Abstract
Here we describe the development and application of miniature integrated microscopes (miniscopes) paired with microendoscopes that allow for the visualization and manipulation of neural circuits in superficial and subcortical brain regions in freely behaving animals. Over the past decade the miniscope platform has expanded to include simultaneous optogenetic capabilities, electrically-tunable lenses that enable multi-plane imaging, color-corrected optics, and an integrated data acquisition platform that streamlines multimodal experiments. Miniscopes have given researchers an unprecedented ability to monitor hundreds to thousands of genetically-defined neurons from weeks to months in both healthy and diseased animal brains. Sophisticated algorithms that take advantage of constrained matrix factorization allow for background estimation and reliable cell identification, greatly improving the reliability and scalability of source extraction for large imaging datasets. Data generated from miniscopes have empowered researchers to investigate the neural circuit underpinnings of a wide array of behaviors that cannot be studied under head-fixed conditions, such as sleep, reward seeking, learning and memory, social behaviors, and feeding. Importantly, the miniscope has broadened our understanding of how neural circuits can go awry in animal models of progressive neurological disorders, such as Parkinson’s disease. Continued miniscope development, including the ability to record from multiple populations of cells simultaneously, along with continued multimodal integration of techniques such as electrophysiology, will allow for deeper understanding into the neural circuits that underlie complex and naturalistic behavior.
Keywords: miniscope, calcium imaging, neuroscience, neural circuits
Introduction
The ability to monitor and modulate the activity of large numbers of neurons in vivo enables neuroscientists to elucidate the correlative and causal links between neural circuit dynamics and behaviors. Recently, optical tools to record and manipulate neural circuit activity in behaving animals have emerged, revolutionizing the study of brain circuit function during a behavior. Optical tools enable precise spatiotemporal visualization and modulation of neural circuits on a physiologically relevant timescale. With genetically encoded activity-sensing fluorescent indicators, particularly calcium indicators such as GCaMP, it is now possible to image neural dynamics in behaving rodents at the cellular level over periods of weeks at ∼10-ms time resolution [1–3]. With genetically encoded opsins, it is now also possible to modulate cellular dynamics with light at the network and single-cell level [4–6]. These optical tools have overcome prior limitations with functional magnetic resonance imaging and multielectrode array technologies, which exhibit poor spatial and/or temporal resolution, require animals to be immobilized and are unable to target specific cell types. The advent of these new optical tools for in vivo calcium imaging has necessitated the development of novel algorithms for translating the noisy fluorescent signals into meaningful representations of neural activity. Several novel applications of matrix factorization combined with appropriate statistical modeling now make it possible to automatically extract neuronal activity with increasing accuracy and minimal human intervention, thereby enabling a scalable analysis of circuit activity at a single-neuron resolution from optical calcium activity signals [7–10].
In this review, we describe developments and advancements in optical tools for imaging neural activity in vivo. We focus on miniaturized microscopy (miniscope) development for recording and manipulating neural circuit activity in freely behaving animals and review neuroscience research that has been enabled with these miniscopes. Specifically, we will review the application of miniscope technology to the study of neural computations underlying stimulus encoding, decision-making, and learning and memory to regulate a diverse array of behavioral outputs, from naturalistic and instinctive behaviors, such as food intake and sleep, to more complex repertoires of behaviors, such as environmental exploration and reward-seeking. These powerful new tools have so far mostly been applied in mice, taking advantage of transgenic mouse lines to target specific cell types and model human disease. Nevertheless, their use has increasingly extended to other less genetically tractable species that serve as important animal models for basic and translational neuroscience research and offer unique species-specific advantages including songbirds, rats, prairie voles and non-human primates (NHPs).
Optical tools for imaging neural activity in vivo
Two-photon microscopy is an established method for functional and morphological neural imaging in vivo. Two-photon microscopy is a laser-scanning technique that allows for optical sectioning to exclude out-of-focus fluorescence, resulting in high-resolution three-dimensional images of depths of up to 1 mm in brain tissue. The high-resolution capabilities of two-photon imaging enable the investigation of subcellular dynamics such as calcium dynamics of dendritic spines [11] and subcellular membrane potential [12]. Two-photon microscopy is also capable of larger-scale imaging and has been routinely used to study cortical network activity [13,14] and has widely been used to interrogate the cortical circuit underpinnings of sensory processing [15–17]. More recently, two-photon microscopy has also been combined with imaging through gradient refractive index (GRIN) lenses to access deep brain regions such as the hypothalamus [18,19].
A major limitation of traditional two-photon imaging when studying neural circuit dynamics is the reliance on a head fixation preparation. While some naturalistic behaviors can be modeled under head-fixed conditions, head fixation can introduce added confounds such as stress [20]. Importantly, many naturalistic behaviors such as social interaction and sleep behaviors are not suited for head fixation. To overcome the limitation of traditional two-photon imaging, efforts have been made to miniaturize two-photon systems by adapting fiber optic technology to enable two-photon imaging of layer 2/3 dendritic calcium transients [21] and small-scale neuronal populations [22] during free behaviors, but these miniaturized two-photon systems are fundamentally limited to small fields of view and/or a low temporal resolution that preclude the study of neuronal activity in hundreds to thousands of neurons under naturalistic conditions.
Fiber optic technology has also been adapted for one-photon imaging of neural dynamics. Flusberg et al. [1] developed a head-mounted miniaturized microscope that enabled cellular-resolution neural recordings as well as hemodynamic measurements in freely behaving rodents. These fiber-based one-photon imaging technologies were however limited by rigid and pixelated fibers, severely constraining animal behaviors and resulting in low-resolution and low-fidelity imaging signals. Fiber photometry is another technique that uses optical fibers to record changes in calcium activity in neural circuits during free behaviors [23]. With fiber photometry, photons from the optical fibers are summed, resulting in a single bulk fluorescent photometry signal. Owing to the small spatial footprint of optical fiber ferrules, implanting multiple optical fibers into different brain regions is relatively straightforward and has been used to collect simultaneous photometry signals from cortical and subcortical brain regions [24]. While fiber photometry-based imaging approaches are low cost and relatively easy to implement, a bulk signal is inherently spatially and temporally lossy, precluding the analysis of cellular-level neuronal dynamics and potentially resulting in the misleading interpretation of brain circuit activity.
Development of a miniaturized microscope for recording cellular-level neural circuit dynamics in freely behaving animals
To overcome the limitations of the aforementioned optical tools and enable neuroscientists to record and monitor large-scale calcium dynamics at single-cell resolution in freely behaving rodents, the integrated miniaturized head-mounted microscope (miniscope) was invented and developed by a team at Stanford University [25]. Miniscopes weigh ∼2 g and, when borne on the cranium of a freely behaving rodent and used in conjunction with GCaMP, allow neuroscientists to visualize activity in hundreds of genetically defined neurons during natural behaviors. When paired with custom-designed optical GRIN lenses that are implanted in the brain, these miniscopes permit neuroscientists to target and image from deep brain regions and from regions inaccessible to other large-scale recording technologies.
Miniscopes utilize a conventional epi-fluorescence microscope design in which the objective serves to both deliver the excitation light to the specimen and collect the fluorescence emitted by the specimen to form an image of the brain region of interest (ROI). Excitation light and emission light are separated by a dichroic mirror assisted by additional excitation and emission bandpass filters for further spectral cleanup (Fig. 1). Miniscopes predominantly use a GRIN optic as an objective as these offer large numerical apertures in a small form factor, which is not readily obtained using discrete optical elements.
Fig. 1.

Design and fabrication of an integrated fluorescence microscope. (a) Computer-assisted design of a first-generation epifluorescence miniscope. (b) Assembled miniscope. Note the fine-threaded turret enabling manual focusing by the rotation of the image sensor mount. Figure reprinted from Ghosh et al. [25].
Building on the success of these early miniscopes, next-generation miniscopes are now commercially available and include enhanced functionality. The nVistaTM system is a single-channel epifluorescence miniscope that has electronic focusing capabilities to enable simultaneous multiplane imaging, thus maximizing the number of recorded cells. The nVokeTM system integrates simultaneous and sequential same field of view optogenetics, allowing researchers to casually link neural circuit activity with behaviors [26]. Finally, the recently developed nVueTM system includes dual-color imaging capabilities, enabling researchers to image from two brain signals simultaneously.
Each of these miniscope systems has a nonintrusive data-acquisition and control box that can be remotely controlled without interfering with the animal’s behavior and/or the ongoing experiment. The nVista, nVoke and nVue boxes are web-enabled enabling direct data-streaming to the cloud (if allowed by the available network capacity and bandwidth). They each comprise Graphics processing unit (GPU)-based video processing, synchronization with external data streams (such as behavioral video streams) and various analog and digital input/output options (Fig. 2).
Fig. 2.

Next-generation miniscope. Control and data-acquisition device for the Inscopix nVista miniscope system allowing remote control via a web-browser-enabled GUI.
Extracting neural signals from noisy optical calcium imaging data
Processing calcium activity movies into estimations of individual neuron activity requires two main steps: (i) identifying the spatial footprints of individual neurons and (ii) computing the time-varying calcium activity signal for each neuron. The most straightforward solution is to visually inspect the movie, manually draw ‘ROIs’ for each neuron and then estimate each neuron’s calcium activity as the mean ROI pixel intensity; however, this ‘manual ROI’ approach has several disadvantages. First, it is time-consuming as careful visual inspection and identification of ROIs can take several hours of expert labor. Second, it does not account for out-of-focus fluorescent signals or leverage understanding of the underlying biosensor kinetics, which can significantly degrade the accuracy of the extracted neural signals. Finally, manual ROI selection is vulnerable to human bias and subjectivity that can both drift over time and vary across people, all of which can potentially confound the estimation of neural activity.
To overcome the limitations of manual ROI selection, several automated algorithms have been developed over the past decade, each improving the accuracy and speed of transforming calcium imaging movies into representations of neural circuit activity. Here, we briefly review and compare the two most common techniques currently used for processing miniscope calcium imaging data, as well as the software products available for their deployment. Note that while the discussion below focuses on algorithms for translating pixel intensities to neural calcium dynamics, we should not forget the importance of preprocessing data to correct for brain motion (which varies dramatically in magnitude depending on the brain region and surgical preparation), as well as deconvolution and event detection [27].
The first majorly successful automated source extraction method for calcium imaging was the development of ‘principal component analysis–independent component analysis (PCA–ICA)’ by Mukamel et al. in 2009 [7]. The method comprises two main steps: first, a PCA to remove shot noise and reduce dimensionality, and second, an (ICA) to identify the spatial footprints and signal dynamics of each neuron. PCA–ICA is computationally efficient and remarkably effective at cell detection without requiring any preconceptions of neurons’ spatial shape or temporal activity kinetics. There is, however, an assumption that neurons’ spatial location and temporal activity are sparse and statistically independent.
PCA–ICA was originally developed for two-photon calcium imaging data, where a high z-resolution minimizes signal contamination from out-of-focus neurons expressing fluorescent calcium indicators. Unfortunately, poorer z-resolution of one-photon microscopy results in a much larger contribution of the fluorescent signal from neurons residing above and below the imaging plane, which acts to confound and obscure the activities of in-focus neurons. To better account for this dynamic background signal in one-photon data, constrained non-negative matrix factorization for microendoscopic imaging (CNMF-E) was proposed in 2018 [8]. In this approach, movies are factorized into four separate matrices: a constant background, a fluctuating background, the neural signals and the residual (Fig. 3a and b). Factorization is achieved via optimization of several model constraints including the following: (i) neuron shapes are roughly spherical and of a certain diameter and (ii) neural activity is well-described by an autoregressive model with exponential decay constants matching our knowledge of biosensor kinetics. Fluctuating background is computed for each neuron individually by considering a ring of pixels greater than the diameter of the neuron to approximate signal contributions from blurred neurons residing above or below the imaging plane. Compared to PCA–ICA, the authors of CNMF-E showed evidence that it yields a higher cell identification recall, with a higher signal-to-noise ratio of the extracted activity traces (Fig. 3d–f). However, these advantages come at a cost: CNMF-E tends to identify more false positives (lower precision) and is computationally slow and expensive compared to PCA–ICA.
Fig. 3.

Comparison of PCA/ICA and CNMF-E for extracting neural signals from miniscope calcium movies. (a) An example frame of the raw data and its four components decomposed by CNMF-E. (b) The mean fluorescence traces of the raw data, the estimated background activity and the background-subtracted data within the segmented area (red box) in (a). (c) The distributions of the variance explained by different components over all pixels; note that estimated background signals dominate the total variance of the signal. (d) The contour plot of all neurons detected by CNMF-E and PCA/ICA superimposed on the correlation image. Green areas represent the components that are only detected by CNMF-E. The components are sorted in decreasing order based on their SNRs (from red to yellow). (e) The spatial and temporal components of 14 example neurons that are only detected by CNMF-E (green areas in d). (f) The SNRs of all neurons detected by both methods. Colors match the example traces shown in (g), which shows the spatial and temporal components of 10 example neurons detected by both methods. Scale bar: 10 s. Figure reprinted from Zhou et al. [8] under the Creative Commons Attribution license. SNR: Signal-to-noise ratio.
While PCA–ICA and CNMF-E both represent critical advances in our ability to automatically detect neurons and extract calcium activity from one-photon miniscope data, there remain limitations in efficiency and accuracy that will require continued innovation to overcome. Recent attempts to improve background estimation and dimensionality reduction for CNMF-E [28], as well as an entirely new method of matrix factorization based on leveraging robust estimation theory and GPU implementation [9] show exciting promise in this regard.
In addition to method innovation, there is a need to standardize and package existing techniques into software tools and products for wide adoption by miniscope users. For those comfortable programming, the CaImAn computational toolbox offers several methods for calcium imaging processing, including a Python version of CNMF-E [10]. CNMF-E was recently implemented in C++ and can be used in the Inscopix Data Processing Software (IDPS) Graphical user interface (GUI), Python application programming interface (API) or Matlab API. IDPS also offers PCA–ICA and several other preprocessing tools (e.g. motion processing) in its GUI and APIs. CIAtah, written in Matlab, is another software package that allows users to access several different algorithms for one-photon calcium imaging with a GUI [29]. As more neuroscientists adopt miniscope technology for investigating the brain, it will be increasingly important to develop easy-to-use software tools for processing, analyzing and managing calcium data so that it can be interpreted without requiring computer science expertise.
Of course, extracting neuronal signals from the noisy optical calcium imaging movies is just the first step toward identifying and interpreting meaningful neural circuit activity patterns. Dimensionality reduction methods [30], generalized linear models [31] and recurring switching dynamical systems [32] are just a few examples of computational techniques that are proving useful for uncovering latent patterns embedded in high-dimensional neural circuit activity data. Analogous methods are being developed and used for quantifying the equally rich behavior video data (for a nice summary of computational ethology methods, see ref. [33]). With miniscopes, it is now possible to combine these datasets and computational techniques together toward a better understanding of how neural circuit dynamics relate and give rise to freely moving behaviors [34].
Scientific applications in rodent models
Features of the miniscope described above have greatly expanded the breadth of neuroscience research questions that can be asked. The ability to monitor the activity of large populations of neurons during naturalistic behaviors has been particularly advantageous as many adaptive and translationally relevant behaviors are not compatible with conditions where movement is restricted. For example, animals cannot sleep under head-fixed conditions, and social behaviors that require dynamic back-and-forth interactions between individuals are severely limited under restrained locomotion [35]. An important additional advantage of the miniscope technology is the ability to repeatedly track the same cells over time, which is critical to understand how experience-induced plasticity can alter behaviors. This feature was first applied to studies of learning and memory [36] but has more recently been applied to studies of homeostatic drive, where it is critical to track changes in neural activity alongside changes in internal states [37]. Finally, the combination of miniscopes with tools to genetically and anatomically define cell types as well as GRIN lenses to bring the fluorescently encoded activity patterns of cells above the surface of the brain is enabling neuroscientists to decode the precise neural circuits that drive complex behavioral outputs [38]. Together, these features are unlocking experiments that were not previously feasible to increase our understanding of neural circuit computations that regulate hunger, thirst, sleep, environment exploration and social behaviors.
Hunger and thirst
Hunger and thirst direct appetitive or consummatory actions toward maintaining body homeostasis at an optimal set point for normal functioning [39]. This set point is not static, as energy needs will change with survival security, internal energy state and desirability of resources consumed [40]. However, how the brain receives and integrates signals to direct consumptive behaviors is not well understood. The application of miniscope technology to this field has been advantageous as it has identified heterogeneous signals within deep brain regions that generate complex modulations of feeding and drinking behaviors.
Cellular resolution imaging of genetically defined neuron populations by miniscopes revealed previously unidentified heterogenous patterns of neuronal activity that regulate consumptive behaviors. For example, when feeding was classified into constituent phases of food-seeking and consumption, miniscope experiments revealed that subpopulations of γ-aminobutyric acid (GABA) neurons in the lateral hypothalamus (LH) encode functionally discrete appetitive and consummatory behaviors (Fig. 4) [41]. Miniscope imaging also revealed that bidirectional modulation within genetically defined cell types is responsible for the varied permutations of consumptive decision-making, as glutamatergic neurons in the median preoptic area (MPOA) bidirectionally respond to water intake [42] and clusters of glutamatergic neurons in the anterior peri-locus coeruleus are activated or inhibited by food and water intake [43]. Single-cell recordings of parabrachial neurons expressing transcription factor Satb2 have also shown that the same neurons can be excited or inhibited by different taste stimuli [44]. Importantly, some miniscope results have challenged earlier findings on hunger and thirst that originated from studies lacking single-cell resolution. For example, several studies observed heterogenous or transient neuronal responses that were not observed in previous photometry studies [45–48]. Together, these results have revealed complex patterns of activity within genetically and anatomically defined circuits that regulate feeding and drinking behaviors.
Fig. 4.

Heterogeneous responses to consumptive behaviors. (a and b) Targeting LH GABAergic neurons to study regulation of appetitive versus consummatory behaviors. (c and d) Cell maps and corresponding example calcium traces showing that LH GABAergic neurons differently encode appetitive versus consummatory behaviors. (e and f) Miniscope imaging revealed that individual LH GABAergic neurons functionally encode either nose-poke responsive cells or lick-responsive cells, with a small (1.9%) overlap in LH GABAergic neurons that encode both. Figure adapted from Jennings et al. [41].
It is critical that neural circuits that control food intake are also capable of suppressing feeding behaviors when it is not immediately relevant for survival. Taking advantage of the miniscope’s capability to repeatedly monitor the activity of neurons under varying metabolic states and environmental conditions, Viskaitis et al. demonstrated that steroidogenic factor 1 (SF1) neurons within the ventral medial hypothalamus (VMH) differentially encode feeding versus threat and that the activity patterns of these neurons can exert opposing regulation over behavioral output. Specifically, the feeding behavior was associated with low activity of these neurons, while a high activity was associated with threatening environments as well as reduction in exploration and feeding [37]. While previous studies had demonstrated a role for SF1-VMH neurons in both feeding and defensive behaviors [49,50] it was unknown how SF1 neurons in the VMH differently encode rewarding versus threatening stimuli to drive opposing behavioral responses. The application of miniscope technology to complex experimental conditions where competing drive states can be assessed therefore has the ability to increase our understanding of how the brain integrates internal and external factors to generate behavioral responses that are adaptive under specific physiological states and environmental context.
Sleep
Sleep is a fundamental biological phenomenon widely observed in the animal kingdom. The regulation of sleep is primarily controlled by a network of nuclei located within the deep brain regions such as the hypothalamus, midbrain and the brainstem. Measuring the activities of these sleep-regulating neurons has been technically challenging due to a mix of different cell types activating at different brain states, neurons located at deep brain regions and the requirement of nonrestrained recording for natural sleep–wake cycle. Moreover, traditional in vivo electrophysiology or photometry cannot clearly distinguish intermingled cell types, and two-photon imaging requires head restraint that is not conducive to sleep. Miniscope imaging overcomes these limitations and can be combined with electroencephalography (EEG) and Electromyography (EMG) to classify the three distinct phases of sleep—wakefulness, rapid eye movement (REM) sleep and non-REM (NREM) sleep [51]. Miniscope imaging has therefore become a powerful tool in studying sleep circuits.
The first application of using miniscope imaging to study sleep was published in 2015 and characterized the activity of glutamatergic, GABAergic and cholinergic pontine neurons in sleep–wake regulation [52]. Thereafter, scientists have applied miniscope imaging to identify the activity of different cell types across different sleep–wake nuclei, such as GABAergic neurons in the ventral tegmental area, neurotensinergic neurons in the midbrain [53,54] and many different inhibitory neurons controlling REM–NREM transitions [55–57]. Recently, it was also used to identify dynamic changes in astrocyte activity across brain states that was distinct from that of neurons (Fig. 5) [58]. This finding not only identifies a role for glial cells in sleep regulation but also highlights a direction of applying miniscope imaging to investigate the role of non-neural cell types in the regulation of behaviors. Neural mechanisms of sleep disorder such as cataplexy were also studied by this approach, and abnormal activity of GABAergic neurons in the amygdala was found to be associated with emotion-induced cataplexy [59]. These findings demonstrate the broad application of miniscope imaging in answering the neural mechanisms of sleep–wake regulation and elucidating the pathological mechanisms of sleep disorders.
Fig. 5.

Miniscope imaging to investigate the role of astrocytes in sleep regulation. (a) Expression of GCaMP6f within cortical astrocytes, scale bar, 20 mm. (b) Schematic of miniscope imaging with EEG and EMG recordings. (c) Maximum projection dF/F images of calcium imaging in astrocytes during wake, NREM and REM sleep. Scale bar, 100 mm. (d) An example of astrocyte calcium dynamics with corresponding EEG, EEG power spectrogram and color-coded brain states. Figure adapted from Ingiosi et al. [58].
Miniscope technology can be used not only to study the sleep circuit (how we sleep) but also to study the function of sleep (why we sleep). Most of the recent efforts have focused on REM sleep, given its significant role in learning, memory and neural plasticity. Zhou et al. found the activity of hippocampal CA1 neurons was greater in REM sleep than NREM sleep and wakefulness, suggesting the activities of these memory-associated neurons are modulated by sleep states [60]. Adult-born neurons in the dentate gyrus (DG) are also important for learning and memory, and miniscope imaging identified a role for these neurons in consolidation of context–shock association during REM sleep [61,62]. Researchers not only examined known memory-associated neurons during sleep but also explored the role of sleep-regulatory neurons in memory formation. Izawa et al. found REM sleep–active melanin concentrating hormone–producing neurons in the LH are involved in active forgetting, illustrating hypothalamic sleep regulators can also be important for memory regulation [63]. Future studies utilizing the miniscope for different behavior tasks or combining with optogenetic approaches may help further understand other unknown functions of sleep.
Learning and memory
Our everyday life involves multiple sensory experiences, including exploring different places and interacting with animate and inanimate objects. Episodic memories reflect the ability to recollect the temporal and spatial context of these experiences. Their formation requires rapid synaptic plasticity within the hippocampus and is gradually consolidated within the hippocampal–entorhinal/perirhinal cortex–neocortex for permanent storage [64]. The consolidation units, named engrams, have been theorized for over a century [64,65] as a functional entity enabling memory storage and retrieval through the dynamic interaction of a large number of neurons. While synaptic plasticity has been studied with electrophysiological techniques established in the 20th century [66], the systematic investigation of large neuronal networks and their modulation has been limited by the lack of supporting technologies until very recently. Notably, the visualization of calcium dynamics in freely moving animals with miniscopes has been largely used to define the neural substrate of engrams, for their unique ability to track the activity of large populations of neurons across time, while the experimental subjects are in a naturalistic environment.
The hippocampus has been among the first and most studied brain areas related to learning and memory. The so-called place cells in dorsal CA1 (dCA1) have been indeed known for several decades [67], but the investigation of the neuronal activity was usually limited to few tens of cells per experimental subject, and therefore, the data were too sparse to study them at the ensemble level. Ziv et al. [36] tracked thousands of neurons over weeks from different mice exploring familiar environments. They found that the dCA1 coding had a day-to-day dynamism at the cellular level while preserving spatial information in the ∼15–25% overlap between coding ensembles from any 2 days. In other hippocampal regions, such as the DG, the use of miniscope associated with machine learning demonstrated that, although each individual neuron carries only a small amount of spatial information, the population activity pattern encodes orientation features such as position, speed and motion direction (Fig. 6) [68]. Further analyses of similar experimental data suggest that the day-scale ensemble dynamics might serve as timestamps for the formation of temporal information and to support the episodic memory [69,70].
Fig. 6.

Miniscopes enable tracking large populations of neurons over weeks. (a) Map of calcium activity in mouse imaged for 45 days. Color code as in (b): of all the active cells in day 1, a fraction (represented by the histogram) was active in the subsequent imaging sessions. Inset, notably, a constant fraction of all neurons detected over time was active each day. (c) Probability for a given neuron to be active (blue data) and to code for a place field (red data) in subsequent sessions declined with time. (d) The centroid shifts of identified cells were stable across days (color code days between sessions). (e–g) Example of place cells found on multiple sessions, ordered by place fields’ centroid positions on day 5 (e), day 20 (f) or day 35 (g) (mean ± SEM, data from four mice). Figure adapted from Ziv et al. [36].
Several recent studies [71,72] employed an activity-dependent cell-labeling technology, the c-fos-tet-tag system [73], to tag neurons that are active during a specific memory formation task, namely considered part of a given engram. The neural activity associated with memory formation induces the early gene c-fos expression, which in the c-fos-tTA transgenic mice induces activity-dependent tTA expression under the control of the c-fos promoter. The system is inactive during the administration of doxycycline that can therefore be used as a switch. In this way, it is possible to know what neurons participated in the memory formation and compare their activity during the time with neighboring neurons that are not part of the engram. Ghandour et al. [71] used this approach to monitor the activity of hundreds of dCA1 neurons during a contextual learning task, while selectively discriminating between engram (c-fos positive) and non-engram cells. By utilizing the selective tagging approach combined with freely behaving calcium imaging, they were able to demonstrate that engram cells were more synchronized than non-engram cells not only during the task but also during the post-learning sleep and retrieval sessions, supporting the role of sleep in memory consolidation.
Using a similar approach, Kitamura et al. [72] labeled neurons in the prefrontal cortex (PFC) that were active during a contextual fear conditioning (CFC) task to show that c-fos increased in PFC neurons early on during the task, but the calcium activity in PFC in response to the CFC shock was much higher 15 days after exposure than 1–2 days after exposure. These data, combined with optogenetic experiments, suggest that the PFC engrams are generated early on during CFC, but they become fully functional during the time. These findings highlight the role of the PFC in the early stages of memory formation and are in contrast with a previous model that hypothesized that remote memory is stored in the cortex by a slow transfer of hippocampal memory, rather than being primed at the time of the memory formation. The ability to track the activity of specific cell populations within complex circuits in freely moving animals greatly enhanced the knowledge of learning and memory mechanisms related to spatial orientation initially and of broader aspects of cognition such as emotions and social behaviors.
Social behavior
Social behavior is highly adaptive and critical for the survival of all sexually reproducing species [74]. At its core, it can be described as any form of communication or interaction with conspecifics; however, the appropriate display of social behaviors often requires the dynamic processing of social cues as well as the subsequent output of complex behavioral repertoires [75]. Moreover, social decisions, such as the decision to cooperate or fight, are context- and stimulus-specific, change across development as well as with experience, and are dependent on internal states, further adding to the complexity of social behaviors [76]. As a result, the neural mechanisms underlying the rapid integration of social cues with internal states to drive complex social interactions are not well understood. The application of miniscope technology to this field has been particularly advantageous to elucidate the neural correlates of social behaviors because its lightweight and flexible tether allows subjects to naturally interact with social conspecifics as well as for individual neurons to be tracked under experimental conditions where changes in social perception are hypothesized to occur.
An obvious difference in the display of social behaviors occurs with the types of behaviors that are directed toward adult conspecifics of the opposite sex [77]. Specifically, upon encountering a conspecific, an individual must first encode the properties of the stimulus (adult versus juvenile or male versus female) before selecting the appropriate behavioral output (prosocial, defensive or non-social/ignore) [78]. Although social stimulus perception is considered a critical first step in the display of appropriate behavioral responses, due to previous methodological limitations in the ability to repeatedly track the activity patterns of neurons under different social contexts, how the brain selectively encodes unique social stimulus properties is not well understood. To circumvent these limitations, Li et al. used miniscope imaging to track the activity of neurons within the medial amygdala (MeA), during naturalistic stimulus investigation [76]. Interestingly, their data revealed that in sexually naive males and females, conspecific social cues were uniquely encoded within the MeA of both sexes, with the majority of neurons showing a selective increase in activity in response to the social stimulus and the remainder of neurons showing either decreases in activity or non-selective responses to the social stimulus. Socially responsive neurons also displayed unique patterns of activity to specific categories of social stimuli, but the activity pattern of individual neurons was not consistent across interaction epochs. Instead, discrimination of social stimuli was encoded at the population level, and the size of the ensemble was inversely correlated with the latency to engage in aggressive or reproductive behaviors [76]. These data indicate that while encoding of social stimuli at the individual neuron level may be noisy, encoding at the population level can accurately represent social information to drive the appropriate behavioral response.
The MeA projects to regions of the hypothalamus that have also been implicated in the regulation of aggression and reproductive behaviors, such as the VMH and the MPOA [79], and studies utilizing miniscope imaging have begun to identify how neurons within these regions encode socially specific stimuli. Within the VMH of male mice, estrogen receptor-1 (Esr1+) expressing neurons showed increases in activity in response to both male and female conspecifics (Fig. 7a–d) and that repeated exposure to male and female conspecifics activated similar neuronal ensembles across trials, indicating that these neurons accurately encode the sex of the intruder. Moreover, cells activated by male and female conspecifics were largely distinct populations of neurons (Fig. 7f and g), and the specificity of neural stimulus encoding increased with social experience [80]. Similar changes in population encoding of social information following sexual experience have also been identified within non-hypothalamic nuclei such as the MeA [76] of mice and the Nucleus Accumbens (NAc) [81] and are associated with changes in behavioral states toward conspecifics. In addition to sexual experience, repeated exposure to social stress can also impact social stimulus encoding as chronic social defeat leads to increased activity of granule cells located within the ventral hippocampus as well as an increase in social avoidance behaviors [82]. Together, these observations enabled by miniscope imaging have revealed a distributed network of brain regions where experience-dependent plasticity underlies dynamic coding of social stimuli to drive adaptive behavioral responses. The identification of plasticity within hypothalamic nuclei was particularly surprising because similar to other instinctive behaviors such as feeding, innate social behaviors such as mating and aggression were thought to be ‘hard-wired’ and follow a labeled line theory. Thus, the ability to repeatedly monitor the activity of cells has provided key insights into the understanding of how social information is represented within the brain.
Fig. 7.
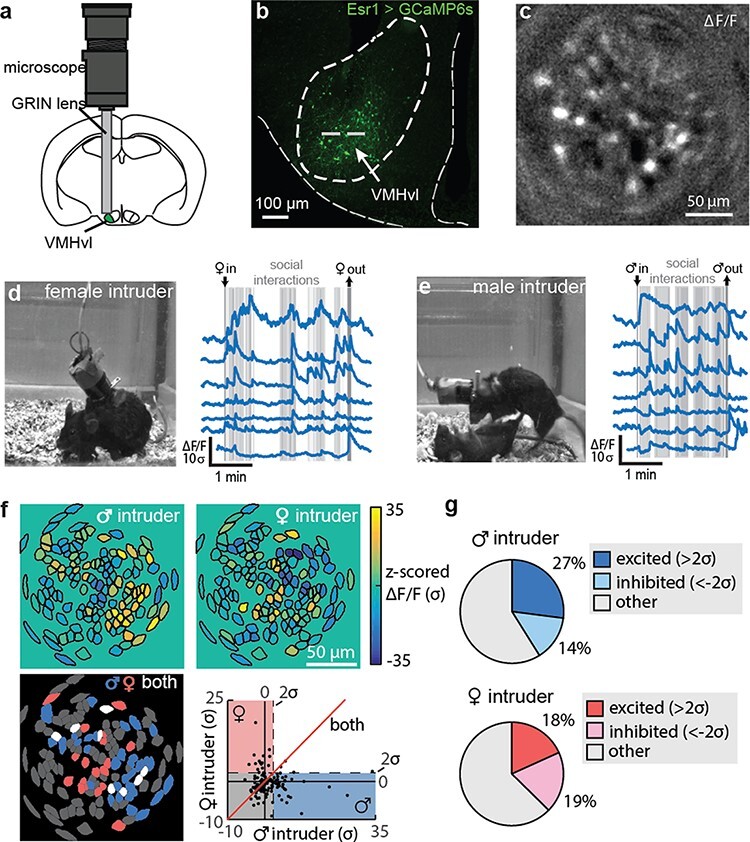
Miniscope imaging during dynamic social interactions. (a) Schematic of miniscope imaging. (b) GCaMP6 expression within VMH Esr1+ cells and approximate imaging plane. (c) Representative image from miniscope recording. (d and e) Representative frame from behavior video (left) and corresponding calcium traces aligned to movie frame (right) from either a (d) reproductive encounter or (e) an aggressive encounter. (f) Cells showing significant changes in activity to either a male or female intruder were largely distinct populations of neurons. (g) Male- and female-preferring cells responded with both increases and decreases in neural activity. Figure adapted from Remedios et al. [80].
In addition to increasing our understanding of how social stimuli are encoded within the brain, the ability to repeatedly track cells over time with miniscope methods has been utilized to disentangle neural correlates underlying social memory formation. In regard to social memory, studies have focused on examining both how the memory of conspecifics are encoded in the brain and the location in space where social encounters have previously occurred. These studies have identified that social memories of conspecifics are stored within the CA1 region of the ventral hippocampus [83], while prelimbic neurons that project to the NAc encode the location of spatial encounters [84]. Within the PFC, neurons located in the prelimbic region [84] and agranular insular cortex [85] are also activated during interactions with social stimuli, regardless of spatial location or stimulus familiarity, suggesting that these neurons may also play a more general role in social processing, perhaps through interactions with downstream regions associated with valence processing, motivation and social decision-making.
A major goal within the social neuroscience field is to identify the neural mechanisms that underlie the complexity of social behaviors that are not only critical for survival but also have large impacts on human mental health. Taking advantage of the ability to study naturalistic behaviors in freely behaving animals, miniscope imaging has greatly increased our understanding of the neural correlates of social behaviors along multiple steps of processing. Specifically, studies to date have identified experience-dependent computational patterns within the amygdala and hypothalamus that are associated with the encoding of specific stimuli, regions of the hippocampus and cortex that regulate social memory, and neuronal ensembles within the NAc that drive social motivation. Importantly, impairment in social functioning is a comorbidity of many psychiatric diseases [86], but the specific circuit disruptions that lead to social behavior abnormalities are not well understood. Applying miniscope imaging to studies of social functioning may thus have a translational impact, especially when combined with genetic models for disorders such as autism and schizophrenia to identify pathophysiological mechanisms associated with aberrant social function [87,88].
Translational applications
In addition to the basic research applications described above, the combination of miniscope imaging with animal models of disease may prove to be a valuable tool for identifying novel biomarkers of disease and screening potential therapeutic compounds. Indeed, recent studies using miniscopes to examine the pathogenesis underlying neurological disorders, such as epilepsy [89] and Parkinson’s disease [90], are providing novel mechanistic insights into circuit abnormalities that lead to aberrant behavior phenotypes associated with these conditions. Moreover, miniscope imaging can easily be combined with pharmacological administration of therapeutic compounds [89–92] to test target engagement and perform predictive efficacy assays, which together with an increased understanding of underlying disease pathology may help to improve the translatability of therapeutic compounds.
Miniscope imaging is beginning to reveal novel mechanistic insights into debilitating neurological disorders, such as epilepsy, in part through the ability to perform large-scale circuit dynamic recordings under conditions that were previously inaccessible with other methodologies. For example, although elevations in hippocampal calcium signaling have long been hypothesized to contribute to increased neuronal excitation and synchronization during seizure as well as seizure-induced brain damage [93], previous technical limitations made it challenging to examine calcium dynamics in freely behaving animals undergoing seizure. By combining miniscope imaging with an established model of epilepsy, kainic acid treatment [94] and assessments of seizure activity such as EEG and behavioral measures, Berdyyeva et al. were able to not only provide evidence supporting the hypothesis that calcium elevations are associated with neural and behavioral signatures of seizure but also identify novel neural signatures that may better characterize the underlying disease pathogenesis [89]. Specifically, they identified waves of calcium activity that occurred well before the onset of motor convulsions (average of 33 minutes prior) and had no consistent EEG phenotype. Thus, it is possible that motor convulsions and EEG signatures typically used to study seizures may reflect the latent expression of central nervous system pathology and may result from seizure propagation rather than initiation [89]. Moreover, when valproate, a commonly prescribed seizure medication [95], was administered prior to seizure induction, it was able to reduce behavioral symptoms of seizure (e.g. motor convulsions) but failed to alleviate aberrant calcium dynamics [89]. This latter finding is of particular importance, given that current epilepsy treatments often fail and one possibility may be that therapeutics are developed to ameliorate the symptoms but fail to target the underlying pathology. Integrating calcium imaging with current assessments of seizures in animal models may therefore improve the translatability of therapeutic compounds.
The ability to repeatedly track the activity dynamics of individual neurons is another advantage of miniscope imaging for translationally relevant studies because it allows for pre- and post-disease states to be compared, for changes in activity dynamics across the transgression of a disease states to be analyzed and for the ability of therapeutic compounds to reverse aberrant circuit dynamics to be assessed. These features may be especially important when trying to understand the pathology underlying progressive neurodegenerative disorders such as Parkinson’s disease where a gradual loss of dopamine leads to motor deficits and the efficacy of current therapeutics to treat these motor deficits degrades over time [96]. A hypothesized mechanism behind motor dysfunctions associated with Parkinson’s disease is that they arise from an imbalance in the activity of direct and indirect spiny projection neurons (SPNs) within the striatum. More specifically, hypo- and hyperactivity of direct and indirect pathway SPNs is thought to contribute to movement inhibition in the Parkinsonian state, while the inverse activity pattern within SPNs is thought to underlie Levodopa (L-DOPA)-induced dyskinesia [97]. However, direct evidence supporting this hypothesis had been lacking due to challenges in repeatedly monitoring the activity of direct and indirect SPNs across critical experimental time points.
By utilizing miniscopes to monitor the activity of direct and indirect SPNs before and after DA depletion with 6-Hydroxydopamine hydrobromide (6-OHDA), a classical model of Parkinson’s disease [96], Parker et al. were able to demonstrate that DA depletion indeed reduced the activity of direct pathway SPNs while increasing the activity of indirect pathway SPNs. In addition, they also demonstrated that the inverse of this activity pattern was true following L-DOPA-induced dyskinesia. Moreover, repeated miniscope imaging also enabled them to identify more complex alterations in striatal activity such as a reduction in activity coupling to motion onset and offset as well as a decrease in spatial activity clustering in indirect SPNs during the Parkisonian state. The opposite impact on motor activity coupling and spatial activity clustering within indirect SPNs occurred following L-DOPA-induced dyskinesia [90]. Together, these neural circuit signatures may act as biomarkers to facilitate the development of next-generation therapeutics that not only aim to restore activity balance within the striatum but also to restore spatiotemporal patterns that are critical for normal motor function. These findings also suggest that therapeutics for chronic neurological conditions also need to target treatment-induced neural adaptations in addition to the underlying neural circuit abnormalities that lead to the onset of the disease.
In addition to models described above, miniscope imaging has also been used with animal models of chronic pain [29], addiction [98,99], narcolepsy [100], anorexia [91], traumatic brain injury [101] and genetic models of social deficits. The combination of miniscope imaging with genetic models of psychiatric disorders is particularly advantageous because it has the potential to connect gene–circuit interactions to aberrant behavioral patterns. For example, by utilizing circuit-selective transgenic tools and a genetic mouse model of maladaptive social behaviors, Kim et al. were able to demonstrate that disruption of the Arp2/3 complex, a cytoskeleton regulator of dendritic spines, specifically within PFC neurons that project to the basolateral amygdala alters neural encoding of social stimuli and leads to disruptions in social behaviors [87]. Thus, they identified how a genetic mutation within a specific component of social circuitry was sufficient to disrupt the proper expression of adaptive social behaviors. Given that many psychiatric disorders are highly heritable [102], understanding how genetic mutations lead to specific circuit abnormalities may facilitate the discovery of novel treatments with possible improved therapeutic efficacy. In addition, the continued development of genetic tools and ability to combine these tools with miniscope technology for functional imaging will also be critical to decode the complex circuit interactions that govern how an individual’s perception of the external world is integrated with internal states to drive adaptive behaviors as well as how disruption of these circuits can lead to maladaptive behaviors associated with central nervous system disorders. Finally, integration of miniscope technology with primate models, which may more closely resemble clinical features of human diseases, may also provide valuable insights into pathology underlying brain disorders [96] and improve the successful translation of drugs into the clinic.
Miniscope imaging in higher-order species
As reviewed above, enormous progress has already been made applying miniscopes toward advancing our understanding of the neural circuit mechanisms underlying a vast array of brain functions and behaviors. The most common animal model species used for this work so far has been the mouse, largely motivated by the availability of transgenic lines (e.g. Cre recombinase lines) and other next-generation genetic and molecular technologies that have enabled imaging and optogenetic manipulation of highly precise cell populations and circuits [103]. While the genetic tractability of a model species is an important consideration for the deployment of these techniques, equally or more important is the relevance of a particular model species for the brain functions and behaviors under study. Furthermore, understanding the similarities and differences in circuit structure and function across a diverse range of species is essential toward identifying common principles of brain function that are more likely translatable to humans [104,105]. It is therefore no surprise that significant effort has been made toward applying miniscopes beyond the mouse, and exciting progress has already been achieved in species ranging from songbirds to rats and prairie voles [81,99,106–111].
Ultimately, to advance our understanding of higher-cognitive function, complex behavior and mental health, a critical need exists to translate the latest miniscope-based technologies to research using NHPs. Historically, the NHP, rhesus macaque, has been the animal model of choice to study human-relevant brain functions due to well-documented similarities between macaques and humans in terms of brain structures and complex behaviors [112,113]. More recently, the common marmoset (Callithrix jacchus), which has long been recognized as an important model for studying human-relevant social behaviors and vocal communications, has emerged as an attractive alternative to the macaque, particularly due to their small size and high reproductive efficiency, making them more amenable to genetic manipulation [114–118]. Indeed, major efforts are now underway toward studying marmoset brain structure and function and applying the latest gene editing technologies toward developing viable genetic models of human brain disease [119,120].
Significant progress has been made applying two-photon calcium imaging in both marmosets and macaques, overcoming several challenges associated with Adeno-associated viruses (AAV)-based expression of genetically encoded calcium indicators in the NHP and imaging artifacts caused by movements of their larger volume brains [121–130]. These studies have in some cases achieved stable cellular-resolution calcium imaging of large populations of neurons over several months of recordings, relying on transparent cranial windows to enable imaging of the gyral cortex to depths of ∼500 μm. Despite this important progress, several limitations of two-photon microscopy in NHPs remain including the following: (i) infection risk of using cranial windows, which also have limited functional lifetimes and require daily maintenance, (ii) cumbersome alignment between the microscope and the brain to maintain a consistent field of view and follow the same neurons across sessions, (iii) inability to image the gyral cortex deeper than ∼500 μm or cortical and subcortical structures positioned deeper in the brain, and (iv) necessity for head restraint, which limits natural behaviors. Head-mounted, one-photon miniscope calcium imaging is poised to overcome these limitations and thereby significantly expand the brain regions and behaviors that can be studied in NHPs.
The first successful demonstration of miniscope calcium imaging in NHP was recently demonstrated in behaving marmosets (Fig. 8) [131]. In this pioneering work, researchers developed and optimized a viral strategy and surgical methods to express GCaMP and chronically implant GRIN prism lenses into deep cortical layers of the primary motor cortex. They relied on an AAV system that utilizes tetracycline-controlled transcriptional activation to drive adequate levels of GCaMP expression while providing a mechanism (via doxycycline administration) to prevent overexpression [130]. They were able to record from hundreds of neurons in deep layers of the primary motor cortex while the marmosets engaged in natural motor behaviors including a seated lever-pulling task and a bidirectional arm-reaching task. As expected from previous electrophysiological studies in the primary motor cortex, neuronal calcium dynamics exhibited activity levels tuned to the direction of the animal’s reach, and the population calcium dynamics could be used to decode the animal’s reach direction on individual trials. Finally, to underscore the possibilities of studying unrestrained behaviors, researchers conducted imaging in marmosets performing a natural, minimally restrained ladder-climbing task. Together, this study confirmed several advantages of this technology as applied in the NHP, including imaging access to deep cortical layers beyond the current depth limits of two-photon imaging through cranial windows, and the ability to perform imaging during more natural, free behaviors as compared to two-photon imaging under head-fixed conditions. This study marked an important first step toward miniscope calcium imaging applications in NHPs and established a foundation upon which to apply similar approaches to the more commonly studied rhesus macaque.
Fig. 8.

Miniscope calcium imaging in behaving marmoset and macaque. (a) Tetracycline-controlled transcriptional activation system (TET-off AAV) used to express GCaMP in the motor cortex of the marmoset. Following lens implantation, miniscope imaging of motor cortical activity was performed in unrestrained animals performing naturalistic motor behaviors (top). Max projection image of in vivo GCaMP fluorescence–the bright colored regions in the image indicate cells that exhibited active calcium dynamics during the recording. Activity traces of example cells showing reach direction selectivity (bottom). Figure adapted from Kondo etal. [131]. (b) Post-mortem GCaMP expression in the premotor cortex of a macaque following injection of the AAV Tet-Off system. Cellular calcium dynamics were imaged bilaterally in the premotor cortices of a macaque performing a reach to reward motor task. Heatmaps depicting reach direction selectivity across the population of recorded cells (top). Stability of imaging allowed longitudinal tracking of cells across sessions that were multiple days apart, thereby allowing study of the relationship between neural activities and motor behaviors over time (bottom). Figure adapted from Bollimunta et al. [132].
Right on the heels of the success achieved in marmosets by Kondo et al., [131] our group recently achieved successful head-mounted microendoscopic calcium imaging in behaving macaques (Fig. 8) [132]. In this study, we demonstrated plug-and-play, head-mounted recordings of cellular-resolution calcium dynamics from large populations of neurons in the dorsal premotor cortex of macaques performing a motor reach task. As in the marmoset, these recordings were performed with the head unrestrained and therefore under relatively natural behavioral conditions. We used the same viral strategy (AAV Tet system) used in marmosets to express GCaMP in macaque premotor cortex and followed similar methods for chronic lens implantation, with the important addition of a cranial chamber surrounding the lens to provide added mechanical protection. Neuronal calcium dynamics exhibited tuning for reach direction and could be used to accurately decode the animal’s motor behavior on individual trials. Importantly, imaging was stable over months, allowing us to track individual neurons and monitor their relationship to motor behaviors over time. Finally, we were able to take advantage of the larger macaque brain to implant multiple lenses and perform simultaneous multi-site imaging from bilateral dorsal premotor cortices.
Together, the work in marmosets and macaques described above has established miniscope calcium imaging as an important new approach for studying the neural circuit mechanisms underlying complex and clinically relevant behaviors in NHPs. In order to take full advantage of this new approach, further developments are warranted to streamline the surgical workflow and improve overall ease of use. Novel viral approaches are needed to expand the options available for cell type–specific targeting, and, critically, longer lenses are needed to access subcortical regions deep within the NHP brain. As researchers increasingly pursue more naturalistic behavioral paradigms, the development of a wireless miniscope for NHPs will be essential. Finally, combining imaging and optogenetics would enable studies testing the causal relationship between neural activity and behaviors [26]. Together with these future developments, miniscope calcium imaging in NHPs will fuel important new insights into the neural circuit mechanisms underlying clinically relevant human behaviors and should help to accelerate the development of effective therapies for complex brain disorders.
Future directions
The decade-long developments in miniscope technology discussed here, from their initial invention and reduction to practice at Stanford University to the use of commercially available systems at hundreds of Neuroscience labs around the world, have progressed our understanding of the relationship between neural circuit dynamics, brain function and behaviors in both health and disease. By recording the activity of neurons during a behavior, the miniscope technology enables scientists to investigate the neural circuit correlates of behaviors, while the integration of optogenetic capabilities with conventional miniscope technology has allowed scientists to probe individual neural circuit elements to investigate the causal relationship between neural circuits and behaviors.
The addition of color-corrected optics and the development of achromatic miniscopes has paved the way to enable two-color imaging and simultaneous recordings of two distinct brain signals. Two-color imaging opens up exciting new neural circuit applications, including recording calcium activity in two distinct cell types, simultaneously imaging calcium activity with static markers that can identify neurons based on projection, activity or genetics, and simultaneously imaging calcium activity and blood flow to investigate neurovascular coupling. By combining green calcium indicators, such as GCaMP, with recently developed red calcium indicators, such as jRGECO1a [133] and XCaMP-R [134], researchers can explore the intricacies of how two populations of neurons interact during free behaviors using a dual-color miniscope system such as nVue.
Another exciting avenue for the development of miniscope technology is integration with other neuroscience tools to better understand brain function. One such tool is electrophysiology, specifically electrophysiological probes integrated with implanted GRIN lenses. While calcium imaging provides cell type–specific activity from genetically defined cells, same-site multiunit activity or local field potentials can add a more temporally precise activity measure of the local network in which the cells are embedded. Additional neuroscience tools that can be paired with miniscope imaging are sophisticated behavioral tracking software, such as DeepLabCut [135] and Syllable [136], that use machine learning techniques to detect nuanced rodent behaviors such as pose estimation and dynamics. The pairing of miniscope imaging with next-generation behavioral tracking will give unprecedented insight into the neural correlates underlying behaviors.
Finally, as the miniscope platform continues to evolve, an exciting application of the technology is to better understand how neural circuit patterns go awry in the context of disease. Tracking the relationship between the activity of specific cell populations and ongoing behaviors over the course of disease progression and in response to therapeutic intervention promises to significantly advance our understanding of disease pathophysiology and therapeutic mechanism of action. This should in turn help to accelerate the development of next-generation circuit-based therapeutics for a wide range of debilitating neurological and neuropsychiatric disorders.
Funding
None declared.
Conflicts of interest statement
Koen Visscher owns Inscopix stock. All authors are employed by Inscopix.
Contributor Information
Alice M Stamatakis, Inscopix Inc., 2462 Embarcadero Way, Palo Alto, CA 94303, USA.
Shanna L Resendez, Inscopix Inc., 2462 Embarcadero Way, Palo Alto, CA 94303, USA.
Kai-Siang Chen, Inscopix Inc., 2462 Embarcadero Way, Palo Alto, CA 94303, USA.
Morgana Favero, Inscopix Inc., 2462 Embarcadero Way, Palo Alto, CA 94303, USA.
Jing Liang-Guallpa, Inscopix Inc., 2462 Embarcadero Way, Palo Alto, CA 94303, USA.
Jonathan J Nassi, Inscopix Inc., 2462 Embarcadero Way, Palo Alto, CA 94303, USA.
Shay Q Neufeld, Inscopix Inc., 2462 Embarcadero Way, Palo Alto, CA 94303, USA.
Koen Visscher, Inscopix Inc., 2462 Embarcadero Way, Palo Alto, CA 94303, USA.
Kunal K Ghosh, Inscopix Inc., 2462 Embarcadero Way, Palo Alto, CA 94303, USA.
References
- 1.Flusberg B A, Nimmerjahn A, Cocker E D, Mukamel E A, Barretto R P J, Ko T H. et al. (2008) High-speed, miniaturized fluorescence microscopy in freely moving mice. Nat. Methods 5: 935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Göbel W and Helmchen F (2007) In vivo calcium imaging of neural network function. Physiology (Bethesda) 22: 358–365. [DOI] [PubMed] [Google Scholar]
- 3.Greenberg D S, Houweling A R, and Kerr J N D (2008) Population imaging of ongoing neuronal activity in the visual cortex of awake rats. Nat. Neurosci. 11: 749–751. [DOI] [PubMed] [Google Scholar]
- 4.Deisseroth K (2015) Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenno L, Yizhar O, and Deisseroth K (2011) The development and application of optogenetics. Annu. Rev. Neurosci. 34: 389–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prakash R, Yizhar O, Grewe B, Ramakrishnan C, Wang N, Goshen I. et al. (2012) Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat. Methods 9: 1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukamel E A, Nimmerjahn A, and Schnitzer M J (2009) Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63: 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P, Resendez S L, Rodriguez-Romaguera J, Jimenez J C, Neufeld S Q, Giovannucci A. et al. (2018) Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife 22: e28728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inan H, Schmuckermair C, Tasci T, Ahanonu B O, Hernandez O, Lecoq J. et al. (2021) Fast and statistically robust cell extraction from large-scale neural calcium imaging datasets, bioRxiv, 2021.03.24.436279. [Google Scholar]
- 10.Giovannucci A, Friedrich J, Gunn P, Kalfon J, Brown B L, Koay S A. et al. (2019) CaImAn an open source tool for scalable calcium imaging data analysis. eLife 8: e38173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Leischner U, Rochefort N L, Nelken I, and Konnerth A (2011) Functional mapping of single spines in cortical neurons in vivo. Nature 475: 501–505. [DOI] [PubMed] [Google Scholar]
- 12.Yang H H, St-Pierre F, Sun X, Ding X, Lin M Z, and Clandinin T R (2016) Subcellular imaging of voltage and calcium signals reveals neural processing in vivo. Cell 166: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sofroniew N J, Vlasov Y A, Hires S A, Freeman J, and Svoboda K (2015) Neural coding in barrel cortex during whisker-guided locomotion. eLife 4: e12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stosiek C, Garaschuk O, Holthoff K, and Konnerth A (2003) In vivo two-photon calcium imaging of neuronal networks. Proc. Natl. Acad. Sci. U.S.A. 100: 7319–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrillo-Reid L, Han S, Yang W, Akrouh A, and Yuste R (2019) Controlling visually guided behavior by holographic recalling of cortical ensembles. Cell 178: 447–457.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cottam J C H, Smith S L, and Häusser M (2013) Target-specific effects of somatostatin-expressing interneurons on neocortical visual processing. J. Neurosci. 33: 19567–19578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J, Hu H, Agmon A, and Svoboda K (2019) Recruitment of GABAergic interneurons in the barrel cortex during active tactile behavior. Neuron 104: 412–427.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossi M A, Basiri M L, McHenry J A, Kosyk O, Otis J M, van den Munkhof H E. et al. (2019) Obesity remodels activity and transcriptional state of a lateral hypothalamic brake on feeding. Science 364: 1271–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Resendez S L, Namboodiri V M K, Otis J M, Eckman L E H, Rodriguez-Romaguera J, Ung R L. et al. (2020) Social stimuli induce activation of oxytocin neurons within the paraventricular nucleus of the hypothalamus to promote social behavior in male mice. J. Neurosci. 40: 2282–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juczewski K, Koussa J A, Kesner A J, Lee J O, and Lovinger D M (2020) Stress and behavioral correlates in the head-fixed method: stress measurements, habituation dynamics, locomotion, and motor-skill learning in mice. Sci. Rep. 10: 12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helmchen F, Fee M S, Tank D W, and Denk W (2001) A miniature head-mounted two-photon microscope: high-resolution brain imaging in freely moving animals. Neuron 31: 903–912. [DOI] [PubMed] [Google Scholar]
- 22.Sawinski J, Wallace D J, Greenberg D S, Grossmann S, Denk W, and Kerr J N D (2009) Visually evoked activity in cortical cells imaged in freely moving animals. Proc. Natl. Acad. Sci. U.S.A. 106: 19557–19562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunaydin L A, Grosenick L, Finkelstein J C, Kauvar I V, Fenno L E, Adhikari A. et al. (2014) Natural neural projection dynamics underlying social behavior. Cell 157: 1535–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim C K, Yang S J, Pichamoorthy N, Young N P, Kauvar I, Jennings J H. et al. (2016) Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. Nat. Methods 13: 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh K K, Burns L D, Cocker E D, Nimmerjahn A, Ziv Y, Gamal A E. et al. (2011) Miniaturized integration of a fluorescence microscope. Nat. Methods 8: 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamatakis A M, Schachter M J, Gulati S, Zitelli K T, Malanowski S, Tajik A. et al. (2018) Simultaneous optogenetics and cellular resolution calcium imaging during active behavior using a miniaturized microscope. Front. Neurosci. 12. doi: 10.3389/fnins.2018.00496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans M H, Petersen R S, and Humphries M D (2020) On the use of calcium deconvolution algorithms in practical contexts, bioRxiv, 871137. [Google Scholar]
- 28.Buchanan E K, Kinsella I, Zhou D, Zhu R, Zhou P, Gerhard F. et al. (2019) Penalized matrix decomposition for denoising, compression, and improved demixing of functional imaging data, bioRxiv, 334706. [Google Scholar]
- 29.Corder G, Ahanonu B, Grewe B F, Wang D, Schnitzer M J, and Scherrer G (2019) An amygdalar neural ensemble that encodes the unpleasantness of pain. Science 363: 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Churchland M M, Cunningham J P, Kaufman M T, Foster J D, Nuyujukian P, Ryu S I. et al. (2012) Neural population dynamics during reaching. Nature 487: 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Driscoll L N, Pettit N L, Minderer M, Chettih S N, and Harvey C D (2017) Dynamic reorganization of neuronal activity patterns in parietal cortex. Cell 170: 986–999.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glaser J, Whiteway M, Cunningham J P, Paninski L, and Linderman S (2020) Recurrent switching dynamical systems models for multiple interacting neural populations. Adv. Neural. Inf. Process. Syst. 33: 14867–14878. [Google Scholar]
- 33.Pereira T D, Shaevitz J W, and Murthy M (2020) Quantifying behavior to understand the brain. Nat. Neurosci. 23: 1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markowitz J E, Gillis W F, Beron C C, Neufeld S Q, Robertson K, Bhagat N D. et al. (2018) The striatum organizes 3D behavior via moment-to-moment action selection. Cell 174: 44–58.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Resendez S L and Stuber G D (2015) In vivo calcium imaging to illuminate neurocircuit activity dynamics underlying naturalistic behavior. Neuropsychopharmacology 40: 238–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziv Y, Burns L D, Cocker E D, Hamel E O, Ghosh K K, Kitch L J. et al. (2013) Long-term dynamics of CA1 hippocampal place codes. Nat. Neurosci. 16: 264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viskaitis P, Irvine E E, Smith M A, Choudhury A I, Alvarez-Curto E, Glegola J A. et al. (2017) Modulation of SF1 neuron activity coordinately regulates both feeding behavior and associated emotional states. Cell Rep. 21: 3559–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamel E J O, Grewe B F, Parker J G, and Schnitzer M J (2015) Cellular level brain imaging in behaving mammals: an engineering approach. Neuron 86: 140–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris R B (1990) Role of set-point theory in regulation of body weight. FASEB J. 4: 3310–3318. [DOI] [PubMed] [Google Scholar]
- 40.Toates F M (1981) The control of ingestive behaviour by internal and external stimuli—a theoretical review. Appetite 2: 35–50. [DOI] [PubMed] [Google Scholar]
- 41.Jennings J H, Ung R L, Resendez S L, Stamatakis A M, Taylor J G, Huang J. et al. (2015) Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell 160: 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmerman C A, Huey E L, Ahn J S, Beutler L R, Tan C L, Kosar S. et al. (2019) A gut-to-brain signal of fluid osmolarity controls thirst satiation. Nature 568: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong R, Xu S, Hermundstad A, Yu Y, and Sternson S M (2020) Hindbrain double-negative feedback mediates palatability-guided food and water consumption. Cell 182: 1589–1605.e22. [DOI] [PubMed] [Google Scholar]
- 44.Jarvie B C, Chen J Y, King H O, and Palmiter R D (2021) Satb2 neurons in the parabrachial nucleus mediate taste perception. Nat. Commun. 12: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C, Navarrete J, Liang-Guallpa J, Lu C, Funderburk S C, Chang R B. et al. (2019) Defined paraventricular hypothalamic populations exhibit differential responses to food contingent on caloric state. Cell Metab. 29: 681–694.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu O, Iwai Y, Kondoh K, Misaka T, Minokoshi Y, and Nakajima K (2019) SatB2-expressing neurons in the parabrachial nucleus encode sweet taste. Cell Rep. 27: 1650–1656.e4. [DOI] [PubMed] [Google Scholar]
- 47.Augustine V, Gokce S K, Lee S, Wang B, Davidson T J, Reimann F. et al. (2018) Hierarchical neural architecture underlying thirst regulation. Nature 555: 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abbott S B G, Machado N L S, Geerling J C, and Saper C B (2016) Reciprocal control of drinking behavior by median preoptic neurons in mice. J. Neurosci. 36: 8228–8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Y, Nedungadi T P, Zhu L, Sobhani N, Irani B G, Davis K E. et al. (2011) Distinct hypothalamic neurons mediate estrogenic effects on energy homeostasis and reproduction. Cell Metab. 14: 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunwar P S, Zelikowsky M, Remedios R, Cai H, Yilmaz M, Meister M. et al. (2015) Ventromedial hypothalamic neurons control a defensive emotion state. eLife 4: e06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown R E, Basheer R, McKenna J T, Strecker R E, and McCarley R W (2012) Control of sleep and wakefulness. Physiol. Rev. 92: 1087–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox J, Pinto L, and Dan Y (2016) Calcium imaging of sleep–wake related neuronal activity in the dorsal pons. Nat. Commun. 7: 10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu X, Li W, Ma Y, Tossell K, Harris J J, Harding E C. et al. (2019) GABA and glutamate neurons in the VTA regulate sleep and wakefulness. Nat. Neurosci. 22: 106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong P, Zhang Z, Barger Z, Ma C, Liu D, Ding X. et al. (2019) Control of non-REM sleep by midbrain neurotensinergic neurons. Neuron 104: 795–809.e6. [DOI] [PubMed] [Google Scholar]
- 55.Chen K-S, Xu M, Zhang Z, Chang W-C, Gaj T, Schaffer D V. et al. (2018) A hypothalamic switch for REM and non-REM sleep. Neuron 97: 1168–1176.e4. [DOI] [PubMed] [Google Scholar]
- 56.Weber F, Hoang Do J P, Chung S, Beier K T, Bikov M, Saffari Doost M. et al. (2018) Regulation of REM and non-REM sleep by periaqueductal GABAergic neurons. Nat. Commun. 9: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blanco-Centurion C, Luo S, Spergel D J, Vidal-Ortiz A, Oprisan S A, Van den Pol A N. et al. (2019) Dynamic network activation of hypothalamic MCH neurons in REM sleep and exploratory behavior. J. Neurosci. 39: 4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ingiosi A M, Hayworth C R, Harvey D O, Singletary K G, Rempe M J, Wisor J P. et al. (2020) A role for astroglial calcium in mammalian sleep and sleep regulation. Curr. Biol. 30: 4373–4383.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Y, Blanco-Centurion C, Bendell E, Vidal-Ortiz A, Luo S, and Liu M (2019) Activity dynamics of amygdala GABAergic neurons during cataplexy of narcolepsy. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou H, Neville K R, Goldstein N, Kabu S, Kausar N, Ye R. et al. (2019) Cholinergic modulation of hippocampal calcium activity across the sleep-wake cycle. eLife 8. doi: 10.7554/eLife.39777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar D, Koyanagi I, Carrier-Ruiz A, Vergara P, Srinivasan S, Sugaya Y. et al. (2020) Sparse activity of hippocampal adult-born neurons during rem sleep is necessary for memory consolidation. Neuron 107: 552–565.e10. [DOI] [PubMed] [Google Scholar]
- 62.Carrier-Ruiz A, Sugaya Y, Kumar D, Vergara P, Koyanagi I, Srinivasan S. et al. (2021) Calcium imaging of adult-born neurons in freely moving mice. STAR Protoc. 2: 100238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Izawa S, Chowdhury S, Miyazaki T, Mukai Y, Ono D, Inoue R. et al. (2019) REM sleep–active MCH neurons are involved in forgetting hippocampus-dependent memories. Science 365: 1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eichenbaum H (2016) Still searching for the engram. Learn Behav. 44: 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tonegawa S, Liu X, Ramirez S, and Redondo R (2015) Memory engram cells have come of age. Neuron 87: 918–931. [DOI] [PubMed] [Google Scholar]
- 66.Bliss T V P and Lømo T (1973) Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J. Physiol. 232: 331–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O’Keefe J and Dostrovsky J (1971) The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 34: 171–175. [DOI] [PubMed] [Google Scholar]
- 68.Murano T, Nakajima R, Nakao A, Hirata N, Amemori S, Murakami A. et al. (2020) Multiple types of navigational information are independently encoded in the population activities of the dentate gyrus neurons. Available at SSRN: https://ssrn.com/abstract=3664712. doi: 10.1101/2020.06.09.141572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mau W, Sullivan D W, Kinsky N R, Hasselmo M E, Howard M W, and Eichenbaum H (2018) The same hippocampal CA1 population simultaneously codes temporal information over multiple timescales. Curr. Biol. 28: 1499–1508.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marks W D, Osanai H, Yamamoto J, Ogawa S K, and Kitamura T (2019) Novel nose poke-based temporal discrimination tasks with concurrent in vivo calcium imaging in freely moving mice. Mol. Brain 12: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghandour K, Ohkawa N, Fung C C A, Asai H, Saitoh Y, Takekawa T. et al. (2019) Orchestrated ensemble activities constitute a hippocampal memory engram. Nat. Commun. 10: 2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kitamura T, Ogawa S K, Roy D S, Okuyama T, Morrissey M D, Smith L M. et al. (2017) Engrams and circuits crucial for systems consolidation of a memory. Science 356: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reijmers L G, Perkins B L, Matsuo N, and Mayford M (2007) Localization of a stable neural correlate of associative memory. Science 317: 1230–1233. [DOI] [PubMed] [Google Scholar]
- 74.O’Connell L A and Hofmann H A (2011) Genes, hormones, and circuits: an integrative approach to study the evolution of social behavior. Front. Neuroendocrinol. 32: 320–335. [DOI] [PubMed] [Google Scholar]
- 75.Chen P and Hong W (2018) Neural circuit mechanisms of social behavior. Neuron 98: 16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Y, Mathis A, Grewe B F, Osterhout J A, Ahanonu B, Schnitzer M J. et al. (2017) Neuronal representation of social information in the medial amygdala of awake behaving mice. Cell 171: 1176–1190.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beach F A (1947) A review of physiological and psychological studies of sexual behavior in mammals. Physiol. Rev. 27: 240–307. [DOI] [PubMed] [Google Scholar]
- 78.Resendez S L and Aragona B J (2013) Aversive motivation and the maintenance of monogamous pair bonding. Rev. Neurosci. 24: 51–60. doi: 10.1515/revneuro-2012-0068/html [DOI] [PubMed] [Google Scholar]
- 79.Keshavarzi S, Sullivan R K P, Ianno D J, and Sah P (2014) Functional properties and projections of neurons in the medial amygdala. J. Neurosci. 34: 8699–8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Remedios R, Kennedy A, Zelikowsky M, Grewe B F, Schnitzer M J, and Anderson D J (2017) Social behaviour shapes hypothalamic neural ensemble representations of conspecific sex. Nature 550: 388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scribner J L, Vance E A, Protter D S W, Sheeran W M, Saslow E, Cameron R T. et al. (2020) A neuronal signature for monogamous reunion. Proc. Natl. Acad. Sci. U.S.A. 117: 11076–11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anacker C, Luna V M, Stevens G S, Millette A, Shores R, Jimenez J C. et al. (2018) Hippocampal neurogenesis confers stress resilience by inhibiting the ventral dentate gyrus. Nature 559: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Okuyama T, Kitamura T, Roy D S, Itohara S, and Tonegawa S (2016) Ventral CA1 neurons store social memory. Science 353: 1536–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murugan M, Jang H J, Park M, Miller E M, Cox J, Taliaferro J P. et al. (2017) Combined social and spatial coding in a descending projection from the prefrontal cortex. Cell 171: 1663–1677.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miura I, Sato M, Overton E T N, Kunori N, Nakai J, Kawamata T. et al. (2020) Encoding of social exploration by neural ensembles in the insular cortex. PLoS Biol. 18: e3000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kennedy D P and Adolphs R (2012) The social brain in psychiatric and neurological disorders. Trends Cogn. Sci. (Regul. Ed.) 16: 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim I H, Kim N, Kim S, Toda K, Catavero C M, Courtland J L. et al. (2020) Dysregulation of the synaptic cytoskeleton in the PFC drives neural circuit pathology, leading to social dysfunction. Cell Rep. 32: 107965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun L, Chen R, Li L, Yuan B, Song K, Pan N. et al. (2020) Visualization and correction of social abnormalities-associated neural ensembles in adult MECP2 duplication mice. Sci. Bull. 65: 1192–1202. [DOI] [PubMed] [Google Scholar]
- 89.Berdyyeva T K, Frady E P, Nassi J J, Aluisio L, Cherkas Y, Otte S. et al. (2016) Direct imaging of hippocampal epileptiform calcium motifs following kainic acid administration in freely behaving mice. Front. Neurosci. 10. doi: 10.3389/fnins.2016.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parker J G, Marshall J D, Ahanonu B, Wu Y-W, Kim T H, Grewe B F. et al. (2018) Diametric neural ensemble dynamics in parkinsonian and dyskinetic states. Nature 557: 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sweeney P, Bedenbaugh M N, Maldonado J, Pan P, Fowler K, Williams S Y. et al. (2021) The melanocortin-3 receptor is a pharmacological target for the regulation of anorexia. Sci. Transl. Med. 13. doi: 10.1126/scitranslmed.abd6434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Griessner J, Pasieka M, Böhm V, Grössl F, Kaczanowska J, Pliota P. et al. (2021) Central amygdala circuit dynamics underlying the benzodiazepine anxiolytic effect. Mol. Psychiatry 26: 534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Berdichevsky E, Riveros N, Sánchez-Armáss S, and Orrego F (1983) Kainate, N-methylaspartate and other excitatory amino acids increase calcium influx into rat brain cortex cells in vitro. Neurosci. Lett. 36: 75–80. [DOI] [PubMed] [Google Scholar]
- 94.Tse K, Puttachary S, Beamer E, Sills G J, and Thippeswamy T (2014) Advantages of repeated low dose against single high dose of kainate in C57BL/6J mouse model of status epilepticus: behavioral and electroencephalographic studies. PLoSOne 9: e96622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kanner A M and Balabanov A (2002) Valproate: a practical review of its uses in neurological and psychiatric disorders. Expert Rev. Neurother 2: 151–165. [DOI] [PubMed] [Google Scholar]
- 96.Duty S and Jenner P (2011) Animal models of Parkinson’s disease: a source of novel treatments and clues to the cause of the disease. Br. J. Pharmacol. 164: 1357–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Albin R L, Young A B, and Penney J B (1989) The functional anatomy of basal ganglia disorders. Trends Neurosci. 12: 366–375. [DOI] [PubMed] [Google Scholar]
- 98.Siciliano C A, Noamany H, Chang C-J, Brown A R, Chen X, Leible D. et al. (2019) A cortical-brainstem circuit predicts and governs compulsive alcohol drinking. Science 366: 1008–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cameron C M, Murugan M, Choi J Y, Engel E A, and Witten I B (2019) Increased cocaine motivation is associated with degraded spatial and temporal representations in IL-NAc neurons. Neuron 103: 80–91.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun Y, Blanco-Centurion C, Bendell E, Vidal-Ortiz A, Luo S, and Liu M (2019) Activity dynamics of amygdala GABAergic neurons during cataplexy of narcolepsy. eLife 8: e48311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tummala S R, Hemphill M A, Nam A, and Meaney D F (2020) Concussion increases CA1 activity during prolonged inactivity in a familiar environment. Exp. Neurol. 334: 113435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brainstorm Consortium, Anttila V, Bulik-Sullivan B, Finucane H K, Walters R K, Bras J, Duncan L. et al. (2018Analysis of shared heritability in common disorders of the brain. Science 360: eaap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Luo L, Callaway E M, and Svoboda K (2018) Genetic dissection of neural circuits: a decade of progress. Neuron 98: 256–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brenowitz E A and Zakon H H (2015) Emerging from the bottleneck: benefits of the comparative approach to modern neuroscience. Trends Neurosci. 38: 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miller C T, Hale M E, Okano H, Okabe S, and Mitra P (2019) Comparative principles for next-generation neuroscience. Front. Behav. Neurosci. 13: 12. doi: 10.3389/fnbeh.2019.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hornung R, Pritchard A, Kinchington P R, and Kramer P R (2020) Reduced activity of GAD67 expressing cells in the reticular thalamus enhance thalamic excitatory activity and varicella zoster virus associated pain. Neurosci. Lett. 736: 135287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Anner P, Passecker J, Klausberger T, and Dorffner G (2020) Ca2+ imaging of neurons in freely moving rats with automatic post hoc histological identification. J. Neurosci. Methods 341: 108765. [DOI] [PubMed] [Google Scholar]
- 108.Roberts T F, Hisey E, Tanaka M, Kearney M G, Chattree G, Yang C F. et al. (2017) Identification of a motor-to-auditory pathway important for vocal learning. Nat. Neurosci. 20: 978–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Markowitz J E, Liberti III W A, Guitchounts G, Velho T, Lois C, and Gardner T J (2015) Mesoscopic patterns of neural activity support songbird cortical sequences. PLoS Biol. 13: e1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Daliparthi V K, Tachibana R O, Cooper B G, Hahnloser R H, Kojima S, Sober S J. et al. (2019) Transitioning between preparatory and precisely sequenced neuronal activity in production of a skilled behavior. eLife 8: e43732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mackevicius E L, Bahle A H, Williams A H, Gu S, Denisenko N I, Goldman M S. et al. (2019) Unsupervised discovery of temporal sequences in high-dimensional datasets, with applications to neuroscience. eLife 8: e38471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Phillips K A, Bales K L, Capitanio J P, Conley A, Czoty P W, ‘t Hart B A. et al. (2014) Why primate models matter. Am. J. Primatol. 76: 801–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roelfsema P R and Treue S (2014) Basic neuroscience research with nonhuman primates: a small but indispensable component of biomedical research. Neuron 82: 1200–1204. [DOI] [PubMed] [Google Scholar]
- 114.Eliades S J and Miller C T (2017) Marmoset vocal communication: behavior and neurobiology. Dev. Neurobiol. 77: 286–299. [DOI] [PubMed] [Google Scholar]
- 115.Miller C T, Freiwald W A, Leopold D A, Mitchell J F, Silva A C, and Wang X (2016) Marmosets: a neuroscientific model of human social behavior. Neuron 90: 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chan A W S (2009) Transgenic primate research paves the path to a better animal model: are we a step closer to curing inherited human genetic disorders? J. Mol. Cell Biol. 1: 13–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shen H (2013) Precision gene editing paves way for transgenic monkeys. Nature 503: 14–15. [DOI] [PubMed] [Google Scholar]
- 118.Izpisua Belmonte J C, Callaway E M, Caddick S J, Churchland P, Feng G, Homanics G E. et al. (2015) Brains, genes, and primates. Neuron 86: 617–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Feng G, Jensen F E, Greely H T, Okano H, Treue S, Roberts A C. et al. (2020) Opportunities and limitations of genetically modified nonhuman primate models for neuroscience research. Proc. Natl. Acad. Sci. U.S.A. 117: 24022–24031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Okano H, Miyawaki A, and Kasai K (2015) Brain/MINDS: brain-mapping project in Japan. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370: 20140310. doi: 10.1098/rstb.2014.0310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Trautmann E M, O’Shea D J, Sun X, Marshel J H, Crow A, Hsueh B. et al. (2021) Dendritic calcium signals in rhesus macaque motor cortex drive an optical brain-computer interface, Nat Commun. 12: 3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Garg A K, Li P, Rashid M S, and Callaway E M (2019) Color and orientation are jointly coded and spatially organized in primate primary visual cortex. Science 364: 1275–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ju N, Jiang R, Macknik S L, Martinez-Conde S, and Tang S (2018) Long-term all-optical interrogation of cortical neurons in awake-behaving nonhuman primates. PLoS Biol. 16: e2005839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li M, Liu F, Jiang H, Lee T S, and Tang S (2017) Long-term two-photon imaging in awake macaque monkey. Neuron 93: 1049–1057.e3. [DOI] [PubMed] [Google Scholar]
- 125.Zeng H-H, Huang J-F, Chen M, Wen Y-Q, Shen Z-M, and Poo M-M (2019) Local homogeneity of tonotopic organization in the primary auditory cortex of marmosets. Proc. Natl. Acad. Sci. U.S.A. 116: 3239–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ebina T, Masamizu Y, Tanaka Y R, Watakabe A, Hirakawa R, Hirayama Y. et al. (2018) Two-photon imaging of neuronal activity in motor cortex of marmosets during upper-limb movement tasks. Nat. Commun. 9: 1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Santisakultarm T P, Kersbergen C J, Bandy D K, Ide D C, Choi S-H, and Silva A C (2016) Two-photon imaging of cerebral hemodynamics and neural activity in awake and anesthetized marmosets. J. Neurosci. Methods 271: 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yamada Y, Matsumoto Y, Okahara N, and Mikoshiba K (2016) Chronic multiscale imaging of neuronal activity in the awake common marmoset. Sci. Rep. 6: 35722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Seidemann E, Chen Y, Bai Y, Chen S C, Mehta P, Kajs B L. et al. (2016) Calcium imaging with genetically encoded indicators in behaving primates. eLife 5: e16178. doi: 10.7554/eLife.16178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sadakane O, Masamizu Y, Watakabe A, Terada S-I, Ohtsuka M, Takaji M. et al. (2015) Long-term two-photon calcium imaging of neuronal populations with subcellular resolution in adult non-human primates. Cell Rep. 13: 1989–1999. [DOI] [PubMed] [Google Scholar]
- 131.Kondo T, Saito R, Otaka M, Yoshino-Saito K, Yamanaka A, Yamamori T. et al. (2018) Calcium transient dynamics of neural ensembles in the primary motor cortex of naturally behaving monkeys. Cell Rep. 24: 2191–2195.e4. [DOI] [PubMed] [Google Scholar]
- 132.Bollimunta A, Santacruz S R, Eaton R W, Xu P S, Morrison J H, Moxon K A. et al. (2021) Head-mounted microendoscopic calcium imaging in dorsal premotor cortex of behaving rhesus macaque, Cell Reports. 35: 109239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman J P. et al. (2016) Sensitive red protein calcium indicators for imaging neural activity. eLife 5: e12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Inoue M, Takeuchi A, Manita S, Horigane S-I, Sakamoto M, Kawakami R. et al. (2019) Rational engineering of XCaMPs, a multicolor GECI suite for in vivo imaging of complex brain circuit dynamics. Cell 177: 1346–1360.e24. [DOI] [PubMed] [Google Scholar]
- 135.Mathis A, Mamidanna P, Cury K M, Abe T, Murthy V N, Mathis M W. et al. (2018) DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21: 1281–1289. [DOI] [PubMed] [Google Scholar]
- 136.Wiltschko A B, Johnson M J, Iurilli G, Peterson R E, Katon J M, Pashkovski S L. et al. (2015) Mapping sub-second structure in mouse behavior. Neuron 88: 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]


