (MRS 0–2) and poor (MRS ≥ 3–5) outcome patients as evaluated at 3 months after stroke. The PFO on the MRS was defined as ≥3 on the basis of what was observed in medical literature.38
Ethical Issues
The study protocol was approved in every participating center from local ethical committee. All patients gave informed consent. The study was in compliance with the Declaration of Helsinki.39
Sample Collection
Whole venous blood was collected in tubes without anticoagulant 24 h after thrombolysis (t1). Tubes were centrifuged at room temperature at 1500g for 15 min, and the supernatants were stored in aliquots at −80 °C until NMR measurements. Samples were analyzed in a unique NMR facility.
NMR Experiments
Serum samples were analyzed using a Bruker 600 MHz spectrometer working at 600.13 MHz proton Larmor frequency equipped with a 5 mm PATXI 1H-13C-15N and 2H decoupling probe. This includes a z axis gradient coil, an automatic tuning-matching, and an automatic and refrigerate sample changer (SampleJet). To stabilize approximately, at a level of ±0.1 K, the sample temperature (310 K), a BTO 2000 thermocouple was employed, and each NMR tube was kept for at least 5 min inside the NMR probe head to equilibrate the acquisition temperature of 310 K.
The analytical preparation of serum samples and their NMR spectra acquisition followed established procedures.13 For each serum specimen, the 1D nuclear Overhauser effect spectroscopy (NOESY) pulse sequence was applied to acquire 1H-NMR spectra. Raw NMR data were multiplied by an exponential function of 0.3 Hz line-broadening factor, before the application of Fourier transform. Phase and baseline distortions were automatically corrected, and transformed spectra were calibrated to the glucose doublet at 5.24 ppm using TopSpin 3.2 (BrukerBioSpin).
Metabolite and Lipid Identification and Quantification
One hundred twenty-eight (J = 128) analytes (18 metabolites and 110 lipoprotein and lipid fractions) were unambiguously identified and quantified using the AVANCE Bruker IVDr (Clinical Screening and In Vitro Diagnostics research, Bruker BioSpin) software using the 1D NOESY NMR spectra.40
For each lipid main class (VLDL, LDL, intermediate-density lipoprotein (IDL), and HDL) and subclass (VLDL-1 to VLDL-5, LDL-1 to LDL-6, and HDL-1 to HDL-4), reported data consist of concentrations of lipids (total cholesterol, free cholesterol, phospholipids, and triglycerides) contained in each fraction. Concentrations of apolipoproteins Apo-A1 and ApoA2 were estimated for the HDL class and each relative subclass, while Apo-B concentrations are calculated for VLDL and IDL classes and all LDL subclasses. A complete list of quantified analytes can be found in Supplementary Tables S1–S4.
Statistical Analysis
Univariate Analysis
The Mann–Whitney–Wilcoxon test41 was used to compare the concentration of the J = 128 analytes (18 metabolites +110 lipoproteins and lipid concentrations) between patient groups (3M-PFO vs 3M-GFO, 3M-nD vs 3M-D) treated with the thrombolytic therapy. The Benjamini–Hochberg method42 was used to correct for multiple testing; analytes for which false discovery rate (FDR) <0.01 are considered statistically significant and discussed in this paper.
Multivariate Exploratory and Classification
Principal Component Analysis (PCA)
PCA43 was applied on all quantified analytes (metabolites + lipoproteins and lipid fractions) from serum samples collected at t1 (24 h post rt-PA, see Figure 1 for an overview), to investigate, in an unsupervised manner, the data structure and highlight the possible presence of metabolite and lipid signatures differentiating (i) patients with the PFO (3M-PFO) from those with the GFO (3M-GFO) and (ii) patients who were alive (3M-nD) from those who were not (3M-D) at 3 months from the AIS. Analysis was performed on data scaled to unit variance.44
Random Forest Modeling
The Random Forest algorithm was employed for sample classification;45 two classification models were built to discriminate 3M-GFO/3M-PFO and 3M-nD/3M-D patients using samples collected at t1 (Table 2). Considering the unbalanced number of subjects in each group to be compared, Random Forest models were built after the two groups to be compared were made of the same size using a resample approach: (20 randomly sampled observations were used for the 3M-D vs 3M-nD comparison; 80 were used for the 3M-GFO vs 3M-PFO); 100 resampling iterations were performed to take into account the (re)sampling variability.
Table 2. Mean Values of Accuracy, Specificity, Sensitivity, and Corresponding 95% CIs of Random Forest Classification Models Built on the Metabolite and Lipid Profiles of Patients Who Suffered AISa.
| model
quality measures |
|||
|---|---|---|---|
| RF model | accuracy mean—95%CI | specificity mean—95%CI | sensitivity mean—95%CI |
| 3M-nD vs 3M-D | 53.8 (51.9, 55.6) | 57.5 (55.6, 59.5) | 50.0 (47.8, 52.2) |
| 3M-GFO vs 3M-PFO | 59.8 (59.1, 60.4) | 60.5 (59.7, 61.4) | 59.1 (58.2, 59.9) |
The classification models considered are (i) 3M-nD patients (alive at 3 months after AIS, n = 226) versus 3M-D patients (deceased at 3 months after AIS, n = 22) and (ii) 3M-GFO patients (with a GFO at 3 months after AIS, n = 166) vs 3M-PFO patients (with a PFO at 3 months after AIS, n = 82).
Accuracy, sensitivity, and specificity of each classification model were calculated according to standard definitions.46 Average values and 95% confidence interval (CI) are calculated over the 100 repetitions.
Network Analysis
Reconstruction of Association Networks
Metabolite–lipid correlation networks were constructed using the probabilistic context likelihood relatedness on correlation (PCLRC) algorithm.22 This algorithm estimates correlation considering the background distribution of correlation (implementing the CLRC approach47) and using resampling to obtain robust estimations.48 The algorithm outputs a J × J matrix P (with J the total number of variables/molecular features) containing the likelihood 0 ≤ pij ≤ 1 of every correlation rij that is used to filter background correlations. In particular, for the correlation rij between two metabolites i and j:
| 1 |
Determining the Significance of Metabolite and Lipid Differential Connectivity
Differences in terms of connectivity among metabolic features in each couple of networks (3M-PFO vs 3M-GFO and 3M-nD vs 3M-D patients) were analyzed 24 h after rt-PA administration (t1).
The connectivity of the i metabolite or i lipid is given by
| 2 |
and differential connectivity is defined as:
| 3 |
where χi1 and χi are the connectivity of metabolite or lipid i estimated from metabolite–lipid association networks calculated from data from conditions 1 and 2, respectively, (i.e., 3M-GFO and 3M-PFO, 3M-nD, and 3M-D, at t1).
The statistical significance of the metabolite and lipid differential connectivity was established using a permutation test (n = 1000) according to a previously described publication.25
Metabolites, lipoproteins, and lipid fractions that were statistically significantly connected (P-value < 0.05 after adjustment for multiple corrections with the Benjamini–Hochberg method) were considered to be related to the specific condition under study (poststroke 3-months impairment/death).
Software
All calculations were performed using R (version 3.6.2). Random Forest was performed using the R “randomForest” package,45 using the default settings. The R code for the PCLRC algorithm and the code to perform differential connectivity analysis are available at link: semantics.systemsbiology.nl under the SOFTWARE tab.
Results and Discussion
Univariate Analysis
The concentrations of blood metabolites, lipoproteins, and lipid fractions were compared between the different patient groups (3M-D vs 3M-nD and 3M-PFO vs 3M-GFO): results are given in Tables 3 and 4, respectively. When comparing patients with the PFO and GFO at 3 months after thrombolytic treatment (Table 3), 18 out of 128 analytes have concentrations significantly different between the two groups (P-value <0.01) of which only 3 are significant after correction for multiple testing (FDR < 0.01). A similar situation is observed for the comparison between patients who survived or died at 3 months after treatment: only 5 out of 128 analytes (Table 4) have concentrations significantly different between the two groups (P-value <0.01); however, these differences are not significant once correction for multiple testing is applied (FDR < 0.01).
Table 3. Results of Univariate Analysis (Mann–Whitney–Wilcoxon Test) for the Comparison of the Concentration Levels of Metabolite and Lipoproteins and Lipid Fractions in Patients with Poor (3M-PFO) and Good (3M-GFO) functional outcome at 3 months after AIS and treatment with rt-PAa.
| patient
groups |
||||||
|---|---|---|---|---|---|---|
| metabolite/lipid | 3M-GFO | 3M-PFO | P-value | FDR | Log2(FC) | trend |
| 3-hydroxybutyrate | 0.10 ± 0.12 | 0.24 ± 0.23 | 0.00003 | 0.0043 | 1.26 | ↑ |
| SubApoB_LDL-3 | 6.70 ± 3.44 | 8.71 ± 3.26 | 0.0001 | 0.0044 | 0.38 | ↑ |
| LDL3_PNb | 121.78 ± 62.42 | 158.41 ± 59.35 | 0.0001 | 0.0044 | 0.38 | ↑ |
| SubFreeChol_LDL-3 | 3.84 ± 1.86 | 4.80 ± 1.58 | 0.0004 | 0.0118 | 0.32 | ↑ |
| SubPhosp_LDL-3 | 6.20 ± 2.97 | 8.04 ± 3.22 | 0.0005 | 0.0118 | 0.37 | ↑ |
| SubChol_LDL-3 | 9.96 ± 6.25 | 13.54 ± 6.12 | 0.0006 | 0.0124 | 0.44 | ↑ |
| acetone | 0.09 ± 0.09 | 0.16 ± 0.13 | 0.0007 | 0.0129 | 0.78 | ↑ |
| phenylalanine | 0.08 ± 0.03 | 0.09 ± 0.03 | 0.0008 | 0.0128 | 0.17 | ↑ |
| SubTrigl_HDL-4 | 3.35 ± 0.99 | 2.91 ± 0.82 | 0.0015 | 0.0208 | –0.21 | ↓ |
| Apo-B100-Apo-A1 | 0.61 ± 0.15 | 0.66 ± 0.14 | 0.0016 | 0.0208 | 0.12 | ↑ |
| LMF_FreeChol_LDL | 27.3 ± 8.91 | 31.43 ± 8.25 | 0.0021 | 0.0236 | 0.20 | ↑ |
| SubFreeChol_VLDL-5 | 1.12 ± 0.51 | 0.87 ± 0.44 | 0.0022 | 0.0236 | –0.36 | ↓ |
| SubPhosp_VLDL-5 | 1.90 ± 0.73 | 1.63 ± 0.50 | 0.0045 | 0.0442 | –0.22 | ↓ |
| SubChol_VLDL-5 | 1.600 ± 0.76 | 1.41 ± 0.71 | 0.0055 | 0.0502 | –0.19 | ↓ |
| LMF_Trigl_LDL | 20.53 ± 5.91 | 23.85 ± 6.02 | 0.0059 | 0.0504 | 0.22 | ↑ |
| citric acid | 0.14 ± 0.04 | 0.16 ± 0.06 | 0.0073 | 0.0565 | 0.19 | ↑ |
| SubTrigl_LDL-6 | 4.51 ± 1.22 | 4.98 ± 1.36 | 0.0075 | 0.0565 | 0.14 | ↑ |
| SubFreeChol_LDL-4 | 2.24 ± 1.85 | 2.97 ± 2.24 | 0.0098 | 0.0683 | 0.41 | ↑ |
| glucose | 6.70 ± 1.78 | 7.40 ± 1.85 | 0.0101 | 0.0683 | 0.14 | ↑ |
| SubTrigl_LDL-2 | 2.30 ± 0.76 | 2.64 ± 0.85 | 0.0111 | 0.0700 | 0.20 | ↑ |
| LMF_Phosp_LDL | 51.69 ± 15.48 | 57.27 ± 16.09 | 0.0134 | 0.0785 | 0.15 | ↑ |
| LMF_ApoB_LDL | 60.2 ± 20.19 | 67.02 ± 21.55 | 0.0141 | 0.0785 | 0.15 | ↑ |
| LDL_PNb | 1094.52 ± 367.12 | 1218.62 ± 391.78 | 0.0141 | 0.0785 | 0.15 | ↑ |
| SubFreeChol_HDL-2 | 1.75 ± 0.5 | 1.98 ± 0.69 | 0.0166 | 0.0859 | 0.17 | ↑ |
| acetic acid | 0.05 ± 0.03 | 0.06 ± 0.04 | 0.0168 | 0.0859 | 0.26 | ↑ |
| SubTrigl_LDL-4 | 1.75 ± 1.25 | 2.26 ± 1.35 | 0.0183 | 0.0900 | 0.37 | ↑ |
| SubTrigl_LDL-3 | 2.64 ± 0.67 | 2.86 ± 0.55 | 0.0199 | 0.0934 | 0.11 | ↑ |
| LDL-Chol | 89.91 ± 29.6 | 98.16 ± 30.33 | 0.0212 | 0.0934 | 0.13 | ↑ |
| LMF_Chol_LDL | 89.91 ± 29.6 | 98.16 ± 30.33 | 0.0212 | 0.0934 | 0.13 | ↑ |
| LDL-HDL-Chol | 1.71 ± 0.63 | 1.94 ± 0.58 | 0.0247 | 0.1044 | 0.18 | ↑ |
| glutamine | 0.64 ± 0.16 | 0.58 ± 0.19 | 0.0256 | 0.1044 | –0.13 | ↓ |
| LDL2_PNb | 183.26 ± 63.07 | 207.11 ± 69.98 | 0.0268 | 0.1044 | 0.18 | ↑ |
| SubApoB_LDL-2 | 10.08 ± 3.47 | 11.39 ± 3.85 | 0.0269 | 0.1044 | 0.18 | ↑ |
| SubPhosp_LDL-2 | 9.87 ± 3.48 | 11.43 ± 3.62 | 0.0329 | 0.1240 | 0.21 | ↑ |
| SubFreeChol_HDL-1 | 3.82 ± 1.82 | 4.15 ± 1.60 | 0.0369 | 0.1345 | 0.12 | ↑ |
| Apo-B100 | 81.59 ± 23.07 | 87.27 ± 23.47 | 0.0477 | 0.1648 | 0.10 | ↑ |
| TPNb | 1483.43 ± 419.52 | 1586.76 ± 426.77 | 0.0477 | 0.1648 | 0.10 | ↑ |
| SubFreeChol_LDL-2 | 6.23 ± 1.93 | 6.9 ± 2.58 | 0.0489 | 0.1648 | 0.15 | ↑ |
In total, 128 blood analytes (18 metabolites and 110 lipoproteins/lipid fractions) were quantified using NMR. Only results for analytes for which the unadjusted raw P-value is <0.05 are reported: median (± MAD, median absolute deviation) in the two groups, raw P-value, Adjusted P-value (FDR, Benjamini–Hochberg false discovery rate), Log2 of the fold change, and trend (↓ decreasing trend, ↑ increasing trend referred to the 3M-PFO group). The concentrations of lipoproteins and lipid fractions are in mg/dL; the concentrations of metabolites are in mmol/L.
Particle number (PN) expressed as nmol/L. Abbreviations: Chol, cholesterol; LMF, lipoprotein main fraction; Phosp, phospholipid; PN, particle number; Sub, subfraction; and Trigl, triglycerides. A table containing results for all 128 analytes can be found in Supplementary Table S1.
Table 4. Results of Univariate Analysis (Mann–Whitney–Wilcoxon Test) for the Comparison of the Concentration Levels of Metabolite and Lipoproteins and Lipid Fractions in the Nondeceased (3M-nD) and Deceased (3M-D) Patients at 3 Months after AIS and Treatment with Rt-PAa.
| patient
groups |
||||||
|---|---|---|---|---|---|---|
| analyte | 3M-nD | 3M-D | P-value | FDR | Log2(FC) | trend |
| SubTrigl_HDL-4 | 3.25 ± 0.99 | 2.46 ± 1.09 | 0.001 | 0.1307 | –0.40 | ↓ |
| SubApoA1_HDL-4 | 69.23 ± 15.68 | 57.14 ± 17.01 | 0.0072 | 0.2418 | –0.28 | ↓ |
| phenylalanine | 0.08 ± 0.03 | 0.10 ± 0.04 | 0.0087 | 0.2418 | 0.32 | ↑ |
| lactic acid | 2.3 ± 1.04 | 3.10 ± 0.89 | 0.0089 | 0.2418 | 0.43 | ↑ |
| citric acid | 0.15 ± 0.05 | 0.20 ± 0.08 | 0.0094 | 0.2418 | 0.38 | ↑ |
| 3-hydroxybutyrate | 0.14 ± 0.16 | 0.26 ± 0.24 | 0.0129 | 0.2758 | 0.95 | ↑ |
| SubPhosp_HDL-4 | 23.31 ± 6.37 | 18.48 ± 7.58 | 0.0207 | 0.3382 | –0.34 | ↓ |
| SubChol_VLDL-5 | 1.50 ± 0.70 | 1.27 ± 0.61 | 0.0253 | 0.3382 | –0.24 | ↓ |
| SubApoA2_HDL-4 | 18.45 ± 4.57 | 14.60 ± 5.40 | 0.0299 | 0.3382 | –0.34 | ↓ |
| SubChol_HDL-1 | 15.45 ± 7.06 | 17.99 ± 8.48 | 0.0307 | 0.3382 | 0.22 | ↑ |
| SubPhosp_VLDL-5 | 1.79 ± 0.66 | 1.54 ± 0.38 | 0.0312 | 0.3382 | –0.21 | ↓ |
| LMF_ApoA2_HDL | 30.81 ± 5.09 | 28.44 ± 5.54 | 0.0362 | 0.3382 | –0.12 | ↓ |
| SubFreeChol_HDL-2 | 1.79 ± 0.53 | 2.24 ± 0.56 | 0.0402 | 0.3382 | 0.32 | ↑ |
| LMF_ApoA1_HDL | 132.35 ± 24 | 121.96 ± 19.09 | 0.0414 | 0.3382 | –0.12 | ↓ |
| Apo-A2 | 30.14 ± 5.15 | 27.89 ± 6.12 | 0.0420 | 0.3382 | –0.11 | ↓ |
| acetone | 0.12 ± 0.11 | 0.17 ± 0.12 | 0.0453 | 0.3382 | 0.44 | ↑ |
| SubChol_HDL-4 | 18.37 ± 5.06 | 14.43 ± 7.81 | 0.0472 | 0.3382 | –0.35 | ↓ |
| SubChol_LDL-5 | 11.14 ± 7.86 | 6.66 ± 6.45 | 0.0482 | 0.3382 | –0.74 | ↓ |
In total, 128 blood analytes (18 metabolites and 110 lipoproteins/lipid fractions) were quantified using NMR. Only results for analytes for which the unadjusted raw P-value is <0.05 are reported: median (± MAD, median absolute deviation) in the two groups, raw P-value, adjusted P-value (FDR, Benjamini-Hochberg false discovery rate), Log2 of the fold change, and trend (↓ decreasing trend, ↑ increasing trend referred to the 3M-D group). The concentrations of lipoproteins and lipid fractions are in mg/dL; the concentrations of metabolites are in mmol/L.
Multivariate Exploratory Analysis
PCA was applied, as a multivariate exploratory approach, on all 128 quantified analytes (metabolites and lipoproteins and lipid fractions) of all available samples, to obtain an overview of the variation in the data and to check for the presence of metabolic signatures among the different groups compared. Results are shown in Figure 2; there is no obvious separation among the samples of patients with the PFO (3M-PFO) and GFO (3M-GFO) and of patients who were alive (3M-nD) or deceased (3M-D) at 3 months from the AIS.
Figure 2.
Scatter plots of PCA. Each dot represents the serum metabolic profile of patients who suffered AIS at 24 h post rt-PA. (A) Analysis of 3M-nD patients (alive at 3 months after AIS, n = 226, green dots) and 3M-D patients (deceased at 3 months after AIS, n = 22, blue dots). (B) Analysis of 3M-GFO patients (with GFO at 3 months after AIS, n = 166, orange dots) and 3M-PFO patients (with PFO at 3 months after AIS, n = 82, purple dots).
Prediction of Patient Outcome with Random Forest
Because PCA analysis was not able to highlight observable differences in serum profiles of the different patient groups (3M-D and 3M-nD; 3M-GFO and 3M-PFO), we used the supervised classification method (Random Forest) to investigate whether the metabolite and lipid profiles could be employed to classify 3M-GFO/3M-PFO patient groups: all classification models (3M-D vs 3M-nD; 3M-GFO vs 3M-PFO) built on metabolite and lipoprotein/lipid fraction concentrations had low discriminating power, as shown in Table 2.
Altogether, the common multivariate analysis of serum profiles of AIS patients proved to be unsuccessful to identify multiple spectral characteristics that are different between 3M-PFO and 3M-GFO or between 3M-nD and 3M-D.
Differential Network Analysis
We built serum metabolite–lipid association networks specific for 3M-PFO/3M-GFO and 3M-nD/3M-D patients at t1 (24 h post rt-PA) with the scope of investigating possible perturbations in patients’ metabolic status that could be captured by differential network analysis: the rationale is that metabolites participating in the same metabolic pathway tend to have higher levels of correlations and connectivity,49 while a significantly decreased metabolite may indicate a reduced role of certain pathways where those metabolites participate.
Differential Analysis of Metabolite–Lipid Association Networks Associated with the Patient FO at 3 Months after Thrombolysis Treatment
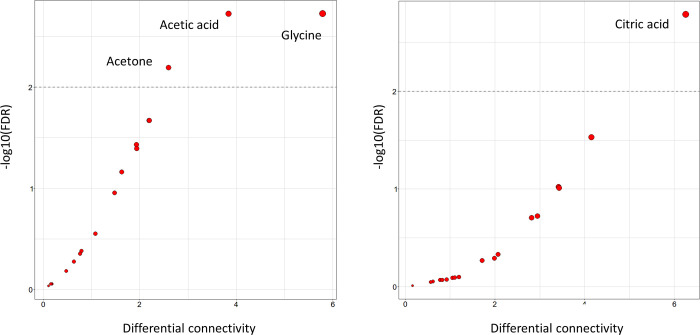
The metabolite and lipid association networks specific to 3M-GFO and 3M-PFO patients are given in Figure 3A,B, respectively, while differential connectivity plots (for metabolites) are reported in Figure 4A. Statistically significant differences in connectivity are reported for glutamine, tyrosine, leucine, lactate, acetone, acetate, and glycine. Results of differential connectivity analysis for lipoprotein and lipid fractions are given in Supplementary Table S3.
Figure 3.
Metabolite–metabolite and metabolite–lipid association networks for (A) 3M-PFO and (B) 3M-GFO patients. Networks are reconstructed using the PCLRC algorithm from serum metabolites and lipid fractions from samples collected 24 h post rt-PA (t1). The two networks show only the nodes (and corresponding edges) that are found to be significantly differentially connected (FDR < 0.05); see eqs 2 and 3, Figure 4, and associated captions. Only edges corresponding to correlations between lipids and metabolites |rij| > 0.6 are shown (eq 1). Note that a differentially connected node can have edges to nondifferentially connected nodes which are, in consequence, also shown. Correlation networks have been reconstructed using the PCLRC algorithm. Nodes are arranged and colored (from pink to purple) according to connectivity (i.e., number of connecting edges, aka degree). Abbreviations are reported as follows: analytes: 3-HB, 3-hydroxybutyrate; Apo, Apolipoproteins; Chol, cholesterol; LMF, lipoprotein main fractions; Phosp, phospholipids; PN, particle number; Sub, subfractions; and Trigl, triglycerides. Amino acids are reported with a three letter code.
Figure 4.
Differential connectivity analysis. Scatter plot of metabolite differential connectivity (see eqs 2 and 3) against statistical significance (P-value corrected (FDR) for multiple testing using the Benjamini–Hochberg approach) obtained by means of permutation test.25 The horizontal lines indicate the 0.01 significance threshold α on the -log(p-val) scale. (A) Comparison of metabolite–metabolite association networks for patients with poor (3M-PFO) and good functional outcome (3M-GFO) at 3 months after AIS and thrombolysis treatment. (B) Comparison of metabolite–metabolite association networks for patients deceased (3M-D) and alive (3M-nD) at 3 months after AIS and thrombolysis treatment. Results of differential connectivity analysis for lipids are given in Supplementary Tables S1 and S2.
The alteration of lactic acid connections observed in patients with impairment could suggest its decreased role in providing substitute energy fuel and in the metabolic pathways of neuroprotection where lactic acid is normally largely involved; in fact, the transition from aerobic to anaerobic glycolysis is enhanced to support the increasing demand of energy. As a result, the production of pyruvate and lactate increases, and this last one can be shuttled to neurons to guarantee neuron protection and survival.50
We observed a reduction of 3-hydroxybutyrate concentration (P-value = 3 × 10–5 and FDR = 0.0043) in patients with the PFO; there are also a decreasing trend for glucose (P-value = 0.0101 and FDR = 0.0683) and an increase of acetone (P-value = 0.0007 and FDR = 0.0129) which are, however, not significant after adjustment.
Changes in the connectivity of leucine, citric acid, acetate, and acetone specific to patients who developed neurological adverse outcomes treated with thrombolysis after the ischemic stroke indicate unbalances in the energy metabolism and oxidative stress-related pathways: alterations in brain energy metabolism are linked to energy deficits associated with ischemia and reperfusion injury, and downregulation of citric acid has been associated with the PFO after AIS,11 while the metabolism of ketone bodies is upregulated to provide alternative energy sources and to maintain free radical homeostasis during ischemia–reperfusion injury.51,52
Increased glutamine connectivity for patients with an adverse outcome suggests alterations in glutamine/glutamate metabolism. Glutamine is a main precursor of glutamate, and both are interconverted among astrocytes and neurons, guaranteeing glutamine homeostasis and glutamate generation and recycling. However, if glutamate generation and recycling are impaired, glutamate can be an excitatory and possibly toxic neurotransmitter, which can lead to glutamate-induced neurotoxicity. Alterations in glutamine/glutamate metabolism have been observed in patients with ischemic stroke, and increasing levels of serum glutamine were associated with compensatory adaptative mechanisms to counteract glutamate-induced neurotoxicity.53
Moreover, we observed increased levels of phenylalanine (P-value = 0.0008 and FDR = 0.0128) in patients with the PFO and differential connectivity of tyrosine. This suggests that phenylalanine and its metabolite tyrosine might be associated with glutamate-induced neurotoxicity, because phenylalanine can suppress the excitatory glutamatergic synaptic transmission.54
Summarizing, the metabolic profiles of 3M-PFO patients are consistent with the hypothesis of deregulated glutamate metabolism and subsequent glutamate-induced neurotoxicity that seems responsible for the worsening of poststroke quality of life.
We observed increased glycine connectivity for 3M-PFO patients, indicating a possible role of glycine in impairment. The role of glycine for AIS is quite controversial. Recent studies demonstrated that lower levels of glycine are deleterious for ischemic neuronal injury, while higher levels of the same metabolite seem to be neuroprotective.55,56
We found altered patterns of connectivity among almost all lipid features (see Supplementary Table S3), indicating that overall alteration and remodulation of lipid metabolism may be involved in poststroke adverse outcome and neurological disabilities. Several studies have explored the relationships between lipidic features and ischemic stroke,6,9,57,58 demonstrating how these molecules play dual roles in the etiology and progression of the disease. Serum cholesterol has been found to be an independent predictor for long-term FOs, and higher serum total cholesterol levels have been associated with better prognosis.59 Triacylglycerols have been significantly associated with ischemic stroke.60
In the metabolite–lipid association network specific to 3M-PFO AIS patients, triglycerides show a decreased connectivity. Because triglycerides are hydrolyzed to fatty acids to provide alternative energy sources, we associated decreased connectivity with alterations in the triglyceride metabolism leading to a decreased role of triacylglycerols in supporting alternative energy fuel in patients who developed a PFO poststroke. This is line with previous observations regarding the general condition of energy failure that characterizes the topology of the association network of 3M-PFO patients. The metabolites and lipids highlighted by the differential network analysis are involved in the mechanisms guaranteeing energy homeostasis and show decreased connectivity in the specific network of patients who developed a PFO 3 months after the ischemic stroke.
Differential Analysis of Metabolite–Lipid Association Networks Associated with Patient Mortality at 3 Months after Thrombolysis Treatment
The metabolite–lipid association networks specific for 3M-nD and 3M-D AIS patients are shown in Figure 5. The networks are strikingly different, with the network built from the samples of patients who did not survive at 3 months after AIS being totally disconnected (Figure 5B). We shall comment that two groups (3M-nD and 3M-D) are different in size (n = 226 and n = 22, respectively), and this may have influenced the reconstruction of the networks. However, n = 22 samples is sufficient to estimate a bivariate correlation >0.5 with 80% power at the α = 0.05 significant level: the two networks shown in Figure 5 are restricted to nodes that are differentially connected and have node–node correlation |rij| > 0.6; thus the difference in the network structure is likely due to underlying biological differences rather than a bias induced by the different sample size.
Figure 5.
Metabolite–metabolite and metabolite–lipid association networks for (A) 3M-nD and (B) 3M-D patients. Networks are reconstructed using the PCLRC algorithm from serum metabolites and lipid fractions from samples collected 24 h post rt-PA (t1). The two networks show only the nodes (and corresponding edges) that are found to be significantly differentially connected (FDR < 0.05); see eqs 2 and 3, Figure 4, and associated captions. Only edges corresponding to correlations between lipids and metabolites |rij| > 0.6 are shown (eq 1). Note that a differentially connected node can have edges to nondifferentially connected nodes which are, in consequence, also shown. Correlation networks have been reconstructed using the PCLRC algorithm. Nodes are arranged and colored (from pink to purple) according to connectivity (i.e., number of connecting edges, aka degree). Abbreviations are reported as follows: analytes: 3-HB, 3-hydroxybutyrate; Apo, Apolipoproteins; Chol, cholesterol; LMF, lipoproteins main fractions; Phosp, phospholipids; PN, particle number; Sub, subfractions; and Trigl, triglycerides. Amino acids are reported with a three letter code.
There is a statistically significant reduction in the connectivity of blood circulating citric acid and acetone and several lipid fractions in the correlation networks of patients who, after 3 months, did not survive the acute cerebral ischemia (Figure 4B; results of differential connectivity analysis for lipoprotein and lipid fractions are given in Supplementary Table S4).
In particular, we observed a loss of structural connections between metabolites and LDL-related fractions. Citric acid showed also higher concentrations (P-value = 0.0094) in deceased patients, although significance disappears after adjustment for multiple corrections (FDR = 0.2418)
As previously discussed, citric acid and ketone bodies are involved in energy metabolism, and their levels can change during and after the brain stroke to restore energy homeostasis. Our results suggest that dysregulations in energy metabolism may be associated also with underlying causes related to an increased risk of death at 3 months from the AIS.
As in the previous case, we observed a statistically significant differential connectivity of several lipoproteins and lipid fractions (see Supplementary Table S4). In particular, we observed a decrease of LDL connectivity (especially VLDL and LDL-5 and LDL-6 subparticles), in the network specific to patients deceased at 3 months after AIS. Lipoproteins and lipids have been associated with IS;61 small dense LDL (sdLDL) and small-sized HDL particles are established risk factors for this disease. It has been shown that AIS is associated with adverse distributions of LDL and HDL subclasses, and short-term mortality is linked to increased levels of sdLDL particles.62 Because sdLDLs are more susceptible to oxidation than larger LDLs, we suggest that sdLDL particles may provide an optimal substrate for rt-PA-induced oxidative action and that alterations in the connectivity patterns of lipid subfractions reflect an increase in the rt-PA-mediated oxidative damage associated with poststroke mortality.
There is evidence that ketone bodies and ketone body metabolism play a role in the pathologic and functional outcomes following stroke,63 and changes in the metabolism of ketone bodies have been observed during stress conditions and reperfusion oxidative stress.64
Ketone bodies are the only circulating substrates in addition to glucose known to contribute significantly to cerebral metabolism; however, the exact role of ketone bodies and the precise mechanisms whereby they can provide protection in ischemic stroke and thus being associated with reduced mortality and/or better FO in AIS patients are largely unknown.64 During ischemic stroke, oxygen levels fall resulting in mitochondria malfunction: this induces a short-term ketosis, and increased reliance on ketone metabolism (which is enzymatically simpler and more efficient than glucose or pyruvate metabolism) seems to be a mechanism of cerebral metabolic adaptation65 and is reported to increase global cerebral blood flow.66,67
Our results indicate strong perturbation in the processes involving serum acetone and VLDL-related subfractions, 3-hydroxybutyrate, and free cholesterol linked to VLDL-1 subfractions. In particular, disruption of the connectivity of acetone and VLDL again suggests remodulation of energy metabolisms.
Overall, the reduction of metabolite–lipid connectivity for 3 M-D patients may suggest alterations in lipid metabolism during cerebral ischemia, which can strongly affect poststroke mortality, as well as the development of poststroke PFO.
Conclusions
In this study, we have analyzed serum circulating metabolites and lipids of patients who suffered AIS with the aim of highlighting molecular mechanisms associated with the patient FO and mortality. Metabolite and lipoprotein and lipid fraction concentrations were measured on patients 24 h treatment with rt-PA using NMR and mortality, and FOs were evaluated at 3 months after stroke.
We applied an integrated top-down system-biology approach deploying standard univariate and multivariate analysis together with machine learning and network analysis.
While standard approaches failed to discriminate between the patient groups, network analysis was successful in detecting marked metabolic differences that could be related to the development of post-AIS PFO and mortality. Although the patterns of dysregulated metabolite and lipoprotein and lipid fraction correlation cannot be used at the patient level as prognostic markers for mortality and FO, this analysis provides a working hypothesis that warrants further investigation at both the molecular and patient levels.
We showed that dysregulations of metabolic mechanisms involving triglycerides, HDL, LDL, and VLDL fractions, and related subfractions), amino acids (leucine, glycine, glutamine, tyrosine, and phenylalanine), organic acids (citric, lactic, and acetic acids), and ketone bodies (acetone, 3-hydroxybutyrate) are associated with patient mortality suggesting a role of energy failure, glutamate-induced neurotoxicity, oxidative stress, and neuroprotection in determining patient survival at 3 months after AIS. Of particular interest is the involvement of lipid and lipid metabolism with patients’ mortality and FO, which warrant further investigation in the light of expanding field of lipidomic research.
Furthermore, ketone bodies emerged as largely involved in the determination of both 3-month outcomes (PFO and mortality) in ischemic stroke treated with thrombolysis, which reinforce already existing evidence. In conclusion, this study affords important information on how metabolite–metabolite and metabolite–lipid association networks of AIS patients differ according to the patient outcomes and highlight the utility of the analysis of biological networks.
Finally, to be translated to predictive biomarkers one should search for markers in blood that can be measured at the patient level and are representative of metabolic perturbation. Our analysis/results, being exploratory in nature, as all network-based analyses, suggest where to search for such markers, that is, to which metabolic pathways to look at.
Acknowledgments
The Biological Markers Associated with Acute Ischemic Stroke (MAGIC) Study was funded by grants from Italian Ministry of Health, 2006 Finalized Research Programs (RFPS-2006-1-336520).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.1c00406.
Table S1: Complete univariate results for the comparison between 3M-PFO and 3M-GFO patient groups; Table S2: Complete univariate results for the comparison between 3M-D and 3M-nD patient groups; Table S3: Complete differential connectivity analysis results for the comparison between 3M-PFO and 3M-GFO patient groups; And Table S4: Complete differential connectivity analysis for the comparison between 3M-D and 3M-nD patient groups (PDF)
Author Contributions
§ C.L. and L.T. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Phipps M. S.; Cronin C. A. Management of Acute Ischemic Stroke. BMJ 2020, 368, l6983. 10.1136/bmj.l6983. [DOI] [PubMed] [Google Scholar]
- Goyal M.; Menon B. K.; van Zwam W. H.; Dippel D. W. J.; Mitchell P. J.; Demchuk A. M.; Dávalos A.; Majoie C. B. L. M.; van der Lugt A.; de Miquel M. A.; Donnan G. A.; Roos Y. B. W. E. M.; Bonafe A.; Jahan R.; Diener H.-C.; van den Berg L. A.; Levy E. I.; Berkhemer O. A.; Pereira V. M.; Rempel J.; Millán M.; Davis S. M.; Roy D.; Thornton J.; Román L. S.; Ribó M.; Beumer D.; Stouch B.; Brown S.; Campbell B. C. V.; van Oostenbrugge R. J.; Saver J. L.; Hill M. D.; Jovin T. G.; Endovascular Thrombectomy after Large-Vessel Ischaemic Stroke: A Meta-Analysis of Individual Patient Data from Five Randomised Trials. Lancet 2016, 387, 1723–1731. 10.1016/S0140-6736(16)00163-X. [DOI] [PubMed] [Google Scholar]
- Albers G. W.; Marks M. P.; Kemp S.; Christensen S.; Tsai J. P.; Ortega-Gutierrez S.; McTaggart R. A.; Torbey M. T.; Kim-Tenser M.; Leslie-Mazwi T.; Sarraj A.; Kasner S. E.; Ansari S. A.; Yeatts S. D.; Hamilton S.; Mlynash M.; Heit J. J.; Zaharchuk G.; Kim S.; Carrozzella J.; Palesch Y. Y.; Demchuk A. M.; Bammer R.; Lavori P. W.; Broderick J. P.; Lansberg M. G. Thrombectomy for Stroke at 6 to 16 Hours with Selection by Perfusion Imaging. N. Engl. J. Med. 2018, 378, 708–718. 10.1056/NEJMoa1713973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira R. G.; Jadhav A. P.; Haussen D. C.; Bonafe A.; Budzik R. F.; Bhuva P.; Yavagal D. R.; Ribo M.; Cognard C.; Hanel R. A.; Sila C. A.; Hassan A. E.; Millan M.; Levy E. I.; Mitchell P.; Chen M.; English J. D.; Shah Q. A.; Silver F. L.; Pereira V. M.; Mehta B. P.; Baxter B. W.; Abraham M. G.; Cardona P.; Veznedaroglu E.; Hellinger F. R.; Feng L.; Kirmani J. F.; Lopes D. K.; Jankowitz B. T.; Frankel M. R.; Costalat V.; Vora N. A.; Yoo A. J.; Malik A. M.; Furlan A. J.; Rubiera M.; Aghaebrahim A.; Olivot J.-M.; Tekle W. G.; Shields R.; Graves T.; Lewis R. J.; Smith W. S.; Liebeskind D. S.; Saver J. L.; Jovin T. G. Thrombectomy 6 to 24 Hours after Stroke with a Mismatch between Deficit and Infarct. N. Engl. J. Med. 2018, 378, 11–21. 10.1056/NEJMoa1706442. [DOI] [PubMed] [Google Scholar]
- Wesley U. V.; Bhute V. J.; Hatcher J. F.; Palecek S. P.; Dempsey R. J. Local and Systemic Metabolic Alterations in Brain, Plasma, and Liver of Rats in Response to Aging and Ischemic Stroke, as Detected by Nuclear Magnetic Resonance (NMR) Spectroscopy. Neurochem. Int. 2019, 127, 113–124. 10.1016/j.neuint.2019.01.025. [DOI] [PubMed] [Google Scholar]
- Jung J. Y.; Lee H.-S.; Kang D.-G.; Kim N. S.; Cha M. H.; Bang O.-S.; Ryu D. H.; Hwang G.-S. 1H-NMR-Based Metabolomics Study of Cerebral Infarction. Stroke 2011, 42, 1282–1288. 10.1161/STROKEAHA.110.598789. [DOI] [PubMed] [Google Scholar]
- Szpetnar M.; Hordyjewska A.; Malinowska I.; Golab P.; Kurzepa J. The Fluctuation of Free Amino Acids in Serum during Acute Ischemic Stroke. Curr. Issues Pharm. Med. Sci. 2016, 29, 151–154. 10.1515/cipms-2016-0031. [DOI] [Google Scholar]
- Wang D.; Kong J.; Wu J.; Wang X.; Lai M. GC-MS-Based Metabolomics Identifies an Amino Acid Signature of Acute Ischemic Stroke. Neurosci. Lett. 2017, 642, 7–13. 10.1016/j.neulet.2017.01.039. [DOI] [PubMed] [Google Scholar]
- Liu P.; Li R.; Antonov A. A.; Wang L.; Li W.; Hua Y.; Guo H.; Wang L.; Liu P.; Chen L.; Tian Y.; Xu F.; Zhang Z.; Zhu Y.; Huang Y. Discovery of Metabolite Biomarkers for Acute Ischemic Stroke Progression. J. Proteome Res. 2017, 16, 773–779. 10.1021/acs.jproteome.6b00779. [DOI] [PubMed] [Google Scholar]
- Kimberly W. T.; Wang Y.; Pham L.; Furie K. L.; Gerszten R. E. Metabolite Profiling Identifies a Branched Chain Amino Acid Signature in Acute Cardioembolic Stroke. Stroke 2013, 44, 1389–1395. 10.1161/STROKEAHA.111.000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.; Zhou K.; Li H.; Dong X.; Tan G.; Chai Y.; Wang W.; Bi X. Potential of Serum Metabolites for Diagnosing Post-Stroke Cognitive Impairment. Mol. BioSyst. 2015, 11, 3287–3296. 10.1039/c5mb00470e. [DOI] [PubMed] [Google Scholar]
- Ke C.; Pan C.-W.; Zhang Y.; Zhu X.; Zhang Y. Metabolomics Facilitates the Discovery of Metabolic Biomarkers and Pathways for Ischemic Stroke: A Systematic Review. Metabolomics 2019, 15, 152. 10.1007/s11306-019-1615-1. [DOI] [PubMed] [Google Scholar]
- Vignoli A.; Ghini V.; Meoni G.; Licari C.; Takis P. G.; Tenori L.; Turano P.; Luchinat C. High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. Ed. Engl. 2019, 58, 968–994. 10.1002/anie.201804736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takis P. G.; Ghini V.; Tenori L.; Turano P.; Luchinat C. Uniqueness of the NMR Approach to Metabolomics. TrAC Trends Anal. Chem. 2019, 120, 115300. 10.1016/j.trac.2018.10.036. [DOI] [Google Scholar]
- Meoni G.; Lorini S.; Monti M.; Madia F.; Corti G.; Luchinat C.; Zignego A. L.; Tenori L.; Gragnani L. The Metabolic Fingerprints of HCV and HBV Infections Studied by Nuclear Magnetic Resonance Spectroscopy. Sci. Rep. 2019, 9, 4128. 10.1038/s41598-019-40028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignoli A.; Orlandini B.; Tenori L.; Biagini M. R.; Milani S.; Renzi D.; Luchinat C.; Calabrò A. S. Metabolic Signature of Primary Biliary Cholangitis and Its Comparison with Celiac Disease. J. Proteome Res. 2019, 18, 1228–1236. 10.1021/acs.jproteome.8b00849. [DOI] [PubMed] [Google Scholar]
- Vignoli A.; Paciotti S.; Tenori L.; Eusebi P.; Biscetti L.; Chiasserini D.; Scheltens P.; Turano P.; Teunissen C.; Luchinat C.; Parnetti L. Fingerprinting Alzheimer’s Disease by 1H Nuclear Magnetic Resonance Spectroscopy of Cerebrospinal Fluid. J. Proteome Res. 2020, 19, 1696–1705. 10.1021/acs.jproteome.9b00850. [DOI] [PubMed] [Google Scholar]
- Vignoli A.; Tenori L.; Giusti B.; Takis P. G.; Valente S.; Carrabba N.; Balzi D.; Barchielli A.; Marchionni N.; Gensini G. F.; Marcucci R.; Luchinat C.; Gori A. M. NMR-Based Metabolomics Identifies Patients at High Risk of Death within Two Years after Acute Myocardial Infarction in the AMI-Florence II Cohort. BMC Med. 2019, 17, 3. 10.1186/s12916-018-1240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Donato S.; Mislang A. R.; Vignoli A.; Mori E.; Vitale S.; Biagioni C.; Hart C.; Becheri D.; Del Monte F.; Luchinat C.; Di Leo A.; Mottino G.; Tenori L.; Biganzoli L. D20 - Serum Metabolomic as Biomarkers to Differentiate Early from Metastatic Disease in Elderly Colorectal Cancer (Crc) Patients. Ann. Oncol. 2016, 27, iv45. 10.1093/annonc/mdw335.20. [DOI] [Google Scholar]
- Rosato A.; Tenori L.; Cascante M.; De Atauri Carulla P. R.; Martins dos Santos V. A. P.; Saccenti E. From Correlation to Causation: Analysis of Metabolomics Data Using Systems Biology Approaches. Metabolomics 2018, 14, 37. 10.1007/s11306-018-1335-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho D.; de la Fuente A.; Mendes P. The Origin of Correlations in Metabolomics Data. Metabolomics 2005, 1, 53–63. 10.1007/s11306-005-1107-3. [DOI] [Google Scholar]
- Saccenti E.; Suarez-Diez M.; Luchinat C.; Santucci C.; Tenori L. Probabilistic Networks of Blood Metabolites in Healthy Subjects As Indicators of Latent Cardiovascular Risk. J. Proteome Res. 2015, 14, 1101–1111. 10.1021/pr501075r. [DOI] [PubMed] [Google Scholar]
- Saccenti E.; Menichetti G.; Ghini V.; Remondini D.; Tenori L.; Luchinat C. Entropy-Based Network Representation of the Individual Metabolic Phenotype. J. Proteome Res. 2016, 15, 3298–3307. 10.1021/acs.jproteome.6b00454. [DOI] [PubMed] [Google Scholar]
- Vignoli A.; Tenori L.; Luchinat C.; Saccenti E. Age and Sex Effects on Plasma Metabolite Association Networks in Healthy Subjects. J. Proteome Res. 2018, 17, 97–107. 10.1021/acs.jproteome.7b00404. [DOI] [PubMed] [Google Scholar]
- Vignoli A.; Tenori L.; Giusti B.; Valente S.; Carrabba N.; Balzi D.; Barchielli A.; Marchionni N.; Gensini G. F.; Marcucci R.; Gori A. M.; Luchinat C.; Saccenti E. Differential Network Analysis Reveals Metabolic Determinants Associated with Mortality in Acute Myocardial Infarction Patients and Suggests Potential Mechanisms Underlying Different Clinical Scores Used To Predict Death. J. Proteome Res. 2020, 19, 949–961. 10.1021/acs.jproteome.9b00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal M.; Saccenti E.; Madsen M. B.; Hansen M. B.; Hyldegaard O.; Skrede S.; Martins Dos Santos V. A. P.; Norrby-Teglund A.; Svensson M. Integrated Univariate, Multivariate, and Correlation-Based Network Analyses Reveal Metabolite-Specific Effects on Bacterial Growth and Biofilm Formation in Necrotizing Soft Tissue Infections. J. Proteome Res. 2020, 19, 688–698. 10.1021/acs.jproteome.9b00565. [DOI] [PubMed] [Google Scholar]
- Suarez-Diez M.; Adam J.; Adamski J.; Chasapi S. A.; Luchinat C.; Peters A.; Prehn C.; Santucci C.; Spyridonidis A.; Spyroulias G. A.; Tenori L.; Wang-Sattler R.; Saccenti E. Plasma and Serum Metabolite Association Networks: Comparability within and between Studies Using NMR and MS Profiling. J. Proteome Res. 2017, 16, 2547–2559. 10.1021/acs.jproteome.7b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori A. M.; Giusti B.; Piccardi B.; Nencini P.; Palumbo V.; Nesi M.; Nucera A.; Pracucci G.; Tonelli P.; Innocenti E.; Sereni A.; Sticchi E.; Toni D.; Bovi P.; Guidotti M.; Tola M. R.; Consoli D.; Micieli G.; Tassi R.; Orlandi G.; Sessa M.; Perini F.; Delodovici M. L.; Zedde M. L.; Massaro F.; Abbate R.; Inzitari D. Inflammatory and Metalloproteinases Profiles Predict Three-Month Poor Outcomes in Ischemic Stroke Treated with Thrombolysis. J. Cereb. Blood Flow Metab. 2017, 37, 3253–3261. 10.1177/0271678X17695572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzitari D.; Giusti B.; Nencini P.; Gori A. M.; Nesi M.; Palumbo V.; Piccardi B.; Armillis A.; Pracucci G.; Bono G.; Bovi P.; Consoli D.; Guidotti M.; Nucera A.; Massaro F.; Micieli G.; Orlandi G.; Perini F.; Tassi R.; Tola M. R.; Sessa M.; Toni D.; Abbate R. MMP9 Variation After Thrombolysis Is Associated With Hemorrhagic Transformation of Lesion and Death. Stroke 2013, 44, 2901–2903. 10.1161/STROKEAHA.113.002274. [DOI] [PubMed] [Google Scholar]
- Fieschi C.; Argentino C.; Lenzi G. L.; Sacchetti M. L.; Toni D.; Bozzao L. Clinical and Instrumental Evaluation of Patients with Ischemic Stroke within the First Six Hours. J. Neurol. Sci. 1989, 91, 311–321. 10.1016/0022-510x(89)90060-9. [DOI] [PubMed] [Google Scholar]
- Tissue Plasminogen Activator for Acute Ischemic Stroke. N. Engl. J. Med. 1995, 333, 1581–1588. 10.1056/NEJM199512143332401. [DOI] [PubMed] [Google Scholar]
- Hacke W.; Kaste M.; Bluhmki E.; Brozman M.; Dávalos A.; Guidetti D.; Larrue V.; Lees K. R.; Medeghri Z.; Machnig T.; Schneider D.; von Kummer R.; Wahlgren N.; Toni D. Thrombolysis with Alteplase 3 to 4.5 Hours after Acute Ischemic Stroke. N. Engl. J. Med. 2008, 359, 1317–1329. 10.1056/NEJMoa0804656. [DOI] [PubMed] [Google Scholar]
- Hacke W.; Donnan G.; Fieschi C.; Kaste M.; von Kummer R.; Broderick J. P.; Brott T.; Frankel M.; Grotta J. C.; Haley E. C.; Kwiatkowski T.; Levine S. R.; Lewandowski C.; Lu M.; Lyden P.; Marler J. R.; Patel S.; Tilley B. C.; Albers G.; Bluhmki E.; Wilhelm M.; Hamilton S.; Association of Outcome with Early Stroke Treatment: Pooled Analysis of ATLANTIS, ECASS, and NINDS Rt-PA Stroke Trials. Lancet 2004, 363, 768–774. 10.1016/S0140-6736(04)15692-4. [DOI] [PubMed] [Google Scholar]
- Wahlgren N.; Ahmed N.; Dávalos A.; Ford G. A.; Grond M.; Hacke W.; Hennerici M. G.; Kaste M.; Kuelkens S.; Larrue V.; Lees K. R.; Roine R. O.; Soinne L.; Toni D.; Vanhooren G. SITS-MOST investigators. Thrombolysis with Alteplase for Acute Ischaemic Stroke in the Safe Implementation of Thrombolysis in Stroke-Monitoring Study (SITS-MOST): An Observational Study. Lancet 2007, 369, 275–282. 10.1016/S0140-6736(07)60149-4. [DOI] [PubMed] [Google Scholar]
- Sulter G.; Steen C.; Jacques de Keyser Use of the Barthel Index and Modified Rankin Scale in Acute Stroke Trials. Stroke 1999, 30, 1538–1541. 10.1161/01.STR.30.8.1538. [DOI] [PubMed] [Google Scholar]
- Broderick J. P.; Adeoye O.; Elm J. Evolution of the Modified Rankin Scale and Its Use in Future Stroke Trials. Stroke 2017, 48, 2007–2012. 10.1161/STROKEAHA.117.017866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Swieten J. C.; Koudstaal P. J.; Visser M. C.; Schouten H. J.; van Gijn J. Interobserver Agreement for the Assessment of Handicap in Stroke Patients. Stroke 1988, 19, 604–607. 10.1161/01.str.19.5.604. [DOI] [PubMed] [Google Scholar]
- Kay R.; Wong K. S.; Perez G.; Woo J. Dichotomizing Stroke Outcomes Based on Self-Reported Dependency. Neurology 1997, 49, 1694–1696. 10.1212/wnl.49.6.1694. [DOI] [PubMed] [Google Scholar]
- World Medical Association Declaration of Helsinki Ethical Principles for Medical Research Involving Human Subjects. Bull World Health Organ 2001, 79, 373–374. [PMC free article] [PubMed] [Google Scholar]
- Jiménez B.; Holmes E.; Heude C.; Tolson R. F.; Harvey N.; Lodge S. L.; Chetwynd A. J.; Cannet C.; Fang F.; Pearce J. T. M.; Lewis M. R.; Viant M. R.; Lindon J. C.; Spraul M.; Schäfer H.; Nicholson J. K. Quantitative Lipoprotein Subclass and Low Molecular Weight Metabolite Analysis in Human Serum and Plasma by 1H NMR Spectroscopy in a Multilaboratory Trial. Anal. Chem. 2018, 90, 11962–11971. 10.1021/acs.analchem.8b02412. [DOI] [PubMed] [Google Scholar]
- Neuhäuser M.Wilcoxon–Mann–Whitney Test. In International Encyclopedia of Statistical Science; Springer, Berlin, Heidelberg, 2011; 1656–1658. 10.1007/978-3-642-04898-2_615. [DOI] [Google Scholar]
- Benjamini Y.; Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Hotelling H. Analysis of a Complex of Statistical Variables into Principal Components. J. Educ. Psychol. 1933, 24, 417–417. 10.1037/h0071325. [DOI] [Google Scholar]
- van den Berg R. A.; Hoefsloot H. C.; Westerhuis J. A.; Smilde A. K.; van der Werf M. J. Centering, Scaling, and Transformations: Improving the Biological Information Content of Metabolomics Data. BMC Genomics 2006, 7, 142. 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Mach. Learn. 2001, 45, 5–32. 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Parikh R.; Mathai A.; Parikh S.; Chandra Sekhar G.; Thomas R. Understanding and Using Sensitivity, Specificity and Predictive Values. Indian J. Ophthalmol. 2008, 56, 45–50. 10.4103/0301-4738.37595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith J. J.; Hayete B.; Thaden J. T.; Mogno I.; Wierzbowski J.; Cottarel G.; Kasif S.; Collins J. J.; Gardner T. S. Large-Scale Mapping and Validation of Escherichia Coli Transcriptional Regulation from a Compendium of Expression Profiles. PLoS Biol. 2007, 5, e8 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahagirdar S.; Saccenti E. On the Use of Correlation and MI as a Measure of Metabolite—Metabolite Association for Network Differential Connectivity Analysis. Metabolites 2020, 10, 171. 10.3390/metabo10040171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T.; Soyer O. S.; Bonhoeffer S. The Evolution of Connectivity in Metabolic Networks. PLoS Biol. 2005, 3, e228 10.1371/journal.pbio.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet C.; Castillo X.; Magistretti P. J.; Hirt L. New Evidence of Neuroprotection by Lactate after Transient Focal Cerebral Ischaemia: Extended Benefit after Intracerebroventricular Injection and Efficacy of Intravenous Administration. Cerebrovasc. Dis. 2012, 34, 329–335. 10.1159/000343657. [DOI] [PubMed] [Google Scholar]
- Au A.Metabolomics and Lipidomics of Ischemic Stroke. In Advances in Clinical Chemistry; Makowski G. S.Elsevier; 2018, 85, 31 −69, 10.1016/bs.acc.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Di Marino S.; Viceconte N.; Lembo A.; Summa V.; Tanzilli G.; Raparelli V.; Truscelli G.; Mangieri E.; Gaudio C.; Cicero D. O. Early Metabolic Response to Acute Myocardial Ischaemia in Patients Undergoing Elective Coronary Angioplasty. Open Heart 2018, 5, e000709 10.1136/openhrt-2017-000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonet D.; Rodríguez M. J.; Fredriksson K.; Bernal F.; Mahy N. In Vivo Neuroprotective Adaptation of the Glutamate/Glutamine Cycle to Neuronal Death. Hippocampus 2004, 14, 586–594. 10.1002/hipo.10188. [DOI] [PubMed] [Google Scholar]
- Kagiyama T.; Glushakov A. V.; Sumners C.; Roose B.; Dennis D. M.; Phillips M. I.; Ozcan M. S.; Seubert C. N.; Martynyuk A. E. Neuroprotective Action of Halogenated Derivatives of L-Phenylalanine. Stroke 2004, 35, 1192–1196. 10.1161/01.STR.0000125722.10606.07. [DOI] [PubMed] [Google Scholar]
- Yao W.; Ji F.; Chen Z.; Zhang N.; Ren S.-Q.; Zhang X.-Y.; Liu S.-Y.; Lu W. Glycine Exerts Dual Roles in Ischemic Injury through Distinct Mechanisms. Stroke 2012, 43, 2212–2220. 10.1161/STROKEAHA.111.645994. [DOI] [PubMed] [Google Scholar]
- Lu Y.; Zhang J.; Ma B.; Li K.; Li X.; Bai H.; Yang Q.; Zhu X.; Ben J.; Chen Q. Glycine Attenuates Cerebral Ischemia/Reperfusion Injury by Inhibiting Neuronal Apoptosis in Mice. Neurochem. Int. 2012, 61, 649–658. 10.1016/j.neuint.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Yang L.; Lv P.; Ai W.; Li L.; Shen S.; Nie H.; Shan Y.; Bai Y.; Huang Y.; Liu H. Lipidomic Analysis of Plasma in Patients with Lacunar Infarction Using Normal-Phase/Reversed-Phase Two-Dimensional Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry. Anal. Bioanal. Chem. 2017, 409, 3211–3222. 10.1007/s00216-017-0261-6. [DOI] [PubMed] [Google Scholar]
- Ding X.; Liu R.; Li W.; Ni H.; Liu Y.; Wu D.; Yang S.; Liu J.; Xiao B.; Liu S. A Metabonomic Investigation on the Biochemical Perturbation in Post-Stroke Patients with Depressive Disorder (PSD). Metab. Brain Dis. 2016, 31, 279–287. 10.1007/s11011-015-9748-z. [DOI] [PubMed] [Google Scholar]
- Pan S.-L.; Lien I.-N.; Chen T. H.-H. Is Higher Serum Total Cholesterol Level Associated with Better Long-Term Functional Outcomes after Noncardioembolic Ischemic Stroke?. Arch. Phys. Med. Rehabil. 2010, 91, 913–918. 10.1016/j.apmr.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Labreuche J.; Deplanque D.; Touboul P.-J.; Bruckert E.; Amarenco P. Association between Change in Plasma Triglyceride Levels and Risk of Stroke and Carotid Atherosclerosis: Systematic Review and Meta-Regression Analysis. Atherosclerosis 2010, 212, 9–15. 10.1016/j.atherosclerosis.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Holmes M. V.; Millwood I. Y.; Kartsonaki C.; Hill M. R.; Bennett D. A.; Boxall R.; Guo Y.; Xu X.; Bian Z.; Hu R.; Walters R. G.; Chen J.; Ala-Korpela M.; Parish S.; Clarke R. J.; Peto R.; Collins R.; Li L.; Chen Z. Group, on behalf of the C. K. B. C. Lipids, Lipoproteins, and Metabolites and Risk of Myocardial Infarction and Stroke. J. Am. Coll. Cardiol. 2018, 71, 620–632. 10.1016/j.jacc.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeljkovic A.; Vekic J.; Spasojevic-Kalimanovska V.; Jelic-Ivanovic Z.; Bogavac-Stanojevic N.; Gulan B.; Spasic S. LDL and HDL Subclasses in Acute Ischemic Stroke: Prediction of Risk and Short-Term Mortality. Atherosclerosis 2010, 210, 548–554. 10.1016/j.atherosclerosis.2009.11.040. [DOI] [PubMed] [Google Scholar]
- Faria M. H. G.; Muniz L. R. F.; de Vasconcelos P. R. L. Ketone Bodies Metabolism during Ischemic and Reperfusion Brain Injuries Following Bilateral Occlusion of Common Carotid Arteries in Rats. Acta Cir. Bras. 2007, 22, 125–129. 10.1590/s0102-86502007000200009. [DOI] [PubMed] [Google Scholar]
- Gibson C. L.; Murphy A. N.; Murphy S. P. Stroke Outcome in the Ketogenic State - a Systematic Review of the Animal Data. J. Neurochem. 2012, 123, 52–57. 10.1111/j.1471-4159.2012.07943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanero S.; Gelderblom M.; Magnus T.; Arumugam T. V. Calorie Restriction and Stroke. Exp. Transl. Stroke Med. 2011, 3, 8. 10.1186/2040-7378-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior M.; Rogawski M. A.; Hartman A. L. Neuroprotective and Disease-Modifying Effects of the Ketogenic Diet. Behav. Pharmacol. 2006, 17, 431–439. 10.1097/00008877-200609000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins M. Diet, Ketones, and Neurotrauma. Epilepsia 2008, 49, 111–113. 10.1111/j.1528-1167.2008.01852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.