Abstract
Neurexins are central to trans-synaptic cell adhesion and signaling during synapse specification and maintenance. The past two decades of human genetics research have identified structural variations in the neurexin gene family, in particular NRXN1 copy number variants (CNVs), implicated in multiple neuropsychiatric and developmental disorders. The heterogeneity and reduced penetrance of NRXN1 deletions, in addition to the pleiotropic, circuit-specific functions of NRXN1, present substantial obstacles to understanding how compromised NRXN1 function predisposes individuals to neuropsychiatric disorders. Here, we provide an updated review of NRXN1 genetics in disease, followed by recently published work using both human induced pluripotent stem cell (iPSC) derived systems and animal models to understand the mechanisms of disease pathophysiology. Finally, we suggest our outlook on how the field should progress to improve our understanding of neurexin mediated disease pathogenesis. We believe that understanding how structural genetic variants in NRXN1 contribute to disease pathophysiology requires parallel approaches in iPSC and mouse model systems, each leveraging their unique strengths — analysis of genetic interactions and background effects in iPSCs and neural circuit and behavioral analysis in mice.
Neurexins
Neurexin protein structure
Neurexins are evolutionarily conserved presynaptic cell adhesion molecules that specify, organize, and maintain synaptic structure and function via diverse binding partnerships with a repertoire of postsynaptic cell adhesion and secreted bridging molecules (see Südhof 2017 for comprehensive review [1]). In mammals, three neurexins (Nrxn 1-3 in mice and NRXN 1-3 in humans) are transcribed by α-promoters and β-promoters, producing distinct long and short isoforms respectively [2]. A newly discovered γ isoform is also produced from a distinct promoter specifically in Nrxn1 [3]. α-neurexins consist of six LNS (laminin-neurexin sex hormone binding globulin) domains with epidermal growth factor (EGF)-like repeats interspersed throughout whereas β-neurexins have a single LNS domain. They share the transmembrane domain and C-terminal tail, which contains a PDZ-binding motif in all isoforms. The extracellular domain of these molecules dictates the specificity of interactions with binding partners including neuroligins, leucine-rich repeat transmembrane proteins (LRRTMs), latrophilins, cerebellins and C1q-like proteins [1]. The intracellular domain of neurexins binds to the calcium/calmodulin-dependent serine protein kinase (CASK) scaffolding molecule, which mediates intracellular complexes important for coupling of Ca2+ channels to synaptic release machinery [4-8].
Neurexin splicing
Though all three neurexin genes are transcribed in the brain, α-neurexins are more abundantly expressed than β-neurexins [9,10,11••]. Extensive alternative splicing at splice sites (SS1-5) in the neurexin genes, in addition to the newly identified splice site (SS6) at α-Nrxn1/3 genes, are predicted to produce over 4000 isoforms, thereby supporting extensive protein and functional diversity at synapses [10,12,13]. This structural diversity dictates binding affinities to postsynaptic cell adhesion and secreted adaptor proteins to control the neurexin ligand interaction network [14-20]. Furthermore, neurexin splicing is differentially regulated depending on the specific cell type and brain region [21], suggesting a mechanism for functional diversification of neurexin function by cellular context.
Context-specific neurexin functions
Neurexins exhibit substantial isoform and splicing diversity, are expressed over broad temporal windows — from late neurogenesis into the adult — and are found across many CNS cell types — from neurons to glia [22-25]. Early observation of pan-α-neurexin constitutive knockout (KO) mice suggested a broad role in mediating neurotransmitter release by coupling Ca2+ influx to presynaptic release sites [7]. Recently, conditional KO of pan-β-neurexins in mice showed presynaptic release deficits resulting from abnormal tonic postsynaptic endocannabinoid signaling [26], highlighting the non-redundant functions of α-neurexins and β-neurexins. Further evidence for functional diversification of each neurexin molecule was observed for neurexin splicing mutants, which showed differential effects on α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR)-mediated and N-methyl-d-aspartate receptor (NMDAR)-mediated excitatory synaptic transmission — Nrxn1 SS4+ selectively enhanced NMDAR-responses, but not AMPAR-responses while Nrxn3 SS4+ induced suppression in AMPAR-responses but not NMDAR-responses [9,27,28]. Finally, depending on the specific cellular context in which neurexin mutations were manipulated and studied, neurexins showed distinct regulatory roles in shaping synapse properties [9,27,28,29••,30]. In summary, these studies highlight the importance of neurexin molecules in specifying and maintaining distinct synapse properties in a circuit-specific manner. Given the protracted expression and dynamic regulation of neurexins [30], further work is needed to confirm whether the synaptogenic period is the peak window of vulnerability to neurexin genetic insults.
Clinically associated exonic deletions in NRXN1 gene
Monoallelic heterozygous CNVs
The human genome encodes three neurexin genes (NRXN1, 2p16.3; NRXN2, 11q13.1; NRXN3, 14q24-q31.1), which span large genomic regions of up to 1.1 Megabase. Copy number variants in all three neurexins have been associated with human disease, although the frequency of NRXN1-related disease far outnumbers those of NRXN2 and NRXN3 structural variants. Rare genomic lesions of NRXN2 and NRXN3 have been identified in autism spectrum disorder (ASD) patients [31,32] and associations of NRXN3 single nucleotide polymorphisms (SNPs) were found in schizophrenia (SCZ) and substance abuse cohorts [33,34]. The genomic region of NRXN1 is particularly susceptible to non-recurrent deletions which are clinically associated with various neurodevelopmental disabilities, including developmental delay (DD), intellectual disability (ID), ASD, SCZ, attention deficit hyperactivity disorder (ADHD), epilepsy, Tourette Syndrome (TS), obsessive compulsive disorder (OCD), and Pitt Hopkins Syndrome 2 (for a comprehensive review, see Dabell et al. [35], Bena et al. [36], Castronovo et al. [37]). Most frequent are heterozygous (monoallelic) exonic NRXN1 deletions, which are significantly enriched in cases versus controls. It is estimated that across these disorders, 0.18% cases versus 0.02% controls exhibit NRXN1 structural variants, which translates to ~ 10-fold risk of developing such disorders for individuals with NRXN1 CNVs [37,38,39••]. Castronovo et al. estimates 9-fold, 36-fold, and 8-fold increases in the frequency of these variants in the cases compared to controls for DD/ID, ASD, and SCZ cohorts, respectively [37]. Interestingly, exonic NRXN1 deletions in clinical cases frequently associate with 5′ deletion of the gene resulting in selective disruption of NRXN1α, although cases do exist showing deletions of NRXN1α and β.
Bialleleic compound heterozygous mutations
While monoallelic deletions are frequently observed in disease-association studies, biallelic deletions with compound heterozygosity have been observed in 11 cases to date [37]. The extreme case of complete NRXN1 null individuals (usually with disruptions in the promoter region and first few exons of the NRXN1α transcripts) were identified with Pitt Hopkins syndrome (PHS), which often presents with severe developmental delay, loss of expressive language, muscle hypotonia, motor stereotypes, social interaction deficits, abnormal sleep-wake cycle and chronic constipation [37]. Further studies requiring additional sample collection and tracing of parental alleles would permit a better description and functional analysis of compound heterozygous NRXN1 mutations and their contribution to PHS.
Based on current clinical genetic data, monoallelic deficiency of NRXN1 alone appears insufficient to cause neuropsychiatric disorder. This has strengthened the hypothesis that ultimate clinical severity and specificity are driven by interactions between the functional effects of NRXN1 CNVs and either genetic or environmental modifiers. In the case of biallelic disruptions associated with PHS, the second hit in the NRXN1 allele may be sufficient, as demonstrated by behavioral changes in core behavioral domains also observed with neuropsychiatric disease.
Understanding NRXN1 CNVs through human iPSC models
To investigate the functional impact of heterozygous NRXN1 CNVs in human neurons, Pak et al. engineered two different NRXN1 α/β heterozygous conditional KO alleles in an isogenic human embryonic stem cell (ESC) background and analyzed the morphology, physiology and gene expression in human induced cortical excitatory neurons [11••]. This study showed that NRXN1 haploinsufficiency in human neurons affected synaptic transmission properties (reduced excitatory synaptic transmission and neurotransmitter release probability) without affecting overall neuronal development (dendritic complexity and synapse morphology) [11••]. Interestingly, these mutant neurons exhibited upregulation of CASK protein, a binding partner of the NRXN1 intracellular domain [4]. In a follow up study, Pak et al. confirmed these electrophysiological and morphological phenotypes in neurons derived from SCZ patients carrying NRXN1 CNVs (2 individuals with α-deletions and 1 individual with α/β deletion) versus paired controls (3 individuals) [40]. The CASK protein upregulation was similarly replicated in these patient neurons. In addition, bulk transcriptomic analysis comparing SCZ-NRXN1 deleted neurons versus controls showed that while intrinsic developmental transcriptomic programs were comparable, genes with post-neurogenesis functions were differentially regulated, consistent with the observed morphological and electrophysiological phenotypes [40]. Importantly, mouse ESC derived induced neurons harboring the same Nrxn1α/β heterozygous mutations showed no electrophysiological phenotypes, suggesting potential species differences in gene dosage effects, with human cortical neurons being more vulnerable to heterozygous loss ofNRXN1 than similar cultures in mouse [40].
Interestingly, using human neurons differentiated from patients with BP, SCZ, and childhood onset SCZ carrying heterozygous NRXN1 deletions (4 cases versus 3 controls), Flaherty et al. profiled total neurexin transcripts and identified a reduction in normal NRXN1α isoforms with misexpression of novel isoforms from the mutant allele [41••]. The altered isoforms, when expressed in a wild type background, reduced spontaneous network activity, while the normal isoforms, when introduced in a disease background, rescued the neuronal firing deficit [41••]. While it is presently unclear how widespread this phenomenon is upon heterozygous loss of NRXN1, it represents a path by which changes in splice isoform alter trans-synaptic binding interactions to contribute to disease.
A central question in these iPSC studies is whether the same genetic NRXN1 deletion generates consistent or distinct phenotypic outcomes depending on the disease background the mutation interacts with. In a neurodevelopmental model of ASD, Avazzedah et al. generated iPSC lines from ASD individuals carrying NRXN1 heterozygous deletions (3 cases and 5 controls) and showed in their differentiated neurons elevated frequency, duration and amplitude of somatic Ca2+ transients [42]. It will be important to test the effects of NRXN1 CNVs in other disease contexts including TS, PHS, and ID in multiple patient-control cohorts to identify potential common pathophysiological mechanisms.
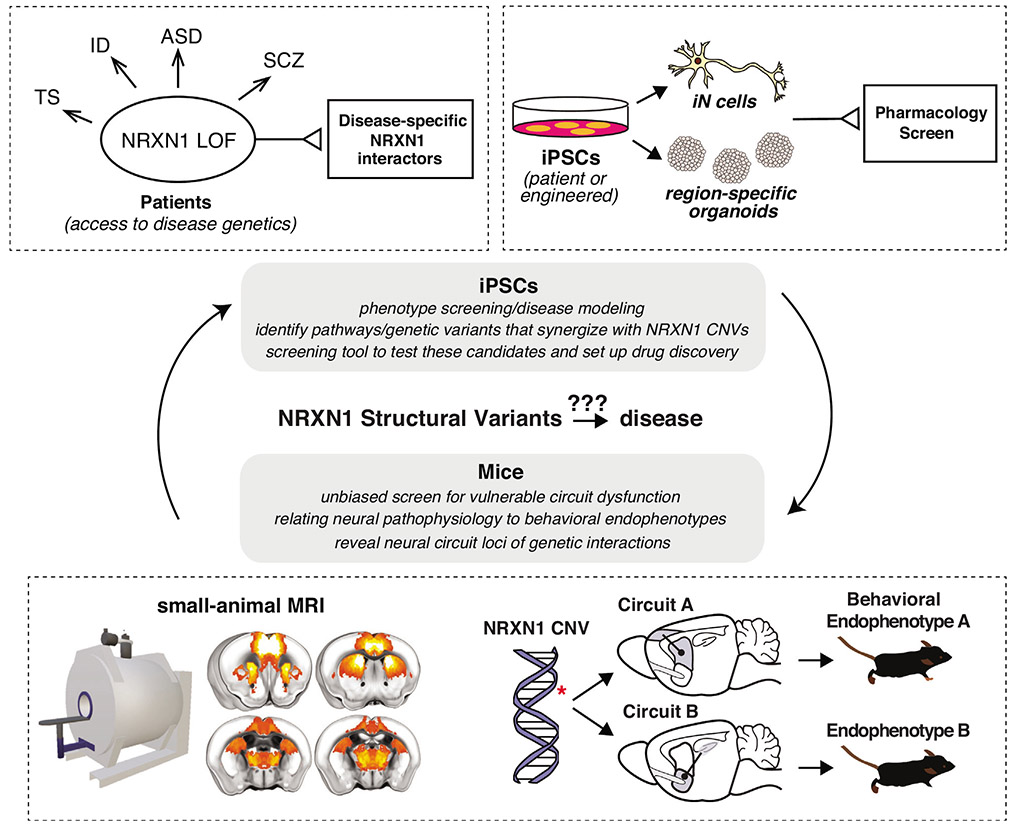
Moving forward, it is imperative to analyze the functional effects of NRXN1 CNVs and how they interact with the genetic backgrounds of different disorders (ID versus ASD versus SCZ versus TS) (Figure 1). Here, the robustness of the observed phenotypes thus far in iPSC-based models provides a solid foundation for dissecting the functional interactions between NRXN1 CNVs and additional genetic modifiers as well as a path toward eventual drug discovery. Second, it is important to expand our analyses to various neuronal and non-neuronal cell types and across multiple developmental time windows at single cell resolution. Here, iPSC-derived brain organoid models represent a tractable system to probe multiple cell types and developmental time points influenced by NRXN1 disruption in brain development.
Figure 1.
Future research directions in understanding phenotypes and mechanisms of NRXN1 copy number variants.
Understanding how NRXN1 CNVs contribute to human disease will require coordinated efforts between iPSCs, where genetic interactions can be explored and high-throughput screens performed, and murine genetic models, where these mutations can be placed in neural circuit and behavioral contexts (Small animal imaging data modified from Canella et al., bioRxiv, 2020; doi: https://doi.org/10.1101/2020.08.05.237958).
Understanding Nrxn1 functional contributions to disease-relevant behavioral dysfunction
Despite the enormous power of iPSC models for exploring early development trajectories and uncovering molecular mechanisms unique to distinct clinical populations, these preparations are limited in their ability to reproduce the cellular diversity and precise connectivity of the brain, as well as to generate its most relevant output, behavior. Rodent and small non-human primate models can fill this gap, with the promise of establishing causal relationships between neurexin-associated changes in neuronal processing and altered behavioral output. Success requires: (1) appropriate modeling of disease-relevant genetic alterations; (2) quantitative analysis of disease-relevant behavioral endophenotypes and (3) higher-throughput methods for observing brain activity (Figure 1).
Neurexin mutations and behavior
Behavioral analyses have been a frequent component of the extensive work on neurexin gene function in rodents. Nevertheless, it is challenging to derive general disease relevant principles from these data as they use a variety of behavioral assays and complex genetic crosses primarily intended to probe neurexin functional redundancy. A smaller set of studies has explicitly sought to model neurexin-associated disease, by looking at the effects of specific mono-allelic and bi-allelic disruption of neurexins on putative disease relevant behavioral endophenotypes (e.g. social or appetitive reward processing, motor control, sensorimotor gating, cognitive flexibility). For Nrxn2α, homozygous KO mice exhibited increased anxiety and decreased sociability, while Nrxn2α heterozygotes exhibited only minor social approach abnormalities [43,44]. For Nrxn1α-associated social behaviors, gene dosage also had distinct effects — homozygote mutants exhibited increased social approach but also heightened aggression, while heterozygotes only had subtle deficits in social preference tasks. Etherton et al. described a range of behavioral control phenotypes including reduced pre-pulse inhibition (commonly observed in schizophrenic patients), increased locomotor activity and elevated grooming [45••]. Furthermore, a robust enhancement in motor learning on the accelerating rotarod was noted, a curious behavioral endophenotype observed in multiple ASD genetic models [46]. Reward processing is another key behavioral domain driving neuropsychiatric disease pathology recently linked to Nrxn1α perturbations. Rats with homozygous mutations in Nrxn1α had male-specific deficits in a simple instrumental learning task for reward [47]. More recently, Alabi et al. employed value-based operant tasks coupled with reinforcement learning behavioral modeling to quantify choice abnormalities in Nrxn1α mutant mice [48••]. Furthermore, they used conditional deletion of Nrxn1α to localize these deficits to disruption within telencephalic excitatory neurons, initiating the challenging task of uncovering pathology-driving neural circuit dysfunction.
Neurexin mutations and physiology
The synaptic function of neurexins has been examined in numerous circuits including brain stem, hippocam-pus, amygdala, prefrontal cortex and cerebellum [7,29••,47,49,51]. These experiments suggest divergent functions based on cellular context, as clearly demonstrated by Nrxn1/2/3 triple KO mice, where cerebellar climbing fibers exhibited altered localization and function while inhibitory connections in prefrontal cortex were absent [29••]. Despite the diversity of neurexin-associated phenotypes observed across circuits, some recurrent trends are apparent — (1) Nrxn1 and Nrxn3 can exert trans-synaptic effects on the composition of postsynaptic glutamate receptors [9,27,28]; (2) Neurexin-Neuroligin networks can modulate the synaptic tone of endocannabinoids [26,50]; (3) both α and β Nrxns support dynamic Ca2+ signals in terminals required for synaptic release [7,8,26,29••]. Regarding physiology of neurexin disease models, there has been a single study of synaptic phenotypes in the hippocampus of Nrxn1α KOs, showing a subtle decrease in excitatory synaptic strength [45••]. Recent work has extended these analyses to striatal circuits, demonstrating startling specificity of Nrxn1α KO phenotypes within a single circuit — prefrontal synapses onto indirect but not direct pathway striatal spiny neurons (iSPNs/dSPNs) have reduced synaptic release, while thalamo-striatal projections have normal synaptic release but lower synaptic NMDAR content on both SPN targets. Interestingly, all of these phenotypes are also observed in Nrxn1α heterozygotes, providing evidence for haploinsufficiency in Nrxn1α mouse models [51].
Other considerations
Despite progress on functional characterization of neurexin mutations, outstanding issues abound. Foremost are considerations about the suitability of current loss-of-function approaches to model the genetics of disease. The use of Nrxn1α null alleles seems reasonable given the 5′ location of the majority of disease-associated NRXN1 CNVs [37], but the default use of homozygous deletions is questionable. While homozygous models may serve as a sensitized genetic state to reveal neural circuits vulnerable to Nrxn1α loss, it is clear that increased focus on heterozygote models as well as splice-altering mutations is warranted [41••,43,44,51]. Another key outstanding question is whether there exist disease-relevant circuits that are vulnerable to neurexin mutations and how to proceed in finding and causally testing such populations. The use of circuit-specific molecular genetic dissection, as with the cortical versus thalamic deletion of Nrxn1α [48••], is a first step in addressing this challenge, but is not amenable to higher throughput scaling and assumes that disease pathology does not arise from dysfunction at multiple circuit nodes. Finally, it is important to consider the utility of both higher and lower model systems, which can provide novel perspectives about how neurexin mutations contribute to circuit pathology. While marmosets have become a potential avenue to analyze single gene mutations in primate brains, flies, fish and worms remain powerful systems for probing genetic interactions and pharmacological modulators, despite the substantial divergence of their neurexin homologs from those of mammals.
Future directions
Two broad questions should guide future studies in understanding how NRXN1 structural variants provide the initiating event for a multitude of brain diseases (Figure 1) — (1) how do the functional outcomes of NRXN1 CNVs interact and synergize with other small effect size variants to impact brain function and are these interactions disease specific? (2) What neural circuits are predisposed to dysfunction with NRXN1 mutation and does the diversity of disease associated with NRXN1 CNVs map to distinct neural circuit perturbations? It is clear that iPSC research could lead the task of dissecting the interplay between genetic background and NRXN1 CNV effects, while simultaneously providing a highly scalable platform for drug discovery. In mice, the task of mapping NRXN1 mutation-associated specific neural circuit dysfunction to behavior can be accomplished via the utilization of improved behavioral modeling, coupled with multi-site high density neural recordings or resting state functional connectivity measures [52,53]. The combined use of these systems will help distinguish whether genetic interactions are acting within individual cells or across neural circuits. We also envision future synergistic interactions between these model systems with mapping of human transcriptomic data to disease-vulnerable neural circuits in mice as a path towards understanding pathophysiological mechanisms and uncovering potential pharmacological tools based on molecular diversity. Ongoing large-scale multi-center efforts, such as the BRAIN Initiative Cell Census Consortium will help better inform experimental designs across species as more complete inventory of brain cell types of the human and rodent developing brain states is achieved [54]. Finally, these basic research directions must be complemented by similar boundary-pushing approaches in the clinical setting. In particular, longitudinal studies that map patient’s developing clinical course with quantifiable changes in structural and functional brain imaging can be coupled with whole genome sequencing of these patients and their immediate family members [55]. These unprecedented datasets will eventually give us a first-hand view of the developmental trajectories associated with genetic alterations.
Acknowledgements
The authors thank Maj Bucan and members of both labs for comments and acknowledge funding support from the National Institute of Mental Health (Pak, R01 MH122519, Fuccillo, R01 MH115030 and R00 MH099243), TAA Young Investigator Award (Fuccillo) and University of Massachusetts Amherst/Institute of Applied Life Sciences start up fund (Pak).
Footnotes
Declarations of interest
None.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
•• of outstanding interest
- 1.Südhof TC: Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell 2017, 171:745–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tabuchi K, Südhof TC: Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics 2002, 79:849–859. [DOI] [PubMed] [Google Scholar]
- 3.Sterky FH, Trotter JH, Lee S-J, Recktenwald CV, Du X, Zhou B, Zhou P, Schwenk J, Fakler B, Südhof TC: Carbonic anhydrase-related protein CA10 is an evolutionarily conserved pan-neurexin ligand. Proc Natl Acad Sci U S A 2017, 114:E1253–E1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hata Y, Butz S, Südhof TC: CASK: a novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J Neurosci 1996, 16:2488–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biederer T, Südhof TC: Mints as adaptors. Direct binding to neurexins and recruitment of munc18. J Biol Chem 2000, 275:39803–39806. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee K, Sharma M, Urlaub H, Bourenkov GP, Jahn R, Südhof TC, Wahl MC: CASK functions as a Mg2+-independent neurexin kinase. Cell 2008, 133:328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Südhof TC: Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature 2003, 423:939–948. [DOI] [PubMed] [Google Scholar]
- 8.Luo F, Sclip A, Jiang M, Südhof TC: Neurexins cluster Ca2+ channels within the presynaptic active zone. EMBO J 2020, 39: e103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Südhof TC: Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell 2013, 154:75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreiner D, Nguyen T-M, Russo G, Heber S, Patrignani A, Ahrné E, Scheiffele P: Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron 2014, 84:386–398. [DOI] [PubMed] [Google Scholar]
- 11.••. Pak C, Danko T, Zhang Y, Aoto J, Anderson G, Maxeiner S, Yi F, Wernig M, Südhof TC: Human neuropsychiatric disease modeling using conditional deletion reveals synaptic transmission defects caused by heterozygous mutations in nrxn1. Cell Stem Cell 2015, 17:316–32826279266 Using conditional gene targeting in isogenic human ESCs, this study demonstrates for the first time the functional consequences of NRXN1 heterozygous loss-of-function in human induced neurons. Most prominent phenotypes described are decreased excitatory neurotransmitter release, decreased synaptic strength and increased CASK protein levels.
- 12.Ullrich B, Ushkaryov YA, Südhof TC: Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron 1995,14:497–507. [DOI] [PubMed] [Google Scholar]
- 13.Treutlein B, Gokce O, Quake SR, Südhof TC: Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc Natl Acad Sci U S A 2014, 111: E1291–E1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boucard AA, Chubykin AA, Comoletti D, Taylor P, Südhof TC: A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron 2005. 48:229–236. [DOI] [PubMed] [Google Scholar]
- 15.Chih B, Gollan L, Scheiffele P: Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron 2006, 51:171–178. [DOI] [PubMed] [Google Scholar]
- 16.Comoletti D, Flynn RE, Boucard AA, Demeler B, Schirf V, Shi J, Jennings LL, Newlin HR, Südhof TC, Taylor P: Gene selection, alternative splicing, and post-translational processing regulate neuroligin selectivity for beta-neurexins. Biochemistry 2006. 45:12816–12827. [DOI] [PubMed] [Google Scholar]
- 17.Ko J, Fuccillo MV, Malenka RC, Südhof TC: LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron 2009, 64:791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siddiqui TJ, Pancaroglu R, Kang Y, Rooyakkers A, Craig AM: LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J Neurosci 2010, 30:7495–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boucard AA, Ko J, Südhof TC: High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J Biol Chem 2012, 287:9399–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uemura T, Lee S-J, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M: Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 2010, 141:1068–1079. [DOI] [PubMed] [Google Scholar]
- 21.Fuccillo MV, Földy C, Gökce Ö, Rothwell PE, Sun GL, Malenka RC, Südhof TC: Single-cell mRNA profiling reveals cell-type-specific expression of neurexin isoforms. Neuron 2015,87:326–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N et al. : An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014, 34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins AK, Paterson C, Wang Y, Hyde TM, Kleinman JE, Law AJ: Neurexin 1 (NRXN1) splice isoform expression during human neocortical development and aging. Mol Psychiatry 2016, 21:701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harkin LF, Lindsay SJ, Xu Y, Alzu’bi A, Ferrara A, Gullon EA, James OG, Clowry GJ: Neurexins 1-3 each have a distinct pattern of expression in the early developing human cerebral cortex. Cereb Cortex 2017, 27:216–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trotter JH, Dargaei Z, Wohr M, Liakath-Ali K, Raju K, Essayan-Perez S, Nabet A, Liu X, Südhof TC: Astrocytic neurexin-1 orchestrates functional synapse assembly. bioRxiv 2020. 10.1101/2020.08.21.262097. [DOI] [Google Scholar]
- 26.Anderson GR, Aoto J, Tabuchi K, Földy C, Covy J, Yee AX, Wu D, Lee S-J, Chen L, Malenka RC et al. : β-neurexins control neural circuits by regulating synaptic endocannabinoid signaling. Cell 2015, 162:593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai J, Aoto J, Südhof TC: Alternative splicing of presynaptic neurexins differentially controls postsynaptic NMDA and AMPA receptor responses. Neuron 2019, 102:993–1008.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aoto J, Földy C, Ilcus SMC, Tabuchi K, Südhof TC: Distinct circuit-dependent functions of presynaptic neurexin-3 at GABAergic and glutamatergic synapses. Nat Neurosci 2015, 18:997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.••. Chen LY, Jiang M, Zhang B, Gokce O, Südhof TC: Conditional deletion of all neurexins defines diversity of essential synaptic organizer functions for neurexins. Neuron 2017, 94:611–625.e428472659 This study uses complete genetic disruption of Neurexins in mice to clearly demonstrate divergent circuit-specific requirements for Neurexin function.
- 30.Trotter JH, Hao J, Maxeiner S, Tsetsenis T, Liu Z, Zhuang X, Südhof TC: Synaptic neurexin-1 assembles into dynamically regulated active zone nanoclusters. J Cell Biol 2019, 218:2677–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gauthier J, Siddiqui TJ, Huashan P, Yokomaku D, Hamdan FF, Champagne N, Lapointe M, Spiegelman D, Noreau A, Lafrenière RG et al. : Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum Genet 2011, 130:563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaags AK, Lionel AC, Sato D, Goodenberger M, Stein QP, Curran S, Ogilvie C, Ahn JW, Drmic I, Senman L et al. : Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am J Hum Genet 2012, 90:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu X, Zhang J, Jin C, Mi W, Wang F, Ma W, Ma C, Yang Y, Li W, Zhang H et al. : Association study of NRXN3 polymorphisms with schizophrenia and risperidone-induced bodyweight gain in Chinese Han population. Prog Neuropsychopharmacol Biol Psychiatry 2013, 43:197–202. [DOI] [PubMed] [Google Scholar]
- 34.Stoltenberg SF, Lehmann MK, Christ CC, Hersrud SL, Davies GE: Associations among types of impulsivity, substance use problems and neurexin-3 polymorphisms. Drug Alcohol Depend 2011, 119:e31–e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dabell MP, Rosenfeld JA, Bader P, Escobar LF, El-Khechen D, Vallee SE, Dinulos MBP, Curry C, Fisher J, Tervo R et al. : Investigation of NRXN1 deletions: clinical and molecular characterization. Am J Med Genet A 2013, 161A:717–731. [DOI] [PubMed] [Google Scholar]
- 36.Béna F, Bruno DL, Eriksson M, van Ravenswaaij-Arts C, Stark Z, Dijkhuizen T, Gerkes E, Gimelli S, Ganesamoorthy D, Thuresson AC et al. : Molecular and clinical characterization of 25 individuals with exonic deletions of NRXN1 and comprehensive review of the literature. Am J Med Genet Part B: Neuropsychiatr Genet 2013, 162:388–403. [DOI] [PubMed] [Google Scholar]
- 37.Castronovo P, Baccarin M, Ricciardello A, Picinelli C, Tomaiuolo P, Cucinotta F, Frittoli M, Lintas C, Sacco R, Persico AM: Phenotypic spectrum of NRXN1 mono- and bi-allelic deficiency: a systematic review. Clin Genet 2020, 97:125–137. [DOI] [PubMed] [Google Scholar]
- 38.Lowther C, Speevak M, Armour CM, Goh ES, Graham GE, Li C, Zeesman S, Nowaczyk MJM, Schultz L-A, Morra A et al. : Molecular characterization of NRXN1 deletions from 19,263 clinical microarray cases identifies exons important for neurodevelopmental disease expression. Genet Med 2017, 19:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.••. Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, Antaki D, Shetty A, Holmans PA, Pinto D et al. : Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 2017, 49:27–3527869829 This study interrogates, in a genome wide manner, the overall disease burden of CNVs in schizophrenia in the largest cohort of patients (21 094) versus controls (20 227) at the time of study, and identifies 2p16.3 (NRXN1) as a rare but genome wide significant single gene CNV for schizophrenia among other loci.
- 40.Pak C, Danko T, Mirabella V, Wang J, Zhang X, Ward T, Grieder S, Vangipuram M, Huang A, Liu Y et al. : Cross-platform validation of neurotransmitter release impairments in schizophrenia patient-derived NRXN1-mutant neurons. bioRxiv 2020. 10.1101/2020.11.03.366617. (Manuscript is currently under revision and will be updated before final publication stage of this review article). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.••. Flaherty E, Zhu S, Barretto N, Cheng E, Deans PJM, Fernando MB, Schrode N, Francoeur N, Antoine A, Alganem K et al. : Neuronal impact of patient-specific aberrant NRXN1α splicing. Nat Genet 2019, 51:1679–169031784728 This study shows that heterozygous loss of NRXN1, depending on the specific location of the deletion, results in differential expression of NRXN1 splice variants. Also, describes, using patient derived iPSC induced neurons, a possible mechanism of disease-related novel NRXN1 isoforms in driving neuronal activity deficits seen in these neurons.
- 42.Avazzadeh S, McDonagh K, Reilly J, Wang Y, Boomkamp SD, McInerney V, Krawczyk J, Fitzgerald J, Feerick N, O’Sullivan M et al. : Increased Ca2+ signaling in NRXN1α+/− neurons derived from ASD induced pluripotent stem cells. Mol Autism 2019, 10:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dachtler J, Glasper J, Cohen RN, Ivorra JL, Swiffen DJ, Jackson AJ, Harte MK, Rodgers RJ, Clapcote SJ: Deletion of α-neurexin II results in autism-related behaviors in mice. Transl Psychiatry 2014, 4:e484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dachtler J, Ivorra JL, Rowland TE, Lever C, Rodgers RJ, Clapcote SJ: Heterozygous deletion of α-neurexin I or α-neurexin II results in behaviors relevant to autism and schizophrenia. Behav Neurosci 2015, 129:765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.••. Etherton MR, Blaiss CA, Powell CM, Südhof TC: Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci U S A 2009, 106:17998–1800319822762 The first study to describe behavioral and electrophysiological phenotypes of Neurexin1a knockout mice, demonstrating a range of disease-relevant endophenotypes and subtle changes to hippocampal excitatory neurotransmission.
- 46.Rothwell PE, Fuccillo MV, Maxeiner S, Hayton SJ, Gokce O, Lim BK, Fowler SC, Malenka RC, Südhof TC: Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell 2014, 158:198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esclassan F, Francois J, Phillips KG, Loomis S, Gilmour G: Phenotypic characterization of nonsocial behavioral impairment in neurexin 1α knockout rats. Behav Neurosci 2015, 129:74–85. [DOI] [PubMed] [Google Scholar]
- 48.••. Alabi OO, Davatolhagh MF, Robinson M, Fortunato MP, Vargas Cifuentes L, Kable JW, Fuccillo MV: Disruption of Nrxn1α within excitatory forebrain circuits drives value-based dysfunction. eLife 2020, 9 This study uses novel value-based operant paradigms and reinforcement learning choice modeling to quantitatively characterize the reward processing deficits of Nrxn1a knockout mice. It also shows that telencephalic excitatory but not thalamic excitatory neuron deletion of Nrxn1a recapitulates the brain-wide knockout phenotype.
- 49.Asede D, Joseph A, Bolton MM: Deletion of NRXN1a impairs long-range and local connectivity in amygdala fear circuit. Transl Psychiatry 2020, 10:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Földy C, Malenka RC, Südhof TC: Autism-associated neuroligin-3 mutations commonly disrupt tonic endocannabinoid signaling. Neuron 2013, 78:498–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davatolhagh MF, Fuccillo MV: Neurexin1α differentially regulates synaptic efficacy within striatal circuits. Cell Rep 2021, 34:108773 10.1016/j.celrep.2021.108773 PMID: 33626349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hultman R, Ulrich K, Sachs BD, Blount C, Carlson DE, Ndubuizu N, Bagot RC, Parise EM, Vu M-AT, Gallagher NM et al. : Brain-wide electrical spatiotemporal dynamics encode depression vulnerability. Cell 2018, 173:166–180.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pagani M, Bertero A, Liska A, Galbusera A, Sabbioni M, Barsotti N, Colenbier N, Marinazzo D, Scattoni ML, Pasqualetti M et al. : Deletion of autism risk gene shank3 disrupts prefrontal connectivity. J Neurosci 2019, 39:5299–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ecker JR, Geschwind DH, Kriegstein AR, Ngai J, Osten P, Polioudakis D, Regev A, Sestan N, Wickersham IR, Zeng H: The BRAIN initiative cell census consortium: lessons learned toward generating a comprehensive brain cell atlas. Neuron 2017, 96:542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hagler DJ, Hatton S, Cornejo MD, Makowski C, Fair DA, Dick AS, Sutherland MT, Casey BJ, Barch DM, Harms Mp et al. : Image processing and analysis methods for the Adolescent Brain Cognitive Development Study. Neuroimage 2019, 202:116091. [DOI] [PMC free article] [PubMed] [Google Scholar]