Abstract
Pax4 is a paired-domain (PD)-containing transcription factor which plays a crucial role in pancreatic β/δ-cell development. In this study, we characterized the DNA-binding and transactivation properties of mouse Pax4. Repetitive rounds of PCR-based selection led to identification of the optimal DNA-binding sequences for the PD of Pax4. In agreement with the conservation of the optimal binding sequences among the Pax family transcription factors, Pax4 could bind to the potential binding sites for Pax6, another member of the Pax family also involved in endocrine pancreas development. The overexpression of Pax4 in HIT-T15 cells dose dependently inhibited the basal transcriptional activity as well as Pax6-induced activity. Detailed domain mapping analyses using GAL4-Pax4 chimeras revealed that the C-terminal region of Pax4 contains both activation and repression domains. The activation domain was active in the embryonic kidney-derived 293/293T cells and embryonal carcinoma-derived F9 cells, containing adenoviral E1A protein or E1A-like activity, respectively but was inactive or very weakly active in other cells including those of pancreatic β- and α-cell origin. Indeed, the exogenous overexpression of type 13S E1A in heterologous cell types could convert the activation domain to an active one. On the other hand, the repression domain was active regardless of the cell type. When the repression domain was linked to the transactivation domain of a heterologous transcription factor, PDX-1, it could completely abolish the transactivation potential of PDX-1. These observations suggest a primary role of Pax4 as a transcriptional repressor whose function may involve the competitive inhibition of Pax6 function. The identification of the E1A-responsive transactivation domain, however, indicates that the function of Pax4 is subject to posttranslational regulation, providing further support for the complexity of mechanisms that regulate pancreas development.
The Pax genes constitute a small family of conserved genes encoding paired box-containing transcription factors. They play a major role in embryonic pattern formation, as illustrated by the dynamic expression patterns during ontogenesis and their association with mouse developmental mutants and human disease syndromes (32). The nine mammalian Pax genes reported to date are classified into five different subclasses according to structural similarities (57). Two Pax genes, Pax4 and Pax6, were recently shown to be essential for endocrine pancreas development (49, 50).
Endocrine cells of the adult pancreas are composed of four major islet cell types, α, β, δ, and PP cells, which synthesize glucagon, insulin, somatostatin, and pancreatic polypeptide, respectively. The development of these four endocrine cell types starts with a common endocrine precursor, and the terminal differentiation leading to the expression of one hormone in each islet cell type occurs late in development (47, 53). During mouse development, Pax6 is first detectable in the pancreatic bud at embryonic day 9 (E9.0); its expression is maintained in a subset of cells throughout development of the pancreas and becomes restricted to the islets of Langerhans in newborn animals (50, 56). Besides being involved in pancreas development, Pax6 also functions as a key regulating molecule for the development of the eye and the central nervous system (16, 32). On the other hand, the expression of Pax4 is detectable in the pancreas at E9.5 in mice and is maintained in cells within the dorsal pancreas at around E10.5 and in the ventral pancreas at around E11.0. In newborn animals, its expression is restricted to the insulin-producing β cells (49). Although Pax4 is also expressed in extrapancreatic tissue such as the ventral spinal cord, the physiological significance of the factor is evident only in the developing pancreas: null mutant mice for Pax4 lack the insulin- and somatostatin-producing cells (49). This contrasts with the phenotypes of the natural or artificial mutants of Pax6, which manifested as partial or total deficiency of glucagon-producing α cells and reduction of β cells (45, 50). Thus, Pax4 was shown to be essential for β/δ-cell development, whereas Pax6 is important for islet cell development with particular commitment to α-cell differentiation.
To understand the functions of transcription factors, it is essential to clarify the target genes of their regulation. However, this is often difficult, especially when null mutations in genes encoding those transcription factors prevent the development of an organ. In terms of the target of its function, Pax6 was shown to bind to the cis-acting element called PISCES (pancreatic islet cell enhancer sequence) that is a common and crucial element in the glucagon, insulin, and somatostatin promoters and thereby activates the expression of those genes (45). In contrast, no information is yet available as to the mechanism of Pax4 involvement in β- and δ-cell development.
In this study, we examined the DNA-binding and transactivation properties of Pax4. We here report an optimal binding sequence for mouse Pax4, which is similar to the previously described Pax6 consensus binding motifs. Indeed, Pax4 binds to the PISCES elements of glucagon, insulin, and somatostatin gene promoters as does Pax6. Also, we have identified in the C-terminal region of Pax4 juxtaposed transactivation and repression domains that can independently function as regulatory modules when linked to a heterologous transcription factor. While the transactivation domain (TAD) can be active in the presence of adenoviral E1A, the repression domain of Pax4 appeared to be active regardless of cell type, suggesting a primary role of Pax4 as a transcriptional repressor.
MATERIALS AND METHODS
Materials.
Restriction enzymes, DNA polymerases, and other modification enzymes were purchased from commercial suppliers (Toyobo, Tokyo, Japan; Takara, Kyoto, Japan; Stratagene, San Diego, Calif.; Promega Biotec, Madison, Wis.; New England Biolabs, Beverly, Mass.), and radioisotopes were from Amersham Japan (Tokyo, Japan). Tissue culture media were purchased from Nacalai Tesque (Tokyo, Japan), and fetal bovine serum was from ICN Biomedicals, Inc. (Costa Mesa, Calif.). Oligonucleotides were synthesized with a DNA/RNA synthesizer (model 394; Applied Biosystems, Tokyo, Japan). Anti-HA (hemagglutinin epitope) monoclonal antibody 12CA5 was purchased from Boehringer Mannheim Co. Ltd. (Tokyo, Japan).
Library screening.
A mouse MIN6 cell-derived cDNA library (25) constructed on the Lambda ZAPII vector (Stratagene) was a kind gift from H. Ishihara (University of Tokyo, Tokyo, Japan). The probe for the screening was a short fragment of Pax4 cDNA corresponding nucleotides 75 to 350 of previously reported partial Pax4 sequence (accession no. Y09584) and was prepared by reverse transcription-PCR (RT-PCR) starting with 1 μg of total RNA of βTC1 cells. For library screening, Escherichia coli XL-1 Blue MRF was infected with phage (7 × 105 plaques in total) and plated on 10 agar plates (150-mm diameter). Library screening followed a standard protocol. After a 16-h incubation at 37°C, phage plates (7 × 104 plaques per 150-mm-diameter plate) were overlaid with Optitran BA-S reinforced nitrocellulose filters (Schleicher & Schuell, Dassel, Germany). Filters were then hybridized with 32P-labeled mouse Pax4 cDNA probe by using standard protocols. Plaques giving positive signals were isolated and subjected to secondary and tertiary screenings to ensure plaque purification. The cDNA inserts from plaque-purified clones were sequenced.
Cell culture and transfection.
The β-cell-derived βTC1 cells, HIT-T15 cells, and the α-cell-derived αTC1 cells were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (FCS), penicillin, and streptomycin. 293/293T (human embryonic kidney-derived) cells and HeLa cells grown in Dulbecco’s modified Eagle’s medium with 10% FCS, penicillin, and streptomycin. CHO-K1 cells (Riken Cell Bank, Tsukuba, Japan) were maintained in F-12–Ham medium supplemented with 10% FCS, penicillin, and streptomycin. Twenty-four hours before transfection experiments, the cells were replated in 100-mm-diameter plates for 293/293T cells and HeLa cells (approximately 3 × 106 cells) or 60-mm-diameter plates for CHO-K1 cells (approximately 106 cells).
Plasmid construction.
A full-length cDNA clone for the mouse Pax6 gene (kindly provided by P. Gruss) (58) was cloned into an expression plasmid pcDNA3 to produce the Pax6 expression vector pcDNA3Pax6. The Pax-responsive luciferase (Luc) reporter construct (Pax)5TKLuc was created by cloning five copies of high-affinity Pax6-binding site (5′-TCGAGAAAATTTTCACGCTTGAGTTCACAGCTCGAG-3′) (12) into the SalI site of TK (thymidine kinase)-Luc (kindly provided by K. Umesono).
Pax paired domain (PD)-glutathione S-transferase (GST) fusion protein preparations.
A full-length cDNA clone for mouse Pax4 and Pax6 genes were used as templates in a PCR with primers selected to amplify the sequence corresponding to amino acids (aa) 1 to 149 for Pax4 and aa 1 to 179 for Pax6. The PCR products were cloned into the BamHI-EcoRI site of pGEX-3T (Pharmacia), and the resulting clones were verified by sequencing. E. coli JM109 was transformed, and the fusion proteins were purified according to the standard protocol, using glutathione-agarose beads (Pharmacia). For electrophoretic mobility shift assay (EMSA) reactions, 50 ng of each fusion protein was used.
DNA-binding site selection.
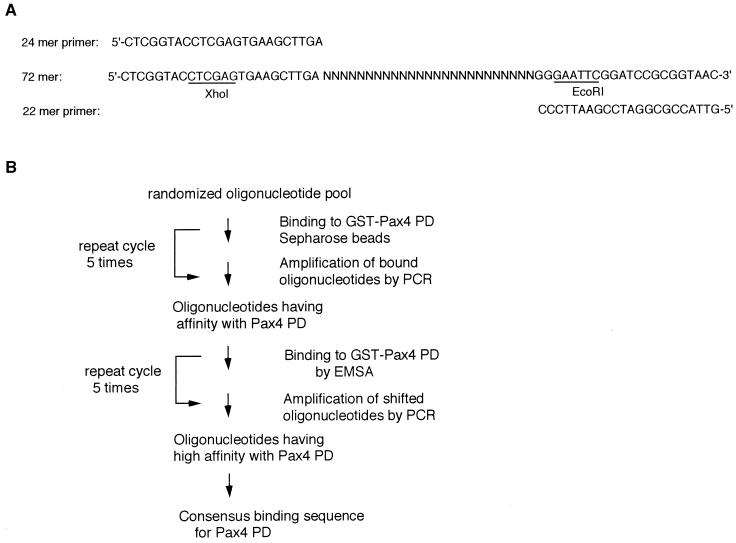
DNA-binding site selection was performed by using a modification of the strategy described by Chittenden et al. (4). A 71-bp oligonucleotide, 5′ CTCGGTACCTCGAGTGAAGCTTGA(N)25GGGAATTCGGATCCGCGGTAAC 3′, was used for the DNA-binding site selection assay. The second strand of this oligonucleotide was synthesized with Klenow polymerase and 22-mer 3′ primer (5′-GTTACCGCGGATCCGAATTCCC-3′) and purified on a 12% polyacrylamide gel in 1× Tris-borate-EDTA buffer. The double-stranded pool was incubated 60 min at 4°C with 20 μl of GST-Pax4 Sepharose beads in 300 μl of binding buffer containing 20 mM Tris (pH 7.4), 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, 1 mg of bovine serum albumin per ml, and 5 μg of poly(dI-dC). The mixture was centrifuged, and the pellet was washed twice in binding buffer. The pellet was resuspended in 30 μl of water, boiled for 3 min, and centrifuged. The eluted oligonucleotides were amplified by PCR using 24-mer 5′ and 22-mer 3′ primers, purified on polyacrylamide gels, and used in a second round of selection. The oligonucleotides selected after the fifth cycle were subjected to five additional rounds of binding and selection, using preparative EMSA performed as described below. The shifted oligonucleotides were excised from the gel, eluted, and amplified by PCR. After the final cycle, the selected oligonucleotides were digested by XhoI and EcoRI, ligated into a Bluescript vector (Stratagene), and sequenced.
DNA sequencing.
DNA sequencing of oligonucleotide inserts in the DNA-binding site selection and PCR products used in the plasmid construction was performed by the dideoxy-chain termination method using an ABI Prism 310 automated sequencer (Applied Biosystems).
EMSA.
The double-stranded oligonucleotide probes were end labeled with T4 polynucleotide kinase and [γ-32P]ATP. Purified fusion proteins (50 ng) or nuclear extracts (5 μg) were preincubated for 20 min at room temperature with 1.5 μg of poly(dI-dC) in a buffer containing 10 mM Tris (pH 7.4), 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 1 mg of bovine serum albumin per ml, and 10% glycerol in a final volume of 20 μl. After addition of the probe (3 × 104 cpm), the samples were incubated for 15 min at room temperature, and then electrophoresis was performed on 5% nondenaturing polyacrylamide gels in 0.5× Tris-borate-EDTA buffer for 90 min at 150 V at 4°C. In the competition studies, a 10-, 100-, or 200-fold molar excess of unlabeled oligonucleotide competitor was added with the probe.
Transient expression of HA-tagged Pax4 protein and detection by Western blot analysis.
A DNA fragment encoding a portion of homophilic influenza virus HA (MYPYDVPDYAS) (3) was linked to the amino terminus of mouse Pax4 cDNA (clone A) and cloned into an expression plasmid pcDNA3 to produce pcDNA3HA-Pax4. The sequence fidelity of DNA fragments was confirmed by sequencing. 293T cells were replated in a 100-mm-diameter dish (approximately 3 × 106 cells) 24 h before transfection. Eight micrograms of pcDNA3HA-Pax4 or the mock vector pcDNA3 was transfected into 293T cells by the lipofection method using LipofectAMINE reagent (Life Technologies, Tokyo, Japan). Forty-eight hours after transfection, the cells were harvested and nuclear extracts were prepared as described previously (43). Ten-microgram aliquots of nuclear extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 5 to 20% gradient gel (Bio-Rad) and electrotransferred to a polyvinylidene difluoride membrane (Immobilon-P; Nihon Millipore, Tokyo, Japan). The membrane was probed with anti-HA monoclonal antibody 12CA5. Immunoreactive bands were detected by autoluminography with an enhanced chemiluminescence detection kit (Amersham).
GAL4 fusion protein reporter gene analyses.
The GAL4 fusion constructs were generated by isolating (by PCR) and introducing appropriate DNA fragments of mouse Pax4, Pax6, and PDX-1 into the KpnI-XbaI sites of a pSG424 plasmid (44) which contained the DNA-binding domain (positions 1 to 147) of GAL4. The GAL4-responsive reporter plasmid (GAL4)5E1bTATA Luc generated from GAL4E1bCAT (minimal promoter containing a TATA box) (28) by replacing the chloramphenicol acetyltransferase reporter with luciferase structural gene was a kind gift from K. Nakajima (Osaka City University School of Medicine, Osaka, Japan). Another GAL4-responsive reporter plasmid, (GAL4)4TKLuc (same as MH100x4-TK-Luc in reference 33, which has four copies of GAL4-binding sites upstream of the TK promoter linked to Luc structural gene) was a kind gift from K. Umesono (Kyoto University, Kyoto, Japan). Two micrograms of each GAL4 fusion plasmid was cotransfected into host cells by the lipofection method along with 2 μg of (GAL4)5E1bTATALuc and 1 μg of plasmid pEFBOSβ-Gal. The luciferase activity from the reporter plasmid was normalized with respect to the β-galactosidase (β-Gal) activity of the cotransfected internal control plasmid pEFBOSβ-Gal.
Nucleotide sequence accession numbers.
The nucleotide sequences of clones A and B can be obtained from GenBank under accession no. AB010557 and AB010558, respectively.
RESULTS
Determination of the optimal binding sequence for the Pax4 PD.
By screening of a mouse β-cell tumor-derived cDNA library (25), two clones, A and B, comprised of 1,295 and 1,310 bp, respectively, were isolated and identified as encoding Pax4. The two cDNA clones had the same 5′ end and used the same polyadenylation site, but only the longer cDNA clone (clone B) had a 15-bp insertion within its 5′ noncoding sequence, which may be derived from an alternative spliced exon. The predicted amino acid sequence of Pax4 is identical to a recently published Pax4 sequence obtained from the same β-cell line via RT-PCR (20). For the expression studies described below, clone A was used.
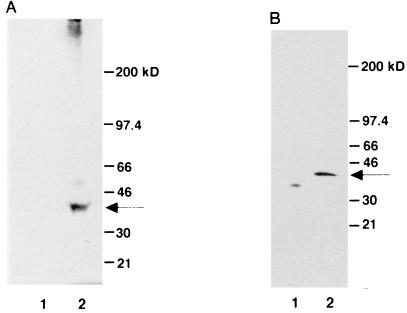
To examine the integrity of the encoded gene product, we allowed Pax4 mRNA to be transcribed and translated in vitro, using a rabbit reticulocyte lysate system. As shown in Fig. 1A, the mouse Pax4 mRNA could be translated into a single protein of 38 kDa in accordance with the size predicted by the nucleotide sequence (data not shown). We also verified that the mouse Pax4 protein could be expressed in mammalian cells by transient transfection. Forty-eight hours after transient transfection of the embryonic kidney-derived 293T cells and CHO-K1 cells, the Pax4 protein with an HA tag (HA-Pax4) could be identified as a single band of 38 kDa in Western blot analysis using an anti-HA monoclonal antibody (Fig. 1B and data not shown).
FIG. 1.
Translation of a 38-kDa protein from a mouse Pax4 mRNA. (A) In vitro translation of mouse Pax4 cDNA. A plasmid containing mouse Pax4 cDNA was isolated from a β-cell-derived cDNA library and subjected to in vitro transcription and translation using a TNT coupled reticulocyte lysate system (Promega). The reaction mixture was incubated in the presence of pCS2 Pax4 (lane 2) or the mock vector pCS2 (54) (lane 1) and then analyzed by SDS-PAGE on a 5 to 20% acrylamide gradient gel (Bio-Rad). (B) Exogenous expression of Pax4 protein in 293T cells. Nuclear extracts were isolated from 293T cells that had been transiently transfected with HA-Pax4 expression plasmid (lane 2) or the control plasmid (lane 1). Aliquots of the extracts were separated by SDS-PAGE, transferred to a membrane, and allowed to react with anti-HA monoclonal antibody 12CA5.
A modified version of the PCR-mediated DNA-binding site selection was used to determine the optimal DNA-binding sequences for Pax4. Oligonucleotides containing 25 consecutive random nucleotides were prepared (Fig. 2A) and allowed to bind to bacterially synthesized fusion proteins of GST and the Pax4 PD. Multiple rounds of selection were performed under the protocol illustrated in Fig. 2B. Following amplification by PCR, the oligonucleotides selected were radiolabeled and used in EMSA to verify the progressive enrichment for Pax4 PD-binding oligonucleotides (data not shown). After the last round of selection, individual oligonucleotides were cloned for DNA sequencing.
FIG. 2.
Strategy for PCR-mediated DNA-binding site selection. (A) DNA sequences of synthetic oligonucleotides. The 72-mer oligonucleotide contained a 25-base consecutive randomized sequence flanked by the defined terminal ends of 24 bases and 22 bases. The 24- and 22-mer oligonucleotides were used as primers for the PCR amplification of the 72-mer oligonucleotide. (B) Protocol of the PCR-mediated binding site selection for Pax4 PD. Each round of selection consisted of incubation of 72-bp oligonucleotide DNA with GST-Pax4 PD for the binding, subsequent purification (with Sepharose beads or with EMSA as shown), and PCR amplification of the bound DNA, which was then carried forward to the next round of selection.
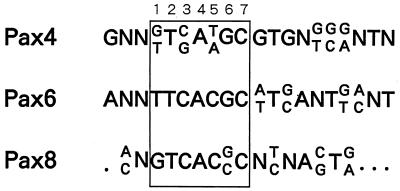
Alignment of nucleotide sequence data revealed the optimal binding sequence for Pax4 PD (Table 1). In terms of the 7-bp core motif (C1 to C7 in Table 1), which shows high conservation among all Pax proteins characterized to date (Fig. 3), clear nonrandom distribution of nucleotides was observed. Additional nucleotides surrounding the core motif also showed some nonrandom distribution. In particular, three consecutive nucleotides located just 3′ downstream of the core motif (positions +1 to +3 in Table 1) were as well conserved as those in the core motif.
TABLE 1.
Summary of sequences selected with the Pax4 PD
| Nucleotide | Sequences selected with the Pax4 PDa
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −4 | −3 | −2 | −1 | C1 | C2 | C3 | C4 | C5 | C6 | C7 | +1 | +2 | +3 | +4 | +5 | +6 | +7 | +8 | +9 | +10 | |
| G | 8 | 26 | 10 | 16 | 25 | 3 | 14 | 6 | 6 | 34 | 7 | 33 | 7 | 23 | 5 | 12 | 11 | 13 | 8 | 4 | 6 |
| A | 4 | 1 | 6 | 11 | 3 | 0 | 0 | 36 | 11 | 0 | 3 | 3 | 0 | 7 | 8 | 6 | 4 | 10 | 8 | 2 | 6 |
| T | 7 | 4 | 10 | 0 | 12 | 40 | 0 | 0 | 21 | 5 | 3 | 4 | 27 | 3 | 8 | 8 | 1 | 3 | 7 | 8 | 4 |
| C | 7 | 4 | 16 | 15 | 3 | 4 | 29 | 1 | 5 | 4 | 30 | 3 | 7 | 5 | 11 | 4 | 14 | 5 | 6 | 13 | 9 |
| Consensusb | G | G/T | T | C/G | A | T/A | G | C | G | T | G | G/T | G/C | G/A | T/C | ||||||
Sequences corresponding to the 25 consecutive random nucleotides were aligned so as to maximize the homology as shown, and the number of each nucleotide in each position was calculated.
Optimal binding sequence, determined by alignment of frequently observed nucleotides.
FIG. 3.
Comparison of the putative Pax4 PD-binding motif with those of other Pax family proteins. The obtained sequence for the optimal binding of Pax4 PD was aligned with the binding consensus sequences for other Pax family proteins. The core motif is boxed. References for the selected binding sites: Pax4 PD, this study (Table 1); Pax6 PD, 12; Pax8 PD, 22.
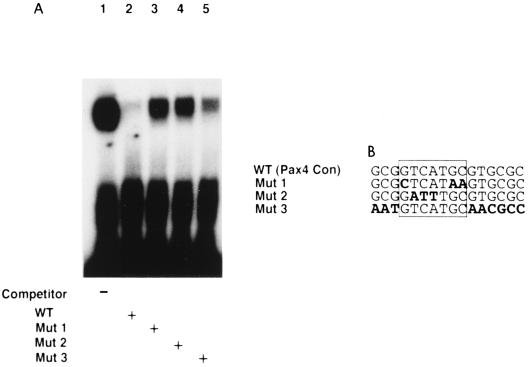
To assess the feasibility and specificity of the obtained binding consensus for Pax4 PD and to determine the nucleotide residues essential for the binding specificity, we performed EMSAs. As shown in Fig. 4A, the binding of Pax4 PD to the consensus binding site was well competed by addition of wild-type competitor (lane 2). Mutations introduced into the core region of the binding site significantly reduced the efficiency of competition (lanes 3 and 4). As expected from the high level of conservation observed within the surrounding sequences of the core region, destruction of those sequences also reduced to some extent the efficiency of competition (lane 5). These results indicate that the Pax4 binding to the proposed sequence is specific and that the specificity depends on the core region and, to some degree, its surrounding sequences.
FIG. 4.
(A) Specificity of Pax4 PD binding to putative recognition sequences. EMSAs were performed with purified GST-Pax4 PD chimeric protein and a labeled double-stranded oligonucleotide probe (5′-GGTGCGCGGTCATGCGTGCGCGACCGCTCCATG-3′) representing the putative binding sequence for Pax4 PD. (B) Mutations introduced into unlabeled competitors. Lengths of the competitors were the same as that of the binding probe (33-mer). Each competitor was added at a 100-fold molar excess. Similar results were obtained in three independent experiments. WT, wild type; Con, consensus.
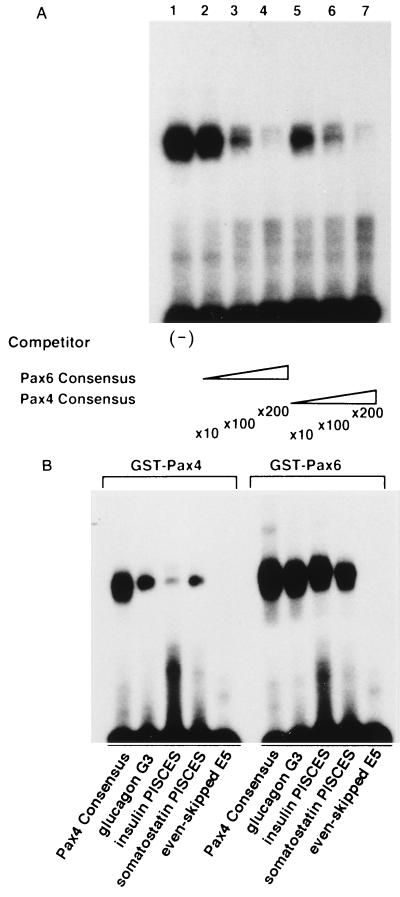
In agreement with the high similarity in the binding consensus between the Pax4 and Pax6 PDs, Pax4 could bind to the Pax6 binding sites. The EMSA results (Fig. 5A) indicated that the binding of Pax4 PD to the Pax4 consensus is well competed by the addition of Pax6 consensus oligonucleotides. Also, Pax4 PD could bind to some of the putative target sequences for Pax6, such as the G3 element (PISCES) of the glucagon gene promoter and PISCES elements in the insulin and somatostatin gene promoters (Fig. 5B) (45), although with lower affinity than Pax6.
FIG. 5.
Conserved DNA-binding specificity for Pax4 PD and Pax6 PD. (A) The Pax6 consensus sequence competes against the Pax4 consensus for binding to Pax4 PD. EMSAs were performed with purified GST-Pax4 PD protein and the oligonucleotide representing the Pax4 PD consensus (see Fig. 4 for sequence) as the binding probe. Unlabeled probe (lanes 5 to 7) or an oligonucleotide representing the Pax6 PD consensus (5′-AAAATTTTCACGCTTGAGTTCACAGCTCGA-3′ [12]; lanes 2 to 4) was added to the binding reactions, and the efficiency of competition was evaluated. (B) Pax4 PD binds to the putative Pax6 target sites. EMSAs were performed with purified GST-Pax4 PD and GST-Pax6 PD. Sequences of the probes used: Pax6 consensus, as described above; glucagon G3 element, 5′-GTAGTTTTTCACGCCTGACTGAGATTGAAGGGT-3′ (45); insulin PISCES (C2 element), 5′-CTTTCTGGGAAATGAGGTGGAAAATGCT CAGCCAA-3′ (45); somatostatin PISCES, 5′-TGATTTTGCGAGGCTAATGGTGCGTAAAAGCACTG-3′ (45); Even-skipped E5, 5′-CCGCACGATTAGCACCGTTCCGCTCAGGCTCGG-3′ (5). Similar results were obtained in three independent experiments.
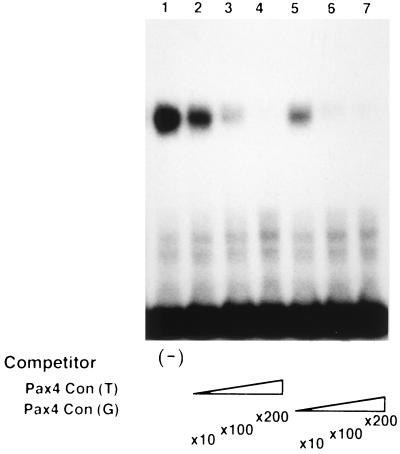
Within the well-conserved core region of the consensus motifs for Pax proteins, the nucleotide residue at position C1 of the Pax6 consensus (T in Fig. 3) differs from that (G) for the other Pax consensus sequences. This difference has been discussed with respect to possible association with the difference in one amino acid residue within the PD (5): the amino acid at position 47, which is Asp in Pax6 but His in Pax2, Pax5, and Pax8. According to crystal structure analysis, the amino acid at position 47 seems to be in contact with the 5′ end of the core in the major groove and may thus determine the preference of the nucleotide residue at position C1 in the core motif, i.e., G when His is at position 47 and T when Asp is at position 47.
In the case of Pax4, the amino acid residue at position 47 is Asp. While the nucleotide residue at position C1 in the Pax4 consensus that we determined was G or T, we further evaluated the possible association between the nucleotide residue at position C1 and the amino acid residue at position 47. We examined the effect of conversion of the G residue at position C1 to the T residue on the efficiency for the Pax4 PD binding. As shown in Fig. 6, Pax4 seems to bind to the sequence with G at position C1 at least as efficiently as to the sequence with T at the same position. Thus, the association in binding preference between the nucleotide residue at position C1 and the amino acid residue at position 47 observed for Pax6 may not be applicable for Pax4. Whereas the amino acid residues at position 42 and 44 were also suggested to be critical for the DNA binding (5), the difference in these residues between Pax4 and Pax6 (Ser versus Ile at position 42 and Lys versus Gln at position 44) may be responsible for this observation.
FIG. 6.
Effect of a single nucleotide substitution (G to T) on the binding affinity for Pax4. Significance of the nucleotide residue at position C1 of the core region (Fig. 3) for Pax4 binding was evaluated by EMSA. The purified GST-Pax4 PD chimeric protein was bound to a γ-32P-labeled double-stranded oligonucleotide probe representing the putative binding sequence for Pax4 PD (5′-GGTGCGCGGTCATGCGTGCGCGACCGCTCCATG-3′). The cold probe (lanes 5 to 7) or its variant in which a single G residue was changed to T (5′-GGTGCGCGTTCATGCGTGCGCGACCGCTCCATG-3′; lanes 2 to 4) was used as a competitor, being added at a 10-fold (lanes 2 and 5), 100-fold (lanes 3 and 6), or 200-fold (lanes 4 and 7) molar excess. No competitor was added to the lane 1 sample. Similar results were obtained in three independent experiments.
Evaluation of transactivation properties of Pax4.
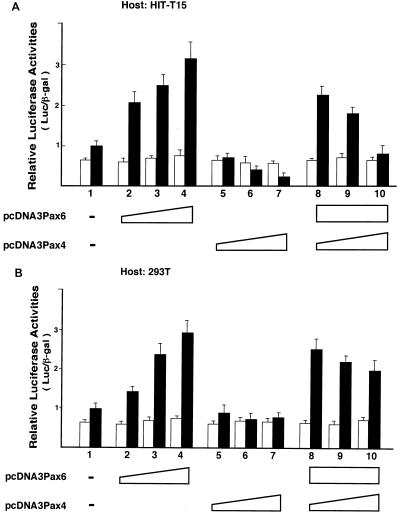
We next evaluated the transactivation properties of Pax4 in reporter gene analyses. An artificial enhancer-promoter reporter construct which contained five copies of the consensus binding sequence for Pax6 linked to the TK gene promoter and a Luc reporter [(Pax)5TKLuc] was used. Prior to this experiment, we allowed the full-length Pax4 and Pax6 to be transiently expressed in 293T cells and verified that both can bind to the Pax6 consensus sequence (data not shown). The reporter plasmid was cotransfected into HIT-T15 cells with various amounts of the Pax4-expressing plasmid (pcDNA3Pax4), the Pax6-expressing plasmid (pcDNA3Pax6), or both expression plasmids. As shown in Fig. 7, Pax6 stimulated the promoter activity containing the Pax6 and Pax4 binding sites in HIT-T15 cells and 293T cells (lanes 2 to 4 in both panels). In contrast, the Pax4 overexpression dose dependently suppressed the same enhancer-promoter in HIT-T15 cells (Fig. 7A, lanes 5 to 7). Also, when the Pax4 expression plasmid was cotransfected into HIT-T15 cells together with a fixed amount of Pax6 expression plasmid and the reporter plasmid, Pax4 dose dependently suppressed the Pax6-induced activation of its target enhancer reporter (Fig. 7A, lanes 8 to 10). Because no changes in the promoter activities were observed with the enhancerless TK-Luc promoter, these suppressive effects of Pax4 depended on its binding to the target sequence. Similar transcription-suppressing effects of Pax4 were observed in βTC1 and HeLa cells (data not shown).
FIG. 7.
Pax4 displays transcriptional repression activity and inhibits Pax6-induced transactivation. The effects of Pax4 and Pax6 on the target promoter were evaluated by reporter gene analyses in HIT-T15 cells (A) and 293T cells (B). The reporter plasmids used were (Pax)5TKLuc, which contained five copies of the Pax6-binding sequence (12) linked to the TK gene promoter (filled bars), and TK-Luc, the enhancerless control (blank bars). Together with the reporter plasmid (Pax)5TKLuc or TKLuc (2 μg), the Pax6 (pcDNA3Pax6) and/or Pax4 (pcDNA3Pax4) expression plasmids were cotransfected into the cells, and the effects of Pax6 and Pax4 on reporter activity were evaluated. In lanes 2 to 4 and 5 to 7, 100, 200, and 500 ng, respectively, of pcDNA3 Pax6 and of pcDNA3 Pax4 were used; in lanes 8 to 10, pcDNA3 Pax4 was added at 100, 200, and 500 ng, respectively, along with a fixed amount (500 ng) of pcDNA3 Pax6. To make the total amount of DNA transfected into the cells the same for all lanes, an appropriate amount of the empty expression vector pcDNA3 was also added. The results were normalized to the β-Gal activity derived from cotransfected pEFBOSβ-Gal and presented as means ± SD of at least three independent experiments.
In contrast to these observations obtained with HIT-T15 cells and other cell types, the transcription-suppressing effects of Pax4 were not evident in embryonic kidney-derived 293T cells (Fig. 7B, lanes 5 to 7). Also, the effect on the action of coexpressed Pax6 was trivial if any (Fig. 7B, lanes 8 to 10). These results thus indicated that Pax4 functions as a transcriptional repressor rather than an activator in most of the cells investigated. Also, when coexpressed with Pax6, it may inhibit the Pax6 function in those cells. This transcription-repressing effect of Pax4 was not evident in 293T cells.
Identification of transactivation and repression domains in the C-terminal region of Pax4.
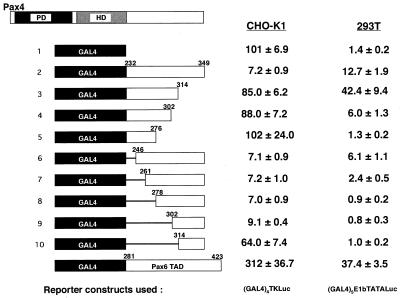
C-terminal portions of Pax family proteins have been shown to constitute a potent transactivation domain (10, 52). Moreover, in the case of Pax2, Pax5/BSAP, and Pax8, there seems to be a region in the extreme C terminus that negatively regulates the function of the adjacent transactivation domain (10). To address the molecular basis of the Pax4 function, we characterized the transactivation property of the C-terminal region of Pax4 by using the Saccharomyces cerevisiae GAL4 fusion protein reporter system. The C-terminal region of Pax4 (aa 232 to 349) or its deleted fragment was fused in frame to the heterologous DNA-binding domain of the GAL4 transcription factor (Fig. 8). These chimeric GAL4-Pax4 fusion proteins were expressed in various cells, and effects on the GAL4 reporter were evaluated. The expression of each fusion protein was verified by EMSA. Neither lack of expression nor greatly reduced expression of the fusion proteins was observed (data not shown).
FIG. 8.
Evaluation of transactivation/repression potential of the C-terminal region of Pax4. Schematic representation of the GAL4-Pax4 chimeras (left) and their transactivation/repression potential in CHO-K1 and 293T cells (right) are shown. The fusion proteins GAL4-Pax4 and GAL4-Pax6 were obtained by fusing the DBD of GAL4 (aa 1 to 147) to the C-terminal region of mouse Pax4 and mouse Pax6, respectively. The deletion endpoints of Pax4 and Pax6 in each GAL4 chimera are indicated. Cells were transfected with 2 μg of a GAL4-responsive reporter plasmid [(GAL4)4TKLuc for CHO-K1 cells or (GAL4)5E1bTATALuc for 293T], 2 μg of GAL4-Pax4 (or GAL4-Pax6) chimera, and 1 μg of an internal control, pEFBOSβ-Gal. Luciferase results were normalized with respect to the transfection efficiency assessed by β-Gal assay. The data are presented as relative activities (means ± SD) to those obtained in the cells cotransfected with the empty expression vector (pcDNA3), arbitrarily set at 100 (for CHO-K1) or 1 (for 293T).
In contrast to the comparable region of Pax6, the C-terminal region of Pax4 revealed transcriptional suppression activity in most of the cell types investigated including CHO-K1, βTC1, αTC1, HeLa, and HIT-T15 cells (Fig. 8 and Table 2). When the entire C-terminal region of Pax4 (aa 232 to 349) was fused to the GAL4 DBD, the promoter activity of the (GAL4)4TKLuc plasmid was reduced by approximately 93% in CHO-K1 cells, 69% in βTC1 cells, and 50% in HIT-T15 cells. Thus, the C-terminal region of Pax4 functions as an active repressor of transcription in these cells.
TABLE 2.
Transactivation and repression properties of GAL4-Pax4 chimeras in various cell typesa
| Constructa | Relative activity (mean ± SD)
|
||
|---|---|---|---|
| GAL4 DBD | GAL4–Pax4 232–349b | GAL4–Pax4 232–314b | |
| (GAL4)5E1bTATALucc | |||
| 293 | 1.11 ± 0.3 | 15.7 ± 1.7 | 35.1 ± 9.9 |
| 293T | 1.44 ± 0.2 | 12.7 ± 1.9 | 42.4 ± 9.4 |
| F9 | 1.07 ± 0.3 | 0.37 ± 0.15 | 11.7 ± 5.4 |
| (GAL4)4TKLucc | |||
| βTC1 | 102 ± 10.5 | 30.9 ± 10.3 | 97.2 ± 16.6 |
| αTC1 | 101 ± 14.2 | 61.0 ± 29.1 | 98.4 ± 15.2 |
| HIT-T15 | 106 ± 23.4 | 49.7 ± 5.7 | 144 ± 34.0 |
| HeLa | 98 ± 9.0 | 15.5 ± 3.5 | 73.7 ± 24.9 |
| CHO-K1 | 101 ± 6.9 | 7.2 ± 0.9 | 85.0 ± 6.2 |
Two micrograms of an expression plasmid (pcDNA, GAL4 DBD, GAL4–Pax4 232–349, or GAL4–Pax4 232–314), 1 μg of pEFBOSβ-Gal, and 2 μg of a GAL4 reporter plasmid [(GAL4)5E1bTATALuc or (GAL4)4TKLuc] were used. The data are presented as activity relative to that obtained in cells transfected with the empty vector (pcDNA3), arbitrarily set at 1 [for (GAL4)5E1bTATALuc] or 100 [for (GAL4)4TKLuc].
Structure is illustrated in Fig. 8.
Structure and origin are described in Materials and Methods.
However, in the case of 293T cells, the Pax4 C-terminal region activates the GAL4 DBD-containing reporter; when the (GAL4)5E1bTATALuc plasmid, which has a relatively low background promoter activity, was used, the activation was more than 10-fold (Table 2 and Fig. 8). Although the fold induction was lower than this, promoter activation by the Pax4 C-terminal region (105 ± 5.6 [mean ± standard deviation {SD} to 157 ± 21.2; approximately 1.5-fold induction) was also observed with the (GAL4)4TKLuc plasmid, which had a relatively high background promoter activity. This pattern of cell-type dependency was consistent with the results obtained with the nonchimeric, wild-type Pax4 overexpression and the Pax6/Pax4-binding-site-containing reporter in those cells (Fig. 7). Thus, the function of the C-terminal region of Pax4 varies depending on the cell types in which it functions. In most cell types, it exerts transcription repression activity but in some, such as 293/293T cells, it can function as a transcriptional activator.
The results of detailed domain mapping analyses are shown in Fig. 8. Deletion in the extreme C terminus (aa 314 to 349) almost perfectly masked the transcription-suppressing activity of the Pax4 C-terminal region in CHO-K1 cells (lines 2 and 3). In 293T cells, the deletion of this region caused further increase in the transactivating activity, suggesting that this region of Pax4 constitutes a transcriptional repression domain and that although the transactivation potential of the C-terminal region can be seen in 293T cells, the repression domain is also active in the 293T cells.
Although the region between aa 314 and 349 was not sufficient, the region between aa 302 and 349 was enough to cause transcriptional suppression in CHO-K1 cells (Fig. 8, lines 9 and 10) when linked to a heterologous DBD and thus constituted an active repression domain. This transrepression activity was also detectable in 293T cells when the (GAL4)4TKLuc plasmid, with a relatively high background promoter activity, was used (105 ± 5.6 to 39.2 ± 6.6 [data not shown]), although it was not as intense as that observed in CHO-K1 cells (101 ± 6.9 to 9.1 ± 0.4 [Fig. 8]). This may be due to some residual transactivation potential derived from the TAD, a part of which was likely included in the region between aa 302 and 349. Alternatively, the efficiency of repression may depend on the cell type. Thus, these observations suggest that the extreme C-terminal region of Pax4 (aa 278 to 349) functions as a transcription repression domain regardless of cell type.
The deletions between aa 232 and 302 had no significant effects in CHO-K1 cells (Fig. 8, lines 2 and 6 to 9). In contrast, the same deletion caused a stepwise reduction of the transcriptional activity in 293T cells; when the region between aa 232 and 278 was deleted, the transactivation potential, which had been detectable in 293T cells but not in CHO-K1 cells, was lost (line 8). Also, a similar stepwise reduction of transcriptional activity was observed in 293T cells when the region between aa 314 and 276 was deleted (lines 3 to 5). Such reduction was not observed in CHO-K1 cells. Thus, the C-terminal region of Pax4 contains a TAD (aa 232 to 314) which functions in a cell-type-dependent manner. Because the deletion between aa 314 and 302 reduced the transactivation potential of the GAL4-fused protein (lines 4 and 3) and the deletion between aa 302 and 314 (lines 9 and 10) masked the transrepression activity, the C-terminal end of the activation domain and N-terminal end of the repression domain reside within this narrow range, between aa 302 and 314. Thus, the transactivation and repression domains may be very closely located or even overlap each other.
Adenoviral E1A product can potentiate the transactivation activity of Pax4.
Among the cells used in our GAL4 reporter experiments, 293 and 293T cells and embryonal carcinoma-derived F9 cells allowed us to detect the TAD of Pax4 (Table 2). The same region of Pax4 did not show comparable transactivation potential in other cell types. The 293 and 293T cells are known to be transformed with adenoviral E1A gene and to continue expressing this viral product (51). Also, F9 cells are known to have intrinsic E1A-like activity (19). Accordingly, we examined the possibility that adenoviral E1A is involved in the difference in the function of the Pax4 C-terminal region. The 12S and 13S fragments of E1A were expressed in CHO-K1 cells by transient transfection of expressing plasmids, and effects on the transactivation potential of Pax4 in these cells were investigated.
As shown in Table 3, exogenous expression of the E1A 12S and 13S fragments increased the transactivation potential of the TAD (aa 232 to 314) of Pax4 (138 to 495%). It did not significantly affect the function of the transrepression domain (aa 278 to 349) of Pax4, in agreement with the observation that the transrepression domain functions in a cell-type-independent manner (Fig. 8). In total, the overall repression activity of the entire C-terminal region (aa 232 to 349) was significantly weakened (from 7.1% up to 60% [Table 3]) by coexpression of the E1A 12S and 13S fragments. Because this was not observed with the expression of E1A 12S only (Table 3), the 13S fragment of E1A would primarily be responsible for the activation of the TAD of Pax4. We also introduced the E1A 12S and 13S expression plasmids into 293 cells, but no further activation of the Pax4 TAD was observed (data not shown). Thus, these observations suggest that the E1A 13S fragment, which is present in 293/293T cells, is involved in the cell-type-dependent activation of the Pax4 TAD.
TABLE 3.
Effects of adenoviral E1A products on the transactivation potential of Pax4a
| Plasmid | Relative activity (mean ± SD [n = 3])
|
||
|---|---|---|---|
| pRSVH2O | pRSV5E1Ac | pRSVJF12d | |
| GAL4 DBD | 104 ± 7.8 | 111 ± 13 | 106 ± 6.6 |
| GAL4–Pax4 232–349b | 7.1 ± 1.2 | 60 ± 20.8 | 17 ± 7.4 |
| GAL4–Pax4 232–314b | 138 ± 28.0 | 495 ± 35.0 | 160 ± 18.5 |
| GAL4–Pax4 278–349b | 10 ± 2.1 | 18 ± 4.2 | 18 ± 3.3 |
One microgram of an E1A expression plasmid (pRSVH2O [empty vector], pRSV5E1A, or pRSVJF12), 1 μg of a GAL4-Pax4 chimera expression plasmid (pcDNA, GAL4 DBD, GAL4–Pax4 232–349, GAL4–Pax4 232–314, or GAL4–Pax4 278–349), 2 μg of (GAL4)4TKLuc, and 1 μg of pEFBOSβ-Gal were transiently transfected into CHO-K1 cells. The data are presented as activity relative with that obtained in cells transfected with the empty vector (pcDNA3), arbitrarily set at 100.
Structure is illustrated in Fig. 8.
Expression plasmid for wild-type E1A including both 12S- and 13S-type E1A.
Expression plasmid for 12S-type E1A.
Pax4 contains a portable transcription repression domain.
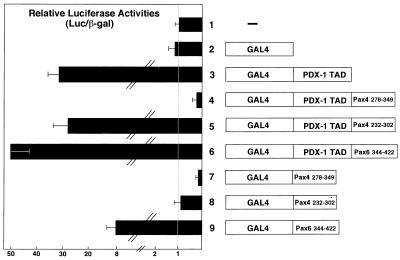
As a step toward characterizing the mechanism underlying the transcriptional repression by Pax4, we examined whether the repression domain of Pax4 can also function in a heterologous transcription factor. The C-terminal 72 aa (aa 278 to 349) of Pax4 were fused to the TAD of PDX-1 (38), and the transactivation (or transrepression) potential of this chimeric protein was evaluated in the GAL4 reporter system (Fig. 9). Comparable expression of the fusion proteins used in this experiment was verified by their DNA-binding characteristics in EMSA (data not shown). As shown in Fig. 9, while the PDX-1 TAD could activate the GAL4 reporter approximately 30-fold (lane 3), the fusion protein containing the same region of PDX-1 TAD and the Pax4 transrepression domain (aa 278 to 349 [lane 4]) suppressed the basal promoter activity of the GAL4 reporter gene by 75%. This did not occur when a different portion of Pax4 with a similar length (lane 5) or a comparable region of Pax6 (lane 6) was linked to PDX-1 TAD. These results thus indicated that the transrepression domain of Pax4 can totally abolish the transactivation potential of PDX-1 TAD and even reveal the fusion protein to be a transcriptional repressor rather than an activator.
FIG. 9.
Pax4 contains a portable transcription repression domain. Shown on the right is a schematic representation of the chimeric proteins used in the assay. The fusion protein construct GAL4–PDX-1 TAD (lane 3) was designed to express the DBD of GAL4 linked to the previously described N-terminal TAD of mouse PDX-1 (aa 1 to 149) (38). The constructs for the GAL4–PDX-1 TAD–Pax4/Pax6 fusion proteins (lanes 4 to 6) encoded the same GAL4 DNA-binding domain and PDX-1 TAD linked to the Pax4 repression domain (aa 278 to 349; lane 4), a part of the Pax4 TAD (aa 232 to 302; lane 5) and an extreme C-terminal region of Pax6 (aa 344 to 422; lane 6), respectively. The constructs used in lanes 7 to 9 were the same as those used in lanes 4 to 6, respectively, except that they lacked the PDX-1 TAD. Each GAL4 fusion protein expression plasmid was cotransfected into CHO-K1 cells with the (GAL4)5E1bTATALuc and pEFBOSβ-Gal plasmid; 48 h after transfection, luciferase and β-Gal assays were performed. The luciferase results were normalized with respect to transfection efficiency, using the results of β-Gal assays. The data are expressed as relative light units (means ± SD of three independent experiments), with transfection of the same amount of an insertionless expression plasmid, arbitrarily set at 1.
DISCUSSION
In this study, we have shown that Pax4 is a potential repressor of transcription. The C-terminal region of the protein contains an active and transferable transrepression domain and a TAD, which was active only in the cells with adenoviral E1A or E1A-like activity. We also determined the optimal binding sequence for Pax4 and found it to be similar to those for other Pax family proteins such as Pax6 (Fig. 3). Indeed, Pax4 binds to the putative Pax6-binding motif (Fig. 5 and data not shown).
Because Pax4 fused to the GAL4 DBD repressed transcription (Fig. 8), Pax4 appears to be able to exert its active transrepression effect even in the absence of positive transcription factors. However, if it is coexpressed with other Pax family proteins and homeodomain (HD)-containing transcription factors which recognize a common binding motif, Pax4 will be a potential antagonist of those proteins with which it may be in competition for binding. In support of this view, we have shown that when Pax4 is coexpressed with Pax6 in HIT-T15 cells, it dose dependently suppressed the Pax6-induced transactivation of the reporter gene (Fig. 7A). Also, Smith et al. (48) have shown that in vitro-transcribed/translated Pax4 binds to the PISCES sequences in insulin, somatostatin, and glucagon genes, to which Pax6 also binds, and that those promoters could be suppressed by exogenous expression of Pax4. During pancreas development, Pax4 seems to be expressed in relatively restricted cell lineages (49), while Pax6 is expressed broadly in the endocrine pancreas (50). A similar rule may also be applicable to the timing of expression; the Pax6 expression seemed to be maintained to adulthood in islets, but Pax4 expression was undetectable after birth (48). Thus, most, if not all, Pax4-expressing cells in the pancreas also would express Pax6, supporting the idea that Pax4 functions as a negative regulator of Pax6 in vivo.
As can be expected from the structural similarity among Pax family proteins, the extreme C-terminal regions in Pax2, Pax5, and Pax8 (collectively referred to as Pax2/5/8) may also function as a repression domain. According to the report by Dörfler and Busslinger, when the extreme C-terminal regions in Pax2/5/8, which corresponds to the aa 315–349 region of Pax4, were removed, their transactivation potentials became increased (10). In contrast to our observations with Pax4 (Fig. 8), the extreme C-terminal regions of Pax2/5/8, when fused to a heterologous transcription factor, failed to function as an active repression domain (10). However, the authors themselves pointed out that this did not totally eliminate the possibility of Pax2/5/8 having an active repression domain as Pax4 does. The C-terminal region of Pax5 with which they evaluated the portability of the transrepression activity corresponded to aa 314 to 349 in Pax4 and was shorter than the portion of Pax4 that we used in the present study (aa 278 to 349 or 302 to 349). Indeed, the comparable region of Pax4 (aa 314 to 349), when linked to the GAL4 DBD, was able to suppress the activity of the target reporter by only 36% (Fig. 8). Thus, it is still possible that a transrepression domain like that identified in Pax4 operates in some other Pax family proteins as well. However, this does not seem to be the case for Pax6; deletion of the comparable C-terminal region in Pax6 even decreased transcriptional activity (52). Although it was recently shown that Pax6 represses transcription of the βB1-crystalline gene, the HD appears to be primarily responsible for the transcriptional suppression (11).
It remains to be clarified how the transrepression domains of Pax4 or other Pax family proteins exert their effects. Because the domain in Pax4 was shown to be transferable to a heterologous transcription factor, it is likely to function by interacting with a certain molecule which can negatively regulate the transactivation potency rather than by some intramolecular events such as physical masking of the activation domain as characterized in C/EBPβ (NF-M) (26). Apart from Pax family proteins, several putative repression domains have been found and analyzed to date. They were identified in the Drosophila and mammalian transcription factors Engrailed, Even-skipped, Krüppel, Hairy/Hes-1, WT1, and Mad/MXI1 (17, 37). Although little is known about the structures and functions of those repression domains, they seem to exert their effects through multiple mechanisms. Note that some repression domains have been shown to recruit a corepressor, a mediator of transcription-repressing activity; Hairy/Hes-1, Mad/MXI1, and basic Krüppel-like factor recruit their specific corepressors, Groucho (37), SIN3A/SIN3B (1), and mCtBP2 (55), respectively. Interestingly, these corepressors seem to be shared by some other transcription factors as well (21, 55). Because the C-terminal repression domain of Pax4 could function regardless of the cell types, the repression domain may interact with an ubiquitously expressed corepressor and thereby achieve its function. Alternatively, the repression domain of Pax4 may exert its effect through direct interaction with a component of the basal transcription machinery such as TATA-binding protein (TBP), as recently suggested in the unliganded thyroid hormone receptor- or Even-skipped-mediated transcriptional repression (13, 27).
In addition to the transferable repression domain, detailed domain mapping analyses allowed us to identify an activation domain which functions in a cell-type-dependent manner. The transactivation potential derived from the domain was evident only in 293/293T cells and F9 cells, which contain adenoviral E1A oncogene products or intrinsic E1A-like activity. Indeed, the exogenous expression of E1A in a heterologous cell line (CHO-K1) could convert the activation domain into an active one, whereas it did not affect the function of the repression domain (Table 3). During revision of this report, Kalousová et al. reported that whereas Pax4 and Pax6 have similar DNA-binding specificities, the Pax4 C-terminal region can activate the GAL4 reporter 2.5-fold less than the comparable region of Pax6 (23). Their experiments used 293 cells, and the relative potency of transactivation by Pax4 to that by Pax6 was consistent with the data that we obtained for 293T cells (Fig. 8). Although they suggested that Pax4 may act as a Pax6 repressor in a passive manner due to competition for binding sites and lower transactivation potential, our present results indicate that Pax4 is an active repressor in most of the cell types.
The adenovirus E1A gene encodes multifunctional products, which are involved in a wide variety of biological events such as transcriptional activation and repression, immortalization, inhibition of cell differentiation, promotion of cell proliferation, and stimulation of DNA synthesis (46, 59). Two major E1A proteins are translated from the differentially spliced 13S and 12S mRNAs, which consist of 289 and 243 amino acid residues, respectively. While the E1A proteins contain three conserved regions, designated as CR1, CR2, and CR3, CR1 and CR2 are included in both 12S and 13S but CR3 is unique to 13S E1A (34). Because 13S but not 12S E1A activated the transactivation potential of the domain (Table 3), CR3 should be essential for the phenomenon.
E1A has been shown to regulate gene transcription through several putative mechanisms. In terms of the transcriptional suppression by E1A, it is known that E1A interacts with p300/CBP and thereby interferes with the association between tissue-specific transcription factors such as c-Myb, MyoD, and p300/CBP (7, 40). Indeed, E1A inhibits insulin gene transcription by sequestering p300 from BETA2/NeuroD, E12/E47, and HEB, which bind to and activate the E-box enhancer in the insulin gene promoter (41). On the other hand, several mechanisms have been identified for E1A-mediated gene activation. First, E1A is known to interact directly with DNA-bound transcription factors such as TBP, USF, ATF2/CRE-BP1, Sp1, and c-Jun via its promoter-targeting region located within CR3 (29). E1A can simultaneously associate with the basal transcription machinery (2), which can lead to the recruiting of sequence-specific transcription factors to the basal transcription components. To investigate whether E1A activates the Pax4 TAD through a similar mechanism, the possible direct interaction between Pax4 and E1A needs to be examined. The observation that the 13S E1A, which encodes CR3, is required for the activation domain of Pax4 to work supports this possibility. Second, E1A can activate transcription by chelating negative regulators of transcription. For example, the E1A-mediated activation of a transcription factor E2F involves neutralization by E1A of the retinoblastoma gene product (pRb), p107, and p130. E1A tightly traps pRb and the two other proteins, which otherwise bind with E2F and silence its function (35). However, since E1A seemed to act not on the transrepression domain but on the TAD (Table 3), a similar mechanism is unlikely to operate for the Pax4 activation.
Even in the absence of adenovirus infection, intrinsic E1A-like activity was shown to be expressed in murine oocytes, developing embryos, and adult ovaries, and this activity has been suggested to be a marker of the undifferentiated cell state (9). In embryonal carcinoma-derived F9 cells, which also has intrinsic E1A 13S-like activity (19), activation of the TAD could be observed. Therefore, if some intrinsic E1A-like activity is expressed during pancreas development, it may modulate the Pax4 function as a repressor of transcription and thus be involved in regulating development.
In the case of ATF2/CRE-BP1, the activation by 13S E1A is suggested to involve the phosphorylation of Thr-69 and Thr-71, located on the activation domain of ATF2 (30). This phosphorylation is considered to be mediated by 13S E1A-activated kinases, possibly mitogen-activated protein kinase (MAPK)/stress-activated protein kinase (SAPK) family serine/threonine kinases (8, 30). During pancreas development, various growth factors such as hepatocyte growth factor, heparin-binding epidermal growth factor-like growth factor (HB-EGF), and activin A and their associated factors are expressed and may exert effects on growth and differentiation (15, 24, 36), and at least some of them were shown to activate MAPK/SAPK family serine threonine kinases (18, 42). Therefore, as is the case with ATF2 activation, the function of Pax4 could also be regulated through activation of the MAPK/SAPK family kinases even in the absence of the E1A-like activity. Support for this possibility comes from the fact that the transactivation domain of Pax4 contains a PXSP sequence, a putative target motif for MAPK family. Also, we found that the activation potential of the C-terminal region of Pax4 did indeed increase in response to serum stimulation (14).
In this study, we used only the PD of Pax4 as a bait for the DNA-binding site selection assay. While HDs are known to bind to AT-rich sequences such as TAAT, the PD is considered to be important primarily for the specificity of DNA binding and has therefore been used in similar experiments for other Pax family proteins such as Pax6 (6, 12). Thus, the use of Pax4 PD allowed us to compare the consensus obtained with those of other Pax family transcription factors reported previously (Fig. 3). However, it has been suggested that the HDs of Pax proteins cooperatively function with PDs and thus play an essential role in achieving high-affinity binding to their target sites (22, 39). In this context, we note the outcome of the binding site selection assay that Smith et al. (48) performed with a truncated Pax4 containing both PD and HD. The consensus that they obtained includes a region (CACC) which can be attributable to the binding with PD as well as an AT-rich region (ATTA) to which the HD of Pax4 should bind. Using the same truncated Pax4 containing both PD and HD, they also showed that Pax4 binds to the insulin gene C2 element with rather high affinity. This contrasts with our present results obtained with the PD of Pax4, revealing rather low affinity for the same site (Fig. 5B, lane 3), thus suggesting that the cooperativity of the HD of Pax4 also contributes to the determination of the binding preference for Pax4.
Although the GAL4-fused chimeric protein containing the entire C-terminal region of Pax4 (GAL4–Pax4 232–349) revealed a strong transactivating potential in 293/293T cells (Table 2), this was not evident when the whole molecule of Pax4 was expressed in the same cells (Fig. 7B). This may be due to an additional transcription repression activity which resides outside of the C-terminal region. Indeed, when GAL4–Pax4 172–349, which contains the entire HD and C-terminal region of Pax4, was expressed in CHO-K1 cells, a repression more profound than that found with GAL4-Pax4 232–349 was observed (14). Similar results were also obtained by Smith et al. (48). While the HD of Pax3 appears to mediate transcriptional suppression by interacting with corepressors such as pRb (60) and HIRA (31), the HD of Pax4 may also function as another transrepression domain.
In summary, our present observations indicated Pax4 to be a potential repressor of transcription. The extreme C-terminal region as well as HD would be responsible for its transcription-repressing activity. Because the optimal binding sequence for Pax4 seemed similar to that for Pax6, it may inhibit the expression of putative Pax6 target genes during pancreas development and thus contribute to β-cell differentiation. Interestingly, Pax4 also has a TAD whose function can be modified by the presence of adenoviral E1A, suggesting the possible implication of posttranslational regulation of the Pax4 function. These observations provide further support for the complexity of the mechanisms underlying pancreas development.
ACKNOWLEDGMENTS
We thank Hisamitsu Ishihara, University of Tokyo Graduate School of Medicine, for kindly providing the MIN6 cDNA library; Peter Gruss, Max Plank Institute, for the mouse Pax6 cDNA plasmid; Kazuhiko Umesono, Kyoto University, for the (GAL4)4TKLuc plasmid (MH100x4-TK-Luc); Koichi Nakajima, Osaka City University School of Medicine, for the (GAL4)5E1bTATALuc plasmid and E1A expression plasmids; and Masahiko Hibi, Osaka University School of Medicine, for the pCS2 plasmid. We also appreciate Koichi Nakajima for helpful suggestions and Masahiko Hibi for comments on the manuscript.
This work was supported in part by a Grant-in-Aid for Scientific Research (to Y.K. and Y.Y.) and a grant from Suzuken Memorial Foundation (to Y.K.).
REFERENCES
- 1.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 2.Boyer T G, Berk A J. Functional interaction of adenovirus E1A with holo-TFIID. Genes Dev. 1993;7:1810–1823. doi: 10.1101/gad.7.9.1810. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y T, Holcomb C, Moore H P H. Expression and localization of two low molecular weight GTP-binding proteins, Rab8 and Rab10, by epitope tag. Proc Natl Acad Sci USA. 1993;90:6508–6512. doi: 10.1073/pnas.90.14.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chittenden T, Livingston D M, Kaelin W G., Jr The T/E1A-binding domain of the retinoblastoma product can interact selectively with a sequence-specific DNA-binding protein. Cell. 1991;65:1073–1082. doi: 10.1016/0092-8674(91)90559-h. [DOI] [PubMed] [Google Scholar]
- 5.Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- 7.Dai P, Akimaru H, Tanaka Y, Hou D X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 8.Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dooley T P, Miranda M, Jones N C, DePamphilis M L. Transactivation of the adenovirus EIIA promoter in the absence of adenovirus E1A protein is restricted to mouse oocytes and preimplantation embryos. Development. 1989;107:945–956. doi: 10.1242/dev.107.4.945. [DOI] [PubMed] [Google Scholar]
- 10.Dörfler P, Busslinger M. C-terminal activating and inhibitory domains determine the transactivation potential of BSAP (Pax-5), Pax-2 and Pax-8. EMBO J. 1996;15:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan M K, Haynes II J I, Cvekl A, Piatigorsky J. Dual roles for Pax-6: a transcriptional repressor of lens fiber cell-specific β-crystallin genes. Mol Cell Biol. 1998;18:5579–5586. doi: 10.1128/mcb.18.9.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 13.Fondell J D, Brunel F, Hisatake K, Roeder R G. Unliganded thyroid hormone receptor α can target TATA-binding protein for transcriptional repression. Mol Cell Biol. 1996;16:281–287. doi: 10.1128/mcb.16.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujitani, Y., and Y. Kajimoto. Unpublished observation.
- 15.Furukawa M, Eto Y, Kojima I. Expression of immunoreactive activin A in fetal rat pancreas. Endocrine J. 1995;42:63–68. doi: 10.1507/endocrj.42.63. [DOI] [PubMed] [Google Scholar]
- 16.Glaser T, Jepeal L, Edwards J G, Young S R, Favor J, Maas R L. Pax6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 17.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 18.Hartley R S, Lewellyn A L, Maller J L. MAP kinase is activated during mesoderm induction in Xenopus laevis. Dev Biol. 1994;163:521–524. doi: 10.1006/dbio.1994.1168. [DOI] [PubMed] [Google Scholar]
- 19.Imperiale M J, Kao H T, Feldman L T, Nevins J R, Strickland S. Common control of the heat shock gene and early adenovirus genes: evidence for a cellular E1A-like activity. Mol Cell Biol. 1984;4:867–874. doi: 10.1128/mcb.4.5.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue H, Nomiyama J, Nakai K, Matsutani A, Tanizawa Y, Oka Y. Isolation of full-length cDNA of mouse PAX4 gene and identification of its human homologue. Biochem Biophys Res Commun. 1998;243:628–633. doi: 10.1006/bbrc.1998.8144. [DOI] [PubMed] [Google Scholar]
- 21.Jimenez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- 23.Kalousová A, Benes V, Paces J, Paces V, Kozmik Z. DNA binding and transactivation properties of the paired and homeobox protein Pax4. Biochem Biophys Res Commun. 1999;259:510–518. doi: 10.1006/bbrc.1999.0809. [DOI] [PubMed] [Google Scholar]
- 24.Kaneto H, Miyagawa J, Kajimoto Y, Yamamoto K, Watada H, Umayahara Y, Hanafusa T, Matsuzawa Y, Yamasaki Y, Higashiyama S, Taniguchi N. Expression of heparin-binding epidermal growth factor-like growth factor during pancreas development. J Biol Chem. 1997;272:29137–29143. doi: 10.1074/jbc.272.46.29137. [DOI] [PubMed] [Google Scholar]
- 25.Katagiri H, Terasaki J, Murata T, Ishihara H, Ogihara T, Inukai K, Fukushima Y, Anai M, Kikuchi M, Miyazaki J, Yazaki Y, Oka Y. A novel isoform of syntaxin-binding protein homologous to yeast Sec1 expressed ubiquitously in mammalian cells. J Biol Chem. 1995;270:4963–4966. doi: 10.1074/jbc.270.10.4963. [DOI] [PubMed] [Google Scholar]
- 26.Kowenz-Leutz E, Twamley G, Ansieau S, Leutz A. Novel mechanism of C/EBP β (NF-M) transcriptional control: activation through derepression. Genes Dev. 1994;15:2781–2791. doi: 10.1101/gad.8.22.2781. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Manley J L. Even-skipped represses transcription by binding TATA binding protein and blocking the TFIID-TATA box interaction. Mol Cell Biol. 1998;18:3771–3781. doi: 10.1128/mcb.18.7.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lillie J W, Green M R. Transcriptional activation by adenovirus E1A protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 29.Liu F, Green M R. Promoter targeting by adenovirus E1a through interaction with different cellular DNA-binding domains. Nature. 1994;368:520–525. doi: 10.1038/368520a0. [DOI] [PubMed] [Google Scholar]
- 30.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magnaghi P, Roberts C, Lorain S, Lipinski M, Scamber P J. HIRA, a mammalian homologue of Saccharomyces cerevisiae transcriptional co-repressors, interacts with Pax3. Nat Genet. 1998;20:74–77. doi: 10.1038/1739. [DOI] [PubMed] [Google Scholar]
- 32.Mansouri A, Hallonet M, Gruss P. Pax genes and their roles in cell differentiation and development. Curr Opin Cell Biol. 1996;8:851–857. doi: 10.1016/s0955-0674(96)80087-1. [DOI] [PubMed] [Google Scholar]
- 33.Minoguchi S, Taniguchi Y, Kato H, Okazaki T, Strobl L J, Zimber-Stroubl U, Bornkamm G W, Honjo T. RBP-L, a transcription factor related to RBP-Jκ. Mol Cell Biol. 1997;17:2679–2687. doi: 10.1128/mcb.17.5.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran E, Mathews M B. Multiple functional domains in the adenovirus E1A gene. Cell. 1987;48:177–178. doi: 10.1016/0092-8674(87)90418-1. [DOI] [PubMed] [Google Scholar]
- 35.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 36.Otonkoski T, Beattie G M, Rubin J S, Lopez A D, Baird A, Hayek A. Hepatocyte growth factor/scatter factor has insulinotropic activity in human fetal pancreatic cells. Diabetes. 1994;43:947–953. doi: 10.2337/diab.43.7.947. [DOI] [PubMed] [Google Scholar]
- 37.Paroush Z, Finley R L, Jr, Kidd T, Wainwright S M, Ingham P W, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with Hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 38.Peshavaria M, Henderson E, Sharma A, Wright C V E, Stein R. Functional characterization of the transactivation properties of the PDX-1 homeodomain protein. Mol Cell Biol. 1997;17:3987–3996. doi: 10.1128/mcb.17.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phelan S A, Loeken M R. Identification of a new binding motif for the paired domain of Pax-3 and unusual characteristics of spacing of bipartite recognition elements on binding and transcription activation. J Biol Chem. 1998;273:19153–19159. doi: 10.1074/jbc.273.30.19153. [DOI] [PubMed] [Google Scholar]
- 40.Puri P L, Avantaggiati M L, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;15:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu Y, Sharma A, Stein R. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol Cell Biol. 1998;18:2957–2964. doi: 10.1128/mcb.18.5.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigues G A, Park M, Schlessinger J. Activation of the JNK pathway is essential for transformation by the Met oncogene. EMBO J. 1997;16:2634–2645. doi: 10.1093/emboj/16.10.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadowski H B, Gilman M Z. Cell-free activation of a DNA-binding protein by epidermal growth factor. Nature. 1993;362:79–83. doi: 10.1038/362079a0. [DOI] [PubMed] [Google Scholar]
- 44.Sadowski I, Ptashne M. A vector for expressing GAL4 (1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539–7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sander M, Neubuser A, Kalamaras J, Ee H C, Martin G R, German M S. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 46.Shenk T, Flint J. Transcriptional and transforming activities of the adenovirus E1A proteins. Adv Cancer Res. 1991;57:47–85. doi: 10.1016/s0065-230x(08)60995-1. [DOI] [PubMed] [Google Scholar]
- 47.Slack J M W. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 48.Smith S B, Ee H C, Conners J R, German M S. Paired-homeodomain transcription factor PAX4 acts as a transcriptional repressor in early pancreatic development. Mol Cell Biol. 1999;19:8272–8280. doi: 10.1128/mcb.19.12.8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The Pax4 gene is essential for differentiation of insulin-producing β cells in the mammalian pancreas. Nature. 1997;386:399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- 50.St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Pax6 is required for differentiation of glucagon-producing α-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 51.Svensson C, Akusjarvi G. Adenovirus 2 early region 1A stimulates expression of both viral and cellular genes. EMBO J. 1984;3:789–794. doi: 10.1002/j.1460-2075.1984.tb01886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang H K, Singh S, Saunders G F. Dissection of the transactivation function of the transcription factor encoded by the eye developmental gene Pax6. J Biol Chem. 1998;273:7210–7221. doi: 10.1074/jbc.273.13.7210. [DOI] [PubMed] [Google Scholar]
- 53.Teitelman G, Alpert S, Polak J M, Martinez A, Hanahan D. Precursor cells of mouse endocrine pancreas coexpress insulin, glucagon and the neuronal proteins tyrosine hydroxylase and neuropeptide Y, but not pancreatic polypeptide. Development. 1993;118:1031–1039. doi: 10.1242/dev.118.4.1031. [DOI] [PubMed] [Google Scholar]
- 54.Turner D L, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 55.Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Krüppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turque N, Plaza S, Radvanyi F, Carriere C, Saule S. PaxQNR/Pax-6, a paired box- and homeobox-containing gene expressed in neurons, is also expressed in pancreatic endocrine cells. Mol Endocrinol. 1994;8:929–938. doi: 10.1210/mend.8.7.7984154. [DOI] [PubMed] [Google Scholar]
- 57.Walther C, Guenet J L, Simon D, Deutsch U, Jostes B, Goulding M D, Plachov D, Balling R, Gruss P. Pax: a murine multigene family of paired box-containing genes. Genomics. 1991;11:424–434. doi: 10.1016/0888-7543(91)90151-4. [DOI] [PubMed] [Google Scholar]
- 58.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 59.Whyte P, Williamson N W, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 60.Wiggan O, Taniguchi-Sidle A, Hamel P A. Interaction of the pRB-family proteins with factors containing paired-like homeodomains. Oncogene. 1998;16:227–236. doi: 10.1038/sj.onc.1201534. [DOI] [PubMed] [Google Scholar]