Abstract
Positive regulation of gene expression by the yeast Saccharomyces cerevisiae transcription factor Yap1p is required for normal tolerance of oxidative stress elicited by the redox-active agents diamide and H2O2. Several groups have provided evidence that a cluster of cysteine residues in the extreme C terminus of the factor are required for normal modulation of Yap1p by oxidant challenge. Deletion of this C-terminal cysteine-rich domain (c-CRD) produces a protein that is highly active under both stressed and nonstressed conditions and is constitutively located in the nucleus. We have found that a variety of different c-CRD mutant proteins are hyperactive in terms of their ability to confer diamide tolerance to cells but fail to provide even normal levels of H2O2 resistance. Although the c-CRD mutant forms of Yap1p activate an artificial Yap1p-responsive gene to the same high level in the presence of either diamide or H2O2, these mutant factors confer hyperresistance to diamide but hypersensitivity to H2O2. To address this discrepancy, we have examined the ability of c-CRD mutant forms of Yap1p to activate expression of an authentic target gene required for H2O2 tolerance, TRX2. When assayed in the presence of c-CRD mutant forms of Yap1p, a TRX2-lacZ fusion gene fails to induce in response to H2O2. We have also identified a second cysteine-rich domain, in the N terminus (n-CRD), that is required for H2O2 but not diamide resistance and influences the localization of the protein. These data are consistent with the idea that the function of Yap1p is different at promoters of loci involved in H2O2 tolerance from promoters of genes involved in diamide resistance.
To grow in the presence of oxygen, cells must be able to deal with reactive oxygen species (ROS) that are produced during metabolism. Aerobes have the ability both to detoxify ROS and to repair macromolecules that are damaged by these highly reactive compounds (26). Owing to the potentially lethal action of ROS, cells maintain constant surveillance of intracellular ROS levels and rapidly activate the expression of loci involved in oxidative stress tolerance (7).
The yeast Saccharomyces cerevisiae has been a useful model for studies of the eukaryotic response to oxidant challenge (15). S. cerevisiae produces a variety of enzymes, such as catalase, superoxide dismutase, and glutathione peroxidase, and small molecules and peptides (glutathione and thioredoxins) that detoxify ROS (reviewed in reference 10). Recent data have provided insight into the regulation of the biosynthesis of these enzymatic activities.
One of the key regulators of oxidative stress tolerance in S. cerevisiae is the Yap1p transcription factor. The YAP1 gene is required for normal tolerance to a wide variety of oxidants and is essential for normal synthesis of a variety of antioxidant activities, including glutathione and glutathione reductase (4, 5, 19, 27). Later studies established that Yap1p-dependent transactivation was markedly enhanced when cells were challenged with oxidants including diamide and H2O2 (13). Western blotting experiments demonstrated that the increased activity of Yap1p was probably due to a posttranslational modification of the factor (14, 24). Mutational analyses have implicated a set of three cysteine residues contained in the C-terminal cysteine-rich domain (c-CRD) of the factor as being required for the normal elevation of Yap1p transactivation upon oxidative stress (14, 24). Cysteine residues have been implicated as likely sensors of the reducing environment of cells in other redox-regulated transcription factors (reviewed in reference 22). Recent studies have implicated the c-CRD in maintaining Yap1p in the cytoplasm until oxidant challenge drives the protein into the nucleus (14, 29). Deletion of the c-CRD produced a mutant Yap1p that appeared to be constitutively localized in the nucleus and conferred a hyperresistance phenotype to diamide (14). These data led to the model that regulation of Yap1p by the redox state of the cell was via oxidant-regulated nuclear localization regulated by the c-CRD of the protein (14, 29).
In this work, we demonstrate that the regulatory role of the Yap1p c-CRD cannot be solely to modulate nuclear localization of the factor. Loss of the c-CRD results in a Yap1p derivative that confers hyperresistance to diamide but hypersensitivity to H2O2. This phenotypic pattern is reproduced by a number of different mutant forms of Yap1p lacking normal c-CRD regions. A second CRD, in the amino terminus of Yap1p (n-CRD), is also required for normal oxidative stress regulation of the protein. Finally, we show that while Yap1p c-CRD mutants activate the expression of a synthetic reporter gene to high levels, these mutant derivatives fail to normally activate the promoter of a gene required for H2O2 tolerance. These data argue that mutants lacking wild-type CRD regions are defective in an additional regulatory function required for activation of transcription at promoters involved in H2O2 resistance.
MATERIALS AND METHODS
Yeast methods.
The yeast strains used in this study were SEY6210 (MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 Mel−), SM12 (MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 Mel− yap1-Δ1::HIS3 ARE-TRP5-lacZ), SM13 (MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 Mel− yap1-Δ2::hisG), YSC6 (MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 Mel- yap1- Δ2::hisG ARE-TRP5-lacZ), and YSC18 (MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 Mel− yap1-Δ2::hisG TRX2-lacZ). YSC6 was constructed by cutting pTEP9 (ARE-TRP5-lacZ) with KpnI (to direct recombination to LEU2) and transforming SM13 to Leu2+. YSC18 was generated by cutting pSC99 containing TRX2-lacZ with NheI and integrating this linear plasmid in the HIS3 gene of SM13. YSC6 and YSC18 were both checked by Southern blotting to confirm proper chromosomal structure of the recombinants. YSC25 was constructed by transforming SEY6210 with SacI-cleaved pGC61 (skn7Δ::TRP1). YSC26 (Δskn7 yap1) was produced by transforming YSC25 with a Asp718-SacI fragment containing yap1-Δ2::hisG-URA3-hisG. Ura3+ transformants were treated with 5-fluoroorotic acid to remove the URA3 gene. Yeast cells were grown either in rich, nonselective medium (yeast extract-peptone-dextrose medium [YPD]), minimal medium (synthetic dextrose [SD]) with required supplements, or SD supplemented with Casamino Acids (20). Transformation was performed by the lithium acetate technique of Ito et al. (9). Assays for β-galactosidase activity were carried out on permeabilized cells as described previously (6) or with a luminescent substrate for detection of low levels of β-galactosidase activity as described by the manufacturer (Clontech). Diamide and H2O2 resistance assays were carried out by spot tests (28).
Plasmids.
The integrating ARE-TRP5-lacZ construct pTEP9 has been described previously (25). Low-copy-number plasmids carrying the wild-type or CSE629AAA forms of YAP1 (pSM58wt and pSMS37, respectively) were constructed previously (24). The YAP1 deletion mutant plasmids pJAW1 and pJAW15 were described previously (25). All PCR products were sequenced in their entirety to ensure that no errors had occurred during amplification.
The integrating TRX2-lacZ plasmid (pSC99) was generated by PCR. A 505-bp fragment of the TRX2 promoter and translation start signals was produced with an upstream primer (GCG AAT TCA TCC AGA CTT TTA CGG GTG GCA) and a downstream primer (GCG GAT CCG TGA CCA TTA TTG ATG TGT TA) corresponding to positions −497 to +8. The resulting product was cleaved with EcoRI-BamHI and cloned into the lacZ fusion plasmid pSEYC102 (3) to produce plasmid pSC99. An EcoRI-SalI TRX2-lacZ fragment was then cloned from pSC99 into pRS303 to form the integrating reporter plasmid.
The low-copy-number YRETRX2-CYC1-lacZ reporter plasmid was generated in two steps. First, a CYC1-lacZ fusion gene was isolated from pCBS1 (8) as a SalI-NruI fragment and cloned into SalI-SmaI-cleaved pRS314 (21). The resulting low-copy-number TRP1 CYC1-lacZ plasmid was designated p314ClZ. To analyze the function of the TRX2 Yap1p response elements (YREs), oligonucleotides corresponding to the YREs at position −181 were generated. The oligonucleotides had the following sequences: −181 top, GAT CCT CTT TTC TTA CTA AGC GCG TTC; −181 bottom, GAT CGA ACG CGC TTA GTA AGA AAA GAG. The underlined residues correspond to the TRX2 YREs. These oligonucleotides were annealed and cloned into BglII-digested p314ClZ.
Fusions between the TRX2 promoter region and CYC1-lacZ were constructed by PCR. All fragments except the −255 to −141 fragment were inserted as BamHI-BglII fragments into the BglII site of p314CIZ. The −255 to −141 fragment was inserted as a SalI-EcoRV fragment into SalI-BglII-filled p314ClZ plasmid. All PCR-generated fragments were sequenced to ensure that no errors had occurred during amplification and cloning.
The alanine scanning mutations were generated by PCR in a two-step procedure as described previously (18). The mutagenic primers used were as follows: C629A, ATG GCA AAG GCA AAA GCC TCA GAA AGA GGG GTT GTC ATC AAT; S630A, ATG GCA AAG GCA AAA TGT GCG GAA AGA GGG GTT GTC ATC AAT; and E631A, ATG GCA AAG GCA AAA TGT TCA GCA AGA GGG GTT GTC ATC AAT. Each alanine mutant generated was cloned as an Asp718-HindIII fragment into pSMS37. These plasmids were then examined for the loss of a SacII restriction site and sequenced to avoid introduction of an unwanted mutation.
Amino-terminal internal deletions were generated by PCR. All PCR amplifications were performed with pSM58wt as a template. The Δ220–243 and Δ220–307 internal YAP1 deletions were constructed by using custom primers (GGA GTT CGT CGA CTT AAT AAC ACA CCA AAC TCC to synthesize the fragment starting at residue 244 and GGA ATT CGT CGA CAG GTA TGT GGA ACA AGG CAA to synthesize the fragment starting at residue 308). The standard M13 reverse-sequencing primer was used as the reverse PCR primer. The PCR products were cloned into pBluescript KSII+ as EcoRI fragments. These deletion fragments were then transferred back into the context of the YAP1 gene by replacing the SalI-KpnI fragment from the Δ220–335 YAP1 deletion mutant described previously (25). The Δ317–335 deletion was constructed by PCR amplification of the 5′ end of the YAP1 gene with custom forward (GGG AGA TCT CCA TGA GTG TGT CTA CCG CCA AGA GGT CGC TGG AT) and reverse (GGA ATT CGT CGA CCA ATG GGA CAT TGC CTT GTT CC) primers. The PCR product was inserted as a BglII-EcoRI fragment into BamHI-EcoRI-cleaved pBluescript KSII+. From this clone, a HpaI-SalI fragment containing the coding sequence for residues 156 to 317 was placed into the same sites in the Δ220–335 YAP1 deletion mutant to generate the complete Δ317–335 YAP1 clone. The C303A mutant was constructed by using a forward primer with a 5′ XhoI restriction site (CGC TCG AGG ATT AAG TGA CGC TAC AGA TTC CTC CAG) and a reverse mutagenic primer with an EcoRI restriction site (GCC GAA TTC TTC GAA GCA AAC TCC GAA ACT TG). The underlined GC identifies the mutant positions in the primer. This PCR product was cloned as a XhoI-EcoRI fragment into pBluescript KSII+ and then subcloned as an Eco47III/BstBI fragment into pUC19-BH3 (25). This recombinant contained the C303A mutant in a YAP1 BamHI-HindIII fragment, which was used to replace the same BamHI-HindIII fragment in pSM58wt to generate the full-length YAP1 C303A.
The wild-type green fluorescent protein (GFP)-YAP1 fusion gene was constructed by a PCR-based method (30). Primers NT3 (AAG TTG TTT CTT AAA CCA TGT CTA AAG GTG AAG AA) and NT4 (CTC TTG GCG GTA GAC ACA CTT TTG TAC AAT TCA TC) were used to amplify the GFP gene from an appropriate template. The underlined segments represent YAP1 sequence flanking the ATG and result in GFP being inserted between codons 1 and 2 of YAP1. This YAP1-GFP chimeric fragment was then annealed to a YAP1 plasmid and the resulting template used in two separate PCR reactions. Additional upstream DNA flanking YAP1 was appended to the YAP1-GFP chimera with primers NT4 and C (GGA ACA AGA GTC CAC), while additional downstream DNA was attached with primers NT3 and D (TGG AGG AAT CTG TAG CGT CA). The products of each of these reactions were purified, mixed, and subjected to a final PCR with primers C and D. This PCR product was transformed into Δyap1 cells along with a YAP1-containing plasmid that had been gapped by restriction enzyme cleavage. This gap removed the YAP1 promoter and N-terminal coding sequences from residues 1 to 63. Recombinants between the PCR product and the gapped plasmid were selected, and the structure of the GFP-YAP1 fusion gene was verified by restriction enzyme digestion and DNA sequence analysis. Mutant forms of this GFP-YAP1 fusion were constructed by replacing the wild-type BamHI-HindIII fragment with the same fragment from a mutant construct of interest.
Construction of a YAP1 mutant library and selection of hyperactive YAP1 alleles.
The c-CRD-encoding region of YAP1 was subcloned as an Asp718-HindIII fragment into pBluescript KSII+ to form pSMS26. A 20-μg portion of this plasmid was then mutagenized with 100 μl of 45% formic acid for 10 min. DNA was recovered by ethanol precipitation, and the carboxy terminus of YAP1 was amplified by PCR with flanking T3 and T7 primers. The PCR product was cleaved with Asp718-HindIII and ligated into the context of wild-type YAP1. The resulting library was amplified in bacteria, and plasmid DNA was generated.
The yeast strain SM12 was transformed with the library and replica plated onto YPD plates containing 2.5 mM diamide. The wild-type YAP1 gene is not able to support growth at this concentration of diamide (data not shown). Survivors were tested for increased expression from the ARE-TRP5-lacZ reporter, and plasmids were recovered. Recovered plasmids were retransformed into SM12 and retested for increased diamide resistance and ARE-TRP5-lacZ expression. The sequence of the carboxy-terminal segment was determined by the University of Iowa DNA Core Facility with a custom oligonucleotide primer.
Western blotting analysis.
Cells were grown in 100 ml of minimal medium to an absorbance at 600 nm (A600) of 0.6, drug was added, and cells were incubated for an additional 1.5 h. Diamide or hydrogen peroxide was added to give final concentrations of 1.5 mM and 1.0 mM, respectively. Cells were harvested, washed, and broken by glass bead lysis in buffer containing 300 mM sorbitol, 100 mM NaCl, 5 mM MgCl2, 10 mM Tris (pH 7.4), and complete protease inhibitors (Boehringer Mannheim). Cell lysates were cleared, and the Bradford protein assay (Bio-Rad) was used to determine the protein concentration of the supernatant. Portions (50 or 100 μg) of protein from each sample were run on an 8% polyacrylamide gel. The proteins were transferred to nitrocellulose, blocked with 2.5% nonfat dry milk in phosphate-buffered saline, and probed with the anti-Yap1p polyclonal antiserum. Horseradish peroxidase-conjugated secondary antibody and the ECL kit (Pierce) were used to visualize immunoreactive protein. The affinity-purified Yap1p antiserum was prepared by passing serum over columns of protein extract prepared from a Δyap1 strain or bacterially purified Yap1p linked to Sepharose beads as specified by the manufacturer (Pierce Amino-link).
Fluorescence microscopy analysis.
Transformants expressing GFP-Yap1p fusions of interest were grown and subjected to oxidative stress as described above. After the desired stress regimen had been imposed, the cells were incubated with 4′,6-diamidino-2-phenylindole (DAPI) to visualize DNA. Cells were then viewed with an Olympus BX60 fluorescence microscope. Images were captured with a Hammamatsu ORCA charge-coupled device camera and processed with Adobe Photoshop 5.0 software.
RESULTS
Alanine-scanning mutagenesis of the CSE629 repeat.
Previous studies from our laboratory (24) and others (14, 29) have implicated cysteine residues in the c-CRD as being critical for the normal response of Yap1p to oxidant challenge. These cysteines in the Yap1p c-CRD are present as three CSE repeat clusters, and our data suggested that the most C-terminal of these clusters (CSE629) played a major role in regulation of Yap1p function (24). However, these mutagenesis experiments involved replacing all three amino acids of the CSE repeats with alanine residues and precluded an individual assessment of the regulatory contribution of each individual amino acid. To explore the necessity of each position in the CSE629 repeat for normal control of Yap1p function, each amino acid was individually replaced with an alanine residue. The resulting single-amino-acid substitution mutants were then assayed for their ability to transactivate a Yap1p-dependent lacZ reporter gene, oxidative stress phenotypes, and steady-state protein levels.
Irrespective of which of the CSE629 residues were replaced with alanine, the resulting mutant protein behaved as a strong constitutive activator of the ARE-TRP5-lacZ reporter gene (Fig. 1). All three of the single-alanine-substitution mutations essentially reproduced the constitutive hyperactive phenotype of the original CSE629AAA allele described previously (24).
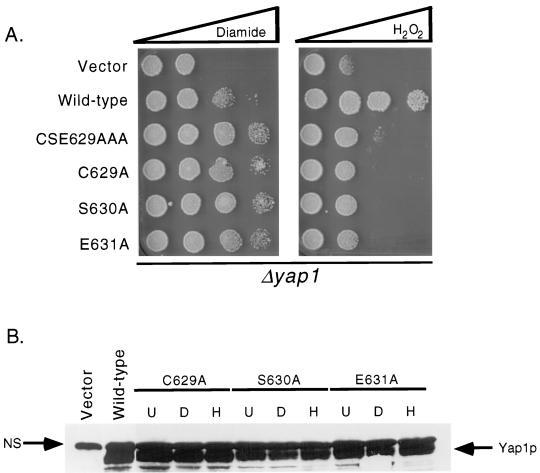
FIG. 1.
Transactivation of ARE-TRP5-lacZ expression by alanine scanning mutations in CSE629. Strain YSC6 (yap1-Δ2::hisG ARE-TRP5-lacZ) was transformed with low-copy-number plasmids expressing the indicated forms of Yap1p. Transformants were grown on SC medium (20) to an A600 of 0.6 and then split into three equal aliquots, which were subjected to diamide- or H2O2-induced oxidative stress or left untreated (No Stress) for 1.5 h. Cells were then processed and β-galactosidase activity was measured as described previously (6). The locations of the basic-region leucine zipper DNA binding domain and the two separable transactivation domains in Yap1p are indicated on the left-hand side of the figure. The drawing represents the Yap1p protein chain, and the numbers indicate the position along the factor.
Along with the analysis of the activation of the Yap1p-dependent reporter gene, each mutant factor was tested for its ability to complement the diamide- and H2O2-hypersensitive phenotypes of a Δyap1 strain. The mutant Yap1p proteins were expressed from low-copy-number plasmids and challenged for growth in the presence of increasing concentrations of oxidants (Fig. 2A). Each of the single-alanine-replacement mutants exhibited the same diamide-hyperresistant and H2O2-hypersensitive phenotypes as those conferred by the CSE629AAA triple-mutant protein. Western blot analyses established that the three single-replacement mutations were all expressed at levels roughly comparable to that of the wild-type factor (Fig. 2B). These data establish that while the cysteine residue at position 629 is necessary for appropriate redox control of Yap1p function, it is not sufficient.
FIG. 2.
Phenotype and expression of alanine-scanning mutations in CSE629. (A) Cells lacking the YAP1 gene (Δyap1) were transformed with low-copy-number plasmids expressing the indicated forms of Yap1p or the vector only (pRS316). Transformants were grown to an A600 of 1, and spots of 1,000 cells were placed on YPD containing diamide or H2O2. Each oxidant was present in a concentration gradient, as indicated by the bar at the top of the figure. (B) Δyap1 cells expressing the indicated forms of Yap1p were grown in the absence of oxidants (U) or challenged for 1.5 h with diamide (D) or H2O2 (H). Protein extracts were prepared, and either 50 μg (wild-type Yap1p) or 100 μg (mutants) of protein was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The gel was then transferred to nitrocellulose and probed with a rabbit anti-Yap1p antiserum (24). Bound antibody was detected by using goat anti-rabbit antibody and chemiluminescence (Pierce).
Random mutagenesis of the Yap1p C terminus.
To gain additional insight into the amino acids required for normal function of the Yap1p C terminus, we generated a collection of random mutations in a restriction fragment encoding this region of the factor (see Materials and Methods for details). This restriction fragment was then reintroduced into an otherwise unaltered YAP1 gene carried on a low-copy-number plasmid. This mutant plasmid pool was transformed into a Δyap1 strain carrying the ARE-TRP5-lacZ gene, and transformants that were hyperresistant to diamide were selected. Cells that exhibited enhanced diamide tolerance were then assayed for their levels of ARE-directed β-galactosidase activity. Plasmids were recovered from colonies that expressed elevated β-galactosidase levels and tested for their ability to retransform both the diamide hyperresistance and the high-level ARE-TRP5-lacZ expression on a fresh version of the original Δyap1 strain. Plasmids that were able to retransform these traits were then subjected to DNA sequence analysis to identify the altered position in the C terminus.
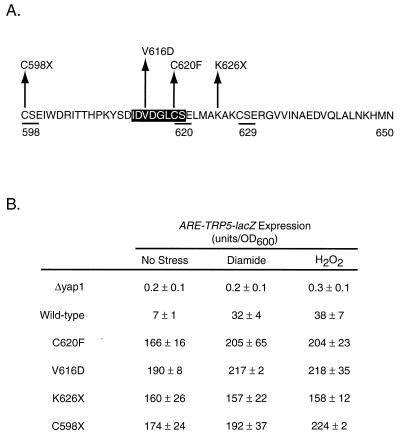
While a large number of mutant plasmids were recovered, we focused our attention on four different mutations, since these served to define the general classes of lesions that would give rise to a hyperactive form of Yap1p. To designate the identity of the mutation recovered, the wild-type residue is listed in the single-letter code followed by the position and finally the residue present in the mutant. The nonsense terminations are designated X in the mutant residue position.
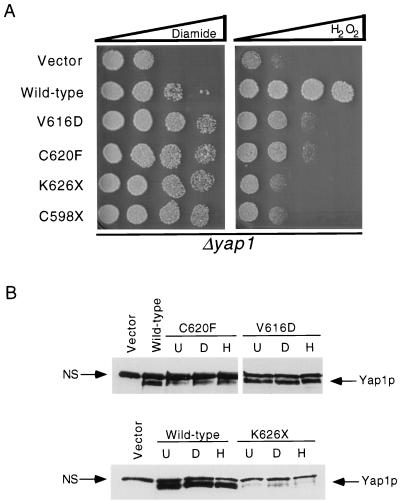
We recovered two distinctly different mutant classes of hyperactive Yap1p: nonsense mutations K626X and C598X and missense mutations V616D and C620F. All four of these mutant alleles led to high-level constitutive expression of the Yap1p-dependent reporter gene and markedly increased diamide resistance above that conferred by the wild-type protein (Fig. 3 and 4). Interestingly, these C-terminal mutant proteins failed to restore normal levels of H2O2 resistance to the Δyap1 strain. All these mutant proteins were detectable by Western blot analysis under control and oxidative stress conditions, indicating that their in vivo behavior could not be explained by a change in expression of the mutant factors relative to the wild-type protein. These data strongly support the idea that the function of the c-CRD of Yap1p varies depending on the oxidative challenge the cell is experiencing.
FIG. 3.
ARE-TRP5-lacZ expression supported by diamide-hyperresistant mutants with mutations in the c-CRD region. (A) The sequence of the C-terminal 53 amino acids of Yap1p is shown in the single-letter amino acid code. The numbers refer to the position of each amino acid along the 650-residue length of Yap1p, and the locations of the three CSE repeats are indicated by underlining. The highlighted region corresponds to a putative nuclear export signal as suggested previously (29). The position and sequence of each mutant analyzed here are indicated above the amino acid sequence. (B) The ability of each mutant to regulate gene expression was assayed by introducing low-copy-number plasmids expressing the indicated forms of Yap1p into a Δyap1 strain containing the ARE-TRP5-lacZ reporter gene, as described for Fig. 1. Levels of ARE-dependent β-galactosidase activity were determined in the unstressed cells (No Stress) and in cells subjected to diamide- or H2O2-induced oxidative stress.
FIG. 4.
Oxidative stress phenotypes and expression level of random mutants with mutations in the c-CRD. (A) Δyap1 cells were transformed with low-copy-number plasmids expressing the indicated forms of Yap1p. Transformants were assayed for their ability to tolerate diamide- or H2O2-induced stress by a spot test assay as described in the legend to Fig. 2. (B) Steady-state protein levels of the indicated Yap1p derivatives were determined by Western blotting with the rabbit anti-Yap1p antiserum as described in the legend to Fig. 2.
An N-terminal cysteine-rich domain regulates Yap1p function.
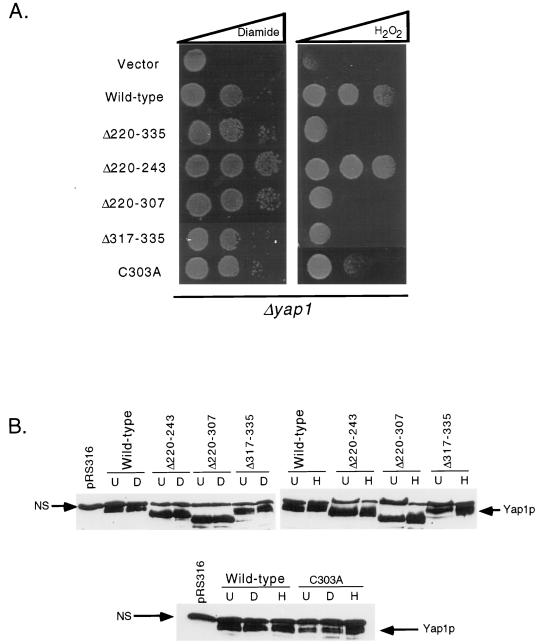
Analysis of an internal-deletion derivative of Yap1p that lacked residues 220 to 335 (Yap1p Δ220–335) indicated that this segment of the protein was required for normal regulation (24, 25). Loss of the region from residues 220 to 335 resulted in a Yap1p derivative that conferred hyperresistance to diamide but failed to provide normal H2O2 resistance. A striking feature of the region of Yap1p from residues 220 to 335 was the presence of the only other three cysteine residues in the protein. Since the cysteine residues in the c-CRD play important roles in regulation of Yap1p, we prepared a series of mutations to examine in more detail the possible role of these N-terminal cysteines in control of Yap1p function. Each mutant factor was expressed from a low-copy-number plasmid and assayed for the ability to complement both the ARE-TRP5-lacZ expression levels and oxidative stress phenotypes of a Δyap1 strain (Fig. 5 and 6).
FIG. 5.
The n-CRD is required for normal redox regulation. The extent of the Yap1p sequence deleted in each construct is indicated by the gap and by the numbers on the left-hand side of the drawing. The other symbols and labeling are as in Fig. 1. ARE-dependent β-galactosidase activity was determined for each construct as described in the legend to Fig. 1.
FIG. 6.
Oxidant resistance and expression profiles of n-CRD mutant proteins. Δyap1 cells were transformed with low-copy-number plasmids expressing the indicated forms of Yap1p. Oxidative stress phenotypes (A) and steady-state protein levels (B) were assayed as described in the legend to Fig. 2.
Three different deletion derivatives were prepared to explore the role of the three cysteine residues at positions 303, 310, and 315, respectively. A small truncation that removed residues 220 to 243 (Yap1p Δ220–243) while maintaining all the cysteines in the N terminus was generated. Yap1p Δ220–243 was strongly derepressed under nonstressed conditions and produced 52 U per optical density unit (A600) of β-galactosidase activity compared to only 7 U/A600 for the wild-type protein (Fig. 5). Yap1p Δ220–243 was still able to confer oxidant-inducible expression on the ARE-TRP5-lacZ fusion, albeit to a lesser extent than was the normal factor. The oxidant resistance profile (Fig. 6) of Yap1p Δ220–243 correlated well with the expression of the ARE reporter gene, with this mutant factor conferring hyperresistance to diamide and even modestly increasing tolerance to H2O2. These data supported the idea that the region of Yap1p from residues 220 to 243 normally acted to repress the activity of the protein.
Further deletion to position 307 generated a mutant regulatory protein (Yap1p Δ220–307) that lacked the most N-terminal of the six cysteine residues normally contained in this factor. This mutant factor behaved nearly indistinguishably from Yap1p Δ220–335, conferring increased diamide-dependent reporter gene expression and diamide tolerance, along with decreased expression in response to H2O2 and depressed tolerance of this oxidant. Finally, a truncation mutant that lacked sequences immediately downstream from the three N-terminal cysteine residues was analyzed. This mutant (Yap1p Δ317–335) behaved normally under both nonstressed and diamide-challenged conditions but failed to respond to H2O2 stress, even though all three amino-terminal cysteine residues were still present. Together, these data argue that this n-CRD is required for normal regulation of Yap1p by oxidative stress even if the c-CRD is fully intact.
Since each of the above-described deletion mutants lacked sequences around the cysteine residues of the n-CRD, we were unable to confirm if these particular amino acids were key players in the function of this N-terminal regulatory region. A site-directed mutation changing cysteine 303 to an alanine was prepared to examine the role played by this amino acid in the response of Yap1p to oxidant challenge. As with the deletion mutations, this substitution mutation was expressed from a low-copy-number plasmid and subjected to the same battery of assays for Yap1p function. Yap1p C303A behaved like the wild-type factor in the absence of stress and upon diamide stress. However, Yap1p C303A was unable to respond to H2O2 challenge, as assayed by either ARE-TRP5-lacZ expression or complementation of H2O2 sensitivity of Δyap1.
Genetic interaction of two negative regulatory domains in Yap1p.
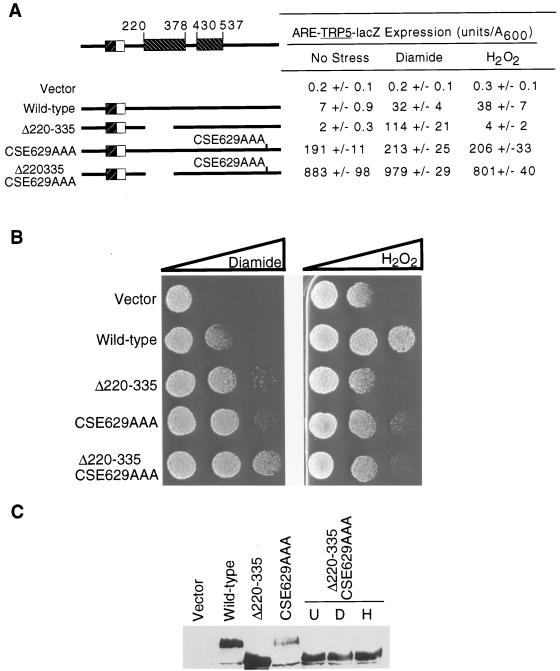
The above data strongly suggested that both the n-CRD and c-CRD are crucial for the normal response to oxidants. To evaluate if these two spatially separate regulatory domains control Yap1p activity through a common regulatory step, we constructed a double-mutant factor lacking the region from residues 220 to 335 and carrying the CSE629AAA allele. This double-mutant factor (Yap1p Δ220–335/CSE629AAA) was introduced into Δyap1 strains carrying the ARE-TRP5-lacZ reporter gene. Plasmids expressing the Δ220–335 or CSE629AAA forms of Yap1p alone were assayed as controls.
Yap1p Δ220–335/CSE629AAA behaved as a factor exhibiting the sum of the characteristics of each of the single mutants (Fig. 7). This double-mutant protein led to three important changes in the properties of Yap1p compared to the single Yap1p CSE629AAA derivative. First, the Yap1p Δ220–335/CSE629AAA exhibited increased basal β-galactosidase activity from the ARE-TRP5-lacZ. Second, the double mutant led to increased resistance to diamide. Third, the Yap1p Δ220–335/CSE629AAA protein accumulated to much higher levels than did the original Yap1p CSE629AAA mutant. Taken together, these data argue that Yap1p receives regulatory information through both its N- and C-terminal regions.
FIG. 7.
The n- and c-CRD regions contribute regulatory information to the redox response of Yap1p. (A) All Yap1p derivatives were introduced on low-copy-number plasmids into the Δyap1 ARE-TRP5-lacZ reporter strain and assayed for ARE-dependent β-galactosidase activity as described in the legend to Fig. 1. (B) Oxidative stress resistance phenotypes of Δyap1 cells carrying low-copy-number plasmids expressing the indicated forms of Yap1p were assayed as described in the legend to Fig. 2. (C) Steady-state protein levels of the double and single CRD mutants were analyzed by Western blotting. This blot was probed with affinity-purified anti-Yap1p antiserum that was depleted for antibodies that recognize the nonspecific protein species. The double CRD mutant was grown in the absence of stress (U) or subjected to oxidative stress by exposure to diamide (D) or H2O2 (H) prior to preparation of protein extracts.
Trafficking information for Yap1p is contained in both the n- and c-CRD regions.
Previous studies have demonstrated that Yap1p subcellular localization changes from primarily cytoplasmic in the absence of stress to nuclear when cells are challenged with either diamide or diethylmaleate (14). We wanted to examine the localization of Yap1p in response to mutations in the n-CRD and H2O2 challenge, two issues that have not been previously explored. To facilitate evaluation of the Yap1p subcellular location, we used PCR to insert the GFP after the ATG codon of the wild-type YAP1 gene. The resulting GFP-Yap1p chimera was carried on a low-copy-number plasmid. Mutant coding sequences were introduced into this fusion gene in vitro and then transformed into a Δyap1 strain carrying the ARE-TRP5-lacZ reporter gene.
Transformants were assayed for their oxidative stress resistance phenotypes, regulation of the ARE-TRP5-lacZ gene, and steady-state protein levels to ensure that the addition of the GFP moiety to the amino terminus did not alter the behavior of either the wild-type or mutant Yap1p derivatives that we constructed. This analysis indicated that the presence of the 30-kDa GFP domain on the amino terminus of these Yap1p derivatives did not alter their previously analyzed behavior (data not shown). We next assessed the subcellular distribution of the wild-type and mutant forms of Yap1p by fluorescence microscopy (Fig. 8).
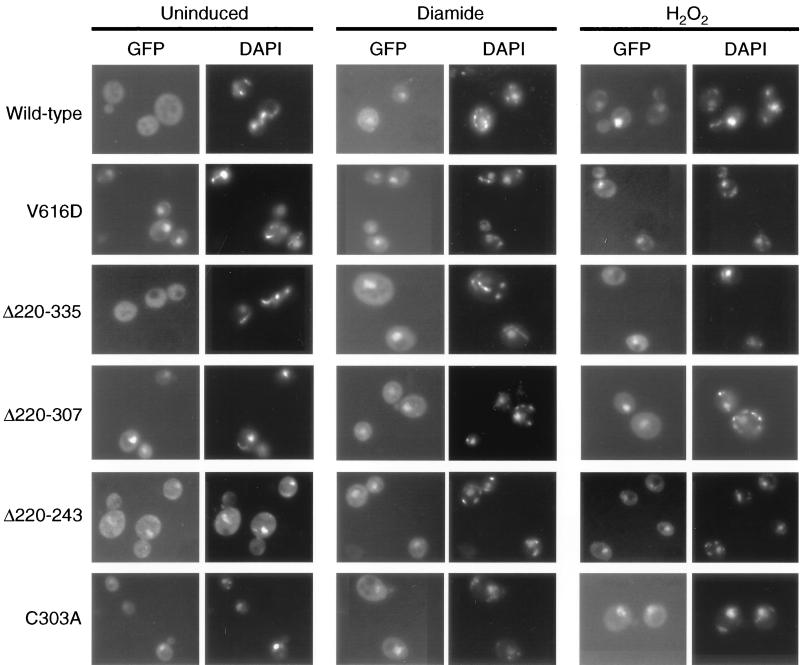
FIG. 8.
Oxidant-specific subcellular localization of Yap1p. Low-copy-number plasmids expressing the indicated alleles of YAP1 as GFP fusion proteins were introduced into Δyap1 cells. Transformants were grown under nonstressed conditions (Uninduced) or subjected to oxidative stress elicited by diamide or H2O2. Living cells were then stained with DAPI and analyzed by microscopy for Yap1p localization (GFP) and DAPI fluorescence.
Wild-type Yap1p is not found in the nucleus in the absence of stress but is clearly localized in this organelle in response to diamide challenge, as seen previously (14, 29). Importantly, H2O2-elicited oxidative stress also caused Yap1p to localize to the nucleus. Irrespective of the agent used to elicit oxidative stress, the subcellular localization of Yap1p changes in response to this stress. Yap1p V616D is constitutively localized in the nucleus, independent of diamide- or H2O2-induced oxidative stress. These data indicated that the failure of Yap1p V616D to normally complement the H2O2-hypersensitive defect of a Δyap1 strain is not due to a failure to localize to the nucleus when cells are treated with this oxidant. Localization of Yap1p C620F was indistinguishable from that of the V616D derivative (data not shown).
Along with these two c-CRD mutant proteins, subcellular localization of four different n-CRD mutant derivatives was examined. Yap1p Δ220–335 was previously found to be inducible by diamide- but not H2O2-elicited oxidative stress (24). One possible explanation for this behavior is the failure of this mutant protein to translocate to the nucleus in response to H2O2 treatment. Analysis of the ability of diamide and H2O2 to induce nuclear accumulation of the Yap1p Δ220–335 mutant showed that this mutant protein entered the nucleus under both conditions, indicating that the H2O2 defect of this factor was not due to defective trafficking. Two smaller n-CRD mutants (Yap1p Δ220–243 and Δ220–307) were also examined for their subcellular distribution. Yap1p Δ220–243 produced 700% more expression of the ARE-TRP5-lacZ relative to the wild-type Yap1p in the absence of oxidative stress (Fig. 5) and was constitutively localized in the nucleus, consistent with the high, oxidant-independent level of gene expression support by this mutant protein. A larger n-CRD deletion derivative lacking sequences from positions 220 to 307 was also constitutively localized to the nucleus but otherwise behaved similarly to the Yap1p Δ220–335 mutant. Note that even though the c-CRD was unaltered in both Yap1p Δ220–243 and Yap1p Δ220–307, these mutant factors were still present in the nucleus in the absence of oxidative stress.
Since these deletion derivatives might have a grossly disturbed protein structure that precludes normal functioning of the c-CRD, we assayed the subcellular localization of a single-amino-acid replacement form of Yap1p (Yap1p C303A) that changed a cysteine residue in the n-CRD to alanine. This mutant derivative was also constitutively localized in the nucleus in an oxidant-independent fashion. Yap1p Δ220–243, Yap1p Δ220–307, and Yap1p C303A have wild-type c-CRD regions but still exhibit constitutive nuclear localization. These data indicate that the n-CRD also contains targeting information that is required for normal functioning of Yap1p.
Differential activation of a TRX2-lacZ reporter gene in response to oxidant exposure.
One complicating feature of the behavior of several mutant Yap1p derivatives characterized in this work and previously (24) was the ability of some mutants (such as Yap1p K626X [Fig. 4]) to strongly activate ARE-TRP5-lacZ expression in the presence of H2O2 but their inability to confer even normal H2O2 resistance on a Δyap1 strain. One hypothesis to explain this discrepancy was that a unique action of the Yap1p C terminus was required at promoters involved in H2O2 tolerance but not at promoters of genes playing a role in diamide resistance. To directly test this idea, we analyzed the ability of several different mutant derivatives of Yap1p to stimulate the expression of a gene involved in H2O2 resistance, TRX2.
TRX2 has previously been shown to be regulated by Yap1p and to be required for resistance to H2O2 but not to diamide (13). A TRX2-lacZ fusion gene was constructed and integrated into the genome of a Δyap1 strain to produce the reporter strain YSC18. This TRX2-lacZ Δyap1 strain was then transformed with plasmids expressing different forms of Yap1p, and levels of TRX2-dependent β-galactosidase in the presence or absence of oxidative stress were determined (Table 1).
TABLE 1.
H2O2-mediated activation of the TRX2 gene requires normal Yap1p function
| YAP1 allelea |
TRX2-lacZ expression (U/A600) withb:
|
||
|---|---|---|---|
| No stress | Diamide | H2O2 | |
| Δyap1 | 3 ± 0.2 | 2 ± 0.3 | 3 ± 0.4 |
| YAP1 | 6 ± 0.5 | 32 ± 3 | 31 ± 1 |
| 2μm YAP1 | 40 ± 5 | 45 ± 12 | 52 ± 12 |
| CSE629AAA | 13 ± 0.4 | 21 ± 1 | 13 ± 1 |
| C629A | 9 ± 0.4 | 9 ± 1 | 8 ± 1 |
| V616D | 16 ± 1 | 19 ± 0.4 | 12 ± 0.3 |
| Δ627–3′UTR | 12 ± 0.6 | 21 ± 1 | 13 ± 1 |
| Δ220–243 | 6 ± 0.1 | 38 ± 1 | 40 ± 6 |
| Δ220–335 | 3 ± 0.1 | 17 ± 0.4 | 8 ± 1 |
| C303A | 4 ± 0.1 | 15 ± 1 | 6 ± 0.1 |
YSC18 cells (relevant genotype: ura3-52 yap1-Δ2::hisG TRX2-lacZ) were transformed with the indicated YAP1 alleles carried on plasmids. In all cases except for the 2μm YAP1 allele, these Yap1p derivatives were expressed from low-copy-number plasmids.
In the presence of a low-copy-number plasmid expressing wild-type Yap1p, TRX2-dependent expression changed from 6 U/A600 under nonstressed conditions to approximately 31 U/A600 when challenged with either diamide or H2O2. Overproduction of Yap1p from a 2μm plasmid led to high-level expression of TRX2, which was no longer responsive to oxidative stress. In the absence of an intact YAP1 structural gene, the TRX2 reporter gene produced low-level enzyme activity, which was not inducible by oxidative stress. These data are consistent with those reported previously (13, 16) and establish that this TRX2-lacZ gene faithfully reflected Yap1p regulation of the endogenous gene.
The expression of the TRX2-lacZ gene was then examined in the presence of an array of different mutant derivatives of Yap1p. Yap1p CSE629AAA was found to be a strong constitutive activator of ARE-TRP5-lacZ expression when challenged with either diamide or H2O2, but although this mutant protein conferred hyperresistance to diamide, it did not normally complement H2O2 tolerance in a Δyap1 background (24). In the absence of oxidative stress, TRX2-lacZ expression in the presence of the Yap1p CSE629AAA allele was elevated to 13 U/A600 compared to 6 U/A600 in the presence of the wild-type protein. However, Yap1p CSE629AAA was able to increase TRX2-lacZ expression to only 21 U/A600 when exposed to diamide and, importantly, completely failed to respond to H2O2 challenge. Two other C-terminal mutants (the C629A and V616D mutants) exhibited the same inability to increase TRX2-lacZ expression after H2O2 exposure, even though each of these mutant factors was able to confer diamide hyperresistance and high-level expression of the ARE-TRP5-lacZ fusion gene.
Along with these C-terminal mutant proteins, several N-terminal mutants were tested for their ability to elevate TRX2-lacZ expression during H2O2-induced oxidative stress. Yap1p Δ220–243 normally conferred H2O2 resistance and strongly induced the expression of the TRX2 reporter gene in the presence of H2O2. Two other N-terminal mutant proteins (Yap1p Δ220–335 and Yap1p C303A) that both failed to normally complement the H2O2-hypersensitive defect of the Δyap1 strain were also incapable of stimulating TRX2-lacZ expression. These data strongly support the idea that the function of Yap1p in response to oxidative stress elicited by H2O2 is not the same as that during diamide stress.
The TRX2 YRE is capable of responding to Yap1p C629A.
To examine if the failure of a c-CRD mutant form of Yap1p to elicit overproduction of TRX2 was due to some property of the YRE in TRX2, we examined the ability of a YRE in the TRX2 promoter to serve as a Yap1p-dependent upstream activator sequence of a heterologous promoter. An oligonucleotide that corresponds to the TRX2 YRE at position −181 in the promoter of this gene was synthesized. This oligonucleotide (YRETRX2) was placed upstream of a CYC1-lacZ promoter in place of the normal CYC1 upstream activator sequence elements. The resulting construct was then introduced into Δyap1 cells along with a low-copy-number plasmid expressing either the wild-type or C629A forms of Yap1p. The vector plasmid alone was included as a control. Transformants were grown in selective media and YRETRX2-dependent β-galactosidase activity determined as described above.
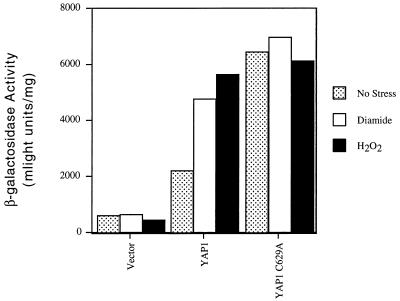
Expression of the YRETRX2-CYC1-lacZ fusion gene was constitutively high in the presence of the Yap1p C629A mutant (Fig. 9). Significantly, in the absence of stress, the expression level of the YRETRX2-CYC1-lacZ reporter gene was approximately 300% greater when the C629A form of Yap1p was present than when the wild-type factor was present. When the TRX2-lacZ was used as the reporter gene in this same comparison (Table 1), only a 50% increase in expression was seen when the C629A and wild-type forms of Yap1p were compared. This experiment suggests that the behavior of this TRX2 YRE is modified by the normal TRX2 promoter context in which it is embedded.
FIG. 9.
A TRX2 YRE is capable of responding to Yap1p C629A when assayed in a heterologous promoter context. An oligonucleotide corresponding to the YRE present at position −181 in the TRX2 promoter (YRETRX2) was inserted into a CYC1-lacZ fusion plasmid in place of the normal CYC1 upstream sequences. This YRETRX2-CYC1-lacZ fusion gene was carried on a low-copy-number plasmid and introduced into a Δyap1 strain along with a second low-copy-number plasmid containing the indicated forms of Yap1p. Transformants were grown and assayed for β-galactosidase activity by using a chemiluminescent substrate as described in Materials and Methods.
The −141 to −61 region of the TRX2 promoter blocks H2O2 induction in the absence of a normal c-CRD region of Yap1p.
To map the region of the TRX2 promoter that requires the function of the Yap1p c-CRD for induction, we fused various segments of the 5′-flanking DNA of TRX2 to a CYC1-lacZ fusion gene lacking its normal upstream activator sequences. These TRX2-CYC1-lacZ fusion genes were introduced into a Δyap1 strain along with a low-copy-number plasmid vector expressing wild-type or C629A mutant forms of Yap1p. β-Galactosidase activities under unstressed and H2O2-challenged conditions were then determined (Fig. 10).
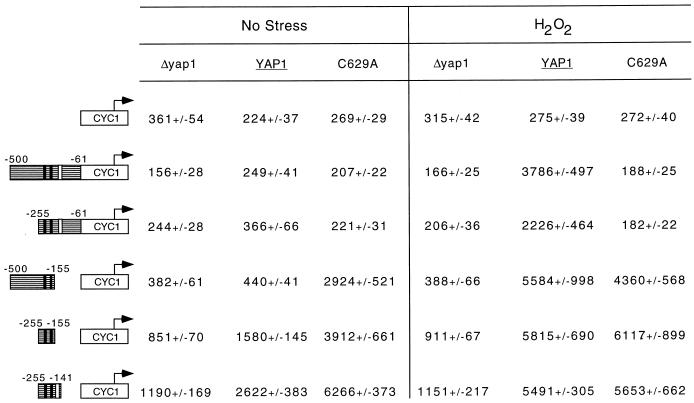
FIG. 10.
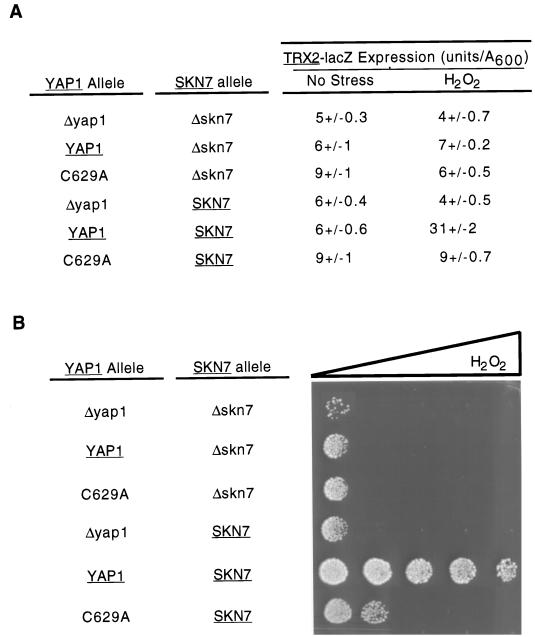
An intact Yap1p c-CRD region is required for normal induction of the TRX2 promoter during H2O2 challenge. Cells lacking the YAP1 structural gene were transformed with a low-copy-number plasmid vector (Δyap1) or the same vector expressing the wild-type or C629A form of Yap1p. Along with these three alleles of YAP1, various fusion genes carrying the indicated segments of the TRX2 promoter region placed upstream of a CYC1-lacZ fusion gene were introduced. The numbers are relative to the start of the TRX2 coding sequence. The solid vertical bars indicate the positions of the two YREs, and the single open box denotes the location of the Skn7p binding site. β-Galactosidase activities were determined by using a chemiluminescent substrate as above.
Fusion genes containing TRX2 upstream DNA from either −500 to −61 or −255 to −61 exhibited very similar behavior. Both these reporter plasmids supported strong H2O2 induction in the presence of wild-type YAP1 but were not induced when the C629A form of Yap1p was expressed. Both of these chimeric TRX2-CYC1-lacZ fusion genes exhibited the same dependence of H2O2 induction on the intact Yap1p c-CRD as was seen for the wild-type TRX2 gene. Two further deletion derivatives that contained TRX2 promoter DNA from −500 to −155 or −255 to −155 were constructed. These two constructs were now able to respond to the presence of the C629A derivative of Yap1p. Even in the absence of H2O2 exposure, the mutant TRX2-CYC1-lacZ fusion genes lacking the −155 to −61 region of TRX2 produced high levels of β-galactosidase activity, suggesting that this portion of TRX2 was responsible for the unique requirement of this gene for the Yap1p c-CRD.
Previous work on the TRX2 promoter demonstrated that a binding site for the transcription factor Skn7p was located between positions −164 and −142 (16). To determine if interaction between Yap1p and Skn7p was involved in the dependence of TRX2 H2O2 activation on the Yap1p c-CRD, two additional experiments were performed. First, a TRX2-CYC1-lacZ fusion gene that contained the Skn7p binding site and extended from −255 to −141 was constructed. Second, a Δskn7 yap1 strain was prepared and transformed with a plasmid expressing wild-type or C629A Yap1p or the empty vector. These transformants were then tested for expression of a TRX2-lacZ reporter gene and H2O2 tolerance.
Inclusion of the Skn7p binding site in the −255 to −141 TRX2-CYC1-lacZ reporter plasmid did not interfere with the ability of the TRX2 promoter segment to support high-level expression in the presence of the C629A form of Yap1p (Fig. 11). This behavior was also seen in the context of the wild-type TRX2 promoter (Table 1). Irrespective of the presence of SKN7, C629A Yap1p was not able to induce TRX2 expression upon H2O2 exposure. Consistent with the results of others (16), loss of either YAP1 or SKN7 blocked the ability of the TRX2-lacZ gene to be induced by H2O2. Phenotypic assays of these strains on H2O2 gradient plates mirrored this defect in H2O2 activation of TRX2 gene expression. Interestingly, Yap1p C629A was able to slightly increase the H2O2 tolerance of a Δyap1 cell but only to a fraction of the resistance shown by a wild-type strain. We conclude from these data that the failure of TRX2 to respond to Yap1p C629A is not dependent on Skn7p and can be rescued by removal of the DNA region between positions −141 and −61.
FIG. 11.
A hyperactive YAP1 allele cannot bypass the Skn7p requirement of H2O2 activation of the TRX2 promoter. A low-copy-number plasmid expressing the indicated forms of YAP1 was introduced into Δyap1 skn7 or Δyap1 cells along with a TRX2-lacZ gene fusion. TRX2-dependent β-galactosidase activity was determined under nonstressed (No Stress) or H2O2-challenged conditions. (B) Aliquots of 1,000 cells of each of the transformants described in panel A were placed on YPD containing a gradient of H2O2 increasing from left to right.
DISCUSSION
Control of the activity of the positive transcriptional regulatory protein Yap1p is one of the key steps modulating the response of S. cerevisiae to oxidative stress. While the action of Yap1p as a positive regulator of gene expression is required for normal tolerance to the oxidants diamide and H2O2, the responses of Yap1p to these oxidants exhibit important differences. These differential responses are most clearly seen in the analysis of mutant forms of Yap1p. A number of lesions that either perturb (C629A) or remove (K626X) the c-CRD in Yap1p produce derivatives that confer hyperresistance to diamide yet fail to provide normal H2O2 tolerance to a Δyap1 strain. These findings strongly suggest that the role of the Yap1p c-CRD is different in the face of the distinct oxidative challenge elicited by either diamide or H2O2. Diamide is believed to shift the pool of glutathione into a predominantly oxidized state (12), while H2O2 is a promiscuous oxidizing agent that acts to damage many different macromolecules in the cell (2). Since the modes of oxidative damage induced by these two agents are not identical, the cellular responses to these stresses are likely to be different. At least part of this selective cellular response to these oxidants is programmed into the oxidant-specific behavior of Yap1p.
The oxidant-specific behavior of the Yap1p c-CRD has important implications for the physiological action of this domain of the protein. Previous work has shown that the c-CRD is involved in the control of Yap1p nuclear localization (14), probably at the level of nuclear export (29). These studies led to the proposal that the Yap1p c-CRD acted to enhance nuclear export in the absence of oxidants but that Yap1p would accumulate in the nucleus upon oxidant challenge (14, 29). Our findings indicate that while c-CRD-regulated localization of Yap1p may be a role of this domain of the factor, it clearly is not the sole activity of this region.
In addition to the already recognized role of the c-CRD in regulation of Yap1p activity, we provide evidence that the amino terminus of Yap1p is also required for the appropriate redox response of the factor. Importantly, the n-CRD also contains information that is required for normal trafficking of Yap1p. Loss of small segments of Yap1p (residues 220 to 243 or 220 to 307) or a single-amino-acid substitution in the n-CRD leads to constitutive nuclear localization of the resulting mutant protein. However, a larger deletion mutant (Yap1p Δ220–335) is normally excluded from the nucleus in the absence of oxidative challenge. These data suggest that the n-CRD contains nuclear targeting information that is normally masked in the absence of oxidative stress. Loss of the sequences between positions 220 and 243 or the C303A mutation unmasks this targeting determinant, and the mutant proteins are found in the nucleus. A larger deletion mutant lacking residues 220 to 335 is found in the nucleus only upon oxidant challenge. This derivative (Yap1p Δ220–335) may lack the targeting information that was unmasked by the two more amino-terminal mutations.
Along with this newly identified role in redox-dependent trafficking of Yap1p, the n-CRD is a key contributor to the ability of the protein to activate transcription of a subset of downstream target genes. A wild-type n-CRD is essential for normal H2O2 tolerance but dispensable for diamide resistance. This finding suggests that genes involved in H2O2 resistance require both CRD regions to be normally activated by Yap1p but that genes required for diamide resistance do not. Previous mutagenesis experiments have identified other substitution mutations in both CRD regions that confer an H2O2 sensitive phenotype on cells and prevent normal activation of TRX2 expression upon H2O2 challenge (23). Interestingly, loss of the n-CRD leads to a H2O2-specific defect in the ability of the resulting mutant Yap1p to activate both the ARE-TRP5-lacZ and the TRX2-lacZ reporter genes whereas c-CRD mutations prevent induction only of TRX2 transcription in the presence of H2O2. These data suggest that Yap1p-mediated transactivation requires normal n-CRD function at all promoters but that c-CRD function is specifically needed to induce expression of genes involved in H2O2 tolerance.
To determine the possible regulatory relationship of these two domains of Yap1p, we constructed a mutant derivative that lacked both the n-CRD (Yap1p Δ220–335) and c-CRD (Yap1p CSE629AAA). This was done to determine if the n-CRD could still influence Yap1p function in the absence of c-CRD function. One possible model explaining the roles of the two CRD regions would be that the n-CRD acts simply to modulate c-CRD function. In the absence of a functional c-CRD, n-CRD mutations would not be expected to influence function of the resulting Yap1p derivative. Contrary to this prediction, our results suggest that both CRD regions play critical roles in Yap1p function and regulation. The mutant lacking both the n-CRD and c-CRD regions (Yap1p Δ220–335/CSE629AAA) is more active than either single mutant. Both of the CRD regions act to repress the factor during conditions of diamide stress, and both are required for Yap1p to normally mediate resistance to H2O2 stress.
The analysis of the c-CRD was complicated by the observation that most c-CRD mutants behaved as constitutive activators of the ARE-TRP5-lacZ reporter gene but failed to normally complement the H2O2 hypersensitivity of the Δyap1 mutant. Our finding that the same c-CRD mutants that drive greater expression of the ARE-TRP5-lacZ reporter than wild-type Yap1p fail to properly regulate transcription of the TRX2 gene clarifies this apparent paradox. The ARE-TRP5-lacZ reporter serves as a model promoter for the response of isolated YREs to Yap1p. This synthetic promoter accurately predicts the diamide resistance phenotype of all Yap1p mutant derivatives that we have analyzed. Yap1p mutants that drive high-level expression of ARE-TRP5-lacZ also confer a strong diamide resistance phenotype on Δyap1 cells. However, this artificial reporter fails to predict the corresponding H2O2 resistance phenotype of the same mutants. When c-CRD mutants are analyzed for their ability to induce transcription of TRX2 in response to H2O2, a marked defect is observed (Table 1).
We believe that these observations suggest that the mode of transactivation of Yap1p at promoters involved in diamide tolerance is different from that at promoters involved in H2O2 resistance. Transcriptional activation of genes involved in diamide tolerance seems to be able to be reduced to the behavior of an isolated YRE. Up-regulation of genes required for H2O2 resistance is more complex and must be examined in the context of the native promoter. This is illustrated by the analysis of the H2O2 induction of the TRX2 promoter by Yap1p. A mutant Yap1p lacking an intact c-CRD region cannot induce TRX2 in the presence of H2O2 unless a region of the TRX2 promoter is removed. The presence of this inhibitory region of TRX2 also prevents the otherwise hyperactive C629A form of Yap1p from driving high-level constitutive expression of TRX2. These data suggest the presence of a binding site for a factor that interacts with the Yap1p c-CRD before activation of TRX2 can occur. This behavior is a unique feature of TRX2, since other Yap1p target genes exhibit dramatic increases in expression if the c-CRD is defective (1, 23). Current work focuses on identification of the factor binding this TRX2 inhibitory site.
Finally, it is important to contrast the physiological significance of tolerance to either diamide or H2O2. While both of these oxidants are useful tools to elicit oxidative stress in the laboratory, H2O2 is arguably the more relevant phenotype to S. cerevisiae cells in particular and fungi in general. Neutrophils, a major host defense against fungal infections, eliminate invading fungal cells through oxidative killing (reviewed in reference 17). While there are several different redox-active chemicals that can be produced by neutrophils, a major oxidant is H2O2 (11). Understanding how fungal cells tolerate H2O2 toxicity will have important implications in the treatment of pathogenic fungal infections.
ACKNOWLEDGMENTS
This work was supported in part by NIH grants GM49825 to W.S.M. and DK25295 to the University of Iowa Diabetes and Endocrinology Center. W.S.M. is an Established Investigator of the American Heart Association. Sean Coleman was the recipient of an NIH predoctoral fellowship (NIH/NIA T32 AG00214).
We thank Chris Grant for helpful discussions and Laura Davis for sharing data prior to publication.
REFERENCES
- 1.Coleman S T, Tseng E, Moye-Rowley W S. Saccharomyces cerevisiae basic region-leucine zipper protein regulatory networks converge at the ATR1 structural gene. J Biol Chem. 1997;272:23224–23230. doi: 10.1074/jbc.272.37.23224. [DOI] [PubMed] [Google Scholar]
- 2.Collinson L P, Dawes I W. Inducibility of the response of yeast cells to peroxide stress. J Gen Microbiol. 1992;138:329–335. doi: 10.1099/00221287-138-2-329. [DOI] [PubMed] [Google Scholar]
- 3.Emr S D, Vassarotti A, Garret J, Geller B C, Takeda M, Douglas M G. The amino terminus of the yeast F1-ATPase beta-subunit precursor functions as a mitochondrial import signal. J Cell Biol. 1986;102:523–533. doi: 10.1083/jcb.102.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant C M, Collinson L P, Roe J-H, Dawes I W. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol Microbiol. 1996;21:739–746. doi: 10.1046/j.1365-2958.1996.6351340.x. [DOI] [PubMed] [Google Scholar]
- 5.Grant C M, MacIver F H, Dawes I W. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr Genet. 1996;29:511–515. doi: 10.1007/BF02426954. [DOI] [PubMed] [Google Scholar]
- 6.Guarente L. Yeast promoter and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 7.Harris E D. Regulation of antioxidant enzymes. FASEB J. 1992;6:2675–2683. doi: 10.1096/fasebj.6.9.1612291. [DOI] [PubMed] [Google Scholar]
- 8.Harshman K D, Moye-Rowley W S, Parker C S. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell. 1988;53:321–330. doi: 10.1016/0092-8674(88)90393-5. [DOI] [PubMed] [Google Scholar]
- 9.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jamieson D J. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 11.Klebanoff S J. Oxygen metabolism and the toxic properties of phagocytes. Ann Intern Med. 1980;93:480–489. doi: 10.7326/0003-4819-93-3-480. [DOI] [PubMed] [Google Scholar]
- 12.Kosower N S, Kosower E M. Formation of disulfides with diamide. Methods Enzymol. 1987;143:264–270. doi: 10.1016/0076-6879(87)43050-4. [DOI] [PubMed] [Google Scholar]
- 13.Kuge S, Jones N. YAP1-dependent activation of TRX2 is essential for the response of S. cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moradas-Ferreira P, Costa V, Piper P, Mager W. The molecular defenses against reactive oxygen species in yeast. Mol Microbiol. 1996;19:651–658. doi: 10.1046/j.1365-2958.1996.403940.x. [DOI] [PubMed] [Google Scholar]
- 16.Morgan B A, Banks G R, Toone W M, Raitt D, Kuge S, Johnston L H. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997;16:1035–1044. doi: 10.1093/emboj/16.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy J W, Friedman H, Bendinelli M. Fungal infections and immune responses. New York, N.Y: Plenum Press; 1993. [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 19.Schnell N, Krems B, Entian K-D. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homolog, is involved in oxygen metabolism. Curr Genet. 1992;21:269–273. doi: 10.1007/BF00351681. [DOI] [PubMed] [Google Scholar]
- 20.Sherman F, Fink G, Hicks J. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1979. [Google Scholar]
- 21.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Y, Oberly L W. Redox regulation of transcriptional activators. Free Radic Biol Med. 1996;21:335–348. doi: 10.1016/0891-5849(96)00109-8. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi T, Miyahara K, Hirata D, Miyakawa T. Mutational analysis of Yap1 protein, an AP-1-like transcriptional activator of Saccharomyces cerevisiae. FEBS Lett. 1997;416:339–343. doi: 10.1016/s0014-5793(97)01233-7. [DOI] [PubMed] [Google Scholar]
- 24.Wemmie J A, Steggerda S M, Moye-Rowley W S. The Saccharomyces cerevisiae AP-1 protein discriminates between oxidative stress elicited by the oxidants H2O2 and diamide. J Biol Chem. 1997;272:7908–7914. doi: 10.1074/jbc.272.12.7908. [DOI] [PubMed] [Google Scholar]
- 25.Wemmie J A, Wu A-L, Harshman K D, Parker C S, Moye-Rowley W S. Transcriptional activation mediated by the yeast AP-1 protein is required for normal cadmium tolerance. J Biol Chem. 1994;269:14690–14697. [PubMed] [Google Scholar]
- 26.Wolff S P, Garner A, Dean R T. Free radicals, lipids and protein degradation. Trends Biochem Sci. 1986;11:27–31. [Google Scholar]
- 27.Wu A, Moye-Rowley W S. GSH1, which encodes δ-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol Cell Biol. 1994;14:5832–5839. doi: 10.1128/mcb.14.9.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu A, Wemmie J A, Edgington N P, Goebl M, Guevara J L, Moye-Rowley W S. Yeast bZIP proteins mediate pleiotropic drug and metal resistance. J Biol Chem. 1993;268:18850–18858. [PubMed] [Google Scholar]
- 29.Yan C, Lee L H, Davis L I. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 1998;17:7416–7429. doi: 10.1093/emboj/17.24.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yon J, Fried M. Precise gene fusion by PCR. Nucleic Acids Res. 1989;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]