Abstract
Transitions from environmental sex determination (ESD) to genotypic sex determination (GSD) require an intermediate step of sex reversal, i.e., the production of individuals with a mismatch between the ancestral genotypic and the phenotypic sex. Among amniotes, the sole well-documented transition in this direction was shown in the laboratory in the central bearded dragon, Pogona vitticeps, where very high incubation temperatures led to the production of females with the male-typical (ZZ) genotype. These sex-reversed females then produced offspring whose sex depended on the incubation temperature. Sex-reversed animals identified by molecular and cytogenetic markers were also reported in the field, and their increasing incidence was speculated as a climate warming-driven transition in sex determination. We show that the molecular and cytogenetic markers normally sex-linked in P. vitticeps are also sex-linked in P. henrylawsoni and P. minor, which points to quite ancient sex chromosomes in this lineage. Nevertheless, we demonstrate, based on a crossing experiment with a male bearded dragon who possesses a mismatch between phenotypic sex and genotype, that the used cytogenetic and molecular markers might not be reliable for the identification of sex reversal. Sex reversal should not be considered as the only mechanism causing a mismatch between genetic sex-linked markers and phenotypic sex, which can emerge also by other processes, here most likely by a rare recombination between regions of sex chromosomes which are normally sex-linked. We warn that sex-linked, even apparently for a long evolutionary time, and sex-specific molecular and cytogenetic markers are not a reliable tool for the identification of sex-reversed individuals in a population and that sex reversal has to be verified by other approaches, particularly by observation of the sex ratio of the progeny.
Keywords: Molecular markers, Reversal, Sex chromosomes, Sex linkage, Vertebrates
Introduction
Amniotes possess a large variability in sex determination. According to comparative studies, genotypic sex determination (GSD) and thus sex chromosomes evolved within amniotes independently around 40 times [Johnson Pokorná and Kratochvíl, 2016; Rovatsos et al., 2016, 2019a]. Some species − among amniotes forming a minority − rely on environmental sex determination (ESD), where sexes do not differ in their genotypes and the sex of an individual is set by environmental conditions during a sensitive developmental period. ESD represents a special case of polyphenism. Once emerged, GSD seems to be very evolutionarily stable at least in some lineages of amniotes, as demonstrated by the stability of sex chromosomes in mammals [Waters et al., 2007; Cortez et al., 2014], birds [Shetty et al., 1999; Zhou et al., 2014], softshell turtles [Rovatsos et al., 2017], iguanas [Gamble et al., 2014; Rovatsos et al., 2014a, b; Altmanová et al., 2018], caenophidian snakes [Rovatsos et al., 2015a], lacertids [Rovatsos et al., 2016, 2019b], varanid lizards [Iannucci et al., 2019; Rovatsos et al., 2019c], and skinks [Kostmann et al., 2021]. While independent evolution of GSD within amniotes is well supported by its phylogenetic distribution and genomic evidence for nonhomology of sex chromosomes among particular amniote lineages (although some lineages co-opted the same genomic regions for the function of sex chromosomes) [Rovatsos et al., 2019a; Straková et al., 2020], independent origins of ESD from the ancestral GSD are still controversial. Some authors propose that transitions in this direction might be rapid and common [Janzen and Paukstis, 1991; Barske and Capel, 2008; Marshall Graves, 2008; Sarre et al., 2011; Ezaz et al., 2017; Pennel et al., 2018]. However, many of the reported GSD to ESD transitions suggested within lacertids, skinks, varanids, and chameleons appeared to be based on the erroneous assignment of GSD species as ESD [Rovatsos et al., 2015b; Hill et al., 2018; Nielsen et al., 2018; Iannucci et al., 2019; Kostmann et al., 2021]. Other authors question the frequent transitions from GSD to ESD based on the phylogenetic distribution of ESD clades and general evolutionary stability of GSD [Pokorná and Kratochvíl, 2009; Gamble et al., 2015; Johnson Pokorná and Kratochvíl, 2016; Straková et al., 2020].
A transition from GSD to ESD requires an intermediate step of sex reversal, i.e., the production of individuals with a mismatch between the ancestral genotypic and the actual phenotypic (gonadal) sex. Such a transition from the ancestral GSD to ESD was unequivocally demonstrated only in the laboratory in the central bearded dragon, Pogona vitticeps [Ahl, 1926] [Holleley et al., 2015]. Sex in this species is determined by female heterogamety (ZZ/ZW sex chromosomes), where the W sex chromosome is cytogenetically easily recognizable due to a large heterochromatic block containing an accumulation of repeats with the AAGG motif [Ezaz et al., 2005; Matsubara et al., 2016]. However, high constant incubation temperatures starting above 32°C feminized genotypic males, i.e., led to the production of females with the male-typical ZZ genotype [Quinn et al., 2007; Holleley et al., 2015]. In the next generation, such sex-reversed females mated with normal ZZ males produced offspring exclusively with the ZZ genotype whose sex depended on the incubation temperature. These laboratory studies demonstrated that the cytogenetically recognizable sex chromosome (here the W) can be lost in a single generation, and therefore, a short time is potentially needed for GSD to ESD transitions [Holleley et al., 2015]. At a population level, the derived ESD could then potentially sweep out GSD and become fixed in a population. Sex-reversed animals identified by molecular and cytogenetic markers were reported in the central bearded dragon also in the field [Holleley et al., 2015; Castelli et al., 2020], and their higher incidence in recent years is thought to be in connection with global warming [Holleley et al., 2015]. While the scenario of a climate change-driven transition in sex determination in the central bearded dragon sounds theoretically appealing, here we show that both cytogenetic and molecular genetic markers used for the demonstration of the occurrence of sex reversal in the field in this species are not sufficient for detection of sex reversals, hence evidence for sex-reversed animals in wild populations is not very strong. More generally, we investigated what evidence is needed for the identification of sex reversal, which has a profound influence on our understanding of the stability of sex determination in nature and points to limitations of the commonly used approaches based on genetic markers.
Materials and Methods
The central bearded dragon is an agamid lizard native to arid central Australia. It reaches approximately 40–60 cm in total length and has a body mass of around 200–500 g. Females typically lay 15–40 eggs per clutch. This species became a popular pet worldwide [Cogger, 1992; De Vosjoli et al., 2016]. During a genetic screening of captive bred central bearded dragons and the previous experiment with sex-reversed individuals [Ehl et al., 2017], we uncovered a male with a mismatch between the phenotypic sex and the genotype: it possessed a female-specific allele in the locus assigned as 'contig C' by Quinn et al. [[2010]], serving as a sex-specific PCR marker previously used for detection of sex-reversed individuals in this species [Quinn et al., 2010; Holleley et al., 2015; Ehl et al., 2017; Castelli et al., 2020]. The W-specific fragment represents an anonymous sequence which differs in 4 single nucleotide polymorphisms (SNPs) from the homologous Z-linked locus [Quinn et al., 2010]. We hypothesized that this individual could represent female-to-male sex reversal and tested this possibility by experimental crosses to uncover inheritance of this putative sex-specific locus. Furthermore, as the W chromosome is well defined in this species and cytogenetics was also used for determination of sex-reversed individuals [Ezaz et al., 2005; Holleley et al., 2015], we performed cytogenetic analyses and determined how the molecular and cytogenetic markers correspond to the phenotypic sex. Additionally, 2 pairs of P. henrylawsoni (PHE) and 1 pair of P. minor (PMI) were analysed as an outgroup.
Experimental Crosses
We mated the male with the PCR W-linked fragment (the genotype assigned further as ZWm, Wm indicates male W-like chromosome) with 3 unrelated females. One of the females possessed the typical ZW genotype, the other 2 females were sex reversed (ZZ sex chromosomes) by the high incubation temperature of 35.5°C [Ehl et al., 2017]. Eggs from these crosses were incubated at the constant temperature of 28°C, the standard incubation temperature which does not lead to thermally-induced sex reversal [Quinn et al., 2010; Holleley et al., 2015; Ehl et al., 2017]. These experimental crosses were done to elucidate the potential effect of the Wm in sex determination and viability of the progeny. Predictions on the sex ratio in the cross between the ZWm male and the ZW female depend on the viability of WWm individuals. In lineages with a degenerated W chromosome, the WW genotype is lethal [Harada and Buss 1981; Watts et al., 2006], while poorly differentiated sex chromosomes allow viable WW offspring [Roco et al., 2015]. Although sex chromosomes in P. vitticeps are cytogenetically easily distinguishable, no Z-specific gene (i.e., a gene missing on the W chromosome) was identified yet suggesting that its Z and W sex chromosomes do not differ substantially in gene content [Georges et al., 2015; Deakin et al., 2016]. Gonadal phenotype in ZWm offspring should differentiate between sex reversal (ZWm offspring incubated at the standard, non-reverting temperature should be females) versus its alternatives (the Wm chromosome does not participate in sex determination as the W and offspring should be ZWm males).
The schematic overview of the experimental crosses in P. vitticeps is presented in online supplementary Figure 1 (for all online suppl. material, see www.karger.com/doi/10.1159/514195).
PCR Genotyping
Approximately 200 μL of blood was collected from the ventral tail vein for cytogenetic approaches and DNA isolation. In individuals who died during/early after hatching the tail tip was collected instead. DNA was extracted using DNeasy Blood & Tissue kit (Qiagen) according to the manufacturer's instructions. For PCR genotyping, we used the protocol by Quinn et al. [[2010]]. This duplex PCR was designed to simultaneously amplify the W-specific fragment present only in females (224 bp, primers F4-F1) and an additional fragment present in both sexes (963 bp, primers E-C) as a positive control.
The W-specific PCR products from 4 individuals were sequenced in order to check the amplification of specific sequences and the presence of female-specific SNPs. The PCR products were purified with QIAquick PCR Purification Kit (Qiagen). Sequencing reactions were performed with BigDye Terminator Cycle Sequencing Kit 1.1 (Applied Biosystems) according to the manufacturer's protocol. Sequence products were purified using DyeEx 2.0 Spin Kit (Qiagen) and sequenced bi-directionally on an Applied Biosystems 3130 Genetic Analyser using 30 cm long capillaries and POP-7 polymer. The obtained sequences were assembled and checked by eye using the SeqMan II version 5.05 module of the Lesergene software package (DNASTAR). Obtained sequences were checked by BLASTN search [Altschul et al., 1990] for similarity with sequences already existing in GenBank. Consensus sequences were aligned in Sequencher 4.1.4 (Gene Codes Corporation) together with reference sequences from the previous study of Quinn et al. [[2010]].
Cytogenetic Analyses
According to previous cytogenetic studies, Z and W are microchromosomes and the W chromosome is highly heterochromatic [Ezaz et al., 2005] and possesses notable accumulations of the AAGG repetitive motif [Matsubara et al., 2016]. Therefore, we applied C-banding visualizing heterochromatin [Sumner, 1972] and FISH with the (AAGG)8 motif as a probe [for detailed methodology see Altmanová et al., 2016; Suwala et al., 2020] to chromosome spreads from 22 individuals representing all experimental groups: parents and offspring of all genotypic combinations (summarized in online suppl. Table 1). Chromosome spreads were obtained by cultivation of whole blood according to our standard protocol [Pokorná et al., 2014]. In all cytogenetically tested individuals (online suppl. Tables 1, 2), we scored at least 10 metaphases per each individual and method.
Phenotypic Sexing
We determined the sex of the experimental animals by gonadal inspection, breeding history, and/or external morphology (enlarged hemipenes). The phenotypic sex can be determined by the presence of an enlarged hemipenes in males. However, according to our experience, the determination of the phenotypic sex based on external morphology is reliable only in older animals with a larger body size. Therefore, we consider determination of sex based on external morphology as reliable only in subadult animals above 200 g. The phenotypic sex determined by external morphology was verified by dissection and examination of gonad morphology in 10 individuals from the ZW female × ZWm male cross (5 males, 5 females).
Results
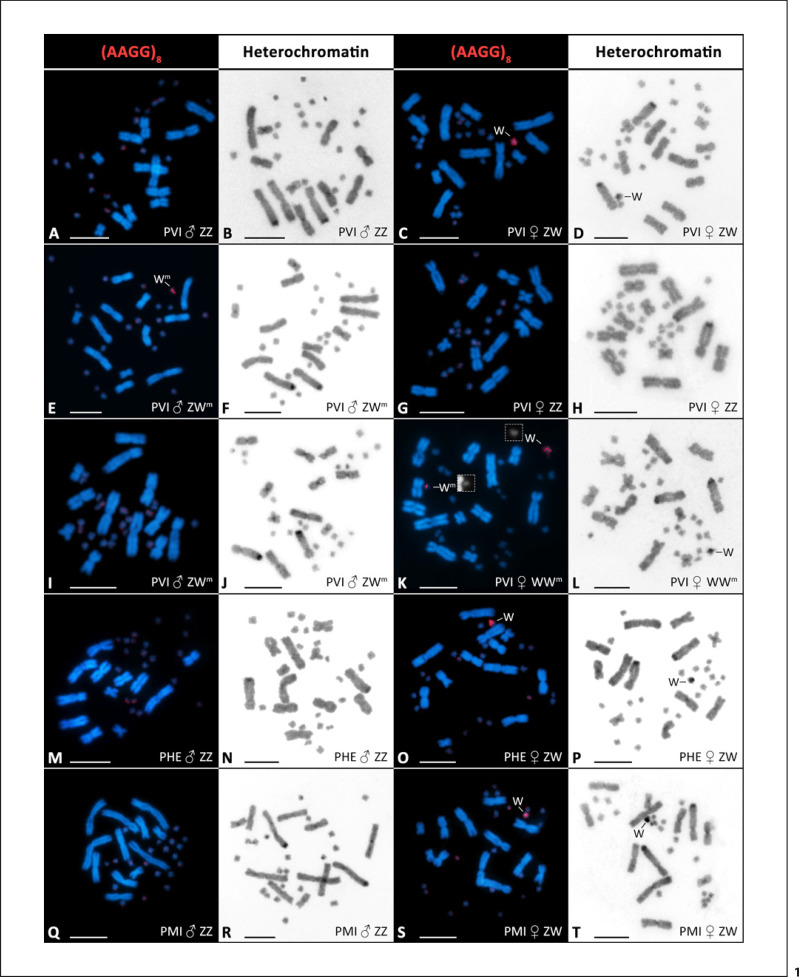
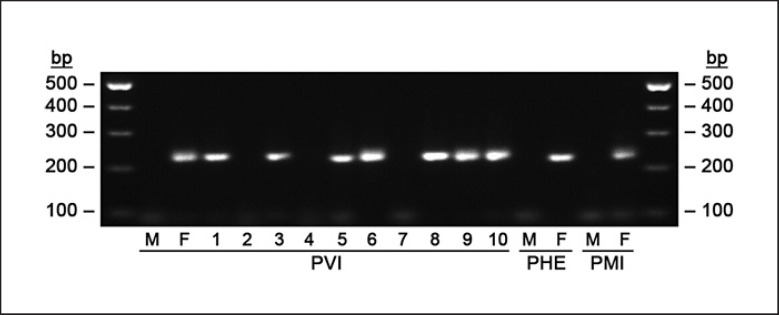
All 22 karyotyped individuals of P. vitticeps (listed in online suppl. Table 1) had 32 chromosomes, consisting of 12 macrochromosomes and 20 microchromosomes, which is congruent with previous studies [Witten, 1983; Ezaz et al., 2005]. We did not observe aneuploidy or chromosomal abnormalities in any tested individual. C-banding revealed heterochromatin in the centromeres of macrochromosomes and at a distal part of the second largest chromosome pair in all individuals, confirming that the method was applied successfully (Fig. 1; online suppl. Fig. 2). A notable heterochromatic accumulation lacking in a control, standard male (Fig. 1B) was observed in a control female on the W chromosome (Fig. 1D) as reported previously by Ezaz et al. [[2005]]. FISH with the probe containing the AAGG repetitive motif detected a weak accumulation of this motif on several microchromosome pairs [similarly to Holleley et al., 2015] and a strong accumulation on the W chromosome (Fig. 1C). This accumulation was absent in a control male (Fig. 1A). As expected, the female-specific PCR marker was amplified only in the standard female and not in the male (Fig. 2). Similarly to P. vitticeps, P. henrylawsoni (2 males and 2 females examined; the results of only 1 pair are shown in Fig. 1M–P) and P. minor (1 male and 1 female; Fig. 1Q–T) possessed 2n = 32 chromosomes. Also heterochromatin and accumulations of the AAGG repetitive motifs were distributed in the same way as in P. vitticeps with apparently strong accumulation on the W chromosomes of females (Fig. 1O, S). The female-specific PCR marker was also amplified only in females of these 2 species (Fig. 2). The results of all studied individuals of P. henrylawsoni and P. minor are summarized in online suppl. Table 2. None of the ZZ females of P. vitticeps which were sex-reversed by high temperature exhibited the female-specific cytogenetic markers (heterochromatic block and AAGG accumulation, Fig. 1G, H'; online suppl. Fig. 2E–H and Table 1) or the female-specific PCR marker (Fig. 2). The male with the W-specific PCR fragment (Fig. 2) lacked the female-specific heterochromatic block (Fig. 1F) but possessed the otherwise female-specific AAGG accumulation, although under closer examination the signal was fainter than in females and the chromosome bearing it was notably smaller than the W chromosome (Fig. 1E).
Fig. 1.
Visualization of the accumulation of AAGG repeats and heterochromatin in selected individuals of P. vitticeps (PVI; A–K), P. henrylawsoni (PHE; M–P), and P. minor (PMI; Q–T). In P. vitticeps, the figure represents successively a standard male ZZ karyotype with no accumulations (A, B), standard female ZW karyotype with AAGG (C) and heterochromatin accumulation (D), karyotype of a ZWm male with AAGG accumulation (E) but without heterochromatinization (F), karyotype of a sex-reversed ZZ female with no accumulations (G, H), karyotype of a ZWm male with no accumulations (I, J), and karyotype of a WWm female with 2 AAGG accumulations (K) and unpaired heterochromatic block (L). In P. henrylawsoni and P. minor, male karyotypes with no accumulations (M, N, Q, R) and female karyotypes with AAGG (O, S) and heterochromatin accumulations (P, T) are shown. Wm in the caption in males reflects the presence of the W-specific marker in PCR, the assignment in females is based on cytogenetics. Boxes in K show W and Wm chromosomes in separated blue channel mode to present their size difference. Scale bars, 10 μm.
Fig. 2.
Results of PCR test with primers for the W-specific fragment in P. vitticeps (PVI), P. henrylawsoni (PHE), and P. minor (PMI) visualized by agarose gel electrophoresis. Standard females (F) of PVI, PHE, and PMI display the W-specific fragment (224 bp), whereas in standard males (M) this fragment is not amplified. PVI individuals 1–10 include: 1) ZWm male; 2) sex-reversed ZZ female; 3, 4) male offspring of sex-reversed females and ZWm male; 5) offspring of unknown sex from the same cross; 6) standard female used in the ZW × ZWm cross, i.e., the mother of the offspring 7–10 depicted here; 7, 8) male offspring of standard female and ZWm male; and 9, 10) female offspring of standard female and ZWm male.
Sex-Reversed ZZ Female × ZWm Male
Among the progeny of the cross between the male carrying the W-specific marker (ZWm) and the 2 ZZ sex-reversed females, exactly 50% out of 40 offspring inherited the W-specific marker obviously from their father, confirming Mendelian inheritance of this locus. As the offspring from this cross were used in another project, there was only 1 offspring carrying the W-specific marker and 1 lacking it from this cross available for reliable sexing and karyotyping (Fig. 2). Both these individuals were males, fertility was confirmed in the male with the W-specific marker (online suppl. Table 1). The male without the W-specific marker had the same cytogenetic characteristics as the standard male (online suppl. Fig. 2A, B), i.e., no microchromosome with heterochromatic and AAGG blocks. The cytogenetic profile of the male carrying the W-specific marker was not identical to his father: he lacked heterochromatin accumulation, but also a notable AAGG accumulation (Fig. 1I, J').
Standard ZW Female × ZWm Male
Among offspring from the cross between the ZWm male with the standard ZW female, 42 out of 57 juveniles possessed the W-specific marker. The ratio of individuals without and with the W-specific marker from this cross was 1:2.8, which is neither statistically different from 1:3 (χ2 = 0.46, df = 1, p = 0.50), which is expected if the WWm genotype is viable, nor from 1:2 (χ2 = 0.63, df = 1, p = 0.43), which is expected for the lethality of the WWm genotype. Nevertheless, the cytogenetic examinations revealed that the WWm genotype is viable (see below). The offspring with the W-specific PCR marker were of both sexes. Determination of phenotypic sex by external morphology was successfully validated in all 10 examined individuals by dissection and gonad inspection. Gonads of 5 animals with the male-typical external morphology were undoubtedly testes, while ovaries were found in all 5 dissected animals with a female-typical external morphology. Three cytogenetically examined males from the cross with the W-specific PCR marker possessed male-typical cytogenetic characteristics, i.e., absence of AAGG and heterochromatin accumulation (online suppl. Fig. 2K–P). In other 3 cytogenetically examined males with the W-specific PCR marker, the examination revealed a prominent AAGG accumulation of the motif on a microchromosome notably smaller than the W chromosome (online suppl. Fig. 2O, Q, S), but no heterochromatic accumulation (online suppl. Fig. 2P, R, T). Among 7 cytogenetically examined individuals from the cross, 1 female and 2 juveniles with undetermined sex had typical female cytogenetic characteristics (online suppl. Fig. 2U–Z). Four females possessed the W chromosome with the strong AAGG accumulation and heterochromatic block together with a smaller chromosome without heterochromatin but with notable AAGG accumulation (Fig. 1K, L; online suppl. Fig. 2A′–F′, summarized in online suppl. Table 1).
Sequence of the Wm Fragment
BLASTN search of the obtained 201 bp long sequences after exclusion of the A20 F1 primer motif found 99–100% cover and 92–97% similarity with P. vitticeps clone C1 sex chromosome anonymous locus genomic sequence [GenBank sequence ID: EU938138.1, published in Quinn et al., 2010]. A partial 46 bp long sequence was determined by Quinn et al. [[2010]] to be informative for the identification of the W-specific fragment with the Z- and W-specific alleles differing in 4 SNPs. Fragments in all 4 individuals sequenced by us, i.e., standard ZW female and 3 males with the amplified W-specific marker, possess the 4 SNPs assigned as female-specific (Fig. 3).
Fig. 3.
Sequences of the W-specific fragment with the male and female reference. P. vitticeps clone C1 sex chromosome anonymous locus genomic sequence (GenBank sequence ID: EU938138.1, compared using BLASTN search), primer F1 used for the amplification of the W-specific fragment, male and female reference sequences differing in 4 SNPs from Quinn et al. [[2010]], and 4 individuals used in the study identical with the female reference sequence are shown. These individuals are a standard ZW female and a male possessing the W-specific marker and the AAGG accumulation involved in ♀ZW × ♂ZWm cross and their offspring, the male possessing the W-specific marker and the AAGG accumulation. The last depicted sequence represents the male possessing the W-specific marker but without the AAGG accumulation originating from the cross between female sex reversal and the male with the W-specific marker (♀ZZ × ♂ZWm).
Discussion
The initial result, stimulating further experiments described in the current study, was the discovery of the amplification of the molecular W-specific marker among the offspring of the thermally-reversed ZZ female with a standard, presumably ZZ, male. Further testing including cytogenetics confirmed that the mother was indeed a reversed ZZ female, whereas the father together with around a half of the offspring displayed markers for the W chromosome (ZWm individuals; summarized in online suppl. Table 1 and Fig. 1, 2, 3). The same results were obtained from the cross of the same male with another sex-reversed ZZ female. The linkage of the PCR and cytogenetic markers to sex seems to be very strong as they are sex-linked not only in P. vitticeps but also in other amphibolurine dragon lizards. The same molecular and cytogenetic W-specific markers developed for P. vitticeps were observed to be W-linked also in P. henrylawsoni and P. minor (this study), and previously, parts of the content of P. vitticeps sex chromosomes mapped to sex chromosomes in P. barbata and Diporiphora nobbi [Ezaz et al., 2009; Quinn et al., 2010]. All of these observations point to the stability and common origin of Pogona and Diporiphora sex chromosomes, which according to the estimated divergence times, evolved around 25 million years ago [Tonini et al., 2016]. Considering this large evolutionary stability of the linkage of the markers to sex, we speculated that the male with the W-specific markers is a reversed ZW individual. We tested this hypothesis by determination of the phenotypic sex in crosses with ZZ and ZW females. However, the results show that the chromosome assigned by us as Wm does not operate in the same way as the W in sex determination: most importantly, the individuals with the ZWm genotype are males.
C-banding and FISH detecting the accumulation of the AAGG motif gave different results. Whereas the conspicuous heterochromatin block is located exclusively on the female-specific W chromosome (and is thus exclusively present in females with either a ZW or WWm genotype), the AAGG accumulation was detected in both sexes − on the female-specific W and on the Wm present in part of males and females. The physical mapping of the sex chromosome content revealed that the Z chromosome is smaller than the W in P. vitticeps [Ezaz et al., 2013]. The Wm chromosome is also obviously smaller than the W chromosome. Therefore, we suggest that the Wm likely originated from the Z by an addition of a small part including the PCR W-specific marker and a part of the AAGG accumulation from the W chromosome. The absence of heterochromatin on the Wm chromosome might reflect different epigenetic dynamics of the Z chromosome, or simply the method of heterochromatin visualization, which is relatively crude and the detection of heterochromatin in a smaller block of repeats might be below the detection limit in comparison with the sensitive PCR and FISH analyses. We cannot exclude that the genetic material was translocated from the W to another microchromosome other than the Z. The Wm would then actually be an autosome. In any case, the linkage between the cytogenetic and PCR markers is loose, as the progeny of the ZWm male had various combinations of presence/absence of these markers (Fig. 1, 2; online suppl. Fig. 2). Additional experiments would be needed to decipher the position of the otherwise W-specific markers in the genome in the family studied by us.
Our finding has important consequences as P. vitticeps has a prominent status among amniote vertebrates. It is the only amniote with a well-supported transition from the ancestral GSD (in amphibolurids stable for around 25 million of years) to ESD. Despite the cytogenetic difference of sex chromosomes and their age, it has been shown in the laboratory that high incubation temperatures override the genotype and produce ZZ females in P. vitticeps [Quinn et al., 2007]. Offspring sex ratio of these sex-reversed females depends on the incubation temperature, suggesting that one generation is sufficient for a GSD-to-ESD transition and the loss of the W chromosome [Holleley et al., 2015]. It was demonstrated that the lack of W chromosome and/or extreme incubation conditions affect morphological and behavioural traits of sex-reversed ZZ females [Holleley et al., 2015; Li et al., 2016; Jones et al., 2020]; however, the influence of sex reversal on fitness under natural conditions is not known. In the laboratory, the females are fully viable and fertile [Holleley et al., 2015]. Sex-reversed animals identified by molecular and cytogenetic markers were reported in the bearded dragon also in the field, and their slightly higher incidence in recent years was put into connection with global warming [Holleley et al., 2015]. In the recent study [Castelli et al., 2020], the mismatch between phenotypic and genotypic sex was assigned by the same molecular marker [Quinn et al., 2010; Holleley et al., 2015; Ehl et al., 2017] (all these studies used the same contig as the sex-specific marker although they differed in the primers used for amplifications of fragments from the same W-linked regions) and was found in 5% out of 534 examined individuals of P. vitticeps covering the whole species range. Notably, all 28 animals with the mismatch were phenotypic females and they were recorded only in the south-western part of the species range. While this clustered distribution is consistent with non-random occurrence of sex reversals in certain environmental conditions at the edge of the species distribution as interpreted by Castelli et al. [[2020]], the spatial clustering can also reflect the geographic spread of a mutation, recombination, or other rearrangement concerning the region of the W chromosome containing the otherwise female-specific marker used for identification of the individuals with the mismatch. The current study discloses that the given marker might not be as reliable as thought for identification of sex-reversed individuals in the field and that the causes of the mismatch between the phenotypic and genotypic sex should be investigated more rigorously. Holleley et al. [[2015]] tested the reliability of the molecular marker by a congruence with cytogenetics, but our current work demonstrates that neither presence of the accumulation of AAGG repeats is reliable. Crossbreeding experiments with the animals assigned as sex-reversed in the field based on genetic markers were not performed. Our study indicates that it can be premature to assign an animal with a mismatch between the phenotypic and genotypic sex as sex-reversed without the necessary experimental crosses. As demonstrated here, even molecular markers linked to sex for many millions of years can be misleading in this respect.
Various techniques were used for genotypic sexing in vertebrates, including analysis of karyotypes, anonymous sex-linked markers, (derived for example from reduced-representation sequencing and microsatellites), and comparison of copy number variation between sexes by quantitative PCR [Cooper et al., 1997; Halverson and Spelman, 2002; Sulandari et al., 2014; Gamble et al., 2015; Rovatsos et al., 2017b]. As the transition from GSD to ESD requires an intermediate step of sex reversal, the reliable detection of sex-reversed individuals is crucial for studies of a turnover of sex determination mechanisms. Individuals with a mismatch between the genotypic and phenotypic sex were quite often reported in the field and were interpreted as sex-reversed individuals and thus as evidence for turnover in the sex determination system (from GSD to ESD or for a turnover in sex chromosomes), a potential for such a turnover or a co-occurrence of GSD and ESD in a population [Shine et al., 2002; Barske and Capel, 2008; Holleley et al., 2015; Hundt et al., 2019; Lambert et al., 2019]. We think that these studies should be taken as good evidence for mismatches between molecular markers and phenotypic sex, not as the documentation of the frequency of environmentally-induced sex reversals in the field. This might seem like a subtle shift but it is important for interpretations and formulations of hypotheses about transitions from GSD to ESD, which require sex reversals. We warn that there are also other mechanisms producing such a mismatch between phenotypic sex and genetic markers, for example, a recombination between sex chromosomes in otherwise non-recombining or rarely recombining regions, translocation of a sex-specific marker to autosomes, translocation of sequences between non-recombining regions of sex chromosomes, or a mutation in a sex-determining gene [Hawkins et al., 1992; Murata et al., 2016; Furman et al., 2020; Xu et al., 2020]. We should learn more about the frequency of events leading to a mismatch between gonadal and genotypic sex and evaluate to what extend are the reports of sex reversals based on given markers reliable. We conclude that individuals should not be assigned as sex-reversed by environmental conditions unless their genotype is carefully examined or a functional analysis of their genetic constitution is evaluated in breeding experiments.
Statement of Ethics
All breeding and experimental procedures were carried out under the supervision and with the approval of the Ethics Committee of the Faculty of Science, Charles University, followed by the Ministry of Education, Youth and Sports (permissions No. 35484/2015–14 and 23,852/2014–11).
Conflict of Interest Statement
We declare no competing interests in relation to the work.
Funding Sources
M.A. was supported by Charles University Research Centre programme 204069, L.K., and M.A. by Czech Science Foundation project No. 20–27236J. The founders did not have any role in the preparation of data or the manuscript.
Author Contribution
All 3 authors designed the project and wrote and approved the manuscript. J.E. performed the work with animals, and M.A. performed the cytogenetic analyses.
Supplementary Material
Supplementary data
Acknowledgement
We would like to express our gratitude to the rock band U2 for inspiration for the title, to Petr Ráb and other colleagues from the Laboratory of Fish Genetics IAPG for their ongoing support, to Jasna Vukić for helpful comments and discussions regarding technical issues, and to Jan Červenka for his valuable insights and help with experimental animal handling.
References
- Altmanová M, Rovatsos M, Kratochvíl L, Johnson Pokorná M. Minute Y chromosomes and karyotype evolution in Madagascan iguanas (Squamata: Iguania: Opluridae) Biol J Linn Soc. 2016;118((3)):618–33. [Google Scholar]
- Altmanová M, Rovatsos M, Johnson Pokorná M, Veselý M, Wagner F, Kratochvíl L. All iguana families with the exception of basilisks share sex chromosomes. Zoology. 2018;126:98–102. doi: 10.1016/j.zool.2017.11.007. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215((3)):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barske LA, Capel B. Blurring the edges in vertebrate sex determination. Curr Opin Genet Dev. 2008;18((6)):499–505. doi: 10.1016/j.gde.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli MA, Georges A, Cherryh C, Rosauer DF, Sarre SS, Contador‐Kelsall I, et al. Evolving thermal thresholds explain the distribution of temperature sex reversal in an Australian dragon lizard. Divers Distrib. 2020 ..in press. [Google Scholar]
- Cogger HG . Reptiles and Amphibians of Australia. Chatswood, New South Wales: Reed Books; 1992. [Google Scholar]
- Cooper SJ, Bull CM, Gardner MG. Characterization of microsatellite loci from the socially monogamous lizard Tiliqua rugosa using a PCR-based isolation technique. Mol Ecol. 1997;6((8)):793–5. doi: 10.1046/j.1365-294x.1997.00242.x. [DOI] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, et al. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508((7497)):488–93. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- Deakin JE, Edwards MJ, Patel H, O'Meally D, Lian J, Stenhouse R, et al. Anchoring genome sequence to chromosomes of the central bearded dragon (Pogona vitticeps) enables reconstruction of ancestral squamate macrochromosomes and identifies sequence content of the Z chromosome. BMC Genomics. 2016;17:447. doi: 10.1186/s12864-016-2774-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vosjoli P, Sommella TM, Mailloux R, Donoghue S, Klingenberg RJ, Cole J. The Bearded Dragon Manual: Expert Advice for Keeping and Caring for a Healthy Bearded Dragon. Lumina Media; 2016. [Google Scholar]
- Ezaz T, Quinn AE, Miura I, Sarre SD, Georges A, Marshall Graves JA. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. 2005;13((8)):763–76. doi: 10.1007/s10577-005-1010-9. [DOI] [PubMed] [Google Scholar]
- Ezaz T, Quinn AE, Sarre SD, O'Meally D, Georges A, Graves JA. Molecular marker suggests rapid changes of sex-determining mechanisms in Australian dragon lizards. Chromosome Res. 2009;17((1)):91–8. doi: 10.1007/s10577-008-9019-5. [DOI] [PubMed] [Google Scholar]
- Ehl J, Vukić J, Kratochvíl L. Hormonal and thermal induction of sex reversal in the bearded dragon (Pogona vitticeps, Agamidae) Zool Anz. 2017;271:1–5. [Google Scholar]
- Ezaz T, Azad B, O'Meally D, Young MJ, Matsubara K, Edwards MJ, et al. Sequence and gene content of a large fragment of a lizard sex chromosome and evaluation of candidate sex differentiating gene R-spondin 1. BMC Genomics. 2013;14:899. doi: 10.1186/1471-2164-14-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaz T, Srikulnath K, Graves JA. Origin of amniote sex chromosomes: an ancestral super-sex chromosome, or common requirements? J Hered. 2017;108((1)):94–105. doi: 10.1093/jhered/esw053. [DOI] [PubMed] [Google Scholar]
- Furman BLS, Metzger DCH, Darolti I, Wright AE, Sandkam BA, Almeida P, et al. Sex chromosome evolution: so many exceptions to the rules. Genome Biol Evol. 2020;12((6)):750–63. doi: 10.1093/gbe/evaa081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T, Geneva AJ, Glor RE, Zarkower D. Anolis sex chromosomes are derived from a single ancestral pair. Evolution. 2014;68((4)):1027–41. doi: 10.1111/evo.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T, Coryell J, Ezaz T, Lynch J, Scantlebury DP, Zarkower D. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol Biol Evol. 2015;32((5)):1296–309. doi: 10.1093/molbev/msv023. [DOI] [PubMed] [Google Scholar]
- Georges A, Li Q, Lian J, O'Meally D, Deakin J, Wang Z, et al. High-coverage sequencing and annotated assembly of the genome of the Australian dragon lizard Pogona vitticeps. Gigascience. 2015;4:45. doi: 10.1186/s13742-015-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada K, Buss EG. The chromosomes of turkey embryos during early stages of parthenogenetic development. Genetics. 1981;98((2)):335–45. doi: 10.1093/genetics/98.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JR, Taylor A, Berta P, Levilliers J, Van der Auwera B, Goodfellow PN. Mutational analysis of SRY: nonsense and missense mutations in XY sex reversal. Hum Genet. 1992;88((4)):471–4. doi: 10.1007/BF00215684. [DOI] [PubMed] [Google Scholar]
- Hill PL, Burridge CP, Ezaz T, Wapstra E. Conservation of sex-linked markers among conspecific populations of a viviparous skink, Niveoscincus ocellatus, Exhibiting genetic and temperature-dependent sex determination. Genome Biol Evol. 2018;10((4)):1079–87. doi: 10.1093/gbe/evy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleley CE, O'Meally D, Sarre SD, Marshall Graves JA, Ezaz T, Matsubara K, et al. Sex reversal triggers the rapid transition from genetic to temperature-dependent sex. Nature. 2015;523((7558)):79–82. doi: 10.1038/nature14574. [DOI] [PubMed] [Google Scholar]
- Hundt P, Liddle E, Nielsen S, Pinto B, Gamble T. Sex chromosomes and sex-specific molecular markers in Indo-Pacific combtooth blennies (Blenniidae, Istiblennius) Mar Ecol Prog Ser. 2019;627:195–200. [Google Scholar]
- Iannucci A, Altmanová M, Ciofi C, Ferguson-Smith M, Milan M, Pereira JC, et al. Conserved sex chromosomes and karyotype evolution in monitor lizards (Varanidae) Heredity (Edinb) 2019;123((2)):215–27. doi: 10.1038/s41437-018-0179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen FJ, Paukstis GL. Environmental sex determination in reptiles: ecology, evolution, and experimental design. Q Rev Biol. 1991;66((2)):149–79. doi: 10.1086/417143. [DOI] [PubMed] [Google Scholar]
- Johnson Pokorná M, Kratochvíl L. What was the ancestral sex-determining mechanism in amniote vertebrates? Biol Rev Camb Philos Soc. 2016;91((1)):1–12. doi: 10.1111/brv.12156. [DOI] [PubMed] [Google Scholar]
- Jones MEH, Pistevos JCA, Cooper N, Lappin AK, Georges A, Hutchinson MN, et al. Reproductive phenotype predicts adult bite-force performance in sex-reversed dragons (Pogona vitticeps) J Exp Zool A Ecol Integr Physiol. 2020;333((4)):252–63. doi: 10.1002/jez.2353. [DOI] [PubMed] [Google Scholar]
- Kostmann A, Kratochvíl L, Rovatsos M. Poorly differentiated XX/XY sex chromosomes are widely shared across skink radiation. Proc Biol Sci. 2021;288((1943)):20202139. doi: 10.1098/rspb.2020.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert MR, Tran T, Kilian A, Ezaz T, Skelly DK. Molecular evidence for sex reversal in wild populations of green frogs (Rana clamitans) PeerJ. 2019;7:e6449. doi: 10.7717/peerj.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Holleley CE, Elphick M, Georges A, Shine R. The behavioural consequences of sex reversal in dragons. Proc R Soc B. 2016;283((1832)):20160217. [Google Scholar]
- Marshall Graves JA. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu Rev Genet. 2008;42:565–86. doi: 10.1146/annurev.genet.42.110807.091714. [DOI] [PubMed] [Google Scholar]
- Matsubara K, O'Meally D, Azad B, Georges A, Sarre SD, Graves JA, et al. Amplification of microsatellite repeat motifs is associated with the evolutionary differentiation and heterochromatinization of sex chromosomes in Sauropsida. Chromosoma. 2016;125((1)):111–23. doi: 10.1007/s00412-015-0531-z. [DOI] [PubMed] [Google Scholar]
- Murata C, Kuroki Y, Imoto I, Kuroiwa A. Ancestral Y-linked genes were maintained by translocation to the X and Y chromosomes fused to an autosomal pair in the Okinawa spiny rat Tokudaia muenninki. Chromosome Res. 2016;24((3)):407–19. doi: 10.1007/s10577-016-9531-y. [DOI] [PubMed] [Google Scholar]
- Nielsen SV, Banks JL, Diaz RE, Jr, Trainor PA, Gamble T. Dynamic sex chromosomes in Old World chameleons (Squamata: Chamaeleonidae) J Evol Biol. 2018;31((4)):484–90. doi: 10.1111/jeb.13242. [DOI] [PubMed] [Google Scholar]
- Pennell MW, Mank JE, Peichel CL. Transitions in sex determination and sex chromosomes across vertebrate species. Mol Ecol. 2018;27((19)):3950–63. doi: 10.1111/mec.14540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorná M, Kratochvíl L. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool J Linn Soc. 2009;156((1)):168–83. [Google Scholar]
- Pokorná M, Rens W, Rovatsos M, Kratochvíl L. A ZZ/ZW sex chromosome system in the thick-tailed gecko (Underwoodisaurus milii; Squamata: Gekkota: Carphodactylidae), a member of the ancient gecko lineage. Cytogenet Genome Res. 2014;142((3)):190–6. doi: 10.1159/000358847. [DOI] [PubMed] [Google Scholar]
- Quinn AE, Georges A, Sarre SD, Guarino F, Ezaz T, Graves JA. Temperature sex reversal implies sex gene dosage in a reptile. Science. 2007;316((5823)):411. doi: 10.1126/science.1135925. [DOI] [PubMed] [Google Scholar]
- Quinn AE, Ezaz T, Sarre SD, Graves JM, Georges A. Extension, single-locus conversion and physical mapping of sex chromosome sequences identify the Z microchromosome and pseudo-autosomal region in a dragon lizard, Pogona vitticeps. Heredity (Edinb) 2010;104((4)):410–7. doi: 10.1038/hdy.2009.133. [DOI] [PubMed] [Google Scholar]
- Roco ÁS, Olmstead AW, Degitz SJ, Amano T, Zimmerman LB, Bullejos M. Coexistence of Y, W, and Z sex chromosomes in Xenopus tropicalis. Proc Natl Acad Sci USA. 2015;112((34)):E4752–61. doi: 10.1073/pnas.1505291112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Kratochvíl L. Molecular sexing applicable in 4000 species of lizards and snakes? From dream to real possibility. Methods Ecol Evol. 2017;8((8)):902–6. [Google Scholar]
- Rovatsos M, Altmanová M, Pokorná M, Kratochvíl L. Conserved sex chromosomes across adaptively radiated Anolis lizards. Evolution. 2014a;68((7)):2079–85. doi: 10.1111/evo.12357. [DOI] [PubMed] [Google Scholar]
- Rovatsos M, Pokorná M, Altmanová M, Kratochvíl L. Cretaceous park of sex determination: sex chromosomes are conserved across iguanas. Biol Lett. 2014b;10((3)):20131093. doi: 10.1098/rsbl.2013.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Vukić J, Lymberakis P, Kratochvíl L. Evolutionary stability of sex chromosomes in snakes. Proc Biol Sci. 2015a;282((1821)):20151992. doi: 10.1098/rspb.2015.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Johnson Pokorná M, Altmanová M, Kratochvíl L. Female heterogamety in Madagascar chameleons (Squamata: Chamaeleonidae: Furcifer): differentiation of sex and neo-sex chromosomes. Sci Rep. 2015b;5:13196. doi: 10.1038/srep13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Vukić J, Altmanová M, Johnson Pokorná M, Moravec J, Kratochvíl L. Conservation of sex chromosomes in lacertid lizards. Mol Ecol. 2016;25((13)):3120–6. doi: 10.1111/mec.13635. [DOI] [PubMed] [Google Scholar]
- Rovatsos M, Farkačová K, Altmanová M, Johnson Pokorná M, Kratochvíl L. The rise and fall of differentiated sex chromosomes in geckos. Mol Ecol. 2019a;28((12)):3042–52. doi: 10.1111/mec.15126. [DOI] [PubMed] [Google Scholar]
- Rovatsos M, Vukić J, Mrugała A, Suwala G, Lymberakis P, Kratochvíl L. Little evidence for switches to environmental sex determination and turnover of sex chromosomes in lacertid lizards. Sci Rep. 2019b;9((1)):7832. doi: 10.1038/s41598-019-44192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Praschag P, Fritz U, Kratochvíl L. Stable Cretaceous sex chromosomes enable molecular sexing in softshell turtles (Testudines: Trionychidae) Sci Rep. 2017;7:42150. doi: 10.1038/srep42150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Rehák I, Velenský P, Kratochvíl L. Shared ancient sex chromosomes in varanids, beaded lizards, and alligator lizards. Mol Biol Evol. 2019c;36((6)):1113–20. doi: 10.1093/molbev/msz024. [DOI] [PubMed] [Google Scholar]
- Sarre SD, Ezaz T, Georges A. Transitions between sex-determining systems in reptiles and amphibians. Annu Rev Genomics Hum Genet. 2011;12:391–406. doi: 10.1146/annurev-genom-082410-101518. [DOI] [PubMed] [Google Scholar]
- Shetty S, Griffin DK, Graves JA. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res. 1999;7((4)):289–95. doi: 10.1023/a:1009278914829. [DOI] [PubMed] [Google Scholar]
- Shine R, Elphick MJ, Donnellan S. Co-occurrence of multiple, supposedly incompatible modes of sex determination in a lizard population. Ecol Lett. 2002;5((4)):486–9. [Google Scholar]
- Straková B, Rovatsos M, Kubička L, Kratochvíl L. Evolution of sex determination in amniotes: Did stress and sequential hermaphroditism produce environmental determination? BioEssays. 2020;42((10)):e2000050. doi: 10.1002/bies.202000050. [DOI] [PubMed] [Google Scholar]
- Sulandari S, Zein MSA, Arida EA, Hamidy A. Molecular sex determination of captive Komodo dragons (Varanus komodoensis) at Gembira Loka Zoo, Surabaya Zoo, and Ragunan Zoo, Indonesia. HAYATI J Biosci. 2014;21((2)):65–75. [Google Scholar]
- Sumner AT. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 1972;75((1)):304–6. doi: 10.1016/0014-4827(72)90558-7. [DOI] [PubMed] [Google Scholar]
- Suwala G, Altmanová M, Mazzoleni S, Karameta E, Pafilis P, Kratochvíl L, et al. Evolutionary variability of W-linked repetitive content in lacertid lizards. Genes. 2020;11((5)):531. doi: 10.3390/genes11050531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonini JFR, Beard KH, Ferreira RB, Jetz W, Pyron RA. Fully-sampled phylogenies of squamates reveal evolutionary patterns in threat status. Biol Conserv. 2016;204((Part A)):23–31. [Google Scholar]
- Waters PD, Wallis MC, Marshall Graves JA. Mammalian sex − Origin and evolution of the Y chromosome and SRY. Semin Cell Dev Biol. 2007;18((3)):389–400. doi: 10.1016/j.semcdb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Watts PC, Buley KR, Sanderson S, Boardman W, Ciofi C, Gibson R. Parthenogenesis in Komodo dragons. Nature. 2006;444((7122)):1021–2. doi: 10.1038/4441021a. [DOI] [PubMed] [Google Scholar]
- Witten GJ. Some karyotypes of Australian agamids (Reptilia: Lacertilia) Aust J Zool. 1983;31:533–40. [Google Scholar]
- Xu L, Irestedt M, Zhou Q. Sequence transpositions restore genes on the highly degenerated W chromosomes of songbirds. Genes. 2020;11((11)):1267. doi: 10.3390/genes11111267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhang J, Bachtrog D, An N, Huang Q, Jarvis ED, et al. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science. 2014;346((6215)):1246338. doi: 10.1126/science.1246338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data