In the crystal, molecules are linked by C—H⋯O hydrogen bonds, forming layers parallel to the (001) plane. These layers of molecules are connected by C—H⋯π interactions along the c-axis direction. Interlayer van der Waals and interhalogen interactions stabilize the packing.
Keywords: crystal structure, tetrahydrofuran rings, C—H⋯O hydrogen bonds, C—H⋯π interactions, Hirshfeld surface analysis, IMDAF reaction, Diels–Alder reaction
Abstract
In the title compound, C24H18Cl3NO3, the tetrahydrofuran rings adopt envelope conformations. In the crystal, C—H⋯O hydrogen bonds connect molecules, generating layers parallel to the (001) plane. These layers are connected along the c-axis direction by C—H⋯π interactions. The packing is further stabilized by interlayer van der Waals and interhalogen interactions. The most important contributions to the surface contacts are from H⋯H (36.8%), Cl⋯H/H⋯Cl (26.6%), C⋯H/H⋯C (18.8%) and O⋯H/H⋯O (11.3%) interactions, as concluded from a Hirshfeld surface analysis.
Chemical context
In recent years, the IMDAF cycloaddition (the intramolecular furan Diels–Alder reaction) in combination with other known reactions in a tandem or sequential manner is pursued for the construction of several important bicyclic or polycyclic compounds, including natural ones (for some reviews on this topic, see: Zubkov et al., 2005 ▸; Takao et al., 2005 ▸; Juhl et al., 2009 ▸; Padwa et al., 2013 ▸; Parvatkar et al., 2014 ▸; Krishna et al., 2021 ▸). Cascade sequences comprising two or more successive [4 + 2] cycloaddition steps are a powerful and frequently used protocol in modern syntheses aimed at constructing cyclohexene derivatives thanks to their exceptional chemoselectivity, regioselectivity, diastereoselectivity, and capability to create more than four chiral centers in a single synthetic step (Criado et al., 2010 ▸, 2013 ▸). It has been shown previously that the Diels–Alder reaction of bis-dienes with derivatives of maleic acid, esters of acetylene dicarboxylic acid and hexafluoro-2-butyne proceeds in all cases diastereo- and chemoselectively and leads, depending on the temperature, to annelated diepoxynaphthalenes of the ‘domino’ or ‘pincer’ type (Borisova et al., 2018a
▸,b
▸; Grudova et al., 2020 ▸; Kvyatkovskaya et al., 2020 ▸, 2021 ▸). In order to expand the limits of the applicability of the IMDAF strategy, we tested in this study dehydrobenzene generated in situ in the role of dienophile. It was demonstrated that the products of the parallel [4 + 2] cycloaddition of two aryne moieties to both the furan fragments of the bis-diene system (Fig. 1 ▸, 1 and 2) prevails over the adduct (3) of the IMDAF reaction (Fig. 1 ▸).
Figure 1.
Synthesis scheme for 2,2,2-trichloro-N,N-bis[(1R,4SR)-1,4-epoxynaphthalen-1(4H)-ylmethyl]acetamide (1).
On the other hand, intermolecular non-covalent interactions organize the molecular aggregates, catalytic intermediates, etc., which play crucial roles for the functional properties of heterocyclic compounds (Gurbanov et al., 2020a ▸,b ▸; Khalilov et al., 2018a ▸,b ▸; Ma et al., 2017a ▸,b ▸, 2020 ▸, 2021 ▸; Mahmudov et al., 2020 ▸; Mizar et al., 2012 ▸). Thus, attached –CCl3 and C=O groups can participate in intermolecular interactions and affect the properties of 1–3.
Structural commentary
In the title compound (1, Fig. 2 ▸), the tetrahydrofuran rings (O19/C11–C14 and O29/C21–C24) adopt envelope conformations with the O atoms as the flaps. The molecular conformation is stabilized by intramolecular C10—H10A⋯O29 and C20—H20A⋯O19 hydrogen bonds and C20—H20B⋯Cl1 and C20—H20B⋯Cl3 interactions (Table 1 ▸).
Figure 2.
The molecule of the title compound 1 with atom-labeling scheme and displacement ellipsoids drawn at the 30% probability level. Hydrogen atoms are shown as spheres of arbitrary radius.
Table 1. Hydrogen-bond geometry (Å, °).
Cg8 is the centroid of the C24A/C25–C28/C28A aromatic ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C10—H10A⋯O29 | 0.97 | 2.35 | 3.074 (2) | 131 |
| C12—H12A⋯O1i | 0.93 | 2.66 | 3.494 (2) | 150 |
| C17—H17A⋯O1ii | 0.93 | 2.51 | 3.427 (3) | 168 |
| C20—H20A⋯O19 | 0.97 | 2.39 | 3.068 (2) | 127 |
| C27—H27A⋯O19iii | 0.93 | 2.51 | 3.438 (3) | 175 |
| C20—H20B⋯Cl1 | 0.97 | 2.55 | 3.1744 (18) | 122 |
| C20—H20B⋯Cl3 | 0.97 | 2.64 | 3.2921 (19) | 125 |
| C13—H13A⋯Cg8iv | 0.93 | 2.90 | 3.633 (2) | 136 |
Symmetry codes: (i) -x+{\script{3\over 2}}, y+{\script{1\over 2}}, -z+{\script{1\over 2}}; (ii) -x+{\script{3\over 2}}, y-{\script{1\over 2}}, -z+{\script{1\over 2}}; (iii) -x+{\script{1\over 2}}, y-{\script{1\over 2}}, -z+{\script{1\over 2}}; (iv) x-{\script{1\over 2}}, -y+{\script{1\over 2}}, z-{\script{1\over 2}}.
Supramolecular features and Hirshfeld surface analysis
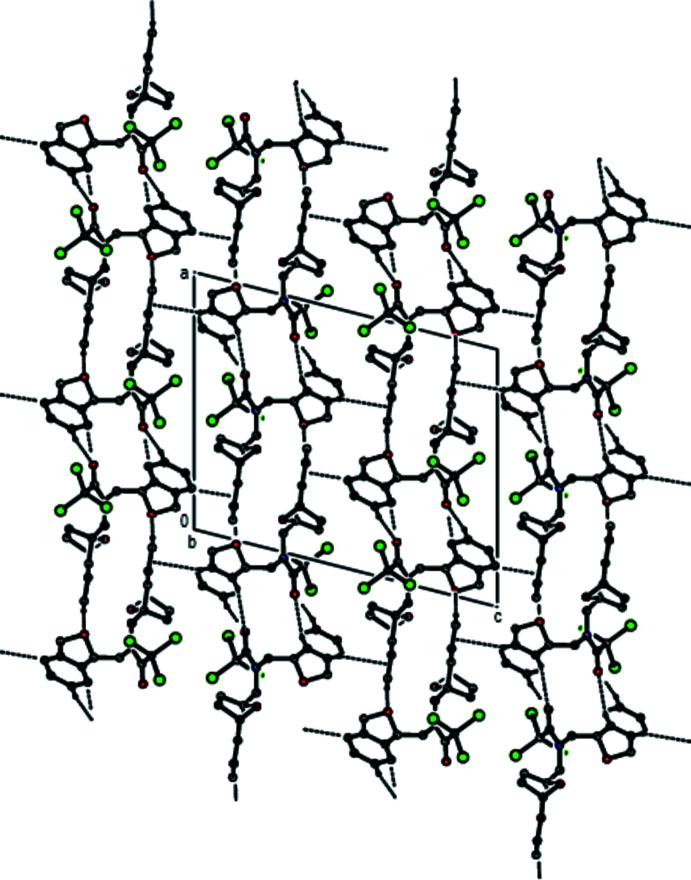
In the crystal, hydrogen bonds of the C—H⋯O type link the molecules, generating layers parallel to the (001) plane (Table 1 ▸; Figs. 3 ▸, 4 ▸, 5 ▸ and 6 ▸). These layers are connected by C—H⋯π interactions (C13—H13A⋯Cg8; Table 1 ▸), where Cg8 is the centroid of the C24A/C25–C28/C28A aromatic ring. The intermolecular interactions in the crystal of the title compound (Table 2 ▸) were quantified using Hirshfeld surface analysis (Spackman & Jayatilaka, 2009 ▸) and the associated two-dimensional fingerprint plots (McKinnon et al., 2007 ▸) were generated. The calculations and visualization were performed using CrystalExplorer17 (Turner et al., 2017 ▸). The three-dimensional Hirshfeld surface mapped over d norm in the range −0.1862 (red) to +1.4233 (blue) a.u. is shown in Fig. 7 ▸. The short and long contacts are indicated as red and blue spots, respectively, on the Hirshfeld surfaces, and contacts with distances approximately equal to the sum of the van der Waals radii are represented as white spots. The Cl⋯H and C—H⋯O interactions, which play a key role in the molecular packing, can be correlated with the bright-red patches near Cl1, Cl2, O1 and O19 and hydrogen atoms H14A and H16A, which highlight their functions as donors and/or acceptors. Fig. 8 ▸ shows the full two-dimensional fingerprint plot (Fig. 8 ▸ a) and those delineated into the major contacts: H⋯H (36.8%, Fig. 8 ▸ b) interactions are the major factor in the crystal packing together with Cl⋯H/H⋯Cl (26.6%, Fig. 8 ▸ c), C⋯H/H⋯C (18.8%, Fig. 8 ▸ d) and O⋯H/H⋯O (11.3%, Fig. 8 ▸ e) interactions representing the next highest contributions. The percentage contributions of other weak interactions are listed in Table 3 ▸.
Figure 3.
A general view of the intermolecular C—H⋯O hydrogen bonds and C—H⋯π interactions (depicted by dashed lines) in the unit cell of the title compound 1. [Symmetry codes: (a)  − x, −
− x, − + y,
+ y,  − z; (b)
− z; (b)  − x, −
− x, − + y,
+ y,  − z; (c)
− z; (c)  − x,
− x,  + y,
+ y,  − z; (d)
− z; (d)  + x,
+ x,  − y,
− y,  + z].
+ z].
Figure 4.
Packing viewed along the a-axis direction with the intermolecular C—H⋯O hydrogen bonds and C—H⋯π interactions depicted by dashed lines.
Figure 5.
Packing viewed along the b-axis direction with the intermolecular C—H⋯O hydrogen bonds and C—H⋯π interactions depicted by dashed lines.
Figure 6.
Packing viewed along the c-axis direction with the intermolecular C—H⋯O hydrogen bonds and C—H⋯π interactions depicted by dashed lines.
Table 2. Summary of short interatomic contacts (Å) in the title compound (1).
| Contact | Distance | Symmetry operation |
|---|---|---|
| Cl1⋯H10A | 3.10 | x, −1 + y, z |
| H20A⋯H25A | 2.44 | {1\over 2} − x, −{1\over 2} + y, {1\over 2} − z |
| H17A⋯O1 | 2.51 | {3\over 2} − x, −{1\over 2} + y, {1\over 2} − z |
| H23A⋯Cl2 | 3.07 | 1 − x, 1 − y, −z |
| C28⋯H16A | 2.96 | −{1\over 2} + x, {1\over 2} − y, −{1\over 2} + z |
| H14A⋯C25 | 2.90 | −{1\over 2} + x, {1\over 2} − y, −{1\over 2} + z |
| H15A⋯H14A | 2.56 | 1 − x, 1 − y, 1 − z |
Figure 7.
Hirshfeld surface of the title molecule 1 mapped with d norm.
Figure 8.
Fingerprint plots showing (a) all intermolecular interactions and resolved into (b) H⋯H, (c) Cl⋯H/H⋯Cl, (d) C⋯H/H⋯C and (e) O⋯H/H⋯O contacts.
Table 3. Percentage contributions of interatomic contacts to the Hirshfeld surface for the title compound (1).
| Contact | Percentage contribution |
|---|---|
| H⋯H | 36.8 |
| Cl⋯H/H⋯Cl | 26.6 |
| C⋯H/H⋯C | 18.8 |
| O⋯H/H⋯O | 11.3 |
| Cl⋯C/C⋯Cl | 4.4 |
| Cl⋯O/O⋯Cl | 0.8 |
| Cl⋯Cl | 0.8 |
| O⋯C/C⋯O | 0.4 |
| C⋯C | 0.1 |
Database survey
A search of the Cambridge Structural Database (CSD version 5.40, update of September 2019; Groom et al., 2016 ▸) for structures having the epoxyisoindole moiety gave ten hits that closely resemble the title compound, viz. 4,5-dibromo-2-[4-(trifluoromethyl)phenyl]hexahydro-3a,6-epoxyisoindol-1(4H)-one (CSD refcode IQOTOA; Mertsalov et al., 2021a ▸), 3-hydroxy-2-{[2-(4-methylbenzene-1-sulfonyl)-2,3,7,7a-tetrahydro-3a,6-epoxyisoindol-6(1H)-yl]methyl}-2,3-dihydro-1H-isoindol-1-one (OMUTAU; Mertsalov et al., 2021b ▸), 2-benzyl-4,5-dibromohexahydro-3a,6-epoxyisoindol-1(4H)-one (OMEMAX; Mertsalov et al., 2021c ▸), 4,5-dibromo-6-methyl-2-phenylhexahydro-3a,6-epoxyisoindol-1(4H)-one (IMUBIE; Mertsalov et al., 2021a ▸), (3aR,6S,7aR)-7a-chloro-2-[(4-nitrophenyl)sulfonyl]-1,2,3,6,7,7a-hexahydro-3a,6-epoxyisoindole (AGONUH; Temel et al., 2013 ▸), (3aR,6S,7aR)-7a-chloro-6-methyl-2-[(4-nitrophenyl)sulfonyl]-1,2,3,6,7,7a-hexahydro-3a,6-epoxyisoindole (TIJMIK; Demircan et al., 2013 ▸), 5-chloro-7-methyl-3-[(4-methyl-phenyl)sulfonyl]-10-oxa-3-azatricyclo[5.2.1.01,5]dec-8-ene (YAXCIL; Temel et al., 2012 ▸), (3aR,6S,7aR)-7a-bromo-2-[(4-methylphenyl)sulfonyl]-1,2,3,6,7,7a-hexahydro-3a,6-epoxyiso-indole (UPAQEI; Koşar et al., 2011 ▸), (3aR,6S,7aR)-7a-bromo-2-methylsulfonyl-1,2,3,6,7,7-hexahydro-3a,6-epoxyisoindole (ERIVIL; Temel et al., 2011 ▸) and tert-butyl 3a-chloroper-hydro-2,6a-epoxyoxireno(e)isoindole-5-carboxylate (MIGTIG; Koşar et al., 2007 ▸).
In the crystal of IQOTOA, the asymmetric unit consists of two crystallographically independent molecules. In both molecules, the pyrrolidine and tetrahydrofuran rings adopt envelope conformations. In the crystal, molecules are linked in pairs by C— H⋯O hydrogen bonds. These pairs form a tetrameric supramolecular motif, leading to molecular layers parallel to the (100) plane formed by C— H⋯π and C—Br⋯π interactions. OMUTAU also crystallizes with two independent molecules in the asymmetric unit. In the central ring systems of both molecules, the tetrahydrofuran rings adopt envelope conformations, the pyrrolidine rings adopt twisted-envelope conformations and the six-membered ring is in a boat conformation. In both molecules, the nine-membered groups attached to the central ring system are essentially planar. In the crystal, strong intermolecular O—H⋯O hydrogen bonds and weak intermolecular C—H⋯O contacts link the molecules, forming a three-dimensional network. In addition, weak π–π stacking interactions between the pyrrolidine rings are observed. OMEMAX again crystallizes with two molecules in the asymmetric unit of the unit cell. In both molecules, the tetrahydrofuran rings adopt envelope conformations with the O atoms as the flaps and the pyrrolidine rings also adopt envelope conformations. In the crystal, molecules are linked by weak C—H⋯O hydrogen bonds, forming sheets lying parallel to the (001) plane. These sheets are connected only by weak van der Waals interactions. In the crystal of IMUBIE, the molecules are linked into dimers by pairs of C—H⋯O hydrogen bonds, thus generating  (18) rings. The crystal packing is dominated by H⋯H, Br⋯H, H⋯π and Br⋯π interactions. In the crystal structures of IQOTOA, OMUTAU, OMEMAX, AGONUH, TIJMIK, YAXCIL, UPAQEI and ERIVIL, the molecules are predominantly linked by C—H⋯O hydrogen bonds, giving various hydrogen-bonding pattern connectivities. In the crystal of AGONUH, the molecules are connected in zigzag chains running along the b-axis direction. In TIJMIK, two types of C—H⋯O hydrogen bonds are found, viz. R
2
2(20) and
(18) rings. The crystal packing is dominated by H⋯H, Br⋯H, H⋯π and Br⋯π interactions. In the crystal structures of IQOTOA, OMUTAU, OMEMAX, AGONUH, TIJMIK, YAXCIL, UPAQEI and ERIVIL, the molecules are predominantly linked by C—H⋯O hydrogen bonds, giving various hydrogen-bonding pattern connectivities. In the crystal of AGONUH, the molecules are connected in zigzag chains running along the b-axis direction. In TIJMIK, two types of C—H⋯O hydrogen bonds are found, viz. R
2
2(20) and  (26) rings, with adjacent rings running parallel to the ac plane. Additionally, C—H⋯O hydrogen bonds form a C(6) chain, linking the molecules in the b-axis direction. In the crystal of ERIVIL, the molecules are connected into
(26) rings, with adjacent rings running parallel to the ac plane. Additionally, C—H⋯O hydrogen bonds form a C(6) chain, linking the molecules in the b-axis direction. In the crystal of ERIVIL, the molecules are connected into  (8) and
(8) and  (14) rings along the b-axis direction. In MIGTIG, the molecules are linked only by weak van der Waals interactions.
(14) rings along the b-axis direction. In MIGTIG, the molecules are linked only by weak van der Waals interactions.
Synthesis and crystallization
CsF (1.7 g, 0.011 mol) was added to 2,2,2-trichloro-N,N-bis(furan-2-ylmethyl)acetamide (0.0022 mol) dissolved in dry CH3CN (20 mL). Then an equivalent of 2-(trimethylsilyl)phenyl trifluoromethanesulfonate (0.54 mL, 0.022 mol) was added to the solution under an argon atmosphere. The mixture was refluxed for 4 h (TLC control). After that, one more portion of 2-(trimethylsilyl)phenyl trifluoromethanesulfonate (0.27 mL, 0.011 mol) and CsF (1.7 g, 0.011 mol) was added to the mixture, repeating all procedures again. After the mixture was cooled, CsF was filtered off through a thin layer of SiO2, and the resulting solution was concentrated under reduced pressure. The residue (brown oil) was separated using column chromatography on silica gel (a mixture EtOAc/hexane = 1/25 as eluent) to give compounds 1–3 in the ratio ∼30/25/45. Single crystals of compound 1 was obtained by slow crystallization from a hexane/EtOAc mixture.
Compound 1: white powder (0.29 g, 0.62 mmol, 28%); Rf 0.50 (‘Sorbfil’ plates for thin-layer chromatography, EtOAc/hexane, 1:4, Sorbfil); m.p. 431.7–433.4 K. 1H NMR (600.2 MHz, CDCl3) δ 7.19–7.24 (4H, m, H-Ar), 7.07 (1H, dd, J = 1.5 and J = 5.6 Hz, H-2′), 7.04 (2H, br dd, J = 2.0 and J = 5.0 Hz, H-3,3′), 6.95–6.99 (4H, m, H-Ar), 6.85 (1H, d, J = 5.6 Hz, H-2), 5.73 (1H, d, J = 1.5 Hz, H-4′), 5.71 (1H, d, J = 1.5 Hz, H-4), 5.11 (1H, d, J = 16.2 Hz, H-1′B), 4.87 (1H, d, J = 16.2 Hz, H-1B), 4.76 (1H, d, J = 15.1 Hz, H-1′A), 4.72 (1H, br d, J = 15.1 Hz, H-1A). 13C NMR (150.9 MHz, CDCl3) d 161.5, 150.2, 149.9, 148.7, 148.2, 145.2, 145.1, 143.3, 143.0, 125.5, 125.3, 125.2, 125.1, 120.5, 120.2, 120.0, 119.6, 94.1, 93.3, 92.1, 82.4, 82.2, 49.1, 45.6. IR νmax/cm−1 (tablet KBr): 2953, 2919, 1702, 1632, 1462, 1410, 1236. HRMS (ESI–TOF): calculated for C24H18Cl3NO4 [M + H]+ 473.0352; found 473.0358.
Refinement details
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. All C-bound H atoms were placed in calculated positions and refined using a riding model, with C—H = 0.93–0.98 Å, and with U
iso(H) = 1.2U
eq(C). Six reflections ( 01, 011, 101, 110, 002 and 200), which were obscured by the beam stop, and nine outliers (343, 253,
01, 011, 101, 110, 002 and 200), which were obscured by the beam stop, and nine outliers (343, 253,  ,1,15, 3,6,11, 15,4,4, 072, 4,6,12,
,1,15, 3,6,11, 15,4,4, 072, 4,6,12,  ,3,22 and 13,6,2) were omitted during the final refinement cycle.
,3,22 and 13,6,2) were omitted during the final refinement cycle.
Table 4. Experimental details.
| Crystal data | |
| Chemical formula | C24H18Cl3NO3 |
| M r | 474.74 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 296 |
| a, b, c (Å) | 15.0134 (6), 8.1336 (3), 18.2841 (6) |
| β (°) | 104.307 (2) |
| V (Å3) | 2163.48 (14) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.45 |
| Crystal size (mm) | 0.34 × 0.18 × 0.14 |
| Data collection | |
| Diffractometer | Bruker Kappa APEXII area-detector diffractometer |
| Absorption correction | Multi-scan (SADABS; Bruker, 2013 ▸) |
| Tmin, Tmax | 0.743, 0.940 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 17833, 4991, 3570 |
| R int | 0.030 |
| (sin θ/λ)max (Å−1) | 0.652 |
| Refinement | |
| R[F2 > 2σ(F 2)], wR(F 2), S | 0.041, 0.118, 1.01 |
| No. of reflections | 4991 |
| No. of parameters | 280 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.30, −0.36 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989021009907/yk2156sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021009907/yk2156Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989021009907/yk2156Isup3.cml
CCDC reference: 2095762
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors’ contributions are as follows. Conceptualization, MA and AB; synthesis, GZM; X-ray analysis, ZA and GZM; writing (review and editing of the manuscript), ZA, GZM and MA; supervision, MA and AB.
supplementary crystallographic information
Crystal data
| C24H18Cl3NO3 | F(000) = 976 |
| Mr = 474.74 | Dx = 1.458 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 15.0134 (6) Å | Cell parameters from 4831 reflections |
| b = 8.1336 (3) Å | θ = 2.8–26.7° |
| c = 18.2841 (6) Å | µ = 0.45 mm−1 |
| β = 104.307 (2)° | T = 296 K |
| V = 2163.48 (14) Å3 | Fragment, colourless |
| Z = 4 | 0.34 × 0.18 × 0.14 mm |
Data collection
| Bruker Kappa APEXII area-detector diffractometer | 3570 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.030 |
| Absorption correction: multi-scan (SADABS; Bruker, 2013) | θmax = 27.6°, θmin = 3.2° |
| Tmin = 0.743, Tmax = 0.940 | h = −19→19 |
| 17833 measured reflections | k = −10→10 |
| 4991 independent reflections | l = −23→23 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.041 | H-atom parameters constrained |
| wR(F2) = 0.118 | w = 1/[σ2(Fo2) + (0.0576P)2 + 0.6103P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.01 | (Δ/σ)max < 0.001 |
| 4991 reflections | Δρmax = 0.30 e Å−3 |
| 280 parameters | Δρmin = −0.36 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.57432 (13) | 0.4170 (2) | 0.17193 (10) | 0.0396 (4) | |
| C2 | 0.53382 (14) | 0.2461 (3) | 0.14061 (12) | 0.0450 (5) | |
| C10 | 0.57234 (13) | 0.6612 (2) | 0.24521 (11) | 0.0386 (4) | |
| H10A | 0.533570 | 0.757230 | 0.231289 | 0.046* | |
| H10B | 0.628106 | 0.679444 | 0.228600 | 0.046* | |
| C11 | 0.59728 (12) | 0.6449 (2) | 0.32991 (11) | 0.0353 (4) | |
| C12 | 0.64935 (13) | 0.7922 (2) | 0.37350 (12) | 0.0445 (5) | |
| H12A | 0.686507 | 0.866559 | 0.356181 | 0.053* | |
| C13 | 0.63075 (14) | 0.7916 (2) | 0.44014 (12) | 0.0473 (5) | |
| H13A | 0.651717 | 0.865687 | 0.479366 | 0.057* | |
| C14 | 0.56784 (13) | 0.6447 (2) | 0.43983 (11) | 0.0417 (4) | |
| H14A | 0.530748 | 0.647835 | 0.477024 | 0.050* | |
| C14A | 0.62536 (12) | 0.4899 (2) | 0.44023 (11) | 0.0390 (4) | |
| C15 | 0.65706 (14) | 0.3678 (3) | 0.49181 (12) | 0.0466 (5) | |
| H15A | 0.643575 | 0.368000 | 0.538795 | 0.056* | |
| C16 | 0.71008 (15) | 0.2436 (3) | 0.47154 (13) | 0.0531 (5) | |
| H16A | 0.731376 | 0.158196 | 0.505157 | 0.064* | |
| C17 | 0.73137 (15) | 0.2450 (3) | 0.40299 (13) | 0.0521 (5) | |
| H17A | 0.767642 | 0.161342 | 0.391117 | 0.062* | |
| C18 | 0.69959 (13) | 0.3700 (2) | 0.35035 (12) | 0.0436 (4) | |
| H18A | 0.714646 | 0.371239 | 0.303977 | 0.052* | |
| C18A | 0.64551 (11) | 0.4905 (2) | 0.36943 (10) | 0.0356 (4) | |
| C20 | 0.42684 (12) | 0.5001 (2) | 0.20562 (10) | 0.0359 (4) | |
| H20A | 0.422213 | 0.498360 | 0.257597 | 0.043* | |
| H20B | 0.404568 | 0.395232 | 0.183106 | 0.043* | |
| C21 | 0.36523 (12) | 0.6346 (2) | 0.16411 (10) | 0.0348 (4) | |
| C22 | 0.37783 (13) | 0.7010 (3) | 0.08860 (11) | 0.0440 (4) | |
| H22A | 0.405680 | 0.647375 | 0.055171 | 0.053* | |
| C23 | 0.34135 (14) | 0.8485 (3) | 0.08083 (13) | 0.0503 (5) | |
| H23A | 0.338084 | 0.920840 | 0.040843 | 0.060* | |
| C24 | 0.30557 (14) | 0.8769 (2) | 0.15056 (12) | 0.0467 (5) | |
| H24A | 0.296146 | 0.991967 | 0.162742 | 0.056* | |
| C24A | 0.22319 (13) | 0.7630 (2) | 0.14421 (10) | 0.0396 (4) | |
| C25 | 0.12998 (14) | 0.7862 (3) | 0.13138 (11) | 0.0485 (5) | |
| H25A | 0.104624 | 0.891139 | 0.125152 | 0.058* | |
| C26 | 0.07468 (15) | 0.6477 (3) | 0.12801 (12) | 0.0556 (6) | |
| H26A | 0.011307 | 0.660262 | 0.118843 | 0.067* | |
| C27 | 0.11209 (14) | 0.4931 (3) | 0.13797 (12) | 0.0558 (6) | |
| H27A | 0.073843 | 0.402457 | 0.136186 | 0.067* | |
| C28 | 0.20793 (13) | 0.4696 (3) | 0.15092 (11) | 0.0448 (4) | |
| H28A | 0.233674 | 0.365028 | 0.157926 | 0.054* | |
| C28A | 0.26130 (12) | 0.6058 (2) | 0.15276 (9) | 0.0350 (4) | |
| N1 | 0.52425 (10) | 0.51733 (18) | 0.20508 (8) | 0.0353 (3) | |
| O1 | 0.65181 (10) | 0.4475 (2) | 0.16809 (9) | 0.0594 (4) | |
| O19 | 0.51694 (8) | 0.64419 (16) | 0.36140 (7) | 0.0389 (3) | |
| O29 | 0.37315 (9) | 0.78716 (15) | 0.20640 (7) | 0.0431 (3) | |
| Cl1 | 0.49921 (4) | 0.13248 (6) | 0.21124 (4) | 0.05856 (17) | |
| Cl2 | 0.61916 (5) | 0.13144 (9) | 0.11317 (5) | 0.0795 (2) | |
| Cl3 | 0.44055 (5) | 0.27068 (8) | 0.06047 (4) | 0.0719 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0400 (10) | 0.0380 (10) | 0.0425 (10) | −0.0010 (8) | 0.0136 (8) | 0.0011 (8) |
| C2 | 0.0436 (11) | 0.0415 (10) | 0.0510 (11) | 0.0025 (8) | 0.0138 (9) | −0.0063 (9) |
| C10 | 0.0366 (10) | 0.0304 (9) | 0.0486 (11) | −0.0056 (7) | 0.0101 (8) | 0.0000 (7) |
| C11 | 0.0276 (8) | 0.0296 (9) | 0.0489 (10) | −0.0020 (7) | 0.0096 (7) | −0.0022 (7) |
| C12 | 0.0393 (11) | 0.0305 (9) | 0.0603 (13) | −0.0053 (8) | 0.0060 (9) | −0.0042 (8) |
| C13 | 0.0465 (12) | 0.0361 (10) | 0.0552 (12) | −0.0006 (9) | 0.0051 (9) | −0.0105 (9) |
| C14 | 0.0358 (10) | 0.0437 (11) | 0.0449 (10) | −0.0001 (8) | 0.0088 (8) | −0.0059 (8) |
| C14A | 0.0309 (9) | 0.0356 (10) | 0.0485 (10) | −0.0048 (7) | 0.0062 (7) | −0.0043 (8) |
| C15 | 0.0417 (11) | 0.0463 (11) | 0.0489 (11) | −0.0066 (9) | 0.0056 (9) | 0.0023 (9) |
| C16 | 0.0491 (12) | 0.0401 (11) | 0.0632 (14) | 0.0007 (9) | 0.0006 (10) | 0.0081 (10) |
| C17 | 0.0450 (12) | 0.0366 (10) | 0.0707 (14) | 0.0091 (9) | 0.0067 (10) | −0.0039 (10) |
| C18 | 0.0382 (10) | 0.0389 (10) | 0.0536 (12) | 0.0009 (8) | 0.0109 (8) | −0.0047 (8) |
| C18A | 0.0266 (8) | 0.0312 (9) | 0.0473 (10) | −0.0040 (7) | 0.0060 (7) | −0.0008 (7) |
| C20 | 0.0323 (9) | 0.0329 (9) | 0.0431 (10) | −0.0024 (7) | 0.0102 (7) | 0.0010 (7) |
| C21 | 0.0346 (9) | 0.0309 (9) | 0.0383 (9) | −0.0023 (7) | 0.0082 (7) | −0.0031 (7) |
| C22 | 0.0381 (10) | 0.0508 (12) | 0.0455 (11) | −0.0010 (9) | 0.0150 (8) | 0.0056 (9) |
| C23 | 0.0445 (11) | 0.0458 (12) | 0.0601 (13) | −0.0024 (9) | 0.0118 (9) | 0.0166 (10) |
| C24 | 0.0445 (11) | 0.0323 (10) | 0.0595 (13) | 0.0040 (8) | 0.0058 (9) | −0.0005 (9) |
| C24A | 0.0406 (10) | 0.0413 (10) | 0.0370 (9) | 0.0020 (8) | 0.0098 (7) | −0.0010 (8) |
| C25 | 0.0463 (12) | 0.0594 (13) | 0.0416 (10) | 0.0136 (10) | 0.0142 (9) | 0.0030 (9) |
| C26 | 0.0345 (10) | 0.0849 (18) | 0.0496 (12) | 0.0021 (11) | 0.0149 (9) | 0.0111 (11) |
| C27 | 0.0412 (11) | 0.0735 (16) | 0.0533 (12) | −0.0185 (11) | 0.0125 (9) | 0.0043 (11) |
| C28 | 0.0438 (11) | 0.0458 (11) | 0.0446 (10) | −0.0064 (9) | 0.0106 (8) | 0.0017 (9) |
| C28A | 0.0346 (9) | 0.0410 (10) | 0.0302 (8) | −0.0022 (7) | 0.0093 (7) | −0.0020 (7) |
| N1 | 0.0325 (8) | 0.0314 (8) | 0.0418 (8) | −0.0030 (6) | 0.0086 (6) | −0.0015 (6) |
| O1 | 0.0475 (9) | 0.0566 (9) | 0.0848 (11) | −0.0086 (7) | 0.0368 (8) | −0.0112 (8) |
| O19 | 0.0279 (6) | 0.0422 (7) | 0.0460 (7) | 0.0007 (5) | 0.0081 (5) | −0.0044 (6) |
| O29 | 0.0429 (8) | 0.0311 (7) | 0.0497 (8) | 0.0011 (6) | 0.0011 (6) | −0.0067 (6) |
| Cl1 | 0.0690 (4) | 0.0354 (3) | 0.0743 (4) | −0.0035 (2) | 0.0234 (3) | 0.0056 (2) |
| Cl2 | 0.0716 (4) | 0.0686 (4) | 0.1080 (6) | 0.0077 (3) | 0.0409 (4) | −0.0319 (4) |
| Cl3 | 0.0757 (4) | 0.0686 (4) | 0.0586 (4) | −0.0006 (3) | −0.0076 (3) | −0.0129 (3) |
Geometric parameters (Å, º)
| C1—O1 | 1.209 (2) | C17—H17A | 0.9300 |
| C1—N1 | 1.351 (2) | C18—C18A | 1.372 (3) |
| C1—C2 | 1.568 (3) | C18—H18A | 0.9300 |
| C2—Cl2 | 1.755 (2) | C20—N1 | 1.472 (2) |
| C2—Cl1 | 1.767 (2) | C20—C21 | 1.510 (2) |
| C2—Cl3 | 1.770 (2) | C20—H20A | 0.9700 |
| C10—N1 | 1.472 (2) | C20—H20B | 0.9700 |
| C10—C11 | 1.506 (3) | C21—O29 | 1.451 (2) |
| C10—H10A | 0.9700 | C21—C22 | 1.537 (3) |
| C10—H10B | 0.9700 | C21—C28A | 1.540 (2) |
| C11—O19 | 1.459 (2) | C22—C23 | 1.312 (3) |
| C11—C18A | 1.538 (2) | C22—H22A | 0.9300 |
| C11—C12 | 1.540 (2) | C23—C24 | 1.519 (3) |
| C12—C13 | 1.316 (3) | C23—H23A | 0.9300 |
| C12—H12A | 0.9300 | C24—O29 | 1.447 (2) |
| C13—C14 | 1.522 (3) | C24—C24A | 1.527 (3) |
| C13—H13A | 0.9300 | C24—H24A | 0.9800 |
| C14—O19 | 1.449 (2) | C24A—C25 | 1.374 (3) |
| C14—C14A | 1.526 (3) | C24A—C28A | 1.394 (3) |
| C14—H14A | 0.9800 | C25—C26 | 1.392 (3) |
| C14A—C15 | 1.371 (3) | C25—H25A | 0.9300 |
| C14A—C18A | 1.400 (3) | C26—C27 | 1.371 (3) |
| C15—C16 | 1.392 (3) | C26—H26A | 0.9300 |
| C15—H15A | 0.9300 | C27—C28 | 1.412 (3) |
| C16—C17 | 1.368 (3) | C27—H27A | 0.9300 |
| C16—H16A | 0.9300 | C28—C28A | 1.362 (3) |
| C17—C18 | 1.400 (3) | C28—H28A | 0.9300 |
| O1—C1—N1 | 123.52 (18) | C18—C18A—C11 | 134.72 (18) |
| O1—C1—C2 | 116.95 (17) | C14A—C18A—C11 | 104.65 (15) |
| N1—C1—C2 | 119.42 (16) | N1—C20—C21 | 114.45 (14) |
| C1—C2—Cl2 | 109.31 (13) | N1—C20—H20A | 108.6 |
| C1—C2—Cl1 | 110.74 (13) | C21—C20—H20A | 108.6 |
| Cl2—C2—Cl1 | 107.36 (11) | N1—C20—H20B | 108.6 |
| C1—C2—Cl3 | 110.99 (14) | C21—C20—H20B | 108.6 |
| Cl2—C2—Cl3 | 107.91 (11) | H20A—C20—H20B | 107.6 |
| Cl1—C2—Cl3 | 110.41 (11) | O29—C21—C20 | 113.09 (14) |
| N1—C10—C11 | 114.17 (14) | O29—C21—C22 | 99.57 (14) |
| N1—C10—H10A | 108.7 | C20—C21—C22 | 120.64 (16) |
| C11—C10—H10A | 108.7 | O29—C21—C28A | 98.57 (13) |
| N1—C10—H10B | 108.7 | C20—C21—C28A | 115.56 (14) |
| C11—C10—H10B | 108.7 | C22—C21—C28A | 106.13 (14) |
| H10A—C10—H10B | 107.6 | C23—C22—C21 | 106.13 (18) |
| O19—C11—C10 | 112.73 (14) | C23—C22—H22A | 126.9 |
| O19—C11—C18A | 98.64 (13) | C21—C22—H22A | 126.9 |
| C10—C11—C18A | 121.53 (15) | C22—C23—C24 | 105.84 (18) |
| O19—C11—C12 | 99.35 (14) | C22—C23—H23A | 127.1 |
| C10—C11—C12 | 115.42 (15) | C24—C23—H23A | 127.1 |
| C18A—C11—C12 | 105.78 (15) | O29—C24—C23 | 100.59 (15) |
| C13—C12—C11 | 106.25 (17) | O29—C24—C24A | 99.27 (15) |
| C13—C12—H12A | 126.9 | C23—C24—C24A | 106.97 (16) |
| C11—C12—H12A | 126.9 | O29—C24—H24A | 115.9 |
| C12—C13—C14 | 105.83 (17) | C23—C24—H24A | 115.9 |
| C12—C13—H13A | 127.1 | C24A—C24—H24A | 115.9 |
| C14—C13—H13A | 127.1 | C25—C24A—C28A | 121.17 (18) |
| O19—C14—C13 | 100.44 (15) | C25—C24A—C24 | 134.57 (19) |
| O19—C14—C14A | 99.30 (14) | C28A—C24A—C24 | 104.24 (16) |
| C13—C14—C14A | 107.32 (15) | C24A—C25—C26 | 117.9 (2) |
| O19—C14—H14A | 115.8 | C24A—C25—H25A | 121.0 |
| C13—C14—H14A | 115.8 | C26—C25—H25A | 121.0 |
| C14A—C14—H14A | 115.8 | C27—C26—C25 | 121.08 (19) |
| C15—C14A—C18A | 121.38 (18) | C27—C26—H26A | 119.5 |
| C15—C14A—C14 | 134.49 (18) | C25—C26—H26A | 119.5 |
| C18A—C14A—C14 | 104.12 (16) | C26—C27—C28 | 120.9 (2) |
| C14A—C15—C16 | 117.8 (2) | C26—C27—H27A | 119.6 |
| C14A—C15—H15A | 121.1 | C28—C27—H27A | 119.6 |
| C16—C15—H15A | 121.1 | C28A—C28—C27 | 117.6 (2) |
| C17—C16—C15 | 121.1 (2) | C28A—C28—H28A | 121.2 |
| C17—C16—H16A | 119.5 | C27—C28—H28A | 121.2 |
| C15—C16—H16A | 119.5 | C28—C28A—C24A | 121.36 (17) |
| C16—C17—C18 | 121.3 (2) | C28—C28A—C21 | 134.20 (17) |
| C16—C17—H17A | 119.4 | C24A—C28A—C21 | 104.43 (15) |
| C18—C17—H17A | 119.4 | C1—N1—C20 | 127.57 (15) |
| C18A—C18—C17 | 117.81 (19) | C1—N1—C10 | 116.44 (15) |
| C18A—C18—H18A | 121.1 | C20—N1—C10 | 115.99 (14) |
| C17—C18—H18A | 121.1 | C14—O19—C11 | 96.05 (13) |
| C18—C18A—C14A | 120.62 (17) | C24—O29—C21 | 96.01 (13) |
| O1—C1—C2—Cl2 | 4.6 (2) | C21—C22—C23—C24 | 0.0 (2) |
| N1—C1—C2—Cl2 | −171.75 (15) | C22—C23—C24—O29 | 33.4 (2) |
| O1—C1—C2—Cl1 | 122.72 (17) | C22—C23—C24—C24A | −69.8 (2) |
| N1—C1—C2—Cl1 | −53.7 (2) | O29—C24—C24A—C25 | 146.2 (2) |
| O1—C1—C2—Cl3 | −114.28 (18) | C23—C24—C24A—C25 | −109.7 (2) |
| N1—C1—C2—Cl3 | 69.3 (2) | O29—C24—C24A—C28A | −35.51 (18) |
| N1—C10—C11—O19 | −67.92 (19) | C23—C24—C24A—C28A | 68.64 (19) |
| N1—C10—C11—C18A | 48.7 (2) | C28A—C24A—C25—C26 | 0.7 (3) |
| N1—C10—C11—C12 | 178.87 (15) | C24—C24A—C25—C26 | 178.8 (2) |
| O19—C11—C12—C13 | 32.80 (19) | C24A—C25—C26—C27 | 0.7 (3) |
| C10—C11—C12—C13 | 153.59 (17) | C25—C26—C27—C28 | −0.9 (3) |
| C18A—C11—C12—C13 | −69.00 (19) | C26—C27—C28—C28A | −0.3 (3) |
| C11—C12—C13—C14 | 0.4 (2) | C27—C28—C28A—C24A | 1.7 (3) |
| C12—C13—C14—O19 | −33.89 (19) | C27—C28—C28A—C21 | −179.31 (18) |
| C12—C13—C14—C14A | 69.4 (2) | C25—C24A—C28A—C28 | −1.9 (3) |
| O19—C14—C14A—C15 | −144.6 (2) | C24—C24A—C28A—C28 | 179.46 (17) |
| C13—C14—C14A—C15 | 111.3 (2) | C25—C24A—C28A—C21 | 178.80 (17) |
| O19—C14—C14A—C18A | 36.13 (17) | C24—C24A—C28A—C21 | 0.19 (18) |
| C13—C14—C14A—C18A | −67.96 (18) | O29—C21—C28A—C28 | −144.1 (2) |
| C18A—C14A—C15—C16 | −0.1 (3) | C20—C21—C28A—C28 | −23.4 (3) |
| C14—C14A—C15—C16 | −179.27 (19) | C22—C21—C28A—C28 | 113.2 (2) |
| C14A—C15—C16—C17 | 1.3 (3) | O29—C21—C28A—C24A | 35.00 (16) |
| C15—C16—C17—C18 | −0.9 (3) | C20—C21—C28A—C24A | 155.77 (15) |
| C16—C17—C18—C18A | −0.6 (3) | C22—C21—C28A—C24A | −67.64 (17) |
| C17—C18—C18A—C14A | 1.8 (3) | O1—C1—N1—C20 | 172.92 (18) |
| C17—C18—C18A—C11 | −179.78 (19) | C2—C1—N1—C20 | −10.9 (3) |
| C15—C14A—C18A—C18 | −1.5 (3) | O1—C1—N1—C10 | −6.9 (3) |
| C14—C14A—C18A—C18 | 177.93 (16) | C2—C1—N1—C10 | 169.28 (16) |
| C15—C14A—C18A—C11 | 179.68 (16) | C21—C20—N1—C1 | −114.5 (2) |
| C14—C14A—C18A—C11 | −0.92 (17) | C21—C20—N1—C10 | 65.2 (2) |
| O19—C11—C18A—C18 | 147.2 (2) | C11—C10—N1—C1 | −104.29 (19) |
| C10—C11—C18A—C18 | 23.7 (3) | C11—C10—N1—C20 | 75.91 (19) |
| C12—C11—C18A—C18 | −110.5 (2) | C13—C14—O19—C11 | 52.63 (15) |
| O19—C11—C18A—C14A | −34.20 (16) | C14A—C14—O19—C11 | −57.06 (15) |
| C10—C11—C18A—C14A | −157.68 (16) | C10—C11—O19—C14 | −174.45 (14) |
| C12—C11—C18A—C14A | 68.14 (18) | C18A—C11—O19—C14 | 55.97 (14) |
| N1—C20—C21—O29 | −76.82 (19) | C12—C11—O19—C14 | −51.72 (15) |
| N1—C20—C21—C22 | 40.7 (2) | C23—C24—O29—C21 | −52.34 (16) |
| N1—C20—C21—C28A | 170.63 (14) | C24A—C24—O29—C21 | 57.01 (16) |
| O29—C21—C22—C23 | −33.12 (19) | C20—C21—O29—C24 | −178.99 (15) |
| C20—C21—C22—C23 | −157.33 (17) | C22—C21—O29—C24 | 51.68 (16) |
| C28A—C21—C22—C23 | 68.77 (19) | C28A—C21—O29—C24 | −56.40 (15) |
Hydrogen-bond geometry (Å, º)
Cg8 is the centroid of the C24A/C25–C28/C28A aromatic ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C10—H10A···O29 | 0.97 | 2.35 | 3.074 (2) | 131 |
| C12—H12A···O1i | 0.93 | 2.66 | 3.494 (2) | 150 |
| C17—H17A···O1ii | 0.93 | 2.51 | 3.427 (3) | 168 |
| C20—H20A···O19 | 0.97 | 2.39 | 3.068 (2) | 127 |
| C27—H27A···O19iii | 0.93 | 2.51 | 3.438 (3) | 175 |
| C20—H20B···Cl1 | 0.97 | 2.55 | 3.1744 (18) | 122 |
| C20—H20B···Cl3 | 0.97 | 2.64 | 3.2921 (19) | 125 |
| C13—H13A···Cg8iv | 0.93 | 2.90 | 3.633 (2) | 136 |
Symmetry codes: (i) −x+3/2, y+1/2, −z+1/2; (ii) −x+3/2, y−1/2, −z+1/2; (iii) −x+1/2, y−1/2, −z+1/2; (iv) x−1/2, −y+1/2, z−1/2.
References
- Borisova, K. K., Kvyatkovskaya, E. A., Nikitina, E. V., Aysin, R. R., Novikov, R. A. & Zubkov, F. I. (2018a). J. Org. Chem. 83, 4840–4850. [DOI] [PubMed]
- Borisova, K. K., Nikitina, E. V., Novikov, R. A., Khrustalev, V. N., Dorovatovskii, P. V., Zubavichus, Y. V., Kuznetsov, M. L., Zaytsev, V. P., Varlamov, A. V. & Zubkov, F. I. (2018b). Chem. Commun. 54, 2850–2853. [DOI] [PubMed]
- Bruker (2013). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Criado, A., Peña, D., Cobas, A. & Guitián, E. (2010). Chem. Eur. J. 16, 9736–9740. [DOI] [PubMed]
- Criado, A., Vilas-Varela, M., Cobas, A., Pérez, D., Peña, D. & Guitián, E. (2013). J. Org. Chem. 78, 12637–12649. [DOI] [PubMed]
- Demircan, A., Temel, E., Kandemir, M. K., Çolak, M. & Büyükgüngör, O. (2013). Acta Cryst. E69, o1628–o1629. [DOI] [PMC free article] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Grudova, M. V., Gil, D. M., Khrustalev, V. N., Nikitina, E. V., Sinelshchikova, A. A., Grigoriev, M. S., Kletskov, A. V., Frontera, A. & Zubkov, F. I. (2020). New J. Chem. 44, 20167–20180.
- Gurbanov, A. V., Kuznetsov, M. L., Demukhamedova, S. D., Alieva, I. N., Godjaev, N. M., Zubkov, F. I., Mahmudov, K. T. & Pombeiro, A. J. L. (2020a). CrystEngComm, 22, 628–633.
- Gurbanov, A. V., Kuznetsov, M. L., Mahmudov, K. T., Pombeiro, A. J. L. & Resnati, G. (2020b). Chem. Eur. J. 26, 14833–14837. [DOI] [PubMed]
- Juhl, M. & Tanner, D. (2009). Chem. Soc. Rev. 38, 2983–2992. [DOI] [PubMed]
- Khalilov, A. N., Asgarova, A. R., Gurbanov, A. V., Maharramov, A. M., Nagiyev, F. N. & Brito, I. (2018a). Z. Kristallogr. New Cryst. Struct. 233, 1019–1020.
- Khalilov, A. N., Asgarova, A. R., Gurbanov, A. V., Nagiyev, F. N. & Brito, I. (2018b). Z. Kristallogr. New Cryst. Struct. 233, 947–948.
- Koşar, B., Demircan, A., Arslan, H. & Büyükgüngör, O. (2011). Acta Cryst. E67, o994–o995. [DOI] [PMC free article] [PubMed]
- Koşar, B., Karaarslan, M., Demir, I. & Büyükgüngör, O. (2007). Acta Cryst. E63, o3323.
- Krishna, G., Grudinin, D. G., Nikitina, E. V. & Zubkov, F. I. (2021). Synthesis, 53, https://doi.org/10.1055/s-0040-1705983.
- Kvyatkovskaya, E. A., Epifanova, P. P., Nikitina, E. V., Senin, A. A., Khrustalev, V. N., Polyanskii, K. B. & Zubkov, F. I. (2021). New J. Chem. 45, 3400–3407.
- Kvyatkovskaya, E. A., Nikitina, E. V., Khrustalev, V. N., Galmés, B., Zubkov, F. I. & Frontera, A. (2020). Eur. J. Org. Chem. pp. 156–161.
- Ma, Z., Gurbanov, A. V., Maharramov, A. M., Guseinov, F. I., Kopylovich, M. N., Zubkov, F. I., Mahmudov, K. T. & Pombeiro, A. J. L. (2017a). J. Mol. Catal. A Chem. 426, 526–533.
- Ma, Z., Gurbanov, A. V., Sutradhar, M., Kopylovich, M. N., Mahmudov, K. T., Maharramov, A. M., Guseinov, F. I., Zubkov, F. I. & Pombeiro, A. J. L. (2017b). Mol. Catal. 428, 17–23.
- Ma, Z., Mahmudov, K. T., Aliyeva, V. A., Gurbanov, A. V., Guedes da Silva, M. F. C. & Pombeiro, A. J. L. (2021). Coord. Chem. Rev. 437, 213859.
- Ma, Z., Mahmudov, K. T., Aliyeva, V. A., Gurbanov, A. V. & Pombeiro, A. J. L. (2020). Coord. Chem. Rev. 423, 213482.
- Mahmudov, K. T., Gurbanov, A. V., Aliyeva, V. A., Resnati, G. & Pombeiro, A. J. L. (2020). Coord. Chem. Rev. 418, 213381.
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. pp. 3814–3816. [DOI] [PubMed]
- Mertsalov, D. F., Alekseeva, K. A., Daria, M. S., Cheshigin, M. E., Çelikesir, S. T., Akkurt, M., Grigoriev, M. S. & Mlowe, S. (2021a). Acta Cryst. E77, 466–472. [DOI] [PMC free article] [PubMed]
- Mertsalov, D. F., Nadirova, M. A., Sorokina, E. A., Vinokurova, M. A., Çelikesir, S. T., Akkurt, M., Kolesnik, I. A. & Bhattarai, A. (2021b). Acta Cryst. E77, 260–265. [DOI] [PMC free article] [PubMed]
- Mertsalov, D. F., Zaytsev, V. P., Pokazeev, K. M., Grigoriev, M. S., Bachinsky, A. V., Çelikesir, S. T., Akkurt, M. & Mlowe, S. (2021c). Acta Cryst. E77, 255–259. [DOI] [PMC free article] [PubMed]
- Mizar, A., Guedes da Silva, M. F. C., Kopylovich, M. N., Mukherjee, S., Mahmudov, K. T. & Pombeiro, A. J. L. (2012). Eur. J. Inorg. Chem. pp. 2305–2313.
- Padwa, A. & Flick, A. C. (2013). Adv. Heterocycl. Chem. 110, 1–41.
- Parvatkar, P. T., Kadam, H. K. & Tilve, S. G. (2014). Tetrahedron, 70, 2857–2888.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Takao, K., Munakata, R. & Tadano, K. (2005). Chem. Rev. 105, 4779–4807. [DOI] [PubMed]
- Temel, E., Demircan, A., Arslan, H. & Büyükgüngör, O. (2011). Acta Cryst. E67, o1304–o1305. [DOI] [PMC free article] [PubMed]
- Temel, E., Demircan, A., Beyazova, G. & Büyükgüngör, O. (2012). Acta Cryst. E68, o1102–o1103. [DOI] [PMC free article] [PubMed]
- Temel, E., Demircan, A., Kandemir, M. K., Çolak, M. & Büyükgüngör, O. (2013). Acta Cryst. E69, o1551–o1552. [DOI] [PMC free article] [PubMed]
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. The University of Western Australia.
- Zubkov, F. I., Nikitina, E. V. & Varlamov, A. V. (2005). Russ. Chem. Rev. 74, 639–669.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989021009907/yk2156sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021009907/yk2156Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989021009907/yk2156Isup3.cml
CCDC reference: 2095762
Additional supporting information: crystallographic information; 3D view; checkCIF report